Abstract
Peptidyl-prolyl isomerase (PIN1) specifically binds and isomerizes the phosphorylated serine/threonine–proline (pSer/Thr–Pro) motif, which results in the alteration of protein structure, function, and stability. The altered structure and function of these phosphorylated proteins regulated by PIN1 are closely related to cancer development. PIN1 is highly expressed in human cancers and promotes cancer as well as cancer stem cells by breaking the balance of oncogenes and tumor suppressors. In this review, we discuss the roles of PIN1 in cancer and PIN1-targeted small-molecule compounds.
Keywords: cancer therapeutics, PIN1, PIN1 inhibitor, proline-directed phosphorylation, prolyl isomerase, tumorigenesis
Introduction
Proline (Pro)-directed serine/threonine (Ser/Thr) phosphorylation is a modification of various signaling pathways. Proline is the unique amino acid with the ability to have either a cis or trans structure, and these isomerizations are catalyzed by peptidylprolyl isomerases (PPIases). The cis–trans isomerization of proline in the phosphorylated Ser/Thr–Pro motif is mediated by PIN1 (Liou et al., 2011). The PPIase superfamily contains FK506-binding proteins (FKBPs), cyclophilins and parvulins. FKBPs and cyclophilins are inhibited by the immunosuppressants FK506/rapamycin and cyclosporine A (CyA). PIN1 is a kind of parvulins and inhibited by juglone. PIN1 is well known PPIase that controls the isomerization of the phosphorylated Ser/Thr–Pro (pSer/Thr–Pro) motif.
PIN1 contains two domains including an WW domain in N-terminal and a PPIase domain in C-terminal (Lu et al., 1996, 1999). The N-terminal WW domain interacts with specific pSer–Pro or pThr–Pro motifs, which are the regulatory phosphorylation sites of substrate proteins (Lu et al., 1996, 1999). After interacting with its substrate, the PPIase domain isomerizes the pSer/Thr–Pro motifs, which affect the function of protein by the conformational changes of target protein (Lu et al., 1999; Lu P. -J. et al., 2002).
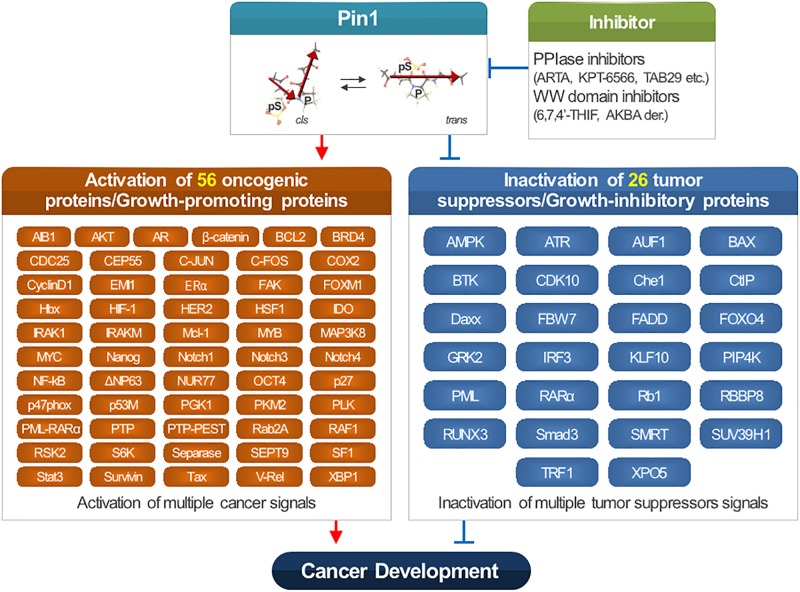
The post-translational modifications of PIN1, containing oxidation, sumoylation, phosphorylation, and ubiquitination, control the PPIase activity and stability of PIN1, and contribute to the high expression and/or activation of Pin1 in cancer development. PIN1 is involved in the cell cycle, synthesized protein folding, and DNA damage responses (Lu et al., 1996). PIN1 is overexpressed in several human cancers (Lee T. H. et al., 2011), including prostate cancer (Ayala et al., 2003; La Montagna et al., 2012), breast cancer (Wulf et al., 2001; Ryo et al., 2002), and oral squamous carcinomas (Miyashita et al., 2003). In cancer patients, a high expression of PIN1 correlates with a poor clinical outcome, lymph node metastasis in non-small cell lung cancer patients, and disease progression in patients with oral squamous carcinoma (Ryo et al., 2001; Ayala et al., 2003; Bao et al., 2004; Suizu et al., 2006). PIN1 overexpression induces chromosome instability and tumorigenesis. PIN1 inactivates and activates more than 26 tumor suppressors and 56 oncogenes, respectively.
In cancer stem cells, multiple PIN1 substrates play an important role. PIN1 regulates the tumorigenesis and expansion of CSCs in leukemia and breast cancer. However, it is not fully understood how PIN1 controls cancer and cancer stem cell development. Several studies have reported that some single nucleotide polymorphisms (SNPs) of the Pin1 gene increases the cancer risk, whereas other variants function as protective factors (Segat et al., 2007; Lu et al., 2009; Li et al., 2013). In this review, we summarize the function of PIN1 in regulating cancer development and small-molecule compounds that exhibit anticancer activities by targeting PIN1.
Transcriptional and Post-Translational Regulation of PIN1 in Cancer
Oncogenes activating E2F transcriptional factor including H-Ras, Her2, p38, and PI3K increase the mRNA expression of Pin1, which appears to activate Pin1 transcription by E2F, considering the existence of the E2F consensus sequence in the Pin1 promoter region (Ryo et al., 2002, 2009; Kamimura et al., 2011). The transcriptional activation of PIN1 is induced by the E2F or by the binding of Notch1 with the Pin1 promoter region (Ryo et al., 2002; Rustighi et al., 2009). In acute myeloid leukemia (AML), oncogenic CCAAT/enhancer binding protein-α ((C/EBPα)-p30) is a dominant negative isoform of the tumor suppressor C/EBPα that is generated by CEBPA mutations. C/EBPα-p30 recruits the E2F transcription factor to bind to the PIN1 pro-moter.
On the contrary, p53 and AP4 act as transcriptional repressors and reduce the Pin1 transcription (Mitchell and Smith, 1988; Jeong et al., 2014). Xbp1 induces the transcription of p53 via HEPN1 and represses E2F1 via NF-κB activation, resulting in reduced Pin1 transcription (Chae et al., 2016). The transcription of PIN1 is repressed by BRCA1, a tumor suppressor gene (MacLachlan et al., 2000). BRCA1 interacts with some proteins to control DNA repair. During cancer development, BRCA1 is often mutated, resulting in the accumulation of DNA damage in cells (Mersch et al., 2015). The mRNA stability of PIN1 is reduced by microRNAs, such as miR-200c (Luo et al., 2014), miR-200b (Zhang et al., 2013) and miR296-5p (Lee et al., 2014) in breast cancer, breast CSCs, and prostate cancer.
Under physiological conditions, the protein activity is generally regulated by post-translational modifications. Post-translational modifications at specific sites, including sumoylation, phosphorylation, ubiquitination, and oxidization, can regulate the PIN1 protein activity and function. The S65, S71, S138, and S16 residues in PIN1 protein sequence are reported as phosphorylation sites (Eckerdt et al., 2005; Rangasamy et al., 2012; Bhaskaran et al., 2013). The PIN1 phosphorylation at Ser16 in the N-terminal WW domain, inhibits the ability of PIN1 to bind with its substrates (Lu P. -J. et al., 2002), and it can be induced by ribosomal S6 kinase 2 (Cho et al., 2012), protein kinase A (Lu K. P. et al., 2002), and aurora kinase A (Lee et al., 2013). The PIN1 phosphorylation at Ser65 in the C-terminal PPIase domain by polo-like kinase (Plk1) (Eckerdt et al., 2005) induces the ubiquitination and stabilization of PIN1. The PIN1 phosphorylation at Ser138 by mixed-lineage kinase 3 induces its nuclear translocation and catalytic activity (Rangasamy et al., 2012). The PIN1 phosphorylation at Ser71 by death-associated protein kinase 1 (DAPK1) can reduce MYC and E2F-mediated oncogenic transformation.
PIN1 sumoylation at Lys6 in the N-terminal WW domain and Lys63 in the C-terminal PPIase domain suppresses its oncogenic function and enzymatic activity (Chen et al., 2013). PIN1 desumoylation at Lys6 and Lys63 by SUMO1/sentrin specific peptidase 1 (SENP1) recovers its substrate-binding and catalytic activity. Under oxidative stress, PIN1 is generally oxidized at Cys113 in the PPIase catalytic site, which can suppress the enzymatic activity of PIN1 (Chen et al., 2015).
PIN1 reduces the degradation of oncogenes and/or growth-promoting regulators, such as β-catenin, AKT, c-fos, cyclin D1, c-Jun, ER, HER2, Hbx, HIF-1, Mcl-1, NF-κB, Nanog, NUR77, PML-RARa, Oct4, Stat3, and Tax (Lu and Zhou, 2007; Gianni et al., 2009; Liao et al., 2009; Moretto-Zita et al., 2010; Lu and Hunter, 2014; Wei et al., 2015). On the contrary, PIN1 induces the degradation of tumor suppressors such as Daxx, FoxO4, Fbw7, GRK2, PML, KLF10, RARa, RUNX3, RBBP8, Smad, SUV39H1, SMRT, and TRF1 (Lu and Zhou, 2007; Lee T. H. etal., 2009; Ryo et al., 2009; de Thé et al., 2012; Lu and Hunter, 2014; Ueberham et al., 2014; Wei et al., 2015). ERα increases the tumor proliferation through regulating the expression of estrogen response element (ERE)-containing genes in breast cancer (Anderson, 2002). PIN1 induces the ERE-binding affinity and transcription activity, and reduces the ERα degradation mediated by E3 ligase E6AP in breast cancer (Rajbhandari et al., 2012, 2014, 2015). Through inhibiting ubiquitination and destabilizing the transcriptional corepressor SMRT, PIN1 increases HER2 activity (Lam et al., 2008; Stanya et al., 2008). PIN1 also increases the activity of NF-κB pathway via inducing the nuclear accumulation of c-Rel, RelA/p65, and v-Rel (Ryo et al., 2003; Fan et al., 2009). Furthermore, it inhibits the p65 ubiquitination mediated by SOCS-1 (Ryo et al., 2003). PIN1 directed NF-κB activation regulates the proliferation of AML, endometrial carcinoma, glioblastoma, and hepatocellular carcinoma (HCC) (Atkinson et al., 2009; Saegusa et al., 2010; Shinoda et al., 2015; Chen et al., 2016). An isoform of p63, ΔNp63 lacking an intact N-terminal transactivational domain is important for cancer development (Murray-Zmijewski et al., 2006). PIN1 reduces the ΔNp63 ubiquitination induced by WWP1 to enhance the proliferation of oral squamous cell carcinoma (Li et al., 2013). PIN1 stabilizes BRD4 protein to increase the migration and proliferation of gastric cancer (Hu et al., 2017). It also upregulates c-Jun, c-Myc, FoxM1, β-catenin, NUR77, and XBP1 (Chen et al., 2012; Helander et al., 2015; Chae et al., 2016; Kruiswijk et al., 2016; Zhu et al., 2016; Csizmok et al., 2018).
PIN1 and Signal Transduction in Cancer
PIN1 is associated with the development of various cancers, including melanoma, breast cancer, gastric cancer, cervical cancer, gallbladder cancer, pancreatic ductal carcinoma, colorectal cancer, prostate cancer, ovarian cancer, non-small cell lung cancer, osteosarcoma, esophageal cancer, hepatitis B virus (HBV)-induced hepatocellular carcinoma, Burkitt lymphoma, and T cell acute lymphoblastic leukemia. PIN1 is reported to activate 56 oncogenes and/or growth-promoting regulators. Also, it is reported to inactivate 26 tumor suppressors and/or growth-inhibitory regulators (Figure 1).
FIGURE 1.
Function of PIN1 in cancer development.
In human breast cancer, PIN1 promotes oncogenesis via the cyclin D1 regulation (Ryo et al., 2001; Wulf et al., 2001). Studies have shown that PIN1 increases cyclin D1 transcription in association with the HER2–HRAS–JNK–AP1, WNT–β-catenin, and NF-κB pathways. PIN1 regulates HER2, NOTCH1, NOTCH3, androgen receptor (AR) and estrogen receptor α (ERα), which are cancer-driving receptors (La Montagna et al., 2012; Rajbhandari et al., 2012). Furthermore, PIN1 regulates AMPK, AKT93, MYC, PKM2, RAF1, SMAD2, SMAD3, STAT3, the RAS family member RAB2A28, FAK, protein tyrosine phosphatase, PTP-PEST, S6K, and SGK1, which act as intracellular signaling modulators (Lee N. Y. et al., 2009; Jo et al., 2015; Chen Y. et al., 2018). PIN1 induces the interaction of non-receptor type 12 (PTP-PEST) with FAK to increase the FAK Tyr397 dephosphorylation, which induces cancer metastasis (Zheng et al., 2009, 2011). PIN1 also promotes epithelial–mesenchymal transition (EMT) of MCF-7 cells by inducing the transcriptional activity of STAT3 and recruiting its transcription coactivator p300 (Lufei et al., 2007). PIN1 induces the cancer metastasis and invasion by activating β-catenin, BRD4, NF-κB, and p53M (Muller et al., 2009; Wang et al., 2012; Zhu et al., 2016; Hu et al., 2017). Overexpression of PIN1 increases the PTOV1 expression as a novel interactome of PIN1, and knockdown of both genes inhibits the expression of β-catenin, cyclin D1, and c-Myc in breast cancer MDA-MB-231 cells (Karna et al., 2019).
Pin1 transgenic mice in mammary glands induces mammary hyperplasia and malignant mammary tumors (Suizu et al., 2006). Pin1-deficient mice inhibit the massive proliferation of breast epithelium in pregnancy through reducing cyclin D1 levels (Liou et al., 2002) and decreases β-catenin expression in breast cancer (Ryo et al., 2001). Pin1 knockout mice show defects in breast development and induces retinal degeneration and neurodegenerative disorder in brain (Fujimori et al., 1999; Liou et al., 2002).
In human liver cancer, PIN1 is associated with the transcription levels of RhoC and RhoA, and co-overexpression of both genes correlates with metastasis and recurrence of HCC (Ng et al., 2019). All-trans retinoic acid (ATRA) is potent PIN1 inhibitor in hepatocellular carcinoma (Liao X. -H. et al., 2017) and co-targeting p53-RS (p53-R249S) with CDK4, c-Myc, or PIN1 is more effective against the treatment of HCC (Liao X. -H. et al., 2017). PIN1 inhibitor (AF-39) significantly suppresses cell proliferation through the XPO5 subcellular distribution and miRNAs biogenesis in HCC cells (Zheng et al., 2019). Inhibition of Pin1 reverses regorafenib resistance in hepatocellular carcinoma (HCC) with reducing EMT, migration and metastasis (Wang et al., 2019).
In pancreatic cancer, PIN1 was highly expressed in pancreatic ductal adenocarcinoma (PDAC) tissues and significantly correlated with the worst outcomes in patients. PIN1 inhibition with specific siRNA or ATRA suppressed tumor growth in PDAC (Chen et al., 2019). Pin1 is overexpressed and correlated with poor prognosis in gastric cancer (Shi et al., 2015). Pin1 inhibition using small molecule inhibitor such as ATRA or short hairpin RNA, reduces cancer development by inhibiting Wnt/β-catenin and PI3K/AKT signaling pathways in gastric cancer (Zhang et al., 2019).
In nasopharyngeal carcinoma (NPC), Pin1 inhibition reduced NPC cell proliferation, colony formation and anchorage-independent growth through the decrease of cyclin D1 expression and the activation of caspase-3 (Xu et al., 2016). Pin1 enhances transcription activity of ATF1 and induces tumorigenesis in NPC (Huang et al., 2016). Using specific siRNA, Pin1-targeted inhibition suppresses transformed properties and prevents cell proliferation in prostate cancer cells (Ryo et al., 2005). In human melanoma metastasis, although the expression of cytoplasmic Pin1 is not associated with primary melanoma clinical outcome, Pin1 expression in cytosol is correlated with poor survival of melanoma patients (Chen X. et al., 2018). In human colorectal cancer, the expression of cytoplasmic Pin1 is importantly correlated with aggressive tumor behaviors and a worse prognosis in colorectal cancer (Pyo et al., 2018).
In osteosarcoma, PIN1 overexpression using adenovirus significantly stimulates MG-63 and U2-OS cell proliferation. Also, PIN1 inhibitor, juglone reduces cell proliferation in osteosarcoma cells (Zhou et al., 2013). In esophageal squamous cell carcinoma (ESCC), increased Pin1 expression is associated with worse outcome of ESCC patients. Also, Pin1 promotes the aggressiveness of ESCC via β-catenin and cyclin D (Lin et al., 2014). In human lung cancer, cancer patients without Pin1 overexpression has longer cancer-related survival than cancer patients with Pin1 overexpression. Pin1 knockdown in H1299 cell reduces cell invasion and migration (Tan et al., 2010).
In metastatic cancer, PIN1 level is considerably higher than that in primary cancer. The TGF-β signaling promotes the metastasis of cancer. PIN1 increases SMAD degradation mediated by E3 ligase Smurf-2 to repress TGF-β signaling (Nakano et al., 2009). In prostate cancer, PIN1 promotes TGF-β-induced metastasis (Matsuura et al., 2010). Inhibiting the phosphorylation of SMAD3 represses the aggressiveness of breast cancer by reducing the interaction with PIN1 (Thomas et al., 2017).
PIN1 is also involved in angiogenesis. It enhances the transcriptional activity and of stability HIF-1α in several cancer cells (Jalouli et al., 2014; Han et al., 2016). PIN1 promotes the VEGF expression mediated by NF-κB in HCC and regulates the transcriptional factors by VEGF including β-catenin and FoxM1 (Wang et al., 2007; Jiang et al., 2015; Shinoda et al., 2015). Overexpression of HIF-1α, VEGF, and Pin1 is correlated to TAM-resistant MCF-7 cell lines (TAMR-MCF-7) (Oh et al., 2010; Lee T. H. et al., 2011). RNA interference of Pin1 inhibits the angiogenesis as well as the growth of prostate cancer. In TAMR-MCF-7 cells, PI3K/p38 signal pathways increase the Pin1 expression through increasing E2F1 (Lee K. Y. et al., 2011).
PIN1 and Signal Transduction in Cancer Stem Cells (CSCs)
Studies have shown a role of PIN1 in stem cells of breast cancer and leukemia (Luo et al., 2014, 2015; Rustighi et al., 2014; Wei et al., 2015). PIN1 induces NOTCH1 cleavage by γ-secretase, leading to enhanced NOTCH1 transcriptional and tumorigenic activities. PIN1 increases NOTCH1 stability to promote self-renewal and metastasis of breast CSCs by reducing the ubiquitin ligase F-box/WD repeat-containing protein 7 (FBXW7)-mediated degradation of NOTCH1 and NOTCH4 (Rustighi et al., 2014). The deletion of Pin1 decreases the NOTCH-induced invasion of T cell acute lymphoblastic leukemia (T-ALL) cells (Franciosa et al., 2016). PIN1 interacts with the AP1 transcription factors JUN and FOS to activate AP1-dependent RAB2A transcription to promote the expansion and tumorigenesis of breast CSCs (Luo et al., 2015). Overexpression of PIN1 converts normal human breast epithelial cells to cells with stem-like and EMT phenotypes, whereas PIN1 silencing reduces the tumorigenesis and self-renewal activity of breast CSCs in primary breast cancer tissue (Luo et al., 2014, 2015; Rustighi et al., 2014). PIN1 is a pivotal target of miR-200c, a key negative regulator of CSC function and EMT (Shimono et al., 2009; Luo et al., 2014). Inhibition of PIN1 induces the degradation of the fusion oncogene promyelocytic leukemia (PML)–retinoic acid receptor-α (PML–RARα) that drives leukemia stem cells (LSCs), and thereby, treats APL without inducing myeloid differentiation (Ito et al., 2008; de Thé and Chen, 2010). PIN1 controls the maintenance of stability of Nanog, octamer-binding protein 4 (OCT4), and MYC (Nishi et al., 2011; Farrell et al., 2013) and is important for the self-renewal of CSCs.
Pin1 Regulates the Cell Death Resistance and Inflammation of Cancer
Pin1 inhibits apoptosis through BAX as proapoptotic factor in human eosinophils (Shen et al., 2009) and regulates death-associated proteins DAXX to promote its degradation in human gliomas (Ryo et al., 2007). Pin1 induces cell death resistance function of BCL-2 and myeloid cell leukemia-1 (MCL-1) as anti-apoptosis factors (Basu and Haldar, 2002; Ding et al., 2008). Pin1 increases the survival of cisplatin-treated cervical cancer cells through Wnt/β-catenin and FoxM1 signaling (Wang et al., 2016). Pin1 increases the tamoxifen resistance upregulating LC-3 in breast cancer (Namgoong et al., 2010). Pin1 inhibit proapoptotic signals and activate antiapoptotic signals which consequently regulates the cell death resistance in cancer cells.
In allergen-injected rat, inhibition of Pin1 decreases the production of GM-CSF (Esnault et al., 2007). Pin1 induces the IL-22-induced proliferation and survival of breast cancer cells by activating c-Jun, and STAT3 (Kim et al., 2014). Pin1 is involved in inflammatory diseases such as non-alcoholic steatohepatitis (NASH) (Nakatsu et al., 2012), atherosclerosis (Paneni et al., 2015), rheumatoid arthritis (Jeong et al., 2009), and biliary cholangitis (Asuri et al., 2018).
Therapeutic Targeting of PIN1
PIN1 is reported to be highly expressed in variety of human cancers, such as hepatic, prostate, lung, colorectal and esophageal cancers. It participates in diverse cancer-associated signaling pathways. Thus, the development of PIN1 inhibitors has been the focus of several research groups (Table 1).
TABLE 1.
PIN1 inhibitors for cancer treatment.
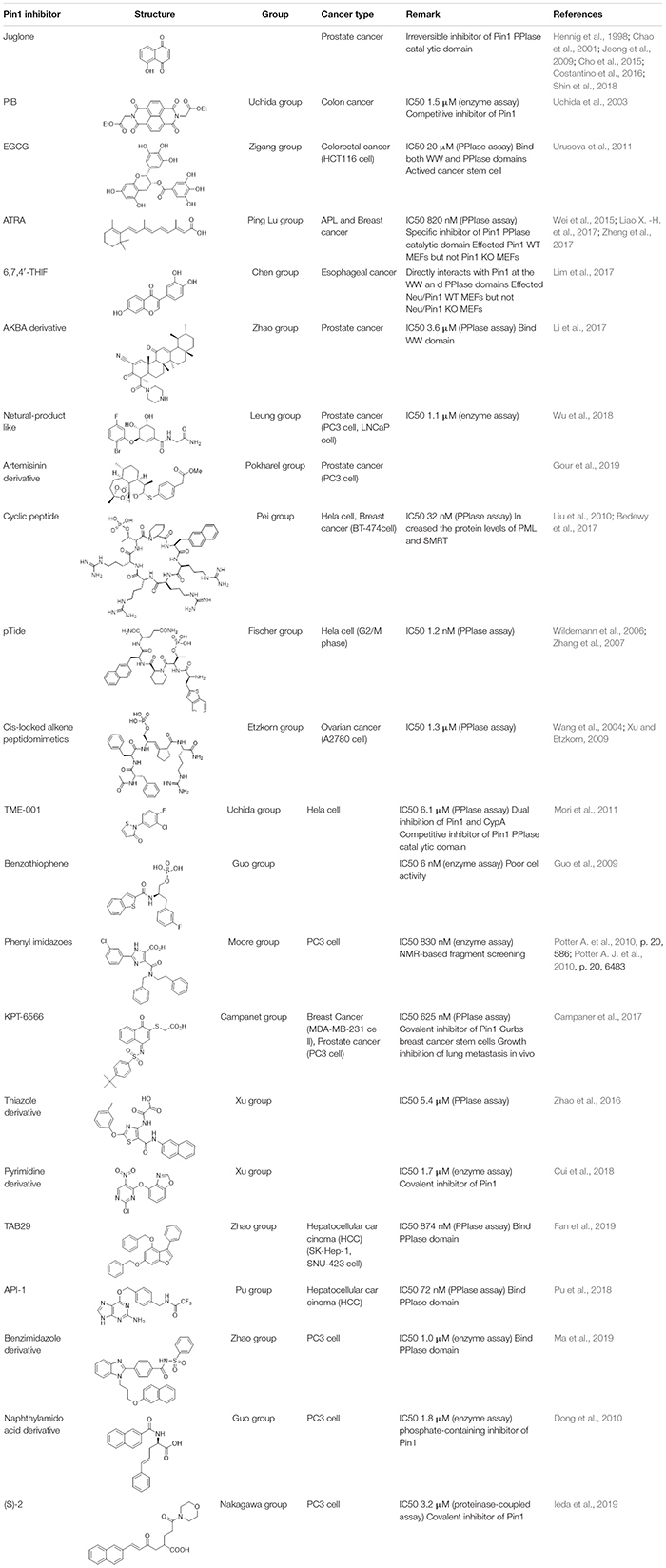 |
The first PIN1 inhibitor discovered by low-throughput screening is juglone. Juglone functions to inhibit the PIN1 PPIase activity in C-terminal catalytic domain, and a high dose of juglone reduces PIN1 protein expression. In addition, juglone has also shown to reduce the prostate cancer cell growth by inhibiting PIN1 activity (Hennig et al., 1998; Chao et al., 2001; Jeong et al., 2009; Costantino et al., 2016; Wang et al., 2017; Shin et al., 2018). Nevertheless, juglone possesses a primarily simple structure that may affect diverse specificity.
A chemically synthesized library containing compounds having a double-ring structure was screened and PiB inhibiting PIN1 (IC50 = 1.5 μM) was identified. Unlike juglone, PiB has been shown to a competitive inhibitor that inhibits the growth of Pin1-containing cells, but not that of Pin1-deficient cells. Furthermore, the inhibition of PIN1 by PiB treatment destabilizes Nanog, transcription factor required for the essential survival of cancer stem cells (Uchida et al., 2003). Uchida et al. identified TME-001 (IC50 = 6.1 μM) for a PIN1 inhibitor by library screening using in vitro enzymatic assay. The results revealed that this compound prevents the growth of HeLa cells (Mori et al., 2011).
Like juglone and PiB, other PIN1 inhibitors have been screened by low-throughput or high-throughput screening. pTide peptide shows PIN1 inhibition at 1.2 nM in vitro, but it is inactive in cells (Wildemann et al., 2006). The specificity of pTide against PIN1 has been shown by the X-ray crystal structure (Zhang et al., 2007). Attachment of an PIN1 octaarginine sequence to the pTide fragment enhances the membrane permeable ability and inhibits the cell growth in cancer (Liu et al., 2010).
A cyclic peptide derivative with increased cell permeable ability repressed the activity of PIN1 (IC50 = 32 nM) and inhibited the BT-474 breast cancer cell proliferation (Liu et al., 2010). Treatment of 100 nM of this peptide in cancer cell lines (HeLa and BT-474) increases the levels of PML and SMRT, and inhibits intracellular PIN1 activity (Liu et al., 2010; Bedewy et al., 2017). A major flavonoid of green tea, epigallocatechin 3 gallate (EGCG) is widely known as chemo-preventive compound for cancer and one of PIN1 inhibitor. Urusova et al. found utilizing X-ray crystal co-structure that EGCG binds to the N-terminal WW as well as C-terminal PPIase domains of PIN1. EGCG inhibits the activity of PIN1 in vitro enzyme assay (IC50 = 20 μM) and reduces JNK signaling pathway, and Bcl-xL and cyclin D1 expression in MEF cells transformed by ErbB2 (Urusova et al., 2011).
Structure-based design induces the identification of cis-locked alkene peptidomimetics as PIN1 inhibitors. They exhibited anti-proliferation activities in A2780 ovarian carcinoma cell line (Wang et al., 2004; Xu and Etzkorn, 2009). Using structure-based drug design, Vernalis and Pfizer develop small molecules. These inhibitors often contain a phosphate or carboxylate as isostere or a phenyl imidazole core, which is required to target the phosphate-binding pocket of the PIN1 protein (Guo et al., 2009; Dong et al., 2010; Potter A. et al., 2010; Potter A. J. et al., 2010; Guo et al., 2014). Pfizer has identified an inhibitor that repressed the PPIase activity of PIN1 at nano-molar concentrations (IC50 = 6 nM) by investigating and exploring the protein crystal structure of PIN1 (Guo et al., 2009; Dong et al., 2010; Guo et al., 2014). Using 900-number fragment library, Vernalis has developed a NMR-based fragment screen to isolate PIN1 inhibitors through the protease-coupled in vitro enzyme assay. A PIN1 inhibitor was synthesized (Potter A. J. et al., 2010) and showed good nanomolar inhibition against PIN1 in vitro (IC50 = 830 nM). However, they are poorly active or inactive in cell lines since the phosphate or carboxylate renders the inhibitors poor cell permeable ability (Guo et al., 2009; Dong et al., 2010; Potter A. et al., 2010; Potter A. J. et al., 2010).
Leung et al. have identified a PIN1 inhibitor from natural-product library using structure-based virtual screening and they show that compound 1 targets PIN1 and interferes the interaction of PIN1 with the NF-κB p65 subunit in cells. Moreover, a natural-product compound induced apoptosis in PC-3 cell lines (Wu et al., 2018). Using the virtual screening analysis, PIN1 protein has been identified as a target of 6,7,4′-trihydroxyisoflavone (6,7,4′-THIF). 6,7,4′-THIF bound to PIN1 protein, but did not bind to the family proteins such as FKBP or cyclophilin A, suggesting a selective and specific binding with PIN1. 6,7,4′-THIF compound was analyzed for specific inhibitory activity for PIN1 using Neu/Pin1 knockout (KO) and Neu/Pin1 wild-type (WT) MEFs. This PIN1 inhibitor affected Neu/Pin1 WT MEF cells, but not Neu/Pin1 KO MEF cells. In addition, the result of a xenograft tumor growth assay in mice utilizing Neu/Pin1 KO and WT MEF cells have been shown similar to the result from the in vitro enzyme assay (Lim et al., 2017).
One of the reported inhibitors of PIN1, aetyl-11-keto-β-boswellic acid (AKBA) derivative has been shown to inhibit the growth of prostate cancer PC-3 (IC50 = 40 nM) and LNCaP (IC50 = 270 nM) cell lines. The compound inhibited the activity of PIN1, to stabilize cyclin D1, which improved anti-proliferative effects of prostate cancer treatment through new mechanisms (Li et al., 2017). In addition to the previous inhibitors of PIN1, thiazole derivatives (IC50 = 5.3 μM), pyrimidine derivatives (IC50 = 1.7 μM), benzimidazle derivatives (IC50 = 1.0 μM), 6-O-benzylguaninie derivative API-1 (IC 50 = 72 nM), and phenylbenzofuran derivative TAB29 (IC50 = 874 nM) have been reported as non-small molecule inhibitors (Zhao et al., 2016; Cui et al., 2018; Pu et al., 2018; Fan et al., 2019; Ma et al., 2019). In particular, in silico virtual screening was performed using the PIN1crystal structures and identified API-1 and TAB29 as small molecules that bind to the PPIase domain. Furthermore, PIN1 inhibition by API-1 and TAB29 upregulates miRNA biogenesis by maintaining the active XPO5 conformation and represses the development of hepatocellular carcinoma (HCC), suggesting that PIN1 mediates miRNA biogenesis mechanism, API-1 can be a drug candidate for therapy for Pin1-overexpressing or extracellular signal-regulated kinase-activated HCC (Pu et al., 2018; Fan et al., 2019).
The Food and Drug Administration (FDA) approved all-trans retinoic acid (ATRA) for acute promyelocytic leukemia (APL) therapy (Wei et al., 2015). ATRA was identified using a mechanism-based high-throughput screening system. ATRA inhibited the activity of PIN1 by binding with the C-terminal catalytic PPIase domain of PIN1. ATRA induces the degradation of PIN1 protein, but also suppresses the oncogenic function by decreasing the expression of cyclin D1. Furthermore, PIN1 inhibition mediated by ATRA induces the degradation of PML-RARA oncoprotein, resulting in anti-proliferative effect in APL cells and mouse models, as well as in humans. Moreover, a slow-release ATRA formulation induces the degradation of PIN1 and decreases tumorigenicity in mice xenograft model of HCC (Wei et al., 2015; Liao P. et al., 2017). Additionally, a combination of ATRA and sorafenib for the HCC treatment decreases the expression of PIN1 protein, increases cancer cell death, and represses the HCC growth compared with sorafenib or ATRA alone. These results provide an important rationale for further PIN1 inhibitor development to increase the therapeutic efficacy of general drug for HCC (Zheng et al., 2017).
A more recent study identifies KPT-6566, a novel PIN1 small molecule inhibitor, possessing high potency (IC50 = 625 nM) and specificity from a drug-like collection of 0.2 million commercial compounds (Campaner et al., 2017). Compounds capable of covalently binding to the C113 residue of the PIN1 catalytic domain were selected by virtual structure-based screening and cytotoxicity testing to select the final compounds. Structurally, the electrophile sulfonyl-acetate moiety of KPT-6566 directly faces the nucleophile sulfur atom of C113. Like ATRA, KPT-6566 also promotes the degradation of PIN1, resulting in the reduction of hyper-phosphorylated pRB and cyclin D1 levels. KPT-6566 increases the apoptosis and decreases the cancer cell proliferation such as pancreatic, lung, prostate, and breast cancers. It showed a better anti-proliferative effect on cancer cell lines than on normal cell lines. Furthermore, treatment with KPT-6566 inhibited the overexpression of Pin1, confirming the reduction of breast cancer stem cells. In addition, in in vivo studies, KPT-6566 has been shown to decrease the lung metastasis in breast cancer mouse models. Currently, KPT-6566 is the only PIN1 inhibitor in the preclinical stage of research.
A study reported by the Pokharel et al. shows that the artemisinin derivatives commonly used as antimalarial drugs are very effective in variety of cancer cell lines to inhibit cancer cell growth. Especially compound 9a, one of the artemisinin derivatives increases anti-proliferative, pro-apoptotic and anti-metastatic effect in PC-3 prostate cancer cells by decreasing the expression of Pin1, cyclin D1, c-Myc, elF4E, and PCNA (Gour et al., 2019).
Irreversible PIN1 inhibitor (S)-2 (IC50 = 3.2 μM), and its derivatives recently designed by Ieda et al. show the inhibition of Pin1 in protease-coupled in vitro assay and the reduction of cyclin D1 expression in PC-3 prostate cancer cell (Ieda et al., 2019).
Conclusion
PIN1 is a well-known PPIase that regulates the cis–trans isomerization of pSer/Thr–Pro, which highlights its importance in the control of Pro-directed phosphorylation. PIN1 regulates protein function via conformational changes of target protein and is associated with the oncogenic pathway activation by controlling tumor suppressors and oncogenes. PIN1 is overexpressed in cancer tissues and CSCs, and correlated with poor clinical outcome in various cancer patients. Inhibition of PIN1 plays an important role in the tumorigenesis and angiogenesis of cancer, thereby providing a new great therapeutic target. Recently, PIN1 inhibitors have been developed elsewhere using structure-based drug designs and natural compounds that inhibit the activity of cancer. PIN1 obviously can be an super attractive target for curing cancer and cancer stem cells.
Author Contributions
All authors designed and wrote this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (NRF-2015M3A9C7030181, NRF-2016M3A9E4947797 and NRF-2017R1D1A1B03034810).
References
- Anderson E. (2002). Progesterone receptors - animal models and cell signaling in breast cancer: the role of oestrogen and progesterone receptors in human mammary development and tumorigenesis. Breast Cancer Res. 4 197–201. 10.1186/bcr452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuri S., McIntosh S., Taylor V., Rokeby A., Kelly J., Shumansky K., et al. (2018). Primary Biliary Cholangitis in British Columbia First Nations: clinical features and discovery of novel genetic susceptibility loci. Liver Int. 38 940–948. 10.1111/liv.13686 [DOI] [PubMed] [Google Scholar]
- Atkinson G. P., Susan E. N., Harrison D. K., Stonecypher M. S., Chen D., Benveniste E. N. (2009). The prolyl isomerase Pin1 regulates the NF-κB signaling pathway and interleukin-8 expression in glioblastoma. Oncogene 28 3735–3745. 10.1038/onc.2009.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala G., Wang D., Wulf G., Frolov A., Li R., Sowadski J., et al. (2003). The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 63 6244–6251. [PubMed] [Google Scholar]
- Bao L., Kimzey A., Sauter G., Sowadski J. M., Lu K. P., Wang D.-G. (2004). Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am. J. Pathol. 164 1727–1737. 10.1016/S0002-9440(10)63731-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Haldar S. (2002). Signal-induced site specific phosphorylation targets Bcl2 to the proteasome pathway. Int. J. Oncol. 21 597–601. [PubMed] [Google Scholar]
- Bedewy W., Liao H., Abou-Taleb N. A., Hammad S. F., Nasr T., Pei D. (2017). Generation of a cell-permeable cycloheptapeptidyl inhibitor against the peptidyl–prolyl isomerase Pin1. Org. Biomol. Chem. 15 4540–4543. 10.1039/C7OB00430C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran N., van Drogen F., Ng H.-F., Kumar R., Ekholm-Reed S., Peter M., et al. (2013). Fbw7α and Fbw7γ collaborate to shuttle cyclin E1 into the nucleolus for multiubiquitylation. Mol. Cell. Biol. 33 85–97. 10.1128/MCB.00288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner E., Rustighi A., Zannini A., Cristiani A., Piazza S., Ciani Y., et al. (2017). A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat. Commun. 8:15772. 10.1038/ncomms15772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae U., Park S.-J., Kim B., Wei S., Min J.-S., Lee J.-H., et al. (2016). Critical role of XBP1 in cancer signalling is regulated by PIN1. Biochem. J. 473 2603–2610. 10.1042/bcj20160482 [DOI] [PubMed] [Google Scholar]
- Chao S.-H., Greenleaf A. L., Price D. H. (2001). Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res. 29 767–773. 10.1093/nar/29.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Chang C.-C., Lee T. H., Luo M., Huang P., Liao P.-H., et al. (2013). SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res. 73 3951–3962. 10.1158/0008-5472.CAN-12-4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Li W., Sultana R., You M.-H., Kondo A., Shahpasand K., et al. (2015). Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer’s disease. Neurobiol. Dis. 76 13–23. 10.1016/j.nbd.2014.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Z., Li L., Wang W. J., Du X. D., Wen Q., He J. P., et al. (2012). Prolyl isomerase Pin1 stabilizes and activates orphan nuclear receptor TR3 to promote mitogenesis. Oncogene 31 2876–2887. 10.1038/onc.2011.463 [DOI] [PubMed] [Google Scholar]
- Chen J.-Y., Lai Y.-S., Tsai H.-J., Kuo C.-C., Yen B. L., Yeh S.-P., et al. (2016). The oncometabolite R-2-hydroxyglutarate activates NF-κB-dependent tumor-promoting stromal niche for acute myeloid leukemia cells. Sci. Rep. 6:32428. 10.1038/srep32428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xu X., Wen X., Xu S., Wang L., Lu W., et al. (2019). Targeting PIN1 exerts potent antitumor activity in pancreatic ductal carcinoma via inhibiting tumor metastasis. Cancer Sci. 110 2442–2455. 10.1111/cas.14085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu X., Deng B., Martinka M., Zhou Y., Lan X., et al. (2018). Cytoplasmic Pin1 expression is increased in human cutaneous melanoma and predicts poor prognosis. Sci. Rep. 8:16867. 10.1038/s41598-018-34906-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wu Y.-R., Yang H.-Y., Li X.-Z., Jie M.-M., Hu C.-J., et al. (2018). Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. 9:883. 10.1038/s41419-018-0844-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. A., Jue S. S., Bae W. J., Heo S. H., Shin S. I., Kwon I. K., et al. (2015). PIN1 inhibition suppresses osteoclast differentiation and inflammatory responses. J. Dent. Res. 94 371–380. 10.1177/0022034514563335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. S., Park S. Y., Kim D. J., Lee S.-H., Woo K.-M., Lee K.-A., et al. (2012). TPA-induced cell transformation provokes a complex formation between Pin1 and 90 kDa ribosomal protein S6 kinase 2. Mol. Cell. Biochem. 367 85–92. 10.1007/s11010-012-1322-y [DOI] [PubMed] [Google Scholar]
- Costantino S., Paneni F., Lüscher T. F., Cosentino F. (2016). Pin1 inhibitor Juglone prevents diabetic vascular dysfunction. Int. J. Cardiol. 203 702–707. 10.1016/j.ijcard.2015.10.221 [DOI] [PubMed] [Google Scholar]
- Csizmok V., Montecchio M., Lin H., Tyers M., Sunnerhagen M., Forman-Kay J. D. (2018). Multivalent interactions with Fbw7 and Pin1 facilitate recognition of c-Jun by the SCFfbw7 ubiquitin ligase. Structure 26 28–39.e2. 10.1016/j.str.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Cui G., Jin J., Chen H., Cao R., Chen X., Xu B. (2018). Synthesis and biological evaluation of pyrimidine derivatives as, novel human Pin1 inhibitors. Bioorg. Med. Chem. 26 2186–2197. 10.1016/j.bmc.2018.03.024 [DOI] [PubMed] [Google Scholar]
- de Thé H., Chen Z. (2010). Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat. Rev. Cancer 10 775–783. 10.1038/nrc2943 [DOI] [PubMed] [Google Scholar]
- de Thé H., Le Bras M., Lallemand-Breitenbach V. (2012). Acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 198 11–21. 10.1083/jcb.201112044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Huo L., Yang J.-Y., Xia W., Wei Y., Liao Y., et al. (2008). Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. 68 6109–6117. 10.1158/0008-5472.CAN-08-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Marakovits J., Hou X., Guo C., Greasley S., Dagostino E., et al. (2010). Structure-based design of novel human Pin1 inhibitors (II). Bioorg. Med. Chem. Lett. 20 2210–2214. 10.1016/j.bmcl.2010.02.033 [DOI] [PubMed] [Google Scholar]
- Eckerdt F., Yuan J., Saxena K., Martin B., Kappel S., Lindenau C., et al. (2005). Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. J. Biol. Chem. 280 36575–36583. 10.1074/jbc.M504548200 [DOI] [PubMed] [Google Scholar]
- Esnault S., Rosenthal L., Shen A. Z., Sedgwick J. J., Szakaly B. R., Sorkness J. (2007). A critical role for Pin1 in allergic pulmonary eosinophilia in rats. J. Allergy Clin. Immunol. 120 1082–1088. 10.1016/j.jaci.2007.06.024 [DOI] [PubMed] [Google Scholar]
- Fan G., Fan Y., Gupta N., Matsuura I., Liu F., Zhou X. Z., et al. (2009). Peptidyl-prolyl isomerase Pin1 markedly enhances the oncogenic activity of the Rel proteins in the nuclear factor-κB family. Cancer Res. 69 4589–4597. 10.1158/0008-5472.CAN-08-4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., He H., Li J., Luo G., Zheng Y., Zhou J.-K., et al. (2019). Discovery of 4,6-bis(benzyloxy)-3-phenylbenzofuran as a novel Pin1 inhibitor to suppress hepatocellular carcinoma via upregulating microRNA biogenesis. Bioorg. Med. Chem. 27 2235–2244. 10.1016/j.bmc.2019.04.028 [DOI] [PubMed] [Google Scholar]
- Farrell A. S., Pelz C., Wang X., Daniel C. J., Wang Z., Su Y., et al. (2013). Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol. Cell. Biol. 33 2930–2949. 10.1128/MCB.01455-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciosa G., Diluvio G., Gaudio F. D., Giuli M. V., Palermo R., Grazioli P., et al. (2016). Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression. Oncogene 35 4741–4751. 10.1038/onc.2016.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori F., Takahashi K., Uchida C., Uchida T. (1999). Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem. Biophys. Res. Commun. 265 658–663. 10.1006/bbrc.1999.1736 [DOI] [PubMed] [Google Scholar]
- Gianni M., Boldetti A., Guarnaccia V., Rambaldi A., Parrella E. (2009). Inhibition of the peptidyl-prolyl-isomerase Pin1 enhances the responses of acute myeloid leukemia cells to retinoic acid via stabilization of RARα and PML-RARα. Cancer Res. 69 1016. 10.1158/0008-5472.CAN-08-2603 [DOI] [PubMed] [Google Scholar]
- Gour R., Ahmad F., Prajapati S. K., Giri S. K., Karna S. K. L., Kartha K. P. R., et al. (2019). Synthesis of novel S-linked dihydroartemisinin derivatives and evaluation of their anticancer activity. Eur. J. Med. Chem. 178 552–570. 10.1016/j.ejmech.2019.06.018 [DOI] [PubMed] [Google Scholar]
- Guo C., Hou X., Dong L., Dagostino E., Greasley S., Ferre R., et al. (2009). Structure-based design of novel human Pin1 inhibitors (I). Bioorg. Med. Chem. Lett. 19 5613–5616. 10.1016/j.bmcl.2009.08.034 [DOI] [PubMed] [Google Scholar]
- Guo C., Hou X., Dong L., Marakovits J., Greasley S., Dagostino E., et al. (2014). Structure-based design of novel human Pin1 inhibitors (III): optimizing affinity beyond the phosphate recognition pocket. Bioorg. Med. Chem. Lett. 24 4187–4191. 10.1016/j.bmcl.2014.07.044 [DOI] [PubMed] [Google Scholar]
- Han H.-J., Kwon N., Choi M.-A., Jung K. O., Piao J.-Y., Ngo H. K. C., et al. (2016). Peptidyl prolyl isomerase PIN1 directly binds to and stabilizes hypoxia-inducible factor-1α. PLoS One 11:e0147038. 10.1371/journal.pone.0147038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander S., Montecchio M., Pilstål R., Su Y., Kuruvilla J., Elvén M., et al. (2015). Pre-Anchoring of Pin1 to Unphosphorylated c-Myc in a fuzzy complex regulates c-Myc activity. Structure 23 2267–2279. 10.1016/j.str.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Christner C., Kipping M., Schelbert B., Rücknagel K. P., Grabley S., et al. (1998). Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by Juglone. Biochemistry 37 5953–5960. 10.1021/bi973162p [DOI] [PubMed] [Google Scholar]
- Hu X., Dong S. H., Chen J., Zhou X. Z., Chen R., Nair S., et al. (2017). Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4. Oncogene 36 5177–5188. 10.1038/onc.2017.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. L., Liao D., Chen H., Lu Y., Chen L., Li H., et al. (2016). The protein level and transcription activity of activating transcription factor 1 is regulated by prolyl isomerase Pin1 in nasopharyngeal carcinoma progression. Cell Death Dis. 7:e2571. 10.1038/cddis.2016.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda N., Itoh K., Inoue Y., Izumiya Y., Kawaguchi M., Miyata N., et al. (2019). An irreversible inhibitor of peptidyl-prolyl cis/trans isomerase Pin1 and evaluation of cytotoxicity. Bioorg. Med. Chem. Lett. 29 353–356. 10.1016/j.bmcl.2018.12.044 [DOI] [PubMed] [Google Scholar]
- Ito K., Bernardi R., Morotti A., Matsuoka S., Saglio G., Ikeda Y., et al. (2008). PML targeting eradicates quiescent leukaemia-initiating cells. Nature 453 1072–1078. 10.1038/nature07016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalouli M., Déry M.-A. C., Lafleur V. N., Lamalice L., Zhou X. Z., Lu K. P., et al. (2014). The prolyl isomerase Pin1 regulates hypoxia-inducible transcription factor (HIF) activity. Cell. Signal. 26 1649–1656. 10.1016/j.cellsig.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Jeong H. G., Pokharel Y. R., Lim S. C., Hwang Y. P., Han E. H., Yoon J.-H., et al. (2009). Novel role of pin1 induction in type II collagen-mediated rheumatoid arthritis. J. Immunol. 183 6689–6697. 10.4049/jimmunol.0901431 [DOI] [PubMed] [Google Scholar]
- Jeong K., Kim S.-J., Oh Y., Kim H., Lee Y.-S., Kwon B.-S., et al. (2014). p53 negatively regulates Pin1 expression under ER stress. Biochem. Biophys. Res. Commun. 454 518–523. 10.1016/j.bbrc.2014.10.101 [DOI] [PubMed] [Google Scholar]
- Jiang L., Yin M., Wei X., Liu J., Wang X., Niu C., et al. (2015). Bach1 represses Wnt/β-Catenin signaling and angiogenesis. Circ. Res. 117 364–375. 10.1161/CIRCRESAHA.115.306829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A. R. A., Yun H. J., Kim J. Y., Lim S.-C., Choi H. J., Kang B. S., et al. (2015). Prolyl isomerase PIN1 negatively regulates SGK1 stability to mediate tamoxifen resistance in breast cancer cells. Anticancer Res. 35 785–794. [PubMed] [Google Scholar]
- Kamimura T., Miyamoto T., Nagafuji K., Numata A., Henzan H., Takase K., et al. (2011). Role of autotransplantation in the treatment of acute promyelocytic leukemia patients in remission: fukuoka BMT Group observations and a literature review. Bone Marrow Transpl. 46 820–826. 10.1038/bmt.2010.207 [DOI] [PubMed] [Google Scholar]
- Karna S. K. L., Ahmad F., Lone B. A., Pokharel Y. R. (2019). Knockdown of PTOV1 and PIN1 exhibit common phenotypic anti-cancer effects in MDA-MB-231 cells. PLoS One 14:e0211658. 10.1371/journal.pone.0211658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kim G., Yun J. K., Yun H. J., Choi H. S. (2014). Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis 35 1352–1361. 10.1093/carcin/bgu044 [DOI] [PubMed] [Google Scholar]
- Kruiswijk F., Hasenfuss S. C., Sivapatham R., Baar M. P., Putavet D., Naipal K. A. T., et al. (2016). Targeted inhibition of metastatic melanoma through interference with Pin1-FOXM1 signaling. Oncogene 35 2166–2177. 10.1038/onc.2015.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Montagna R., Caligiuri I., Maranta P., Lucchetti C., Esposito L., Paggi M. G., et al. (2012). Androgen receptor serine 81 mediates Pin1 interaction and activity. Cell Cycle 11 3415–3420. 10.4161/cc.21730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P. B., Burga L. N., Wu B. P., Hofstatter E. W., Lu K. P., Wulf G. M. (2008). Prolyl isomerase Pin1 is highly expressed in Her2-positive breast cancer and regulates erbB2 protein stability. Mol. Cancer 7:91. 10.1186/1476-4598-7-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Lin F. C., Hsu T. I., Lin J. T., Guo J. H., Tsai C. H., et al. (2014). MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim. Biophys. Acta 1843 2055–2066. 10.1016/j.bbamcr.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Lee K. Y., Jeong Woon L., Hyun Jeong N., Jeong-Hyun S., Youngsup S., Keon Wook K. (2011). PI3-Kinase/p38 kinase-dependent E2F1 activation is critical for Pin1 induction in tamoxifen-resistant breast cancer cells. Mol. Cells 32 107–111. 10.1007/s10059-011-0074-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N. Y., Choi H.-K., Shim J.-H., Kang K.-W., Dong Z., Choi H. S. (2009). The prolyl isomerase Pin1 interacts with a ribosomal protein S6 kinase to enhance insulin-induced AP-1 activity and cellular transformation. Carcinogenesis 30 671–681. 10.1093/carcin/bgp027 [DOI] [PubMed] [Google Scholar]
- Lee T. H., Chen C.-H., Suizu F., Huang P., Schiene-Fischer C., Daum S., et al. (2011). Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol. Cell 42 147–159. 10.1016/j.molcel.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Tun-Kyi A., Shi R., Lim J., Soohoo C., Finn G., et al. (2009). Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat. Cell Biol. 11 97–105. 10.1038/ncb1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-C., Que J., Chen Y.-C., Lin J.-T., Liou Y.-C., Liao P.-C., et al. (2013). Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A–Bora complex. J. Cell Sci. 126 4862–4872. 10.1242/jcs.121368 [DOI] [PubMed] [Google Scholar]
- Li C., Chang D. L., Yang Z., Qi J., Liu R., He H., et al. (2013). Pin1 modulates p63α protein stability in regulation of cell survival, proliferation and tumor formation. Cell Death Dis. 4:e943. 10.1038/cddis.2013.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Li L., Wang S., Li X., Ma T., Liu D., et al. (2017). Design and synthesis of novel 2-substituted 11-keto-boswellic acid heterocyclic derivatives as anti-prostate cancer agents with Pin1 inhibition ability. Eur. J. Med. Chem. 126 910–919. 10.1016/j.ejmech.2016.09.089 [DOI] [PubMed] [Google Scholar]
- Liao P., Zeng S. X., Zhou X., Chen T., Zhou F., Cao B., et al. (2017). Mutant p53 gains its function via c-Myc activation upon CDK4 phosphorylation at serine 249 and consequent PIN1 binding. Mol. Cell 68 1134–1146. 10.1016/j.molcel.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.-H., Zhang A. L., Zheng M., Li M.-Q., Chen C. P., Xu H., et al. (2017). Chemical or genetic Pin1 inhibition exerts potent anticancer activity against hepatocellular carcinoma by blocking multiple cancer-driving pathways. Sci. Rep. 7:43639. 10.1038/srep43639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Wei Y., Zhou X., Yang J. Y., Dai C., Chen Y. J., et al. (2009). Peptidyl-prolyl cis/trans isomerase Pin1 is critical for the regulation of PKB/Akt stability and activation phosphorylation. Oncogene 28 2436–2445. 10.1038/onc.2009.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T.-G., Lee S.-Y., Duan Z., Lee M.-H., Chen H., Liu F., et al. (2017). The prolyl isomerase Pin1 is a novel target of 6,7,4′-trihydroxyisoflavone for suppressing esophageal cancer growth. Cancer Prev. Res. 10 308–318. 10.1158/1940-6207.CAPR-16-0318 [DOI] [PubMed] [Google Scholar]
- Lin F. C., Lee Y. C., Goan Y. G., Tsai C. H., Yao Y. C., Cheng H. C., et al. (2014). Pin1 positively affects tumorigenesis of esophageal squamous cell carcinoma and correlates with poor survival of patients. J. Biomed. Sci. 21:75. 10.1186/s12929-014-0075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou Y. C., Ryo A., Huang H. K., Lu P. J., Bronson R., Fujimori F., et al. (2002). Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. U.S.A. 99 1335–1340. 10.1073/pnas.032404099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou Y.-C., Zhou X. Z., Lu K. P. (2011). Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 36 501–514. 10.1016/j.tibs.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Y., Kao H.-Y., Pei D. (2010). Membrane permeable cyclic peptidyl inhibitors against human peptidylprolyl isomerase Pin1. J. Med. Chem. 53 2494–2501. 10.1021/jm901778v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Hu Z., Wei S., Wang L.-E., Liu Z., El-Naggar A. K., et al. (2009). A novel functional variant (-842G>C) in the PIN1 promoter contributes to decreased risk of squamous cell carcinoma of the head and neck by diminishing the promoter activity. Carcinogenesis 30 1717–1721. 10.1093/carcin/bgp171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. P., Hanes S. D., Hunter T. (1996). A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380 544–547. 10.1038/380544a0 [DOI] [PubMed] [Google Scholar]
- Lu K. P., Liou Y.-C., Zhou X. Z. (2002). Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 12 164–172. 10.1016/S0962-8924(02)02253-5 [DOI] [PubMed] [Google Scholar]
- Lu K. P., Zhou X. Z. (2007). The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 8 904–916. 10.1038/nrm2261 [DOI] [PubMed] [Google Scholar]
- Lu P.-J., Zhou X. Z., Liou Y.-C., Noel J. P., Lu K. P. (2002). Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J. Biol. Chem. 277 2381–2384. 10.1074/jbc.c100228200 [DOI] [PubMed] [Google Scholar]
- Lu P.-J., Zhou X. Z., Shen M., Lu K. P. (1999). Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283 1325–1328. 10.1126/science.283.5406.1325 [DOI] [PubMed] [Google Scholar]
- Lu Z., Hunter T. (2014). Prolyl isomerase Pin1 in cancer. Cell Res. 24 1033–1049. 10.1038/cr.2014.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufei C., Koh T. H., Uchida T., Cao X. (2007). Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene 26 7656–7664. 10.1038/sj.onc.1210567 [DOI] [PubMed] [Google Scholar]
- Luo M.-L., Gong C., Chen C.-H., Hu H., Huang P., Zheng M., et al. (2015). The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via Erk signaling activation. Cell Rep. 11 111–124. 10.1016/j.celrep.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.-L., Gong C., Chen C.-H., Lee D. Y., Hu H., Huang P., et al. (2014). Prolyl isomerase Pin1 acts downstream of miR200c to promote cancer stem–like cell traits in breast cancer. Cancer Res. 74 3603–3616. 10.1158/0008-5472.CAN-13-2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Huang M., Li A., Zhao F., Li D., Liu D., et al. (2019). Design, synthesis and biological evaluation of benzimidazole derivatives as novel human Pin1 inhibitors. Bioorg. Med. Chem. Lett. 29 1859–1863. 10.1016/j.bmcl.2018.11.045 [DOI] [PubMed] [Google Scholar]
- MacLachlan T. K., Somasundaram K., Sgagias M., Shifman Y., Muschel R. J., Cowan K. H., et al. (2000). BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J. Biol. Chem. 275 2777–2785. 10.1074/jbc.275.4.2777 [DOI] [PubMed] [Google Scholar]
- Matsuura I., Chiang K.-N., Lai C.-Y., He D., Wang G., Ramkumar R., et al. (2010). Pin1 promotes transforming growth factor-β-induced migration and invasion. J. Biol. Chem. 285 1754–1764. 10.1074/jbc.M109.063826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersch J., Jackson M. A., Park M., Nebgen D., Peterson S. K., Singletary C., et al. (2015). Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 121 269–275. 10.1002/cncr.29357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. J., Smith S. L. (1988). Effects of the chitin synthetase inhibitor plumbagin and its 2-demethyl derivative juglone on insect ecdysone 20-monooxygenase activity. Experientia 44 990–991. 10.1007/BF01939896 [DOI] [PubMed] [Google Scholar]
- Miyashita H., Mori S., Motegi K., Fukumoto M., Uchida T. (2003). Pin1 is overexpressed in oral squamous cell carcinoma and its levels correlate with cyclin D1 overexpression. Oncol. Rep. 10 455–461. 10.3892/or.10.2.455 [DOI] [PubMed] [Google Scholar]
- Moretto-Zita M., Jin H., Shen Z., Zhao T., Briggs S. P., Xu Y. (2010). Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc. Natl. Acad. Sci. U.S.A. 107 13312–13317. 10.1073/pnas.1005847107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Hidaka M., Lin Y.-C., Yoshizawa I., Okabe T., Egashira S., et al. (2011). A dual inhibitor against prolyl isomerase Pin1 and cyclophilin discovered by a novel real-time fluorescence detection method. Biochem. Biophys. Res. Commun. 406 439–443. 10.1016/j.bbrc.2011.02.066 [DOI] [PubMed] [Google Scholar]
- Muller P. A. J., Caswell P. T., Doyle B., Iwanicki M. P., Tan E. H., Karim S., et al. (2009). Mutant p53 drives invasion by promoting integrin recycling. Cell 139 1327–1341. 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F., Lane D. P., Bourdon J. C. (2006). p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 13 962–972. 10.1038/sj.cdd.4401914 [DOI] [PubMed] [Google Scholar]
- Nakano A., Koinuma D., Miyazawa K., Uchida T., Saitoh M., Kawabata M., et al. (2009). Pin1 down-regulates transforming growth factor-β (TGF-β) signaling by inducing degradation of Smad proteins. J. Biol. Chem. 284 6109–6115. 10.1074/jbc.M804659200 [DOI] [PubMed] [Google Scholar]
- Nakatsu Y., Otani Y., Sakoda H., Zhang J. (2012). Role of Pin1 protein in the pathogenesis of nonalcoholic steatohepatitis in a rodent model. J. Biol. Chem. 287 44526–44535. 10.1074/jbc.M112.397133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong G. M., Khanal P., Cho H. G., Lim S. C., Oh Y. K., Kang B. S. (2010). The prolyl isomerase Pin1 induces LC-3 expression and mediates tamoxifen resistance in breast cancer. J. Biol. Chem. 285 23829–23841. 10.1074/jbc.M109.092874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L., Kwan V., Chow A., Yau T. C., Poon R. T., Pang R., et al. (2019). Overexpression of Pin1 and rho signaling partners correlates with metastatic behavior and poor recurrence-free survival of hepatocellular carcinoma patients. BMC Cancer 19:713. 10.1186/s12885-019-5919-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M., Akutsu H., Masui S., Kondo A., Nagashima Y., Kimura H., et al. (2011). A distinct role for Pin1 in the induction and maintenance of pluripotency. J. Biol. Chem. 286 11593–11603. 10.1074/jbc.M110.187989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J., Kim O., Lee J. S., Kim J.-A., Kim M. R., Choi H. S., et al. (2010). Inhibition of angiogenesis by quercetin in tamoxifen-resistant breast cancer cells. Food Chem. Toxicol. 48 3227–3234. 10.1016/j.fct.2010.08.028 [DOI] [PubMed] [Google Scholar]
- Paneni F., Costantino S., Castello L., Battista R., Capretti G., Chiandotto S., et al. (2015). Targeting prolyl-isomerase Pin1 prevents mitochondrial oxidative stress and vascular dysfunction: insights in patients with diabetes. Eur. Heart J. 36 817–828. 10.1093/eurheartj/ehu179 [DOI] [PubMed] [Google Scholar]
- Potter A., Oldfield V., Nunns C., Fromont C., Ray S., Northfield C. J., et al. (2010a). Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution. Bioorg. Med. Chem. Lett. 20 6483–6488. 10.1016/j.bmcl.2010.09.063 [DOI] [PubMed] [Google Scholar]
- Potter A. J., Ray S., Gueritz L., Nunns C. L., Bryant C. J., Scrace S. F., et al. (2010b). Structure-guided design of α-amino acid-derived Pin1 inhibitors. Bioorg. Med. Chem. Lett. 20 586–590. 10.1016/j.bmcl.2009.11.090 [DOI] [PubMed] [Google Scholar]
- Pu W., Li J., Zheng Y., Shen X., Fan X., Zhou J.-K., et al. (2018). Targeting Pin1 by inhibitor API-1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology 68 547–560. 10.1002/hep.29819 [DOI] [PubMed] [Google Scholar]
- Pyo J. S., Son B. K., Oh I. H. (2018). Cytoplasmic Pin1 expression is correlated with poor prognosis in colorectal cancer. Pathol. Res. Pract. 214 1848–1853. 10.1016/j.prp.2018.09.018 [DOI] [PubMed] [Google Scholar]
- Rajbhandari P., Finn G., Solodin N. M., Singarapu K. K., Sahu S. C., Markley J. L., et al. (2012). Regulation of estrogen receptor α N-terminus conformation and function by peptidyl prolyl isomerase Pin1. Mol. Cell. Biol. 32 445–457. 10.1128/MCB.06073-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajbhandari P., Ozers M. S., Solodin N. M., Warren C. L., Alarid E. T. (2015). Peptidylprolyl isomerase Pin1 directly enhances the DNA binding functions of estrogen receptor α. J. Biol. Chem. 290 13749–13762. 10.1074/jbc.M114.621698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajbhandari P., Schalper K. A., Solodin N. M., Ellison-Zelski S. J., Ping Lu K., Rimm D. L., et al. (2014). Pin1 modulates ERα levels in breast cancer through inhibition of phosphorylation-dependent ubiquitination and degradation. Oncogene 33 1438–1447. 10.1038/onc.2013.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy V., Mishra R., Sondarva G., Das S., Lee T. H. J., Bakowska C., et al. (2012). Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proc. Natl. Acad. Sci. U.S.A. 109 8149–8154. 10.1073/pnas.1200804109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustighi A., Tiberi L., Soldano A., Napoli M., Nuciforo P., Rosato A., et al. (2009). The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat. Cell Biol. 11 133–142. 10.1038/ncb1822 [DOI] [PubMed] [Google Scholar]
- Rustighi A., Zannini A., Tiberi L., Sommaggio R., Piazza S., Sorrentino G., et al. (2014). Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol. Med. 6 99–119. 10.1002/emmm.201302909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A., Hirai A., Nishi M., Liou Y.-C., Perrem K., Lin S.-C., et al. (2007). A suppressive role of the Prolyl isomerase pin1 in cellular apoptosis mediated by the death-associated protein Daxx. J. Biol. Chem. 282 36671–36681. 10.1074/jbc.m704145200 [DOI] [PubMed] [Google Scholar]
- Ryo A., Liou Y.-C., Wulf G., Nakamura M., Lee S. W., Lu K. P. (2002). PIN1 Is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22 5281–5295. 10.1128/MCB.22.15.5281-5295.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A., Nakamura M., Wulf G., Liou Y.-C., Lu K. P. (2001). Pin1 regulates turnover and subcellular localization of β-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3 793–801. 10.1038/ncb0901-793 [DOI] [PubMed] [Google Scholar]
- Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y.-C., Wulf G., et al. (2003). Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12 1413–1426. 10.1016/S1097-2765(03)00490-8 [DOI] [PubMed] [Google Scholar]
- Ryo A., Uemura H., Ishiguro H., Saitoh T., Yamaguchi A., Perrem K. (2005). Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin. Cancer Res. 11 7523–7531. 10.1158/1078-0432.ccr-05-0457 [DOI] [PubMed] [Google Scholar]
- Ryo A., Wulf G., Lee T. H., Lu K. P. (2009). Pinning down HER2-ER crosstalk in SMRT regulation. Trends Biochem. Sci. 34 162–165. 10.1016/j.tibs.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Saegusa M., Hashimura M., Kuwata T. (2010). Pin1 acts as a modulator of cell proliferation through alteration in NF-κB but not β-catenin/TCF4 signalling in a subset of endometrial carcinoma cells. J. Pathol. 222 410–420. 10.1002/path.2773 [DOI] [PubMed] [Google Scholar]
- Segat L., Pontillo A., Annoni G., Trabattoni D., Vergani C., Clerici M., et al. (2007). PIN1 promoter polymorphisms are associated with Alzheimer’s disease. Neurobiol. Aging 28 69–74. 10.1016/j.neurobiolaging.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Shen Z.-J., Esnault S., Schinzel A., Borner C., Malter J. S. (2009). The peptidyl-prolyl isomerase Pin1 facilitates cytokine-induced survival of eosinophils by suppressing Bax activation. Nat. Immunol. 10 257–265. 10.1038/ni.1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Chen L., Ji J., Cai Q., Yu Y., Liu B., et al. (2015). Pin1 is overexpressed and correlates with poor prognosis in gastric cancer. Cell Biochem. Biophys. 71 857–864. 10.1007/s12013-014-0274-0 [DOI] [PubMed] [Google Scholar]
- Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., et al. (2009). Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138 592–603. 10.1016/j.cell.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. R., Bae H. S., Kim B. S., Yoon H. I., Cho Y. D., Kim W. J., et al. (2018). PIN1 is a new therapeutic target of craniosynostosis. Hum. Mol. Genet. 27 3827–3839. 10.1093/hmg/ddy252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K., Kuboki S., Shimizu H., Ohtsuka M., Kato A., Yoshitomi H., et al. (2015). Pin1 facilitates NF-κB activation and promotes tumour progression in human hepatocellular carcinoma. Br. J. Cancer 113 1323–1331. 10.1038/bjc.2015.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanya K. J., Liu Y., Means A. R., Kao H.-Y. (2008). Cdk2 and Pin1 negatively regulate the transcriptional corepressor SMRT. J. Cell Biol. 183 49–61. 10.1083/jcb.200806172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suizu F., Ryo A., Wulf G., Lim J., Lu K. P. (2006). Pin1 regulates centrosome duplication, and its overexpression induces centrosome amplification, chromosome instability, and oncogenesis. Mol. Cell. Biol. 26 1463–1479. 10.1128/MCB.26.4.1463-1479.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Zhou F., Wan J., Hang J., Chen Z., Li B., et al. (2010). Pin1 expression contributes to lung cancer: prognosis and carcinogenesis. Cancer Biol. Ther. 9 111–119. 10.4161/cbt.9.2.10341 [DOI] [PubMed] [Google Scholar]
- Thomas A. L., Lind H., Hong A., Dokic D., Oppat K., Rosenthal E., et al. (2017). Inhibition of CDK-mediated Smad3 phosphorylation reduces the Pin1-Smad3 interaction and aggressiveness of triple negative breast cancer cells. Cell Cycle 16 1453–1464. 10.1080/15384101.2017.1338988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T., Takamiya M., Takahashi M., Miyashita H., Ikeda H., Terada T., et al. (2003). Pin1 and Par14 Peptidyl prolyl isomerase inhibitors block cell proliferation. Chem. Biol. 10 15–24. 10.1016/S1074-5521(02)00310-1 [DOI] [PubMed] [Google Scholar]
- Ueberham U., Rohn S., Ueberham E., Wodischeck S., Hilbrich I., Holzer M., et al. (2014). Pin1 promotes degradation of Smad proteins and their interaction with phosphorylated tau in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 40 815–832. 10.1111/nan.12163 [DOI] [PubMed] [Google Scholar]
- Urusova D. V., Shim J.-H., Kim D. J., Jung S. K., Zykova T. A., Carper A., et al. (2011). Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1. Cancer Prev. Res. 4 1366–1377. 10.1158/1940-6207.CAPR-11-0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu K., Wang X. F., Sun D. J. (2017). Juglone reduces growth and migration of U251glioblastoma cells and disrupts angiogenesis. Oncol. Rep. 38 1959–1966. 10.3892/or.2017.5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ray P. S., Sim M. S., Zhou X. Z., Lu K. P., Lee A. V., et al. (2012). FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-κB signaling. Oncogene 31 4798–4802. 10.1038/onc.2011.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang N., Han Q., Lu W., Wang L., Yang D., et al. (2019). Pin1 inhibition reverses the acquired resistance of human hepatocellular carcinoma cells to Regorafenib via the Gli1/Snail/E-cadherin pathway. Cancer Lett. 444 82–93. 10.1016/j.canlet.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Liu Z., Shi F., Wang J. (2016). Pin1 modulates chemo-resistance by up-regulating FoxM1 and the involvements of Wnt/β-catenin signaling pathway in cervical cancer. Mol. Cell. Biochem. 413 179–187. 10.1007/s11010-015-2651-4 [DOI] [PubMed] [Google Scholar]
- Wang X. J., Xu B., Mullins A. B., Neiler F. K., Etzkorn F. A. (2004). Conformationally locked Isostere of PhosphoSer-cis-Pro inhibits Pin1 23-fold better than PhosphoSer-trans-Pro isostere. J. Am. Chem. Soc. 126 15533–15542. 10.1021/ja046396m [DOI] [PubMed] [Google Scholar]
- Wang Z., Banerjee S., Kong D., Li Y., Sarkar F. H. (2007). Down-regulation of Forkhead box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 67 8293–8300. 10.1158/0008-5472.can-07-1265 [DOI] [PubMed] [Google Scholar]
- Wei S., Kozono S., Kats L., Nechama M., Li W., Guarnerio J., et al. (2015). Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat. Med. 21 457–466. 10.1038/nm.3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildemann D., Erdmann F., Alvarez B. H., Stoller G., Zhou X. Z., Fanghänel J., et al. (2006). Nanomolar inhibitors of the peptidyl prolyl Cis/Trans isomerase Pin1 from combinatorial peptide libraries. J. Med. Chem. 49 2147–2150. 10.1021/jm060036n [DOI] [PubMed] [Google Scholar]
- Wu K.-J., Zhong H.-J., Yang G., Wu C., Huang J.-M., Li G., et al. (2018). Small molecule Pin1 inhibitor blocking NF-κB signaling in prostate cancer cells. Chem. Asian J. 13 275–279. 10.1002/asia.201701216 [DOI] [PubMed] [Google Scholar]
- Wulf G. M., Ryo A., Wulf G. G., Lee S. W., Niu T., Petkova V., et al. (2001). Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20 3459–3472. 10.1093/emboj/20.13.3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. G., Etzkorn F. A. (2009). Pin1 as an anticancer drug target. Drug News Perspect. 22 399–407. 10.1358/dnp.2009.22.7.1381751 [DOI] [PubMed] [Google Scholar]
- Xu M., Cheung C. C., Chow C., Lun S. W., Cheung S. T., Lo K. W. (2016). Overexpression of PIN1 enhances cancer growth and aggressiveness with cyclin D1 induction in EBV-associated nasopharyngeal carcinoma. PLoS One 11:e0156833. 10.1371/journal.pone.0156833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang B., Gao J., Wang X., Liu Z. (2013). Regulation of the microRNA 200b (miRNA-200b) by transcriptional regulators PEA3 and ELK-1 protein affects expression of Pin1 protein to control anoikis. J. Biol. Chem. 288 32742–32752. 10.1074/jbc.M113.478016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Daum S., Wildemann D., Zhou X. Z., Verdecia M. A., Bowman M. E., et al. (2007). Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem. Biol. 2 320–328. 10.1021/cb7000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yu W., Zheng M., Liao X., Wang J., Yang D., et al. (2019). Pin1 inhibition potently suppresses gastric cancer growth and blocks PI3K/AKT and Wnt/β-catenin oncogenic pathways. Mol. Carcinog. 58 1450–1464. 10.1002/mc.23027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Cui G., Jin J., Chen X., Xu B. (2016). Synthesis and Pin1 inhibitory activity of thiazole derivatives. Bioorg. Med. Chem. 24 5911–5920. 10.1016/j.bmc.2016.09.049 [DOI] [PubMed] [Google Scholar]
- Zheng M., Xu H., Liao X.-H., Chen C. P., Zhang A. L., Lu W., et al. (2017). Inhibition of the prolyl isomerase Pin1 enhances the ability of sorafenib to induce cell death and inhibit tumor growth in hepatocellular carcinoma. Oncotarget 8 29771–29784. 10.18632/oncotarget.15967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Pu W., Li J., Shen X., Zhou Q., Fan X., et al. (2019). Discovery of a prenylated flavonol derivative as a Pin1 Inhibitor to suppress hepatocellular carcinoma by modulating microRNA biogenesis. Chem. Asian J. 14 130–134. 10.1002/asia.201801461 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Xia Y., Hawke D., Halle M., Tremblay M. L., Gao X., et al. (2009). FAK phosphorylation by ERK primes Ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Mol. Cell 35 11–25. 10.1016/j.molcel.2009.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Yang W., Xia Y., Hawke D., Liu D. X., Lu Z. (2011). Ras-induced and extracellular signal-regulated kinase 1 and 2 phosphorylation-dependent isomerization of protein tyrosine phosphatase (PTP)-PEST by PIN1 promotes FAK dephosphorylation by PTP-PEST. Mol. Cell. Biol. 31 4258–4269. 10.1128/MCB.05547-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Park B. H., Park J. H., Jang K. Y., Park H. S., Wagle S., et al. (2013). Overexpression of the prolyl isomerase PIN1 promotes cell growth in osteosarcoma cells. Oncol. Rep. 29 193–198. 10.3892/or.2012.2112 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Zhang H., Lang F., Liu G., Gao D., Li B., et al. (2016). Pin1 promotes prostate cancer cell proliferation and migration through activation of Wnt/β-catenin signaling. Clin. Transl. Oncol. 18 792–797. 10.1007/s12094-015-1431-7 [DOI] [PubMed] [Google Scholar]