Abstract
A greater understanding of factors causing cancer initiation, progression and evolution is of paramount importance. Among them, the serine/threonine phosphatase PPM1D, also referred to as wild-type p53-induced phosphatase 1 (Wip1) or protein phosphatase 2C delta (PP2Cδ), is emerging as an important oncoprotein due to its negative regulation on a number of crucial cancer suppressor pathways. Initially identified as a p53-regulated gene, PPM1D has been afterwards found amplified and more recently mutated in many human cancers such as breast cancer. The latest progress in this field further reveals that selective inhibition of PPM1D to delay tumor onset or reduce tumor burden represents a promising anti-cancer strategy. Here, we review the advances in the studies of the PPM1D activity and its relevance to various cancers, and recent progress in development of PPM1D inhibitors and discuss their potential application in cancer therapy. Consecutive research on PPM1D and its relationship with cancer is essential, as it ultimately contributes to the etiology and treatment of cancer.
Keywords: PPM1D, phosphatase, inhibitor, p53, cancer
Introduction
PPM1D, also known as (PPM1D, PPMID, PP2Cδ or IDDGIP), is a wild-type p53-induced phosphatase 1 (Wip1). The PPM1D gene is located at 17q23. The complete protein encoded by PPM1D (605 amino acids) is a member of the PP2C family of Ser/Thr protein phosphatase and a negative regulator of cellular stress response pathway. The expression of this gene is induced in a p53-dependent manner in response to various environmental threats, and plays an important role in cell stress, cell cycle regulation, DNA damage repair and tumor cell metabolism. At present, PPM1D has been found to be amplified and overexpressed in various tumors and is currently considered to be an oncogene. This article reviews the relationship between PPM1D and cancer as well as advances in studies of its inhibitors as anti-cancer agents.
In 1997, Fiscella M et al. exposed a large number of animal-derived cells including myeloid leukemia, lymphoblastoid, colon carcinoma, lung carcinoma, fibroblasts etc. to 6.3 Gy of ionizing radiation, extracted RNA and cDNA, and found a new gene named PPM1D. This gene is induced by ionizing radiation in a p53-dependent manner. It is largely induced in the presence of wild-type p53, but is not sensitive to mutant p53. In addition, their studies demonstrate its homology with type 2C protein phosphatase, including dependence on Mg2+ and relative insensitivity to okadaic acid. PPM1D gene transcription is activated rapidly and briefly after ionizing radiation, and the expressed protein is located in the nucleus [1]. Only 3 years later, the structure and expression of mouse PPM1D gene were also determined. PPM1D has been located near p53 gene on chromosome 11. Mouse PPM1D gene consists of 6 exons, spanning 36 kb of DNA length. Compared with human PPM1D (605 amino acids), mouse PPM1D protein is composed of 598 amino acids and their identity and similarity reach 83% and 86%, respectively. The authors observed mouse embryos and various adult tissues including mammary gland, uterus, ovary, adrenal gland, skin and testis in different periods and found that PPM1D mRNA seems to be universally expressed in all tissues and has very high level of expression in testis. Although the expression level fluctuates during development, PPM1D is also expressed at all embryonic stages [2]. In order to further determine the normal biological function of PPM1D in mammalian organisms, the authors obtained PPM1D+/−, PPM1D−/− and PPM1D+/+ through gene recombination. The body weight, fertility, life span and tumor of the mice were observed for 2 years. The results showed that the male mice with PPM1D−/− genotype had smaller body size, atrophic reproductive organs, reduced fertility and life span, and that their 2-year survival rate was less than 20%, while the survival rate of female mice with PPM1D−/−, all PPM1D+/− and PPM1D+/+ mice was about 80% in the same period. Moreover, PPM1D−/− male mice also showed increased susceptibility to pathogens and weakened T and B cell functions, resulting in death of all infected PPM1D−/− male mice within 11 days after influenza virus infection, while the survival rate of PPM1D+/− heterozygote mice infected with virus at the same time was as high as 80%. However, it is noteworthy that PPM1D−/− mice did not show any signs of tumor [3]. Is this just a coincidence?
The NIH Mammalian Gene Collection (MGC) program published related articles in the same year to sequence and verify the complete Open Reading Frame (ORF) clone, which contains a non-redundant set of > 9,000 human and > 6,000 mouse genes. Candidate full ORF clones of another 7,800 individuals and 3,500 mouse genes were also identified [4] and provide a basis for the follow-up research of PPM1D. In the same year Bulavin et al. first studied the relationship between PPM1D and tumor. This study measured PPM1D mRNA content in human embryonic fibroblasts, breast cancer cells, ovarian cancer cells, non-small cell lung cancer cells, renal cancer, T lymphocyte leukemia cell lines, etc. and determined the high expression of PPM1D in breast cancer cell lines BT474 and MCF7. This work not only found a breakthrough for the relationship between PPM1D and cancer, but also established the central position of breast cancer in PPM1D and tumor-related research [5]. The research on the relationship between PPM1D and the occurrence and development of tumor has been started since then, and we will make a systematic review for this.
Breast Cancer
Bulavin et al. found that genes encoding PPM1D (17q22/q23) were amplified in human breast tumor cell lines and about 11% of primary breast tumors, most of which carried wild-type p53. Inactivation of p38MAPK (product of mapk14) in vivo through overexpression of PPM1D accelerates tumor formation [5–7]. Moreover, a new amplification region of breast cancer was found on chromosome 17q23. In addition to PPM1D, the amplification of ERBB2, RPS6KB1, TBX2 and ZNF217 were also found [7, 8]. Interestingly, PPM1D amplification is related to ERBB2 expression. Some researchers have proposed that PPM1D phosphatase plays a role in MKK6/p38 MAPK signaling pathway to promote ErbB2-driven breast tumor occurrence [9]. Meanwhile, some scholars found that 35% of PPM1D mRNA was up-regulated in invasive breast cancer samples. The overexpression of PPM1D was negatively correlated with the overexpression of p-p38 MAPK, suggesting that PPM1D overexpression eliminated the steady-state balance maintained by p38-p53-PPM1D pathway [10]. Inhibiting BRCA1 expression can effectively reduce PPM1D expression, thus enhancing the activity of p38MAPK and effectively improving cell survival rate [11]. In a cohort study from Brazil, the author tried to find out the relationship between long-term clinical efficacy and gene variation by using gene expression chips of tumor samples from 24 patients with invasive ductal breast cancer, and followed up the patients for at least 5 years. They demonstrated that the up-regulation of B3GNT7, PPM1D, TNKS2, PHB and GTSE1 genes in different breast cancer patients was related to the poor prognosis of patients [12]. Other scholars have introduced next-Generation Sequencing (NGS) into clinical diagnosis, and found that gene mutation including PPM1D may lead to increased risk of bilateral breast cancer (BBC) [13]. Other scholars have evaluated the genetic risk of breast cancer, put forward the important role of single nucleotide polymorphisms (SNPs) in cancer diagnosis, and proposed that PPM1D gene mutation will increase the genetic risk of breast cancer [14]. Kim et al. performed NGS analysis on blood samples of patients suspected of having hereditary breast cancer. For patients with PPM1D mutation in blood, routine sequencing was used to check PPM1D mutation in tumor tissue samples. The results showed that in 719 patients with breast cancer, the mutation rate was less than 0.3%, with truncating mutations in exon 6 as the main mutation, which may be related to previous chemotherapy [15]. Regarding the genetic mutation of breast cancer, Mahdavi et al. pointed out that the important reason for the occurrence of genetic breast cancer is the mutation of susceptible genes, including BRCA1, BRCA2, TP53, CHEK2, PTEN, ATM and PPM1D. These mutations are crucial to early onset and the increased risk of familial breast cancer, and lead to 90% of hereditary breast cancer cases [16]. With the rise of research on special types of breast cancer in recent years, triple negative breast cancer (TNBCs) has become the target of conquest. Quist et al. have adopted 5 independent cohort studies with a total of 1,168 patients. Four genes EXO1, TP53BP2, FOXM1 and RSU1 have been identified to be related to genomic instability, malignant growth or therapeutic response. In TNBC MC6 cell line, inhibition of PPM1D can increase the sensitivity of the cancer cells to platinum drugs [17]. Our team also conducted relevant research on diabetes-associated breast cancer and observed that high glucose (HG) promotes the expression of PPM1D through PKC-GSK3β and oxidative stress ROS-NF-KB pathway, thus inhibiting p53 function and promoting the proliferation, migration and invasion of breast cancer cells [18].
In another cohort study of clinical trials, 245 patients with invasive breast cancer were observed and analyzed by in situ hybridization and RT-PCR. It was found that breast cancer samples from clinical patients highly expressed estrogen receptor and progesterone receptor, and PPM1D amplification was significantly correlated with HER2, TOP2A and CCND1 amplification [7]. Some researchers have further studied the relationship between estrogen signal transduction and PPM1D expression. MCF7 cells were treated with different concentrations of estradiol (E2) (1, 10, 100 nM) and for 4, 8, 16, 24 hours. The results showed that the expression of PPM1D reached a peak at 16 hours under the treatment of 10 nM E2. Immunoprecipitation technique confirmed the direct binding of estrogen receptor (ER)α to PPM1D promoter. In addition, the cDNA sequence of PPM1D transcribed by adenovirus caused overexpression of PPM1D, inactivating p53 through dephosphorylation and promoting tumor proliferation [19]. Other studies have confirmed that the expression of PPMID in breast cancer can be independent of p53 regulation, and breast cancer can also show malignant process without p53 mutation. In this study, 201 specimens of primary invasive ductal carcinoma were examined. The results of follow-up for 10 years showed that PPM1D (+) itself is a prognostic factor for breast cancer. The expression of PPM1D could reduce the 10-year survival rate of patients from more than 70% to less than 50%, and what’s more, the 10-year survival rate of patients with PPM1D(+)p21(−) is lower (nearly 30%) [20].
Esophageal cancer
PPM1D overexpression predicts poor prognosis in esophageal squamous cell carcinoma (ESCC). Li et al. studied 101 patients with ESCC and 1: 1 matched control group. Immunohistochemical staining showed that the positive expression of PPM1D protein in tumor tissues of patients with ESCC accounts for 70 cases (69.3%), while the control group accounts for only 15 cases (14.9%), with significant difference. Moreover, the expression of PPM1D mRNA in tumor patients is also significantly higher than that of the control group. The expression of PPM1D in patients with metastatic ESCC is significantly higher than that in patients with non-metastatic ESCC. COX risk model analysis indicates that lymph node metastasis and high expression of PPM1D are independent prognostic factors of ESCC. Follow-up data show that the 5-year survival rate of patients with high expression of PPM1D in tumors is less than 20%, while that of patients with negative PPM1D is about 50%, suggesting that PPM1D may be a new marker for metastasis and prognosis of ESCC patients [21]. In addition, PPMID was also used to evaluate autophagic effect after paclitaxel treatment of esophageal adenocarcinoma cells [22].
Intriguingly, most recently Yokoyama et al. analyzed 157 cases of physiologically normal oesophageal epithelia (PNE), 12 cases of esophageal epithelial dysplasia and 519 cases of esophageal squamous cell carcinoma. It was found that smoking and drinking contributed the most to gene mutation, and age itself was also a risk factor. People over the age of 76 are more likely to develop esophageal cancer. The comparative analysis of normal esophageal epithelium and esophageal carcinoma indicated that the increase of TP53 gene expression and the decrease of PPM1D and NOTCH1 gene expression were statistically significant. Compared to the mutations of esophageal cancer, the normal esophageal epithelia has obvious overrepresentation of NOTCH1 and PPM1D mutations. These mutations can be acquired before late puberty (as early as infancy) and increase significantly with heavy smoking and drinking [23].
Colon cancer
DNA damage can increase PPM1D content and initiate ATM/CHK2, ATR/CHK1 as well as p38-MAPK-induced p53 inhibition to facilitate damage repair. Kleiblova et al. found that PPM1D gene mutation exists in colon cancer, which lead to the persistent suppression of p53 and the occurrence of tumors [24]. Oliva et al. also confirmed that overexpression of PPM1D inhibited CHK2’s ability to detect and repair cell cycle damage in colon cancer cells, leading to malignant progression of cancer [25]. Some scholars have pointed out that PPM1D can activate downstream p38MAPK and JNK signaling pathways independent of p53 in colon cancer cells [26].
Li, Wang, Bai and Peng et al. analyzed the expression of PPM1D protein and mRNA in colorectal cancer and normal tissues. Their results show unanimously that PPM1D protein and mRNA levels in colorectal cancer tissues are significantly higher than those in normal control group and are correlated with lymph node metastasis, Dukes stage, histological grade, 5 year survival rate and liver metastasis [27–30]. Studies on the related mechanisms indicate that p53-dependent PPM1D-KPNA2-AKT/GSK-3beta pathway and NF-kappa B-PPM1D-mTOR-P21 pathway are new pathways for molecular regulation of colon cancer [28, 29]. Using lentiviral shRNA to reduce the expression of PPM1D in RKO cells, Yin et al. demonstrated that inhibition of PPM1D expression effectively inhibited the proliferation and colony formation, and that the cell cycle arrested at the G0/G1 phase and a large number of tumor cells accumulated in G1 phase [31]. Moreover, Suman et al. demonstrated that the deletion of PPM1D inhibited the development of radiation-induced intestinal tumors [32].
Apart from interfering with the development of colon cancer, PPM1D also contributes to the drug resistance of colon cancer. Xia added 50 nmo/L PPM1D siRNA to 5-FU, oxaliplatin and doxorubicin-induced drug resistance model, successfully reducing IC50 of 5-FU-, oxaliplatin- and doxorubicin-resistant strains from 56.88 to 25.32 μmol/L, 43.60 to 18.74 μmol/L, 2.13 to 0.88 μmol/L, respectively [33]. P53-negative tumors are more likely to develop resistance to antineoplastic drugs. However, some scholars have found that overexpression of PPM1D in p53-negative tumors makes them sensitive to chemotherapeutics and protects normal tissues from side effects of anticancer therapy [34].
Hematological Tumors
In clonal hematopoiesis of leukemia, mutations in TP53 and PPM1D seem to lead to clonal growth, which may result in subsequent malignant tumors [35]. In the early stage of tumorigenesis, CDK6 promotes tumorigenesis through regulating transcriptional response in a stage-specific manner. In hematopoietic stem cells, CDK6 inhibits the function of p53 gene by binding to PPM1D [36]. Coombs et al. found that myelodysplastic syndrome was related to mutation of PPM1D and that chemotherapy could cause mutation of PPM1D and TP53. However, for the treatment of hematological tumors, patients with PPM1D mutation are more likely to need growth factor therapy [37]. Xie et al. focused on TCGA database to study gene mutations of tumors in the blood system. Seventy-seven specific mutations were found through analysis of blood source-related genes in 2728 individuals. Among them, PPM1D mutations were associated with hematological diseases such as myelodysplastic syndrome, leukemia and lymphoma [38]. Previous studies indicated that PPM1D gene was amplified and overexpressed in leukemia. Kamada et al. found that PPM1D expression increased in nucleus and cytoplasm of human promyelocytic leukemia cell line HL-60 when it differentiated into neutrophils. PPM1D inhibitor can increase the proportion of HL-60 differentiating into neutrophils, and also induce G1 cell cycle arrest in HL-60 cells. Their results suggest that PPM1D may be a potential therapeutic target for hematopoietic diseases, including leukemia [39]. Other researchers have confirmed that arsenic trioxide (ATO) can activate Chk2/p53 and p38MAPK/p53 pathways by inhibiting the function of PPM1D, leading to apoptosis of acute promyelocytic leukemia cells. Its role has been verified by using PPM1D siRNA [40].
Thyroid cancer
Pekova et al. studied 113 children’s thyroid samples, including 30 benign lesions and 83 thyroid cancer samples. Mutant genes in thyroid cancer were identified by Next Generation Gene Sequencing, and mutations were found in genes including PPM1D [41]. Another study included 301 thyroid patients, including 119 males and 182 females, with a median age of 34 years. Analysis with PCR and Sanger sequencing revealed that EIF1AX, PPM1D and CHEK2 are the three most important new mutation genes [42, 43].
Sarcoma
Osteosarcoma (OS) is a primary malignant bone tumor with high incidence. After analyzing the expression of PPM1D in 18 pairs of osteosarcoma tissues and control tissues, Long et al. found that the expression of PPM1D in osteosarcoma was significantly higher than that in control tissues, and that PPM1D mRNA was highly expressed in U2OS and MG63 cells [44]. A miRNA microarray analysis of cancer tissues and adjacent non-cancer tissues from 62 OS patients showed that 29 miRNA were up-regulated and 26 miRs were down-regulated, of which miR-499a-5p was down-regulated the most. In addition, expression levels of PPM1D mRNA and protein in osteosarcoma tissue are higher than those in non-cancerous tissue. Targetscan predicts that PPM1D is regulated by miR-499a-5p. In order to further confirm the regulatory relationship between miR-499a-5p and PPM1D, they transfected miR-499a-5p mimics into osteosarcoma cell MG-63, and found that it can down-regulate the expression of PPM1D mRNA and protein through Akt/GSK-3β signaling, thus inhibiting tumor cell proliferation [45]. Stole et al. also found that the intervention of PPM1D can reduce the cell viability of Ewing sarcoma, showing the target effect of PPM1D in Ewing sarcoma treatment [46].
Lung cancer
Yang et al. performed immunohistochemical determination of PPM1D in 60 non-small cell lung cancer (NSCLC) tissues and 20 normal lung tissues, and found PPM1D expression in 38 lung cancer tissues, but negative expression in normal tissues. This difference is related to tumor size (3 cm as the boundary) and differentiation degree. It was also observed that the expression of PPM1D was negatively correlated with the expression of p38MAPK, p53 and p16 [47]. Zhao et al. also observed this phenomenon. Among 117 cases of NSCLC, 81 cases expressed PPM1D, but there was no expression or only weak expression in 15 normal lung tissues. The data of follow-up for more than 6 years show that the 6-year survival rate of lung cancer patients with high PPM1D expression is less than 20%, while the survival rate of lung cancer patients with low PPM1D expression is more than 40%. COX risk model regression analysis shows that the difference in survival rate is mainly related to PPM1D protein expression, lymph nodes metastasis and pathological stage [48]. Yang also observed a significant increase in PPM1D mRNA expression in NSCLC tissues, which is significantly correlated with tumor grade, tumor size T stage, clinical stage, and lymph node metastasis [49], suggesting PPM1D has independent evaluation significance for lung cancer [49–51].
Not only can PPM1D be expressed in lung cancer tissues, but it can also mediate the regulation of amyloid protein-binding protein 2 (APPBP2) on lung cancer tissues as an intermediate molecule of pathway. Gong et al. observed an ectopic expression of APPBP2, PPM1D and SPOP in human NSCLC tissues. The results suggest that APPBP2 promotes the progression of NSCLC by regulating PPM1D and SPOP signaling pathways [52].
In view of the significance of PPM1D in the prognosis of lung cancer, researchers began to study the intervention of PPM1D. Gu et al. studied the role of mir-16 in lung cancer and confirmed that PPM1D was the target gene of mir-16 in lung cancer cell line A459. Transfection of mir-16 mimics significantly inhibited the expression of wild-type PPM1D, and reduced the proliferation of A459 cells and promoted cell apoptosis [53]. PPM1D snRNA treatment in A459 and H1299 cells could also reduce the proliferation of tumor cells and induce cell cycle arrest in G0/G1 phase [54].
Ovarian cancer
Previous studies have confirmed that PPM1D gene amplification is closely related to ovarian cancer [55, 56]. Phroah et al. studied 3236 patients with ovarian cancer and found that 0.37% of them had chimeric mutations in PPM1D. However, PPM1D mutation was detected in the blood of 1827 patients after chemotherapy. The authors believe that these PPM1D mutations are related to chemotherapy, but not to the susceptibility of primary tumors [57]. Akbari et al. observed 1,295 cases of ovarian cancer and concluded that in the absence of a family history of cancer, PPM1D chimeric mutation could make women more susceptible to ovarian cancer [58].
Feng et al. transfected PPM1D siRNA into ovarian cancer cell SKOV3 and found that after PPM1D was inhibited, the percentage of tumor cell apoptosis increased, the expression of P53 increased, and the Bax/bcl2 ratio also increased [59]. However, Yin et al. have reached the opposite conclusion. Inhibition of PPM1D expression with shRNA increased the proliferation, migration and invasion ability of Hey A8 and A2780 cells. Moreover, overexpression of PPM1D can inhibit proliferation, migration and invasion of ovarian cancer cells in SKOV3 and OVCA433 cells and mouse metastatic tumor models. At the same time, they substantiated that PPM1D can inhibit ovarian cancer metastasis through negative regulation of p-ATM, p-Akt and Snail. This mechanism is independent of the previous regulatory mechanism involving p53, and may provide new ideas for the treatment of ovarian cancer [60]. Furthermore, Hirata et al. confirmed that miR-21, which located at the same gene position 17q23–25 as PPM1D may also participate in the pathogenesis of ovarian cancer [61].
PPM1D not only can be used as a gene target for ovarian cancer, but also plays an important role in relieving drug resistance of ovarian cancer. Ali proposed a tumor drug resistance mechanism model. In sensitive cells, cisplatin (CDDP) induces PPM1D nuclear rejection and proteasome degradation, thus allowing CDDP to induce Chk1 and p53 activation and eventually initiate cell apoptosis. However, in chemoresistant cells, Akt activation and overexpression stabilize PPM1D, leading to sustained PPM1D expression and nuclear localization, which inhibits Chk1 and p53 activity and reduces apoptosis, thus contributing to CDDP resistance in cancer cells. PPM1D, as a target molecule of AKT, plays an important role in eliminating chemotherapeutic drug resistance [62, 63]. Tsuyoshi confirmed that Saikosaponin-d (Ssd) can induce cancer cell death and sensitize chemoresistant cancer cells to anti-cancer drugs by down-regulating PPM1D and increasing the phosphorylation of checkpoint protein kinase (Chk) 1, cell division cycle 25C (Cdc25c) and cyclin dependent kinase 1 (Cdk1) [64].
Pancreatic cancer
The study of PPM1D in pancreatic cancer is relatively few. Wu et al. found that the expression of PPM1D protein and mRNA in human pancreatic cancer tissues was significantly higher than that in normal pancreas. The expression of PPM1D in pancreatic cancer tissues was related to tumor size (2cm as the boundary), case classification, lymph node metastasis and vascular invasion. Further mechanism study showed that the expression of PPM1D mRNA and protein in PANC-1 and MIA Paca-2 cell lines was higher. High expression of PPM1D can significantly promote the proliferation, invasion and migration of cancer cells through wnt/beta-catenin pathway. Down-regulation of PPM1D expression with PPM1D siRNA significantly increased aspp2 and p38MAPK/p53, and promoted apoptosis of cancer cells. Experiments in nude mice also confirmed that PPM1D siRNA could significantly reduce tumors, suggesting that PPM1D is a carcinogenic gene of pancreatic cancer [65]. Other studies indicated the independent predictive effect of PPM1D on pancreatic cancer patients and that the 3-year survival rate of patients with high PPM1D expression was 0%, while the 10-year survival rate of patients with low PPM1D expression was 10% [66].
Glioma
Dodgshum et al. observed PPM1D mutation in a 3-year-old patient with glioblastoma multiforme [67]. The deletion of Rb gene inactivation of p38MAPK through PPM1D is a new biological behavior of glioma [68]. Wang et al. found that PPM1D is expressed in glioma cell line U87-MG. After silencing PPM1D and treating the cell with TMZ, they showed that PPM1D gene silencing can better inhibit TMZ-induced cell proliferation and induce cell apoptosis and cell cycle arrest, and that PIK3R1/AKT pathway plays a role in various functions of glioma cells [69].
Gastric Cancer
It has been reported that PPM1D is highly expressed in glands of gastric cancer tissues, while almost no PPM1D is expressed in normal gastric mucosa. High expression of PPM1D is associated with lymph node metastasis, distant metastasis and vascular invasion. Survival analysis showed that the 5-year survival rate of patients with PPM1D positive expression is about 40%, while that of patients with PPM1D negative expression is over 70%. Therefore, the expression of PPM1D in cancer tissues could be used as an indicator of poor prognosis of gastric cancer patients [70]. Fuku et al. proposed that the expression of PPM1D in human gastric cancer tissues is related to the size of tumors and the dephosphorylation of Chk2. In addition, carrying out experiments in HEK293 and MKN-74 gastric cancer cells, the authors suggest that ionizing radiation (IR) can induce PPM1D up-regulation and inhibition of CHK2, leading to down-regulation of p53. This pathway provides a new way for the treatment of gastric cancer [71].
Nasopharyngeal Carcinoma
The expression of PPM1D in nasopharyngeal carcinoma is significantly higher than that in normal tissues. Its positive expression is significantly correlated with T stage, lymph node metastasis, clinical stage, tumor differentiation and radiotherapy response. The 5-year survival rate of PPM1D positive patients is less than 40%, while that of PPM1D negative patients is more than 60%. Transfection of PPM1D siRNA into nasopharyngeal carcinoma CNE2 cells significantly reduces the proliferation, invasion and migration of tumor cells, increases apoptosis, and promotes protein expressions of p53 and p16. It is suggested that PPM1D may participate in the development of nasopharyngeal carcinoma by regulating p53-p16 pathway [72].
Liver Cancer
The expression of PPM1D in hepatocellular carcinoma is significantly higher than that in normal liver tissues. Immunohistochemistry showed that PPM1D protein is highly expressed in hepatocellular carcinoma tissues, but hardly expressed in normal tissues. The expression of PPM1D in hepatocellular carcinoma is associated with family history, tumor size, alpha-fetoprotein (α-FP), TNM stage and recurrence. However, it is not significantly correlated with age, gender, portal vein invasion, lymph node metastasis, hepatitis B virus (HBV) infection and alcohol intake. The 3-year survival rate of patients with high expression of PPM1D is 0%, while that of patients with low expression of PPM1D is close to 40%. These results suggest that high expression of PPM1D is associated with poor clinical prognosis of patients with hepatocellular carcinoma [73]. Wang et al. reported that microRNA-29c was down-regulated in hepatocellular carcinoma tissues in 50.6% patients of 255 cases, while PPM1D was up-regulated in 45.4% of these tissues, and the down-regulation of microRNA-29c was negatively correlated with the up-regulation of PPM1D. Ectopic overexpression of microRNA-29c significantly inhibited cell proliferation and induced apoptosis of HepG2 cells and G1 cell cycle arrest. In contrast, knockdown of miR-29c greatly enhanced the proliferation of HepG2 cells and inhibited cell apoptosis. These studies confirmed that the target of biological effect of miR-29c is PPM1D, and proposed that miR-29c can be used as an intervention target for liver cancer [74]. In addition, other scholars have proposed that the synergistic anticancer effect of miR-29a and arsenic trioxide may provide new opportunities for the treatment of liver cancer by reducing the dose and side effects of arsenic trioxide [75].
Renal Carcinoma and Bladder Cancer
The expression of PPM1D protein and mRNA in human renal cell carcinoma is significantly higher than that in normal kidney tissues. The high expression of PPM1D is related to the depth of invasion T stage, Fuhrman grade, lymph node metastasis and distant metastasis. PPM1D shRNA can inhibit the proliferation, migration and invasion of 786-O and RLC-310 renal cancer cells, while overexpression of PPM1D promotes the growth and invasion of these cells in vitro. Survival analysis showed that the 5-year survival rate of patients with high expression of PPM1D in renal cell carcinoma was 0%, while that of patients with low expression of PPM1D was more than 20%. Regression analysis showed that the depth of T3+T4, Fuhrman grade of G3-G4, distant metastasis and high expression of PPM1D are risk factors for poor prognosis of renal cancer patients [76, 77].
Wang et al. transfected PPM1D ShRNA into 5367 and T24 cells via lentiviral vectors. shRNA-mediated PPM1D knockdown significantly inhibited cell growth and colony formation in bladder cancer cell lines 5637 and T24, increased the proportion of G0/G1 phase cells, and decreased the proportion of S and G2 phase cells. These results suggest that PPM1D is a target molecule for bladder cancer treatment [78]. In the treatment of advanced or metastatic bladder cancer, cisplatin resistance often occurs. Lin et al. found that overexpression of HIPK2 in bladder cancer TR4 cells can sensitize chemoresistant bladder cancer cells to cisplatin by regulating the expression of PPM1D [79].
Prostate Cancer
The expression of PPM1D in prostate cancer is significantly higher than that in benign prostatic hyperplasia (BPH) control group, and the expression of PPM1D is related to Gleason score, T stage and lymph node infiltration. Kaplan-Meier curve analysis showed that the 10-year survival rate of PPM1D positive prostate cancer patients was about 50%, while that of PPM1D negative prostate cancer patients was more than 90%. COX risk model regression analysis showed that the difference of survival rate was mainly related to PPM1D protein expression, Gleason score and T stage. In order to further study the mechanism of PPM1D in prostate cancer, Jiao et al. transfected prostate cancer cell lines CP-3 and LNCaP with PPM1D siRNA and found that PPM1D knockdown inhibits the proliferation, migration and invasion of PC-3 and LNCaP cells [80]. Song et al. reported that PPM1D protein expression in prostate cancer LNCaP cells increased significantly after exposure to irradiation. LNCaP cells were continuously exposed to 10Gy radiation for 2–4 hours, p53 content was reduced, p38-p, c-Jun-p, JNK-p, ATR-p, MKK4-p were all inhibited, while PPM1D inhibitor CCT00793 could reverse the inhibition of the above molecules. Further studies have confirmed that PPM1D directly interacts with BAX and dephosphorylates it. Overexpression of PPM1D and BAX in BAX deficient cells can greatly reduce cell apoptosis, reflecting the downregulation of BAX activity by PPM1D [81].
In addition to the various tumors mentioned above, a large number of studies have shown that PPM1D inhibition can also become a new way to treat oral cancer/laryngeal cancer [82], neuroblastoma [83, 84] and melanoma [85].
PPM1D Inhibition as a promising strategy for cancer treatment
PPM1D is amplified and overexpressed in a number of human tumors. Based on Mouse genetic studies and data from RNAi-mediated depletion of PPM1D in cancer cell lines, PPM1D was proposed as an attractive pharmacological target [86–88]. Recently, significant progress has been made in the design of PPM1D inhibitors. On the basis of the PPM1D p53 substrate, peptide inhibitors were designed and further developed to produce a cyclic thioether peptide, which has an in vitro Ki of 110 nM against PPM1D [89]. In spite of a valuable tool compound, the two phosphoric acid moieties on the peptide strictly limit cell entry and the binding of PPM1D. Hence, peptide-based inhibitors have not been further studied to determine their antiproliferative effect. As an alternative to rational design, some labs have screened and developed small molecule inhibitors of Wip1 phosphatase [18, 90–92]. Nevertheless, the majority of these compounds lack the efficacy, target specificity or bioavailability required to encourage further research. Here, we summarize the currently available proof-of-principle small molecule inhibitors and attempt to give a prospect on PPM1D targeted therapeutic strategies in the future.
M321237 and CCT-007093
Compound M321237 was identified through screening chemical libraries based on its ability to repress PPM1D protein phosphatase activity in vitro [93]. Cell viability assay demonstrated that M321237 sensitizes breast cancer MCF-7 cells to doxorubicin. In vivo studies showed that M321237 reduced tumor volumes in xenograft models; nonetheless, the selectivity of M321237 to PPM1D was never validated.
Similar screening methods resulted in identification of CCT007093, which has an in vitro PPM1D protein phosphatase IC50 value of only 8.4 μM [92]. CCT007093 suppresses cell viability in p53-proficient tumor cells carrying amplified PPM1D [92]. Alternatively, CCT007093 inhibits UV-induced apoptosis in skin keratinocytes through blocking activation of JNK, suggesting that the inhibitor is less specific to WIP1 [94]. Additionally, CCT007093 was shown to inhibit cell proliferation irrespective of the presence of WIP1 in U2OS cells, confirming its off-target effect [95]. In addition, CCT007093 treatment does not affect levels of p53-pS15 and γH2AX, both of which are well-established substrates of WIP1 [95]. Finally, CCT-007093 is a strong Michael acceptor that may produce adverse irreversible target inhibition [92]. These data indicate that CCT007093 does not impede WIP1 in cells and emphasize the urgent need to verify the specificity of small molecule inhibitors in cell models, including the CRISPR/Cas9-mediated target gene knockout.
SPI-001 and SL-176
Compared with aforementioned compounds, small molecule perhydrophenanthrene SPI-001 and its analogue SL-176 were identified as non-competitive inhibitors of recombinant WIP1 (IC50 = 110 and 86.9 nM, respectively) [91, 96]. In addition, the specificity of SPI-001 to PPM1D protein phosphatase was determined to be about 50 times higher than that of another PP2C phosphatase PPM1A [91]. Both SPI-001 and SL-176 inhibit the proliferation in human breast cancer MCF7 cells overexpressing wild-type (WT) PPM1D in a dose-dependent manner [96]. In human colorectal cancer HCT-116 cells expressing truncated PPM1D, SPI-001 treatment does not affect cell proliferation, nonetheless combination treatment with SPI-001 and doxorubicin enhances cell growth inhibition by increasing p53 phosphorylation at Ser15 [97]. In summary, SPI-001 and SL-176 are promising lead compounds but further analysis is needed to test their specificity, efficiency and pharmacokinetics in organismal levels [91].
GSK2830371
A particularly intriguing PPM1D protein phosphatase inhibitor with high selectivity was identified recently by combining biochemical and biophysical screens, which employed suppression of PPM1D enzymatic activity and high-affinity binding as readouts, respectively [90]. Firstly, a number of capped amino acids (CAA) were identified (in vitro IC50: 10–20 nM) [90]. The lead CAA compounds are characterized as non-competitive, allosteric PPM1D inhibitors, which bind to a conformationally flexible flap domain that is implicated in substrate conjugation. This is a non-conserved sequence among homologous PPM1D phosphatase family members, assuming that it provides selectivity for PPM1D inhibitors. The peptide properties of these lead compounds led to poor cell permeability and consequently stimulated the further exploration of structure-activity relationship that produces GSK2830371. It has an in vitro IC50 of 6 nM against PPM1D and in vivo activity against a B-cell lymphoma xenograft model, despite a fairly aggressive oral treatment regimen (150 mg/kg, three times a day for two weeks) [90]. It is noteworthy that GSK2830371 also rapidly reduces the level of PPM1D protein in cancer cells through a mechanism that has not been fully described. Moreover, cell proliferation studies showed that GSK2830371 effectively inhibited the proliferation of cancer cells carrying PPM1D amplification while retaining WT p53, e.g. some breast cancer, neuroblastoma, and hematological cancer cell lines [84, 90, 95, 98, 99]. Notably, U2OS-PPM1D-KO cells knocked out by CRISPR/Cas9 did not respond to GSK2830371, which further confirmed its specificity to PPM1D at the cellular level [95]. Inhibition of PPM1D by GSK2830371 up-regulates expression of p53 target genes such as CDKN1A, PUMA, and BAX, leading to cell cycle arrest, but is not enough to induce cell death [90, 95, 98, 100]. Of note, these studies also indicate that GSK2830371 is orally bioavailable. Nevertheless, the relatively low stability in blood could limit its clinical application. Further optimization of GSK2830371 is expected to develop a small molecule PPM1D inhibitor with better pharmacokinetic properties.
Compound 23
Recently, we identified a 1,5-diheteroarylpenta-1,4-dien-3-one (Compound 23, or C23) as an effective PPM1D protein phosphatase inhibitor via cell based screening assay for growth inhibition and activity of a series of curcumin mimics [18]. In structure, CCT007093 and these compounds have a similar central pentacarbon monoketone linker and two same aromatic terminal rings. These curcumin mimics are structurally different from CCT007093 in that they have a distinctive nitrogen containing moiety in the terminal heteroaromatic rings. The growth inhibition assay demonstrated that C23 has a remarkable cytotoxicity on MCF-7 cells (IC50: 0.98 μM) but not MCF-12A cells compared to CCT007093. The docking simulation for binding mode of C23 to the PPM1D phosphatase domain shows that C23 is buried into the catalytic site consisting of Asp105, Arg110, Arg243, Arg258, Arg259, Gln265, Phe268, Asp314, Arg364, Asp366, and Asn367. The carbonyl oxygen atom and the nitrogen atom in C23 heteroaromatic ring form two hydrogen bonds with side chains of Arg258 and Asp105, which leads to a stable binding model of C23. In addition, C23 has a good selectivity versus two other Ser/Thr phosphatases, i.e. PP2Cβ and PP2A.
In addition to the direct inhibition of PPM1D phosphatase activity, C23 suppresses high glucose induction of PPM1D expression through heat shock protein 27 (HSP27) induction and subsequent inhibition of ROS/NF-kB pathway. The water solubility and bioavailability of C23 are still insufficient. To improve its aqueous solubility and bioavailability, we developed PLGANPs of C23, which could significantly inhibit the growth of breast cancer MCF-7 xenografts in diabetic nude mice. We suggest that C23 can be developed as a unique therapeutic agent for breast cancer patients with diabetes.
Conclusions and future perspectives
PPM1D phosphatase is an important negative regulator of p53 pathway and DNA damage response. Overexpressed PPM1D damages p53 function and promotes tumorigenesis, generally in concert with activation of other oncogenes. Amplification, overexpression or mutation of PPM1D are closely related to many human tumors. In contrast, loss of PPM1D dramatically postpones cancer development in mice and depletion of PPM1D by genetic approach reactivates p53 and hinders proliferation in p53-proficient cancers. Until recently, the selective inhibition of PPM1D phosphatase is still a main challenge, and the lack of specific small molecule inhibitors limits the development of wip1 as a pharmacological target for cancer treatment. Because the structure of PPM1D is still unknown, the potential inhibitors of PPM1D have been identified through high throughput screening of a large number of chemical libraries. In the past decade, several compounds have been developed to antagonize the activity of PPM1D; nonetheless, only two of these inhibitors i.e., GSK2830371 and C23, shows high specificity to PPM1D and encouraging results in preclinical analysis. Notably, GSK2830371 is bioavailable by oral administration, and its ability to inhibit the growth of cancer cells in vivo has been confirmed in xenograft models. At the same time, GSK2830371 is promptly inactivated in plasma, limiting its further clinical application. The water solubility and bioavailability of C23 still need to be improved. Thus, further development of GSK2830371 and C23 derivatives with better pharmacokinetic properties is extremely desirable. In addition, solving the three-dimensional structure of PPM1D can stimulate the development of more specific inhibitors. The existing studies show that the inhibition of PPM1D will be the most effective in cancers with WT p53 and amplification or mutations of PPM1D. Therefore, it is important to determine the status of TP53 and PPM1D in tumor for predicting the treatment outcome of PPM1D inhibitors. Although mice are well tolerated to loss of PPM1D, there is new evidence that a lack of PPM1D in the immune system induces an inflammatory environment [101]. In view of these newly discovered physiological effects of PPM1D, it is important to address the potential side effects of temporary suppression of PPM1D during treatment interventions.
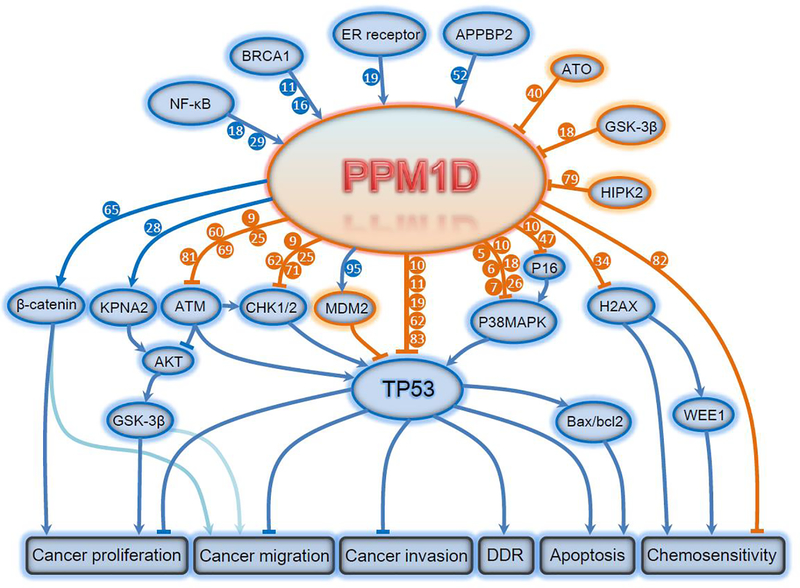
Figure 1. Targets and functional consequences of PPM1D signaling.
The expression and function of PPM1D is regulated by upstream NF-kB, BRCA1, ER receptor, APPBP2, etc. PPM1D phosphatase directly dephosphorylates target proteins including KPNA2, ATM, Chk1/2, Mdm2, p53, p38 MAPK, p16, H2AX and p16, leading to inhibition of apoptosis and promotion of tumorigenesis, invasion, migration and chemoresistance. AKT, AKT Serine/Threonine Kinase; APPBP2, Amyloid Beta Precursor Protein Binding Protein 2; ATM, Ataxia Telangiectasia Mutated; ATO, Arsenic Trioxide; BAX, BCL2 Associated X; BCL2, BCL2 Apoptosis Regulator; BRCA1, BRCA1 DNA Repair Associated; CHK1/2, Checkpoint Kinase 1/2; DDR, DNA Damage Response; ER, Estrogen Receptor; GSK-3β, Glycogen Synthase Kinase 3 Beta; H2AX, H2A Histone Family Member X; HIPK2, Homeodomain Interacting Protein Kinase 2; KPNA2, Karyopherin Subunit Alpha 2; MDM2, Murine Double Minute 2; NF-κB, Nuclear Factor Kappa B; P38 MAPK, P38 Mitogen Activated Protein Kinases; PPM1D, Protein Phosphatase, Mg2+/Mn2+ Dependent 1D; TP53, Tumor Protein P53; WEE1, WEE1 G2 Checkpoint Kinase.
Table 1.
PPM1D in cancers
| Cancer type | Amplification | mRNA upregulation | Protein upregulation | Mutation | Reference |
|---|---|---|---|---|---|
| Breast cancer | + | + | + | + | 5–7,10–11,13 |
| Esophageal cancer | − | + | + | + | 21,22,23 |
| Colon cancer | − | + | + | + | 24,26,27 |
| Hematological tumor | + | − | − | + | 35,37,39 |
| Thyroid cancer | − | − | − | + | 41,42 |
| sarcoma | − | + | + | − | 44,45 |
| Lung cancer | − | + | + | + | 48,49,53 |
| Ovarian cancer | + | + | + | + | 56,57,58,59,61 |
| Pancreatic cancer | − | + | + | − | 65 |
| glioma | + | + | − | + | 67,69 |
| Gastric cancer | − | − | + | − | 70,71 |
| Nasopharyngeal carcinoma | − | + | + | − | 72 |
| Liver cancer | − | + | + | − | 73,74,75 |
| Renal carcinoma | − | + | + | − | 76 |
| Bladder cancer | − | + | + | − | 77, |
| Prostate cancer | − | + | + | − | 80,81 |
| Oral cancer | − | − | + | − | 82 |
| neuroblastoma | − | + | + | − | 83,84 |
| melanoma | − | + | + | − | 85 |
Table 2.
PPM1D inhibitors
| Compound name | Structure | IC50 cell proliferation | IC50 phosphatase activity | Animal study | Animal model | Shortcomings | Reference |
|---|---|---|---|---|---|---|---|
| M321237 | 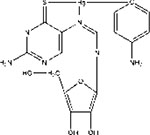 |
− | 0.5μM /MCF7 | + | Transgenic mice/Xenograft | selectivity | 93 |
| CCT007093 | 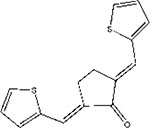 |
− | 8.4μM /MCF7 | + | Knockout mice | selectivity | 94 |
| SPI-001 | 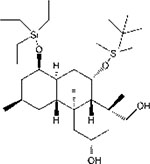 |
26.9μM /MCF7 | 86.9 nM /MCF7 | − | − | Need more test | 91,96 |
| SL-176 | 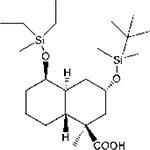 |
7.4μM /MCF7 | 110 nM /MCF7 | − | − | Need more test | 96 |
| GSK2830371 |  |
− | 6.0 nM /MCF7 | + | Xenograft | Low stability in blood | 90,98 |
| C23 | 0.98μM /MCF7 | − | + | Xenograft | Water solubility and bioavailability | 18 |
Highlights.
PPM1D, an oncoprotein, negatively regulates several cancer suppressor pathways
PPM1D has been found amplified and mutated in many human cancers
Selective inhibition of PPM1D represents a promising anti-cancer strategy
Recent progress in PPM1D’s relevance to cancers and development of PPM1D inhibitors
Acknowledgments
Funding: This work was supported in part by NIH-NIMHD U54MD007598, NIH/NCI1U54CA14393, U56 CA101599–01; Department-of-Defense Breast Cancer Research Program grant BC043180, NIH/NCATS CTSI UL1TR000124 to J.V. Vadgama, and Accelerating Excellence in Translational Science Pilot Grants G0812D05, NIH/NCI SC1CA200517 to Y. Wu. Research reported in this publication was partially supported by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under award number S21 MD000103. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This study was also partially supported by the National Natural Science Foundation of China (No. 81500488), the China Scholarship Council (CSC (2018) 10038) and Wuhan Young and Middle-aged Medical Backbone Talent Training Project (Wuhan Municipal Commission of Health and Family Planning (2019) 27), China.
Footnotes
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O’Connor PM, Appella E, Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner, Proc. Natl. Acad. Sci. U. S. A 94(12) (1997) 6048–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Choi J, Appella E, Donehower LA, The structure and expression of the murine wildtype p53-induced phosphatase 1 (Wip1) gene, Genomics 64(3) (2000) 298–306. [DOI] [PubMed] [Google Scholar]
- [3].Choi J, Nannenga B, Demidov ON, Bulavin DV, Cooney A, Brayton C, Zhang Y, Mbawuike IN, Bradley A, Appella E, Donehower LA, Mice deficient for the wild-type p53-induced phosphatase gene (Wip1) exhibit defects in reproductive organs, immune function, and cell cycle control, Mol Cell Biol 22(4) (2002) 1094–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA, Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences, Proceedings of the National Academy of Sciences of the United States of America 99(26) (2002) 16899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace AJ Jr., Appella E, Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity, Nat Genet 31(2) (2002) 210–5. [DOI] [PubMed] [Google Scholar]
- [6].Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A, The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours, Breast cancer research and treatment 95(3) (2006) 257–63. [DOI] [PubMed] [Google Scholar]
- [7].Lambros MB, Natrajan R, Geyer FC, Lopez-Garcia MA, Dedes KJ, Savage K, Lacroix-Triki M, Jones RL, Lord CJ, Linardopoulos S, Ashworth A, Reis-Filho JS, PPM1D gene amplification and overexpression in breast cancer: a qRT-PCR and chromogenic in situ hybridization study, Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 23(10) (2010) 1334–45. [DOI] [PubMed] [Google Scholar]
- [8].Sinclair CS, Rowley M, Naderi A, Couch FJ, The 17q23 amplicon and breast cancer, Breast cancer research and treatment 78(3) (2003) 313–22. [DOI] [PubMed] [Google Scholar]
- [9].Demidov ON, Kek C, Shreeram S, Timofeev O, Fornace AJ, Appella E, Bulavin DV, The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis, Oncogene 26(17) (2007) 2502–6. [DOI] [PubMed] [Google Scholar]
- [10].Yu E, Ahn YS, Jang SJ, Kim MJ, Yoon HS, Gong G, Choi J, Overexpression of the wip1 gene abrogates the p38 MAPK/p53/Wip1 pathway and silences p16 expression in human breast cancers, Breast cancer research and treatment 101(3) (2007) 269–78. [DOI] [PubMed] [Google Scholar]
- [11].Chock K, Allison JM, Elshamy WM, BRCA1-IRIS overexpression abrogates UV-induced p38MAPK/p53 and promotes proliferation of damaged cells, Oncogene 29(38) (2010) 5274–85. [DOI] [PubMed] [Google Scholar]
- [12].Canevari RA, Marchi FA, Domingues MA, de Andrade VP, Caldeira JR, Verjovski-Almeida S, Rogatto SR, Reis EM, Identification of novel biomarkers associated with poor patient outcomes in invasive breast carcinoma, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 37(10) (2016) 13855–13870. [DOI] [PubMed] [Google Scholar]
- [13].Tedaldi G, Tebaldi M, Zampiga V, Danesi R, Arcangeli V, Ravegnani M, Cangini I, Pirini F, Petracci E, Rocca A, Falcini F, Amadori D, Calistri D, Multiple-gene panel analysis in a case series of 255 women with hereditary breast and ovarian cancer, Oncotarget 8(29) (2017) 47064–47075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mahdi KM, Nassiri MR, Nasiri K, Hereditary genes and SNPs associated with breast cancer, Asian Pacific journal of cancer prevention : APJCP 14(6) (2013) 3403–9. [DOI] [PubMed] [Google Scholar]
- [15].Kim B, Won D, Lee ST, Choi JR, Somatic mosaic truncating mutations of PPM1D in blood can result from expansion of a mutant clone under selective pressure of chemotherapy, PloS one 14(6) (2019) e0217521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mahdavi M, Nassiri M, Kooshyar MM, Vakili-Azghandi M, Avan A, Sandry R, Pillai S, Lam AK, Gopalan V, Hereditary breast cancer; Genetic penetrance and current status with BRCA, Journal of cellular physiology 234(5) (2019) 5741–5750. [DOI] [PubMed] [Google Scholar]
- [17].Quist J, Mirza H, Cheang MCU, Telli ML, O’Shaughnessy JA, Lord CJ, Tutt ANJ, Grigoriadis A, A Four-gene Decision Tree Signature Classification of Triple-negative Breast Cancer: Implications for Targeted Therapeutics, Molecular cancer therapeutics 18(1) (2019) 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wu K, Yu X, Huang Z, Zhu D, Yi X, Wu YL, Hao Q, Kemp KT 2nd, Elshimali Y, Iyer R, Nguyen KT, Zheng S, Chen G, Chen QH, Wang G, Vadgama JV, Wu Y, Targeting of PP2Cdelta By a Small Molecule C23 Inhibits High Glucose-Induced Breast Cancer Progression In Vivo, Antioxid Redox Signal 30(17) (2019) 1983–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Han HS, Yu E, Song JY, Park JY, Jang SJ, Choi J, The estrogen receptor alpha pathway induces oncogenic Wip1 phosphatase gene expression, Molecular cancer research : MCR 7(5) (2009) 713–23. [DOI] [PubMed] [Google Scholar]
- [20].Inoue Y, Yamashita N, Kitao H, Tanaka K, Saeki H, Oki E, Oda Y, Tokunaga E, Maehara Y, Clinical Significance of the Wild Type p53-Induced Phosphatase 1 Expression in Invasive Breast Cancer, Clinical breast cancer 18(4) (2018) e643–e650. [DOI] [PubMed] [Google Scholar]
- [21].Li K, Liu Y, Xu S, Wang J, PPM1D Functions as Oncogene and is Associated with Poor Prognosis in Esophageal Squamous Cell Carcinoma, Pathology oncology research : POR (2018). [DOI] [PubMed] [Google Scholar]
- [22].Adams O, Janser FA, Dislich B, Berezowska S, Humbert M, Seiler CA, Kroell D, Slotta-Huspenina J, Feith M, Ott K, Tschan MP, Langer R, A specific expression profile of LC3B and p62 is associated with nonresponse to neoadjuvant chemotherapy in esophageal adenocarcinomas, PloS one 13(6) (2018) e0197610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S, Age-related remodelling of oesophageal epithelia by mutated cancer drivers, Nature 565(7739) (2019) 312–317. [DOI] [PubMed] [Google Scholar]
- [24].Kleiblova P, Shaltiel IA, Benada J, Sevcik J, Pechackova S, Pohlreich P, Voest EE, Dundr P, Bartek J, Kleibl Z, Medema RH, Macurek L, Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint, J Cell Biol 201(4) (2013) 511–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F, The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase, Oncogene 26(10) (2007) 1449–58. [DOI] [PubMed] [Google Scholar]
- [26].Park JY, Song JY, Kim HM, Han HS, Seol HS, Jang SJ, Choi J, p53-Independent expression of wild-type p53-induced phosphatase 1 (Wip1) in methylmethane sulfonate-treated cancer cell lines and human tumors, The international journal of biochemistry & cell biology 44(6) (2012) 896–904. [DOI] [PubMed] [Google Scholar]
- [27].Li ZT, Zhang L, Gao XZ, Jiang XH, Sun LQ, Expression and significance of the Wip1 proto-oncogene in colorectal cancer, Asian Pacific journal of cancer prevention : APJCP 14(3) (2013) 1975–9. [DOI] [PubMed] [Google Scholar]
- [28].Wang P, Zhao Y, Liu K, Liu X, Liang J, Zhou H, Wang Z, Zhou Z, Xu N, Wip1 cooperates with KPNA2 to modulate the cell proliferation and migration of colorectal cancer via a p53-dependent manner, Journal of cellular biochemistry (2019). [DOI] [PubMed] [Google Scholar]
- [29].Bai F, Zhou H, Fu Z, Xie J, Hu Y, Nie S, NF-kappaB-induced WIP1 expression promotes colorectal cancer cell proliferation through mTOR signaling, Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 99 (2018) 402–410. [DOI] [PubMed] [Google Scholar]
- [30].Peng TS, He YH, Nie T, Hu XD, Lu HY, Yi J, Shuai YF, Luo M, PPM1D is a prognostic marker and therapeutic target in colorectal cancer, Experimental and therapeutic medicine 8(2) (2014) 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yin H, Yan Z, Liang Y, Liu B, Su Q, Knockdown of protein phosphatase magnesium-dependent 1 (PPM1D) through lentivirus-mediated RNA silencing inhibits colorectal carcinoma cell proliferation, Technology in cancer research & treatment 12(6) (2013) 537–43. [DOI] [PubMed] [Google Scholar]
- [32].Suman S, Moon BH, Thakor H, Fornace AJ Jr., Datta K, Wip1 abrogation decreases intestinal tumor frequency in APC(Min/+) mice irrespective of radiation quality, Radiation research 182(3) (2014) 345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xia ZS, Wu D, Zhong W, Lu XJ, Yu T, Chen QK, Wip1 gene silencing enhances the chemosensitivity of human colon cancer cells, Oncology letters 14(2) (2017) 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Clausse V, Goloudina AR, Uyanik B, Kochetkova EY, Richaud S, Fedorova OA, Hammann A, Bardou M, Barlev NA, Garrido C, Demidov ON, Wee1 inhibition potentiates Wip1-dependent p53-negative tumor cell death during chemotherapy, Cell death & disease 7 (2016) e2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Silver AJ, Jaiswal S, Clonal hematopoiesis: Pre-cancer PLUS, Advances in cancer research 141 (2019) 85–128. [DOI] [PubMed] [Google Scholar]
- [36].Bellutti F, Tigan AS, Nebenfuehr S, Dolezal M, Zojer M, Grausenburger R, Hartenberger S, Kollmann S, Doma E, Prchal-Murphy M, Uras IZ, Hollein A, Neuberg DS, Ebert BL, Ringler A, Mueller AC, Loizou JI, Hinds PW, Vogl C, Heller G, Kubicek S, Zuber J, Malumbres M, Farlik M, Villunger A, Kollmann K, Sexl V, CDK6 Antagonizes p53-Induced Responses during Tumorigenesis, Cancer discovery 8(7) (2018) 884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, Arcila ME, Ladanyi M, Tallman MS, Levine RL, Berger MF, Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes, Cell stem cell 21(3) (2017) 374–382 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L, Age-related mutations associated with clonal hematopoietic expansion and malignancies, Nature medicine 20(12) (2014) 1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kamada R, Kudoh F, Yoshimura F, Tanino K, Sakaguchi K, Inhibition of Ser/Thr phosphatase PPM1D induces neutrophil differentiation in HL-60 cells, Journal of biochemistry 162(4) (2017) 303–308. [DOI] [PubMed] [Google Scholar]
- [40].Yoda A, Toyoshima K, Watanabe Y, Onishi N, Hazaka Y, Tsukuda Y, Tsukada J, Kondo T, Tanaka Y, Minami Y, Arsenic trioxide augments Chk2/p53-mediated apoptosis by inhibiting oncogenic Wip1 phosphatase, The Journal of biological chemistry 283(27) (2008) 18969–79. [DOI] [PubMed] [Google Scholar]
- [41].Pekova B, Dvorakova S, Sykorova V, Vacinova G, Vaclavikova E, Moravcova J, Katra R, Vlcek P, Sykorova P, Kodetova D, Vcelak J, Bendlova B, Somatic genetic alterations in a large cohort of pediatric thyroid nodules, Endocrine connections (2019) 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Alzahrani AS, Murugan AK, Qasem E, Alswailem MM, AlGhamdi B, Moria Y, Al-Hindi H, Absence of EIF1AX, PPM1D, and CHEK2 mutations reported in Thyroid Cancer Genome Atlas (TCGA) in a large series of thyroid cancer, Endocrine 63(1) (2019) 94–100. [DOI] [PubMed] [Google Scholar]
- [43].Integrated genomic characterization of papillary thyroid carcinoma, Cell 159(3) (2014) 676–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Long X, Lin XJ, P65-mediated miR-590 inhibition modulates the chemoresistance of osteosarcoma to doxorubicin through targeting wild-type p53-induced phosphatase 1, Journal of cellular biochemistry 120(4) (2019) 5652–5665. [DOI] [PubMed] [Google Scholar]
- [45].Liu J, Huang L, Su P, Song T, Zhang W, Fan J, Liu Y, MicroRNA-499a-5p inhibits osteosarcoma cell proliferation and differentiation by targeting protein phosphatase 1D through protein kinase B/glycogen synthase kinase 3beta signaling, Oncology letters 15(4) (2018) 4113–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stolte B, Iniguez AB, Dharia NV, Robichaud AL, Conway AS, Morgan AM, Alexe G, Schauer NJ, Liu X, Bird GH, Tsherniak A, Vazquez F, Buhrlage SJ, Walensky LD, Stegmaier K, Genome-scale CRISPR-Cas9 screen identifies druggable dependencies in TP53 wild-type Ewing sarcoma, The Journal of experimental medicine 215(8) (2018) 2137–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yang S, Dong S, Qu X, Zhong X, Zhang Q, Clinical significance of Wip1 overexpression and its association with the p38MAPK/p53/p16 pathway in NSCLC, Molecular medicine reports 15(2) (2017) 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhao M, Zhang H, Zhu G, Liang J, Chen N, Yang Y, Liang X, Cai H, Liu W, Association between overexpression of Wip1 and prognosis of patients with non-small cell lung cancer, Oncology letters 11(4) (2016) 2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yang H, Gao XY, Li P, Jiang TS, PPM1D overexpression predicts poor prognosis in non-small cell lung cancer, Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 36(3) (2015) 2179–84. [DOI] [PubMed] [Google Scholar]
- [50].Lindskog C, Edlund K, Mattsson JS, Micke P, Immunohistochemistry-based prognostic biomarkers in NSCLC: novel findings on the road to clinical use?, Expert review of molecular diagnostics 15(4) (2015) 471–90. [DOI] [PubMed] [Google Scholar]
- [51].Satoh N, Maniwa Y, Bermudez VP, Nishimura K, Nishio W, Yoshimura M, Okita Y, Ohbayashi C, Hurwitz J, Hayashi Y, Oncogenic phosphatase Wip1 is a novel prognostic marker for lung adenocarcinoma patient survival, Cancer science 102(5) (2011) 1101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gong H, Liu F, Liu X, Min S, Wu N, Liu Y, Han S, Zhang Y, Hu Y, Wang X, APPBP2 enhances non-small cell lung cancer proliferation and invasiveness through regulating PPM1D and SPOP, EBioMedicine 44 (2019) 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gu Y, Wang XD, Lu JJ, Lei YY, Zou JY, Luo HH, Effect of mir-16 on proliferation and apoptosis in human A549 lung adenocarcinoma cells, International journal of clinical and experimental medicine 8(3) (2015) 3227–33. [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang C, Chen Y, Wang M, Chen X, Li Y, Song E, Liu X, Kim S, Peng H, PPM1D silencing by RNA interference inhibits the proliferation of lung cancer cells, World journal of surgical oncology 12 (2014) 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bonache S, Esteban I, Moles-Fernandez A, Tenes A, Duran-Lozano L, Montalban G, Bach V, Carrasco E, Gadea N, Lopez-Fernandez A, Torres-Esquius S, Mancuso F, Caratu G, Vivancos A, Tuset N, Balmana J, Gutierrez-Enriquez S, Diez O, Multigene panel testing beyond BRCA1/2 in breast/ovarian cancer Spanish families and clinical actionability of findings, Journal of cancer research and clinical oncology 144(12) (2018) 2495–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, Gao Q, Marchio C, Geyer FC, Savage K, Parry S, Fenwick K, Tamber N, Mackay A, Dexter T, Jameson C, McCluggage WG, Williams A, Graham A, Faratian D, El-Bahrawy M, Paige AJ, Gabra H, Gore ME, Zvelebil M, Lord CJ, Kaye SB, Ashworth A, Reis-Filho JS, PPM1D is a potential therapeutic target in ovarian clear cell carcinomas, Clinical cancer research : an official journal of the American Association for Cancer Research 15(7) (2009) 2269–80. [DOI] [PubMed] [Google Scholar]
- [57].Pharoah PDP, Song H, Dicks E, Intermaggio MP, Harrington P, Baynes C, Alsop K, Bogdanova N, Cicek MS, Cunningham JM, Fridley BL, Gentry-Maharaj A, Hillemanns P, Lele S, Lester J, McGuire V, Moysich KB, Poblete S, Sieh W, Sucheston-Campbell L, Widschwendter M, Whittemore AS, Dork T, Menon U, Odunsi K, Goode EL, Karlan BY, Bowtell DD, Gayther SA, Ramus SJ, PPM1D Mosaic Truncating Variants in Ovarian Cancer Cases May Be Treatment-Related Somatic Mutations, Journal of the National Cancer Institute 108(3) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Akbari MR, Lepage P, Rosen B, McLaughlin J, Risch H, Minden M, Narod SA, PPM1D mutations in circulating white blood cells and the risk for ovarian cancer, Journal of the National Cancer Institute 106(1) (2014) djt323. [DOI] [PubMed] [Google Scholar]
- [59].Feng Y, Liu F, Du Z, Zhao D, Cheng J, Guo W, Wip1 regulates SKOV3 cell apoptosis through the p38 MAPK signaling pathway, Molecular medicine reports 15(6) (2017) 3651–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yin S, Wang P, Yang L, Liu Y, Wang Y, Liu M, Qi Z, Meng J, Shi TY, Yang G, Zang R, Wip1 suppresses ovarian cancer metastasis through the ATM/AKT/Snail mediated signaling, Oncotarget 7(20) (2016) 29359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hirata Y, Murai N, Yanaihara N, Saito M, Urashima M, Murakami Y, Matsufuji S, Okamoto A, MicroRNA-21 is a candidate driver gene for 17q23–25 amplification in ovarian clear cell carcinoma, BMC cancer 14 (2014) 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ali AY, Kim JY, Pelletier JF, Vanderhyden BC, Bachvarov DR, Tsang BK, Akt confers cisplatin chemoresistance in human gynecological carcinoma cells by modulating PPM1D stability, Molecular carcinogenesis 54(11) (2015) 1301–14. [DOI] [PubMed] [Google Scholar]
- [63].Ali AY, Abedini MR, Tsang BK, The oncogenic phosphatase PPM1D confers cisplatin resistance in ovarian carcinoma cells by attenuating checkpoint kinase 1 and p53 activation, Oncogene 31(17) (2012) 2175–86. [DOI] [PubMed] [Google Scholar]
- [64].Tsuyoshi H, Wong VKW, Han Y, Orisaka M, Yoshida Y, Tsang BK, Saikosaponin-d, a calcium mobilizing agent, sensitizes chemoresistant ovarian cancer cells to cisplatin-induced apoptosis by facilitating mitochondrial fission and G2/M arrest, Oncotarget 8(59) (2017) 99825–99840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wu B, Guo BM, Kang J, Deng XZ, Fan YB, Zhang XP, Ai KX, PPM1D exerts its oncogenic properties in human pancreatic cancer through multiple mechanisms, Apoptosis : an international journal on programmed cell death 21(3) (2016) 365–78. [DOI] [PubMed] [Google Scholar]
- [66].Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, Kosuge T, Kanai Y, Hosoda F, Imoto I, Ohki M, Inazawa J, Hirohashi S, Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome, Cancer science 98(3) (2007) 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Dodgshun AJ, Sexton-Oates A, Saffery R, Sullivan MJ, Biallelic FANCD1/BRCA2 mutations predisposing to glioblastoma multiforme with multiple oncogenic amplifications, Cancer genetics 209(12) (2016) 53–6. [DOI] [PubMed] [Google Scholar]
- [68].Seoane M, Iglesias P, Gonzalez T, Dominguez F, Fraga M, Aliste C, Forteza J, Costoya JA, Retinoblastoma loss modulates DNA damage response favoring tumor progression, PloS one 3(11) (2008) e3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang P, Ye JA, Hou CX, Zhou D, Zhan SQ, Combination of lentivirus-mediated silencing of PPM1D and temozolomide chemotherapy eradicates malignant glioma through cell apoptosis and cell cycle arrest, Oncology reports 36(5) (2016) 2544–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ma D, Zhang CJ, Chen ZL, Yang H, Prognostic value of PPM1D in 800 gastric cancer patients, Molecular medicine reports 10(1) (2014) 191–4. [DOI] [PubMed] [Google Scholar]
- [71].Fuku T, Semba S, Yutori H, Yokozaki H, Increased wild-type p53-induced phosphatase 1 (Wip1 or PPM1D) expression correlated with downregulation of checkpoint kinase 2 in human gastric carcinoma, Pathology international 57(9) (2007) 566–71. [DOI] [PubMed] [Google Scholar]
- [72].Sun GG, Zhang J, Ma XB, Wang YD, Cheng YJ, Hu WN, Overexpression of Wild-Type p53-Induced Phosphatase 1 Confers Poor Prognosis of Patients with Nasopharyngeal Carcinoma, Pathology oncology research : POR 21(2) (2015) 283–91. [DOI] [PubMed] [Google Scholar]
- [73].Li GB, Zhang XL, Yuan L, Jiao QQ, Liu DJ, Liu J, Protein phosphatase magnesium-dependent 1delta (PPM1D) mRNA expression is a prognosis marker for hepatocellular carcinoma, PloS one 8(3) (2013) e60775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang B, Li D, Sidler C, Rodriguez-Juarez R, Singh N, Heyns M, Ilnytskyy Y, Bronson RT, Kovalchuk O, A suppressive role of ionizing radiation-responsive miR-29c in the development of liver carcinoma via targeting WIP1, Oncotarget 6(12) (2015) 9937–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Meng XZ, Zheng TS, Chen X, Wang JB, Zhang WH, Pan SH, Jiang HC, Liu LX, microRNA expression alteration after arsenic trioxide treatment in HepG-2 cells, Journal of gastroenterology and hepatology 26(1) (2011) 186–93. [DOI] [PubMed] [Google Scholar]
- [76].Liu S, Qi L, Han W, Wan X, Jiang S, Li Y, Xie Y, Liu L, Zeng F, Liu Z, Zu X, Overexpression of wip1 is associated with biologic behavior in human clear cell renal cell carcinoma, PloS one 9(10) (2014) e110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang ZP, Chen SY, Tian Y, Wild-type p53-induced phosphatase 1 is a prognostic marker and therapeutic target in bladder transitional cell carcinoma, Oncology letters 13(2) (2017) 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wang W, Zhu H, Zhang H, Zhang L, Ding Q, Jiang H, Targeting PPM1D by lentivirus-mediated RNA interference inhibits the tumorigenicity of bladder cancer cells, Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas 47(12) (2014) 1044–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lin J, Zhang Q, Lu Y, Xue W, Xu Y, Zhu Y, Hu X, Downregulation of HIPK2 increases resistance of bladder cancer cell to cisplatin by regulating Wip1, PloS one 9(5) (2014) e98418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jiao L, Shen D, Liu G, Jia J, Geng J, Wang H, Sun Y, PPM1D as a novel biomarker for prostate cancer after radical prostatectomy, Anticancer research 34(6) (2014) 2919–25. [PubMed] [Google Scholar]
- [81].Song JY, Ryu SH, Cho YM, Kim YS, Lee BM, Lee SW, Choi J, Wip1 suppresses apoptotic cell death through direct dephosphorylation of BAX in response to gamma-radiation, Cell death & disease 4 (2013) e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang L, Mosel AJ, Oakley GG, Peng A, Deficient DNA damage signaling leads to chemoresistance to cisplatin in oral cancer, Molecular cancer therapeutics 11(11) (2012) 2401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Baxter EW, Milner J, p53 Regulates LIF expression in human medulloblastoma cells, Journal of neuro-oncology 97(3) (2010) 373–82. [DOI] [PubMed] [Google Scholar]
- [84].Richter M, Dayaram T, Gilmartin AG, Ganji G, Pemmasani SK, Van Der Key H, Shohet JM, Donehower LA, Kumar R, WIP1 phosphatase as a potential therapeutic target in neuroblastoma, PLoS One 10(2) (2015) e0115635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Pandolfi S, Montagnani V, Penachioni JY, Vinci MC, Olivito B, Borgognoni L, Stecca B, WIP1 phosphatase modulates the Hedgehog signaling by enhancing GLI1 function, Oncogene 32(40) (2013) 4737–47. [DOI] [PubMed] [Google Scholar]
- [86].Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJ Jr., Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway, Nat Genet 36(4) (2004) 343–50. [DOI] [PubMed] [Google Scholar]
- [87].Shreeram S, Hee WK, Demidov ON, Kek C, Yamaguchi H, Fornace AJ Jr., Anderson CW, Appella E, Bulavin DV, Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase, J Exp Med 203(13) (2006) 2793–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Demidov ON, Timofeev O, Lwin HN, Kek C, Appella E, Bulavin DV, Wip1 phosphatase regulates p53-dependent apoptosis of stem cells and tumorigenesis in the mouse intestine, Cell Stem Cell 1(2) (2007) 180–90. [DOI] [PubMed] [Google Scholar]
- [89].Hayashi R, Tanoue K, Durell SR, Chatterjee DK, Jenkins LM, Appella DH, Appella E, Optimization of a cyclic peptide inhibitor of Ser/Thr phosphatase PPM1D (Wip1), Biochemistry 50(21) (2011) 4537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Gilmartin AG, Faitg TH, Richter M, Groy A, Seefeld MA, Darcy MG, Peng X, Federowicz K, Yang J, Zhang SY, Minthorn E, Jaworski JP, Schaber M, Martens S, McNulty DE, Sinnamon RH, Zhang H, Kirkpatrick RB, Nevins N, Cui G, Pietrak B, Diaz E, Jones A, Brandt M, Schwartz B, Heerding DA, Kumar R, Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction, Nat Chem Biol 10(3) (2014) 181–7. [DOI] [PubMed] [Google Scholar]
- [91].Yagi H, Chuman Y, Kozakai Y, Imagawa T, Takahashi Y, Yoshimura F, Tanino K, Sakaguchi K, A small molecule inhibitor of p53-inducible protein phosphatase PPM1D, Bioorg Med Chem Lett 22(1) (2012) 729–32. [DOI] [PubMed] [Google Scholar]
- [92].Rayter S, Elliott R, Travers J, Rowlands MG, Richardson TB, Boxall K, Jones K, Linardopoulos S, Workman P, Aherne W, Lord CJ, Ashworth A, A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D, Oncogene 27(8) (2008) 1036–44. [DOI] [PubMed] [Google Scholar]
- [93].Belova GI, Demidov ON, Fornace AJ Jr., Bulavin DV, Chemical inhibition of Wip1 phosphatase contributes to suppression of tumorigenesis, Cancer Biol Ther 4(10) (2005) 1154–8. [DOI] [PubMed] [Google Scholar]
- [94].Lee JS, Park JR, Kwon OS, Kim H, Fornace AJ Jr., Cha HJ, Off-target response of a Wip1 chemical inhibitor in skin keratinocytes, J Dermatol Sci 73(2) (2014) 125–34. [DOI] [PubMed] [Google Scholar]
- [95].Pechackova S, Burdova K, Benada J, Kleiblova P, Jenikova G, Macurek L, Inhibition of WIP1 phosphatase sensitizes breast cancer cells to genotoxic stress and to MDM2 antagonist nutlin-3, Oncotarget 7(12) (2016) 14458–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ogasawara S, Kiyota Y, Chuman Y, Kowata A, Yoshimura F, Tanino K, Kamada R, Sakaguchi K, Novel inhibitors targeting PPM1D phosphatase potently suppress cancer cell proliferation, Bioorg Med Chem 23(19) (2015) 6246–9. [DOI] [PubMed] [Google Scholar]
- [97].Kozakai Y, Kamada R, Kiyota Y, Yoshimura F, Tanino K, Sakaguchi K, Inhibition of C-terminal truncated PPM1D enhances the effect of doxorubicin on cell viability in human colorectal carcinoma cell line, Bioorg Med Chem Lett 24(24) (2014) 5593–5596. [DOI] [PubMed] [Google Scholar]
- [98].Chen Z, Wang L, Yao D, Yang T, Cao WM, Dou J, Pang JC, Guan S, Zhang H, Yu Y, Zhao Y, Wang Y, Xu X, Shi Y, Patel R, Zhang H, Vasudevan SA, Liu S, Yang J, Nuchtern JG, Wip1 inhibitor GSK2830371 inhibits neuroblastoma growth by inducing Chk2/p53-mediated apoptosis, Sci Rep 6 (2016) 38011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kojima K, Maeda A, Yoshimura M, Nishida Y, Kimura S, The pathophysiological significance of PPM1D and therapeutic targeting of PPM1D-mediated signaling by GSK2830371 in mantle cell lymphoma, Oncotarget 7(43) (2016) 69625–69637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Sriraman A, Radovanovic M, Wienken M, Najafova Z, Li Y, Dobbelstein M, Cooperation of Nutlin-3a and a Wip1 inhibitor to induce p53 activity, Oncotarget 7(22) (2016) 31623–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Uyanik B, Grigorash BB, Goloudina AR, Demidov ON, DNA damage-induced phosphatase Wip1 in regulation of hematopoiesis, immune system and inflammation, Cell Death Discov 3 (2017) 17018. [DOI] [PMC free article] [PubMed] [Google Scholar]