Abstract
Sterol regulatory element–binding protein 2 (SREBP2) is the master transcription factor that regulates cholesterol metabolism. SREBP2 activation is regulated by SREBP chaperone SCAP. Here we show that ring finger protein 5 (RNF5), an endoplasmic reticulum-anchored E3 ubiquitin ligase, mediates the Lys-29–linked polyubiquitination of SCAP and thereby activates SREBP2. RNF5 knockdown inhibited SREBP2 activation and reduced cholesterol biosynthesis in human hepatoma cells, and RNF5 overexpression activated SREBP2. Mechanistic studies revealed that RNF5 binds to the transmembrane domain of SCAP and ubiquitinates the Lys-305 located in cytosolic loop 2 of SCAP. Moreover, the RNF5-mediated ubiquitination enhanced an interaction between SCAP luminal loop 1 and loop 7, a crucial event for SREBP2 activation. Notably, an overexpressed K305R SCAP variant failed to restore the SREBP2 pathway in SCAP-deficient cell lines. These findings define a new mechanism by which an ubiquitination-induced SCAP conformational change regulates cholesterol biosynthesis.
Keywords: cholesterol, E3 ubiquitin ligase, endoplasmic reticulum (ER), lipid metabolism, polyubiquitin chain, post-translational modification (PTM), ring finger protein 5 (RNF5), SREBP chaperone (SCAP), sterol regulatory element-binding protein (SREBP)
Introduction
Cholesterol is a crucial component of cellular membranes and serves as a precursor of steroid hormones and bile acids (1–3). Despite being a life-sustaining molecule, a high level of cholesterol in blood has also been linked to cardiovascular diseases (4). Hence, to maintain a delicate cholesterol balance, mammalian cells have evolved an intricate feedback mechanism featuring sterol regulatory element–binding protein 2 (SREBP2)3 and its sterol-sensing chaperone, SCAP (5, 27).
SREBP2 regulates the transcription of key genes involved in cholesterol biosynthesis and import, such as HMGCS, HMGCR, and LDLR. The precursor of SREBP2 is synthesized on the endoplasmic reticulum (ER) membrane and requires constant binding to SCAP to avoid rapid degradation (6). In addition, SCAP also mediates the COPII-dependent ER-to-Golgi transport of SREBP2. When the intramembrane cholesterol level decreases, the SCAP-SREBP2 complex is transported to the Golgi, where SREBP2 is proteolytically activated to release its N-terminal transcriptionally matured form (5, 27). When the intramembrane cholesterol level becomes sufficient, cholesterol binds to SCAP luminal loop 1 to promote its dissociation from luminal loop 7. This dissociation alters SCAP's conformation and blocks access to the COPII vesicle, thereby inhibiting SREBP2 activation in the Golgi (7).
We previously identified heat shock protein 90 (Hsp90) as a chaperone of SCAP and showed that its inhibition led to the rapid degradation of SCAP via a proteasome-dependent pathway (8). Although others have also described the proteasomal degradation of SCAP (9, 10), the E3 ubiquitin ligase regulating SCAP degradation is unknown.
In this study, we set out to search for the E3 ubiquitin ligase that modulates SCAP by analyzing SCAP-interacting proteins and identified RING finger protein 5 (RNF5), an ER-anchored E3 ubiquitin ligase that participates in ER-associated protein degradation (11). RNF5 regulates the stability of mutant cystic fibrosis transmembrane regulator (12), glutamine carrier proteins (13), and stimulator of interferon genes (STING), which is the central modulator of antiviral immune response (14).
The functions of RNF5 in lung, muscle, intestine, cancer, and immune cells have been studied; however, its role in hepatocytes and cholesterol metabolism is unknown. Here, we report that RNF5 plays an important role in SCAP-mediated SREBP2 activation and elucidated the molecular basis underlying the RNF5-dependent regulation of cholesterol biosynthesis.
Results
RNF5 interacts with SCAP on the ER membrane
To search for the E3 ligase that regulates SCAP, we examined the existing SCAP-interacting proteins and the related E3 ligases (Fig. S1A). FBW7 and RNF20 regulate the stability of matured SREBP in the nucleus (15, 16), whereas AMFR/gp78, TRC8, and RNF5 are associated with insulin-induced genes (Insigs), which link the E3 ligases to their substrates (17–19). Insigs also bind to SCAP in a sterol-dependent manner (20, 21), so it is likely that AMFR/gp78, TRC8, or RNF5 is also associated with SCAP. Intriguingly, we noticed a conserved sequence between SCAP and the RNF5-interacting domain of STING (Fig. S1B) (14), which prompted us to examine the interaction between RNF5 and SCAP.
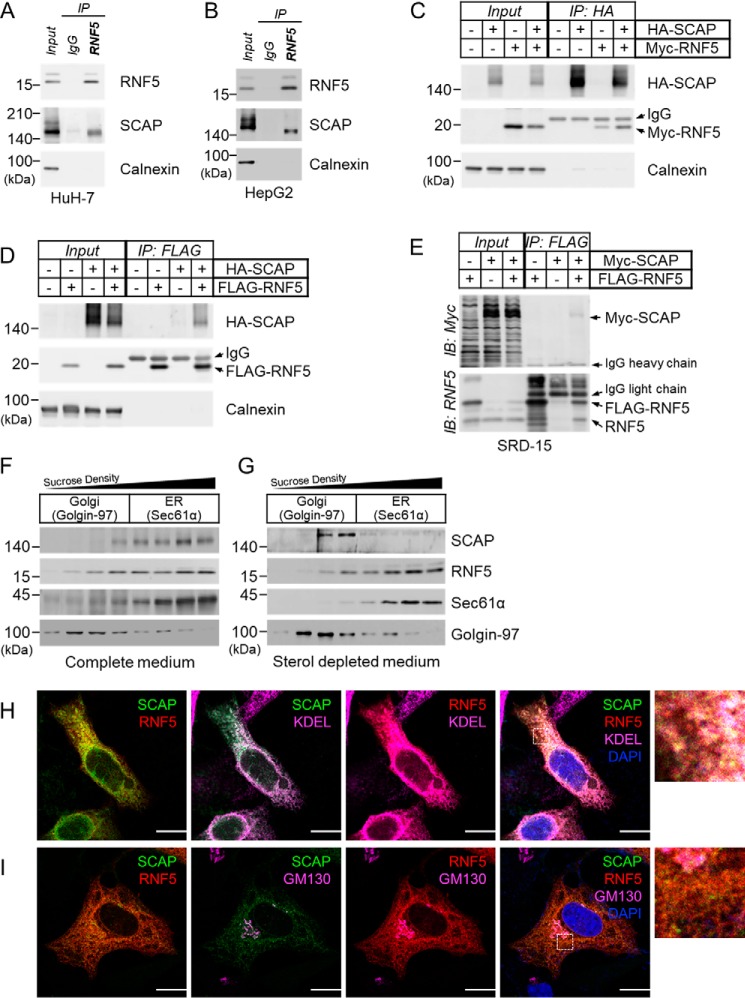
To determine whether RNF5 is associated with SCAP, we performed anti-RNF5 immunoprecipitation (IP) using human hepatoma cell lines HepG2 and HuH-7. We found that SCAP was co-precipitated with RNF5 in both HuH-7 and HepG2 cells (Fig. 1, A and B). In addition, interactions between HA-SCAP and Myc-RNF5, HA-SCAP and FLAG-RNF5, and FLAG-SCAP and Myc-RNF5 were confirmed by anti-HA, anti-FLAG, and anti-Myc IP experiments, respectively (Fig. 1, C and D; Fig. S1C). These results demonstrate a clear interaction between RNF5 and SCAP.
Figure 1.
RNF5 interacts with SCAP on the ER membrane. A and B, anti-RNF5 IP experiments were performed using whole cell lysates prepared from (A) HuH-7 and (B) HepG2 cells. C and D, HEK293 cells were transfected with indicated combinations of plasmid and cultured for 48 h and then harvested for (C) anti-HA-SCAP IP and (D) anti–FLAG-RNF5 IP to examine RNF5-SCAP interaction. E, Insig-deficient SRD-15 cells were transfected with Myc-SCAP and/or FLAG-RNF5 expression plasmids and cultured for 48 h. The cells were then harvested for anti-FLAG-RNF5 IP to determine RNF5-SCAP interaction. F and G, HuH-7 cells were cultured in complete DMEM supplemented with 10% (v/v) FBS (F) or sterol-depleted medium supplemented with 5% LPDS (G) for 16 h and then harvested for subcellular fractionation. Sec61α and Golgi-97 represents a marker for the ER and the Golgi apparatus, respectively. H and I, HeLa cells were co-transfected with EGFP-SCAP and mCherry-RNF5 expression plasmids and cultured for 2 days in compete medium. Localization of EGFP-SCAP (green), mCherry-RNF5 (red), and ER (KDEL) or Golgi (GM130) markers (magenta) were then analyzed by confocal microscopy. Nuclei were stained with DAPI (blue). Enlarged images enclosed by dotted squares were shown in the right. Images for individual channels were shown in Fig. S2, A and B. Scale bar, 10 μm.
We noticed that Insig1 was quite easily detected in HuH-7 but nearly undetectable in HepG2 cells, whereas Insig2 was highly expressed in HepG2 but nearly undetectable in HuH-7 cells (Fig. S1D). Because RNF5-SCAP interaction was found in both cell lines, it is suggested that either both Insigs play a complementary role in RNF5-SCAP interaction or Insigs are not involved. To address this, we performed IP experiments using an Insig-deficient Chinese hamster ovarian (CHO) cell line, SRD-15 (22). We found that Myc-SCAP was co-precipitated with FLAG-RNF5 even in the absence of Insigs (Fig. 1E). This indicates that the RNF5-SCAP interaction is independent of Insigs.
To define the subcellular localization of the RNF5-SCAP complex, we conducted cell fractionation by sucrose-gradient ultracentrifugation. We found that, as described in previous studies (12, 14), RNF5 existed predominantly in the ER fractions with SCAP in HuH-7cells (Fig. 1F). Sterol-dependent ER-to-Golgi movement plays a key role in SCAP function; however, whether RNF5 can also be transported to the Golgi is unknown. To address this, we performed cell fractionation experiments on HuH-7 cells cultured in sterol-depleted or sterol-loaded medium. We found that, when cells were cultured in sterol-depleted medium, SCAP was detected predominantly in the Golgi fractions, whereas RNF5 remained in the ER fractions (Fig. 1G).
To further validate the RNF5-SCAP interaction in the ER, we next performed immunofluorescence staining using antibodies to ER markers (KDEL or calnexin) and to Golgi markers (GM130 or Golgin-97) on HeLa, HEK293, or HuH-7 cells co-transfected with EGFP-SCAP and mCherry-RNF5. We found that when HeLa cells were cultured in complete medium supplemented with 10% FBS, both SCAP and RNF5 co-localized predominantly with an ER marker (KDEL), not with a Golgi marker (GM130) (Fig. 1, H and I). Similarly, in HuH-7 and HEK293 cells cultured in complete medium, RNF5 and a majority of SCAP were found in the ER (Fig. S2). On the other hand, incubating cells in cholesterol-depleted medium caused translocation of SCAP, but not RNF5, to the Golgi (Fig. S2F), which is consistent with our fractionation experiments (Fig. 1G).
These findings indicate that RNF5 interacted with SCAP on the ER membrane but was not co-transported with SCAP to the Golgi when cellular sterol level becomes insufficient.
RNF5 knockdown inhibits SREBP2 activation to block cholesterol biosynthesis
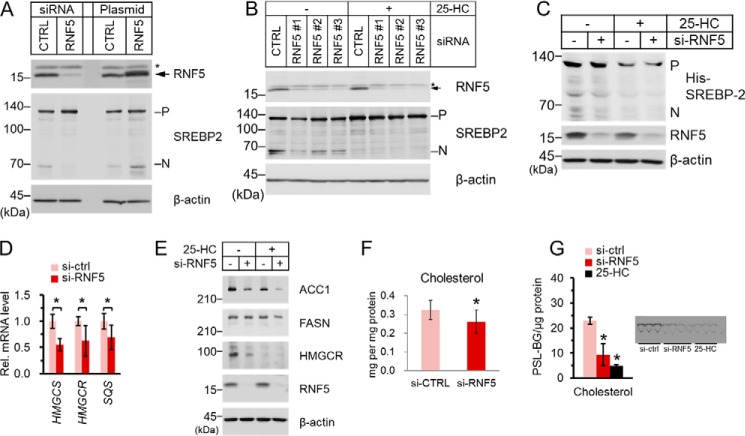
To investigate the function of RNF5, we performed siRNA knockdown (KD) experiments in HuH-7 cells. First, we examined whether RNF5 KD rescued proteasomal degradation of SCAP degradation induced by Hsp90 inhibitor 17-AAG treatment (8). However, we found that although MG132 rescued SCAP from 17-AAG–induced degradation (Fig. S3A), RNF5 KD had no effect on the SCAP protein level (Fig. S3B). This result suggests that RNF5 is not involved in the ubiquitin-proteasomal degradation of SCAP. This finding is similar to that in a previous report showing that RNF145, an E3 ubiquitin ligase, ubiquitinated SCAP to inhibit SREBP2 rather than inducing SCAP degradation (23).
Interestingly, we found that RNF5 KD decreased the matured form of SREBP2, whereas the overexpression of RNF5 had the opposite effect (Fig. 2A). We also examined another three different RNF5 siRNAs and found that RNF5 KD decreased the amount of matured SREBP2 (Fig. 2B). Notably, when the cells were treated with 25-hydroxylcholesterol (25-HC), a strong inhibitor of SREBP processing, RNF5 KD no longer reduced the protein level of matured SREBP2 (Fig. 2B). In addition, to rule out the influence of SREBP self-regulation, we showed that RNF5 KD decreased the matured form of overexpressed His-tagged SREBP2 whose transcription was controlled by a cytomegalovirus promoter (Fig. 2C). These findings suggest that RNF5 positively regulates SREBP2 activation.
Figure 2.
RNF5 knockdown inhibits SREBP2 activation to block cholesterol biosynthesis. A, HuH-7 cells were transfected with control (sc-37007) or RNF5-specific siRNA (sc-95209) or transfected with pcDNA3.1 empty vector or pcDNA-RNF5 expression plasmid. The cells were cultured for 32 h and then switched to sterol-depleted medium and cultured for another 16 h and then harvested for immunoblot analysis. B, HuH-7 cells were transfected with a control siRNA (assay ID: 12935300) or three RNF5-specific siRNA targeting different locations (assay ID: HSS155076, HSS155077, HSS155078). The cells were cultured for 32 h and then switch to sterol-depleted medium supplemented with 1 μg/ml 25-HC or with equal volume of ethanol and cultured for another 16 h and then harvested for immunoblot analysis. In A and B, arrows indicate RNF5 protein bands, whereas asterisks indicate nonspecific protein bands. C, HEK293 cells were reverse transfected with His-SREBP2 expression plasmid and cultured for 24 h. The cells were then transfected with siRNA as described in (A) and then switched to fresh medium as described in (B). The cells were then harvested for immunoblot analysis. A–C, the precursor (P) form and the nuclear (N) form of SREBP2 was detected by anti-SREBP2 (RS004) antibody that recognizes the N-terminal domain of SREBP2. D, HuH-7 cells were transfected with siRNA and cultured as described in (A). The cells were then harvested for total RNA extraction followed by reverse transcription and quantitative PCR. E, HuH-7 cells were transfected with siRNA as described in (A) and switched to fresh medium as described in (B). The cells were then harvested for immunoblot analysis. F, HuH-7 cells were treated as described in (D). The cells were then harvested for quantification of cellular cholesterol. G, HuH-7 cells were transfected with siRNA and cultured as described in (A). A group of cells was mock transfected and then cultured in sterol-depleted medium containing 1 μg/ml 25-HC. De novo cholesterol biosynthesis was determined as described under “Experimental procedures.” The Bar chart shows the signal of [14C]-labeled cholesterol normalized by cellular protein level. The image showing cholesterol bands shows the [14C]-cholesterol bands visualized by TLC. The unit of radiolabeled signal is presented as PSL-BG, which stands for photostimulated luminescence minus background. Data shown in D and F were pooled from three independent experiments performed in triplicate (n = 9), whereas data shown in (G) were pooled from two independent experiments performed in triplicate (n = 6). In D, F, and G, data are presented as mean ± S.D. Asterisks indicate difference between groups were significant as determined by two-tailed unpaired Student's t-tests (p < 0.05).
We then examined the effect of RNF5 KD on SREBP2-target gene expression. We found that the mRNA levels of HMGCS, HMGCR, and SQS were significantly down-regulated by RNF5 KD (Fig. 2D). The protein level of HMGCR as well as acetyl-CoA carboxylase 1, an SREBP1 target gene was also drastically reduced by RNF5 KD, whereas the protein level of fatty acid synthase was less affected by RNF5 KD (Fig. 2E). Although 25-HC treatment also led to the proteasomal degradation of HMGCR (24, 25), this event was independent of the presence of RNF5 (Fig. 2E).
To determine the physiological outcome of RNF5 KD, we measured the total cellular cholesterol content of HuH-7 cells. We found that total cholesterol in the cells was significantly decreased by about 20% in RNF5 KD cells (Fig. 2F). Moreover, using [14C] radioisotope-labeled acetic acid, we quantified the amount of de novo synthesized cholesterol in HuH-7 cells. De novo cholesterol synthesis was reduced by more than 60% in RNF5 KD cells, whereas treatment of 25-HC, a strong inhibitor of SREBP processing and an HMGCR degrader, led to even more significant suppression of cholesterol synthesis (Fig. 2G).
RNF5 mediates formation of Lys-29–linked polyubiquitin chain on SCAP
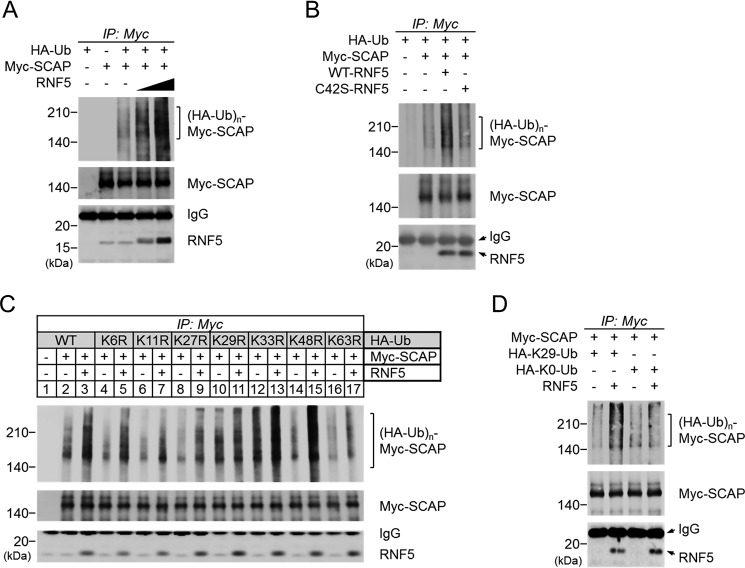
To understand how RNF5 affects SREBP2 activation by interacting with SCAP, we first examined whether RNF5 mediated the ubiquitination of SCAP. Cells of the SCAP-deficient CHO cell line SRD13A (6) were transfected with HA-ubiquitin, Myc-SCAP, and increasing amounts of RNF5 expression plasmids to perform anti-Myc IP. We found an RNF5-dependent increase of polyubiquitinated SCAP (Fig. 3A). This polyubiquitination was not observed when a C42S mutant RNF5 that had lost its E3 activity (14) was transfected (Fig. 3B). These results indicate that RNF5 is an E3 ubiquitin ligase that mediates the polyubiquitination of SCAP.
Figure 3.
RNF5 mediates Lys-29–linked polyubiquitination of SCAP. A, SRD-13A cells were seeded in 6-well plates and cultured until cell density reached 90% confluent. The cells were then switched to Opti-MEM and transfected with 0.5 μg/well HA-ubiquitin (Ub), 0.5 μg/well Myc-SCAP, and/or 0.5 or 1.0 μg/well RNF5 expression plasmid and incubated for 2 h. The total amount of transfected plasmid was adjusted to 2 μg/well using pcDNA3.1 empty vector. The cells were then switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 20 h. The cells were then switched to fresh medium supplemented with 5% LPDS and 10 μm MG132 and cultured for another 2 h. The cells were then harvested for anti-Myc-SCAP IP. B, SRD-13A cells were transfected with 1.0 μg/well HA-Ub, 0.5 μg/well Myc-SCAP and/or 0.5 μg/well WT or mutant RNF5 expression plasmid and then cultured for IP experiment as described in (A). C and D, SRD-13A cells were transfected with 0.5 μg/well Myc-SCAP and WT or 1.0 μg/well mutant HA-Ub in the presence or absence of 0.5 μg/well RNF5 expression plasmid and then cultured for IP experiment as described in (A).
It has been reported that RNF5 induced the Lys-48–linked polyubiquitination of STING, leading to its degradation (14). On the other hand, AMFR/gp78 induced the Lys-27–linked polyubiquitination of STING to activate downstream signaling (18). It was essential to determine which kind of polyubiquitination was induced by RNF5 on SCAP. To this end, we generated seven HA-ubiquitin plasmids that carried an arginine mutation at lysine residue Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, or Lys-63 to perform SCAP ubiquitination experiments. We found that RNF5 failed to induce the polyubiquitination of SCAP when Lys-29 of ubiquitin was mutated (Fig. 3C). This indicates that Lys-29 was crucial for RNF5-induced SCAP polyubiquitination, whereas other lysine residues were dispensable.
To confirm this finding, we generated two other HA-ubiquitin mutant plasmids, one with all of the lysines mutated to arginine (Lys-0) and the other with all but the Lys-29 lysine residue mutated to arginine (Lys-29). We found that RNF5 induced Lys-29–ubiquitin to form a polyubiquitin chain on SCAP, whereas no polyubiquitination was observed with Lys-0–ubiquitin (Fig. 3D). Taking these findings together, we revealed that RNF5 mediated the Lys-29–linked polyubiquitination of SCAP.
RNF5 ubiquitinates Lys-305 located in cytosolic loop 2 of SCAP
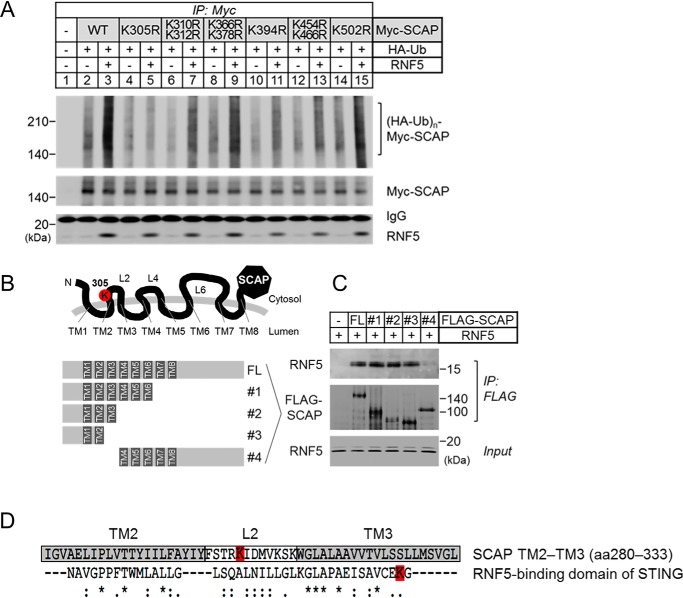
Next, we began to identify the site where RNF5-induced polyubiquitination occurred. There are 18 cytosolic lysine residues in human SCAP that were possible ubiquitination sites (Fig. S4A). We generated 14 SCAP mutants that carried an arginine mutation at one or two lysine residues located in the cytosolic loops or the N-terminal or C-terminal cytosolic domains of SCAP. We found that arginine mutation at Lys-305 located in cytosolic loop 2 of SCAP drastically impaired RNF5-induced polyubiquitination, whereas mutations of Lys-310 and Lys-312 in the same loop had no effect (Fig. 4A). Mutation of lysine in cytosolic loop 4 and loop 6 did not affect the RNF5-induced polyubiquitination of SCAP.
Figure 4.
RNF5 ubiquitinates lysine Lys-305 located in cytosolic loop 2 of SCAP. A, SRD-13A cells were seeded in 6-well plates and cultured until cell density reached 90% confluent. The cells were then switched to Opti-MEM and transfected with 0.5 μg/well HA-ubiquitin (Ub) and 1.0 μg/well WT or mutant Myc-SCAP in the presence or absence of 0.5 μg/well RNF5 expression plasmid and incubated for 2 h. The total amount of transfected plasmid was adjusted to 2 μg/well using pcDNA3.1 empty vector. The cells were then switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 20 h and then switched to fresh medium supplemented with 5% LPDS and 10 μm MG132 and cultured for another 2 h. The cells were then harvested for anti-Myc-SCAP IP. B, schematic presentation of the FLAG-SCAP plasmids used in (C). FL indicates full-length of SCAP, whereas plasmid no.1 to 4 are truncated from either C-terminal or N-terminal end of SCAP. TM indicates transmembrane domain, whereas L indicates loops pointed to the cytosol. Lysine residue Lys-305 on loop 2 is highlighted in red. C, SRD-13A cells were seeded as described in (A) and transfected with 2 μg/well empty vector or WT or truncated FLAG-SCAP plasmids in Opti-MEM and incubated for 2 h. The cells were then switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 22 h. The cells were then harvested for anti-FLAG-SCAP IP. D, alignment of RNF5-binding region of SCAP and STING (14). Gray boxes indicate the amino acid sequences of transmembrane domains TM2 and TM3. White box indicates the amino acid sequence of cytosolic loop 2. The lysine residues that were ubiquitinated by RNF5 in SCAP and STING are highlighted in red.
Arginine mutation at the Lys-10 lysine residue located in the N-terminal cytosolic domain and other lysine residues located in the C-terminal cytosolic domain had no influence on RNF5-induced polyubiquitination of SCAP (Fig. S4B). Collectively, these results demonstrate that Lys-305 lysine residue is essential for the RNF5-mediated polyubiquitination of SCAP.
Because lysine Lys-305 is located in loop 2 flanked by transmembrane domains TM2 and TM3 (Fig. S4A), we examined whether RNF5 binds to nearby regions of loop 2. We generated full-length, TM1–6, TM1–3, TM1–2, and TM4–C-terminal truncated versions of FLAG-SCAP (Fig. 4B) to study their binding to RNF5 by anti-FLAG IP. We found that RNF5 was co-precipitated with full-length SCAP as well as TM1–6, TM1–3, and TM1–2 of SCAP (Fig. 4C, full-length (FL) and no. 1–3), but not with TM4-C-terminal of SCAP (Fig. 4C, no. 4). We also noticed that the amino acid sequence of SCAP TM2–3 shares 58% similarity with RNF5-binding domain of STING (Fig. 4D). Taking these findings together, we revealed that RNF5 binds to a region near loop 2 and ubiquitinates Lys-305 of SCAP.
K305R mutation impairs SCAP-mediated SREBP activation
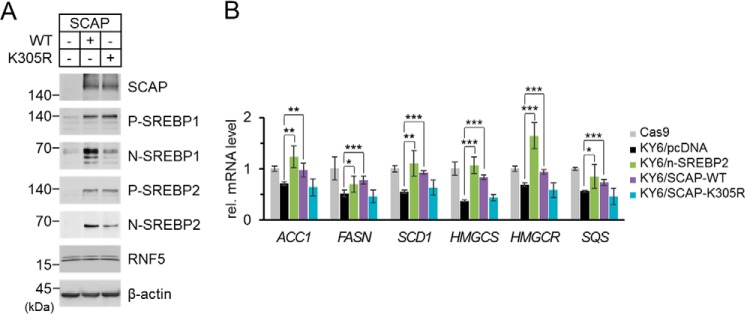
To study the relationship between RNF5-induced SCAP polyubiquitination and RNF5-mediated SREB2 activation, we transfected WT or K305R mutant SCAP into SCAP-deficient SRD-13A cells and examined the effect on SREBP activation. We found that the expression levels of WT and K305R SCAP were quite comparable and they had similar abilities to stabilize precursor SREBPs (Fig. 5A). Intriguingly, judging from the amount of N-terminal matured form of SREBPs, although WT SCAP rescued SREBP activation, K305R mutation of SCAP drastically impaired SREBP activation (Fig. 5A). To determine the physiological outcome of K305R mutation in SCAP, we generated a SCAP-knockout (KO) HEK293 cell line using the CRISPR/Cas9 system following a previously published method (26). SCAP was undetectable in the SCAP-KO HEK293 cell line, and the protein level of SREBP was drastically reduced (Fig. S5A), which led to 30–50% down-regulation of SREBP target genes compared with that in a control cell line that expressed Cas9 (Fig. S5B). We named the SCAP-KO and Cas9 control HEK293 cell lines KY6 and Cas9, respectively. When we transfected matured SREBP2, or WT or K305R mutant SCAP into KY6 cells, we found that exogenous SREBP2 and WT SCAP, but not K305R-mutant SCAP, restored SREBP target gene expression to a level comparable with that of Cas9 cells (Fig. 5B). These experiments demonstrate that Lys-305 polyubiquitination of SCAP is crucial for SREBP2 activation.
Figure 5.
K305R mutation impairs SCAP-mediated SREBP activation. A, SRD-13A cells were seeded in 6-well plates and cultured until cell density reached to 90% confluent. The cells were then switched to Opti-MEM and transfected with 2 μg/well empty vector or WT or K305R mutant SCAP and incubated for 2 h. The cells were switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 22 h. The cells were then harvested for immunoblot analysis. P-SREBP and N-SREBP indicate the precursor form and the N-terminal processed form of SREBP, respectively. B, SCAP-deficient HEK293 cell line KY6 was established by CRISPR/Cas9 system as described previously (26). A control cell line Cas9 was established in parallel by transfecting the cells with only the Cas9 plasmid without a guide RNA. Cas9 and KY6 cells were seeded in 12-well plates and cultured until cell density reached 50% confluent. The cells were then switched to Opti-MEM and transfected with 1 μg/well empty vector, N-terminal matured form of SREBP2, WT SCAP, or K305R mutant SCAP expression plasmids and incubated for 2 h. The cells were switched to DMEM supplemented with 10% FBS, 5 μg/ml cholesterol, 1 mm sodium mevalonate, and 20 μm sodium oleate and cultured for 22 h. The cells were switched again to DMEM supplemented with 5% LPDS, 50 μm sodium mevalonate, and 12.5 μm fluvastatin and cultured for 16 h. The cells were then harvested for total RNA extraction followed by reverse transcription and quantitative PCR. Data were pooled from two independent experiments performed in triplicate and presented as mean ± S.D. (n = 6). Values between groups were analyzed by two-tailed unpaired Student's t-tests. Asterisks indicate that the difference between groups were significant. *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
RNF5-dependent polyubiquitination enhances luminal loop 1 and loop 7 interaction of SCAP
SREBP activation is tightly regulated by the interaction between cholesterol and SCAP. When the cholesterol level in the ER membrane rises, cholesterol binds to luminal loop (L1) of SCAP, causing the dissociation of L1 from L7. The separation of L1 and L7 results in a conformational change that hides the ER exit signal “MELADL” sequence and allows Insig binding of SCAP; both actions lead to ER retention of SCAP-SREBP complex and obstruction of SREBP processing (27).
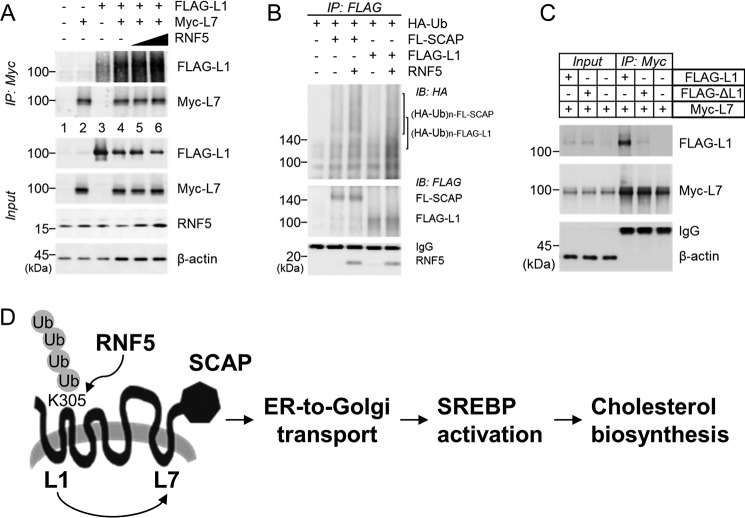
We noticed that Lys-305 is located at the junction of TM2 and loop2 and hypothesized that the ubiquitination of Lys-305 helped SCAP to assume an L1-L7–bound conformation. To test this assumption, we took advantage of an experimental system developed by the Brown and Goldstein laboratory (7). SRD-13A cells were transfected with FLAG-L1 and Myc-L7, and L1-L7 binding was assessed by anti-Myc IP. We observed L1-L7 interaction as expected (Fig. 6A, lanes 1–4). Intriguingly, co-transfection of RNF5 dose-dependently enhanced L1-L7 binding (Fig. 6A, lanes 4–6). This finding supports our hypothesis, so we went on and examined whether ubiquitination was involved in the regulation of L1-L7 interaction. We found that RNF5 interacted with FLAG-L1 and mediated its polyubiquitination (Fig. 6B). Moreover, when we used K305R-L1 mutant (FLAG-ΔL1) to perform the experiment, the mutant form of L1 failed to bind to L7 as its WT counterpart (Fig. 6C). Taking these data into account, we propose that RNF5 facilitates SREBP2 activation by mediating the polyubiquitination of SCAP at K305, thereby securing L1-L7 interaction, which is critical to ER-to-Golgi transport of the SCAP-SREBP protein complex (Fig. 6D).
Figure 6.
RNF5-dependent polyubiquitination enhances luminal loop 1 and loop 7 interaction of SCAP. A, SRD-13A was seeded in 6-well plates and cultured until cell density reached 90% confluent. The cells were switched to Opti-MEM and transfected with 0.5 μg/well FLAG-SCAP(amino acids 2–472) (L1) and/or 0.5 μg/well Myc-SCAP(amino acids 496–1279) (L7) and 0.5, 0.75, 1.0 μg/well RNF5 expression plasmids and incubated for 2 h. The cells were then switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 22 h and then harvested for anti-Myc IP. B, SRD-13A were cultured as described in (A) and transfected with 1.0 μg/well HA-ubiquitin and/or 0.5 μg/well FLAG-SCAP or FLAG-L1 in the presence or absence of 0.5 μg/well RNF5 expression plasmids and incubated for 2 h. The cells were then switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 22 h and then harvested for anti-Myc IP. C, SRD-13A were as described in (A) and transfected with 1.0 μg/well Myc-L7 and/or 1.0 μg/well FLAG-L1 or FLAG-L1 K305R mutant plasmids and incubated for 2 h. The cells were then switched to DMEM/Ham's F-12 supplemented with 5% LPDS and cultured for 22 h and then harvested for anti-Myc IP. D, schematic illustration of the proposed mechanism. RNF5 induces polyubiquitination of SCAP at Lys-305, promoting loop 1–loop 7 interaction, which is a crucial event for SREBP-mediated cholesterol biosynthetic pathway.
Discussion
Cholesterol homeostasis is regulated by a delicate feedback inhibition mechanism in which excessive cholesterol inactivates SREBP2 to shut down cholesterol biosynthesis and import via binding to SCAP (27). A fundamental aspect of this regulation is the ability of SCAP to change conformation in response to cholesterol binding (7). In this report, we show that RNF5, an ER-bound E3 ubiquitin ligase, contributes to SCAP conformational change via inducing Lys-29–linked polyubiquitination.
Zhang et al. (23) reported that RNF145, an LXR-responsive ER-anchored E3 ubiquitin ligase, induced the polyubiquitination of SCAP to inhibit cholesterol biosynthesis. RNF145 induced the polyubiquitination of cytosolic loop 6 of SCAP, presumably blocking access of the “MELADAL” sequence to COPII vesicle, and thereby preventing ER-to-Golgi transport. Interestingly, we found that RNF5 had the opposite function. RNF5 induced the polyubiquitination of cytosolic loop 2 of SCAP to promote interaction between SCAP luminal loop 1 and loop 7, a crucial event for cholesterol biosynthesis (7).
We also identified that RNF5 induced Lys-29–linked polyubiquitination. The biochemical and structural features of Lys-29–linked ubiquitination have been studied in detail in recent years. For example, Michel et al. (28) found that human HECT E3 ligases UBE3C and AREL1 assemble Lys-48/Lys-29– and Lys-11/Lys-33–linked ubiquitin chains that adopt open and dynamic conformations. Moreover, Kristariyanto et al. (29) determined the crystal structures of Lys-29–linked di-ubiquitin and its complex with NZF1 domain of TRABID, a Lys-29/Lys-33–specific deubiquitinating enzyme. They found that the flexibility and linkage-specific binding were achieved by using the hydrophobic patch on only one of the ubiquitin moieties. Therefore, it is plausible to assume that Lys-29 linkage is involved in mediating protein-protein interaction. Indeed, it was reported that SMURF1 ubiquitinates axin through Lys-29–linked polyubiquitin chains to disrupt its interaction with the Wnt coreceptors LRP5/6 to repress Wnt/β-catenin signaling (30). SMURF1 also mediates the Lys-29–linked polyubiquitination of UVRAG, disrupting UVRAG-RUBCN binding to promote autophagosome maturation (31). TRAF4, an E3 ubiquitin ligase that is highly expressed in prostate cancer, regulates metastasis by inducing Lys-27/Lys-29–polyubiquitination of TrkA kinase to facilitate its interactions with downstream proteins (32). In a study of Parkinson's disease, WSB1 was shown to mediate the Lys-27/Lys-29 polyubiquitination of LRRK2 to promote LRRK2 aggregation, leading to neuronal protection (33).
In this report, we show that RNF5-induced Lys-29 polyubiquitination led to conformational change of SCAP, which promoted cholesterol biosynthesis. Although we present evidence of enhanced loop 1–loop 7 interaction of SCAP, it is possible that protein-protein interaction is also regulated by RNF5. Lys-29 polyubiquitin chains on SCAP may serve as a platform for the recruitment of COPII vesicle proteins to facilitate ER-to-Golgi transport; alternatively, they may hinder Insig-SCAP binding and thereby facilitate ER exit. In addition, it is possible that RNF5 might also regulate SCAP in other organelles. Indeed, we were able to detect RNF5 in both ER and Golgi fractions, whereas Zhong et al. (14) reported that RNF5 was presented in both ER and mitochondria fractions. Of note, this is one of the two reports showing that nondegradative ubiquitination of SCAP modulates cholesterol biosynthesis.
Interestingly, Zhang et al. (23) found that RNF145 suppresses SREBP2 activation. In addition, studies also showed that RNF145 mediates polyubiquitination and proteasomal degradation of HMGCR (42, 43). These studies clearly demonstrate the role of RNF145 in inhibiting cholesterol biosynthesis, whereas we found here that RNF5 promotes SREBP2 activation and cholesterol biosynthesis. Whether RNF5 and RNF145 coordinate to form a ubiquitin coding system as an additional mechanism to regulate cholesterol homeostasis requires further investigation.
RNF145 is expressed in liver and the expression of RNF145 is induced by LXR ligand and high-cholesterol feeding-responsive gene, indicating that RNF145 serves as an alternative feedback inhibition pathway to control cholesterol biosynthesis (23). We also investigated whether RNF5 expression was also regulated by a similar mechanism. We examined the effect of 25-HC, a potent LXR agonist, on RNF5 expression. However, 25-HC treatment failed to alter RNF5 expression (Fig. S6). This implies that RNF5 regulation of cholesterol might be involved in pathways other than lipid metabolism. RNF5 is involved in the regulation of altered metabolism fueled by glutamine in response to ER stress-inducing chemotherapy in breast cancer (13). In addition, RNF5 mediates the clearance of the inflammatory-inducing protein S100A8. In a murine model of DSS-induced colitis, RNF5 KO mice exhibited more severe disease symptoms than did the WT littermates (34). Moreover, RNF5 is also involved in the control of unfolded protein response signaling in response to anti-tumor immune therapy (35). These studies reveal the link between RNF5 and cancer survival, ER stress, unfolded protein response, and inflammatory response. Notably, SREBP activation also plays critical roles in tumorigenesis, ER stress response, and inflammation (9, 36–38). Whether the RNF5 regulation of SREBP is involved in these pathways awaits further examination.
Finally, as mentioned earlier, RNF5 modulates the proteasomal degradation of STING, the central mediator of antiviral type I IFN signaling. Zhong et al. (14) found that when viruses infect cells they trigger ER-to-mitochondrion translocation of RNF5. In the mitochondria, RNF5 mediates the Lys-48–linked polyubiquitination of STING, leading to the proteasome-dependent degradation of STING and the suppression of type I IFN expression. It was proposed that viruses use this mechanism to evade host immune surveillance. Interestingly, York et al. (39) found that inhibition of the cholesterol biosynthetic pathway via the deletion of SCAP or SREBP2 led to the activation of type I IFN signaling. Our finding in this paper may provide an important link between these pathways. It seems plausible that, when viral infection triggers the translocation of RNF5 out of the ER membrane, it would cause the inhibition of SREBP2 and thereby activate type I IFN signaling. This would counteract the viral strategy of using RNF5 against host immunity. We are currently investigating this possibility.
In conclusion, we identified a mechanism that is crucial for the regulation of cholesterol homeostasis. We elucidated that RNF5-induced Lys-29–linked polyubiquitination of SCAP activates SREBP2 to promote cholesterol biosynthesis. We present evidence showing that the formation of Lys-29–linked polyubiquitin chain alters the SCAP conformation to modulate lipid metabolism. These findings provide insights into a ubiquitination-dependent pathway that controls SCAP, an important mediator in both lipid and immune homeostasis.
Experimental procedures
Cell lines and culture medium
HEK293, HeLa, HepG2, and HuH-7 cells were maintained in DMEM supplemented with 10% (v/v) FBS. SRD-15, an Insig-deficient line derived from CHO) cells were maintained in DMEM/Ham's F-12 supplemented 5% (v/v) FBS. SRD-13A, an SCAP-deficient line derived from CHO was maintained in DMEM/Ham's F-12 supplemented with 5% (v/v) FBS, 5 μg/ml cholesterol, 1 mm sodium mevalonate, and 20 μm sodium oleate. SRD-15 cells were originated from the laboratory of Russell A. DeBose-Boyd (22) and SRD-13A cells were originated from the Brown and Goldstein Laboratory (6). In some experiments, cells were cultured in sterol-depleted medium supplemented with 50 μm sodium mevalonate, 12.5 μm fluvastatin, and 5% (v/v) lipoprotein-deficient serum (LPDS) in the absence or presence of 1 μg/ml 25-hydroxylcholesterol (25-HC) for indicated periods to manipulate SREBP processing. KY6, an SCAP-deficient stable line derived from HEK293 cells was established and maintained following a method described previously (26). All media were supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin. All cells were grown in a humidified incubator at 37 °C and 5% CO2.
Plasmids and siRNA transfection
The coding sequence of human RNF5 (NM_006913) was cloned into p3×FLAG-CMV-7.1 vector (Sigma-Aldrich) or a previously described pcDNA-2×vector (8) to construct FLAG-RNF5 and Myc-RNF5 expression plasmids, respectively. The coding sequence of human SCAP (NM_012235) was cloned into pcDNA-2×vector to construct Myc-SCAP expression plasmid. A synthesized oligonucleotide encoding three consecutive hemagglutinin (HA) tag was cloned into pcDNA3.1/Hygro(+) vector (Thermo Fisher Scientific) to construct pcDNA-3×HA vector. Coding sequence of human SCAP and human ubiquitin monomer (NM_001281716) was cloned into pcDNA-3×HA vector to construct HA-SCAP and HA-ubiquitin expression plasmid, respectively. Point mutations in RNF5, SCAP, and ubiquitin plasmids were generated by inverse PCR using KOD-Plus Mutagenesis Kit (Toyobo). Stealth RNAi siRNA Negative Control, Med GC (code: 12935300) and RNF5 Stealth siRNA were purchased from Thermo Fisher Scientific (assay ID: HSS155076, HSS155077, HSS155078), and control siRNA-A and RNF5 siRNA were purchased from Santa Cruz Biotechnology (codes: sc-37007 and sc-95209, respectively). Transfection of plasmids was performed using Lipofectamine 3000 Reagent, whereas transfection of siRNA was performed using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific) following manufacturer's protocols.
Antibodies
Monoclonal anti-RNF5, anti-SREBP1, and anti-Insig1 antibodies were purchased from Santa Cruz Biotechnology (codes: sc-81716, sc-13551, sc-390504). Monoclonal anti-FLAG, anti–β-actin, polyclonal anti–c-Myc antibodies were purchased from Sigma-Aldrich (codes: F1804, A1978, C3956). Anti-calnexin, anti-golgin-97, and anti-STING antibodies were purchased from Cell Signaling Technologies (codes: 2433, 13192, 13647). Rabbit anti-Sec61α antiserum was purchased from EMD Millipore (code: 07-204). Rabbit anti-SREBP2 antiserum was produced as described previously (40). Mouse monoclonal anti-HA antibody was purchased from BioLegend (code: 901501). Mouse anti-GFP antibody was purchased from Clontech (code: 632381). Rabbit monoclonal anti-SCAP antibody was purchased from Bethyl Laboratory (code: A303-553A). Purified mouse anti-GM130 (clone 35) mAb (code: 610822) and mouse anti-KDEL mAb (code: M181-3) were obtained from BD Transduction Laboratories and Medical & Biological Laboratories, respectively. Alexa Fluor 647–conjugated goat anti-rabbit IgG antibody (code: A21245) was purchased from Thermo Fisher Scientific. Peroxidase-conjugated affinity-purified donkey anti-mouse IgG and peroxidase-conjugated affinity purified donkey anti-rabbit IgG were purchased from Jackson ImmunoResearch Laboratories (codes: 715-035-151, 111-035-144).
Immunoprecipitation
Immunoprecipitation was performed following previously described methods with minor modifications (8). Briefly, anti-RNF5 and isotype matched control antibody were used with Protein G Sepharose 4 Fast Flow (GE Healthcare Life Sciences) to perform anti-RNF5 IP. Anti-HA-tag, Anti-DDDDK-tag, and Anti-Myc-tag mAb-Magnetic Beads (codes: M132-11, M185-11, M047-11) were used to perform anti-HA, anti-FLAG, and anti-Myc IP, respectively, following manufacturer's protocols.
Subcellular fractionation
HuH-7 cells were cultured for subcellular fractionation following a method described previously (8). In one experiment, cell was cultured in sterol-depleted medium for 16 h before harvest.
Real-time quantitative PCR and immunoblot analysis
RNA extraction, reverse transcription, real-time quantitative PCR, and immunoblot analysis were carried out as described previously. The sequences for quantitative PCR are listed in Table S1.
Confocal microscopy
The localization of RNF5 and SCAP was determined by confocal microscopy following a protocol modified from a method described previously (41). Briefly, HeLa, HuH-7, and HEK293 cells were seeded on glass coverslips and cultured for 16 h. The cells were then co-transfected with EGFP-SCAP and mCherry-RNF5 expression plasmids using Lipofectamine 3000 Reagent. The cells were switch to fresh medium 12 h post transfection and cultured for 1–2 days. The cells were then fixed with 4% (w/v) paraformaldehyde in PBS for 15 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. After blocking with 5% FBS (in PBS) for 1 h, specimens were incubated with anti-KDEL (1:500), anti-GM130 (1:500), anti-calnexin (1:1000), or anti-Golgin-97 (1:1000) in 2% FBS for 1.5 h, followed by incubation with Alexa Flour 647–conjugated goat anti-mouse or anti-rabbit IgG antibodies (1:600) for 1 h. Nuclei were stained with DAPI for 15 min. The specimens were washed extensively, and mounted on a glass slide with ProLong Gold Antifade Mount (Thermo Fisher Scientific). Images were obtained using a ZEISS LSM 800 confocal laser scanning microscope (ZEISS) equipped with a Plan-Apochromat 63x/1.40 Oil DIC M27 objective lens or Plan-Apochromat 40x/1.40 Oil DIC M27 objective lens. Images were processed with a Zen software (Zeiss).
Measurement of cellular and de novo biosynthesis of cholesterol
The cellular cholesterol content in HuH-7 cells was measured as described previously (8). The experimental procedures for quantifying de novo synthesized cholesterol were modified from a previous method (41). In brief, HuH-7 cells were seeded in 6-well plates at a density of 3 × 105 cells per well for 24 h. The cells were then transfected with control or RNF5-specific siRNA and grown for 24 h. The cells were then switched to sterol-depleted medium with or without the addition of 1 μg/ml 25-HC. After growing for another 16 h, the cells were switched to fresh DMEM supplemented with 5% LPDS and 1.6 μCi/ml [14C]acetate and incubated for 6 h. The cells were then harvested and washed twice with PBS. The cell pellets were mixed with 1 ml of 8 N potassium hydrate and 1 ml of ethanol and then heated at 100 °C for 2 h. The mixtures were extracted twice with 2 ml of petroleum ether to collect the cholesterol containing fractions. The cholesterol extracts were then analyzed by TLC on Silica Gel 60 (Merck) and quantified with a FLA3000 imaging system (Fujifilm). The de novo biosynthesis of cholesterol was determined by normalizing the signal of [14C]cholesterol by the total cellular protein level.
Statistical analysis
All experiments were performed at least three times on different days. For qPCR and cholesterol quantification experiments, data from at least two independent experiments performed in triplicates (n = 3) are pooled for statistical analysis. Data are presented as means ± S.D., and significant difference was determined by computing p values between control and treated groups using a two-tailed equal variance Student's t test where the null hypothesis was rejected when p value is less than 0.05. Statistical analysis was performed using Microsoft Excel software.
Author contributions
Y.-C. K. and R. S. conceptualization; Y.-C. K. data curation; Y.-C. K. formal analysis; Y.-C. K. and Y. T. validation; Y.-C. K. investigation; Y.-C. K. visualization; Y.-C. K., T. M., M. S., and Y. Y. methodology; Y.-C. K. writing-original draft; Y.-C. K. and R. S. writing-review and editing; M. S., Y. Y., and R. S. resources; R. S. supervision; R. S. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Brown at the New South Wales University for helpful discussion about this work. We thank Enago for English language review.
This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (S) 15H05781, Cross-ministerial Strategic Innovation Promotion Program Grant 14533567, Japanese Agency for Medical Research and Development Grant 16gm0910008h0001 (to R. S.), and a JSPS KAKENHI Grant-in-Aid for JSPS Fellows 17F17100 (to Y.-C. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Table S1.
- SREBP
- sterol regulatory element-binding protein
- 25-HC
- 25-hydroxylcholesterol
- CHO
- Chinese hamster ovarian
- ER
- endoplasmic reticulum
- HMGCR
- 3-hydroxy-3-methylglutaryl-CoA reductase
- HMGCS
- 3-hydroxy-3-methylglutaryl-CoA synthase
- Insig
- insulin-induced gene
- IP
- immunoprecipitation
- KD
- knockdown
- LPDS
- lipoprotein-deficient serum
- RING
- really interesting new gene
- RNF
- RING finger protein
- SCAP
- SREBP cleavage-activating protein
- SQS
- squalene synthase
- STING
- stimulator of interferon genes.
References
- 1. Simons K., and Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- 2. Payne A. H., and Hales D. B. (2004) Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 25, 947–970 10.1210/er.2003-0030 [DOI] [PubMed] [Google Scholar]
- 3. Chiang J. Y. (2013) Bile acid metabolism and signaling. Compr. Physiol. 3, 1191–1212 10.1002/cphy.c120023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu D., Cai Y., Qin R., Graffy J., Holman D., Zhao Z., and Simmons D. (2018) Total/high density lipoprotein cholesterol and cardiovascular disease (re)hospitalization nadir in type 2 diabetes. J. Lipid Res. 59, 1745–1750 10.1194/jlr.P084269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Radhakrishnan A., Goldstein J. L., McDonald J. G., and Brown M. S. (2008) Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 8, 512–521 10.1016/j.cmet.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rawson R. B., DeBose-Boyd R., Goldstein J. L., and Brown M. S. (1999) Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274, 28549–28556 10.1074/jbc.274.40.28549 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y., Lee K. M., Kinch L. N., Clark L., Grishin N. V., Rosenbaum D. M., Brown M. S., Goldstein J. L., and Radhakrishnan A. (2016) Direct demonstration that loop1 of Scap binds to loop7: A crucial event in cholesterol homeostasis. J. Biol. Chem. 291, 12888–12896 10.1074/jbc.M116.729798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuan Y. C., Hashidume T., Shibata T., Uchida K., Shimizu M., Inoue J., and Sato R. (2017) Heat shock protein 90 modulates lipid homeostasis by regulating the stability and function of sterol regulatory element-binding protein (SREBP) and SREBP cleavage-activating protein. J. Biol. Chem. 292, 3016–3028 10.1074/jbc.M116.767277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng C., Ru P., Geng F., Liu J., Yoo J. Y., Wu X., Cheng X., Euthine V., Hu P., Guo J. Y., Lefai E., Kaur B., Nohturfft A., Ma J., Chakravarti A., and Guo D. (2015) Glucose-mediated N-glycosylation of SCAP is essential for SREBP-1 activation and tumor growth. Cancer Cell 28, 569–581 10.1016/j.ccell.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asano L., Watanabe M., Ryoden Y., Usuda K., Yamaguchi T., Khambu B., Takashima M., Sato S. I., Sakai J., Nagasawa K., and Uesugi M. (2017) Vitamin D metabolite, 25-hydroxyvitamin D, regulates lipid metabolism by inducing degradation of SREBP/SCAP. Cell Chem. Biol. 24, 207–217 10.1016/j.chembiol.2016.12.017 [DOI] [PubMed] [Google Scholar]
- 11. Kuang E., Qi J., and Ronai Z. (2013) Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem. Sci. 38, 453–460 10.1016/j.tibs.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., and Cyr D. M. (2006) Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 26, 571–582 10.1016/j.cell.2006.06.041 [DOI] [PubMed] [Google Scholar]
- 13. Jeon Y. J., Khelifa S., Ratnikov B., Scott D. A., Feng Y., Parisi F., Ruller C., Lau E., Kim H., Brill L. M., Jiang T., Rimm D. L., Cardiff R. D., Mills G. B., Smith J. W., Osterman A. L., Kluger Y., and Ronai Z. A. (2015) Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell 27, 354–369 10.1016/j.ccell.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhong B., Zhang L., Lei C., Li Y., Mao A. P., Yang Y., Wang Y. Y., Zhang X. L., and Shu H. B. (2009) The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30, 397–407 10.1016/j.immuni.2009.01.008 [DOI] [PubMed] [Google Scholar]
- 15. Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., and Ericsson J. (2005) Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 10.1016/j.cmet.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 16. Lee J. H., Lee G. Y., Jang H., Choe S. S., Koo S. H., and Kim J. B. (2014) Ring finger protein 20 regulates hepatic lipid metabolism through protein kinase A-dependent sterol regulatory element binding protein1c degradation. Hepatology 60, 844–857 10.1002/hep.27011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jo Y., Sguigna P. V., and DeBose-Boyd R. A. (2011) Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 286, 15022–15031 10.1074/jbc.M110.211326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Q., Liu X., Cui Y., Tang Y., Chen W., Li S., Yu H., Pan Y., and Wang C. (2014) The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41, 919–933 10.1016/j.immuni.2014.11.011 [DOI] [PubMed] [Google Scholar]
- 19. Rolland T., Taan M., Charloteaux B., Pevzner S. J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., Kamburov A., Ghiassian S. D., Yang X., Ghamsari L., Balcha D., et al. (2014) A proteome-scale map of the human interactome network. Cell 159, 1212–1226 10.1016/j.cell.2014.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., and Brown M. S. (2002) Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110, 489–500 10.1016/S0092-8674(02)00872-3 [DOI] [PubMed] [Google Scholar]
- 21. Yabe D., Brown M. S., and Goldstein J. L. (2002) Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 12753–12758 10.1073/pnas.162488899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee P. C., Sever N., and Debose-Boyd R. A. (2005) Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J. Biol. Chem. 280, 25242–25249 10.1074/jbc.M502989200 [DOI] [PubMed] [Google Scholar]
- 23. Zhang L., Rajbhandari P., Priest C., Sandhu J., Wu X., Temel R., Castrillo A., de Aguiar Vallim T. Q., Sallam T., and Tontonoz P. (2017) Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP. eLife 6, e28766 10.7554/eLife.28766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A. (2003) Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 10.1016/s1097-2765(02)00822-5 [DOI] [PubMed] [Google Scholar]
- 25. Jo Y., and DeBose-Boyd R. A. (2010) Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit. Rev. Biochem. Mol. Biol. 45, 185–198 10.3109/10409238.2010.485605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shao W., Machamer C. E., and Espenshade P. J. (2016) Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner. J. Lipid Res. 57, 1564–1573 10.1194/jlr.M069583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown M. S., Radhakrishnan A., and Goldstein J. L. (2018) Retrospective on cholesterol homeostasis: The central role of Scap. Annu. Rev. Biochem. 87, 783–807 10.1146/annurev-biochem-062917-011852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michel M. A., Elliott P. R., Swatek K. N., Simicek M., Pruneda J. N., Wagstaff J. L., Freund S. M., and Komander D. (2015) Assembly and specific recognition of K29- and K33-linked polyubiquitin. Mol. Cell 58, 95–109 10.1016/j.molcel.2015.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kristariyanto Y. A., Abdul Rehman S. A., Campbell D. G., Morrice N. A., Johnson C., Toth R., and Kulathu Y. (2015) K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of K29 polyubiquitin. Mol. Cell 58, 83–94 10.1016/j.molcel.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fei C., Li Z., Li C., Chen Y., Chen Z., He X., Mao L., Wang X., Zeng R., and Li L. (2013) Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/β-catenin signaling. Mol. Cell Biol. 33, 4095–4105 10.1128/MCB.00418-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng X., Jia Y., Zhang Y., Ma F., Zhu Y., Hong X., Zhou Q., He R., Zhang H., Jin J., Piao D., Huang H., Li Q., Qiu X., and Zhang Z. (2019) Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 15, 1130–1149 10.1080/15548627.2019.1570063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh R., Karri D., Shen H., Shao J., Dasgupta S., Huang S., Edwards D. P., Ittmann M. M., O'Malley B. W., and Yi P. (2018) TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J. Clin. Invest. 128, 3129–3143 10.1172/JCI96060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nucifora F. C. Jr., Nucifora L. G., Ng C. H., Arbez N., Guo Y., Roby E., Shani V., Engelender S., Wei D., Wang X. F., Li T., Moore D. J., Pletnikova O., Troncoso J. C., Sawa A., Dawson T. M., Smith W., Lim K. L., and Ross C. A. (2016) Ubiqutination via K27 and K29 chains signals aggregation and neuronal protection of LRRK2 by WSB1. Nat. Commun. 7, 11792 10.1038/ncomms11792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fujita Y., Khateb A., Li Y., Tinoco R., Zhang T., Bar-Yoseph H., Tam M. A., Chowers Y., Sabo E., Gerassy-Vainberg S., Starosvetsky E., James B., Brown K., Shen-Orr S. S., Bradley L. M., Tessier P. A., and Ronai Z. A. (2018) Regulation of S100A8 stability by RNF5 in intestinal epithelial cells determines intestinal inflammation and severity of colitis. Cell Rep. 24, 3296–3311.e6 10.1016/j.celrep.2018.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y., Tinoco R., Elmén L., Segota I., Xian Y., Fujita Y., Sahu A., Zarecki R., Marie K., Feng Y., Khateb A., Frederick D. T., Ashkenazi S. K., Kim H., Perez E. G., et al. (2019) Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5−/− mice. Nat. Commun. 10, 1492 10.1038/s41467-019-09525-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim J. Y., Garcia-Carbonell R., Yamachika S., Zhao P., Dhar D., Loomba R., Kaufman R. J., Saltiel A. R., and Karin M. (2018) ER stress drives lipogenesis and steatohepatitis via caspase-2 activation of S1P. Cell 175, 133–145.e15 10.1016/j.cell.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J. H., Phelan P., Shin M., Oh B. C., Han X., Im S. S., and Osborne T. F. (2018) SREBP-1a-stimulated lipid synthesis is required for macrophage phagocytosis downstream of TLR4-directed mTORC1. Proc. Natl. Acad. Sci. U.S.A. 115, E12228–E12234 10.1073/pnas.1813458115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oishi Y., Spann N. J., Link V. M., Muse E. D., Strid T., Edillor C., Kolar M. J., Matsuzaka T., Hayakawa S., Tao J., Kaikkonen M. U., Carlin A. F., Lam M. T., Manabe I., Shimano H., Saghatelian A., and Glass C. K. (2017) SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 25, 412–427 10.1016/j.cmet.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. York A. G., Williams K. J., Argus J. P., Zhou Q. D., Brar G., Vergnes L., Gray E. E., Zhen A., Wu N. C., Yamada D. H., Cunningham C. R., Tarling E. J., Wilks M. Q., Casero D., Gray D. H., et al. (2015) Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 163, 1716–1729 10.1016/j.cell.2015.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato R., Miyamoto W., Inoue J., Terada T., Imanaka T., and Maeda M. (1999) Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J. Biol. Chem. 274, 24714–24720 10.1074/jbc.274.35.24714 [DOI] [PubMed] [Google Scholar]
- 41. Miyata S., Inoue J., Shimizu M., and Sato R. (2015) Xanthohumol improves diet-induced obesity and fatty liver by suppressing sterol regulatory element-binding protein (SREBP) activation. J. Biol. Chem. 290, 20565–20579 10.1074/jbc.M115.656975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang L. Y., Jiang W., Tian N., Xiong Y. N., Liu J., Wei J., Wu K. Y., Luo J., Shi X. J., and Song B. L. (2018) Ring finger protein 145 (RNF145) is a ubiquitin ligase for sterol-induced degradation of HMG-CoA reductase. J. Biol. Chem. 293, 4047–4055 10.1074/jbc.RA117.001260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Menzies S. A., Volkmar N., van den Boomen D. J., Timms R. T., Dickson A. S., Nathan J. A., and Lehner P. J. (2018) The sterol-responsive RNF145 E3 ubiquitin ligase mediates the degradation of HMG-CoA reductase together with gp78 and Hrd1. eLife 7, e40009 10.7554/eLife.40009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.