Abstract
Human astrovirus (HAstV) constitutes a major cause of acute gastroenteritis in children. The viral 5′ and 3′ untranslated regions (UTR) have been involved in the regulation of several molecular mechanisms. However, in astrovirues have been less characterized. Here, we analyzed the secondary structures of the 5′ and 3′ UTR of HAstV, as well as their putative target sites that might be recognized by cellular factors. To our knowledge, this is the first bioinformatic analysis that predicts the HAstV 5′ UTR secondary structure. The analysis showed that both the UTR sequence and secondary structure are highly conserved in all HAstVs analyzed, suggesting their regulatory role of viral activities. Notably, the UTRs of HAstVs contain putative binding sites for the serine/arginine-rich factors SRSF2, SRSF5, SRSF6, SRSF3, and the multifunctional hnRNPE2 protein. More importantly, putative binding sites for PTB were localized in single-stranded RNA sequences, while hnRNPE2 sites were localized in double-stranded sequence of the HAstV 5′ and 3′ UTR structures. These analyses suggest that the combination of SRSF proteins, hnRNPE2 and PTB described here could be involved in the maintenance of the secondary structure of the HAstVs, possibly allowing the recruitment of the replication complex that selects and recruits viral RNA replication templates.
Electronic supplementary material
The online version of this article (10.1007/s11033-018-4498-8) contains supplementary material, which is available to authorized users.
Keywords: RNA virus, Replication, SR proteins, Stem-loop RNA structures
Introduction
Human astroviruses (HAstVs) are a major cause of acute gastroenteritis in children, the elderly and in immunocompromised subjects [1], therefore constituting an important problem public health. Recently, new members of the Astroviridae family have been associated with neurologic disorders in humans, and several reports have suggested the possibility of a cross-species transmission that could eventually become a zoonotic problem [2, 3].
HAstVs virions are non-enveloped and contain a positive-sense, single-stranded polyadenylated genome of approximately 7 kb. The genome is organized into three open reading frames (ORFs), ORF1a, ORF1b, and ORF2, flanked by 5′ and 3′ untranslated regions (UTRs) of 80–100 nucleotides (nt) [4]. A VPg protein covalently linked to the 5′ terminus of the genome is essential for viral infectivity [5]. While ORF1a encodes four nonstructural proteins, one of which is a putative serine-protease (nsP1a3), ORF1b codes for the viral RNA-dependent RNA polymerase (nsP1b) and ORF2 encodes for the capsid conforming proteins [6]. It has been suggested that the proteins encoded in the C-terminal domain of ORF1a and the complete ORF1b are involved in viral replication [7].
Regulatory 5′ and 3′ UTRs are found in single stranded positive RNA viruses (ssRNA+). These UTRs have been implicated in viral translation and replication mainly because they contain sequences and RNA secondary structures that are recognized by cellular and viral proteins involved in these processes [8–11]. The roles of several of these proteins in the regulation of both the initiation of translation and the synthesis of the viral genomic RNAs have been described (see Tables 1, 2). However, a full understanding of the precise mechanism underlying the function of all the viral and cellular proteins involved remains incomplete. Very little is known about translation and replication mechanisms of the HAstVs, and even less is known about how the astroviral UTRs are involved in such processes. In spite of this, only one report exists on the RNA secondary structure of the 3′ end of HAstV-1, which is similar to those of other RNA viruses [12]. Moreover, nothing is known about the possible RNA secondary structure of the 5′ UTR of any HAstVs, or its possible role in the regulation of viral processes such as translation or replication, which has been well documented in other positive-stranded RNA viruses.
Table 1.
Cellular proteins bound to 5′ untranslated region of ss+RNA viruses
| Virus | Protein | Viral process | Reference |
|---|---|---|---|
| Poliovirus | PCBP | Replication | [13] |
| Initial stability and translation | |||
| PTB | Translation | [14–17] | |
| La | |||
| eIF-2 | |||
| p100 | |||
| p54 | |||
| p48/38 | |||
| Dengue | La | Stabilization of the replication complex | [18] |
| Calicivirus | eIF4E, eIF3 | Translation | [19] |
| PTB La, PCBP1/2, hnRNPL, Nucleolin, DDX3 | Translation and/or replication | [16] | |
| hnRNP A1 | Replication | [20] | |
| Hepatitis C | PTB, La autoantigen, nucleolin, EIF2Bγ | Translation | [21] |
| NFAR | Replication | [22] | |
| Bovine viral diarrhea virus | NFAR | Translation and replication | [23] |
| Human astrovirus serotype 8 | PTB | Replication | [This review] |
Table 2.
Cellular proteins that bound to 3′ untranslated region of ss + RNA viruses
| Virus | Protein | Viral process | Reference |
|---|---|---|---|
| Dengue | EF1-α | Replication | [18, 24–31] |
| La | |||
| PTB | |||
| PABP | |||
| NF90 | Replication | [32] | |
| DHX9 | |||
| NF45 | |||
| YB1 | Translational repression | [33] | |
| hnRNP A1 | Unknown | [33] | |
| hnRNP A2/B2 | |||
| hnRNP Q | |||
| hnRNP H | |||
| Caprin-1 | Antiviral INF-β response | [34–36] | |
| DDX6 | |||
| USP10 | |||
| G3PB1 and G3PB2 | |||
| P100 | Unknown | [37] | |
| PDI | Unknown | [38] | |
| Calreticulin | |||
| Calicivirus | La | Replication | [39] |
| PTB | |||
| PAPB | |||
| 110, 97, 75, 50 and 42 kDa | Unknown | [40] | |
| PTB | Replication | [20, 41] | |
| PCBP1 and 2 | |||
| HnRNP A1 | |||
| PABP, PTB, PCBP | Replication | [42] | |
| La, DDX3, HuR, hnRNP A1, hnRNP A1/B2 and hnRNP A/B | |||
| Nucleolin | Replication | [43] | |
| Poliovirus | PABP | Replication | [44] |
| hnRNP C | Replication | [45, 46] | |
| Nucleolin | Replication | [47] | |
| hnRNP A1 La, Nucleolin hnRNP K and hnRNP C | Antiviral response | [48] | |
| Hepatitis C | PTB, hnRNPC, La,HuR,NFAR | Replication | [21, 22, 49] |
| Bovine viral diarrhea virus | NFAR | Translation replication | [23] |
| Human astrovirus serotype 8 | PTB | Replication | [50] |
| hnRNPE2 | Unknown | [This review] |
A general mechanism followed by positive-sense ssRNA viruses to replicate their genome consists of the following. After the release of the RNA viral genome into the cytoplasm, it is translated to produce non-structural viral proteins that, along with cellular factors, assemble viral replication complexes (VRCs) that participate in negative-stranded RNA synthesis. This RNA is used as a template to synthesize a large copy of new positive-sense RNA virus, which is followed by additional rounds of translation/replication to form new viral particles. The balance between positive and negative stranded RNA synthesis is disproportionate, which means that there are more positive RNA viral molecules than negative-stranded RNA viral molecules, which depends on the RNA viruses [51].
Several single-stranded positive animal RNA viruses, such as poliovirus, hepatitis C, West-Nile, coronavirus, dengue, calicivirus family members and others, have been used as a model to study the role of the UTRs in many processes during viral infections [8, 9, 21, 52]. It has been suggested that the interaction between the UTRs and cellular factors is important during the viral replication cycle. For example, heterogeneous nuclear ribonucleoproteins (hnRNPs), such as polypyrimidine tract-binding protein (PTB), also known as hnRNP1, is an RNA binding protein involved in multiple aspects of cellular mRNA metabolism, including splicing regulation, polyadenylation, 3′ end formation, translation of RNAs, RNA localization and stability. Additionally, PTB binding to viral RNAs is by far the best characterized to date [14, 21, 24, 25, 41, 49, 50, 53, 54].
Other proteins include the La auto-antigen (La), which is a 47 kDa nuclear RNA-phosphoprotein that associates with transcripts of RNA polymerase III [55, 56], and has also been associated with protein transport from the nucleus to the cytoplasm and their re-localization to the cytoplasm and has been observed in several RNA viral infections [57] and might play a role in positive and negative-strand RNA synthesis [15, 18, 26, 38, 49, 58]. Additionally, La protein could be involved in the interplay between viral and host proteins to determine the essential switch from translation to replication in the viral life cycle as described in Hepatitis C virus [59].
PCBP2 (poly (rC)-binding protein 2), also known as hnRNPE2, appears to be a multifunctional protein, is one of the major cellular poly(rC)-binding proteins, binds to the UTRs of several positive RNAs viruses and has been shown to function in both, RNA translation and replication [57, 60–62]. Translation elongation factor 1-α (EF1-alpha) is an abundant multifunctional protein, a GTP-binding protein that catalyzes the binding of aminoacyl-transfer RNAs to the ribosome, is involved in nucleocytoplasmic trafficking and might be involved in viral replication [8, 9, 24, 63].
Finally, some members of the SR (serine/arginine-rich) protein family [64, 65] could participate in the viral replicative cycle because they often contain one or more RNA recognition motifs, and they have been classically described as regulators in precursor (pre)-mRNA splicing [66, 67]. More recently, the shuttling of SR proteins acting as adaptors for mRNA export and as regulators for translation in the cytoplasm has been described [68]. In this review, we explore the conserved similarities of the available sequences of the 5′ and 3′ UTRs of HAstVs, and aim to describe the presence of several putative binding sites for different cellular factors. We also consider whether these RNA–protein interactions might be involved in the several viral functions of human astrovirus.
Comparisons of the human astrovirus 5′ and 3′ untranslated region secondary structures
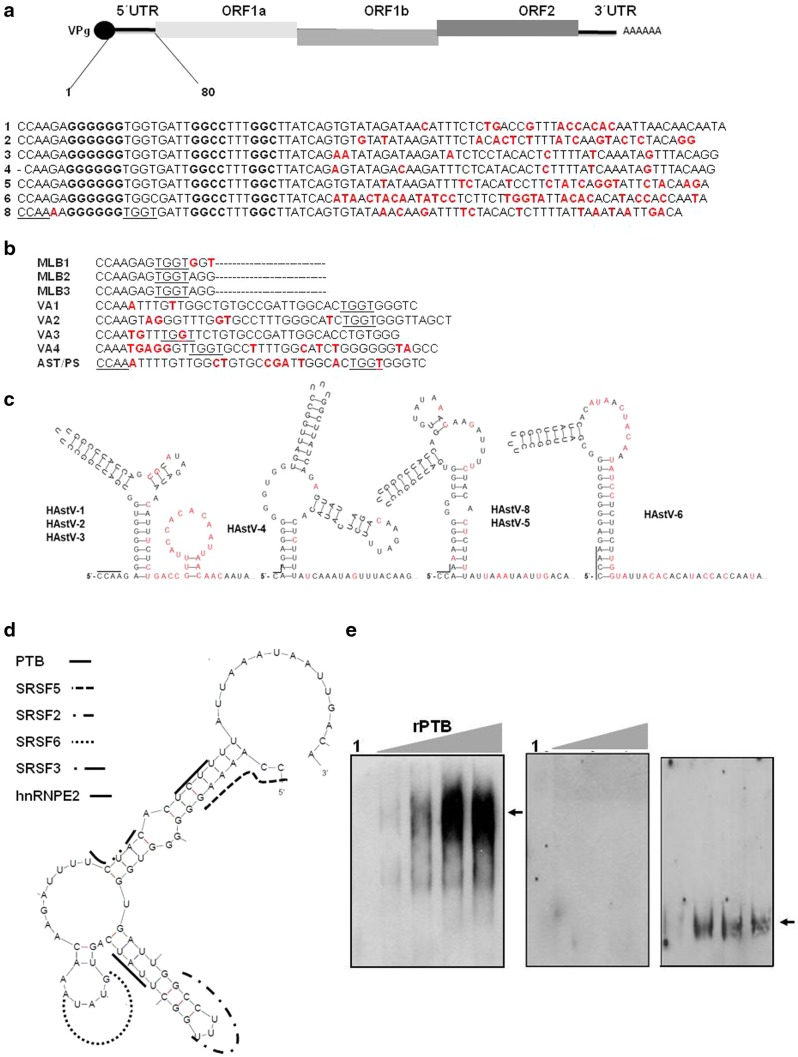
The genome of RNA viruses is extremely versatile. To maximize the efficiency of the genome, RNA viruses regulate translation and RNA synthesis through a switch between these two stages, which is triggered by the communication between the UTRs via both RNA–RNA and RNA–protein interactions [69]. These UTRs contain sequences recognized by cellular and viral proteins that participate in many viral functions. It has been suggested that the UTRs of several RNA viruses, such as dengue, human calicivirus, norovirus, West Nile, enterovirus, Hepatitis C virus and others, are involved in the translation and replication [16, 19, 21, 27, 69]. To explore a possible link between the sequence conservation and functional significance in classical HAstV, we carried out multiple sequence alignments of the 5′ UTRs of all the classical HAstVs and novel astrovirus (known as non-classical astrovirus) sequences available in the GenBank [2, 3]. The analysis revealed that the 5′ UTR contains several conserved motifs identical among all HAstV serotypes and novel astroviruses. For example, CCAA (position 1–4) residues are present in both classical viruses and novel serotypes, except for the first C which is absent in HAstV-4. The adenine present in position 5 in classical HAstVs is replaced by G/T in almost all novel astrovirus, with the exception of VA1 and AST/PS. Meanwhile the GGGGG and GGCC motifs localized around position 7 and 21, respectively, depending on the serotype (indicated in bold in Fig. 1a) are present only in classical viruses. Additionally, another sequence is conserved between the former motifs, TGGT residues are common in all classical HAstV (position nt 13–16), except in HAstV-6 where the last T residue changes to C, as well in VA and AST/PS genotypes. In MLB 1–3 clade viruses (position nt 8–11) as well in VA1-4 clade virus this motif is present but in different position (underlined). This position variability might have arisen as a function of UTR length variability: MLB1-3 (14 nt), VA1-4 (36–40 nt). Another conserved motif TTT was found in all human astrovirus and the VA1-4 clade, this is localized in position 25 and absent in MLB1-3 clade (Fig. 1b). This high degree of sequence conservation among the different classical and novel astrovirus serotypes supports the possible involvement of this region in important viral functions. The sequence similarities in both virus types may result of recombination events between human and animal strains, as previously suggested [2, 3], suggesting that they share similar mechanisms such UTRs-proteins interaction-mediated replication.
Fig. 1.
Sequences alignment, secondary structures predictions and putative binding sites for cellular proteins in the 5′ Untranslated region of HAstVs. a Alignment of the human astrovirus. 5’UTR sequences 1–8 (Accession Nos. Z25771, L13745, AF141381, AY720891, DQ028633, GQ495608.1, AF260508 respectively) except HAstV-7, which the 5′UTR is not available. b Alignment of novel astrovirus sequences available MLB1-3 (Accession Nos. FJ222451, NC_016155.1, JX857870.1), VA1-VA4 (Accession Nos. FJ973620.1, GQ502193.2, JX857868.1, JX857869.1), and AST/PS (Accesion No. GQ891990.1) The nucleotide position used to aligment MLB1-3 (nt 1–14 length) For VA1 nt 1–38, VA2 nt 1–42, VA3 nt 1–36, VA4 nt 1–49 and AST/PS nt 1–39. Red marked nucleotides correspond to sequence differences. c Predicted RNA secondary structures of the 5′UTR of human astrovirus 1–8 determined by the MFold software release, version 3.2 [70]. For secondary structure modeling and alignments, the following nucleotide positions were used: HAstV-1 nt 1–86; HAstV-2 nt 1–82, HAstV-3 nt 1–85; HAstV-4 nt 1–84; HAstV-5 nt 1–84, HAstV-6 nt 1–83 and HAstV–8 nt 1–81. d Localization of the putative binding sites for different cell factors in the HAstV-8 5′UTR RNA structure. e Mobility shift assay carried out with biotin labeled HAstV-8 5′UTR RNA incubated with 0.50, 0.75, and 1 µg of recombinant His-PTB protein (left panel), 0.5, 0.75, and 1 µg of BSA (middle panel) and heterologous RNA with 0.50, 0.75, and 1 µg His-PTB (right panel). Free probe is shown in lane 1 and PTB complexes formation is indicated by arrow. The RNA–protein complexes were detected with chemiluminiscent nucleic acid detection module (Pierce) following the conditions described by the manufacturer
From our analysis above and considering the proposal that 5′ and 3′ UTR might be required elements for viral replication and RNA synthesis [27], we set to establish whether the 5′ UTR of the HAstV region could be structurally conserved. Therefore we generated their predicted secondary structure. This is of upmost importance since no single report exists on the possible 5′ UTR RNA secondary structure (classical or novel astroviruses), neither on the importance of the 5′ UTRs in the regulation of viral processes such as translation or replication, as has been documented for other positive-stranded RNA viruses.
The analysis of classical astroviruses revealed that their secondary structures are very similar, with free energies (∆G) ranging from − 16 (HAstV-5) to − 12.1 kcal, except for HAstV6 whose predicted secondary structure has a ∆G of − 24.30 kcal. As observed above, compared to the rest of HAstVs, the 5′ UTR of HAstV-8 exhibits A to G change in position 5, and greater sequence conservation is observed towards the 5′ ends of the UTRs than towards their 3′ ends (nt 55–80) (Fig. 1a). These features correlate with the secondary structure modeling results. In general, the 5′ UTR RNA structures showed two main loops connected by a large and a small hairpin (Fig. 1c). The main differences observed in the aforementioned regions showed major nucleotide changes. Depending on the serotype, the first loop was localized around nucleotides 8/10 (HAstV-3, 4, 5, 8) or 10/14 (HAstV-1, 6), and the second loop was localized around nucleotides 55/84 (Fig. 1c). To our knowledge, this is the first report describing the possible secondary structure of the 5′ UTR from HAstV.
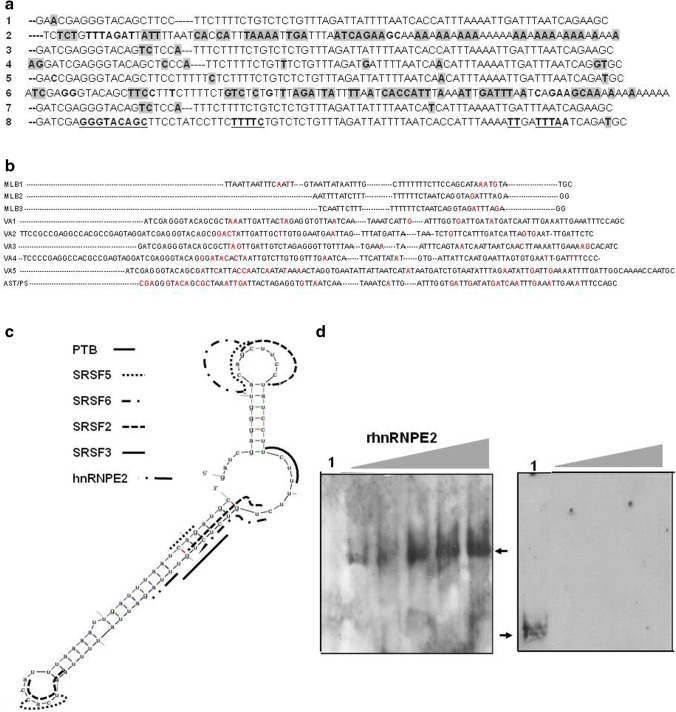
The pioneering work of Willcocks and Carter proposed the HAstV-1 3′ UTR secondary structure [12]. Recently, we described the predicted structures of the 3′ UTR of all HastV sequences available to date [50]. Our analysis showed that this region is highly conserved, complying with a double hairpin-loop secondary structure that is linked by a single-stranded RNA spacer. Following the approach used for the 5′ UTR; we performed multiple sequence alignment of the 3′ UTRs of all classical HAstVs and non- classical astrovirus available in the GenBank. The comparison revealed that the 3′ UTR contains several conserved motifs. For example, a GGGTACAGCG motif was located in both virus groups. For classical HAstV the sequence appears around positions 6739/6680, except in HAstV-2, and in non-classical astrovirus the motif is localized depending of the clade group: in MLB1-3 these motifs are absent, but in clade VA1-5, including AST/PS, the motif appears in 6406/6502. The TTT/TTTTA motif was localized around positions 6743/6801 on classical HAstV depending on the serotype (Fig. 2a). Similarly, conserved TT motifs appear on non-classical astrovirus, in MLB 1–3 are present within the first nucleotides, and VA1, 3, 5 and AST/PS clade the motif appears in different positions. Another slightly conserved sequence among no classical clade is the TTT motif, that changes the last T for G/A (VA1-VA2), or changes the middle T for A (VA4–VA5), and in AST/PS the T in the first position is changed for A (Fig. 2b). The phylogenetic analysis carried out with 3′ UTR sequences showed that classical and novel astrovirus are divergent, in which the clade VA1-VA5 and AST/PS for example, is less related with the classical astrovirus than the MLB1-3 clade as previously reported [2, 3]. The biological significance of sequence conservation between classical and non-classical astrovirus, particularly the GGGTACAGCG motif shared in classical and clade VA1-VA5 and AST/PS, remains to be uncovered, although it could be speculated that it might be involved in global replication process.
Fig. 2.
Sequence alignments of the 3′UTR sequence and putative binding sites for cellular proteins in the 3′ untranslated region of HAstVs. a Alignment of the human astrovirus 3′UTR sequences 1, 3, 4, 5, 6, 7, and 8. The accession numbers of Genbank as mentioned before (Genbank HAstV-7 accession number Y08632). The following nucleotide positions were used: HAstV-1 nt 6733–6813; HAstV-3 nt 6733–6815; HAstV-4 nt 6643–6723; HAstV-5 nt 66,677–6762; HAstV-6 nt 6665–6757 HAstV-7 nt 2403–2484 (nt numbering differs because of the partiality of the sequence); HAstV-8 nt 6674–6759. Bold and shaded nucleotides correspond to sequence differences. Gaps are noted with hyphens. GGGTACAG and TT/TTTA conserved elements are bold and underlined. b Alignment of novel astrovirus 3′ UTR sequences MLB1-3, VA1-VA5 and AST/PS The accession numbers of Genbank as mentioned before and included VA5 (Accession No KJ656124.1). The following nucleotide positions to aligment were used: MLB1 nt 6114–6171, MLB2 nt 6081–6119, MLB3 nt 6087–6124, VA1 nt 6489–6586, VA2 nt 6415–6531, VA3 nt 6487–6581, VA4 nt 6404–6518, VA5 nt 6401–6519, and AST/PS nt 6489–6584. Red marked nucleotides correspond to sequence differences. c Localization of the putative binding sites for the different cell factors in the HAstV-8 3′UTR RNA structure. d Mobility shift assay carried out with biotin labeled HAstV-8 3′UTR RNA incubated with 0.50, 0.75, and 1 µg of recombinant His-hnRNPE2 protein (left panel), or 0.5, 0.75, and 1 µg of BSA (middle panel). The heterologous RNA used was the same to Fig. 1e. Free probe is shown in lane 1 and hnRNPE2 complexes formation is indicated by arrow. The RNA–protein complexes were detected with Chemiluminiscent Nucleic Acid Detection Module (Pierce) following the conditions described by the manufacturer
Several works revealed that the integrity of the secondary structure of the UTRs of different single-stranded, positive sense RNA viruses is very important for the recruitment of both cellular and viral factors that are constituents of the replication complex (see Tables 1, 2). This complex selects and recruits viral RNA replication templates to perform synthesis of the minus-strand RNA [8] and participates in other functions, such as viral viability, RNA stability, translation initiation, and intracellular localization [62, 71–74]. Thus, this structural conservation in all HAstVs agrees and supports the possible implication of the biological functions proposed [12, 73].
Putative binding sites for cellular factors of Human Astrovirus 5′ and 3′ untranslated regions
As mentioned previously, several reports showed that cellular and viral proteins can bind to the UTRs in positive-sense RNA viruses and play important roles in many viral processes. Using various search engines, such as ESE Finder [75, 76], ESR search [77, 78] EBI-EMBL and RBPmap [79], we analyzed the 5′ and 3′ UTRs of HAstVs available looking for the target sites of cellular factors.
In silico analysis carried out with the 5′ UTR sequence showed putative binding sites for SRSF2 (GGCCTTTG), and SRSF5 (ACAGG) (Fig. 1d). Although the analysis was carried out for all classical HAstV, for the sake of clarity the sites were depicted on the secondary structure HAstV-8 only. In the case of HAstV-1 (CCACACA) and HAstV-8 (CCAAAAG) the putative binding sites were selected among the highest scores of the search engines used. The SRSF6 putative binding site (TGTATA) was identified in all 5′ UTRs analyzed except for HAstV-3 and 4 which lack this putative binding site. As before, the SRSF6 putative binding site in HAstV-2 (TGTGTA) also corresponds to the highest score in the ESEFinder software. Whereas the putative binding site (TCTAC) for SRSF3, previously known as SRp20, was observed in HAstV-2, 4, 5 and 8, using highest scores with RBPmap software, other SRSF3 binding motifs were observed in HAstV-1, 3 and 6.
hnRNPE2 (TTAT) putative binding sites are present in all serotype analyzed. Except for HAstV-3 a, the same holds for TIA1 (ATTTTCT) (Figs. S1a, b). This brings into question the role of SR proteins in the localization and infectivity of the viral particles and in the RNA secondary structure stabilization for the recruitment of the replication complex. More studies will be necessary to elucidate the participation of the SR proteins in the replicative cycle of astrovirus [64, 66, 77–79].
It has been described that the SR protein family, recently known as SRSF (SR splicing factor) [64, 65], often contains one or more RNA recognition motifs. They have been classically described as regulators of precursor (pre)-mRNA splicing [66, 67], but SRSF proteins have been involved in a series of transcription-related activities, including transcriptional elongation [80], RNA transport [81, 82], nonsense-mediated decay enhancement [28], translation-stimulation [83], genome stability maintenance [84] and cell cycle progression [85]. Furthermore, SRSF3 is required for efficient IRES-mediated translation in poliovirus through interacting with the PCBP2/hnRNPE2 protein, which directly binds to the poliovirus IRES. This probably results in either the direct or indirect recruitment of the translation complex to the viral RNA through other protein–protein or protein–RNA associations [86, 87]. So far, this is the only evidence that SRSF proteins are involved in translation of an RNA virus.
In addition to SR proteins, we found PTB/hnRNP1 putative binding sites in all viruses studied. Interestingly, in HAstV-1, -2, -3, -4.-6 and -8, the strong motif for PTB (TTCT) was localized in the double-stranded linker between the two hairpin loops that form the 5′ UTR, but not in HAstV-5 (Fig. S1a). However, we found a consensus motif TCTT located in a compromising RNA position (63–66 nt). Many reports have demonstrated the interaction of PTB (Table 1) with a viral 5′ UTR, suggesting its participation in viral translation or replication as mentioned previously [16, 54, 88–91]. To corroborate the presence of PTB binding site in the HAstV-8 5′ UTR (nt 61–64), mobility shift assays were performed with different amounts of recombinant PTB (rPTB). The results show its ability to interact with the strong motif in the 5′ UTR (Fig. 1e).
Likewise, the remaining HAstV 5′ UTRs contain putative binding site for PTB/hnRNP I and hnRNPE2/PCBP2, and more importantly these putative binding sites for both cellular proteins are located in double stranded RNA sequences suggesting a functional role of this RNA–protein interaction, such as RNA chaperones that could remodel RNA secondary structures to conform RNA–protein complexes, possibly involved in astrovirus replication process by mediating the contact of the ends of the viral genome as successfully seen in several single-stranded positive animal RNA viruses. Additionally, the putative binding site found to SRSF3 in this region suggests it may interact with hnRNE2 as previously reported for poliovirus.
A great deal of evidence has been published that describes the interaction of cellular proteins with the 5′ UTR. For example, picornaviruses interact with PCBP2/hnRNPE2, La, eEF1A, SRSF3 and PTB [15, 60, 86, 89], and some of these cellular proteins have also been observed to bind to the 5′ end of Norwalk virus (La, hnRNPL, PCBP2 and PTB) [90] and feline calicivirus (PTB) [16, 54]. In the case of the viral bovine diarrhea virus and Hepatitis C, the binding of NFAR proteins (and other cellular factors) at both UTRs is responsible for mediating the contacts between the 5′ and 3′ ends of the RNA [22, 23] (Table 1). There is no previous study on the 5′ UTR that could suggest a possible role in any viral function. In addition to the 3′ UTR, we think that this structure might be necessary for the recruitment of the viral replication complex. This includes the viral RNA polymerase (nsP1b) and the viral nsP1a-4 protein, which has been co-localized with the viral RNA containing a VPg covalently linked to the 5′ terminus in the rough endoplasmic membranes of infected cells [5, 7].
The analysis in silico carried out with 3′ UTRs of human astrovirus revealed putative binding sites for the SRSF proteins showed affinity motifs for SRSF2 (TGTCTCTG), SRSF5 (TCAGA), and except for HAstV-3 that lacks such motif, SRSF6 (TACAGC). Additionally, putative binding sites were found for SRSF3 (CTCTGTT), hnRNPE2 (TTAG) and TIA1 (TTATTTT); interestingly these protein recognition motifs are also present in the 5′ UTR of most HAstV (Figs. 2c, S2a, b). The few variations at this respect are the lack of hnRNPE2, SRSF3 and TIA1 in HAstV-2, and the lack of SRFS6 in HAstV-3 and -7. As described for the 5′ UTRs, the binding sites localized in the 3′ UTRs correspond to the highest scores obtained in all search engines used.
The possible recruitment of SRSF proteins to the 3′ UTR in astrovirus leads to the speculation that SRSF proteins could determine the localization and infectivity of the viral particles produced, probably because they function in RNA transport and nuclear export [86]. However, they could also help in the stabilization of RNA secondary structure and the recruitment of the replication complex. In this analysis, we found PTB/hnRNP1 putative binding sites in all astroviruses studied, except in HAstV-2. The TCTT motif is present in the 3′ UTR HAstV-8 structure (nt 6700–6704) is recognized by PTB, protein found in infected CaCo2 cell extracts. Published data from our group described that PTB/hnRNP1 knockdown in CaCo2 cells affects HAstV-8 viral RNA replication, suggesting that it is required for this viral function. Additionally, UV cross-linked assays with uninfected and infected CaCo2 cell extracts showed that other six proteins of 35, 40, 45, 50, 52, and 75 kDa from both cell extracts were bound to the 3′ UTR [50]. Preliminary results using mobility shift assays revealed that recombinant hnRNPE2 is able to interact with the 3′ UTR (nt 6717–6720/ TTAG) of HAstV-8 (Fig. 2d). Experiments exploring whether SRSF or TIA1 proteins are components of the astrovirus replication complex are currently being carried out in our laboratory.
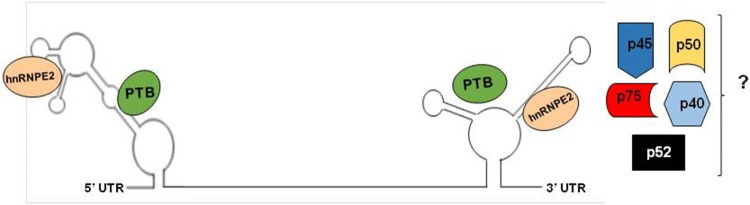
Interestingly, both human astrovirus UTRs have one PTB binding site as observed in preliminary in silico analysis carried out in our lab. Additionally, it is possible that hnRNPE2 (PCPB2) and SRSF3 might be involved (Fig. 3). hnRNPE2 is capable of establishing independent protein–protein interactions both with PTB and SRSF3 [86, 92, 93]. PTB-hnRNPE2 and SRSF3-hnRNPE2 interactions might occur during astrovirus replication as has been observe in poliovirus and hepatitis C [61, 86, 87]. PTB proteins could also interact with each other and with the ends of the viral RNA to form a circular ribonucleoprotein (RNP) complex as reported previously [20, 44, 69, 74].
Fig. 3.
Schematic representation of the interaction of HAstVs UTR regions with cellular factors. The 5′ and 3′ UTR of astroviral RNA binds to PTB and hnRNPE2 (a.k.a. PCBP2), proteins. Additionally, the 3′ UTR binds to five others proteins p40, p45, p50, p52 and p75, the identity of these proteins has to be determined. The hnRNPE2 would be interacting with UUAT motif (nt 31–34) in 5′ UTR or UUAG motif in 3′ UTR (nt 6717–6720)
Conclusions
Our findings suggest the probable mode of action of hnRNPs and SRSF proteins that bind to human astrovirus UTRs and their involvement in the viral life cycles. This is similar to RNA chaperones, which maintain RNA structure in a conformation that favors viral RNA replication. These proteins play important roles in assembling the viral RNA replication complex, selecting and recruiting viral RNA replication templates, and other processes. Even when it has been proposed that circularization does not depend on the 5′ and 3′UTR protein interactions, multiple reports suggest that, in addition to viral proteins, the binding of cellular proteins in trans is required to promote or facilitate viral replication. The question is how the interplay of the human astrovirus 5′ and 3′ UTRs maximize the efficacy of the genome via either RNA–protein (PTB, hnRNPE2, and/or SRSF) or RNA–RNA interactions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Bert L. Semler (University of California, Irvine) for the pET22b-PCBP2 plasmid. This work was partly supported by Secretaría de Investigación y Posgrado (SIP) of Instituto Politécnico Nacional (IPN) Grants 20151007, 20160939, 20170853, 20180182 (M. De N-O.), and by Consejo Nacional de Ciencia y Tecnología (CONACyT) Grants 127557-M and 236104 (J.V.). M.C.S was a CONACyT (747789) and BEFI SIP-IPN (reg. 16783) fellowships recipient. M. De N-O., J.S.B. and L.G.M. received fellowships from Comisión de Operación y Fomento a las Actividades Académicas (COFAA), and M. De N-O, J.S.B received Estímulo al Desempeño de los Investigadores (EDI) from IPN. Additionally, M. De N-O, C.V.V., J.S.B and J.V. received fellowships Sistema Nacional de Investigadores (SNI) CONACyT.
Author contributions
M. De N-O, C.V.V, J.S.B, J.V. and L.G.M., wrote the introduction, RNA structural analysis, comparison between the 5′ and 3′ UTR sections and the cellular protein interactions with other RNA viruses. M.C.S and W.E.H made the table containing the proteins that bind to the 5′ UTR and 3′ UTR respectively. M.C.S prepared the Figs. 1c, e, 2d and 3. All authors discussed the manuscript contents and agreed with this paper.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Mendez E, Murillo A, Velázquez R, Burnham A, Arias C. Replication cycle of astrovirus. In: Schultz-Cherry S, editor. Astrovirus research; essential ideas, everyday impacts. New York: Springer; 2013. pp. 19–45. [Google Scholar]
- 2.La Vu D, Cordey S, Brito F, Kaiser L. Novel human astroviruses: novel human diseases? J Clin Virol. 2016;82:56–63. doi: 10.1016/j.jcv.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Krishna T. Novel human astroviruses: challenges for developing countries. Virus Dis. 2014;25:208–214. doi: 10.1007/s13337-014-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendez E, Arias CF. Astroviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 5. Philadelphia: Lippincott Williams & Willkins; 2007. pp. 981–997. [Google Scholar]
- 5.Fuentes C, Bosch A, Pintó RM, Guix S. Identification of human astrovirus genome-linked protein (VPg) essential for virus infectivity. J Virol. 2012;86:10070–10078. doi: 10.1128/JVI.00797-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monroe SS, Jiang B, Stine SE, Koopmans M, Glass RI. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J Virol. 1993;67:3611–3614. doi: 10.1128/jvi.67.6.3611-3614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guix S, Caballero S, Bosch A, Pinto RM. Human astrovirus C-terminal nsP1a protein is involved in RNA replication. Virology. 2005;333:124–131. doi: 10.1016/j.virol.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 8.Ahlquist P, Noueiry AO, Lee WM, Kushner BD, Dye BT. Host factors in positive strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai MM. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 10.Edgil D, Harris E. End-to-end communication in the modulation of translation by mammalian RNA virus. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray D, Wu B, White KA. A second functional RNA domain in the 5ʹ UTR of the Tomato bushy stunt virus genome: intra- and interdomain interactions mediate viral RNA replication. RNA. 2003;9:1232–1245. doi: 10.1261/rna.5630203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willcocks MM, Carter MJ. The 3′ terminal sequence of a human astrovirus. Arch Virol. 1992;124:279–289. doi: 10.1007/BF01309809. [DOI] [PubMed] [Google Scholar]
- 13.Barton DJ, O´Donnell BJ, Flanegan JB. 5′cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutiérrez- EAL, De Nova-Ocampo M, Racaniello VR, Del Angel RM. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J Virol. 1997;71:3826–3833. doi: 10.1128/jvi.71.5.3826-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrenfel E, Gebhard JG. Interaction of cellular proteins with the poliovirus 5′ noncoding region. Arch Virol Suppl. 1994;9:269. doi: 10.1007/978-3-7091-9326-6_27. [DOI] [PubMed] [Google Scholar]
- 16.Alhatlani B, Vashist S, Goodfllow I. Functions of the 5′ and 3′ ends of calicivirus genomes. Virus Res. 2015;206:134–143. doi: 10.1016/j.virusres.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller AA, Semler BL. Stem-loop structure synergy in binding cellular proteins to the 5′ noncoding region of poliovirus RNA. Virology. 1995;206:923–934. doi: 10.1006/viro.1995.1015. [DOI] [PubMed] [Google Scholar]
- 18.García-Montalvo BM, Medina F, del Angel RM. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of Dengue 4 virus RNA. Virus Res. 2004;102:141–150. doi: 10.1016/j.virusres.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberté J, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez ME, Vashist S, Ureña L, Goodfellow I, Chavez P, Mora HJE, Cancio C, Garrido E, Gutierrez-Escolano AL. Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNPA1. J Virol. 2013;87:11371–11387. doi: 10.1128/JVI.03433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris D, Zhang Z, Chaubey B, Pandey NV. Identification of cellular factors associated with the 3′ nontranslated region of the hepatitis C virus genome. Mol Cell Proteom. 2006;5:1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Isken O, Baroth M, Grassmann WC, Weinlich S, Ostareck HD, Ostareck-Lederer A, Behrens SE. Nuclear factors are involved in hepatitic C virus RNA replication. RNA. 2007;13:1675–1692. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isken O, Grassmann CW, Yu H, Behrens SE. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Nova-Ocampo M, Villegas-Sepulveda N, Del Angel RM. Translation elongation factor-1 alfa, La and PTB interact with the 3′untranslated region of Dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 25.Agis-Juarez RA, Galvan I, Medina F, Daikoku T, Padmanabhan R, Ludert JE, del Angel RM. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J Gen Virol. 2009;90:2893–2901. doi: 10.1099/vir.0.013433-0. [DOI] [PubMed] [Google Scholar]
- 26.Yocupicio-Monroy RM, Padmanabhan R, Medina F, del Angel RM. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology. 2007;357:29–40. doi: 10.1016/j.virol.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 27.Yu L, Nomaguchi M, Padmanabhan R, Markoff L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology. 2008;374:170–185. doi: 10.1016/j.virol.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 29.Anwar A, Leong KM, Ng ML, Chu JJ, Garcia-Blanco MA. The polypyrimidine tract binding protein is required for efficient dengue virus propagation and associates with the viral replication machinery. J Biol Chem. 2009;284:17021–17029. doi: 10.1074/jbc.M109.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paranjape SM, Harris E. Control of dengue virus translation and replication. Curr Top Microbiol Immun. 2010;338:15–34. doi: 10.1007/978-3-642-02215-9_2. [DOI] [PubMed] [Google Scholar]
- 31.Zeng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomila RC, Martin GW, Gehrke L. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS ONE. 2011;6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 34.Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a Dengue virus non-coding RNA. PLoS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Feng L, Pan W, Shi X, Dai J. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem Biophys Res Commun. 2015;456:327–332. doi: 10.1016/j.bbrc.2014.11.080. [DOI] [PubMed] [Google Scholar]
- 36.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011;8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei Y, Huang Y, Zhang H, Yu L, Zhang M, Dayton A. Functional interaction between cellular p100 and the dengue virus 3′ UTR. J Gen Virol. 2011;9:796–806. doi: 10.1099/vir.0.028597-0. [DOI] [PubMed] [Google Scholar]
- 38.Yocupicio-Monroy RM, Medina F, Reyes-del Valle J, del Angel RM. Cellular proteins from human monocytes bind to dengue 4 virus minus-strand 3′ untranslated region RNA. J Virol. 2003;77:3067–3076. doi: 10.1128/JVI.77.5.3067-3076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutiérrez-Escolano AL, Vázquez-Ochoa M, Escobar-Herrera J, Hernández-Acosta J. La, PTB and PABP proteins bind to the 3′ untranslated region of Norwalk virus genomic RNA. Bioch. Biophys Res Commun. 2003;311:759–766. doi: 10.1016/j.bbrc.2003.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandoval-Jaime C, Gutiérrez-Escolano AL. Cellular proteins mediate 5′-3′ end contacts of Norwalk virus genomic RNA. Virology. 2009;387:322–330. doi: 10.1016/j.virol.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Bailey D, Karakasiliotis I, Vashist S, Chung LM, Rees J, McFadden N, Benson A, Yarovinsky F, Simmonds P, Goodfellow I. Functional analysis of RNA structures present at the 3′ extremity of the murine norovirus genome: the variable polypirimidine tract plays a role in viral virulence. J Virol. 2010;84:2859–2870. doi: 10.1128/JVI.02053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vashist S, Ureña L, Chaudhry Y, Goodfellow I. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J Virol. 2012;86:11977–11990. doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancio-Lonches C, Yocupicio-Monroy M, Sandoval-Jaimes C, Galvan-Mendoza I, Ureña L, Vashist S, Goodfellow I, Salas-Benito J, Gutierrez-Escolano AL. Nucleolin interacts with the feline calicivirus 3′ untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replications. J Virol. 2011;85:8056–8068. doi: 10.1128/JVI.01878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunner JE, Nguyen JHC, Roehl HH, Ho TV, Swiderek KM, Semler BL. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J Virol. 2005;79:3254–3266. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roehl HH, Semler BL. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand. RNA J Virol. 1995;69:2954–2961. doi: 10.1128/jvi.69.5.2954-2961.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waggoner S, Sarnow P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J Virol. 1998;72:6699–6709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustin KE, Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, Das S. HuR displaces polypyrimidine tract binding protein to facilitate La binding to the 3′ untranslated region and enhances hepatitis C virus replication. J Virol. 2015;89:11356–11371. doi: 10.1128/JVI.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espinosa-Hernandez W, Velez-Uriza D, Valdes J, Velez C, Salas-Benito J, Martínez-Contreras R, García M, Salas M, Vega-Almeida T, De Nova-Ocampo M. PTB binds to the 3′ Untranslated region of the Human astrovirus type 8: A possibke role in viral replication. PLoS ONE. 2014;9:e113113. doi: 10.1371/journal.pone.0113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagy DP, Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol. 2012;10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rios MP, Romero LC, Berzal H. The cis-acting replication element of the hepatitis C virus genome recruits host factors that influence viral replication and translation. Sci Rep. 2016;6:25729,1–15. doi: 10.1038/srep25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auweter DS, Allai THF. Structure-function relationships of the polypyrimidine tract binding protein. Cell Mol Life Sci. 2008;65:516–527. doi: 10.1007/s00018-007-7378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karakasiliotis I, Vashist S, Bailey D, Abente JE, Green YK, Roberts OL, Sosnovtsev VS, Goodfellow GI. Polypyrimidine tract binding protein functions as a negative regulator of feline calicivirus translation. PloS ONE. 2010;5:1–17. doi: 10.1371/journal.pone.0009562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woling LS, Cedervall T. The La protein. Ann Rev Bioch. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 56.Gottlieb E, Stelitz JA. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bedard KM, Semler BL (2004) Regulation of picornavirus gene expression. Microbes Infect.702–713. 10.1016/j.micinf.2004.03.001 [DOI] [PubMed]
- 58.Meerovitch K, Svitkin YV, Lee HS, Lejbkowics F, Kenan DJ, Chan EKL, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ray U, Saumitra D. Interplay between NS3 protease and human La protein regulates translation-replication switch of hepatitis C virus. Sci Rep. 2011;1:1–9. doi: 10.1038/srep00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhen L, Dong X, Li Y, Zhang Q, Kim C, Song Y, Kang L, Liu Y, Wu K, Wu J. PolyC-binding protein 1 interacts with 5′-untranslated region of enterovirus 71 RNA in membrane-associated complex to facilitate viral replication. PLoS ONE. 2014;9:e87491. doi: 10.1371/journal.pone.0087491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang LY, Jeng KS, Lai MC. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol. 2011;85:7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flynn RA, Martin L, Spitale CR, Do TB, Sagan MS, Zarnegar B, Qu K, Khavari AP, Quake RS, Sarnow P, Chang YH. Dissecting noncoding and pathogen RNA-protein interactomes. RNA. 2015;21:135–143. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li D, Wei T, Abbott MC, Harrich D. The unexpected roles of eukaryotic translation elongation factors in RNA Virus replication and pathogenesis. Microbiol Mol Biol Rev. 2013;77:253–265. doi: 10.1128/MMBR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai MC, Peng TY, Tarn WY. Functional interplay between viral and cellular SR proteins in control of post-transcriptional gene regulation. FEBS J. 2009;276:1517–1526. doi: 10.1111/j.1742-4658.2009.06894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manley LJ, Krainer RA. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/S1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shepar JP, Hertel JK. The SR protein family. Genome Biol. 2009 doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong S, Proteins SR. Binders, regulators, and connectors of RNA. Mol Cells. 2017;40:1–9. doi: 10.14348/molcells.2017.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgil D, Polacek C, Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol. 2006;80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol. 2006;80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernandez-Miragall O, Lopez de Quinto S, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 2009;139:172–182. doi: 10.1016/j.virusres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 73.Monceyron C, Grinde B, Jonassen TO. Molecular characterisation of the 3′-end of the astrovirus genome. Arch Virol. 1997;142:699–706. doi: 10.1007/s007050050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villordo SM, Gamarnik A. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;15:3568. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Human Mol Gen. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 77.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 78.Goren A, Ram O, Amit M, Keren H, Lev-Maor G. Comparative analysis identifies exonic splicing regulatory sequences-the complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 79.Paz I, Kosti I, Ares MJr, Cline M, Mandel-Gutfreund Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42:W361–W367. doi: 10.1093/nar/gku406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fededa JP, Kornblihtt AR. A splicing regulator promotes transcriptional elongation. Nat Struc Mol Biol. 2008;15:779–781. doi: 10.1038/nsmb0808-779. [DOI] [PubMed] [Google Scholar]
- 81.Huang Y, Steitz JA. SRprises along messenger’s journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 82.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Nat Acad Sci. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 85.Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fitzgerald KD, Semler BL. Poliovirus infections induces the co-localization of cellular protein SRp20 with TIA-1, a cytoplasmic stress granule protein. Virus Res. 2013;176:223–231. doi: 10.1016/j.virusres.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Florez PM, Sessions OM, Wagner EJ, Gromeier M, García-Blanco MA. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J Virol. 2005;79:6172–6179. doi: 10.1128/JVI.79.10.6172-6179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gutierrez-Escolano AL, Uribe-Brito Z, del Angel RM, Jiang X. Interaction of cellular proteins with the 5′ end of Norwalk virus genomic RNA. J Virol. 2000;74:8558–8562. doi: 10.1128/JVI.74.18.8558-8562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang P, Lai MC. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim KJ, Hahm B, Kim KY, Choi M, Jang KS. Protein–protein interaction among hnRNPs shuttling between nucleous and cytoplasm. J Mol Biol. 2000;298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 93.Caputi M, Zahler AM. SR proteins and hnRNP H regulated the splicing of the HIV-1 tev specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.