Currently, domesticated dogs and cats are the most cherished companion animals for humans, and concerns about their health and well-being are therefore important. In this context, the gut microbiota plays a crucial role in maintaining and promoting host health. However, despite the social relevance of domesticated dogs and cats, their intestinal microbial communities are still far from being completely understood. In this study, the taxonomical composition of canine and feline gut microbiota was explored at genus and bifidobacterial species levels, allowing classification of these microbial populations into distinct gut community state types at either of the two investigated taxonomic levels. Furthermore, the reconstruction of core gut microbiota coupled with covariance network analysis based on bifidobacterial internally transcribed spacer (ITS) profiling revealed differences in the bifidobacterial compositions of canine and feline gut microbiota, suggesting that particular bifidobacterial species have developed a selective ability to colonize a specific host.
KEYWORDS: gut microbiota, bifidobacteria, metagenomics, dog, cat, gut microbiota
ABSTRACT
During the course of evolution, dogs and cats have been subjected to extensive domestication, becoming the principal companion animals for humans. For this reason, their health care, including their intestinal microbiota, is considered of considerable importance. However, the canine and feline gut microbiota still represent a largely unexplored research area. In the present work, we profiled the microbiota of 23 feline fecal samples by 16S rRNA gene and bifidobacterial internally transcribed spacer (ITS) approaches and compared this information with previously reported data from 138 canine fecal samples. The obtained data allowed the reconstruction of the core gut microbiota of the above-mentioned samples coupled with their classification into distinct community state types at both genus and species levels, identifying Bacteroides, Fusobacterium, and Prevotella 9 as the main bacterial components of the canine and feline gut microbiota. At the species level, the intestinal bifidobacterial gut communities of dogs and cats differed in terms of both species number and composition, as emphasized by a covariance analysis. Together, our findings show that the intestinal populations of cats and dogs are similar in terms of genus-level taxonomical composition, while at the bifidobacterial species level, clear differences were observed, indicative of host-specific colonization behavior by particular bifidobacterial taxa.
IMPORTANCE Currently, domesticated dogs and cats are the most cherished companion animals for humans, and concerns about their health and well-being are therefore important. In this context, the gut microbiota plays a crucial role in maintaining and promoting host health. However, despite the social relevance of domesticated dogs and cats, their intestinal microbial communities are still far from being completely understood. In this study, the taxonomical composition of canine and feline gut microbiota was explored at genus and bifidobacterial species levels, allowing classification of these microbial populations into distinct gut community state types at either of the two investigated taxonomic levels. Furthermore, the reconstruction of core gut microbiota coupled with covariance network analysis based on bifidobacterial internally transcribed spacer (ITS) profiling revealed differences in the bifidobacterial compositions of canine and feline gut microbiota, suggesting that particular bifidobacterial species have developed a selective ability to colonize a specific host.
INTRODUCTION
The gastrointestinal tract (GIT) of mammals is home to a complex and intricate community of microorganisms collectively representing the gut microbiota. The long-lasting interplay between this microbial ecosystem and its hosts has ensured the establishment of a symbiotic relationship from which both parties benefit (1). The host offers a suitable environment for microbial growth, while bacterial cells contribute to certain physiological host functions, affecting absorption and metabolism of nutrients, promoting protection against pathogens, and stimulating the immune system (2, 3), thereby implicating the GIT as the most important interface between host and microorganisms. Culture-dependent and culture-independent methods coupled with biochemical and physiological investigations purport bifidobacteria as a group of microorganisms that positively affect the host by exerting health-promoting effects (4, 5). Members of the Bifidobacterium genus are prominent constituents of the gut microbiota of various animals that provide parental care to their offspring, including warm-blooded mammals and social insects (6, 7). It is worth noting that these commensal bacteria are among the first colonizers of the neonatal gut, thus modulating metabolic and immune activities of the host following its birth. In this context, it is widely accepted that bifidobacteria contribute to various biological processes in their human host, such as maturation of the immune system, induction of mucus layer production, and degradation of nondigestible diet- and host-derived glycans to produce nutrition for the host’s enterocytes (8).
Domesticated dogs and cats are the most popular human companion animals. Their domestication began many centuries ago, and these animals have since gained an increasingly important role within the social structure of a family, even being considered genuine family members (2). In this context, health care for dogs and cats has substantially increased over time, with recent attention also including the impact of canine and feline intestinal microbiota on health and well-being (2, 9). Nevertheless, the taxonomical composition and functional investigations of the intestinal microbial communities of dogs and cats is still far from being fully characterized. In particular, only a couple of studies have reported detailed insights into the composition of the bifidobacterial populations harbored by the canine and feline intestine (7, 10). Indeed, most of the published research limited their investigations to culture-dependent methods, which is commonly known to be restricted by medium and growth conditions (11–13). Furthermore, other publications have reported on the variation of bifidobacterial relative abundance following administration of prebiotics and/or probiotics to dogs and cats, while not including a general overview of the autochthonous species-level bifidobacterial community that may be present in the intestines of these animal companions (14, 15). In the present study, 16S rRNA gene sequencing was exploited to assess the overall taxonomical composition of the gut microbiota of dogs and cats, involving 23 feline stool samples specifically collected for this study and 138 canine fecal samples retrieved from a previous study (10). Moreover, a genus-specific investigation aimed at dissecting the bifidobacterial community of the same fecal samples was performed by means of bifidobacterial internally transcribed spacer (ITS) profiling, an approach that relies on sequencing of the ITS hypervariable region for taxonomic reconstruction of the bifidobacterial population down to the subspecies level (7, 16).
RESULTS AND DISCUSSION
Characterization of the feline and canine intestinal microbial community by means of 16S rRNA-based microbial profiling.
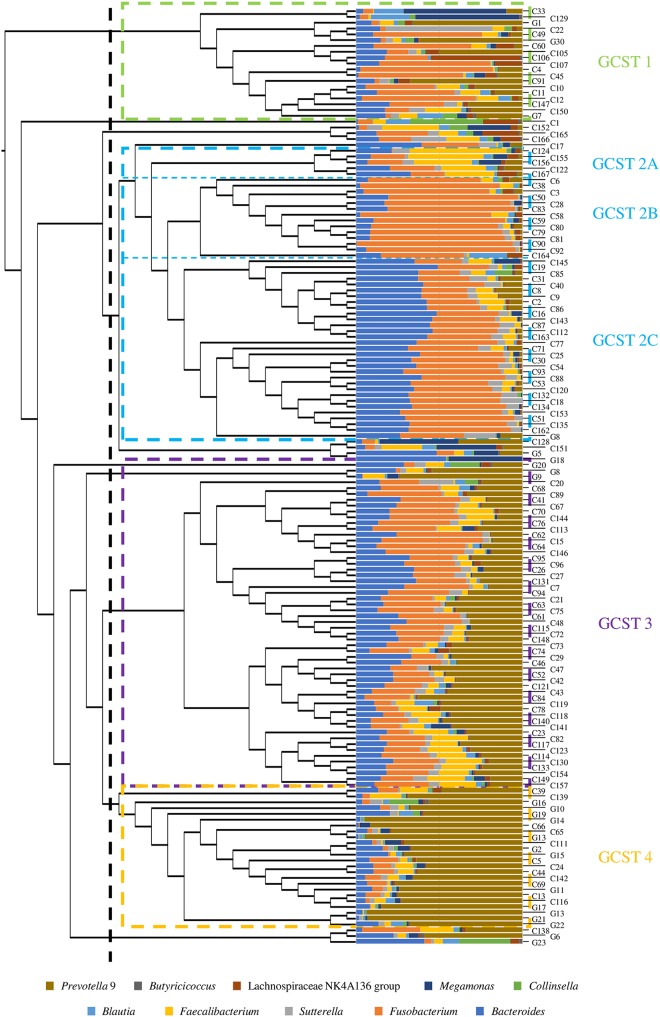
To investigate the taxonomical composition of the intestinal bacterial community of the mammalian species Canis lupus familiaris and Felis catus silvestris, a total of 161 fecal samples, composed of 23 feline fecal samples and 138 canine stool samples, were analyzed, representing 7 and 46 distinct feline and canine breeds, respectively, and corresponding to 161 different animals (see Table S1 in the supplemental material). In detail, feline fecal samples were specifically collected for this study and subjected to 16S rRNA gene sequencing, as previously described (17). Conversely, the 16S rRNA microbial profiling data of the canine fecal samples were obtained by reanalyzing the 16S rRNA gene sequences from a previous study that followed the same experimental procedures used for feline samples processed here (10). In this case, as all feline fecal samples belonged to cats that followed a commercial food-based diet, only fecal samples corresponding to the commercial food-fed dogs were taken into consideration to prevent any bias, as previously discussed (10). Illumina sequencing generated a total of 8,732,180 sequenced reads with an average of 53,902 reads per sample. Quality and chimera filtering then generated a total of 8,052,497 filtered sequence reads with an average of 49,706 reads per sample (see Table S2). The generated data were used to evaluate bacterial biodiversity through alpha diversity analysis based on the observed exact sequence variants (ESVs). Assessment of the two rarefaction curves corresponding to the averages observed for the two mammalian groups revealed differences in the biodiversity of canine and feline gut microbiota. In detail, Student’s t test statistical analysis (t value < 0.05) revealed a significantly higher level of diversity among the feline fecal bacterial community than among that harbored by canine fecal samples (see Fig. S1a), in line with what had previously been observed (18). Additionally, 16S rRNA gene sequencing data were used to reconstruct the canine and feline core gut microbiota, defined as the ensemble of microbial genera that are shared across samples of a defined cohort (19). To identify the core intestinal bacterial communities of dogs and cats, only bacterial genera that were identified in at least 80% of collected samples and with an average relative abundance of >0.1% were considered. Interestingly, while the canine gut microbiota counted 21 bacterial genera fulfilling these two “core” inclusion criteria, the feline core intestinal community consisted of 33 microbial taxa. In detail, the core gut microbiota of dogs is dominated by Fusobacterium (average relative abundance of 24.32%), Bacteroides (13.66%), and Prevotella 9 (a subcluster of the Prevotellaceae family inferred from available in silico data from the SILVA database) (15.18%), as reported previously (10). Conversely, in the feline core intestinal population, Prevotella 9 was predominant with an average relative abundance of 28.92% followed by Bacteroides (8.32%), Romboutsia (5.28%), and Clostridium sensu stricto 3 (5.32%). Members of the genus Fusobacterium, despite being part of the feline core gut microbiota, displayed a markedly reduced relative abundance (2.34%) compared to that of the canine group (Fig. S1b and c), indicating that the Fusobacterium genus may have undergone specific adaptations in order to colonize the Canis lupus familiaris intestine. These taxonomical differences were further substantiated by a hierarchical clustering analysis based on 16S rRNA microbial profiling of the canine and feline fecal samples (Fig. 1). Indeed, even though the generated cladogram did not display a clear separation of the collected samples according to these two mammalian groups, most of the feline fecal samples clustered together. Particularly, the cladogram revealed the presence of four main clusters, one of which may be separated into three different subclusters. These findings allowed separation of the collected fecal samples into six distinct gut community state types (GCSTs), each dominated by a minimum of one to a maximum of three bacterial genera with an average relative abundance of >10%: Lactobacillus (GCST 1); Bacteroides, Fusobacterium, and Faecalibacterium (GCST 2A); Fusobacterium (GCST 2B); Fusobacterium and Bacteroides (GCST 2C); Fusobacterium, Prevotella 9, and Bacteroides (GCST 3); and Prevotella 9 (GCST 4) (Fig. 1). Although six subclusters were identified, three groups were shown to contain the largest number of samples, i.e., clusters 2C, 3, and 4, which represent 31, 53, and 26 samples, respectively. This suggests that Fusobacterium, Prevotella 9, and Bacteroides are the major players of the canine and feline fecal samples. Clustering of the collected samples according to abundance variation of certain microbial genera was further corroborated by beta-diversity analysis performed by means of Bray-Curtis distance matrix and represented by principal-coordinate analysis (PCoA) (see Fig. S2). This PCoA clustering analysis was confirmed by a permutational multivariate analysis of variance (PERMANOVA) when the various GCSTs were compared (P value < 0.05). Interestingly, the latter analysis reveals partial overlap between GCST 2C and 3, probably due to their similar microbial compositions.
FIG 1.
Classification into distinct compositional types of the gut microbiota of dogs and cats. The cladogram on the left displays the hierarchical clustering of the collected samples based on their average 16S rRNA gene microbial profiles. The bar plot in the center depicts the abundance of the 10 most representative microbial taxa of the considered samples. The numbers on the right correspond to the names of collected samples. The colored dotted rectangles separate samples into the six identified gut community state types (GCSTs) whose names are reported on the right with the same GCST color code.
Distribution of bifidobacteria in the intestinal microbial community of dogs and cats.
16S rRNA microbial profiling data were exploited to evaluate the relative abundance of the Bifidobacterium genus within the canine and feline gut microbiota. Interestingly, while the feline gut microbiota showed an average relative bifidobacterial abundance of 0.46% with a prevalence of 95.65%, the canine intestinal microbial community only possessed an average relative bifidobacterial abundance of 0.20% with a lower prevalence level (74.64%). Bifidobacterial species have been reported to be universally distributed across the mammalian species, including both cats and dogs, suggesting that these intestinal commensals have evolved a broad host colonization potential (6, 7). In this context, the observed lower prevalence level of bifidobacterial species in the gut microbiota of dogs (compared to that found in cats) may be a consequence of the limited sensitivity of the 16S rRNA gene-based microbial profiling (16). To overcome this limitation, ITS bifidobacterial profiling was performed by means of genus-specific primers that target the hypervariable ITS region of bifidobacteria, allowing in-depth phylotype-level community profiling, according to a previously described protocol (16). Illumina sequencing of the bifidobacterial ITS region generated a total of 1,771,641 sequenced reads, which when quality and chimera filtered resulted in a total of 1,686,399 filtered reads (see Table S3). Beta-diversity analysis performed by means of principal-coordinate analysis (PCoA) through the Bray-Curtis distance matrix revealed clustering of the data sets based on host species, i.e., Felis silvestris catus and Canis lupus familiaris, indicating that the gut microbiota of these two domesticated animals each contain a specific bifidobacterial community (see Fig. S3). Furthermore, the obtained ITS data revealed differences in the number of bifidobacterial species harbored by the intestinal tracts of dogs and cats. Indeed, canine fecal samples appear to contain a higher number of known bifidobacterial species with an average relative abundance of ≥0.01% than the feline gut microbiota (50 and 37 bifidobacterial species, respectively). Furthermore, ITS data were used to reconstruct the bifidobacterial core gut microbiota of the two animal companion groups. Specifically, the bifidobacterial core gut microbiota of cats and dogs were obtained considering only those bifidobacterial species with a prevalence of ≥80%, leading to the identification of 13 core taxa in dogs, of which Bifidobacterium breve (average relative abundance, 18.11%), Bifidobacterium magnum (13.17%), Bifidobacterium longum subsp. longum (11.30%), Bifidobacterium adolescentis (5.82%), Bifidobacterium bifidum (3.86%), and Bifidobacterium animalis subsp. lactis (3.45%) coupled with a novel putative bifidobacterial species, i.e., Bifidobacterium pseudolongum_new_subsp. (15.28%) (see below) as the most prevalent and the most abundant bifidobacterial species of the canine gut microbiota (see Table S4). Conversely, using the same criteria, just seven bifidobacterial species represented the bifidobacterial core gut microbiota of cats, i.e., B. adolescentis (24.61%), B. breve (9.18%), Bifidobacterium pseudolongum_new_subsp. (8.18%), B. longum subsp. longum (6.87%), B. dentium (4.29%), B. animalis subsp. lactis (2.68%), and Bifidobacterium crudilactis (1.17%). Interestingly, all species that belong to the bifidobacterial core gut microbiota of cats, with the exception of B. crudilactis, were also part of the canine bifidobacterial core gut microbiota (Table S4), suggesting that these bifidobacterial species are able to adapt and colonize the intestine of either of these two hosts. Of note, B. longum subsp. longum and B. adolescentis have been detected in the intestinal communities of several mammalian species (7), while B. animalis subsp. lactis and B. dentium have also been reported to enjoy a cosmopolitan lifestyle (20, 21). On the other hand, the high prevalence of bifidobacterial taxa that are typically associated with the human gut, i.e., B. breve and B. catenulatum, may be the result of closer or more frequent physical contact between a dog and its owner compared to that between cats and humans (10, 22). Indeed, cats generally live a solitary existence and tend to be more linked to their territory than to their owners (23). In this context, the closer relationship between dogs and humans may have promoted colonization of the above-mentioned bifidobacterial species in the canine intestinal tract, presumably through horizontal transfer events. Moreover, statistical analyses were performed in order to evaluate if differences may occur in the bifidobacterial population based on sex, breed, or age in the feline and canine gut microbiota. A hierarchical clustering analysis of the bifidobacterial ITS microbial profiling data revealed that fecal samples did not cluster in separate groups based on animal breed, indicating that this parameter does not impact the bifidobacterial population (see Fig. S4). In addition, no significant differences were observed in the bifidobacterial community of feline fecal samples based on sex, whereas two bifidobacterial species, i.e., Bifidobacterium pseudocatenulatum and Bifidobacterium_new_taxa_9 (see below for the classification of Bifidobacterium new taxa), exhibited a significantly higher relative abundance in female canine fecal samples than in their male counterparts (P value < 0.05) (see Table S5). This suggests that sex may influence the relative abundance of certain minority bifidobacterial species in the canine intestine. Finally, analysis of variance (ANOVA) was applied to evaluate possible differences in the bifidobacterial compositions when samples were grouped based on age, as previously reported, i.e., puppy (0 to 8 months old), junior (9 to 24 months old), adult (25 to 96 months old), and senior (>97 months old) (10). Interestingly, no significant differences were observed for feline fecal samples, while six bifidobacterial taxa significantly varied in canine fecal samples according to the above-mentioned age groups (P value < 0.05) (see Table S6). However, only two of the latter bifidobacterial species, i.e., Bifidobacterium_new_taxa_60 and Bifidobacterium thermacidophilum subsp. porcinum, exhibited an average relative abundance of >0.05% in at least one age group (Table S6). Altogether, these results suggest that the fecal bifidobacterial population of dogs, in contrast to what was observed for cats, may be subject to variations based on age and/or sex.
Investigation of putative novel members of the Bifidobacterium genus.
Bifidobacterial ITS profiling showed the presence of ESVs with an identity level of <93% compared to the currently classified bifidobacterial species, suggesting the presence of putative novel bifidobacterial taxa in the gut microbiota of dogs and cats. In-depth analysis of these putative new taxa was performed by employing a previously described customized database (7, 16). The latter contains the ITS sequences of publicly available bifidobacterial (sub)species as well as the ITS sequences corresponding to all other nonbifidobacterial members of the Bifidobacteriaceae family in order to avoid misclassification of ITS sequences. This database also included 89 different putative new (sub)species, identified as previously described (7). Interestingly, canine and feline fecal samples were characterized by average relative abundances of 26.19% and 26.82% of putative novel bifidobacterial species, respectively, compared to the overall bifidobacterial community. Remarkably, 67 ESVs were classified as representative of a putative novel taxon of the Bifidobacterium genus classified as Bifidobacterium pseudolongum_new_subsp., showing a prevalence of >80% in the gut microbiota of both dogs and cats, suggesting that this taxon has evolved the ability to colonize the intestinal tracts of these two mammalian species. Moreover, the canine bifidobacterial core gut microbiota included two other putative novel species, designated here Bifidobacterium_new_taxa_60 and Bifidobacterium_new_taxa_65 (Table S4). Conversely, feline fecal microbiota possessed Bifidobacterium_new_taxa_50 and Bifidobacterium_new_taxa_55, two putative novel bifidobacterial species that are phylogenetically related to B. pseudocatenulatum, with a high average relative abundance (7.59% and 3.74%, respectively) but with lower prevalence (65.22% and 26.09%, respectively). Isolation and subsequent phenotypical and genomic characterization of these putative novel taxa will be important to decipher their biological relevance in the canine and feline gut.
Covariance of bifidobacterial species among the gut microbiota of dogs and cats.
ITS bifidobacterial profiling data sets were then exploited to evaluate the cooccurrence and coexclusion of known and putative novel bifidobacterial species across the fecal samples of dogs and cats by means of a Pearson correlation index. Force-driven networks were constructed using the statistically significant (P value < 0.05, with P values corrected for multiple comparisons) data obtained by considering the bifidobacterial species with an average relative abundance of ≥0.3% and ≥0.05% in case of canine and feline fecal samples, respectively. The different relative abundance cutoffs used to define the covariance analyses were chosen to have a comparable number of bifidobacterial species for the two mammalian groups.
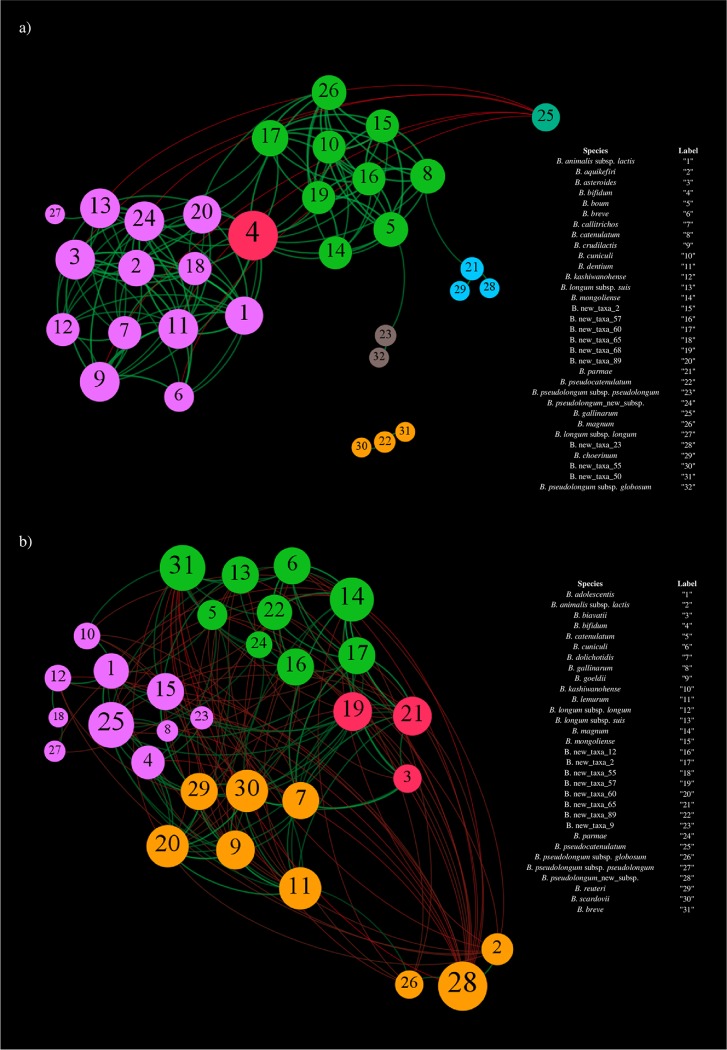
The force-driven network generated by modularity analysis by means of Gephi software for bifidobacterial species harbored by the feline gut microbiota highlighted positive interactions among all the considered bifidobacterial species, except for B. gallinarum, thus supporting the notion of extensive coevolution among bifidobacterial members of the intestinal microbial community of cats (Fig. 2a). Conversely, coexclusion was observed between B. gallinarum and six other bifidobacterial taxa, including Bifidobacterium asteroides, B. breve, B. crudilactis, B. dentium, B. longum subsp. suis, and B. longum_new_subsp. Probably, when present, the high relative abundance of B. gallinarum (average of 17.96%) may play a role in excluding other bifidobacterial species from the colonization of the feline intestinal tract, suggesting a dominant ecological behavior of B. gallinarum in the cat’s gut microbiota. In this context, despite the fact that B. gallinarum was originally isolated from a chicken cecum, its extensive presence in the fecal microbiota of cats may emphasize the cosmopolitan ecological feature of certain bifidobacterial species that may have been prompted by specific predatory/dietary habits, thus corroborating previous reports (7, 24). Furthermore, the constructed force-driven covariance network showed the separation of bifidobacterial species into two main clusters and three minor clusters, all characterized by a high degree of cooccurrence among the microbial taxa within a single cluster. Outside this particular case, very few cooccurrence relationships were detected among bifidobacterial species belonging to different clusters. This observation indicates that bifidobacteria may have evolved specific trophic interactions between cocolonizers. Noteworthy, the bifidobacterial species of the two main clusters positively covaried with B. bifidum, suggesting social behavior of this particular bifidobacterial species. The ability of B. bifidum to create trophic relationships with other bifidobacterial species was previously observed in humans (25). In this context, B. bifidum contains genes encoding glycosyl hydrolases that are predicted to degrade both diet- and host-derived complex glycans, thus allowing the release of partially degraded glycans and subsequent establishment of cross-feeding activities by other bifidobacterial species (26). Therefore, as observed in humans, when present, B. bifidum also appears to exhibit social behavior within the feline gut microbiota by creating a network of metabolic interactions with other bifidobacterial species.
FIG 2.
Covariance network of the most abundant bifidobacterial species of feline and canine fecal microbiota. (a) Force-driven network obtained by considering the bifidobacterial species with an average relative abundance of ≥0.05% of the feline fecal samples. (b) Force-driven network constructed by considering the bifidobacterial taxa with an average relative abundance of ≥0.3% of the canine fecal samples. In both cases, the force-driven networks exhibit the predicted c-variances with a P value of <0.05 between the profiled bifidobacterial species. Moreover, the node size is proportional to the number of covariances, while the node color indicates the different observed clusters.
On the other hand, the force-driven covariance network constructed using profiles of the bifidobacterial species constituting the gut microbiota of dogs showed the formation of four clusters predicted by modularity analysis by Gephi software, with three unclustered species (Fig. 2b). In this context, while only positive interactions were observed among bifidobacterial species within a single cluster, various coexclusion relationships were identified among bifidobacterial taxa belonging to different clusters. Specifically, B. pseudolongum_new_subsp. was shown to display a coexclusion interaction with 17 of the considered bifidobacterial species while exhibiting just two positive occurrences with B. animalis subsp. lactis and B. pseudolongum subsp. globosum. Possibly, as observed here for B. gallinarum in the feline gut microbiota, B. pseudolongum_new_subsp. has evolved the ability to colonize the canine gut and successfully compete with other bifidobacterial species in this ecological niche.
Bifidobacterial phylogenetics-based clustering of the canine and feline fecal samples.
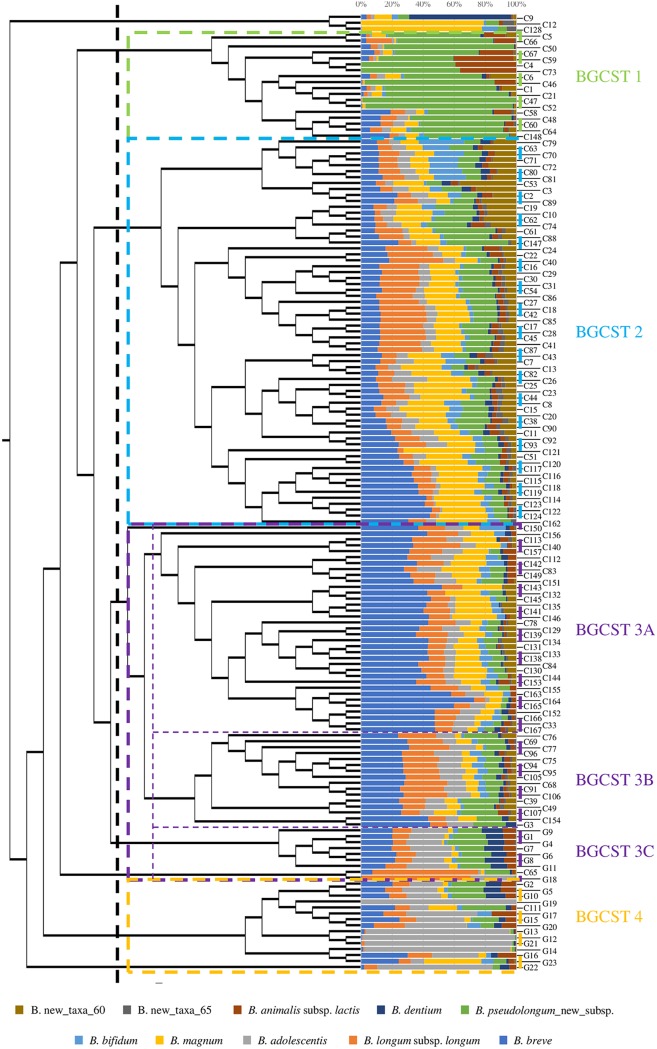
To evaluate if a GCST-based clustering of the collected samples is not only observed at the genus level but also at the species level, the bifidobacterial ITS profiling data were subjected to hierarchical clustering analysis. The generated cladogram highlighted the division of fecal samples into four main monophyletic clusters, named bifidobacterial gut community state types (BGCSTs) (Fig. 3). Furthermore, detailed analysis revealed the presence of three monophyletic subclusters for one of the main BGCSTs. Therefore, when considering bifidobacterial taxa with an average relative abundance of >10%, the six clusters are dominated by B. pseudolongum_new_subsp. (BGCST 1); B. breve, B. longum subsp. longum, B. pseudolongum_new_subsp., and B. magnum (BGCST 2); B. breve, B. longum subsp. longum, and B. magnum (BGCST 3A); B. breve, B. longum subsp. longum, and B. pseudolongum_new_subsp. (BGCST 3B); B. breve, B adolescentis, and B. pseudolongum_new_subsp. (BGCST 3C); and B. adolescentis (BGCST 4). Interestingly, BGCST 3C and 4 are exclusively represented by feline fecal samples with the exception of two canine fecal samples, while BGCSTs 1, 2, 3A, and 3B encompassed only canine fecal samples (Fig. 3). Furthermore, beta-diversity analysis performed by means of Bray-Curtis distance matrix showed a clear separation of the collected fecal samples according to the above-mentioned clusters, a finding that was also confirmed by PERMANOVA (P value < 0.05) (see Fig. S5), thus corroborating the presence of recurrent bifidobacterial combinations among the individual canine and feline gut microbiota assemblies. Interestingly, PCoA revealed a partial overlap of samples belonging to the BGCST 2, 3A, and 3B subclusters. Altogether, these results suggest that the intestinal communities of dogs and cats harbor distinct bifidobacterial combinations that can be assigned to six different BGCST clusters.
FIG 3.
Classification of the canine and feline gut microbiota into distinct types based on their bifidobacterial community. The cladogram on the left depicts the hierarchical clustering of the considered fecal samples based on their average bifidobacterial profiles. The bar plot in the center shows the abundance of the 10 most representative bifidobacterial species of the collected samples, whose names are reported on the right. The colored dotted rectangles divide samples into the eight identified bifidobacterial gut community state types (BGCSTs), whose designations are reported on the right with the same BGCST color code.
Covariance of bifidobacterial species and other major microbial players of the canine and feline gut microbiota.
To investigate the covariance interactions among bifidobacterial species and the other most abundant microbial genera of the canine and feline gut microbiota, a force-driven network was built. Specifically, this cooccurrence analysis was performed involving those bacterial genera with an average relative abundance of >1% of the gut microbiota of dogs and cats and by normalizing the bifidobacterial ITS profiling data of each sample for the Bifidobacterium genus relative abundance obtained from the 16S rRNA gene sequencing. As revealed by Gephi modularity analysis, bifidobacteria did not cluster together in a single group, but as was observed as described above, bifidobacterial species established positive interactions among each other, supporting once again the assumption of trophic behavior between various bifidobacterial members of the canine and feline gut microbiota (see Fig. S6). Conversely, positive interactions were established between certain bifidobacterial species and one of the most abundant microbial genera, i.e., Prevotella 9. The latter, in humans, has been associated with a carbohydrate- and fiber-based diet. Indeed, species belonging to the Prevotella genus are able to degrade a wide range of plant-derived carbohydrates facilitated by specific glycosyl hydrolase-encoding genes in their genomes (27–29). In the same manner, bifidobacterial species were reported to have evolved genetic strategies to get access to diet- and host-derived glycans (30, 31). In this context, the cooperation observed between bifidobacterial taxa and Prevotella 9 may be the result of syntrophic interactions between members of these two saccharolytic microbial genera. Conversely, coexclusion connections were observed between bifidobacterial species and two other main players of the canine and feline gut microbiota, corresponding to Fusobacterium and Bacteroides. Specifically, the latter are known to be functionally involved in the degradation of protein (32). Indeed, both Fusobacterium and Bacteroides have been related to a high fat- and protein-based diet (33–35). In this context, the proteolytic features of these microbial taxa may have contributed to the negative correlation with bifidobacterial species known to be saccharolytic microorganisms.
Conclusions.
From the establishment of their domestication, dogs and cats have played a special role as family members, and for this reason, their health is as important to us as our own. An increasing number of scientific studies have dissected the feline and canine gut microbiota, as it is believed to directly impact host health status. However, the possible presence of distinct community composition types at both genus and species levels as well as the autochthonous bifidobacterial community of the canine and feline gut microbiota had not yet been investigated. In the present study, the application of 16S rRNA gene profiling allowed the reconstruction of the canine and feline gut microbiota as well as the identification of GCSTs. Both analyses revealed that Bacteroides, Fusobacterium, and Prevotella 9 dominated the canine and feline gut microbiota, suggesting that these microbial taxa are major players of the intestinal communities of the two above-mentioned companion animals. Moreover, ITS profiling revealed differences in the bifidobacterial compositions of canine and feline fecal samples both in terms of number and species of bifidobacteria as indicated by the reconstruction of the corresponding bifidobacterial core gut microbiota and covariance network analysis. This observation was further corroborated by a hierarchical clustering analysis showing a clear separation of the collected fecal samples according to the two considered mammalian species. Our results therefore show that at the genus level, the intestinal populations of cats and dogs are similar in terms of taxonomical composition, though clear differences are observed when the data are scrutinized at the bifidobacterial species level, indicative of extensive adaptation of the bifidobacterial species to colonize a specific host.
MATERIALS AND METHODS
Ethical statement.
This study was performed in compliance with the rules, regulations, and recommendations of the ethical committee of the University of Parma. All animal procedures were carried out in accordance with national guidelines (Decreto legislativo 26/2014).
Sample collection and DNA extraction.
For the purpose of the present study, a total of 138 canine stool samples and 23 feline fecal samples were collected through a collaboration with several Italian dog and cat breeders in the north and center of Italy (see Table S1 in the supplemental material). To be enrolled in the study, animals had to be healthy without having been treated with probiotics or drugs during the 6 months prior to sample collection. Moreover, breed, age, weight, and sex were noted for each sample (Table S1). Stool samples were collected immediately after defecation, kept on ice, and shipped to the laboratory under frozen conditions where they were preserved at –20°C until they were processed. DNA extraction form each sample was performed using the QIAmp DNA Stool minikit according to the manufacturer’s instructions (Qiagen, Germany).
16S rRNA gene sequencing.
Partial 16S rRNA gene sequences were amplified from extracted DNA using primer pair Probio_Uni (5′-CCTACGGGRSGCAGCAG-3′) and Probio_Rev (5′-ATTACCGCGGCTGCT-3′), targeting the V3 region of the 16S rRNA gene sequence (17). Illumina adapter overhang nucleotide sequences were added to the partial 16S rRNA gene-specific amplicons and to the generated ITS amplicons of approximately 200 bp, which were further processed using the 16S metagenomic sequencing library preparation protocol (part number [no.] 15044223 rev. B; Illumina). Amplifications were carried out using a Verity thermocycler (Applied Biosystems). The integrity of the PCR amplicons was analyzed by electrophoresis on a 2200 Tape Station instrument (Agilent Technologies, USA). DNA products obtained following PCR-mediated amplification of the 16S rRNA gene sequences were purified by means of a magnetic purification step involving Agencourt AMPure XP DNA purification beads (Beckman Coulter Genomics GmbH, Bernried, Germany) in order to remove primer dimers. DNA concentration of the amplified sequence library was determined by a fluorometric Qubit quantification system (Life Technologies, USA). Amplicons were diluted to a concentration of 4 nM, and 5-μl quantities of each diluted DNA amplicon sample were mixed to prepare the pooled final library. 16S rRNA gene sequencing was performed using an Illumina MiSeq sequencer with MiSeq reagent kit v3 chemicals. Each step of the library preparation was performed using HiPure molecular biology-grade water (GE Healthcare, USA). In addition, a negative control was sequenced in order to verify that any contamination did not occur during the amplification and sequencing phases. Briefly, the negative control was processed as a normal sample (see above), but HiPure molecular biology grade-water was used instead of a DNA sample. Following sequencing, the .fastq files were processed using QIIME2 software with the SciKit-learn classifier (36). Paired-end reads were merged, and quality control retained sequences with a length between 140 and 400 bp and mean sequence quality score of >20, while sequences with homopolymers of >7 bp and mismatched primers were omitted. To calculate downstream diversity measures, alpha- and beta-diversity (Bray-Curtis), 16S rRNA operational taxonomic units (OTUs) were defined at 100% sequence homology using DADA2 (37), therefore generating exact sequence variants (ESVs). ESVs not encompassing at least two sequences of the same sample were removed. All reads were classified to the lowest possible taxonomic rank using QIIME2 (36, 38) and a reference data set from the SILVA database (39). PCoA representations of beta-diversity were performed using QIIME2 (36, 38).
Bifidobacterial ITS sequencing.
Partial ITS sequences were amplified from extracted DNA using the primer pair Probio-bif_Uni (5′-CTKTTGGGYYCCCKGRYYG-3′) and Probio-bif_Rev (5′-CGCGTCCACTMTCCAGTTCTC-3′), which targets the spacer region between the 16S rRNA and the 23S rRNA genes within the rRNA locus (16). Illumina adapter overhang nucleotide sequences were added to the partial ITS amplicons, which were further processed employing the 16S metagenomic sequencing library preparation protocol (part no. 15044223 rev. B; Illumina). PCR amplifications and library preparation, including the negative control, were performed as described above for the 16S rRNA microbial profiling analyses. Following sequencing, the .fastq files were processed using a custom script based on the QIIME software suite (38). Paired-end read pairs were assembled to reconstruct the complete Probio-bif_Uni/Probio-bif_Rev amplicons. Quality control retained sequences with a length between 100 and 400 bp and mean sequence quality score of >20, while sequences with homopolymers of >7 bp in length and mismatched primers were removed. To calculate downstream diversity measures, alpha- and beta-diversity (Bray-Curtis), ITS operational taxonomic units (OTUs) were defined at 100% sequence homology using uclust (40), generating ESVs. All reads were classified to the lowest possible taxonomic rank using QIIME2 (36, 38) and a reference data set, i.e., an updated version of the bifidobacterial ITS database (16).
Statistical analyses and network representations.
PERMANOVAs were carried out with QIIME2 software (36). ANOVA, Student’s t test, and covariance statistical analyses were performed with SPSS software (IBM SPSS Statistics for Windows, version 22.0; IBM Corp., Armonk, NY, USA), and the subsequent force-driven network representations were obtained with the Gephi software (41). Network clusters were predicted through the modularity statistical function of the Gephi software. The software MeV v.4.9.0 was used to construct a hierarchical clustering based on the Pearson correlation distance matrix.
Data availability.
16S rRNA gene and bifidobacterial ITS profiling data were deposited in the SRA database with the accession number PRJNA579980 for feline fecal samples and PRJNA504009 for canine stool samples.
Supplementary Material
ACKNOWLEDGMENTS
This work was primarily funded by the EU Joint Programming Initiative–A Healthy Diet for a Healthy Life (JPI HDHL; http://www.healthydietforhealthylife.eu/) to D.V.S. (in conjunction with Science Foundation Ireland [SFI], grant number 15/JP-HDHL/3280). D.V.S. is member of APC microbiome Ireland, funded by SFI through the Irish Government’s National Development Plan (grant numbers SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2). The study was supported by Fondazione Cariparma, under TeachInParma Project (to D.V.S.). G.A. is supported by Fondazione Cariparma, Parma, Italy. GenProbio srl provided financial support to the Laboratory of Probiogenomics.
This research benefited from the HPC (High Performance Computing) facility of the University of Parma, Italy. We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Suchodolski JS. 2011. Companion animals symposium: microbes and gastrointestinal health of dogs and cats. J Anim Sci 89:1520–1530. doi: 10.2527/jas.2010-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grześkowiak Ł, Endo A, Beasley S, Salminen S. 2015. Microbiota and probiotics in canine and feline welfare. Anaerobe 34:14–23. doi: 10.1016/j.anaerobe.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickard JM, Zeng MY, Caruso R, Nunez G. 2017. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sanchez B, Margolles A. 2017. Bifidobacteria and their health-promoting effects. Microbiol Spectr 5:BAD-0010-2016. doi: 10.1128/microbiolspec.BAD-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottacini F, van Sinderen D, Ventura M. 2017. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem J 474:4137–4152. doi: 10.1042/BCJ20160756. [DOI] [PubMed] [Google Scholar]
- 6.Bunesova V, Vlkova E, Rada V, Killer J, Musilova S. 2014. Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Benef Microbes 5:377–388. doi: 10.3920/BM2013.0081. [DOI] [PubMed] [Google Scholar]
- 7.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, van Sinderen D, Ventura M. 2017. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Redfern A, Suchodolski J, Jergens A. 2017. Role of the gastrointestinal microbiota in small animal health and disease. Vet Rec 181:370. doi: 10.1136/vr.103826. [DOI] [PubMed] [Google Scholar]
- 10.Alessandri G, Milani C, Mancabelli L, Mangifesta M, Lugli GA, Viappiani A, Duranti S, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M. 2019. Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ Microbiol 21:1331–1343. doi: 10.1111/1462-2920.14540. [DOI] [PubMed] [Google Scholar]
- 11.Strompfova V, Laukova A. 2014. Isolation and characterization of faecal bifidobacteria and lactobacilli isolated from dogs and primates. Anaerobe 29:108–112. doi: 10.1016/j.anaerobe.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Masuoka H, Shimada K, Kiyosue-Yasuda T, Kiyosue M, Oishi Y, Kimura S, Ohashi Y, Fujisawa T, Hotta K, Yamada A, Hirayama K. 2017. Transition of the intestinal microbiota of cats with age. PLoS One 12:e0181739. doi: 10.1371/journal.pone.0181739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuoka H, Shimada K, Kiyosue-Yasuda T, Kiyosue M, Oishi Y, Kimura S, Yamada A, Hirayama K. 2017. Transition of the intestinal microbiota of dogs with age. Biosci Microbiota Food Health 36:27–31. doi: 10.12938/bmfh.BMFH-2016-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinna C, Vecchiato CG, Bolduan C, Grandi M, Stefanelli C, Windisch W, Zaghini G, Biagi G. 2018. Influence of dietary protein and fructooligosaccharides on fecal fermentative end-products, fecal bacterial populations and apparent total tract digestibility in dogs. BMC Vet Res 14:106. doi: 10.1186/s12917-018-1436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabbioni A, Ferrario C, Milani C, Mancabelli L, Riccardi E, Di Ianni F, Beretti V, Superchi P, Ossiprandi MC. 2016. Modulation of the bifidobacterial communities of the dog microbiota by zeolite. Front Microbiol 7:1491. doi: 10.3389/fmicb.2016.01491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. 2014. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 17.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 19.Salonen A, Salojarvi J, Lahti L, de Vos WM. 2012. The adult intestinal core microbiota is determined by analysis depth and health status. Clin Microbiol Infect 18 Suppl 4:16–20. doi: 10.1111/j.1469-0691.2012.03855.x. [DOI] [PubMed] [Google Scholar]
- 20.Lugli GA, Mancino W, Milani C, Duranti S, Mancabelli L, Napoli S, Mangifesta M, Viappiani A, Anzalone R, Longhi G, van Sinderen D, Ventura M, Turroni F. 2019. Dissecting the evolutionary development of the species Bifidobacterium animalis through comparative genomics analyses. Appl Environ Microbiol 85:e02086-18. doi: 10.1128/AEM.02806-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turroni F, van Sinderen D, Ventura M. 2011. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol 149:37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, van Sinderen D, Ventura M. 2018. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci 75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Driscoll CA, Macdonald DW, O'Brien SJ. 2009. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci U S A 106 Suppl 1:9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watabe J, Benno Y, Mitsuoka T. 1983. Bifidobacterium gallinarum sp. nov.: a new species isolated from the ceca of chickens. Int J Syst Bacteriol 33:127–132. doi: 10.1099/00207713-33-2-127. [DOI] [Google Scholar]
- 25.Duranti S, Lugli GA, Milani C, James K, Mancabelli L, Turroni F, Alessandri G, Mangifesta M, Mancino W, Ossiprandi MC, Iori A, Rota C, Gargano G, Bernasconi S, Di Pierro F, Sinderen D, Ventura M. 2019. Bifidobacterium bifidum and the infant gut microbiota: an intriguing case of microbe-host co-evolution. Environ Microbiol 21:3683–3695. doi: 10.1111/1462-2920.14705. [DOI] [PubMed] [Google Scholar]
- 26.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki K, Hirase T, Kojima Y, Flint HJ. 2005. Medium- to large-sized xylo-oligosaccharides are responsible for xylanase induction in Prevotella bryantii B14. Microbiology 151:4121–4125. doi: 10.1099/mic.0.28270-0. [DOI] [PubMed] [Google Scholar]
- 29.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 30.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2016. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. 2018. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol 26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Butowski CF, Thomas DG, Young W, Cave NJ, McKenzie CM, Rosendale DI, Bermingham EN. 2019. Addition of plant dietary fibre to a raw red meat high protein, high fat diet, alters the faecal bacteriome and organic acid profiles of the domestic cat (Felis catus). PLoS One 14:e0216072. doi: 10.1371/journal.pone.0216072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt M, Unterer S, Suchodolski JS, Honneffer JB, Guard BC, Lidbury JA, Steiner JM, Fritz J, Kolle P. 2018. The fecal microbiome and metabolome differs between dogs fed bones and raw food (BARF) diets and dogs fed commercial diets. PLoS One 13:e0201279. doi: 10.1371/journal.pone.0201279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media, 17 to 20 May 2009, San Jose, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene and bifidobacterial ITS profiling data were deposited in the SRA database with the accession number PRJNA579980 for feline fecal samples and PRJNA504009 for canine stool samples.