Graphical abstract
Keywords: Protein stability change, Integrated prediction, Machine learning
Abstract
Protein mutations can lead to structural changes that affect protein function and result in disease occurrence. In protein engineering, drug design or and optimization industries, mutations are often used to improve protein stability or to change protein properties while maintaining stability. To provide possible candidates for novel protein design, several computational tools for predicting protein stability changes have been developed. Although many prediction tools are available, each tool employs different algorithms and features. This can produce conflicting prediction results that make it difficult for users to decide upon the correct protein design. Therefore, this study proposes an integrated prediction tool, iStable 2.0, which integrates 11 sequence-based and structure-based prediction tools by machine learning and adds protein sequence information as features. Three coding modules are designed for the system, an Online Server Module, a Stand-alone Module and a Sequence Coding Module, to improve the prediction performance of the previous version of the system. The final integrated structure-based classification model has a higher Matthews correlation coefficient than that of the single prediction tool (0.708 vs 0.547, respectively), and the Pearson correlation coefficient of the regression model likewise improves from 0.669 to 0.714. The sequence-based model not only successfully integrates off-the-shelf predictors but also improves the Matthews correlation coefficient of the best single prediction tool by at least 0.161, which is better than the individual structure-based prediction tools. In addition, both the Sequence Coding Module and the Stand-alone Module maintain performance with only a 5% decrease of the Matthews correlation coefficient when the integrated online tools are unavailable. iStable 2.0 is available at http://ncblab.nchu.edu.tw/iStable2.
1. Introduction
When the amino acid of a protein is changed, it may affect the structural stability, hydrogen bonding, activity, etc., of the protein and then may affect protein function and may even cause disease [1], [2], [3]. In protein engineering, drug design, and the optimization of industrial processes, mutations are often used to increase protein stability or maintain its stability while altering protein properties [4], [5], [6]. Single amino acid mutation could change the structural stability of a protein by making a smaller free energy change (ΔG, or dG) after folding, while the difference in folding free energy change between wild type and mutant protein (ΔΔG, or ddG) is often considered as an impact factor of protein stability changes [7], [8], [9]. Many studies use computer science, mathematics, statistics and other methods to make predictions [10]. The main methods are divided into four categories: (a) physical potential, which investigates molecular dynamics to simulate protein folding trajectories and obtain accurate structures and relative free energy levels [11], [12]; (b) statistical potential, where statistical analysis is performed on the substitutions, occurrences and pairing frequencies of the 20 amino acid to make predictions from the database of known protein structures [13], [14], [15], [16]; (c) empirical potential, which uses physical energy terms, statistical energy terms and structural descriptors for the difference of free energy [17], [18], [19]; and (d) machine learning methods, such as support vector machine (SVM), neural network (NN), decision tree, and random forest (RF) and which are currently used to solve bioinformatics problems, could be used to make predictions by extensive training of relevant data in the field [20], [21], [22], [23]. The features used with machine learning in different research fields will be different, but are not restricted by knowledge of any one particular research field.
Many studies have used machine learning to construct prediction models for the effects of single point mutations on protein stability. The features used can be mainly divided into two types based on the prediction methods that employ them: (i) Sequence-based methods consider point mutations and upstream and downstream amino acids. Then, the features and physiochemical properties of amino acids, the position-specific scoring matrix (PSSM) and other features are extracted to construct prediction models. This type of tool usually takes a protein sequence as input and include examples such as I-Mutant2.0 [24], MUpro [21], iPTREE-STAB [25], INPS [26] and EASE-MM [27]; (ii) Structure-based methods use 3D protein-structure information to extract features such as secondary structures, chemical composition, and interatomic interactions. These tools usually take a 3D protein structure (PDB) as input and include examples such as I-Mutant2.0, CUPSAT [28], PoPMuSiC [29], [30], [31], SDM [32], [33], mCSM [34], MAESTRO [35] and AUTO-MUTE2.0 [36], [37]. However, prediction models that use structure-based methods typically perform better than sequence-based methods.
Each tool performs its predictions with different characteristics and algorithms, and consequently the respective performances are also different. The same input may have conflicting results across different tools, so it would be difficult for the user to decide upon the correct protein design. If the outputs of the prediction tools are integrated through machine learning methods, the user is provided with a higher accuracy prediction than through the use of a single tool, which may alleviate the user's potential concerns. Integrated methods have been successfully applied in other fields, such as in predicting miRNA binding sites with mirMeta, which integrates five different tools [38], as well as in predicting protein-protein interactions [39] and phosphorylation sites [40]. However, there are only a few integrated tools for predicting protein stability changes, such as DUET [41], which integrates the prediction tools mCSM and SDM developed by their own team and only makes integrated predictions for structural information. iStable [9] can use sequence or structure information as inputs for prediction and integrates the results from five prediction tools.
iStable integrates tools developed by different teams to combine a greater number of diverse predicting and feature coding methods. However, it is difficult to integrate tools from different sources. The inputs and outputs of the tools don't have uniform formats, execution time of prediction tools and operating system of stand-alone functionality are different. In addition, if the integrated predicting tool provides services through a web server, our system may encounter unpredictable situations. For example, if the website interface changes or the web server or Internet experiences connectivity issues, the prediction results from Internet-connected tools cannot be obtained, which would affect the performance of iStable. In terms of the predictive model, it can be seen from the literature that iStable focuses on training and testing classification models and comparing the results with other tools [9]. For regression models, the prediction of ddG is weak, even without the integration of sequence-based tools. To improve the above shortcomings, we propose a new system architecture and classification and regression model; the improved system is named iStable 2.0.
In this study, we selected 11 prediction tools based on execution time for integration: the sequence-based tools I-Mutant2.0, MUpro and iPTREE-STAB and the structure-based tools I-Mutant2.0, CUPSAT, PoPMuSiC, AUTO-MUTE2.0, SDM, DUET, mCSM, MAESTRO and SDM2 [33]. The results of the prediction tools are integrated through a machine learning approach, which constructs classification and regression models with structure-based and sequence-based inputs. The system is divided into three modules: Online Server Module (OSM), Stand-alone Module (SAM), and Sequence Coding Module (SCM). The Online Server Module is responsible for obtaining the results of the prediction tools from a web server. The Stand-alone Module is responsible for sending commands and obtaining the results from stand-alone prediction tools. The Sequence Coding Module is responsible for extracting features from protein sequences and performing encoding. With three module integration, iStable 2.0 can improve the MCC (Matthews correlation coefficient) of single structure-based prediction tools from 0.547 to 0.708, and the PCC (Pearson correlation coefficient) of the regression model can be increased from 0.669 to 0.714. The MCC of the sequence-based classification model was 0.652, and the regression model PCC was 0.695, which are better than those of the individual structure-based prediction tools. Without the Internet-dependent OSM, the MCC calculated from use of only the SAM and SCM models drops to only 5% of that from the operation of the three modules. From the feature analysis, it is found that the importance of SCM is increased without OSM. Therefore, iStable 2.0 provides predictions with stable performance. The iStable 2.0 web server is freely available at http://ncblab.nchu.edu.tw/iStable2.
2. Materials and methods
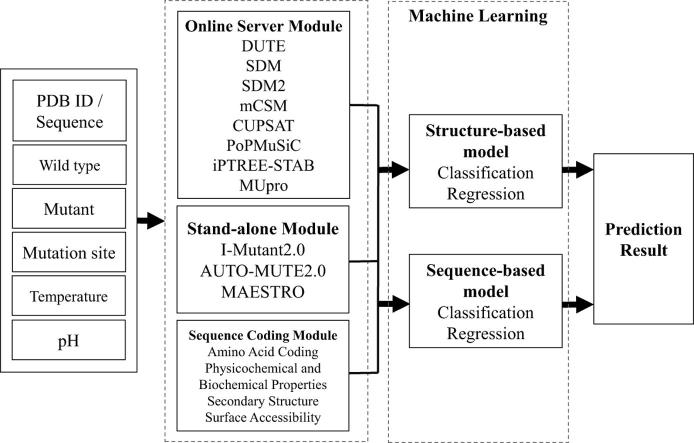
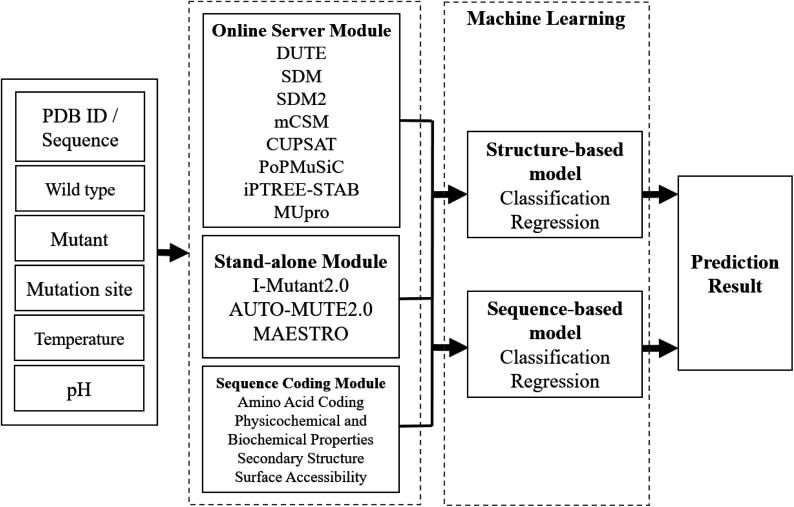
We propose the system architecture shown in Fig. 1. These features were obtained from the predicted results of the integrated tools when they were input the information about the protein mutation point. The system provides two types of models, one that uses protein structure as the input and the other that uses protein sequences, constructs binary classification models that predict stability and instability after mutation, and uses regression models to predict ddG after mutation.
Fig. 1.
System architecture.
2.1. Compilation of single-mutation datasets
The training data set was obtained from I-Mutant [24] and PoPMuSiC [30]; I-Mutant contained 1948 mutation sites in 58 proteins, and PoPMuSiC contained 2648 mutation sites in 131 proteins. The combined dataset contained a total of 4596 single point mutations, which had been obtained as experimental results in protein stability change experiments with environmental conditions such as pH and temperature. To evaluate the performance of the prediction model and compare it with other methods, we prepared an independent test set from data obtained from ProTherm (thermodynamic database for proteins and mutants, last updated in 2013) [42], containing 2869 single point mutations in 81 proteins. Each single point mutation from I-Mutant, PoPMuSiC, and ProTherm has five types of information: 1) protein data bank (PDB) ID, which contains 3D structure information for proteins, 2) the site of the mutation position and residue with the native and mutant proteins, 3) the temperature used in the experiment, 4) the pH used in the experiment, and 5) the free energy change between the wild-type and mutant protein (ΔΔG, or ddG). All the data with the same information from type 1 to type 5 is defined redundancy, and which is defined contradiction with the same information from type 1 to type4. However, the training and independent test data had redundant and contradictory data that could have led to biases in training and evaluation. After removing the contradictory and redundant data, the training data set contained 3568 mutations and was named S3568. The independent test data set ultimately contained 1372 mutations. To prevent the model from learning with the independent test samples in the training stage, we removed data from the independent test set that was found in the training test set. Finally, 630 mutations remained, and the resulting independent test set was named S630.
2.1.1. Definitions of positive and negative data
To construct the classification model, we defined stabilizing data with a ddG value ≥0 as positive (+) and destabilizing data with a ddG value less than 0 as negative (−). Because PopMuSiC’s data used the opposite definition to the above, we inverted the signs of the ddG values for its 2648 mutation sites.
2.1.2. Correction of sequence information
Because the position of the mutation point is derived from the absolute position of the PDB, the prediction tools that use the sequence as input will produce errors. Therefore, we corrected the position of the mutation site with the relative position such that the first amino acid of the sequence obtained from PDB was defined as position 1.
2.2. Selection strategy for integration of prediction tools
We chose single mutation-point protein predicting tools and an execution time of less than 5 min for integration. If the tool has web server and stand-alone functionality, we may prioritize integration of the stand-alone functionality. Table S1 lists the prediction tools published between 2005 and 2018. This study integrated I-Mutant2.0, MUpro, and iPTREE-STAB to construct the protein sequence-based prediction model, and the additional9 prediction tools, including I-Mutant2.0, CUPSAT, PoPMuSiC, AUTO-MUTE2.0, SDM, DUET, mCSM, MAESTRO and SDM2, were used to construct the structure-based prediction model. INPS, EASE-MM, ELASPIC [43], TopologyNet [44], TML [45], and DynaMut [46] were also investigated, but each required between five minutes and several hours of execution time, so they were not included in the integration.
2.3. Feature encoding
This study divides feature encoding into two types: prediction tool results and protein sequence information. The prediction tool results refer to the encoding of the output of the 11 prediction tools as a vector of features. The protein sequence information contains the amino acid composition surrounding the mutation site and the physicochemical properties and secondary structure of the protein. The integrated prediction tools already contain the relevant features for using protein sequence information; however, to maintain our prediction tool reliability when the integrated online tools are not available, the protein sequence information would still be needed in order to ensure prediction performance.
2.3.1. Predictor result features
We encoded the results of the prediction tools that were integrated into the system. The encoding method for nondigital values is shown below. A decrease of free energy change (ddG) in the value is encoded as 0, and an increase of free energy change (ddG) is encoded as 1. Other numerical values, such as ddG, predictive confidence score and RSA (relative surface accessibility), are used directly. The details of the features for each prediction tool are shown in Tables S2 and S3.
2.3.2. Sequence coding features
Four types of features were extracted from the protein sequence, including concerning the mutation point and the 9 amino acids up- and downstream of the site. Because no direct structural information for the protein sequences were available, we obtained the predicted surface accessibility and secondary structure through NetSurfP [47].
2.3.2.1. Amino acid coding (AAC)
We use one-hot encoding to represent the 20 amino acids as one 20-dimensional vector each, and gaps are represented as a 20-zero value vector. The encoding is shown in Table S4. We encoded the mutant and wild-type amino acids and the 9 upstream and downstream amino acids in this manner.
2.3.2.2. Physicochemical and biochemical properties (PBP)
We encoded the physicochemical and biochemical properties of the amino acids using the scheme proposed by William et al. [48] and Mathura et al. [49] for a total of 10 properties: Polarity, Secondary structure, Molecular size or volume, Codon diversity, Electrostatic charge, Hydrophobicity, Side chain length, α-helix propensity, Number of codons, and β-strand propensity. The detailed values of this encoding are shown in Tables S5 and S6. An amino acid is represented by 10 values, and we encoded the mutant and wild-type amino acids and the 9 upstream and downstream amino acids in this manner. We also calculated the numerical differences between the attributes of the wild-type and mutant amino acids.
2.3.2.3. Surface accessibility (SA)
Solvent accessibility is related to the spatial arrangement of the amino acids during the process of protein folding [50]. It has proven useful for protein–protein interactions, fold recognition [51], intrinsic disorder [52], DNA-binding protein prediction [53], protein-ligand binding sites prediction [54], [55] and so on. We encoded Buried as 00001 and Exposed as 00100; we also encoded the values of the relative surface accessibility (RSA), the absolute surface accessibility (ASA), and the Z-score for the elative surface accessibility and calculated the frequencies of Buried and Exposed in the protein sequence. Information on the wild-type amino acids and the 9 upstream and downstream amino acids was encoded in this manner.
2.3.2.4. Secondary structure (SS)
From NetSurfP, the probability of the amino acid being in an α-helix, β-strand or Coil were obtained. We used the one-hot encoding method to encode the highest value among the three to the structure: H (α-helix) was encoded to 010, S (β-strand) was encoded to 100, and C (Coil) was encoded to 001. Next, we calculated the frequency of the three secondary structures in the entire protein. The wild-type amino acids and the 9 amino acids up- and downstream were encoded in manner.
2.4. Model generation
According to the input protein information, we divided the prediction model into a structure-based model and a sequence-based model and constructed classification and regression models for each. The structure-based model only integrates the tools that use protein sequences and structures as input. The sequence-based model only integrates the tools that use protein sequences as input. Both models use our prepared sequence features. We used Weka [56] and XGBoost [57] to construct the classification and regression models and the training S3568 data with 10-fold cross-validation to determine the best parameters for the training model. Finally, we used the independent test data S630 to compare the results with the different tools.
2.5. Model evaluation
Correct predictions of positive and negative data have different meanings because the effects of mutations are not always detrimental to protein function. One of the purposes of predicting protein stability changes is to identify the mechanisms of structural stability change upon single amino acid mutation; another goal is to apply this knowledge to protein design to modify proteins into more stable and thermal-tolerant forms. Since it is equally important to understand the mechanisms underlying stabilizing and destabilizing mutations, we expect an integrated predictor to make correct predictions in both cases. Since the minority result could be the right answer, we want to prove that iStable 2.0, with training, would know right from wrong and not just pick the majority answer. Accuracy (Acc), sensitivity (Sn), specificity (Sp), and the Matthews correlation coefficient (MCC) were used to evaluate the predictive ability of each system, calculated as follows:
and
where TP, FP, FN and TN are the true positives, false positives, false negatives, and true negatives, respectively. Sn and Sp represent the ratio of true positives to the number of all correctly classified items and of true negatives to the number of all incorrectly classified items, respectively. Acc is the overall accuracy of prediction, and the MCC is a measure of the quality of the classifications, whose value may range between −1 (an inverse prediction) and +1 (a perfect prediction), with 0 denoting a random prediction [9].
2.6. Description of system modules
As shown in Fig. 1, the process of transferring the input information to the structure-based model and sequence-based model is divided among three modules: Online Server Module (OSM), Sequence Coding Module (SCM), and Stand-alone Module (SAM). Online Server Module (OSM): This module handles prediction tools that operate using web server functionality and have an execution time of no >5 min. The prediction tools CUPSAT, PoPMuSiC, SDM, DUET, mCSM, SDM2, MUpro and iPTREE-STAB are classified into this module. Stand-alone Module (SAM): This module handles stand-alone prediction tools, that is, those that do not require web access. The prediction tools in this group are AUTO-MUTE2.0, MAESTRO and I-Mutant2.0. Sequence Coding Module (SCM): This module handles protein sequence coding, including one-hot encoding of the amino acids and the physicochemical and biochemical properties of the amino acids and of the secondary structure and solvent accessibility information from NetSurfP. Stand-alone Module & Sequence Coding Module: The Online Server Module depends on the Internet; the servers linked to the tools in the OSM are required for their operation. If the OSM module fails due to a lack of Internet access, the system's performance will be affected. This study solves this problem through the SAM and the SCM. The two modules will work on if the OSM module is not functioning.
3. Results and discussion
3.1. Performance of machine learning algorithms in the training set
To select the appropriate algorithm for predicting sequence and structure point mutations, we used the S3568 training data to evaluate the performance of 85 classifiers and 39 regression methods with 10-fold cross-validation. Tables S8 and S9 show the top 10 methods sorted by performance. XGBoost is the best performing method in the structure-based classification model, with an Sn of 0.758, an Sp of 0.964, an Acc of 0.908 and an MCC of 0.758. XGBoost is also the best performing method in the sequence-based classification model, with an Sn of 0.670, an Sp of 0.953, an Acc of 0.877, and an MCC of 0.672, which. The PCC of XGBoost reached 0.864 and 0.818, having the best performance in the structure-based and sequence-based regression models, respectively. Finally, we constructed four prediction models with the XGBoost algorithm.
3.2. Training set performance
3.2.1. Evaluation of the structure-based classification model
Table 1 shows the performance of iStable2.0_PDB and the integrated structure-based tools in training data S3568. iStable2.0_PDB has an MCC of 0.758 on training set S3568, outperforming all other integrated prediction tools. A ranking of feature importance (Table S9) shows that AUTO-TUTE2.0_RF/TR, which have the best performance with S3568, rank third and fourth. The performance of MAESTRO in the S3568 data set ranks 11th among the 14 methods; however, it is ranked first and second in terms of feature importance. Therefore, we nevertheless chose to integrate tools that had low performance because the performance could be complemented through machine learning. The three sequence-based tools, I-Mutant2.0_SEQ, iPTREE-STAB and MUpro, are also no less important than the structure-based tools.
Table 1.
Performance of the classifier and regression models with structure-based and sequence-based tools with S3568.
| Model | Method | Classification |
Regression | |||
|---|---|---|---|---|---|---|
| Sn | Sp | Acc | MCC | PCC | ||
| Structure-based | iStable2.0_PDB | 0.758 | 0.964 | 0.908 | 0.758 | 0.864 |
| DUET | 0.499 | 0.891 | 0.787 | 0.421 | 0.655 | |
| SDM | 0.566 | 0.748 | 0.700 | 0.293 | 0.474 | |
| SDM2 | 0.562 | 0.744 | 0.696 | 0.286 | 0.485 | |
| mCSM | 0.298 | 0.955 | 0.781 | 0.354 | 0.638 | |
| CUPSAT | 0.513 | 0.798 | 0.723 | 0.305 | 0.188 | |
| I-Mutant2.0_PDB | 0.650 | 0.922 | 0.850 | 0.601 | 0.689 | |
| PoPMuSiC | 0.333 | 0.936 | 0.776 | 0.347 | 0.626 | |
| AUTO-MUTE2.0_SVM | 0.802 | 0.848 | 0.840 | 0.560 | 0.716 | |
| AUTO-MUTE2.0_RF/TR | 0.604 | 0.979 | 0.879 | 0.676 | 0.725 | |
| MAESTRO | 0.457 | 0.850 | 0.746 | 0.322 | 0.566 | |
| Sequence-based | iStable2.0_SEQ | 0.670 | 0.953 | 0.877 | 0.672 | 0.820 |
| iPTREE-STAB | 0.537 | 0.945 | 0.837 | 0.550 | 0.484 | |
| I-Mutant2.0_SEQ | 0.565 | 0.919 | 0.825 | 0.525 | 0.625 | |
| MUpro_SVM | 0.599 | 0.906 | 0.825 | 0.531 | – | |
| MUpro_NN | 0.536 | 0.924 | 0.821 | 0.509 | – | |
3.2.2. Evaluation of the sequence-based classification model
Table 1 shows the performance of iStable2.0_SEQ and the integrated sequence-based tools in training data S3568. The integration of iStable2.0_SEQ results in greater values of Sn and Sp compared to those obtained from integrating iPTREE-STAB, I-Mutant2.0_SEQ and MUpro. From the feature importance rankings shown in Table S10, it can be seen that the importance of ddG from tools I-Mutant2.0_SEQ and iPTREE-STAB rank third and fourth, respectively. It is worth mentioning that ddG has a higher importance in the classification model, and the difference in the physicochemical properties of the wild-type and mutant amino acids is of great importance.
3.2.3. Evaluation of structure-based regression model
Table 1 also shows the performance of the structure-based regression models in training data S3568. The maximum PCC of AUTO-MUTE2.0_TR is 0.725 with S3568, while integration of iStable2.0_PDB in the regression model results in a PCC of 0.864, a difference of 0.139. The ddG from tools iPTREE-STAB, MAESTRO, DUET and mCSM is shown in Table S11 to be of high importance when iStable2.0_PDB is integrated in the regression model; these tools were not integrated in the previous version of iStable and may be used to improve prediction performance.
3.2.4. Evaluation of the sequence-based regression model
As shown in Table 1, iStable2.0_SEQ_Regression has a PCC of 0.820 with S3568, which is better than the PCC of 0.625 with I-Mutant2.0_SEQ. In Table S12, I-Mutant2.0_SEQ and the ddG of iPTREE-STAB rank first and second in terms of feature importance. In addition, the difference in the hydrophobicity property of the wild-type and mutant amino acids is also an important feature.
3.3. Performance of the classification model with and without the OSM module
The performance of the prediction models with and without the features of the OSM module is shown in Table 2. iStable2.0_PBD_SASC's MCC is 0.747 with the training data set, which is higher than the MCC of the best integrated tool, AUTO-TUTE2.0_RF (0.676). The difference in the MCCs of iStable2.0_PBD_SASC and iStable2.0_PBD was 0.011 when the model was trained in S3568 and 0.019 when the model was tested with S630. iStable2.0_PBD_SASC's MCC is 0.689 with independence test set. The sequence classification model, iStable2.0_SEQ_SASC, has an MCC of 0.625 with training set S3568 and 0.603 with independent test set S630. The difference in the MCCs between iStable2.0_SEQ_SASC and iStable2.0_SEQ with training set S3568 is 0.047, and the MCC of iStable2.0_SEQ_SASC is reduced by 0.049 with independent test S630. In Figs. S1 and S2, the SCM module with iStable2.0_PBD uses 400 of the 883 features, and the SCM module iStable2.0_PBD_SASC uses 442. The feature with the greatest increase is AAC feature, while the MCC is only reduced by 0.011. From Table S17, there are 883 features in the SCM module with iStable2.0_SEQ. iStable2.0_SEQ uses 508 features, and the number of features used in iStable2.0_SEQ_SASC is increased to 638, most of which are AAC features. In addition, iStable2.0_SEQ only integrates i-Mutant2.0_SEQ, but it can nevertheless maintain accuracy. Table S15 shows the feature importance ranking for features with an f-score >100. The number of important features of iStable2.0_SEQ_SASC is greater than that of iStable2.0_SEQ.
Table 2.
Impact evaluation of OSM for prediction performance from the classification model.
| Model | Method | S3568 |
S630 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sn | Sp | Acc | MCC | Sn | Sp | Acc | MCC | |||
| Structure-based | iStable2.0_PDB | 0.758 | 0.964 | 0.908 | 0.758 | 0.718 | 0.953 | 0.892 | 0.708 | |
| iStable2.0_PBD_SASC* | 0.740 | 0.964 | 0.904 | 0.747 | 0.687 | 0.955 | 0.886 | 0.689 | ||
| Sequence-based | iStable2.0_SEQ | 0.670 | 0.953 | 0.877 | 0.672 | 0.644 | 0.953 | 0.873 | 0.652 | |
| iStable2.0_SEQ_SASC* | 0.640 | 0.941 | 0.861 | 0.625 | 0.620 | 0.938 | 0.856 | 0.603 | ||
SASC: indicate the models used SAM and SCM only.
3.4. Performance of the regression model with and without the OSM module
As shown in Table 3, the PCC of iStable2.0_PDB_Regression_SASC is only reduced by 0.05 (the PCC of iStable2.0_PDB_Regression is 0.714) when only integrating AUTO-MUTE2.0, I-Mutant2.0 and MAESTRO with S630. The PCC of iStable2.0_SEQ_Regression_SASC decreased by 0.044 (the PCC of iStable2.0_SEQ_Regression is 0.695) without iPTREE-STAB and MUpro. Tables S15 and S16 show that predicting ddG based on sequence or structure is a more difficult problem, so the number of important features required will been increasing when OSM module does not work. In addition, Figs. S3 and S4 shows that iStable2.0_PDB_Regression uses 461 of the 883 features in the SCM module, while iStable2.0_PDB_Regression_SASC uses 647. Table S17 shows that iStable2.0_SEQ_Regression uses 452 of the 883 features in the SCM, while the number of features used by iStable2.0_SEQ_Regression_SASC has increased to 618.
Table 3.
Impact evaluation of OSM for prediction performance from the regression model.
| Model | Method | S3568 | S630 |
|---|---|---|---|
| PCC | PCC | ||
| Structure-based | iStable2.0_PDB_Regression | 0.864 | 0.714 |
| iStable2.0_PDB_Regression_SASC* | 0.861 | 0.709 | |
| Sequence-based | iStable2.0_SEQ_Regression | 0.820 | 0.695 |
| iStable2.0_SEQ_Regression_SASC* | 0.821 | 0.651 |
SASC: indicate the models used SAM and SCM only.
3.5. Independent test performance
Table 4 shows the comparison of the performance of the iStable 2.0 classification model with that of the individual methods with independent test S630. The performance of iStable based on each integration tool has prediction results if some of the tools absent could decrease iStable performance. The results for SDM and mCSM were obtained from DUET. The highest Sn for predicting stability after a mutation is obtained with SDM (0.620) among the individual integrated tools, which can be improved to 0.718 through integration. S630 is an imbalanced data set containing 143 stable and 467 unstable data, so we used the MCC since it could objectively evaluate performance, unlike the ACC. The accuracy of iStable 2.0 based on the integrated strategy is better than that of I-Mutant2.0_PDB, which has the highest accuracy among the single method tools. The MCC of the former and latter are 0.708 and 0.547, respectively; meanwhile, if the number of positive and negative voting meets even, then the prediction result is set to stable, Majority Voting only can reach a MCC of 0.493 (Table S18). In addition, we compared the performance of the previous version with that of the current version and the MCC increased by 0.068. Among the sequence-based tools, the best MCC is obtained with I-Mutant2.0_SEQ (0.491). After integration in iStable 2.0, the MCC increases to 0.652, which is also superior to the performances of the unconsolidated tools, such as EASE-MM and INPS.
Table 4.
Performance of classifiers and regression with structure-based and sequence-based models with S630.
| Model | Tool | Classification |
Regression | |||
|---|---|---|---|---|---|---|
| Sn | Sp | Acc | MCC | PCC | ||
| Structure-based | iStable2.0_PDB | 0.718 | 0.953 | 0.892 | 0.708 | 0.714 |
| iStable_PDB | 0.744 | 0.901 | 0.860 | 0.640 | 0.665 | |
| DUET | 0.405 | 0.906 | 0.776 | 0.358 | 0.458 | |
| SDM | 0.620 | 0.392 | 0.451 | 0.010 | 0.349 | |
| SDM2 | 0.497 | 0.771 | 0.700 | 0.256 | 0.352 | |
| mCSM | 0.239 | 0.953 | 0.768 | 0.285 | 0.447 | |
| CUPSAT | 0.442 | 0.82 | 0.722 | 0.266 | 0.274 | |
| I-Mutant2.0_PDB | 0.571 | 0.929 | 0.837 | 0.547 | 0.669 | |
| PoPMuSiC | 0.344 | 0.901 | 0.757 | 0.291 | 0.424 | |
| AUTO-MUTE2.0_SVM | 0.245 | 0.981 | 0.790 | 0.370 | 0.520 | |
| AUTO-MUTE2.0_RF/TR | 0.350 | 0.981 | 0.817 | 0.473 | 0.534 | |
| MAESTRO | 0.417 | 0.807 | 0.706 | 0.227 | 0.329 | |
| Sequence-based | iStable2.0_SEQ | 0.644 | 0.953 | 0.873 | 0.652 | 0.695 |
| iStable_SEQ | 0.702 | 0.903 | 0.849 | 0.611 | – | |
| iPTREE-STAB | 0.350 | 0.970 | 0.810 | 0.443 | 0.496 | |
| I-Mutant2.0_SEQ | 0.509 | 0.927 | 0.819 | 0.491 | 0.546 | |
| MUpro_SVM | 0.264 | 0.923 | 0.752 | 0.247 | – | |
| MUpro_NN | 0.245 | 0.934 | 0.756 | 0.248 | – | |
| EASE-MM | 0.693 | 0.732 | 0.722 | 0.384 | 0.541 | |
| INPS | 0.472 | 0.857 | 0.757 | 0.343 | 0.449 | |
The best PCC for predicting ddG among the structure-based regression models is 0.669 for I-Mutant2.0_PDB. The PCC of iStable 2.0 increased by 0.045 to 0.714, which was also 0.049 higher than the previous version. Among the sequence-based tools, the best PCC is obtained with I-Mutant2.0_SEQ (0.546). After integration in iStable 2.0, the PCC increases to 0.695, which is also superior to those of the unconsolidated tools, such as EASE-MM and INPS.
3.6. Performance of different thresholds
To explore the feasibility of the regression prediction, which is replaced by the classification model, we converted the predicted ddG value into a binary classification of stable or unstable and compared the resulting accuracy with the results from the classification model. The conversion rules are as follows. We defined the prediction result as stable when ddGpred ≧ threshold and as unstable when ddGpred < threshold, where threshold = {−0.5, −0.1, 0, 0.1, 0.5}. Table 5 shows the performance of the regression model conversion and that of the classification for the five thresholds. Using the threshold converts the structure-based and sequence-based regression models to classification models, which biases the performance for the prediction results. The Sp is highest when the threshold value is set to 0.5, and the highest Sn is obtained with the threshold set to −0.5. The balanced performance with structure-based and sequence-based regression model at threshold of zero, but their MCC of regression model with thresholds at zero are not superior to the classification model. Therefore, the classifier model is more accurate than the regression model in solving classification problem, and so the final classification model is used in our prediction system, and the performance of ROC curve was shown in Fig. S5.
Table 5.
Comparison of the performance of models with different thresholds tested with S630.
| Model | Method | Sn | Sp | Acc | MCC |
|---|---|---|---|---|---|
| Structure-based | iStable2.0_PDB | 0.718 | 0.953 | 0.892 | 0.708 |
| Threshold: 0.5 | 0.451 | 0.960 | 0.894 | 0.475 | |
| Threshold: 0.1 | 0.601 | 0.953 | 0.876 | 0.613 | |
| Threshold: 0 | 0.577 | 0.949 | 0.852 | 0.590 | |
| Threshold:−0.1 | 0.608 | 0.932 | 0.841 | 0.585 | |
| Threshold: −0.5 | 0.684 | 0.887 | 0.802 | 0.590 | |
| Sequence-based | iStable2.0_PDB | 0.644 | 0.953 | 0.873 | 0.652 |
| Threshold: 0.5 | 0.476 | 0.951 | 0.889 | 0.468 | |
| Threshold: 0.1 | 0.601 | 0.945 | 0.870 | 0.595 | |
| Threshold: 0 | 0.613 | 0.942 | 0.857 | 0.607 | |
| Threshold: −0.1 | 0.593 | 0.929 | 0.832 | 0.569 | |
| Threshold:−0.5 | 0.609 | 0.898 | 0.776 | 0.539 |
3.7. Performance under different experimental conditions
Most of the prediction tools do not allow input for temperature and pH. To objectively evaluate the performance of the tools, we observed their accuracies at various temperatures and pH ranges with the S630 independent test set. Table 6 shows the performance of all the tools for nine evaluation items [9], where most of the mutation data in the training set are in the range of pH 6 ~ 8 and temperature ≦ 37 °C. The structural classification model of iStable 2.0 has higher accuracy than the individual tools for the three temperature intervals at pH 6 ~ 8. In addition, the performance of iStable 2.0 is greatly improved compared to the previous version of iStable in the 37 ~ 65 and >65 temperature intervals at pH 6 ~ 8.
Table 6.
Evaluation of prediction results with data from pH-temperature ranges by accuracy.
| pH | ≦6 |
6 ~ 8 |
>8 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | ≦37 | 37 ~ 65 | >65 | ≦37 | 37 ~ 65 | >65 | ≦37 | 37 ~ 65 | >65 |
| iStable2.0_PDB | 0.857 | 0.870 | 1.000 | 0.936 | 0.837 | 0.929 | 0.556 | 1.000 | 1.000 |
| iStable_PDB | 0.898 | 0.844 | 1.000 | 0.900 | 0.741 | 0.571 | 0.722 | 0.800 | 1.000 |
| DUET | 0.755 | 0.766 | 1.000 | 0.829 | 0.660 | 0.714 | 0.667 | 0.900 | 0.000 |
| SDM | 0.367 | 0.636 | 0.969 | 0.819 | 0.558 | 0.500 | 0.500 | 0.800 | 1.000 |
| SDM2 | 0.673 | 0.662 | 0.938 | 0.758 | 0.605 | 0.500 | 0.500 | 0.800 | 0.500 |
| mCSM | 0.694 | 0.779 | 1.000 | 0.833 | 0.626 | 0.786 | 0.722 | 0.800 | 0.000 |
| CUPSAT | 0.796 | 0.857 | 0.969 | 0.740 | 0.619 | 0.571 | 0.278 | 0.700 | 0.000 |
| I-Mutant2.0_PDB | 0.837 | 0.909 | 1.000 | 0.890 | 0.748 | 0.714 | 0.278 | 0.900 | 0.000 |
| PoPMuSiC | 0.694 | 0.740 | 1.000 | 0.822 | 0.653 | 0.643 | 0.611 | 0.700 | 0.000 |
| AUTO-MUTE2.0_SVM | 0.714 | 0.818 | 1.000 | 0.904 | 0.639 | 0.643 | 0.278 | 0.600 | 0.000 |
| AUTO-MUTE2.0_RF | 0.878 | 0.870 | 1.000 | 0.911 | 0.653 | 0.714 | 0.278 | 0.600 | 0.000 |
| MAESTRO | 0.714 | 0.753 | 1.000 | 0.694 | 0.694 | 0.643 | 0.278 | 0.800 | 0.500 |
| iStable2.0_SEQ | 0.878 | 0.844 | 1.000 | 0.911 | 0.830 | 0.857 | 0.500 | 0.900 | 1.000 |
| iStable_SEQ | 0.878 | 0.857 | 1.000 | 0.922 | 0.769 | 0.500 | 0.722 | 0.900 | 1.000 |
| iPTREE-STAB | 0.755 | 0.844 | 1.000 | 0.872 | 0.667 | 0.786 | 0.722 | 0.900 | 0.000 |
| I-Mutant2.0_SEQ | 0.796 | 0.896 | 1.000 | 0.865 | 0.741 | 0.571 | 0.333 | 1.000 | 0.000 |
| MUpro_SVM | 0.714 | 0.727 | 0.969 | 0.883 | 0.585 | 0.571 | 0.278 | 0.500 | 0.000 |
| MUpro_NN | 0.714 | 0.753 | 0.969 | 0.883 | 0.585 | 0.500 | 0.278 | 0.600 | 0.000 |
| EASE-MM | 0.816 | 0.766 | 1.000 | 0.687 | 0.701 | 0.571 | 0.556 | 0.900 | 0.500 |
| INPS | 0.776 | 0.597 | 0.844 | 0.808 | 0.755 | 0.643 | 0.500 | 0.800 | 1.000 |
4. Conclusion
iStable 2.0 successfully integrates sequence- and structure-based tools to improve the predictive performance of protein stability changes, which compare to various machine learning methods and prediction tools. In the evaluations of the training and test sets, it was found that these tools provide predicted results of protein stability using predicted ddG values and have high PCC and low MCC performance. According to our experimental results obtained from converting regression to classification, we found that training of both regression and classification models was necessary. In addition, there are some issues which should be considered when we adopt an integrated approach: 1) different input and output formats from different tools, 2) how to determine which tools should be integrated, 3) how to improve the performance of the integrated system, and 4) how to maintain system performance when the integration system fails. Majority Voting is a simple and intuitive integration method; this strategy is often used by biologists for many prediction tools. However, the predicted performance of iStable 2.0 using machine learning integration is better than the Majority Voting method because majority vote cannot consider the confidence score in the prediction results from different prediction tools. Our integration strategy only considers the execution time of the integrated tools but not the performance in order to complete the prediction of the integration calculation within a limited time, and from the feature analysis, the integration tools with low performance also provide contribution to the model. We additionally trained models that relied on the Stand-alone Module (SAM) and Sequence Coding Module (SCM). The integrated tools that cannot grasp the computing status are divided into an Online Server Module (OSM) so when access cannot be obtained by the integrated tools, the system performance will depend on SAM and SCM. iStable 2.0 is more effective at predicting point mutations between pH 6 ~ 8 than any integrated tools. However, each tool has its own advantages, such as a certain temperature, pH range, or protein type. Determining how to integrate the strengths of each tool into a model to enhance the performance will be a further improvement.
CRediT authorship contribution statement
Chi-Wei Chen: Methodology, Software, Writing - original draft. Meng-Han Lin: Data curation, Investigation, Validation, Writing - original draft. Chi-Chou Liao: Writing - original draft. Hsung-Pin Chang: Methodology, Writing - review & editing, Supervision. Yen-Wei Chu: Conceptualization, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by (a) Ministry of Science and Technology under grant number 108-2321-B-005-008 and 108-2634-F-005-002. (b) National Chung Hsing University and Chung-Shan Medical University under grant number NCHU-CSMU-10811.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.02.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Tokuriki N., Tawfik D.S. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19(5):596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Stefl S., Nishi H., Petukh M., Panchenko A.R., Alexov E. Molecular mechanisms of disease-causing missense mutations. J Mol Biol. 2013;425(21):3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yue P., Li Z., Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353(2):459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Frokjaer S., Otzen D.E. Protein drug stability: a formulation challenge. Nat Rev Drug Discovery. 2005;4(4):298. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 5.Rathi P.C., Jaeger K.-E., Gohlke H. Structural rigidity and protein thermostability in variants of lipase A from Bacillus subtilis. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0130289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom J.D., Labthavikul S.T., Otey C.R., Arnold F.H. Protein stability promotes evolvability. Proc Natl Acad Sci. 2006;103(15):5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DePristo M.A., Weinreich D.M., Hartl D.L. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6(9):678. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 8.Tokuriki N., Stricher F., Serrano L., Tawfik D.S. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4(2) doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Chi-Wei, Lin Jerome, Chu Yen-Wei. iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinf. 2013;14(S2) doi: 10.1186/1471-2105-14-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montanucci L., Capriotti E., Frank Y., Ben-Tal N., Fariselli P. DDGun: an untrained method for the prediction of protein stability changes upon single and multiple point variations. BMC Bioinf. 2019;20(14):335. doi: 10.1186/s12859-019-2923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 12.Pitera J.W., Kollman P.A. Exhaustive mutagenesis in silico: multicoordinate free energy calculations on proteins and peptides. Proteins Struct Funct Bioinf. 2000;41(3):385–397. [PubMed] [Google Scholar]
- 13.Thomas P.D., Dill K.A. Statistical potentials extracted from protein structures: how accurate are they? J Mol Biol. 1996;257(2):457–469. doi: 10.1006/jmbi.1996.0175. [DOI] [PubMed] [Google Scholar]
- 14.Carter C.W., Jr, LeFebvre B.C., Cammer S.A., Tropsha A., Edgell M.H. Four-body potentials reveal protein-specific correlations to stability changes caused by hydrophobic core mutations. J Mol Biol. 2001;311(4):625–638. doi: 10.1006/jmbi.2001.4906. [DOI] [PubMed] [Google Scholar]
- 15.Topham C.M., Srinivasan N., Blundell T.L. Prediction of the stability of protein mutants based on structural environment-dependent amino acid substitution and propensity tables. Protein Eng. 1997;10(1):7–21. doi: 10.1093/protein/10.1.7. [DOI] [PubMed] [Google Scholar]
- 16.Gilis D., Rooman M. Prediction of stability changes upon single-site mutations using database-derived potentials. Theor Chem Acc. 1999;101(1–3):46–50. [Google Scholar]
- 17.Bordner A., Abagyan R. Large-scale prediction of protein geometry and stability changes for arbitrary single point mutations. Proteins Struct Funct Bioinf. 2004;57(2):400–413. doi: 10.1002/prot.20185. [DOI] [PubMed] [Google Scholar]
- 18.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320(2):369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 19.Yin S., Ding F., Dokholyan N.V. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15(12):1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Capriotti E., Fariselli P. Casadio R: A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 2004;20(suppl_1):i63–i68. doi: 10.1093/bioinformatics/bth928. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct Funct Bioinf. 2006;62(4):1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 22.Huang L.-T., Gromiha M.M., Ho S.-Y. Sequence analysis and rule development of predicting protein stability change upon mutation using decision tree model. J Mol Model. 2007;13(8):879–890. doi: 10.1007/s00894-007-0197-4. [DOI] [PubMed] [Google Scholar]
- 23.Kourou K., Exarchos T.P., Exarchos K.P., Karamouzis M.V., Fotiadis D.I. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucl Acids Res. 2005;33(Web Server):W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L.-T., Gromiha M.M., Ho S.-Y. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics. 2007;23(10):1292–1293. doi: 10.1093/bioinformatics/btm100. [DOI] [PubMed] [Google Scholar]
- 26.Fariselli P., Martelli P.L., Savojardo C., Casadio R. INPS: predicting the impact of non-synonymous variations on protein stability from sequence. Bioinformatics. 2015;31(17):2816–2821. doi: 10.1093/bioinformatics/btv291. [DOI] [PubMed] [Google Scholar]
- 27.Folkman L., Stantic B., Sattar A., Zhou Y. EASE-MM: sequence-based prediction of mutation-induced stability changes with feature-based multiple models. J Mol Biol. 2016;428(6):1394–1405. doi: 10.1016/j.jmb.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucl Acids Res. 2006;34(suppl_2):W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilis D., Rooman M. PoPMuSiC, an algorithm for predicting protein mutant stability changes. Application to prion proteins. Protein Eng. 2000;13(12):849–856. doi: 10.1093/protein/13.12.849. [DOI] [PubMed] [Google Scholar]
- 30.Dehouck Y., Grosfils A., Folch B., Gilis D., Bogaerts P., Rooman M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009;25(19):2537–2543. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- 31.Dehouck Y., Kwasigroch J.M., Gilis D., Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinf. 2011;12(1):151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worth C.L., Preissner R., Blundell T.L. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucl Acids Res. 2011;39(suppl_2):W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandurangan A.P., Ochoa-Montaño B., Ascher D.B., Blundell T.L. SDM: a server for predicting effects of mutations on protein stability. Nucl Acids Res. 2017;45(W1):W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pires D.E., Ascher D.B., Blundell T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2013;30(3):335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laimer J., Hofer H., Fritz M., Wegenkittl S., Lackner P. MAESTRO-multi agent stability prediction upon point mutations. BMC Bioinf. 2015;16(1):116. doi: 10.1186/s12859-015-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masso M., Vaisman I.I. AUTO-MUTE: web-based tools for predicting stability changes in proteins due to single amino acid replacements. Protein Eng Des Sel. 2010;23(8):683–687. doi: 10.1093/protein/gzq042. [DOI] [PubMed] [Google Scholar]
- 37.Masso Majid, Vaisman Iosif I. AUTO-MUTE 2.0: a portable framework with enhanced capabilities for predicting protein functional consequences upon mutation. Adv Bioinf. 2014;2014:1–7. doi: 10.1155/2014/278385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue B., Lipps D., Devineni S. Integrated strategy improves the prediction accuracy of miRNA in large dataset. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0168392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia J.-F., Zhao X.-M., Huang D.-S. Predicting protein–protein interactions from protein sequences using meta predictor. Amino Acids. 2010;39(5):1595–1599. doi: 10.1007/s00726-010-0588-1. [DOI] [PubMed] [Google Scholar]
- 40.Wan J., Kang S., Tang C., Yan J., Ren Y., Liu J. Meta-prediction of phosphorylation sites with weighted voting and restricted grid search parameter selection. Nucl Acids Res. 2008;36(4) doi: 10.1093/nar/gkm848. e22–e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires D.E., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucl Acids Res. 2014;42(W1):W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bava K.A., Gromiha M.M., Uedaira H., Kitajima K., Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucl Acids Res. 2004;32(suppl_1):D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witvliet D.K., Strokach A., Giraldo-Forero A.F., Teyra J., Colak R., Kim P.M. ELASPIC web-server: proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity. Bioinformatics. 2016;32(10):1589–1591. doi: 10.1093/bioinformatics/btw031. [DOI] [PubMed] [Google Scholar]
- 44.Cang Z., Wei G.-W. TopologyNet: Topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLoS Comput Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cang Z., Wei G.-W. Analysis and prediction of protein folding energy changes upon mutation by element specific persistent homology. Bioinformatics. 2017;33(22):3549–3557. doi: 10.1093/bioinformatics/btx460. [DOI] [PubMed] [Google Scholar]
- 46.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucl Acids Res. 2018;46(W1):W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen B., Petersen T.N., Andersen P., Nielsen M., Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9(1):51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atchley W.R., Zhao J., Fernandes A.D., Drüke T. Solving the protein sequence metric problem. Proc Natl Acad Sci. 2005;102(18):6395–6400. doi: 10.1073/pnas.0408677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkatarajan M.S., Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical–chemical properties. Mol Model Annual. 2001;7(12):445–453. [Google Scholar]
- 50.Hartlmüller C., Göbl C., Madl T. Prediction of protein structure using surface accessibility data. Angew Chem Int Ed. 2016;55(39):11970–11974. doi: 10.1002/anie.201604788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S., Zhang C., Liang S., Zhou Y. Fold recognition by concurrent use of solvent accessibility and residue depth. Proteins Struct Funct Bioinf. 2007;68(3):636–645. doi: 10.1002/prot.21459. [DOI] [PubMed] [Google Scholar]
- 52.He B., Wang K., Liu Y., Xue B., Uversky V.N., Dunker A.K. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19(8):929. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 53.Hu J., Zhou X., Zhu Y.-H., Yu D.-J., Zhang G. TargetDBP: Accurate DNA-Binding Protein Prediction via Sequence-based Multi-View Feature Learning. IEEE/ACM Trans Comput Biol Bioinf. 2019 doi: 10.1109/TCBB.2019.2893634. [DOI] [PubMed] [Google Scholar]
- 54.Chen K., Mizianty M.J., Kurgan L. Prediction and analysis of nucleotide-binding residues using sequence and sequence-derived structural descriptors. Bioinformatics. 2011;28(3):331–341. doi: 10.1093/bioinformatics/btr657. [DOI] [PubMed] [Google Scholar]
- 55.Hu J., Li Y., Zhang M., Yang X., Shen H.-B., Yu D.-J. Predicting protein-DNA binding residues by weightedly combining sequence-based features and boosting multiple SVMs. IEEE/ACM Trans Comput Biol Bioinf. 2016;14(6):1389–1398. doi: 10.1109/TCBB.2016.2616469. [DOI] [PubMed] [Google Scholar]
- 56.Frank E., Hall M., Trigg L., Holmes G., Witten I.H. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20(15):2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 57.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. ACM; 2016. Xgboost: A scalable tree boosting system; pp. 785–794. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.