Summary
KCNQ1, also known as Kv7.1, is a voltage-dependent K+ channel that regulates gastric acid secretion, salt and glucose homeostasis and heart rhythm. Its functional properties are regulated in a tissue-specific manner through co-assembly with beta subunits KCNE1-5. In non-excitable cells KCNQ1 forms a complex with KCNE3, which suppresses channel closure at negative membrane voltages that otherwise would close it. Pore opening is regulated by the signaling lipid PIP2. Using cryo-EM we show that KCNE3 tucks its single membrane spanning helix against KCNQ1, at a location that appears to lock the voltage sensor in its depolarized conformation. Without PIP2, the pore remains closed. Upon addition, PIP2 occupies a site on KCNQ1 within the inner membrane leaflet, which triggers a large conformational change that leads to dilation of the pore’s gate. It is likely that this mechanism of PIP2 activation is conserved among Kv7 channels.
Graphical Abstract
In Brief
Cryo-EM structures of the human voltage-dependent potassium channel KCNQ1 in complex with its ancillary subunit KCNE3, in the presence or absence of the PIP2 activating cofactor, provide insight into how PIP2 binding leads to channel opening.
Introduction
KCNQ1 (also known as Kv7.1 or KvLQT1) is widely distributed throughout different organs such as heart (Barhanin et al., 1996; Sanguinetti et al., 1996), colon (Preston et al., 2010; Schroeder et al., 2000), inner ear (Warth and Bleich, 2000), testis (Manderfield and George, 2008), kidney (Zheng et al., 2007) and stomach (Grahammer et al., 2001), and plays essential roles in human physiology (Abbott et al., 2014; Jespersen et al., 2005). KCNQ1 is the pore-forming subunit of the slow-delayed rectifier potassium channel (IKs) in the human heart and therefore is vital for maintaining cardiac rhythm (Barhanin et al., 1996; Sanguinetti et al., 1996). KCNQ1—in complex with KCNE3—provides the driving force for proximal tubular and intestinal sodium absorption, gastric acid secretion and intestinal cAMP-induced chloride secretion (Al-Hazza et al., 2016; Julio-Kalajzic et al., 2018; Nakano and Shimizu, 2016; Preston et al., 2010). Mutations in the kcnq1 gene or pharmacological disruption of KCNQ1 channel function cause human diseases such as long-QT syndrome, short-QT syndrome, atrial fibrillation, diabetes mellitus and deafness (Nakano and Shimizu, 2016; Peroz et al., 2008; Thevenod, 2002).
KCNEs (KCNE1-5, also called minK-related peptides, MiRPs) are single transmembrane-spanning beta-subunits of potassium channels that modulate KCNQ1 functional properties and cellular localization (Abbott et al., 2014; McCrossan and Abbott, 2004; Roura-Ferrer et al., 2010). To fulfill its diverse roles in both excitable and nonexcitable cells, KCNQ1 is always found in association with one or more KCNE subunits in vivo. All five KCNE subunits can interact with KCNQ1, resulting in channel complexes with a wide range of functional properties (Van Horn et al., 2011). For example, KCNE1 right-shifts the voltage dependence of KCNQ1 activation (that is, causes the channel to activate at more positive voltages), slows its activation and deactivation, increases the single channel conductance and eliminates inactivation. KCNE4 on the other hand is an inhibitor of KCNQ1 activity (Grunnet et al., 2002).
KCNE3 alters the voltage sensitivity of KCNQ1 such that the current-voltage (I-V) relationship is almost linear over physiological membrane potentials (Barro-Soria et al., 2015; Schroeder et al., 2000). A linear I-V relationship contrasts the more typical non-linear, step function-like I-V curve exhibited by most other Kv channels, including KCNQ1 in the absence of KCNE3. Linearity of the KCNQ1-KCNE3 I-V curve gives rise to K+ conductivity in non-excitable cells such as polarized epithelial cells of the colon, small intestine and airways. KCNE3 is thought to stabilize the S4 segment of KCNQ1 voltage sensors in an activated state, which in turn locks the channel open at negative voltages that would otherwise cause it to close (Barro-Soria et al., 2015; Barro-Soria et al., 2017). In addition, a plausible mechanism has been proposed, suggesting that D54 and D55 of KCNE3 stabilize the KCNQ1 S4 in its activated conformation through electrostatic interactions with arginines on S4 (Barro-Soria et al., 2015; Choi and Abbott, 2010). However, a concrete structural description of how KCNE3 does this is still missing.
The channel activity of KCNQ1 is also regulated by the signaling lipid phosphatidylinositol 4,5-bisphosphate (PIP2) (Loussouarn et al., 2003). PIP2 functions as a cofactor of KCNQ1 and is required for the activation of KCNQ1 homo-tetramers as well as its complexes with different KCNEs (Zaydman and Cui, 2014). PIP2 likely acts by coupling the voltage sensor domain to the pore domain of KCNQ1 because in the absence of PIP2 the voltage sensors move but the pore does not open (Zaydman et al., 2013). Our previous structural study offered a hypothesis for the site of PIP2 binding (Sun and MacKinnon, 2017). Now we test this hypothesis and in addition describe the conformational changes that ensue once PIP2 binds.
Results and Discussion
Structure of the hKCNQ1EM-KCNE3-CaM complex
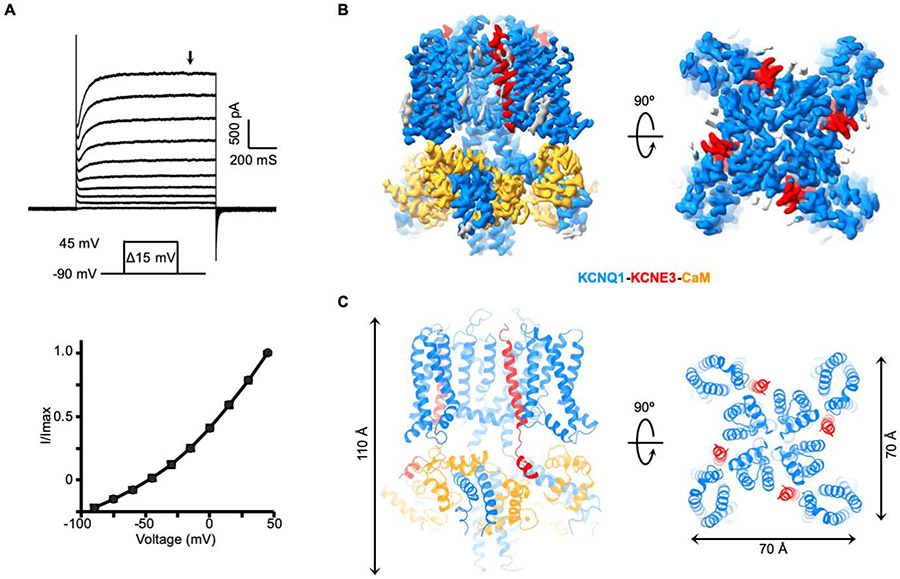
We first carried out structural studies of the “PIP2-free” hKCNQ1EM-KCNE3 complex together with calmodulin (CaM), which is likely a structurally obligate subunit of KCNQ1 (Ghosh et al., 2006; Sachyani et al., 2014; Shamgar et al., 2006). The N-terminal and C-terminal loops of human KCNQ1 were truncated to obtain a biochemically stable construct (hereafter called hKCNQ1EM). Full-length KCNE3 was tagged with mCherry fluorescence protein at its N terminus to monitor the intactness of the protein complex during purification. CaM was also overexpressed, although endogenous CaM from the HEK293S GnTl− cells used for protein expression may already be sufficient (Figure S1A) (Goehring et al., 2014). The resulting hKCNQ1EM-KCNE3-CaM complex has an almost linear activation curve in electrophysiological recordings from Chinese Hamster Ovary (CHO) cells, manifesting functional features similar to those described in previous reports (Gao et al., 2008; Kroncke et al., 2016; Mazhari et al., 2002; Van Horn et al., 2011) (Figure 1A).
Figure 1. Function validation and structure determination of hKCNQ1EM-KCNE3-CaM complex.
(A) Voltage family current trace of the hKCNQ1EM-KCNE3-CaM complex recorded in CHO cells (top) and the I-V curve of the channel activation plotted at the time point indicated by the black arrow.
(B) Side and top view of the hKCNQ1EM-KCNE3-CaM cryo-EM density map. KCNQ1, KCNE3 and CaM are colored in blue, red and orange, respectively.
(C) Structure model of the hKCNQ1EM-KCNE3-CaM complex oriented and colored as in (B).
The structure of the “PIP2-free” hKCNQ1EM-KCNE3-CaM complex was determined at a resolution of 3.1 Å using single-particle cryo-EM analysis with C4 symmetry imposed (Figure S1B-S1D, Table S1). Structural studies were carried out in the presence of 4 mM EGTA. The final cryo-EM map allowed de novo model building (Figure 1B and 1C). In the presence of KCNE3 the architecture of human KCNQ1-CaM is similar to that of the Xenopus complex (Sun and MacKinnon, 2017) and thus we use the same nomenclature for secondary structure elements (Figure S2A). The channel complex has dimensions ~70 Å x 70 Å x 110 Å (Figure 1C). KCNQ1 adopts a “decoupled” configuration with activated voltage sensors and a closed pore as expected in the absence of PIP2. The interface between KCNQ1 and CaM is conserved between the two species (Figure S2C). The overall RMSD between the human and Xenopus KCNQ1-CaM monomer is about 1.1 Å (Figure S2B).
The cryo-EM density permitted building of a model of KCNE3 that included the transmembrane helix, a cytosolic helix and the connecting loop (amino acids 53 to 99). The remainder of KCNE3 was not resolved, presumably owing to structural disorder; the cryo-EM density of the cytoplasmic helix is weaker than that of the transmembrane helix. Of note, the structural model of KCNE3 in the complex deviates from an NMR structure of KCNE3 in the absence of KCNQ1 with an RMSD between the two structures of 7.6 Å (Figure S2F) (Kroncke et al., 2016) The structural differences undoubtedly reflect the separate contexts in which these structures were determined, one by itself and the other in a complex with the channel.
Four KCNE3 molecules are bound to one KCNQ1 tetramer. The number of KCNE subunits that can associate with the KCNQ1 tetramer has been debated (Chen et al., 2003; Morin and Kobertz, 2008; Murray et al., 2016; Nakajo et al., 2010; Plant et al., 2014). Previous investigations into the stoichiometry between KCNQ1 and KCNE1 (a paralog of KCNE3) have proposed either a fixed KCNQ1:KCNE1 stoichiometry of 4:2 or the possibility of multiple stoichiometries ranging from 4:1 to 4:4. During the structural reconstruction of the hKCNQ1EM-KCNE3-CaM complex we carried out 3D refinement without imposing symmetry restraints; the results support a 4:4 stoichiometry (Figure S1B), thus demonstrating that a 4:4 stoichiometry can occur.
Modulation of KCNQ1 by KCNE3
To document the influence of KCNE3 on the structure of KCNQ1 we also determined the cryo-EM structure of hKCNQ1EM-CaM in the absence of KCNE3 at a resolution of 3.1-Å (Figure S3, Table S1). When aligning the pore domain of these two structures, we observe a ~7-degree counterclockwise rotation of the voltage sensors relative to the pore in the KCNE3-bound complex, when viewed from the extracellular side (Figure S2E). This rotation moves the voltage sensor domains of KCNQ1 slightly away from the pore domain, allowing space for the KCNE3 subunits.
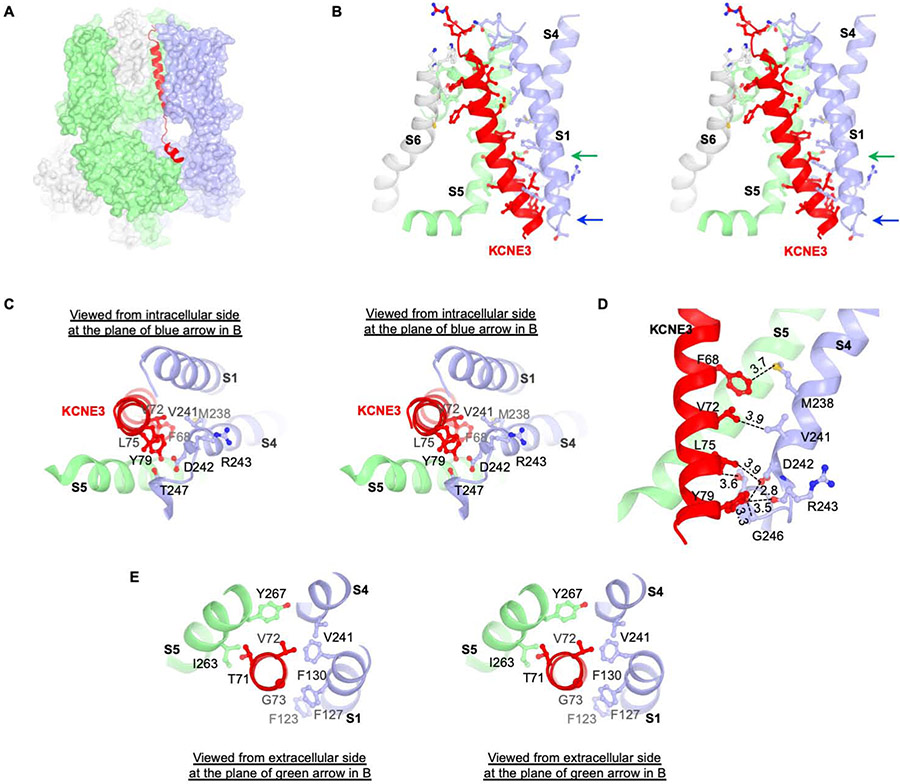
The interface between KCNE3 and KCNQ1-CaM consists of two parts: cytosolic interactions between KCNE3 and CaM (Figure S2D) and transmembrane interactions between KCNE3 and KCNQ1 (Figure 2A). At the cytosolic interface, KCNE3 lies on the top of a surface shaped by two neighboring CaM subunits Figure S2D). At the transmembrane interface KCNE3 is tucked into a cleft formed by three KCNQ1 subunits. The mainly hydrophobic surface of this cleft is lined by S6 from one subunit, S5 and the pore helix from a second subunit, and S1 and S4 from a third subunit (Figure 2B). The extracellular half of KCNE3 lies in-between S1 and S6, consistent with previous biochemical data showing that residues flanking the extracellular end of the KCNE1 transmembrane helix could be crosslinked to S1 and S6 of KCNQ1 by introducing disulfide bonds (Figure S2G). Therefore, we expect that KCNE1 and KCNE3 occupy a similar location in the complex. We note, however, that some homologous residue pairs that could be strongly crosslinked in a previous study are quite far away in the hKCNQ1EM-KCNE3-CaM complex (Chung et al., 2009). While it is possible that KCNE1 and KCNE3 can bind differently to KCNQ1, we think it is also likely that crosslinks sometimes occur between distant residues (Figure S2G).
Figure 2. Interaction between KCNQ1-CaM and KCNE3.
(A) Interaction between KCNQ1 and KCNE3. KCNE3 is shown as ribbons and KCNQ1-CaM is shown as surface representation. KCNE3 interacts with three protomers of KCNQ1-CaM colored in green, light blue and grey, respectively. The same color code is used in panel (B), (C) and (D).
(B) Stereo view of the interaction between KCNQ1 and KCNE3 in the transmembrane region. Side chains of residues on the interface are shown as stick and balls.
(C) Stereo view from intracellular side of the KCNQ1 and KCNE3 interface at the plane indicated by blue arrow in (B).
(D) Interactions between S4 of KCNQ1 and transmembrane helix of KCNE3. The distances between potential interaction residues (within 4 Å) are labeled. Main chain of R243 is shown in order to demonstrate the potential interaction between Y79 and main chain of R243.
(E) Stereo view from extracellular side of the TVG region and its binding site in KCNQ1 at the plane indicated by green arrow in (B).
The position of KCNE3 suggests a plausible explanation for why it prevents closure of KCNQ1 at negative voltages, thus rendering the I-V relation approximately linear. We observe that KCNE3 directly contacts the bottom of S4 when the voltage sensor adopts its up (depolarized) conformation (Figure 2C). If the contact stabilizes S4 in this conformation, then it would diminish the tendency for S4 to move downward toward the cytoplasm at hyperpolarizing membrane voltages. Likewise, the position of KCNE3 might destabilize S4 in its downward conformation. In either case, KCNE3 seems well poised to favor a depolarized conformation of the voltage sensor. This explanation is consistent with a voltage clamp fluorometry study, which showed that KCNE3 primarily affects the movement of S4 to modify the voltage-dependent gating of KCNQ1 (Barro-Soria et al., 2015). However, the molecular details of the locking mechanism we describe are distinct from an earlier model, in which D54 and D55 of KCNE3 were proposed to interact with R228 or R237 on S4 (Barro-Soria et al., 2015; Choi and Abbott, 2010). In the structure, we observe that distances between D54 and R228 of 19.7 Å, between D55 and R228 of 27.4 Å, between D54 and R237 of 25.6 Å and between D55 and R237 of 30.6 Å are too great to mediate the proposed electrostatic interactions.
Melman and colleagues showed that it is possible to modify KCNE3 so that its influence on KCNQ1 gating is similar to that of KCNE1 (Melman et al., 2001; Melman et al., 2002). The modification they made was mutation of amino acids Thr71, Val72 and Gly73 to the corresponding amino acids – Phe, Thr, Leu – found in KCNE1. In the structure, these amino acids reside deep inside the membrane, where KCNE3 interacts with helices S1, S4 and S5 (Figure 2E). How this segment of the KCNE transmembrane helix confers specific gating properties onto KCNQ1 is unclear.
Structure of hKCNQ1EM-KCNE3-CaM in the presence of PIP2
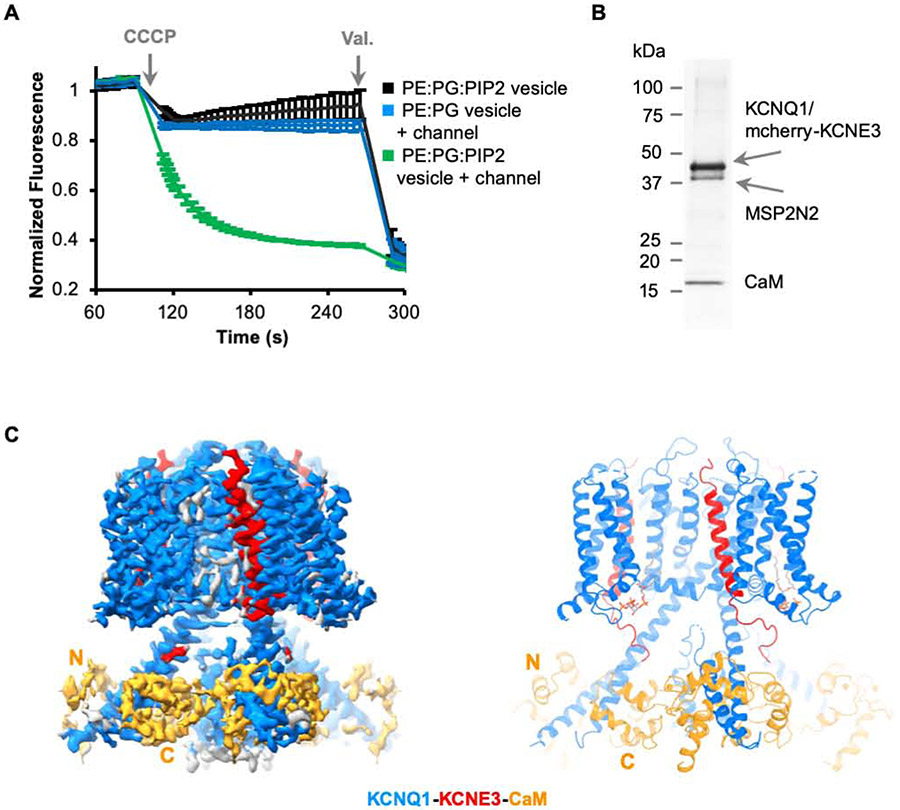
Because KCNE3 modifies KCNQ1 such that voltage-dependent closure does not occur across the physiological range of voltages, the KCNQ1-KCNE3-CaM channel essentially functions as a ligand-gated ion channel, with the ligand being the signaling lipid PIP2. To demonstrate PIP2 activation of hKCNQ1EM-KCNE3-CaM we used a liposome flux assay (Su et al., 2016). Proteoliposomes were reconstituted with a high concentration of K+ (160 mM KCl) and then diluted into K+-free solution with ionic strength held constant with 160 mM NaCl. Fluorescence decrease due to K+ efflux was observed only when both hKCNQ1EM-KCNE3-CaM and PIP2 were present in the vesicles, indicating that the purified channel complex is activated by PIP2 in vitro (Figure 3A).
Figure 3. Structure of hKCNQ1EM-KCNE3-CaM complex in the presence of PIP2.
(A) Liposome based flux assay. Recordings (n=3) from PE:PG:PIP2 alone, PE:PG with hKCNQ1EM-KCNE3-CaM and PE:PG:PIP2 with hKCNQ1EM-KCNE3-CaM vesicles are plotted in black, blue and green traces.
(B) SDS-PAGE showing the reconstitution of hKCNQ1EM-KCNE3-CaM complex in nanodiscs containing PIP2 using MSP2N2 as the scaffold protein.
(C) Cryo-EM map and structure model of hKCNQ1EM-KCNE3-CaM in the presence of PIP2. KCNQ1, KCNE3 and CaM are colored in blue, red and orange, respectively.
We determined a cryo-EM structure of hKCNQ1EM-KCNE3-CaM with PIP2 in nanodiscs at an overall resolution of 3.9 Å (Figure 3C and Figure S4, Table S1) (Denisov and Sligar, 2016; Nath et al., 2007). hKCNQ1EM-KCNE3-CaM was first purified in detergent and then reconstituted into lipid nanodiscs containing soybean polar extract and brain PIP2 (Figure 3B). Following data collection and image analysis the map showed the highest quality density in the transmembrane domain and in cytoplasmic helices HA and HB, allowing side chain assignment in these regions. Because the density in most other areas, including the CaM N-lobe, was too poor to ensure accurate sequence registration, we initiated model building by docking the model of the PIP2-free structure into the map. It was immediately evident that the PIP2-bound hKCNQ1EM-KCNE3-CaM channel exhibited large conformational changes relative to the PIP2 structure (Figure 1B and 3C).
PIP2-binding site and conformational changes induced by PIP2
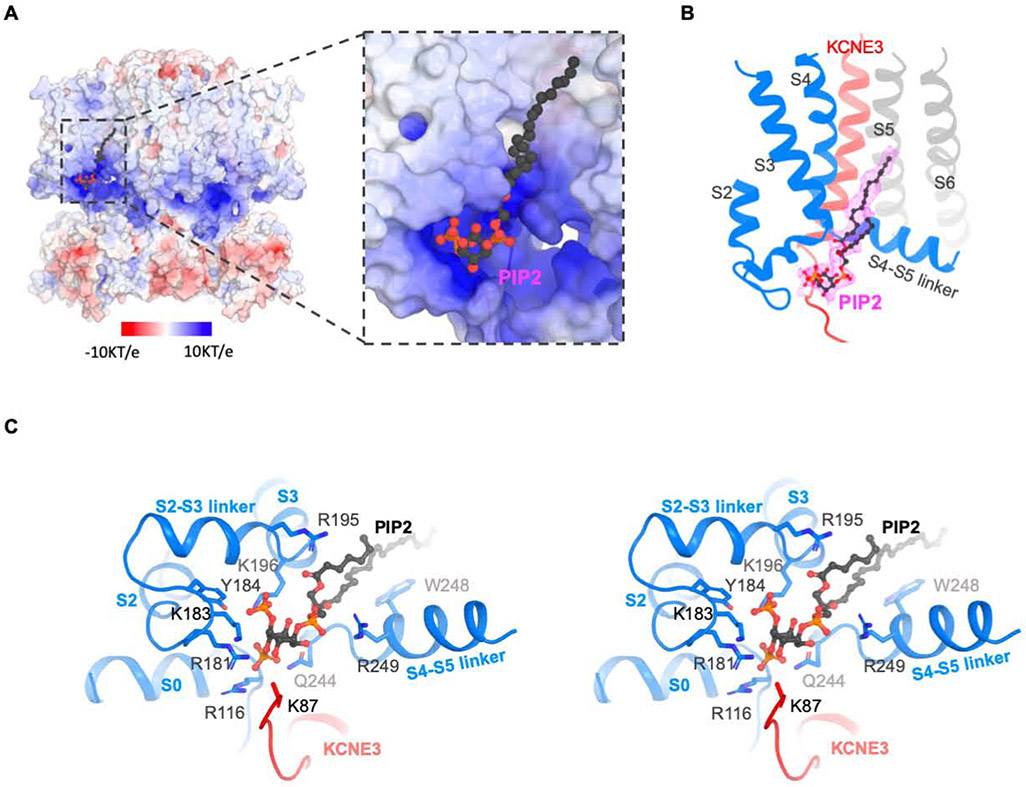
Density consistent with PIP2 is observed near a loop that connects S4 to the S4-S5 linker of KCNQ1 (Figure 4B). The binding site contains multiple amino acids with positive-charged side chains contributed by S0, the S2-S3 loop and the S4-S5 linker of KCNQ1 and possibly more distant interactions with amino acids from KCNE3 (Figure 4A and Figure 4C). The location of PIP2 binding is consistent with biochemical and mutational studies that concluded PIP2 interacts with these same regions (Choveau et al., 2018; Eckey et al., 2014; Kasimova et al., 2015; Telezhkin et al., 2013; Zaydman et al., 2013). Several studies implicated also the importance of the junction between S6 and the HA helix, the HB helix and the linker connecting these helices of KCNQ1 (Choveau et al., 2018; Tobelaim et al., 2017; Zaydman et al., 2013). The residues at the junction between S6 and HA may be important for the PIP2-induced conformational changes discussed below. However, the sites on HB or the linker connecting the HA and HB helices are not easily explained by our cryo-EM structure. KCNE3 also comes in close proximity to PIP2, suggesting that it might influence PIP2 regulation (Li et al., 2011).
Figure 4. PIP2 binding site of the hKCNQ1EM-KCNE3-CaM channel complex.
(A) Surface charge representation (−10KT/e to 10 KT/e) of the PIP2 binding site. The surface potential is calculated using APBS plugin in PyMOL. PIP2 binding site is zoomed in on the right. PIP2 molecule is shown as balls and sticks.
(B) Cryo-EM density of PIP2 in the complex. Only the surrounding region of PIP2 binding site are shown in cartoon for clarity. KCNE3 is colored in red and two neighboring KCNQ1 subunits are colored in grey and blue, respectively.
(C) Stereo view of the PIP2 binding site. The side chains of residues within 4 A of PIP2 are shown as sticks. PIP2 is shown as balls and sticks.
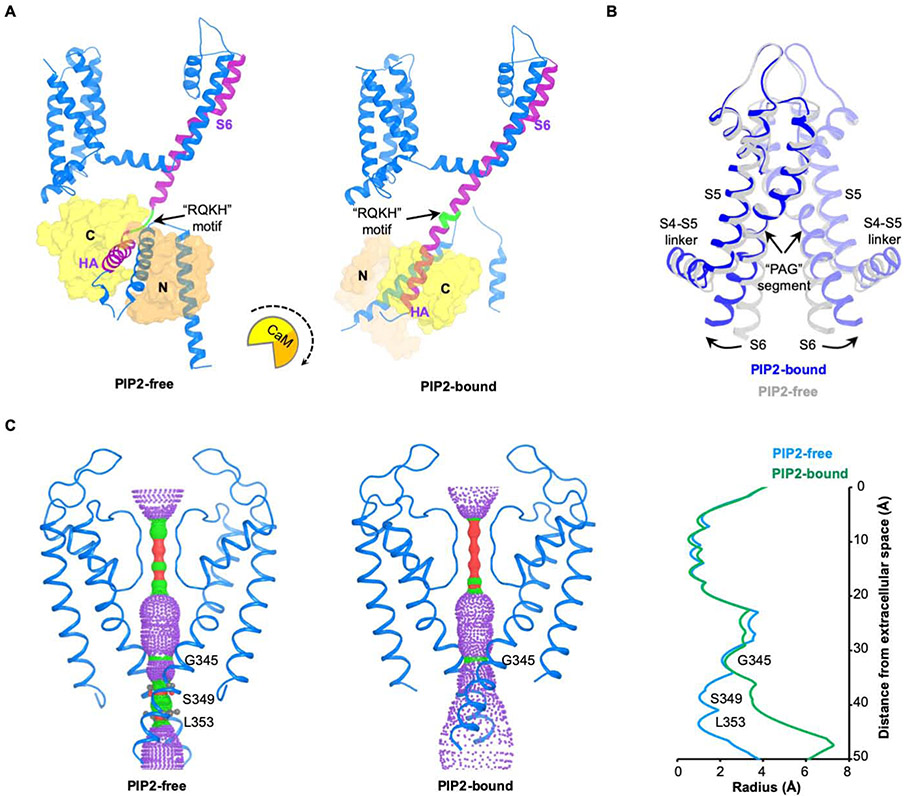
The binding of PIP2 leads to large conformational changes in KCNQ1 (Figure 5). The S6 and HA helices, which form a helix-loop-helix structure in the “PIP2-free” state, become a single continuous long helix (Figure 5A). This transition causes CaM to rotate almost 180 degrees, release its interaction with the voltage sensor, and move towards the central axis of the molecule. The linker connecting S6 to the HA helices (here called the “RQKH motif”) undergoes a structural rearrangement from a loop to a helix. The RQKH motif is absolutely conserved in sequence among KCNQ1 orthologs and paralogs (KCNQ1-5) (Figure S5E), suggesting the possibility of a similar PIP2-gating mechanism for the entire KCNQ family.
Figure 5. Conformational changes induced by PIP2.
(A) Conformational change of the channel complex in one KCNQ1-CaM protomer. The “RQKH” motif that undergoes structural rearrangement from a loop to a helix is colored in green. S6 and HA helixes of KCNQ1 are colored in purple. CaM is shown as surface with its N-lobe in orange and C-lobe in yellow. The rotational motion of CaM associated with the conformational change is indicated by a cartoon with a dash arrow.
(B) Overlay of the pore domains of PIP2-free and PIP2-bound structures showing the conformational change in the ion conducting pathway. The PIP2-free state is colored in grey and PIP2-bound state in blue. The S6 helix bends outward upon PIP2 binding at the point of PAG segment.
(C) Left: view of ion conducting pathways for PIP2-free and PIP2-bound states with front and back subunits excluded for clarity. Right: radius of the pore calculated using HOLE program. The amino acids restricting the pore are labeled.
Correlated with the above conformational changes, the C-terminal half of S6 bends at the conserved “PAG” segment in the presence of PIP2, which has been identified as an important gating element (Figure 5B) (Boulet et al., 2007; Seebohm et al., 2007). The bend causes the cytoplasmic pore entrance to dilate (Figure 5C). The region near Ser349, proposed to form the inner gate, expands from ~1 Å to >3.5 Å (Figure 5C) (Sun and MacKinnon, 2017). Similar opening mechanisms through bending of S6 have also been observed in other channels (Hite and MacKinnon, 2017; Hite et al., 2017; Lee and MacKinnon, 2018). The narrowest region of the ion conducting pathway remains ~2.5 Å radius. Since a hydrated K+ ion is larger than that (radius ~ 4 Å), we are left to wonder whether the pore can dilate further, or whether K+ can diffuse across this region because the side chains are sufficiently dynamic. This situation of ‘a little narrower than expected’ has occurred so repeatedly in various ion channels under conditions that should render conductivity, we wonder whether the diffusion of ions through a sufficiently dynamic region is analogous to the diffusion of molecular oxygen to heme in hemoglobin (Hansen et al., 2011; Lee and MacKinnon, 2018; Park and MacKinnon, 2018; Perutz and Mathews, 1966; Takayanagi et al., 2013; Whorton and MacKinnon, 2013).
Because we determined the PIP2-free structure in detergent micelles (digitonin) and the PIP2-bound structure in nanodiscs, we repeated the structural comparison under conditions in which the only difference is the presence or absence of PIP2. To this end we determined the structure of hKCNQ1EM-KCNE3-CaM in nanodiscs without PIP2 at a resolution of 6.2 Å. The “PIP2-free” hKCNQ1EM-KCNE3-CaM model docks almost perfectly into the map (Figure S5A). Thus, PIP2 drives the conformational change. We also examined the effects of PIP2 on KCNQ1 in the absence of KCNE3 and found that it produced the same conformational change (Figure S5B). To better appreciate the magnitude of conformational changes caused by the binding of PIP2, we made movies connecting the PIP2-free and PIP2-bound structures of the hKCNQ1EM-KCNE3-CaM complex (Movie S1 and S2).
The PIP2-mediated conformational change helps to rationalize the functional effects of certain disease-causing mutations, for example R555H (Figure S5D). R555 is located on the HC helix of KCNQ1, which interacts with the CaM C-lobe in the PIP2-bound state. In cellular electrophysiological studies this mutant channel exhibits markedly reduced IKs current without altering channel trafficking (Aromolaran et al., 2014). The structures suggest that R555H might disrupt interactions between HC and CaM that are important for stabilization of the open KCNQ1-KCNE1 complex. The reduced channel activity then leads to a long-QT syndrome.
Conclusion
KCNQ1 is a voltage-dependent K+ channel of physiological importance. In non-excitable cells it forms a complex with KCNE3 that suppresses channel closure at negative membrane voltages that otherwise would close it. Conductivity of the pore is largely regulated by the signaling lipid PIP2: in the absence of PIP2 the channel closes and in its presence it opens. This paper addresses two central questions. First, how does KCNE3 prevent voltage-dependent closure of KCNQ1? And second, how does PIP2 open the KCNQ1-KCNE3-CaM complex? The answer to the first question is, KCNE3 tucks its single membrane spanning helix against KCNQ1 in such a way that it appears to stabilize the up conformation of the voltage sensor, locking it in its ‘open’ configuration. The answer to the second question is, binding of PIP2 orchestrates global conformational changes that open the pore. The PIP2 binding site involves S0, the S2-S3 loop, the S4-S5 linker of KCNQ1 and possibly the juxtamembrane region of KCNE3. Through its interaction with these regions, PIP2 brings about a large conformational change in the cytoplasmic domain and opening of the ion conducting pathway through expansion of the S6 helices.
Star Methods
Lead Contact and Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact (Roderick MacKinnon: mackinn@rockefeller.edu) with a completed Materials Transfer Agreement.
Experimental Model and Subject Details
Cell lines
Chinese hamster ovary (CHO)-K1 cells were cultured in DMEM/F-12 medium (GIBCO) supplemented with 10% fetal bovine serum at 37°C. S f9 cells were cultured in Sf-900 II SFM medium (GIBCO) at 27°C. HEK293S GnTI− cells were cultured in Freestyle 293 medium supplemented with 2% fetal bovine serum at 37°C.
Method Details
Cloning, expression and purification of hKCNQ1EM-KCNE3-CaM and hKCNQ1EM-CaM
DNAs encoding human KCNQ1 (Homo sapiens: NP_000209.2), KCNE3 (Homo sapiens: NP_005463) and CaM (Homo sapiens: NP_001734.1) were synthesized by Genewiz Inc. To improve the biochemical and thermal stability of KCNQ1, the N-terminal and C-terminal regions were truncated, leaving a construct (hKCNQ1EM) with residues 76-620. EcoR I and Xho I sites were used for cloning the hKCNQ1EM construct into a BacMam expression vector with a C-terminal green fluorescent protein (GFP)-6xHis tag linked by a preScission protease site. Full-length KCNE3 (Homo sapiens: NP_005463) was cloned into a BacMam expression vector with an IL2 signal peptide followed by a mCherry2 fluorescence protein tag. CaM was cloned into a BacMam expression vector without any tags (Figure S1A).
Recombinant baculoviruses of hKCNQ1EM, KCNE3 and CaM were generated separately using the Bac-to-Bac system according to manufacturer's instructions (Invitrogen). Then P3 virus mixture of hKCNQ1EM:CaM (9:1) or hKCNQ1EM:KCNE3:CaM (5:5:1) was used for transduction of HEK293S GnTI− cells for protein expression. Briefly, for 1L cultures of HEK293S GnTI− cells (1.5-3x106 cells/mL) in Freestyle 293 media (Gibco) supplemented with 2% FBS (Gibco), about 100 ml P3 virus mixture was used. Infected cells were incubated at 37°C overnight, and protein expression was induced by adding 10 mM sodium butyrate. Cells were cultured at 30°C for another 48-60 hrs before harvest (Goehring et al., 2014).
4L of cell pellet was resuspended in 75 ml lysis buffer (20 mM HEPES pH7.4, 150 mM KCl, 4 mM EGTA, 2 μM leupepetin, 1 μM pepstatin, 100 μM PMSF, 3 μM E-64 and 1 μg/mL aprotinin), and then solubilized by adding 70 ml detergent mixture LMNG/CHS (8:1 mass ratio, 2% stock solubilized in 20 mM HEPES pH7.4, 150 mM KCl) to a final concentration of 1 % while stirring at 4°C for 2 hrs. Solubilized hKCNQ1EM-CaM or hKCNQ1EM-KCNE3-CaM was separated from the insoluble fraction by high-speed centrifugation (38,000g for 1 hr), and incubated with 4 ml CNBR-activated sepharose beads (GE healthcare) coupled with 4 mg high-affinity GFP nanobodies (GFP-NB) (Kirchhofer et al., 2010). The GFP tag was cleaved by preScission protease 2hr or overnight at 4°C, and hKCNQ1EM-CaM or hKCNQ1EM-KCNE3-CaM complex was further purified by size-exclusion chromatography with a Superose 6, 10/300 GL column (GE Healthcare) equilibrated with 20 mM HEPES pH7.4, 150 mM KCl, 4 mM EGTA, 0.06% Digitonin. The peak fractions were pooled, concentrated to 5-6 mg/ml using a 100-kDa MWCO centrifugal device (Ambion) and immediately used for cryo-EM grid preparation.
Reconstitution of Nanodiscs
To prepare the lipid stock, soybean polar lipid extract (Soy-PL, Avanti) or Soy-PL:brain PIP2 (Avanti) mixture (1:1 mass ratio) dissolved in chloroform was dried as a thin film in a glass tube using an argon stream. Next, the lipid film was solubilized in 20 mM HEPES pH 7.4 with 0.8% DDM/CHS mixture (DDM:CHS=5:1), resulting in a final concentration of 10 mM lipid as calculated from the average molecular weight of the lipid species.
Protein complex was reconstituted with MSP2N2 and Soy-PL or mixture of Soy-PL and brain PIP2 in a final ratio of 1:3:450, which was prepared in the following manner (Denisov and Sligar, 2016). hKCNQ1EM-CaM or hKCNQ1EM-KCNE3-CaM eluted from the GFP-NB column was mixed with the DDM-CHS solubilized lipid, which was incubated for 30min at 4 degree to form mixed lipid-detergent micelles. Next, MSP2N2 was added to the protein-lipid mixture for another 30min incubation before an equal volume of freshly activated bio-beads SM2 were added to remove the detergent from the system. The resulted mixture was incubated at 4°C overnight with constant rotation. Bio-beads were removed, and the reconstituted mixture was further purified using size-exclusion chromatography with a Superose 6, 10/300 GL column (GE Healthcare) equilibrated with 20 mM HEPES pH7.4, 150 mM KCl and 4 mM EGTA. The peak fractions were pooled, concentrated to 5-6 mg/ml using a 100-kDa MWCO centrifugal device (Ambion). For cryo-EM grid preparation, the concentrated protein sample is mixed with 120 mM Fos-Choline-8 at a volume ratio of 1:50 immediately before freezing.
Electrophysiology
Chinese hamster ovary (CHO) cells, cultured in DMEM-F12 (Gibco) with 10% FBS and L-glutamine, were transfected with the hKCNQ1EM and KCNE3 expression plasmid (1:2 mass ratio) with lipofectamine 3000 reagent (Thermo Fisher Scientific). A medium exchange was carried out after 4hr incubation with lipofectamine reagent. 48 hrs following transfection, the media was replaced with bath solution and experiments were performed at room temperature using whole-cell patch clamp techniques with polished borosilicate glass pipettes with resistance between 2-4 MΩ. All recordings were carried out using pClamp10.5 software (Molecular Devices), an Axopatch 200B amplifier (Molecular Devices), and an Axon digidata 1550 digitizer (Molecular Devices). Data were filtered at 1 kHz and digitized at 10 kHz.
The bath solution contains 10 mM HEPES-Na pH 7.2, 20 mM KCl, 130 mM NaCl, 5 mM glucose, 2 mM MgCl2 and 1 mM CaCl2, and the pipette solution was 10 mM HEPES-Na pH 7.2, 50 mM KF, 100 mM KCl, 5 mM EGTA. Data was processed using Origin 9 (OriginLab Corp.).
Flux assay
Lipid mixtures, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-snglycero-3-phospho-(1'-rac-glycerol) (POPG) at a 3:1 ratio (w/w) or POPE, POPG and PI(4,5)P2 at a 3:1:0.1 ratio were dried under an argon stream, and placed in a vacuum chamber overnight. Dried lipids were suspended by sonication in buffer containing 20 mM Tris pH 8 and 160 mM KCl. DDM was added and mixed with the lipid suspension for 1 hour at room temperature. The final DDM concentration was 10 mM and the lipid concentration 10 mg/ml. Purified channel complex was added to the lipid/DDM mixture at a protein-to-lipid ratio of 1:250 (w/w). After 30-min incubation at room temperature, SM2 biobeads (Bio-Rad) were added to remove DDM by incubating the mixture at 4 °C overni ght.
Proteoliposome vesicles were collected, briefly sonicated and diluted 25 times in buffer containing 20 mM Tris pH 8 and 160 mM NaCl and prior to the assay. 6 μl vesicles was mixed with 6 μl ACMA solution (7.5 μM) and 12 μl buffer (20 mM Tris pH 8 and 160 mM NaCl). Fluorescence was recorded every 5 seconds (λEX = 410 nm, λEM = 490 nm) using a plate reader (Tecan Infinite M1000). After the ACMA fluorescence stabilized, K+ flux was initiated by addition of 6 μl CCCP (40 μM). At the end of the assay, 2 μM valinomycin was added to release K+ from all vesicles and yield a minimum baseline fluorescence.
Cryo-EM analysis
Cryo-EM grids were prepared with a Vitrobot Mark IV (FEI). Quantifoil R1.2/1.3 holey carbon gold grids were glow-discharged for 15s. Then 3.6 μL of 5-6 mg/ml protein sample was pipetted onto the grids, which were blotted for 2s under blot force 1 at 100% humidity and frozen in liquid nitrogen cooled liquid ethane. The grids were loaded onto a 300 keV Titan Krios (FEI) with a K2 direct electron detector (Gatan).
Images of all datasets except the hKCNQ1EM-KCNE3-CaM with PIP2 were recorded with SerialEM (Mastronarde, 2005) in super-resolution mode with a super resolution pixel size of 0.515 Å and a defocus range of −1.1 to −2.4 μm. Data were collected with a dose rate of 8 electrons per physical pixel per second, and images were recorded with a 12.5 s exposure and 250 ms subframes (50 total frames). The hKCNQ1EM-KCNE3-CaM with PIP2 was collected on Krios #2 equipped with a Cs corrector, energy filter and K2 detector in Simons Electron Microscopy Center, NYSBC. Images were recorded in super-resolution mode with a super resolution pixel size of 0.545 Å and a defocus range of −1.1 to −2.2 μm. Dose rate is set at 8 electrons per physical pixel per second, and images were recorded with a 10 s exposure using 200 ms subframes.
Super-resolution image stacks were gain-normalized, binned by 2 with Fourier cropping, and corrected for beam-induced motion using MotionCor2 (Zheng et al., 2017). Contrast transfer function parameters were estimated from motion-corrected summed images without dose-weighting using GCTF (Zhang, 2016). All subsequent processing was performed on motion-corrected summed images with dose weighting. About 3,000 particles were manually picked and processed with reference-free 2D classification in RELION to generate a template (Zivanov et al., 2018), which was used for particle picking using Gautomatch (written by Kai Zhang). Auto-picked particles were visually examined to remove false positives and were further cleaned up by multiple rounds of 2D classification in cryoSPARC (Punjani et al., 2017).
For hKCNQ1EM-CaM and hKCNQ1EM-KCNE3-CaM datasets, ab initio reconstruction was carried out in cryoSPARC asking for three classes. The best class is further refined by cryoSPARC followed with local refinement in RELION. For hKCNQ1EM-KCNE3-CaM with PIP2 dataset, the best classes from cryoSPARC were refined in RELION followed by 3D classification without alignment, resulting in one class with continuous density for all transmembrane helixes. Then, particles from the best class were used for Bayesion polishing (Scheres, 2012) followed by another round 2D classification, 3D refinement and 3D classification without alignment in RELION. Next, the classes with good densities were combined for 3D refinement.
Model building and refinement
Either RELION or auto_sharpen of the Phenix program (Terwilliger et al., 2018) was used for map sharpening. Models were built in Coot (Emsley et al., 2010). First, a homology model was generated using the I-TASSER server (Yang et al., 2015; Yang and Zhang, 2015) and docked into the cryo-EM map. From this starting point manual rebuilding were carried out. The structural model was refined using phenix.real_space_refine (Afonine et al., 2013) with secondary structure restraints and Coot iteratively. Protein structure quality was monitored using the Molprobity server (Chen et al., 2010). The resulting hKCNQ1EM-CaM structure includes KCNQ1 residues 104-218, 223-396 and 506-568 and CaM residues 6-149. The resulting hKCNQ1EM-KCNE3-CaM structure includes KCNQ1 residues 104-218, 222-396 and 506-569, KCNE3 residues 53-99 and CaM residues 6-149. The structure of hKCNQ1EM-KCNE3-CaM with PIP2 includes hKCNQ1 residues 104-218, 225-387, 506-537 and 542-562, KCNE3 residues 53-92 and CaM residues 6-149.The pore radii were calculated using HOLE (Smart et al., 1996). Figures were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC.) and UCSF Chimera (Pettersen et al., 2004).
Quantification and Statistical Analysis
All reported resolutions in cryoEM studies are based upon the 0.143 Fourier Shell Correlation criterion. Error bars in Figure 3A represent standard error of the mean for three independent experiments.
Data and Code Availability
Data Resources
The three-dimensional cryo-EM density maps for hKCNQ1EM-CaM, hKCNQ1EM-KCNE3-CaM and hKCNQ1EM-KCNE3-CaM with PIP2 have been deposited in the EM Database under the accession code EMDB: EMD-20965, EMD-20966 and EMD-20967, and the coordinates for the structure have been deposited in Protein Data Bank under accession code PDB: 6UZZ, 6V00 and 6V01.
Supplementary Material
Figure S1. Structure determination of hKCNQ1EM-KCNE3-CaM complex. Related to Figure 1.
(A) Construct design of KCNQ1, KCNE3 and CaM used for structural studies.
(B) Flowchart of hKCNQ1EM-KCNE3-CaM structure determination. The steps in blue are carried out in cryoSPARC and ones in green in RELION.
(C) FSC of the final map.
(D) Local resolution of the channel complex calculated by Blocres software.
Figure S2. Structural feature of the human KCNQ1 and KCNE3. Related to Figure 2.
(A) A cartoon showing the secondary structure of KCNQ1, CaM and KCNE3.
(B) Comparison of protomers between human and frog KCNQ1-CaM complex
(C) Comparison of KCNQ1 and CaM interactions between frog and human KCNQ1 complexes.
(D) The cytoplasmic interface between KCNE3 and CaM. CaM (with HA and HB helixes) is shown as surface with ribbons and two subunits are colored in green and light blue. The cytoplasmic helix of KCNE3 is colored in red with sidechains of interface residues shown.
(E) KCNE3 induced conformational changes viewed from extracellular side. hKCNQ1-CaM with or without KCNE3 is colored in blue and grey, respectively.
(F) Structural comparison of KCNE3 structure determined by cryo-EM in the presence of KCNQ1 (red) and by NMR without KCNQ1 (grey).
(G) Comparison of the interaction between KCNE1 and KCNE3 to KCNQ1. Top: sequence alignment between KCNE1 and KCNE3. Middle: interaction between KCNE3 and KCNQ1 at the extracellular side. Bottom: crosslinking residue pairs in KCNE1 and their distances (in Å) at homologous positions in the KCNQ1-KCNE3 structure.
Figure S3. Structure determination of hKCNQ1EM-CaM complex. Related to Figure 2.
(A) Flowchart of hKCNQ1EM-CaM structure determination. The steps in blue are carried out in cryoSPARC and ones in green in RELION.
(B) Structural model of hKCNQ1EM-CaM complex.
(C) FSC of the final map
(D) Local resolution of the channel complex calculated by Blocres software.
(E) Left: view of ion conducting pathways for PIP2-free and PIP2-bound states with front and back subunits excluded for clarity. Right: radius of both of hKCNQ1EM-CaM (purple) and hKCNQ1EM-KCNE3-CaM (blue) pore calculated using HOLE program.
Figure S4. Structure determination of hKCNQ1EM-KCNE3-CaM complex in the presence of PIP2. Related to Figure 3.
(A) Flowchart of structure determination for PIP2-bound hKCNQ1EM-KCNE3-CaM complex. The steps in blue are carried out in cryoSPARC and ones in green in RELION.
(B) FSC of the final map.
(C) Local resolution of the final map calculated by Blocres software.
Figure S5. Analysis of the hKCNQ1EM-KCNE3-CaM complex in the presence of PIP2. Related to Figure 5.
(A) PIP2-free hKCNQ1EM-KCNE3-CaM structure (solved in detergent) docked into hKCNQ1EM-KCNE3-CaM map solved in nanodics without PIP2. The dashed box shows the S6-loop-HA helix docked into the cryo-EM map of hKCNQ1EM-KCNE3-CaM from nanodics.
(B) PIP2-bound hKCNQ1EM-KCNE3-CaM structure docked into hKCNQ1EM-CaM map that is determined in nanodiscs with PIP2. KCNE3 is not shown.
(C) 12 of the most populated 2D classes of hKCNQ1EM-KCNE3-CaM with C8-PIP2 in detergent.
(D) The interface between HC helix bundle and CaM viewed from intracellular side. CaM is shown as orange surface and KCNQ1 as blue ribbon.
(E) Sequence alignment of the RQKH motif among KCNQ1 orthologs and paralogs (human KCNQ1, mouse KCNQ1, frog KCNQ1, human KCNQ2-5).
Movie S1. PIP2 induced conformational changes in a protomer. Related to Figure 5.
Movie S2. Pore dilation of the KCNQ1 channel induced by PIP2. Related to Figure 5.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM/F-12 medium | GIBCO | Cat# 11330-032 |
| SF-900 II SFM medium | GIBCO | Cat# 10902-088 |
| L-Glutamine (100x) | GIBCO | Cat# 25030-081 |
| Pen Strep | GIBCO | Cat# 15140-122 |
| Grace’s insect medium | GIBCO | Cat# 11605-094 |
| Freestyle 293 medium | GIBCO | Cat# 12338-018 |
| Fetal bovine serum | GIBCO | Cat# 16000-044 |
| Lipofectamine 3000 transfection reagent | Invitrogen | Cat# 11668019 |
| Cellfectin II reagent | Invitrogen | Cat# 10362100 |
| Lauryl Maltose Neopentyl Glycol (LMNG) | Anatrace | Cat# NG310 |
| Cholesteryl hemisuccinate | Anatrace | Cat# CH210 |
| Digitonin | Millipore Sigma | Cat# 300410 |
| Critical Commercial Assays | ||
| CNBR-activated sepharose beads | GE Healthcare | Cat# 17-0430-01 |
| Superose 6, 10/300 GL | GE Healthcare | Cat# 17-5172-01 |
| Deposited Data | ||
| hKCNQ1EM-CaM density map | This study | EMD-20965 |
| hKCNQ1EM-CaM atomic model | This study | PDB6UZZ |
| hKCNQ1EM-KCNE3-CaM density map | This study | EMD-20966 |
| hKCNQ1EM-KCNE3-CaM atomic model | This study | PDB 6V00 |
| hKCNQ1EM-KCNE3-CaM with PIP2 density map | This study | EMD-20967 |
| hKCNQ1EM-KCNE3-CaM with PIP2 atomic model | This study | PDB 6V01 |
| Experimental Models: Cell Lines | ||
| CHO-K1 | ATCC | Cat# CRL-9618 |
| Sf9 | ATCC | Cat# CRL-1711 |
| HEK293S GnTI− | ATCC | Cat# CRL-3022 |
| Recombinant DNA | ||
| Synthesized human KCNQ1 cDNA | Genewiz. Inc | N/A |
| Synthesized Calmodulin cDNA | Genewiz. Inc | N/A |
| Synthesized KCNE3 cDNA | Genewiz. Inc | N/A |
| Software and Algorithms | ||
| SerialEM | Mastronarde, 2005 | http://bio3d.colorado.edu/SerialEM |
| RELION | Scheres, 2012 | http://www2.mrc-lmb.cam.ac.uk/relion |
| EMAN2 | Tang et al., 2007 | http://blake.bcm.edu/emanwiki/EMAN2 |
| Blocres | Cardone et al., 2013 | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera |
| PHENIX | Afonine et al., 2013 | https://www.phenix-online.org |
| COOT | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| HOLE | Smart et al., 1996 | http://www.holeprogram.org/ |
| PyMOL | Molecular Graphics System, Version 1.8 Schrodinger, LLC | http://www.pymol.org |
| Other | ||
| Quantifoil R1.2/1.3 400 mesh gold holey carbon grids | SPI supplies | 4240G-XA |
Highlights:
Structures of human KCNQ1-CaM and KCNQ1-KCNE3-CaM complexes without PIP2
Structural mechanism of KCNQ1 modulation by KCNE3
Structure of human KCNQ1-KCNE3-CaM with PIP2
Gating mechanism of KCNQ1 by PIP2
Acknowledgments
We thank M. Ebrahim and J. Sotiris at The Rockefeller University Evelyn Gruss Lipper Cryo-electron Microscopy Resource Center and L.Yen at the New York Structural Biology Center for help with cryo-EM data collection, Y. C. Hsiung for help with mammalian cell culture, members of the MacKinnon lab for helpful discussions, and C. Zhao and G. Mitta for critical reading of the manuscript. J.S. is funded by NIH (HL143037). This work was also supported by NIH (GM43949). R.M. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no conflict of interest.
References
- Abbott GW, Tai KK, Neverisky DL, Hansler A, Hu Z, Roepke TK, Lerner DJ, Chen Q, Liu L, Zupan B, et al. (2014). KCNQ1, KCNE2, and Na+-coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability. Sci Signal 7, ra22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Adams PD, and Urzhumtsev A (2013). Bulk-solvent and overall scaling revisited: faster calculations, improved results. Acta Crystallogr D Biol Crystallogr 69, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hazza A, Linley J, Aziz Q, Hunter M, and Sandle G (2016). Upregulation of basolateral small conductance potassium channels (KCNQ1/KCNE3) in ulcerative colitis. Biochem Biophys Res Commun 470, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aromolaran AS, Subramanyam P, Chang DD, Kobertz WR, and Colecraft HM (2014). LQT1 mutations in KCNQ1 C-terminus assembly domain suppress IKs using different mechanisms. Cardiovasc Res 104, 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, and Romey G (1996). K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 384, 78–80. [DOI] [PubMed] [Google Scholar]
- Barro-Soria R, Perez ME, and Larsson HP (2015). KCNE3 acts by promoting voltage sensor activation in KCNQ1. Proc Natl Acad Sci U S A 112, E7286–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro-Soria R, Ramentol R, Liin SI, Perez ME, Kass RS, and Larsson HP (2017). KCNE1 and KCNE3 modulate KCNQ1 channels by affecting different gating transitions. Proc Natl Acad Sci U S A 114, E7367–E7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet IR, Labro AJ, Raes AL, and Snyders DJ (2007). Role of the S6 C-terminus in KCNQ1 channel gating. J Physiol 585, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, and Abbott GW (2010). A shared mechanism for lipid- and beta-subunit-coordinated stabilization of the activated K+ channel voltage sensor. FASEB J 24, 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choveau FS, De la Rosa V, Bierbower SM, Hernandez CC, and Shapiro MS (2018). Phosphatidylinositol 4,5-bisphosphate (PIP2) regulates KCNQ3 K(+) channels by interacting with four cytoplasmic channel domains. J Biol Chem 293, 19411–19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung DY, Chan PJ, Bankston JR, Yang L, Liu G, Marx SO, Karlin A, and Kass RS (2009). Location of KCNE1 relative to KCNQ1 in the I(KS) potassium channel by disulfide cross-linking of substituted cysteines. Proc Natl Acad Sci U S A 106, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, and Sligar SG (2016). Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckey K, Wrobel E, Strutz-Seebohm N, Pott L, Schmitt N, and Seebohm G (2014). Novel Kv7.1-phosphatidylinositol 4,5-bisphosphate interaction sites uncovered by charge neutralization scanning. J Biol Chem 289, 22749–22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Xiong Q, Sun H, and Li M (2008). Desensitization of chemical activation by auxiliary subunits: convergence of molecular determinants critical for augmenting KCNQ1 potassium channels. J Biol Chem 283, 22649–22658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Nunziato DA, and Pitt GS (2006). KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ Res 98, 1048–1054. [DOI] [PubMed] [Google Scholar]
- Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, and Gouaux E (2014). Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahammer F, Herling AW, Lang HJ, Schmitt-Graff A, Wittekindt OH, Nitschke R, Bleich M, Barhanin J, and Warth R (2001). The cardiac K+ channel KCNQ1 is essential for gastric acid secretion. Gastroenterology 120, 1363–1371. [DOI] [PubMed] [Google Scholar]
- Grunnet M, Jespersen T, Rasmussen HB, Ljungstrom T, Jorgensen NK, Olesen SP, and Klaerke DA (2002). KCNE4 is an inhibitory subunit to the KCNQ1 channel. J Physiol 542, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SB, Tao X, and MacKinnon R (2011). Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, and MacKinnon R (2017). Structural Titration of Slo2.2, a Na(+)-Dependent K(+) Channel. Cell 168, 390–399 e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite RK, Tao X, and MacKinnon R (2017). Structural basis for gating the high-conductance Ca(2+)-activated K(+) channel. Nature 541, 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen T, Grunnet M, and Olesen SP (2005). The KCNQ1 potassium channel: from gene to physiological function. Physiology (Bethesda) 20, 408–416. [DOI] [PubMed] [Google Scholar]
- Julio-Kalajzic F, Villanueva S, Burgos J, Ojeda M, Cid LP, Jentsch TJ, and Sepulveda FV (2018). K2P TASK-2 and KCNQ1-KCNE3 K(+) channels are major players contributing to intestinal anion and fluid secretion. J Physiol 596, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimova MA, Zaydman MA, Cui J, and Tarek M (2015). PIP(2)-dependent coupling is prominent in Kv7.1 due to weakened interactions between S4-S5 and S6. Sci Rep 5, 7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. (2010). Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–138. [DOI] [PubMed] [Google Scholar]
- Kroncke BM, Van Horn WD, Smith J, Kang C, Welch RC, Song Y, Nannemann DP, Taylor KC, Sisco NJ, George AL Jr., et al. (2016). Structural basis for KCNE3 modulation of potassium recycling in epithelia. Sci Adv 2, e1501228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, and MacKinnon R (2018). Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures. Science 360, 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zaydman MA, Wu D, Shi J, Guan M, Virgin-Downey B, and Cui J (2011). KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc Natl Acad Sci U S A 108, 9095–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loussouarn G, Park KH, Bellocq C, Baro I, Charpentier F, and Escande D (2003). Phosphatidylinositol-4,5-bisphosphate, PIP2, controls KCNQ1/KCNE1 voltage-gated potassium channels: a functional homology between voltage-gated and inward rectifier K+ channels. EMBO J 22, 5412–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manderfield LJ, and George AL Jr. (2008). KCNE4 can co-associate with the I(Ks) (KCNQ1-KCNE1) channel complex. FEBS J 275, 1336–1349. [DOI] [PubMed] [Google Scholar]
- Mazhari R, Nuss HB, Armoundas AA, Winslow RL, and Marban E (2002). Ectopic expression of KCNE3 accelerates cardiac repolarization and abbreviates the QT interval. J Clin Invest 109, 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrossan ZA, and Abbott GW (2004). The MinK-related peptides. Neuropharmacology 47, 787–821. [DOI] [PubMed] [Google Scholar]
- Melman YF, Domenech A, de la Luna S, and McDonald TV (2001). Structural determinants of KvLQT1 control by the KCNE family of proteins. J Biol Chem 276, 6439–6444. [DOI] [PubMed] [Google Scholar]
- Melman YF, Krummerman A, and McDonald TV (2002). KCNE regulation of KvLQTI channels: structure-function correlates. Trends Cardiovasc Med 12, 182–187. [DOI] [PubMed] [Google Scholar]
- Nakano Y, and Shimizu W (2016). Genetics of long-QT syndrome. J Hum Genet 61, 51–55. [DOI] [PubMed] [Google Scholar]
- Nath A, Atkins WM, and Sligar SG (2007). Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069. [DOI] [PubMed] [Google Scholar]
- Park E, and MacKinnon R (2018). Structure of the CLC-1 chloride channel from Homo sapiens. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peroz D, Rodriguez N, Choveau F, Baro I, Merot J, and Loussouarn G (2008). Kv7.1 (KCNQ1) properties and channelopathies. J Physiol 586, 1785–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, and Mathews FS (1966). An x-ray study of azide methaemoglobin. J Mol Biol 21, 199–202. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Preston P, Wartosch L, Gunzel D, Fromm M, Kongsuphol P, Ousingsawat J, Kunzelmann K, Barhanin J, Warth R, and Jentsch TJ (2010). Disruption of the K+ channel beta-subunit KCNE3 reveals an important role in intestinal and tracheal Cl− transport. J Biol Chem 285, 7165–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Roura-Ferrer M, Sole L, Oliveras A, Dahan R, Bielanska J, Villarroel A, Comes N, and Felipe A (2010). Impact of KCNE subunits on KCNQ1 (Kv7.1) channel membrane surface targeting. J Cell Physiol 225, 692–700. [DOI] [PubMed] [Google Scholar]
- Sachyani D, Dvir M, Strulovich R, Tria G, Tobelaim W, Peretz A, Pongs O, Svergun D, Attali B, and Hirsch JA (2014). Structural basis of a Kv7.1 potassium channel gating module: studies of the intracellular c-terminal domain in complex with calmodulin. Structure 22, 1582–1594. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, and Keating MT (1996). Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature 384, 80–83. [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Waldegger S, Fehr S, Bleich M, Warth R, Greger R, and Jentsch TJ (2000). A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature 403, 196–199. [DOI] [PubMed] [Google Scholar]
- Seebohm G, Strutz-Seebohm N, Birkin R, Dell G, Bucci C, Spinosa MR, Baltaev R, Mack AF, Korniychuk G, Choudhury A, et al. (2007). Regulation of endocytic recycling of KCNQ1/KCNE1 potassium channels. Circ Res 100, 686–692. [DOI] [PubMed] [Google Scholar]
- Shamgar L, Ma L, Schmitt N, Haitin Y, Peretz A, Wiener R, Hirsch J, Pongs O, and Attali B (2006). Calmodulin is essential for cardiac IKS channel gating and assembly: impaired function in long-QT mutations. Circ Res 98, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376. [DOI] [PubMed] [Google Scholar]
- Su Z, Brown EC, Wang W, and MacKinnon R (2016). Novel cell-free high-throughput screening method for pharmacological tools targeting K+ channels. Proc Natl Acad Sci U S A 113, 5748–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, and MacKinnon R (2017). Cryo-EM Structure of a KCNQ1/CaM Complex Reveals Insights into Congenital Long QT Syndrome. Cell 169, 1042–1050 e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi M, Kurisaki I, and Nagaoka M (2013). Oxygen entry through multiple pathways in T-state human hemoglobin. J Phys Chem B 117, 6082–6091. [DOI] [PubMed] [Google Scholar]
- Telezhkin V, Thomas AM, Harmer SC, Tinker A, and Brown DA (2013). A basic residue in the proximal C-terminus is necessary for efficient activation of the M-channel subunit Kv7.2 by PI(4,5)P(2). Pflugers Arch 465, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Sobolev OV, Afonine PV, and Adams PD (2018). Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D Struct Biol 74, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenod F (2002). Ion channels in secretory granules of the pancreas and their role in exocytosis and release of secretory proteins. Am J Physiol Cell Physiol 283, C651–672. [DOI] [PubMed] [Google Scholar]
- Tobelaim WS, Dvir M, Lebel G, Cui M, Buki T, Peretz A, Marom M, Haitin Y, Logothetis DE, Hirsch JA, et al. (2017). Competition of calcified calmodulin N lobe and PIP2 to an LQT mutation site in Kv7.1 channel. Proc Natl Acad Sci U S A 114, E869–E878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn WD, Vanoye CG, and Sanders CR (2011). Working model for the structural basis for KCNE1 modulation of the KCNQ1 potassium channel. Curr Opin Struct Biol 21, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth R, and Bleich M (2000). K+ channels and colonic function. Rev Physiol Biochem Pharmacol 140, 1–62. [DOI] [PubMed] [Google Scholar]
- Whorton MR, and MacKinnon R (2013). X-ray structure of the mammalian GIRK2- betagamma G-protein complex. Nature 498, 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015). The I-TASSER Suite: protein structure and function prediction. Nat Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, and Zhang Y (2015). I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43, W174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaydman MA, and Cui J (2014). PIP2 regulation of KCNQ channels: biophysical and molecular mechanisms for lipid modulation of voltage-dependent gating. Front Physiol 5, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaydman MA, Silva JR, Delaloye K, Li Y, Liang H, Larsson HP, Shi J, and Cui J (2013). Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc Natl Acad Sci U S A 110, 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Verlander JW, Lynch IJ, Cash M, Shao J, Stow LR, Cain BD, Weiner ID, Wall SM, and Wingo CS (2007). Cellular distribution of the potassium channel KCNQ1 in normal mouse kidney. Am J Physiol Renal Physiol 292, F456–466. [DOI] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Structure determination of hKCNQ1EM-KCNE3-CaM complex. Related to Figure 1.
(A) Construct design of KCNQ1, KCNE3 and CaM used for structural studies.
(B) Flowchart of hKCNQ1EM-KCNE3-CaM structure determination. The steps in blue are carried out in cryoSPARC and ones in green in RELION.
(C) FSC of the final map.
(D) Local resolution of the channel complex calculated by Blocres software.
Figure S2. Structural feature of the human KCNQ1 and KCNE3. Related to Figure 2.
(A) A cartoon showing the secondary structure of KCNQ1, CaM and KCNE3.
(B) Comparison of protomers between human and frog KCNQ1-CaM complex
(C) Comparison of KCNQ1 and CaM interactions between frog and human KCNQ1 complexes.
(D) The cytoplasmic interface between KCNE3 and CaM. CaM (with HA and HB helixes) is shown as surface with ribbons and two subunits are colored in green and light blue. The cytoplasmic helix of KCNE3 is colored in red with sidechains of interface residues shown.
(E) KCNE3 induced conformational changes viewed from extracellular side. hKCNQ1-CaM with or without KCNE3 is colored in blue and grey, respectively.
(F) Structural comparison of KCNE3 structure determined by cryo-EM in the presence of KCNQ1 (red) and by NMR without KCNQ1 (grey).
(G) Comparison of the interaction between KCNE1 and KCNE3 to KCNQ1. Top: sequence alignment between KCNE1 and KCNE3. Middle: interaction between KCNE3 and KCNQ1 at the extracellular side. Bottom: crosslinking residue pairs in KCNE1 and their distances (in Å) at homologous positions in the KCNQ1-KCNE3 structure.
Figure S3. Structure determination of hKCNQ1EM-CaM complex. Related to Figure 2.
(A) Flowchart of hKCNQ1EM-CaM structure determination. The steps in blue are carried out in cryoSPARC and ones in green in RELION.
(B) Structural model of hKCNQ1EM-CaM complex.
(C) FSC of the final map
(D) Local resolution of the channel complex calculated by Blocres software.
(E) Left: view of ion conducting pathways for PIP2-free and PIP2-bound states with front and back subunits excluded for clarity. Right: radius of both of hKCNQ1EM-CaM (purple) and hKCNQ1EM-KCNE3-CaM (blue) pore calculated using HOLE program.
Figure S4. Structure determination of hKCNQ1EM-KCNE3-CaM complex in the presence of PIP2. Related to Figure 3.
(A) Flowchart of structure determination for PIP2-bound hKCNQ1EM-KCNE3-CaM complex. The steps in blue are carried out in cryoSPARC and ones in green in RELION.
(B) FSC of the final map.
(C) Local resolution of the final map calculated by Blocres software.
Figure S5. Analysis of the hKCNQ1EM-KCNE3-CaM complex in the presence of PIP2. Related to Figure 5.
(A) PIP2-free hKCNQ1EM-KCNE3-CaM structure (solved in detergent) docked into hKCNQ1EM-KCNE3-CaM map solved in nanodics without PIP2. The dashed box shows the S6-loop-HA helix docked into the cryo-EM map of hKCNQ1EM-KCNE3-CaM from nanodics.
(B) PIP2-bound hKCNQ1EM-KCNE3-CaM structure docked into hKCNQ1EM-CaM map that is determined in nanodiscs with PIP2. KCNE3 is not shown.
(C) 12 of the most populated 2D classes of hKCNQ1EM-KCNE3-CaM with C8-PIP2 in detergent.
(D) The interface between HC helix bundle and CaM viewed from intracellular side. CaM is shown as orange surface and KCNQ1 as blue ribbon.
(E) Sequence alignment of the RQKH motif among KCNQ1 orthologs and paralogs (human KCNQ1, mouse KCNQ1, frog KCNQ1, human KCNQ2-5).
Movie S1. PIP2 induced conformational changes in a protomer. Related to Figure 5.
Movie S2. Pore dilation of the KCNQ1 channel induced by PIP2. Related to Figure 5.
Data Availability Statement
Data Resources
The three-dimensional cryo-EM density maps for hKCNQ1EM-CaM, hKCNQ1EM-KCNE3-CaM and hKCNQ1EM-KCNE3-CaM with PIP2 have been deposited in the EM Database under the accession code EMDB: EMD-20965, EMD-20966 and EMD-20967, and the coordinates for the structure have been deposited in Protein Data Bank under accession code PDB: 6UZZ, 6V00 and 6V01.