Significance
Because the B cell receptor (BCR) is central to the development and activation of normal B cell subpopulations, its function is tightly controlled. We show that a novel tumor suppressor, KLHL14, is a subunit of a ubiquitin ligase that associates with the endoplasmic reticulum-associated protein degradation (ERAD) machinery. KLHL14 promotes ubiquitylation of BCR subunits and decreases the stability of immature BCR glycoforms in the endoplasmic reticulum, thereby reducing BCR levels. Conversely, loss of KLHL14 promotes BCR-dependent NF-κB activation and survival. These findings explain why KLHL14 inactivation occurs selectively in DLBCLs belonging to the MCD genetic subtype, since these tumors rely on BCR-dependent NF-κB signaling for survival and are highly sensitive to the BTK inhibitor ibrutinib, which blocks this pathway.
Keywords: KLHL14, DLBCL, B cell receptor, NF-κB
Abstract
The KLHL14 gene acquires frequent inactivating mutations in mature B cell malignancies, especially in the MYD88L265P, CD79B mutant (MCD) genetic subtype of diffuse large B cell lymphoma (DLBCL), which relies on B cell receptor (BCR) signaling for survival. However, the pathogenic role of KLHL14 in DLBCL and its molecular function are largely unknown. Here, we report that KLHL14 is in close proximity to the BCR in the endoplasmic reticulum of MCD cell line models and promotes the turnover of immature glycoforms of BCR subunits, reducing total cellular BCR levels. Loss of KLHL14 confers relative resistance to the Bruton tyrosine kinase (BTK) inhibitor ibrutinib and promotes assembly of the MYD88-TLR9-BCR (My-T-BCR) supercomplex, which initiates prosurvival NF-κB activation. Consequently, KLHL14 inactivation allows MCD cells to maintain NF-κB signaling in the presence of ibrutinib. These findings reinforce the central role of My-T-BCR–dependent NF-κB signaling in MCD DLBCL and suggest that the genetic status of KLHL14 should be considered in clinical trials testing inhibitors of BTK and BCR signaling mediators in DLBCL.
Diffuse large B cell lymphoma (DLBCL), the most common and aggressive form of non-Hodgkin lymphoma, has been subdivided by gene expression profiling into germinal center B cell-like (GCB), activated B cell-like (ABC), and unclassified subgroups (1). Multiplatform genomic analysis further subdivided these subgroups into four genetic subtypes that share multiple genomic aberrations, termed MCD (MYD88L265P/CD79B mutations), BN2 (BCL6 translocation/NOTCH2 mutation), N1 (NOTCH1 mutation), and EZB (EZH2 mutation/BCL2 translocation) (2). The survival of DLBCL patients following standard R-CHOP immunochemotherapy is influenced by these molecular distinctions, with the least favorable outcomes associated with the MCD subset of ABC DLBCL (2).
The viability of ABC but not GCB DLBCL cells depends on NF-κB activation via chronic active B cell receptor (BCR) signaling (3), which is initiated by interaction of the BCR with self-antigens (4). Accordingly, clinical responses to the BTK inhibitor ibrutinib, which blocks the BCR-dependent NF-κB pathway, occur preferentially in ABC DLBCL. Tumors with the MCD genetic subtype had a particularly high 80% response rate (5). Ibrutinib is also effective in another aggressive lymphoma, primary central nervous system lymphoma (PCNSL), which is also enriched for the MYD88L265P and CD79B mutations as well as other MCD-defining genetic aberrations (6).
MYD88L265P forms a complex with IRAK kinases in ABC DLBCL to promote NF-κB and JAK-STAT signaling (7). Furthermore, MYD88L265P associates with TLR9 in ABC cells and, unexpectedly, with the BCR to form the My-T-BCR supramolecular complex (8). The My-T-BCR nucleates prosurvival NF-κB signaling on the surface of endolysosomes by attracting the CBM (CARD11-BCL10-MALT1) complex, IκB kinase (IKK), and NF-κB transcription factor subunits. The My-T-BCR is readily detected in ibrutinib-sensitive MCD cells and is disrupted by ibrutinib, suggesting that it may be a critical target of this drug (8).
Genetic analysis revealed recurrent mutations of the KLHL14 gene in DLBCL, often in ABC tumors of the MCD genetic subtype (2) and in PCNSL (6, 9). KLHL14 (also known as Printor) (10) belongs to the Kelch-like family of proteins that can serve as subunits of Cullin-RING ubiquitin ligase (CRL) complex (reviewed in ref. 11). Kelch proteins have an amino-terminal BTB domain, which binds to the Cullin3 subunit of CRL ligases, and six carboxyl-terminal Kelch repeats, which mediate substrate recognition. KLHL14 is highly expressed in immune tissues, especially in B cells (12, 13), but its function is poorly understood. Whereas homozygous deletion of Klhl14 is embryonic lethal in mice, Klhl14 heterozygous mice are viable but have alterations in the self-renewing B-1 B cell lineages, thus revealing a role for KLHL14 in controlling B cell differentiation and/or function (13). These mice have reduced numbers of B-1a B cells, which have polyreactive and autoreactive BCRs, but increased numbers of B-1b B cells, which mediate memory responses to T independent type 2 antigens such as polysaccharides.
The mechanism by which KLHL14 regulates the genesis of normal and malignant B cells is unknown. Herein, we use proteomic and functional genomic methods to explore the relationship between KLHL14 and known oncogenic mechanisms in ABC DLBCL cells, revealing an unexpected control mechanism governing oncogenic BCR signaling.
Results
KLHL14 Is a Tumor Suppressor in ABC DLBCL.
We previously identified KLHL14 mutations by genomic analysis of 574 DLBCL biopsy samples (2). KLHL14 mutations were most prevalent in ABC DLBCL (10.8%) and were highly enriched in tumors belonging to the MCD genetic subtype (29.6%) (Fig. 1A). In a broader survey of published whole exome and genome sequencing data from patients with lymphoid malignancies (n = 5,692) (SI Appendix), KLHL14 mutations were most prevalent in DLBCL (4.7%) and in two other aggressive lymphoma entities, PCNSL (17.8%) and primary cutaneous lymphoma (PCL, 9.7%). PCNSL and PCL have the same histology as DLBCL but arise initially in the central nervous system and skin, respectively, rather than lymph nodes. Both entities typically have the ABC gene expression phenotype and the MCD genotype (6), suggesting that KLHL14 mutations may play a specific pathogenetic role in this genetic subtype.
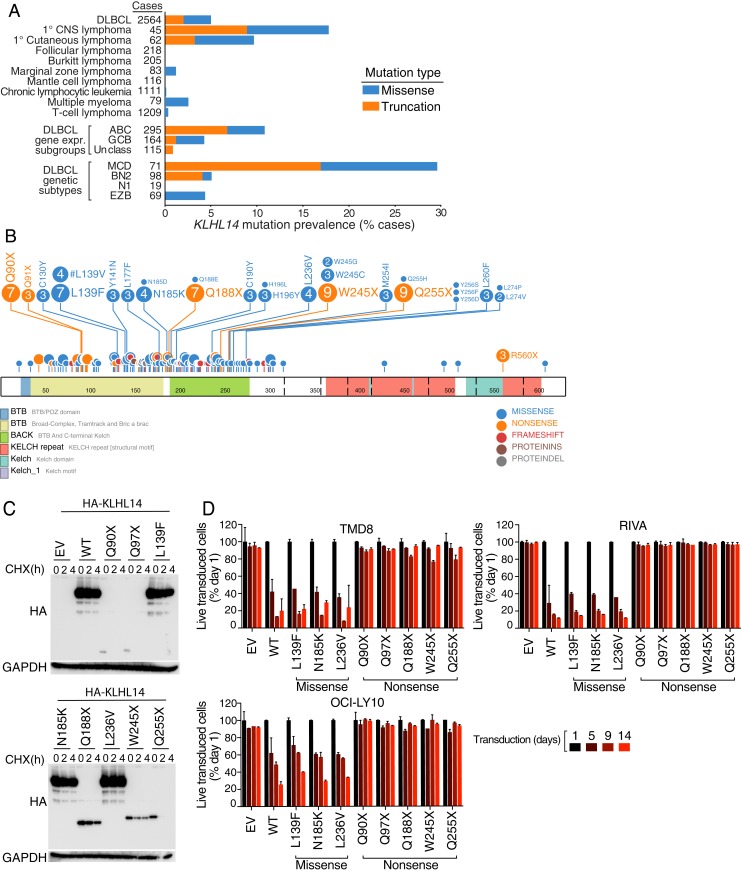
Fig. 1.
KLHL14 is a tumor suppressor in ABC DLBCL. (A) Percentage of KLHL14 mutations in DLBCL patients (n = 574) and in patients with lymphoid malignancies (n = 5,692). (B) Schematic representation of KLHL14 domain, organization, and mutations in DLBCL. (C) Western blot analysis of whole cell lysates, from TMD8 cells retrovirally transduced with cDNAs encoding an empty vector (EV), hemagglutinin (HA)-tagged version of KLHL14 wild-type (KLHL14WT) or eight lymphoma-derived KLHL14 mutant isoforms. Cells were treated with 50 μg/mL of cycloheximide (CHX) for the indicated time points before cell lysis. (D) FACS analysis of TMD8, RIVA, and OCI-LY10 cells retrovirally transduced with cDNAs encoding EV, KLHL14WT, or eight KLHL14 mutant isoforms along with the LYT2 (mouse CD8a) surface marker. The percent viable LYT2+ (KLHL14 expressing) cells was analyzed on the indicated posttransduction days. Error bars represent SD of triplicates, and data are representative of three independent experiments.
Although KLHL14 mutations are introduced by aberrant somatic hypermutation in DLBCL (2), those that are acquired by MCD tumors are likely to be drivers rather than passengers: the majority cluster in the BTB domain with hotspots at Gln90, Gln188, Trp245, and Gln255, each acquiring nonsense substitutions (Fig. 1B). We compared the ratio of truncation to missense KLHL14 mutations in DLBCL with the median ratio in other genes with similar mutational loads in DLBCL (mutation count ≥100 per sample). KLHL14 sustained truncating mutations significantly more often than other genes (odds ratio = 2.4 and P value = 0.0012), consistent with a tumor suppressor function.
To test whether these mutations affect KLHL14 protein abundance, we transduced TMD8, an ABC DLBCL cell line model of the MCD genetic subtype, with hemagglutinin (HA)-tagged versions of eight lymphoma-derived KLHL14 mutant isoforms along with KLHL14 wild-type (KLHL14WT). Immunoblot analysis revealed very low levels of KLHL14 BTB nonsense mutant isoforms relative to KLHL14WT and to KLHL14 isoforms with BTB missense substitutions (Fig. 1C). We assessed KLHL14 protein stability by treatment with cycloheximide and observed that the truncation isoforms were considerably less stable than the missense isoforms (Fig. 1C).
To investigate the functional consequences of KLHL14 mutations on ABC DLBCL cell proliferation and survival, we infected a panel of ABC cell lines (TMD8, RIVA, and OCI-LY10) with retroviruses coexpressing various KLHL14 isoforms and the LYT2 (mouse CD8a) surface marker. We then measured the fraction of viable, LYT2+ (KLHL14 expressing) cells over time by flow cytometry. Expression of KLHL14WT or KLHL14 BTB missense isoforms (L139F, N185K, and L236V) was toxic while KLHL14 BTB nonsense isoforms (Q90X, Q97X, Q188X, W245X, and Q255X) were not (Fig. 1D). These results support a loss-of-function tumor suppressor role for KLHL14 in ABC DLBCL.
KLHL14-Dependent Ubiquitylation and Down-Regulation of BCR Subunits.
We next tested whether KLHL14 assembles a Cullin-RING ubiquitin ligase (CRL), using its BTB domain to bind Cullin3 (10, 12, 14). Immunoblot analysis of KLHL14 immunoprecipitates detected its association with Cullin3 in TMD8 lysates (Fig. 2A). In TMD8 cells transduced with Flag epitope-tagged KLHL14 isoforms, anti-Flag immunoprecipitates from cells expressing KLHL14WT or KLHL14Q255X contained Cullin3, as assessed by immunoblot analysis, but Cullin3 did not associate with KLHL14Q97X (Fig. 2A). Whereas KLHL14Q255X is truncated in the BACK domain, leaving the BTB domain intact, the KLHL14Q97X disrupts the BTB domain. This suggests that the KLHL14 BTB is necessary and sufficient for Cullin3 association, as is the case for CRL ubiquitin ligases formed by other Kelch-like proteins. By performing an in vitro ubiquitylation assay, we found that KLHL14WT promoted autoubiquitylation, and KLHL14Q255X mutant lacking the Kelch domain, albeit containing the intact BTB domain, was sufficient to catalyze autoubiquitylation to a greater degree (Fig. 2B).
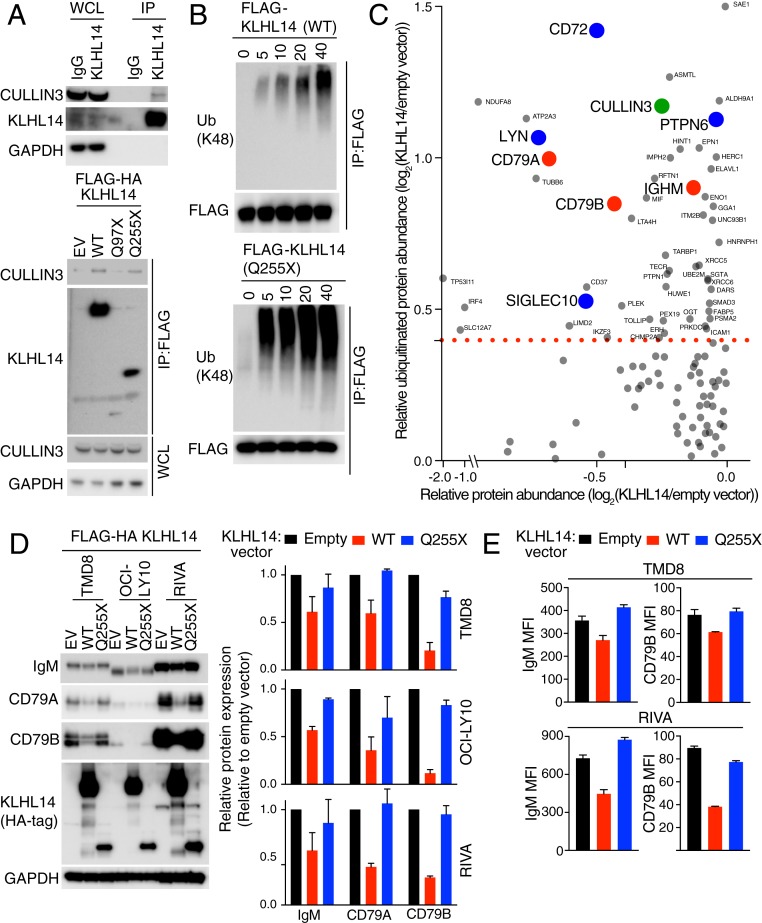
Fig. 2.
KLHL14-dependent ubiquitylation and down-regulation of BCR subunits. (A, Top) Western blot analysis of immunoprecipitated endogenous KLHL14 in TMD8 cells. IgG antibody immunoprecipitates = negative control; IP, immunoprecipitated; WCL, whole cell lysates. (A, Bottom) Western blot analysis of immunoprecipitated empty vector (EV), FLAG-HA-tagged KLHL14WT, two KLHL14 mutant isoforms in TMD8 cells. (B) In vitro ubiquitylation reaction of immunopurified FLAG-tagged KLHL14WT (Top) and KLHL14Q255X mutant (Bottom). Ubiquitin reaction buffer contains E1, E2 (UbcH5c), ATP, and ubiquitin. Ub, ubiquitin. (C) Mass-spectrometry measurements of relative protein abundance versus relative ubiquitinated protein abundance upon KLHL14 expression in TMD8 cells. BCR subunits, BCR-associated negative regulators, and Cullin3 are labeled red, blue, and green, respectively. (D, Left) Western blot analysis of whole cell lysates from TMD8, OCI-LY10, and RIVA cells retrovirally transduced with cDNAs encoding EV, FLAG-HA-tagged KLHL14WT, or KLHL14Q255X mutant. (D, Right) Quantification of BCR protein levels. (E) FACS analysis of surface IgM and CD79B expression on TMD8 and RIVA cells retrovirally transduced with cDNAs encoding EV, KLHL14WT, or KLHL14Q255X mutant coexpressing the LYT2 surface marker. Error bars represent SD of triplicates, and data are representative of three independent experiments.
To identify the proteins that are ubiquitylated and down-regulated by KLHL14, we conducted a proteomewide assessment of ubiquitinated proteins using diglycine remnant profiling and SILAC (stable isotope labeling with amino acids in cell culture)-based mass spectrometry (15, 16). Diglycine remnants are peptides that contain lysine residues from the ubiquitinated protein that are conjugated to GG residues of ubiquitin (K-ε-GG peptides), thereby enabling global quantification of ubiquitin attachment sites. SILAC mass spectrometry of total protein abundance and K-ε-GG peptides were used to compare TMD8 cells transduced with KLHL14WT under the control of a doxycycline-inducible promoter with control TMD8 cells transduced with an empty vector following doxycycline induction for 12 h. We identified 2,214 proteins whose expression levels were decreased upon KLHL14 expression and 491 unique ubiquitylation sites derived from 354 proteins that were more abundant in KLHL14-expressing cells than in control cells (Fig. 2C and SI Appendix, Tables S1 and S2). KLHL14-expressing cells had increased ubiquitinated Cullin3, suggesting autoubiquitination of a KLHL14-containing CRL ubiquitin ligase. Notably, the BCR subunits CD79A, CD79B, and IgM were ubiquitinated by ectopic KLHL14 expression as were the BCR-associated negative regulators LYN, PTPN6, CD72, and SIGLEC10. The protein abundance of CD79A and CD79B was concomitantly decreased, suggesting a role for KLHL14 in regulating chronic active BCR signaling, the hallmark of ABC DLBCL. Of note, the KLHL14-induced ubiquitylation of IgM occurs on lysine K292 in its endoplasmic reticulum (ER) luminal domain, whereas CD79A and CD79B ubiquitylation occurred on lysines K225 and K219, respectively, in their cytoplasmic domains.
We validated the proteomic data by assessing the abundance of IgM, CD79A, and CD79B upon induction of KLHL14WT in the ABC cell lines TMD8, OCI-LY10, and RIVA. As a control, we expressed KLHL14Q255X, which associates with Cullin3 but lacks the Kelch repeats that mediate substrate engagement in CRL ubiquitin ligases. Compared with cells transduced with empty vector or KLHL14Q255X, expression of KLHL14WT decreased the abundance of each BCR subunit, with the greatest effect on CD79B levels (Fig. 2D). Moreover, surface IgM and CD79B levels were decreased in cells ectopically expressing KLHL14WT but not KLHL14Q255X (Fig. 2E).
KLHL14 and BCR Subunits Are in Close Proximity in ABC DLBCL Cells.
KLHL14 has previously been localized to the ER in neuronal cells and interacts with the ER protein TorsinA (10). We therefore carried out immunofluorescence microcopy to determine the subcellular localization of KLHL14 in ABC cell lines. In TMD8 cells transduced with BioID2-HA-tagged KLHL14, anti-BioID2 staining was confined to a cytoplasmic compartment that colocalized with LRMP, a resident ER protein (17) (Fig. 3A), suggesting that KLHL14 is localized to the cytoplasmic face of the ER. Consistently, endogenous immunoprecipitation of KLHL14 showed its interaction with p97/VCP protein, an ATPase enzyme that associates and extracts ER-associated protein degradation (ERAD) substrates from the ER membrane to cytosol for protein degradation (SI Appendix, Fig. S1A) (18). The IgM component of the endogenous BCR was localized to both the plasma membrane and to the LRMP+ compartment (Fig. 3B), likely representing immature IgM isoforms in the ER.
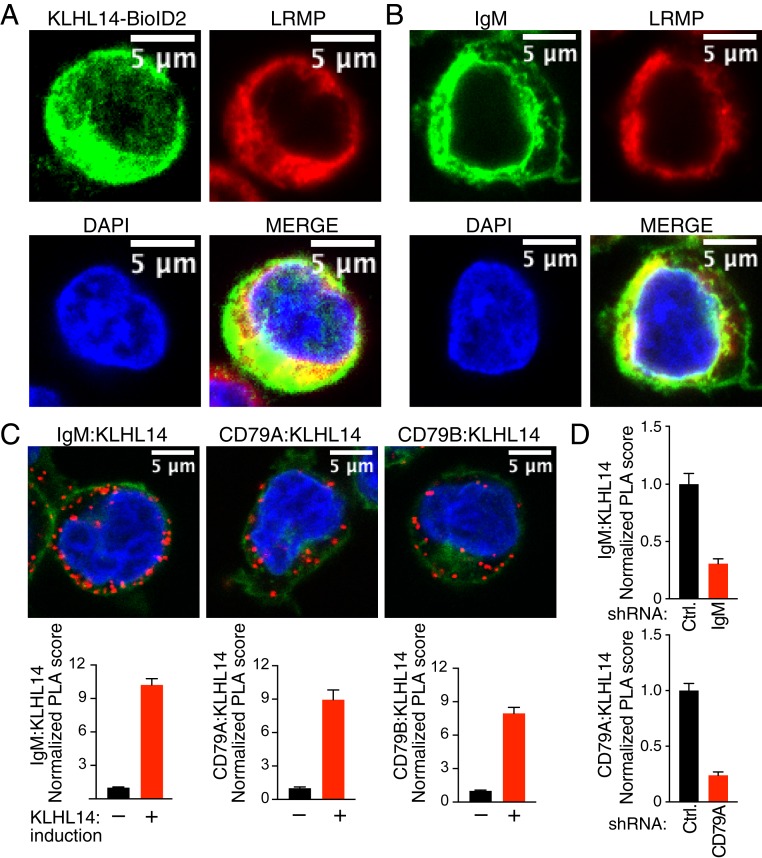
Fig. 3.
KLHL14 and BCR subunits are in close proximity in ABC DLBCL cells. (A) Confocal images of Tet repressor-expressing TMD8 cells retrovirally transduced with cDNAs encoding BioID2-HA-tagged KLHL14. Cells were treated with 1 μg/mL of doxycycline (DOX) for 24 h and stained as indicated. (B) Confocal images of TMD8 cells stained as indicated. (C, Top) Confocal images of PLAs (red) showing interaction of induced KLHL14 with IgM (C, Left), CD79A (C, Middle), and CD79B (C, Right) in Tet repressor-expressing TMD8 cells retrovirally transduced with cDNAs encoding BioID2-HA-tagged KLHL14 and treated with DOX (1 μg/mL) for 24 h. (Bottom) Normalized PLA score for each PLA pair. (D) Normalized PLA score after shRNA knockdown of the indicated genes (see Materials and Methods for detail). Error bars represent SEM, and data are representative of three independent experiments.
To test whether KLHL14 and BCR components interacts within cells, we used the proximity ligation assay (PLA), which detects proteins within tens of nanometers of each other (8, 19). We stably expressed BioID2-HA-KLHL14 in TMD8 cells under the control of a doxycycline-inducible promoter and performed a PLA using anti-BioID2 antibodies with antibodies to BCR subunits. PLAs for KLHL14-IgM, KLHL14-CD79A, and KLHL14-CD79B produced fluorescent cytoplasmic puncta in a doxycycline-inducible fashion (Fig. 3C). The KLHL14-IgM and KLHL14-CD79A PLA scores of doxycycline-treated cells were diminished by the knockdown of IgM or CD79A, respectively, further supporting the specificity of these assays (Fig. 3D). Thus, KLHL14 and BCR subunits are in close proximity, presumably at the ER membrane.
KLHL14 Decreases Stability of the Immature BCR Glycoforms in the ER.
Next, we investigated the role of KLHL14 in regulating the levels of BCR subunits in the ER. The steady state subcellular distribution of BCR in TMD8 cells was measured by a lectin purification method that separates proteins based on N-linked glycan composition. Relatively immature, ER-resident glycoproteins have mannose and glucose residues and can be fractionated by Pisum sativum agglutinin (PSA) lectin pulldown (SI Appendix, Fig. S1B) (20, 21). More mature glycoproteins containing galactose residues, which are usually added in the trans-Golgi, can be fractionated by Ricinus communis agglutinin I (RCA) lectin pulldown (20, 21). To elucidate the subcellular location of KLHL14 and the BCR, we performed both PSA and RCA pulldowns on total cellular lysates. Endoglycosidase H (EndoH) treatment was used as an additional control for lectin specificity in that it cleaves high mannose oligosaccharides associated with glycoproteins in the ER. These proteins become resistant to EndoH treatment after further sugar modification upon transport to the medial Golgi compartment (SI Appendix, Fig. S1B) (22). All BCR subunits were detected in both the PSA- and RCA-bound fractions and the RCA-bound fraction was completely EndoH resistant (SI Appendix, Fig. S1C). The PSA fraction contained all three BCR subunits whose mobility was predominantly EndoH sensitive (SI Appendix, Fig. S1C), suggesting enrichment of proteins in the ER.
Using this lectin-based fractionation method, we measured the protein stability of immature (ER) and mature (Golgi) BCR components upon ectopic KLHL14 expression or knockout (KO). Induction of KLHL14 expression by doxycycline shortened the half-lives of the IgM, CD79A, and CD79B subunits of the BCR in the ER fraction (Fig. 4A and SI Appendix, Fig. S1D). In the Golgi fraction, the BCR subunits had longer half-lives than in the ER fraction and expression of KLHL14 had little if any effect. Consistent with these observations, knockout of KLHL14 significantly extended the half-lives of BCR in the ER fraction while those in the Golgi fraction were not affected (Fig. 4B and SI Appendix, Fig. S1E). The effect of KLHL14 ectopic expression and knockout was specific for the BCR subunits because the half-life of calnexin, an ER-resident protein, was not affected (SI Appendix, Fig. S1 F and G).
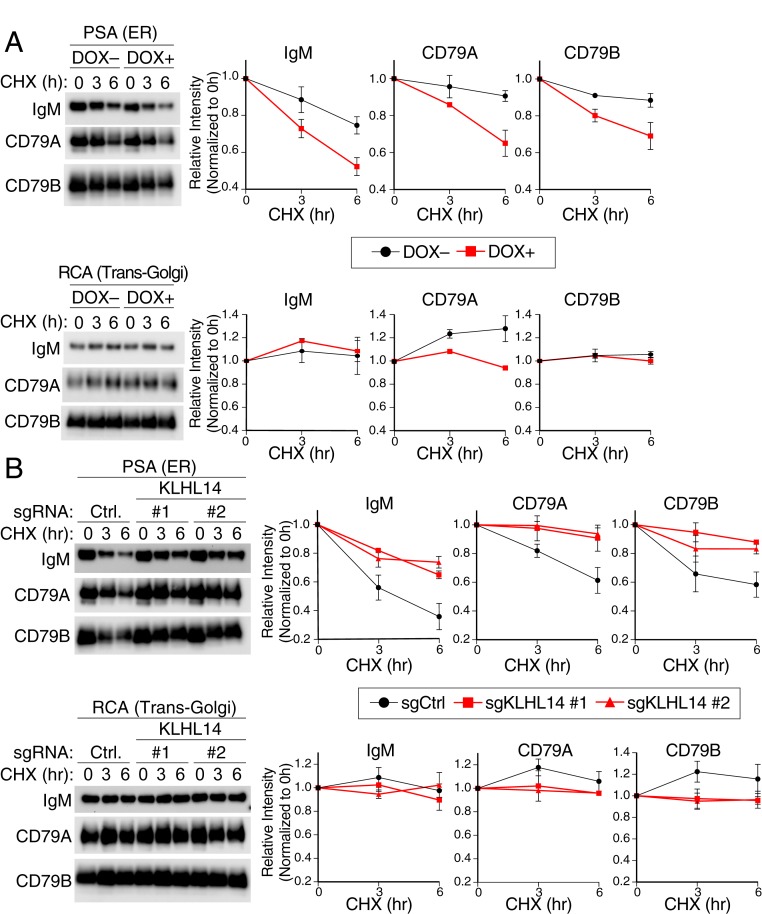
Fig. 4.
KLHL14 decreases stability of the immature BCR glycoforms in the ER. (A, Left) Western blot analysis of PSA (ER associated, Top)- and RCA (Golgi associated, Bottom)-bound fractions of BCR in Tet repressor-expressing TMD8 cells retrovirally transduced with cDNAs encoding BioID2-HA-tagged KLHL14. Cells were pretreated with DOX (1 μg/mL) for 16 h and treated with CHX (50 μg/mL) for the indicated time points before lysis. Right, quantification of BCR protein levels for PSA (ER associated, Top)- and RCA (Golgi associated, Bottom)-bound fractions. (B, Left) Western blot analysis of PSA (ER associated, Top)- and RCA (Golgi associated, Bottom)-bound fractions of BCR in TMD8-Cas9 cells lentivirally transduced with indicated sgRNAs. Cells were treated with CHX (50 μg/mL) for the indicated time points before lysis. (B, Right) Quantification of BCR protein levels for PSA (ER associated, Top)- and RCA (Golgi associated, Bottom)-bound fractions. Ctrl, nontargeting control. Error bars represent SD of triplicates, and data are representative of three independent experiments.
KLHL14 Alters BCR-Dependent NF-κB Signaling in ABC DLBCL.
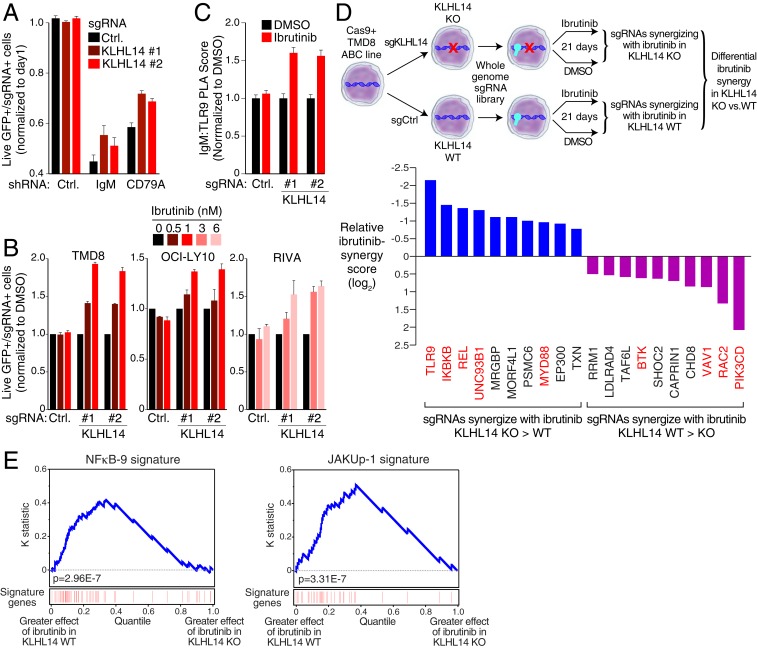
Given that KLHL14 negatively regulates BCR protein levels, we investigated whether knockout of KLHL14 would alter the sensitivity of ABC cells to inhibition of BCR-dependent NF-κB signaling. Knockdown of IgM and CD79A was toxic to TMD8 ABC DLBCL cells as previously reported (Fig. 5A) (3). Knockout of KLHL14 decreased this toxicity, suggesting that inactivation of KLHL14 promotes cell survival when BCR signaling is inhibited. Bruton tyrosine kinase (BTK) couples BCR signaling to intracellular calcium release and NF-κB activation and consequently the BTK inhibitor ibrutinib kills ABC DLBCL cells (3). We therefore investigated whether KLHL14 inactivation alters ibrutinib sensitivity in ABC cells. For this, we infected a panel of Cas9-expressing ABC cell lines (TMD8, RIVA, and OCI-LY10) exhibiting no genetic KLHL14 mutations with lentiviruses coexpressing a GFP-puromycin fusion protein along with a single guide RNA (sgRNA) targeting KLHL14 or a nontargeting control sgRNA. Following puromycin selection for stable integration, these cells were mixed with GFP-negative cells expressing a nontargeting control sgRNA at a ∼1:6 ratio. Cells were cultured for 9 d in DMSO or ibrutinib (0.5 to 6 nM), and the relative abundance of GFP-positive cells was tracked by flow cytometry over time. Two sgRNAs targeting KLHL14 conferred a competitive advantage in ibrutinib-treated cultures while the control sgRNA did not (Fig. 5B), suggesting that KLHL14 inactivation promotes BCR-dependent NF-κB signaling and thus renders ABC DLBCL cells relatively ibrutinib resistant.
Fig. 5.
KLHL14 alters BCR-dependent NF-κB signaling in ABC DLBCL. (A) FACS analysis of TMD8-Cas9 cells lentivirally transduced with indicated sgRNAs coexpressing a BFP-neomycin fusion protein along with indicated shRNAs coexpressing, GFP (see Materials and Methods for details). (B) FACS analysis of TMD8-, OCI-LY10-, and RIVA-Cas9 cells lentivirally transduced with sgRNA targeting KLHL14 or a nontargeting control, coexpressing a GFP-puromycin fusion protein. Cells were mixed at an ∼1:6 ratio with GFP, negative cells infected with a nontargeting control gRNA, and cultured for 9 d in DMSO, or indicated concentration of ibrutinib. (C) Normalized PLA score for IgM-TLR9 in TMD8- Cas9 cells lentivirally transduced with indicated sgRNAs treated with DMSO or ibrutinib, (5 nM) for 16 h. (D, Top) Schematic representation of CRISPR screen for ibrutinib synergy, in KLHL14 WT and KLHL14 KO TMD8 cells. (D, Bottom) Relative ibrutinib synergy score. My-T-BCR, NF-κB, and proximal BCR signaling regulators are colored red. (E) Kolmogorov–Smirnov test of gene expression signature enrichment in KLHL14 WT and KO cells. Genes were ranked based on the difference in digital gene expression (DGE) between the effect of 24-h ibrutinib (2 nM) treatment (DGEIbrutinib − DGEDMSO) in KLHL14 WT and KO cells, as indicated. The position of genes in the NF-κB-9 and JAKUp-1 signatures in the ranked gene list is shown in red. In A and B, error bars represent SD of triplicates. In C, error bars represent SEM. Data are representative of three independent experiments except in D and E.
In the subset of ABC DLBCL belonging to the MCD genetic subtype, prosurvival NF-κB signaling is coordinated by the My-T-BCR multiprotein supercomplex, which is comprised of the BCR, TLR9, MYD88 and associated signaling mediators (8). Knockout of KLHL14 increased My-T-BCR abundance in the presence of ibrutinib compared with DMSO-treated cells, as assessed by an IgM-TLR9 PLA (Fig. 5C). These findings suggest that increased My-T-BCR assembly in KLHL14-deficient cells might increase NF-κB signaling, thereby promoting resistance to ibrutinib.
To test this hypothesis and to understand generally how KLHL14 inactivation alters the response to ibrutinib, we performed a genomewide CRISPR-Cas9 screen in cells with and without KLHL14 knockout, in the presence or absence of ibrutinib (Fig. 5D). First, we stably expressed a KLHL14 sgRNA or a control sgRNA in Cas9-expressing TMD8 cells, creating KLHL14 KO cells and KLHL14 WT cells, respectively. KLHL14 KO and WT lines were then transduced with the Brunello sgRNA library, which contains 77,441 sgRNAs targeting four unique positions in most protein-coding genes (n = 19,114) in the human genome (23). After 1 wk of culture to allow for sgRNA-mediated genomic deletions, cells were divided and either cultured in a low concentration of ibrutinib (1 nM) or DMSO for 14 additional days. This concentration of ibrutinib was chosen to be sublethal, allowing us to identify sgRNAs that synergize with ibrutinib in killing these cells. Analysis of sgRNA abundance in DMSO-treated KLHL14 WT and KO cultures at day 21 versus day 0 revealed that the essential genes in ABC DLBCL cells (8) are not altered by KLHL14 inactivation in the absence of ibrutinib (SI Appendix, Tables S3 and S4).
To investigate the effect of KLHL14 KO on the ibrutinib response, we calculated an ibrutinib synergy score for each sgRNA as the ratio of its abundance in ibrutinib to its abundance in DMSO at day 21 (Log2 ratio ibrutinib versus DMSO). We then computed a relative ibrutinib synergy score as the ratio of these values in KLHL14 KO versus WT cells (Materials and Methods). Notably, in a ranking of genes by this relative ibrutinib sensitization metric, the top 10 genes with a greater effect in KLHL14 KO than WT cells when inactivated included TLR9, MYD88, and UNC93B1, encoding a chaperone that is required for the localization of TLR9 to the endolysosome. Also, these genes included IKBKB, encoding IκB kinase, and REL, encoding the NF-κB subunit c-REL. Conversely, the top 10 genes that sensitized to ibrutinib to a greater extent in KLHL14 WT than KO cells when inactivated encoded several signaling mediators that are engaged by proximal BCR signaling, namely PI3 kinase δ (PIK3CD), the small GTPase RAC2 and its binding partner VAV1, and BTK. The synergy of BTK knockout with ibrutinib is most likely due to increased lethality of the drug at early time points when only one allele of BTK is knocked out. Thus, this ibrutinib sensitization screen revealed that BCR signaling in cells with and without KLHL14 is fundamentally distinct, with KLHL14 KO cells relying more on My-T-BCR–dependent NF-κB signaling and relatively less on proximal BCR-mediated PI3 kinase signaling.
To investigate this hypothesis further, we profiled gene expression changes in KLHL14 WT and KO cells in the presence or absence of ibrutinib. An analysis of gene expression signatures revealed that these isogenic cell lines differed in their transcriptional response to ibrutinib (SI Appendix, Table S5). The NF-κB-9 gene expression signature, which reflects NF-κB signaling in ABC DLBCL cells (3), was down-regulated by ibrutinib treatment of KLHL14 WT cells, as expected, but was significantly less down-regulated by ibrutinib in KLHL14 KO cells using a Kolmogorov–Smirnov gene set enrichment test (24) (P = 2.96E-7, Fig. 5E). Among the NF-κB target that was less down-regulated by ibrutinib in KLHL14 KO cells was IL6, encoding a cytokine that stimulates JAK1 following engagement of its receptor. Accordingly, a signature of JAK1 signaling in ABC DLBCL (25) was also more down-regulated by ibrutinib in KLHL14 WT than KO cells (P = 3.31E-7, Fig. 5E). These results fit well with the observation that My-T-BCR–dependent NF-κB activation is relatively more essential for cell survival in ibrutinib-treated KLHL14 KO than WT cells.
Discussion
KLHL14 is a tumor suppressor gene that is recurrently mutated in mature B cell malignancies (2, 9). Our genetic survey of all major lymphoma diagnostic categories revealed that KLHL14 inactivation is remarkably limited to the MCD subtype of nodal DLBCL and to MCD-like primary extranodal lymphomas such as PCNSL and PCL. We demonstrated that KLHL14 binds Cullin3 and showed that it can function as a CRL ubiquitin ligase. Indeed, proteomic analysis revealed KLHL14-dependent ubiquitination of the BCR subunits IgM, CD79A, and CD79B, all of which decreased in abundance upon expression of KLHL14. KLHL14 was found in close proximity with a subpopulation of BCRs in the ER and selectively promoted the turnover of BCR subunits in the ER. KLHL14 loss promoted the survival of ABC DLBCL cell line models in the presence of ibrutinib. Moreover, our functional genomic studies revealed an unexpected dichotomy between the survival signaling in KLHL14 WT and KO cells in the presence of ibrutinib. In low concentrations of ibrutinib, KLHL14 KO cells were particularly reliant on activation of NF-κB by TLR9 and MYD88, which are components of the My-T-BCR supercomplex (8). Accordingly, loss of KLHL14 promoted assembly of the My-T-BCR and maintained expression of NF-κB signature genes under the selective pressure of ibrutinib. These results have implications for both the pathogenesis of lymphomas and the deployment of therapies targeting BCR-dependent NF-κB activation.
The selective tumor-suppressor function of KLHL14 in MCD-like human lymphomas is paralleled by the narrow and selective functions of KLHL14 during normal mouse B cell differentiation. Heterozygous deficiency of Klhl14 does not alter mature follicular B cell numbers but does alter the abundance of B-1 B cell subpopulations. Specifically, these Klhl14-deficient mice have decreased numbers of the B-1a subpopulation, but increased numbers of B-1b cells. Thus, KLHL14 exerts a negative function on both the differentiation of B-1b cells and on the formation of MCD DLBCL tumors. The B-1a and B-1b subpopulations have distinct developmental origins, utilize distinct repertoires of Ig variable regions, and respond differentially to T independent antigens (reviewed in ref. 26). The human equivalent of B-1b cells has not been identified but could potentially represent an extrafollicular cell of origin for MCD DLBCL.
In several respects, the BCR ubiquitylation that we describe here is distinct from previously described ubiquitylation of cell surface BCRs, which contributes to BCR internalization, endocytic sorting, degradation, and antigen presentation and signaling (27–30). Specifically, KLHL14 regulated the turnover of immature BCR subunit glycoforms in the ER rather than mature BCRs on the plasma membrane. Furthermore, the pattern of BCR ubiquitylation that we observed is unusual in that it targets both the cytoplasmic and luminal domains of BCR subunits: KLHL14 increased ubiquitylation of IgM in its ER luminal domain, whereas CD79A and CD79B were ubiquitinylated in their cytoplasmic domains. A clue to this unexpected observation lies in the interaction of KLHL14 with p97 and TorsinA (10), both of which participate in the ERAD mechanism. TorsinA is a resident ER protein that associates with the ERAD subunits p97, Derlin-1, and VIMP (31). TorsinA is thought to function as an ATP-dependent chaperone for misfolded proteins, facilitating their retrotranslocation by the ERAD machinery into the cytoplasm where they are ubiquitylated and degraded by proteasomes. Several E3 ligases have been implicated in ERAD processes, each targeting specific sets of misfolded proteins (reviewed in ref. 32). We speculate that KLHL14 may be an ERAD-associated E3 ligase that is devoted to quality control of BCR subunits in the ER. This would account for the ubiquitylation of BCR subunits on both ER luminal and cytoplasmic domains since ERAD ubiquitylation occurs during retrotranslocation of proteins from the ER to the cytoplasm. This putative mechanism is reminiscent of previous studies reporting that deficiency of Sel1L, a component of the ERAD machinery, impairs pre-BCR degradation and elevates surface pre-BCR levels in pre-B cells, thereby promoting pre-BCR signaling and the proliferation of large pre-B cells (33). Whether this KLHL14-dependent mechanism senses misfolding, abnormal glycosylation, or incomplete assembly of BCR subunits in the ER remains to be elucidated.
Our functional studies help to explain why KLHL14-inactivating mutations are selectively acquired by MCD DLBCL and by MCD-like extranodal lymphomas. These tumors are characterized by the My-T-BCR protein supercomplex, in which MYD88, TLR9, the BCR, and numerous colocated proteins coordinately activate IκB kinase and downstream NF-κB prosurvival signaling (8). The viability of MCD cells is not only maintained by NF-κB, but also by PI3 kinase signaling, which is initiated by proximal BCR signaling (8, 34, 35). Our CRISPR screens of KLHL14 WT and KO cells suggest that KLHL14 subtly shifts the balance of this survival signaling. Under the pressure of sublethal treatment with ibrutinib, the viability of KLHL14 KO cells relied to a greater extent than WT cells on TLR9 and MYD88, a functional dependency that was explained by greater My-T-BCR formation and NF-κB activity in the KO cells. Under the same conditions, KLHL14 WT cells had a greater dependence than KO cells on PI3 kinase δ, VAV1, and its binding partner, the small GTPase RAC2. In normal B cells, BCR engagement by antigen activates Src-family kinases, which in turn activate PI3 kinase and RAC2 via VAV1 (36). These observations lead us to hypothesize that KLHL14 mutations are selected by MCD lymphomas because they promote My-T-BCR–dependent NF-κB signaling, which distinctively characterizes this genetic subtype. It is unclear at present whether the increased abundance of BCR subunits in KLHL14 KO cells is itself sufficient to drive My-T-BCR formation or whether KLHL14 alters signaling and/or trafficking of the My-T-BCR components by some other mechanism.
What influence might KLHL14 inactivation have on the response of DLBCL tumors to BTK inhibitors and other drugs targeting the BCR-dependent NF-κB pathway? Although KLHL14 deficiency fosters ibrutinib resistance in vitro, this effect is only partial and relative to KLHL14 WT cells. Hence KLHL14-deficient tumors may still respond to ibrutinib. Indeed, ibrutinib monotherapy elicited objective responses in ≥80% of MCD DLBCL tumors with both MYD88L265P and CD79B mutations (5) and in PCNSL (37), both of which frequently inactivate KLHL14. Whether KLHL14-inactivating mutations are an accurate biomarker of response to BTK inhibitors should be tested in prospective trials of these drugs in aggressive lymphomas.
Materials and Methods
Cell Culture, Reagents, and Antibodies for Biochemical Methods.
TMD8, OCI-Ly10, and RIVA cells engineered to express an ecotropic retroviral receptor and tetracycline (Tet) repressor (3) or doxycycline-inducible Cas-9 (8) were grown in advanced RPMI medium 1640 containing 5% fetal bovine serum (FBS) (Tet and Tet derivatives tested, Atlanta Biologics) and 1% penicillin/streptomycin and 1% l-glutamine (Invitrogen). For SILAC, cells were grown in RPMI media lacking arginine and lysine (Thermo Fisher Scientific) and containing 10% heat-inactivated dialyzed fetal calf serum (FCS) (Sigma-Aldrich), 100 U/mL penicillin/100 mg/mL streptomycin (Life Technologies). The media were supplemented with heavy (13C615N4-arginine and 13C615N2-lysine), medium-heavy (13C6-arginine and D4-lysine) or regular/light (12C614N4-arginine and 12C614N2-lysine) amino acids (Cambridge Isotope Laboratories). HEK293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing FBS and 1% penicillin/streptomycin and 1% l-glutamine. All cell lines were maintained in a humidified 5% CO2 incubator at 37 °C and tested for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza). The following drugs were used: cycloheximide (Sigma Aldrich; 50 μg/mL final concentration), ibrutinib (Selleckchem; 0.5 to 6 nm final concentration), doxycycline hyclate (Sigma-Aldrich; 200 ng/mL to 1 μg/mL final concentration), puromycin (Sigma-Aldrich; 0.5 μg/mL to 1 μg/mL final concentration), and neomycin/G418 (Corning; 0.5 mg/mL final concentration).
All antibodies were diluted with 1:1,000 ratios unless otherwise specified. The following antibodies were used: anti-HA (Cell Signaling Technology, catalog no. 3724), anti-GAPDH (Cell Signaling Technology, catalog no. 2118, 1:3,000), anti-Cullin3 (Bethyl, catalog no. A301-109A), anti-KLHL14 (MBS, catalog no. 2526408, 1:300), anti-FLAG (Sigma, catalog no. F7425), anti-IgM (Santa Cruz Biotechnology, catalog no. sc-53347), anti-CD79A (Santa Cruz Biotechnology, catalog no. sc-20064,1:500), anti-CD79B (Santa Cruz Biotechnology, catalog no. sc-53210), anti-rabbit IgG (Cell Signaling Technology, catalog no. 2729), anti-rabbit IgG HRP-linked (Cell Signaling Technology, catalog no. 7074, 1:5,000), and anti-mouse IgG HRP-linked (Cell Signaling Technology, catalog no. 706, 1:5,000). KLHL14 antibody has been validated in multiple cell lines utilizing knockout (gRNAs) as well as overexpression.
The following agarose beads were used: anti-FLAG-M2 affinity gel (Sigma, A2220) and Pierce protein A agarose (Thermo Scientific, catalog no. 20333).
Plasmids and Cloning.
Human KLHL14 cDNA isolated from human tonsil B cells was subcloned into pcDNA3.1-FLAG-streptavidin vector. The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to generate KLHL14 cancer mutants. For retrovirus production, HA-tagged KLHL14 wild type and cancer mutants were subcloned into the retroviral vector pBMN-Ires-Lyt2. cDNAs encoding FLAG-HA-tagged KLHL14 wild type and two cancer mutants (Q97X and Q255X) were subcloned into the retroviral vector pMSCV. BioID2-HA-tagged KLHL14 and FLAG-tagged KLHL14 were subcloned into the retroviral vector pRetroCMV/TO (38). The vector pRSMX-Puro-GFP coexpressing an shRNA targeting SC4, IgM, and CD79A was previously described (3). Lentivirus encoding gRNAs targeting human KLHL14 or control gRNA were subcloned into pLKO-Puro-GFP vector or PLKO-Puro vector (8). pLKO-Neo-BFP vector is made by replacing hU6-sgRNA-Puro-GFP in pLKO-Puro-GFP vector with hU6-sgRNA-Puro-BFP from pMJ117 vector (39) and puromycin cassette with neomycin cassette using Gibson cloning techniques.
The target sequences to knockout human KLHL14 were:
sgKLHL14#1: GCACCGAGATCTGGTCGTTG
sgKLHL14#2: GGTGCTCGAGTACCTCTACA
sgCtrl: TAAAGCAGAAGAATATACAG.
Western Blot, Quantification of Protein, and Immunoprecipitation.
Extract preparation, immunoprecipitation, and immunoblotting were carried out as previously described (40). In brief, RIPA-lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% Nonidet P-40) supplemented with protease and phosphatase inhibitors was used for immunoblotting. Protein concentrations were quantified using a Pierce BCR protein assay kit according to the manufacturer’s protocol. Equal amounts of protein (10 to 15 μg) were loaded into NuPAGE 4 to 12% Bis-Tris Protein Gels (Thermo Fisher Scientific). Bands quantification was performed using ImageJ software and plotted using Prism8. For endogenous protein immunoprecipitation, Nonidet P-40 lysis buffer (15 mM Tris pH 7.4, 1 mM EDTA, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40) containing protease inhibitors and phosphatase inhibitors was used. Cleared supernatants were incubated with 20 μL of Pierce protein A agarose with 2 μg of anti-rabbit IgG or anti-KLHL14 antibody per each sample for 2 h at 4 °C. For anti-FLAG immunoprecipitation, cleared supernatants were incubated with 15 μL of anti-FLAG agarose. After four washes with Nonidet P-40 lysis buffer, the immunoprecipitates were eluted with 2× lithium dodecyl sulfate (LDS) sample buffer (Invitrogen).
Lectin Affinity Chromatography and Endoglycosidase Digestion.
Pisum sativum agglutinin (Vector Laboratories, catalog no. AL2053), Ricinus communis Agglutinin I (Vector Laboratories, catalog no. AL1083) coupled to agarose beads were washed in Nonidet P-40 lysis buffer. A total of 100 × 106 cells for each sample were lysed in Nonidet P-40 lysis buffer and incubated with 40 μL of RCA beads for 2 h at 4 °C. Subsequently, the supernatant was incubated with 40 μL of PSA beads for 2 h at 4 °C. After incubation, beads were washed five times in Nonidet P-40 lysis buffer and either directly suspended in 2× LDS sample buffer or resuspended with 40 μL of Tris-NaCl (15 mM Tris pH 7.4, 150 mM NaCl) buffer containing EndoH (New England Biolabs; 1 μL for each reaction). EndoH digestion was performed according to the manufacturer’s protocol and reaction was stopped by boiling in 2× LDS sample buffer.
In Vitro Ubiquitylation Assay.
The ubiquitylation of KLHL14 was performed in an ubiquitin reaction buffer containing 20 μL of 50 mM Tris pH 7.4, 5 mM MgCl2, 2 mM ATP (Roche), 10 ng/μL of E1 (Boston Biochem), 10 ng/μL of E2-UbcH5c (Boston Biochem), 50 ng/μL ubiquitin (Boston Biochem), and purified FLAG-tagged KLHL14 WT and KLHL14 Q255X from FLAG immunoprecipitation in HEK293T cells. The reaction was incubated at 37 °C for indicated time points, eluted in 4× LDS sample buffer, and subjected to Western blot analysis.
Transient Transfections and Virus Production/Transduction.
Lentivirus and retrovirus production/transduction have been previously described (3, 40). HEK239FT or HEK293T cells were transfected using Trans-IT 293T (Mirus) with a plasmid mixture of gag and pol, a mutant ecotropic env, and retroviral vectors for retrovirus production, and pPAX2, pMD2.g, and lentiviral vectors for lentivirus production. Lentivirus supernatants were concentrated with Lenti-X concentrator (Clontech) and used for infection as previously described (8).
FACS Analysis.
Flow cytometry was performed on BD FACS Calibur using CellQuest Pro version 6.0 and analyzed with FlowJo version 9. For Lyt2 monitoring viability assay, HA-tagged KLHL14 wild type and cancer mutants were retrovirally transduced using pBMN-Ires-Lyt2 vector into ABC cells (TMD8, RIVA, and OCI-LY10) expressing ecotropic retroviral receptor. Cells were split every 2 to 3 d and Lyt2 levels were followed for 14 d by staining with Alexa Fluor 647 conjugate anti-mouse CD8a (BD, catalog no. 557682). For detection of surface IgM and CD79B expression, cells were stained with anti-mouse CD8a and PE anti-human IgM (Biolegend, catalog no. 314508) or PE anti-human CD79B (Biolegend, catalog no. 341404), and Lyt2-positive cells were gated to measure change in surface BCR expression 3 d posttransduction. All staining was performed according to manufacturers’ protocols. For investigating shRNA effects, TMD8-Cas9 cells were infected with lentiviruses encoding gRNAs targeting human KLHL14 using pLKO-Neo-BFP vector and selected with neomycin in the presence of doxycycline. These cells were transduced with pRSMX-Puro-GFP vector expressing an shRNA against human SC4 (control), IgM, and CD79A. The number of infected cells was determined by measuring GFP levels the next day (day 1). Two days posttransduction, the number of viable GFP-positive cells on day 3 was calculated in relation to day 1 to normalize for transduction efficiency.
Growth Competition Assay.
TMD8-, RIVA-, or OCI-LY10-Cas9 cells were infected with lentiviruses encoding gRNAs targeting KLHL14 or control gRNA using pLKO-Puro-GFP vector or PLKO-Puro vector and selected with puromycin. GFP-positive and KLHL14 gRNA-positive cells were then mixed at a 1:6 ratio with GFP-negative and control gRNA-positive cells. These mixed cells were grown for 9 d in the presence of DMSO or ibrutinib (0.5 to 6 nM) with 1 μg/mL of doxycycline and replenished with fresh media and drugs every 2 to 3 d. The GFP-positive cells were quantified using flow cytometry. The percentage of GFP-positive and gRNA-positive cells treated with ibrutinib was normalized to that of cells treated with DMSO, which was set to 1.
Immunofluorescence Microscopy and PLA.
TMD8 cells expressing BioID2-HA-tagged KLHL14 under the Tet repressor were treated with doxycycline for 24 h and plated, permeabilized, and fixed as previously described (8). Cells were blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 30 min at room temperature. Cells were then incubated with mouse anti-BioID2 (Novus Biologicals, NBP2-59941, 1:100) and rabbit anti-LRMP (Abcam, catalog no. 202418, 1:400) antibody overnight at 4 °C. Cells were washed twice for 20 min in a large volume of 1% BSA in PBS and incubated with anti-rabbit F(ab′)2 conjugated to Alexa Fluor 555 (Cell Signaling Technology, catalog no. 4413, 1:300) and anti-mouse F(ab′)2 conjugated to Alexa Fluor 488 (Cell Signaling Technology, catalog no. 4408, 1:300) for 1 h at 37 °C. Cells were washed twice for 20 min and mounted with Prolong Diamond mounting media (Invitrogen). For colocalization of IgM and LRMP, TMD8 cells were stained with mouse anti-IgM (Santa Cruz Biotechnology, catalog no. sc-53347, 1:50) and rabbit anti-LRMP antibody (Abcam, catalog no. 202418, 1:400). All antibodies were diluted in 1% BSA in PBS. A PLA experiment was performed as previously described (8) using the same TMD8 cells expressing BioID2-HA-tagged KLHL14. Anti-IgM (Santa Cruz Biotechnology, catalog no. sc-53347, 1:100), anti-CD79A (Santa Cruz Biotechnology, catalog no. sc-20064, 1:100), anti-CD79B (Santa Cruz Biotechnology, catalog no. sc-53210, 1:100), and anti-BioID2 (Novus Biologicals, catalog no. NBP2-59941, 1:100) were used for each PLA pair. To confirm the specificity of each PLA pair, cells were transduced with either control shRNA (SC4) or shRNAs targeting CD79A or IgM, selected with puromycin plus doxycycline 2 d posttransduction, and subjected to PLA as described above. For TLR9-IgM PLA pair, anti-IgM (Jackson Immunoresearch, catalog no. 109-005-229 132466, 1:1,000) and anti-TLR9 (Cell Signaling Technology, catalog no. 2254S, 1:200) were used as previously described (8). The PLA score was determined by normalizing the number of PLA puncta counted in each sample to the number of PLA puncta counted in the control sample, which was set to 1. All images were acquired on a Zeiss LSM 880 confocal microscope and prepared with NIH ImageJ/FIJI software version 2.0.0-rc-65/1.5ls (41). PLA spots were counted in cell lines using Blobfinder version 3.2 (42).
Global Protein Expression Profiling.
For full proteome analysis, TMD8 cells with an ecotropic receptor were labeled using SILAC media for 2 wk. FLAG-HA-tagged KLHL14 (WT) or empty vector was retrovirally transduced using pMSCV vector and selected with puromycin. After 2 d of puromycin selection, 5 × 106 cells were lysed in 50 μL of modified Nonidet P-40 lysis buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40, 5 mM NaF, 1 mM Na3VO4, sodium deoxycholate [0.1%], Complete Protease Inhibitor Mixture, EDTA-free [Roche]). The cell lysates were mixed in a 1:1:1 ratio and analyzed as described previously (43).
Enrichment of Diglycine Remnant Peptides for Ubiquitination Analysis.
For diglycine remnant profiling, FLAG-tagged KLHL14 was retrovirally transduced using pRetroCMV/TO vector into TMD8 cells expressing an ecotropic retroviral receptor and Tet repressor. Infected cells were selected with puromycin and labeled using SILAC media for 2 wk before expansion to 150 × 106 cells. Cells were treated with doxycycline (1 μg/mL) for 12 h before cell lysis.
For antibody-based enrichment of peptides with diglycine remnants on lysine residues after tryptic digestion, the PTMScan Ubiquitin Remnant Motif Kit was used following the instructions of the manufacturer (Cell Signaling Technology). Briefly, an equal number of cells was lysed in 10 mL of urea lysis buffer (20 mM Hepes pH 8.0, 9 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate) at room temperature. The cleared lysates were mixed in a 1:1 ratio and reduced with dithiothreitol (DTT) and alkylated with iodoacetamide (IAA). After bringing the urea concentration to 2 M with 20 mM Hepes, pH 8.0 protein digestion with trypsin (Promega) was conducted overnight. The resulting peptides were purified with Sep-Pak C18 cartridges (Waters Corporation) and lyophilized. Following immunoprecipitation with the Ubiquitin Remnant Motif Antibody Bead Conjugate, the enriched diglycine remnant peptides were eluted from the bead matrix under acidic conditions, purified with C18 microtips, and vacuum dried.
For Fig. 2C, when multiple ubiquitylated sites were identified per protein, the most abundant one was used for each protein.
Mass Spectrometry and Data Analysis.
The peptide mixtures were analyzed on a Q Exactive HF orbitrap mass spectrometer (Thermo Fisher Scientific) coupled online to an Ultimate 3000 RSLC system (Dionex). After washing on a trap column (ReproSil-Pur 120 C18-AQ, 5 µm; Dr. Maisch GmbH; 100 µm × 5 cm; self-packed) the samples were separated on an analytical column (ReproSil-Pur 120 C18-AQ, 3 μm; Dr. Maisch GmbH; 350 × 0.075 mm; self-packed) using a 70-min linear solvent gradient of 2 to 40% B (80% acetonitrile [ACN], 0.1% formic acid [FA]) versus A (0.1% FA in water) at a flow rate of 300 nL·min–1. Eluting peptides were analyzed in a data-dependent acquisition scheme using a top 20 tandem mass spectrometry (MS/MS) method. After a survey scan (resolution 120,000 full width at half maximum [FWHM] at m/z 200, automatic gain control [AGC] target value 1 × 106, maximum injection time 40 ms) the 20 most abundant precursor peptide ions were selected for higher-energy collisional dissociation (HCD) with a normalized energy (NCE) setting of 28% and an isolation window of m/z 1.4. Fragment ions were analyzed with a resolution setting of 35,000 FWHM at m/z 200, an AGC target value of 1 × 105, and a maximum injection time of 64 ms.
MS raw data were analyzed with the MaxQuant software (version 1.6.5.0, Max Planck Institute of Biochemistry [MPI of Biochemistry]) (44). Fragment ion spectra were searched using the UniProtKB/Swiss-Prot reference database of human proteins (date: February 2019) including 245 common contaminants via the integrated Andromeda search engine (45). The mass tolerances for precursor and fragment ions were set to 6 and 20 ppm after initial recalibration, respectively. Diglycine remnants on lysine residues, oxidation of methionine, and acetylation on the protein N terminus were considered as variable modifications. Carbamidomethylation of cysteine was defined as a fixed modification. Minimal peptide length was set to seven amino acids, allowing up to two missed cleavages. Both on the peptide and protein level, the false discovery rate (FDR) was set to 1% using a forward-and-reverse decoy database search approach. SILAC multiplicity was set to triple labeling (Lys+0/Arg+0, Lys+4/Arg+6, Lys+8/Arg+10) requiring at least two ratio counts for peptide quantitation. Additionally, the “re-quantify” option of MaxQuant was enabled.
Further data evaluation was carried out using the Perseus software (version 1.6.0.7, MPI of Biochemistry) (46). After removal of potential contaminants and hits to the decoy database, identified ubiquitination sites were filtered for a localization probability of at least 75%.
RNA-Seq.
TMD8-Cas9 cells stably expressing sgRNA targeting KLHL14 or a nontargeting control were treated with DMSO or ibrutinib (2 nM) for 24 h. Total RNA was extracted using RNeasy Mini Kit (QIAGEN, catalog no. 74104) and RNA libraries were generated with TruSeq Stranded mRNA Library Prep (illumina) according to the manufacturer’s protocol. Pooled libraries were sequenced on HiSeq4000 using pair-end sequencing (2 × 75 bp reads). To calculate Digital Gene Expression (DGE) values, reads were aligned using STAR software and total numbers of reads were counted using HTSeq software. Normalization of counts was performed as previously described (47) and normalized counts were Log2 transformed to generate final DGE values. All of the RNA-seq gene expression datasets have been deposited in Gene Expression Omnibus (GEO) under accession GSE141143.
Genome-Wide Scale CRISPR-Cas9 Screen.
TMD8-Cas9 cells were infected with lentiviruses encoding gRNAs targeting KLHL14 or control gRNA using pLKO-Neo-BFP vector. Upon neomycin selection (0.5 mg/mL), biological replicates were transduced with the genome-wide Brunello sgRNA library (23) such that an average of 500 copies of each sgRNA (500× coverage) was present after puromycin selection (0.5 μg/mL). Library virus preparation, titration, and transduction have been previously described (8, 48). Cells were harvested for day 0 time point and replicates were treated with DMSO or ibrutinib (1 nM) with 200 ng/mL of doxycycline, counted, and passaged every 2 d with fresh media containing drugs and doxycycline until day 21. Cells were collected at day 21 and genomic DNA was isolated using QIAamp DNA Blood Maxi kits. Library amplification, sequence alignment and counts, and CRISPR score calculation were carried out as previously described (8, 48). In brief, sgRNA sequence was isolated and amplified by a two-step PCR using Extaq (Takara) for 18 cycles of amplification. PCR products were size selected using an eGel (Invitrogen) and the resulting amplicons were quantitated by Qubit HS (Thermo Fisher Scientific), pooled, and sequenced using a high output single-read cycle read flow cell. Libraries were multiplexed using indexes with the previously described primers (8) and BaseSpace sequence hub was used for demultiplexing and quality measurement. Sequences were aligned to the sgRNA library using Bowtie 2 version 2.2.9 and custom scripts as previously described (8). For the first PCR, the following primers were used: forward 5′-AGGGGACCCAGAGAGGGCC-3′ and reverse 5′-GCCAATTCCCACTCCTTTCAAGACC-3′. For the second PCR, unique pairs of secondary PCR primers (48) were used to amplify each sample.
The relative ibrutinib synergy score was calculated by normalizing the raw sequencing counts of each CRISPR screen sample by the total read counts of that sample and then normalized to 40,000,000 before adding 1 to remove zero count gRNAs. Low read count gRNAs were then filtered (≤40) and the Log2 fold change of each gRNA was calculated as either ibrutinib day 21/DMSO day 21 or DMSO day 21/day 0. Next, all gRNA ratios were averaged per gene. The average gene level metric for the control (KLHL14 WT) ibrutinib-treated score was then subtracted from KLHL14 KO ibrutinib-treated score, resulting in the “relative ibrutinib synergy score.” The top 10 differentially synergistic genes are shown for each condition (Fig. 5D).
Data Availability.
The datasets generated are included in the supplementary information. All of the RNA-seq gene expression datasets have been deposited in Gene Expression Omnibus (GEO) under accession GSE141143.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
Footnotes
The authors declare no competing interest.
Data deposition: All of the RNA-seq gene expression datasets have been deposited in Gene Expression Omnibus (GEO) www.ncbi.nlm.nih.gov/geo (accession no. GSE141143).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921187117/-/DCSupplemental.
References
- 1.Alizadeh A. A., et al. , Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Schmitz R., et al. , Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 378, 1396–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis R. E., et al. , Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463, 88–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young R. M., et al. , Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc. Natl. Acad. Sci. U.S.A. 112, 13447–13454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson W. H., et al. , Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat. Med. 21, 922–926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young R. M., et al. , Taming the heterogeneity of aggressive lymphomas for precision therapy. Annu. Rev. Cancer Biology 3, 429–455 (2019). [Google Scholar]
- 7.Ngo V. N., et al. , Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phelan J. D., et al. , A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 560, 387–391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vater I., et al. , The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia 29, 677–685 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Giles L. M., Li L., Chin L. S., Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis. J. Biol. Chem. 284, 21765–21775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X., et al. , Kelch-like proteins: Physiological functions and relationships with diseases. Pharmacol. Res. 148, 104404 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G., Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S., et al. , Kelch-like protein 14 promotes B-1a but suppresses B-1b cell development. Int. Immunol. 30, 311–318 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Xu B. J., et al. , Quantitative analysis of the secretome of TGF-beta signaling-deficient mammary fibroblasts. Proteomics 10, 2458–2470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Udeshi N. D., Mertins P., Svinkina T., Carr S. A., Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens T. W., et al. , Jaw1, A lymphoid-restricted membrane protein localized to the endoplasmic reticulum. J. Immunol. 153, 682–690 (1994). [PubMed] [Google Scholar]
- 18.Ye Y., Meyer H. H., Rapoport T. A., The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Söderberg O., et al. , Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Brouns G. S., de Vries E., Neefjes J. J., Borst J., Assembled pre-B cell receptor complexes are retained in the endoplasmic reticulum by a mechanism that is not selective for the pseudo-light chain. J. Biol. Chem. 271, 19272–19278 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Lehrman M. A., Oligosaccharide-based information in endoplasmic reticulum quality control and other biological systems. J. Biol. Chem. 276, 8623–8626 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Freeze H. H., Kranz C., Endoglycosidase and glycoamidase release of N-linked glycans. Curr. Protoc. Protein Sci. 62, 12.4.1–12.4.25 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Lam L. T., et al. , Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-kappaB pathways in subtypes of diffuse large B-cell lymphoma. Blood 111, 3701–3713 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas K. M., B-1 lymphocytes in mice and nonhuman primates. Ann. N. Y. Acad. Sci. 1362, 98–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veselits M., et al. , Igβ ubiquitination activates PI3K signals required for endosomal sorting. J. Exp. Med. 214, 3775–3790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drake L., McGovern-Brindisi E. M., Drake J. R., BCR ubiquitination controls BCR-mediated antigen processing and presentation. Blood 108, 4086–4093 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M., et al. , Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J. Immunol. 179, 4435–4443 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Satpathy S., et al. , Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol. Syst. Biol. 11, 810 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nery F. C., et al. , TorsinA participates in endoplasmic reticulum-associated degradation. Nat. Commun. 2, 393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Z., Brodsky J. L., Protein quality control in the secretory pathway. J. Cell Biol. 218, 3171–3187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji Y., et al. , The Sel1L-Hrd1 endoplasmic reticulum-associated degradation complex manages a key checkpoint in B cell development. Cell Rep. 16, 2630–2640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul J., et al. , Simultaneous inhibition of PI3Kδ and PI3Kα induces ABC-DLBCL regression by blocking BCR-dependent and -independent activation of NF-κB and AKT. Cancer Cell 31, 64–78 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Pongas G. N., Annunziata C. M., Staudt L. M., PI3Kδ inhibition causes feedback activation of PI3Kα in the ABC subtype of diffuse large B-cell lymphoma. Oncotarget 8, 81794–81802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arana E., et al. , Activation of the small GTPase Rac2 via the B cell receptor regulates B cell adhesion and immunological-synapse formation. Immunity 28, 88–99 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Lionakis M. S., et al. , Inhibition of B Cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell 31, 833–843 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngo V. N., et al. , A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Adamson B., et al. , A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi J., et al. , Loss of KLHL6 promotes diffuse large B-cell lymphoma growth and survival by stabilizing the mRNA decay factor roquin2. Nat. Cell Biol. 20, 586–596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allalou A., Wählby C., BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 94, 58–65 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Corso J., et al. , Elucidation of tonic and activated B-cell receptor signaling in Burkitt’s lymphoma provides insights into regulation of cell survival. Proc. Natl. Acad. Sci. U.S.A. 113, 5688–5693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Cox J., et al. , Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Ceribelli M., et al. , A druggable TCF4- and BRD4-dependent transcriptional network sustains malignancy in blastic plasmacytoid dendritic cell neoplasm. Cancer Cell 30, 764–778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webster D. E., Roulland S., Phelan J. D., Protocols for CRISPR-cas9 screening in lymphoma cell lines. Methods Mol. Biol. 1956, 337–350 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated are included in the supplementary information. All of the RNA-seq gene expression datasets have been deposited in Gene Expression Omnibus (GEO) under accession GSE141143.