Introduction
Since their discovery 40 years ago, Ebola viruses (in the following: EBOV; family Filoviridae, genus Ebolavirus) continue to emerge unpredictably and cause Ebola virus disease (EVD) in humans and susceptible animals in tropical Africa (Leroy et al. 2004; Feldmann and Geisbert 2011). The scale of the current epidemic in West Africa demonstrates the impact that a single spillover event can have (Baize et al. 2014; Gire et al. 2014). Meanwhile, the reservoir(s) and ecology of EBOV remain largely unknown (Groseth et al. 2007; Feldmann and Geisbert 2011), hampering prediction of future outbreaks.
To date, the only laboratory-confirmed sources of human EVD outbreaks were infected great apes and duikers (Leroy et al. 2004). However, these species are unlikely reservoirs as high mortality rates rule out an indefinite infection chain (Leroy et al. 2004; Bermejo et al. 2006; Wittmann et al. 2007). Scientists are therefore searching for other hosts where EBOV circulate without major negative effects; fruit bats have received the most research attention and are frequently referred to as the reservoir for African EBOV (Centers for Disease Control and Prevention 2014b; O’Shea et al. 2014; World Health Organization 2014). We review current evidence and highlight that fruit bats may not represent the main, or the sole, reservoir. We discuss evidence implicating insectivorous bats and reiterate that bats themselves might not be the ultimate reservoir for EBOV. Knowing which species are involved will facilitate an understanding of factors allowing spillover to susceptible human and wildlife populations (Viana et al. 2014; Plowright et al. 2015).
The Current Hypothesis of Fruit Bats as Reservoir: A Story of Chinese Whispers?1
Although a number of potential reservoirs have been considered since EBOV were first detected in the mid-1970s, the hypothesis of fruit bats as EBOV reservoir has been dominant for over a decade (Swanepoel et al. 1996; Leirs et al. 1999; Leroy et al. 2005; Pourrut et al. 2005; Olson et al. 2012). Although evidence suggests EBOV ecology involves fruit bats, the case for sustained circulation remains largely unconfirmed and epidemiological links between fruit bats and human index-cases is sparse.
Evidence of Exposure to EBOV and Outcome of Infection
The discovery of viral RNA in 13 specimens of Epomops franqueti, Hypsignathus monstrosus, and Myonycteris torquata collected during the EVD outbreak investigations in Gabon, 2003, entrenched fruit bats as the likely reservoir. However, the virus itself could not be isolated from these samples and despite an intensive search, it has not been possible to generate viral sequences from bats captured since. EBOV-specific antibodies were found in 16 samples, but not in PCR-positive specimens. Rather, there was a shift in the proportion of PCR- and seropositive individuals over a 5-month period: viral prevalence in Mbomo soon after onset of the outbreak was 22.6%, and no bats exhibited antibodies. Five months later, viral prevalence declined to 2.2%, and antibody prevalence had increased to 7.5%. Thus, fruit bats at the beginning of the outbreak seemed not to have had previous EBOV exposure and appeared being able to clear infections (Leroy et al. 2005).
Several studies confirmed EBOV-specific antibodies in certain populations of Eidolon helvum, Epomophorus gambianus, Rousettus aegyptiacus, Micropteropus pusillus, Epomops franquetti, and H. monstrosus (Pourrut et al. 2007, 2009; Hayman et al. 2010, 2012; Ogawa et al. 2015); seroprevalence was generally low but varies by site and population (Fig. 1). Sampling effort has targeted EVD outbreak areas, but screening has also been done in a number of countries and regions where human outbreaks have not been observed, suggesting EBOV may be circulating in areas where human outbreaks have not been documented (e.g., Olival et al. 2013; Ogawa et al. 2015). Live seropositive specimens suggest bats of these species are exposed to EBOV and survive infections, confirming what was shown by experimental infections of Epomophorus wahlbergi (Swanepoel et al. 1996). However, the patchy pattern of seropositive bat populations across the predicted EBOV-niche in Africa and the inability to find further EBOV sequences despite intensive efforts suggest EBOV are not widely and generally present in these populations (Pigott et al. 2014).
Figure 1.
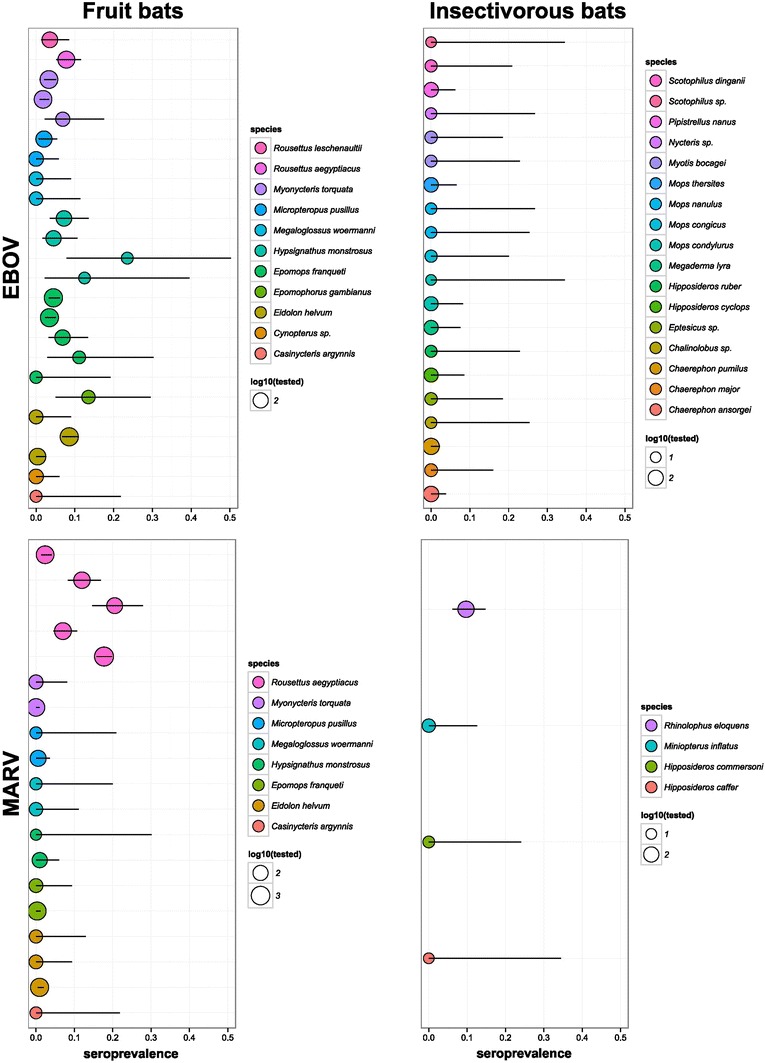
EBOV and MARV seroprevalence in bats. In each panel, each line represents a study. Circle size is proportional to the number of individuals that were tested. Bars represent 95% confidence intervals on proportions.
Knowledge About Shedding and Transmission
PCR-positive fruit bat specimens were collected soon after the onset of the human outbreak when shedding and transmission rates in bats are expected to be high. However, the only PCR-positive organs were liver and spleen; levels of viral RNA were low, and no live virus was isolated. In other blood-filled organs (heart, liver, kidneys), no viral RNA was detected (Leroy et al. 2005). This raises questions about the ability of the virus to shed in bat bodily fluids. One theory of how great apes and other animals who do not hunt bats are infected is via fecal contamination of their food or habitat. Although experimental infection of E. wahlbergi resulted in fecal shedding (Swanepoel et al. 1996), evidence of EBOV fecal shedding has not been described in wild bat populations. The low number of EBOV-positive bats detected in the wild has limited our understanding of shedding and transmission.
Epidemiological Evidence for Zoonotic Transmission
No fruit bat hunter has been reported as index-case, despite widespread hunting across Africa (Mickleburgh et al. 2009; Kamins et al. 2011). The only proposed epidemiological link between fruit bats and an outbreak relies on limited evidence from the Luebo-2007 outbreak (Leroy et al. 2009); it was suggested that the first person to succumb to EVD (a 4-year-old child) was infected via sweat by her father, who had bought fruit bat meat from the local market and was presumed to be the index-case. The father did not fall ill or show typical signs of EVD, nor were any of the hunters or villagers involved in the annual 3-week mass-hunting and butchering of migrating fruit bats among the first to succumb to the virus. While evidence of asymptomatic infections is mounting, individuals are currently only presumed infectious when symptomatic (Leroy et al. 2000; Becquart et al. 2010; Schoepp et al. 2014). Whether zoonotic transmission resulted from fruit bat bushmeat purchased by the father or via the exposure of the 4-year-old child to an alternative zoonotic source remains unclear. It was not possible to isolate EBOV from any wildlife in the region, although a second human outbreak occurred 1 year later, and high genetic similarity between EBOV strains from these human outbreaks suggests the virus had persisted undetected in local wildlife between outbreaks rather than in migrating fruit bats (Grard et al. 2011).
Evidence of Link Between Fruit Bats and Marburg Viruses
Evidence for a filovirus–fruit bat link is stronger for Marburg virus (MARV), although knowledge gaps regarding the full host range and circulation also remain for this filovirus (Swanepoel et al. 2007; Towner et al. 2009; Amman et al. 2012; Paweska et al. 2015; reviewed in Olival and Hayman 2014). Virological studies focused on R. aegyptiacus inhabiting East African caves where MARV outbreaks occurred, found live, healthy specimens of R. aegyptiacus to be MARV PCR and seropositive. Population PCR prevalence up to 13.3% was recorded and in contrast to EBOV, live MARV was isolated from wild bat spleens and livers (Towner et al. 2009; Amman et al. 2014). However, virus was not detected in feces or urine collected from infected specimens or the cave floor (Amman et al. 2012). Laboratory experimental subcutaneous infection of R. aegyptiacus identified a number of PCR-positive tissues including salivary glands in asymptomatic bats and viral loads detected in oral and rectal swabs are consistent with biting as a mode of bat–bat transmission (Amman et al. 2015; Paweska et al. 2015). However, the period during which the virus could be isolated was limited to a few days, and no transmission from the infected specimens to naïve, in-contact conspecifics could be induced.
MARV antibodies were detected in other fruit (E. helvum, E. franqueti, H. monstrosus, and M. pusillus), and insectivorous species (Rhinolophus eloquens) indicating that this filovirus is a multihost-parasite (Swanepoel et al. 2007; Pourrut et al. 2009) (Fig. 1). Insectivorous species (R. eloquens, Miniopterus inflatus, and Hipposideros spp.) were also PCR positive for MARV; in one outbreak, PCR prevalence in R. eloquens was similar (7/197; 3.6%) to that of R. aegyptiacus (4/127; 3.1%) tested in the same cave (Swanepoel et al. 2007). While accompanying serology suggested relatively higher viral exposure in R. aegyptiacus (20.5% vs 9.7% for R. eloquens); these findings suggest insectivorous bats may play an underappreciated role in MARV ecology. Such multispecies infection within a single cave suggests that transmission may occur between co-roosting species; however, further research is needed to confirm this and in which direction and to which extent such cross-species infection occurs. Indeed, in at least one MARV-cave, R. aegyptiacus is the only chiropteran inhabitant, which may suggest another insectivorous host is not necessary for the circulation or occurrence of MARV in a cave (Amman et al. 2012). MARV outbreaks have primarily been linked to miners’ or tourists’ entry into bat caves; to date, no bat-hunter or researcher has been recorded as an index case (Centers for Disease Control and Prevention 2014a). The seasonality of human outbreaks coincides with rapid seasonal increases in viral prevalence in juvenile R. aegyptiacus bats; these biannual pulses of infection are believed to be required for the virus to persist in this population (Amman et al. 2012; Hayman 2015).
Sampling Biases in the Search for the Ebola Reservoir
Several potential reservoirs have been sampled and subjected to experimental infections, especially in the early days of EBOV research (Swanepoel et al. 1996; Leirs et al. 1999; Pourrut et al. 2005; Olson et al. 2012). For example, as many primates as fruit bats had been sampled by the mid-2000s; however, since viral sequences were generated from fruit bats in 2005, the fruit bat reservoir hypothesis has very understandably driven an enormous sampling bias in the wildlife tested (Fig. 2). This observed bias may be exacerbated by a publication bias, as negative results are difficult to publish and not well documented. Peer-reviewed literature studies report that more than 800 individuals of E. franqueti (and more than 700 for the other two main suspected fruit bat species combined) have been analyzed and fruit bats represent the overwhelming majority of recent sampling efforts (Table 1). In contrast, insectivorous bats have received less research attention, yet evidence originating from a relatively small number of specimens suggests they also play a role in EBOV ecology. Less than 1000 African insectivorous bats representing approximately 20 species (in many cases, species determinations were not precise) have been analyzed (Table 1). Among them, Mops condylurus specimens were found to harbor EBOV-specific antibodies, suggesting exposure (Pourrut et al. 2009). Two insectivorous bat species, M. condylurus and Tadarida pumila (i.e., Chaerephon pumilus), survived experimental infections while displaying high viremia (Swanepoel et al. 1996). Closely related Mops trevori were present in the cotton factory where spillover was presumed to have occurred in Sudan in 1976 and 1979 (Francis et al. 1978; Pattyn 1978; Smith 1978; Baron et al. 1983). It is hypothesized that the current EVD epidemic was started with a 2-year-old boy’s contact to a high-density colony of M. condylurus, although the evidence supporting this claim remains largely circumstantial as no EBOV-positive wildlife species were detected in the region following the outbreak (Saéz et al. 2015). Collectively, these results indicate that insectivorous bats are involved in EBOV ecology and possibly an EBOV source for humans. Aspects of insectivorous bat ecology may be informative for predicting future EVD outbreaks (Pigott et al. 2014).
Figure 2.
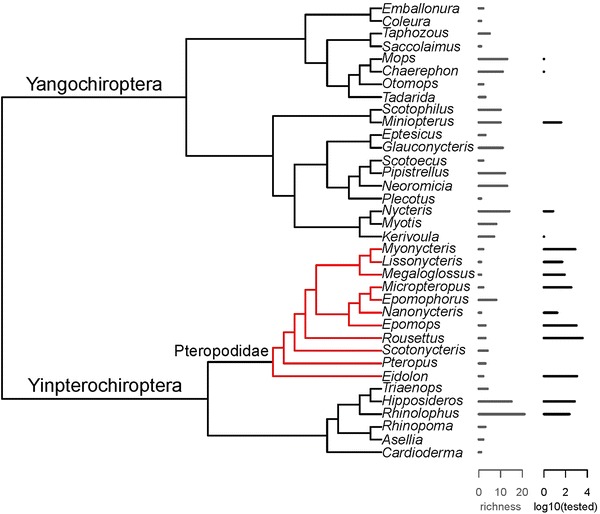
Cladogram of bat genera occurring in countries predicted to belong to the EBOV zoonotic niche, bat species richness and EBOV testing intensity. This cladogram was created using PhyML 3.0 (Guindon et al. 2010) and derived from the analysis of short barcode sequences (partial mitochondrial cytochrome c oxidase subunit 1) and may therefore not accurately represent the relationships between genera: Red branches on the phylogeny represent genera in the family Pteropodidae. Note that genera for which no barcode was available are not included (pteropodids: Casinycteris, Hypsignathus, Plerotes; other bats: Cistugo, Cloeotis, Laephotis, Lavia, Mimetillus, Mormopterus, Myopterus, Myzopoda, Nycticeinops, Platymops, Sauromys); altogether these genera only account for 18/221 species (8%) and 128/7672 of the bats tested for EBOV (21 Casinycteris and 127 Hypsignathus; 2%). The list of countries predicted to belong to the EBOV zoonotic niche was obtained from (Pigott et al. 2014).
Table 1.
Sampling Effort Targeting Wild African Bats Where EV PCR or Serological Tests Were Performed.
| Fruit bats | Insectivorous bats | Sampling country | References | ||
|---|---|---|---|---|---|
| PCR | Serology | PCR | Serology | ||
| 0/1 | DRC | Germain (1978) | |||
| 0/27 | 0/413 | DRC, Cameroon | Breman et al. (1999) | ||
| 0/125 | 0/414 | DRC | Leirs et al. (1999) | ||
| 0/132 | 0/7 | CAR | Morvan et al. (1999) | ||
| 13/279 | 16/192 | Gabon, RC | Leroy et al. (2005)a | ||
| 40/1390 | Gabon, RC, Senegal | Pourrut et al. (2007)a | |||
| 92/2123 | 3/24 | Gabon, RC | Pourrut et al. (2009)a | ||
| 1/265 | Ghana | Hayman et al. (2010) | |||
| 10/85 | Ghana | Hayman et al. (2012) | |||
| 0/111 | 0/58 | Guinea | Saéz et al. (2015) | ||
| 0/367 | 64/748 | Zambia | Ogawa et al. (2015) | ||
Number positive/number tested. Positive results highlighted in bold.
DRC Democratic Republic of Congo, RC Republic of Congo, CAR Central African Republic.
aThese publications report partially the same specimens and tests.
Future Directions
The emerging picture of EBOV ecology is one of multiple host species, with a blend of carrier-, resistant-, stuttering chain-, and multiplier hosts involved (Viana et al. 2014; Plowright et al. 2015), although it is unlikely that efforts have sampled the full range of potential hosts (Lahm et al. 2007; Mandl et al. 2015). Interestingly, such a complex viral ecology would not be without precedent. Other zoonotic viruses indeed exist, which are able to infect multiple, phylogenetically distant hosts (e.g., lymphocytic choriomeningitis virus in domestic mice and hamsters; Albariño et al. 2010), including some primary reservoirs of which are bats (e.g., SARS and MERS coronaviruses in bats and small carnivores and bats and camels, respectively; Chan et al. 2015).
Despite valiant efforts and large-scale sampling of an impressive number of taxa, followed by a decade of more targeted sampling of fruit bats following the discovery of viral RNA in a number of species (Leroy et al. 2005), the evidence for a fruit bat reservoir is still far from decisive. Bats are evidently involved in EBOV ecology and may represent the best place to begin studying EBOV circulation. However, it remains possible that bats are intermediate hosts occasionally exposed via another intermediate host or unknown reservoir. Viral emergence might be more related to environmental factors and other hosts than bats themselves. The combination of ecological factors determining the occurrence of outbreaks has not been identified (Pigott et al. 2014), and there is little agreement on if and how movement of EBOV occurs between the large distances observed between outbreaks (Leroy et al. 2004; Walsh et al. 2005; Biek et al. 2006; Wittmann et al. 2007). Data on seroprevalence in fruit and insectivorous bats across Africa may help targeting hot zones; a shared database including negative, otherwise difficult to publish, results would be helpful toward this end, and consensus and validation of serology methods in different bat species will be critical (discussed in more detail in: Olival and Hayman 2014). Similarly, studies on bat EBOV infection and immunity could help in understanding how long bats are infected and potentially infectious, and the duration and possible role of antibodies in Ebola resistance. Rapid detection of human and wildlife outbreaks remains a cornerstone in the prevention of large outbreaks and will allow timely sampling of ecological conditions and potential reservoirs that could help us understand EBOV ecology and ultimately the development of intervention strategies.
Acknowledgments
The authors thank Bodil Jensen for support and discussions. JFG was supported by an NSF Graduate Research Fellowship (DGE-1142336), the Canadian Institutes of Health Research’s Strategic Training Initiative in Health Research’s Systems Biology Training Program; an NSERC Vanier Canada Graduate Scholarship (CGS), a long-term Research Grant from the German Academic Exchange Service (DAAD-91525837-57048249), a Quebec Centre for Biodiversity Science Excellence Award, and a Graduate Research Mobility Award from McGill University.
Footnotes
This children’s game also goes by whisper down the lane or telephone, depending on the country where it is played. While the pioneering studies discussed below were well done and careful to state that their evidence did not confirm the sole or ultimate reservoir, this message of restraint has been lost in some recent reviews and popular media. Our goal here is to reiterate the original message and highlight future directions.
Siv Aina J. Leendertz and Jan F. Gogarten contributed equally to this article.
References
- Albariño CG, Palacios G, Khristova ML, Erickson BR, Carroll SA, Comer JA, et al. High diversity and ancient common ancestry of lymphocytic choriomeningitis virus. Emerging Infectious Diseases. 2010;16:1093–1100. doi: 10.3201/eid1607.091902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathogens. 2012;8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Jones ME, Sealy TK, Uebelhoer LS, Schuh AJ, Bird BH, et al. Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus) Journal of Wildlife Diseases. 2015;51:113–124. doi: 10.7589/2014-08-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Nyakarahuka L, McElroy AK, Dodd KA, Sealy TK, Schuh AJ, et al. Marburg virus resurgence in Kitaka Mine bat population after extermination attempts, Uganda. Emerging Infectious Diseases. 2014;20:1761–1764. doi: 10.3201/eid2010.140696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba NF, et al. Emergence of Zaire Ebola virus disease in Guinea—preliminary report. New England Journal of Medicine. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- Baron RC, McCormick JB, Zubeir OA. Ebola virus disease in southern Sudan: hospital dissemination and intrafamilial spread. Bulletin of the World Health Organization. 1983;61:997–1003. [PMC free article] [PubMed] [Google Scholar]
- Becquart P, Wauquier N, Mahlakõiv T, Nkoghe D, Padilla C, Souris M, et al. High prevalence of both humoral and cellular immunity to Zaire Ebolavirus among rural populations in Gabon. PLoS ONE. 2010;5:e9126. doi: 10.1371/journal.pone.0009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo M, Rodriguez-Teijeiro JD, Illera G, Barroso A, Vila C, Walsh PD. Ebola outbreak killed 5000 gorillas. Science. 2006;314:1564–1564. doi: 10.1126/science.1133105. [DOI] [PubMed] [Google Scholar]
- Biek R, Walsh PD, Leroy EM, Real LA. Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathogens. 2006;2:e90. doi: 10.1371/journal.ppat.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breman JG, Johnson KM, van der Groen G, Robbins CB, Szczeniowski MV, Ruti K, et al. A search for Ebola virus in animals in the Democratic Republic of the Congo and Cameroon: ecologic, virologic, and serologic surveys, 1979–1980. Ebola Virus Study Teams. The Journal of Infectious Disease. 1999;179(Suppl 1):S139–147. doi: 10.1086/514278. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2014a). Chronology of Marburg hemorrhagic fever outbreaks http://www.cdc.gov/vhf/marburg/resources/outbreak-table.html. Accessed on March 24, 2015
- Centers for Disease Control and Prevention (2014b). Ebola virus disease—transmission http://www.cdc.gov/vhf/ebola/transmission/index.html. Accessed on 10.02.2015
- Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle east respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clinical Microbiology Reviews. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Geisbert TW. Ebola haemorrhagic fever. The Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis D, Smith D, Highton R, Simpson D, Lolik P, Deng IM, et al. (1978) Ebola fever in the Sudan, 1976: epidemiological aspects of the disease. Ebola Virus Haemorrhagic Fever :100–104
- Germain M. Collection of mammals and arthropods during the epidemic of haemorrhagic fever in Zaire. Ebola virus haemorrhagic fever. Amsterdam: Elsevier; 1978. pp. 185–189. [Google Scholar]
- Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard G, Biek R, Tamfum JJ, Fair J, Wolfe N, Formenty P, et al. Emergence of divergent Zaire ebola virus strains in Democratic Republic of the Congo in 2007 and 2008. Journal of Infectious Diseases. 2011;204(Suppl 3):S776–784. doi: 10.1093/infdis/jir364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Feldmann H, Strong JE. The ecology of Ebola virus. Trends in Microbiology. 2007;15:408–416. doi: 10.1016/j.tim.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hayman DT. Biannual birth pulses allow filoviruses to persist in bat populations. Proceedings of the Royal Society of London B: Biological Sciences. 2015;282:20142591. doi: 10.1098/rspb.2014.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DT, Emmerich P, Yu M, Wang L-F, Suu-Ire R, Fooks AR, et al. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS ONE. 2010;5:e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DT, Yu M, Crameri G, Wang LF, Suu-Ire R, Wood JL, et al. Ebola virus antibodies in fruit bats, Ghana, West Africa. Emerging Infectious Diseases. 2012;18:1207–1209. doi: 10.3201/eid1807.111654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamins AO, Restif O, Ntiamoa-Baidu Y, Suu-Ire R, Hayman DT, Cunningham AA, et al. Uncovering the fruit bat bushmeat commodity chain and the true extent of fruit bat hunting in Ghana, West Africa. Biological Conservation. 2011;144:3000–3008. doi: 10.1016/j.biocon.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm SA, Kombila M, Swanepoel R, Barnes RF. Morbidity and mortality of wild animals in relation to outbreaks of Ebola haemorrhagic fever in Gabon, 1994–2003. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101:64–78. doi: 10.1016/j.trstmh.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Leirs H, Mills JN, Krebs JW, Childs JE, Akaibe D, Woollen N, et al. Search for the Ebola virus reservoir in Kikwit, Democratic Republic of the Congo: reflections on a vertebrate collection. Journal of Infectious Diseases. 1999;179(Suppl 1):155–163. doi: 10.1086/514299. [DOI] [PubMed] [Google Scholar]
- Leroy E, Baize S, Volchkov V, Fisher-Hoch S, Georges-Courbot M, Lansoud-Soukate J, et al. Human asymptomatic Ebola infection and strong inflammatory response. The Lancet. 2000;355:2210–2215. doi: 10.1016/S0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Epelboin A, Mondonge V, Pourrut X, Gonzalez J-P, Muyembe-Tamfum J-J, et al. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector-Borne and Zoonotic Diseases. 2009;9:723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Leroy EM, Rouquet P, Formenty P, Souquiere S, Kilbourne A, Froment JM, et al. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- Mandl JN, Ahmed R, Barreiro LB, Daszak P, Epstein JH, Virgin HW, et al. Reservoir host immune responses to emerging zoonotic viruses. Cell. 2015;160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickleburgh S, Waylen K, Racey P. Bats as bushmeat: a global review. Oryx. 2009;43:217–234. doi: 10.1017/S0030605308000938. [DOI] [Google Scholar]
- Morvan JM, Deubel V, Gounon P, Nakoune E, Barriere P, Murri S, et al. Identification of Ebola virus sequences present as RNA or DNA in organs of terrestrial small mammals of the Central African Republic. Microbes and Infection. 1999;1:1193–1201. doi: 10.1016/S1286-4579(99)00242-7. [DOI] [PubMed] [Google Scholar]
- O’Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DT, Luis AD, et al. Bat flight and zoonotic viruses. Emerging Infectious Diseases. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Miyamoto H, Nakayama E, Yoshida R, Nakamura I, Sawa H, et al. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. Journal of Infectious Diseases. 2015 doi: 10.1093/infdis/jiv063. [DOI] [PubMed] [Google Scholar]
- Olival KJ, Hayman DT. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6:1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival KJ, Islam A, Yu M, Anthony SJ, Epstein JH, Khan SA, et al. Ebola virus antibodies in fruit bats, Bangladesh. Emerging Infectious Diseases Journal. 2013;19:270. doi: 10.3201/eid1902.120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson SH, Reed P, Cameron KN, Ssebide BJ, Johnson CK, Morse SS, et al. Dead or alive: animal sampling during Ebola hemorrhagic fever outbreaks in humans. Emerging Health Threats Journals. 2012 doi: 10.3402/ehtj.v5i0.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn S. Ebola virus haemorrhagic fever. Amsterdam: Elsevier/North-Holland; 1978. [Google Scholar]
- Paweska JT, van Vuren PJ, Fenton KA, Graves K, Grobbelaar AA, Moolla N, et al. Lack of Marburg virus transmission from experimentally infected to susceptible in-contact Egyptian fruit bats. Journal of Infectious Diseases. 2015 doi: 10.1093/infdis/jiv132. [DOI] [PubMed] [Google Scholar]
- Pigott DM, Golding N, Mylne A, Huang Z, Henry AJ, Weiss DJ, et al. Mapping the zoonotic niche of Ebola virus disease in Africa. Elife. 2014 doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, et al. Ecological dynamics of emerging bat virus spillover. Proceedings of the Royal Society B: Biological Sciences. 2015;282:20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourrut X, Delicat A, Rollin PE, Ksiazek TG, Gonzalez JP, Leroy EM. Spatial and temporal patterns of Zaire Ebolavirus antibody prevalence in the possible reservoir bat species. Journal of Infectious Diseases. 2007;196:S176–S183. doi: 10.1086/520541. [DOI] [PubMed] [Google Scholar]
- Pourrut X, Kumulungui B, Wittmann T, Moussavou G, Délicat A, Yaba P, et al. The natural history of Ebola virus in Africa. Microbes and Infection. 2005;7:1005–1014. doi: 10.1016/j.micinf.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez JP, et al. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infectious Diseases. 2009;9:159. doi: 10.1186/1471-2334-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saéz AM, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Düx A, et al. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Molecular Medicine. 2015;7:17–23. doi: 10.15252/emmm.201404792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp RJ, Rossi CA, Khan SH, Goba A, Fair JN. Undiagnosed acute viral febrile illnesses, Sierra Leone. Emerging Infectious Diseases. 2014;20:1176. doi: 10.3201/eid2007.131265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DIH. Ebola haemorrhagic fever, 1976. Bulletin of the World Health Organization. 1978;56:247–270. [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R, Leman PA, Burt FJ, Zachariades NA, Braack L, Ksiazek TG, et al. Experimental inoculation of plants and animals with Ebola virus. Emerging Infectious Diseases. 1996;2:321. doi: 10.3201/eid0204.960407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R, Smit SB, Rollin PE, Formenty P, Leman PA, Kemp A, et al. Studies of reservoir hosts for Marburg virus. Emerging Infectious Diseases. 2007;13:1847. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, Carroll SAR, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathogens. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana M, Mancy R, Biek R, Cleaveland S, Cross PC, Lloyd-Smith JO, et al. Assembling evidence for identifying reservoirs of infection. Trends in Ecology & Evolution. 2014;29:270–279. doi: 10.1016/j.tree.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh PD, Biek R, Real LA. Wave-like spread of Ebola Zaire. PLoS Biology. 2005;3:e371. doi: 10.1371/journal.pbio.0030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann TJ, Biek R, Hassanin A, Rouquet P, Reed P, Yaba P, et al. Isolates of Zaire Ebolavirus from wild apes reveal genetic lineage and recombinants. Proceedings of the National Academy of Sciences USA. 2007;104:17123–17127. doi: 10.1073/pnas.0704076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2014) Ebola virus disease—fact sheet. Accessed Oct 2, 2015. No. 103


