Significance
Stomatal cell fate and patterning, which are regulated by key transcriptional factors and intercellular communications, are critical for plant growth and survival. The known regulators of stomatal development do not appear to have microRNAs (miRNAs) regulating them. Thus, it remains elusive as to whether and how miRNAs are involved in stomatal development. This study identifies stomatal lineage miRNAs including developmental stage-specific miRNAs. Genetic analysis shows that stomatal lineage miRNAs positively or negatively regulate stomatal formation and patterning. Moreover, biological processes modulated by stomatal lineage miRNAs reveal previously unknown regulatory pathways in stomatal development, indicating that miRNAs function as a critical element of stomatal development. These results provide a resource for guiding the study of stomatal development.
Keywords: PHO2, stomatal development, stomatal lineage miRNA
Abstract
Stomata in the plant epidermis play a critical role in growth and survival by controlling gas exchange, transpiration, and immunity to pathogens. Plants modulate stomatal cell fate and patterning through key transcriptional factors and signaling pathways. MicroRNAs (miRNAs) are known to contribute to developmental plasticity in multicellular organisms; however, no miRNAs appear to target the known regulators of stomatal development. It remains unclear as to whether miRNAs are involved in stomatal development. Here, we report highly dynamic, developmentally stage-specific miRNA expression profiles from stomatal lineage cells. We demonstrate that stomatal lineage miRNAs positively and negatively regulate stomatal formation and patterning to avoid clustered stomata. Target prediction of stomatal lineage miRNAs implicates potential cellular processes in stomatal development. We show that miR399-mediated PHO2 regulation, involved in phosphate homeostasis, contributes to the control of stomatal development. Our study demonstrates that miRNAs constitute a critical component in the regulatory mechanisms controlling stomatal development.
Control of cell lineage and patterning plays a crucial role in the development of multicellular organisms (1–3). Transcription factors can act as master modulators of cell fate specification (2, 4). Environmental factors, including positional cues and neighboring cells, have also been shown to affect cell fate during development (5, 6), indicating developmental flexibility and regulatory complexity in cellular decision making. Genetic reprogramming, including epigenetic regulation and posttranslational modification, consists of multilayers of control and plays a crucial role in the development of cell lineage and patterning (7, 8).
Stomata are microscopic pores formed by a pair of guard cells (GCs) on the plant epidermis. They govern gas exchange and water loss between plants and the atmosphere. Stomatal functions are tightly regulated as they are critical for photosynthesis and responses to environmental changes. They also play a role in the global ecosystem, affecting atmospheric carbon levels and the global water cycle (9). Stomatal density and distribution affect the functional efficiency of stomata; thus, stomatal development is strictly controlled by developmental and environmental cues to ensure precise stomatal lineage and patterning. For instance, pathogen infection and high temperature reduce stomatal density (10, 11).
Stomatal stem cells undergo a series of asymmetric and symmetric cell divisions to form mature GCs. Meristemoid mother cells (MMCs) undergo asymmetric division to produce meristemoids (stomatal entry) that can then undergo additional asymmetric divisions before developing into guard mother cells (GMCs) (commitment). GMCs undergo a single symmetric cell division, and the resultant GCs then differentiate (differentiation) (SI Appendix, Fig. S1A). The stomatal lineage is sequentially regulated by three basic helix–loop–helix (bHLH) transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA (12). Stomatal patterning is regulated by intracellular and intercellular communications involving a mitogen-activated protein (MAP) kinase pathway and small peptide/ligand signaling (13). In addition, environmental cues are integrated into the developmental program, modulating stomatal cell fate and patterning and aiding in environmental adaptation (11, 14).
MicroRNAs (miRNAs) are 21- to 24-nucleotide (nt), small noncoding RNAs that posttranscriptionally regulate gene expression (15, 16). Primary miRNA transcripts are processed via premiRNAs into mature miRNAs through sequential cleavages by the DICER-LIKE1 (DCL1) protein complex. Mature miRNAs are methylated by HUA ENHANCER1 (HEN1) and loaded into ARGONAUTE (AGO) in the RNA-induced silencing complex (RISC), which in turn represses the target gene(s) through mRNA cleavage and/or translation inhibition in a sequence-specific manner (17). Genetic and miRNA profiling analyses have revealed that cell type-specific miRNAs are implicated in animal development including cell lineage and patterning (18, 19). In plants, studies have identified cell type-specific miRNAs that regulate embryogenesis and root development (20, 21), and miRNA-deficient mutants, such as dcl1 and ago1, display changes in stomatal density and patterning, suggesting that miRNAs play a role in stomatal development (22, 23). However, miRNAs have not been identified that target the known regulators of stomatal lineage control.
Here, we report the profiles of developmental stage-specific miRNAs and their predicted targets in stomatal lineage cells. We have developed a transgenic system in which GFP-slicer–defective AGO1 (GFP-AGO1DAH) was expressed in Arabidopsis using the promoters of the stomatal stage-specific marker genes, thereby allowing for the isolation of AGO1-associated miRNAs in a developmental stage-specific manner. Small RNA-sequencing (RNA-seq) analysis has revealed the dynamic expression patterns of these miRNAs during stomatal development. The predicted target genes serve as a resource for guiding the study of stomatal development. In addition, we show that miR399-mediated PHO2 regulation, involved in phosphate homeostasis, also contributes to the control of stomatal development. Overall, our results show that miRNAs play a crucial role in regulating stomatal development, contributing to developmental robustness and plasticity.
Results
A System for miRNA Profiling in Stomatal Lineage Cells.
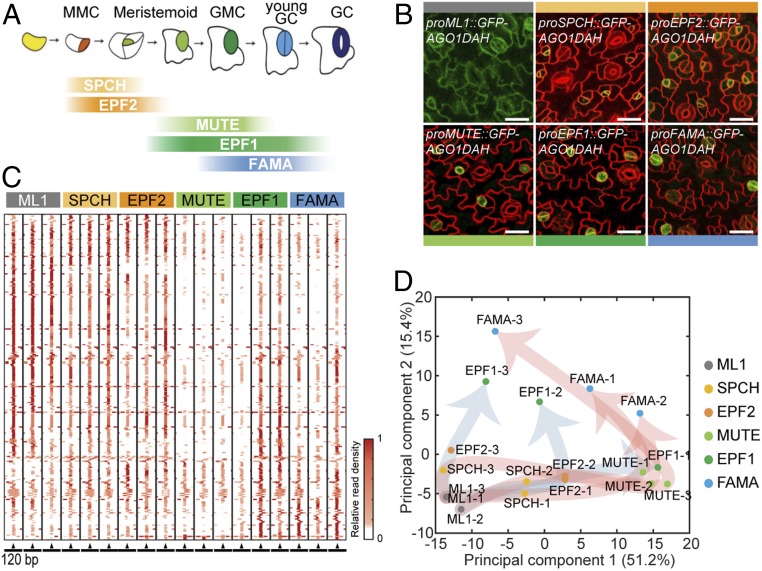
To conduct expression profiling of developmental stage-specific miRNAs in stomatal lineage cells, we expressed AGO1 fused to GFP (GFP-AGO1) in Arabidopsis using the promoters of the stomatal stage-specific marker genes SPCH, MUTE, FAMA, EPF1, and EPF2. The three bHLH transcription factors SPCH, MUTE, and FAMA are expressed predominantly in the stomatal lineage in a developmental stage-specific manner, regulating sequential stomatal cell fate transitions; stomatal entry, commitment, and differentiation, respectively (Fig. 1A and SI Appendix, Fig. S1A). EPF2 and EPF1 are peptide ligands expressed in stomatal lineage cells that regulate stomatal patterning (Fig. 1A and SI Appendix, Fig. S1A) (24, 25). For comparison, we used the ML1 promoter that drives expression in all epidermal cells (26). For effective isolation of AGO1–miRNA complexes, we used AGO1DAH, a slicer-defective AGO1 that stabilizes the association of AGO1 with miRNA but does not affect the maturation and binding specificity of miRNAs to AGO1 (27).
Fig. 1.
Profiling of stomatal lineage miRNAs. (A) A diagram showing an expression window of each developmental stage-specific marker in the stomatal lineage. (B) Confocal images of transgenic plants expressing GFP-AGO1DAH under the control of the promoters of the marker genes. AGO1DAH from epidermal and GCs was immunoprecipitated to isolate stomatal lineage miRNAs associated with AGO1. Promoters used: ML1 (epidermal cells), SPCH (MMC, Meristemoid), EPF2 (Meristemoid), MUTE (Meristemoid, GMC), EPF1 (GMC, young GC), and FAMA (GMC, young GC). Cell outlines are visualized using FM4-64. (Scale bars, 20 μm.) (C) The heatmap of MIR gene sequences shows the position of AGO1-associated 21-nt small RNAs isolated from stomatal lineage cells, at each developmental stage, harboring AGO1DAH driven by the promoter of the marker genes. MIR genomic sequences are presented in the 120-bp window with mature miRNA sequences placed in the center (arrowhead). The abundance of aligned reads was normalized to the total mapped reads in the individual samples and further normalized to the maximum abundance among 120-bp window across the six stages. (D) Principal-component analysis (PCA) of 266 AGO1-associated miRNAs in stomatal lineage cells indicates divergence in the miRNA populations of the SPCH–MUTE–FAMA (red arrows) and EPF2–EPF1 (blue arrows) paths.
GFP expression patterns in transgenic plant harboring each promoter::GFP-AGO1DAH construct were validated by confocal microscopy (Fig. 1B and SI Appendix, Fig. S1 B–F) and were consistent with previous findings (Fig. 1A) (24, 25, 28–30). proSPCH::GFP-AGO1DAH was mainly detected in MMCs and initial meristemoids (SI Appendix, Fig. S1B). proMUTE::GFP-AGO1DAH was detected in meristemoids that had undergone asymmetric division and, at lower levels, in young GMCs (SI Appendix, Fig. S1C). The expression of proFAMA::GFP-AGO1DAH was restricted to GMCs and young GCs (SI Appendix, Fig. S1D). proEPF2::GFP-AGO1DAH was detected in MMCs and their early descendants (SI Appendix, Fig. S1E), and proEPF1::GFP-AGO1DAH in late meristemoids, GMCs, and young GCs (SI Appendix, Fig. S1F). There were no changes in the accumulation levels of endogenous miRNAs in GFP-AGO1DAH plants, suggesting that its introduction not affect the production of endogenous miRNAs (SI Appendix, Fig. S2). These results indicate that the molecular constructs were suitable for the isolation of stomatal lineage miRNAs.
AGO1-Associated miRNA Profiles of Stomatal Lineage Cells.
After immunoprecipitation of GFP-AGO1DAH from each transgenic line followed by small RNA extraction, we performed small RNA-seq analysis. The GFP-AGO1DAH immunoprecipitates were verified by Western blot using anti-GFP antibody (SI Appendix, Fig. S3A). Small RNA-seq of each sample yielded 10.4 million total reads on average. Reads between 15 and 48 nt were analyzed further: 39.2% were 21 nt long, typical of AGO1-associated small RNAs (SI Appendix, Fig. S3B). Alignment of these reads to the Arabidopsis genome revealed that of the 427 annotated MIR gene sequences, 266 (62.3%) were found to be expressed the cells at one or more stomatal developmental stage. To assess the nature of the small RNAs aligned to the MIR genes, we examined where the reads accumulated. Within a 120-bp window centered on the 266 MIR genomic sequences, the majority of the small RNAs were found to accumulate at the center of the window where the mature miRNA sequences are positioned (Fig. 1C), suggesting that the RNAs represent mature miRNAs that are presumably functional in RISC.
To compare the miRNA profiles among lineage-specific cells, we performed principal-component analysis (PCA) and found a clear separation between the MUTE–FAMA and EPF2–EPF1 stages (Fig. 1D). Stomatal lineage cells at the SPCH stage are precursors to those at the MUTE stage (29), suggesting that the SPCH–MUTE–FAMA and EPF2–EPF1 stages possess distinct miRNA profiles to a certain extent. Moreover, miRNA profiles at the FAMA and EPF1 stages are more heterogeneous compared to those at the SPCH, EPF2, and MUTE stages, as indicated by the larger variability among the replicates of the FAMA or EPF1 stage (Fig. 1D). The mRNA profiles at the SPCH stage were previously shown to be more heterogeneous compared to those at FAMA and MUTE stages (2), suggesting there may be distinct mRNA and miRNA regulatory programs in stomatal lineages.
Stomatal Lineage miRNA Dynamics.
Dynamic expression of miRNAs during lineage progression may contribute to the regulation of cell fate and patterning. We therefore analyzed miRNAs that are differentially expressed (DE miRNAs) in at least at one stage of stomatal development (SPCH, MUTE, FAMA, EPF2, and/or EPF1 stages) and identified 224 DE miRNAs (Dataset S1). We assessed the expression of the DE miRNAs during the SPCH–MUTE–FAMA and EPF2–EPF1 developmental paths, classifying the DE miRNAs into three major groups. Two groups displayed higher expression during stomatal entry (SPCH and/or EPF2 stage) or stomatal differentiation (FAMA and/or EPF1 stage) in the two development paths (Fig. 2A). The third group of DE miRNAs showed either higher or lower expression during stomatal commitment (MUTE stage), compared with the SPCH and FAMA stages.
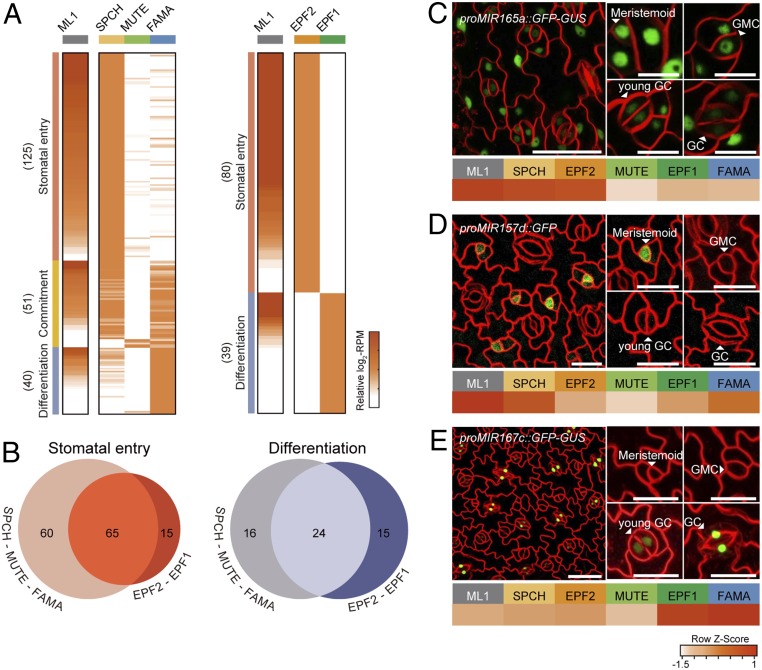
Fig. 2.
miRNA expression profiles in the SPCH–MUTE–FAMA and EPF2–EPF1 paths during stomatal development. (A) The heatmaps show clusters of DE miRNAs expressed in the stomatal lineage cells. DE miRNAs of the SPCH–MUTE–FAMA (Left) and EPF2–EPF2 (Right) paths were grouped into three developmental stages (stomatal entry, differentiation, and commitment) and two developmental stages (stomatal entry and differentiation), respectively. The color bars of the heatmaps represent the gradient scale of relative log2-RPM values for each DE miRNA, which was normalized to the minimum and maximum log2-RPMs across SPCH, MUTE, and FAMA stages (A, Left) as well as EPF2 and EPF1 stages (A, Right). The numbers in parentheses indicate number of miRNAs at each stage. (B) Venn diagrams show overlapping and distinct DE miRNAs between the SPCH–MUTE–FAMA and EPF2–EPF1 paths at the stomatal entry or differentiation stage. (C–E) The heatmaps show the expression levels of miR165a-3p (C), miR157d (D), and miR167c-3p (E) in each of the stomatal lineage cells. Representative confocal images of the epidermis of at least three independent proMIR165a::GFP-GUS (C), proMIR157d::GFP (D), and proMIR167c::GFP-GUS (E) transgenic plants. The arrows indicate cells at each stage of stomatal lineage progression. GC, guard cell; GMC, guard mother cell. Cell outlines are visualized by FM4-64. (Scale bars, 20 μm.)
Approximately two-thirds of the DE miRNAs in each path belonged to the stomatal entry group (Fig. 2A), among which 65 miRNAs overlapped between the 125 (52%) and 80 (81.3%) DE miRNAs from the SPCH–MUTE–FAMA and EPF2–EPF1 paths, respectively (Fig. 2 B, Left). This overlap is consistent with the high similarity in miRNA profiles between the SPCH and EPF2 stages, revealed by PCA (Fig. 1D). In the stomatal differentiation group, 24 miRNAs overlapped between the 40 (60%) and 39 (61.5%) DE miRNAs from the two paths, respectively (Fig. 2 B, Right). These results indicate that miRNA expression patterns are more distinct during differentiation than the stomatal entry stage.
Validation of Stage-Specific miRNAs.
To validate DE miRNA expression in the three major groups, we generated transgenic plants in which GFP reporters were driven by selected DE miRNA gene promoters. miRNA profiling showed that miR165a and miR157d belong to the stomatal entry group with high expression in the epidermis and at the SPCH stage, which gradually decreases as stomatal development proceeds from the entry to differentiation stages. GFP fluorescence from proMIR165a::GFP-GUS plants was detected in epidermal cells and was reduced in stomatal lineage cells (Fig. 2C and SI Appendix, Fig. S4A), while GFP fluorescence from proMIR157d::GFP plants was stomatal lineage-specific and enriched in meristemoids (Fig. 2D and SI Appendix, Fig. S4B). Confocal imaging of proMIR167c::GFP-GUS plants showed preferential GFP expression at the differentiation stages (EPF1 and FAMA), consistent with miR167c being part of the stomatal differentiation group, based on small RNA-seq data (Fig. 2E and SI Appendix, Fig. S4C). Overall, these results show that miRNAs identified from stomatal lineage cells are dynamically expressed during development.
Developmental miRNAs Regulate Stomatal Development.
To determine whether developmental stage-specific DE miRNAs play a role in stomatal development in vivo, we selected three miRNAs (miR829, miR861, and miR3932) that have not studied for functions in stomatal development, misexpressed them in Arabidopsis plants, and analyzed stomatal phenotypes. miR829 (EPF2 stage in the EPF2–EPF1 path) and miR3932 (SPCH stage in the SPCH–MUTE–FAMA path) appear to be preferentially expressed during stomatal entry (Fig. 3 A and B, Lower), whereas miR861 (EPF1 and FAMA stages in the EPF2–EPF1 and SPCH–MUTE–FAMA paths, respectively) seems to be preferentially expressed during differentiation stages (Fig. 3 C, Lower).
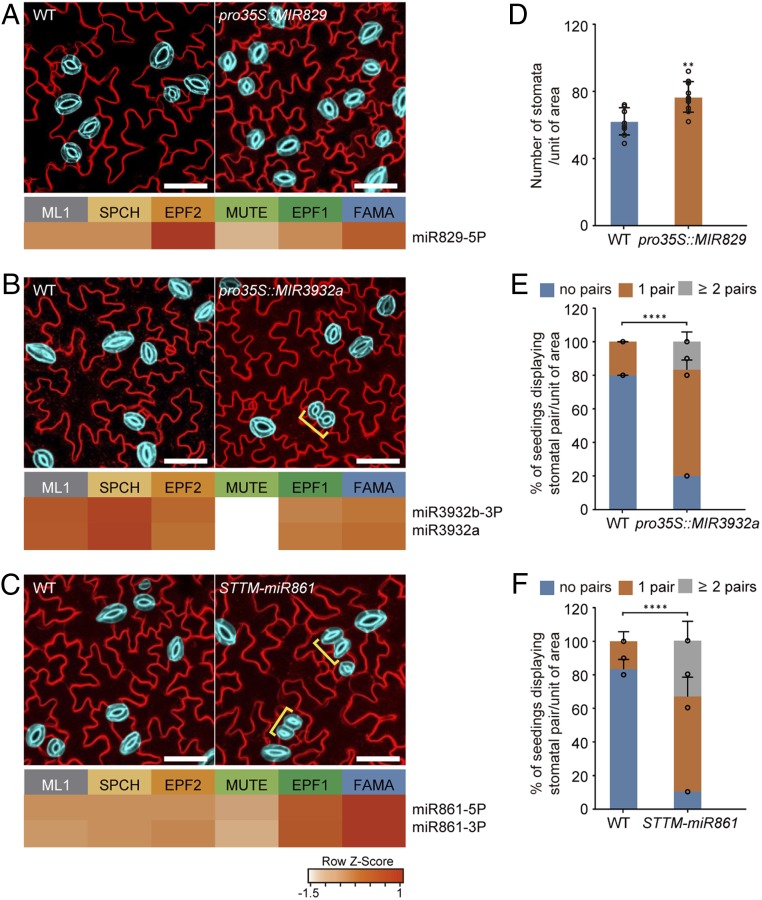
Fig. 3.
Stomatal lineage miRNAs modulate stomatal formation and patterning. (A–C) Stomatal development phenotypes of transgenic plants in which stage-specific miRNAs were overexpressed or down-regulated. The heatmaps show the expression levels of miR829-5p (A), miR3932 (B), and miR861 (C) in the stomatal lineage cells. Representative confocal images of stomata of at least three independent Col-0 (WT), pro35S::MIR829 (A), pro35S::MIR3932a (B), and STTM-miR861 (C) transgenic plants. Mature GCs are highlighted in blue for elucidation, and the brackets indicate stomatal pairs. Cell outlines are visualized by FM4-64. (Scale bar, 50 μm.) (D) Stomatal density in pro35S:MIR829 transgenic plants compared to WT. The number of GCs per unit area (780 × 780 μm2) was scored from at least 10 seedlings for each line. Error bars represent mean ± SD. Two-sided Student’s t test P values; **P < 0.01. (E and F) Numbers of stomatal pairs in pro35S:MIR2932 and STTM-MIR861 plants compared to WT. Percentage of plants having stomatal pairs per area (780 × 780 μm2) in cotyledons of 10-d-old seedlings. Error bars represent mean ± SD calculated from at least 10 seedlings. Two-sided Student’s t test P values; ****P < 0.0001.
Overexpression of miR829 and miR3932 led to altered stomatal development. In pro35S::MIR829 plants, the number of stomata was significantly increased (Fig. 3 A and D and SI Appendix, Figs. S5A and S6A). In contrast, pro35S::MIR3932 plants displayed an increase in the number of stomatal pairs composed of two stomata without changes in stomatal density (Fig. 3 B and E and SI Appendix, Figs. S5B and S6B). Since the overexpressions of the two entry stage-preferential miRNAs, miR829 and miR3932, resulted in distinct stomatal phenotypes, they seem to play differing roles during stomatal development possibly by modulating stomatal fate acquisition or intercellular signaling. Overexpression of miR861 caused no alteration in stomatal development (SI Appendix, Fig. S7 A and B). To inhibit miR861, we used the short tandem target mimic (STTM) strategy, which causes the degradation of target small RNAs (31). STTM-miR861 transgenic plants increased the number of stomatal pairs (Fig. 3 C and F and SI Appendix, Figs. S5C and S6C). Based on the expression pattern, miR861 may suppress a target gene(s) that positively regulates terminal differentiation of stomata and/or symmetric cell division of GCs. Our results suggest that these miRNAs provide an additional layer of regulation in cell fate control and stomatal development.
Target mRNA and Genetic Pathway Predictions.
To predict potential mRNA targets of the DE miRNAs, we used previously reported stomatal lineage transcriptome data to identify genes that are differentially expressed (DEGs) during at least at one stage of stomatal development. For each of the three major groups of DE miRNAs, we selected potential target DEG mRNAs that satisfy the following two criteria: 1) they contain complementary sequences to a corresponding DE miRNA, and 2) they show anticorrelating expression patterns with an miRNA during stomatal lineage progression (r less than −0.5). We identified a total of 868 putative mRNA targets, involving 553 expressed during stomatal entry, 264 during commitment, and 105 during the differentiation (Fig. 4A and Dataset S2).
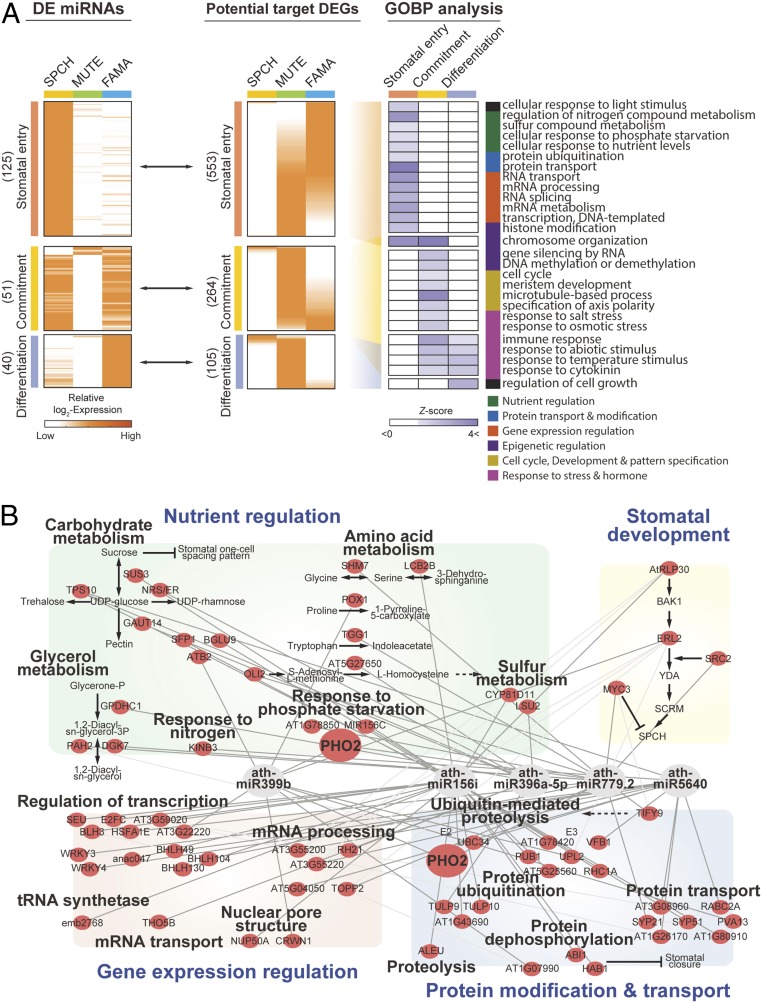
Fig. 4.
Predicted cellular pathways modulated by stomatal lineage miRNAs and anticorrelated DEGs. (A) Anticorrelation between stomatal lineage miRNAs and their predicted target DEGs. GO enrichment-based cellular activities at SPCH, MUTE, and FAMA stages are shown. The numbers in parentheses indicate number of miRNAs at each stage. (B) Regulatory cellular networks consisted of hub DE miRNAs and their predicted target mRNAs at the stomatal entry stage. The gray and light gray lines indicate the predicted miRNA-target gene and known protein–protein interactions, respectively.
We performed an enrichment analysis of gene ontology biological processes (GOBPs) to examine cellular processes associated with the predicted target genes for DE miRNAs in each of the three major groups (Fig. 4A and Dataset S3). The putative target genes of the stomatal entry DE miRNAs were strongly associated with gene expression regulation (mRNA processing and transcription) and protein modification/transport such as transcription factors, histone-modifying enzymes, and protein ubiquitination-related proteins (Fig. 4A and Dataset S4). This result suggests that these DE miRNAs may be involved in stomatal cell fate specification through transcriptional programing and posttranslational modification of key components required for the maintenance of stemness (32). Genes involved in nutrient regulation were also predicted to be modulated by stomatal entry DE miRNAs (Fig. 4A), implying they might contribute to the regulation of stomatal development at the stomatal entry stage. The predicted target genes in the stomatal commitment group were associated with epigenetic regulation (chromatin organization and gene silencing) and development (cell cycle and meristem development) (Fig. 4A and Dataset S4). Since nucleosome-remodeling factors play a critical role in the transition from stem cell state to differentiation (33), miRNAs may play a role in the commitment to GC fate. The predicted target genes for stomatal entry DE miRNAs were associated with light responses that affect the development of stomatal entry cells, whereas those for stomatal differentiation DE miRNAs were associated with defense/temperature responses that regulate stomatal movements (34, 35) (Fig. 4A).
To identify key DE miRNAs, we built a network model describing the regulatory relationship between the DE miRNAs and their putative targets. We identified 10 hub DE miRNAs that putatively regulate a large number of the predicted target genes in the network model (P < 0.01). We focused on the DE miRNAs and their predicted target genes in the stomatal entry group that had the largest number of hub DE miRNAs (i.e., five hubs; Dataset S5). A subnetwork for the stromal entry group describes the regulatory relationship of the five hub miRNAs with the predicted target genes involved in nutrient regulation, gene expression regulation, and protein modification/transport (Fig. 4B). These results identify key miRNA candidates that act in the regulation of stomatal entry, as well as their targets, associated with the regulation of nutrient homeostasis and gene expression. Our results will help develop the current model for stomatal lineage control and guide the study of stomatal development.
MiR399-Mediated PHO2 Regulation Controls Stomatal Development.
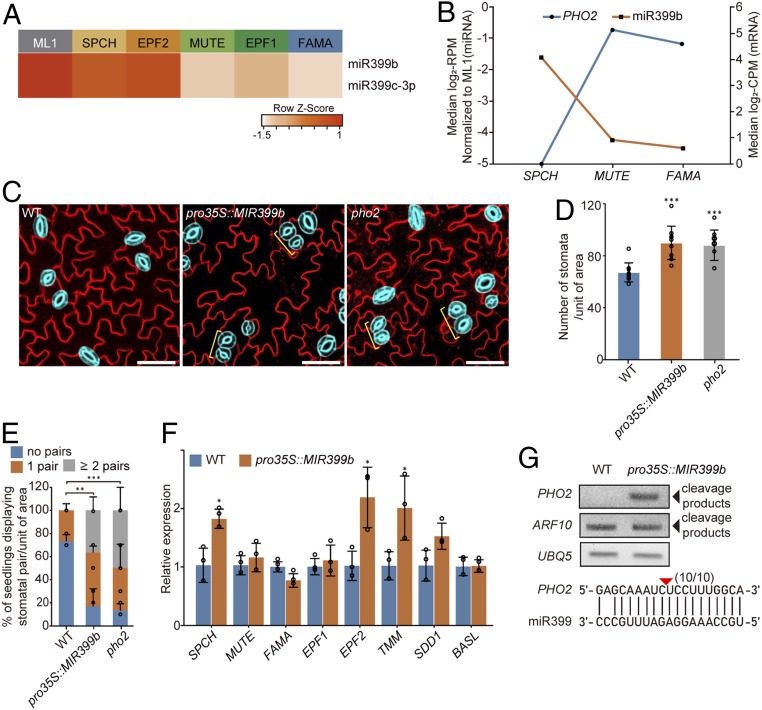
MiR399, a hub DE miRNA in the stomatal entry group, is conserved across plant species and plays a role in phosphate homeostasis by modulating the ubiquitin-conjugating E2 enzyme PHO2, which regulates the PHO1 and PHT1 phosphate transporters (36). The MIR399 family is composed of six members (MIR399a–f). MiR399b and miR399c were classified into the stomatal entry group in both the SPCH–MUTE–FAMA and EPF2–EPF1 paths (Fig. 5A). They are highly expressed in epidermal cells, and their expression during the stomatal lineage progression is most abundant at the SPCH and EPF2 stages followed by a gradual decline (Fig. 5A). The stomatal transcriptome data generated by Adrian et al. (2) show that the expression of PHO2 is highly up-regulated at the MUTE and FAMA stages compared to the SPCH stage. The expression pattern of miR399 in the stomatal lineage is anticorrelated with that of PHO2 (Fig. 5B), implying that miR399-mediated PHO2 regulation may contribute to the control of stomatal development in addition to its role in nutrient homeostasis.
Fig. 5.
miR399-mediated regulation of E3 ubiquitin ligase PHO2 guides stomatal development. (A) The heatmap shows the expression levels of miR399b and miR399c-3p in the stomatal lineage cells. (B) Anticorrelation in expression levels of miR399b and PHO2 during stomatal development. (C) Representative confocal images show stomatal development phenotypes of at least three independent WT, pro35S:MIR399, and pho2 plants. Mature GCs are highlighted in blue for elucidation, and the brackets indicate stomatal pairs. Cell outlines are visualized by FM4-64. (Scale bar, 50 μm.) (D) Stomatal density is increased in pro35S:MIR399 and pho2 plants compared to WT plants. The number of GCs per unit area (780 × 780 μm2) in cotyledons of 10-d-old seedlings. Error bars represent mean ± SD calculated from at least 10 plants. Two-sided Student’s t test P values: ***P < 0.001. (E) Numbers of stomatal pairs in pro35S:MIR399 and pho2 plants compared to WT. Percentage of plants having stomatal pairs per unit area (780 × 780 μm2) in cotyledons of 10-d-old seedlings. Error bars represent mean ± SD calculated from at least 10 seedlings. Two-sided Student’s t test P values; **P < 0.01; ***P < 0.001. (F) Expression levels of the key regulators of stomatal development in 4-d-old WT and pro35S:MIR399 seedlings. The expression levels were normalized to ACTIN2. Error bars represent mean ± SD calculated from three independent biological repeats. Two-sided Student’s t test P values; *P < 0.05. (G) miRNA399-guided 3′ cleavage products of PHO2 mRNA were detected in pro35S:MIR399 plants. The arrowheads indicate the miRNA-guided cleavage products. ARF10 and UBQ5 were used as internal controls. The arrow above the sequences indicates the cleavage site verified from 10 out of 10 clones sequenced.
To test this hypothesis, we generated transgenic plants overexpressing miR399b (pro35S::MIR399b) in which PHO2 expression was largely reduced (SI Appendix, Fig. S5 D and E), resulting in an increase in the number of stomatal pairs and a few stomatal clusters composed of more than two stomata, as well as an increase in stomatal density (Fig. 5 C–E and SI Appendix, Fig. S6D). Furthermore, we found that the levels of SPCH, EPF2, and TMM transcripts were substantially increased in pro35S::MIR399b plants (Fig. 5F). The altered expression levels of the stomatal lineage regulators could be either a result of their up-regulation via miR399 and/or an increased number of stomatal cells expressing these genes. A PHO2 null mutant displayed stomatal pairs similar in the pro35S::MIR399b plants (Fig. 5 C–E), and the cleavage products of PHO2 mRNA were present at high levels in the pro35S::MIR399 plants (Fig. 5G), supporting that miR399-mediated PHO2 regulation plays a role in proper stomatal development.
Discussion
Although two previous studies have implicated miRNAs in stomatal lineage determination (22, 37), the extent to which miRNA regulation plays a role in stomatal development has remained unclear. In this study, we demonstrate that stomatal lineage miRNAs are dynamically expressed during stomatal development and are critical for stomatal formation, in concert with the bHLH transcription factors SPCH, MUTE, and FAMA, the MAP kinase pathway, and small peptide/ligand signaling cascade (12). We have also identified the putative target mRNAs regulated by the developmental stage-specific miRNAs expressed in stomatal lineage cells, implicating an array of genes and pathways in stomatal development.
Our analysis identified 224 DE miRNAs in the stomatal lineage (Fig. 2A), slightly more than one-half of the annotated miRNAs in Arabidopsis, suggesting that miRNA dynamics and function may be crucial for stomatal development. Nearly two-thirds of the 224 DE miRNAs are implicated in the initiation of cell fate specification, belonging to the stomatal entry group. Stomatal lineage cells initially acquire their cell fate through the conversion of protodermal cells to MMCs and meristemoids, all of which possess stem cell-like activity, divide asymmetrically, and have self-renewing properties (38). In this regard, they are distinct from other cell types in the epidermis. Systemic changes in miRNA expression levels during entry into stomatal development may be part of the genetic program specifying stomatal stem cells. It is unclear how a protodermal cell is selected for stomatal lineage. SPCH has been suggested to be a master regulator in this process (39). It is expressed in MMCs and meristemoids and binds directly to ∼8,000 genes, including those involved in cell fate specification (39), implying that many events occurs during entry into the stomatal lineage. Forty-three MIRNA genes are included among the SPCH-binding genes, 33 of which are stomatal lineage miRNAs identified in our analysis, suggesting that SPCH-regulated miRNAs may contribute to stomatal lineage initiation. The miR171, miR394, and miR156 genes bound by SPCH have been shown to play a role in cell differentiation and patterning of meristem maintenance and gynoecium development (40–43). Since they belong to the stomatal entry group, a subset of stomatal lineage miRNAs may directly participate in the specification of stomatal stem cells.
Our PCA analysis reveals that the SPCH–MUTE–FAMA and EPF2–EPF1 paths have distinct miRNA populations (Fig. 1D) with certain miRNAs shared between the two (Fig. 2 A and B). Evidence indicates miRNAs have an impact on intercellular transcriptional heterogeneity that ultimately affects cell fate (44, 45). It is possible that stomatal lineage cells undergo a series of transitions in which cells with different properties, and populations of DE miRNAs, transiently exist and progress in the lineage, influencing cell fate and patterning. Transcriptome analysis, including single-cell RNA-seq, and visualization of stomatal lineage cells may address whether subtly different cell populations contribute to stomatal development.
Stomatal density and patterning are influenced by the environment (46–48). For example, increased temperature and high CO2 levels result in a low stomatal density through regulation by SPCH (11, 14). Certain plant species residing in dry or salty environments develop clustered stomatal complexes (49) and drought and salt stresses induce stomatal clustering, suggesting that stomatal patterning may contribute to environmental adaptation (49). Posttranscriptional regulation by miRNAs is one of the mechanisms underlying developmental plasticity in plants that facilitates adaptation to environmental changes. Overexpression or knockdown of DE miRNAs resulted in altered stomata number and patterning (Fig. 3), suggesting that environmental changes could alter miRNA expression, which in turn modulates stomatal development to better adapt to the environmental changes.
Overexpression of miR829 resulted in increased stomatal density (Fig. 3 A and D), whereas overexpression of miR3932 and knockdown of miR861 both resulted in an increase in the number of stomatal pairs, implying that miR3932 is a negative regulator, and miR829 and miR861 are positive regulators of stomatal patterning. NEK5 (AT3G20860) is one of the putative target genes for miR829 (Dataset S2), encoding a NIMA-related serine/threonine kinase (Neks). Several Nek family members play a role in cell cycle control (50), and NEK5 functions in the G2/M transition during mouse oocyte maturation (51). The precise control of stomatal cell division is critical for stomatal development, and several cell cycle regulators function in the regulation of stomatal development (52–55). Misexpression of CDC10 TARGET 1 (CDT1) and CELL DIVISION CONTROL PROTEIN 6 (CDC6), DNA replication licensing components, and B1-TYPE CYCLIN-DEPENDENT KINASE CDKB1;1 resulted in altered stomatal density by modulating the production of satellite meristemoid at the stomatal entry stage (52, 54). It might be possible that miR829 contributes to stomatal development by regulating NEK5-mediated cell division at the stomatal entry stage.
miR861 is predicted to target several genes including NAP1 (AT2G35110), a component of the SCAR/WAVE complex, which regulates trichome morphogenesis in Arabidopsis (56). Although the function of NAP1 for stomatal development has not yet been reported in Arabidopsis, two SCAR/WAVE complex components, LPL2 and LPL3, modulate epidermal cell morphogenesis including stomatal density and shape in rice (57), suggesting that the SCAR/WAVE complex may play a role in stomatal development. None of the predicted targets for miR3932 has been implicated in development or cell fate control and patterning.
Nutrient homeostasis is critical for plant growth and development. miRNAs play a role in this process by regulating genes involved in nutrient transport and assimilation (58). However, the relationship between nutrition and stomatal development is largely unknown. Our study revealed that miR399, which negatively regulates the ubiquitin-conjugating E2 enzyme PHO2 involved in phosphate homeostasis, controls stomatal development (Fig. 5). PHO2 mediates the degradation of the phosphate transporter PHO1, which is responsible for phosphate loading into the xylem to maintain Pi homeostasis in plants (59). PHO2 is known to interact with PHOSPHATE TRANSPORTER1 (PHT1) proteins in the postendoplasmic reticulum and mediate PHT1 degradation by ubiquitination, resulting in the modulation of Pi acquisition (60). A subset of PHT1 genes is differentially expressed in stomatal lineage cells. PHT1;1 and PHT1;2 transcripts are enriched at the early stage of stomatal development, while PHT1;4 and PHT1;5 transcripts are enriched at the differentiation stage of stomatal development based on previously reported transcriptome data (2). The level of the inorganic phosphate (Pi) transporter, PiT1 is critical for the regulation of cell division and cell proliferation in mammalian cells (61). Thus, this result suggests that the phosphate level may contribute to the regulation of stomatal cell lineage. Furthermore, when plants have higher stomatal density and conductance, a larger root with enhanced phosphate uptake is produced (62), which is attributable to demand for water. Given that pho2 mutants display enhanced phosphate uptake (59) and root-to-shoot translocation and that phosphate starvation leads to up-regulation of miRNA399 (63), our results together with the previous studies suggest that phosphate uptake controlled by the miR399-PHO2 (and possibly phosphate transporters) module could coordinate the regulation of stomatal development.
Photosynthesis relies on optimal gas exchange and water use, which requires efficient allocation of leaf surface space to stomata. Plants control stomatal density and pattern to maintain the one-cell spacing rule and possess greater maximum stomatal conductance for optimal gas exchange and water use (64, 65). Furthermore, altered stomatal development imposed by misexpression of SPCH, EPF, or TMM leads to changes in photosynthetic mesophyll tissues (66). The presence of a regulatory pathway by miRNAs, independent of the known regulators of stomatal development, may indicate a developmental strategy of plants for the efficient epidermal architecture to ensure optimal gas exchange and water use for photosynthesis.
Materials and Methods
Plant materials and growth conditions, plasmid construction, plant transformation, microscopy, RNA immunoprecipitation and small RNA isolation, Western blot analysis, total RNA extraction, RT-qPCR analysis, stomatal phenotype analysis, small RNA library sequencing and data analysis, identification of DE miRNAs, identification of potential target DEGs for DE miRNAs, functional enrichment analysis of GOBPs, construction of miRNA-target mRNA regulatory network, and RNA ligase-mediated rapid amplification of 5′ cDNA ends are descried in SI Appendix, Materials and Methods.
Data Availability.
Data are available in the paper, in SI Appendix, and at the Gene Expression Omnibus database under accession number GSE140918.
Supplementary Material
Acknowledgments
We thank Myeong Min Lee for critical reading of the manuscript, Jiyul Jung for the pho2 seeds, Juan Dong for the ML1p::YFP-RCI2A and SPCHp::SPCH-GFP transgenic plants, Dominique C. Bergmann for the MUTEp::nucGFP transgenic plants, Yunde Zhao for the MIR mutant seeds, and Seung-Jin Lee for the technical support. This work was supported by Grant IBS-R013-G2 from the Institute for Basic Science and in part by a grant from National Research Foundation (2019R1A2C3007376) and start-up funds from DGIST to J.M.K., and in part by Grant IBS‐R013‐D1 from the Institute for Basic Science.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE140918).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919722117/-/DCSupplemental.
References
- 1.Semrau S., et al. , Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 8, 1096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian J., et al. , Transcriptome dynamics of the stomatal lineage: Birth, amplification, and termination of a self-renewing population. Dev. Cell 33, 107–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shulse C. N., et al. , High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 27, 2241–2247.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurdon J. B., “Cell fate determination by transcription factors” in Current Topics in Developmental Biology, Wassarman P. M., Ed. (Academic Press, 2016), pp. 445–454. [DOI] [PubMed] [Google Scholar]
- 5.Scheres B., Plant cell identity. The role of position and lineage. Plant Physiol. 125, 112–114 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephenson R. O., Rossant J., Tam P. P. L., Intercellular interactions, position, and polarity in establishing blastocyst cell lineages and embryonic axes. Cold Spring Harb. Perspect. Biol. 4, a008235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawakatsu T., et al. , Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2, 16058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann D. C., Lukowitz W., Somerville C. R., Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Heimann M., Reichstein M., Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 451, 289–292 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Dutton C., et al. , Bacterial infection systemically suppresses stomatal density. Plant Cell Environ. 42, 2411–2421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau O. S., et al. , Direct control of SPEECHLESS by PIF4 in the high-temperature response of stomatal development. Curr. Biol. 28, 1273–1280.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillitteri L. J., Dong J., Stomatal development in Arabidopsis. Arabidopsis Book 11, e0162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau O. S., Bergmann D. C., Stomatal development: A plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139, 3683–3692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engineer C. B., et al. , Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513, 246–250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y., Jia T., Chen X., The “how” and “where” of plant microRNAs. New Phytol. 216, 1002–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S., Castillo-González C., Yu B., Zhang X., The functions of plant small RNAs in development and in stress responses. Plant J. 90, 654–670 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Mi S., et al. , Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wienholds E., et al. , MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Hwang H. W., Mendell J. T., MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94, 776–780 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vashisht D., Nodine M. D., MicroRNA functions in plant embryos. Biochem. Soc. Trans. 42, 352–357 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Breakfield N. W., et al. , High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res. 22, 163–176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jover-Gil S., et al. , The microRNA pathway genes AGO1, HEN1 and HYL1 participate in leaf proximal-distal, venation and stomatal patterning in Arabidopsis. Plant Cell Physiol. 53, 1322–1333 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Yang K., Jiang M., Le J., A new loss-of-function allele 28y reveals a role of ARGONAUTE1 in limiting asymmetric division of stomatal lineage ground cell. J. Integr. Plant Biol. 56, 539–549 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Hara K., Kajita R., Torii K. U., Bergmann D. C., Kakimoto T., The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara K., et al. , Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Sessions A., Weigel D., Yanofsky M. F., The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Carbonell A., et al. , Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24, 3613–3629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacAlister C. A., Ohashi-Ito K., Bergmann D. C., Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Pillitteri L. J., Sloan D. B., Bogenschutz N. L., Torii K. U., Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Ohashi-Ito K., Bergmann D. C., Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang G., et al. , Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58, 118–125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D., Bu F., Zhang W., The role of ubiquitination in regulating embryonic stem cell maintenance and cancer development. Int. J. Mol. Sci. 20, 2667 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Signolet J., Hendrich B., The function of chromatin modifiers in lineage commitment and cell fate specification. FEBS J. 282, 1692–1702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcázar R., Reymond M., Schmitz G., de Meaux J., Genetic and evolutionary perspectives on the interplay between plant immunity and development. Curr. Opin. Plant Biol. 14, 378–384 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Hronková M., et al. , Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J. Exp. Bot. 66, 4621–4630 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Chiou T. J., et al. , Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18, 412–421 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutter C., Schöb H., Stadler M., Meins F. Jr, Si-Ammour A., MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19, 2417–2429 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le J., Zou J., Yang K., Wang M., Signaling to stomatal initiation and cell division. Front. Plant Sci. 5, 297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau O. S., et al. , Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345, 1605–1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulze S., Schäfer B. N., Parizotto E. A., Voinnet O., Theres K., LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 64, 668–678 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Knauer S., et al. , A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Garcia D., A miRacle in plant development: Role of microRNAs in cell differentiation and patterning. Semin. Cell Dev. Biol. 19, 586–595 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Xing S., et al. , SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 75, 566–577 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Wang N., et al. , Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 10, 95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambardella G., et al. , The impact of microRNAs on transcriptional heterogeneity and gene co-expression across single embryonic stem cells. Nat. Commun. 8, 14126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casson S., Gray J. E., Influence of environmental factors on stomatal development. New Phytol. 178, 9–23 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Lake J. A., Woodward F. I., Response of stomatal numbers to CO2 and humidity: Control by transpiration rate and abscisic acid. New Phytol. 179, 397–404 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Qi X., Torii K. U., Hormonal and environmental signals guiding stomatal development. BMC Biol. 16, 21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gan Y., et al. , Stomatal clustering, a new marker for environmental perception and adaptation in terrestrial plants. Bot. Stud. 51, 325–336 (2010). [Google Scholar]
- 50.Fry A. M., O’Regan L., Sabir S. R., Bayliss R., Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125, 4423–4433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y.-Y., et al. , NEK5 regulates cell cycle progression during mouse oocyte maturation and preimplantation embryonic development. Mol. Reprod. Dev. 86, 1189–1198 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Castellano M. d. M., Boniotti M. B., Caro E., Schnittger A., Gutierrez C., DNA replication licensing affects cell proliferation or endoreplication in a cell type–specific manner. Plant Cell 16, 2380–2393 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han S.-K., et al. , MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev. Cell 45, 303–315.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Boudolf V., et al. , B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang K., et al. , A conserved but plant-specific CDK-mediated regulation of DNA replication protein A2 in the precise control of stomatal terminal division. Proc. Natl. Acad. Sci. U.S.A. 116, 18126–18131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basu D., et al. , DISTORTED3/SCAR2 is a putative arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17, 502–524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou W., et al. , Homologs of SCAR/WAVE complex components are required for epidermal cell morphogenesis in rice. J. Exp. Bot. 67, 4311–4323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paul S., Datta S. K., Datta K., miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 6, 232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu T.-Y., et al. , PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24, 2168–2183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang T.-K., et al. , Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25, 4044–4060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byskov K., et al. , Regulation of cell proliferation and cell density by the inorganic phosphate transporter PiT1. Cell Div. 7, 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hepworth C., Turner C., Landim M. G., Cameron D., Gray J. E., Balancing water uptake and loss through the coordinated regulation of stomatal and root development. PLoS One 11, e0156930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujii H., Chiou T.-J., Lin S.-I., Aung K., Zhu J.-K., A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15, 2038–2043 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Nadeau J. A., Sack F. D., Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 (2002). [DOI] [PubMed] [Google Scholar]
- 65.de Boer H. J., et al. , Optimal allocation of leaf epidermal area for gas exchange. New Phytol. 210, 1219–1228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dow G. J., Berry J. A., Bergmann D. C., Disruption of stomatal lineage signaling or transcriptional regulators has differential effects on mesophyll development, but maintains coordination of gas exchange. New Phytol. 216, 69–75 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the paper, in SI Appendix, and at the Gene Expression Omnibus database under accession number GSE140918.