Abstract
Bile acids (BAs) are important regulators of metabolism and energy balance, but excess BAs cause cholestatic liver injury. The histone methyltransferase, Mixed-lineage leukemia-4 (MLL4), is a transcriptional coactivator of the BA-sensing nuclear receptor, Farnesoid-X-Receptor (FXR), and epigenetically upregulates FXR targets important for the regulation of BA levels, Small Heterodimer Partner (SHP) and Bile Salt Export Pump (BSEP). MLL4 expression is aberrantly downregulated and BA homeostasis is disrupted in cholestatic mice, but the underlying mechanisms are unclear. Here, we examined whether elevated microRNA-210 (miR-210) in cholestatic liver promotes BA-induced pathology by inhibiting MLL4 expression. MiR-210 was the most highly elevated miR in hepatic SHP-downregulated mice with elevated hepatic BA levels. MLL4 was identified as a direct target of miR-210 and overexpression of miR-210 inhibited MLL4, and subsequently, BSEP and SHP expression, resulting in defective BA metabolism and hepatotoxicity with inflammation. MiR-210 levels were elevated in cholestatic mouse models, and in vivo silencing of miR-210 ameliorated BA-induced liver pathology and decreased hydrophobic BA levels in a MLL4-dependent manner. In gene expression studies, SHP inhibited miR-210 expression by repressing a transcriptional activator, Kruppel-like factor-4 (KLF4). In primary biliary cholangitis/cirrhosis (PBC) patients, hepatic levels of miR-210 and KLF4 were highly elevated, whereas nuclear levels of SHP and MLL4 were reduced. In conclusion, hepatic miR-210 is physiologically regulated by SHP, but is elevated in cholestatic mice and PBC patients, promoting BA-induced liver injury in part by targeting MLL4. The miR-210/MLL4 axis is a potential target for the treatment of BA-associated hepatobiliary disease.
Keywords: FXR, SHP, BSEP, FGF19, PBC patients
Introduction
Bile acids (BAs) are amphipathic steroids that facilitate absorption of lipid nutrients, but also function as signaling molecules that profoundly impact metabolism and energy balance (1–3). Due to their detergent-like properties, hepatic BA levels must be tightly controlled. Transcriptional regulation, primarily by the nuclear receptor, Farnesoid-X-Receptor (FXR, NR1H4), plays a crucial role in maintaining BA homeostasis (4). Deficiencies in adaptive responses to elevated BA levels result in accumulation of BAs in the liver, and BA-mediated liver injury, which can further progress to fibrosis, cirrhosis, and hepatocellular carcinoma (5, 6). The molecular pathological mechanisms by which BA homeostasis is disrupted in the cholestatic liver remain unclear.
The orphan nuclear receptor SHP is transcriptionally induced by BA-activated FXR, together with its transcriptional coactivator, Mixed-Lineage Leukemia 3/4 (MLL3/4) (7, 8), and plays a critical role in BA metabolism (9). MLL3 (KMT2C) and its closest homolog MLL4 (KMT2D) are known epigenetic regulators of BA metabolism (7, 8, 10). SHP does not have a DNA binding domain but acts as a co-repressor for numerous transcriptional factors (11, 12). SHP links elevated hepatic BA levels to epigenetic repression of genes involved in BA synthesis and reuptake, including Cyp7a1, Cyp8b1 and Ntcp, by recruiting repressive histone modifying proteins to these genes (13–15). The gene regulatory functions and protein stability of SHP are enhanced by a late fed-state intestinal hormone, Fibroblast Growth Factor-19 (FGF19, mouse FGF15) (15–17). SHP also regulates hepatic functions in part via transcriptional control of microRNAs (miRs) (18–20). MiRs are small non-coding RNAs that have emerged as powerful post-transcriptional negative gene regulators (21). MiRs are aberrantly expressed in numerous diseases, including metabolic disorders and cancer, and serve as promising potential therapeutic targets and/or diagnostic markers (22). Aberrantly expressed miRs have been identified in alcoholic (23, 24) and non-alcoholic fatty liver diseases (19, 25) but whether this aberrant expression in cholestatic liver causes defective regulation of BA metabolism is not known.
Here, we show that miR-210, normally repressed by SHP, is aberrantly elevated in BA- and drug-induced cholestatic mice, resulting in disruption of BA balance, in part by inhibiting MLL4 and, subsequently, Bsep and Shp expression. Remarkably, in primary biliary cholangitis/cirrhosis (PBC) patients, hepatic miR-210 levels were highly elevated, whereas levels of nuclear SHP and MLL4 were reduced, suggesting the potential of targeting the miR210-MLL4 axis for treatment of BA-associated hepatobiliary disease.
Experimental Procedures
Animal experiments.
Viruses were injected into mice i.v. via the tail vein. To generate liver-specific SHP-knockdown (SHP-LKD) mice, 8 week old male SHP-floxed mice (SHP f/f) were injected with 100 μl of 2 × 1011 genome copies/body weight (Vector Biolabs) AAV-TBG-Cre (or -GFP) 6 weeks before sacrifice. For overexpression of SHP, 100 μl of 0.5–1 × 109 active viral particles of adenovirus expressing SHP (or GFP as a control) was injected into C57BL/6J mice 3 weeks before sacrifice. For overexpression of miR-210, 100 μl of 5–8 X 109 IU/ml of lentivirus expressing pre-miR-210 was injected; for the miR-210 downregulation, 100 μl of 5–8 X 109 IU/ml of lentivirus expressing shRNA miR-210 was injected; and for the MLL4 downregulation, 100 μl of 1X109 IU/ml lentivirus expressing MLL4 shRNA was injected, in each case into C57BL/6 mice 2 weeks before sacrifice. To induce cholestasis, mice were treated by gavage with 75 mg/kg ANIT (olive oil) for 24 h or fed chow supplemented with 1% cholic acid (CA)-chow for 1 week. Cholestasis was also induced by bile duct ligation (BDL) or sham-operation in mice for 24 h (26). To examine the effects of obeticholic acid (OCA), an FXR agonist, on ANIT-induced cholestasis (5, 27), mice were treated with vehicle or 10 mg/kg of OCA, i.p., and after five days, with 35 mg/kg ANIT by gavage daily for 2 days. With the exception of refeeding experiments, mice were fasted for 4 to 5 h before sacrifice. All animal use and biosafety protocols were approved by the Institutional Animal Use and Care and Biosafety Committees of the University of Illinois at Urbana-Champaign and were in accordance with NIH Guidelines.
MicroRNA-seq analysis.
RNA was isolated from the liver of mice using the miRNeasy kit (Qiagen). The cDNA library was sequenced using an Illumina HiSeq2000 (Illumina, San Diego, CA) to produce paired-end 100 bp reads. The microRNA-seq data was processed using the miARma-seq analysis pipeline. After the initial inspection of reads quality using FastQC, residual 3’ adapters reads were trimmed (Cutadapt) and reads shorter than 15 nt were discarded and reads were mapped to the mouse genome (mm10) using Bowtie1. FeatureCounts was used to quantify the number of mapped reads in each known microRNA (miRBase). Differentiation expression analysis was performed by edgeR and microRNAs with at least one count per million (CPM) in at least two samples were included in the analysis.
Measurement of BA levels and composition.
Total BA levels from the liver, intestine, plasma, and gallbladder were measured with a BA assay kit (MAK309, Sigma-Aldrich). BAs were extracted from tissues in 0.4N perchloric acid and individual bile acid levels were determined using the 5500 QTRAP LC-MS/MS system (Sciex, Framingham, MA) in the Metabolomics Center, University of Illinois at Urbana-Champaign.
Liver histology and hepatotoxicity.
Fixed liver sections were stained with hematoxylin and eosin (H&E) or F4/80 antibody was detected by IHC and imaged as described (16). Plasma ALT levels were measured using colorimetric analysis kits (MAC052, Sigma-Aldrich).
Lentiviral production.
Lentiviruses expressing pre-miR-210 or anti-miR-210 were prepared by transient transfection of 293T cells (2X107 cells) with the respective target plasmid (7.5 μg), the packaging plasmid psPAX2 (5.6 μg) and the VSV-G envelope protein-coding plasmid pMD2G (1.9 μg).
Luciferase reporter assays.
DNA fragments containing the miR-210 promoter (−270 to −581 bp) with the WT or mutated KLF4 binding motif were inserted into the pGL3-luc. A reporter plasmid containing the Mll4 3’-UTR (219 bp) containing the miR-210 seed sequence or its mutated sequence was inserted into the pmirGLO vector (Promega, E1330) using site-directed mutagenesis (Agilent, 200521). Hepa1c1c7 and HEK293 cells were transfected with 200 ng luciferase reporter plasmids containing the Mll4 3’-UTR, 100 ng of CMV β-gal plasmid, 50–150 ng pre-miR-210 or anti-miR-210. Hepa1c1c7 cells were transfected with luciferase vectors containing the miR-210 promoter and 5–50 ng KLF4 or 5–50 ng SHP expression plasmids. Luciferase activities were normalized to β-galactosidase activities.
ChIP, re-ChIP, and Co-IP assays.
ChIP and re-ChIP assays were performed as described (11) with primer sequences shown in Supplemental Table S1. Co-IP assays using liver whole cell extracts were performed as described (12).
Human PBC patient liver samples.
Frozen unidentifiable liver samples of 15 normal individuals and 15 PBC patients were obtained from the Liver Tissue Procurement and Distribution System. Ethical approval was not required. Hepatic mRNA and protein levels were determined by RT-qPCR and IB, respectively.
IB analysis.
IB analysis of proteins in whole cell extracts of liver or cultured cells was performed as described (12).
Subcellular localization.
Liver tissues were minced and resuspended in hypotonic buffer (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2% NP40, 1 mM EDTA and 5% sucrose) and lysed by homogenization. Nuclei were pelleted by centrifugation at 4500 x g for 3 min through a 10% sucrose cushion.
RT-qPCR.
Total RNA was isolated using TRIzol (Invitrogen) or RNeasy mini kits (Qiagen), cDNA was synthesized, and RT-qPCR was performed with the iCycler iQ (Bio-Rad) using primers listed in supplemental Table S1. The mRNA amounts were normalized to 36B4 mRNA. Primers for RT-qPCR of miR-210 were purchased (Thermo Fisher, Inc).
Statistical analysis.
Data analysis was performed using GraphPad Prism 7 (GraphPad software). Statistical significance was determined by the Student’s two-tailed t-test or one- or two-way ANOVA with the False Discovery Rate post-test for single or multiple comparisons as appropriate. P-values < 0.05 were considered statistically significant.
Results
MicroRNA-210 is the most highly elevated microRNA in SHP-LKD mice.
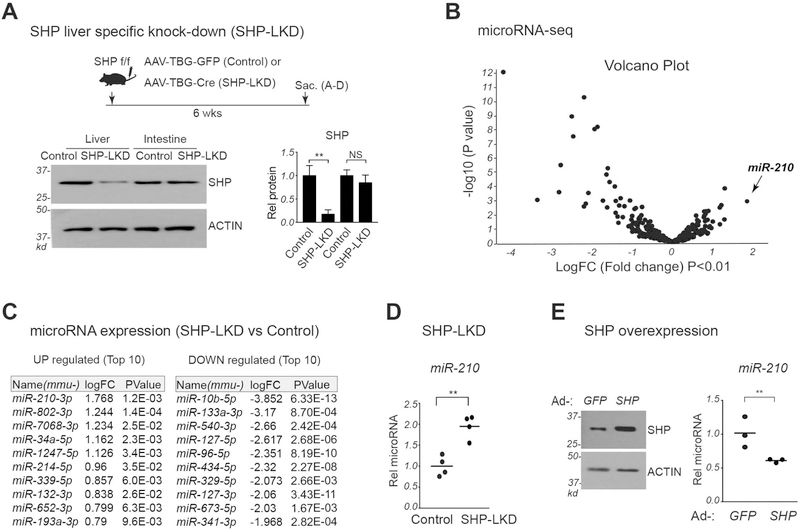
Hepatic SHP-regulated miRs were identified by microRNA-seq analysis of livers from control and SHP-LKD mice (Fig. 1A). Since SHP represses its target genes, we focused on miRs upregulated in SHP-LKD mice. Levels of several hepatic miRs were elevated in the SHP-LKD mice, including the known SHP target, miR-34a (18), but miR-210 was the most highly elevated (Fig. 1B, C). Confirming this result, hepatic miR-210 levels were increased about 2-fold in SHP-LKD mice (Fig. 1D). Notably, hepatic BA levels were elevated over 2-fold in SHP-LKD mice (Supplemental Fig. S1). Conversely, liver-specific overexpression of SHP decreased miR-210 levels (Fig. 1E). This inverse correlation between SHP and miR-210 levels suggests that SHP may regulate hepatic BA levels in part by inhibiting miR-210 expression.
Figure 1. MiR-210 is the most highly elevated hepatic miR in SHP-LKD mice.
(A-D) To specifically downregulate (knock down) hepatic SHP (SHP-LKD), AAV-TBG-Cre, or AAV-TBG-GFP as a control, was injected into SHP-floxed (SHP f/f) mice. Six weeks after injection, the mice were fasted for 4 h and sacrificed. (A) Experimental approach (top) and liver and intestinal SHP protein levels measured by immunoblotting (bottom). (B) Global expression changes in microRNA expression displayed in a volcano plot. (C) List of miRs with levels up-regulated (left panel) or down-regulated (right panel) by 1.5 fold or more with P values <0.05. (D) Relative levels of miR-210 with the mean control level set to 1. (E) C57BL/6 mice were injected with adenovirus expressing GFP or SHP and sacrificed 3 weeks later. SHP levels detected by IB (left) and relative miR-210 levels with the mean GFP value set to 1 (right). (A, D, E) Statistical significance was determined by the Student’s t-test, standard deviation (SD, n=5), ** p < 0.01.
Overexpression of miR-210 in mice results in defective BA regulation and cholestatic liver injury.
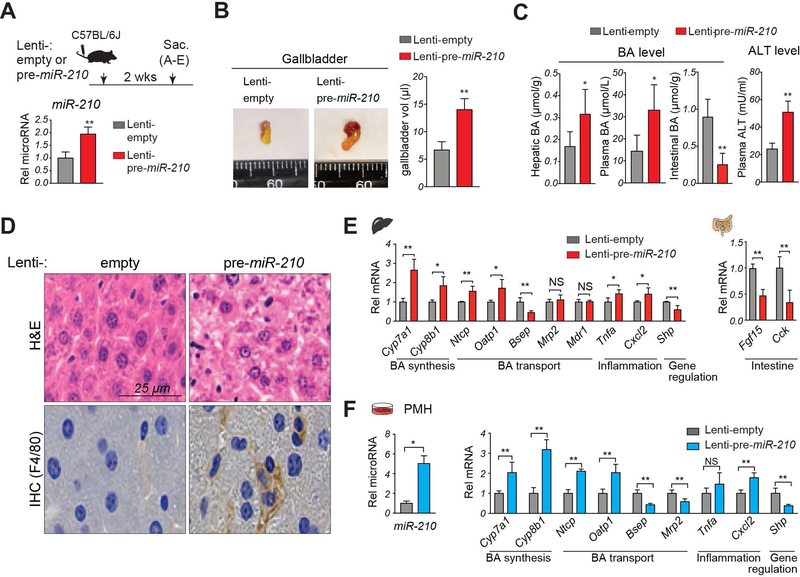
To determine whether miR-210 played a role in regulation of BA metabolism by SHP, we examined the effects of about a 2-fold overexpression of pre-miR-210 in C57BL/6 mice (Fig. 2A), which was similar to the increase in SHP-LKD mice (Fig. 1D). In these mice, gallbladder size and volume were increased (Fig. 2B), hepatic and plasma BA levels increased, intestinal BA levels decreased, and plasma levels of ALT increased (Fig. 2C). Further, pathological changes were observed, including hepatocellular ballooning and increased hepatic inflammation (Fig. 2D), and mRNA levels of BA synthetic genes, Cyp7a1 and Cyp8b1, and transporters, Ntcp, Oatp1 and Bsep, and of pro-inflammatory genes, Tnfα and Cxcl2, and intestinal genes, Fgf15 and Cck, were significantly changed, whereas Mrp2 and Mdr1 were not (Fig. 2E). Somewhat surprising was the increase in gallbladder volume by miR-210 overexpression despite decreased Bsep expression. However, expression of other export transporters, Mrp2 and Mdr1, were unchanged (Fig. 2E) and expression of Cck, which stimulates gallbladder emptying, was inhibited (Fig. 2E), and intestinal BA levels were decreased, all of which are consistent with the increased gallbladder size. Intriguingly, expression of Shp (Fig. 2E) was decreased after overexpression of miR-210, suggesting that a pathway involving SHP and miR-210 may form a feedback loop to regulate BA levels.
Figure 2. Overexpression of miR-210 results in defective BA regulation, hepatoxicity and inflammation.
(A-E) C57BL/6 mice were injected with either lenti-pre-miR-210 or control lenti-empty and the mice were sacrificed after 2 weeks. (A) Experimental approach (top). Relative levels of miR-210 with the Lenti-empty value set to 1 (bottom). (B) Representative images (left) of gallbladders and gallbladder volumes (right). (C) Hepatic, plasma, and intestinal BA and plasma ALT levels. (D) Liver sections stained with H&E (top) and by IHC with F4/80 antibody to detect macrophages (bottom). (E) Levels of the indicated mRNAs. (F) PMH were infected with either empty lentivirus (Lenti-empty) or lentivirus expressing the miR-210 precursor (Lenti-pre-miR-210) for 3 days. Relative levels of miR-210 and the indicated mRNAs with the Lenti-empty levels set to 1. (A-F) Statistical significance was determined by the Student’s t-test, SD (n=4–5), * p < 0.05, ** p < 0.01, NS, not significant.
Since lentiviruses target multiple tissues, we examined the effects of overexpression of miR-210 in primary mouse hepatocytes (PMH) to determine if the miR-210 effects were cell autonomous. Consistent with the in vivo data, overexpression of pre-miR-210 in hepatocytes led to increased miR-210 levels and changes in gene expression (Fig. 2F), indicating that the effects of overexpression of miR-210 on BA metabolic gene expression are hepatocyte cell autonomous. These findings, together, suggest that increased hepatic miR-210 levels in normal mice can cause defective BA metabolism and cholestatic pathology, including hepatotoxicity and inflammation.
Inverse correlation between hepatic miR-210 and MLL4 levels.
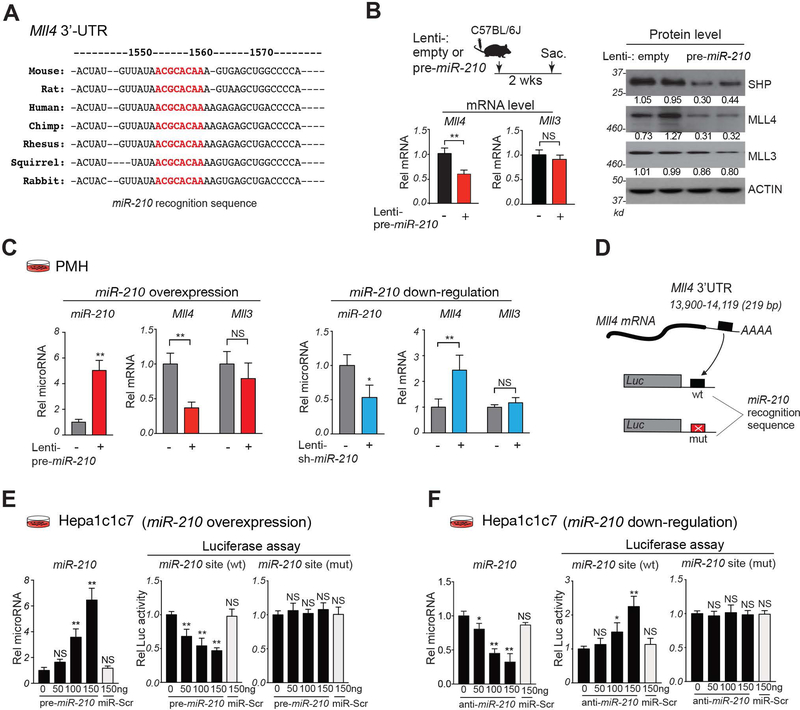
Target scan analysis was performed to potential targets of miR-210 in BA regulation. Mouse MLL4, but not its closest homolog MLL3, contains a miR-210 recognition sequence in its 3’-UTR, which is highly conserved in mammals, including humans (Fig. 3A). MLL4 was particularly interesting because it is a known FXR coactivator in induction of Shp and Bsep (7, 10). Protein and mRNA levels of MLL4, but not those of MLL3, were reduced by 50% by overexpression of miR-210 in mice (Fig. 3B). Similarly, in PMH, overexpression of miR-210 decreased expression of MLL4, but not MLL3 and conversely, downregulation of miR-210 specifically increased MLL4 expression (Fig. 3C). These results demonstrate that MLL4 and miR-210 levels are inversely correlated, suggesting that MLL4 may be a direct target of miR-210.
Figure 3. MiR-210 directly targets the MLL4 3’-UTR.
(A) MiR-210 recognition sequences (red) in the 3’-UTR of mammalian Mll4 mRNAs. (B) C57BL/6 mice were treated as described in the Fig. 2A legend. Experimental approach (top, left). Relative levels of Mll3 and Mll4 mRNAs with lenti-empty values set to 1 (bottom, left) and protein levels (right). (C) PMH were infected Lenti-empty (−), Lentiviruses expressing pre-miR-210 (left) or sh-miR-210 (right) for 3 days. Relative levels of miR-210 and Mll4 and Mll3 mRNA with the control RNA value set to 1. (D-F) (D) Schematic of the luciferase constructs containing WT or mutated miR-210 sites in the Mll4 3’-UTR. Hepa1c1c7 cells were co-transfected with indicated luciferase plasmids and oligonucleotides for pre-miR-210 (E), antisense-miR-210 (F), or control scrambled RNA (miR-Scr), and 48 h later, luciferase and β-galactosidase activities were measured. (E,F) Relative levels of miR-210 (left) and relative luciferase activity normalized to β-galactosidase activity (right). (B-F) Statistical significance was determined by the Student’s t-test (B-C) or one-way ANOVA (E, F), SD (n=4–5), * p < 0.05, ** p < 0.01, NS, not significant.
The Mll4 3’-UTR contains a functional binding site for miR-210.
To determine whether the miR-210 recognition sequence in Mll4 is functional, the Mll4 3’-UTR containing either the WT or a mutated sequence was inserted into a luciferase reporter (Fig. 3D). Expression of pre-miR-210, but not control RNA, in Hepa1c1c7 cells increased miR-210 levels and decreased luciferase activity in vectors with the WT, but not mutated, sequence (Fig. 3E). Conversely, treatment with anti-miR-210, but not control RNA, decreased miR-210 levels and increased luciferase activity in vectors with the WT, but not mutated, miR-210 sequence (Fig. 3F). Similar results were observed in human HEK293 cells (Supplemental Fig. S2). These results suggest that MLL4 is a direct target of miR-210.
MiR-210 attenuates FXR induction of Shp and Bsep mediated by MLL4.
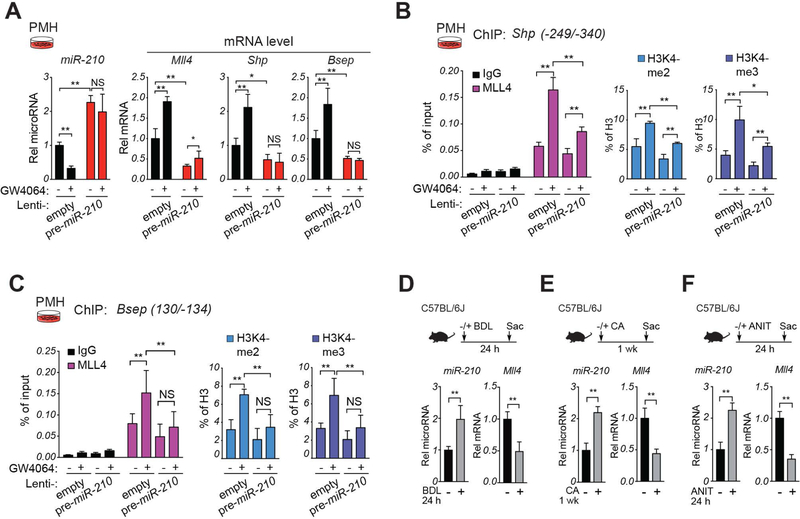
Since MLL4 coactivates BA activated FXR induction of Shp and Bsep (7, 10) and miR-210 inhibits expression of MLL4 (Fig. 3), we examined whether miR-210 blunts FXR-mediated induction. Treatment of PMH with an FXR agonist, GW4064, decreased miR-210 levels and increased mRNA levels of Mll4, Shp and Bsep (Fig. 4A). GW4064 increased occupancy of MLL4 and levels of gene-activating methylated histones, H3K4-me2 and H3K4-me3, at the Shp (Fig. 4B) and Bsep (Fig. 4C) promoters. In each case, these GW4064-mediated effects were blunted by overexpression of pre-miR-210. These results indicate that epigenetic coactivation of FXR induction of BA metabolic genes by MLL4 is blocked by overexpression of miR-210.
Figure 4. (A-C) MiR-210 blocks MLL4-mediated FXR-mediated induction of Shp and Bsep.
(A-C) PMH were infected either empty lentivirus or lentivirus expressing pre-miR-210 and after 3 days were treated with vehicle or GW4064 (200 nM) for 6 h. (A) Levels of miR-210 and the indicated mRNAs detected by RT-qPCR. (B,C) Occupancy of MLL4 and levels of H3K4-me2, and H3K4-me3 at the Shp (B) or Bsep (C) promoters determined by ChIP assay. (A-C) The mean and standard deviation of the mean are shown. (D-F) In cholestatic mouse models, hepatic miR-210 levels are elevated and MLL4 levels decreased. Cholestatic mice were induced by bile duct ligation (BDL) for 24 h (D), by feeding 1% CA-chow for 7 days (E), or by treatment with 75 mg/kg ANIT by gavage for 24 h (F). Hepatic miR-210 and Mll4 mRNA levels, as indicated. Statistical significance was determined by two-way ANOVA (A-C) or Student’s t-test (D-F), SD (n=4–5), * p < 0.05, ** p < 0.01, NS, not significant.
Hepatic miR-210 and MLL4 levels are inversely correlated in cholestatic mice.
Hepatic MLL4 expression is substantially downregulated and histone H43K4-me3 levels at Bsep are decreased in cholestatic mice (10). Since miR-210 inhibits MLL4 expression, we examined whether miR-210 is upregulated in cholestatic mice. Cholestatic liver injury was induced by bile duct ligation, feeding CA-chow for 1 week, or by treatment with α-naphthyl isothiocyanate (ANIT), a chemical inducer of intrahepatic cholestasis (28). In each of these models, hepatic miR-210 levels were increased and Mll4 mRNA levels were decreased (Fig. 4D-F). Interestingly, treatment with obeticholic acid (OCA), a potent FXR agonist and an FDA-approved therapeutic agent for cholestatic liver disease (5, 27), largely reversed the ANIT-induced increases in miR-210 levels and decreases in Mll4, Shp, and Bsep expression (Supplemental Fig. S3). These results indicate that hepatic miR-210 levels are elevated in cholestatic mouse models, which may contribute to decreased MLL4 expression, and that activation of FXR in cholestatic mice can reverse the increase in miR-210 levels and subsequently, expression of Mll4, Shp and Bsep.
Systemic silencing of elevated miR-210 largely restored MLL4 expression and ameliorated liver injury in cholestatic mice.
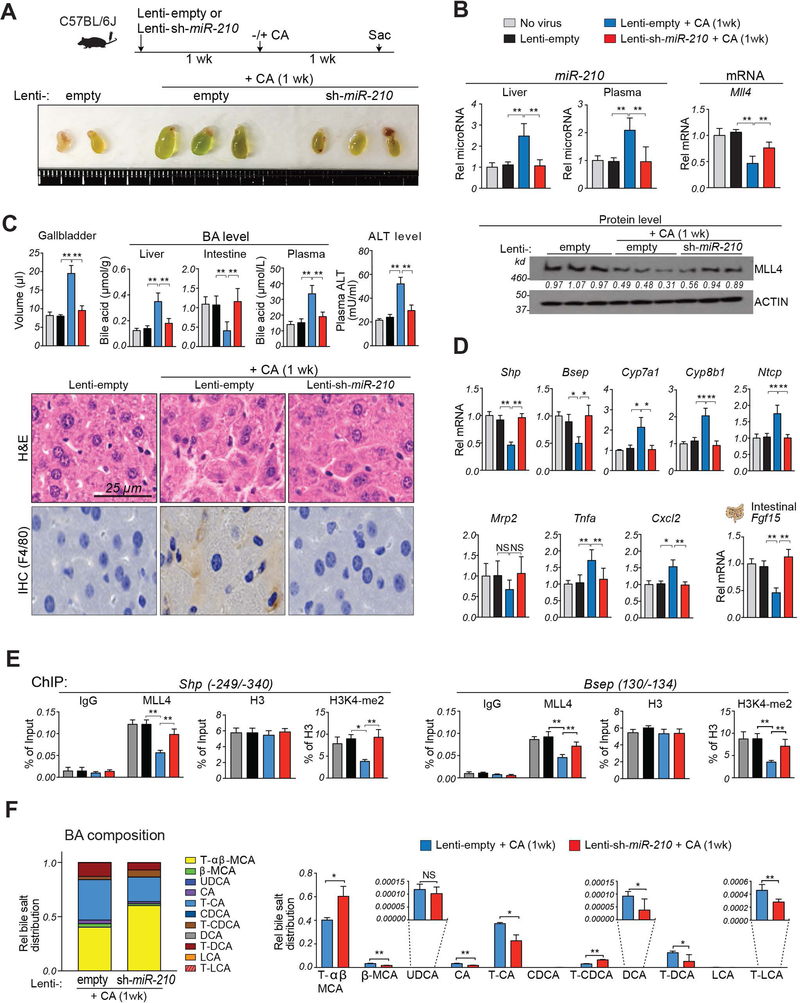
To examine whether reducing the elevated miR-210 levels in cholestatic mice would have beneficial effects, miR-210 was downregulated by lentiviral expression in mice fed 1% CA-chow for 1 week. In these mice, gallbladders were more greenish and increased in size (Fig. 5A). Both hepatic and plasma miR-210 levels were increased, suggesting that plasma miR-210 might serve as a diagnostic marker for liver disease, and mRNA and protein levels of MLL4 were decreased (Fig. 5B). Further, hepatotoxicity was evident with increased hepatic and plasma BA levels and plasma ALT levels (Fig. 5C, top) and hepatocyte ballooning with increased inflammation (Fig. 5C, bottom).
Figure 5. Downregulation of miR-210 in cholestatic mice reverses low MLL4 levels, defective BA metabolism, and liver injury.
C57BL/6 mice were either not infected (no virus) or injected with lentivirus expressing miR-210 shRNA (Lenti-sh-miR-210) or empty lentivirus (Lenti-empty) and 1 week later, as indicated, were fed either a normal or CA-chow diet for 1 week. (A) Experimental approach (top) and images of gallbladders (bottom). (B) Levels of miR-210 in the liver and plasma (top, left) and levels of Mll4 mRNA (top, right) and protein (bottom) (C) Gallbladder volumes and levels of BAs in the liver, intestine, and plasma and ALT in the plasma (top). Liver sections stained with H&E (bottom, upper panels) or by IHC with F4/80 antibody (bottom, lower panels). (D) Levels of mRNAs of the indicated genes. (E) Occupancy of MLL4 or levels of histone H3 or H3K4-me2 at the Shp promoter (left) or Bsep promoter (right). (F) BA composition analysis: C57BL/6 mice were injected with lentivirus expressing shRNA for miR-210 or empty lentivirus and 4 weeks later, the mice were fed CA-chow for 1 week. BA composition (left) of total BA pool and fractional amounts of selected BAs in the BA pool (right). Statistical significance was determined by the two-way ANOVA (B-F), standard error of the mean (SEM, n=4–5), * p < 0.05, ** p < 0.01, NS, not significant.
Expression of Shp and Bsep and hepatic genes involved in BA synthesis, transport and inflammation was altered in a manner consistent with increased hepatic BA levels and inflammation, and intestinal expression of Fgf15, which inhibits hepatic BA synthesis (29), was decreased (Fig. 5D). Notably, each of these pathological outcomes in BA-overloaded mice was ameliorated by miR-210 downregulation (Fig. 5B-D). These results, together, demonstrate that decreased MLL4 levels and pathological changes in cholestatic liver (10) are, at least in part, dependent on elevated miR-210.
The decreased hepatic Shp expression in C57BL/6 mice after feeding CA-chow for 1 week (Fig. 5D) is consistent with decreased Shp expression in cholestatic C57BL/6 mice induced by bile duct ligation (30), but inconsistent with increased Shp expression in 129×1/SvJ mice fed with CA-chow for 1 week (31). The 129×1/SvJ mice have a profound defect in response to inflammatory stimulus (32) and C57BL/6 mice are more susceptible to liver damage than 129×1/SvJ mice (33), so the differences in hepatic responses to BA-induced toxicity, including Shp expression, are likely a strain-specific effect.
To determine whether the decreased MLL4 levels in the cholestatic liver affected the MLL4 action at Shp and Bsep, we examined the occupancy of MLL4 and methylated histone H3K4-me2 levels at promoter regions in mice fed CA-chow for 1 week. Occupancy of MLL4 and histone H3K4-me2 levels at Shp and Bsep promoters were decreased in these mice, but these effects were blunted by downregulation of miR-210 (Fig. 5E). These results demonstrate that reduction of elevated miR-210 in the cholestatic liver restored MLL4 expression and subsequently, epigenetic induction of Shp and Bsep.
BA composition is an important determinant of BA-induced hepatotoxicity (34). We, therefore, examined whether downregulation of miR-210 in mice fed CA-chow for 1 week altered the BA composition (Fig. 5F, left). In these mice, approximately equal relative amounts of hydrophilic BAs, predominantly the taurine conjugates of α- and β-muricholic acid (MCA), and hydrophobic BAs, predominantly taurine conjugates of CA and deoxycholic acid (DCA), were present (Fig. 5F, left). Notably, downregulation of miR-210 increased the relative levels of α- and β-MCA and decreased the levels of more toxic DCA and CA (Fig. 5F, right). These results suggest that overall decreases in BA levels particularly hydrophobic BAs, contribute to the beneficial effects of miR-210 downregulation in the BA-overloaded mice.
MLL4 is important for beneficial effects in cholestatic mice of miR-210 downregulation.
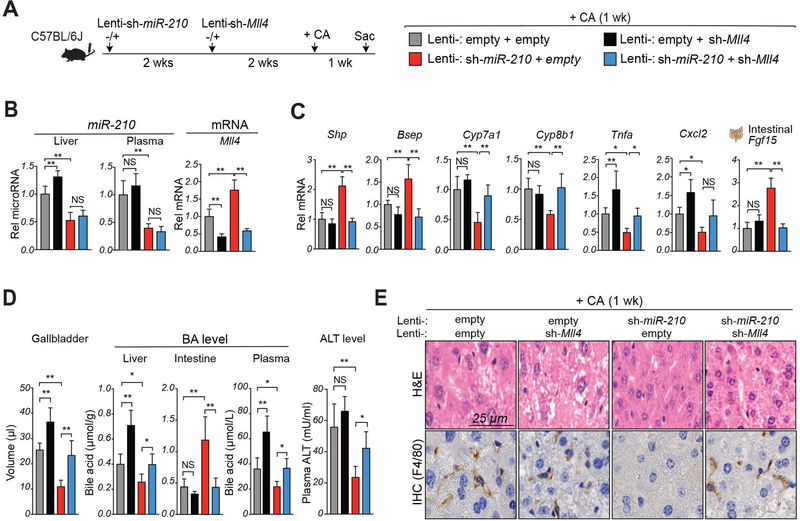
To determine the relative importance of MLL4, among the multiple potential targets of miR-210, in mediating miR-210 on BA metabolism, MLL4 was downregulated after downregulation of miR-210 followed by feeding a CA-chow for 1 week (Fig. 6A, left). Similar to the results of the studies in Fig. 5, downregulation of miR-210 altered gene expression (Fig. 6B, C), gallbladder volume (Fig. 6D), plasma BA and ALT levels (Fig. 6D), and liver histology (Fig. 6E), in a manner suggesting amelioration of hepatotoxicity. Remarkably, all these beneficial effects observed in the cholestatic liver after systemic silencing of miR-210 were blunted by downregulation of MLL4 (Fig. 6B-E). These results indicate that beneficial effects of downregulation of miR-210 in BA-challenged mice are largely dependent on MLL4.
Figure 6. Beneficial effects of downregulation of miR-210 in cholestatic mice are largely dependent on MLL4.
Mice were injected with empty lentivirus (Lenti-empty) or lenti-virus expressing miR-210 shRNA (Lenti-sh-miR-210), 2 weeks later mice were injected with empty lentivirus (Lenti-empty) or lentivirus expressing shRNA for Mll4 (Lenti-sh-Mll4), and 2 weeks later the mice were fed CA-chow for 1 week. (A) Experimental approach (left) and legends showing viral injection groups (right). (B) Levels of miR-210 in liver and plasma (left) and hepatic Mll4 mRNA (right) (C) Levels of mRNAs of the indicated genes. (D) Gallbladder volume (n=5), levels of BAs in the liver, intestine, and plasma and ALT in the plasma. (E) Liver sections stained with H&E (upper panels) or by IHC with F4/80 antibody (lower panels). Statistical significance was determined by two-way ANOVA (B, C, E), SD (n=4–5), * p < 0.05, ** p < 0.01, NS, not significant.
SHP inhibits transcriptional activation of miR-210 by Kruppel-like factor-4 (KLF4).
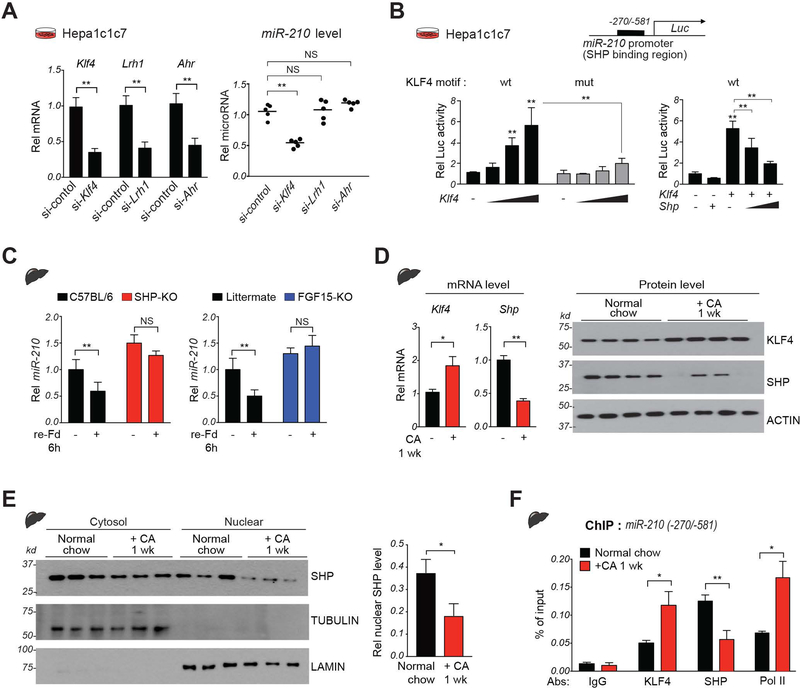
To determine how SHP directly represses miR-210 expression, we first analyzed SHP binding in published hepatic SHP ChIP-seq data (11). SHP binding was increased at the promoter region of the miR-210 gene after FGF19 treatment (Supplemental Fig. S4A), which was confirmed in ChIP assays showing that FGF19 treatment increased SHP binding at the miR-210 promoter (Supplemental Fig. S4B). SHP lacks a DNA binding domain (11, 12) and cannot bind directly to the miR-210 promoter. Within the SHP binding region, several sites for potential binding partners were identified using JASPAR motif analysis, including KLF4 and AhR (Supplemental Fig. S5). Since KLF4 motifs had the highest scores and AhR is a known transcriptional partner of SHP (12), we initially focused on these two factors, compared to a known SHP binding partner LRH-1 (35).
Downregulation of KLF4, but not AhR nor LRH-1, resulted in decreases in miR-210 levels in Hepa1c1c7 cells (Fig. 7A). Further, overexpression of KLF4 increased luciferase activity of a reporter containing the WT, but not mutant, KLF4 motif in miR-210 (Fig. 7B, left) and overexpression of SHP inhibited the KLF4-mediated increase in luciferase activity (Fig. 7B, right). These results suggest that KLF4 is a previously unknown transcriptional activator of miR-210 and that its transactivation is inhibited by SHP.
Figure 7. SHP directly inhibits hepatic expression of miR-210 in part by repressing KLF4.
(A) Hepa1c1c7 cells were transfected with siRNAs as indicated for 48 h. Levels of the indicated mRNAs (left) or miR-210 (right). (B) Hepa1c1c7 cells were transfected with luciferase vectors containing WT or mutated KLF4 motifs in the miR-210 promoter and with KLF4 and SHP expression vectors as indicated. Luciferase activity was normalized to that of β-galactosidase. (C) C57BL/6 (WT), SHP-KO, and FGF15-KO mice were refed for 6 h after fasting overnight and hepatic miR-210 levels were determined. (D-F) C57BL/6 mice were fed normal chow or CA-chow for 1 week. (D) Levels of hepatic Klf4 and Shp mRNA (left) or protein levels (right). (E) Nuclear localization studies: Levels of the indicated proteins in the nuclear and cytosolic fractions in liver extracts determined by IB. (F) Occupancy of indicated proteins at miR-210 promoter. Mean with SD (A-E) or SEM (F) is plotted. Statistical significance was determined by Student’s t-test (A, D, E), one-way ANOVA (B) or two-way ANOVA (C, F), n=5. * p < 0.05, ** p < 0.01. NS, not significant.
Hepatic expression of miR-210 is physiologically regulated by SHP and FGF15.
The nuclear localization and gene regulatory function of SHP are increased by FGF15/19 signaling in the late fed-state (12, 15, 16, 36), which provides a physiological model to study regulation of miR-210 by SHP. Notably, hepatic miR-210 levels in WT mice were decreased after refeeding for 6 h after overnight fasting, but these decreases were blunted in both SHP-KO and FGF15-KO mice (Fig. 7C). These results indicate that both SHP and FGF15 are physiological regulators of miR-210 in the late fed-state. Supporting these data, refeeding dramatically increased the interaction of SHP with KLF4 and increased SHP occupancy in KLF4-bound chromatin at the miR-210 promoter (Supplemental Fig. S6). These results suggest that SHP inhibits KLF4-mediated transactivation of miR-210 under physiological conditions in mice.
Altered expression of KLF4 and SHP in cholestatic mice contributes to elevated miR-210 expression.
We next examined whether the increased miR-210 levels in mice fed CA-chow for 1 week (Fig. 4E) are associated with altered expression of KLF4 and SHP. Hepatic mRNA and protein levels increased for KLF4 and decreased for SHP in these mice (Fig. 7D). Notably, SHP levels, particularly nuclear levels, were reduced in the BA-overloaded cholestatic mice (Fig. 7E). Consistent with these results, occupancy of KLF4 and RNA polymerase II, a marker of active transcription, was increased at the miR-210 promoter, while SHP occupancy was decreased (Fig. 7F). These results indicate that increased hepatic expression of KLF4 and decreased levels of SHP, in particular nuclear SHP, in mice chronically fed CA-chow contribute to elevated miR-210 expression.
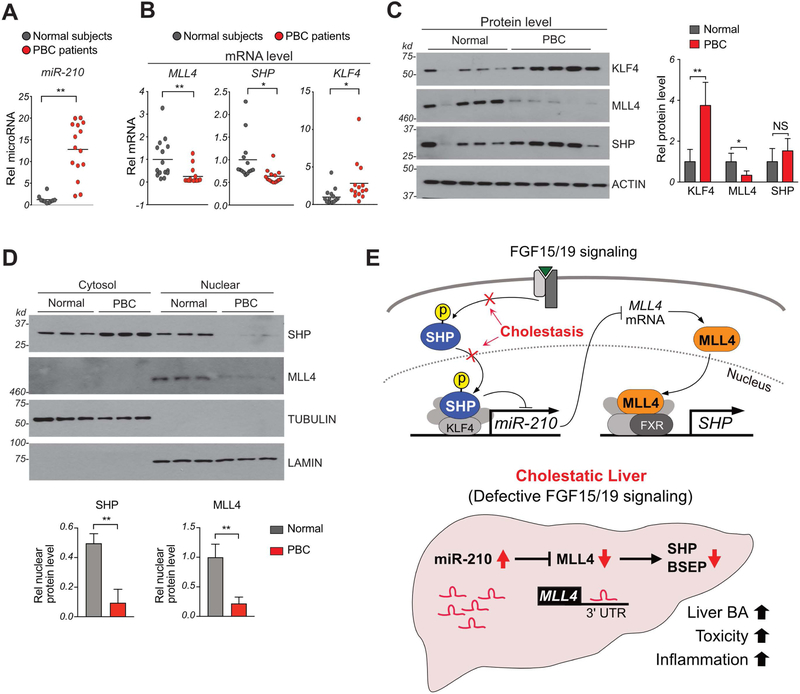
In PBC patients, hepatic miR-210 and KLF4 levels are increased and nuclear levels of SHP and MLL4 are decreased.
To determine whether the studies in cholestatic mice above are relevant to humans, we examined the livers of PBC patients. Hepatic miR-210 levels were increased more than 10-fold in PBC patients (Fig. 8A). Hepatic mRNA levels of KLF4 were also markedly increased, whereas those of MLL4 and SHP were decreased (Fig. 8B). Hepatic protein levels of KLF4 were increased and those of MLL4 were substantially reduced (Fig. 8C) while protein levels of SHP were not significantly changed in contrast to the decrease in mRNA levels (Fig. 8C). MLL4 was detected only in the nucleus, but remarkably, in PBC patients, SHP was detected predominantly in the cytoplasm, while it was detected at similar levels in the cytoplasm and nucleus in normal individuals (Fig. 8D). Thus, although the PBC patients have normal levels of SHP protein, functional nuclear levels are dramatically reduced. These results suggest that the hepatic miR-210/MLL4 axis is likely dysregulated in PBC patients and further support the findings in cholestatic mice that increased KLF4 expression and decreased nuclear levels of SHP contribute to elevated miR-210 expression, and subsequently, downregulation of MLL4 (Fig. 8E).
Figure 8. Hepatic miR-210, MLL4, SHP, and KLF4 levels are altered in PBC patients.
(A,B) Levels of miR-210 (A) and mRNAs (B) of the indicated genes in liver samples of 15 PBC patients and 15 normal individuals. (C) Livers extracts from 3 each of 15 patients or of 15 normal individuals were randomly pooled into 5 samples. (n=5). Levels of the indicated with proteins detected by IB. (D) Livers extracts from 5 each of 15 patients or 15 normal individuals were randomly pooled into 3 samples (n=3). The pooled liver extracts were fractionated into nuclear and cytosolic fractions. Levels of the indicated with proteins. (A-D) Mean and SD is plotted. Statistical significance was determined by Student’s t-test, *p < 0.05, **p < 0.01. NS, not significant. (E) Model: Regulation of BA levels by a SHP-miR-210-MLL4 axis triggered by FGF15/19 signaling. In normal liver, FGF15/19-activated SHP via phosphorylation increases nuclear localization of SHP (16, 42), inhibits KLF4 and miR-210 expression and consequently, increases expression of MLL4, a direct target of miR-210. MLL4, then, acts as a coactivator of BA-activated FXR and induces SHP expression, which further upregulates MLL4 via inhibition of miR-210. In contrast, in cholestatic liver (bottom), defective FGF15/19 signaling (28) impairs nuclear localization of SHP, which increases KLF4 activity and miR-210 expression. The elevated miR-210, then, inhibits MLL4 expression with decreased SHP expression, resulting in increased hepatic BA levels, toxicity, and inflammation.
Discussion
In this study, we identify miR-210 as a post-transcriptional regulator that disrupts BA balance and promotes cholestatic liver pathology in mice. Hepatic miR-210, normally repressed by SHP, is aberrantly elevated in cholestatic mice. Intriguingly, elevated miR-210 levels were not limited to cholestatic mouse models but were also highly elevated in livers of PBC patients. MiR-210 inhibits expression of the methyltransferase, MLL4, by directly binding to the 3’-UTR of Mll4 mRNA, which blocks MLL4-mediated epigenetic induction of Shp and Bsep genes. Decreased expression of Shp and Bsep contributes to defective BA metabolism and cholestatic liver pathology, including hepatotoxicity and inflammation.
In cholestasis, impairment of bile flow from the liver leads to abnormal accumulation of BAs, causing liver damage and inflammation (5, 6). Overexpression of miR-210 in normal mice caused elevated liver and plasma BA levels and pathological changes in the liver, whereas systemic downregulation of miR-210 in cholestatic mice ameliorated BA-associated liver pathologies. These findings indicate that elevated miR-210 in cholestatic mice disrupts BA balance and promotes liver injury. Notably, silencing of miR-210 in cholestatic mice increased hydrophilic MCA levels and decreased toxic hydrophobic BA levels, particularly taurine conjugates of DCA and CA, which is consistent with decreased expression of Cyp8b1, a key determinant of the hydrophobicity of the BA pool (1, 34). These results suggest that decreased BA hydrophobicity, as well as, decreased hepatic BA levels, contributes to beneficial effects of downregulation of miR-210 in cholestatic mice.
MLL4 is a transcriptional coactivator of FXR and plays a critical role in epigenetic induction of Bsep and Shp and thus, regulation of hepatic BA levels (8, 10). MLL4 expression was dramatically downregulated in bile duct ligated cholestatic mice by unknown mechanisms (10). Our studies suggest that elevated miR-210 in cholestatic mice decreases MLL4 expression and subsequently, epigenetic induction of Bsep and Shp. Overexpression of pre-miR-210 in PMH blunted FXR-mediated induction of these genes. Although MLL3 is the closest homolog of MLL4 and these proteins have redundant functions in BA regulation (7, 8), MLL3 was not a direct target of miR-210. Consistent with these results, beneficial effects observed by downregulation of miR-210 in cholestatic mice were markedly diminished after downregulation of MLL4.
The present study adds further complexity to FXR-related networks regulating BAs by showing that SHP-regulated miR-210 inhibits expression of MLL4, which targets SHP and BSEP. Nearly every aspect of BA synthesis, transport and metabolism is regulated transcriptionally by FXR-dependent pathways, including induction of SHP, FGF19, β-KL (37), the essential coreceptor for FGF19, MAFG, a transcriptional repressor of BA synthesis (38), and LSD1, a SHP-interacting epigenetic repressor of BA synthesis (13). BA levels are further regulated via FGF19 signaling-induced post-translational modifications of FXR and SHP (15, 16, 28, 39). Recently, a role for the FXR-induced RNA binding protein, ZFP36L1, in posttranscriptional regulation of Cyp7a1 was also shown (40). This remarkable complexity in the regulation emphasizes the importance of the need for tight physiological regulation of BA levels in response to hormonal, nutritional and circadian cues.
KLF4 was identified as a new transcriptional activator of miR-210 and its activity was repressed by SHP. KLF4, thus, has a role in BA metabolism in addition to its roles in proliferation, apoptosis, and stem cell biology (41). FGF15/19 signaling, which activates SHP via phosphorylation (15, 16), is likely important for its nuclear localization and the SHP-mediated repression of miR-210 since the feeding-mediated inhibition of miR-210 was absent in FGF15-KO, as well as, SHP-KO mice. These findings from previous (16, 42) and current studies, together, reveal an intriguing positive feedback loop in which BA-activated FXR, together with its coactivator MLL4, induces Shp (7), which leads to increased MLL4 expression via inhibition of KLF4-mediated transactivation of miR-210, resulting in sustained expression of SHP. In contrast, in cholestatic liver, impaired FGF15/19 signaling (28) results in defective nuclear localization of SHP and subsequently, increased the activity of KLF4 and miR-210 expression. The elevated miR-210 levels inhibit expression of MLL4, which further inhibits FXR-mediated induction of SHP, and thus, forms a negative feedback loop that reinforces reduced SHP function and elevated miR-210 levels (Model, Fig. 8E).
A surprising finding in the present study is that the nuclear levels of SHP were dramatically reduced in PBC patients and significantly decreased in cholestatic mice, which may contribute to the pathological mechanisms in this severe liver disease. Nuclear localization and activity of SHP in mice requires FGF15/19 signaling-induced posttranslational modifications, including phosphorylation (15, 16). In PBC patients, hepatic expression of β-KL, the essential co-receptor of FGF19 (29), was dramatically reduced, p-ERK levels were reduced (28), and circulating FGF19 levels increased (43), indicative of impaired FGF19 signaling. Thus, impaired FGF19 signaling in PBC patients may result in defective nuclear localization of SHP, which together with elevated KLF4 expression, in part, explains elevated miR-210 expression.
In conclusion, we demonstrate that elevated miR-210 in cholestatic liver disrupts BA balance and promotes liver injury in part by inhibiting MLL4. Importantly, miR-210 levels are highly elevated and MLL4 expression is reduced in PBC patients, suggesting that the miR-210/MLL4 axis is dysregulated in these patients. Treatment with UDCA and norUDCA are first line treatment for cholestatic liver disease, but FXR, FXR-induced FGF19, and BA transporters have received great attention as pharmacological targets for cholestatic liver diseases (5, 27). Elevated hepatic miR-210 in PBC patients, thus, may provide a new therapeutic target for BA-associated hepatobiliary diseases.
Supplementary Material
Acknowledgments
We thank Bo Kong and Grace Guo at Rutgers University for providing liver samples of FGF15-KO mice, Kristina Schoonjans at Ecole Polytechnique Fédérale de Lausanne for providing SHP-floxed mice, and H. Eric Xu at Van Andel Research Institute for recombinant FGF19. We also thank the Liver Tissue Cell Distribution System, University of Minnesota (NIH Contract # HHSN276201200017C) for providing liver specimens of PBC patients.
Financial Support
This study was supported by an American Heart Association Scientist Development Award (16SDG27570006) to YK and grants from the National Institutes of Health (DK062777 and DK095842) to JK.
List of Abbreviations
- BA
bile acid
- FXR
Farnesoid X Receptor
- SHP
Small Heterodimer Partner
- FGF15/19
Fibroblast Growth Factor-15/19
- miR
microRNA
- MLL3/4
Mixed-lineage leukemia3/4
- BSEP
Bile Salt Export Pump
- Cyp7a1
cholesterol 7α hydroxylase
- Cyp8b1
sterol 12α hydroxylase
- KLF4
Kruppel-like factor-4
- PBC
primary biliary cirrhosis
- KO
knockout
- PMH
primary mouse hepatocytes
- LKD
liver-specific knockdown
- β-KL
β-Klotho
- AhR
aromatic hydrocarbon receptor
- LRH-1
liver receptor homolog-1
- 3’-UTR
3’-untranslated region
- ALT
alanine aminotransferase
- ChIP
chromatin immunoprecipitation
- BDL
bile duct ligation
- CA
cholic acid
- ANIT
α-naphthyl isothiocyanate
- OCA
obeticholic acid
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Chiang JY. Bile acid metabolism and signaling. Comprehensive Physiology 2013;3:1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab 2013;17:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev 2014;66:948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol 2012;13:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trauner M, Fuchs CD, Halilbasic E, Paumgartner G. New therapeutic concepts in bile acid transport and signaling for management of cholestasis. Hepatology 2017;65:1393–1404. [DOI] [PubMed] [Google Scholar]
- 6.Wagner M, Trauner M. Recent advances in understanding and managing cholestasis. F1000Res 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DH, Lee J, Lee B, Lee JW. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Mol Endocrinol 2009;23:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DH, Rhee JC, Yeo S, Shen R, Lee SK, Lee JW, Lee S. Crucial roles of mixed-lineage leukemia 3 and 4 as epigenetic switches of the hepatic circadian clock controlling bile acid homeostasis in mice. Hepatology 2015;61:1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta 2011;1812:893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, Suchy FJ. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 2011;300:G771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YC, Byun S, Zhang Y, Seok S, Kemper B, Ma J, Kemper JK. Liver ChIP-seq analysis in FGF19-treated mice reveals SHP as a global transcriptional partner of SREBP-2. Genome Biol 2015;16:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YC, Seok S, Byun S, Kong B, Zhang Y, Guo G, Xie W, et al. AhR and SHP regulate phosphatidylcholine and S-adenosylmethionine levels in the one-carbon cycle. Nat Commun 2018;9:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YC, Fang S, Byun S, Seok S, Kemper B, Kemper JK. FXR-induced lysine-specific histone demethylase, LSD1, reduces hepatic bile acid levels and protects the liver against bile acid toxicity. Hepatology 2015;62:220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol 2007;27:1407–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seok S, Kanamaluru D, Xiao Z, Ryerson D, Choi SE, Suino-Powell K, Xu HE, et al. Bile acid signal-induced phosphorylation of small heterodimer partner by protein kinase Czeta is critical for epigenomic regulation of liver metabolic genes. J Biol Chem 2013;288:23252–23263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DH, Kwon S, Byun S, Xiao Z, Park S, Wu SY, Chiang CM, et al. Critical role of RanBP2-mediated SUMOylation of Small Heterodimer Partner in maintaining bile acid homeostasis. Nature Communications 2016;7:12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, et al. Bile acid signaling pathways increase stability of Small Heterodimer Partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev 2009;23:986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem 2010;285:12604–12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu T, Choi SE, Kim DH, Seok S, Suino-Powell KM, Xu HE, Kemper JK. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor beta-Klotho. Proc Natl Acad Sci U S A 2012;109:16137–16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Y, Lu S, Zhao J, Wang L. Nuclear Receptor SHP: A Critical Regulator of miRNA and lncRNA Expression and Function. Nucl Receptor Res 2017;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 22.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203–222. [DOI] [PubMed] [Google Scholar]
- 23.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 2013;10:542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satishchandran A, Ambade A, Rao S, Hsueh YC, Iracheta-Vellve A, Tornai D, Lowe P, et al. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology 2018;154:238–252 e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SE, Fu T, Seok S, Kim DH, Yu E, Lee KW, Kang Y, et al. Elevated microRNA-34a in obesity reduces NAD levels and SIRT1 activity by directly targeting NAMPT. Aging cell 2013;12:1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Woolbright BL, Zhao W, Wang Y, Matye D, Hagenbuch B, Jaeschke H, et al. Sortilin 1 Loss-of-Function Protects Against Cholestatic Liver Injury by Attenuating Hepatic Bile Acid Accumulation in Bile Duct Ligated Mice. Toxicol Sci 2018;161:34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beuers U, Trauner M, Jansen P, Poupon R. New paradigms in the treatment of hepatic cholestasis: from UDCA to FXR, PXR and beyond. J Hepatol 2015;62:S25–37. [DOI] [PubMed] [Google Scholar]
- 28.Byun S, Kim DH, Ryerson D, Kim YC, Sun H, Kong B, Yau P, et al. Postprandial FGF19-induced phosphorylation by Src is critical for FXR function in bile acid homeostasis. Nature Communications 2018;9:2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kliewer SA, Mangelsdorf DJ. Bile Acids as Hormones: The FXR-FGF15/19 Pathway. Dig Dis 2015;33:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, Pope C, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology 2017;66:1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinal C, Tohkin M, Miyata M, Ward J, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 2000;102:731–744. [DOI] [PubMed] [Google Scholar]
- 32.White P, Liebhaber SA, Cooke NE. 129X1/SvJ mouse strain has a novel defect in inflammatory cell recruitment. J Immunol 2002;168:869–874. [DOI] [PubMed] [Google Scholar]
- 33.Wu R, Wu X, Wang H, Fang X, Li Y, Gao L, Sun G, et al. Strain differences in arsenic-induced oxidative lesion via arsenic biomethylation between C57BL/6J and 129X1/SvJ mice. Sci Rep 2017;7:44424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann AF, Hagey LR. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J Lipid Res 2014;55:1553–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci U S A 2005;102:9505–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YC, Byun S, Seok S, Guo G, Xu HE, Kemper B, Kemper JK. Small Heterodimer Partner and Fibroblast Growth Factor 19 Inhibit Expression of NPC1L1 in Mouse Intestine and Cholesterol Absorption. Gastroenterology 2018;156:1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu T, Kim YC, Byun S, Kim DH, Seok S, Suino-Powell K, Xu HE, et al. FXR primes the liver for intestinal FGF15 signaling by transient induction of betaKlotho. Mol Endocrinol 2015;30:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Aguiar Vallim TQ, Tarling EJ, Ahn H, Hagey LR, Romanoski CE, Lee RG, Graham MJ, et al. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell Metab 2015;21:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab 2009;10:392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarling EJ, Clifford BL, Cheng J, Morand P, Cheng A, Lester E, Sallam T, et al. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest 2017;127:3741–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer 2006;6:11–23. [DOI] [PubMed] [Google Scholar]
- 42.Kim YC, Seok S, Byun S, Kong B, Zhang Y, Guo G, Xie W, et al. AhR and SHP regulate phosphatidylcholine and S-adenosylmethionine levels in the one-carbon cycle. Nat Commun 2018;9:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wunsch E, Milkiewicz M, Wasik U, Trottier J, Kempinska-Podhorodecka A, Elias E, Barbier O, et al. Expression of hepatic Fibroblast Growth Factor 19 is enhanced in Primary Biliary Cirrhosis and correlates with severity of the disease. Sci Rep 2015;5:13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.