SUMMARY
Brown adipose tissue (BAT) activity protects animals against hypothermia and represents a potential therapeutic target to combat obesity. The transcription factor early B cell factor-2 (EBF2) promotes brown adipocyte differentiation, but its roles in maintaining brown adipocyte fate and in stimulating BAT recruitment during cold exposure were unknown. We find that the deletion of Ebf2 in adipocytes of mice ablates BAT character and function, resulting in cold intolerance. Unexpectedly, prolonged exposure to cold restores the thermogenic profile and function of Ebf2 mutant BAT. Enhancer profiling and genetic assays identified EBF1 as a candidate regulator of the cold response in BAT. Adipocyte-specific deletion of both Ebf1 and Ebf2 abolishes BAT recruitment during chronic cold exposure. Mechanistically, EBF1 and EBF2 promote thermogenic gene transcription through increasing the expression and activity of ERRα and PGC1α. Together, these studies demonstrate that EBF proteins specify the developmental fate and control the adaptive cold response of brown adipocytes.
Graphical Abstract
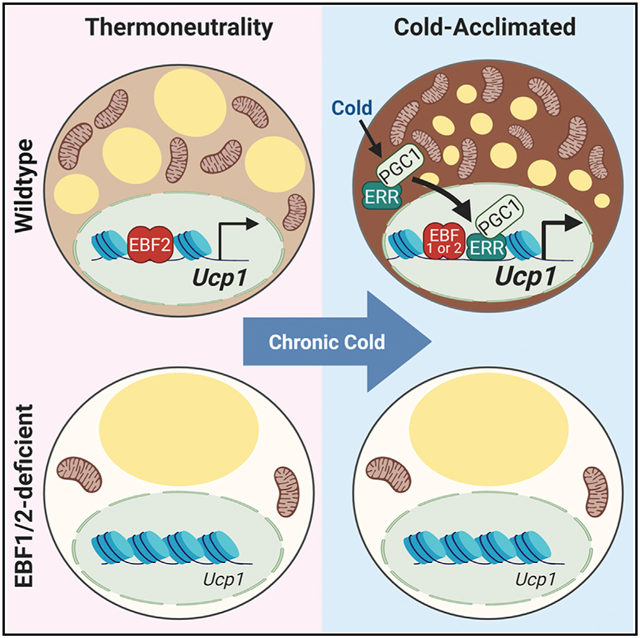
In Brief
Angueira et al. show that early B cell factors (EBFs) control both basal and cold-induced thermogenic activity in brown adipocytes. EBF2 is required to maintain BAT fate under basal conditions, ensuring a capacity for thermogenesis upon cold challenge. BAT recruitment during chronic cold exposure requires EBF1 or EBF2 activity.
INTRODUCTION
Brown adipose tissue (BAT) thermogenesis protects animals from hypothermia during cold exposure and is associated with a resistance to obesity (Harms and Seale, 2013; Yoneshiro et al., 2013). Brown adipocytes are packed with mitochondria that contain uncoupling protein-1 (UCP1). When activated, UCP1 dissipates the mitochondrial membrane potential, which creates a driving force to burn large amounts of glucose and fatty acids, generating heat as a byproduct. The therapeutic potential of BAT to reduce metabolic disease has motivated great interest in defining the pathways that control the development and function of brown adipocytes.
BAT depots develop prenatally in mice, providing newborns with an increased capacity for non-shivering thermogenesis. Cold exposure is the major physiological activator of BAT thermogenesis. Cold-exposure-induced β-adrenergic signaling in brown adipocytes drives lipolysis, substrate oxidation, and uncoupled respiration (Cannon and Nedergaard, 2004). β-adrenergic signaling also increases the expression of Ucp1 and mitochondrial genes in adipocytes to augment BAT capacity.
Distinct transcriptional pathways regulate the development and thermogenic activation of brown adipocytes in response to stimulation. The p38 mitogen-activated protein kinase (MAPK) signaling cascade plays a critical role in mediating thermogenic gene activation in response to β-adrenergic agonists (Cao et al., 2004). p38 phosphorylates activating transcription factor 2 (ATF2) and peroxisome proliferator-activated receptor gamma (PPARγ) coactivator 1α (PGC1α) to increase the transcription of Ucp1 and other mitochondrial genes. Other transcription factors also promote thermogenic gene induction in response to β-adrenergic agonists, including the estrogen-related receptors (ERRs), cAMP response element-binding protein (CREB), ZFP516, and interferon reglatory factor-4 (IRF4) (Ahmadian et al., 2018; Brown et al., 2018; Dempersmier et al., 2015; Emmett et al., 2017; Kong et al., 2014; Kozak et al., 1994; Yubero et al., 1994).
We previously identified the transcription factor early B cell factor-2 (EBF2) as an important regulator of brown fat cell commitment. EBF2 expression is enriched in brown relative to white fat precursor cells and adipocytes (Rajakumari et al., 2013; Wang et al., 2014). Ectopic expression of EBF2 in white fat precursors or muscle cells promotes brown adipocyte differentiation (Rajakumari et al., 2013; Stine et al., 2015). Overexpression of EBF2 or deletion of the EBF repressor ZFP423 in the white adipose tissue (WAT) of mice stimulates browning and suppresses the development of obesity (Shao et al., 2016; Stine et al., 2015). Whole-body knockout (KO) of Ebf2 impairs fetal brown fat development (Rajakumari et al., 2013; Wang et al., 2014). However, EBF2 is expressed in various cell types, precluding studies addressing the physiologic role of EBF2 in regulating BAT activity of adult mice.
Here, we define a critical requirement for EBF2 in establishing and maintaining the thermogenic character of brown adipocytes under basal conditions. Deletion of Ebf2 in adipocytes of mice ablated BAT activity, causing cold intolerance. Interestingly, sustained cold exposure restored the thermogenic profile and function of Ebf2 mutant BAT. Enhancer activity analyses identified an enrichment of DNA-binding motifs for EBFs and ERRs in mutant BAT in response to cold exposure, implicating a potential role for the related EBF family member, EBF1. Deletion of EBF1 alone in adipocytes had minor effects on BAT. However, concurrent deletion of EBF1 and EBF2 caused a complete loss of brown fat thermogenic fate and ablated BAT recruitment during chronic cold exposure. Mechanistic studies showed that EBF1 or EBF2 enhances the activity and expression of the cold-induced transcription factors ERRα and PGC1α to stimulate Ucp1 transcription. Together, these studies demonstrate that EBF proteins control basal and cold-induced thermogenic gene programing in adipocytes.
RESULTS
Adipocyte EBF2 Controls Adaptive Thermogenic Responses
To evaluate the role of EBF2 in regulating the thermogenic programing of adipocytes, we generated adipocyte-specific Ebf2 mutant mice using the Adipoq-Cre driver strain (Ebf2ΔAdipoq). Ebf2 expression was decreased in the BAT of Ebf2ΔAdipoq mice but was unaffected in the stromal-vascular fraction (SVF) of BAT, which contains brown preadipocytes (Figures 1A and S1A). By contrast, Myf5-Cre, which expresses in embryonic brown fat progenitor cells, decreased Ebf2 expression in the BAT SVF (Figure 1A). At late stages of fetal development, the BAT of Ebf2ΔAdipoq mice had higher lipid content and reduced eosinophilic (mitochondrial) staining compared to the BAT from control mice (Figure 1B). Ebf2ΔMyf5 and Ebf2ΔAdipoq mutant BAT at this stage expressed greatly reduced levels of Ucp1 and the mitochondrial marker Cytochrome C (Cycs), along with elevated levels of the white adipocyte marker Resistin (Retn), relative to wild-type (WT) controls (Figure S1). Additionally, BAT from Ebf2ΔAdipoq mice displayed a near-complete loss of UCP1 expression and lower levels of many mitochondrial respiratory chain proteins (Figure 1C). Conversely, the complex V protein ATP5a, which is normally expressed at low levels in brown relative to white fat, was upregulated in Ebf2 mutant BAT (Figure 1C). Transmission electron microscopy analyses showed that the BAT of Ebf2ΔAdipoq mice had greatly diminished mitochondrial density and larger lipid droplets (Figure 1D).
Figure 1. Adipocyte EBF2 Controls BAT Thermogenic Gene Expression and Activity.
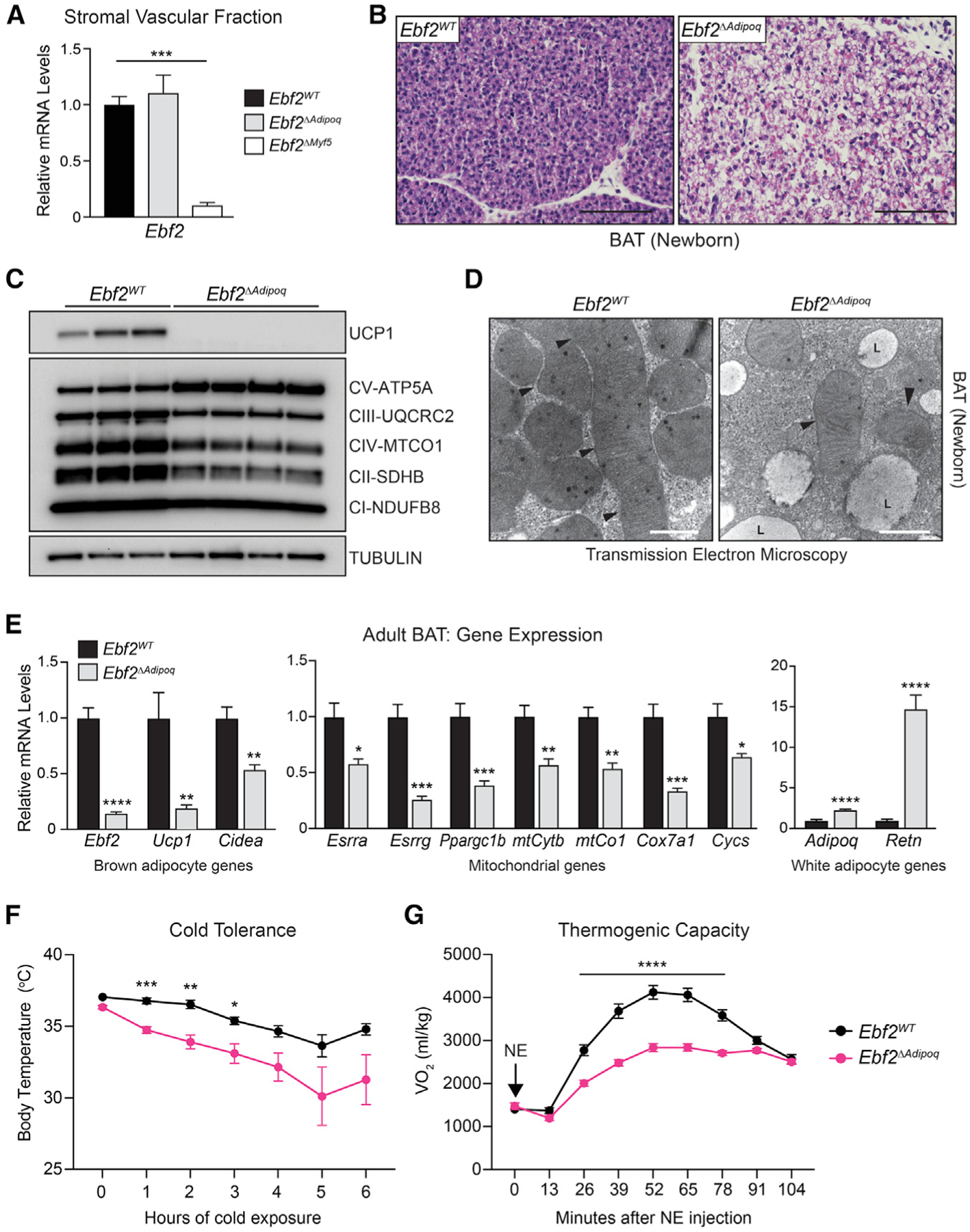
(A) Ebf2 mRNA levels in the stromal-vascular fraction of BAT from 3–4-week-old Ebf2WT, Ebf2ΔMyf5, and Ebf2ΔAdipoq mice (n = 3–5 mice per group; mean ± SEM).
(B) Hematoxylin and eosin staining of BAT from
P0.5 Ebf2WT and Ebf2ΔAdipoq mice (scale bar, 100 μm).
(C) Western blot analysis of UCP1 and mitochondrial respiratory chain components in BAT from P0.5 Ebf2WT and Ebf2ΔAdipoq mice.
(D) Electron micrographs of BAT from P0.5 Ebf2WT and Ebf2ΔAdipoq mice (scale bar, 500 nm; arrow-head: mitochondria; L: lipid droplet).
(E) Relative mRNA levels of indicated genes in BAT from 8- to 10-week-old Ebf2WT and Ebf2ΔAdipoq mice housed at room temperature (RT; 24°C) (n = 5 mice per group; mean ± SEM).
(F) Cold-tolerance test in RT-acclimated, 8- to 10-week-old Ebf2WT and Ebf2ΔAdipoq mice (n = 8–10 mice per group; mean ± SEM).
(G) Volume of O2 (VO2) consumed following norepinephrine (NE) treatment of Ebf2WT and Ebf2ΔAdipoq mice housed at RT (n = 7–10 mice per group; mean ± SEM).
The BAT of adult (8- to 10-week-old) Ebf2ΔAdipoq mice also expressed lower levels of thermogenic and mitochondrial genes, including a ~80% reduction of Ucp1 relative to that in control animals (Figure 1E). Moreover, Ebf2 mutant BAT expressed diminished levels of several transcriptional regulators of mitochondrial biogenesis, including Esrra, Esrrg, and Ppargc1b (Figure 1E). White-fat-enriched genes, including Adipoq and Retn, were upregulated in Ebf2 mutant BAT (Figure 1E).
Upon acute exposure to 4°C, Ebf2ΔAdipoq mutant mice experienced a greater decline in body temperature compared to control mice, indicative of cold intolerance (Figure 1F). We further assessed the thermogenic capacity of control and mutant mice in response to norepinephrine (NE). NE augmented oxygen consumption by ~4-fold in control animals, and this response was markedly blunted in Ebf2ΔAdipoq mice (Figure 1G). Altogether, these results demonstrate that adipocyte-EBF2 is required to establish and maintain the thermogenic function of brown adipocytes.
An EBF2-Independent Pathway Mediates Chronic Cold-Induced BAT Recruitment
Mice housed at room temperature (RT; 22°C) experience mild cold stress, characterized by constitutive BAT activation (Feldmann et al., 2009). To determine if EBF2 regulates BAT fate under basal (unstimulated) conditions, we housed Ebf2WT and Ebf2ΔAdipoq mice at 30°C, near their thermoneutral point. At this temperature, Ebf2 mutant BAT displayed an even more pronounced loss of thermogenic character, compared to the effects at 22°C. This included a dramatic morphologic conversion of BAT to WAT-like tissue, characterized by increased lipid deposition, a near-complete loss of Ucp1 mRNA and protein expression, and diminished levels of mitochondrial genes (Figures 2A–2C). Ucp1 mRNA levels were reduced by ~16-fold in Ebf2 mutant versus control BAT in thermoneutral-housed mice, compared to a ~3-fold reduction observed at RT (Figure 2B).
Figure 2. EBF2 Is Not Required for Chronic Cold-Induced Thermogenic BAT Recruitment.
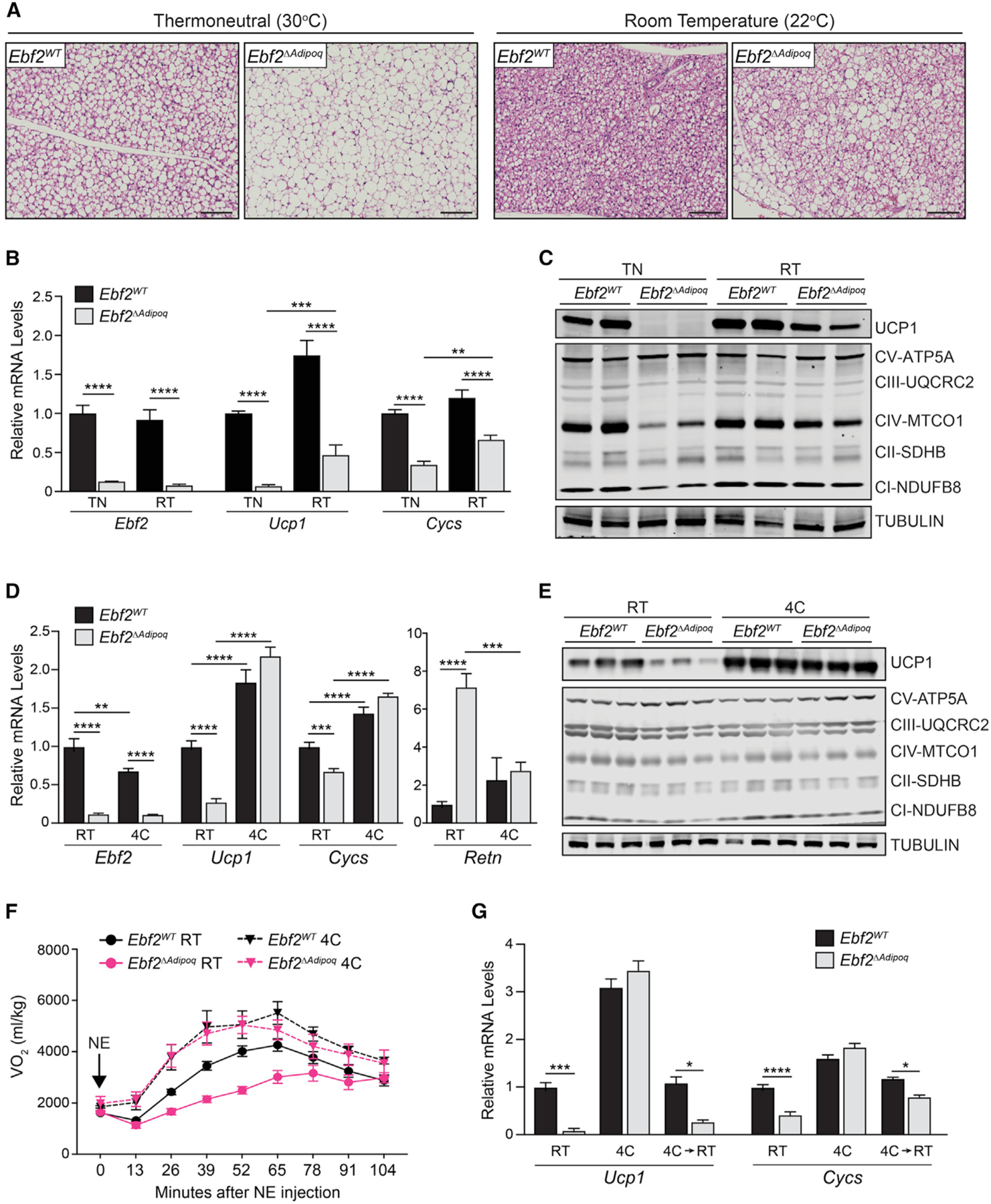
(A) Hematoxylin and eosin staining of BAT in Ebf2WT and Ebf2ΔAdipoq mice housed at RT or 30°C (thermoneutrality [TN]) for 1 month (scale bar, 100 μm).
(B) Relative mRNA levels of Ebf2, Ucp1, and Cycs in BAT from Ebf2WT and Ebf2ΔAdipoq mice housed at RT or TN (n = 5–6 mice per group; mean ± SEM).
(C) Western blot analysis of UCP1 and mitochondrial respiratory chain components in BAT from Ebf2WT and Ebf2ΔAdipoq mice housed at RT or TN. Tubulin was used for loading control.
(D) Relative mRNA levels of indicated genes in BAT from Ebf2WT and Ebf2ΔAdipoq mice housed at RT or 4°C for 1 week (n = 9–11 mice per group; mean ± SEM).
(E) Western blot analysis of UCP1 and mitochondrial respiratory chain components in BAT from Ebf2WT and Ebf2ΔAdipoq mice housed at RT or 4°C for 1 week.
(F) VO2 consumed following NE treatment of Ebf2WT and Ebf2ΔAdipoq mice housed at RT or 4°C for 1 week (n = 5–7 mice per group; mean ± SEM).
(G) Relative mRNA levels of Ucp1 and Cycs in BAT from Ebf2WT and Ebf2ΔAdipoq mice housed at (1) RT, (2) 4°C for 1 week, or (3) 4°C for one week, then RT for 1 week (4C/RT) (n = 3–6 mice per group; mean ± SEM).
The capacity for moderate cold exposure to increase the thermogenic profile of Ebf2 mutant BAT led us to examine the effects of chronic exposure to more severe cold. Following exposure to 4°C for 1 week, the BAT of both Ebf2WT and Ebf2ΔAdipoq mice assumed the histological appearance of thermogenically active tissue, including depleted lipid content and dense eosinophilic staining (Figure S2A). Ucp1 and Cycs were induced to similarly high expression levels in control and Ebf2 mutant BAT (Figure 2D). UCP1 and mitochondrial proteins were also expressed at equivalent levels in control and Ebf2 mutant BAT following cold exposure (Figure 2E). Finally, cold exposure normalized Retn expression in Ebf2 mutant BAT (Figure 2D).
We next assessed if long-term cold exposure normalized the thermogenic capacity of Ebf2ΔAdipoq mice. As shown above, NE increased oxygen consumption to a much greater extent in control than in Ebf2 mutant animals housed at 22°C (RT) (Figure 2F). Following housing at 4°C for 7 days, NE raised oxygen consumption to similarly high levels in control and mutant animals (Figure 2F). There were no statistically significant differences in oxygen consumption between cold-acclimated control and mutant mice at any time points. To assess if one bout of cold exposure permanently restored the thermogenic program in Ebf2-mutant BAT, we cold exposed mice for 1 week and then re-acclimated them to RT for another week. The BAT of the “re-warmed” Ebf2ΔAdipoq mice (4C→RT) expressed lower levels of thermogenic genes than did control mice, similar to the pattern observed in cold-naive mice (Figure 2G). These findings show that sustained cold exposure enables Ebf2-independent thermogenic gene expression and function in BAT.
In addition to driving BAT recruitment, cold exposure elicits the development of UCP1+ multilocular “beige” adipocytes in WAT depots. Adipocyte Ebf2 deficiency caused a marked decrease in beige adipocyte formation and Ucp1 expression in inguinal WAT (iWAT) during cold exposure (Figures S2B–S2D). The loss of Ebf2 in adipocytes did not affect either basal or b3-adrenergic agonist-stimulated lipolysis in epididymal adipose tissue (Figure S2E). Moreover, adipocyte Ebf2 deficiency did not affect body weight or composition in mice housed at RT (Figure S2F).
The Classic Thermogenic Program in Adipocytes Is Dispensable for High-Fat-Diet-Induced Thermogenesis
High-fat or cafeteria (high fat, high sugar) diets increase energy expenditure, though the contribution of BAT thermogenesis or UCP1 to this process is debated (Anunciado-Koza et al., 2008; Feldmann et al., 2009; Kazak et al., 2017; Kontani et al., 2005; Ma et al., 1988; Rothwell and Stock, 1979). To test if the increase in energy expenditure induced by a high-fat diet (HFD) depends on EBF2 action in adipocytes, we housed Ebf2WT and Ebf2ΔAdipoq mice at 30°C (to exempt them from cold stress) and fed them a HFD (Figure S3A). Under these experimental conditions, the BAT of mutant mice expressed greatly reduced levels of UCP1 and mitochondrial genes and higher levels of Retn compared to controls (Figure S3B). Ebf2 mutant animals also displayed a severe reduction in NE-stimulated oxygen consumption (Figure S3C). Despite this deficit in BAT activity, Ebf2 mutant mice gained similar amounts of body weight as the control animals (Figure S3D). Control and Ebf2 mutant animals also had similar blood glucose and insulin levels under both fasting and re-fed conditions (Figures S3E and S3F). Daily energy expenditure during HFD feeding was equivalent between control and mutant mice (Figure S3G). During the switch from normal chow to HFD, oxygen consumption increased and the respiratory exchange ratio decreased to a similar extent in control and mutant animals (Figures S3H–S3J). Overall, these findings indicate that the EBF2-driven BAT thermogenic program (including UCP1) is not essential for HFD-induced thermogenesis.
EBF Activity Controls Basal and Adaptive Thermogenic Gene Expression
To investigate the mechanism responsible for Ebf2-independent BAT recruitment, we performed unbiased gene expression profiling of BAT from Ebf2WT and Ebf2ΔAdipoq mice at RT and following cold exposure (Figure 3A). Clustering analysis identified a large set of genes (light green cluster) that were induced by cold to a similar extent in control and Ebf2 mutant BAT. This cluster was enriched for genes involved in oxidative metabolism and mitochondrial activity, including Ucp1, Ppargc1b, Esrra, and Esrrg (Figures 3B and 3C). To identify EBF2- and cold-regulated enhancers, we profiled the genome-wide levels of H3K27-acetylation (H3K27ac) in BAT. Genomic regions with reduced levels of H3K27ac in Ebf2 mutant versus control BAT at RT were enriched for the EBF binding motif, suggesting that EBF2 functions as a transcriptional activator at many of these sites (Figure 3D). Regions with higher H3K27ac levels in control BAT during cold exposure (cold induced) were enriched with binding motifs for the ERRs, thyroid hormone receptor (TR), and IRX3 (Figure 3E). Of note, the cold-induced H3K27ac regions in Ebf2 mutant BAT were enriched for an EBF motif (Figure 3F), leading us to consider whether cold engages the activity of another EBF family member.
Figure 3. EBF Activity Controls Basal and Adaptive Thermogenic Gene Expression.
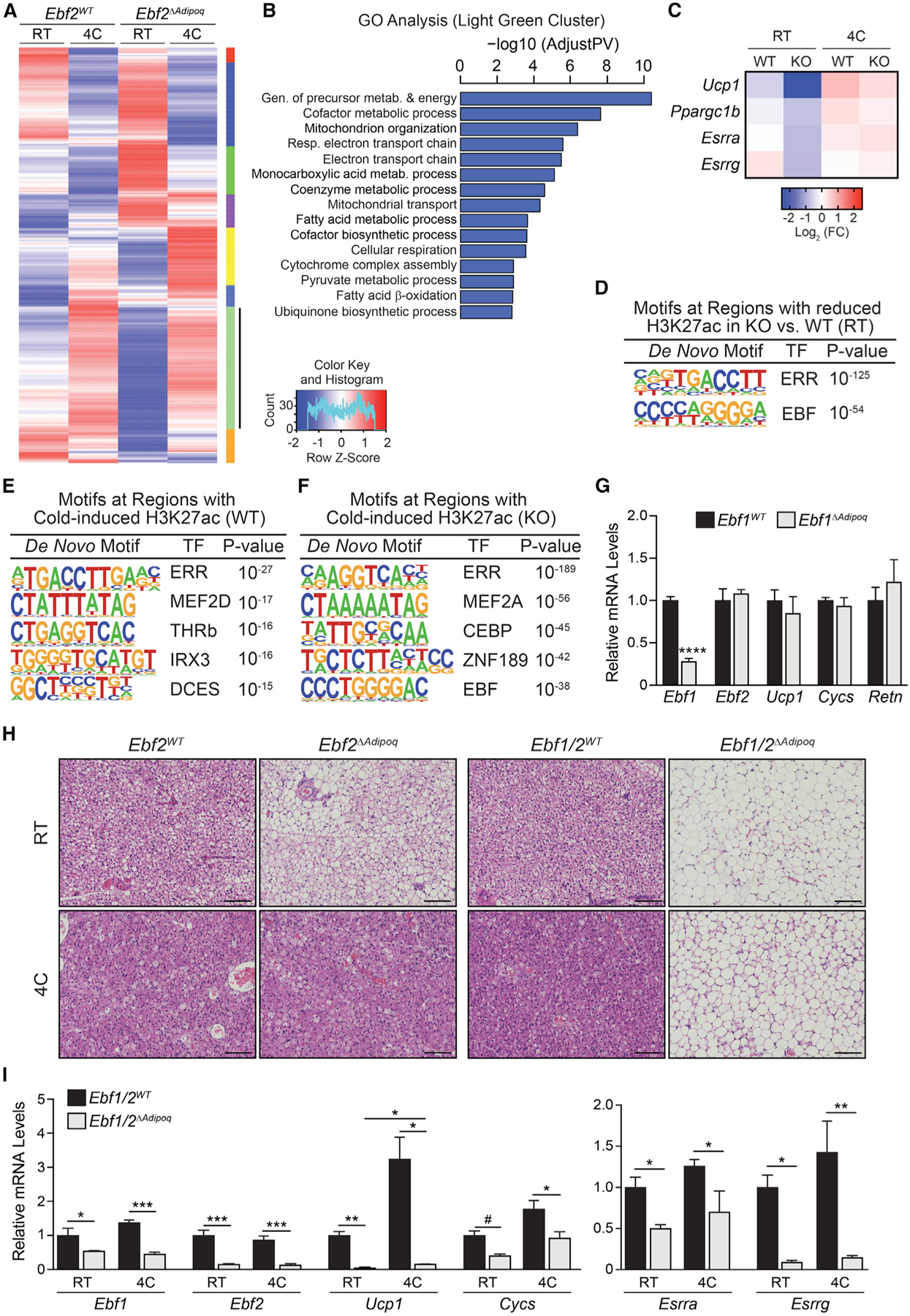
(A) Clustering analysis of gene expression in BAT from Ebf2WT and Ebf2ΔAdipoq mice housed at RT or 4°C for 1 week (n = 3 mice per group).
(B) Gene Ontology analysis of genes in the light green cluster.
(C) Heatmaps of selected genes from light green cluster in (A). Mean log2 fold change (FC).
(D) De novo motif analysis of regions with decreased H3K27ac levels in Ebf2 mutant versus control (WT) BAT of mice housed at RT.
(E and F) De novo motif analysis of regions that display increased H3K27ac levels during cold exposure in BAT from (E) Ebf2WT and (F) Ebf2ΔAdipoq mice.
(G) Relative mRNA levels of Ebf1, Ebf2, and other indicated genes in BAT from Ebf1WT and Ebf1ΔAdipoq mice housed at RT (n = 4 mice per group; mean ± SEM).
(H) Hematoxylin and eosin staining of BAT from Ebf2WT, Ebf2ΔAdipoq, Ebf1/2WT, and Ebf1/2ΔAdipoq mice housed at RT or 4°C for 1 week (scale bar, 100 μm).
(I) Relative mRNA levels of indicated genes in BAT from Ebf1/2WT (control) and Ebf1/2ΔAdipoq (double knockout [DKO]) mice housed at RT or 4°C for 1 week (n = 3–4 mice per group; mean ± SEM).
EBF1 is highly expressed in adipocytes and is known to regulate white adipocyte differentiation (Akerblad et al., 2002; Griffin et al., 2013; Jimenez et al., 2007). To determine if EBF1 regulates brown adipocyte fate, we generated adipocyte-specific Ebf1 mutant (Ebf1ΔAdipoq) mice. The BAT of Ebf1ΔAdipoq mice did not display an altered expression of Ebf2, Ucp1, Cycs, or Retn at RT (Figure 3G). Additionally, the levels of UCP1 and mitochondrial proteins were unaffected by the loss of Ebf1 (Figure S4A). At thermoneutrality (TN; 30°C), the BAT of Ebf1 mutant mice displayed a modest reduction in Ucp1 expression and elevated Retn levels (Figure S4B). To determine if EBF1 compensates for the loss of EBF2 to mediate BAT recruitment during cold exposure, we generated adipocyte-specific Ebf1/Ebf2 double KO (Ebf1/2ΔAdipoq) mice. At RT, the concurrent loss of Ebf1 and Ebf2 resulted in a greater reduction of Ucp1 and an increase in Retn expression in BAT, as compared with the loss of Ebf2 alone (Figure S4C). As observed before, control and Ebf2 single-mutant BAT adopted the characteristics of thermogenically active tissue following chronic cold exposure, including lipid depletion and dense eosinophilic staining (Figure 3H). By contrast, Ebf1/2 double-mutant BAT retained a white-fat-like morphology and did not lose its lipid stores during cold exposure (Figure 3H). The induction of Ucp1 expression in response to cold was nearly absent in Ebf1/2 double-mutant BAT (Figures 3I and S4D). Moreover, cold failed to normalize the expression of Esrra or Esrrg in the BAT of Ebf1/2 double KO (DKO) animals (Figure 3I). These results demonstrate that EBF1 or EBF2 is required for cold-stimulated BAT recruitment.
EBF2 Cooperates with PGC1α and ERRα to Promote Ucp1 Expression
Lastly, we sought to determine the mechanism by which EBF proteins control the cold-induced expression of Ucp1. The −6 kb Ucp1 enhancer is occupied by EBF2, ERRα, and the adipocyte-determination factor PPARγ in BAT (Figure 4A) (Emmett et al., 2017; Shapira et al., 2017). Motif analyses of cold-induced enhancers in BAT identified a high enrichment of ERR binding sites, raising the hypothesis that EBFs cooperate with ERRα and its co-activator partner PGC1α to stimulate transcription (Figures 3D–3F). The −6 kb Ucp1 enhancer contains putative DNA binding sites for EBF2 and ERR (Figure 4B). In reporter gene assays, we found that either EBF2 or EBF1, while having little activity on their own, synergized with PGC1α or PGC1β to strongly activate transcription from the −6 kb Ucp1 enhancer (Figures 4C and S4E). The addition of ERRα further enhanced the transcriptional activity of EBF2 and PGC1α (Figures 4C and S4E). EBF1 and EBF2 were also able to synergize with PGC1β to activate the −6 kb Ucp1 enhancer (Figures S4F and S4G).
Figure 4. EBF2 Cooperates with ERRα and PGC1α to Promote Ucp1 Transcription.
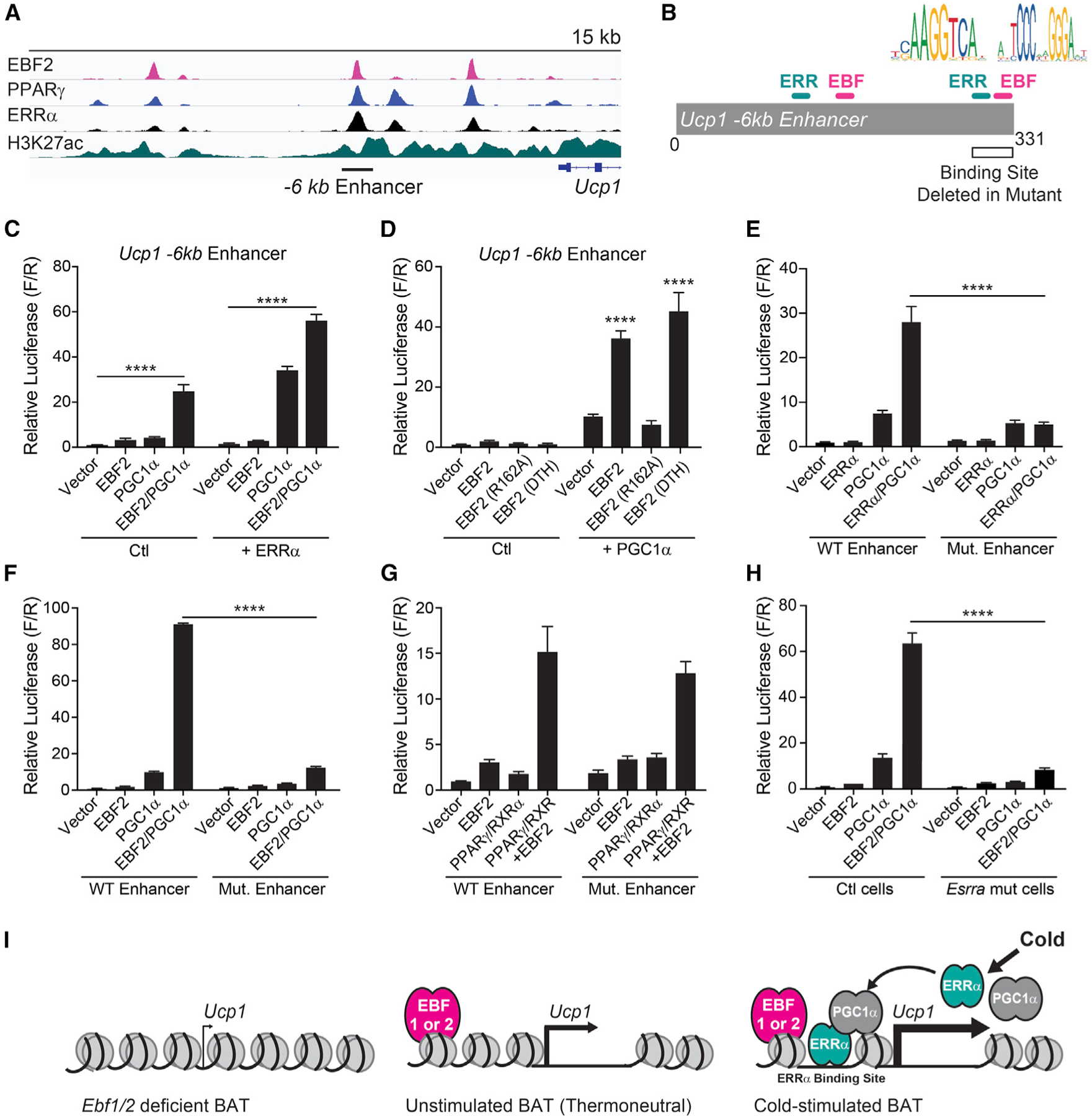
(A) Chromatin immunoprecipitation sequencing (ChIP-seq) tracks of indicated transcription factors and histone modifications at the Ucp1 locus (Emmett et al., 2017; Shapira et al., 2017).
(B) Schematic depicting Ucp1 −6 kb reporter construct and putative EBF and ERR binding sites deleted in mutant reporter.
(C and D) Transcriptional activity of the Ucp1 −6 kb enhancer in NIH 3T3 cells upon expression of (C) EBF2, PGC1α, and/or ERRα; (D) EBF2, EBF2 R162A (DNA-binding mutant) or EBF2 DTH (C-terminal domain mutant); and/or PGC1α (n = 3 replicates per group; mean ± SEM).
(E–G) Transcriptional activity of the WT or mutant (Mut.) Ucp1 −6 kb enhancer in NIH 3T3 cells upon expression of (E) ERRα and/or PGC1α (n = 3); (F) EBF2 and/or PGC1α (n = 3); or (G) EBF2 and/or PPARγ/RXRα (n = 9) (mean ± SEM).
(H) Transcriptional activity of the Ucp1 −6 kb enhancer in control (ctl) or Esrra-deficient NIH 3T3 cells upon expression of EBF2 and/or PGC1α (n = 3 replicates per group; mean ± SEM).
(I) Model of EBF function at thermogenic genes (i.e., Ucp1) in the context of cold exposure.
To determine the nature of this cooperativity, we tested the transcriptional activity of various mutant forms of EBF2. EBF2 R162A, which contains a mutation in the DNA-binding domain, was unable to cooperate with PGC1α to activate transcription (Figure 4D). Deletion of the C-terminal serine/proline transactivation domain did not affect EBF2 transcriptional activity (Figure 4D). To test if EBF2 enhances the function of an ERR/PGC1α complex, we mutated a candidate ERR-binding site within the −6 kb Ucp1 enhancer (Figure 4B). Deletion of this site eliminated the ability of ERRα/PGC1α to activate transcription, validating it as a functional ERRα site (Figure 4E). Mutation of this site also largely abolished the capacity for EBF2 or EBF1 to cooperate with PGC1α and stimulate transcription (Figures 4F and S4H). Notably, EBF2 still efficiently cooperated with PPARγ to activate transcription from the mutant enhancer, suggesting distinct sites for EBF2 action (Figure 4H). To determine if ERRα expression was required for the transcriptional activity of EBF2 and PGC1α, we performed transcription assays in Esrra mutant cells (Figures S4I and S4J). In the absence of ERRα, EBF2 was unable to cooperate with PGC1α or efficiently promote transcription from the Ucp1 enhancer (Figure 4I). Altogether, these results indicate that EBF2 enhances the transcriptional activity of an ERRα/PGC1α complex to drive Ucp1 transcription (model; Figure 4I).
DISCUSSION
This study reveals that EBF proteins control the thermogenic gene program in adipocytes during development and in response to environmental cold. Adipocyte-EBF2 is essential for maintaining brown fat fate in mice housed under standard conditions, ensuring proper thermogenic function and tolerance upon cold challenge. Chronic cold exposure recruited BAT activity through a pathway that depended on either EBF1 or EBF2.
Non-shivering thermogenesis in BAT has been suggested to counteract weight gain. While the EBF2-driven thermogenic program in adipocytes was essential for cold tolerance in mice, this program (including UCP1 expression) was dispensable for HFD-induced thermogenesis. We found that the HFD induced an equivalent increase in oxygen consumption and shift to higher fatty acid utilization in control and Ebf2 mutant mice. Several studies have described UCP1-independent forms of energetic uncoupling in adipocytes, which could mediate HFD-induced thermogenesis and/or compensate for a loss of UCP1 (Anunciado-Koza et al., 2008; de Meis, 2003; de Meis et al., 2006; Flachs et al., 2013; Granneman et al., 2003; Ikeda et al., 2017; Kazak et al., 2015, 2017; Long et al., 2016; Ukropec et al., 2006). In particular, a creatine cycling pathway in adipocytes of mice counteracts HFD-induced weight gain (Kazak et al., 2015, 2017, 2019). We speculate that EBF2 controls the UCP1-based pathway in adipocytes without influencing the activity of other thermogenic pathways, such as the creatine cycle.
Cold exposure activates BAT and elicits the development of thermogenic UCP1+ (beige) adipocytes in WAT. While chronic cold exposure rescued BAT activity, it did not restore beige adipocyte formation in Ebf2ΔAdipoq mice. This may be due to lower expression and/or activity of other EBFs in WAT versus BAT. Alternatively, WAT browning may rely more on EBF2 due to the lower levels of sympathetic innervation and adrenergic stimulation in WAT compared to BAT. Of note, cold-adapted Ebf2 mutant mice displayed the same thermogenic capacity as WT animals despite a near-complete absence of UCP1 expression in WAT. This result suggests that BAT is the predominant source of adrenergically stimulated thermogenesis during cold exposure. Alternatively, the level of acute NE-stimulated thermogenesis could be limited by other factors, such as oxygen delivery or blood flow, precluding the ability to detect a contribution from beige fat.
Genomic and transcriptional studies indicate that EBF proteins promote thermogenic gene transcription through cooperation with members of the ERR and PGC1 protein families, which are known to regulate the BAT cold response (Brown et al., 2018; Emmett et al., 2017; Gantner et al., 2016; Kleiner et al., 2012). Our data support a model in which EBF binding to thermogenic gene enhancers such as Ucp1 facilitates the binding and/or transcriptional function of ERR/PGC1α (Figure 4I). We previously showed that EBF2 associates with the BAF chromatin-remodeling complex to increase PPARγ activity during brown adipocyte differentiation. A similar mechanism may enhance ERR/PGC1α activity at thermogenic genes during cold exposure. In support of this, BRG1, a member of the BAF chromatin-remodeling complex, is recruited to brown adipocyte enhancers in response to β-agonist treatment (Abe et al., 2015). In addition to regulating the transcriptional activity of ERR/PGC1, EBF1 and EBF2 promote the expression of Esrra, Esrrg, and Ppargc1b, establishing a feed-forward loop to drive the BAT cold response. It is yet unknown whether cold enhances the chromatin binding or remodeling activity of EBFs.
In summary, our data highlight the critical function of EBF proteins in regulating the basal and adaptive thermogenic gene programs in BAT. Identification of the factors that stimulate EBF expression and/or activity in adipocytes may thus reveal new approaches to increase BAT activity for the treatment of obesity and related disease.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Patrick Seale (sealep@pennmedicine.upenn.edu). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were housed under the care of University of Pennsylvania University Laboratory Animal Resources (ULAR), which provides both basic husbandry and veterinary care. Animals were raised at room temperature on standard chow with a 12-hour light/dark cycle. The following strains were obtained from the Jackson Laboratory: AdipoqCre (strain name: B6 B6;FVB-Tg(Adipoq-cre)1Evdr/J, RRID:IMSR_JAX:010803) (Eguchi et al., 2011). The following stock was generated by Patrick Seale: Ebf2 loxP/loxP (Shapira et al., 2017). The following stock was generated by Rudolph Grosschedl: Ebf1 loxP/loxP (Györy et al., 2012). Experiments performed at P0.5 and embryonic time points were conducted on male and female mice. Experiments at adult time points were performed in male mice between the ages of 8 to 12 weeks at the onset of the experiment. All mouse housing and husbandry occurred at RT (22°C) unless specified otherwise. For chronic cold exposure, 8- to 10-week-old mice were pair-housed in cages at 4°C. For thermoneutral acclimation, 4to 5-week old mice were housed at 30°C for one month.
METHOD DETAILS
Thermogenic Capacity Assays and Metabolic Phenotyping
For thermogenic capacity assays, CLAMS metabolic chambers (Columbus Instruments) were warmed to 33°C. Mice were placed in chambers and sedated with 75 mg/kg Nembutal. Following 1 round of reading (13 minutes), mice were injected with 1 mg/kg NE (Sigma A9512–1G) subnuchally and VO2 was measured until mice woke up (Kissig et al., 2017). For basal metabolic phenotyping, mice were placed into CLAMS metabolic chambers warmed to 30°C. Average hourly light/dark oxygen consumption and RER measurements were obtained using Cal-R software (Mina et al., 2018). 45% HFD was obtained from Research Diets [D12451].
Cold Tolerance Tests
8- to 10-week-old room temperature acclimated mice were singly housed in pre-chilled cages at 4°C, with rectal temperatures measured hourly for 6 hours.
Body Composition Analysis
Body weight and body composition were measured in ad-lib fed conscious mice using EchoMRI 3-in-1 system nuclear magnetic resonance spectrometer (Echo Medical Systems, Houston, TX) to determine whole body lean and fat mass.
Lipolysis Assay
Fresh eWAT depots were dissected from ad libitum-fed mice and put into FluoroBrite DMEM. The depots were then cut into 4–6 small similar-sized pieces and transferred into 150 μL FluoroBrite DMEM supplemented with 2% FA-free BSA in 96-well plates for 30 min for pre-incubation. To analyze basal lipolysis, tissues were transferred into 150 μL of fresh FluoroBrite DMEM supplemented with 2% FA-free BSA for 60 min. Then, the tissues were transferred and pre-incubated for 30 min in 150 μL FluoroBrite DMEM supplemented with 2% FA-free BSA in the presence of 10 μM CL 316,243. To analyze stimulated lipolysis, tissues were transferred into 150 μL fresh FluoroBrite DMEM supplemented with 2% FA-free BSA and 10 μM of CL 316,243 for another 60 min. Glycerol content was analyzed by combining 5 μL of supernatant and 200 μL of Free Glycerol Reagent and incubating for 15 min at room temperature before measuring absorbance at 540 nm. At the end of the experiment, the tissue pieces were delipidated by CHCL3 extraction and solubilized in 0.3 N NaOH/0.1% SDS at 65°C overnight. Protein content was determined using Pierce BCA Protein assay. Results are expressed as μmol of glycerol per mg of tissue protein.
Isolation of stromal vascular fraction (SVF) from brown adipose tissue (BAT)
BAT SVF was isolated as previously described (Rajakumari et al., 2013). Briefly, BAT was minced and placed into digestion medium (DMEM, Collagenase D: 6.1mg/ml (Roche), Dispase II: 2.4 mg/ml (Roche) and placed at 37°C with constant agitation at 200 rpm for 45 min. Cells were filtered through a 100 μm filter into an equal volume of complete medium (DMEM/10% FBS). Cells were pelleted at 400 g for 4 min.
Histology
Tissues were fixed in 4% PFA overnight, washed in PBS, dehydrated in ethanol, paraffin-embedded and sectioned. Sections were stained with hematoxylin and eosin. Images were captured on a Keyence inverted microscope. For transmission electron microscopy, BAT was fixed in 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) overnight at 4°C. Thin sections were stained with uranyl acetate and lead citrate and analyzed with a JEOL 1010 electron microscope.
RNA Extraction, qRT-PCR, and RNA-sequencing analyses
Total RNA was extracted from using TRIzol (Invitrogen) combined with Purelink RNA columns (Fisher) and quantified using a Nano-drop. mRNA was reverse transcribed into cDNA using the ABI High-Capacity cDNA Synthesis kit (ABI). Real-time PCR was performed on an ABI7900HT PCR machine using SYBR green fluorescent dye (Applied Biosystems). Fold changes were calculated using the ΔΔCT method, with Tata Binding Protein (Tbp) mRNA serving as a normalization control. RNaseq libraries were prepared using the NEB Next Ultra RNA Library Prep Kit and sequenced on a NovaSeq 6000 (PE150) (Novogene). RNA-seq reads were aligned to UCSC mm9 genome using STAR aligner (Dobin et al., 2013) with an option, “–outSAMstrandField intronMotif–outFilterMultimapNmax 1.” Mitochondrial reads were filtered out to avoid sequencing depth bias due to mitochondrial abundance. Then, reads aligned to genes were counted using featureCounts (Liao et al., 2014). Differential gene expression analysis was performed using edgeR (Robinson et al., 2010). Hierarchical clustering was performed to identify distinct functional modules of genes using Ward’s criterion and Pearson correlation as a similarity measure. Gene ontology analysis was done using EnrichR (Chen et al., 2013).
Chromatin Immunoprecipitation (ChIP) and ChIP-Sequencing Analysis
ChIP analysis was performed as previously described (Emmett et al., 2017). Briefly, frozen BAT was thawed and minced in 1% form-aldehyde for 20 min at room temperature and quenched with the addition of 2.5 M glycine. BAT was sonicated in (50 mM HEPES pH 7.5, 155 mM NaCl, 1 mM EDTA, 1.1% Triton X-100, 0.11% Sodium-deoxycholate, 0.1% SDS) using probe sonication (30 s on 30 s off, 3 times at amplitude 10, then 30 s on 30 s off, 3 times at amplitude 15) at 4°C (Fischer Scientific, FB705 Sonic Dismembrator). Primary antibodies were added for overnight incubation at 4°C with rotation. Primary antibody used was 1 μg per sample anti-histone H3K27Ac (ab4729). Protein A Sepharose beads (GE healthcare) were then added for 4 hours at 4°C. Samples were washed twice with sonication buffer (50 mM HEPES pH7.5, 155 mM NaCl, 1 mM EDTA, 1.1% Triton X-100, 0.11% Sodium-deoxycholate, 0.1% SDS), once with sonication buffer with 500mM NaCL, once with wash buffer (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Sodium-deoxycholate, 1 mM EDTA), and twice with TE (1 mM EDTA, 20 mM Tris at pH 8). Samples were then eluted from beads with warm 100 mM NaHCO3/1% SDS buffer. Samples were reverse cross-linked overnight at 65°C with RNase A and then treated with proteinase K and column purified (Clontech NucleoSpin). Libraries were prepared using the NEB Next Ultra DNA Library Prep Kit and sequenced on a Illumina Hiseq 4000 (PE150) (Novogene). ChIP-sequencing reads were aligned to UCSC mm9 genome using STAR aligner with an option, “–alignSJDBoverhangMin 999–alignIntronMax 1–alignMatesGapMax 2000–outFilterMultimapNmax 1–alignEndsType EndToEnd–outFilterMismatchNoverLmax 0.05.” Before downstream analysis, aligned read-pairs were down-sampled to 1/3 for each sample. H3K27ac peaks were called using “findPeaks” in Homer (Heinz et al., 2010) with an option, “-nfr -tbp 0.” Specifically, “-nfr” option was used to center peaks at nucleosome free regions as a putative transcription factor binding sites. All the genomics regions of 200 bp around the peak centers were pooled, merged, and resized to 200 bp to prepare a master enhancer set as an anchor for comparative analysis. Then, H3K27ac tag counts were measured in these regions across all the samples. Differential analysis was done using edgeR to identify genotype or environment-dependent enhancers. De novo motif search was performed using Homer to predict potential transcription factors that are responsible for the enhancer regulation.
Western blotting
Adipose tissue samples were homogenized in RIPA (50mM Tris HCl, 150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS) in a TissueLyser (QIAGEN) at 4°C. Samples were separated on 4%–12% Bis-Tris NuPage gels (Invitrogen) and transferred to nitrocellulose membranes for western blot analysis on Odyssey Li-Cor Machine. Primary antibodies used were anti-UCP1 (1:1000; R&D Systems, MAB6158), anti-TUBULIN (1:5000; Sigma, DM1A), and total-rodent OXPHOS antibody cocktail (1:5000; Abcam, 110143).
Cell Culture & Transcription Assays
NIH 3T3 cells (ATCC) were grown in 10% BCS/DMEM. The plasmid containing Ucp1 −6kb enhancer in pGL4 was described before (Emmett et al., 2017). The start of the enhancer (331 bases total) is located 5964 bases upstream of the Ucp1 start site. De novo motif analyses were performed using JASPAR. The Ucp1–6kb mutant enhancer was constructed by digesting the Ucp1–6kb enhancer with AvrII (NEB) and BglII (NEB), and inserting a 51 bp annealed oligonucleotide synthesized with the ERR binding site deleted. pRL-CMV was used for internal normalization of luciferase assays. The EBF2 R162A DNA binding mutant was generated by site-directed PCR mutagenesis and designed using homology from EBF1 (Fields et al., 2008). The C-terminal domain mutant form of EBF2 was generated by PCR to encode only the N-terminal 421 amino acids of the wild-type protein (by analogy to EBF1; Boller et al., 2016). Expression plasmids (pcDNA-PGC1α, pcDNA-ERRα, pcDNA-EBF2, pcDNA-EBF1, pcDNA-EBF2, pcDNA-EBF3, pcDNA-PPARγ, and pcDNA-RXRα) were described before (Emmett et al., 2017; Rajakumari et al., 2013). Plasmids were transfected into NIH 3T3 cells with Lipofectamine 2000 (Invitrogen). 48 hours following transfection, cells were harvested in passive lysis buffer and dual luciferase assay was performed (Promega E1910) with a Synergy plate reader (BioTek). For CRISPR experiments, gRNA constructs were designed using CRISPOR and cloned into lentiCRISPR v2 as described previously (Haeussler et al., 2016; Shapira et al., 2017). Two gRNAs against each experimental target were co-infected to create mutant cell lines.
QUANTIFICATION AND STATISTICAL ANALYSIS
No power calculations were performed prior to initiation of the study. No mice were omitted from the study. All individual data points were plotted to assay normality. Two sample t tests were performed where comparisons between two groups were being assayed. One way ANOVAs with pairwise comparisons corrected with the Holm Sidak method were performed where comparisons between more than two groups were being assayed. One-way repeated-measure ANOVAs with pairwise comparisons corrected with the Holm Sidak method were performed where comparisons between more than two groups and repeated-measures were being assayed. # indicates p value < 0.1; * indicates p value < 0.05, ** indicates p value < 0.01, *** indicates p value < 0.001, **** indicates p value < 0.0001.
DATA AND CODE AVAILABILITY
The accession number for the high throughput sequencing datasets (ChiPseq and RNaseq) reported in this paper is GEO: GSE144188.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rb anti Histone H3 (acetyl K27) | Abcam | RRID:AB_2118291 |
| Ms anti Human/Mouse UCP1 | R&D | RRID: AB_10572490 |
| Ms anti Alpha tubulin | Sigma | RRID:AB_477583 |
| Ms anti Total Oxphos Rodent Antibody Cocktail | Abcam | RRID:AB_2629281 |
| Anti-rabbit IgG IRDye 800CW | Li-Cor | RRID:AB_621848 |
| Anti-mouse IgG IRDye 800CW | Li-Cor | RRID:AB_621847 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BCS | ATCC | Cat#30–2030 |
| HEPES 1M | Thermo Fisher | Cat#15630080 |
| 16% Paraformaldehyde | EMS | Cat#15710 |
| Protein-A Sepharose CL-4B Beads | GE Healthcare | Cat#17–0780-01 |
| PCR Master Mix, Power SYBR Green | Applied Biosystems | Cat#4367659 |
| Lipofectamine 2000 | Invitrogen | Cat#11668027 |
| NEBNext Ultra II DNA Library Prep with Sample Purification Beads | NEB | Cat#E7103 |
| NEB Next Ultra RNA Library Prep Kit | NEB | Cat#E7530 |
| DMEM | MediaTech | Cat#MT10–017-CV |
| Dispase II | Roche | Cat#4942078001 |
| Collagenase D | Roche | Cat# 11088882001 |
| TRIzol | Invitrogen | Cat#15596018 |
| Purelink RNA Mini columns | Invitrogen | Cat#LT-12183018 |
| ABI High-Capacity cDNA Synthesis kit | Applied Biosystems | Cat#4368813 |
| FluoroBrite DMEM | Thermo Fisher | Cat# A1896701 |
| CL-316,243 | Sigma Aldrich | Cat#C5976 |
| Free Glycerol Reagent | Sigma Aldrich | Cat#F6428 |
| Bovine Serum Albumin, Fraction V, Fatty Acid Free | Roche | Cat#03117057001 |
| Pierce BCA Protein Assay | Thermo Fischer | Cat#23225 |
| Deposited Data | ||
| RNA sequencing data | GEO repository | GSE144188 |
| H3K27Ac ChIP Sequencing data | GEO repository | GSE144188 |
| Experimental Models: Organisms/Strains | ||
| Adipoq-Cre | The Jackson Laboratory | RRID:IMSR_JAX:010803 |
| Ebf2 fl/fl | Dr. Patrick Seale | N/A |
| Ebf1 fl/fl | Dr. Rudolph Grosschedl | N/A |
| Recombinant DNA | ||
| pcDNA-PGC1 α, and pcDNA-ERRa | Dr. Mitchell Lazar | N/A |
| pcDNA-EBF2, pcDNA-EBF1, pcDNA-EBF2, pcDNA-EBF3, pcDNA-PPARγ, pGL4-Ucp1 −6, and pcDNA-RXRα | Dr. Patrick Seale | N/A |
| lentiCRISPR v2 | Addgene (F. Zhang) | RRID:Addgene_52961 |
| pMD2.G | Addgene (D. Trono) | RRID:Addgene_12259 |
| psPAX2 | Addgene (D. Trono) | RRID:Addgene_12260 |
| Software and Algorithms | ||
| GraphPad Prism 7 | Graphpad (www.graphpad.com/scientific-software/prism/ | RRID:SCR_002798 |
| UCSC Genome Browser | UCSC (http://genome.ucsc.edu/) | RRID:SCR_005780 |
| Morpheus (Heatmaps) by the Broad Institute | https://software.broadinstitute.org/morpheus/ | RRID:SCR_017386 |
| Homer | http://homer.ucsd.edu/ | RRID:SCR_010881 |
| Star Aligner | https://github.com/alexdobin/STAR | RRID:SCR_015899 |
| ToppGene | https://toppgene.cchmc.org/ | RRID:SCR_005726 |
Highlights.
Adipocyte EBF2 maintains BAT thermogenic character and function under basal conditions
EBF1 or EBF2 is required for chronic cold-induced BAT recruitment
EBF1 or EBF2 cooperate with ERRa and PGC1a to promote Ucp1 transcription
EBF activity controls basal and adaptive thermogenic gene programing in adipocytes
ACKNOWLEDGMENTS
We thank Marine Adlanmerini for thoughtful discussion. We are grateful to Dr. Rudolf Grosschedl for generously providing the Ebf1 floxed mice. We thank Lan Cheng for expert tissue processing and histological staining. This work was supported by NIH grants DK10300802 to P.S., DK120062 to A.R.A., and DK43806 to M.A.L. We also acknowledge support from the Mouse Metabolic Phenotyping Core and Functional Genomics Core of the Penn Diabetes Research Center (DK P3019525) and the Cox Medical Research Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.023.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abe Y, Rozqie R, Matsumura Y, Kawamura T, Nakaki R, Tsurutani Y, Tanimura-Inagaki K, Shiono A, Magoori K, Nakamura K, et al. (2015). JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun 6, 7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian M, Liu S, Reilly SM, Hah N, Fan W, Yoshihara E, Jha P, De Magalhaes Filho CD, Jacinto S, Gomez AV, et al. (2018). ERRγ Preserves Brown Fat Innate Thermogenic Activity. Cell Rep. 22, 2849–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerblad P, Lind U, Liberg D, Bamberg K, and Sigvardsson M (2002). Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol. Cell. Biol 22, 8015–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anunciado-Koza R, Ukropec J, Koza RA, and Kozak LP (2008). Inactivation of UCP1 and the glycerol phosphate cycle synergistically increases energy expenditure to resist diet-induced obesity. J. Biol. Chem 283, 27688–27697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schübeler D, and Grosschedl R (2016). Pioneering Activity of the C-Terminal Domain of EBF1 Shapes the Chromatin Landscape for B Cell Programming. Immunity 44, 527–541. [DOI] [PubMed] [Google Scholar]
- Brown EL, Hazen BC, Eury E, Wattez JS, Gantner ML, Albert V, Chau S, Sanchez-Alavez M, Conti B, and Kralli A (2018). Estrogen-Related Receptors Mediate the Adaptive Response of Brown Adipose Tissue to Adrenergic Stimulation. iScience 2, 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, and Nedergaard J (2004). Brown adipose tissue: function and physiological significance. Physiol. Rev 84, 277–359. [DOI] [PubMed] [Google Scholar]
- Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, and Collins S (2004). p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol 24, 3057–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meis L (2003). Brown adipose tissue Ca2+-ATPase: uncoupled ATP hydrolysis and thermogenic activity. J. Biol. Chem 278, 41856–41861. [DOI] [PubMed] [Google Scholar]
- de Meis L, Arruda AP, da Costa RM, and Benchimol M (2006). Identification of a Ca2+-ATPase in brown adipose tissue mitochondria: regulation of thermogenesis by ATP and Ca2+. J. Biol. Chem 281, 16384–16390. [DOI] [PubMed] [Google Scholar]
- Dempersmier J, Sambeat A, Gulyaeva O, Paul SM, Hudak CS, Raposo HF, Kwan HY, Kang C, Wong RH, and Sul HS (2015). Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell 57, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, and Rosen ED (2011). Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett MJ, Lim HW, Jager J, Richter HJ, Adlanmerini M, Peed LC, Briggs ER, Steger DJ, Ma T, Sims CA, et al. (2017). Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, and Nedergaard J (2009). UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209. [DOI] [PubMed] [Google Scholar]
- Fields S, Ternyak K, Gao H, Ostraat R, Akerlund J, and Hagman J (2008). The ‘zinc knuckle’ motif of Early B cell Factor is required for transcriptional activation of B cell-specific genes. Mol. Immunol 45, 3786–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachs P, Rossmeisl M, Kuda O, and Kopecky J (2013). Stimulation of mitochondrial oxidative capacity in white fat independent of UCP1: a key to lean phenotype. Biochim. Biophys. Acta 1831, 986–1003. [DOI] [PubMed] [Google Scholar]
- Gantner ML, Hazen BC, Eury E, Brown EL, and Kralli A (2016). Complementary Roles of Estrogen-Related Receptors in Brown Adipocyte Thermogenic Function. Endocrinology 157, 4770–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Burnazi M, Zhu Z, and Schwamb LA (2003). White adipose tissue contributes to UCP1-independent thermogenesis. Am. J. Physiol. Endocrinol. Metab 285, E1230–E1236. [DOI] [PubMed] [Google Scholar]
- Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, and Rosen ED (2013). Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J. Biol. Chem 288, 35925–35939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györy I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E, and Grosschedl R (2012). Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 26, 668–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, and Seale P (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med 19, 1252–1263. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, Shinoda K, Chen Y, Lu X, Maretich P, et al. (2017). UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med 23, 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez MA, Akerblad P, Sigvardsson M, and Rosen ED (2007). Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol. Cell. Biol 27, 743–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, Vetrivelan R, Lu GZ, Laznik-Bogoslavski D, Hasenfuss SC, et al. (2015). A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Lu GZ, Jedrychowski MP, Bare CJ, Mina AI, Kumari M, Zhang S, Vuckovic I, Laznik-Bogoslavski D, et al. (2017). Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab. 26, 660–671.e663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Rahbani JF, Samborska B, Lu GZ, Jedrychowski MP, Lajoie M, Zhang S, Ramsay LC, Dou FY, Tenen D, et al. (2019). Ablation of adipocyte creatine transport impairs thermogenesis and causes diet-induced obesity. Nat Metab 1, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissig M, Ishibashi J, Harms MJ, Lim HW, Stine RR, Won KJ, and Seale P (2017). PRDM16 represses the type I interferon response in adipocytes to promote mitochondrial and thermogenic programing. EMBO J. 36, 1528–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner S, Mepani RJ, Laznik D, Ye L, Jurczak MJ, Jornayvaz FR, Estall JL, Chatterjee Bhowmick D, Shulman GI, and Spiegelman BM (2012). Development of insulin resistance in mice lacking PGC-1α in adipose tissues. Proc. Natl. Acad. Sci. USA 109, 9635–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, et al. (2014). IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 158, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontani Y, Wang Y, Kimura K, Inokuma KI, Saito M, Suzuki-Miura T, Wang Z, Sato Y, Mori N, and Yamashita H (2005). UCP1 deficiency increases susceptibility to diet-induced obesity with age. Aging Cell 4, 147–155. [DOI] [PubMed] [Google Scholar]
- Kozak UC, Kopecky J, Teisinger J, Enerbäck S, Boyer B, and Kozak LP (1994). An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol. Cell. Biol 14, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Bateman LA, Lin H, Kamenecka T, Lokurkar IA, Lou J, Rao RR, Chang MR, Jedrychowski MP, et al. (2016). The Secreted Enzyme PM20D1 Regulates Lipidated Amino Acid Uncouplers of Mitochondria. Cell 166, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SW, Foster DO, Nadeau BE, and Triandafillou J (1988). Absence of increased oxygen consumption in brown adipose tissue of rats exhibiting “cafeteria” diet-induced thermogenesis. Can. J. Physiol. Pharmacol 66, 1347–1354. [DOI] [PubMed] [Google Scholar]
- Mina AI, LeClair RA, LeClair KB, Cohen DE, Lantier L, and Banks AS (2018). CalR: A Web-Based Analysis Tool for Indirect Calorimetry Experiments. Cell Metab. 28, 656–666.e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, and Seale P (2013). EBF2 determines and maintains brown adipocyte identity. Cell Metab 17, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell NJ, and Stock MJ (1979). A role for brown adipose tissue in diet-induced thermogenesis. Nature 281, 31–35. [DOI] [PubMed] [Google Scholar]
- Shao M, Ishibashi J, Wang Q, Kusminski CM, MacPherson KA, Vishvanath L, Spurgin SB, Hepler C, Holland WL, Seale P, et al. (2016). Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab. 23, 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SN, Lim HW, Rajakumari S, Sakers AP, Ishibashi J, Harms MJ, Won KJ, and Seale P (2017). EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. 31, 660–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine R, Shapira SN, Ishibashi J, Lim HW, Harms M, Won KJ, and Seale P (2015). EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol. Metab 5, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukropec J, Anunciado RP, Ravussin Y, Hulver MW, and Kozak LP (2006). UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1−/− mice. J. Biol. Chem 281, 31894–31908. [DOI] [PubMed] [Google Scholar]
- Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, and Seale P (2014). Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl. Acad. Sci. USA 111, 14466–14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, Iwanaga T, and Saito M (2013). Recruited brown adipose tissue as an anti-obesity agent in humans. J. Clin. Invest 123, 3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubero P, Manchado C, Cassard-Doulcier AM, Mampel T, Viñas O, Iglesias R, Giralt M, and Villarroya F (1994). CCAAT/enhancer binding proteins alpha and beta are transcriptional activators of the brown fat uncoupling protein gene promoter. Biochem. Biophys. Res. Commun 198, 653–659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the high throughput sequencing datasets (ChiPseq and RNaseq) reported in this paper is GEO: GSE144188.


