Abstract
Pantothenate kinase (PanK) is the first enzyme in the coenzyme A (CoA) biosynthetic pathway. The differential expression of the four-active mammalian PanK isoforms regulates CoA levels in different tissues and PANK2 mutations lead to Pantothenate Kinase Associated Neurodegeneration (PKAN). The molecular mechanisms that potentially underly PKAN pathophysiology are investigated in a mouse model of CoA deficiency in the central nervous system (CNS). Both PanK1 and PanK2 contribute to brain CoA levels in mice and so a mouse model with a systemic deletion of Pank1 together with neuronal deletion of Pank2 was generated. Neuronal Pank2 expression in Pank1−/−Syn-Pank2−/− double knockout mice decreased starting at P9-11 triggering a significant brain CoA deficiency. The depressed brain CoA in the Pank1−/−Syn-Pank2−/− mice correlates with abnormal forelimb flexing and weakness that, in turn, contributes to reduced locomotion and abnormal gait. Biochemical analysis reveals a reduction in short-chain acyl-CoAs, including acetyl-CoA and succinyl-CoA. Comparative gene expression analysis reveals that the CoA deficiency in brain is associated with a large elevation of Hif3a transcript expression and significant reduction of gene transcripts in heme and hemoglobin synthesis. Reduction of brain heme levels is associated with the CoA deficiency. The data suggest a response to oxygen/glucose deprivation and indicate a disruption of oxidative metabolism arising from a CoA deficiency in the CNS.
Keywords: pantothenate kinase, PKAN, neurodegeneration, coenzyme A, mouse model
1. Introduction
Pantothenate kinase-associated neurodegeneration (PKAN) is an inherited disorder characterized by brain iron accumulation, progressive dystonia, dysarthria and Parkinson-like features [1-3]. PKAN is associated with mutations in the human pantothenate kinase 2 (PANK2) gene that inactivate or often substantially reduce activity of the expressed pantothenate kinase 2 (PanK2) protein [2-4]. PanK2 is one of four active PanK isoforms that initiate coenzyme A (CoA) biosynthesis [5]. CoA deficiency in the central nervous system (CNS) is thought to underlie the movement disorder and neurodegeneration in PKAN, although CoA deficiency has not been formally demonstrated in human PKAN patients. CoA is cell-autonomous and non-invasive probes for CoA are not available. The view that CoA deficiency is causative in PKAN is supported by the discovery that missense mutations in the human COASY gene that encodes the CoA synthase are also associated with neurological deterioration with a progressive movement disorder and iron accumulation in the basal ganglia [6]. In addition, a mouse model with neuron-specific degradation of CoA exhibits reduced motor coordination [7]. This result indicates that neuronal CoA deficiency likely contributed to development of a movement disorder but only 15% CoA reduction was achieved in the model and the phenotype was relatively mild. Pertubation of CoA homeostasis as a disease-relevant factor is also supported by research in other animal models. The PanK in Drosophila is encoded by the fumble (fbl) gene and a dPANK/fbl hypomorphic mutant has reduced whole body CoA and locomotor dysfunction [8]. Zebrafish have two pank genes and knockdown of pank2 results in developmental defects of the nervous and vascular systems [9]. The few studies that included CoA measurements in PKAN patient fibroblasts and/or cultured neurons derived from induced pluripotent stem cells (iPSCs) are controversial. CoA levels in iPSC-derived neurons from PKAN patients were similar to CoA levels in iPSC-derived neurons from non-diseased, unrelated subjects [10], whereas in a separate study, mitochondrial CoA was reduced in fibroblasts and iPSC-derived neurons obtained from PKAN patients compared to those from control subjects [11]. Our understanding of the pathological mechanisms associated with a substantial CoA deficiency in the CNS has been hampered by the lack of a tractable mouse model with phenotypic characteristics that resemble the human disease.
CoA is an essential cofactor that has a role in energy production, lipid synthesis and metabolism [5]. A general view of the processes most affected by reduction of PANK2 expression in cultured human cells includes dysfunctional mitochondria with overproduction of reactive oxygen species, aberrant mitochondrial iron metabolism and impaired function of electron transport components [12-15]. However, gene inactivation of Pank2 in mice did not result in brain pathology nor impaired movement, but azoospermia was evident and male mice were infertile [16]. Notably, Pank2 expression is particularly high in mouse testes [17]. Pank2 deletion reduced CoA levels in brain during the postnatal period, but in adult mice loss of Pank2 expression did not result in reduced brain CoA levels [18]. Both Pank1 and Pank3 genes are also expressed in the murine CNS and the activities of PanK1 and PanK3 likely adapted to compensate for loss of Pank2 expression in the adult. A report of a PKAN-like disease phenotype induced by a ketogenic diet in Pank2−/− animals [19] did not include CoA measurements and the phenotype was not reproducible in our laboratory. These observations indicate an adaptive cooperativity among the PanKs in mice that together work to regulate and maintain CoA homeostasis. Derivation of a Pank1−/− mouse pointed out a unique role for PanK1 protein in maintaining liver CoA and glucose homeostasis [20]. Pank1 expression is particularly high in murine liver [21] and the Pank1−/− mouse developed a mild hypoglycemia during an extended fast that was associated with a 20-25% reduction in fatty acid oxidation capacity [20]. By contrast, liver CoA levels were the same as wild-type in Pank2−/− [18] and Pank3−/− (unpublished, S. Jackowski) mice. The phenotypes of the deletions of each of the three mouse Pank genes indicate regulatory complexity and PanK crosstalk in maintaining tissue CoA levels.
One explanation for the difficulty in developing a PKAN mouse model may be that the relative expression levels of the murine Pank genes in the CNS are not equivalent to those in the human CNS. Both Pank1 and Pank2 are important for early development in mice [18] and are expressed at the same levels in mouse brain [21]. Analysis of whole-transcriptome gene expression data from human PKAN brain revealed a gene set that belongs to a co-expression module reflecting neuronal signatures in the basal ganglia [22]. Based on this analysis, together with the earlier result of neuronal-specific CoA reduction in association with reduced motor coordination [7], we reasoned that neuron-specific loss of Pank2, in addition to systemic loss of Pank1 expression, would reduce brain CoA sufficiently in mice to elicit an enduring CoA deficiency in the CNS. We generated a Pank1−/−Syn-Pank2−/− mouse model with systemic deletion of Pank1 together with neuron-specific deletion of Pank2 (Syn-Pank2) as driven by the synapsin-Cre transgene. The Pank1−/−Syn-Pank2−/− mouse model represents not only the results of a reduction of Pank2 expression in neurons, but also a loss of Pank1 expression in neurons plus additional cell types in the brain, such as glial cells. Indeed, the Pank1−/−Syn-Pank2−/− mice consistently developed a CoA-deficiency in the late postnatal period that correlated with a sustained forelimb dysfunction and gait abnormality. Comparative investigation of the Pank1−/−Syn-Pank2−/− mice pre- and post-development of the phenotype pointed toward selective alterations in gene expression that, together with biochemical analysis, revealed a heme deficit in brain as well as aspects associated with neurodegenerative diseases.
2. Material and methods
2.1. Generation of Pank1−/−Pank2f/fSyn-Cre+/0 (aka Pank1−/−Syn-Pank2−/−) Mice
All procedures were performed according to protocol 323 which was approved by the St. Jude Children's Research Hospital Institutional Animal Care and Use Committee. Generation of the Pank1−/− and the Pank2f/f mice was reported previously [18, 20]. B6.Cg-Tg(Syn1-cre) 671Jxm/J mice (The Jackson Laboratory) expressing Cre recombinase as controlled by the neuronal synapsin1 (Syn) promoter were crossed with Pank2f/f mice to generate a deletion within the Pank2f/f gene by recombination of the first and third loxP sites, resulting in the loss of exon 3. After ≥6 generations of intercrossing the Pank1−/− with the Pank2f/f and the Syn-Cre+/0 mice, Pank1−/−Pank2f/fSyn-Cre0/0 males and Pank1+/−Pank2f/fSyn-Cre+/0 females were obtained and were bred together to obtain the Pank1−/−Pank2f/fSyn-Cre+/0 (Pank1−/−Syn-Pank2−/−) conditional double knockout progeny used in this study that had deletion of Pank1 systemically and selective deletion of Pank2, in addition to deletion of Pank1, in neurons (Fig. S1). The Syn-Cre transgene was maintained as hemizygous in female breeders. Wild-type matched control animals for the study were generated by mating Pank1+/− Pank2f/fSyn-Cre+/0 males and females together to yield Pank1+/+Pank2f/fSyn-Cre0/0 pups. Pank1+/+Pank2f/fSyn-Cre0/0 males and females were then bred to yield the wild-type matched control mice. PCR analysis was used to genotype tail, toe and organ biopsies using primers (Table S1) described previously [18, 20]. Tail genotyping was performed at P4. A 332-bp product indicated the presence of the Pank2f/f allele and a 285-bp product indicated the presence of the Cre gene. Deletion of the Pank2 allele was confirmed in brains by the presence of a 176-bp product while only the Pank2f/f allele (332-bp) was evident in liver (Fig. S1). Incomplete deletion of the Pank2f/f allele in brain was indicated by coincidence of both the floxed PCR product (332 bp) and the deleted PCR product (176 bp). Cell types other than neurons contributed to the residual undeleted Pank2f/f signal in brain. In Figure S1, the PCR products indicating the unfloxed Pank1+/+ and Pank2+/+ alleles are shown for reference. Genotyping primers and PCR conditions are listed in Supplemental Table 1.
2.2. Animal Studies
Mice were maintained at a room temperature of 72° ± 2°F, humidity of 50%±10%, and a 14-hr light, 10-hr dark cycle, with the dark cycle starting at 20:00 hr. Water and chow (Lab diet 5013) were supplied ad libitum. Blood glucose levels were measured via tail snips using a glucometer (Freestyle). Blood was collected from euthanized animals, plasma or serum was prepared and stored frozen until analysis. Organs, including liver, forebrain, hindbrain and spinal cord, were quickly excised from euthanized animals and immediately flash frozen in liquid nitrogen (LN2) or immersed in RNAlater® (Qiagen) overnight prior to freezing. Forebrain and hindbrain regions were identified according to ‘Paxinos and Franklin's “The Mouse Brain in Stereotaxic Coordinates” [23]. Forebrain included the olfactory system, cortex, and hypothalamus.
Locomotor activity was assessed using an open field test. Individual mice were placed in an open rectangular arena (36.8 cm × 43.2 cm) for 5 min during the light phase and motor activity was evaluated using a video tracking system provided by HVS Image with associated 2100 Plus software (San Diego, CA, USA). Each subject was placed in the center of the arena under standard overhead lighting and the total distance traveled and the percentage of time in motion were recorded. The spontaneous activity of each subject in a novel open field provided a general measure of motor function of the Pank1−/−Syn-Pank2−/− mice, at P19-21. The behaviors of age-matched wild-type control mice were quantified and compared.
Forelimb function was assessed by measuring forelimb grip strength. The wild-type and Pank1−/−Syn-Pank2−/− mice at P19-21 were subjected to grip strength test using the UGO Basile grip strength meter. The force applied to pull the trapeze bar and the grip force was measured in grams and compared with age-matched WT mice of the same genetic lineage.
2.3. PanK Antibodies and Immunodetection
The pan-reactive anti-PanK antibody that recognized all mouse PanK isoforms was purified from rabbit polyclonal antiserum that was raised against pure full-length recombinant mouse Pank1β. Anti-PanK2 rabbit polyclonal antiserum was raised against a synthetic peptide (GESADSEARRRDPLRRR). The PanK2 peptide antigen was prepared by coupling to keyhole limpet hemocyanin at the amino terminus. Immunization of rabbits and collection of antisera for both antibodies were performed by Rockland Immunochemicals, Inc. according to their standard schedule. Antisera were purified by affinity chromatography on Affi-Gel cross-linked either with the recombinant Pank1β protein (for the pan-reactive anti-PanK antibody) or with the PanK2-specific peptide as described previously [24]. The PanK specificities of both antibodies were confirmed by immunoblotting serial dilutions of the purified antibody preparations against purified recombinant mouse Pank1β, PanK2 and PanK3 proteins. In tissue lysates, the pan-reactive anti-PanK antibody recognized a non-specific protein migrating near the 62 kDa size marker, slightly slower than PanK1α on SDS-PAGE, that became evident upon validation using tissues from Pank2−/− mice [17]. Specificities of the PanK antibodies were validated previously [25].
Pre-frozen brain tissue (150 mg) tissues from WT and Pank1−/−Syn-Pank2−/− mice was homogenized in 500 μl low salt lysis buffer (1% NP40, 50 mM Tris-HCl, 5 mM EDTA, pH 8) plus Protease Inhibitor Cocktail (Roche). Homogenates were incubated on ice for 1 hr, then centrifuged at 10,000g at 4°C for 10 min. Aliquots of the supernatants were removed for protein quantification (Bio-Rad). In preparation for pre-clearing the brain supernatant, Protein A Sepharose beads, 50 μl of 50% suspension (Millipore), were preincubated with 5 μl normal rabbit serum in 100 μl low salt lysis buffer at 4°C for 1 hr then washed. Brain supernatants (15 mg protein) were added to the washed beads in 1 ml of low salt buffer and rotated at 4°C for 1 hr for pre-clearing. In preparation for immunoprecipitation, purified anti-PanK antibody (10 μg) was pre-incubated with 100 μl of 50% Protein A-Sepharose beads in low salt lysis buffer at 4°C for 1 hr. For immunoprecipitation, the pre-cleared supernatant was added to the anti-PanK antibody plus beads and rotated at 4°C overnight. Beads were collected, washed 3 times in low salt lysis buffer, and then finally resuspended in 100 μl 1xLDS sample buffer, heated at 95 °C for 10 min, and the supernatant was loaded onto pre-cast 10% Nupage Bis-Tris Gels (Invitrogen). Proteins were fractionated by electrophoresis at 200 V for 2.5 hr according to manufacturer's directions and then transferred using an XCell II Blot module (Invitrogen) to Immobilon-FL membranes (Millipore) for 1 hr. Membranes were blocked in 5% dry milk at room temp for 1 hr. Western blotting was performed using either the pan-reactive anti-PanK primary antibody or the anti-PanK2 primary antibody at 1 μg/ml in TBS-T with incubation overnight at 4°C. Membranes were washed with TBS-T for 15 min 3 times and then incubated with Conformation-Specific Mouse anti-Rabbit IgG (Cell Signaling #3678s) followed by incubation with Alexa Fluor 546 Goat anti-Mouse IgG (Life Technology) for 15 min each. Fluorescence was measure using a Typhoon FLA9500 (GE Healthcare), then quantified using ImageQuant TL software (GE Healthcare). Each brain lysate (20 μg) was fractionated by SDS-PAGE and stained with Coomassie Brilliant Blue to confirm equivalent protein input for the immunoprecipitation assays.
2.4. Tissue CoA Levels
CoA was extracted from flash frozen tissues from matched control wild-type and Pank1−/−Syn-Pank2−/− mice, derivatized and quantified following high-pressure liquid chromatography as described previously [26, 27]. Briefly, organs, including liver, forebrain, hindbrain and spinal cord, were quickly excised from euthanized animals and immediately flash frozen in liquid Nitrogen prior to freezing. Forebrain and hindbrain regions were identified as described [23]. Total CoA was determined using 20-50 mg of tissue (liver, spinal cord, forebrain or hindbrain) homogenized in 2 ml of 1 mM KOH. The pH was adjusted to 12.0 with 0.25 M KOH, and incubated at 55°C for 2 hours. The pH of the sample was re-adjusted to 8.0, and the samples were derivatized with monobromobimane and analyzed by HPLC equipped with a fluorescent detector. The quantified CoA-bimane data were normalized to wet weights (mg) of each sample.
2.5. Measuring CoA Species with HPLC/MS/MS
Frozen Hindbrain (30-40 mg) from matched control wild-type and Pank1−/−Syn-Pank2−/− mice was homogenized in 2 ml of methanol and 1 ml of water. After homogenizing, 1 ml of chloroform and 400 pmol of [13C]acetyl-CoA (Sigma) were added and incubated on ice for 15 min. Chloroform (1.5 ml) and water (1.2 ml) were added, and centrifuged at 2000 × g for 10 min. The top layer was loaded on a 2-(2-pyridyl)ethyl solid phase extraction column (Supleco) which was equilibrated with 1 ml of 50% methanol:2% acetic acid. The column was washed with 2 × 1 ml 50% methanol:2% acetic acid and 1 ml water. CoA and its thioesters were eluted with 2 × 1 ml of [95% ethanol + 50 mM ammonium formate], and dried under nitrogen. The samples were re-suspended 200 μl of [90% methanol + 15 mM ammonium hydroxide] and filtered through a Spin-X centrifuge tube (0.22 um Cellulose Acetate, Costar).
Acyl-CoA was analyzed using a Shimadzu Prominence UFLC attached to a QTrap 4500 equipped with a Turbo V ion source (Sciex). Samples were injected onto an Acquity UPLC HSS T3, 1.8uM, 2.1 x 150 mm column at 40 °C (Waters) using a flow rate of 0.2 ml/min. Solvent A was 10 mM ammonium acetate pH 6.8, and Solvent B was [95% acetonitrile + 10 mM ammonium acetate pH 6.8]. The HPLC elution program was the following: starting solvent mixture of 95% A:5% B, 0 to 2 min isocratic with 5% B; 2 to 20 min linear gradient 100% B; 20 to 25 min isocratic with 100% B; 25 to 27 min linear gradient to 5% B; 27 to 31 min isocratic with 5% B. The QTrap 4500 was operated in the positive mode, and the ion source parameters were: ion spray voltage, 5500 V; curtain gas, 15 psi; temperature, 400 °C; collision gas, medium; ion source gas 1, 15 psi; ion source gas 2, 20 psi; declustering potential, 60 V; and collision energy, 45 V. The MRM transition for CoA species is listed in Table S3. [13C]acetyl-CoA was used to calculate relative abundance of the acyl-CoAs in tissue. The system was controlled by the Analyst® software (Sciex) and analyzed with MultiQuant™ 3.0.2 software (Sciex).
2.6. Glucose, Lactate and β-Hydroxybutyrate Measurements
Glucose, lactate and β-hydroxybutyrate were measured in plasma from matched control and Pank1−/−Syn-Pank2−/− mice that was collected with heparin using the GM7 Micro-Stat Analyzer (Analox Instruments, London, UK). The Analox kits for glucose (GMRD-002A), lactate (GMRD-090) and 3-hydoxybutyrate (GMRD-135) were used for the analysis according to the manufacturer's instructions.
2.7. NAD+, NADH and ATP measurements
NAD, NADH and NAD/NADH levels in the hindbrain of matched control wild-type and Pank1−/−Syn-Pank2−/− mice were determined according to instructions provided with the NAD/NADH colorimetric assay kit from Abcam. Approximately 20 mg of tissue was homogenized in 400 μl of extraction buffer and the supernatant was passed twice through a 10 kDa-cutoff spin column to remove enzymes consuming NADH. The supernatant was assayed according to the manufacturer’s instructions. NADH and NAD were measured at 450 nm using a Spectramax plate reader.
ATP was extracted from 30-40 mg of hindbrain from matched control wild-type and Pank1−/−Syn-Pank2−/− mice by homogenizing the tissue in 80% methanol and 300 pmol of 13C10-15N5-ATP (Millipore Sigma) was added. The samples were incubated at −80 °C for 4 hours and spun at 4000 x g for 10 minutes. The supernatant was vacuum dried and analyzed by mass spectrometry. Samples were resuspended in water and spun through a Spin-X Centrifuge Tube Filter (0.22 μm Cellulose Acetate, Costar). ATP was analyzed using a Shimadzu Prominence UFLC attached to a QTrap 4500 equipped with a Turbo V ion source (Sciex). Samples were injected onto an XSelect® HSS C18, 2.5 μm, 3.0 x 150 mm column using a flow rate of 0.3 ml/min. Solvent A was [100 mM ammonium formate, pH 5.0 + 2% acetonitrile + 0.1% tributylamine (TBA)], and Solvent B was [95% acetonitrile + 50 mM ammonium formate, pH 6.3 + 0.1% TBA]. The HPLC program was the following: starting solvent mixture of 4% B, 0 to 1 min isocratic with 4% B; 2 to 12 min linear gradient to 100% B; 12 to 20 min isocratic with 100% B; 20 to 22 min linear gradient to 4% B; 22 to 25 min isocratic with 4% B. The QTrap 4500 was operated in the negative mode, and the ion source parameters were: ion spray voltage, −4500 V; curtain gas, 25 psi; temperature, 450 °C; collision gas, medium; ion source gas 1, 25 psi; and ion source gas 2, 40 psi. The MRM transition for ATP was 506.0 / 159 m/z and 13C10-15N5-ATP was 521.0 / 159 m/z both with a declustering potential, −80 V and collision energy, −40 V. The system was controlled by the Analyst® software (Sciex) and analyzed with MultiQuant™ 3.0.2 software (Sciex).
2.8. RNAseq Analysis
RNA was isolated from back left brain of 10 and 20 day old matched control wild-type and Pank1−/−Syn-Pank2−/− male and female mice by the Triazole method as described earlier [28] and washed with ethanol thrice. Each sample was assessed for RNA quality and integrity using a 2100 Bioanalyzer Instrument (Agilent). RNA sequencing was done using the Illumina transcriptome sequencing platform. The sequence files were aligned to the reference mouse genome (Ensembl Mus musculus GRCm38 sequence and annotation files) using STAR. STAR was run with the --quantMode GeneCounts option to tabulate the number of reads per gene while mapping each sample. The second read strand aligned with RNA count (column 4) was used for analysis because strand-specific RNA-seq was performed. Differential gene expression using the count data was analyzed via DESeq2 using the R statistical computing environment.
2.9. Real-time Reverse Transcriptase Quantitative PCR (RT-qPCR)
RNA was extracted, cDNAs were prepared by reverse transcription, and RT-qPCR was performed in triplicate as described previously [29]. The Taqman rodent glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Applied Biosystems) was used as the normalization control. All of the values were compared using the ΔCT method [29] and the amount of target cDNA (2−ΔCT) was calculated relative to GAPDH. The relative expression data shown in Fig. 4 were calculated by comparing the ΔCT values of the Pank1−/−Syn-Pank2−/− samples with the values of age-matched control wild-type samples, to obtain 2−ΔΔCT values and setting the wild-type values at 1.0. The primers are listed in Supplemental Table 2.
Fig. 4.
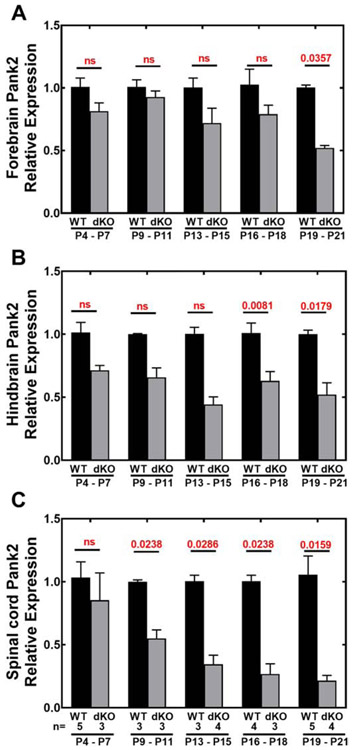
Relative expression of Pank2 in Pank1−/−Syn-Pank2−/− (dKO) compared to wild-type (WT) tissues. RNA was isolated from each dKO and WT tissue at postnatal days P4-P7, P9-P11, P13-P15, P16-P18, P19-P21. The numbers of mice in each group are indicated below panel C. Transcripts were quantified by RT-qPCR as described in Experimental Procedures. A. Relative expression of Pank2 in forebrain; B. Relative expression of Pank2 in hindbrain, C. Relative expression of Pank2 in spinal cord. The data represent the mean ± SEM from triplicate determinations with independent samples and statistical significance was calculated using the non-parametric t-test comparing age matched WT and dKO mice, the values for which are shown in red.
2.10. Heme and Iron Measurements
Tissues (30 – 40 mg) from WT and Pank1−/−Syn-Pank2−/− mice were homogenized in 4 ml 80% acetonitrile:20% 1.7 M hydrochloric acid, and incubated on ice for 30 min. Sample was centrifuged at 5000 x g for 20 minutes to pellet debris. Supernatant was removed and dried under nitrogen overnight.
Heme was analyzed with a Waters e2695 separation module equipped with a Waters 2489 UV/Vis detector controlled by Empower 3 software. Sample was fractionated using a Gemini C18 3 μm column (150 × 4.60 mm) with a flow rate of 1.0 ml/min. Solvent A was [30% acetonitrile + 0.05% trichloroacetic acid], and Solvent B was [90% acetonitrile + 0.05% trichloroacetic acid]. The HPLC program was the following: starting solvent mixture of 100% A: 0% B, 0 to 15 min linear gradient from 0% B to 60% B, 15 to 20 min linear gradient from 60% B to 100% B, 20 to 30 min isocratic with 100% B, 30 to 32 min linear gradient from 100% to 0%, and 32 to 40 min isocratic with 0% B. The UV/Vis detector was set at 400 nm. Area under the heme peak was compared to a standard curve of hemin to determine the amount of heme in a sample and normalized to mg of tissue wet weight. Hemin from Sigma was used as standard.
Iron was measured using the Abcam iron assay kit (ab83366) following the manufacturer’s instructions. Approximately 50 mg of mouse brain tissue was homogenized in the iron assay buffer. Iron was measured following addition of the iron probe and absorbance was measured at 593 nm.
2.11. Statistical analysis
Either the parametric unpaired t-test or nonparametric unpaired (Mann-Whitney) t-test was performed on the data to evaluate significance. All the data were checked for normality using the Shapiro-Wilk normality test. If the data passed the normality test (with an alpha level of 0.05), then the parametric t-test was used, otherwise the nonparametric t-test was used as indicated in each figure legend.
3. Results
3.1. Reduction of Pank2 expression in brain and spinal cord
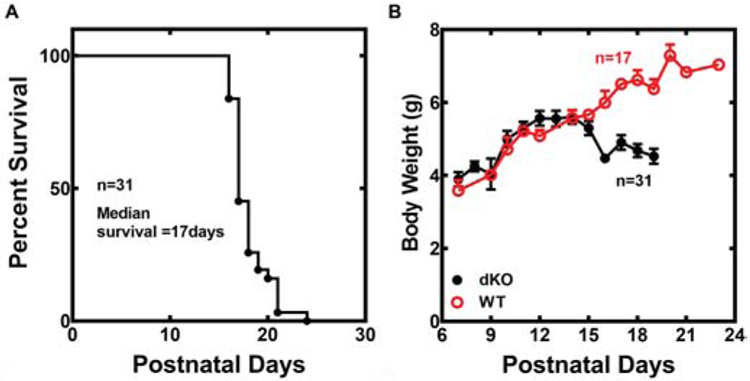
The Pank1−/−Syn-Pank2−/− animals had a median lifespan of 17 days (Fig. 1A) and survival extended from P15 to P23 in the population. Animals lost weight in conjunction with the appearance of a forelimb phenotype and then stabilized at about 10% lesser weight 2-3 days prior to death. Tissues were taken from euthanized animals before and immediately after the weight loss for comparative analyses that are described below. Matched wild-type animals (Pank1+/+Pank2f/f) originating from Pank1+/−Pank2f/f males and females lacking the Syn-Cre transgene continued to gain weight and developed normally through the postnatal period and subsequently (Fig. 1B). Male and female Pank1−/−Syn-Pank2−/− animals had the same average lifespan, and very similar timelines for weight loss and phenotype development.
Fig. 1.
Survival and growth of Pank1−/− Syn-Pank2−/− (dKO) mice. A. Kaplan-Meier survival plot of the dKO mice. Male and female dKO mice have the same lifespan and the data represent both genders. B. Body weights of wild-type (WT) and dKO mice.
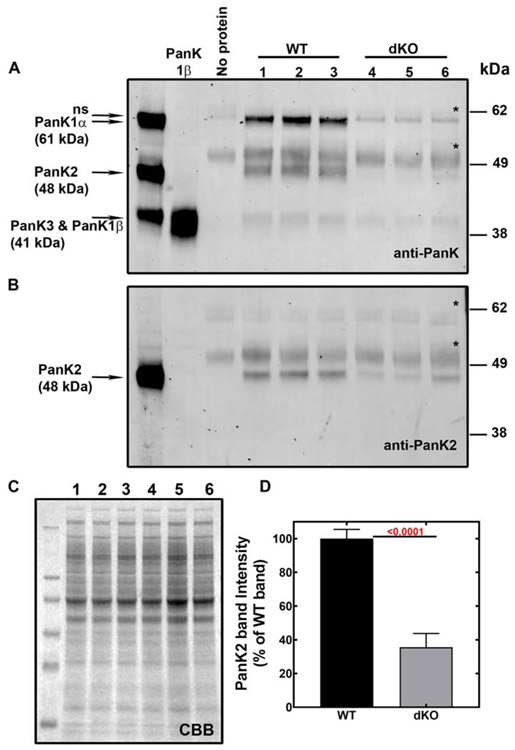
Loss of Pank1 protein and reduction of PanK2 protein expression was confirmed in the Pank1−/−Syn-Pank2−/− brains age P19-20 (Fig. 2). The PanK proteins are in generally low abundance and so relative amounts of the PanK proteins in brain lysates were determined by immunoprecipitation followed by immunoblotting. A pan-reactive polyclonal rabbit antibody, designated as anti-PanK, was raised against full-length recombinant mouse PanK1β and was found to recognize all PanK isoforms (Fig. S2, [25]). When the brain immunoprecipitates were probed with the PanK antibody, PanK1α and PanK2 proteins were dominant in wild-type brain while PanK3 and possibly PanK1β were present at significantly lower levels (Fig. 2A). Equal protein input for the immunoprecipitation of each experimental sample labeled 1-6 is shown in the Coomassie-stained gel (Fig. 2C). A second antibody that specifically recognized mouse PanK2 protein was developed (Fig. S2) and used for immunoblotting. The amounts of PanK2 protein were noticeably reduced by about 60% in the dKO brains when the brain immunoprecipitates were probed with the anti-PanK2 antibody (Fig. 2B and 2D) and the residual PanK2 protein that was detected was likely due to expression in non-neuronal cell types. PanK1α, the dominant PanK1 isoform expressed in wild-type brain (Fig. 3A, inset), was absent in the Pank1−/−Syn-Pank2−/− brain as expected. The molecular sizes of PanK1β and PanK3 are the same (41 kDa) and so could not be distinguished on the blot (Fig. 2A), but the band intensity was lower in the dKO samples compared to wild-type, consistent with the expected absence of Pank1β protein expression. Two non-specific protein bands (*) at about 61.5 kDa and 50 kDa were evident on the blots in the absence of added lysate and were associated with the immunobeads. Altogether, the data demonstrated reduction of PanK1 and PanK2 in brain, without compensation by elevation of PanK3 protein expression.
Fig. 2.
Immunoprecipitation and immunoblots of wild-type (WT) and Pank1−/− Syn-Pank2−/− (dKO) brains. Forebrain lysates were prepared from dKO and age-matched WT animals at P19-P20. Numbered lanes 1-3 contain immunoprecipitates from WT brains (n=3), and numbered lanes 4-6 contain immunoprecipitates from dKO brains (n=3). A. PanK proteins were immunoprecipitated from the lysates using a pan-reactive anti-PanK antibody, fractionated by SDS-PAGE and immunoblotted with the same anti-PANK antibody as described in Materials and methods. The far-left lane contains lysates from HEK 293T cells that overexpressed mouse PanK1α (61 kDa), PanK2 (48 kDa) and PanK3 (41 kDa) proteins as indicated and the 2nd lane from left contains purified recombinant mouse PanK1β (41 kDa). Pank1β and PanK3 protein standards have the same molecular size and were loaded onto gels separately. The 3rd lane from left contains Protein A-Sepharose beads alone without lysate. Non-specific immunereactive band (ns) detected at 55 kDa (immunoglobin) are indicated with an asterisk (*). B. Immunoblot with anti-PanK2 antibody following the identical immunoprecipitation described in panel A of WT and dKO forebrain lysates age P19-P20. C. The brain lysates (20 μg protein per lane) that corresponded to the numbered immunoprecipitates in panels A and B stained with Coomassie Brilliant Blue (CBB) to indicate equivalent protein input in the immunoprecipitation assay. D. Quantification of the PanK2 protein in the dKO forebrains levels as % of WT forebrains shown in panel B. The immunoprecipitation-immunoblot assay was performed independently with brain lysates from 6 mice of each genotype and the data represent the mean ± SEM. Statistical significance values (shown in red) was calculated using the parametric t-test comparing the band intensities of the WT and the dKO.
Fig. 3.
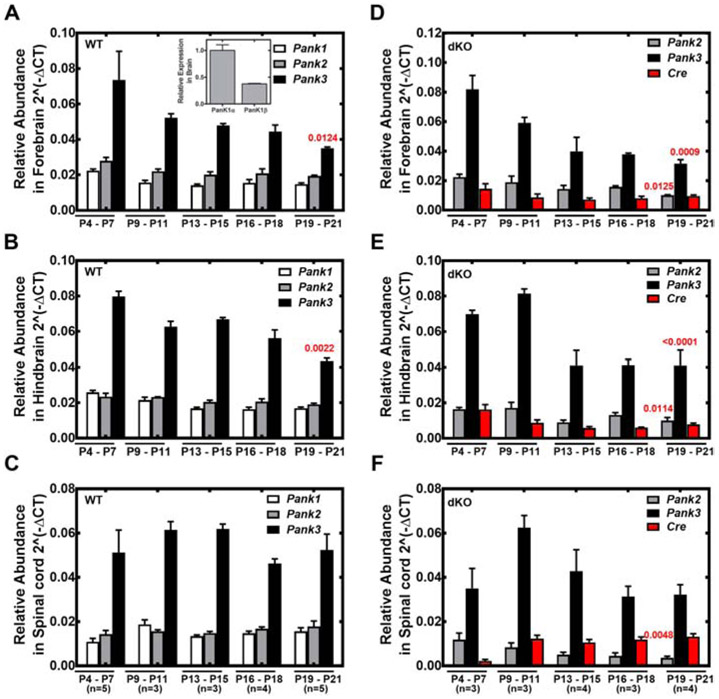
Relative abundance of Pank1, Pank2 and Pank3 gene transcripts during postnatal development of wild-type (WT) mice and Pank1−/− Syn-Pank2−/− (dKO) mice. RNA was extracted from mice at P4-P7, P9-P11, P13-P15, P16-P18, P19-P21 from forebrain, hindbrain and spinal cord. RT-qPCR analysis was performed as described in Materials and methods. The numbers of mice in each group are indicated below panels C and F in parenthesis. A-C. Relative abundance of Pank1, Pank2 and Pank3 transcripts in WT mice. D-F. Relative abundance of Pank2, Pank3 and Cre transcripts in dKO mice. A, D. Forebrain; B,E. Hindbrain; C,F. Spinal cord. Panel A, Inset. Relative abundance of Pank1α and Pank1β in brain. Data represent the mean ± SEM from triplicate determinations with independent samples. Statistical significance values (shown in red) were calculated using the nonparametric t-test comparing the Pank values between P4-11 and P13-21.
3.2. Analysis of PanK expression during postnatal development
Expression of Pank1, Pank2 and Pank3 transcripts in wild-type forebrain, hindbrain and spinal cord during postnatal development (P4-P21) were determined (Fig. 3A-C). Pank3 transcripts were the most abundant in the CNS throughout this period, and the expression levels of Pank1 and Pank2 were equivalent to each other and 2- to 5-fold less compared to Pank3. Pank1α transcripts were consistently in greater abundance relative to Pank1β transcripts (inset Fig. 3A). Overall there was a trending decrease in brain Pank3 transcripts through the postnatal period until weaning age at P21 when Pank3 expression was reduced about 20% in brain. Although Pank3 transcripts were most abundant, the relative amount of PanK3 protein was less compared to the PanK1α protein in the immunoblots (Fig. 2), suggesting a greater turnover rate for the PanK3 protein compared to PanK1α and PanK2 proteins. By comparison, the relative abundance of Pank2, Pank3 and Cre transcripts were monitored in the Pank1−/−Syn-Pank2−/− brain and spinal cord (Fig. 3D-F). The synapsin1-driven Cre transgene was maintained as hemizygous in the breeding scheme and its expression was evident through the postnatal period in forebrain and hindbrain, and became more prominent in spinal cord during progression through the postnatal period. Pank2 transcripts of Pank1−/−Syn-Pank2−/− mice decreased starting at P9-11 and thereafter in forebrain, hindbrain and spinal cord (Fig. 4A-C). Loss of Pank2 expression was most noticeable in spinal cord where the level was reduced to about 15% of wild-type levels at P21 (Fig. 4C). Loss of Pank2 expression in forebrain and hindbrain was more variable and ultimately decreased about 50% by P21 (Fig. 4A and 4B). Pank3 expression in the Pank1−/−Syn-Pank2−/− animals was not significantly different from wild-type levels during postnatal development and Pank1 expression was absent (Fig. 3D-F). The lower levels of Pank2 transcripts that remained in the Pank1−/−Syn-Pank2−/− brain and spinal cord tissues represented the contributions of non-neuronal cell types within the CNS and possibly incomplete deletion by the synapsin1-driven Cre recombinase activity. Pank1 was the most abundant isoform in the liver followed by Pank3 and then Pank2 (Fig. S3). A minor, but significant decrease in Pank2 expression in liver was noted although Cre transcript expression in liver of the Pank1−/−Syn-Pank2−/− animals was below the limits of detection by RT-qPCR (data not shown). The levels of the housekeeping gene GAPDH were constant in the different postnatal days in forebrain, hindbrain, spine and liver in both matched control wild-type and Pank1−/−Syn-Pank2−/− mice as shown in Figure S8. Taken together, these data indicate that Pank2 expression became depressed starting at about P9-11 and continued to decrease until about P15 in the brain and P20 in the spinal cord.
3.3. CoA levels in brain and spinal cord
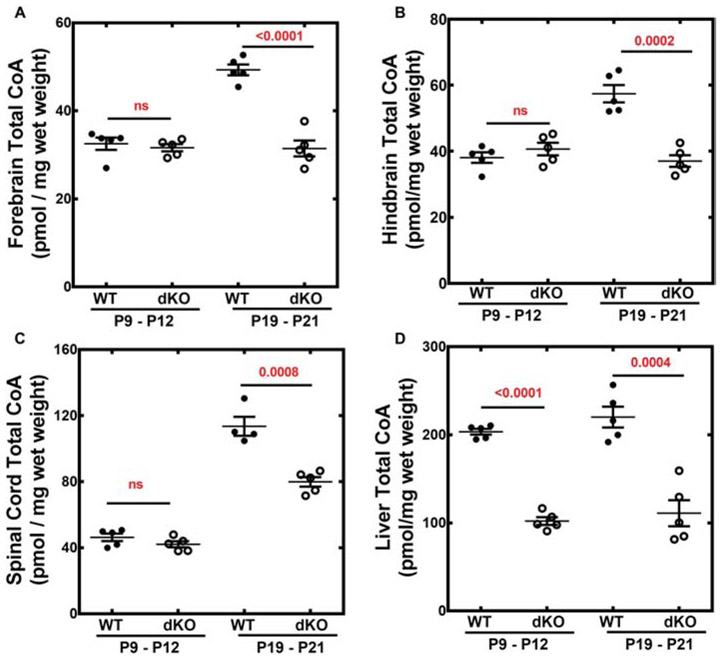
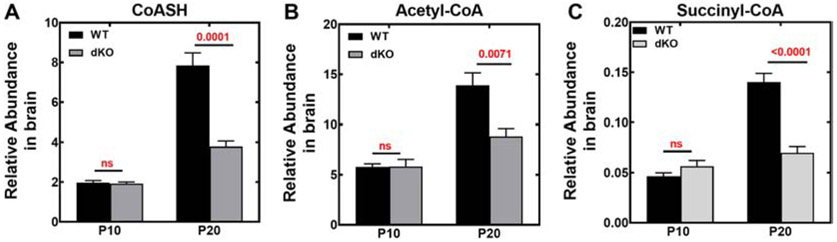
Total CoA levels in wild-type brain and spinal cord increased during postnatal development between P9-12 and P19-21 (Fig. 5A-C). Forebrain CoA increased about 40%, hindbrain CoA increased almost 30% and spinal cord CoA increased 3-fold during wild-type murine development. In contrast, total CoA levels in matched control wild-type liver were consistently about 200 pmol/mg wet weight through the pre-weaning period (Fig. 5D). Liver total CoAs in adult wild-type mice average about 100-120 pmol/mg wet weight [25] and the higher levels during the postnatal period may be due, at least in part, to the ketogenic nature of the pre-weaning diet. At P9-12, the total CoAs in the Pank1−/−Syn-Pank2−/− brains were the same as found in wild-type animals but failed to increase during the period between P9-P12 and P19-21. At the later postnatal time, the Pank1−/−Syn-Pank2−/− brain and spinal cord total CoA levels were ≥ 30% lower compared to wild-type levels. In contrast, the total CoA levels in the Pank1−/−Syn-Pank2−/− livers were reduced about 45% at both the earlier P9-12 and the later P19-21, similar to the levels for the systemic Pank1−/− mouse during the early postnatal period [18]. Analysis of the free CoA (i.e., CoASH) and CoA thioester distribution in the brains of male and female matched control wild-type and Pank1−/−Syn-Pank2−/− mice using mass spectrometry revealed substantial depression of the free CoA (Fig. 6A) and CoA short-chain thioesters at the late stage postnatal period (Fig. 6B-C). However, at the earlier P10 period, the Pank1−/−Syn-Pank2−/− mice of both genders had free CoA and CoA thioester levels equivalent with those in the matched wild-type mice. At the later time, acetyl-CoA levels were 25-40% lower, depending on the gender (Fig. 6B), and succinyl-CoA levels (Fig. 6C) were about 50% lower for both genders in the Pank1−/−Syn-Pank2−/− animals at P19-21. Acetyl-CoA and succinyl-CoA are the two CoA thioesters that are intrinsic components of the tricarboxylic acid cycle that is central to intermediary metabolism and mitochondrial energy production, and succinyl-CoA is also an essential precursor for heme biosynthesis. Acetyl-CoA is the most abundant CoA thioester and has a central role in multiple metabolic and signaling pathways that can impact synaptic function. Additional short-chain CoA thioesters in the Pank1−/−Syn-Pank2−/− brain were depressed at the late postnatal stage and the relative levels are shown in Fig. S4. In contrast, the intermediate- and long-chain CoA thioester levels were not different from wild-type or trended higher in the Pank1−/−Syn-Pank2−/− brains compared to wild-type levels at the late stage of the pre-weaning period as shown in the complete acyl-CoA profiles (Fig. S4). The free CoA levels (Fig. 6A) showed a substantial reduction for both Pank1−/−Syn-Pank2−/− genders at the later stage during the pre-weaning period, but not at the earlier stage. Note that the data in Fig. 6 are not quantitative absolute values but were determined relative to an internal [13C]acetyl-CoA standard and underestimate the true free CoA levels. Nevertheless, the differences between the relative free CoA levels in brains and spinal cords of wild-type and Pank1−/−Syn-Pank2−/− mice as determined by HPLC/MS/MS are substantial and significant. Thus, the reduction in total CoA in the P19-21 Pank1−/−Syn-Pank2−/− brains (Fig. 5) was reflected in multiple CoA species, including free CoA, and the short-chain acyl-CoAs which include acetyl-CoAs and succinyl-CoAs.
Fig. 5.
Total CoA from Pank1−/−Syn-Pank2−/− (dKO) and wild-type (WT) tissues. Total CoA was extracted and quantified as described in Materials and methods from tissues of 5 dKO and 5 WT mice at P9-12 and P19-P21, with the exception of WT spinal cord at P19-21 with n=4. A. Forebrain total CoA; B. Hindbrain total CoA; C. Spinal cord total CoA; D. Liver total CoA. The mean values ± SEM are indicated and statistical significance was calculated using the parametric t-test comparing age matched WT and dKO mice, the values for which are shown in red.
Fig. 6.
CoASH, acetyl-CoA and succinyl-CoA levels. CoASH (free CoA) and CoA thioesters in hindbrain of P10 and P21 wild-type (WT) and Pank1−/−Syn-Pank2−/− (dKO) mice were quantified relative to the [13C]acetyl CoA internal standard using mass spectrometry. n=6 mice per group, 3 males and 3 females. A. CoASH; B. Acetyl-CoA; C. Succinyl-CoA. The mean values ± SEM are indicated and statistical significance was calculated using parametric t-test comparing age-matched WT and dKO mice, the values for which are shown in red.
Blood chemistries revealed lower glucose and lactate levels and elevated ketones in the Pank1−/−Syn-Pank2−/− mice at P19-21 mice compared to age-matched wild-type animals (Fig. S5A-C). The Pank1−/−Syn-Pank2−/− glucose values were similar to those obtained in the Pank1−/− mouse [20], indicating that the changes largely reflected the loss of Pank1 expression in the liver. These data suggested that the lower circulating glucose may have contributed to the phenotype of the Pank1−/−Syn-Pank2−/− animals.
Altered NADH-related activities were observed in cultured neuronal cells derived from PKAN patient fibroblasts [10] and NADH levels were reduced in the livers of Pank1−/−Pank2−/− mice [18]. So we measured the NAD and NADH levels as indicators of metabolic imbalance in brains of the Pank1−/−Syn-Pank2−/− animals and found no significant difference from the levels in matched control wild-type mice at P19-P21, although a non-significant but trending imbalance in the NAD/NADH ratio was observed earlier during development at P9-12 (Fig. S6A-C). Brain ATP levels in the Pank1−/−Syn-Pank2−/− animals also trended toward lower values at P9-P12, but were not significantly different from wild-type values (Fig. S6D). By P19-21, both the NAD/NADH ratios and ATP levels in the Pank1−/−Syn-Pank2−/− brains became normalized in comparison with the levels in wild-type brains, suggesting that any imbalance in these metabolic indicators preceded the development of the forelimb deformity and gait abnormality (see below).
3.4. Phenotype of Pank1−/−Syn-Pank2−/− mouse model
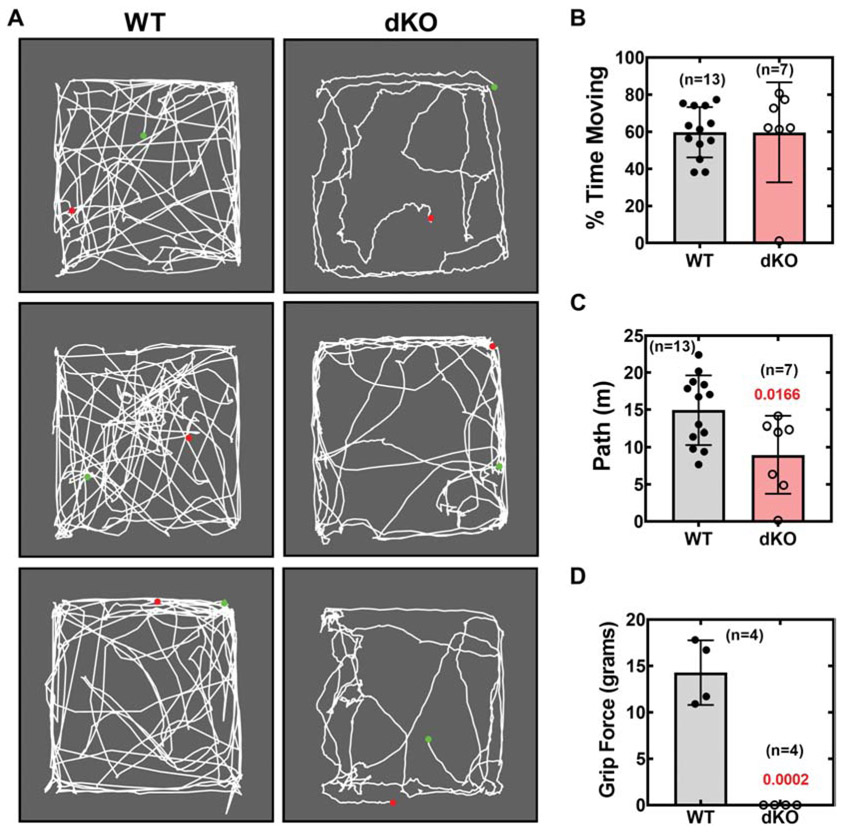
The lack of an increase in brain CoA correlated with development of an overt forelimb deformity associated with weakness. An abnormal dystonia-like flexing of the forelimb and wrist was evident after weight loss in 100% of the Pank1−/−Syn-Pank2−/− animals and this feature prevented the mice from having a normal gait (Fig. 7 and video Fig. S7). The forelimb deformity of the Pank1−/−Syn-Pank2−/− mice was not associated with any anatomical evidence of muscle damage nor did examination of the brain, spinal cord and dorsal root ganglion using hematoxylin and eosin staining indicate any gross abnormalities. Throughout the neuraxis there were no regions of reduced density nor any evidence of pyknotic nuclei that would suggest ongoing cell death (data not shown). About 5% of the longer-lived Pank1−/−Syn-Pank2−/− animals exhibited dragging of the hind limbs prior to death in addition to the forelimb phenotype. The Pank1−/−Syn-Pank2−/− animals were less active compared to matched wild-type mice as demonstrated in an open field test (Fig. 7A). Quantification of the data obtained from the video record of the open field test indicated that the distance traveled by the Pank1−/−Syn-Pank2−/− animals was less compared to the matched wild-type animals at P20 (Fig. 7C) although the per cent movement was not significantly different (Fig 7B). Note that intermittent shaking was associated with pauses in Pank1−/−Syn-Pank2−/− stationary animals as was evident from the mapping pattern of the movement (Fig. 7A) and was included in the calculation of per cent movement. Forelimb grip strength was not measurable in the late postnatal Pank1−/−Syn-Pank2−/− mice (Fig. 7D). These features were unlike the Pank1−/− mice that did not show any overt behavioral phenotype and had a normal lifespan, indicating that the CoA deficiency in brain and spinal cord, in conjunction with a mild metabolic imbalance associated with systemic Pank1 deletion, resulted in a movement disorder.
Fig. 7.
Evaluation of locomotor activity and forelimb grip strength. The open field test was performed on wild-type (WT) and Pank1−/−Syn-Pank2−/− (dKO) mice, ages P19-P20. Mice were allowed to roam freely in a 16.5 X 14.5-inch open arena for 5 minutes. Movement of each individual mouse was recorded with a video camera, with start position indicated by the green dot and stop position indicated by the red dot. The distance traveled and percentage of time spent moving were quantified using Field2100 software. A. Video record for 3 WT and 3 dKO animals. B. Calculated values for percentage (%) of 5-min trial time spent moving. C. Calculated values for distance traveled by the WT and dKO mice in 5 minutes. D. Forelimb grip strength test for WT and dKO mice at P19. The numbers of mice in each group are indicated on the graphs. The data represent the mean values for each genotype ± SEM and statistical significance was calculated using the parametric t-test comparing age-matched WT and dKO mice, the values for which are shown in red.
3.5. Gene Expression Analysis
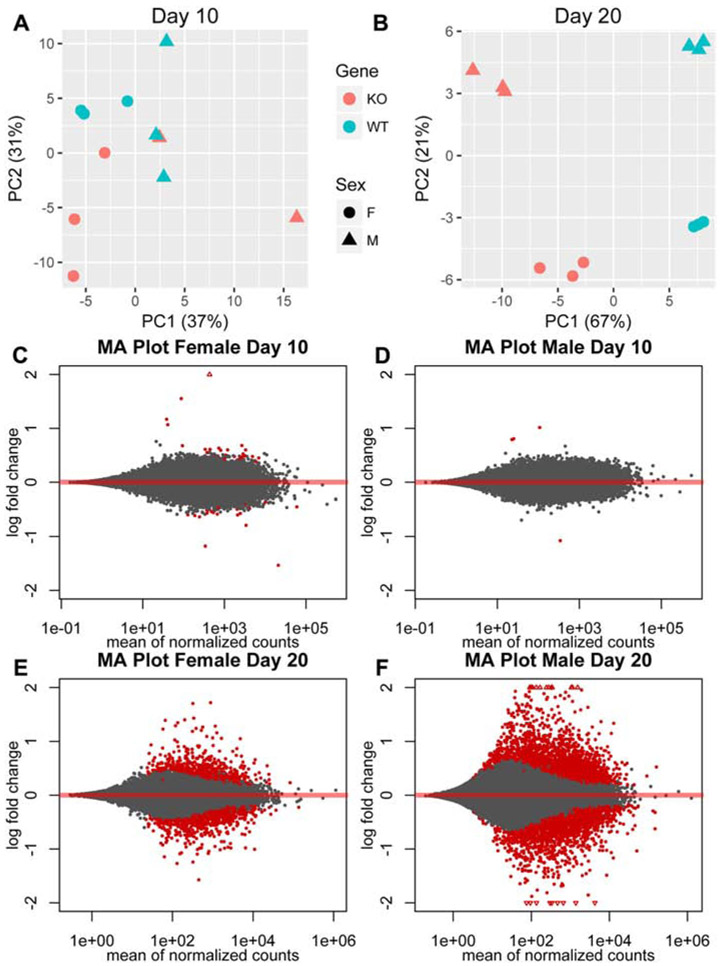
Brain gene expression analysis by RNAseq was performed at P10 and P20 and results from the Pank1−/−Syn-Pank2−/− mice were compared with those from gender- and age-matched wild-type mice of the same genetic lineage. The brain gene expression profiling of the Pank1−/−Syn-Pank2−/− mouse model represents not only the results of a reduction of Pank2 expression in neurons, but also a loss of Pank1 expression in neurons plus additional cell types in the brain, such as glial cells. Comparison was made with matched control wild-type brain in order to include those non-neuronal cell types that may contribute to the phenotype of the mouse model. Principal component analysis (PCA) of the data assessed the global change in gene profiles and evaluated the robustness of the experiment. The wild-type and Pank1−/−Syn-Pank2−/− mice of either gender did not segregate into distinct groups at P10, indicating that there were few differences between their gene expression profiles at that point of postnatal development (Fig. 8A). On the other hand, four distinct data sets, segregated according to genotype and gender, were evident after analysis for day P20, and indicated significant differences in gene expression profiles between the [wild-type female], [wild-type male], [Pank1−/−Syn-Pank2−/− female] and [Pank1−/−Syn-Pank2−/− male] brains (Fig. 8B). The two different genotypes diverged primarily in the first principle component, which explained 67% of the variance. The two genders diverged primarily in the second principle component, which explained 21% of the variance. The clear segregation of the groups based on genotype and gender, and the high amount of variance explained by the first and second principle components, are consistent with robust differences between the genotypes and genders at P20. MA plot analysis, an application of a Bland-Altman plot for visual representation of genomic data, compared the Pank1−/−Syn-Pank2−/− with wild-type mice for each gender at day 10 and day 20 and corroborated the PCA analysis. Relatively few changes in gene expression were observed between the Pank1−/−Syn-Pank2−/− and wild-type mice at P10 (Fig. 8C-D). A significant increase in differential gene expression was found at P20, particularly for the male Pank1−/−Syn-Pank2−/− mice (Fig. 8E-F). These gene expression data have been deposited in NCBI’s Gene Expression Omnibus [30] and are accessible through GEO series accession number GSE130705 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130705).
Fig. 8.
Differential gene expression analysis of Pank1−/−Syn-Pank2−/− (dKO) compared to wild-type (WT) mice. RNAseq analysis was performed using RNA extracted from 3 males and 3 females each for dKO and WT mice at postnatal days P9-P12 or P19-P21. A. Principle component analysis for gene expression at P9-P12. B. Principle component analysis for gene expression at P19-21. Blue indicates WT, red indicates dKO, circle (●) indicates female, triangle (▲) indicates male. C. MA plot of gene expression for female dKO mice compared to female WT mice at P9-P12. D. MA plot of gene expression for male dKO mice compared to male WT mice at P9-P12. E. MA plot of gene expression for female dKO mice compared to female WT mice at P19-P21. F. MA plot of gene expression of male dKO mice compared to male WT mice at P19-P21. Statistically significant differences between WT and dKO gene expression profiles were observed at P19-20 for both sexes.
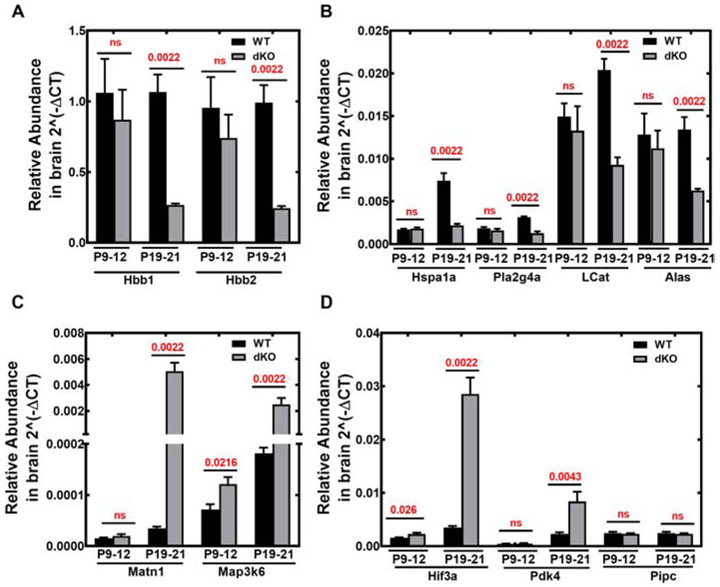
The top ten differentially expressed genes within the day P20 Pank1−/−Syn-Pank2−/− brains included Alas, Hbb1, Hbb2, Hspa1a, Pla2g4a, Lcat, Matn, Map3k6, Pdk4, and Hif3a. These candidate genes identified by RNA-seq were confirmed and quantified by RT-qPCR using specific primer sets (Table S2; Fig. 9). The genes involved in the pathway of hemoglobin synthesis, Hbb1, Hbb2, and Alas were downregulated (Fig. 9A-B). Among other genes that were downregulated we confirmed reduced expression of the heat shock protein Hsp70 (Hspa1a), phospholipase A2 (Pla2g4a) and lecithin cholesterol acyl transferase (Lcat) (Fig. 9B). The genes that were upregulated were matrilin1 cartilage matrix protein (Matn), mitogen activated protein kinase 6 (Map3k6), hypoxia inducible factor 3a (Hif3a), and pyruvate dehydrogenase kinase 4 (Pdk4) (Fig. 9C-D). The Ppic (cyclophilin C) gene was included in the data set as a negative control that showed no change between the Pank1−/−Syn-Pank2−/− and wild-type brains (Fig. 9D). The Hbb1, Hbb2, and Alas genes were among the most highly expressed in brain, and the greatest increases occurred in the expression of Hif3a (7-fold elevation), Map3k6 (10-fold elevation) and Matn1 (126-fold elevation). Elevation of pyruvate dehydrogenase kinase 4 (Pdk4) expression is characteristic of CoA deficiency induced by treatment with HoPan, a PanK inhibitor [31], and Pdk4 expression increased about 4-fold in the Pank1−/−Syn-Pank2−/− brains compared to matched wild-type samples. Analysis of gene pathway and network enrichment identified the aminolevulinic acid synthase (Alas) and beta-hemoglobin subunit genes (Hbb1 and Hbb2) which were down-regulated, and the hypoxia-inducible factor 3a (Hif3a) gene which was up-regulated, and altogether this combination suggested a decreased molecular capacity to carry oxygen. In general, the downregulated gene transcripts represented potential causative factors for phenotype development, whereas several of the upregulated gene transcripts represented possible adaptive responses to stress elicited by oxygen or glucose deprivation.
Fig. 9.
Real time-qPCR evaluation of top 10 differential brain transcripts identified by RNAseq. Expression levels of candidate genes were quantified for Pank1−/−Syn-Pank2−/− (dKO) and matched wild-type (WT) brains using RT-qPCR as described in Materials and methods. RNA was isolated from brains of 3 males and 3 females each for dKO and WT mice at P9-P12 or P19-P21. A. Hbb1 and Hbb2 encode proteins in hemoglobin synthesis. B. Hspa1a, Pla2g4a, LCat and Alas genes encode proteins for heat-shock protein 70, an isoform of phospholipase A2, lecithin-cholesterol acyltransferase, and aminolevulinic acid synthase, respectively. C. Matn and Ma3k6 encode matrilin cartilage protein and a mitogen activated protein kinase, respectively. D. Hif3a, Pdk4 and Pipc encode hypoxia inducible factor 3a, pyruvate dehydrogenase kinase 4, and cyclophilin C, respectively. Pipc was included in the analysis as a negative control. The data represents the combined data from 3 males and 3 females, the mean ± SEM of triplicate determinations with independent samples and statistical significance was calculated using non-parametric t-test comparing age matched WT and dKO mice, the values for which are shown in red.
Based on these results, we measured the transcript levels of the same ten differentially expressed genes in a separate, longer-lived mouse model in which both the Pank1 and Pank2 genes were deleted in neurons only, thereby restricting the CoA deficiency to neurons. The Syn Pank1−/−Syn-Pank2−/− animals have a median survival of 52 days, substantially reduced expression of both Pank1 and Pank2 in brain (Fig. S9A), reduced brain CoA and a substantial movement disorder [25], The Hspa1a and Lcat transcripts were reduced significantly, and the Hif3a transcripts were elevated significantly, whereas the Hbb transcripts trended lower in Syn Pank1−/−Syn-Pank2−/− animals compared to matched control wild-type animals but the differences were not significant (Fig. S9C-F). These data suggested that the systemic deletion of Pank1 in all brain cell types, e.g., glia as well as neurons, in the current mouse model that is described in this report, may contribute to and possibly account for the reductions in Pla2g4a, Alas, Matn1, Map3k6, and Pdk4 transcripts.
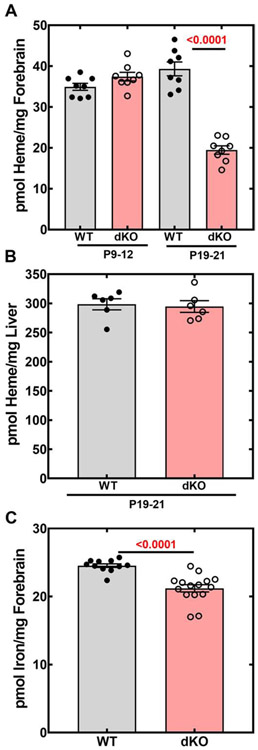
3.6. Heme and Iron Levels
The gene expression profiling, together with the data indicating a reduced amount of succinyl-CoA, suggested a heme deficit in the brains of the Pank1−/−Syn-Pank2−/− mice. Heme levels were determined by an HPLC method in brains and livers of Pank1−/−Syn-Pank2−/− and matched control wild-type mice at the earlier (P9-12) and later (P19-21) postnatal stages (Fig. 10A-B). The amount of heme was not significantly different in the Pank1−/−Syn-Pank2−/− brains at the early stage, but heme was reduced about 50% in the Pank1−/−Syn-Pank2−/− mice at the later stage that corresponded with the appearance of the movement disorder (Fig. 10A). By contrast, liver heme levels were equivalent in mice of both genotypes at P19-21 (Fig. 10B), indicating that the heme deficit was not global and was restricted to the brain. Similarly, the heme levels in the separate, longer-lived Syn-Pank1−/−Syn-Pank2−/− mouse model were also significantly lower at P45 (Fig. S9B) when the animals demonstrated a clear deficit in locomotion [25].
Fig. 10.
Heme levels in brain and liver; iron levels in brain. Heme was measured in forebrain and liver by HPLC from wild-type (WT) and Pank1−/−Syn-Pank2−/− (dKO) mice as described in Materials and methods. Iron levels from the forebrain in the WT and dKO mice at P19-21 were assayed as described in Materials and methods. A. Heme levels in forebrain, n=8 mice in each group. B. Heme levels in liver, n= 6 mice in each group. C. Iron levels in brain, n=15 for WT and n=11 for dKO mice at P19-21. The data represent the mean values for each genotype ± SEM and statistical significance was calculated using the parametric t-test comparing age matched WT and dKO mice for the heme assay and the nonparametric t-test for the iron measurement, the values for which are shown in red.
Iron is sequestered within the heme porphyrin ring (Fig. 11) and brain iron accumulation is a characteristic of PKAN disease [1], so iron levels were measured in the P19-21 forebrains. Total iron levels were about 12% lower in the Pank1−/−Syn-Pank2−/− mice compared to matched wild-type mice (Fig. 10C). This result contrasts with the MRI imaging results of PKAN patients that show an accumulation of iron in the basal ganglia [1].
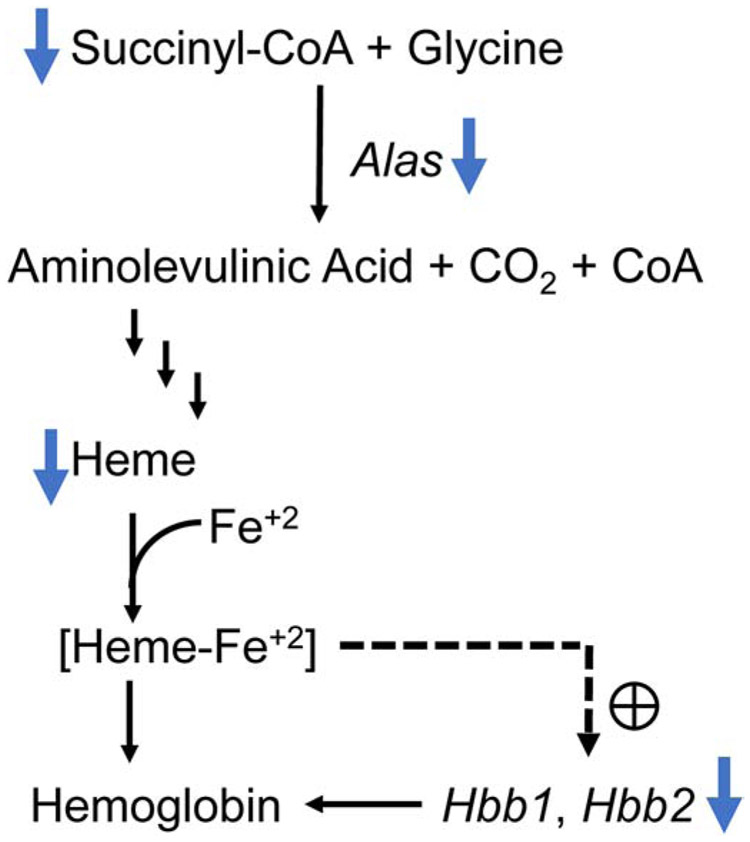
Fig. 11.
Pathway depicting the effect of reduced succinyl-CoA on heme and hemoglobin synthesis. Lower levels of succinyl CoA and heme were observed in the brains of the Pank1−/−Syn-Pank2−/− mice together with fewer transcripts for Alas, Hbb1 and Hbb2. A decrease in succinyl CoA, together with reduced expression of the aminolevulinic acid synthase encoded by the Alas gene leads to decreased amount of the metabolic intermediate, aminolevulinic acid, which, in turn, leads to decreased synthesis of heme. [Heme-Fe+2] is a positive regulator of transcription of Hbb1 and Hbb2 that yield the β subunits of hemoglobin. Depression of this pathway results in reduced active hemoglobin in the brain. The blue bold arrows indicate the observed results in the Pank1−/−Syn-Pank2−/− mice compared to the age-matched controls, the dashed arrow and the plus sign indicate positive regulation of [heme-Fe+2] on Hbb1 and Hbb2 expression.
4. Discussion
The generation of the Pank1−/−Syn-Pank2−/− mice enabled the evaluation of brain CoA deficiency. The Pank1−/−Syn-Pank2−/− mice had similar CoA levels compared to matched wild-type littermates at P9-P12, but had reduced CoA in the brain and spinal cord at postnatal days P19-P21, the time when the Pank1−/−Syn-Pank2−/− mice consistently developed a forelimb deformity similar to dystonia with little to no grip strength (Fig. 7A) and abnormal gait (Fig. S7 video). The wild-type brain CoA increased between P9 and P19, but the Pank1−/−Syn-Pank2−/− brain CoA levels failed to increase showing that the mice were unable to meet the demand for CoA synthesis during brain development. This mouse model exhibits some characteristics of PKAN but differences between mouse and human postnatal development, lifespan, metabolism and tissue gene expression mean that the model does not recapitulate all aspects of PKAN disease. The CoA deficiency in the CNS is severe in the model due to systemic deletion of Pank1 expression in addition to loss of Pank2 expression in neurons, and likely more severe compared to PKAN patients. To the best of our knowledge, human PKAN patients do not carry activity-reducing mutations in PANK1 as well as PANK2, and the lifespan of a typical PKAN patient extends beyond the first signs of disease in childhood into late adolescence or early adulthood. By contrast, the Pank1−/−Syn-Pank2−/− animals expire before weaning and prior to adolescence, as referenced by sexual maturity which occurs at age 6-8 weeks. The neuronal deletion of Pank2 in the Pank1−/−Syn-Pank2−/− mouse also impacted the spinal cord and no reports of spinal cord involvement or pathology in PKAN patients have been published. Nevertheless, the Pank1−/−Syn-Pank2−/− mouse model of brain CoA deficiency reveals measurable differences at the molecular level, together with a clear phenotype of motor dysfunction, that are consistent with aspects of PKAN clinical features and post mortem PKAN brain pathology that suggests cellular hypoxic or ischemic injury [32].
The Pank1−/−Syn-Pank2−/− mice did not accumulate brain iron which is a hallmark of the human disease, although the amount of iron accumulation in the brains of PKAN patients is variable [33] and dependent on disease progression [34]. By the time clear neurologic features are present in PKAN, iron may be visible only later in the disease course [35], suggesting that the molecular events, including heme deficiency, that are associated with the CoA deficiency and motor dysfunction in the Pank1−/−Syn-Pank2−/− model may precede an elevation of brain iron. Indeed, a recent publication indicated that the adult Pank2−/− mouse model had about 27% higher iron, as determined by inductively coupled plasma mass spectrometry, but in the globus pallidus only [36], whereas our measurements were made using whole brain. Unfortunately, the adult Pank2−/− mouse model does not exhibit any problems with motor skills nor any CoA deficiency [17] so it is not clear whether iron accumulation contributes to PKAN disease or is an epiphenomenon. Iron deposition is not unique to PKAN and has been observed in the brain with normal aging and in many chronic neurologic disorders at the later stages, including Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis [37-41].
The correspondence between the dysfunctional locomotion and reduced CoA in the Pank1−/−Syn-Pank2−/− brain and spinal cord suggests that the alterations in brain gene expression and heme production that accompanied the phenotype are informative for understanding the consequences of CoA deficiency. Very few differences in gene expression were found in comparative analysis of wild-type and Pank1−/−Syn-Pank2−/− brain transcripts at P10 (Fig. 8). On the other hand, significant changes in a subset of transcripts were observed in the P20 brain, in both males and females. Reduction of Hbb1 and Hbb2 expression in the Pank1−/−Syn-Pank2−/− mice (Fig. 9) was similar to reduction of Hbb gene expression in other neurodegenerative diseases like Alzheimer’s and Parkinson’s disease [42, 43]. A 50% reduction in the Alas (aminolevulinic acid synthase) transcript was also observed in the Pank1−/−Syn-Pank2−/− brain compared to the wild type brain (Fig. 9) and succinyl-CoA, the precursor to heme, was depressed 50% in the Pank1−/−Syn-Pank2−/− brain (Fig. 6). Measurement of heme levels confirmed a deficit in synthesis in the Pank1−/−Syn-Pank2−/− brain (Fig. 10). An association between reduced brain CoA, reduced brain heme and a movement disorder was confirmed in a separate mouse model, the Syn-Pank1−/−Syn-Pank2−/− model (Fig. S9) which has deletions of both the Pank1 and Pank2 genes restricted to neurons only [25]. Altogether, these data indicate that reduced brain CoA had a major impact on the pathway for synthesis of brain hemoglobin, suggesting reduced oxygen availability and/or reduced redox regulation. Note that the earlier Pank knockout mouse models, i.e., the Pank2−/− model and the Pank1−/−Pank2−/− double systemic knockout, developed brain CoA deficiencies by P10 [18] but did not exhibit a consistent movement disorder because these animals had not yet reached the developmental milestone of consistent locomotion which occurs at P15 [44]. The CoA returned to wild-type levels later during development of the Pank2−/− animals [17] and the Pank1−/−Pank2−/− double systemic knockout animals died between P10-P17, so a correlation between movement, brain CoA deficiency and heme reduction is not possible to evaluate in those earlier models.
As depicted in Fig. 11, succinyl-CoA combines with glycine to yield aminolaevulinic acid mediated by the Alas protein, the rate-limiting step leading to heme [45] which, in turn, positively regulates globin transcription [46]. Heme is the porphyrin cofactor that binds iron and is the prosthetic group that activates the hemoglobin. Oxygen binds to the heme iron in the reduced or ferrous Fe+2 state and hemoglobin expression supports neuronal mitochondrial activity [47]. Heme-containing proteins also include catalase, peroxidases, prostaglandin synthase, guanylate cyclase, nitric oxide synthase, and the microsomal and mitochondrial cytochromes. Thus, heme proteins are involved in mitochondrial redox regulation and cycle between the Fe+2 and Fe+3 states which, in turn, can affect ion-channel gating and neuronal membrane excitability [48]. Reduced hemoglobin mRNA has been observed in the frontal cortex of patients with Alzheimer’s disease [49] and double-labeling immunofluorescence confocal microscopy in Parkinson’s disease showed reduced hemoglobin alpha and beta chain [50]. It has been recognized that insufficient levels of heme may be a factor in mitochondrial and neuronal decay [51] and PKAN patient fibroblasts have a reduced heme content [12, 14]. The results from this study indicate that heme and hemoglobin deficiencies may be a pathological feature of brain CoA deficiency.
Additional genes whose expressions were altered significantly in the postnatal late-stage Pank1−/−Syn-Pank2−/− brains pointed toward cellular stress and the response to cellular stress. Hif3a encodes hypoxia-inducible factor 3α which is a member of the HIF transcription factor family that regulates the cellular adaptive responses to oxygen or glucose deprivation. Hif3a transcript expression was elevated almost 10-fold in the late-stage Pank1−/−Syn-Pank2−/− brains, suggesting that an ischemia-like condition could be associated with CoA deficiency. However, our efforts to demonstrate Hif3a protein levels in the late-stage Pank1−/−Syn-Pank2−/− brains were negative (data not shown). Recent reports show that transient expression of human HIF1A protein drives HIF3A expression in human endothelial cells [52] and that the HIF1A protein is stabilized by deacetylation via SIRT1 [53]. We speculate that the Hif3a transcript elevation in the Pank1−/−Syn-Pank2−/− brains may reflect an upstream event resulting from acetyl-CoA deficiency. We hypothesize that reduced acetyl-CoA levels, as found in the Pank1−/−Syn-Pank2−/− brains, would result in reduced HIF1A acetylation and thereby stabilize the HIF1A protein. Indeed, reduced CoA has been shown to affect global protein acetylation in Drosophila and human cell models [54]. Deacetylated, stabilized HIF1A could theoretically promote Hif3a transcript expression. Interestingly, Hif3a transcripts were upregulated in cortical neurons when treated with the anti-Alzheimer iron chelator drug M30 [55], suggesting a link between Hif3a expression and the response to disruption of iron homeostasis.
Hypoxic or ischemic injury within the globus pallidus of PKAN patients was indicated by pathological findings of multiple infarcts and multiple proteinaceous aggregates enriched in apolipoprotein E (ApoE) [32, 56]. Hspa1a encodes Hsp70 protein which is a molecular chaperone and reduced Hspa1a expression in the late postnatal Pank1−/−Syn-Pank2−/− brains (Fig. 9) indicates a potential issue with protein folding. Hspa1a is expressed at low levels in the adult mouse brain [57] and its expression is induced in response to cerebral ischemia [58]. Overexpression studies indicated that Hsp70 can suppress neuropathology and improve motor function in a mouse model of spinocerebellar ataxia type 1 (SCA1) [59]. Reduced Hspa1 expression leads to accumulation of protein aggregates such as ApoE in brain [60, 61]. ApoE is the major cholesterol carrier that plays an important role in injury repair in the brain [62]. Expression of the LCAT gene, encoding lecithin-cholesterol acyl transferase, was substantially reduced in the late postnatal Pank1−/−Syn-Pank2−/− brains (Fig. 9). The LCAT protein catalyzes the esterification of free cholesterol on circulating apolipoproteins such as ApoE to yield mature high-density lipoprotein [63]. Reduced LCAT expression would impede the esterification of 24-hydroxy-cholesterol, the most abundant cholesterol species in the CNS, and, in turn, reduce the stability of ApoE proteins. 24-Hydroxycholesteryl esters are reduced in Parkinson’s disease [64], amyotrophic lateral sclerosis [65] and Alzheimer’s disease [43] and our data are suggestive of the early stages of possible ApoE instability associated with reduced expression of Hsp70 and reduced LCAT activity. In the brains of the late-stage postnatal Pank1−/−Syn-Pank2−/− mice we also measured a decrease in Pla2g4a expression. Pla2g4a encodes a cytosolic phospholipase that catalyzes the first step in the formation of eicosanoid mediators and inflammation [66, 67]. Interestingly, Pla2g4a expression and the associated microRNA Mir-145-5P have been found to be responsive to the long non-coding RNA SNHG14 during cerebral ischemic stroke [68], forging another potential link between CoA depletion and ischemia.
Additional selective changes in gene expression were associated with brain CoA deficiency in the Pank1−/−Syn-Pank2−/− mouse model. Map3k6 expression is upregulated 10-fold in the late-postnatal Pank1−/−Syn-Pank2−/− dKO brains (Fig. 9). The gene encodes a mitogen activated serine threonine protein kinase involved in signal transduction. Map3k6 is involved in neuronal cell death mediated by endoplasmic reticulum stress in amyotrophic lateral sclerosis [69, 70]. Map3k6 transcripts are also upregulated 60-fold in liver in association with severe CoA deficiency in HoPan-treated mice [31]. The upregulated Matn1 gene encodes the matrilin cartilage protein that is involved in the formation of the filamentous network in the extracellular matrix of various tissues. Increased matrilin levels in circulating plasma are believed to lead to thrombosis [71] and we speculate that elevated Matn1 expression may be associated with the infarct formation in the globus pallidus that has been observed in PKAN patients [72]. The upregulation of Pdk4 expression in the late postnatal Pank1−/−Syn-Pank2−/− brains is a hallmark of CoA deficiency [31]. Pdk4 encodes pyruvate dehydrogenase kinase 4 which inactivates pyruvate dehydrogenase by phosphorylation [73]. An increase in Pdk4 is considered an adaptive response to glucose deprivation as observed in a chemical knockout of CoA production in liver [31]. Taken together, the gene expression data from the late postnatal Pank1−/−Syn-Pank2−/− brains are consistent not only with with several features of CoA deficiency but also selected aspects of the pathologies of several neurodegenerative diseases, including PKAN, and may indicate mechanistic commonality.
5. Conclusions
The conditional knockout of Pank2 in neurons, in conjunction with systemic knockout of Pank1 in neurons plus additional cell types, elicited a CoA deficiency in the murine CNS that correlated with a phenotype with some aspects resembling PKAN disease. A deficit in brain heme levels was revealed following assessment of the CoA molecular species distribution and evaluation of gene expression changes in the late postnatal period. Additional changes in gene expression pointed toward similarities with other neurodegenerative diseases. The model provides an informative platform that furthers our understanding of the molecular pathophysiology associated with CoA deficiency in the murine CNS.
Supplementary Material
Fig. S1. Conditional deletion of the Pank2 gene in brain.
Fig. S2. Immunoblot characterization of pan-reactive anti-PanK and specific anti-PanK2 antibodies.
Fig. S3. Relative abundance of Pank1, Pank2 and Pank3 transcripts in Pank1−/−Syn-Pank2−/− and wild-type liver.
Fig. S4. Relative abundance of CoA thioesters in Pank1−/−Syn-Pank2−/− and wild-type hindbrain at P21.
Fig. S5. Plasma metabolite levels in Pank1−/−Syn-Pank2−/− and wild-type mice.
Fig. S6. Hindbrain NAD, NADH and ATP measurements.
Fig. S7. Video - Pank1−/−Syn-Pank2−/− gait.
Fig S8. GAPDH measurements.
Fig S9. RT-qPCR in WT and Pank1−/−Syn-Pank2−/− mice.
Table S1. Primers for genotyping Pank1−/−Syn-Pank2−/− mice.
Table S2. Primers for RT-qPCR.
Table S3. Acyl-CoA mrm values
Highlights.
Novel mouse with systemic deletion of Pank1 and neuronal deletion of Pank2 was derived as a model for Pantothenate Kinase Associated Neurodegeneration.
CoA deficiency in the CNS developed during the late postnatal, preweaning stage and correlated with a consistent movement disorder.
Free CoA and short-chain CoA thioesters, including acetyl-CoA and succinyl-CoA, were reduced.
Gene expression and biochemical analyses revealed reduced heme synthesis in brain due to CoA deficiency.
Alteration of brain gene expression profile was similar to alterations associated with other neurodegenerative disorders.
Acknowledgments
We thank Drs. Jerold Rehg (Department of Pathology, St. Jude Children’s Research Hospital, Memphis, TN, USA) and Richard J. Smeyne (Department of Neuroscience, Thomas Jefferson University, Philadelphia, PA, USA) for pathology assessments. We thank Lois Richmond for animal handling and grip tests, Katie Creed for open field tests, Karen Miller for RT-qPCR, Jina Wang for immunoprecipitation and immunoblotting, and Caroline Pate for plasma and brain metabolite measurements.
Funding
The main body of this work was funded by National Institutes of Health RO1 GM062896 and the American Lebanese Syrian Associated Charities. The supplemental data from the Syn-Pank1−/−Syn-Pank2−/− mouse model was supported by CoA Therapeutics, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
S.J. is a member of the Scientific Advisory Board of CoA Therapeutics, Inc. and a member of the Scientific and Medical Advisory Board of the NBIA Disorders Association. The other authors declare no conflicts of interest with the contents of this article.
References
- [1].Hogarth P, Kurian MA, Gregory A, Csanyi B, Zagustin T, Kmiec T, Wood P, Klucken A, Scalise N, Sofia F, Klopstock T, Zorzi G, Nardocci N, Hayflick SJ, Consensus clinical management guideline for pantothenate kinase-associated neurodegeneration (PKAN), Mol Genet Metab, 120 (2017) 278–287. [DOI] [PubMed] [Google Scholar]
- [2].Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ, A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome, Nat Genet, 28 (2001) 345–349. [DOI] [PubMed] [Google Scholar]
- [3].Zhang YM, Rock CO, Jackowski S, Biochemical properties of human pantothenate kinase 2 isoforms and mutations linked to pantothenate kinase-associated neurodegeneration, J Biol Chem, 281 (2006) 107–114. [DOI] [PubMed] [Google Scholar]
- [4].Kotzbauer PT, Truax AC, Trojanowski JQ, Lee VM, Altered neuronal mitochondrial coenzyme A synthesis in neurodegeneration with brain iron accumulation caused by abnormal processing, stability, and catalytic activity of mutant pantothenate kinase 2, J Neurosci, 25 (2005) 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Leonardi R, Zhang YM, Rock CO, Jackowski S, Coenzyme A: back in action, Prog Lipid Res, 44 (2005) 125–153. [DOI] [PubMed] [Google Scholar]
- [6].Dusi S, Valletta L, Haack TB, Tsuchiya Y, Venco P, Pasqualato S, Goffrini P, Tigano M, Demchenko N, Wieland T, Schwarzmayr T, Strom TM, Invernizzi F, Garavaglia B, Gregory A, Sanford L, Hamada J, Bettencourt C, Houlden H, Chiapparini L, Zorzi G, Kurian MA, Nardocci N, Prokisch H, Hayflick S, Gout I, Tiranti V, Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation, Am J Hum Genet, 94 (2014) 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shumar SA, Fagone P, Alfonso-Pecchio A, Gray JT, Rehg JE, Jackowski S, Leonardi R, Induction of Neuron-Specific Degradation of Coenzyme A Models Pantothenate Kinase-Associated Neurodegeneration by Reducing Motor Coordination in Mice, PLoS One, 10 (2015) e0130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rana A, Seinen E, Siudeja K, Muntendam R, Srinivasan B, van der Want JJ, Hayflick S, Reijngoud DJ, Kayser O, Sibon OC, Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration, Proc Natl Acad Sci U S A, 107 (2010) 6988–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zizioli D, Tiso N, Guglielmi A, Saraceno C, Busolin G, Giuliani R, Khatri D, Monti E, Borsani G, Argenton F, Finazzi D, Knock-down of pantothenate kinase 2 severely affects the development of the nervous and vascular system in zebrafish, providing new insights into PKAN disease, Neurobiol Dis, 85 (2016) 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Arber C, Angelova PR, Wiethoff S, Tsuchiya Y, Mazzacuva F, Preza E, Bhatia KP, Mills K, Gout I, Abramov AY, Hardy J, Duce JA, Houlden H, Wray S, iPSC-derived neuronal models of PANK2-associated neurodegeneration reveal mitochondrial dysfunction contributing to early disease, PLoS One, 12 (2017) e0184104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Alvarez-Cordoba M, Fernandez Khoury A, Villanueva-Paz M, Gomez-Navarro C, Villalon-Garcia I, Suarez-Rivero JM, Povea-Cabello S, de la Mata M, Cotan D, Talaveron-Rey M, Perez-Pulido AJ, Salas JJ, Perez-Villegas EM, Diaz-Quintana A, Armengol JA, Sanchez-Alcazar JA, Pantothenate Rescues Iron Accumulation in Pantothenate Kinase-Associated Neurodegeneration Depending on the Type of Mutation, Mol Neurobiol, 56 (2019) 3638–3656. [DOI] [PubMed] [Google Scholar]
- [12].Santambrogio P, Dusi S, Guaraldo M, Rotundo LI, Broccoli V, Garavaglia B, Tiranti V, Levi S, Mitochondrial iron and energetic dysfunction distinguish fibroblasts and induced neurons from pantothenate kinase-associated neurodegeneration patients, Neurobiol Dis, 81 (2015) 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Campanella A, Privitera D, Guaraldo M, Rovelli E, Barzaghi C, Garavaglia B, Santambrogio P, Cozzi A, Levi S, Skin fibroblasts from pantothenate kinase-associated neurodegeneration patients show altered cellular oxidative status and have defective iron-handling properties, Hum Mol Genet, 21 (2012) 4049–4059. [DOI] [PubMed] [Google Scholar]
- [14].Orellana DI, Santambrogio P, Rubio A, Yekhlef L, Cancellieri C, Dusi S, Giannelli SG, Venco P, Mazzara PG, Cozzi A, Ferrari M, Garavaglia B, Taverna S, Tiranti V, Broccoli V, Levi S, Coenzyme A corrects pathological defects in human neurons of PANK2-associated neurodegeneration, EMBO Mol Med, 8 (2016) 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Poli M, Derosas M, Luscieti S, Cavadini P, Campanella A, Verardi R, Finazzi D, Arosio P, Pantothenate kinase-2 (Pank2) silencing causes cell growth reduction, cell-specific ferroportin upregulation and iron deregulation, Neurobiol Dis, 39 (2010) 204–210. [DOI] [PubMed] [Google Scholar]
- [16].Kuo YM, Duncan JL, Westaway SK, Yang H, Nune G, Xu EY, Hayflick SJ, Gitschier J, Deficiency of pantothenate kinase 2 (Pank2) in mice leads to retinal degeneration and azoospermia, Hum Mol Genet, 14 (2005) 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Leonardi R, Zhang YM, Lykidis A, Rock CO, Jackowski S, Localization and regulation of mouse pantothenate kinase 2, FEBS Lett, 581 (2007) 4639–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Garcia M, Leonardi R, Zhang YM, Rehg JE, Jackowski S, Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism, PLoS One, 7 (2012) e40871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brunetti D, Dusi S, Giordano C, Lamperti C, Morbin M, Fugnanesi V, Marchet S, Fagiolari G, Sibon O, Moggio M, d'Amati G, Tiranti V, Pantethine treatment is effective in recovering the disease phenotype induced by ketogenic diet in a pantothenate kinase-associated neurodegeneration mouse model, Brain, 137 (2014) 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Leonardi R, Rehg JE, Rock CO, Jackowski S, Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state, PLoS One, 5 (2010) e11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dansie LE, Reeves S, Miller K, Zano SP, Frank M, Pate C, Wang J, Jackowski S, Physiological roles of the pantothenate kinases, Biochem Soc T, 42 (2014) 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bettencourt C, Forabosco P, Wiethoff S, Heidari M, Johnstone DM, Botia JA, Collingwood JF, Hardy J, Consortium UKBE, Milward EA, Ryten M, Houlden H, Gene co-expression networks shed light into diseases of brain iron accumulation, Neurobiol Dis, 87 (2016) 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Franklin KBJ, Paxinos G, Paxinos and Franklin's The mouse brain in stereotaxic coordinates, 2013.
- [24].Lykidis A, Murti KG, Jackowski S, Cloning and characterization of a second human CTP:phosphocholine cytidylyltransferase, J Biol Chem, 273 (1998) 14022–14029. [DOI] [PubMed] [Google Scholar]
- [25].Sharma LK, Subramanian C, Yun MK, Frank MW, White SW, Rock CO, Lee RE, Jackowski S, A therapeutic approach to pantothenate kinase associated neurodegeneration, Nature Communications, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Frank MW, Subramanian C, Rock CO, Jackowski S, Quantification of Coenzyme A in Cells and Tissues, J Vis Exp, (2019). [DOI] [PubMed] [Google Scholar]
- [27].Zano SP, Pate C, Frank M, Rock CO, Jackowski S, Correction of a genetic deficiency in pantothenate kinase 1 using phosphopantothenate replacement therapy, Mol Genet Metab, 116 (2015) 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chomczynski P, A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples, Biotechniques, 15 (1993) 532–534, 536-537. [PubMed] [Google Scholar]
- [29].Winer J, Jung CK, Shackel I, Williams PM, Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro, Anal Biochem, 270 (1999) 41–49. [DOI] [PubMed] [Google Scholar]
- [30].Edgar R, Domrachev M, Lash AE, Gene Expression Omnibus: NCBI gene expression and hybridization array data repository, Nucleic Acids Res, 30 (2002) 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang YM, Chohnan S, Virga KG, Stevens RD, Ilkayeva OR, Wenner BR, Bain JR, Newgard CB, Lee RE, Rock CO, Jackowski S, Chemical knockout of pantothenate kinase reveals the metabolic and genetic program responsible for hepatic coenzyme A homeostasis, Chem Biol, 14 (2007) 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Woltjer RL, Reese LC, Richardson BE, Tran H, Green S, Pham T, Chalupsky M, Gabriel I, Light T, Sanford L, Jeong SY, Hamada J, Schwanemann LK, Rogers C, Gregory A, Hogarth P, Hayflick SJ, Pallidal neuronal apolipoprotein E in pantothenate kinase-associated neurodegeneration recapitulates ischemic injury to the globus pallidus, Molecular Genetics and Metabolism, 116 (2015) 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Amaral LL, Gaddikeri S, Chapman PR, Roy R, Gaddikeri RS, Marussi VH, Bag AK, Neurodegeneration with Brain Iron Accumulation: Clinicoradiological Approach to Diagnosis, J Neuroimaging, 25 (2015) 539–551. [DOI] [PubMed] [Google Scholar]
- [34].Nassif D, Pereira JS, Spitz M, Capitao C, Faria A, Neurodegeneration with brain iron accumulation: A case report, Dement Neuropsychol, 10 (2016) 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gregory A, Hayflick SJ, Pantothenate Kinase-Associated Neurodegeneration, in: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A (Eds.) GeneReviews((R)), Seattle (WA), 1993. [PubMed] [Google Scholar]
- [36].Jeong SY, Hogarth P, Placzek A, Gregory AM, Fox R, Zhen D, Hamada J, van der Zwaag M, Lambrechts R, Jin H, Nilsen A, Cobb J, Pham T, Gray N, Ralle M, Duffy M, Schwanemann L, Rai P, Freed A, Wakeman K, Woltjer RL, Sibon OC, Hayflick SJ, 4'-Phosphopantetheine corrects CoA, iron, and dopamine metabolic defects in mammalian models of PKAN, EMBO Mol Med, (2019) e10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stankiewicz J, Panter SS, Neema M, Arora A, Batt CE, Bakshi R, Iron in chronic brain disorders: imaging and neurotherapeutic implications, Neurotherapeutics, 4 (2007) 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moller HE, Bossoni L, Connor JR, Crichton RR, Does MD, Ward RJ, Zecca L, Zucca FA, Ronen I, Iron, Myelin, and the Brain: Neuroimaging Meets Neurobiology, Trends Neurosci, 42 (2019) 384–401. [DOI] [PubMed] [Google Scholar]
- [39].Ingrassia R, Garavaglia B, Memo M, DMT1 Expression and Iron Levels at the Crossroads Between Aging and Neurodegeneration, Front Neurosci, 13 (2019) 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Faissner S, Gold R, Progressive multiple sclerosis: latest therapeutic developments and future directions, Ther Adv Neurol Disord, 12 (2019) 1756286419878323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ammal Kaidery N, Ahuja M, Thomas B, Crosstalk between Nrf2 signaling and mitochondrial function in Parkinson's disease, Mol Cell Neurosci, 101 (2019) 103413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].La Marca V, Maresca B, Spagnuolo MS, Cigliano L, Dal Piaz F, Di Iorio G, Abrescia P, Lecithin-cholesterol acyltransferase in brain: Does oxidative stress influence the 24-hydroxycholesterol esterification?, Neurosci Res, 105 (2016) 19–27. [DOI] [PubMed] [Google Scholar]
- [43].Benussi L, Ghidoni R, Dal Piaz F, Binetti G, Di Iorio G, Abrescia P, The level of 24-Hydroxycholesteryl Esters is an Early Marker of Alzheimer's Disease, J Alzheimers Dis, 56 (2017) 825–833. [DOI] [PubMed] [Google Scholar]
- [44].Hill JM, Lim MA, Stone MM, Developmental Milestones in the Newborn Mouse, in: Gozes I (Ed.) Neuropeptide Techniques, Humana Press, Totowa, NJ, 2008, pp. 131–149. [Google Scholar]
- [45].Burch JS, Marcero JR, Maschek JA, Cox JE, Jackson LK, Medlock AE, Phillips JD, Dailey HA Jr., Glutamine via alpha-ketoglutarate dehydrogenase provides succinyl-CoA for heme synthesis during erythropoiesis, Blood, 132 (2018) 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tahara T, Sun JY, Nakanishi K, Yamamoto M, Mori H, Saito T, Fujita H, Igarashi K, Taketani S, Heme positively regulates the expression of beta-globin at the locus control region via the transcriptional factor Bach1 in erythroid cells, Journal of Biological Chemistry, 279 (2004) 5480–5487. [DOI] [PubMed] [Google Scholar]
- [47].Singhal NK, Alkhayer K, Shelestak J, Clements R, Freeman E, McDonough J, Erythropoietin Upregulates Brain Hemoglobin Expression and Supports Neuronal Mitochondrial Activity, Mol Neurobiol, 55 (2018) 8051–8058. [DOI] [PubMed] [Google Scholar]
- [48].Todorovic SM, Jevtovic-Todorovic V, Redox Regulation of Neuronal Voltage-Gated Calcium Channels, Antioxid Redox Sign, 21 (2014) 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vanni S, Zattoni M, Moda F, Giaccone G, Tagliavini F, Haik S, Deslys JP, Zanusso G, Ironside JW, Carmona M, Ferrer I, Kovacs GG, Legname G, Hemoglobin mRNA Changes in the Frontal Cortex of Patients with Neurodegenerative Diseases, Front Neurosci, 12 (2018) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ferrer I, Gomez A, Carmona M, Huesa G, Porta S, Riera-Codina M, Biagioli M, Gustincich S, Aso E, Neuronal hemoglobin is reduced in Alzheimer's disease, argyrophilic grain disease, Parkinson's disease, and dementia with Lewy bodies, J Alzheimers Dis, 23 (2011) 537–550. [DOI] [PubMed] [Google Scholar]
- [51].Atamna H, Killilea DW, Killilea AN, Ames BN, Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging, P Natl Acad Sci USA, 99 (2002) 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Janaszak-Jasiecka A, Bartoszewska S, Kochan K, Piotrowski A, Kalinowski L, Kamysz W, Ochocka RJ, Bartoszewski R, Collawn JF, miR-429 regulates the transition between Hypoxia-Inducible Factor (HIF)1A and HIF3A expression in human endothelial cells, Sci Rep, 6 (2016) 22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Joo HY, Yun M, Jeong J, Park ER, Shin HJ, Woo SR, Jung JK, Kim YM, Park JJ, Kim J, Lee KH, SIRT1 deacetylates and stabilizes hypoxia-inducible factor-1alpha (HIF-1alpha) via direct interactions during hypoxia, Biochem Biophys Res Commun, 462 (2015) 294–300. [DOI] [PubMed] [Google Scholar]
- [54].Siudeja K, Srinivasan B, Xu L, Rana A, de Jong J, Nollen EA, Jackowski S, Sanford L, Hayflick S, Sibon OC, Impaired Coenzyme A metabolism affects histone and tubulin acetylation in Drosophila and human cell models of pantothenate kinase associated neurodegeneration, EMBO Mol Med, 3 (2011) 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Avramovich-Tirosh Y, Bar-Am O, Amit T, Youdim MB, Weinreb O, Up-regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-target genes in cortical neurons by the novel multifunctional iron chelator anti-Alzheimer drug, M30, Curr Alzheimer Res, 7 (2010) 300–306. [DOI] [PubMed] [Google Scholar]
- [56].Suzuki N, Gradin K, Poellinger L, Yamamoto M, Regulation of hypoxia-inducible gene expression after HIF activation, Exp Cell Res, 356 (2017) 182–186. [DOI] [PubMed] [Google Scholar]
- [57].Tebbenkamp AT, Borchelt DR, Analysis of chaperone mRNA expression in the adult mouse brain by meta analysis of the Allen Brain Atlas, PLoS One, 5 (2010) e13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Soriano MA, Ferrer I, Rodriguezfarre E, Planas AM, Expression of C-Fos and Inducible Hsp-70 Messenger-Rna Following a Transient Episode of Focal Ischemia That Had Nonlethal Effects on the Rat-Brain, Brain Res, 670 (1995) 317–320. [DOI] [PubMed] [Google Scholar]
- [59].Cummings CJ, Sun YL, Opal P, Antalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY, Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice, Human Molecular Genetics, 10 (2001) 1511–1518. [DOI] [PubMed] [Google Scholar]
- [60].Lu RC, Tan MS, Wang H, Xie AM, Yu JT, Tan L, Heat shock protein 70 in Alzheimer's disease, Biomed Res Int, 2014 (2014) 435203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jinwal UK, Akoury E, Abisambra JF, O'Leary JC 3rd, Thompson AD, Blair LJ, Jin Y, Bacon J, Nordhues BA, Cockman M, Zhang J, Li P, Zhang B, Borysov S, Uversky VN, Biernat J, Mandelkow E, Gestwicki JE, Zweckstetter M, Dickey CA, Imbalance of Hsp70 family variants fosters tau accumulation, FASEB J, 27 (2013) 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Giau VV, Bagyinszky E, An SS, Kim SY, Role of apolipoprotein E in neurodegenerative diseases, Neuropsychiatr Dis Treat, 11 (2015) 1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jonas A, Lecithin cholesterol acyltransferase, Biochim Biophys Acta, 1529 (2000) 245–256. [DOI] [PubMed] [Google Scholar]
- [64].Di Natale C, Monaco A, Pedone C, Tessitore A, De Mase A, Tedeschi G, Netti PA, Abrescia P, The level of 24-hydroxycholesteryl esters decreases in plasma of patients with Parkinson's disease, Neurosci Lett, 672 (2018) 108–112. [DOI] [PubMed] [Google Scholar]
- [65].Abdel-Khalik J, Yutuc E, Crick PJ, Gustafsson JA, Warner M, Roman G, Talbot K, Gray E, Griffiths WJ, Turner MR, Wang Y, Defective cholesterol metabolism in amyotrophic lateral sclerosis, J Lipid Res, 58 (2017) 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Law MH, Cotton RG, Berger GE, The role of phospholipases A2 in schizophrenia, Mol Psychiatry, 11 (2006) 547–556. [DOI] [PubMed] [Google Scholar]
- [67].Wei J, Hemmings GP, A study of a genetic association between the PTGS2/PLA2G4A locus and schizophrenia, Prostaglandins Leukot Essent Fatty Acids, 70 (2004) 413–415. [DOI] [PubMed] [Google Scholar]
- [68].Qi X, Shao M, Sun H, Shen Y, Meng D, Huo W, Long non-coding RNA SNHG14 promotes microglia activation by regulating miR-145-5p/PLA2G4A in cerebral infarction, Neuroscience, 348 (2017) 98–106. [DOI] [PubMed] [Google Scholar]
- [69].Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H, ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1, Genes Dev, 22 (2008) 1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hattori K, Naguro I, Runchel C, Ichijo H, The roles of ASK family proteins in stress responses and diseases, Cell Commun Signal, 7 (2009) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Foradori MJ, Chen Q, Fernandez CA, Harper J, Li X, Tsang PC, Langer R, Moses MA, Matrilin-1 is an inhibitor of neovascularization, J Biol Chem, 289 (2014) 14301–14309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kruer MC, The neuropathology of neurodegeneration with brain iron accumulation, Int Rev Neurobiol, 110 (2013) 165–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sugden MC, Holness MJ, Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs, Am J Physiol Endocrinol Metab, 284 (2003) E855–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Conditional deletion of the Pank2 gene in brain.
Fig. S2. Immunoblot characterization of pan-reactive anti-PanK and specific anti-PanK2 antibodies.
Fig. S3. Relative abundance of Pank1, Pank2 and Pank3 transcripts in Pank1−/−Syn-Pank2−/− and wild-type liver.
Fig. S4. Relative abundance of CoA thioesters in Pank1−/−Syn-Pank2−/− and wild-type hindbrain at P21.
Fig. S5. Plasma metabolite levels in Pank1−/−Syn-Pank2−/− and wild-type mice.
Fig. S6. Hindbrain NAD, NADH and ATP measurements.
Fig. S7. Video - Pank1−/−Syn-Pank2−/− gait.
Fig S8. GAPDH measurements.
Fig S9. RT-qPCR in WT and Pank1−/−Syn-Pank2−/− mice.
Table S1. Primers for genotyping Pank1−/−Syn-Pank2−/− mice.
Table S2. Primers for RT-qPCR.
Table S3. Acyl-CoA mrm values