Abstract
Background
Adrenal cortical carcinoma (ACC) is a rare cancer with a variable prognosis. Several prognostic factors of ACC have been previously reported, but a proteomic analysis has not yet been performed. This study aimed to investigate prognostic biomarkers for ACC using a proteomic approach.
Methods
We used reverse-phase protein array data from The Cancer Proteome Atlas, and identified differentially expressed proteins in metastatic ACCs. Multivariate Cox regression analysis adjusted by age and staging was used for survival analysis, and the C-index and category-free net reclassification improvement (cfNRI) were utilized to evaluate additive prognostic value.
Results
In 46 patients with ACC, cyclin B1, transferrin receptor (TfR1), and fibronectin were significantly overexpressed in patients with distant metastasis. In multivariate models, high expression of cyclin B1 and TfR1 was significantly associated with mortality (hazard ratio [HR], 6.13; 95% confidence interval [CI], 1.02 to 36.7; and HR, 6.59; 95% CI, 1.14 to 38.2; respectively), whereas high fibronectin expression was not (HR, 3.92; 95% CI, 0.75 to 20.4). Combinations of high cyclin B1/high TfR1, high cyclin B1/high fibronectin, and high TfR1/high fibronectin were strongly associated with mortality ([HR, 13.72; 95% CI, 1.89 to 99.66], [HR, 9.22; 95% CI, 1.34 to 63.55], and [HR, 18.59; 95% CI, 2.54 to 135.88], respectively). In reclassification analyses, cyclin B1, TfR1, fibronectin, and combinations thereof improved the prognostic performance (C-index, 0.78 to 0.82–0.86; cfNRI, all P values <0.05).
Conclusion
In ACC patients, the overexpression of cyclin B1, TfR1, and fibronectin and combinations thereof were associated with poor prognosis.
Keywords: Adrenocortical carcinoma, Protein array analysis, Proteomics, Prognosis
INTRODUCTION
Adrenal cortical carcinoma (ACC) is a rare cancer, with an annual incidence of about 1–2 per million [1]. ACC has a poor prognosis, with a 5-year overall survival rate of less than 40% for all cancers and 10% for metastatic cancers [2]. However, even in metastatic ACC, the prognosis is quite variable, with reported survival ranging from a few months to more than 10 years, suggesting the heterogeneity of these tumors [3]. Tumor, node, metastasis (TNM) staging has been used as a predictor of survival and has been modified to improve its prognostic power; currently, the eighth edition of TNM staging and European Network for the Study of Adrenal Tumors (ENSAT) staging systems are used [4,5,6]. Nevertheless, a significant proportion of patients with localized disease according to TNM staging experience recurrence after surgery (up to 70% within 3 years), which is the only curative treatment to date [2]. Tumor grade, as assessed by mitotic count and proliferation indices such as the Ki-67 index, has shown good predictive value for localized ACC [7]. However, tumor grade still has the limitation of poor reproducibility [8]. Molecular studies have focused on several potential driver genes, but their independent prognostic values are controversial [9,10]. Recently, distinct molecular subgroups have been identified via pan-genomic approaches using DNA copy number, mRNA expression, miRNA expression, and DNA methylation, and these subgroups have shown associations with different survival outcomes [11,12]. Moreover, this classification demonstrated high prognostic value when combining with clinical staging and tumor grade [13]. However, the pan-genomic approach is quite complex and not easy to use in clinical practice. In addition, pan-genomic biomarkers had limited value in prognostic performance for metastatic ACC [13].
Few studies have investigated the use of a proteomic approach to predict the prognosis of ACC. Moreover, protein markers can be useful in clinical practice since they can be straightforwardly analyzed using immunohistochemistry or Western blots. Therefore, we aimed to investigate prognostic biomarkers for ACCs using a proteomic approach.
METHODS
Study subjects and data source
In this study, ACC datasets from The Cancer Proteome Atlas (TCPA) database were extracted from the cBioPortal website (http://www.cbioportal.org/) using The Cancer Genome Atlas (TCGA) provisional dataset [14]. We obtained data from reverse-phase protein arrays (RPPA), an antibody-based quantitative method assessing multiple protein markers in a cost-effective, sensitive, and high-throughput manner [14]. RPPA data that had been normalized using the z-score, as provided in cBio-Portal, were used. Among the 91 ACC patients in the TCGA database, we included 46 patients with RPPA data. Clinical data, including staging (ENSAT), treatment, and survival information, were also obtained from the same source [5].
Differential protein expression analysis
Differential protein expression analysis of the RPPA data was performed using the Perseus software (http://www.perseus-framework.org). Differentially expressed proteins (DEPs) in metastatic and non-metastatic ACCs were identified, and the false-discovery rate (FDR) was controlled using the Benjamini-Hochberg method. The list of DEPs was limited to proteins showing a fold-change of equal to or greater than +2, or equal to or less than −2, and an FDR lower than 0.05. Protein expression above the median value was defined as high expression.
Statistical analysis
Kaplan-Meier analysis and the log-rank test were used to construct the survival curve. Cox proportional-hazards regression analyses were used to calculate the hazard ratios (HRs) of DEPs for all-cause mortality. Age and staging were adjusted in the multivariable Cox regression analyses. To determine the incremental discriminative value of DEPs additive to age and staging for prognosis, we calculated C-statistics and the category-free net reclassification improvement (cfNRI). In addition, we performed a subgroup analysis of non-metastatic patients. All P values were two-sided, and P values of lower than 0.05 were considered to indicate statistical significance in all analyses. The statistical analyses were performed using STATA software version 13 (StataCorp, College Station, TX, USA; https://www.stata.com) and R version 3.6.1 (R Foundation, Vienna, Austria; https://www.r-project.org).
Ethical statement
Written informed consent by subjects was waived due to the public database. All data were anonymized, and this study received Institutional Review Board (IRB) approval at Seoul National University Hospital (IRB No. E-1910-005-1067).
RESULTS
Baseline characteristics and clinical outcomes
The baseline characteristics of the participants are shown in Table 1. Their mean age was 47.7±14.4 years, and 18 (39.1%) were male. Thirty-five (76.1%) were white, two (4.4%) were non-white, and the others' racial information was unknown. The number of patients with ENSAT stage I, II, III, and IV disease was two (4.3%), 26 (56.5%), 10 (21.7%), and eight (17.4%), respectively. Complete resection was performed in 33 (71.7%) patients, of whom 11 experienced recurrence. Thirty-two patients (69.6%) received adjuvant chemotherapy after surgery. The patients' mean Weiss score was 5.9±2.0.
Table 1. Baseline Characteristics of Adrenal Cortical Carcinoma Patients (n=46) in the TCPA Database.
| Characteristic | No metastasis (n = 38) | Metastasis (n = 8) | Total (n = 46) | P valuea |
|---|---|---|---|---|
| Age, yr | 47.6±14.8 | 48.0±13.1 | 47.7±14.4 | 0.948 |
| Male sex | 18 (47.4) | 0 | 18 (39.1) | 0.015 |
| Race | 0.440 | |||
| White | 27 (71.1) | 8 (100) | 35 (76.1) | |
| Black | 1 (2.6) | 0 | 1 (2.2) | |
| Asian | 1 (2.6) | 0 | 1 (2.2) | |
| Unknown | 9 (23.7) | 0 | 9 (19.6) | |
| Survival (alive) | 30 (78.9) | 2 (25.0) | 32 (69.6) | 0.006 |
| Follow-up, yr | 3.3 (1.3–5.3) | 2.2 (1.2–3.7) | 3.1 (1.3–5.3) | 0.041 |
| Staging (ENSAT) | <0.001 | |||
| I | 2 (5.3) | 0 | 2 (4.3) | |
| II | 26 (68.4) | 0 | 26 (56.5) | |
| III | 10 (26.3) | 0 | 10 (21.7) | |
| IV | 0 | 8 (100) | 8 (17.4) | |
| Complete resection | 32 (84.2) | 1 (12.5) | 33 (71.7) | <0.001 |
| Chemotherapy | 25 (65.8) | 7 (87.5) | 32 (69.6) | 0.403 |
| Recurrence after complete resectionb | 11 (34.4) | 0 | 11 (33.3) | 1.000 |
| Weiss scorec | 5.8±1.7 | 6.1±3.0 | 5.9±2.0 | 0.503f |
| Mitotic countd | ||||
| Count/HPF | 9.8±8.2 | 33.0±39.9 | 13.6±18.8 | 0.589f |
| No. of >20/HPF | 2 (7.7) | 2 (28.6) | 4 (12.1) | 0.190 |
| Cortisol secretione | 15 (42.9) | 5 (62.5) | 20 (46.5) | 0.440 |
| Differentially expressed proteins | ||||
| High cyclin B1 | 16 (42.1) | 7 (87.5) | 23 (50.0) | 0.047 |
| High TfR1 | 17 (44.7) | 6 (75.0) | 23 (50.0) | 0.243 |
| High fibronectin | 15 (39.5) | 8 (100) | 23 (50.0) | 0.004 |
| High cyclin B1+TfR1 | 11 (28.9) | 6 (75.0) | 17 (37.0) | 0.038 |
| High cyclin B1+fibronectin | 7 (18.4) | 7 (87.5) | 14 (30.4) | <0.001 |
| High TfR1+fibronectin | 8 (21.1) | 6 (75.0) | 14 (30.4) | 0.006 |
| High cyclin B1+TfR1+fibronectin | 7 (18.4) | 6 (75.0) | 13 (28.3) | 0.004 |
Values are expressed as mean±standard deviation, number (%), or median (interquartile range).
TCPA, The Cancer Proteome Atlas; ENSAT, European Network for the Study of Adrenal Tumors; HPF, high power field; TfR1, transferrin receptor.
aP value for a comparison between patients with metastasis and without metastasis; bThe denominator was patients with complete resection; cThe available number was 32 because of missing values; dThe available number was 33 because of missing values; eThe available number was 43 because of missing values; fThe Mann-Whitney test was used, because the Weiss score and mitotic count did not follow normal distributions.
Differential expression of proteins
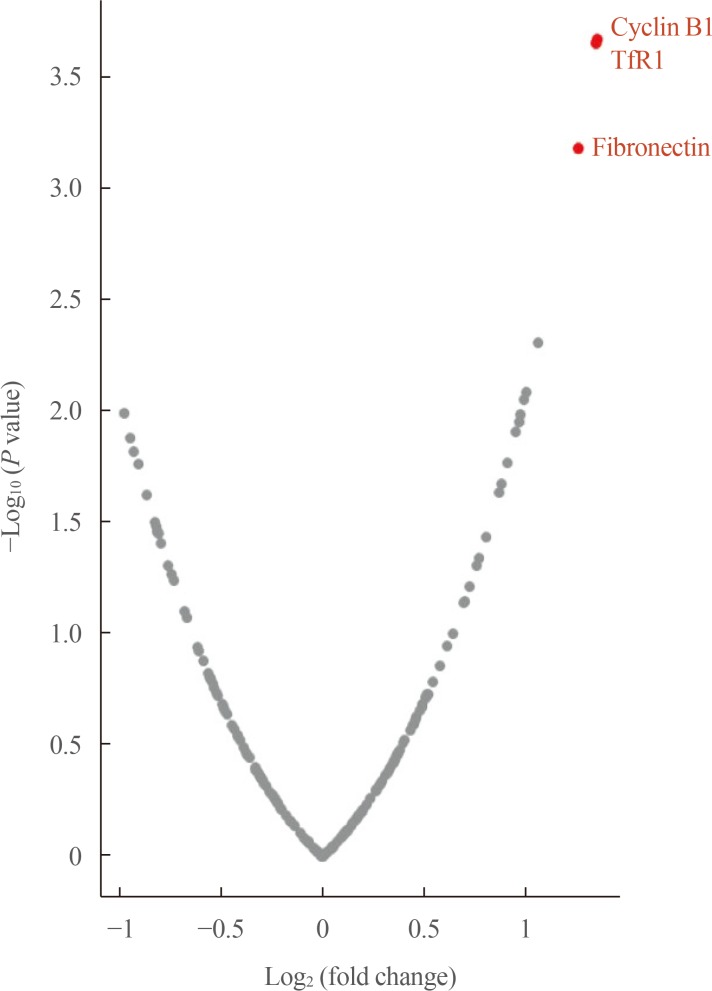
The expression levels of 198 proteins in the RPPA data are shown in Supplemental Table S1. In patients with distant metastasis, three proteins were found to be upregulated (fold-change ≥2)—CCNB1 (cyclin B1), TFRC (transferrin receptor [TfR1]), and FN1 (fibronectin)—and none was found to be downregulated (fold-change ≤−2) A volcano plot of DEPs according to metastasis is presented in Fig. 1.
Fig. 1. Volcano plot of differentially expressed proteins in adrenal cortical carcinoma patients according to metastasis. TfR1, transferrin receptor.
Survival analyses
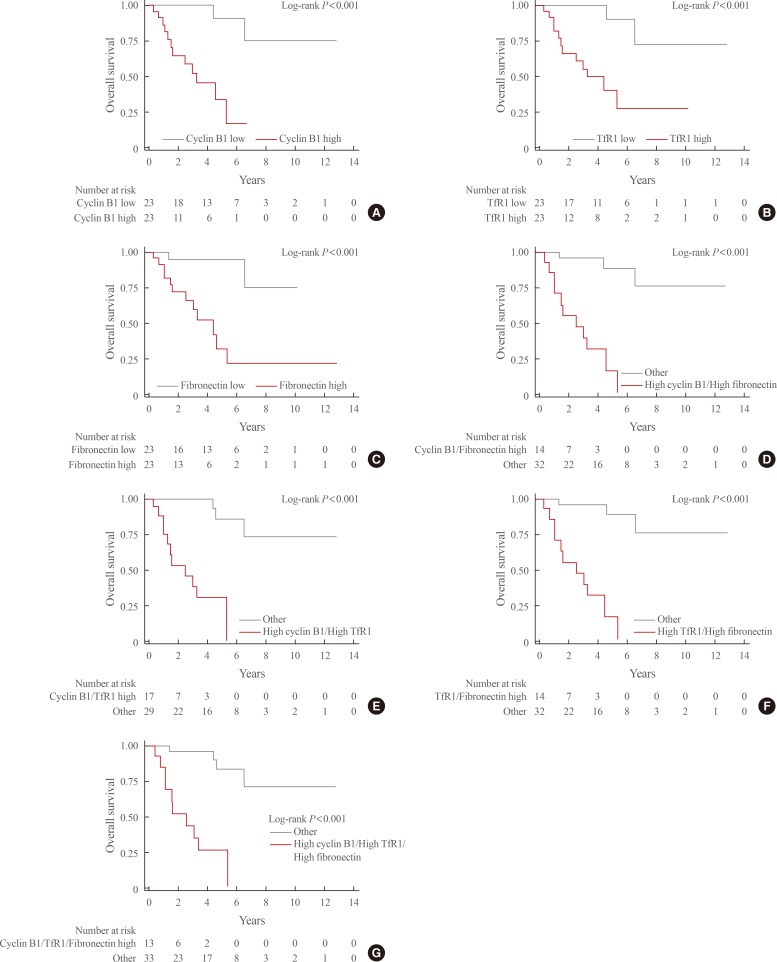
During the median follow-up period of 3.1 years (interquartile range, 1.3 to 5.3), mortality occurred in 14 patients (31.4%). Kaplan-Meier curves for each of the above-mentioned DEPs and combinations thereof are shown in Fig. 2. The survival rate was significantly lower in patients with high expression levels of each of these three proteins than in those with low expression levels (overall log-rank P value <0.001 for all). In addition, significantly lower survival rates were seen for all combinations of these proteins (high cyclin B1/high TfR1, high cyclin B1/high fibronectin, high TfR1/high fibronectin, high cyclin B1/high TfR1/high fibronectin; overall log-rank P value <0.001 for all).
Fig. 2. Kaplan-Meier curves for overall survival based on the expression status of differentially expressed proteins. (A) Cyclin B1. (B) Transferrin receptor (TfR1). (C) Fibronectin. (D) Cyclin B1 and TfR1. (E) Cyclin B1 and fibronectin. (F) TfR1 and fibronectin. (G) Cyclin B1, TfR1, and fibronectin.
In the unadjusted Cox regression analyses, high expression of each protein and all combinations thereof were significantly associated with mortality (Table 2). In multivariate Cox regression analyses adjusted for age and staging, high expression levels of cyclin B1 and TfR1 were significantly associated with high mortality. However, the expression level of fibronectin failed to predict mortality in the multivariate Cox regression model. All combinations of each protein expression signature (high cyclin B1/high TfR1, high cyclin B1/high fibronectin, high TfR1/high fibronectin, and high cyclin B1/high TfR1/high fibronectin) significantly predicted mortality. The HRs for mortality of combinations involving each DEP were higher than those of each protein individually.
Table 2. Cox Regression Analysis of Differentially Expressed Proteins for Overall Survival.
| Variable | Univariate | Multivariatea | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (per 10-year increment) | 1.35 (0.91–2.02) | 0.140 | - | - |
| Staging (ENSAT)b | 11.82 (2.49–56.09) | 0.002 | - | - |
| High cyclin B1 | 11.40 (2.47–52.59) | 0.002 | 6.13 (1.02–36.65) | 0.047 |
| High TfR1 | 8.76 (1.94–39.54) | 0.005 | 6.59 (1.14–38.23) | 0.036 |
| High fibronectin | 8.71 (1.93–39.35) | 0.005 | 3.92 (0.75–20.43) | 0.105 |
| High cyclin B1+high TfR1c | 22.23 (4.59–107.61) | <0.001 | 13.72 (1.89–99.66) | 0.010 |
| High cyclin B1+high fibronectinc | 19.44 (4.22–89.57) | <0.001 | 9.22 (1.34–63.55) | 0.024 |
| High TfR1+high fibronectinc | 21.48 (4.57–100.95) | <0.001 | 18.59 (2.54–135.88) | 0.004 |
| High cyclin B1+high TfR1+high fibronectinc | 15.43 (4.05-58.83) | <0.001 | 8.41 (1.54-45.90) | 0.014 |
HR, hazard ratio; CI, confidence interval; ENSAT, European Network for the Study of Adrenal Tumors; TfR1, transferrin receptor.
aMultivariate analyses are adjusted by age and staging; bThe categorization of I+II vs. III+IV was applied using the ENSAT system; cThe reference group is other subjects, such as those except patients with both high cyclin B1 and high TfR1.
Reclassification analyses of individual DEPs and combinations thereof for overall survival
C-statistics and the cfNRI were used to evaluate the prognostic values of the DEPs additive to age and staging (Table 3). The C-index of the reference model (age and staging) was 0.78, and it increased when each protein (high cyclin B1, high TfR1, and high fibronectin) was added, to 0.82, 0.83, and 0.82, respectively. The C-index increased further when combinations of the proteins were considered (high cyclin B1/high TfR1, high cyclin B1/high fibronectin, high TfR1/high fibronectin, and high cyclin B1/high TfR1/high fibronectin), which yielded C-index values of 0.84, 0.86, 0.86, and 0.86, respectively. In addition, the cfNRI values were also significant for each protein, individually and in combination with others (P value <0.05 for all). The event and nonevent cfNRIs were both positive in all models.
Table 3. Reclassification Analyses of the Individual Differentially Expressed Proteins and Combinations Thereof in Addition to Age and Stage for Overall Survival.
| C-index (95% CI) | cfNRI (95% CI) | P value (cfNRI) | Event cfNRI | Nonevent cfNRI | |
|---|---|---|---|---|---|
| Ref (age+stage)a | 0.78 (0.62–0.94) | - | - | - | - |
| Ref+high cyclin B1 | 0.82 (0.72–0.92) | 1.03 (0.53–1.52) | <0.001 | 0.71 | 0.31 |
| Ref+high TfR1 | 0.83 (0.71–0.94) | 1.03 (0.53–1.52) | <0.001 | 0.71 | 0.31 |
| Ref+high fibronectin | 0.82 (0.70–0.94) | 0.74 (0.16–1.32) | 0.012 | 0.43 | 0.31 |
| Ref+high cyclin B1+high TfR1 | 0.84 (0.76–0.93) | 0.84 (0.34–1.34) | 0.001 | 0.71 | 0.13 |
| Ref+high cyclin B1+high fibronectin | 0.86 (0.78–0.93) | 0.88 (0.34–1.43) | 0.001 | 0.57 | 0.31 |
| Ref+high TfR1+high fibronectin | 0.86 (0.78–0.95) | 1.03 (0.53–1.52) | <0.001 | 0.71 | 0.31 |
| Ref+high cyclin B1+high TfR1+high fibronectin | 0.86 (0.78–0.93) | 0.88 (0.34–1.43) | 0.001 | 0.57 | 0.31 |
CI, confidence interval; cfNRI, category-free net reclassification improvement; TfR1, transferrin receptor.
aAge was analyzed in terms of 10-year increments; for stage, the categorization of I+II vs. III+IV was applied using the European Network for the Study of Adrenal Tumors (ENSAT) system.
Subgroup analysis of non-metastatic ACC patients
We performed a subgroup analysis of non-metastatic patients (Table 4). Some combinations of DEPs (high cyclin B/high TfR1 and high TfR1/high fibronectin) showed significant associations with mortality, while other combinations (high cyclin B1/high fibronectin and high cyclin B1/high TfR1/high fibronectin) showed near-significant associations with mortality.
Table 4. Cox Regression Analysis of Differentially Expressed Proteins for Overall Survival in Non-Metastatic Patients (n=38).
| Variable | Univariate | Multivariatea | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (per 10-year increment) | 1.52 (0.89–2.62) | 0.126 | - | - |
| Staging (ENSAT)b | 7.85 (1.33–46.04) | 0.022 | - | - |
| High cyclin B1 | 7.75 (1.51–39.86) | 0.014 | 5.46 (0.80–37.27) | 0.083 |
| High TfR1 | 11.27 (1.38–92.35) | 0.024 | 6.70 (0.70–63.77) | 0.098 |
| High fibronectin | 5.91 (1.19–29.43) | 0.030 | 3.60 (0.63–20.66) | 0.150 |
| High cyclin B1+high TfR1c | 30.19 (3.45–264.24) | 0.002 | 21.69 (1.58–298.63) | 0.021 |
| High cyclin B1+high fibronectinc | 16.20 (3.07–85.37) | 0.001 | 9.25 (0.96–88.87) | 0.054 |
| High TfR1+high fibronectinc | 29.22 (3.48–245.26) | 0.002 | 84.15 (2.39–2961.35) | 0.015 |
| High cyclin B1+high TfR1+high fibronectinc | 16.20 (3.07–85.37) | 0.001 | 9.25 (0.96–88.87) | 0.054 |
HR, hazard ratio; CI, confidence interval; ENSAT, European Network for the Study of Adrenal Tumors; TfR1, transferrin receptor.
aMultivariate analyses were adjusted by age and staging; bThe categorization of I+II vs. III+IV was applied using ENSAT system; cThe reference group is other subjects, such as those except patients with both high cyclin B1 and high TfR1.
DISCUSSION
In the present study, we identified three proteins (cyclin B1, TfR1, and fibronectin) that were highly expressed in metastatic ACC in the TCPA database. With a median follow-up of 3.1 years, high expression of each of these three proteins was associated with a poor survival rate. Subjects with high expression levels of a combination of two DEPs were at a higher risk for mortality than those with a high expression levels of only one DEP. Cyclin B1, TfR1, and all combinations of the three DEPs showed meaningful prognostic performance independent of age and staging. Moreover, among non-metastatic patients, combinations of these three DEPs showed significant or near-significant associations with mortality. The reason for the non-significance of fibronectin alone needs to be elucidated, but the small number of patients may have contributed to this finding. In addition, the C-index and cfNRI values of high cyclin B1/high TfR1/high fibronectin were the same as those of high cyclin B1/high fibronectin. This phenomenon may be attributed to the similar number of subjects with high expression of all three DEPs and with high cyclin B1 and high fibronectin expression.
We conducted a proteomic analysis to identify prognostic markers for ACC. Several studies have explored prognostic markers for ACC, but most studies have reported clinical or pathological prognostic markers [5,7]. Molecular studies revealed that mutations in several potential driver genes such as CTNNB1 and TP53 may serve as prognostic markers [10,15]. Nevertheless, the independent prognostic value of these markers is controversial [9,10]. Recently, pan-genomic approaches—encompassing DNA copy number, mRNA expression, miRNA expression, and DNA methylation—classified ACC patients into distinct molecular subgroups, which showed different survival outcomes [11,12,13]. Despite the good performance of pan-genomic prognostic markers in previous studies, the relevant analyses are quite complex to perform, and their prognostic values were insufficient in advanced ACC (stage IV) [13]. To overcome these limitations, our present study utilized a proteomic approach, which has not been investigated in previous research on ACC. In this study, the expression status of three DEPs had prognostic value in addition to age and staging, as assessed by C-index and cfNRI. In addition, the highest C-index of our prediction model was quite high (0.86), which is similar to the result of a previous study [13], given that we included stage IV patients. Therefore, the DEPs that we identified can be applied as relatively simple and powerful prognostic markers in ACC.
The three identified DEPs—cyclin B1, TfR1, and fibronectin—have been studied as prognostic markers for other cancers. Cyclin B1 is a regulatory protein that plays an important role in mitosis by forming a complex with Cdk1 [16]. It acts as a switch-like manner in the decision to progress from G2 to the M phase in the cell. In many cancers, such as breast, cervical, colorectal, esophageal, lung, and prostate cancers, high expression levels of cyclin B1 have been found [17,18,19,20,21] and are associated with a poor prognosis [17,21]. In addition, the use of small interfering RNA that targeted cyclin B1 downregulated tumor proliferation and enhanced sensitivity to systemic chemotherapy [22]. Moreover, in a previous study using ACC cell lines (H295R and SW-13 cells), the mechanism of mitotane was found to involve the cyclin B1/Cdk complex [23].
TfR1 is a transmembrane glycoprotein that imports iron-bound transferrin into cells by endocytosis [24]. It has also been reported that TfR1 is overexpressed in many cancers, such as breast, prostate, colon, liver, lung, brain, ovarian, and hematologic cancers [25,26,27,28,29,30,31]. This high expression of TfR1 in cancer is explained by the high iron requirements of cancer cells, as well as by the regulatory role of TfR1 in anti-apoptotic regulation, reactive oxygen species production, and mitochondrial respiration [30,32,33]. In addition, as potential TfR1 inhibitors, curcumin, antibody A24 or JST-TFR09, and miR-320 have been studied in several cancers [34,35,36,37].
Fibronectin is a glycoprotein of the extracellular matrix that plays a role in cell growth, differentiation, migration, and wound healing [38]. It has also been reported that abnormal expression of fibronectin promotes invasion and migration of cancers, including breast, ovarian, prostate, lung, and colon cancers [39,40,41,42,43]. In addition, fibronectin has been reported to play a role in reducing the response of cancer to cytotoxic therapy [44,45]. In vitro studies of lung, pancreatic neuroendocrine, and breast cancers have shown that several molecules (e.g., PP2, PF-04554878, pUR4B, and AdF512v1) that inhibit fibronectin and its upstream or downstream pathways reduced its effects on tumor cell proliferation, migration, and angiogenesis [42,46,47,48].
As discussed above, previous studies have suggested that all three of the proteins (cyclin B1, TfR1, and fibronectin) identified as DEPs in our study may be prognostic markers in several cancers. However, to the best of our knowledge, no study has yet demonstrated an association of these proteins with mortality in ACC patients. Accordingly, these proteins may also be useful novel markers of poor prognosis in ACC patients.
There are several limitations of our study. The number of subjects was small, and our results were not validated in other cohorts. Thus, the prognostic power of these three DEPs remains to be confirmed. In addition, the RPPA data of ACC patients from the TCPA, which were used as the data source in our study, contained information on a small number of proteins. Therefore, it is possible that other protein markers might predict survival in ACC patients. Advanced proteomics techniques, such as gas or liquid chromatography tandem mass spectrometry, might enable the identification of more prognostic protein markers in ACC patients.
Taken together, the overexpression of cyclin B1, TfR1, and fibronectin proteins may have prognostic value for ACC patients. Further studies need to validate these three proteins as prognostic markers in another ACC cohort.
ACKNOWLEDGMENTS
This study was supported by the Korean Endocrine Hypertension-Adrenal Study Group from the Korean Endocrinology Society. This work was presented in abstract form at the 2019 Endocrine Society in New Orleans, USA.
Footnotes
CONFLICTS OF INTEREST: No potential conflict of interest relevant to this article was reported.
- Conception or design: S.J.M., J.H.K., S.H.K., C.S.S.
- Acquisition, analysis, or interpretation of data: S.J.M., J.H.K.
- Drafting the work or revising: S.J.M., J.H.K.
- Final approval of the manuscript: S.J.M., J.H.K., S.H.K., C.S.S.
SUPPLEMENTARY MATERIAL
Protein Expression Data and Differentially Expressed Protein Analysis
References
- 1.Allolio B, Fassnacht M. Clinical review: adrenocortical carcinoma: clinical update. J Clin Endocrinol Metab. 2006;91:2027–2037. doi: 10.1210/jc.2005-2639. [DOI] [PubMed] [Google Scholar]
- 2.Jouinot A, Bertherat J. Management of endocrine disease: adrenocortical carcinoma: differentiating the good from the poor prognosis tumors. Eur J Endocrinol. 2018;178:R215–R230. doi: 10.1530/EJE-18-0027. [DOI] [PubMed] [Google Scholar]
- 3.Else T, Kim AC, Sabolch A, Raymond VM, Kandathil A, Caoili EM, et al. Adrenocortical carcinoma. Endocr Rev. 2014;35:282–326. doi: 10.1210/er.2013-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLellis RA, Lloyd RV, Heitz PU, Eng C. Pathology and genetics of tumours of endocrine organs. 3rd ed. Lyon: IARC Press; 2004. [Google Scholar]
- 5.Fassnacht M, Johanssen S, Quinkler M, Bucsky P, Willenberg HS, Beuschlein F, et al. Limited prognostic value of the 2004 International Union Against Cancer staging classification for adrenocortical carcinoma: proposal for a revised TNM classification. Cancer. 2009;115:243–250. doi: 10.1002/cncr.24030. [DOI] [PubMed] [Google Scholar]
- 6.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th ed. Chicago: Springer; 2017. Chapter 76, Adrenal cortical carcinoma. [Google Scholar]
- 7.Beuschlein F, Weigel J, Saeger W, Kroiss M, Wild V, Daffara F, et al. Major prognostic role of Ki67 in localized adrenocortical carcinoma after complete resection. J Clin Endocrinol Metab. 2015;100:841–849. doi: 10.1210/jc.2014-3182. [DOI] [PubMed] [Google Scholar]
- 8.Papathomas TG, Pucci E, Giordano TJ, Lu H, Duregon E, Volante M, et al. An international Ki67 reproducibility study in adrenal cortical carcinoma. Am J Surg Pathol. 2016;40:569–576. doi: 10.1097/PAS.0000000000000574. [DOI] [PubMed] [Google Scholar]
- 9.Heaton JH, Wood MA, Kim AC, Lima LO, Barlaskar FM, Almeida MQ, et al. Progression to adrenocortical tumorigenesis in mice and humans through insulin-like growth factor 2 and β-catenin. Am J Pathol. 2012;181:1017–1033. doi: 10.1016/j.ajpath.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldmann J, Patsalis N, Fendrich V, Langer P, Saeger W, Chaloupka B, et al. Clinical impact of TP53 alterations in adrenocortical carcinomas. Langenbecks Arch Surg. 2012;397:209–216. doi: 10.1007/s00423-011-0868-6. [DOI] [PubMed] [Google Scholar]
- 11.Assie G, Letouze E, Fassnacht M, Jouinot A, Luscap W, Barreau O, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46:607–612. doi: 10.1038/ng.2953. [DOI] [PubMed] [Google Scholar]
- 12.Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016;29:723–736. doi: 10.1016/j.ccell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assie G, Jouinot A, Fassnacht M, Libe R, Garinet S, Jacob L, et al. Value of molecular classification for prognostic assessment of adrenocortical carcinoma. JAMA Oncol. 2019;5:1440–1447. doi: 10.1001/jamaoncol.2019.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach AE, Nucera C, Lam QT, Nguyen A, Dias-Santagata D, Sadow PM. Genomic and immunohistochemical analysis in human adrenal cortical neoplasia reveal beta-catenin mutations as potential prognostic biomarker. Discoveries (Craiova) 2015;3:e40. doi: 10.15190/d.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Urano T, Miki Y, Moriya T, Akahira J, Ishida T, et al. Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2007;98:644–651. doi: 10.1111/j.1349-7006.2007.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang A, Yoshimi N, Ino N, Tanaka T, Mori H. Overexpression of cyclin B1 in human colorectal cancers. J Cancer Res Clin Oncol. 1997;123:124–127. doi: 10.1007/BF01269891. [DOI] [PubMed] [Google Scholar]
- 19.Nozoe T, Korenaga D, Kabashima A, Ohga T, Saeki H, Sugimachi K. Significance of cyclin B1 expression as an independent prognostic indicator of patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:817–822. [PubMed] [Google Scholar]
- 20.Zhao M, Kim YT, Yoon BS, Kim SW, Kang MH, Kim SH, et al. Expression profiling of cyclin B1 and D1 in cervical carcinoma. Exp Oncol. 2006;28:44–48. [PubMed] [Google Scholar]
- 21.Egloff AM, Weissfeld J, Land SR, Finn OJ. Evaluation of anticyclin B1 serum antibody as a diagnostic and prognostic biomarker for lung cancer. Ann N Y Acad Sci. 2005;1062:29–40. doi: 10.1196/annals.1358.005. [DOI] [PubMed] [Google Scholar]
- 22.Androic I, Kramer A, Yan R, Rodel F, Gatje R, Kaufmann M, et al. Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer. 2008;8:391. doi: 10.1186/1471-2407-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerquetti L, Sampaoli C, Amendola D, Bucci B, Misiti S, Raza G, et al. Mitotane sensitizes adrenocortical cancer cells to ionizing radiations by involvement of the cyclin B1/CDK complex in G2 arrest and mismatch repair enzymes modulation. Int J Oncol. 2010;37:493–501. doi: 10.3892/ijo_00000698. [DOI] [PubMed] [Google Scholar]
- 24.Aisen P. Transferrin receptor 1. Int J Biochem Cell Biol. 2004;36:2137–2143. doi: 10.1016/j.biocel.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Rychtarcikova Z, Lettlova S, Tomkova V, Korenkova V, Langerova L, Simonova E, et al. Tumor-initiating cells of breast and prostate origin show alterations in the expression of genes related to iron metabolism. Oncotarget. 2017;8:6376–6398. doi: 10.18632/oncotarget.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horniblow RD, Bedford M, Hollingworth R, Evans S, Sutton E, Lal N, et al. BRAF mutations are associated with increased iron regulatory protein-2 expression in colorectal tumorigenesis. Cancer Sci. 2017;108:1135–1143. doi: 10.1111/cas.13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kindrat I, Tryndyak V, de Conti A, Shpyleva S, Mudalige TK, Kobets T, et al. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis. Oncotarget. 2016;7:1276–1287. doi: 10.18632/oncotarget.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, Zhang J, Song F, Tian M, Shi B, Jiang H, et al. EGFR regulates iron homeostasis to promote cancer growth through redistribution of transferrin receptor 1. Cancer Lett. 2016;381:331–340. doi: 10.1016/j.canlet.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Rosager AM, Sorensen MD, Dahlrot RH, Hansen S, Schonberg DL, Rich JN, et al. Transferrin receptor-1 and ferritin heavy and light chains in astrocytic brain tumors: expression and prognostic value. PLoS One. 2017;12:e0182954. doi: 10.1371/journal.pone.0182954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017;36:4089–4099. doi: 10.1038/onc.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Z, Wang H, Xia J, Yang Y, Jin Z, Xu H, et al. Decreased ferroportin promotes myeloma cell growth and osteoclast differentiation. Cancer Res. 2015;75:2211–2221. doi: 10.1158/0008-5472.CAN-14-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jian J, Yang Q, Huang X. Src regulates Tyr(20) phosphorylation of transferrin receptor-1 and potentiates breast cancer cell survival. J Biol Chem. 2011;286:35708–35715. doi: 10.1074/jbc.M111.271585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong SM, Hwang S, Seong RH. Transferrin receptor regulates pancreatic cancer growth by modulating mitochondrial respiration and ROS generation. Biochem Biophys Res Commun. 2016;471:373–379. doi: 10.1016/j.bbrc.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 34.Yang C, Ma X, Wang Z, Zeng X, Hu Z, Ye Z, et al. Curcumin induces apoptosis and protective autophagy in castration-resistant prostate cancer cells through iron chelation. Drug Des Devel Ther. 2017;11:431–439. doi: 10.2147/DDDT.S126964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callens C, Moura IC, Lepelletier Y, Coulon S, Renand A, Dussiot M, et al. Recent advances in adult T-cell leukemia therapy: focus on a new anti-transferrin receptor monoclonal antibody. Leukemia. 2008;22:42–48. doi: 10.1038/sj.leu.2404958. [DOI] [PubMed] [Google Scholar]
- 36.Shimosaki S, Nakahata S, Ichikawa T, Kitanaka A, Kameda T, Hidaka T, et al. Development of a complete human IgG monoclonal antibody to transferrin receptor 1 targeted for adult T-cell leukemia/lymphoma. Biochem Biophys Res Commun. 2017;485:144–151. doi: 10.1016/j.bbrc.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 37.Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp Hematol. 2009;37:245–255. doi: 10.1016/j.exphem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115(Pt 20):3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 39.Nam JM, Onodera Y, Bissell MJ, Park CC. Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer Res. 2010;70:5238–5248. doi: 10.1158/0008-5472.CAN-09-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitra AK, Sawada K, Tiwari P, Mui K, Gwin K, Lengyel E. Ligand-independent activation of c-Met by fibronectin and α(5)β(1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30:1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Z, Zhou Z, Shi X, Wang J, Wu X, Sun D, et al. EDB fibronectin specific peptide for prostate cancer targeting. Bioconjug Chem. 2015;26:830–838. doi: 10.1021/acs.bioconjchem.5b00178. [DOI] [PubMed] [Google Scholar]
- 42.Meng XN, Jin Y, Yu Y, Bai J, Liu GY, Zhu J, et al. Characterisation of fibronectin-mediated FAK signalling pathways in lung cancer cell migration and invasion. Br J Cancer. 2009;101:327–334. doi: 10.1038/sj.bjc.6605154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei PL, Kuo LJ, Huang MT, Ting WC, Ho YS, Wang W, et al. Nicotine enhances colon cancer cell migration by induction of fibronectin. Ann Surg Oncol. 2011;18:1782–1790. doi: 10.1245/s10434-010-1504-3. [DOI] [PubMed] [Google Scholar]
- 44.Eke I, Storch K, Krause M, Cordes N. Cetuximab attenuates its cytotoxic and radiosensitizing potential by inducing fibronectin biosynthesis. Cancer Res. 2013;73:5869–5879. doi: 10.1158/0008-5472.CAN-13-0344. [DOI] [PubMed] [Google Scholar]
- 45.Pontiggia O, Sampayo R, Raffo D, Motter A, Xu R, Bissell MJ, et al. The tumor microenvironment modulates tamoxifen resistance in breast cancer: a role for soluble stromal factors and fibronectin through β1 integrin. Breast Cancer Res Treat. 2012;133:459–471. doi: 10.1007/s10549-011-1766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francois RA, Maeng K, Nawab A, Kaye FJ, Hochwald SN, Zajac-Kaye M. Targeting focal adhesion kinase and resistance to mTOR inhibition in pancreatic neuroendocrine tumors. J Natl Cancer Inst. 2015;107:djv123. doi: 10.1093/jnci/djv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hielscher A, Ellis K, Qiu C, Porterfield J, Gerecht S. Fibronectin deposition participates in extracellular matrix assembly and vascular morphogenesis. PLoS One. 2016;11:e0147600. doi: 10.1371/journal.pone.0147600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez MV, Rivera AA, Viale DL, Benedetti L, Cuneo N, Kimball KJ, et al. A tumor-stroma targeted oncolytic adenovirus replicated in human ovary cancer samples and inhibited growth of disseminated solid tumors in mice. Mol Ther. 2012;20:2222–2233. doi: 10.1038/mt.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein Expression Data and Differentially Expressed Protein Analysis