Abstract
Ocimum is one of the most revered medicinally useful plants which have various species. Each of the species is distinct in terms of metabolite composition as well as the medicinal property. Some basil types are used more often as an aromatic and flavoring ingredient. It would be informative to know relatedness among the species which though belong to the same genera while exclusively different in terms of metabolic composition and the operating pathways. In the present investigation the similar effort has been made in order to differentiate three commonly occurring Ocimum species having the high medicinal value, these are Ocimum sanctum, O. gratissimum and O. kilimandscharicum. The parameters for the comparative analysis of these three Ocimum species comprised of temporal changes in number leaf trichomes, essential oil composition, phenylpropanoid pathway genes expression and the activity of important enzymes. O. gratissimum was found to be richest in phenylpropanoid accumulation as well as their gene expression when compared to O. sanctum while O. kilimandscharicum was found to be accumulating terpenoid. In order to get an overview of this qualitative and quantitative regulation of terpenes and phenylpropenes, the expression pattern of some important transcription factors involved in secondary metabolism were also studied.
Subject terms: Plant molecular biology, Secondary metabolism
Introduction
Ocimum is one of the highly important medicinal plants used worldwide in the traditional medicinal systems. It is commonly known as ‘basil’ but is popular as ‘Tulsi’ in India. Not only in medicine, basil also has religious importance1,2. Ocimum belongs to family Lamiaceae and subfamily Nepetoideae which contains strongly fragrant plants marked by the presence of essential oils comprising of sesquiterpenes, monoterpenes and phenylpropanoids. Eugenol, methylchavicol and linalool are the major compounds found in most basil species, however, the composition of every chemical constituent differ in different species or varieties1. Another feature in the plants of Lamiaceae family is the presence of oil glands called glandular trichomes on the leaf surface. Mainly there are two types of oil glands3 that is, peltate and capitate. The glands are differentiated by the number and size of head cells occurring on the basil leaf surface, and the phenylpropenes are exclusively synthesized in the peltate glands4. Surplus glandular trichomes in plants and their association with essential oil biosynthesis in numerous plants including peppermint5–14, lima bean15–18, and tomato19–21 have been talked about, which has improved our comprehension of isoprenoid production in plants. The general phenylpropanoid pathway produces a wide range of secondary metabolites originating from the shikimate pathway intermediates. However, the role and impact of this gene family need to be established22. Phenylpropanoids contribute to both abiotic and biotic plant stress responses. Apart being the indicators of stress, they also help in acquiring resistance against pests23. They support the incursion of fresh habitation24 and help in providing the biochemical sources for successful reproduction25. Phenylpropenes form a collection of small phenolic molecules which result from the general phenylpropanoid biosynthetic pathway, and are the main flavoring factors in a lot of spices and herbs, like- nutmeg, cloves, cinnamon, pimento, allspice, tarragon and basil. Besides this, phenylpropenes (like eugenol, methyleugenol etc.) are the key constituents of the protective mechanism in plants and even help in attracting the pollinators for pollination9. One of the most important reservoirs of phenylpropenes is the ‘Holy basil’ whose whole genome has recently been sequenced. Thus, an effort to study the temporal expression of phenylpropanoid pathway genes in O. sanctum (OS), the study was designed so as to compare the expression pattern of another phenylpropene rich species, O. gratissimum (OG) and a terpene producing species, O. kilimandscharicum (OK). The differences in oil composition, trichome number and also the expression of the genes governing the phenylpropanoid biosynthesis as well as the expression pattern of transcription factors involved in secondary metabolism were studied in these three Ocimum species. Since the synthesis of secondary metabolites as well as the transcripts of respective biosynthesis pathway genes is tightly regulated at various levels26, therefore, transcription factors governing the process of transcription have also been investigated. Characteristics of the three species are discussed here. O. sanctum (tenuiflorum), is a grand sacred medicinal plant of India, commonly known as holy basil or Tulsi. Basically there are two types of varieties (i) Green leaves tulsi plant- Sri/Rama Tulsi and (ii) Purple leaves tulsi plant- Krishna/ Shyama Tulsi27. Traditional medical practitioners widely use O. sanctum to treat various ailments like bronchial asthma, bronchitis, dysentery, malaria, arthritis, diarrhea, chronic fever, skin diseases, insect bite, etc. In addition, it acquires antifertility, antidiabetic, anticancer, antimicrobial, antifungal, cardioprotective, hepatoprotective, antiemetic, analgesic, antispasmodic, diaphoretic and adaptogenic activities28–30. Holy basil possesses a highly-flavored and a bit pungent savor in comparison to other basils because of a phenylpropanoid, eugenol and a sesquiterpenoid, beta caryophyllene present in the leaf essential oil. The application of essential oil has demonstrated the reduction in inflammation, joint pains as well as body rashes29. O. gratissimum (wild basil, tree basil or clove basil) is a perennial herb, aromatic in nature and found to occur in tropical regions, like- India and West Africa31. It is popular in India as Vana tulsi (Hindi) or Vriddhutulsi (Sanskrit)32. Though pungent in taste, it is helpful in treating brain, liver, heart and spleen related ailments, as well as strengthen the gums and eliminates foul breath smell33. It has also been demonstrated to acquire diaphoretic, laxative, antidiarrhoeal, antipyretic, anti-inflammatory, immunostimulatory, cardiovascular, hepatoprotective, antidiabetic, wound healing, antihypertensive, analgesic and anticonvulsant effects29,34. O. gratissimum contain essential oil with a high percentage of eugenol. A variation is found in the chemical composition of the oil and 6 chemotypes based on oil composition variation have been reported. Chief components present in the O. gratissimum essential oils are- eugenol, citral, thymol, geraniol, ethyl cinnamate and linalool35. Essential oil finds application in food, flavor, beverages, dental preparations and detergents etc29. O. kilimandscharicum Guerke, known as ‘Kapoori Tulsi’ (Hindi) or ‘Camphor Basil’ (English) is a foreign west African species native of East Africa which was brought to India and Turkey. This plant attracted attention as a source of camphor36. This species has a strong but less pleasant flavor. The plant has carminative, stimulant, antipyretic, anti-fungal and anti-bacterial properties. Conventionally, in East Africa, plant extracts of O. kilimandscharicum were employed to treat many diseases like cold, cough, diarrhea, measles, abdominal pains as well as it had inhibitory action against mosquitoes and pest37. The leaves of the plant are aromatic, antiviral, thermogenic, acrid, anti-bacterial, appetizing, insecticidal, and ophthalmic36. The color of essential oil is light yellow and owes a strong camphory odour, used widely in perfumery, flavor and pharmaceutical industries.
Temporal variation in the essential oil yield has been attributed to the environmental factors; however, till now there has been no investigation conducted to observe the status of essential oil pathway gene expression and/ or the expression of the regulatory elements, like- the transcription factors. Additionally, the differences among the species of Ocimum have only been established on the basis of essential oil composition and constituents. Therefore, this study was planned to give a comparative overview of the three Ocimum species based on the effect on essential oil yield with time as well as the differences in biochemical constituents like- chlorophyll, phenolics, anthocyanin and activity of some phenylpropanoid pathway enzymes viz. PAL (phenylalanine ammonia lyase), C4H (cinnamic acid 4-hydroxylase), 4CL (4-coumarate CoA ligase) and CAD (cinnamyl alcohol dehydrogenase). Simultaneously, the essential oil profiling and trichome density measurement was also carried out every month to see the effect of time over them.
Results
Chemoprofiling of essential oil
Aiming at the investigation of the seasonal variation, the leaves of O. sanctum, O. gratissimum and O. kilimandscharicum were collected in triplicates throughout one year in the months of July, August, September, October, November, and December 2018. The yields of the leaf essential oils varied between 0.29 and 0.87% (w/ w) for O. sanctum, 0.11 to 1.3% (w/w) for O. gratissimum and between 0.1 and 0.73% (w/w) for O. kilimandscharicum (Table 1). Total compounds which are represented in metabolic profile were those having peak area percent greater than and equal to 0.01 were considered for metabolic profiling analysis (39 compounds from O. sanctum, 47 from O. gratissimum and 22 from O. kilimandscharicum). As evident from Fig. 1, abundant essential oil constituents (d-camphor, D-limonene, camphene, and caryophyllene) in O. kilimandscharicum were found to be present in higher amounts from July to October and thereafter the contents of these constituents started decreasing. Similar pattern of essential oil constituents (eugenol, β-elemene, caryophyllene and methyleugenol) was also observed in the oil profiling from the leaves of O. sanctum. However, no major effect of time and environmental factors was observed in O. gratissimum essential oil constituent profiling (eugenol, β-ocimene, germacrene-D and caryophyllene) (Fig. 1). Eugenol was identified to be present in the highest concentration in the leaf essential oils of both O. sanctum and O. gratissimum while camphor constituted the major proportion of the O. kilimandscharicum oil. As per the oil profiling (Supplementary Table 3) of the Ocimum varieties taken in the present study O. sanctum and O. gratissimum were found to be rich in phenylpropenes while O. kilimandscharicum was rich in terpenes. The essential oil constituents were categorized under five major class of compounds namely, phenylpropenes, monoterpenes, monoterpene alcohols, sesquiterpenes and sesquiterpenoid alcohols. The array of essential oil constituents was highest in O. gratissimum followed by O. sanctum and was lowest in O. kilimandscharicum (Supplementary Table 3).
Table 1.
Essential oil yield of the three Ocimum species for six months.
| Months | O. sanctum (%) | O. gratissimum (%) | O. kilimandscharicum (%) |
|---|---|---|---|
| July | 0.41 ± 0.11 | 0.64 ± 0.08 | 0.48 ± 0.14 |
| August | 0.35 ± 0.05 | 0.83 ± 0.08 | 0.73 ± 0.17 |
| September | 0.68 ± 0.11 | 1.12 ± 0.11 | 0.59 ± 0.06 |
| October | 0.87 ± 0.03 | 1.30 ± 0.40 | 0.42 ± 0.07 |
| November | 0.47 ± 0.04 | 0.35 ± 0.10 | 0.26 ± 0.07 |
| December | 0.29 ± 0.07 | 0.11 ± 0.01 | 0.10 ± 0.02 |
Data are means ± SD (at least three replicates).
Figure 1.
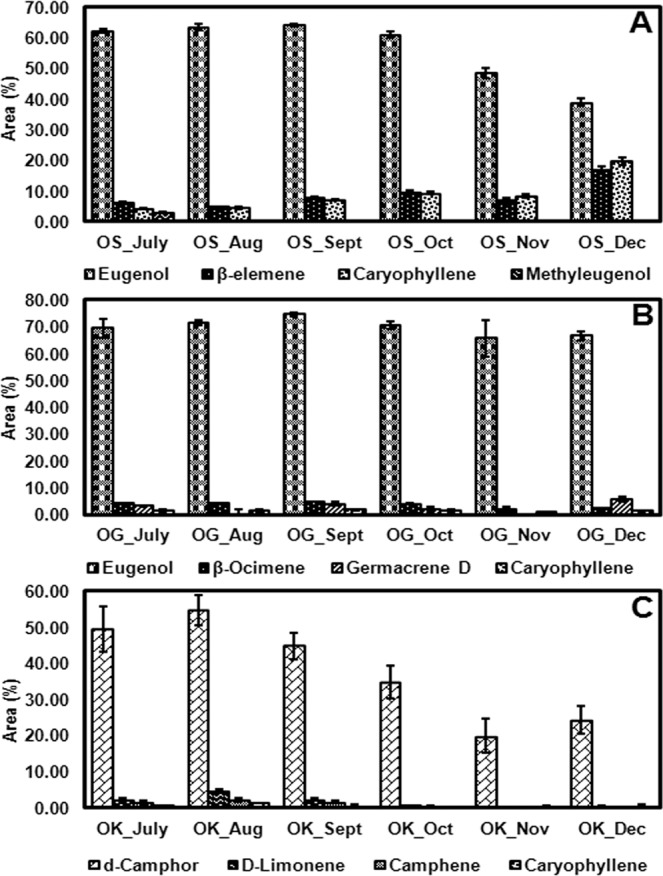
Abundant essential oil constituents from six months oil profiling of (A) Ocimum sanctum (OS) (B) Ocimum gratissimum (OG) and, (C) Ocimum kilimandscharicum (OK). Data are given as means ± SD (at least three independent replicates).
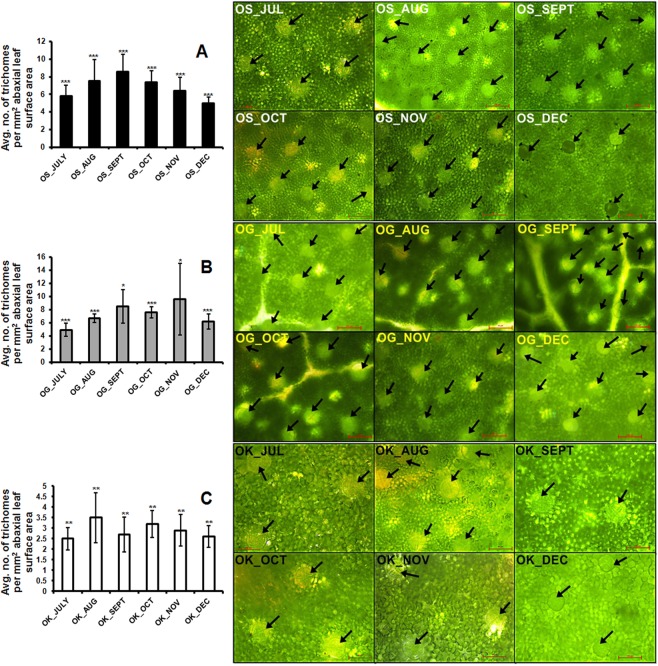
Glandular trichome density
Trichome density in terms of average number of trichomes per mm2 leaf area were recorded from the abaxial surface of leaves in all the six months from July to December in a population size of 10 plants each month for O. sanctum, O. gratissimum as well as O. kilimandscharicum (Fig. 2). Highest effect of variation was found to be observed in O. sanctum species where the trichome density gradually increased from July to September and decreased thereafter onwards October. In O. gratissimum also the trichome density increased from July to September but fluctuated in October and November and finally got decreased in the month of December. However O. kilimandscharicum was found to be having least temporal variation on its trichome densities. Trichome densities were observed to be highest during the months of September, November, and August in O. sanctum, O. gratissimum and O. kilimandscharicum, respectively. O. gratissimum and O. sanctum were found to be having more number of peltate glandular trichomes in comparison to O. kilimandscharicum.
Figure 2.
Trichome densities and their representative microscopic images for six months in three panels for (A) Ocimum sanctum (OS) (B) Ocimum gratissimum (OG) and, (C) Ocimum kilimandscharicum (OK). Statistical analysis was performed using HSD tukey test, standard weighted-means analysis at *P < 0.01, **P < 0.05 and ***P < 0.1 levels of significance. Arrows in the figure are pointing the glandular trichomes on the abaxial leaf surface; Scale bar = 100 µm. Error bars represent ± SD (at least ten independent plants).
Phenylpropanoid pathway gene expression studies
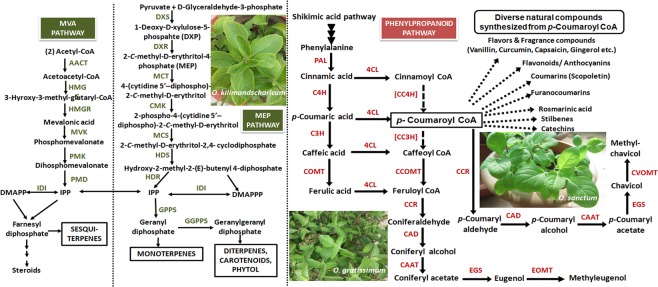
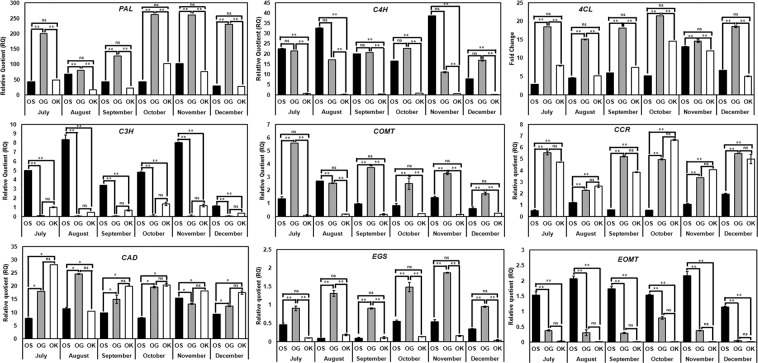
Quantitative real-time PCR (qRT–PCR) analysis was performed to assess the transcript levels of nine important phenylpropanoid pathway genes in glandular trichomes at different developmental stages collected throughout one year in the months of July, August, September, October, November, and December 2018. The nine genes taken in the present study are- phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4- coumarate CoA ligase (4CL), p-coumarate 3-hydroxylase (C3H), caffeic acid O- methyltransferase (COMT), cinnamoyl CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), eugenol synthase (EGS), and the eugenol O- methyltransferase (EOMT). Analysis was performed by designing the primers from the genes reported in the NCBI database as well as from transcriptome and the whole genome sequencing data of O. sanctum reported recently38–40 (Supplementary Table 1). Figure 3 shows the detailed pathway leading to biosynthesis of terpenes and phenylpropenes in the glandular trichomes of the three Ocimum species as earlier discussed by Iijima et al.41 The results reveal that overall there is a higher expression of phenylpropanoid pathway genes in O. gratissimum as compared to O. sanctum and O. kilimandscharicum. In the succession of gene expression, the highest expression in terms of relative quotient (RQ) was observed in PAL followed by CAD, C4H, 4CL, C3H, CCR, COMT, EOMT and EGS (Fig. 4).
Figure 3.
Pathways leading to the generation of terpenes and phenylpropenes in Ocimum sp. giving an overview of the MVA (mevalonate) and MEP (2-C-methyl-D-erythritol-4-phosphate) pathways localized in the cytosol and the plastids, respectively. DXS, DXP synthase; DXR, DXP reductoisomerase; MCT, 2-C-methyl-D-erythritol-4-(cytidyl-5-diphosphate) transferase; CMK, CDP-ME kinase; MCS, CMEPP synthase; HDS, HMBPP synthase; HDR, hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; IDI, IPP isomerase; AACT (acetoacetyl-CoA thiolase), HMGS (HMG synthase), HMGR (HMG reductase), MVK, MVA kinase; PMK, phosphome-valonate kinase; PMD, MVA diphosphate decarboxylase; and GGPPS, GGPP synthase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; C3H, p-coumarate 3-hydroxylase; COMT, caffeoyl O-methyl transferase; CC4H, cinnamoyl-CoA 4-hydroxylase; CC3H, p-coumaroyl- CoA 3-hydroxylase; CCOMT, caffeoyl-CoA O-methyl transferase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; CAAT, coniferyl alcohol acetyl transferase; EGS, eugenol synthase; EOMT, eugenol O-methyl transferase and CVOMT, chavicol O-methyl transferase. The enzymes for 4- and 3-hydroxylation of CoA esters have not been demonstrated yet and hence [CC4H] and [CC3H] are indicated in brackets to indicate their probable role (Gang et al.56) [adapted from Iijima et al.41 and Rastogi et al.38].
Figure 4.
Relative expression levels of PAL, C4H, 4CL, C3H, COMT, CCR, CAD, EGS and EOMT transcripts in O. sanctum (OS), O. gratissimum (OG) and O. kilimandscharicum (OK) during six months starting from July’ 2018 till December’ 2018 determined by Real-time RT-PCR. Data are means ± SD (for at least three replicates), and the y-axis represents the relative quotient (RQ). Statistical analysis was performed using HSD tukey test, standard weighted-means analysis at P < 0.01* and P < 0.05** levels of significance while ‘ns’ means non-significant.
Expression of PAL gene transcripts was found to be highest in the months of October and November in all the three Ocimum species. In O. sanctum, the expression was PAL transcripts was highest in the month of November while in O. kilimandscharicum it was highest in October and in O. gratissimum the expression was nearly equal in October and November. In the month of December, the transcripts of PAL specifically decreased in O. sanctum and O. kilimandscharicum while there was not much decrease in the O. gratissimum’s PAL gene expression. The expression of C4H gene showed quite a uniform pattern from July to December. C4H gene was found to be expressed highest in O. sanctum in the month of November. The expression of C4H in O. gratissimum was comparable with that of its expression in O. sanctum and was highest in the month of October. The expression pattern of C4H transcripts in all the three species of Ocimum was more or less consistent. The 4CL transcript expression was found to be highest in the months of October and November. In the present study, highest 4CL transcript expressing species was O. gratissimum in the month of October. In O. sanctum as well as the O. gratissimum expression pattern of 4CL gradually increased from July to November, but in case of O. sanctum got decreased in December with not much effect in O. gratissimum. The expression of C3H transcript was found to be throughout from the month of July upto November significantly dropped down in December in all the Ocimum species in the present study. O. sanctum showed the highest expression of C3H transcript followed by very low expression O. kilimandscharicum and relatively very low in O. gratissimum. C3H transcript was among the lowest expressing one in O. gratissimum which had maximum high expressing phenylpropanoid gene transcripts. The expression pattern of C3H transcripts in O. sanctum and O. kilimandscharicum was analogous to C4H with an exception of O. gratissimum. O. gratissimum showed the highest expression of COMT gene transcript. The expression was by far highest during the month of July and fairly in August, September, October and November, but gradually decreases during December. Expression pattern of COMT transcript in O. sanctum was also comparable to O. gratissimum while O. kilimandscharicum showed very low expression of transcript. The expression of CCR gene transcript was found to be highest in O. kilimandscharicum which was comparable to O. gratissimum and was least in O. sanctum. CCR expression first increase upto October in O. kilimandscharicum and decreased thereupon, but the decrease was not very significant. However, the expression pattern of CCR transcript in O. gratissimum was more or less constant while showed an increasing pattern in case of O. sanctum. The CAD gene transcript expression was almost similar and uniform in all the three Ocimum species throughout all the six months with an exceptionally high expression observed in O. gratissimum during the month of July. O. gratissimum showed the highest expression of the EGS gene transcript with a gradual increase of expression from July and upto highest in the month of November, thereafter decreasing in December. O. sanctum followed O. gratissimum in EGS transcript expression but in the similar temporal series. O. kilimandscharicum was found to be having very less expression of EGS gene transcript as compared to the other two Ocimum species. The expression of EOMT gene transcript was exceptionally high in O. sanctum, found to be increasing from the month July to November. The highest expression of the transcript was found to be observed during the month of November and thereafter decreases, onwards December. The other two Ocimum species showed the expression of EOMT transcript in very trace levels.
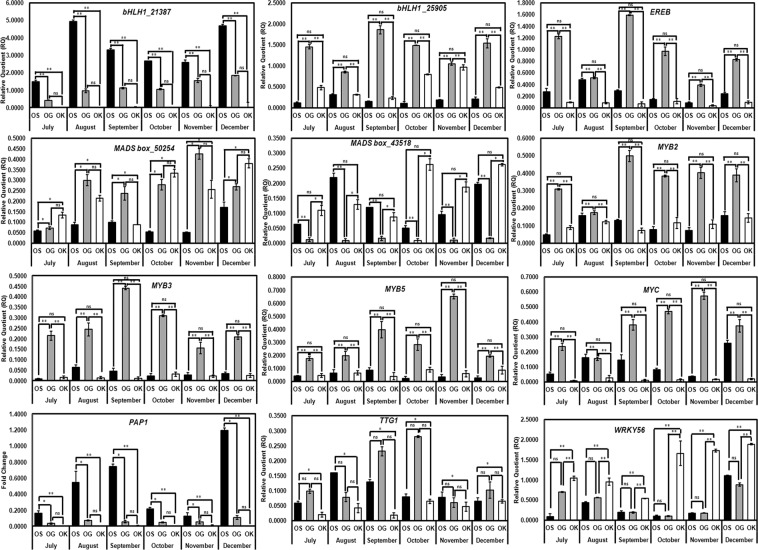
Expression profiling of secondary metabolism related transcription factors (TFs)
Expression pattern of transcription factors pertaining to the secondary metabolism were also studied in the present investigation. Based on the equivalent transcript abundance of digital gene expression data of prior transcriptome analysis39 comparing O. sanctum and O. basilicum, some of the TFs involved in the secondary metabolism were sorted for present analysis. The TFs selected for the expression studies were- bHLH1, EREB, MADS box, MYB2, MYB3, MYB5, MYC, PAP1, and TTG1. Analysis was carried out using the primers designed from the comparative transcriptome sequencing data of O. sanctum and O. basilicum reported recently by Rastogi et al.39 (Supplementary Table 2). Only primer sequence of TF WRKY was designed from the O. basilicum ‘TrichOME Database’ (http://www.planttrichome.org/trichomedb/estbyspecies_detail.jsp?species=Ocimum%20basilicum) (Supplementary Table 2) because the annotations were lacking WRKY which is considered to be an important TF involved in regulation of secondary metabolism pathways42. In case of bHLH1 and MADS BOX TFs the two- two transcripts were selected as these transcript sequences were non-overlapping and were showing equivalent transcript abundance in digital gene expression data of O. sanctum and O. basilicum leaf transcriptomes. As evident from the Fig. 5, out of the 12 TFs selected in the present study, the maximum number of TFs i.e., 8 was expressed higher in O. gratissimum followed by O. sanctum and O. kilimandscharicum. Now considering the expression pattern of each TF it was observed that two non- overlapping transcripts of bHLH1 (bHLH1_21387 and bHLH1_25905) showed an entirely different pattern of expression with respect to each other. Expression of bHLH1_21387 was found to be highest in O. sanctum but there was no temporal pattern of expression. Expression of bHLH1_21387 in O. sanctum increased from July to August, gradually decreased in September with nearly no change in expression during October and November and a sudden increase in the month of December. The expression of bHLH1_21387 in O. sanctum during the December month was comparable to its expression in August. On the contrary, expression of bHLH1_25905 was highest in O. gratissimum but a specific temporal pattern of expression was also not observed, while in case of O. kilimandscharicum there was a gradual increase in bHLH1_25905 transcript expression from the month of July till November and got decreased December onwards. O. sanctum showed the lowest expression of bHLH1_25905 transcript and also not much of significant difference was seen in the expression pattern in all the six months. Observing the expression pattern of EREB transcript it was found to be expressing highest in O. gratissimum during the month of September and decreased thereafter. In case of O. sanctum EREB expression increased from July to August which also got decreased upon maturity, while in O. kilimandscharicum there was a uniform expression pattern of this transcript throughout all the six months. Considering the expression pattern of another non-overlapping TF transcripts MADS box_50254 and MADS box_43518 contrasting results were obtained. In case of MADS box_50254 transcript, expression was highest in O. gratissimum which gradually increased from July upto highest in November and decreased in December. Antagonistically, expression of MADS box_50254 got increased from July upto the month of December in both O. kilimandscharicum and O. sanctum. Interestingly, MADS box_43518 transcript showed the similar expression pattern as of MADS box_50254 in case of O. kilimandscharicum and O. sanctum, while a uniform expression pattern but very low transcript abundance of MADS box_43518 was observed in O. gratissimum. Analyzing the expression pattern of the three transcripts of MYB TF family members, MYB2, MYB3 and MYB5, it was observed that expression of all the three MYB transcripts was highest in O. gratissimum and more or less equivalent pattern in O. kilimandscharicum and O. sanctum. Expression of MYB3 and MYB5 transcripts in O. gratissimum increased from young stage (July) and then decreased upon maturity (December) while MYB2 expression increased from July to September and was nearly constant in the successive months upto December. However, the expression pattern of the three MYB family TFs in O. kilimandscharicum and O. sanctum was nearly temporally unaffected. Next, the MYC transcript expression was also found to be highest in O. gratissimum which increased from July upto November and got reduced in December, while its expression increased in case of O. sanctum from July to December and was least as well as unaffected in O. kilimandscharicum. Expression study data of PAP1 TF transcript reveals O. sanctum to be highest PAP1 expressing Ocimum species among the three whose expression increases as the plant matures and found to be highest in during the month of December. PAP1 expression in other two Ocimum species, O. gratissimum and O. kilimandscharicum was very low as compared to O. sanctum and also no noticeable temporal effect was observed in PAP1 expression pattern as there laid almost evenly distributed transcript abundance throughout the six months. A canonical temporal expression pattern of TTG1 TF transcript was observed in case of O. gratissimum and O. sanctum while in case of O. kilimandscharicum the expression increased from July to October and thereafter remained constant in November and December. The order of expression of TTG1 was highest in O. gratissimum followed by O. sanctum and then O. kilimandscharicum. The last studied TF for expression study was WRKY56 which demonstrated the same expression pattern in all the three Ocimum species i.e., the expression increases as the plant ages. The highest expression was observed in O. kilimandscharicum followed by the other two, O. gratissimum and O. sanctum.
Figure 5.
Relative expression levels of bHLH_21387, bHLH_25905, EREB, MADS box_50254, MADS box_43518, MYB2, MYB3, MYB5, MYC, PAP1, TTG1 and WRKY56 transcription factors in O. sanctum (OS), O. gratissimum (OG) and O. kilimandscharicum (OK) during six months starting from July’ 2018 till December’ 2018 determined by Real-time RT-PCR. Data are means ± SD (for at least three replicates), and the y-axis represents the relative quotient (RQ). Statistical analysis was performed using HSD tukey test, standard weighted-means analysis at *P < 0.01 and **P < 0.05 levels of significance while ‘ns’ means non-significant.
Enzyme activity of some important phenylpropanoid pathway enzymes
Crude protein extracts obtained from young leaves of the mature plant were assayed for activity for the key phenylpropanoid pathway enzymes in leading to the phenylpropenes, as well as for an intermediate enzyme that might also be involved in phenylpropenes biosynthesis. These enzymes included phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate: CoA ligase (4CL), and cinnamyl alcohol dehydrogenase (CAD). Figure 6 shows the activity of the enzymes per mg protein. The order of activity for the four enzymes in the three Ocimum species was highest in O. gratissimum followed by O. sanctum and least in O. kilimandscharicum.
Figure 6.
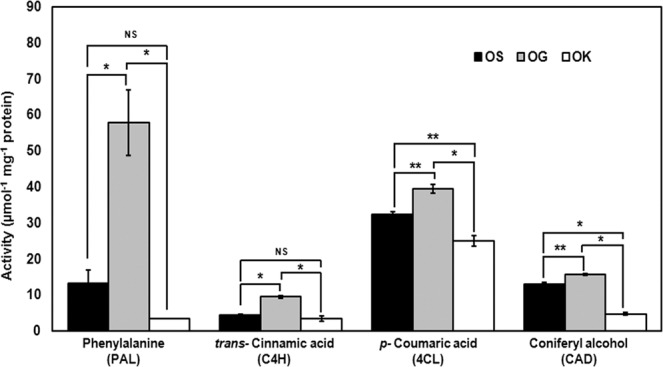
Enzyme activity of PAL, C4H, CAD quantified in terms of product formed and of 4CL in terms of substrate utilized represented per mg of the total plant protein from O. sanctum (OS), O. gratissimum (OG) and O. kilimandscharicum (OK) at maturity. L-Phenylalanine, trans- cinnamic acid, p- coumaric acid and coniferyl alcohol were used as the substrates for PAL, C4H, 4CL and CAD, respectively. Activities are given as means of two independent assays ± SD (standard deviation). Statistical analysis was performed using HSD tukey test, standard weighted-means analysis at *P < 0.01 and **P < 0.05 levels of significance while ‘ns’ means non-significant.
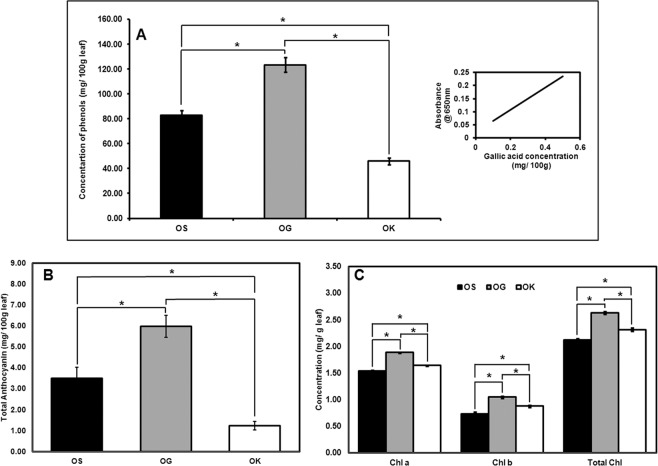
Total phenolic content, anthocyanin content and chlorophyll content
Total phenolics, anthocyanin and chlorophyll contents were estimated from the leaves of the mature plant of all the three Ocimum species in order to know their variability in terms of these three constituents in order to get some correlation of the these constituents composition with that of the gene expression profiling of phenylpropanoid pathway as well as transcription factors.
The phenolics and the pigment analysis results as shown in the Fig. 7 proved O. gratissimum to be richest of all, the total phenolics, anthocyanin as well as chlorophyll pigments.
Figure 7.
The content of (A) total phenols, (B) anthocyanin and, (C) chlorophyll determined in mature plant for comparing the three Ocimum species, O. sanctum (OS), O. gratissimum (OG) and O. kilimandscharicum (OK). Data are means ± SD (for at least three replicates). Statistical analysis was performed using HSD tukey test, standard weighted-means analysis at *P < 0.01 and **P < 0.05 levels of significance.
Discussion
On comparing the oil yields and the trichome densities of the three Ocimum species, it was observed that as the trichome density decreases upon the plant maturity, the oil yield also decreases (Table 1). The essential oil profiling showed that both O. sanctum and O. gratissimum were rich in eugenol which is a phenylpropene compound whereas O. kilimandscharicum was rich in camphor, a well-known terpenoid (Fig. 1 and Supplementary Table 3). As also reported by Joshi43 eugenol was the principal constituent of O. gratissimum and O. sanctum44 while camphor in case of O. kilimandscharicum45. Oil yield of O. gratissimum (1.3%) was highest followed by O. sanctum (0.87%) and O. kilimandscharicum (0.73%) and similar was the pattern of peltate glandular trichome densities (Fig. 2) except in O. kilimandscharicum. O. kilimandscharicum did not show much difference in the trichome distribution throughout the six months and also had the least number of trichomes per mm2 leaf area and also least oil yield as compared to O. sanctum and O. gratissimum. Since the apical leaves were used for the trichome density studies, the number of trichomes increased upto the optimum metabolite expression stage (July to October) and thereafter the number decreased to leaf expansion. While studying the changes in leaf trichomes and epicuticular flavonoids during leaf development in birch taxa, Valkama et al.46 also concluded that rapid decline in the density of leaf trichomes due to growth dilution in expanding leaves, as the total number of trichomes per leaf remained constant and neither the development nor shedding of trichomes at later growth stage occurs. In another study, Adebooye et al.47 also described that the morphology and density of trichomes and stomata of Trichosanthes cucumerina also got affected by leaf age, densities decrease as leaf age increases. Tozin et al.48 while analyzing the density of glandular trichome density and essential oil profile of inflorsences and leaves of Lippia origanoides Kunth belonging to family verbenaceae also reported that higher essential oil yield in the inflorescences as compared to the leaves. In a separate investigation by Werker49, it was demonstrated that trichomes remain functional in mature leaves, contrastingly Gairola et al.50 also reported that at leaf maturity, the functional role of trichomes becomes less important and they therefore senesce or wither. All of the differences in the trichome densities of the three Ocimum species could be related to genetic or physiological or evolutionary mechanisms operating within the genus, which need further investigation. Essential oil of the genus Ocimum is a secondary metabolites reservoir and has been suggested to be correlated with the chromosome numbers of species as well as the oil yield39,51. The chromosome numbers of O. sanctum var. CIM Ayu (2n = 16), O. gratissimum is higher (2n = 40) and O. kilimandscharicum (2n = 38)39,52,53 also support the earlier report of correlation between essential oil yield and chromosome number.
In secondary metabolism, phenylpropanoid biosynthesis is one of the most important pathways as it leads to synthesis of a large group of natural products22,54. The core phenylpropanoid pathway involves three enzymes, PAL, C4H, and 4CL. PAL is the first enzyme in the pathway that catalyzes the conversion of L -phenylalanine to trans-cinnamic acid. Subsequently, C4H, which is a member of cytochrome P450 super-family, hydroxylates trans-cinnamic acid into para-coumaric acid. Lastly, the formation of p-coumaroyl CoA from p-coumaric acid takes place by the reaction catalyzed by 4CL leading to the production of hydroxycinnamic acids, monolignols/lignin, coumarins, benzoic acids, stilbenes, anthocyanins and flavonoids22,55. In Ocimum species, the phenylpropanoid pathway is an important one as it leads to the synthesis of many commercially important phenylpropenes like- eugenol, methyleugenol, chavicol, methylchavicol in the leaf essential oil38,39,44,56. The phenylpropenes synthesized in plant aerial parts helps in plant defense against herbivores and pathogens56 and are also imperative in human diet57,58. Biosynthesis of these phenylpropenes is localized in the specialized glands known as glandular trichomes on the surface of leaves3,56.
PAL is an important enzyme as it is a link between primary and secondary metabolism. It is also a key regulatory enzyme in the phenolics biosynthesis59 as high activity of PAL is usually associated with the accumulation of phenolic compounds in fruit tissues of several species60. Highest expression of PAL (Fig. 4) also correlated with the highest enzyme activity (Fig. 6), highest total phenol and anthocyanin contents (Fig. 7A,B) in O. gratissimum followed by O. sanctum and O. kilimandscharicum. Expression of the PAL, C4H and 4CL transcripts in all the three Ocimum species first increased and then decreased as the plant attained maturity. The expression was optimal at the duration of high oil yield respective to each of the three Ocimum species. Xu et al.61 have also proven that the expression level of GbPAL from Ginkgo biloba was lowest at the beginning of leaf growth, increased gradually, decreased thereafter and subsequently increased further and then remained relatively constant. The results obtained at the metabolic, transcript and protein level support the hypothesis62 that the PAL, C4H, and 4CL genes or gene families irrespective of their variable sizes, represent a case of tight regulation, possibly mediated through large structural and functional similarity in TF binding with their promoters63–66. The real-time expression results of the seven TFs (bHLH1_25905, EREB, MADS box_50254, MYB3, MYB5, MYC and TTG1) out of twelve TFs studied (Fig. 5) in the present investigation also show a similar expression pattern as of the PAL, C4H and 4CL transcripts in the three Ocimum species are also in coherence with the above hypothesis. As also suggested by Koopmann et al.67, the results of the present study indicate to the possibility of the formation of a true multienzyme complex by PAL, C4H and 4CL enzymes.
The activity of C3H in biosynthesis of lignin and many other phenylpropanoid pathway products in plants has been well documented, however, conditions suitable for assay of the enzyme explicitly, still remain unclear. Although p-coumarate acts as the substrate of C3H but its significant activity towards other para hydroxylated substrates cannot be ignored67. Franke et al.68 in their work revealed CYP98A3 is encoded by REF8 gene which is required for the synthesis of wild-type lignin precursors and sinapate esters in Arabidopsis. Gang et al.69 have reported that the differential production of meta hydroxylated phenylpropanoids in sweet basil is controlled by the activities of specific acyltransferases and hydroxylases found in the peltate glandular trichomes and leaves. In the present investigation, the expression pattern of C3H transcript in the three Ocimum species did not show a specific trend. In case of O. sanctum and O. gratissimum, the expression first increased, then decreased, again increased and finally decreased while it was nearly constant in case of O. kilimandscharicum (Fig. 4). Till date the activity of C3H was considered essential for the lignin biosynthesis in plants70,71 but the possibilities of synthesizing other compounds may not be overlooked. As also reported in our previous work38 a new function of 4CL gene was explored towards eugenol biosynthesis rather than considering its conventional involvement in lignin biosynthesis. Hence, a further investigation is required to finally prove its role in the synthesis and/or regulation of phenylpropenes biosynthesis and to evidence the highest transcript expression in O. sanctum. Expression of next gene transcript, COMT was found to be decreased as the plant attained maturity in case of O. gratissimum and O. sanctum whereas in O. kilimandscharicum there was no major change in expression pattern was observed (Fig. 4). It has already been discussed that there is decrease in essential oil metabolites as the plant attains maturity. This nature of expression of COMT gene transcript expression may be elucidated by the experiments carried out by Gang et al.56 where the role of COMT in phenylpropene biosynthesis was evident when the northern blot showed high expression of COMT in glandular trichomes as compared to the whole leaf while studying the relative abundance of mRNA in the peltate glandular trichomes.
The activity of CCR and CAD enzymes till date has been attributed to lignin biosynthesis72–74. The CCR and CAD gene encode the enzymes which catalyze the first and last steps of lignin monomer biosynthesis, respectively and are closely related members of the short-chain dehydrogenase/reductase (SDR) superfamily75. Thus, the constant and increasing patterns of CCR and CAD transcripts expression in the all the three Ocimum species could be correlated with the plant aging (Fig. 4).
EGS and EOMT are the terminal genes which encode for the enzymes responsible for the synthesis of eugenol and methyleugenol from the coniferyl acetate and eugenol as the substrates, respectively76 as also shown in Fig. 3. Expression of EGS transcript was found to be expressed highest in O. gratissimum followed by O. sanctum and vice versa in case of EOMT expression pattern while, O. kilimandscharicum showed very low expression of EGS and even lesser expression of EOMT transcript (Fig. 4). As evident from the Fig. 1, O. gratissimum and O. sanctum had high percentages of eugenol content in their leaf essential oil; hence the high expression of EGS in both the two species gets justified. Gang et al.77 while characterizing the phenylpropene O-methyltransferases from sweet basil with 13 phenolic acid substrates demonstrated that EOMT1 enzyme gave 100% activity with eugenol as a substrate, simultaneously it also gave 29%, 26% and 24% activities with guaicol, isoeugenol and chavicol as substrates. Essential oil profile of O. sanctum and O. gratissimum (Supplementary Table 3) also confirms the eugenol and iso-eugenol presence in high percentages and hence, the high expression of these two gene transcripts in the two Ocimum species may be correlated. Since these two enzymes, EGS and EOMT are localized in the glandular trichomes of leaf, the expression of the transcript decreases as the leaf expands upon maturity.
Transcription factors (TFs) play a dominant role in gene regulation of all plant growth and development aspects, inclusive of secondary metabolism. Recent years, have added to the number of transcription factors involved in plant secondary metabolism regulation. However, the possibility of existence of other mechanisms regulating specific pathways cannot be overruled. Several families of TFs have been ascribed to be regulators of plant secondary metabolism but a few important ones with equivalent digital gene expression from the comparative O. sanctum and O. basilicum transcriptome sequencing data39 were sorted for the temporal expression studies of three Ocimum species. These include two non-overlapping transcripts of bHLH1 (bHLH_21387 and bHLH_25905), MADS box (MADS box_50254 and MADS box_43518) each and single transcripts of EREB, MYB2, MYB3, MYB5, MYC, PAP1, and TTG1 (Fig. 5). MADS box proteins, the MYB and bHLH (basic-helix-loop-helix) families have significantly expanded in the past 100–600 million years and are extensively reviewed78. TFs generally form complexes in order to regulate the metabolic pathways as evident by several examples. It has been reported that the MYB and bHLH TFs function cooperatively and flavonoid biosynthesis is one of the best best-studied pathway of the combinatorial gene regulation by interactions between the two79. Gonzalez et al.80 demonstrated the regulation of the anthocyanin biosynthetic pathway by transcriptional complex formation of TTG1 (transparent testa glabra1)/ bHLH/ MYB TFs in Arabidopsis seedlings. Not only was this TF complex, MYC also suggested to be involved in regulation of anthocyanin biosynthesis in Perilla frutescens81, a member of the same lamiaceae family to which Ocimum belongs. Considering the diverse functions of these TFs it becomes extremely difficult to give explanations to the intricate role of bHLH, MYB, MYC and TTG1 TFs. PAP1 (production of anthocyanin pigment1) being well described for its involvement in the anthocyanin biosynthetic pathway82 but Sekhon et al.83 and Pourtau et al.84 have also reported PAP1 and PAP2 to be involved in senescence induced by pollination prevention in maize and sugar application in Arabidopsis, respectively. Hence, the expression pattern of PAP1 in O. sanctum may be due to some stress induced at the onset of winter season in November and December as Ocimum is a plant of tropics. But no specific trends in the expression pattern of O. gratissimum and O. kilimandscharicum were observed which might be due to the fact that O. sanctum might be comparatively more susceptible to cold climate which needs experimental confirmation. Contrastingly, MADS which is an acronym for the four founder proteins MCM1 (from Saccharomyces cerevisiae), AGAMOUS (from Arabidopsis), DEFICIENS (from Antirrhinum), and SRF (a human protein), on which the definition of this gene family is based. The network of these MADS box genes is not only imperative in contributing to floral organ identity but also in floral meristem identity85. The increasing temporal pattern of expression of MADS box gene transcript in the three Ocimum species of the present study supports the fact of involvement of MADS box gene in flower organ development as meristem. The ERF (ethylene response factor) family formerly known as EREBP (ethylene-responsive element binding proteins) is attributed to regulation of biological processes related to plant growth, metabolism, development, and response to abiotic and biotic stresses86. Since the EREB transcript reveals an increasing and further decreasing temporal expression pattern in the three Ocimum species under the investigation, it might be involved in some of the biological process active during the plant development and slows down as the plant attains maturity. WRKY proteins comprise a large family of TFs which imperative to developmental and defense response in plants, hence the higher expression of WRKY transcript in the three Ocimum species towards the plant maturity might be due to the plant response against the abiotic stresses. The phenylpropenes found in glandular trichomes of Ocimum basilicum play an important in plant resistance against herbivores56. Simultaneously, Valkama et al.46 suggested that during the birch leaf development, the amount of osmiophilic material (phenolics containing o-dihydroxy groups) declines, however 20–40% of cells in aged trichomes possess it.
This study provides a comparative description in trichome number and expression pattern of important genes of phenylpropanoid biosynthesis pathway as well as the transcription factors involved in the secondary metabolism with respect to the differential accumulation and regulation of essential oil metabolites and their composition among three Ocimum species. The final number of trichomes is Ocimum leaf is ascertained at the young stage and does not change during leaf development. On the contrary, the trichome density as well as phenylpropens tends to decline with leaf age. Since the basils are susceptible to winter season, the expression pattern of the genes and transcription factors discussed herewith may be due to some abiotic (cold) or biotic (insects, fungal pathogens etc.) which often attack the plant during this season. A very scarce literature is available to infer the interaction of such abiotic and biotic plant stress exerted over the plant. Present investigation in light of trichomes as well as gene expression studies could be exploited for genetically improving the essential oil biosynthesis in Ocimum species which are becoming highly desirable for fragrance, flavor and pharmaceutical industries.
Methods
Plant material
Leaf tissues from the 3 month plants after seed sowing [O. gratissimum, var: CIM-Ayu of O. sanctum L44., and O. kilimandscharicum] were collected monthly in the year 2018 in triplicates from the research field at the CSIR-Central Institute of Medicinal and Aromatic from July till December.
Extraction and analysis of essential oil
Hydro-distillation of collected plant leaves was conducted in a Clevenger-type apparatus for two hours. 1 µl of 1:10 pentane diluted essential oil was injected in GC-MS (Agilent Technologies 7980 A gas chromatograph system with the 5977 A mass selective detector) for analysis. The HP5-MS column with dimension 30 m × 250 µm having film thickness 0.25 µm was used for obtaining the peak separation in the chromatogram. Helium in a split ratio of 10:1 and flow rate of 1 ml/min was used as the carrier gas. The running condition for the samples was 40° for 5 min as initial hold, subsequently 150 °C at the flow rate of 3 °C/min, followed by a ramping of 5 °C/min until the temperature reaches 200 °C and finally a hold for 10 min after the temperature reaches 300 °C with a ramp rate of 10 °C/min. Mass spectrometry was conducted at 230 °C as a transfer line and ion source temperature while, 150 °C as quadrupole temperature, 70 eV ionization potential and 50 to 550 atomic mass units scan range. Version 2.0 g of NIST/EPA/NIH mass spectral library was used for compound identification (Agilent Technologies, Palo Alto, CA, USA). The relative abundance of particular constituent was considered as the area percent.
Analysis of trichome density
Microscopic analysis was carried out from the leaves of fourth nodes of the plant shoot-tips. Trichomes on the leaves were photographed under an inverted fluorescence microscope (Nikon Eclipse Ti-S) on 20 × magnification with auto exposure of 15 milliseconds and analog gain of 2.0X at a 100 µm scale bar. Thereafter all the images captured were examined with the help of NIS- Elements F (version 4.0) imaging software for the estimation of trichome density as the total number of trichomes within an area of 1.0 mm2 on the leaf abaxial side in a sample size of 10 plants per month from each of the three Ocimum species. Significant differences among populations between months of the collection were calculated using Tukey–HSD post hoc test at p > 0.01, p > 0.05, p > 0.1.
Trichome and RNA isolation
Young leaves of all the three Ocimum species (O. sanctum, O. gratissimum and O. kilimandscharicum) were used to isolate the glandular trichomes following the method used by Rastogi et al.38. The total RNA was isolated from the isolated glandular trichomes using Spectrum Plant Total RNA Kit (Sigma). 2 µg of total RNA was used for the cDNA synthesis via Revert Aid Premium First Strand cDNA Synthesis Kit (Thermo).
Quantitative RT–PCR analysis
Quantitative realtime PCR was performed by the protocol given by Rastogi et al.38 which utilized SYBR Green chemistry (Thermo). The Primer Express Software version 2.0 (Applied Biosystems) was used for the designing of gene-specific primers and were ordered from Integrated DNA Technologies, India (Supplementary Table 1 and 2). The experiment was conducted in ‘7900HT Fast Real Time PCR System’ (Applied Biosystems) with five biological replicates, and the reaction specificity was evaluated by analyzing the melting curve. The parameters of the thermal cycling were: 50 °C for 2 min (initial hold); 95 °C for 10 min (initial denaturation); and 40 amplification cycles (95 °C for 15 s; and 60 °C for 1 min). Subsequently additional steps (60 °C for 15 s, 95 °C for 15 s and 37 °C for 2 min) were followed to get the dissociation curve. Actin of O. sanctum (details provided in the Supplementary Table 1) was used as an endogeneous control to quantify the relative mRNA levels38,87. ∆∆Ct method was used for relative quantification of gene transcripts through Sequence Detection System (SDS) software version 2.2.1. As a result of real-time PCR, the Ct (threshold cycle) values thus obtained were used to calculate ∆Ct value (target-endogenous control). Thereafter, ∆∆Ct was calculated for the quantification by determining the fold difference in gene expression [∆Ct target – ∆Ct calibrator]. Finally, 2−∆∆CT was determined as relative quotient (RQ).
Total phenols, anthocyanin chlorophyll estimation
Anthocyanin content was estimated following the protocol of Neff and Chory88. Anthocyanin quantification was carried out by incubating 1 g leaf samples (grounded in liquid nitrogen) overnight in 150 ml of with 1% HCl acidified methanol in triplicates. Further 100 ml of distilled water and 250 ml chloroform was added to separate anthocyanins from chlorophylls. Absorbance at 530 nm and 657 nm were recorded to determine total anthocyanins using a spectrophotometer (Elico). Relative amount of anthocyanin per gram leaf sample was calculated by subtraction of absorbance at 657 nm from the absorbance at 530 nm.
Total phenolic content was determined by Folin- Ciocalteu method using gallic acid as phenolic standard89. About 100 mg powdered leaf samples of O. sanctum, O. kilimandscharium and O. gratissimum were extracted with 0.5 µl of 80% ethanol in triplicates. The extract was centrifuged for 20 min and the supernatant was collected. The supernatants were evaporated to dryness and dissolved in 0.5 ul of water. Different aliquots of the dissolved extracts were pipetted (2–20 µl) into the micro-centrifuge tubes. The volume of each micro-centrifuge tube was made up to 300 µl final volume with the double distilled water. About 50 µl of Folin-Ciocalteau reagent was added into each tube. After 3 min, 200 µl of 20% Na2CO3 solution was added into the each tube and mixed thoroughly. Each tube was now placed in boiling water for exactly one min. The samples were cooled and measured at 650 nm absorbance using micro-titer plate. The concentration of total phenols was estimated using standard curve and expressed as mg phenols/100 g materials.
Chlorophyll extraction was performed by the protocol given by Sadasivam and Manickam89. About 1 g powdered leaf samples of O. sanctum, O. kilimandscharium and O. gratissimum were extracted with 80% chilled acetone in triplicates till the residues turned colorless. The supernatant was collected into the volumetric flask and the final volume was made upto 100 ml with 80% of chilled acetone. The extracted solutions were measured as 645 nm, 663 nm and 652 nm absorption against 80% acetone as blank. The amount of chlorophyll present in the extract mg chlorophyll per g tissue was calculated using the following equations:
where, A = absorbance at specific wavelengths,
V = final volume of chlorophyll extract in 80% acetone
W = fresh weight of tissue extracted
Enzyme assays
Young leaves were used to prepare the soluble protein extracts. Whole leaves of individual species were weighed and grinded in liquid nitrogen in triplicates. The extraction was carried out in ice-chilled protein extraction buffer (10:1, w/v), containing 50 mM BisTris [2-[bis(hydroxyethyl)amino]-2-(hydroxymethyl)-1-propane- 1,3-diol] HCl, pH 8.0, 14 mM β-mercaptoethanol, and 10% (w/v) glycerol followed by an incubation of 30 min on ice. The ground mixture was then centrifuged at 4 °C for 20 min at 14,000 g to get the protein extract as a clarified supernatant which was transferred to a new tube. The Bradford method90 was used to quantify the concentration of protein in the extract. The protein thus isolated was used for the assay of PAL (phenylalanine ammonia lyase), C4H (cinnamate-4-hydroxylase), 4CL (4-coumarate: CoA ligase) and CAD (cinnamyl alcohol dehydrogenase) enzymes.
PAL, C4H, 4CL, and CAD activities were measured following the procedures described by Gang et al.56, Misra et al.91, Rastogi et al.38, Fu et al.74, respectively. Enzyme assay for each enzyme was set in reaction volume of 1 ml containing 1 mg of the plant protein extracted. There were two controls taken for this enzyme assay, one included all reaction components except the protein and another had all reaction components except substrate. Reactions were incubated for 2 hours at 30 °C and after that it was ended by the addition of 50 µl 6 N HCl. The product was extracted twice by adding equal volume of ethylacetate, vortexing, and centrifuging at 14,000 g for 5 min, followed by evaporation of organic phase in vacuum. Product identification was verified by gradient high-performance liquid chromatography (HPLC) (LCMS-2010 EV, Shimadzu) as described by Proestos and Komaitis92. The mobile phase consisted of eluent A [2% (v/v) acetic acid/water] and eluent B [methanol: acetonitrile (50:50 v/v)]. Extracts were separated on Symmetry C-18 column (5 µm, 4.6 × 250 mm) using a gradient program: 10% B (0–5 min.), 40% B (5–25 min.), 45% B (25–35 min.), 55% B (35–40 min.) and finally 10% B (40–45 min.) at the flow rate of 1 ml/min. Column effluent was monitored at wavelengths of 254 nm, 280 nm, 320 nm and the product was recognized by spectral scans using the photodiode-array detector followed by comparing retention time and UV spectrum with that of genuine standards. In case of 4CL enzyme assay, the activity was measured in terms of substrate utilization rather than product formation due to the commercial unavailability of p-coumaroyl CoA standard.
Statistical analysis
One‐Way Analysis of Variance (ANalysis Of VAriance) with post-hoc Tukey HSD (Honestly Significant Difference) test93 was used for performing all the statistical analysis used in the study at *P < 0.01 and **P < 0.05 levels of significance with ‘ns’ meaning non-significant.
Supplementary information
Acknowledgements
This study was funded by HCP-007, BSC-0203 and YSS/2014/001011.
Author contributions
S.R., S.S., A.K. and R.K. performed the experiments; A.K.S. conceived and designed the study, S.R. and R.K. analyzed and interpreted the results; A.K.S. and S.R. wrote the manuscript. All the authors have read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-61957-5.
References
- 1.Darrah, H. H. The cultivated basils. Buckeye Printing Company, Independence, MO (1980).
- 2.Meyers, M. Basil An Herb Society of America Guide. The Herb Society of America, Kirtland, Ohio (2003).
- 3.Gang DR, Simon J, Lewinsohn E, Pichersky E. Peltate Glandular Trichomes of Ocimum basilicum L. (Sweet Basil) Contain High Levels of Enzymes Involved in the Biosynthesis of Phenylpropenes. J. Herbs Spices Med. P. 2002;9(2):189–195. doi: 10.1300/J044v09n02_27. [DOI] [Google Scholar]
- 4.Deschamps C, Simon JE. Phenylpropanoid biosynthesis in leaves and glandular trichomes of basil (Ocimum basilicum L.) Methods Mol. Biol. 2010;643:263–273. doi: 10.1007/978-1-60761-723-5_18. [DOI] [PubMed] [Google Scholar]
- 5.McCaskill D, Croteau R. Monoterpene and sesquiterpene biosynthesis in glandular trichomes of peppermint (Mentha x piperita) rely exclusively on plastid-derived isopentenyl diphosphate. Planta. 1995;197:49–56. doi: 10.1007/BF00239938. [DOI] [Google Scholar]
- 6.McCaskill D, Croteau R. Strategies for bioengineering the development and metabolism of glandular tissues in plants. Nat. Biotechnol. 1999;17:31–36. doi: 10.1038/5202. [DOI] [PubMed] [Google Scholar]
- 7.McConkey ME, Gershenzon J, Croteau RB. Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint. Plant. Physiol. 2000;122:215–223. doi: 10.1104/pp.122.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange BM, et al. Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc. Natl Acad. Sci. USA. 2000;97:2934–2939. doi: 10.1073/pnas.97.6.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner GW, Gershenzon J, Croteau RB. Distribution of peltate glandular trichomes on developing leaves of peppermint. Plant. Physiol. 2000;124:655–663. doi: 10.1104/pp.124.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner GW, Gershenzon J, Croteau RB. Distribution of peltate glandular trichomes on developing leaves of peppermint. Plant. Physiol. 2000;124:665–679. doi: 10.1104/pp.124.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmoud SS, Croteau RB. Menthofuran regulates essential oil biosynthesis in peppermint by controlling a downstream monoterpene reductase. Proc. Natl Acad. Sci. USA. 2003;100:14481–14486. doi: 10.1073/pnas.2436325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahmoud SS, Williams M, Croteau R. Cosuppression of limonene-3-hydroxylase in peppermint promotes accumulation of limonene in the essential oil. Phytochem. 2004;65(5):547–554. doi: 10.1016/j.phytochem.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Ringer KL, Davis EM, Croteau R. Monoterpene metabolism. Cloning, expression, and characterization of (−)-isopiperitenol/(−)-carveol dehydrogenase of peppermint and spearmint. Plant. Physiol. 2005;137:863–872. doi: 10.1104/pp.104.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyatt DC, et al. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc. Natl Acad. Sci. USA. 2007;104:5360–5365. doi: 10.1073/pnas.0700915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartram S, Jux A, Gleixner G, Boland W. Dynamic pathway allocation in early terpenoid biosynthesis of stress-induced lima bean leaves. Phytochem. 2006;67:1661–1672. doi: 10.1016/j.phytochem.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Pinto DM, et al. Ozone degrades common herbivore induced plant volatiles: does this affect herbivore prey location by predators and parasitoids? J. Chem. Ecol. 2007;33:683–694. doi: 10.1007/s10886-007-9255-8. [DOI] [PubMed] [Google Scholar]
- 17.Behnam S, Farzaneh M, Ahmadzadeh M, Tehrani AS. Composition and antifungal activity of essential oils of Mentha piperita and Lavendula angustifolia on post-harvest phytopathogens. Commun. Agric. Appl. Biol. Sci. 2006;71:1321–1326. [PubMed] [Google Scholar]
- 18.Bertomeu JM, Arrillaga I, Ros R, Segura J. Up-regulation of 1- deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant. Physiol. 2006;142:890–900. doi: 10.1104/pp.106.086355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, et al. The tomato homolog of coronatine-insensitive1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant. Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enfissi EM, et al. Metabolic engineering of the mevalonate and nonmevalonate isopentyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant. Biotechnol. J. 2005;3:17–27. doi: 10.1111/j.1467-7652.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- 21.Fridman E, et al. Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant. Cell. 2005;17:1252–1267. doi: 10.1105/tpc.104.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogt T. Phenylpropanoid Biosynthesis. Mol. Plant. 2010;3(1):2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 23.La Camera S, et al. Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2004;198:267–284. doi: 10.1111/j.0105-2896.2004.0129.x. [DOI] [PubMed] [Google Scholar]
- 24.Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM. Allelopathy and exotic plant invasion: from molecules and genes in species interaction. Sci. 2003;301:1377–1380. doi: 10.1126/science.1083245. [DOI] [PubMed] [Google Scholar]
- 25.Dudareva N, Pichersky E, Gershenzon J. Biochemistry of plant volatiles. Plant. Physiol. 2004;135:1893–1902. doi: 10.1104/pp.104.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang CQ, et al. Transcriptional regulation of plant secondary metabolism. J. Integr. Plant. Biol. 2012;54(10):703–712. doi: 10.1111/j.1744-7909.2012.01161.x. [DOI] [PubMed] [Google Scholar]
- 27.Anonymous. Wealth of India. Publication and Information Directorate; New Delhi, India: CSIR, 79-89 (1991).
- 28.Prakash P, Gupta N. Therapeutic uses of Ocimum sanctum linn (tulsi) with a note on eugenol and its pharmacological actions: A short review. Indian. J. Physiol. Pharmacol. 2005;49(2):125–131. [PubMed] [Google Scholar]
- 29.Bhasin M. Ocimum- Taxonomy, medicinal potentialities and economic value of essential oil. J. Bios. 2012;1:48–50. [Google Scholar]
- 30.Kalita J, Khan ML. Commercial potentialities of essential oil of Ocimum members growing in North East India. Int. J. Pharm. Life Sci. 2013;4(4):2559–2567. [Google Scholar]
- 31.Nadkarni, K. M. Indian Materia Medica, 3rd ed. Popular Prakashan Pvt Ltd: India (1999).
- 32.Prajapati, N. D., Purohit, S. S., Sharma, A. K. & Kumar, T. Agro’s dictionary of medicinal plants. 1st ed. Agrobios: India (2003).
- 33.Cohen MM. Tulsi - Ocimum sanctum: A herb for all reasons. J. Ayurveda Integr. Med. 2014;5(4):251–259. doi: 10.4103/0975-9476.146554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabhu KS, Lobo R, Shirwaikar AA, Shirwaikar A. Ocimum gratissimum: A Review of its chemical, pharmacological and ethnomedicinal properties. Open. Complement. Med. J. 2009;1:1–15. doi: 10.2174/1876391X00901010001. [DOI] [PubMed] [Google Scholar]
- 35.Orwa, C., Muta, A., Kindt, R., Jamnads, R. & Simons, A. Agrofrestre Datbase:a tre refrence and selction guide version 4.0 (htp:/ww.orldagrofrestry.org/aftredb/) (2009).
- 36.Gill D, Soni N, Sagar B, Raheja S, Agrawal S. Ocimum kilimandscharicum: A systematic review. J. Drug. Deliv. Ther. 2012;2(3):45–52. [Google Scholar]
- 37.Kokwaro, J. O. Medicinal plants of East Africa. Kampala: East African Literature Bureau. 384 (1976).
- 38.Rastogi S, et al. 4-coumarate: CoA ligase partitions metabolites for eugenol biosynthesis. Plant. Cell Physiol. 2013;54(8):1238–1252. doi: 10.1093/pcp/pct073. [DOI] [PubMed] [Google Scholar]
- 39.Rastogi S, et al. De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genom. 2014;15:588. doi: 10.1186/1471-2164-15-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rastogi S, et al. Unravelling the genome of Holy basil: an “incomparable” “elixir of life” of traditional Indian medicine. BMC Genom. 2015;16:413. doi: 10.1186/s12864-015-1640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iijima Y, et al. The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant. Physiol. 2004;136(3):3724–3736. doi: 10.1104/pp.104.051318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, et al. Transcriptome Analysis of Medicinal Plant Salvia miltiorrhiza and Identification of Genes Related to Tanshinone Biosynthesis. PLoS ONE. 2013;8(11):e80464. doi: 10.1371/journal.pone.0080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi RK. Chemical composition, in vitro antimicrobial and antioxidant activities of the essential oils of Ocimum gratissimum, O. sanctum and their major constituents. Ind. J. Pharm. Sci. 2013;75(4):457–462. doi: 10.4103/0250-474X.119834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal RK, et al. High yielding eugenol rich oil producing variety of Ocimum sanctum-‘CIM-Ayu’. JMAPS. 2003;25:746–747. [Google Scholar]
- 45.Kashyap CP, Kaur R, Arya V, Kumar V. Therapeutic Potency of Ocimum kilimandscharicum Guerke - A Review. Glob. J. Pharm. 2011;5(3):191–200. [Google Scholar]
- 46.Valkama E, Salminen J, Koricheva J, Pihlaja K. Changes in leaf trichomes and epicuticular flavonoids during leaf development in three birch taxa. Ann. Bot. 2004;94:233–242. doi: 10.1093/aob/mch131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adebooye OC, Hunsche M, Noga G, Lankes C. Morphology and density of trichomes and stomata of Trichosanthes cucumerina (Cucurbitaceae) as affected by leaf age and salinity. Turk. J. Bot. 2012;36:328–335. [Google Scholar]
- 48.Tozin LR, Marques MO, Rodrigues TM. Glandular trichome density and essential oil composition in leaves and inflorescences of Lippia origanoides Kunth (Verbenaceae) in the Brazilian Cerrado. An. Acad. Bras. Cienc. 2015;87(2):943–953. doi: 10.1590/0001-3765201520140376. [DOI] [PubMed] [Google Scholar]
- 49.Werker E. Trichome diversity and development. Adv. Bot. Res. 2000;31:1–35. doi: 10.1016/S0065-2296(00)31005-9. [DOI] [Google Scholar]
- 50.Gairola S, Naidoo Y, Bhatt A, Nicholas A. An investigation of the foliar trichomes of Tetradenia riparia (Hochst.) Codd [Lamiaceae]: an important medicinal plant of Southern. Africa. Flora. 2009;204:325–330. doi: 10.1016/j.flora.2008.04.002. [DOI] [Google Scholar]
- 51.Lavania UC, et al. Autopolyploidy differentially influences body size in plants, but facilitate enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant. J. 2012;71(4):539–549. doi: 10.1111/j.1365-313X.2012.05006.x. [DOI] [PubMed] [Google Scholar]
- 52.Carović-Stanko K, et al. Genetic relations among basil taxa (Ocimum L.) based on molecular markers, nuclear DNA content, and chromosome number. Plant. Syst. Evol. 2009;285(1):13–22. [Google Scholar]
- 53.Bose RB, Choudhury JK. A Comparative Study of the Cytotaxonomy, Pallynology, Physiology of ‘Diploid’ and ‘Polyploid’ Plants of Ocimum kilimandscharicum Guerke and their Yield of Raw Material and Volatile Contents. Caryologia: Int. J. Cytol. Cytosystem Cytogen. 1962;15(2):435–454. doi: 10.1080/00087114.1962.10796070. [DOI] [Google Scholar]
- 54.Turgut-Kara N, Çakır O. Comparative phylogenetic analysis of phenylpropanoid metabolism genes of legume plants. Plant. Omics J. 2015;8(1):55–61. [Google Scholar]
- 55.Dixon RA, et al. The phenylpropanoid pathway and plant defense a genomics perspective. Mol. Plant. Pathol. 2002;3:371–390. doi: 10.1046/j.1364-3703.2002.00131.x. [DOI] [PubMed] [Google Scholar]
- 56.Gang DR, et al. An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant. Physiol. 2001;125:539–555. doi: 10.1104/pp.125.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nahrstedt A. The significance of secondary metabolites for interaction between plants and insects. Planta Med. 1990;55:333–338. doi: 10.1055/s-2006-962023. [DOI] [PubMed] [Google Scholar]
- 58.Prasad NS, Raghavendra R, Lokesh BR, Naidu KA. Spice phenolics inhibit human PMNL 5-lipoxygenase. Prostaglandins Leukot. Essent. Fat. Acids. 2004;70:521–528. doi: 10.1016/j.plefa.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 59.Jones DH. Phenylalanine ammonia-lyase: Regulation of its induction, and its role in plant development. Phytochem. 1984;23:l349–1359. doi: 10.1016/S0031-9422(00)80331-3. [DOI] [Google Scholar]
- 60.Cheng WG, Breen PJ. Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Amer Soc. Hort. Sci. 1991;116(5):865–869. doi: 10.21273/JASHS.116.5.865. [DOI] [Google Scholar]
- 61.Xu F, et al. Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr. J. Biotech. 2008;7(6):721–729. [Google Scholar]
- 62.Batz O, Logemann E, Reinold S, Hahlbrock K. Extensive reprogramming of primary and secondary metabolism by fungal elicitor or infection in parsley cells. Biol. Chem. 1998;379:1127–1135. doi: 10.1515/bchm.1998.379.8-9.1127. [DOI] [PubMed] [Google Scholar]
- 63.Hahlbrock K, et al. Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc. Natl Acad. Sci. USA. 1995;92:4150–4157. doi: 10.1073/pnas.92.10.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C. Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant. Physiol. 1997;113:729–738. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Logemann E, Parniske M, Hahlbrock K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl Acad. Sci. USA. 1995;92:5905–5909. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant. Physiol. 1997;113:755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koopmann E, Logemann E, Hahlbrock K. Regulation and Functional Expression of Cinnamate 4-Hydroxylase from Parsley. Plant. Physiol. 1999;119:49–55. doi: 10.1104/pp.119.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franke R, et al. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant. J. 2002;30(1):33–45. doi: 10.1046/j.1365-313X.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- 69.Gang DR, Beuerle T, Ullmann P, Werck-Reichhart D, Pichersky E. Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant. Physiol. 2002;130(3):1536–1544. doi: 10.1104/pp.007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schoch G, et al. CYP98A3 from Arabidopsis thaliana is a 3’-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem. 2001;276:36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Deng Z, Gao S, Sun X, Tang K. A new gene coding for p-coumarate 3-hydroxylase from Ginkgo biloba. Russ J Plant Physiol. 2008;55(1):82–92. doi: 10.1134/S102144370801010X. [DOI] [Google Scholar]
- 72.Chabannes, M. et al. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J28(3), 257–270 (2001). [DOI] [PubMed]
- 73.Lauvergeat V, et al. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochem. 2001;57(7):1187–1195. doi: 10.1016/S0031-9422(01)00053-X. [DOI] [PubMed] [Google Scholar]
- 74.Fu C, et al. Downregulation of Cinnamyl Alcohol Dehydrogenase (CAD) leads to improved saccharification efficiency in Switchgrass. BioEnergy Res. 2011;4(3):153–164. doi: 10.1007/s12155-010-9109-z. [DOI] [Google Scholar]
- 75.Pan H, et al. Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis. Plant Cell. 2014;26(9):3709–3727. doi: 10.1105/tpc.114.127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koeduka T, Baiga TJ, Noel JP, Pichersky E. Biosynthesis of t-Anethole in Anise: Characterization of t-Anol/Isoeugenol Synthase and an O-Methyltransferase Specific for a C7-C8 Propenyl Side Chain. Plant Physiol. 2009;149:384–394. doi: 10.1104/pp.108.128066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gang DR, et al. Characterization of Phenylpropene O-Methyltransferases from Sweet Basil: Facile Change of Substrate Specificity and Convergent Evolution within a Plant O-Methyltransferase Family. Plant Cell. 2002;14:505–519. doi: 10.1105/tpc.010327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 79.Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 81.Saito K, Yamazaki M. Biochemistry and molecular biology of the late-stage of biosynthesis of anthocyanin: lessons from Perilla frutescens as a model plant. New Phytol. 2002;155:9–23. doi: 10.1046/j.1469-8137.2002.00440.x. [DOI] [PubMed] [Google Scholar]
- 82.Qiu J, et al. Metabolic engineering of the phenylpropanoid pathway enhances the antioxidant capacity of Saussurea involucrate. PLoS One. 2013;8(8):e70665. doi: 10.1371/journal.pone.0070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sekhon RS, et al. Transcriptional and metabolic analysis of senescence induced by preventing pollination in Maize. Plant Physiol. 2012;159:1730–1744. doi: 10.1104/pp.112.199224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A. Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta. 2006;224:556–568. doi: 10.1007/s00425-006-0243-y. [DOI] [PubMed] [Google Scholar]
- 85.Theissen G, et al. A short history of MADS-box genes in plants. Plant Mol Biol. 2000;42:115–149. doi: 10.1023/A:1006332105728. [DOI] [PubMed] [Google Scholar]
- 86.Wu C, Avila CA, Goggin FL. The ethylene response factor Pti5 contributes to potato aphid resistance in tomato independent of ethylene signaling. J Exp Bot. 2014;66(2):559–570. doi: 10.1093/jxb/eru472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bansal S, et al. HMG-CoA reductase from Camphor Tulsi (Ocimum kilimandscharicum) regulated MVA dependent biosynthesis of diverse terpenoids in homologous and heterologous plant systems. Sci Rep. 2018;8(1):3547. doi: 10.1038/s41598-017-17153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neff MM, Chory J. Genetic Interactions between Phytochrome A, Phytochrome B, and Cryptochrome 1 during Arabidopsis Development. Plant Physiol. 1998;118(1):27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sadasivam, S. & Manickam, A. Biochemical methods. 2nd Edition, New age International, New Delhi (1996).
- 90.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 91.Misra A, Chanotiya CS, Gupta MM, Dwivedi UN, Shasany AK. Characterization of cytochrome P450 mono-oxygenases isolated from trichome enriched fraction of Artemisia annua L. leaf. Gene. 2012;510:193–201. doi: 10.1016/j.gene.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 92.Proestos C, Komaitis M. Analysis of Naturally Occurring Phenolic Compounds in Aromatic Plants by RP-HPLC Coupled to Diode Array Detector (DAD) and GC-MS after Silylation. Foods. 2013;2:90–99. doi: 10.3390/foods2010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haynes, W. Tukey’s Test. In: Dubitzky, W., Wolkenhauer, O., Cho, K.H. & Yokota, H. (eds) Encyclopedia of Systems Biology. Springer, New York, NY (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.