Summary
Background
Obesity is associated with an increased risk of developing clear cell renal cell carcinoma (ccRCC) but, paradoxically, with improved oncologic outcomes. As the biologic mechanisms underlying this association are poorly understood, our study aimed to identify transcriptomic differences in primary tumor or peritumoral adipose tissue between obese and normal weight patients.
Methods
After confirming the inverse association between body mass index (BMI ≥ 30 vs < 30 kg/m2) and mortality in three independent clinical cohorts including 453 metastatic ccRCC patients treated with first-line VEGF-directed therapy from the Phase III COMPARZ clinical trial, 152 predominately early stage ccRCC patients from the Cancer Genome Atlas (TCGA), and a Memorial Sloan Kettering (MSK) observational cohort of 203 metastatic ccRCC patients treated with targeted immunotherapy using the Kaplan–Meier method and analyzed by log-rank test, we evaluated and validated primary tumor transcriptomic differences. We also compared gene expression differences in peritumoral adipose tissue from a prospectively collected cohort of patients with non-metastatic ccRCC. Wilcoxon rank-sum test was used to analyze significant differences in gene expression between groups and false discovery correction was performed using the Benjamini-Hochberg procedure.
Findings
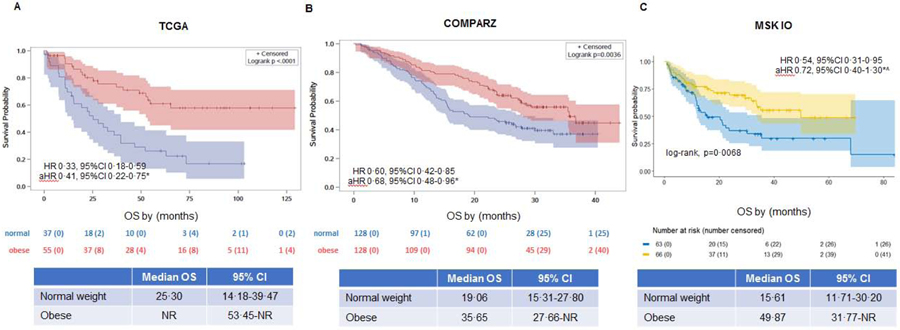
A lower mortality rate was observed in obese patients versus normal weight patients in the in the locally advanced The Cancer Genome Atlas cohort (median OS not-reached (NR), 95%CI 53.45-NR vs 25.30 mos, 95%CI 14.18–39.47, respectively; adjusted-Hazard Ratio (aHR) 0·41, 95%CI 0·22–0·75), and COMPARZ trial cohort of metastatic ccRCC treated with tyrosine kinase inhibitors (median OS 35.65, 95%CI 27.66-NR vs 19.06, 95%CI 15.31–27.80, respectively; aHR 0·68, 95%CI 0·48–0·96). In a cohort of metastatic ccRCC patients treated with immunotherapy, the inverse association with OS was no longer significant after adjustment for International Metastatic Database Consortium risk score (median OS 49.87, 95%CI 31.77-NR vs. 15.61, 95%CI 11.71–30.20, respectively; aHR 0·72, 95%CI 0·40–1·30). Tumors of obese patients were characterized by increased angiogenesis expression (false discovery rate-adjusted p-value=0·01) but showed no significant differences in immune cell programs. We found increased peritumoral adipose tissue inflammation (p<0.05) in obese versus normal weight patients.
Interpretation
Tumor microenvironmental differences in both the tumor and peritumoral adipose tissue by BMI may contribute to the apparent survival advantage experienced by obese ccRCC patients. The complex interplay between the ccRCC tumor and peritumoral adipose tissue microenvironment may have clinical relevance and warrants further investigation.
Funding
Ruth L. Kirschstein Research Service Award, ACSO YIA, Ludwig Center MSK, Weiss Family Kidney Research Fund, Novartis, The Sidney Kimmel Center for Prostate and Urologic Cancers and the NIH/NCI to MSKCC through the Cancer Center Support Grant.
Introduction
Obesity, defined by a body mass index (BMI) ≥ 30 kg/m2, is an established risk factor for developing clear cell renal cell carcinoma (ccRCC)1 but exhibits a counterintuitive association with prognosis.2 We and others found that obese patients with localized ccRCC treated with nephrectomy survive longer than those who are normal weight, a phenomenon known as the obesity paradox.1, 3 We also observed the same pattern among metastatic ccRCC patients treated with targeted therapy.4 McQuade, et al.5 recently reported the obesity paradox among patients with metastatic melanoma treated with immunotherapy, and others have also demonstrated this finding in cohorts of patients with mixed solid tumors treated with immunotherapy.6, 7 Most recently, DeGiorgi, et al.8 found that among metastatic RCC patients treated with ≥ 2nd line nivolumab, those who were normal weight experienced worse overall survival compared to those with higher BMI. Why obese ccRCC patients do better than patients who are normal weight regardless of treatment type is not known. Initial mechanistic insights suggest it may be due to BMI-related differences in the tumor transcriptome.3 We previously showed that the adverse metabolic oncogene, fatty acid synthase (FASN), was downregulated in the tumors of obese vs. normal weight patients. Recently, Wang et al. speculated that the adipocyte-derived hormone leptin in the tumor microenvironment of obese colorectal cancer patients may alter T-cell function thereby improving the response to systemic immunotherapy.6
In this report, we compared angiogenic and immunologic transcriptomic patterns of tumor and peritumoral adipose tissue in obese and normal weight ccRCC patients to shed light on putative mechanisms underlying the obesity paradox.
Methods
Study Design and Participants
We analyzed data from the COMPARZ phase III clinical trial (enrolled from August, 2008, and September, 2011) which demonstrated non-inferiority of the tyrosine kinases pazopanib to sunitinib in treatment-naïve patients with metastatic ccRCC.9 Formalin-fixed paraffin-embedded (FPPE) tumor blocks were collected at baseline from 453 ccRCC patients with sufficient tissue available for RNA microarray and gene mutation calling. 375 patients had microarray, pre-treatment body mass index (BMI), progression-free survival (PFS), and overall survival (OS) available for analyses (Table 1). After excluding overweight patients (n=119), our final cohort consisted of 256 patients. The cohort used in this retrospective study was institutional review board (IRB) approved by participating institutions and registered with ClinicalTrials.gov with the number NCT00720941.
Table 1:
Clinicopathologic characteristics of COMPARZ clinical trial cohort
| N=375 | |
|---|---|
| Age, mean (SD) | 62 (14) |
| Male, n (%) | 282 (75·2) |
| BMI, mean (SD) | 27·49 (7·67) |
| BMI category, n (%) | |
| Normal | 128 (34·13) |
| Overweight | 119 (31·73) |
| Obese | 128 (34·13) |
| MSKCC risk group, n (%) | |
| Favorable | 102 (27·2) |
| Intermediate | 227 (60·53) |
| Poor | 32 (8·53) |
| Treatment, n (%) | |
| Sunitinib | 192 (51·2) |
| Pazopanib | 183 (48·8) |
BMI=body mass index. SD=standard deviation.
In the ccRCC the Cancer Genome Atlas (TCGA) Cohort, we evaluated 152 patients with advanced (American Joint Committee on Cancer, AJCC stage III and IV) ccRCC treated by nephrectomy (Table 2). Clinicopathologic data and tumor specimens (year of collection 1998–2010) were retrospectively collected from multiple institutions after appropriate IRB approval. Primary tumors were previously transcriptomically profiled by the Cancer Genome Atlas (TCGA) and published in 2013.10 Additional pre-operative clinicopathologic variables, including BMI, at the time of nephrectomy were abstracted from each contributing institution. For the majority of cases, data on subsequent recurrences and systemic therapy received if recurrent were not available. After exclusion of overweight patients (n=59), our final cohort consisted of 93 patients.
Table 2:
Clinicopathologic characteristics of TCGA AJCC stage III/IV cohort
| N=152 | |
|---|---|
| Age, mean (SD) | 62 (14) |
| Male, n (%) | 99 (65·1) |
| BMI, mean (SD) | 28·1 (7) |
| BMI category, n (%) | |
| Normal | 38 (25) |
| Overweight | 59 (38·8) |
| Obese | 55 (36·2) |
| AJCC stage, n (%) | |
| III | 88 (57·9) |
| IV | 64 (42·1) |
TCGA=clear cell renal cancer The Cancer Genome Atlas. BMI=body mass index. SD=standard deviation.
The MSK Peritumoral Adipose Tissue Cohort was a Memorial Sloan Kettering (MSK) IRB approved study which enrolled patients from May 2015 to August 2017. We prospectively obtained primary tumor and peritumoral fat specimens from 62 non-metastatic ccRCC patients at the time of nephrectomy (appendix p 19). Peritumoral fat was harvested immediately adjacent (perinephric near, PNN, n=59) and on the opposite unaffected pole (perinephric away, PNA, n=25) from the tumor (n=55). Clinicopathologic variables including BMI at time of nephrectomy were abstracted from the clinical record. After exclusion of overweight patients (n=18 for PNN, n=8 for PNA, n=18 for tumor), our final cohort included n=41 PNN, n=17 PNA, n=37.
In the MSK immunotherapy cohort, we conducted an IRB-approved retrospective observational study to investigate the association between pre-treatment BMI and overall survival (OS) among 203 metastatic ccRCC patients treated with immunotherapy between July 2011 and August 2018 (Table 3). After exclusion of overweight patients (n=74), our final cohort for OS analysis was n=129. RNA and DNA samples were not available for analysis in this cohort. Clinicopathologic variables were abstracted from the clinical record.
Table 3:
Clinicopathologic characteristics of MSK immunotherapy cohort.
| N=203 | |
|---|---|
| Age, mean (SD) | 62 (13) |
| Male, n (%) | 151 (74·0) |
| BMI, mean (SD) | 26·9 (6·3) |
| BMI category, n (%) | |
| Normal | 63 (31·0) |
| Overweight | 74 (36·5) |
| Obese | 66 (32·5) |
| IMDC risk group, n (%) | |
| Favorable | 35 (17·9) |
| Intermediate | 128 (65·3) |
| Poor | 33 (16·8) |
| Unknown | 7 |
| IO Treatment Line, n (%) | |
| First line | 48 (23·6) |
| ≥Second line | 155 (76·4) |
IO=immunotherapy. BMI=body mass index. SD=standard deviation.
Procedures
In the COMPRAZ clinical trial, RNA was extracted by AltheaDx in 2013 according to Qiagen RNAeasy FFPE kit with a modified deparaffinization step. Gene expression profiles were derived via Affymetrix GeneChip HTA 2.0 (Affymetrix) RNA microarray data was normalized to log2 value. Probes without corresponding gene symbol found were excluded from further analysis. For genes matched with multiple probes, the probe with maximum Median Absolute Deviation is chosen for representing the expression of the gene. The log2 normalized expression values were used in subsequent analyses. For differentially expressed (DEG) and individual gene analyses, the quantile normalization values were used.
DNA from the primary tumors and matched normal tissue were extracted using DNA easy kit (Qiagen) according to standard protocol and subjected to analysis. Germline mutations were ruled out by analysis of adjacent non-tumoral tissue or normal germline for every sample. Samples from 377 patients with adequate DNA yields were extracted and sequenced using the 410 oncogene panel MSK Mutation Profiling of Actionable Targets (IMPACT) assay. A minimum of 40 ng of DNA was required for next-generation sequencing. MSK-IMPACT is a hybridization capture-based next-generation sequencing assay for targeted deep sequencing (approx. 500x) of all exons and selected introns of 410 oncogenes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies using Illumina HiSeq 2500 (Illumina Inc, San Diego, CA, USA).11 Our analysis focused on the three most clinically relevant mutations in ccRCC which include PBRM1, BAP1, and TP53.12 Rates of VHL mutation calling are challenging; however, large published mutational analyses have demonstrated that VHL is an early and essential event in the pathogenesis of ccRCC.13, 14 Therefore, an analysis of VHL mutation frequency by BMI groups would not be appropriate in this cohort of patients with ccRCC. Total mutational count was calculated as the total number of somatic mutations.
Publicly-available RNAseq data for the TCGA cohort, including tumor and normal adjacent kidney, were downloaded from the NIH Genomic Data Commons (https://gdc.cancer.gov). For the peritumoral fat cohort, tissue samples were stored in RNALater at 4°C, until RNA was extracted. Total RNA was isolated from tumor and perinephric fat tissues using the RNeasy Mini Kit (Qiagen) and sequenced. The methods for RNA extraction and processing for the TCGA are documented in TCGA, Nature, 2013.10 Tumor and fat specimens with > 500 ng of RNA and RNA integrity number (RIN) > 6.0 were sequenced using RNA poly-A capture. Quality control (QC) of RNAseq using principal components analysis did not demonstrate any clusters or similarity (data not shown). RNAseq raw read sequences were aligned against human genome assembly hg19 (Feb.2009/GRCh37, https://genome.ucsc.edu/cgi-bin/hgGateway?db=hg19) by STAR 2-pass15 alignment. RNAseq gene level count values were computed by using the R package GenomicAlignments16 over aligned reads with UCSC KnownGene17 in hg19 as the base gene model. The Union counting mode was used and only mapped paired reads were considered. Fragments per kilobase million (FPKM) values were then computed from gene level counts by using fpkm function from the R package “DESeq2.”18
Differentially expressed gene (DEG) results were used for gene-set enrichment analyses (GSEA)(19) and building gene set enrichment plots against the Molecular Signatures Database (MSigDB) Hallmark gene sets (http://software.broadinstitute.org/gsea/msigdb) through the R package clusterProfiler.20 The R package “limma” (version 3.29.0) was used for microarray data DEG analysis.21 Limma returns empirical Bayes moderated-t p-values and adjusted P-values (Q-value) to correct for multiple comparisons testing using the Benjamini-Hochberg method to control the false discovery rate (FDR). For the RNAseq cohort, the DEG analysis was performed through DESeq2 R package. In brief, the package DESeq2 provides methods to test for differential expression between conditions by use of negative binomial generalized linear models; the estimates of dispersion and logarithmic fold changes incorporate data-driven prior distributions. Given the raw count data and gene model used, DESeq2 normalized the raw count data by sample specific size factor and took covariates, if any, into account while testing for significant differences in gene expression between conditions with multiple test correction through FDR. The hallmark gene sets include angiogenesis and hypoxia gene signatures. Two additional previously published and validated angiogenesis signatures22, 23 were utilized to measure an overall angiogenesis score (appendix p 14).
Single-Sample GSEA(24) (ssGSEA) was utilized for immune deconvolution analyses to estimate the abundance of immune cell types, T-cell infiltration score (TIS), Immune Infiltration Score (IIS), and fraction of immune cells (ImmuneScore). ssGSEA takes the sample gene expression values as the input and computes an overexpression measure for the given gene list of immune cell type relative to all other genes in the transcriptome. Marker genes of immune cell types for ssGSEA were obtained from Bindea et al.25 and Senbabaoglu et al.26 Infiltration levels for different immune cell types were quantified using the ssGSEA implementation by the R package “gsva.”27 The R package “estimate”28 was used to infer the fraction of stromal and immune cells (ImmuneScore) in tumor samples based on given gene expression profile in FPKM or normalized log2 transformed values. ssGSEA scores for each individual immune cell type were used to calculate total T-cell Infiltration Score (TIS) and Immune Infiltration Score (IIS) as previously describe by Senbabaoglu et al.26
DEG results were used in performing IPA analyses (QIAGEN Inc.) with the Ingenuity Knowledge Base as the reference set.29 IPA was used to evaluate differences in canonical pathways that are predicted to change based on gene expression. IPA analysis was used rather than GSEA due to the smaller cohort sample size in the peritumoral fat cohort. Filters used in picking up DEG genes for IPA include: mean expression > 10, fold change > 20%, and P value < 0.05. The number of DEG genes after filtering was 2517, 675, and 629 for PNN, PNA and tumor, respectively.
We derived an immune cytolytic score (CYT) based on the geometric mean expression of two key cytolytic effectors, granzyme A (GZMA) and perforin (PRF1).(30) Previously published signatures of immune cell function were utilized to assess differences in T-cell function (Teff score22: includes CD8A, EOMES, PRF1, IFNG, and CD274), ImmuneCheckpoint (includes CD274, CTLA4, HAVCR2, LAG3, PDCD1, PDCD1LG2, TIGIT), and myeloid expression (Myeloid score22: includes IL6, CXCL1, CXCL2, CXCL3, CXCL8, PTGS2), and up-regulated Macrophage M1 and M2 signals described as M1_Up and M2_Up signatures by Chung et al.(31)
PD-L1 expression quantification in FFPE samples from the COMPARZ trial was performed using IHC per previously validated protocols and as previously described.32 Briefly, patients were categorized as PD-L1/B7H1 positive when any tumor cell positivity was detected (H-score (HS) > 0).
Outcomes
Overall survival was defined as the time from treatment initiation (COMPARZ, MSK IO) or surgery (TCGA) and date of death because of any cause or censoring on the day of the last follow-up visit. Progression-free survival (COMPARZ) was defined as the time interval between treatment initiation and the earliest date of either disease progression or death of any cause. Pre-treatment (COMPARZ, MSK IO) or pre-surgical BMI (TCGA, peritumoral adipose tissue cohort) was calculated and categorized into normal weight (BMI < 25 kg/m2), overweight (25kg/m2 ≤ BMI < 30kg/m2), and obese (BMI ≥ 30kg/m2) in accordance with World Health Organization criteria.33
Statistical Analysis
Differences in patient characteristics were tested using Fisher’s exact test and Chi-Square test (SAS 9.4, SAS Institute Inc., Cary, NC, USA) and the Wilcoxon rank- sum/Wilcoxon-Mann-Whitney test. SAS 9.4 & R package “ggpubr” version 0.1.5 performs Wilcoxon rank-sum test and reports p-value in the plot for continuous variables (such as immune feature ssGSEA scores and total mutation count) after Z-score normalization between sample groups. False discovery correction was performed using the Benjamini-Hochberg procedure for reporting multiple test adjusted p-values was used for GSEA analysis. Survival curves were calculated according to the Kaplan–Meier method and analyzed by log-rank test (SAS 9.4). The TCGA cohort survival curves were adjusted for stage and grade as these were AJCC stage III and IV where the COMPARZ and MSK IO cohorts were all stage IV and did not require stage or grade adjustment. The hazard ratio (HR) estimates and 95% confidence intervals were determined by the Cox proportional hazards regression modeling (SAS 9.4). Statistical significance was set at p < 0·05.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
Results
Kaplan–Meier plots suggest superior overall survival (OS) for obese vs. normal weight patients (BMI ≥ 30 vs < 25 kg/m2) in the TCGA (adjusted hazard ratio (aHR) 0·41, 95%CI 0·22–0·75) and COMPARZ clinical trial (aHR 0·68, 95%CI 0·48–0·96) after adjustment for stage/grade and IMDC, respectively (Figures 1a–b). The number of deaths in each cohort for obese vs normal weight were 19/55 (34·5%) and 28/37 (75·7%) for TCGA, 57/128 (44·5%) and 76/128 (59·4%) in COMPARZ, and 25/66 (37·9%) and 36/63 (57·4%) for MSK IO, respectively. The OS and PFS (Figure 1c and appendix p 1, respectively) advantage among obese patients was observed in all patients treated with first-line TKIs (pazopanib or sunitinib) in the COMPARZ clinical trial. The inverse association of OS and BMI was strongest in the sunitinib-treated arm (appendix p 2). In the MSK IO cohort, the inverse association with OS (HR 0·54, 95%CI 0·31–0·95) was no longer significant after adjustment for IMDC risk score (aHR 0·72, 95%CI 0·40–1·30) (Figure 1c). These findings were unchanged when the overweight category was included in the analysis or BMI as a continuous variable (appendix p 3-4, 21). Notably, the association between BMI and OS was not significantly different between men and women in a model including IMDC risk score (p>0.05) (appendix p 21).
Figure 1. (A-C): Obesity is associated with improved survival outcomes in the TCGA, COMPARZ, and MSK immunotherapy immunotherapy (IO) cohorts.
Unadjusted Kaplan-Meier curves demonstrating differences in OS between normal and obese patients in the (A) TCGA, (B) COMPARZ, and (C) MSK immunotherapy (IO) cohorts. HR=hazard ratio. aHR = adjusted HR. CI=confidence interval. OS=overall survival. TCGA=The Cancer Genome Atlas clear cell kidney cancer. MSK=Memorial Sloan Kettering. NR=not reached.
*TCGA cohort was adjusted for stage and grade. COMPARZ and MSK IO cohorts adjusted for IMDC alone.
^The association of BMI with overall survival in the MSK IO cohort was no longer statistically significant after adjustment for IMDC. No significant association with OS after adjustment for age and sex (aHR 0·60, 95%CI 0·34–1·08).
After confirming the obesity paradox in our cohorts, we assessed transcriptomic differences in tumors between obese and normal weight BMI groups in the COMPARZ cohort. Tumors from obese patients in the COMPARZ cohort demonstrated significant enrichment in hypoxia, TGF-beta, epithelial mesenchymal transition (EMT), and angiogenesis signaling programs (appendix p 5). Enrichment in TGF-beta, hedgehog, notch and EMT suggest activation of wound healing pathways among obese patients. Obese patients also showed an enrichment of metabolic pathways (e.g., adipogenesis, glycolysis, and fatty acid metabolism).
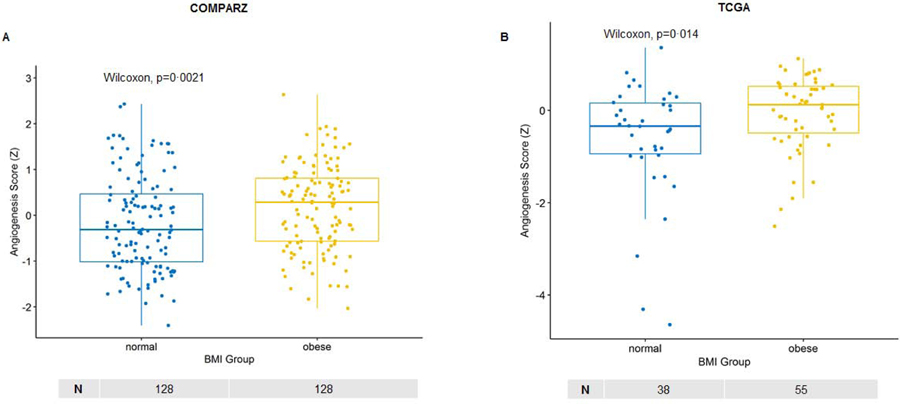
Notably, tumors of obese patients appeared to have distinct angiogenic differences compared to normal weight patients. Higher angiogenesis scores among obese patients found on GSEA-derived Hallmark gene set angiogenesis signature were further validated using two independently published and validated angiogenesis signatures with minimal overlap in the number of genes per signature as each signature is likely representing a different aspect of angiogenesis biology (appendix p 14); one signature by Masiero et al.23 (Figure 2a–b, appendix p 14) and the other signature recently utilized by McDermott, et al.22 to evaluate predictors of response to anti-VEGF, immunotherapy and combination therapy in the IMmotion150 phase II clinical trial.
Figure 2. (A-B): Obese patients demonstrate higher angiogenesis score in the primary tumor compared to normal weight patients.
Box plots demonstrating differences in ssGSEA angiogenesis scores among obese and normal weight patients in the (A) COMPARZ cohort and (B) KIRC TCGA cohorts. The angiogenesis RNA signature was derived from Masiero M, et al. (Cancer Cell, 2013). ssGSEA=single-sample gene set enrichment analysis. TCGA=The Cancer Genome Atlas clear cell kidney cancer. Z=z-score. Wilcoxon=Wilcoxon–Mann–Whitney test. Sample number per group indicated below each graph.
As obesity is associated with a state of chronic systemic inflammation(34), we hypothesized that obese patients harbor increased local inflammation in the primary tumor. Surprisingly, we found that the tumors of obese patients exhibited downregulation of IFN-gamma and no significant change in other inflammatory pathways (INF-alpha and inflammatory response) (appendix p 5). Therefore, tumors arising in an obesogenic environment do not appear to harbor increased local inflammation, but instead have increased activation of angiogenic, metabolic and wound healing pathways.
To further characterize differences in the tumor immune microenvironment by BMI, we utilized immune deconvolution35 using published immune cell signatures.25 Overall, there were no statistically significant differences in total immune infiltration (IIS, ImmuneScore), macrophage, neutrophil, overall myeloid immune cell, or T-cell infiltration score (TIS) between obese and normal weight patients in the COMPARZ cohort. We found that obese patients had higher proportions of infiltrating plasmacytoid dendritic cell (pDCs) infiltration (appendix p 7) but this was not validated in the TCGA cohort (appendix p 15). In a separate cohort of nephrectomy patients with advanced RCC (7 normal weight and 16 obese) analyzed using flow cytometry, we did not find differences in the percent CD45+ of effector immune cells, NK or macrophage populations by BMI status (appendix p 17-18).
Although there were no differences in the overall immune infiltration in the tumor among obese vs normal weight patients, some differences in RNAseq-derived ImmuneCheckpoint signature and PD-L1 IHC were evident in our analyses of COMPARZ. Specifically, obese patients had lower expression of various immune checkpoint molecules (ImmuneChekpoint) (appendix p 8) and decreased PD-L1 tumoral expression by immunohistochemistry (appendix p 9). In the TCGA cohort, there was no difference in ImmuneCheckpoint expression among obese patients (p=0.32) and PD-L1 (appendix p 15).
Finally, because tumor mutational burden has been heavily studied as a biomarker for potential response to immunotherapy,36 we examined whether mutations differed in obese and normal weight patients in COMPARZ. We found no differences in total mutational burden (appendix p 16) or frequency of PBRM1, BAP1 or TP53 mutations in the tumors of obese vs. normal weight patients.
Given the surprising findings of a lack of differences in tumoral immune infiltration in obese patients compared to normal weight patients we hypothesized that the obese adipose tissue surrounding the kidney tumor (i.e. peritumoral fat) may harbor a distinct immunologic milieu possibly contributing to their paradoxical response to immunotherapy. As such, we performed RNAseq and immune deconvolution on tumor and peritumoral fat specimens to characterize possible fat-tumor interactions. Supplementary Figure 5h (appendix p 13) summarizes ingenuity pathway analysis results which suggest that the tumor and the peritumoral fat (near and away from the tumor) among obese patients have higher expression of canonical inflammatory signatures (e.g., Th1/Th2 pathways, CD28 signaling in T-helper cells, dendritic cell maturation). We then used immune deconvolution to further characterize the immune microenvironment of peritumoral fat near (PNN) and away (PNA) from the tumor (appendix p 10). Obese patients demonstrated higher immune infiltration scores (IIS) in the peritumoral fat regardless of proximity to the tumor (appendix p 11). Compared to fat away from the tumor, peritumoral fat near the tumor demonstrated more significant immune infiltration (appendix p 12) among obese as compared with normal weight patients.
As hypoxia is a hallmark of dysregulated adipose tissue in obesity, we assessed hypoxia in peritumoral fat specimens and its potential relationship to immune infiltration. Within peritumoral fat, we found higher hypoxia gene expression scores near the tumor and hypoxia was correlated with higher total immune infiltration (IIS) and type 1 macrophage (M1) infiltration scores (appendix p 12). There was no difference in hypoxia, immune infiltration or macrophage scores within the tumor of obese and normal weight patients.
Discussion
In the present study, we investigated the potential mechanisms underlying the inverse association between BMI and survival that is observed in clinical cohorts of localized and metastatic ccRCC patients regardless of treatment. Our observations suggest that compared to the tumors of normal weight patients, those of obese patients harbor ccRCC tumors with higher angiogenesis scores. Contrary to our initial hypothesis, obese patients did not harbor increased inflammation within their primary tumors. However, tumors of obese patients in the COMPARZ cohort had lower immune checkpoint expression than tumors from normal weight patients and no difference in gene mutational profile or tumor mutational burden. Finally, our analyses of peritumoral fat revealed higher hypoxia and inflammation close to the primary tumor. Taken together, our initial mechanistic findings suggest that differences in the tumor microenvironment may underlie the apparent survival advantage of obese vs. normal weight ccRCC observed in clinical cohorts.
Survival analyses demonstrated improved survival among patients treated with TKI therapy, after adjustment for IMDC risk score, with the most significant association being noted among patients treated with sunitinib. These findings are consistent with a recent report where patients with higher angiogenesis scores are more likely to benefit from sunitinib as compared to pazopanib.37 Notably, the inverse association of BMI with immunotherapy in the MSK IO cohort was attenuated after adjustment for IMDC risk score. The IMDC risk score model encompasses a composite of some “host” factors that indirectly reflect systemic inflammatory effects of the cancer (e.g., anemia, neutrophilia, and thrombocytosis) which may overlap with mechanisms surrounding the association of immunotherapy outcomes and BMI.
Using three published and validated angiogenesis signatures, our finding that tumors of obese patients harbor higher angiogenesis scores than normal weight patients is in line with recent studies demonstrating that obese patients are more likely to have ClearCode34 molecular subtype A (ccA, associated with improved prognosis compared to ccB) which demonstrate enrichment in genes involved in angiogenesis, beta-oxidation pathway, organic acid metabolism, fatty acid metabolism, and pyruvate metabolism.38 Furthermore, patients with high angiogenesis scores exhibit improved survival outcomes with sunitinib compared to other VEGF-directed therapy, immunotherapy or the combination.22, 37 Visceral adipose tissue in obese patients is characterized by adipocyte hypertrophy which leads to regions of hypoxia and subsequent increases in angiogenesis and immune cell infiltration.39 We noted increased hypoxia scores closest to the primary tumor (PNN) and increased expression of canonical inflammatory pathways. Adipocytes produce angiogenic factors which may alter the underlying biology of tumors locally (adipocyte-tumor interface) and systemically.40 Furthermore, leptin secreted by adipocytes induces activation, proliferation, and migration of endothelial cells by upregulating VEGF and VEGFR-2.41 Therefore, we hypothesize that (1) obesity may create an environment that facilitates ccRCC growth via angiogenesis while simultaneously making these tumors more susceptible to TKIs and (2) increased angiogenesis could enhance local drug delivery. Similar findings have been demonstrated among obese metastatic colorectal patients who have been treated with anti-VEGF agents (e.g. bevacizumab, ramucirumab).42 Therefore, differences in angiogenesis expression may partially explain the superior survival outcomes among obese patients receiving TKI therapy.
Obesity-associated inflammation in other solid malignancies has been shown to cause tumor progression and therapeutic failure.43 Chronic inflammation in visceral adipose tissue leads to immune cell infiltration, extracellular matrix remodeling and eventually fibrosis. We did not find increased overall immune cell infiltration in ccRCC tumors from obese and normal weight patients. Higher expression of plasmacytoid dendritic cells was noted in the COMPARZ cohort but this was not validated in the TCGA cohort. James, et al.44, using a mouse model with intrarenal RCC, demonstrated that diet-induced obesity (DIO) lead to increased expression of tumor-suppressive dendritic cells (DC) and accelerated tumor growth which lead to poor response to DC-directed immunotherapy. Further exploration into the impact of obesity on DC cell function are required given these counterintuitive findings. In the COMPARZ cohort, we found lower expression of immune checkpoint molecules (ImmuneCheckpoint and PD-L1 IHC staining) in the tumors of obese patients. These findings are counter to recent findings from Wang, et al. where DIO in tumor-bearing mice cause increased CD8+ T-cell infiltration and T-cell dysfunction as measured by increased immune checkpoint molecule expression (Lag3, Tim3, PD-1).6 However, these results differed when human specimens were analyzed where obese colorectal cancer tumor specimens demonstrated lower CD8+ T-cell infiltration and melanoma cancer tumor specimens demonstrated higher CD8+ T-cell infiltration and immune checkpoint expression.6 Therefore, the underlying mechanism by which obesity alters the tumor microenvironment may differ based on the origin of each cancer. Similar to colorectal cancer, ccRCC arises in close proximity to visceral adipose tissue (i.e., peritumoral fat) and how fat-tumor interactions affect response to immunotherapy are not known.
Notably, the peritumoral fat of obese patients demonstrated increased immune infiltration and hypoxia, both of which were more pronounced near the tumor. The potential significance of the interaction between peritumoral adipose tissue inflammation and ccRCC tumors is unclear. In ovarian, breast and prostate cancer there has been significant cross-talk noted between the primary tumor and adipocytes. Studies such as this require further exploration in ccRCC. Our hypothesis generating study suggests that although the primary tumors of obese patients did not exhibit higher levels of immune infiltration to explain their response to immune checkpoint blockade, the surrounding peritumoral fat did. We speculate that the peritumoral adipose tissue may act as an immune reservoir of activated cells that are made available for mobilization upon administration of various systemic therapies (appendix p 13). Therefore, peritumoral fat may act locally to affect the biology and ultimately survival of ccRCC.
Our study is not without limitations. This study is limited to patients with ccRCC as less is known about the association of obesity with non-clear cell RCC. As this was exploratory analysis of microenvironmental difference between obese and normal weight patients, we did not fully adjust for all clinical co-variates associated with clinical response. We used bulk immune deconvolution to estimate immune cell composition within primary tumors and results were not validated using multiplex IHC. However, we validated our results across multiple cohorts, using different gene expression platforms (microarray and RNAseq) and flow cytometry. We utilized published and validated gene signatures for our analysis and did not seek to generate novel signatures in this study as each angiogenic signature may represent a different aspect of anagenesis biology. We analyzed results from a single area of primary tumors and recognize the possibility that there is significant heterogeneity present in ccRCC.45 We recognize that we are correlating primary tumor specimens with the treatment of metastatic disease which may not be representative of the microenvironment of metastatic lesions.46 Our MSK immunotherapy cohort consisted primarily of patients receiving second line immunotherapy and therefore cannot be generalized to patients treated in the first line setting. Finally, while BMI is a good marker for visceral adiposity, it may not reflect other body composition components (e.g., muscle mass).47
In summary, our findings lend biologic support to the obesity paradox which is observed among ccRCC patients regardless of treatment. We detected differences in the tumor microenvironment in obese compared to normal weight patients. Specifically, the tumors of obese ccRCC patients harbored increased angiogenic programs and did not demonstrate significant differences in overall inflammation. Interestingly it’s the peritumoral fat of obese patients which showed increased inflammation and hypoxia condition compared to normal weight patients. Although there is no direct clinical translatability at this time, differences in transcriptomic pathways associated with obesity and other body composition features should be further investigated so that they may be leveraged to improve outcomes among patients with ccRCC. Future studies should focus on the utility of body size (e.g., BMI) or body composition measures as a predictive factor in combination with clinicopathologic and tumor-specific features (e.g., mutational status) and further explore mechanisms of fat-tumor cross-talk that can be exploited to improve patient outcomes.
Supplementary Material
Research in context.
Evidence before this study
An inverse association between body mass index (BMI) and overall survival has been observed among patients with renal cell carcinoma (RCC) across several cohorts including a recent meta-analysis (pooled HR 0·57, 95%CI 0·43–0·76). We searched PubMed for publications from database inception to June 22, 2019 for studies in English assessing primary tumor and peritumoral adipose tissue gene expression differences to understand the mechanisms underlying the obesity paradox in clear cell RCC (ccRCC). We used the search terms, “obesity kidney cancer”, “obesity renal cell carcinoma gene expression”, and “obesity kidney cancer genomic.” The inverse association between BMI and cancer-specific mortality among 2,119 patients with ccRCC treated with nephrectomy (HR 0·59, 95%CI 0·42–0·83) and among 1,975 patients metastatic ccRCC treated with targeted therapy (HR 0·84, 95%CI 0·73–0·95) where obese patients survived longer than normal weight patients was reported. Among a smaller subset of patients in the previously mentioned localized (n=126) and metastatic ccRCC (n=61) cohorts, expression of the metabolic oncogene fatty acid synthase (FASN) was noted to be significantly lower in the primary tumors of obese patients compared to normal weight. Another study evaluated the association of a previously published and validated 34-gene signature (ClearCode34-identified molecular subtype) with comorbidities in 282 patients with ccRCC and noted that obese patients were more likely to harbor ccA tumors (compared to ccB which denote a more aggressive phenotype) (48% vs. 34%, p=0.02). Recently, a report of 313 metastatic RCC patients treated with ≥ 2nd-line nivolumab reported shorter survival among normal weight patients receiving immunotherapy (HR 1·59, 95% CI 1·10–2·30). A recent study demonstrated that altered PD-1-mediated T-cell dysfunction may be partially driven by leptin which is increased among obese patients. Lastly, emerging evidence points to the importance of understanding microenvironmental differences in primary RCC tumors (e.g., angiogenesis and inflammation) to personalize treatment selection. We did not find any studies evaluating differences in primary tumor or peritumoral fat gene expression pathways between obese or non-obese patients as it relates to the obesity paradox in ccRCC.
Added value of this study
Findings from this study lend biologic support to the obesity paradox in ccRCC. Specifically, gene expression differences in angiogenic and inflammatory programs within the tumor microenvironment (tumor and peritumoral adipose tissue) may help explain the survival advantage experienced by obese as compared to normal weight patients.
Implications of all the available evidence
The obesity paradox observed in ccRCC patients may be driven by differences in tumor microenvironment biology. The cross-talk between the tumor and peritumoral adipose tissue is incompletely understood but may provide new, clinically relevant insights for ccRCC patients. This study highlights the need to evaluate the clinical relevance of host factors, such as body size, and their potential contribution to tumor biology, prognosis and treatment selection. Future studies are required to evaluate the cross-talk between peritumoral adipose tissue and RCC, impact of visceral fat inflammation on outcomes in patients receiving immunotherapy, and therapeutic interventions to address adverse components of body size (e.g., increased visceral fat).
Acknowledgements
Ruth L. Kirschstein Research Service Award (grant T32CA082088 [AS]), American Society of Clinical Oncology Young Investigator Award (AS), Ludwig Center MSK (AAH, FK, HF), Weiss Family Kidney Research Fund (AAH), Novartis (RJM, MHV), the Sidney Kimmel Center for Prostate and Urologic Cancers and the National Institutes of Health/National Cancer Institute to Memorial Sloan Kettering Cancer Center through the Cancer Center Support Grant, award number (grant P30 CA008748 [AAH, RJM, MHV]).
M.H. Voss reports receiving commercial research grants from BMS, Pfizer and Genentech, is a consultant/advisory board member for Alexion Pharmaceuticals, Calithera Biosciences, Corvus Pharmaceuticals, Exelixis, Eisai, Natera, Novartis, and Pfizer, and has received other remuneration from Eisai, Takeda, and Novartis. T.A. Chan reports receiving commercial research grants from Illumina, BMS, Eisai, AstraZeneca, and An2H, has ownership interest (including stock, patents, etc.) in Gritstone Oncology, and is a consultant/advisory board member for Gritstone Oncology, Illumina, and Bristol Myers. R.J. Motzer reports receiving commercial research grants from Bristol-Myers Squibb, Pfizer, Novartis, Genentech/Roche, and Eisai and is a consultant/advisory board member for Genentech, Pfizer, Merck, Novartis, and Incyte. A.J. Dannenberg is a member of the scientific advisory boards of SynDevRx and Quentis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
No potential conflicts of interest were disclosed by the other authors.
Data sharing
Novartis is committed to sharing access to patient-level data and supporting clinical documents from eligible studies with qualified external researchers. These requests are reviewed and approved by an independent review panel on the basis of scientific merit. All data provided are anonymized to respect the privacy of patients who have participated in the trial in line with applicable laws and regulations. This trial data availability is according to the criteria and process described on www.clinicalstudydatarequest.com.
Contributor Information
Alejandro Sanchez, Department of Surgery, Division of Urology, Memorial Sloan Kettering Cancer Center, New York, NY.
Helena Furberg, Department of Epidemiology & Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY.
Fengshen Kuo, Immunogenomics & Precision Oncology Platform, Memorial Sloan Kettering Cancer Center, New York, NY.
Lynda Vuong, Immunogenomics & Precision Oncology Platform, Memorial Sloan Kettering Cancer Center, New York, NY.
Yasser Ged, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY.
Sujata Patil, Department of Epidemiology & Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY.
Irina Ostrovnaya, Department of Epidemiology & Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY.
Stacey Petruzella, Department of Epidemiology & Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY.
Albert Reising, Novartis Oncology, New York, NY.
Parul Patel, Novartis Oncology, New York, NY.
Roy Mano, Department of Surgery, Division of Urology, Memorial Sloan Kettering Cancer Center, New York, NY.
Jonathan Coleman, Department of Surgery, Division of Urology, Memorial Sloan Kettering Cancer Center, New York, NY.
Paul Russo, Department of Surgery, Division of Urology, Memorial Sloan Kettering Cancer Center, New York, NY.
Catherine H. Liu, Department of Medicine, Weill Cornell Medical College, New York, New York
Andrew J. Dannenberg, Department of Medicine, Weill Cornell Medical College, New York, New York
Robert Motzer, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY.
Martin H. Voss, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY
A. Ari Hakimi, Department of Surgery, Division of Urology, Memorial Sloan Kettering Cancer Center, New York, NY.
References
- 1.Choi Y, Park B, Jeong BC, et al. Body mass index and survival in patients with renal cell carcinoma: a clinical-based cohort and meta-analysis. Int J Cancer. 2013;132(3):625–34. [DOI] [PubMed] [Google Scholar]
- 2.Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K. Body Fatness and Cancer--Viewpoint of the IARC Working Group. N Engl J Med. 2016;375(8):794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakimi AA, Furberg H, Zabor EC, et al. An epidemiologic and genomic investigation into the obesity paradox in renal cell carcinoma. J Natl Cancer Inst. 2013;105(24):1862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albiges L, Hakimi AA, Xie W, et al. Body Mass Index and Metastatic Renal Cell Carcinoma: Clinical and Biological Correlations. J Clin Oncol. 2016;34(30):3655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McQuade JL, Daniel CR, Hess KR, et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. Lancet Oncol. 2018;19(3):310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Aguilar EG, Luna JI, et al. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat Med. 2019;25(1):141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortellini A, Bersanelli M, Buti S, et al. A multicenter study of body mass index in cancer patients treated with anti-PD-1/PD-L1 immune checkpoint inhibitors: when overweight becomes favorable. J Immunother Cancer. 2019;7(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Giorgi U, Procopio G, Giannarelli D, et al. Association of Systemic Inflammation Index and Body Mass Index with Survival in Patients with Renal Cell Cancer Treated with Nivolumab. Clin Cancer Res. 2019;25(13):3839–46. [DOI] [PubMed] [Google Scholar]
- 9.Motzer RJ, Hutson TE, Cella D, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med. 2013;369(8):722–31. [DOI] [PubMed] [Google Scholar]
- 10.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voss MH, Reising A, Cheng Y, et al. Genomically annotated risk model for advanced renal-cell carcinoma: a retrospective cohort study. Lancet Oncol. 2018;19(12):1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turajlic S, Xu H, Litchfield K, et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell. 2018;173(3):581–94.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–7. [DOI] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence M, Huber W, Pages H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbloom KR, Armstrong J, Barber GP, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D670–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med. 2018;24(6):749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masiero M, Simoes FC, Han HD, et al. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell. 2013;24(2):229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95. [DOI] [PubMed] [Google Scholar]
- 26.Senbabaoglu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung W, Eum HH, Lee HO, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choueiri TK, Figueroa DJ, Fay AP, et al. Correlation of PD-L1 tumor expression and treatment outcomes in patients with renal cell carcinoma receiving sunitinib or pazopanib: results from COMPARZ, a randomized controlled trial. Clin Cancer Res. 2015;21(5):1071–7. [DOI] [PubMed] [Google Scholar]
- 33.Semlitsch T, Stigler FL, Jeitler K, Horvath K, Siebenhofer A. Management of overweight and obesity in primary care-A systematic overview of international evidence-based guidelines. Obes Rev. 2019. [DOI] [PMC free article] [PubMed]
- 34.Deng T, Lyon CJ, Bergin S, Caligiuri MA, Hsueh WA. Obesity, Inflammation, and Cancer. Annu Rev Pathol. 2016;11:421–49. [DOI] [PubMed] [Google Scholar]
- 35.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandal R, Chan TA. Personalized Oncology Meets Immunology: The Path toward Precision Immunotherapy. Cancer Discov. 2016;6(7):703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakimi AA, Voss MH, Kuo F, et al. Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear cell Renal Cell Cancer - Data from a Randomized Phase III Trial. Cancer Discov. 2019. [DOI] [PMC free article] [PubMed]
- 38.Haake SM, Brooks SA, Welsh E, et al. Patients with ClearCode34-identified molecular subtypes of clear cell renal cell carcinoma represent unique populations with distinct comorbidities. Urol Oncol. 2016;34(3):122.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quail DF, Dannenberg AJ. The obese adipose tissue microenvironment in cancer development and progression. Nat Rev Endocrinol. 2019;15(3):139–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukumura D, Incio J, Shankaraiah RC, Jain RK. Obesity and Cancer: An Angiogenic and Inflammatory Link. Microcirculation. 2016;23(3):191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Perez RR, Lanier V, Newman G. Leptin’s Pro-Angiogenic Signature in Breast Cancer. Cancers (Basel). 2013;5(3):1140–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Himbert C, Ose J, Nattenmuller J, et al. Body Fatness, Adipose Tissue Compartments, and Biomarkers of Inflammation and Angiogenesis in Colorectal Cancer: The ColoCare Study. Cancer Epidemiol Biomarkers Prev. 2019;28(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James BR, Tomanek-Chalkley A, Askeland EJ, Kucaba T, Griffith TS, Norian LA. Diet-induced obesity alters dendritic cell function in the presence and absence of tumor growth. J Immunol. 2012;189(3):1311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearce OMT, Delaine-Smith RM, Maniati E, et al. Deconstruction of a Metastatic Tumor Microenvironment Reveals a Common Matrix Response in Human Cancers. Cancer Discov. 2018;8(3):304–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown JC, Caan BJ, Prado CM, et al. Body Composition and Cardiovascular Events in Patients With Colorectal Cancer: A Population-Based Retrospective Cohort Study. JAMA Oncol. 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.