Abstract
Semen modifies the expression of genes related to immune function along the porcine female internal genital tract. Whether other pathways are induced by the deposition of spermatozoa and/or seminal plasma (SP), is yet undocumented. Here, to determine their relative impact on the uterine and tubal transcriptomes, microarray analyses were performed on the endocervix, endometrium and endosalpinx collected from pre-ovulatory sows 24 h after either mating or artificial insemination (AI) with specific ejaculate fractions containing spermatozoa or sperm-free SP. After enrichment analysis, we found an overrepresentation of genes and pathways associated with sperm transport and binding, oxidative stress and cell-to-cell recognition, such as PI3K-Akt, FoxO signaling, glycosaminoglycan biosynthesis and cAMP-related transcripts, among others. Although semen (either after mating or AI) seemed to have the highest impact along the entire genital tract, our results demonstrate that the SP itself also modifies the transcriptome. The detected modifications of the molecular profiles of the pre/peri-ovulatory endometrium and endosalpinx suggest an interplay for the survival, transport and binding of spermatozoa through, for instance the up-regulation of the Estrogen signaling pathway associated with attachment and release from the oviductal reservoir.
Subject terms: Microarray analysis, Animal breeding
Introduction
A successful establishment and maintenance of pregnancy is highly dependent on uterine receptivity1. Such receptivity implies a reciprocal communication between gametes and the uterine or oviductal epithelia is a key factor for fertilization, embryo development and pregnancy success2,3. Understanding the modifications of female genital molecular profiles triggered by seminal components that interact with the internal genital lining during the peri-ovulatory period could be of great importance to overcome problems associated with fertility. Several studies have described the transcriptome changes in the pig uterus and oviduct motivated by the presence of gametes in the female genital tract4.
Semen deposited into the female genital tract elicits an immediate inflammatory response5. It also activates modifications of genes involved in immune mechanisms6 that will promote a state of immune tolerance to paternal antigens in the female7. Sperm presence in the oviduct elicits different transcriptomic responses by modulating specific signaling pathways depending on which sex chomosome the spermatozoa contain4. Moreover, spermatozoa are suggested to modulate the uterine milieu against oxidative stress to protect spermatozoa and embryos from ROS-induced damage8. Presence of spermatozoa and the following sperm-oocyte interactions influence gene expression in the ampullar-isthmic section of the oviduct, changes that would regulate several processes associated with fertilization and embryo development9.
However, despite spermatozoa modifying the uterine and/or oviductal environment, there is emerging evidence that the SP interaction with the female genital tract also plays a role in the different events that take place surrounding ovulation and fertilization. The SP has the potential to accelerate ovulation by modifying the endocrine-immune-cytokine network in pre-ovulatory follicles; leading to a reduction in the interval between the LH peak and ovulation time-point10. The SP has also been shown to promote subtle changes in the uterine gene expression associated with sperm transport and protection11. Furthermore, the SP seems to elicit changes in the expression of certain genes in the oviductal sperm reservoir: the utero-tubal junction12.
The study presented here aimed to determine which genes and signaling pathways are expression-modified along the peri-ovulatory female pig genital tract (endocervix, endometrium and endosalpinx) 24 h after the entry of semen and/or sperm-free SP. Since the pig ejaculate is expelled in easily identifiable fractions5, where the 1st 10 mL of the sperm-rich fraction (SRF) hereby named P1-fraction contains around 25% of the total spermatozoa of the entire ejaculate mainly immersed in fluid from the epididymis cauda5, comparisons were made for the entire ejaculate or this P1-fraction either containing spermatozoa and seminal plasma (Mating: entire ejaculate vs P1-AI: cauda epididymides sperm-peak fraction) or their sperm-free counterparts (SP-Ejac vs SP-P1).
Results
Differential gene expression is induced by semen (M or P1-AI) and sperm-free SP (SP-Ejac or SP-P1) through the entire female genital tract
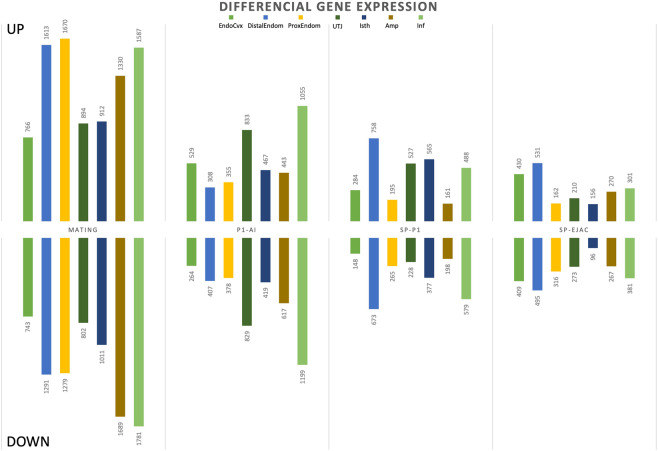
Following statistical analysis of the data provided by the Affymetrix microarray, we found that an exposure to semen modified the expression of a range of transcripts (1,509–3,369 (M) and 715–2,255 (P1-AI) transcripts) while sperm-free SP treatment altered fewer genes (SP-P1: 359–1,431; SP-Ejac: 252–1026) depending on the section of the genital tract assessed (Table 1). Figure 1 shows the numbers of genes differentially expressed in the mucosal samples collected along the sow internal genital tract. Overall, the highest differences were found in the M-group, compared to the rest of the experimental groups among all mucosal segments, with the exception of the utero-tubal junction (UTJ) where the total number of differentially expressed genes was similar between M- and P1-AI semen groups (1,696 vs. 1,662 genes, respectively).
Table 1.
Differential expression (up- and down-regulation) among treatments of total genes included in the microarrays (25,470) in the different segments of the internal genital tract of sows after mating (M) or artificial deposition of the sperm-peak portion P1 (P1-AI); sperm-free seminal plasma of from P1-fraction P1 (SP-P1) or of the whole ejaculate (SP-Ejac).
| M | M (%) | P1-AI | P1-AI (%) | SP-P1 | SP-P1 (%) | SP-Ejac | SP-Ejac (%) | |
|---|---|---|---|---|---|---|---|---|
| EndoCvx | 1509 | 5.9 | 793 | 3.1 | 432 | 1.7 | 839 | 3.3 |
| DistEndom | 2905 | 11.4 | 715 | 2.8 | 1431 | 5.6 | 1026 | 4 |
| ProxEndom | 2949 | 11.6 | 733 | 2.9 | 460 | 1.8 | 478 | 1.9 |
| UTJ | 1696 | 6.7 | 1662 | 6.5 | 755 | 3 | 483 | 1.9 |
| Isth | 1923 | 7.6 | 886 | 3.5 | 942 | 3.7 | 252 | 1 |
| Amp | 3028 | 11.9 | 1060 | 4.2 | 359 | 1.4 | 537 | 2.1 |
| Inf | 3369 | 13.2 | 2255 | 8.9 | 1068 | 4.2 | 682 | 2.7 |
All treatments were compared to Control (AI with 50 mL of BTS).
Figure 1.
Differential expression (up- and down-regulation) of annotated genes in the mucosa of the internal genital tract of sows after Mating or artificial deposition of the sperm-peak portion P1 (P1-AI); sperm-free seminal plasma of the whole ejaculate (SP-Ejac) or of pooled P1 (SP-P1). All treatments were compared to Control (AI with 50 mL of BTS). The numbers represent the number of differentially expressed genes (p-value < 0.05). Endocervix (EndoCvx), endometrium (distal: DistEndom or proximal: ProxEndom) and endosalpinx from 4 different segments; the utero-tubal junction (UTJ), isthmus (Isth), ampulla (Amp) and infundibulum (Inf).
Commonly altered genes among mucosal segments within each experimental group
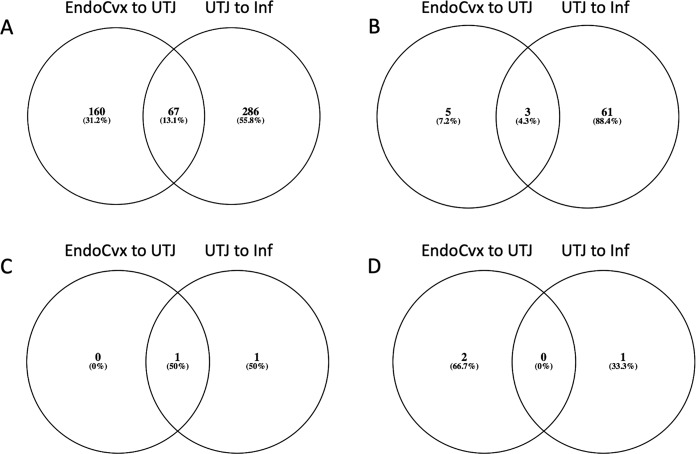
Each treatment induced transcriptomic changes that included a particular subset of genes common to all mucosal segments (the number of these common genes is shown in the Venn diagrams in Fig. 2). The M-treatment induced the largest number of altered genes common to all mucosal segments (19 genes UP- & 47 genes DOWN), followed by P1-AI (2 genes UP- & 1 genes DOWN), and SP-P1 (1 gene UP- & 0 genes DOWN). We did not find altered genes common within all mucosal segments of the SP-Ejac group. The names of the genes represented in the Venn diagrams are shown in Table 2.
Figure 2.
Venn diagrams depicting common genes among mucosal segments of the internal genital tract of the sow, after the various treatments: M (A), P1-AI (B), SP-P1 (C) and SP-Ejac (D).
Table 2.
Identification of the common genes among all tissues represented in Venn diagrams from Fig. 2.
| M | P1-AI | SP-P1 | SP-Ejac | ||||
|---|---|---|---|---|---|---|---|
| UP (19) | DOWN (48) | UP (2) | DOWN (1) | UP (1) | DOWN (0) | UP (0) | DOWN (0) |
| ABCA9, FAM107A, FIGF, FMO1, FMO2, GAB1, CCDC68, MICAL3,NDRG4, SLC2A12, LOC106504056, EZH1, PPIP5K1, DECR2, ALDH6A1, CHDH, LOC100522894, GLUL, NCOA2 | LOC100523107, FAM98A, CALR, VIMP, MIR671, EMC3, CALU, CPQ, SNX31, TMEM263, SEC. 23A, TPM4, RAI14, ARF4, TCEAL8, OSTC, B4GALT1, LOC100514264, RCN1, DNAJB11, ABHD2, GPX8, TSPAN6, HSPA13, FKBP4, KDELR3, TRPC5, LPPR5, FRD1, FAM69A, LOC100624484, SNAI2, REXO2, TDG, OLFML3, FAM19A4, HTR2A, CLRN3, DLGAP2, SRPX2, OLFM3, ENPEP, TAC3, PLAUR, VCAN, FGFBP1, HTRA4 |
AOAH LOC102162715 |
KIT | NMNAT1 | |||
Mating (M) or artificial deposition of the sperm-peak portion P1 (P1-AI); sperm-free seminal plasma of from P1-fraction P1 (SP-P1) or of the whole ejaculate (SP-Ejac). All treatments were compared to Control (AI with 50 mL of BTS).
Biological meaning of the differentially expressed transcripts
Analysis of functional categories
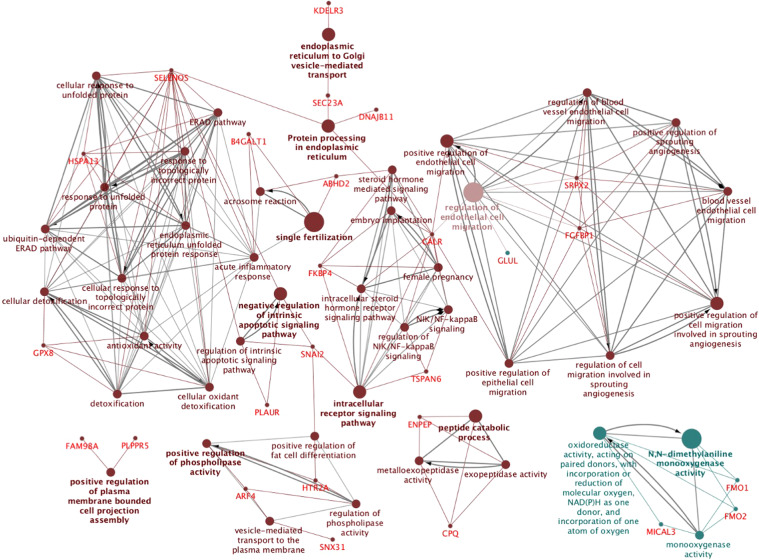
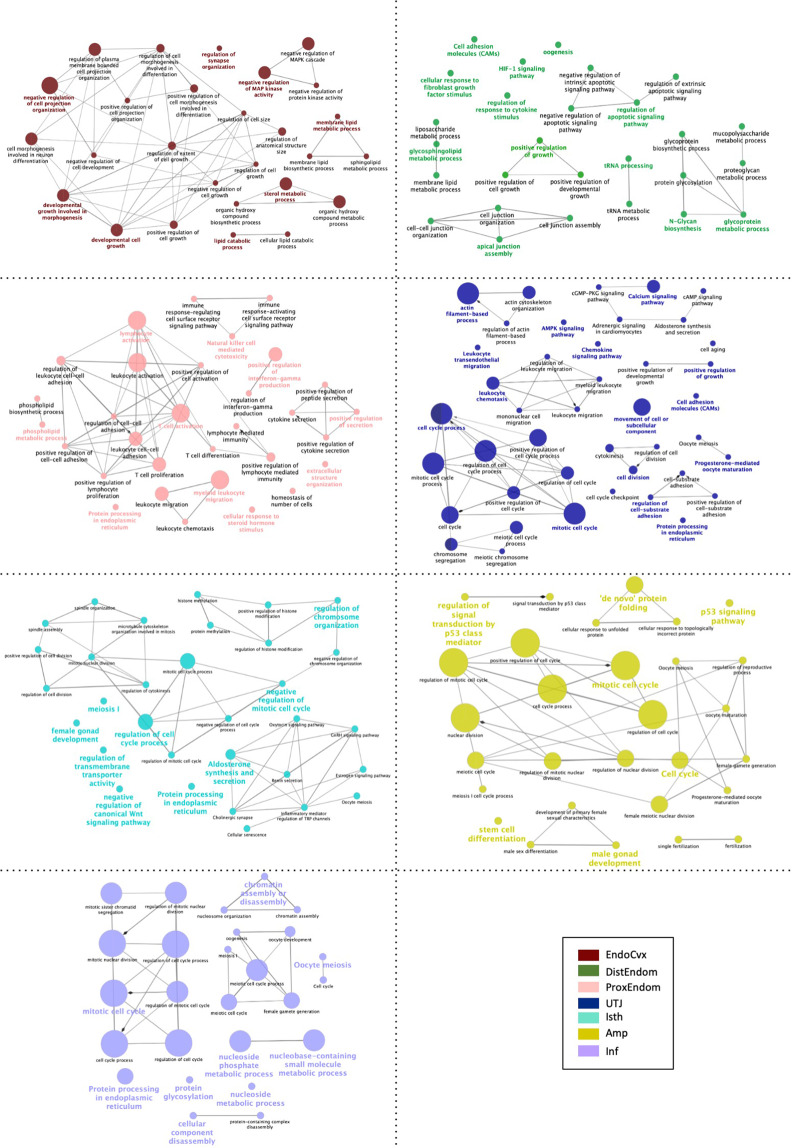
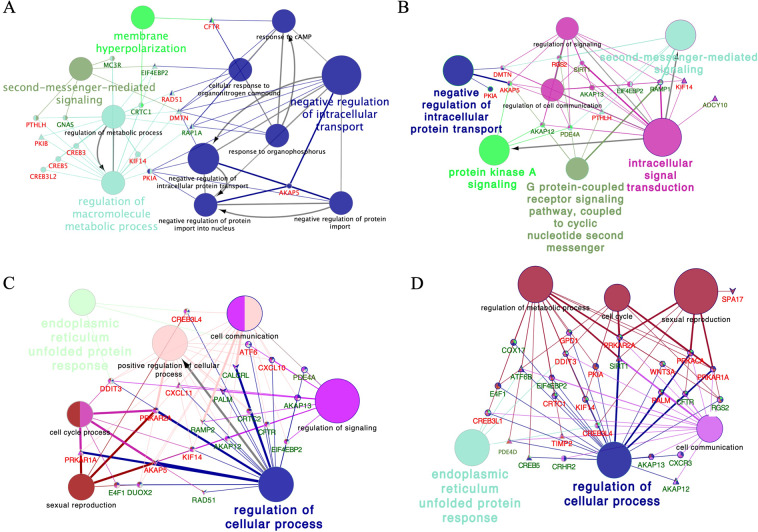
Since the M-group induced the largest number of transcriptomic changes among all tissues relative to the rest of the experimental groups, we performed an overall representation of the most significantly altered biological terms along the entire internal genital tract of the sow, where we found evidence of overrepresentation of processes involved in reproduction such as: single fertilization, acrosome reaction, steroid biosynthesis, and female pregnancy, amongst others (Fig. 3). Figure 4 shows the functional categories of altered genes in the P1-AI group from all mucosal segments analyzed (Endocervix (EndoCvx) to Infundibulum (Inf)) where many cellular processes were consistently represented. Interestingly, the oviduct segments (Isthmus utero-tubal junction (Isth) to Infundibulum (Inf)) identified a high number of GO terms associated with gamete function, oocyte meiosis, male and female gonadal development, male sex differentiation, etc. The most relevant GO terms found in the sperm-free SP groups are shown in Table 3, where the largest number of alterations are found in the SP-Ejac group, while the SP-P1 group identified functions related with sperm survival and apoptosis. Additional gene ontology (GO) analysis of altered cAMP-related transcripts in all experimental groups along the oviduct is presented in Fig. 5A–D (A, UTJ); (B, Isth); (C, Ampulla (Amp)); (D, Inf), depicting the consistent overrepresentation of cAMP-related genes throughout, highly relevant to sperm motility, the main functional mechanism for sperm release from the oviduct reservoir pending fertilization.
Figure 3.
Schematic representation of common altered transcripts among all mucosal tissues collected from the M group. The analysis of overrepresented functional categories was performed using the Cytoscape v3.0.0 application ClueGo v2.0.3. Terms are functionally grouped based on shared genes (kappa score) and are shown in different colors. The size of the nodes indicates the degree of significance, where the biggest nodes correspond to highest significance. The following ClueGo parameters were used: GO tree levels, 3–6 (first level = 0); minimum number of genes, 1; P-value correction, Benjamini-Hochberg, terms with P < 0.05, GO term connection restriction (kappa score), 0.4; GO term grouping, initial group size of 1. The resulting network was modified; that is, some redundant and non-informative terms were deleted and the network manually rearranged.
Figure 4.
Schematic representation of selected altered transcripts in the mucosae of segments of the internal genital tract of the sows in P1-AI group; Endocervix (EndoCvx: (A)), Endometrium (DisEndom: (B), ProEndom: (C)), Utero-tubal Junction (UTJ: (D)), Isthmus (Isth: (E)), Ampulla (Amp: (F)) and Infundibulum (Inf: (G)). The analysis of overrepresented functional categories was performed using the Cytoscape v3.0.0 application ClueGo v2.0.3. Terms are functionally grouped based on shared genes (kappa score) and are shown in different colors. The size of the nodes indicates the degree of significance, where the biggest nodes correspond to highest significance. The following ClueGo parameters were used: GO tree levels 3–6 (first level = 0); minimum number of genes 1; minimum percentage of genes 8, 5P-value correction, Benjamini-Hochberg, terms with P < 0.05, GO term connection restriction (kappa score) 0.4; GO term grouping, GO term fusion; initial group size of 3. The resulting network was modified; that is, some redundant and non-informative terms were deleted and the network manually rearranged.
Table 3.
Biological function of differentially expressed genes (Up & Down regulated) in sperm-free groups among the entire female genital tract of the sow.
| Genes | Tissue | Group | Expression | Biological role |
|---|---|---|---|---|
| GAPDH | Inf | SP-P1 | DOWN | Inition of apoptosis |
| ATM | DistEndom | SP-P1; SP-Ejac | UP | Phosphorylation of key proteins involved in DNA repair and apoptosis |
| Isth | SP-P1 | UP | ||
| IFN-alpha9 | ProxEndom | SP-P1 | DOWN | Activation of immune system |
| ProxEndom | SP-Ejac | UP | ||
| PRLR | ProxEndom | SP-P1; SP-Ejac | DOWN | Prosurvival factor for spermatozoa by inhibiting sperm capacitation through suppression of SRC kinase activation and stimulation of AKT |
| BCL6 | DistEndom | SP-Ejac | DOWN | Modulation of IL-4 and lead the differentiation of naive helper T-cells |
| CDKN2D | DistEndom | SP-Ejac | DOWN | Prevents the activation of CDK kinases |
| G6PC2 | DistEndom | SP-Ejac | DOWN | Catalyzes the final steps of gluconeogenic and glycogenolytic pathways |
| KRAS | DistEndom | SP-Ejac | UP | Controls cell proliferation |
| PRKAB1 | DistEndom | SP-Ejac | UP | Activated protein kinase subunit beta-1 |
| ANAPC13 | DistEndom | SP-Ejac | UP | Controls the G1 phase of the cell cycle |
| HDAC2 | DistEndom | SP-Ejac | UP | Involved in cell cycle progression |
| COL5A3 | ProxEndom | SP-Ejac | DOWN | Collagen alpha-3(V) chain, of the low abundance fibrillar collagens |
| CSF3 | ProxEndom | SP-Ejac | DOWN | Act in hematopoiesis by controlling the production, differentiation, and function of 2 related white cell populations of the blood, the granulocytes and the monocytes-macrophages |
| TSC2 | ProxEndom | SP-Ejac | DOWN | Stimulate GTPases. Interacts with Hsp70 and Hsp90 |
| CHST7 | EndoCvx, Dist endom | SP-Ejac | DOWN | Important determinant of sulfated GAGs expression and the associated function of chondroitin sulfates as regulators of many biologic processes |
| CHST3 | Endo cvx | SP-Ejac | DOWN | Related with GAGs as well as CHST7 |
| CHST12 | DistEndom | SP-Ejac | DOWN | Related with GAGs as well as CHST7 |
| DSE | EndoCvx | SP-Ejac | DOWN | Induce T lymphocytes |
| E2F3 | DistEndom | SP-Ejac | DOWN | Transcription factor, control cycle |
| STAG2 | DistEndom | SP-Ejac | UP | Part of the cohesion comples to hold the chromatid togheter |
| FGFR1 | ProxEndom | SP-Ejac | DOWN | Indirect activation of the gamma isoforms of phospholipase C (PLCγ) |
| PPP2R5D | ProxEndom | SP-Ejac | UP | Negative control of cell growth and division |
Sperm-free seminal plasma of from P1-fraction P1 (SP-P1) or of the whole ejaculate (SP-Ejac). All treatments were compared to Control (AI with 50 mL of BTS).
Figure 5.
Schematic representation of CAMP-related transcripts in the mucosa of the segments of the sow oviduct [Utero-tubal junction (UTJ: (A)), Isthmus (Isth: (B)), Ampulla (Amp: (C)) and Infundibulum (Inf: (D))] after mating (M: Elipse); Artificial insemination of the sperm-peak fraction (P1-AI: Triangle), seminal plasma from the sperm-peak fraction (SP-P1: Hexagon); or seminal plasma from the entire ejaculate (SP-Ejac: V). The analysis of overrepresented functional categories was performed using the Cytoscape v3.0.0 application ClueGo v2.0.3. Terms are functionally grouped based on shared genes (kappa score) and are shown in different colors. The size of the nodes indicates the degree of significance, where the biggest nodes correspond to highest significance. The following ClueGo parameters were used: GO tree levels, 1–4 (first level = 0); minimum number of genes, 1; P-value correction, Benjamini-Hochberg, terms with P < 0.05, GO term connection restriction (kappa score), 0.4; GO term grouping, initial group size of 1. The resulting network was modified; that is, some redundant and non-informative terms were deleted and the network manually rearranged.
Pathway analysis
We selected among differently expressed biological pathways depending on their statistical significance (P < 0.05) in regards to the different treatments and mucosal segments of the internal genital tract (Tables 4–7). We found many altered pathways with important roles in reproductive-related processes such as the phosphatidylinositol 3-kinase (PI3K-Akt) pathway, which was altered in the M-group (distal and proximal endometria (DistEndom, ProxEndom) and Amp) and also in the SP-Ejac (ProxEndom) and SP-P1 (ProxEndom and Amp) groups (Tables 4, 6 and 7), the FoxO signaling pathway, modified by M (Amp and Inf), P1-AI (Inf), SP-Ejac (DistEndom) and SP-P1 (Inf) (Tables 3–6), or the N-glycans biosynthesis, that was enriched after M (UTJ, Amp and Inf), P1-AI (EndoCvx, Isth and Inf), and SP-P1 (EndoCvx) (Tables 4, 5 and 7). The Estrogen signaling pathway appeared to be mainly altered after the sperm-free SP treatment SP-P (UTJ and Isth) (Table 7). The Supplementary figures 1 to 14 depict a detailed view of the up- and down-regulation of genes according to Biological and Molecular functions. In general, for biological functions, Binding and Catalytic activity were constantly overrepresented in all mucosal segments, independently of the experimental group. In the molecular function analysis, the cellular process of biological regulation, the response to stimulus developmental process and metabolic processes were overrepresented, independ of both the mucosal segment and experimental group considered.
Table 4.
List of the most significant differently expressed biological pathways (P < 0.05) examined with KEGG database in mating (M), in comparison with the Control group.
| PATHWAY ID | PATHWAY NAME | ENRICHMENT SCORE | P-VALUE | GENES ALTERED% | GENE LIST | |
|---|---|---|---|---|---|---|
| EndoCvx | ||||||
| 1 | KEGG_103 | Steroid biosynthesis | 6.7 | 0.001 | 41.7 | CYP51, FAXDC2, MSMO1, NSDHL, SQLE |
| 2 | KEGG_8 | Glycosaminoglycan biosynthesis | 5.6 | 0.003 | 33.3 | CHST14, CHST3, CHSY1, CHSY3, DSE |
| 3 | KEGG_34 | Focal adhesion | 5.3 | 0.005 | 14.7 | ACTB, CAV1, CAV2, CDC42, COL1A2, COL3A1, COL6A3, COL6A3, ERBB2, HGF, IGF1R, ITGA2, ITGB6, MAPK1, ROCK2, THBS1, THBS3, VEGFB, ZYX |
| 4 | KEGG_49 | Adherens junction | 5 | 0.007 | 19.6 | ACTB, CDC42, CSNK2A2, CSNK2B, ERBB2, IGF1R, INSR, MAPK1, SNAI2 |
| DistEndom | ||||||
| 1 | KEGG_34 | Focal adhesion | 10.6 | 0.0002 | 25.2 | CCND2, COL11A1, COL1A2, COL3A1, COL4A1, COL4A6, COL5A2, COL6A3, COL6A3, COL6A3, ERBB2, FLT1, IGF1, IGF1R, ITGA1, ITGA2, ITGA5, ITGA8, ITGAV, ITGB6, LAMB1, MAPK1, MAPK8, PDGFC, PDGFD, SHC1, THBS1, THBS3, TLN2, VCL, VEGFA, VEGFB, ZYX |
| 2 | KEGG_213 | PI3K-Akt signaling pathway | 5.4 | 0.004 | 17.9 | CCND2, COL11A1, COL1A2, COL3A1, COL4A1, COL4A6, COL5A2, COL6A3, COL6A3, COL6A3, CREB3L1, CREB3L2, CREB5, CSF1R, CSF3, FGF17, FGF2, FGF20, FGFR1, FLT1, G6PC2, IGF1, IGF1R, IL7, ITGA1, ITGA2, ITGA5, ITGA8, ITGAV, ITGB6, LAMB1, MAPK1, MYB, PDGFC, PDGFD, PIK3AP1, PPP2R5D, STK11, THBS1, THBS3, VEGFA, VEGFB |
| 3 | KEGG_8 | Glycosaminoglycan biosynthesis | 5.2 | 0.005 | 40 | CHST12, CHST7, CHSY1, CHSY3, CSGALNACT2, DSE |
| 4 | KEGG_46 | Estrogen signaling pathway | 3.7 | 0.02 | 26.9 | CALM1, CREB3L1, CREB3L2, CREB5, FKBP4, HBEGF, HSPA2, MAPK1, MMP2, MMP9, PLCB4, PRKACA, SHC1 |
| 5 | KEGG_49 | Adherens junction | 3.2 | 0.03 | 21.7 | ERBB2, FGFR1, IGF1R, MAPK1, PTPN1, PVRL1, SNAI2, SORBS1, TCF7L2, VCL |
| ProxEndom | ||||||
| 1 | KEGG_34 | Focal adhesion | 8.8 | 0.0001 | 20.3 | BRAF, CCND2, COL1A2, COL3A1, COL4A1, COL5A2, COL6A3, COL6A3, COMP, CTNNB1, FIGF, FLT1, HGF, IGF1, IGF1R, ITGA1, ITGA2, ITGA8, KDR, LAMB1, MYLPF, PIK3R5, SHC1, TLN2, TNR, VEGFA |
| 2 | KEGG_213 | PI3K-Akt signaling pathway | 7.8 | 0.0004 | 16.5 | ANGPT2, CCND2, COL1A2, COL3A1, COL4A1, COL5A2, COL6A3, COMP, CREB3L1, CREB3L2, CREB3L4, CREB5, CSF1R, CSF3, EFNA2, FGF17, FGF21, FGF5, FGFR1, FIGF, FLT1, G6PC2, GH1, HGF, IGF1, IGF1R, IL7, ITGA1, ITGA2, ITGA8, KDR, LAMB1, PIK3AP1, PIK3R5, STK11, TNR, VEGFA |
| 3 | KEGG_98 | Rap1 signaling pathway | 6.3 | 0.001 | 17.5 |
ANGPT2, BRAF, CALM1, CSF1R, CTNNB1, EFNA2, FGF17, FGF21, FGF5, FGFR1, FIGF, FLT1, GNAS, GRIN2B, HGF, IGF1, IGF1R, KDR, PIK3R5, PLCB4, RAPGEF2, SIPA1L1 TLN2, VEGFA |
| 4 | KEGG_46 | Estrogen signaling pathway | 5.6 | 0.003 | 21.3 | CALM1, CREB3L1, CREB3L2, CREB3L4, CREB5, FKBP4, GNAS, HSPA2, KCNJ3, MMP9, PIK3R5, PLCB4, SHC1 |
| 5 | KEGG_105 | Cell adhesion molecules | 5.4 | 0.004 | 18.7 | CDH15, CLDN11, CLDN2, CLDN5, ITGA8, NRXN2, NTNG1, PVRL1, SDC1, SDC2, SDC4, SELL, SELP, SIGLEC1, SLA-8, VCAM1, VCAN |
| 6 | KEGG_165 | Mucin type O-Glycan biosynthesis | 4.5 | 0.01 | 28.6 | GALNTL5, GCNT1, GCNT3, GCNT4, ST3GAL1, ST6GALNAC1 |
| 7 | KEGG_8 | Glycosaminoglycan biosynthesis | 4.2 | 0.01 | 36.4 | B3GNT2, B4GALT1, CHST4, ST3GAL1 |
| UTJ | ||||||
| 1 | KEGG_103 | Steroid biosynthesis | 8.7 | 0.0007 | 21.4 | CYP51, FAXDC2, LIPA, MSMO1, NSDHL, SQLE |
| 2 | KEGG_110 | N-glycans biosynthesis | 4.8 | 0.008 | 21.6 | ALG13, B4GALT1, B4GALT3, DDOST, DPM3, MAN2A1, MGAT4B, STT3A |
| 3 | KEGG_8 | Glycosaminoglycan biosynthesis | 3.6 | 0.02 | 26.7 | B4GALT7, CHST12, CHSY3, DSE |
| Isth | ||||||
| 1 | KEGG_59 | Cell cycle | 6.7 | 0.001 | 20.3 | ANAPC13, ATM, CCNB2, CCND2, CDC20, CDC25C, EP300, ESPL1, FZR1, ORC4, PTTG1, RB1, STAG2, TP53, TTK |
| 2 | KEGG_34 | Focal adhesion | 3.5 | 0.03 | 13.8 | BRAF, CCND2, COL4A1, COL5A2, CTNNB1, ELK1, FIGF, HGF, IGF1R, LAMA4, MYLK2, PAK1, PTEN, SHC1, SOS2, SRC, VEGFB |
| 4 | KEGG_49 | Adherens junction | 3.2 | 0.03 | 17.4 | CTNNB1, EP300, IGF1R, SNAI2, SORBS1, SRC, SSX2IP, WASL |
| KEGG_98 | Rap1 signaling pathway | 3.2 | 0.04 | 13.1 | ADORA2A, BRAF, CTNNB1, FIGF, GNAI2, GNAO1, HGF, IGF1R, LPAR2, MAGI3, MAP2K6, MAPK14, P2RY1, PLCB4, RAPGEF6, RASGRP2, SRC, VEGFB | |
| Amp | ||||||
| 1 | KEGG_59 | Cell cycle | 12.4 | 0.0004 | 32.4 | ABL1, ANAPC13, BUB3, CCNA1, CCNB2, CCNB3, CCNE2, CCNH, CDC20, CDC23, CDC25B, CDC25C, CDC27, CDK1, CHEK1, E2F5, EP300, HDAC2, MCM4, MDM2, PTTG1, TTK, YWHAH, YWHAZ |
| 2 | KEGG_66 | FoxO signaling pathway | 6.4 | 0.001 | 23.7 | AKT2, ARAF, BCL2L11, BCL6, CCNB2, CCNB3, CHUK, EP300, FOXO3, IGF1R, INSR, KRAS, MAPK1, MAPK8, MDM2, PCK2, PIK3R5, PLK4, RAG1, RAG2, STAT3, USP7 |
| 3 | KEGG_195 | HIF-1 signaling pathway | 5.3 | 0.004 | 23.4 | AKT2, EGLN1, EGLN3, EIF4E, EP300, ERBB2, HK1, HKDC1, IFNGR1, IGF1R, INSR, MAPK1, NFKB1, PDHB, PIK3R5, RELA, STAT3, TCEB1 |
| 4 | KEGG_103 | Steroid biosynthesis | 4.5 | 0.01 | 41.7 | CYP24A1, HSD17B7, MSMO1, NSDHL, SQLE |
| 5 | KEGG_213 | PI3K-Akt signaling pathway | 3.8 | 0.02 | 17 | AKT2, BCL2L1, BCL2L11, CCNE2, CHUK, COL5A3, CRTC2, DDIT4, EFNA1, EIF4E, FGF19, FN1, FOXO3, GHR, GNB5, GNG11, HGF, IGF1R, INSR, ITGA11, ITGB4, KRAS, LPAR1, LPAR2, LPAR6, MAPK1, MDM2, NFKB1, PCK2, PIK3R5, PPP2R2A, RELA, TNC, TNR, TSC2, VEGFB, YWHAH, YWHAZ |
| 6 | KEGG_110 | N-glycans biosynthesis | 3.4 | 0.03 | 24.3 | ALG14, ALG5, B4GALT1, DAD1, DDOST, DOLPP1, MAN2A2, RPN1, STT3A |
| Inf | ||||||
| 1 | KEGG_59 | Cell cycle | 7.3 | 0.0006 | 28.4 | ANAPC13, BUB3, CCNB2, CCNB3, CCNE2, CDC20, CDC23, CDC25B, CDC25C, CDC27, CDK1, CDK6, CDKN2D, E2F3, EP300, FZR1, HDAC2, PTTG1, TGFB2, TTK, YWHAH |
| 2 | KEGG_49 | Adherens junction | 7.1 | 0.0008 | 32.6 | CDC42, CSNK2B, CTNNA3, EP300, ERBB2, IGF1R, IQGAP1, MAP3K7, MAPK1, MET, PTPN1, PVRL4, SNAI2, SORBS1, SSX2IP, TCF7L1 |
| 3 | KEGG_195 | HIF-1 signaling pathway | 6.7 | 0.001 | 27.3 | ANGPT1, CAMK2D, EGLN1, EGLN3, EIF4E, EP300, ERBB2, GAPDH, HK1, IFNGR1, IGF1, IGF1R, LTBR, MAPK1, MKNK2, NFKB1, NOS2, PDHB, RPS6, STAT3, TEK |
| 4 | KEGG_66 | FoxO signaling pathway | 5.8 | 0.002 | 24.7 | BCL2L11, BCL6, CAT, CCNB2, CCNB3, CDKN2D, CHUK, EP300, FOXO4, GABARAP, IGF1, IGF1R, KRAS, MAPK1, PLK4, PRKAB1, PRMT1, SIRT1, SOS2, STAT3, TGFB2, TNFSF10, USP7 |
| 5 | KEGG_110 | N-glycans biosynthesis | 4.7 | 0.008 | 29.7 | ALG14, ALG2, ALG5, DAD1, DDOST, DOLPP1, MAN1A1, MAN2A2, MGAT2, RPN1, STT3A |
| 6 | KEGG_103 | Steroid biosynthesis | 4.1 | 0.01 | 25.5 | CYP24A1, LIPA, MSMO1, NSDHL, SQLE |
*Upregulated genes are marked in bold; the unmarked were downregulated.
Endocervix (EndoCvx), Endometrium (DistEndom, ProxEndom), and endosalpinx from Utero-tubal Junction (UTJ), Isthmus (Isth), Ampulla (Amp) and Infundibulum (Inf).
Table 7.
List of the most significant differently expressed biological pathways (P < 0.05) examined with KEGG database in artificial deposition of sperm-free seminal plasma (SP) from entire ejaculate (SP-Ejac) group, in comparison with the Control group.
| PATHWAY ID | PATHWAY NAME | ENRICHMENT SCORE | P-VALUE | GENES ALTERED% | GENE LIST | |
|---|---|---|---|---|---|---|
| EndoCvx | ||||||
| 1 | KEGG_105 | Glycosaminoglycan biosynthesis | 3.5 | 0.03 | 20 | CHST3, CHST7, DSE |
| DistEndom | ||||||
| 1 | KEGG_66 | FoxO signaling pathway | 4.1 | 0.01 | 10.8 | ATG12, ATM, BCL6, CDKN2D, G6PC2, HOMER2, KRAS, MAPK10, PRKAB1, PRKAB2 |
| 2 | KEGG_59 | Cell cycle | 3.5 | 0.02 | 10.8 | ANAPC13, ANAPC5, ATM, CDKN2D, E2F3, HDAC2, SMC3, STAG2 |
| 3 | KEGG_105 | Glycosaminoglycan biosynthesis | 3.3 | 0.03 | 20 | CHST7, CHST12, CHST15 |
| ProxEndom | ||||||
| 1 | KEGG_213 | PI3K-Akt signaling pathway | 3.6 | 0.02 | 5.6 | COL5A3, CSF3, FGFR1, IFN-ALPHA-9, JAK1, MCL1, OSM, PKN3, PPP2R5D, PRLR, TN-X, TSC2 |
*Upregulated genes are marked in bold; the unmarked were downregulated.
Endocervix (EndoCvx), Endometrium (DistEndom, ProxEndom).
Table 6.
List of the most significant differently expressed biological pathways (P < 0.05) examined with KEGG database in artificial deposition of sperm-free seminal plasma (SP) from P1-fraction (SP-P1) group, in comparison with the Control group.
| PATHWAY ID | PATHWAY NAME | ENRICHMENT SCORE | P-VALUE | GENES ALTERED% | GENE LIST | |
|---|---|---|---|---|---|---|
| EndoCvx | ||||||
| 1 | KEGG_105 | Cell adhesion molecules | 3.7 | 0.02 | 5.5 | CD8B, SLA-1, SLA-DOA, SLA-DQB1, VCAM1 |
| 2 | KEGG_110 | N-glycans biosynthesis | 3.5 | 0.03 | 8.1 | ALG13, ALG5, B4GALT3 |
| DistEndom | ||||||
| 1 | KEGG_133 | Mismatch repair | 4.9 | 0.007 | 31.2 | AKT2, ATM, BCL2L1, CSF2RB, IRAK1, MYD88, PPP3CB, RELA, RIPK1, TNFRSF1A |
| 2 | KEGG_56 | Apoptosis | 4.7 | 0.082 | 19.2 | LIG1, MSH2, MSH3, PMS2, SSBP1 |
| ProxEndom | ||||||
| 1 | KEGG_213 | PI3K-Akt signaling pathway | 5.6 | 0.003 | 8 | ANGPT1, ANGPT2, CDK6, COL1A2, COL3A1, COL5A2, COL5A3, COL6A3, EPO, FLT1, IFN-ALPHA-9, ITGA5,, LAMA4, LPAR3, PGF, PIK3R5, PPP2CB, PRLR |
| 2 | KEGG_34 | Focal adhesion | 3.6 | 0.02 | 8.1 | COL1A2, COL3A1, COL5A2, COL5A3, COL6A3, FLT1, ITGA5, LAMA4, PGF, PIK3R5 |
| 3 | KEGG_195 | HIF-1 signaling pathway | 3.4 | 0.03 | 9.1 | EPO, NPPA, PIK3R5, TFRC |
| UTJ | ||||||
| 1 | KEGG_139 | GnRH signaling pathway | 5.1 | 0.006 | 8.8 | ATF4, GNRHR2, GRB2, MAP2K1, SOS2 |
| 2 | KEGG_46 | Estrogen signaling pathway | 4.8 | 0.008 | 8.1 | ATF4, GRB2, MAP2K1, SHC1, SOS2 |
| Isth | ||||||
| 1 | KEGG_34 | Focal adhesion | 5.6 | 0.003 | 12.2 | AKT3, BRAF, FYN, IBSP, ITGA2B, ITGA4, JUN, LAMA4, MRLC2, MYLK, MYLK2, PTEN, SHC4, SOS2, VCL |
| 2 | KEGG_98 | Rap1 signaling pathway | 5.5 | 0.004 | 11.7 |
AKT3, ARAP3, BRAF, CNR1, FGF17, FGF9, GNAO1, GNAQ INSR, ITGA2B, KRIT1, LCP2, P2RY1, RAPGEF6, RASGRP3, RGS14 |
| 3 | KEGG_66 | FoxO signaling pathway | 5.1 | 0.005 | 12.9 | AKT3, ATM, BRAF, CDKN1A, CDKN2B, FOXO3, INSR, PRKAA2, PTEN, SIRT1, SMAD4, SOS2 |
| 4 | KEGG_213 | PI3K-Akt signaling pathway | 4.9 | 0.007 | 9.8 | AKT3, CDKN1A, CRTC2, FGF17, FGF9, FOXO3, GHR, GNG13, IBSP, IL2RG, INSR, ITGA2B, ITGA4, JAK2, LAMA4, OSM, PRKAA2, PTEN, SOS2, SYK, TP53, YWHAQ |
| 5 | KEGG_46 | Estrogen signaling pathway | 3.9 | 0.02 | 13.1 | AKT3, FKBP4, GNAO1, GNAQ, JUN, MMP9, SHC4, SOS2 |
| 6 | KEGG_59 | Cell cycle | 3.8 | 0.02 | 12.2 | ATM, CCNH, CDKN1A, CDKN2B, RB1, SMAD4, STAG2, TP53, YWHAQ |
| 7 | KEGG_252 | mTOR signaling pathway | 3.2 | 0.03 | 13.3 | AKT3, BRAF, PRKAA2, PTEN, RICTOR, RRAGC |
| 8 | KEGG_49 | Adherens junction | 3.1 | 0.04 | 13 | FYN, INSR, SMAD4, VCL, WASF1, WASL |
| Inf | ||||||
| 1 | KEGG_195 | HIF-1 signaling pathway | 4.2 | 0.01 | 15.6 | ANGPT4, EGLN2, ENO3, GAPDH, IFNGR1, IL-6, LTBR, MTOR, PIK3R2, PLCG1, RELA, TEK |
*Upregulated genes are marked in bold; the unmarked were downregulated.
Endocervix (EndoCvx), Endometrium (DistEndom, ProxEndom), and endosalpinx from Utero-tubal Junction (UTJ), Isthmus (Isth) and Infundibulum (Inf).
Table 5.
Most significant differently expressed biological pathways (P < 0.05) examined with KEGG database in artificial insemination of P1 fraction (P1-AI) group in comparison with the Control group.
| PATHWAY ID | PATHWAY NAME | ENRICHMENT SCORE | P-VALUE | GENES ALTERED% | GENE LIST | |
|---|---|---|---|---|---|---|
| EndoCvx | ||||||
| 1 | KEGG_103 | Steroid biosynthesis | 6.1 | 0.002 | 12.2 | SQLE, MSMO1, CYP51 |
| 2 | KEGG_110 | N-glycans biosynthesis | 4 | 0.01 | 10.8 | ALG8, B4GALT3, DPM2, UT8 |
| 3 | KEGG_8 | Glycosaminoglycan biosynthesis | 3.3 | 0.03 | 18.1 | B4GALT3, FUT8 |
| DistEndom | ||||||
| 1 | KEGG_91 | Rap1 signaling pathway | 5.6 | 0.003 | 7.3 | ARAP3, KIT, PLCB4, FGF17, FGF9, FGFR1, FLT4, ITGAL, SKAP1, CALM1 |
| ProxEndom | ||||||
| 1 | KEGG_103 | Steroid biosynthesis | 5.7 | 0.0002 | 20.7 | TM7SF2, LIPA, FAXDC2, CYP51 |
| 2 | KEGG_91 | Rap1 signaling pathway | 3.1 | 0.04 | 9.5 |
FGFR1, KIT, ANGPT2, FGFR2, SIPA1L2, GNAS, ITGAL, PLCB1, IGF1 EFNA4, RAPGEF2, LPAR3, LPAR2 |
| 3 | KEGG_250 | Glycosaminoglycan degradation | 3 | 0.05 | 20 | GUSB, HEXA, NAGLU |
| UTJ | ||||||
| 1 | KEGG_91 | Rap1 signaling pathway | 6.5 | 0.001 | 12.4 |
FIGF, ARAP2, IGF1R LPAR1, KIT, RRAS, INSR, AKT3, ITGAL, MAP2K1, PLCB4, TLN1, IGF1, FGF1, SRC, SKAP1, CTNND1 |
| 2 | KEGG_49 | Adherens junction | 5.7 | 0.003 | 17.4 | CSNK2A2, IGF1R, INSR, SORBS1, SRC, WASF1 |
| 3 | KEGG_139 | GnRH signaling pathway | 3.3 | 0.03 | 12.3 | FSHB, CAMK2G, MAP2K1, PLCB4, GNRHR2, SRC, MAP3K3 |
| 4 | KEGG_34 | Focal adhesion | 3.2 | 0.04 | 9.7 | FIGF, LAMA4, VAV2, IGF1R, THBS3, ITGA8, AKT3, PAK7, MAP2K1, TLN1, IGF1, SRC |
| Isth | ||||||
| 1 | KEGG_110 | N-glycans biosynthesis | 4.2 | 0.01 | 21.6 | ALG8, B4GALT1, DAD1, DPM1, MAN2A1, MGAT2, MOGS, STT3A |
| 2 | KEGG_252 | mTOR signaling pathway | 4.1 | 0.01 | 20 | AKT3, IGF1, MLST8, PIK3CD, PRKAA2, PRKCA, RRAGC, ULK1, VEGFA |
| Inf | ||||||
| 1 | KEGG_66 | FoxO signaling pathway | 5.8 | 0.002 | 25.8 | BCL6, BRAF, CCNB3, CCNG2, CDKN2D, EP300, FOXO3, GADD45G, IGF1, IGF1R, INSR, IRS1, MAPK10, MDM2, PCK2, PIK3R2, PLK3, PLK4, PRMT1, SIRT1, SOS2, STK11, TGFBR1, TNFSF10 |
| 2 | KEGG_59 | Cell cycle | 4 | 0.01 | 24.3 | BUB3, CCNB3, CCNH, CDC45, CDK1,CDKN2D, E2F1, E2F4, E2F5, EP300, ESPL1, GADD45G, MDM2, PTTG1, TP53, YWHAG, YWHAH, YWHAQ |
| 3 | KEGG_110 | N-glycans biosynthesis | 3.4 | 0.03 | 24.5 | ALG13, DAD1, DDOST, DOLPP1, DPM3, MAN1A1, MGAT4A, MGAT5, MOGS, STT3A |
| 4 | KEGG_195 | HIF-1 signaling pathway | 3 | 0.04 | 22.1 | ARNT, CAMK2D, EGLN1, EIF4E, EP300, EPO, GAPDH, IGF1, IGF1R, INSR, LTBR, NFKB1, PDHB, PDHB, PIK3R2, PLCG1, TEK, VHL |
*Upregulated genes are marked in bold; the unmarked were downregulated.
Endocervix (EndoCvx), Endometrium (DistEndom, ProxEndom), and endosalpinx from Utero-tubal Junction (UTJ), Isthmus (Isth), Infundibulum (Inf).
Discussion
Although the effects of semen and SP on the porcine uterus have been previously characterized13, the present study provides, to the best of our knowledge, the first description of a genome-wide gene expression analysis of specific endo-cervical, endometrial and endosalpingeal responses to semen or SP (either of the entire ejaculate or of the sperm-peak fraction of the boar ejaculate). We have compared the portion of the ejaculate first contacting the internal genital tract of the sow5,14 with the non-exposed control sows in the same period of the oestrous cycle: peri-ovulation. Considering the transcriptomic effects we detected during the pre-ovulatory oestrus can continue over ovulation and even be visible during the progesterone-dominated stages of the oestrus cycle/early pregnancy, a follow-up study should include experimental stimulation during the post-ovulatory oestrus.
Differences were seen among the various exposures (sperm-containing or solely seminal plasma). Despite these differences were clear, we should not forget that there are other factors that could play roles in the effects, and preliminarily appear as confounding factors. For instance, the entry into oestrus, and the use of manual detection of oestrus behavior both cause elevations of female oxytocin in peripheral blood15. Mating dramatically increases this oxytocin release16 while manipulation of uterus and its cervix causes intermediate responses17. The study, however, was focused on those aspects solely vinculated to the entry and interaction of semen on/with the female internal genital tract. We separated the whole ejaculate (natural mating) from AI with the sperm-peak fraction in a fixed volume, the 1st 10 mL of the sperm-rich fraction (SRF) hereby defined as P1, which contains 25% of the total number of spermatozoa of the entire ejaculate5,18,19. Using such fractionation, we were able to use the same volumen and relative sperm concentration from P1 (containing spermatozoa), derived from ejaculates of 2 boar siblings, whose sperm quality was very similar in between.
The retrieval of mucosal samples 24 hours after semen or SP-only exposure explores the period between semen deposition and the colonization of the sperm reservoir at the UTJ, including sperm transport towards the oviduct. The enrichment analysis of the collected mucosae revealed an overrepresentation of several genes and pathways associated with many reproductive-related processes relevant for the colonization of the sperm reservoirs such as: sperm transport and survival, antioxidant activity, sperm binding and steroid biosynthesis, amongst others.
The phosphatidylinositol 3-kinase (PI3K-Akt) signaling pathway, was one of the most significant altered pathways found in the endometria (Dist and Prox) as well as in the ampulla after mating. This pathway was also altered after SP-Ejac (ProxEndom) and after SP-P1 (ProxEndom and Isth). The PI3K-Akt signaling pathway plays a pivotal role in numerous reproductive processes as an important regulator of cell proliferation and survival, being widely studied due to its participation in endometrial receptivity in several species including the pig20,21. Although the PI3K-Akt signaling pathway is considered strongly associated to uterine decidualization in invasive-implantation species, there is evidence that in porcine (a non-invasive-implantation species), the endometrium undergoes a continuous cycle of changes during pregnancy, including events such as cellular remodeling as well as maintaining hormonal responsiveness and secretory function by the uterine epithelium22. These processes prepare the genital tract mucosae for fertilization, embryo development and implantation23. Interestingly, we found 39 altered transcripts related to the PI3K-Akt signaling pathway in the endometrial samples within the Mating group, suggesting that just 24 hours after semen deposition, the mechanisms that would lead to building up a receptive endometrial environment for embryo survival and implantation are already activated. One of the most interesting genes associated with this pathway is the colony stimulation factor 1 and its receptor (the CSF1-CSF1R complex), that were up-regulated in the endometrial, ampullar and infundibular mucosae after mating. The CSF1-CSF1R complex has been widely related to many biological processes, such as protein phosphorylation, cell proliferation, cell migration, cellular response to macrophage colony-stimulating factor stimulus, cell-cell junction maintenance and cellular response to cytokine stimulus24. Here, we provide evidence that this complex is overexpressed within the entire female reproductive tract after mating, thus triggering the activation of several cellular processes.
Additionally, the FoxO signaling pathway, regulated by the PI3K-Akt signaling pathway25,26, is overrepresented in the upper segments of the oviduct (ampulla and infundibulum) mostly by upregulation. The FoxO transcription factors regulate the expression of genes that are involved in apoptosis, oxidative stress resistance, DNA repair, cell cycle transitions and differentiation27,28 and play important roles in reproductive processes by increasing cellular antioxidant activity29–31. All of these processes are relevant to the site of fertilization, with the presence of immunologically foreign-spermatozoa (as well as a high rate of epithelial cell turnover32). Specially, FOXO3 and FOXO4, which were up-regulated in our experiment by Mating or AI with the P1-semen fraction, have been reported to provide cell protection against ROS-induced damage by directly increasing their quantities of manganese superoxide dismutase (MnSOD)33,34. Insemination triggers an inflammatory cascade in the internal female genital tract during the peri-ovulatory period6,35; an inflammatory process intimately related to the action of reactive oxygen species (ROS)36. Spermatozoa in general, but those of boar in particular, are highly vulnerable to oxidative stress due to the high polyunsaturated acid content in their membranes37. Therefore, we hypothesize that FoxOs are physiologically activated in the oviduct in response to semen exposure in order to overcome the negative impact of oxygen-induced damage surrounding the inflammatory cascade. Once the spermatozoa reach the oviduct, they elicit changes in gene expression to promote a favorable environment by influencing the release of many oviduct secretions38. We identified several genes encoding oviduct-specific proteins with important roles in sperm binding, sperm protection, smooth muscle contractility or gamete-oviduct interactions, suggesting that entry of semen leads to the activation of oviduct mechanisms that trigger these processes. There are several reports studying the mechanisms that might be responsible for retaining spermatozoa in the oviductal reservoir of the sow38, although the molecular interactions that influence this phenomenon as yet unclear. Considerable novel evidence supports the involvement of glycans in oviductal sperm colonization39 which includes the role played N- and O-linked glycans found in oviduct epithelial cells that, by binding specific receptors found in the sperm head membrane40, aid establishing the tubal reservoir in the pig41. Sperm binding to oviduct cells via glycans have been even proposed as additional biomarkers for semen analysis42. At 24 hours after semen deposition, ST3GAL5, MAN1C1, B4GALT1 and MAN2A1, which encode enzymes that modify already syntesized N-linked and/or O-linked glycans by adding or removing a single monosaccharide, were upregulated in the Isthmic segment after Mating and P1-AI treatments respectively. Such findings support the prevailing concept that pig spermatozoa, being stored for up to 30–36 hours in the oviductal reservoir, maintain viability and fertilizing capacity until ovulation takes place12,43,44. In addition, various carbohydrate sulfotransferases genes (CHST7, CHST3 and CHST12), involved in the catalysis of the sulfation of glycosaminoglycans (GAGs)45 were also overrepresented in our study. The non-sulfated GAG hyaluronan is produced by oviduct epithelial cells to act multipurposely, being related to the capture of spermatozoa during their storage in the isthmus reservoir by adhering to sperm surface membrane, to sperm capacitation, acrosome reaction and fertilization32,38,46,47. We identified mostly downregulated targets implicated in the synthesis of HA or its receptors in the Infundibular segment; HAS2, HYAL2, VCAN, HMMR, SPAM1 after Mating, and of VCAN, HAPLN1, HABP4 in the P1-AI group. Similarly, we observed many genes encoding binding-molecules such as annexin were either up or downregulated in our study. Ignotz et al.48 reported in the cow, that oviductal annexin could act as possible oviduct receptors for sperm surface proteins and thus serve to hold the spermatozoa in the oviductal reservoir during the pre-ovulatory period. In the pig, annexins have also been isolated from oviductal epithelial cells probably also involved in sperm-oviduct interactions by acting as sperm-binding proteins49. In our study, ANXA5, a major constituent of the members of the annexin family, was downregulated after Mating. In addition, we found upregulated genes that encode proteins that have heparin affinity such as PRELP, LPL, POSTN, ABI3BP, FGF10 after Mating; and PTN, PRELP, VEGFA after P1-AI exposure. There is evidence that sulfated GAGs, as heparin or heparan-sulfate, produced by the oviductal epithelium50 are powerful inhibitors of sperm binding to oviductal cells47 and so, concerted interplay between various GAGs can be involved when sperm release from the pig oviductal reservoir occurs43,44.
What role can then the seminal plasma play? The capacity of porcine testicular spermatozoa to bind the oviduct epithelium can be advanced by components of the fluid of the cauda epididymides51, i.e. the fluid prevailing in the P1-fraction. Seminal plasma spermadhesins are also related to sperm binding to the epithelium52 act prior to/during capacitation53,54, during the sperm release from the oviductal reservoir55 and even to fertilization, particularly during binding to the zona-pellucida56, commonly acting via their heparin-binding domain57,58. The fact that we have found some altered transcripts associated with either sperm binding or detachment from the oviductal reservoir is consistent with the general concept that the sperm release from oviductal epithelium is a gradual process that restrict the number of sperm in the site of fertilization to control polyspermy59 and maintains sperm in a protective state until they are fertilization-competent18, so it seems plausible to find active mechanisms of either attach or detachment during the peri-ovulatory period, including the action of progesterone and CatSper60 during sperm capacitation, illustrated by the experimental blockade of sperm release inhibition caused by inhibition of the progesterone receptor that activates CatSper61. The reason why these results were not so evident in the P1-AI group may be related to the number of spermatozoa deposited into the reproductive tract, which is approximately 30 times lower than those during natural mating62. In addition, another mechanism that has been implicated in sperm detachment from the oviductal reservoir is the contractility of the myosalpinx smooth muscle63. There is extensive evidence that concerted contractions of the myometrium and the myosalpinx are probably primarily responsible for moving sperm and oocytes to the fertilization point1,18,63. In this context, we found many downregulated genes encoding cAMP-binding proteins which are the major responsible of inhibiting smooth muscle contractility to maintain uterine quiescence during pregnancy64. In the smooth muscle cells of the myometrium or myosalpinx, cAMP functions as a second messenger that activates protein kinase-A (PKA)65, a multifunctional kinase that phosphorylates downstream targets to suppresses contractility by decreasing the intracellular concentration of free Ca2+ and inhibiting myosin light chain kinase activity66,67. Interestingly, regulatory subunits of protein Kinase A, such as PRKAR1A, were downregulated in the Ampulla and Infundibulum in the M and P1-AI groups, and PRKAR2A was downregulated in Ampulla after Mating and in the Infundibulum in both M and P1-AI groups.
Moreover, cyclic nucleotide phosphodiesterases (PDEs), which are degradation enzymes known for their role in cAMP breakdown, were mainly upregulated. Specifically, PDE-4 isoforms were detected. These have been widely studied in several species, and catalyze the hydrolysis and inactivation of cAMP68,69, lowering its concentration in the smooth muscle cells. Based on our findings, we can clearly hypothesize that the presence of spermatozoa in the oviduct could be influencing myosalpinx motility. These effects were more obvious after Mating (PDE-4 up-regulated in Isthmus, Ampulla and Infundibulum) followed by P1-AI (PDE-4 up-regulated in Infundibulum) and SPP1 (PDE-4 up-regulated in Isthmus). These differences could be explained by the fact that when natural mating takes place, the presence of a boar (separate from insemination itself) induces central oxytocin release and thus increases smooth muscle motility in the sow16.
Of note, the SP groups showed the lowest changes in terms of overall transcriptional modifications. In the pig, as in other mammals, there is little likelihood that the SP reaches the oviduct after semen deposition because the utero-tubal junction acts as a physiological barrier38. Therefore, the evidence presented in this report supports the hypothesis that spermatozoa themselves and not the SP are the major agent responsible for the molecular changes elucidated in the pig oviduct.
However, SP molecules can support sperm survival along the female genital tract and thus, enhance reproductive success70. In addition, previous studies suggest that seminal estrogen has a positive effect on sperm transport and fertilization and is also responsible for rapid sperm elimination from the uterus through the cervix in the pig71. It has also been reported that sperm binding to the oviductal cells is triggered by high concentrations of estrogen within the oviduct72,73. Interestingly, the SP-P1 group showed an overrepresentation of the Estrogen signaling pathway in our study. Of particular interest was the up-regulation of HSD17B8 in the UTJ, suggesting that the SP may play a role in sperm attachment and release from the oviduct reservoir via estrogen signaling regulation. It is known that exosomes present in SP bind to the sperm head membrane and improve membrane integrity74. Also, it has been stated that proteins present in SP are involved in the immune and inflammatory response in the uterus75. Moreover, there is evidence that mRNAs and miRNAs are heavily involved in boar sperm response to environmental stimuli, apoptosis, and metabolic activities76. However, the components of SP responsible of the alterations found in this study are yet to be determined.
In conclusion, our study underlies that boar spermatozoa and/or SP have the ability to modify molecular patterns in the endocervix, endometrium and endosalpinx during the peri-ovulatory period, supporting the idea that these contribute to the development of a receptive environment to prepare for successful fertilization, as well as for an adequate milieu for zygote/early embryo development.
Materials and Methods
Ethics statement of the interventional study
All animal husbandry and experimental handling was performed in compliance with the European Community (Directive 2010/63/EU) and current Swedish legislation (SJVFS 2017:40), following the Reduction principles of the 3Rs on animal experimentation (Replacement, Reduction and Refinement) while maintaining enough numbers of biological replicates from distinct animals, to obtain sufficiently reliable enough estimates of variation among samples within conditions to distinguish true differences between conditions. This applied to both treatment and control conditions. The experiments were approved in advance by the “Regional Committee for Ethical Approval of Animal Experiments” (Linköpings Djurförsöksetiska nämnd) in Linköping, Sweden (permits no. 75–12 and no. ID1400).
Experimental design
Changes in the general transcriptome profile of the pre/peri-ovulatory porcine endocervix, endometrium (distal and proximal) and endosalpinx (UTJ, isthmus, ampulla and infundibulum), were studied 24 h after the deposition of semen (i.e. spermatozoa and SP) or of sperm-free SP. Twenty sows displaying standing oestrus in the presence of a boar were equally and at-random allotted to a Control group (n = 4), where females were cervically infused with 50 mL of the protein-free extender “Beltsville Thawing Solution” (BTS)77 or a series of treatment groups; including either entry of semen (spermatozoa and the accompanying SP) either by mating (M, whole ejaculate) or AI of the sperm-peak fraction e.g. the first 10 mL of the sperm-rich fraction (P1) (P1-AI, contained 25% of the total sperm numbers of the ejaculates of the boars used which were ~40 billion spermatozoa) or counterparts holding solely the sperm-free SP (entire SP; SP-Ejac) or SP from the P1 fraction (SP-P1). For the M group (n = 4), the sows were each mated with a single male; in the P1-AI (n = 4) females were cervically artificially inseminated with the ejaculated P1-sperm-peak fraction (extended to 50 mL with BTS) while for SP-Ejac (n = 4) sows were cervically infused with sperm-free SP of the entire ejaculate (50 mL) and finally, in the SP-P1 (n = 4), females were cervically infused with sperm-free SP harvested from ejaculated P1-fractions (pool, 50 mL). Samples from endocervix (EndoCvx), endometrium (distal: DistEndom or proximal: ProxEndom) and endosalpinx from 4 different segments; the utero-tubal junction (UTJ), isthmus (Isth), ampulla (Amp) and infundibulum (Inf) were surgically retrieved under general anesthesia, 24 h after the interventions (Control/Treatments).
Animal management
Pigs of Swedish Landrace breed were recruited from a controlled breeding farm, as weaned sows (parity 1–3, n = 20) and young mature boars (9–11 months, n = 5) of good semen quality (<100 mL volume, >60 × 109 total sperm number, >70% sperm motility, and >75% morphologically normal-looking spermatozoa, controlled weekly)6. Throughout all experiments, animals were handled carefully to avoid any unnecessary stress. The animals were individually kept in separate pens at the Translational Medicine Centre (TMC/CBR-3) of Linköping University under controlled temperature and light regimes (12 h:12 h light/dark cycle). Pigs were fed with commercial feedstuff (Lantmännen, Stockholm, Sweden) according to national standards78, provided with water ad libitum and receiving the same management.
Semen collection and SP harvesting
Ejaculates, collected using the gloved-hand method, and with at least 70% motile and 75% morphologically normal-looking spermatozoa immediately after collection, were used for the experiments. The SP was harvested either from the whole ejaculate or from the sperm-peak P1-fraction after double centrifugation at 1,500 × g for 10 min, and microscopically checked for presence of spermatozoa. The harvested sperm-free crude SP was stored at −20 °C until used.
Handling of sows
Detection of estrus was performed during snout-to-snout contact of females with the presence of mature boars while applying back-pressure to test for standing estrus reflex twice daily, beginning one day after weaning. When sows showed standing estrus reflex they were used in the experiments, considered to be on the first day of behavioral estrus. Sows were cervically inseminated/infused using disposable conventional AI-catheters (Minitüb, Munich, Germany).
Collection of genital tract samples
All sows were subjected to mid-ventral laparotomies to collect the tissue samples 24 h after mating/inseminations (pre-/peri-ovulation period), as described by Alvarez-Rodriguez6. Briefly, sows were sedated by the i.m. administration of a mixture of 5 mg dexmeditomedine (Dexdomitor, Orion Pharma Animal Health, Sollentuna, Sweden) and 100 mg tiletamine hydrochloride/zolazepam hydrochloride (Zoletil vet, Virbac A/S, Kolding, Denmark) followed by anaesthesia induced i.v. with sodium thiopental (Abbot Scandinavia AB, Solna, Sweden, 7 mg/kg bw), and maintained with isoflurane (3.5–5%, Baxter Medical AB, Kista, Sweden) administered via a tracheal cuffed tube by a close-circuit PVC-ventilator (Servo ventilator 900 D, SIEMENS-ELEMA AB, Solna, Sweden). The number of pre-ovulatory follicles or eventual new ovulations per sow was 22.30 ± 7.29 (mean +− standard deviation) pre-ovulatory follicles, without significant differences between sow-groups. Peripheral blood was collected for estradiol 17ß (E2) and progesterone ELISA analyses in plasma, which confirmed the animals were in pre-/peri-ovulatory estrus (mean ± SD pg/mL for P4 = 0.77 ± 0.35 in all sows; and for E2 ranging 376.50 ± 27.76–294.20 ± 80.24 in all experimental groups, e.g., without significant differences). Segments of the female genital tract segments were immediately retrieved and stored at –80 °C in RNAlater (Ambion, Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania) until analysed.
Transcriptome analysis
Analyses of the tissue samples was done as described by Alvarez-Rodriguez6. In brief, total RNA was isolated from tissue samples using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and quality assessment was performed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. The RNA integrity number (RIN) values obtained were in the range of 8 to 10, which guarantied the homogeneity and high quality of the samples. Equal amounts of total RNA (250 ng) from each sample were used to make cDNA using GeneChip® Whole Transcript Plus reagent kit (Affymetrix, Santa Clara, CA, USA) following the manufacturer protocol. cDNA was then hybridized and loaded on the array chip (GeneChip® Porcine Gene 1.0 ST Array, Affymetrix Inc., 3420 Central Expressway, Santa Clara, CA 95051, USA), incubated at 45 °C under 60 rotations per min, for 16 h. The hybridized cartridge array chip was then unloaded and subjected to washing and staining using a GeneChip® Fluidics Station 450 (Affymetrix), to be finally scanned using the Affymetrix GeneChip® scanner GCS3000.
Analysis of microarray data and bioinformatics
The intensity data of each array chip was processed using the robust multi-array average (RMA) normalization, computing average intensity values by background adjustment, quantile normalization among arrays and finally log2 transformation for extracting the expression values of each transcript in the probe set, as implemented in the official Transcriptome Analysis Console (TAC; version 4.0) from Affymetrix. The statistical analysis of the normalized gene expression data was performed using a linear model using the empirical Bayes’ approach as implemented in the package “limma” was used to calculate differentially expressed transcripts using a Benjamini-Hochberg (q < 0.05) correction to control for multiple testing to control type-I errors79 and a fold change (FC) > 1 or <−1.
Enrichment analysis
Data was then analyzed using the PANTHER Classification System for gene ontology (GO) terms80. The differentially expressed genes (p < 0.05) were screened for additional molecular functions with the protein knowledge base of the UniProt Consortium81 to confirm they were correctly classified as different groups. An additional analysis of altered pathways was performed using Partek Genomics Suite 7.0 (Partek) using the KEGG database82. Graphical illustration of overrepresented GO terms and pathways was produced with the Cytoscape v3.0.0 application ClueGO v2.0.383.
Supplementary information
Acknowledgements
Alejandro Vicente-Carrillo, Anders Delleskog, Dan Linghammar, and Rebecca Katajaama are acknowledged for their assistance during pig handling, semen collection, and surgery. We also thank Mohammad Atikuzzaman, Heli Venhoranta, Annette Molbaek and Åsa Schippert, for expert assistance when running the microarrays. We appreciate the kind support provided by Mihaela Martis (BILS, Sweden) regarding quality check controls and the proper use of bioinformatic tools and senior bioinformatician Dr Pedro Madrigal (Stem Cell Institute, University of Cambridge) for expert advice on the experimental design. Open access funding provided by Linköping University.
Author contributions
Conceptualization, H.R.-M., D.W. and M.A.R.; methodology, M.A., M.A.R. and H.R.-M.; software, M.A.R. and C.A.M.; validation, M.A.R., C.A.M. and D.W.; data curation, M.A.R. and C.A.M.; writing—original draft preparation, M.A.R. and C.A.M..; writing—review and editing, D.W. and H.R.-M.; visualization, M.A.R.; supervision, D.W. and H.R.-M.; project administration, H.R.-M.; funding acquisition, H.R.-M and M.A.R.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: M. Alvarez-Rodríguez and C. A. Martinez.
Supplementary information
is available for this paper at 10.1038/s41598-020-60810-z.
References
- 1.Langendijk P, Soede NM, Kemp B. Uterine activity, sperm transport, and the role of boar stimuli around insemination in sows. Theriogenology. 2005;63:500–513. doi: 10.1016/j.theriogenology.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Georgiou AS, et al. Modulation of the oviductal environment by gametes. J. Proteome Res. 2007;6:4656–4666. doi: 10.1021/pr070349m. [DOI] [PubMed] [Google Scholar]
- 3.Maillo V, et al. Oviductal response to gametes and early embryos in mammals. Reproduction. 2016;152:R127–41. doi: 10.1530/REP-16-0120. [DOI] [PubMed] [Google Scholar]
- 4.Alminana C, et al. The battle of the sexes starts in the oviduct: modulation of oviductal transcriptome by X and Y-bearing spermatozoa. BMC Genomics. 2014;15:293. doi: 10.1186/1471-2164-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Martinez H, et al. The physiological roles of the boar ejaculate. Soc. Reprod. Fertil. Suppl. 2009;66:1–21. [PubMed] [Google Scholar]
- 6.Alvarez-Rodriguez M, Atikuzzaman M, Venhoranta H, Wright D, Rodriguez-Martinez H. Expression of immune regulatory genes in the porcine internal genital tract is differentially triggered by spermatozoa and seminal plasma. Int. J. Mol. Sci. 2019;20:1–20. doi: 10.3390/ijms20030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson SA, Sharkey DJ. The role of semen in induction of maternal immune tolerance to pregnancy. Semin. Immunol. 2001;13:243–254. doi: 10.1006/smim.2000.0320. [DOI] [PubMed] [Google Scholar]
- 8.Huang VW, et al. Cell membrane proteins from oviductal epithelial cell line protect human spermatozoa from oxidative damage. Fertil. Steril. 2013;99:1444–1452.e3. doi: 10.1016/j.fertnstert.2012.11.056. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Ubeda R, et al. Oviductal Transcriptome Is Modified after Insemination during Spontaneous Ovulation in the Sow. PLoS One. 2015;10:e0130128. doi: 10.1371/journal.pone.0130128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waberski D, et al. Seminal plasma modulates the immune-cytokine network in the porcine uterine tissue and pre-ovulatory follicles. PLoS One. 2018;13:e0202654. doi: 10.1371/journal.pone.0202654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JC, et al. Seminal plasma induces global transcriptomic changes associated with cell migration, proliferation and viability in endometrial epithelial cells and stromal fibroblasts. Hum. Reprod. 2014;29:1255–1270. doi: 10.1093/humrep/deu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atikuzzaman M, et al. Conserved gene expression in sperm reservoirs between birds and mammals in response to mating. BMC Genomics. 2017;18:98. doi: 10.1186/s12864-017-3488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson SA. Seminal fluid signaling in the female reproductive tract: lessons from rodents and pigs. J. Anim. Sci. 2007;85:36–44. doi: 10.2527/jas.2006-578. [DOI] [PubMed] [Google Scholar]
- 14.Wallgren M, Saravia F, Rodriguez-Martinez H. The Vanguard sperm cohort of the boar ejaculate is overrepresented in the tubal sperm reservoir in vivo. J. Reprod. Dev. 2010;56:68–72. doi: 10.1262/jrd.09-125k. [DOI] [PubMed] [Google Scholar]
- 15.Yun J, Björkman S, Oliviero C, Soede N, Peltoniemi O. The effect of farrowing duration and parity on preovulatory follicular size and oxytocin release of sows at subsequent oestrus. Reprod. Domest. Anim. 2018;53:776–783. doi: 10.1111/rda.13170. [DOI] [PubMed] [Google Scholar]
- 16.Langendijk P, Bouwman EG, Schams D, Soede NM, Kemp B. Effects of different sexual stimuli on oxytocin release, uterine activity and receptive behavior in estrous sows. Theriogenology. 2003;59:849–861. doi: 10.1016/s0093-691x(02)01157-3. [DOI] [PubMed] [Google Scholar]
- 17.Franczak A, Ciereszko R, Kotwica G. Oxytocin (OT) action in uterine tissues of cyclic and early pregnant gilts: OT receptors concentration, prostaglandin F2α secretion, and phosphoinositide hydrolysis. Anim. Reprod. Sci. 2005;88:325–339. doi: 10.1016/j.anireprosci.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Martinez H, et al. Boar spermatozoa in the oviduct. Theriogenology. 2005;63:514–535. doi: 10.1016/j.theriogenology.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Saravia F, Wallgren M, Nagy S, Johannisson A, Rodríguez-Martínez H. Deep freezing of concentrated boar semen for intra-uterine insemination: Effects on sperm viability. Theriogenology. 2005;63:1320–1333. doi: 10.1016/j.theriogenology.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Veillette A, et al. Regulation of the PI3-K/Akt survival pathway in the rat endometrium. Biol. Reprod. 2013;88:79. doi: 10.1095/biolreprod.112.107136. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, et al. Infertility caused by retardation of follicular development in mice with oocyte-specific expression of Foxo3a. 2007;209:199–209. doi: 10.1242/dev.02667. [DOI] [PubMed] [Google Scholar]
- 22.Franczak A, Zglejc-Waszak K, Martyniak M, Waszkiewicz EM, Kotwica G. Peri-conceptional nutritional restriction alters transcriptomic profile in the peri-implantation pig embryos. Anim. Reprod. Sci. 2018;197:305–316. doi: 10.1016/j.anireprosci.2018.08.045. [DOI] [PubMed] [Google Scholar]
- 23.Fabi F, Asselin E. Expression, activation, and role of AKT isoforms in the uterus. Reproduction. 2014;148:R85–95. doi: 10.1530/REP-14-0270. [DOI] [PubMed] [Google Scholar]
- 24.Ovadia S, Insogna K, Yao G-Q. The cell-surface isoform of colony stimulating factor 1 (CSF1) restores but does not completely normalize fecundity in CSF1-deficient mice. Biol. Reprod. 2006;74:331–336. doi: 10.1095/biolreprod.105.045047. [DOI] [PubMed] [Google Scholar]
- 25.Biggs WH, 3rd, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 26.Sedding DG. FoxO transcription factors in oxidative stress response and ageing–a new fork on the way to longevity? Biol. Chem. 2008;389:279–283. doi: 10.1515/BC.2008.033. [DOI] [PubMed] [Google Scholar]
- 27.Carter ME, Brunet A. FOXO transcription factors. Curr. Biol. 2007;17:R113–4. doi: 10.1016/j.cub.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Essers MAG, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goertz MJ, et al. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis Find the latest version: Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelosi E, et al. Constitutively active Foxo3 in oocytes preserves ovarian reserve in mice. Nat. Commun. 2013;4:1843. doi: 10.1038/ncomms2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto H, et al. Lower FOXO3 mRNA expression in granulosa cells is involved in unexplained infertility. J. Obstet. Gynaecol. Res. 2017;43:1021–1028. doi: 10.1111/jog.13307. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Martinez H, et al. Involvement of oviduct in sperm capacitation and oocyte development in pigs. Reprod. Suppl. 2001;58:129–145. [PubMed] [Google Scholar]
- 33.Kops GJPL, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 34.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 35.O’Leary S, Jasper MJ, Warnes GM, Armstrong DT, Robertson SA. Seminal plasma regulates endometrial cytokine expression, leukocyte recruitment and embryo development in the pig. Reproduction. 2004;128:237–247. doi: 10.1530/rep.1.00160. [DOI] [PubMed] [Google Scholar]
- 36.Hussain T, et al. Oxidative Stress and Inflammation: What Polyphenols Can Do for Us? Oxid. Med. Cell. Longev. 2016;2016:7432797. doi: 10.1155/2016/7432797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko EY, Sabanegh ESJ, Agarwal A. Male infertility testing: reactive oxygen species and antioxidant capacity. Fertil. Steril. 2014;102:1518–1527. doi: 10.1016/j.fertnstert.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Martinez H. Role of the oviduct in sperm capacitation. Theriogenology. 2007;68(Suppl 1):S138–46. doi: 10.1016/j.theriogenology.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Miller DJ. Regulation of Sperm Function by Oviduct Fluid and the Epithelium: Insight into the Role of Glycans. Reprod. Domest. Anim. 2015;50(Suppl 2):31–39. doi: 10.1111/rda.12570. [DOI] [PubMed] [Google Scholar]
- 40.Machado SA, et al. LewisX-containing glycans on the porcine oviductal epithelium contribute to formation of the sperm reservoir. Biol. Reprod. 2014;91:140. doi: 10.1095/biolreprod.114.119503. [DOI] [PubMed] [Google Scholar]
- 41.Kadirvel G, et al. Porcine Sperm Bind to Specific 6-Sialylated Biantennary Glycans to Form the Oviduct Reservoir. Biol. Reprod. 2012;87:147. doi: 10.1095/biolreprod.112.103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winters RA, et al. Porcine sperm binding to oviduct cells and glycans as supplements to traditional laboratory semen analysis. J. Anim. Sci. 2018;96:5265–5275. doi: 10.1093/jas/sky372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tienthai P, Johannisson A, Rodríguez-Martínez H. Sperm capacitation in the porcine oviduct. Anim. Reprod. Sci. 2004;80:131–146. doi: 10.1016/S0378-4320(03)00134-9. [DOI] [PubMed] [Google Scholar]
- 44.Hunter RHF, Rodriguez-Martinez H. Capacitation of mammalian spermatozoa in vivo, with a specific focus on events in the Fallopian tubes. Mol. Reprod. Dev. 2004;67:243–250. doi: 10.1002/mrd.10390. [DOI] [PubMed] [Google Scholar]
- 45.Miyata S, Kitagawa H. Formation and remodeling of the brain extracellular matrix in neural plasticity: Roles of chondroitin sulfate and hyaluronan. Biochimica et Biophysica Acta - General Subjects. 2017;1861:2420–2434. doi: 10.1016/j.bbagen.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Martinez H, et al. The ubiquitous hyaluronan: functionally implicated in the oviduct? Theriogenology. 2016;86:182–186. doi: 10.1016/j.theriogenology.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Gualtieri R, et al. Bovine oviductal monolayers cultured under three-dimension conditions secrete factors able to release spermatozoa adhering to the tubal reservoir in vitro. Theriogenology. 2013;79:429–435. doi: 10.1016/j.theriogenology.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Ignotz GG, Cho MY, Suarez SS. Annexins are candidate oviductal receptors for bovine sperm surface proteins and thus may serve to hold bovine sperm in the oviductal reservoir. Biol. Reprod. 2007;77:906–913. doi: 10.1095/biolreprod.107.062505. [DOI] [PubMed] [Google Scholar]
- 49.Teijeiro JM, Ignotz GG, Marini PE. Annexin A2 is involved in pig (Sus scrota) sperm-oviduct interaction. Mol. Reprod. Dev. 2009;76:334–341. doi: 10.1002/mrd.20958. [DOI] [PubMed] [Google Scholar]
- 50.Tienthai P, Kjellén L, Pertoft H, Suzuki K, Rodriguez-Martinez H. Localisation and quantitation of hyaluronan and sulphated glycosaminoglycans in the tissues and intraluminal fluid of the pig oviduct. Reprod Fert Dev. 2000;12:173–182. doi: 10.1071/rd00034. [DOI] [PubMed] [Google Scholar]
- 51.Peña S, Summers P, Gummow B, Paris DBBP. Oviduct binding ability of porcine spermatozoa develops in the epididymis and can be advanced by incubation with caudal fluid. Theriogenology. 2015;83:1502–1513. doi: 10.1016/j.theriogenology.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 52.Ardon F, et al. Dynamics of Bovine Sperm Interaction with Epithelium Differ Between Oviductal Isthmus and Ampulla. Biol. Reprod. 2016;95:90. doi: 10.1095/biolreprod.116.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lefebvre R, Suarez SS. Effect of capacitation on bull sperm binding to homologous oviductal epithelium. Biol. Reprod. 1996;54:575–582. doi: 10.1095/biolreprod54.3.575. [DOI] [PubMed] [Google Scholar]
- 54.Gualtieri R, Talevi R. In vitro-cultured bovine oviductal cells bind acrosome-intact sperm and retain this ability upon sperm release. Biol. Reprod. 2000;62:1754–1762. doi: 10.1095/biolreprod62.6.1754. [DOI] [PubMed] [Google Scholar]
- 55.Suarez SS. Mammalian sperm interactions with the female reproductive tract. Cell Tissue Res. 2016;363:185–194. doi: 10.1007/s00441-015-2244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calvete JJ, et al. Monoclonal Antibodies against Boar Sperm Zona Pellucida-Binding Protein AWN-1. Characterization of a Continuous Antigenic Determinant and Immunolocalization Of AWN Epitopes in Inseminated Sows. Biol. Reprod. 1997;57:735–742. doi: 10.1095/biolreprod57.4.735. [DOI] [PubMed] [Google Scholar]
- 57.Calvete JJ, et al. Mapping the heparin-binding domain of boar spermadhesins. FEBS Lett. 1996;379:207–211. doi: 10.1016/0014-5793(95)01513-2. [DOI] [PubMed] [Google Scholar]
- 58.Rodríguez-Martínez H, Iborra A, Martínez P, Calvete JJ. Immunoelectronmicroscopic imaging of spermadhesin AWN epitopes on boar spermatozoa bound in vivo to the zona pellucida. Reprod. Fertil. Dev. 1998;10:491–497. doi: 10.1071/rd98111. [DOI] [PubMed] [Google Scholar]
- 59.Coy P, et al. Oviduct-specific glycoprotein and heparin modulate sperm-zona pellucida interaction during fertilization and contribute to the control of polyspermy. Proc. Natl. Acad. Sci. USA. 2008;105:15809–15814. doi: 10.1073/pnas.0804422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vicente-Carrillo, A., Álvarez-Rodríguez, M. & Rodríguez-Martínez, H. The CatSper channel modulates boar sperm motility during capacitation. Reprod. Biol. 17 (2017). [DOI] [PubMed]
- 61.Machado, S. A., Sharif, M., Wang, H., Bovin, N. & Miller, D. J. Release of Porcine Sperm from Oviduct Cells is Stimulated by Progesterone and Requires CatSper. Sci. Rep. 9 (2019). [DOI] [PMC free article] [PubMed]
- 62.Knox RV. Artificial insemination in pigs today. Theriogenology. 2016;85:83–93. doi: 10.1016/j.theriogenology.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez-Martinez H, Einarsson S. Spontaneous contractility of isolated muscle layers of the pig oviductal isthmus. Acta Physiol. Scand. 1982;115:325–330. doi: 10.1111/j.1748-1716.1982.tb07085.x. [DOI] [PubMed] [Google Scholar]
- 64.Chanrachakul B, Broughton Pipkin F, Khan RN. Contribution of coupling between human myometrial beta2-adrenoreceptor and the BK(Ca) channel to uterine quiescence. Am. J. Physiol. Cell Physiol. 2004;287:C1747–52. doi: 10.1152/ajpcell.00236.2004. [DOI] [PubMed] [Google Scholar]
- 65.Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway. Differential expression,regulation, and subcellular localization of subunits of PKA. Front. Biosci. 2000;5:D678–93. doi: 10.2741/skalhegg. [DOI] [PubMed] [Google Scholar]
- 66.Morgado M, Cairrao E, Santos-Silva AJ, Verde I. Cyclic nucleotide-dependent relaxation pathways in vascular smooth muscle. Cell. Mol. Life Sci. 2012;69:247–266. doi: 10.1007/s00018-011-0815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yulia A, Singh N, Lei K, Sooranna SR, Johnson MR. Cyclic AMP Effectors Regulate Myometrial Oxytocin Receptor Expression. Endocrinology. 2016;157:4411–4422. doi: 10.1210/en.2016-1514. [DOI] [PubMed] [Google Scholar]
- 68.Oger S, et al. Anti-inflammatory and utero-relaxant effects in human myometrium of new generation phosphodiesterase 4 inhibitors. Biol. Reprod. 2004;70:458–464. doi: 10.1095/biolreprod.103.023051. [DOI] [PubMed] [Google Scholar]
- 69.Mehats C, et al. PDE4 as a target in preterm labour. BMC Pregnancy Childbirth. 2007;7(Suppl 1):S12. doi: 10.1186/1471-2393-7-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaczmarek MM, et al. Seminal plasma affects prostaglandin synthesis in the porcine oviduct. Theriogenology. 2010;74:1207–1220. doi: 10.1016/j.theriogenology.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 71.Troedsson MHT, et al. Components in seminal plasma regulating sperm transport and elimination. Anim. Reprod. Sci. 2005;89:171–186. doi: 10.1016/j.anireprosci.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 72.Suarez S, Redfern K, Raynor P, Martin F, Phillips DM. Attachment of boar sperm to mucosal explants of oviduct in vitro: possible role in formation of a sperm reservoir. Biol. Reprod. 1991;44:998–1004. doi: 10.1095/biolreprod44.6.998. [DOI] [PubMed] [Google Scholar]
- 73.Lamy J, et al. Steroid hormones regulate sperm-oviduct interactions in the bovine. Reproduction. 2017;154:497–508. doi: 10.1530/REP-17-0328. [DOI] [PubMed] [Google Scholar]
- 74.Du J, et al. Boar seminal plasma exosomes maintain sperm function by infiltrating into the sperm membrane. Oncotarget. 2016;7:58832–58847. doi: 10.18632/oncotarget.11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Druart X, de Graaf S. Seminal plasma proteomes and sperm fertility. Anim. Reprod. Sci. 2018;194:33–40. doi: 10.1016/j.anireprosci.2018.04.061. [DOI] [PubMed] [Google Scholar]
- 76.Dai, D.-H. et al. Exploration of miRNA and mRNA Profiles in Fresh and Frozen-Thawed Boar Sperm by Transcriptome and Small RNA Sequencing. Int. J. Mol. Sci. 20 (2019). [DOI] [PMC free article] [PubMed]
- 77.Pursel VG, Johnson LA. Freezing of boar spermatozoa: fertilizing capacity with concentrated semen and a new thawing procedure. J. Anim. Sci. 1975;40:99–102. doi: 10.2527/jas1975.40199x. [DOI] [PubMed] [Google Scholar]
- 78.Simonsson A. Näringsrekommendationer och fodermedelstabeller till svin (Nutrient and metabolizable energy recommendations for swine. In: Swedish. Sweden: Publisher: Swedish University of Agricultural Sciences, SLU Rapporter Husdjur, p. 75, (1991).
- 79.Benjamini, Y. & Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B57, 289–300 (1995).
- 80.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.UniProt: a hub for protein information Nucleic Acids Res. 2015;43:D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.