Abstract
Nanopore analysis is an emerging technique that enables the investigation of the conformation of a single peptide or protein molecule. Briefly, a pore is inserted into a membrane under voltage clamp conditions. When a molecule interacts with the pore there is a change in the current, I, for a time, T. Small unfolded molecules can translocate the pore whereas folded or large molecules tend to simply bump into the pore and then diffuse away. Therefore, the parameters, I and T, are dependent on the conformation of the molecule at the instant at which it encounters the pore. Thus, multiple conformations can be detected simultaneously in a single sample. As well, the analysis can be performed under dilute conditions so that folding or dimerization of a peptide can be followed in real time, which is generally difficult to study for proteins that are prone to aggregate. In this report, we describe our initial analysis of (1) Aβ peptides, which are deposited as amyloid plaques in Alzheimer disease, (2) α-synuclein, which is implicated in Parkinson disease and (3) prion proteins whose misfolding is evident in transmissable spongiform encephalopathies. In each case conformational information can be obtained which may help in understanding the early steps in the misfolding pathways.
Keywords: Aβ-peptides, nanopore analysis, prions, protein misfolding, α-synuclein
Origins
Nanopore analysis arose from a combination of three different disciplines. First, the Coulter Counter is an elegant device that was invented in 1947.1,2 An automated version is now found in most hospital laboratories for measuring complete blood counts, but it can also detect bacteria or viruses in biological samples and particulate matter for pharmaceutical or industrial applications.3 A small hole of about 1 μm was developed in the side of a thin glass tube which was immersed in an electrolyte bath. The sample was placed in the tube so that the level was higher than that in the bath and the hydrostatic pressure would drive liquid from the tube into the bath. Electrodes were placed in the two chambers so that upon application of a voltage, a constant current would be recorded. However, if a particle passed through the hole there would be a resistive pulse as the current transiently dropped. The number of pulses in a given time yielded an estimate of the concentration of particles. The technique could also be used for estimating the relative size of bacteria and viruses since the magnitude of the current drop is proportional to the volume of the pore occluded by the particle.4 Nanopore analysis (see Figure 1) uses a similar principle except that the analytes are usually driven through the pore by an electric field rather than hydrostatic pressure. As well, the measured current is many orders of magnitude smaller and the size of the pore is measured in nanometers.
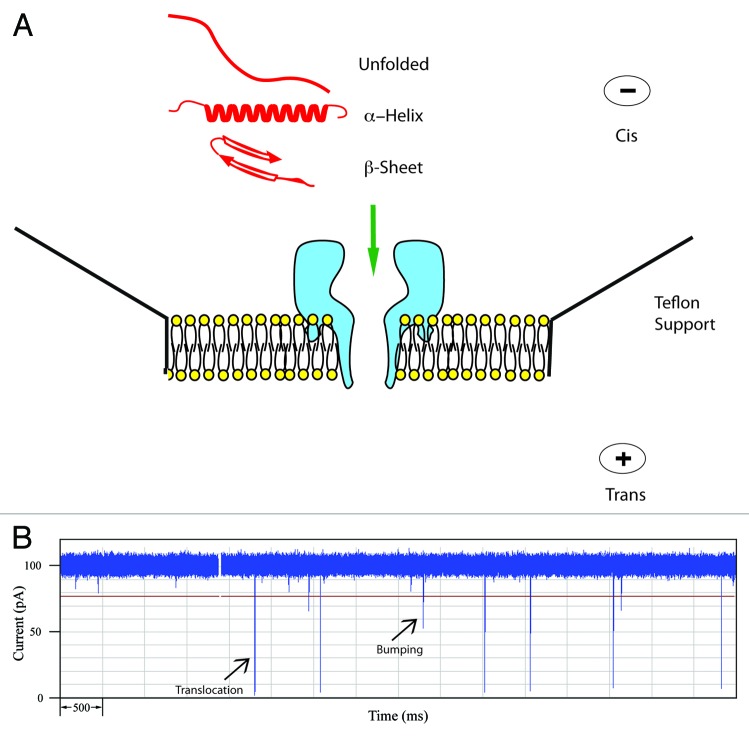
Figure 1. (A) Schematic of the α-hemolysin pore embedded in a lipid membrane. Unfolded and simple α-helical or β-sheet forming peptides can readily translocate. (B) Typical current trace of Aβ1−40 recorded for 10 sec. The open pore current is 100 pA and each spike represents an event where a single peptide interacts with the pore. Typically for a peptide, large spikes are due to translocations and short spikes are bumping events.
Physiologists routinely manipulate currents in the pA range for patch clamp experiments.5 Briefly, a drawn out pipette tip is clamped onto the membrane of an individual cell such that individual pores or ion channels can be interrogated. In one type of experiment, for example, a drug is added which allows calcium ions to flow into the cell causing a small current which is measured by the patch clamp amplifier.6 The event time may be very short so the response time of the amplifier must be less than a millisecond. The final piece of the puzzle was provided by microbiologists.
Many pathogens produce toxins which alter the permeability of cell membranes.7 For example, antibiotics such as Gramicidin insert into the membrane and allow sodium ions to enter which eventually leads to cell lysis.8 A particularly vicious example, from the human perspective, is the toxin α-hemolysin (α-HL) from Staphylococcus aureus which is a common member of the bacterial community found on the skin.9,10 However, if it enters the blood stream the toxin punches holes in red blood cells causing cell lysis and the resulting inflammatory response can lead to multi-organ failure and death. The molecular basis for this behavior was established once the crystal structure of the toxin was solved.9,10 The protein is heptameric and can self-assemble into a membrane resulting in a pore with a minimum diameter of about 1.5 nm (see Figure 1a). In most situations the pore is not gated and allows the translocation of ions, oligosaccharides, peptides and single-stranded DNA. For these reasons the α-HL pore has found widespread use in nanopore analysis. (More recently, the use of other bacterial pores such as Aerolysin have been described as well as solid state pores elaborated in silicon nitride membranes. However this mini-review will be confined to the use of α-HL).11-15
The Basic Technique
A typical experimental set-up is shown in Figure 1a.16-18 A lipid bilayer is painted over a small (150 μM) aperture in a teflon support. Then an electrolyte with high conductance (usually 1 M KCl) is added to both sides of the membrane and a dilute solution of the α-HL is added to one side, usually the cis-side. It should be noted that the pore is asymmetric but it self-assembles in a unique orientation such that the large opening or vestibule is always on the side to which the α-HL was added. Thus the topology and dimensions of the pore are very reproducible. When a voltage is applied across the pore, the transport of ions gives rise to a constant current; coincidentally, at 100 mV the current is – 100 pA in a buffer containing 1 M KCl. With this arrangement of the electrodes, negatively-charged molecules will be electrophoretically driven from the cis-side toward the pore although neutral molecules may also diffuse toward the pore. Interaction of a molecule with the pore will cause a resistive pulse or decrease in the current, I, for a time, T. These events are monitored by a patch clamp amplifier and a typical current trace is shown in 1b.
Each spike is the signal from a single molecule interacting with the pore. The negatively-charged peptides (see Table 1) are driven toward the pore and generally give rise to two types of events: translocations, in which the molecule passes through the pore, and bumping events where the molecule briefly interacts with the vestibule of the pore and then diffuses away. For many proteins, a third type of event called intercalation has also been identified. In this case, a negatively-charged loop or terminus penetrates the vestibule but the bulk of the protein does not allow complete translocation and eventually the protein will diffuse back to the cis-side.19-21Figure 1b shows a current trace for the Alzheimer peptide, Aβ1–40 and two types of events are readily distinguished. The larger spikes with I = - 95 pA are typical of a mostly α-helical conformation.22,23 There are also spikes with smaller current blockades (-20 to -30 pA) which are typical of bumping events. More details concerning these tentative assignments and methods for the processing of this data will be described below.
Table 1. Peptides and proteins used in this work.
| protein or peptide | net charge | peptide sequence or NCBI accession number |
|---|---|---|
| Aβ(1–42) | -3 | DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV IA |
| Aβ(1–40) | -3 | DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV |
| Aβ(1–40) (D23N) | -2 | DAEFRHDSGYEVHHQKLVFFAENVGSNKGAIIGLMVGGVV |
| Wild type α-synuclein | -9 | P37840.1 |
| α-synuclein (A30P) | -9 | P37840.1 |
| α-synuclein (E46K) | -7 | P37840.1 |
| α-synuclein (A53T) | -9 | P37840.1 |
| Bovine PrP (25–242) | +8 | NP_851358.1 |
| Bovine PrP (T194A) | +8 | NP_851358.1 |
| Human PrP (23–231) | +9 | NP_001082180.1 |
| Human PrP (90–231) | +3 | NP_001082180.1 |
NCBI: National center for Biotechnology Information, US, National Library for Medicine
From DNA to Protein
The initial focus of nanopore technology was on nucleic acids with the ultimate goal being rapid DNA sequencing. Pioneering studies by Deamer, Branton, Bayley, Akeson, Meller and colleagues demonstrated that simple sequences such as poly(rU), poly(rA), poly(dC), and poly(dT) could be distinguished on the basis of the parameters I and T.24-30 More recently, with the aid of a modified α-HL pore it was shown that the four individual nucleotides of DNA could be distinguished.31 Thus, by attaching an exonuclease to the vestibule of the pore to cleave off sequential bases, it may be possible to sequence a single DNA molecule.32 However, there are many unresolved issues such as ensuring that each cleaved nucleotide actually enters the pore. It remains to be seen if such a “nanopore DNA sequencer” can compete with conventional technology.
As was the case with nucleic acids, early work with peptides showed that different sequences gave different values for I and T.33-36 However, sequencing peptides and proteins with nanopores will likely be even more challenging because there are 20 common amino acids some of which are chemically very similar. There is also the problem that neutral peptides cannot be driven through the pore. On the other hand, it became clear that small changes in the amino acid sequence could give rise to large changes in the event profiles. For example, both I and T increase for α−helical peptides of increasing length, β−sheet structures tend to give smaller values of I and T compared with α−ηϵλιχϵσ, random coils yield small values of T, and some tetrapeptides give unique event signatures.22,23,37 Thus, although it is not possible to identify with certainty a particular structure based upon the values of I and T tentative assignments can be made by comparison with previous results. Perhaps of more importance to the present discussion, was the ability of the pore to distinguish between multiple conformations in a single sample. For example, in the absence of metal ions Zn-finger peptides, prion peptides from the octarepeat region and myelin basic protein will readily translocate; but in the presence of Zn(II) or Cu(II) the peptides fold into a compact structure and only bumping events are observed.38-40 These results highlight the capacity of the pore to interrogate the conformation of the peptide at the instant at which it interacts with the pore.
Protein Misfolding
Alzheimer disease (AD) is the most widespread of a number of neurodegenerative protein misfolding and aggregation diseases which also include Parkinson disease (PD), Huntington disease (HD), the “prion” diseases such as Creutzfeldt-Jakob Disease (CJD), and amyotrophic lateral sclerosis (ALS).41 In all cases, post-mortem analysis of brain tissue shows the presence of amyloid fibrils, plaques or Lewy bodies which consist of protein aggregates. Surprisingly, the proteins show no obvious sequence or structural homology; e.g., Aβ, which is derived from amyloid precursor protein (APP), in AD; α−synuclein in PD; hungtingtin protein in HD; prion protein in CJD; and superoxide dismutase in ALS. The fact that the proteins are not in their native conformations has led to the hypothesis that they are all “protein misfolding diseases.”42,43
In AD, for example, the amyloid plaques are composed of aggregates of Aβ1–40 and Aβ1–42 which are cleaved from APP by specific peptidases called secretases. The Aβ peptides can oligomerize through β-sheet formation into low molecular weight precursors or protofibrils which then aggregate further into the amyloid fibrils.43,44 There is considerable debate whether the protein aggregates themselves are pathogenic leading to cell death or whether a misfolded protein intermediate is the primary cause. There is currently preference for the latter model since disease progression is correlated with the formation of soluble protofibrils rather than the burden of the amyloid plaques.46-49 Misfolded proteins are normally sequestered or neutralized by cellular defense mechanisms which include the chaperone, proteasome and/or autophagosome responses. Thus one pathogenic possibility is that these responses are affected during AD such that normal protein turnover, which is essential for cell survival, cannot function.50,51 The change in protein turnover and/or clearance undoubtedly accounts for the inverse relation between central nervous system Aβ burden (based on imaging) and plasma levels of the corresponding Aβ species.52 Another possibility is that the initial misfolded Aβ structures bind Cu(II) which can lead to an increase in toxic and potentially lethal reactive oxygen species.53,54 The amyloid plaques on the other hand are relatively inert and are extracellular so that they cannot interfere with protein turnover. Whatever the mechanism, the presence and subsequent misfolding of Aβ peptides plays a central role. Similar arguments can be applied to PD, HD, CJD and ALS.
Nanopore analysis of misfolding
It is perhaps surprising that there is no definitive structure for any of the misfolded proteins. The problem is that at high concentrations they inevitably aggregate which precludes the use of NMR or X-ray crystallography. (One intriguing method to overcome this would be to co-crystallize an antibody-peptide complex in which the antibody prevents an aggregate from forming.)55,56 Nanopore analysis is ideally suited for studying peptides which can adopt multiple conformations since each molecule is interrogated individually. It is also very sensitive since in theory a single molecule can be detected. Therefore, the experiments can be performed at relatively low concentrations so that aggregates will only form slowly if at all. These advantages will be illustrated with three examples. Details of the peptides/proteins and their net charge are shown in Table 1.
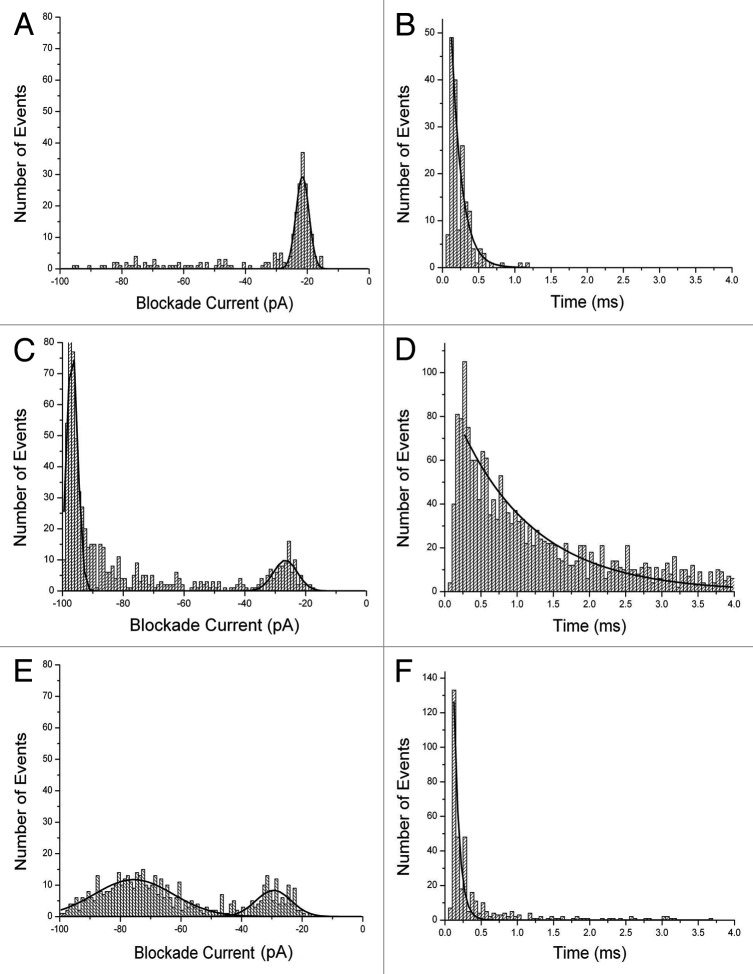
First, the results for Aβ1–42, Aβ1–40 and mutant Aβ1–40 (D23N) are shown in Figure 2. Each peptide was dissolved in 50% trifluoroethanol (TFE) and stored at -20°C before adding an aliquot to the analysis buffer (final concentration 10 μM). The final concentration of TFE was 0.5% but control experiments demonstrated that this concentration had no significant effect on the membrane or pore. The blockade events (see Figure 1) have been converted into histograms of blockade current (I, right) and blockade times (T, left). Aβ1–42 gives events which are mostly of short duration and small blockade current. The event parameters are summarized in Table S1. Previous work20-23 has shown that events with small values of I and T are not due to translocations. Thus, this profile is typical of bumping events due to the molecule being too large to pass through the pore. Similarly, it is known that α-helices and simple two-stranded β-sheets can translocate through the pore.22,23 Thus the implication is that the peptide forms multimers upon storage in 50% TFE. For Aβ1–40 the profile is very different with the majority of events having large I and long T which as mentioned above is typical of a long, mostly α-helical conformation which is translocating through the pore.22 Further evidence for translocation was obtained by measuring the value of T as a function of applied voltage. It was found that t = 3.9, 2.4 and 1.1 ms at 75, 100 and 125 mV respectively. In other words the time decreases as the voltage increases since the peptide is being driven through the pore by the electric field. There are also some bumping events but the distribution is broader than for Aβ1–42. For Aβ1–40 (D23N) the profile is different yet again; most events form a broad Gaussian distribution with an I value of about -75 pA but of short duration. This behavior is typical of a peptide adopting a simple folded β-sheet.23
Figure 2. Event histograms for Aβ peptides. Blockade current and blockade time histograms for Aβ1−42 (A) and (B), Αβ1–40 (C) and (D), and mutant Aβ1–40 D23N (E) and (F). Stock solutions of peptides (rPeptide, Bogart GA) were dissolved in 50% TFE at 1 mg/ml and 10 μL was added to the cis-side of the nanopore chamber which contained 1 M KCl, 10 mM HEPES, pH 7.8. The applied voltage was 100 mV. Further details of the experimental set up have been described previously.12,20-22 (See also Supplementary Information).
It should be noted that the event profiles are constant during the course of an experiment (typically lasting 2–4 h). In other words, once the peptide has been added to the analysis buffer there is no further conformational change or oligomerization. The results themselves may be of relevance to AD since it would appear that Aβ1–42 aggregates more readily than Aβ1–40 and it is known that the presence of Aβ1–42 appears to be more toxic to cells than Aβ1–40.57 On the other hand, the familial AD mutation Aβ1–40 (D23N) forms a β-sheet conformation which does not readily aggregate;57 consistent with the theory that it is a misfolded intermediate which is most toxic to cells rather than the aggregates themselves.46-49
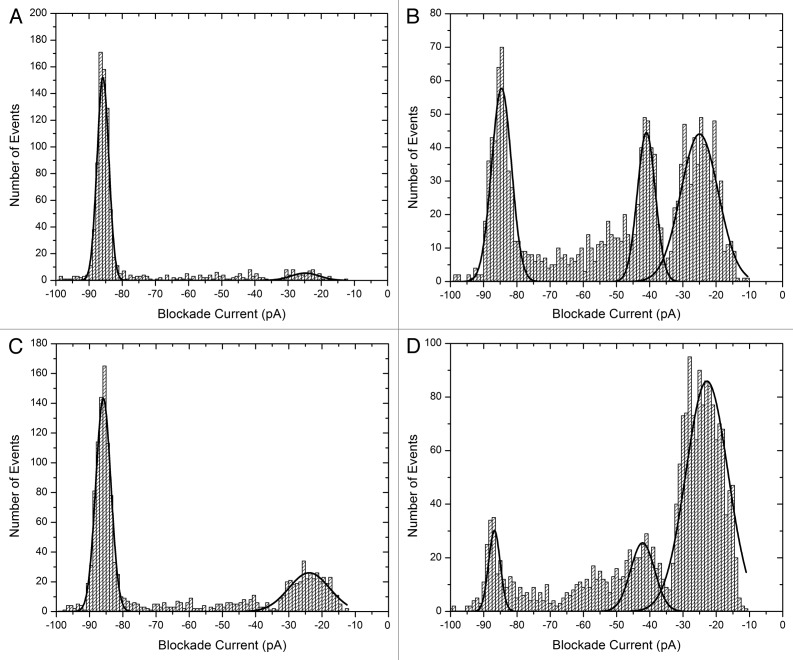
The second example is α-synuclein (Table 1), an intrinsically disordered protein of 140 amino acids which misfolds to form Lewy bodies in PD.59-61 Current blockade histograms of the wild type and three mutant proteins which are implicated in familial forms of the disease, are shown in Figure 3.62 The calculated event parameters are summarized in Table S2. The wild type (Fig. 3a) has a major peak centered at about -86 pA, and a bumping peak at about -25 pA. The peak at -86 pA is almost certainly due to translocation events since the average time decreases as the voltage increases (0.61 and 0.49 ms at 75 and 100 mV respectively). In other words, the protein is being electrophoretically driven through the pore as would be expected for a negatively-charged intrinsically disordered protein. The A30P mutation (Fig. 3b) has a smaller proportion of translocation events, a larger proportion of bumping events and a third peak at about -41 pA, probably due to a folded intermediate, is now evident. For E46K (Fig. 3c) about 67% of the events are translocations which is slightly less than for the wild type. For A53T (Fig. 3d) yet another profile is observed with the majority of events being bumping at – 23pA. It was anticipated that the mutants would show a higher propensity to misfold or aggregate and this appears to be true for A30P and A53T. However, the profile for E46K suggests that it prefers an unfolded conformation like the wild type. Therefore, the molecular mechanism for disease causation is probably different for different mutations. Further work is required to fully understand the misfolding pathways.
Figure 3. Blockade current histograms for α-synuclein. (A) wild type, (B) mutant A30P, (C) mutant E46K and (D) mutant A53T. The peptides (rPeptide) were dissolved in 10 mM TRIS-HCl, pH 7.4 at 1 mg/ml and 10 μL was added to the cis-side of the nanopore chamber which contained 1 M KCl, 10 mM HEPES, 1 mM EDTA, pH 7.8. The applied voltage was 100 mV.
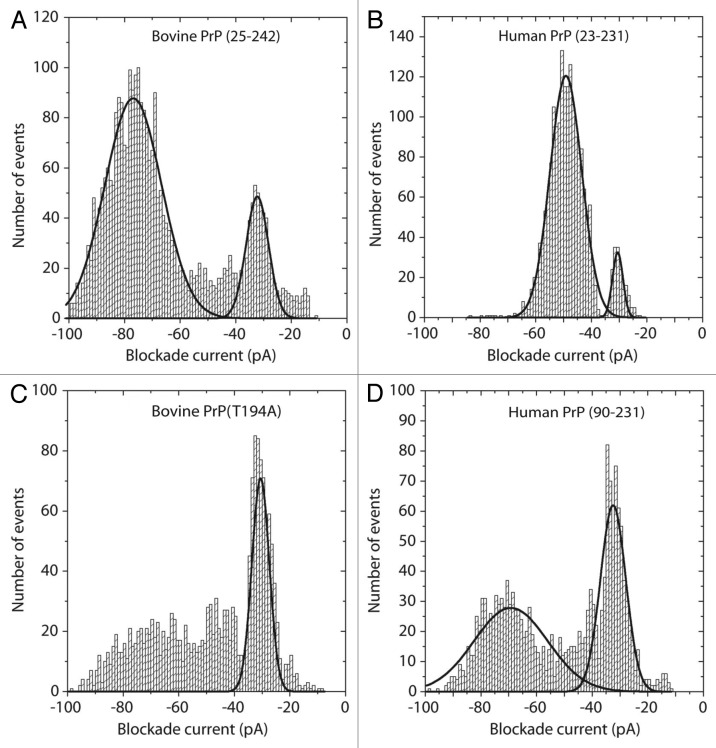
The final example is the prion protein of about 220 amino acids which misfolds to cause a wide variety of transmissible spongiform encephalopathies (TSE).63-65 Perhaps the most intriguing feature of these diseases is the existence of prion strains which arise from different misfolded conformations of identical protein sequences.66,67 Different strains show varying levels of infectivity within the same species and slightly different pathologies. As shown in Figure 4, nanopore analysis can be used to investigate the conformational flexibility of prion proteins. In these experiments, a 10 μl aliquot of the protein was pretreated with 1.5 M gaunadinium-HCl in order to favor the conversion to a different conformation before dilution into 1 ml of the normal 1 M KCl buffer. The blockade current histogram for full length bovine PrP (Fig. 4a) reveals two peaks; a bumping peak at -30 pA and a larger peak at about -80 pA. As has been discussed elsewhere, for many proteins it is difficult to distinguish between intercalation and translocation because the blockade time of these events is little changed by changes in the voltage.21 By comparison, the full length human PrP (Fig. 4b) has a small bumping peak and a major peak at about -50pA. Even small (10–20 amino acid) peptides give translocation events with blockade currents greater than -60 pA and thus, the events at -50 pA for the 208 amino acid human protein must surely be due to intercalation i.e., a loop of the protein transiently enters the pore but the remainder of the molecule is too tightly folded to allow translocation. Further evidence for intercalation is provided by voltage studies since the event time does not change significantly (0.096, 0.092, 0.093 and 0.083 ms at 50, 100, 150 and 200 mV respectively). It is also noticeable that the current distributions for the full length human PrP are narrower than that for the full length bovine PrP. One possible explanation is that the bovine protein has more conformational flexibility and thus is interacting with the pore in different orientations. For the mutant bovine PrP (T194A) the event profile (Fig. 4c) is easily distinguished from the wild type because there are far fewer intercalation events. Thus a single amino acid change in a 200 amino acid protein results in conformational changes which are readily distinguished by the nanopore. Finally, the human PrP protein which is lacking the disordered N-terminus also yields a unique profile (Fig. 4d). Compared with the full length human PrP, there is no peak at -50 pA and the bumping peak at -30 pA is more prominent. Thus, it would appear that the presence of the N-terminus alters the conformation of the C-terminus. We have shown previously that addition of prion-specific antibodies eliminates most events because the antibody/prion complex diffuses to the pore very slowly.68 Thus, epitope specific antibodies could be used to delineate which epitopes are exposed under particular conditions. These results are expected to clarify the nature of conformational differences between prion strains.
Figure 4. Blockade current histograms for prion proteins. (A) Bovine PrP (25–242), (B) human PrP (23–231), (C) mutant bovine PrP (T194A) and (D) Human PrP (90–231). The proteins were obtained from Jena Bioscience, and dissolved in 10 mM TRIS-HCl, 0.1 mM EDTA pH 8.0 at 1 mg/ml. Twenty μL was added to 8.6 μL of 5 M Guanadinium-HCl and incubated at 21°C for 1 h. The whole aliquot was then added to the cis-side of the nanopore chamber which contained 1 M KCl, 10 mM TRIS-HCl pH 7.8. The applied voltage was 100 mV
Future directions
The examples presented above demonstrate that conformational changes can be readily examined with the α-HL pore for peptides and proteins ranging in size from 40 to 220 amino acids. Thus the α-HL pore with a diameter of 1.5 nm is suitable for studying the factors that can either induce or prevent the initial stages in the misfolding pathway. These factors include metal ions, particularly Cu(II) which appears to be involved in most neurodegenerative diseases. The interaction of Cu(II) with prions and prion peptides using nanopore analysis has already been reported and studies with Aβ peptides and α-synuclein are now underway.39 Similarly, small molecules or peptides that prevent misfolding can be analyzed; for example, dopamine which inhibits misfolding of α-synuclein.69 In order to study the later stages of misfolding i.e the formation of oligomers and aggregates as well as larger proteins such as SOD, implicated in ALS, it might be more appropriate to use solid state pores.14,15 These can be fabricated in silicon nitride membranes with diameters in the range of 5–20 nm, allowing the translocation of large proteins or aggregates. Since the blockade current is proportional to the volume of the pore occluded by the protein, it is possible to estimate the size of oligomers/aggregates as well as to distinguish unbound protein from antibody-protein complexes. Recent experiments have shown that folded proteins give smaller blockade currents than the corresponding random coil but it is not clear that different conformations would yield different signatures since the occluded volumes would be very similar.70 There are also problems with the reproducibility of solid state pores since it is very difficult to control the diameter during fabrication and the chemistry of the internal surfaces can be quite variable. One elegant solution is to coat the pore with a lipid.71 This treatment not only makes the pore chemically inert which prevents permanent blockages but also the structure of the internal surface becomes reproducible. In this way it was possible to demonstrate single molecular events for Aβ oligomers and fibrils although no detailed analysis was performed.
In conclusion, many neurodegenerative diseases involve protein misfolding into insoluble fibrils or amyloid plaques. The misfolding processes, particularly the first steps, are very difficult to study by conventional techniques because at high concentrations the proteins inevitably aggregate. Nanopore analysis, being a single molecule technique, is an attractive alternative because it can be performed at very low concentrations.
Supplementary Material
Glossary
Abbreviations:
- AD
Alzheimer disease
- PD
Parkinson disease
- HD
Huntington disease
- CJD
Creutzfeldt-Jacob disease
- ALS
Amyotrophic lateral sclerosis
- α-HL
α-hemolysin
- TFE
Trifluoroethanol
Note
Supplemental Material can be found at: www.landesbiocience.com/journals/pri/article/6.2/18665/
Acknowledgments
Financial support was provided by NSERC.
References
- 1.Coulter WH. Means for counting particles in a fluid. USA patent 2,656.508, 765-772.
- 2.Bayley H, Martin CR. . Resistive-pulse sensing – from microbes to molecules. Chem Rev 2000; 100:2575 - 94; http://dx.doi.org/ 10.1021/cr980099g; PMID: 11749296 [DOI] [PubMed] [Google Scholar]
- 3.Davis RE, Green RE. . Automatic platelet counting with the Couler particle counter. J Clin Pathol 1967; 20:777 - 9; http://dx.doi.org/ 10.1136/jcp.20.5.777; PMID: 5602990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubitschek HE. . Electronic counting and sizing of bacteria. Nature 1958; 182:234 - 5; http://dx.doi.org/ 10.1038/182234a0; PMID: 13577794 [DOI] [PubMed] [Google Scholar]
- 5.Allard B. Patch-clamp methods and protocols. In: Molnar P, Hickman JJ, ed(s) Methods in Molecular Biology. 403. Totowa NJ, Humana Press, 2007: 3-303. [Google Scholar]
- 6.Williams AJ, West DJ, Sitsapesan R. . Light at the end of the Ca(2+)-release channel tunnel: structures and mechanisms involved in ion translocation in ryanodine receptor channels. Q Rev Biophys 2001; 34:61 - 104; http://dx.doi.org/ 10.1017/S0033583501003675; PMID: 11388090 [DOI] [PubMed] [Google Scholar]
- 7.Gilbert RJ. . Pore-forming toxins. Cell Mol Life Sci 2002; 59:832 - 44; http://dx.doi.org/ 10.1007/s00018-002-8471-1; PMID: 12088283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace BA. . Gramicidin channels and pores. Annu Rev Biophys Biophys Chem 1990; 19:127 - 57; http://dx.doi.org/ 10.1146/annurev.bb.19.060190.001015; PMID: 1694667 [DOI] [PubMed] [Google Scholar]
- 9.Gouaux E. . alpha-Hemolysin from Staphylococcus aureus: an archetype of beta-barrel, channel-forming toxins. J Struct Biol 1998; 121:110 - 22; http://dx.doi.org/ 10.1006/jsbi.1998.3959; PMID: 9615434 [DOI] [PubMed] [Google Scholar]
- 10.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. . Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 1996; 274:1859 - 66; http://dx.doi.org/ 10.1126/science.274.5294.1859; PMID: 8943190 [DOI] [PubMed] [Google Scholar]
- 11.Parker MW, Buckley JT, Postma JPM, Tucker AD, Leonard K, Pattus F, et al. . Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states. Nature 1994; 367:292 - 5; http://dx.doi.org/ 10.1038/367292a0; PMID: 7510043 [DOI] [PubMed] [Google Scholar]
- 12.Stefureac RI, Waldner L, Howard P, Lee JS. . Nanopore analysis of a small 86-residue protein. Small 2008; 4:59 - 63; http://dx.doi.org/ 10.1002/smll.200700402; PMID: 18058890 [DOI] [PubMed] [Google Scholar]
- 13.Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, van der Goot FG, Auvray L, et al. . Dynamics of unfolded protein transport through an aerolysin pore. J Am Chem Soc 2011; 133:2923 - 31; http://dx.doi.org/ 10.1021/ja1073245; PMID: 21319816 [DOI] [PubMed] [Google Scholar]
- 14.Dekker C. . Solid-state nanopores. Nat Nanotechnol 2007; 2:209 - 15; http://dx.doi.org/ 10.1038/nnano.2007.27; PMID: 18654264 [DOI] [PubMed] [Google Scholar]
- 15.Wanunu M, Sutin J, Meller A. . DNA profiling using solid-state nanopores: detection of DNA-binding molecules. Nano Lett 2009; 9:3498 - 502; http://dx.doi.org/ 10.1021/nl901691v; PMID: 19585985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bezrukov SM. . Ion channels as molecular coulter counters to probe metabolite transport. J Membr Biol 2000; 174:1 - 13; http://dx.doi.org/ 10.1007/s002320001026; PMID: 10741427 [DOI] [PubMed] [Google Scholar]
- 17.Bayley H, Cremer PS. . Stochastic sensors inspired by biology. Nature 2001; 413:226 - 30; http://dx.doi.org/ 10.1038/35093038; PMID: 11557992 [DOI] [PubMed] [Google Scholar]
- 18.Martin CR, Siwy ZS. . Chemistry. Learning nature’s way: biosensing with synthetic nanopores. Science 2007; 317:331 - 2; http://dx.doi.org/ 10.1126/science.1146126; PMID: 17641190 [DOI] [PubMed] [Google Scholar]
- 19.Mohammad MM, Prakash S, Matouschek A, Movileanu L. . Controlling a single protein in a nanopore through electrostatic traps. J Am Chem Soc 2008; 130:4081 - 8; http://dx.doi.org/ 10.1021/ja710787a; PMID: 18321107 [DOI] [PubMed] [Google Scholar]
- 20.Meng H, Detillieux D, Baran C, Krasniqi B, Christensen C, Madampage C, et al. . Nanopore analysis of tethered peptides. J Pept Sci 2010; 16:701 - 8; http://dx.doi.org/ 10.1002/psc.1289; PMID: 20814890 [DOI] [PubMed] [Google Scholar]
- 21.Christensen C, Baran C, Krasniqi B, Stefureac RI, Nokhrin S, Lee JS. . Effect of charge, topology and orientation of the electric field on the interaction of peptides with the α-hemolysin pore. J Pept Sci 2011; 17:726 - 34; http://dx.doi.org/ 10.1002/psc.1393; PMID: 21766390 [DOI] [PubMed] [Google Scholar]
- 22.Stefureac R, Long YT, Kraatz HB, Howard P, Lee JS. . Transport of α-helical peptides through α-hemolysin and aerolysin pores. Biochemistry 2006; 45:9172 - 9; http://dx.doi.org/ 10.1021/bi0604835; PMID: 16866363 [DOI] [PubMed] [Google Scholar]
- 23.Goodrich CP, Kirmizialtin S, Huyghues-Despointes BM, Zhu A, Scholtz JM, Makarov DE, et al. . Single-molecule electrophoresis of β-hairpin peptides by electrical recordings and Langevin dynamics simulations. J Phys Chem B 2007; 111:3332 - 5; http://dx.doi.org/ 10.1021/jp071364h; PMID: 17388500 [DOI] [PubMed] [Google Scholar]
- 24.Bayley H. . Sequencing single molecules of DNA. Curr Opin Chem Biol 2006; 10:628 - 37; http://dx.doi.org/ 10.1016/j.cbpa.2006.10.040; PMID: 17113816 [DOI] [PubMed] [Google Scholar]
- 25.Rhee M, Burns MA. . Nanopore sequencing technology: nanopore preparations. Trends Biotechnol 2007; 25:174 - 81; http://dx.doi.org/ 10.1016/j.tibtech.2007.02.008; PMID: 17320228 [DOI] [PubMed] [Google Scholar]
- 26.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. . The potential and challenges of nanopore sequencing. Nat Biotechnol 2008; 26:1146 - 53; http://dx.doi.org/ 10.1038/nbt.1495; PMID: 18846088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deamer DW, Akeson M. . Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol 2000; 18:147 - 51; http://dx.doi.org/ 10.1016/S0167-7799(00)01426-8; PMID: 10740260 [DOI] [PubMed] [Google Scholar]
- 28.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. . Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J 1999; 77:3227 - 33; http://dx.doi.org/ 10.1016/S0006-3495(99)77153-5; PMID: 10585944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. . Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci U S A 2000; 97:1079 - 84; http://dx.doi.org/ 10.1073/pnas.97.3.1079; PMID: 10655487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. . Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A 1996; 93:13770 - 3; http://dx.doi.org/ 10.1073/pnas.93.24.13770; PMID: 8943010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nano 2009 [DOI] [PubMed] [Google Scholar]
- 32.Deamer D. . Nanopore analysis of nucleic acids bound to exonucleases and polymerases. Annu Rev Biophys 2010; 39:79 - 90; http://dx.doi.org/ 10.1146/annurev.biophys.093008.131250; PMID: 20192777 [DOI] [PubMed] [Google Scholar]
- 33.Sutherland TC, Long Y, Stefureac R, Bediako-Amoa I, Kraatz H, Lee JS. . Structure of peptides investigated by nanopore analysis. Nano Lett 2004; 4:1273 - 7; http://dx.doi.org/ 10.1021/nl049413e [DOI] [Google Scholar]
- 34.Movileanu L, Schmittschmitt JP, Scholtz JM, Bayley H. . Interactions of peptides with a protein pore. Biophys J 2005; 89:1030 - 45; http://dx.doi.org/ 10.1529/biophysj.104.057406; PMID: 15923222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bikwemu R, Wolfe AJ, Xing X, Movileanu L. . Facilitated translocation of polypeptides through a single nanopore. J Phys 2010; 22:454177 - 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Movileanu L. . Squeezing a single polypeptide through a nanopore. Soft Matter 2008; 4:925 - 31; http://dx.doi.org/ 10.1039/b719850g [DOI] [PubMed] [Google Scholar]
- 37.Zhao Q, Jayawardhana DA, Wang D, Guan X. . Study of peptide transport through engineered protein channels. J Phys Chem B 2009; 113:3572 - 8; http://dx.doi.org/ 10.1021/jp809842g; PMID: 19231820 [DOI] [PubMed] [Google Scholar]
- 38.Stefureac RI, Lee JS. . Nanopore analysis of the folding of zinc fingers. Small 2008; 4:1646 - 50; http://dx.doi.org/ 10.1002/smll.200800585; PMID: 18819138 [DOI] [PubMed] [Google Scholar]
- 39.Stefureac RI, Madampage CA, Andrievskaia O, Lee JS. . Nanopore analysis of the interaction of metal ions with prion proteins and peptides. Biochem Cell Biol 2010; 88:347 - 58; http://dx.doi.org/ 10.1139/O09-176; PMID: 20453935 [DOI] [PubMed] [Google Scholar]
- 40.Baran C, Smith GST, Bamm VV, Harauz G, Lee JS. . Divalent cations induce a compaction of intrinsically disordered myelin basic protein. Biochem Biophys Res Commun 2010; 391:224 - 9; http://dx.doi.org/ 10.1016/j.bbrc.2009.11.036; PMID: 19903451 [DOI] [PubMed] [Google Scholar]
- 41.Lansbury PT, Lashuel HA. . A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature 2006; 443:774 - 9; http://dx.doi.org/ 10.1038/nature05290; PMID: 17051203 [DOI] [PubMed] [Google Scholar]
- 42.Chiti F, Dobson CM. . Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 2006; 75:333 - 66; http://dx.doi.org/ 10.1146/annurev.biochem.75.101304.123901; PMID: 16756495 [DOI] [PubMed] [Google Scholar]
- 43.Schnabel J. . Protein folding: The dark side of proteins. Nature 2010; 464:828 - 9; http://dx.doi.org/ 10.1038/464828a; PMID: 20376124 [DOI] [PubMed] [Google Scholar]
- 44.Fezoui Y, Teplow DB. . Kinetic studies of amyloid β-protein fibril assembly. Differential effects of alpha-helix stabilization. J Biol Chem 2002; 277:36948 - 54; http://dx.doi.org/ 10.1074/jbc.M204168200; PMID: 12149256 [DOI] [PubMed] [Google Scholar]
- 45.Hickman SE, Allison EK, El Khoury J. . Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci 2008; 28:8354 - 60; http://dx.doi.org/ 10.1523/JNEUROSCI.0616-08.2008; PMID: 18701698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, et al. . A specific amyloid-β protein assembly in the brain impairs memory. Nature 2006; 440:352 - 7; http://dx.doi.org/ 10.1038/nature04533; PMID: 16541076 [DOI] [PubMed] [Google Scholar]
- 47.Mucke L. . Neuroscience: Alzheimer’s disease. Nature 2009; 461:895 - 7; http://dx.doi.org/ 10.1038/461895a; PMID: 19829367 [DOI] [PubMed] [Google Scholar]
- 48.Hardy J. . The amyloid hypothesis for Alzheimer’s disease: a critical reappraisal. J Neurochem 2009; 110:1129 - 34; http://dx.doi.org/ 10.1111/j.1471-4159.2009.06181.x; PMID: 19457065 [DOI] [PubMed] [Google Scholar]
- 49.Castellani RJ, Lee H-G, Siedlak SL, Nunomura A, Hayashi T, Nakamura M, et al. . Reexamining Alzheimer’s disease: evidence for a protective role for amyloid-beta protein precursor and amyloid-beta. J Alzheimers Dis 2009; 18:447 - 52; PMID: 19584435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobson CM. . Protein folding and misfolding. Nature 2003; 426:884 - 90; http://dx.doi.org/ 10.1038/nature02261; PMID: 14685248 [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. . Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 2010; 141:1146 - 58; http://dx.doi.org/ 10.1016/j.cell.2010.05.008; PMID: 20541250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, et al. . Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol 2006; 59:512 - 9; http://dx.doi.org/ 10.1002/ana.20730; PMID: 16372280 [DOI] [PubMed] [Google Scholar]
- 53.Curtain CC, Ali F, Volitakis I, Cherny RA, Norton RS, Beyreuther K, et al. . Alzheimer’s disease amyloid-beta binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J Biol Chem 2001; 276:20466 - 73; http://dx.doi.org/ 10.1074/jbc.M100175200; PMID: 11274207 [DOI] [PubMed] [Google Scholar]
- 54.Bush AI, Masters CL, Tanzi RE. . Copper, β-amyloid, and Alzheimer’s disease: tapping a sensitive connection. Proc Natl Acad Sci U S A 2003; 100:11193 - 4; http://dx.doi.org/ 10.1073/pnas.2135061100; PMID: 14506299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paramithiotis E, Pinard M, Lawton T, LaBoissiere S, Leathers VL, Zou WQ, et al. . A prion protein epitope selective for the pathologically misfolded conformation. Nat Med 2003; 9:893 - 9; http://dx.doi.org/ 10.1038/nm883; PMID: 12778138 [DOI] [PubMed] [Google Scholar]
- 56.Polymenidou M, Moos R, Scott M, Sigurdson C, Shi YZ, Yajima B, et al. . The POM monoclonals: a comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One 2008; 3:e3872; http://dx.doi.org/ 10.1371/journal.pone.0003872; PMID: 19060956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez OL, Kuller LH, Mehta PD, Becker JT, Gach HM, Sweet RA, et al. . Plasma amyloid levels and the risk of AD in normal subjects in the Cardiovascular Health Study. Neurology 2008; 70:1664 - 71; http://dx.doi.org/ 10.1212/01.wnl.0000306696.82017.66; PMID: 18401021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grabowski TJ, Cho HS, Vonsattel JP, Rebeck GW, Greenberg SM. . Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Ann Neurol 2001; 49:697 - 705; http://dx.doi.org/ 10.1002/ana.1009; PMID: 11409420 [DOI] [PubMed] [Google Scholar]
- 59.Trojanowski JQ, Lee VM. . Parkinson’s disease and related synucleinopathies are a new class of nervous system amyloidoses. Neurotoxicology 2002; 23:457 - 60; http://dx.doi.org/ 10.1016/S0161-813X(02)00065-7; PMID: 12428717 [DOI] [PubMed] [Google Scholar]
- 60.Chandra S, Gallardo G, Fernández-Chacón R, Schlüter OM, Südhof TC. . Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell 2005; 123:383 - 96; http://dx.doi.org/ 10.1016/j.cell.2005.09.028; PMID: 16269331 [DOI] [PubMed] [Google Scholar]
- 61.Volles MJ, Lansbury PT Jr.. . Zeroing in on the pathogenic form of alpha-synuclein and its mechanism of neurotoxicity in Parkinson’s disease. Biochemistry 2003; 42:7871 - 8; http://dx.doi.org/ 10.1021/bi030086j; PMID: 12834338 [DOI] [PubMed] [Google Scholar]
- 62.Conway KA, Harper JD, Lansbury PT. . Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med 1998; 4:1318 - 20; http://dx.doi.org/ 10.1038/3311; PMID: 9809558 [DOI] [PubMed] [Google Scholar]
- 63.Prusiner SB. . Shattuck lecture--neurodegenerative diseases and prions. N Engl J Med 2001; 344:1516 - 26; http://dx.doi.org/ 10.1056/NEJM200105173442006; PMID: 11357156 [DOI] [PubMed] [Google Scholar]
- 64.Cobb NJ, Surewicz WK. . Prion diseases and their biochemical mechanisms. Biochemistry 2009; 48:2574 - 85; http://dx.doi.org/ 10.1021/bi900108v; PMID: 19239250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morillas M, Vanik DL, Surewicz WK. . On the mechanism of α-helix to β-sheet transition in the recombinant prion protein. Biochemistry 2001; 40:6982 - 7; http://dx.doi.org/ 10.1021/bi010232q; PMID: 11389614 [DOI] [PubMed] [Google Scholar]
- 66.Morales R, Abid K, Soto C. . The prion strain phenomenon: molecular basis and unprecedented features. Biochim Biophys Acta 2007; 1772:681 - 91; PMID: 17254754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collinge J. . Medicine. Prion strain mutation and selection. Science 2010; 328:1111 - 2; http://dx.doi.org/ 10.1126/science.1190815; PMID: 20508117 [DOI] [PubMed] [Google Scholar]
- 68.Madampage CA, Andrievskaia O, Lee JS. . Nanopore detection of antibody prion interactions. Anal Biochem 2010; 396:36 - 41; http://dx.doi.org/ 10.1016/j.ab.2009.08.028; PMID: 19699704 [DOI] [PubMed] [Google Scholar]
- 69.Latawiec D, Herrera F, Bek A, Losasso V, Candotti M, Benetti F, et al. . Modulation of alpha-synuclein aggregation by dopamine analogs. PLoS One 2010; 5:e9234; http://dx.doi.org/ 10.1371/journal.pone.0009234; PMID: 20169066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stefureac RI, Trivedi D, Marziali A, Lee JS. . Evidence that small proteins translocate through silicon nitride pores in a folded conformation. J Phys Condens Matter 2010; 22:454133 - 44; http://dx.doi.org/ 10.1088/0953-8984/22/45/454133; PMID: 21339619 [DOI] [PubMed] [Google Scholar]
- 71.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, et al. . Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotechnol 2011; 6:253 - 60; http://dx.doi.org/ 10.1038/nnano.2011.12; PMID: 21336266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.