Abstract
In our previous study by Gupta et al, dominant T-cell epitopes of SARS CoV-N(N) protein were predicted by software. The spectrum of interferon (IFN)-γ responses of Balb/c mice immunized against two different forms of SARS CoV-N plasmid was then analyzed. A cluster of dominant T-cell epitopes of SARS CoV-N protein was found in the N-terminus (amino acids 76–114). On the basis of this study, four different plasmids were constructed: (i) DNA encoding the unmodified N (p-N) or N70−122 (p-N70−122) as an endogenous cytoplasmic protein or (ii) DNA encoding a lysosome-associated membrane protein (LAMP) chimera with N (p-LAMP/N) or N70−122 (p-LAMP/N70−122). The immune responses of mice to these four constructs were evaluated. The results showed marked differences in the responses of the immunized mice. A single priming immunization with the p-LAMP/N construct was sufficient to elicit an antibody response. Enzyme-linked immunospot (ELISpot) assay indicated that p-LAMP/N70−122 and p-LAMP/N plasmids both elicited a greater IFN-γ response than p-N. p-N and p-N70−122 constructs induced low or undetectable levels of cytokine secretion. We also found that the p-LAMP/N70−122 construct promoted a long-lasting T-cell memory response without an additional boost 6 months after three immunizations. These findings show that DNA vaccines, even epitope-based DNA vaccines using LAMP as chimera, can elicit both humoral and cellular immune responses.
Keywords: SARS CoV, N, LAMP, cluster of dominant T-cell epitopes, DNA vaccine, ELISpot
Introduction
Endogenous antigens encoded by conventional naked DNA plasmids are processed by antigen-presenting cells and presented only to CD8+ T cells through the formation of peptide/major histocompatibility complex class I (MHC I) complexes. Therefore, CD4+ T cells cannot be effectively activated in this model. CD4+ T cell activation is vital for the efficient priming of CD8+ T cells, B lymphocytes and long-term immune memory.1, 2, 3, 4, 5, 6 A weak point in the current theory is that DNA vaccines, when encoding a cytoplasmic protein, usually fail to elicit a sufficient, if any, CD4+ T response, whereas MHC I–cytotoxic T-lymphocyte (CTL) responses are usually preferable. This preference usually leads to the inability of the host immune system to mount greater immune responses and to maintain long-term memory.
The lysosome is one of the most important components of the multilaminar vesicular compartment (MHC class II-processing compartment) in the exogenous antigen-processing pathway.7, 8, 9 Lysosome-associated membrane protein (LAMP) can target and bind the endosome/lysosome through the LAMP transmembrane/cytoplasmic domain. The luminal domain of LAMP is then integrated into the lysosome.10, 11, 12, 13, 14, 15, 16, 17 Consequently, one strategy commonly adopted by our group 18, 19, 20, 21, 22, 23, 24, 25 is to insert the target DNA into LAMP as a LAMP/antigen chimera. In addition, many other laboratories have reported that LAMP targeting could greatly enhance the immune response against a number of antigens.26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 It has been postulated that DNA plasmids encoding endogenous antigen could take advantage of LAMP and be carried directly into MHC class II-processing compartment, where the fusion protein of interest could be digested and presented by MHC II molecules. This strategy could thus boost the CD4+ T-lymphocyte response and, in turn, enhance the immune responses mentioned above.
Our previous work showed that the LAMP–SARS Coronavirus nucleocapsid construct (p-LAMP/N) elicits the strongest T-cell responses when compared with the SARS CoV-N alone (p-N) or a glutathione S-transferase–N fusion protein. A dominant epitope cluster appeared to be located within the N terminus of SARS CoV-N.23 Hence, we were interested to find out whether a sequence containing these epitopes only would suffice to elicit a T-cell response and, if not, whether it was possible to make a LAMP fusion construct to enhance the immune response. In this paper, we will explore the above-mentioned possibilities.
Results
Expression of N, LAMP/N, N70−122 and LAMP/N70−122 in transfected cells
Several DNA plasmids encoding N, N70−122 or LAMP protein chimeras were constructed in the p43 vector backbones to evaluate N protein expression in transfected cells. Expression of the protein encoded by the DNA plasmid constructs in transfected COS-7 or 293 cells was confirmed by fluorescence microscopy and flow cytometry for proteins containing the enhanced green fluorescent protein (EGFP) tag, as well as by western blot.
The levels of EGFP-tagged N70−122 and LAMP/N70−122 expression in transfected 293 or COS-7 cells were analyzed by fluorescence microscopy (Figure 1a) and flow cytometry (Figure 1b). No significant difference in the fluorescence intensity of the transfected cells was observed, suggesting similar synthesis and translation rates for the two EGFP-tagged molecules.
Figure 1.
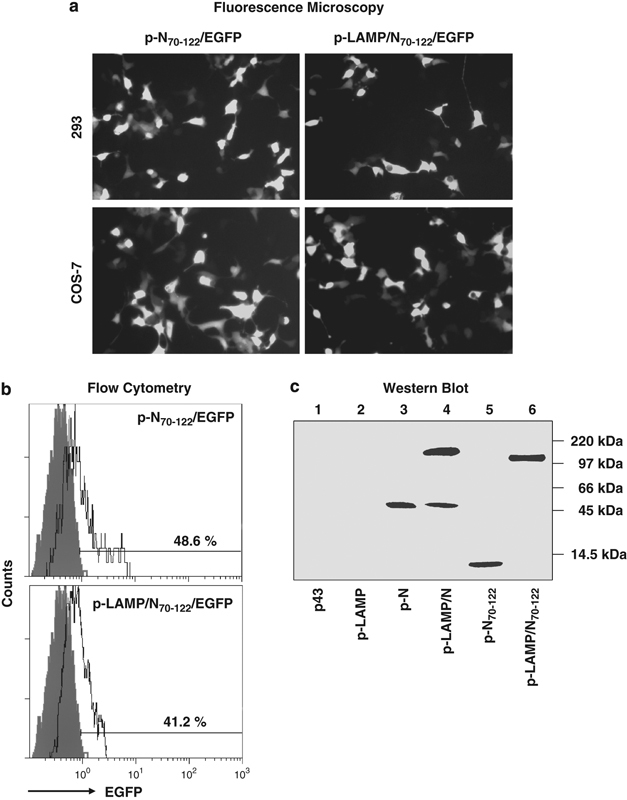
Validation of SARS-N and N70−122 protein expression. 293 or COS-7 cell lines were transfected with different DNA plasmids. (a) and (b) Enhanced green fluorescent protein (EGFP) expression was directly observed by Fluorescence microscopy and flow cytometry. (c) Western blot analysis of transfected 293 cells blotted with anti-N70−159 polyclonal antibody. The positions of the protein markers in kDa are indicated on the right. LAMP, lysosome-associated membrane protein.
Western blot using the anti-SARS-N70−169 polyclonal antibody confirmed the presence of the approximately 160 kDa LAMP/N protein and 45 kDa N protein at comparable levels in transfected 293 cells. Protein degradation of N was detected in the LAMP chimera-transfected cells. LAMP/N70−122 was present at about twofold greater concentrations than the unmodified N70−122 (Figure 1c) by Bio-Rad ChemiDoc scanning (Bio-Rad, Hercules, CA, USA).
Greater concentrations of the LAMP chimera forms of proteins have been observed frequently in cell extracts,18, 21 presumably because of the increased expression of the associated LAMP, a major cellular structural protein, or because of decreased proteolytic degradation as a consequence of the association with the heavily glycosylated and protease-resistant LAMP.13
Antibody responses to repeated immunizations with the SARS CoV-N DNA constructs
Mice were immunized three times (on day 1 and by boosting immunizations on days 21 and 42) by subcutaneous injection of the DNA plasmid. An initial evaluation of the immune response was conducted by measuring antibody binding to the intact N protein 2 weeks after each immunization. An antibody response to LAMP/N immunogen was detected after the first immunization, and strong responses to both N and LAMP/N were detected after the second immunization. A low antibody response to LAMP/N70−122 was observed after the third immunization, compared with both LAMP/N and N-immunized mice. However, the response of mice immunized with LAMP/N70−122 was considerably greater than the response of those immunized with N70−122, which did not differ from either the immunogen control group or the normal mouse serum (Figure 2).
Figure 2.
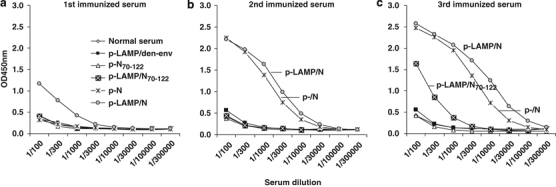
Humoral immune responses. (a–c) Mice were injected on days 1, 21 and 42 with 50 μg of DNA plasmid. Samples were collected 2 weeks after each immunization. Pooled serum from individual groups of mice was serially diluted, and SARS-N-specific total IgG was measured by enzyme-linked immunosorbent assay. LAMP, lysosome-associated membrane protein.
Peptide-specific IFN-γ, IL-2 and IL-4 ELISpot responses to a dominant cluster of T-cell epitopes
We previously reported that the major interferon (IFN)-γ T-cell response of mice immunized with N DNA constructs was to a cluster of N overlapping peptides encompassing amino acids 76–114 from the N-amino terminus, with an approximately twofold greater response to LAMP/N compared with N.23 Sequential studies of this epitope cluster as a potential epitope-based DNA vaccine have shown a remarkable level of T-cell activation by the fragment constructed in the form of a LAMP chimera. Mice were immunized with constructs encoding the intact N and the N70−122 sequence, both as unmodified sequences and as LAMP chimeras. The experimental protocol, as described in Materials and methods, included immunization with p-N and p-LAMP/N as positive controls and, for comparison with previous experiments, with p-N70−122 and p-LAMP/N70−122 for analysis of the dominant epitope cluster as an N antigen and with a p-LAMP/dengue-env construct as a negative-control immunogen.
Enzyme-linked immunospot (ELISpot) assay responses after two immunizations (on day 40) were analyzed with overlapping peptides spanning the N epitope cluster region, amino acids 70–122, and with a yellow fever virus peptide as a negative-control peptide. The IFN-γ response results for the positive-control immunizations with p-N and p-LAMP/N were similar to those reported earlier,23 with strong T-cell responses to LAMP/N immunization that were approximately twofold greater than N immunization (Figure 3a-1). It is noted, however, that a response to p-N70−122 lacking the LAMP sequences was not detected and was equivalent to that of the negative control (p-LAMP/den-env). In contrast, strong responses to the p-LAMP/N70−122 construct were detected, comparable to immunization with the intact N protein. Moreover, the p-LAMP/N70−122-immunized mice showed strong interleukin (IL)-2 and IL-4 responses, similar to those elicited by p-LAMP/N, whereas very little response was observed in the p-N and p-N70−122-immunized mice (Figures 3a-2 and a-3).
Figure 3.
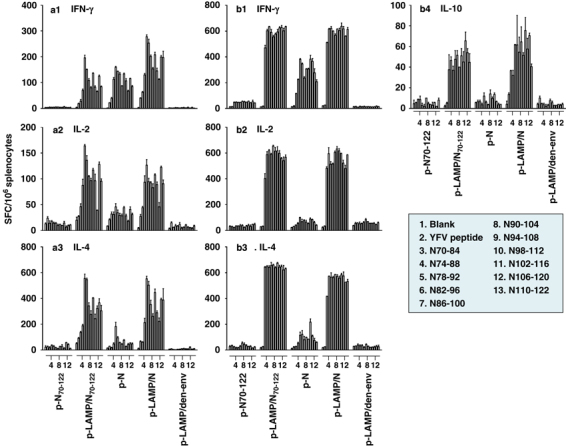
The lysosome-associated membrane protein (LAMP) chimera enhances the antigen-specific cytokine responses. Mice were injected twice (a-1–a-3) and three times (b-1–b-4) with 50 μg of DNA plasmid. Antigen-specific splenocyte cytokine secretions were detected by enzyme-linked immunospot assay. The splenocytes were stimulated with overlapping peptides spanning the SARS-N epitope cluster region, amino acids 70–122, and with a yellow fever virus (YFV) peptide as a negative control. Each result was performed in three times individual experiment.
Enhanced IFN-γ, IL-2, IL-4 and IL-10 T-cell responses after three times of LAMP/N70−122 or LAMP/N immunizations
An additional (second boost) immunization on day 42 with p-LAMP/N and p-LAMP/N70−122 resulted in dramatic cytokine production measured on day 60 (Figure 3b), with the most dramatic results observed for IFN-γ, IL-2, IL-4 and IL-10. Again, almost no IFN-γ, IL-2, IL-4 or IL-10 response was displayed by the p-N70−122-immunized mice, and very little response other than IFN-γ was detected for the p-N-immunized mice. In addition, there was a marked broadening of the enhanced response to the LAMP chimera constructs that included all of the peptides encompassing the cluster of epitopes, and this was presumably because of the bystander activation.
The requirement for LAMP chimera constructs to induce memory T-cell responses
Memory T-cell responses were measured by ELISpot at week 30 in mice treated with the same initial immunization protocol with or without an additional boost injection at week 27. After the boost injection at week 27, strong memory T-cell IFN-γ responses were induced by p-LAMP/N, p-LAMP/N70−122 and p-N and, in contrast, no significant response to p-N70−122 was observed (Figure 4a-2). Without the boost immunization, there were relatively weak but significant memory responses to the LAMP chimera constructs, compared with no response to constructs lacking LAMP (Figure 4a-1).
Figure 4.
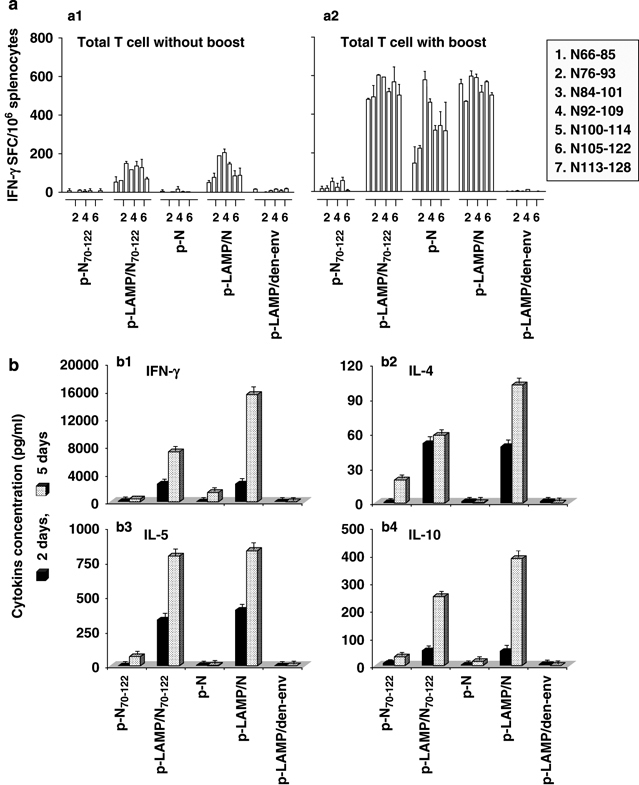
Cytokine responses of memory T cells. Mice were injected three times with 50 μg of DNA plasmid. (a) After 21 weeks, mice were treated with the same initial immunization protocol with (a-2) or without (a-1) an additional boost injection. Memory T-cell interferon (IFN)-γ secretions were determined by enzyme-linked immunospot assay. (b) At week 30, the splenocytes were cultured in the culture medium alone or stimulated with overlapping peptides spanning the SARS-N epitope cluster region, amino acids 66–128, for 2 or 5 days. The cytokine levels (b-1, IFN-γ; b-2, interleukin (IL)-4; b-3, IL-5; b-4, IL-10) in the cultured supernatant were analyzed by enzyme-linked immunosorbent assay. Each result was performed in three times individual experiment. LAMP, lysosome-associated membrane protein.
The splenocytes of immunized mice lacking an additional boost injection at week 30 were cultured in medium alone or with a pool of overlapping peptides spanning the SARS-N epitope cluster region (amino acids 66–128) for 2 or 5 days. The cultured supernatants were respectively collected. The cytokine levels in the supernatant were then determined by enzyme-linked immunosorbent assay (ELISA). At 2 days, greater memory T-cell cytokine secretion, including IFN-γ, IL-4, IL-5 and IL-10, was observed with the p-LAMP/N and p-LAMP/N70−122 constructs in contrast to an almost undetectable response to p-N70−122 and p-N. The strongest memory T-cell cytokine secretion was observed in response to the p-LAMP/N and p-LAMP/N70−122 constructs at 5 days (Figure 4b).
Discussion
Endogenous antigens are processed in the cytoplasm and initiate CTL activation. In contrast, exogenous antigens are degraded in the lysosome of antigen-presenting cells and induce antibody-mediated humoral immune responses and CD4+ T-cell activation and secretion of cytokines to facilitate CTL activation and differentiation. Therefore, the achievement of a desired response by controlling the delivery pathways is indispensable in vaccine development and immunotherapy.39 In addition, the effective activation of CD4+ T cells is critical for generating adaptive immune responses.1, 2, 3, 4, 5 The studies discussed above used a recently developed approach that provides an additional necessary protein element required for antigen trafficking to vesicular lysosomal sites where MHC II antigen presentation takes place. As shown in the results, the DNA vaccine of SARS CoV-N alone definitely elicited the host immune response, and this was most evident for B cells. However, stronger T-cell responses were observed when LAMP fusion DNA vaccines were applied.18 This is consistent with previous studies in which HIV Gag-LAMP elicited superior antibody and CD4+ and CD8+ cellular responses.18 This increase in the CD8+ T-cell response could be attributed to the increased response of CD4+ T cells. This study, together with previous studies, provides indirect evidence that the trafficking of DNA-encoded antigens to the LAMP endosomal/lysosomal vesicular compartments is an important factor affecting the function of genetic vaccines. Whatever the mechanism may be, the effect is evident. In addition to stronger T-cell responses (Figure 3), stronger long-term memory T-cell responses (Figure 4) were also observed, even with the p-LAMP/ N70−122 construct, whereas almost no response was detected to the p-N construct. These results reveal the special advantage of LAMP fusion DNA vaccines, which have a much longer effect than conventional DNA vaccines.
Another finding of this study confirmed our initial prediction that the T-cell dominant epitope cluster of SARS CoV-N resides mainly in the N-terminal 76–114 amino acids of this protein (Figure 5). It is interesting that even though this dominant epitope cluster resides in this area, the DNA SARS CoV-N70−122 vaccine alone failed to elicit immune responses in either B cells or T cells. However, when the fragment was fused to LAMP, dramatic immune responses were observed. The extent of the T-cell response was comparable to that observed with p-LAMP/N, and was stronger than that detected with p-N. Weaker B cell responses were also evident (Figure 2). There are some possible explanations for this result. The major B cell epitope cluster may not be located within the N terminus of SARS CoV-N. This hypothesis is supported by the fact that the p-LAMP/N construct elicited a B-cell response equivalent to the p-N construct and to the findings reported by Zhao et al.40
Figure 5.
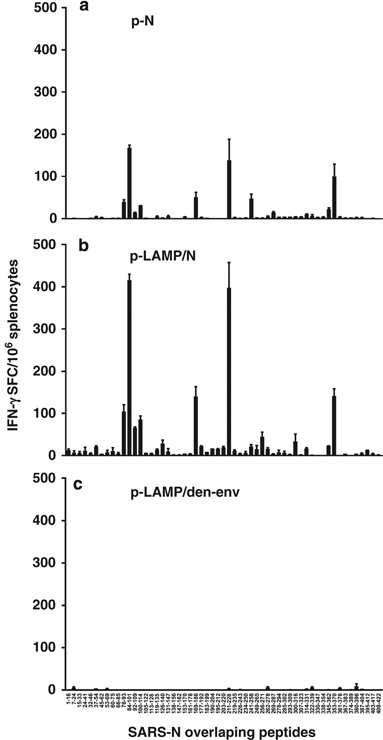
Confirmation of dominant T-cell epitopes cluster of SARS-N protein. (a–c) Mice were killed 21 days after the second immunization with the DNA immunogens and splenocytes were analyzed by enzyme-linked immunospot assay for antigen-specific interferon (IFN)-γ secretion. The splenocytes were stimulated with whole SARS-N overlapping peptides. Plasmid p-LAMP/den-env was used as the immunogen control. LAMP, lysosome-associated membrane protein.
Taken together, this study not only showed that the use of LAMP as a chimera can elicit both humoral and cellular immune responses, even with an epitope-based cluster DNA vaccine, but it also emphasized the significance of targeting DNA-encoded vaccine antigens to the MHC II-processing compartments for induction of long-term immunological memory. These findings are of great value in understanding anti-SARS-CoV immunity and also in designing epitope-based DNA vaccines against the disease.
Materials and methods
Plasmids
The SARS CoV-N cDNA was prepared from a Singapore clinical isolate (GenBank ID, AY307165), as previously described.23 The total N 1–1269 bp sequence and a 208–366 bp fragment were cloned into the p43 vector,41 which contains the adeno-associated virus inverted terminal repeat sequences flanking the expression elements (cytomegalovirus promoter and bovine growth hormone polyadenylation signal). The complete N gene fragment was amplified by PCR using the following primers:
5′-CGGCTAGCATGTCTGATAATGGACCCCAATC-3′
5′-GCGGTACCTTATGCCTGAGTTGAATCAGCAG-3′
5′-CGCTCGAGATGTCTGATAATGGACCCCAATC-3′
5′-CGGAATTCTGCCTGAGTTGAATCAGCAGAA-3′
The 208–366 bp gene region was amplified using the primers:
5′-GCGCTAGCATGGGCCAGGGCGTTCCAATCAACACCAA-3′
5′-CGGGTACCTTA GAATTCAAGTGAAGCTTCTGGGCCAGTT-3′
5′-GCCTCGAGATGGGCCAGGGCGTTCCAATCAACACCAA-3′
5′-CGGAATTC GATATCAAGTGAAGCTTCTGGGCCAGTT-3′
The amplified fragments of native N and N70−122 were inserted into the NheI and KpnI sites of the p43 vector and termed p-N and p-N70−122, respectively. The LAMP chimera constructs, p-LAMP/N and p-LAMP/N70−122, were cloned by inserting the sequences encoding N and N70−122 of SARS CoV between the LAMP luminal domain and the transmembrane/cytoplasmic domains at the XhoI and EcoRI sites, respectively. The vaccination plasmids were produced by transforming Escherichia coli DH5α, and were then purified to remove endotoxin (Qiagen, Valencia, CA, USA). These plasmids were also constructed to carry EGFP, p-N70−122/EGFP and p-LAMP/N70−122/EGFP, which were introduced at the EcoRI and EcoRV sites, for expedient analysis of protein expression (Figure 6).
Figure 6.
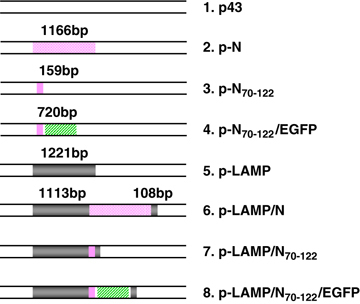
Schematic representations of DNA constructs. p43, a vector with a CMV promoter and BGH polyadenylation signal; N, SARS CoV nucleocapsid protein; N70−122, a cluster of dominant T-cell epitopes of SARS CoV-N protein; EGFP, enhanced green fluorescence protein; LAMP, lysosome-associated membrane protein.
Protein expression analysis
Enhanced green fluorescent protein-tagged N70−122 and LAMP/N70−122 protein expressions were studied in human 293 and COS-7 monkey kidney cell lines. Cells were seeded in 6-well plates (1 × 106 cells per well or 5 × 105 cells per well) during log-phase growth and transfected with the EGFP-labeled N70−122 and LAMP chimera plasmid DNA (4 μg) using FuGENE 6 transfection reagent (Roche, Indianapolis, IN, USA), according to the manufacturer's instructions. EGFP expression was observed 24–48 h after transfection by fluorescence microscopy and flow cytometry.
Western blot analysis of protein expression was analyzed using 293 cells. Cells were seeded in 6-well plates (2 × 106 cells per well) and transfected with plasmid DNA lacking the EGFP tag using the same transfection reagent. The cells were harvested 48 h after transfection and disrupted with CelLytic M Cell Lysis Reagent premixed with Protease Inhibitor Cocktail (Sigma-Aldrich, Inc., St Louis, MO, USA) for 15 min on ice. Cellular debris was then removed by centrifugation,18 and the protein concentration was determined with a BCA kit (Pierce, Rockford, IL, USA). Samples were resolved in 4–15% gradient polyacrylamide ready gels (Bio-Rad) and electrotransferred onto an Immobilon-P membrane (Millipore, Bedford, MA, USA). The membrane was blocked in Tris–HCl-buffered saline (TBS) containing 5% nonfat dried milk. The blot was probed with mouse anti-SARS-N70−169 polyclonal antibody (Genesis Biotech Inc., Taipei, Taiwan) at a 1:1000 dilution for 2 h and then incubated with peroxidase-conjugated goat anti-mouse IgG secondary antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA) at a 1:5000 dilution for 1 h. The bands were visualized by ECL Plus Western Blotting Detection reagents and exposed to Hyperfilm ECL (Amersham Biosciences UK Ltd, Buckinghamshire, UK). Bands were scanned by Bio-Rad ChemiDoc.
Animals and immunization
Five groups of 6–8-week-old female Balb/c mice (Charles Rivers, Wilmington, MA, USA; each group contained 30 mice) were immunized subcutaneously at the base of the tail with 50 μg of the specified endotoxin-free DNA plasmid (p-N, p-LAMP/N, p-N70−122, p-LAMP/N70−122 and p-LAMP/den-env as an immunogen control) diluted in phosphate-buffered saline (PBS). The mice were boosted twice every 3 weeks with the same plasmid. Blood samples were collected by tail vein bleeding of the immunized mice 2 weeks after each immunization. The immunized mice were killed on day 40 or 60.
Furthermore, to analyze the immune recall response, five groups of mice (each group contained 10 mice) were immunized as described above. After an interval of 21 weeks (at week 27), the immunized mice received an additional boost with the same DNA plasmid or with PBS alone. The immunized mice were killed after a 3-week interval (Figure 7).
Figure 7.
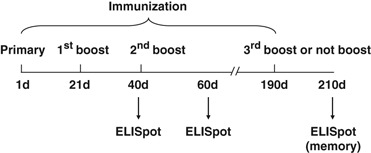
Immunization schedule.
Antibody determination in vaccinated mice
The 96-well ELISA plates (Nunc., Rochester, NY, USA) were coated with recombinant N protein (NIH Biodefense and Emerging Infections Research Resources Repository, NIAID). The protein was diluted in 0.1 M sodium carbonate–bicarbonate buffer (PH 9.5) at a concentration of 5 μg ml–1, and 100 μl was added to each well and incubated overnight at 4 °C. The coating solution was then discarded and the plates were washed three times with PBS containing 0.05% Tween-20 washing buffer (PBST). Immunized serum samples were prepared in eight (threefold) serial dilutions in PBST with 0.1% bovine serum albumin starting at 1:100. One hundred microliters of each dilution was added to each well and the plates were incubated for 2 h at 37 °C. The serum antibody was captured by the coated N protein and detected by incubation with 100 μl of horseradish peroxidase-conjugated goat anti-mouse IgG antibody (1:1000) for 1 h at 37 °C. Color was developed with Turbo tetramethylbenzone substrate solution (BD PharMingen, San Diego, CA, USA) by incubation for 15 min at room temperature. The reaction was stopped by the addition of 50 μl of 2 M sulfuric acid and the absorbance was measured at 450 nm in a TECAN microplate reader (SafireII, San Jose, CA, USA).
Preparation of splenocytes for assaying the T-cell-mediated cellular immune responses of immunized mice
Single cell suspensions depleted of red blood cells were prepared from freshly isolated immunized mouse splenocytes in culture medium (RPMI-1640 medium supplemented with 5% v/v fetal bovine serum, 100 units ml–1 penicillin/streptomycin, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, and 0.01 M HEPES buffer). Splenocytes were counted and resuspended at 1 × 107 cells ml–1 in culture medium for T-cell-mediated assays.
ELISpot assays
Cellular immune responses to N, N70−122 and LAMP chimeras were assessed by multiple cytokine ELISpot assays according to the manufacturer's protocol (BD Pharmingen). Briefly, ELISpot plates were coated overnight at 4 °C with cytokine-specific capture antibody (rat anti-mouse) diluted in sterile PBS at 5 μg ml–1. After blocking for 2 h with RPMI-1640 (Gibco, Invitrogen Co., Carlsbad, CA, USA) containing 10% fetal bovine serum at room temperature, 1 × 106 splenocytes per well were stimulated with the overlapping N synthetic peptides at a final concentration of 10 μg ml–1 (Sin-pep, NIH Biodefense and Emerging Infections Research Resources Repository, NIAID) for 16–40 h at 37 °C in a 5% CO2 incubator. Negative controls were medium alone or a yellow fever virus peptide, respectively. Con A (5 μg ml–1) was used as a positive control. The plates were washed with H2O and PBST and incubated for 2 h at room temperature with 2 μg ml–1 of the relevant biotinylated rat anti-mouse cytokine antibody. After washing with PBST, the plates were incubated with streptavidin–horseradish peroxidase at a 1:100 dilution for 1 h. The reaction was developed with AEC substrate (Calbiochem, San Diego, CA, USA) and was stopped by washing with water. The plates were air dried and the cytokine tracks of antigen-specific cell amplification were counted (ImmunoSpot, CTL Cellular Technology Ltd, Shaker Heights, OH, USA). The frequencies of the cytokine-producing cells were calculated by subtracting the number of detected spots in the absence of antigen from those obtained in the presence of the relevant antigen (performed in duplicate). All results are expressed as the mean number of SFCs per 1 × 106 splenocytes.
Cytokine production by immunized splenocytes assessed by ELISA analysis
Single splenocyte suspensions from immunized mice were prepared as above, and 1 × 107 splenocytes per ml per well were cultured in a 12-well plate (Nunc.) with medium alone or with a final concentration of 20 μg ml–1 of the overlapping peptide pool (N66−122). After incubation for 2 or 5 days at 37 °C in 5% CO2, the culture supernatants were collected for detection of secreted cytokines as measured by OptEIA ELISA kits (BD Pharmingen).
Statistical analyses
The experimental data were obtained from two completely independent mouse immunizations and two different libraries of synthetic peptides. All ELISpot data are expressed as means±s.d. All graphs and unpaired t-test analyses were generated using GraphPad Prism Version 4.0a for Macintosh (GraphPad software, San Diego, CA, USA).
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This study was supported by grants from the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, Department of Health and Human Services, contract no. HHSN266200400085C of USA, and the National High-tech R&D Program (863 Program) of China (No. 2006AA02A237). We thank Chikhlikar PR, Paul Tan, Vladimir Perovic, Keun OK Jung, Garg S, Milton Maciel Jr, Betty Hart and Delores Henson for their excellent technical assistance. The support of the NIH AIDS Research and Reference Reagent Program in providing the SARS-N peptides and purified SARS-N protein is gratefully acknowledged.
Glossary
- SARS
severe acute respiratory syndrome
- SARS CoV
SARS-associated coronavirus
- N
nucleocapsid protein
- LAMP
lysosome-associated membrane protein
- ELISpot
enzyme-linked immunospot assay
- MHC
major histocompatibility complex
- p-N
p43-SARS CoV-N construct
- p-LAMP/N
p43-LAMP SARS CoV-N construct
- p-N70−122
p43-SARS CoV-N70−122 epitopes cluster construct
- p-LAMP/N70−122
p43-LAMP SARS CoV-N70−122 epitopes cluster construct
- p-LAMP/den-env
p43-LAMP Dengue virus envelope construct.
Contributor Information
K Yang, Email: yangkunkun@fmmu.edu.cn.
J T August, Email: taugust@jhmi.edu.
References
- 1.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 2.Rocha B, Tanchot C. Towards a cellular definition of CD8+ T-cell memory: the role of CD4+ T-cell help in CD8+ T-cell responses. Curr Opin Immunol. 2004;16:259–263. doi: 10.1016/j.coi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Janssen EM, Lemmens EE, Wolfe T. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;2003:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 4.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 5.Khanolkar A, Badovinac VP, JT H. CD8 T cell memory development: CD4 T cell help is appreciated. Immunol Res. 2007;39:94–104. doi: 10.1007/s12026-007-0081-4. [DOI] [PubMed] [Google Scholar]
- 6.Sun JC, Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming of memory CD8+ T cells after acute infection. Nat Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JW, Murphy TL, Willingham MC, Pastan I, JT A. Identification of two lysosomal membrane glycoproteins. J Cell Biol. 1985;101:85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/S0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M. Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J Biol Chem. 1991;266:21327–21330. [PubMed] [Google Scholar]
- 11.Hunziker W, Simmen T, Honing S. Trafficking of lysosomal membrane proteins in polarized kidney cells. Nephrologie. 1996;17:347–350. [PubMed] [Google Scholar]
- 12.Peters PJ, Neefjes JJ, Oorschot V, Ploegh HL, Geuze HJ. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 1991;349:669–676. doi: 10.1038/349669a0. [DOI] [PubMed] [Google Scholar]
- 13.Guarnieri FG, Arterburn LM, Penno MB, Cha Y, August JT. The motif Tyr-X-X-hydrophobic residue mediates lysosomal membrane targeting of lysosome-associated membrane protein 1. J Biol Chem. 1993;268:1941–1946. [PubMed] [Google Scholar]
- 14.Kleijmeer MJ, Morkowski S, Griffith JM, Rudensky AY, Geuze HJ. Major histocompatibility complex class II compartments in human and mouse B lymphoblasts represent conventional endocytic compartments. J Cell Biol. 1997;139:639–649. doi: 10.1083/jcb.139.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geuze HJ. The role of endosomes and lysosomes in MHC class II functioning. Immunol Today. 1998;19:282–287. doi: 10.1016/S0167-5699(98)01269-9. [DOI] [PubMed] [Google Scholar]
- 16.Drake JR, Lewis TA, Condon KB, Mitchell RN, Webster P. Involvement of MIIC-like late endosomes in B cell receptor-mediated antigen processing in murine B cells. J Immunol. 1999;162:1150–1155. [PubMed] [Google Scholar]
- 17.Turley SJ, Inaba K, Garrett WS, Ebersold M, Unternaehrer J, Steinman RM. Transport of peptide-MHC class II complexes in developing dendritic cells. Science. 2000;288:522–527. doi: 10.1126/science.288.5465.522. [DOI] [PubMed] [Google Scholar]
- 18.Marques ETAJ, Chikhlikar P, de Arruda LB, Leao IC, Lu Y, Wong J. HIV-1 p55Gag encoded in the lysosome-associated membrane protein-1 as a DNA plasmid vaccine chimera is highly expresse, traffics to the major histocompatibility class II compartment, and elicit enhanced responses. J Biol Chem. 2003;278:37926–37936. doi: 10.1074/jbc.M303336200. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Raviprakash K, Leao IC. Dengue 2 PreM-E/LAMP chimera targeted to the MHC class II compartment elicits long-lasting neutralizing antibodies. Vaccine. 2003;21:2187–2198. doi: 10.1016/S0264-410X(03)00009-4. [DOI] [PubMed] [Google Scholar]
- 20.De Arruda LB, Chikhlikar PR, August JT. DNA vaccine encoding human immunodeficiency virus-1 Gag, targeted to the major histocompatibility complex II compartment by lysosomal-associated membrane protein, elicits enhanced long-term memory response. Immunology. 2004;112:126–133. doi: 10.1111/j.1365-2567.2004.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chikhlikar PR, Barros dAL, Agrawal S. Inverted terminal repeat sequences of adeno-associated virus enhance the antibody and CD8+ responses to a HIV-1 p55Gag/LAMP DNA vaccine chimera. Virology. 2004;323:220–232. doi: 10.1016/j.virol.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 22.Anwar A, Chandrasekaran A, Ng ML. West Nile premembrane-envelope genetic vaccine encoded as a chimera containing the transmembrane and cytoplasmic domains of a lysosome-associated membrane protein: increased cellular concentration of the transgene product, targeting to the MHC II compartment, and enhanced neutralizing antibody response. Virology. 2005;332:66–77. doi: 10.1016/j.virol.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Gupta V, Tabiin TM, Sun K, Chandrasekaran A, Anwar A, Yang K. SARS coronavirus nucleocapsid immunodominant T-cell epitope cluster is common to both exogenous recombinant and endogenous DNA-encoded immunogens. Virology. 2006;347:127–139. doi: 10.1016/j.virol.2005.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arruda LB, Sim D, Chikhlikar PR. Dendritic cell-lysosomal-associated membrane protein (LAMP) and LAMP-1-HIV-1 gag chimeras have distinct cellular trafficking pathways and prime T and B cell responses to a diverse repertoire of epitopes. J Immunol. 2006;177:2265–2275. doi: 10.4049/jimmunol.177.4.2265. [DOI] [PubMed] [Google Scholar]
- 25.Chikhlikar P, Arruda LBd, Maciel M, Lewis P, August T, Marques ETA. DNA encoding an HIV-1 Gag/human lysosome-associate membrane protein-1 elicits a broad cellular immune response in Rhesus macaques. PloS ONE. 2006;1:e135. doi: 10.1371/journal.pone.0000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowell JF, Ruff AL, Guarnieri FG, Staveley-O'Carroll K, Lin X, Tang J. Lysosome-associated membrane protein- 1-mediated targeting of the HIV-1 envelope protein to an endosomal/lysosomal compartment enhances its presentation to MHC class II-restricted T cells. J Immunol. 1995;155:1818–1828. [PubMed] [Google Scholar]
- 27.Wu TC, Guarnieri FG, Staveley-O'Carroll KF, Viscidi RP, Levitsky HI, Hedrick L. Engineering an intracellular pathway for major histocompatibility complex class II presentation of antigens. Proc Natl Acad Sci USA. 1995;92:11671–11675. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruff AL, Guarnieri FG, Staveley-O'Carroll K, Siliciano RF, August JT. The enhanced immune response to the HIV gp160/LAMP chimeric gene product targeted to the lysosome membrane protein trafficking pathway. J Biol Chem. 1997;272:8671–8678. doi: 10.1074/jbc.272.13.8671. [DOI] [PubMed] [Google Scholar]
- 29.Nair SK, Boczkowski D, Morse M, Cumming RI, Lyerly HK, Gilboa E. Induction of primary carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes in vitro using human dendritic cells transfected with RNA. Nat Biotechnol. 1998;16:364–369. doi: 10.1038/nbt0498-364. [DOI] [PubMed] [Google Scholar]
- 30.Bonini C, Lee SP, Riddell SR, Greenberg PD. Targeting antigen in mature dendritic cells for simultaneous stimulation of CD4+ and CD8+ T cells. J Immunol. 2001;166:1665250–1665257. doi: 10.4049/jimmunol.166.8.5250. [DOI] [PubMed] [Google Scholar]
- 31.Raviprakash K, Marques E, Ewing D, Lu Y, Phillips I. Synergistic neutralizing antibody response to a dengue virus type 2 DNA vaccine by incorporation of lysosome-associated membrane protein sequences and use of plasmid expressing GM-CSF. Virology. 2001;290:74–82. doi: 10.1006/viro.2001.1136. [DOI] [PubMed] [Google Scholar]
- 32.Su Z, Vieweg J, Weizer AZ, Dahm P, Yancey D. Enhanced induction of telomerase-specific CD4(+) T cells using dendritic cells transfected with RNA encoding a chimeric gene product. Cancer Res. 2002;62:5041–5048. [PubMed] [Google Scholar]
- 33.Dobano C, Rogers WO, Gowda K, Doolan DL. Targeting antigen to MHC class I and class II antigen presentation pathways for malaria DNA vaccines. Immunol Lett. 2007;111:92–102. doi: 10.1016/j.imlet.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Kang TH, Lee JH, Bae HC, Noh KH, Kim JH, Song CK. Enhancement of dendritic cell-based vaccine potency by targeting antigen to endosomal/lysosomal compartments. Immunol Lett. 2006;106:126–134. doi: 10.1016/j.imlet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Peng S, Trimble C, Ji H, He L, Tsai Y-C, Macaes B. Characterization of HPV-16 E6 DNA vaccines employing intracellular targeting and intercellular spreading strategies. J Biomed Sci. 2005;12:689–700. doi: 10.1007/s11373-005-9012-3. [DOI] [PubMed] [Google Scholar]
- 36.Fassnacht M, Lee J, Milazzo C, Boczkowski D, Su Z, Nair S. Induction of CD4+ and CD8+ T-cell responses to the human stromal antigen, fibroblast activation protein: implication for cancer immunotherapy. Clin Cancer Res. 2005;11:5566–5571. doi: 10.1158/1078-0432.CCR-05-0699. [DOI] [PubMed] [Google Scholar]
- 37.Cheng W-F, Hung C-F, Hsu K-F, Chai C-Y, He L, Ling M. Enhancement of sindbis virus self-replicating RNA vaccine potency by targeting antigen to endosomal/lysosomal compartments. Hum Gene Ther. 2001;12:235–252. doi: 10.1089/10430340150218387. [DOI] [PubMed] [Google Scholar]
- 38.Ji H, Wang T-L, Chen C-H, Sara I, Hung C-F, Lin K-Y. Targeting human papillomavirus type 16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine human papillomavirus type 16 E7-expressing tumors. Hum Gene Ther. 1999;10:2727–2740. doi: 10.1089/10430349950016474. [DOI] [PubMed] [Google Scholar]
- 39.Kwon YJ, Standley SM, Goodwin AP, Gillies ER, Frechet JMJ. Directed antigen presentation using polymeric microparticulate carriers degradable at lysosomal pH for controlled immune responses. Mol Pharmaceutics. 2005;2:83–91. doi: 10.1021/mp0498953. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Huang Q, Wang W, Zhang Y, Lv P, Gao XM. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kessler PD, Podsakoff GM, Chen X, McQuiston SA, Colosi PC, Matelis LA. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Nat Acad Sci USA. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]


