Abstract
Purpose
Abnormal lipoprotein and amino acid profiles are associated with insulin resistance and may help to identify this condition. The aim of this study was to create models estimating skeletal muscle and whole-body insulin sensitivity using fasting metabolite profiles and common clinical and laboratory measures.
Material and Methods
The cross-sectional study population included 259 subjects with normal or impaired fasting glucose or type 2 diabetes in whom skeletal muscle and whole-body insulin sensitivity (M-value) were measured during euglycemic hyperinsulinemic clamp. Muscle glucose uptake (GU) was measured directly using [18F]FDG-PET. Serum metabolites were measured using nuclear magnetic resonance (NMR) spectroscopy. We used linear regression to build the models for the muscle GU (Muscle-insulin sensitivity index [ISI]) and M-value (whole-body [WB]-ISI). The models were created and tested using randomly selected training (n = 173) and test groups (n = 86). The models were compared to common fasting indices of insulin sensitivity, homeostatic model assessment—insulin resistance (HOMA-IR) and the revised quantitative insulin sensitivity check index (QUICKI).
Results
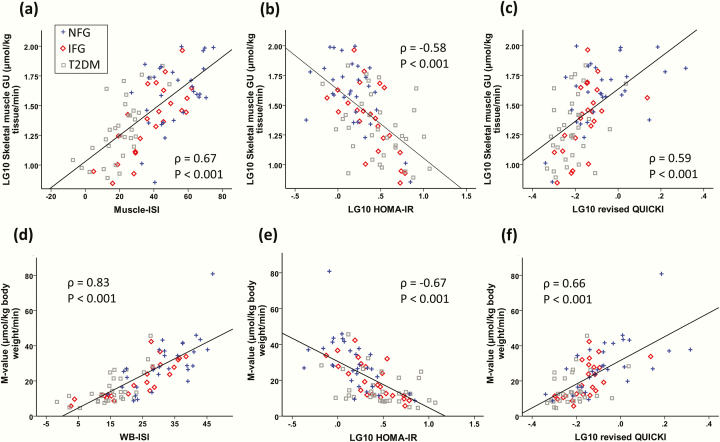
WB-ISI had higher correlation with actual M-value than HOMA-IR or revised QUICKI (ρ = 0.83 vs −0.67 and 0.66; P < 0.05 for both comparisons), whereas the correlation of Muscle-ISI with the actual skeletal muscle GU was not significantly stronger than HOMA-IR’s or revised QUICKI’s (ρ = 0.67 vs −0.58 and 0.59; both nonsignificant) in the test dataset.
Conclusion
Muscle-ISI and WB-ISI based on NMR-metabolomics and common laboratory measurements from fasting serum samples and basic anthropometrics are promising rapid and inexpensive tools for determining insulin sensitivity in at-risk individuals.
Keywords: insulin sensitivity, skeletal muscle, glucose uptake, positron emission tomography, type 2 diabetes, metabolomics
Surrogate insulin sensitivity indices are important for clinical studies where direct measurements would be too laborious and expensive. These indices have been usually validated against the euglycemic-hyperinsulinemic clamp study [1], which is the gold-standard method to measure whole-body insulin sensitivity (M-value). The euglycemic-hyperinsulinemic clamp [1] method is based on the glucose infusion rate, where glucose is kept steady at 5 mmol/L and insulin is infused at a constant rate, most commonly 40 mU/m2 body surface area/minute. Skeletal muscle is the main site of glucose disposal during euglycemic hyperinsulinemia and the key organ in postprandial glucose disposal [2]. Positron emission tomography (PET) in combination with 18F-fluorine-labeled deoxyglucose ([18F]FDG) and euglycemic-hyperinsulinemic clamp allows direct measurement of skeletal muscle insulin sensitivity independently of endogenous glucose production.
Recently, blood branch-chained amino acids (BCAAs), glutamine and glycine have been shown to robustly associate with prediabetes and type 2 diabetes (T2DM) and predict the risk of T2DM [3]. In addition, multiple studies have demonstrated that lipoprotein particle profiles vary according to the degree of glucose tolerance and insulin sensitivity [4–8] and can predict worsening of glycemia and incident T2DM [9–13]. Therefore, we hypothesized that serum metabolomics profiling could provide incremental information about the insulin resistance state in addition to insulin and glucose measurements, which form the basis of commonly used fasting surrogate indices of insulin sensitivity, such as the homeostatic model assessment—insulin resistance (HOMA-IR) [14] or revised quantitative insulin sensitivity check index (QUICKI) which include also fasting free fatty acids (FFAs) [15].
In the present study, we aimed to create models for predicting skeletal muscle insulin sensitivity (Muscle-ISI) and whole-body insulin sensitivity (WB-ISI) by using routine clinical and biochemical measurements and nuclear magnetic resonance (NMR)-metabolomics and compare the predictive performance of the new insulin sensitivity models to HOMA-IR and revised QUICKI. In addition, we aimed to evaluate associations of metabolites with muscle and whole-body insulin sensitivity to better understand the underlying metabolic patterns related to insulin resistance.
1. Participants and Methods
A. Participants
The study population consisted of 259 volunteers, who had participated the cross-sectional CMgene study (ClinicalTrials.gov: NCT03310502) which aims to investigate associations between tissue metabolism, blood biomarkers, and genetic variation. The subjects of CMgene were recruited from separate PET study projects investigating tissue metabolism using PET at Turku PET Centre (Turku, Finland). Subjects of this study consists of persons who participated in studies of muscle and M-value during euglycemic-hyperinsulinemic clamp [16–25] and had a fasting serum sample collected for metabolomics (Table 1). Recruitment for this study was done from 2002 to 2016. The population consisted of individuals with normal fasting glucose (NFG), impaired fasting glucose (IFG), or type 2 diabetes (T2DM). T2DM and IFG were defined by the American Diabetes Association criteria, T2DM by fasting glucose ≥ 7.0 mmol/L, 2-hour glucose ≥ 11.1 mmol/L or glycated hemoglobin A1c (HbA1c) ≥ 6.5% and IFG by fasting glucose between 5.6 and 6.9 mmol/L [26]. Oral glucose tolerance test (OGTT) was not available for all participants, hence it was not used to define the prediabetes group in our study. Seventy-five percent of the subjects with T2DM had diabetes medication but only 2 used thiazolidinedione medication and none was using insulin. HbA1c was 6.6% (5.9%–7.7%) among T2DM subjects in the training group and 6.4% (5.8%–7.3%) in T2DM subjects of the test group. Persons with a condition that could affect metabolism (alcohol or narcotics abuse, liver failure, or cancer) were excluded from the study. All participants gave a written informed consent. The study protocol was approved by the Ethics Committee of the Hospital District of Southwest Finland.
Table 1.
Characteristics of the Study Participants
| Training | Test | |||||||
|---|---|---|---|---|---|---|---|---|
| All | NFG | IFG | T2DM | All | NFG | IFG | T2DM | |
| n (females/males) | 173 (97/76) | 61 (23/38) | 52 (21/31) | 60 (32/28) | 86 (43/43) | 30 (8/22) | 23 (14, 9) | 33 (21, 12) |
| Age, years | 55 (47, 68) | 47 (39, 60) | 66 (53, 71) | 60 (52, 67) | 54 (45, 65) | 45 (38, 54) | 53 (43, 65) | 60 (52, 69) |
| BMI, kg/m2 | 28.4 (24.6, 32.2) | 25.2 (23.0, 28.7) | 27.6 (25.1, 30.8) | 31.3 (28.6, 33.9) | 28.7 (24.6, 31.6) | 25.8 (23.2, 30.4) | 28.1 (24.8, 35.0) | 29.5 (27.4, 32.0) |
| Fasting glucose, mmol/L | 5.8 (5.3, 6.4) | 5.3 (5.1, 5.4) | 5.9 (5.8, 6.3) | 6.9 (6.1, 8.5) | 5.8 (5.3, 6.5) | 5.2 (4.9, 5.3) | 5.9 (5.7, 6.3) | 7.0 (6.2, 7.5) |
| Fasting insulin, μU/mL | 7 (5, 10) | 5 (4, 7) | 8 (6, 10) | 9 (6, 13) | 8 (6, 12) | 7 (4, 9) | 9 (6. 14) | 10 (6, 18) |
| M-value, µmol/kg body weight/min | 20.0 (11.7, 32.7) | 33.1 (22.7, 40.9) | 21.0 (12.7, 30.2) | 11.0 (8.5, 16.5) | 18.3 (11.5, 28.6) | 27.4 (19.0, 37.7) | 17.4 (11.5, 29.2) | 11.9 (8.7, 21.4) |
| Muscle GU, µmol/kg tissue/min | 29.3 (16.4, 55.4) | 58.0 (31.7, 68.9) | 28.5 (19.7, 52.7) | 16.1 (12.0, 26.5) | 27.6 (16.8, 48.0) | 41.2 (26.7, 64.7) | 26.6 (13.0, 42.5) | 19.8 (12.9, 30.6) |
Data represented as median (interquartile range).
Abbreviations: IFG, impaired fasting glucose; M-value, whole-body insulin sensitivity; Muscle GU, skeletal muscle glucose uptake; NFG, normal fasting glucose; T2DM, type 2 diabetes.
B. Study design
The PET studies were performed after an overnight fast. The consumption of alcohol and caffeine was prohibited 12 hours before the study, and subjects were instructed to avoid strenuous physical activity 24 hours before the study. The subjects were lying in a supine position throughout the euglycemic-hyperinsulinemic clamp and PET scanning. Two cannulas were inserted: one in an antecubital vein for the infusion of glucose and insulin and the injection of [18F]FDG, and the other in the opposite upper extremity radial artery or antecubital vein, which was warmed with a heating pillow to arterialize venous blood. Plasma glucose was maintained at euglycemia (~5 mmol/L) by a primed and then continuous insulin infusion at 40 mU/body surface area m2/min and 20% glucose infusion based on plasma glucose measurements taken every 5 to 10 minutes [1]. The rates of M-value were calculated from steady state and reported as the average of 3 separate 20-minute intervals, starting after reaching euglycemia (median 60 minutes from the start of insulin infusion). [18F]FDG was injected at approximately 80 minutes (interquartile range, 61–93 minutes, 78 minutes on average) from the start of insulin infusion, and dynamic scans were performed to get images of femoral or upper arm regions as previously described [27, 28]. The timing when muscle was scanned varied according the original PET research protocol. Plasma radioactivity was measured from arterial or arterialized blood samples.
C. Measurement of skeletal muscle glucose uptake
[18F]FDG was synthesized using a modified method of Hamacher et al [29]. PET-scanners ECAT 931/08 (Siemens Molecular Imaging, Inc., Knoxville, TN), GE Advance, PET/CT Discovery VCT, and PET/CT Discovery 690 (General Electric Medical Systems, Milwaukee, WI) were used. The scanners were cross-calibrated against the same VDC-404 Dose calibrator (COMECER Netherlands, Joure, the Netherlands) to ensure the consistency of the results. All data obtained were corrected for dead time, decay, and measured photon attenuation. The Bayesian iterative reconstruction algorithm, using median root prior with iterations and a Bayesian coefficient of 0.3, was used for image processing when possible [30]. Skeletal muscle glucose uptake (GU) was measured by drawing regions of interest (ROI) to quadriceps femoris (n = 236) or deltoid (n = 23). Magnetic resonance imaging or computed tomography images were used as references for outlining the regions.
Skeletal muscle GU rates were calculated by graphically analyzing plasma and tissue time-activity curves to quantify the fractional phosphorylation rate (Ki) for [18F]FDG [31, 32]. The GU rates were calculated by multiplying Ki by the plasma glucose concentration and dividing by the tissue density and a lumped constant. A lumped constant corrects for the differences in transportation and phosphorylation of [18F]FDG and glucose. A lumped constant value of 1.2 for skeletal muscle was used [27]. Muscle insulin resistance was defined by having skeletal muscle GU rate lower than 33 μmol/kg tissue/min and whole-body insulin resistance by M-value less than 21 µmol/kg body weight/min [33].
D. Biochemical analyses
Plasma glucose was determined in duplicate by the glucose oxidase method (Analox GM7 or GM9, Analox Instruments, London, UK). Serum insulin concentration was measured by a double antibody radioimmunoassay (Phadeseph Insulin RIA kit, Pharmacia & Upjohn, Uppsala, Sweden), fluoroimmunometric assay (AutoDELFIA, PerkinElmer Inc, Turku, Finland) or automatized electro-chemiluminescence immunoassay (Cobas 8000, Roche Diagnostics GmbH, Mannheim, Germany). Alanine aminotransferase and aspartate aminotransferase were measured using automatized enzymatic method (Cobas 8000, Roche Diagnostics GmbH, Mannheim, Germany) and serum FFA concentration using an enzymatic assay (ACS-ACOD, Wako Chemicals GmbH, Neuss, Germany).
E. Serum NMR metabolomics
Metabolic biomarkers were quantified from serum from 259 individuals at fasting using high-throughput proton NMR metabolomics (Nightingale Health Ltd, Helsinki, Finland; University of Eastern Finland, Kuopio, Finland). This method provides simultaneous quantification of routine lipids, fatty acid (FA) composition, and various low-molecular metabolites including amino acids, ketone bodies, and gluconeogenesis-related metabolites in molar concentration units and lipoprotein subclass profiling with lipid concentrations within 14 subclasses. The 14 lipoprotein subclass sizes were defined as follows: extremely large very-low-density lipoprotein (VLDL) with particle diameters from 75 nm upwards and a possible contribution of chylomicrons, 5 VLDL subclasses (average particle diameters of 64.0 nm, 53.6 nm, 44.5 nm, 36.8 nm, and 31.3 nm), intermediate-density lipoprotein (IDL) (28.6 nm), 3 low-density lipoprotein (LDL) subclasses (25.5 nm, 23.0 nm, and 18.7 nm), and 4 high-density lipoprotein (HDL) subclasses (14.3 nm, 12.1 nm, 10.9 nm, and 8.7 nm). The following components of the lipoprotein subclasses were quantified: phospholipids, triglycerides, cholesterol, free cholesterol, and cholesteryl esters. The mean size for VLDL, LDL, and HDL particles were calculated by weighting the corresponding subclass diameters with their particle concentrations. Concentrations below the detection limits were considered as 0. Details of the experimentation and applications of the NMR metabolomics platform have been described previously [34–36].
F. Statistical methods and mathematical modeling
Two regression models were constructed to predict skeletal muscle GU and M-value (Tables 2 and 3). We call these models Muscle-ISI and WB-ISI, respectively. The regression models were built using linear model with lasso regularization [37]. The data was randomly divided into training data (total n = 173; NFG = 61, IFG = 52, T2DM = 60) and test data (total n = 86; NFG = 30, IFG = 23, T2DM = 33) [38] (Table 1) using R function sample. Both models were built using the training data. Independent test data were used for testing.
Table 2.
Coefficients in the Final (Muscle-ISI) Linear Regression Model for Skeletal Muscle Glucose Uptake (GU) Prediction
| Feature | Coefficient | Normalized Coefficient |
|---|---|---|
| Intercept | 62.0 | 38.2 |
| BMI, kg/m2 | -0.888 | -5.33 |
| Insulin, mU/L | -0.171 | -1.22 |
| Free fatty acids, mmol/L | -6.13 | -1.45 |
| Triglycerides in large HDL, mmol/L | 185 | 2.62 |
| Acetoacetate, mmol/L | -81.6 | -3.47 |
| LG10 Phospholipids in small HDL, mmol/L | -22.6 | -1.42 |
| LG10 Ratio of triglycerides to phosphoglycerides | -36.7 | -6.19 |
| LG10 Alanine, mmol/L | -46.7 | -3.77 |
| LG10 Glycine, mmol/L | 37.9 | 3.16 |
| Glycemic status (0 = NFG, 1 = IFG, 2 = T2DM) | -6.45 | -5.41 |
Estimate for skeletal muscle GU is calculated as a sum of the intercept and each subsequent row where the row values are obtained by multiplying value of the variable by the coefficient. Normalized coefficients describe how much variation in each variable contribute to the model. All metabolite and biochemical measurements were from fasting samples (glucose from plasma and others from serum).
Abbreviations: BMI, body mass index; HDL, high-density lipoprotein; IFG, impaired fasting glucose; NFG, normal fasting glucose; T2DM, type 2 diabetes.
Table 3.
Coefficients in the Final (WB-ISI) Linear Regression Model for Whole-Body Insulin Sensitivity (M-value) Prediction
| Feature | Coefficient | Normalized Coefficient |
|---|---|---|
| Intercept | 149 | 23.4 |
| Age, years | -0.0502 | -0.66 |
| BMI, kg/m2 | -0.754 | -4.52 |
| Glucose, mmol/L | -0.777 | -1.09 |
| Insulin, mU/L | -0.154 | -1.08 |
| Free fatty acids, mmol/L | -3.35 | -0.79 |
| Fraction of cholesterol esters of total lipids in medium HDL (%) | 0.373 | 1.02 |
| Alanine, mmol/L | -16.6 | -1.01 |
| Acetoacetate, mmol/L | -46.1 | -1.93 |
| LG10 Ratio of triglycerides to phosphoglycerides | -16.7 | -2.81 |
| LG10 Ratio of saturated FAs to total FAs | -53.2 | -1.31 |
| LG10 Glycine, mmol/L | 23.2 | 1.91 |
| LG10 Valine, mmol/L | -6.48 | -0.60 |
| LG10 Acetate, mmol/L | 6.23 | 0.61 |
| Glycemic status (0 = NFG, 1 = IFG, 2 = T2DM) | -2.85 | -2.40 |
Estimate for M-value is calculated as a sum of the intercept and each subsequent row where the row values are obtained by multiplying value of the variable by the coefficient. Normalized coefficients describe how much variation in each variable contribute to the model. All metabolite and biochemical measurements were from fasting samples (glucose from plasma and others from serum).
Abbreviations: BMI, body mass index; FA, fatty acid; HDL, high-density lipoprotein.
Features for the final models were selected using the training data. Features used in model building are listed in Supplemental Table 1 [39]. For both models, the feature ranking was first determined by creating 100 models using 10-fold cross-validation and defining frequency of each feature in the models. Features with frequency larger than 90% were then selected into the final model. Potential technical confounders (time of the clamp study or skeletal muscle scan) were evaluated by testing if they were significant predictors when added to the models.
The models were tested with the independent testing data, which were not used for model building. The models Muscle-ISI, WB-ISI, HOMA-IR and revised QUICKI were evaluated with Spearman correlations by comparing the estimated values with the measured values. HOMA-IR was calculated as fasting plasma glucose (mmol/L) * fasting serum insulin (mU/L) / 22.5 [14] and revised QUICKI as 1/(log plasma glucose (mmol/L) + log serum insulin (mU/L) + log serum FFAs (mmol/L)) [15].
Receiver-operating characteristic (ROC) analysis was used to find optimal cutoffs for Muscle-ISI, WB-ISI, HOMA-IR, and revised QUICKI for identifying skeletal muscle or whole-body insulin resistance and comparing discrimination performance of these indices. Youden index, which puts equal weight on sensitivity and specificity [40], was used to determine the cutoff points for Muscle-ISI, WB-ISI, HOMA-IR and revised QUICKI from the training data. The performance of Muscle-ISI and WB-ISI cutoff points was compared to HOMA-IR and revised QUICKI cutoff points by using McNemar’s test. Comparison between the actual skeletal muscle GU and M-value measurement and estimates from Muscle-ISI and WB-ISI were done using Wilcoxon signed rank test.
Associations between circulating metabolites and M-value and skeletal muscle insulin-stimulated GU were tested using Spearman correlations. The mathematical modeling and statistical analysis were done using R statistical computing environment (version 3.4) [41]. The R package glmnet [42] was used for linear regression models with lasso regularization, heatmaps were created with the package gplots [43], and areas under the curve (AUCs) in ROC analysis were compared using the package pROC [44]. Correlation coefficients were compared using package WRS [45].
2. Results
A. Prediction of skeletal muscle glucose uptake and whole-body insulin sensitivity
The regression equations Muscle-ISI and WB-ISI for prediction of skeletal muscle GU and M-value are described in Tables 2 and 3. WB-ISI had higher correlation with the actual M-value than HOMA-IR or revised QUICKI in the test group, whereas correlation between Muscle-ISI and muscle GU was not significantly stronger than those between muscle GU and HOMA-IR or revised QUICKI (Tables 4 and 5, Fig. 1). The models gave significantly higher values than the actual skeletal muscle GU and M-value measurements in IFG subjects (Table 6). Medications are a possible error source which could lower the performance of predictive indices. Muscle-ISI and WB-ISI had a significant correlation with the actual measurement among subgroups of subjects with metformin (n = 19) or statin therapy (n = 17) in the test group whereas correlations of HOMA-IR and revised QUICKI with muscle GU and M-value were mostly not significant (Supplemental Tables 2–5) [39]. Furthermore, the models performed well among persons without metformin (correlation Muscle-ISI vs skeletal muscle GU ρ = 0.58, P < 0.001, WB-ISI vs M-value ρ = 0.80, P < 0.001) or statin medication (correlation Muscle-ISI vs skeletal muscle GU ρ = 0.65, P < 0.001, WB-ISI vs M-value ρ = 0.83, P < 0.001). Clamp or skeletal muscle study timings were not significant predictors of M-value or skeletal muscle GU when added to WB-ISI or Muscle-ISI models.
Table 4.
Spearman Correlation Between Measured and Predicted Skeletal Muscle GU Values and P Values for Comparisons of Correlation Coefficients Between Different Indices
| Spearman Correlation | P Value | P Value | |||
|---|---|---|---|---|---|
| Muscle-ISI | HOMA-IR | revQUICKI | Muscle-ISI vs HOMA-IR | Muscle-ISI vs revQUICKI | |
| Training | 0.80 ** | -0.56 * | 0.61 ** | <0.001 | 0.003 |
| Test | 0.67 ** | -0.58 ** | 0.59 ** | 0.280 | 0.357 |
Abbreviations: HOMA-IR, homeostatic model assessment—insulin resistance; Muscle-ISI, muscle insulin sensitivity; revQUICKI, revised quantitative insulin sensitivity check index.
**P < 0.001; *P < 0.01.
Table 5.
Spearman Correlation Between Measured and Predicted Whole-Body Insulin Sensitivity (M-value) and P Values for Comparisons of Correlation Coefficients Between Different Indices
| Spearman correlation | P value | P value | |||
|---|---|---|---|---|---|
| WB-ISI | HOMA-IR | revQUICKI | WB-ISI vs HOMA-IR | WB-ISI vs revQUICKI | |
| Training | 0.86 * | -0.66 * | 0.70 * | <0.001 | <0.001 |
| Test | 0.83 * | -0.67 * | 0.66 * | 0.017 | 0.020 |
Abbreviations: HOMA-IR, homeostatic model assessment—insulin resistance; Muscle-ISI, muscle insulin sensitivity; revQUICKI, revised quantitative insulin sensitivity check index; WB-ISI, whole-body insulin sensitivity.
*P < 0.001.
Figure 1.
Spearman correlations between skeletal muscle glucose uptake (GU) with Muscle-ISI (a), HOMA-IR (b) and revised QUICKI (c) and whole-body insulin sensitivity (M-value) with WB-ISI (d), HOMA-IR (e), and revised QUICKI (f).
Table 6.
Actual and Estimated Values for Whole-Body Insulin Sensitivity (M-value) and Skeletal Muscle Glucose Uptake (GU) in the Test Group
| NFG | IFG | T2DM | |
|---|---|---|---|
| Actual M-value, μmol/kg body weight/min | 27.4 (19.0, 37.7) | 17.4 (11.5, 29.2) | 11.9 (8.7, 21.4) |
| Estimated M-value, µmol/kg body weight/min | 31.5 (23.1, 39.1) | 26.4 (14.2, 30.5)* | 16.0 (11.7, 19.2) |
| Actual skeletal muscle GU, µmol/kg tissue/min | 41.2 (26.7, 64.7) | 26.6 (13.0, 42.5) | 19.8 (12.9, 30.6) |
| Estimated skeletal muscle GU, μmol/kg tissue/min | 51.3 (40.5, 65.4) | 41.2 (28.4, 49.8)* | 22.4 (13.3, 28.3) |
Data represented as median (interquartile range). *P < 0.05 compared to the actual measurement.
Abbreviations: BMI, body mass index; HDL, high-density lipoprotein; IFG, impaired fasting glucose; NFG, normal fasting glucose; T2DM, type 2 diabetes.
Metabolite predictors of insulin resistance in the models included glucose, FFAs, triglycerides to phosphoglycerides ratio, fraction of saturated fatty acids (FAs) from total FAs, small HDL phospholipids, large VLDL triglycerides, total serum triglycerides, alanine, valine, acetoacetate, and omega-3 FAs (Tables 2 and 3). On the contrary, markers of insulin sensitivity included large HDL triglycerides, fraction of cholesterol esters to total lipids in medium HDL, glycine, and acetate.
B. Correlations between insulin sensitivity with metabolites and fatty acids
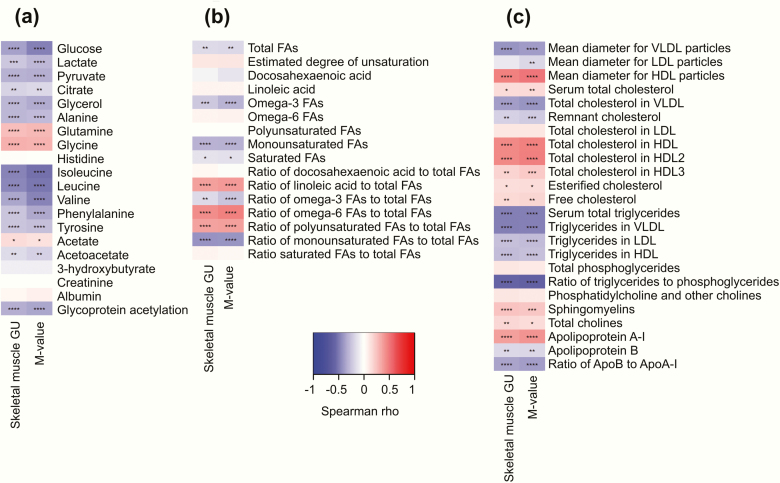
Correlation testing in the whole population showed positive correlations between insulin sensitivity and glycine and glutamine, whereas alanine, isoleucine, leucine, valine, phenylalanine and tyrosine had negative correlation with insulin sensitivity (Fig. 2a). Metabolites related to glycolysis correlated negatively with insulin sensitivity. Monounsaturated and omega-3 FA concentrations were negatively correlated with insulin sensitivity, while linoleic acid, omega-6 FAs, and total polyunsaturated FAs were positively correlated (Fig. 2b).
Figure 2.
Correlation heatmaps between skeletal muscle glucose uptake (GU) and whole-body insulin sensitivity (M-value) with metabolite (a), fatty acid (b), and lipoprotein measures (c). Abbreviation: FAs, fatty acids. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
C. Correlations between insulin sensitivity with lipoprotein subclass measures
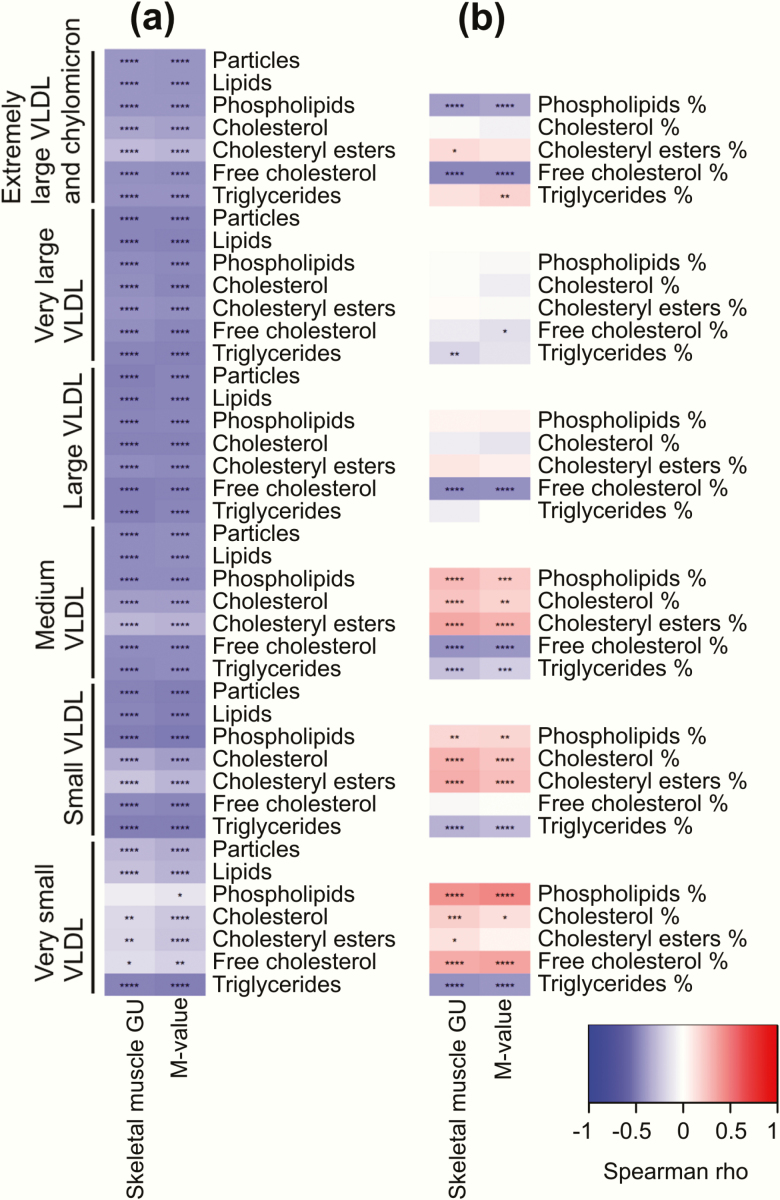
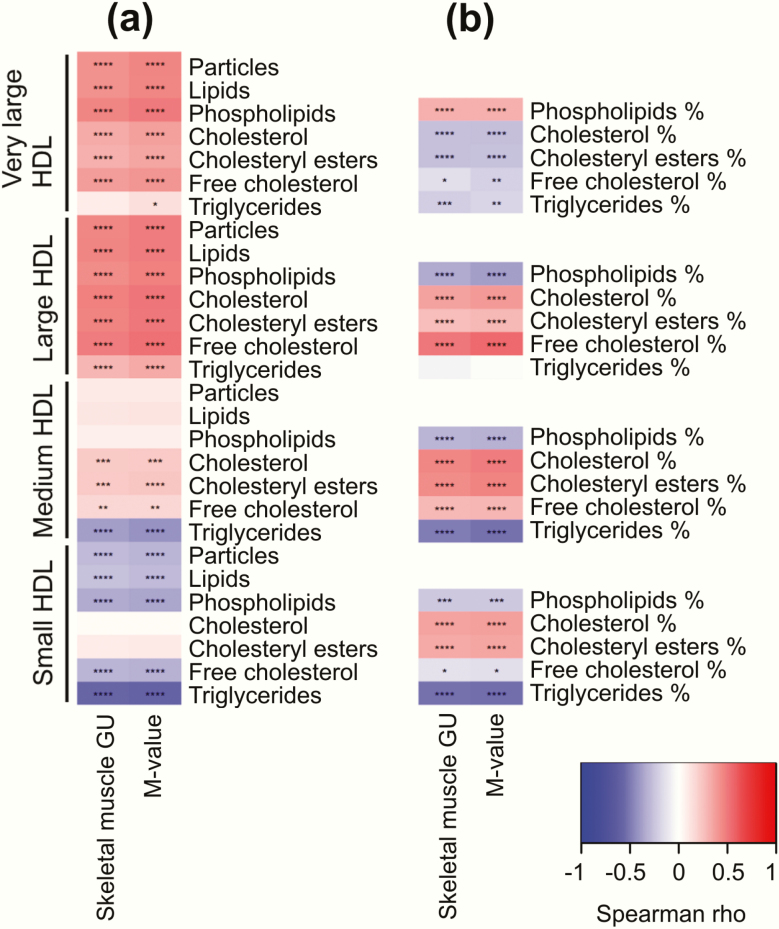
Lipids typical of lipoprotein particle membranes such as sphingomyelins, cholines, and free cholesterol were positively correlated with insulin sensitivity, whereas triglycerides were negatively correlated (Fig. 2c). More detailed inspection of lipoprotein particle composition showed that triglycerides were negatively correlated with insulin sensitivity across all the studied lipoprotein subclasses (Figs. 3–5), except large and very large HDL (Fig. 5).
Figure 3.
Correlation heatmaps between skeletal muscle glucose uptake (GU) and whole-body insulin sensitivity (M-value) with different lipoprotein subclasses: VLDL particles and lipids (a) and VLDL lipid fractions (b). ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
Figure 5.
Correlation heatmaps between skeletal muscle glucose uptake (GU) and whole-body insulin sensitivity (M-value) with different lipoprotein subclasses: HDL particles and lipids (a) and HDL lipid fractions (b). ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
All lipids in VLDL subclasses were negatively correlated with insulin sensitivity; the correlations were strongest among the larger subclasses (Fig. 3a). Further examination of the VLDL subclass lipid fractions revealed that the fractions of free cholesterol and phospholipids to total lipids in the very large VLDL were negatively correlated with insulin sensitivity and free cholesterol and phospholipid fraction in the very small VLDL were positively correlated with insulin sensitivity (Fig. 3b).
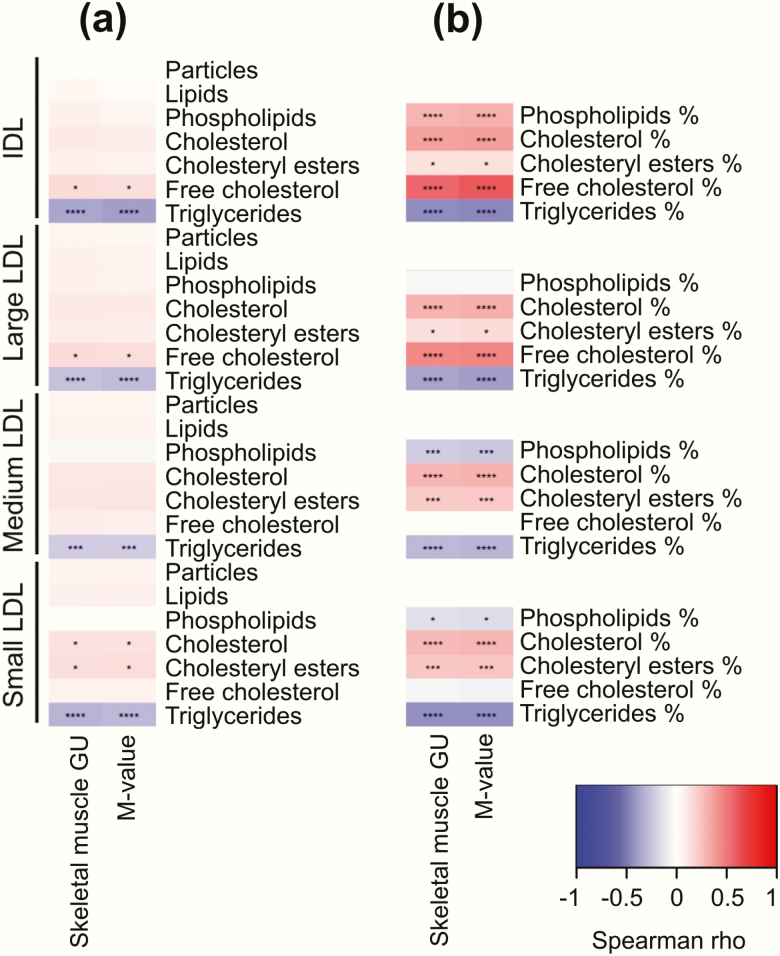
Concentrations of LDL and IDL subclass lipids showed only weak correlations with insulin sensitivity apart the triglycerides (Fig. 4a). However, fraction of phospholipids to total lipids in medium and small LDL were negatively correlated with insulin sensitivity similarly to triglyceride fraction and fraction of cholesterol to total lipids had a positive correlation to insulin sensitivity (Fig. 4b).
Figure 4.
Correlation heatmaps between skeletal muscle glucose uptake (GU) and whole-body insulin sensitivity (M-value) with different lipoprotein subclasses: IDL and LDL particles and lipids (a) and IDL and LDL lipid fractions (b). ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
Fraction of triglycerides to total lipid content in very large HDL particles had negative correlation with insulin sensitivity (Fig. 4b). In contrast to large HDL particles, concentration of small HDL particles was negatively correlated with insulin sensitivity (Fig. 5a). Evaluation of small HDL lipids showed that fractions of phospholipids, free cholesterol, and triglycerides to total lipids were negatively correlated with insulin sensitivity, while fractions of total cholesterol and cholesterol esters correlated positively (Fig. 5b).
D. Identifying insulin resistant subjects
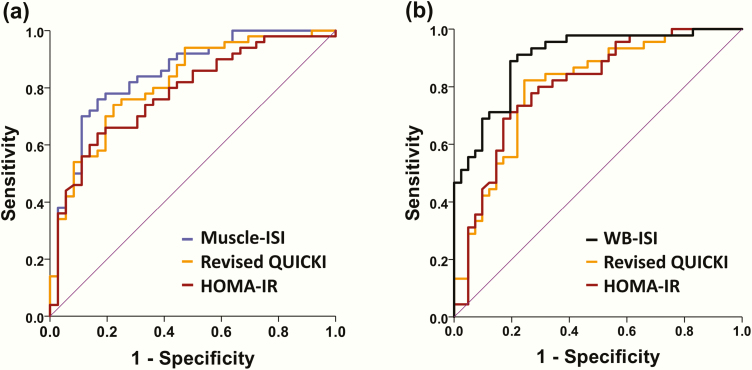
Muscle-ISI and WB-ISI showed very good performance to discriminate between insulin resistant and sensitive individuals (Fig. 6). AUC for Muscle-ISI was 0.85 and for WB-ISI 0.90 in the test group. Also, HOMA-IR (AUC 0.78 for muscle and 0.80 for whole-body insulin resistance) and revised QUICKI (0.82 and 0.80) had good performance in these tasks. WB-ISI was to superior to HOMA-IR and revised QUICKI for detecting whole-body insulin resistance (P = 0.018 and 0.004) in this group, whereas Muscle-ISI was not significantly better than HOMA-IR or revised QUICKI to determine skeletal muscle insulin resistance (P = 0.107 and 0.395) (Fig. 6).
Figure 6.
Receiver-operating characteristic (ROC) curve comparing sensitivity and specificity of (a) Muscle-ISI, HOMA-IR, and revised QUICKI for determining skeletal muscle insulin resistance in the test group and sensitivity and specificity of (b) WB-ISI, HOMA-IR, and revised QUICKI for determining whole-body insulin resistance. Area under the ROC curve for predicting skeletal muscle insulin resistance: Muscle-ISI 0.85, HOMA-IR 0.78, and revised QUICKI 0.82; Area under the ROC curve for predicting whole-body insulin resistance: WB-ISI 0.90, HOMA-IR 0.80, and revised QUICKI 0.80.
We used ROC analysis among training subjects to find optimal cutoff points for Muscle-ISI, WB-ISI, HOMA-IR, and revised QUICKI for distinguishing between persons with and without skeletal muscle and whole-body insulin resistance by using the previously published thresholds for skeletal muscle and M-value as reference standards [33]. The observed cutoffs to determine muscle insulin resistance were 41.2 for Muscle-ISI (87% sensitivity and 84% specificity), 1.83 for HOMA-IR (76% and 76%), and 0.751 (83% and 75%) for revised QUICKI. For determining whole-body insulin resistance, we found cutoffs 23.6 for WB-ISI (86% sensitivity and 88% specificity), 1.83 for HOMA-IR (81% and 80%) and 0.751 for revised QUICKI (89% and 78%). We then tested the performance of these cutoffs among test subjects. We found that the cutoff for Muscle-ISI had 82% sensitivity and 72% specificity, the cutoff for HOMA-IR had 74% sensitivity and 67% specificity, while revised QUICKI had 86% sensitivity and 56% specificity to detect skeletal muscle insulin resistance. The cutoff for Muscle-ISI tended to have higher specificity than revised QUICKI (P = 0.070) in this task. The cutoff for WB-ISI had 86% sensitivity and 81% specificity, the cutoff for HOMA-IR had 80% sensitivity and 68% specificity, and the cutoff for revised QUICKI had 89% sensitivity and 54% specificity for determining whole-body insulin resistance. WB-ISI had higher specificity than revised QUICKI (P = 0.002).
3. Discussion
In this study, we created Muscle-ISI and WB-ISI models for predicting skeletal muscle GU and M-value in a cohort spanning a wide range of body adiposity and insulin sensitivity. WB-ISI performed better than HOMA-IR or revised QUICKI in predicting M-value and may thus be useful for determining whether a person has insulin resistance when more accurate methods are too costly, time-consuming, laborious or ethically not feasible. The surrogate index for skeletal muscle insulin sensitivity was created and validated against the direct measurement of skeletal muscle insulin-stimulated GU and may be useful especially in research settings where muscle insulin sensitivity is of interest, but direct measurement is not feasible.
To build Muscle-ISI and WB-ISI we used linear model with lasso (least absolute shrinkage and selection operator) [37]. Lasso is a regression method, which does simultaneous variable selection and regularization. While typical variable selection methods such as principal component analysis create new variables based on the original ones, lasso uses the original variables with regularized coefficients. Models built using lasso have only a few original features, which makes them easy to interpret. For example, the coefficient −0.888 for body mass index (BMI) in model Muscle-ISI (Table 2) indicates that larger BMI is related to lower skeletal muscle GU.
Insulin, glucose and glycemic status appeared in the models for skeletal muscle GU and M-value, which is not surprising considering the compensatory insulin secretion and raising glucose levels in the insulin resistant state. FFA level in the models is a marker of adipose tissue lipolysis. FFAs are implicated to have a causal role in insulin resistance of various tissues [46]. Moreover, BMI is present in both models, which is unsurprising given that obesity is likely a causal factor for insulin resistance and precedes other metabolic disturbances [47, 48].
An increase in VLDL particles and lipids, especially in large, very large, and extremely large VLDL and chylomicrons, was correlated with a decrease in skeletal muscle and whole-body insulin sensitivity. These findings agree with earlier reports connecting larger VLDL particles with lower insulin sensitivity, worse glycemic control and higher T2DM risk [4–6, 8–13]. A likely cause for the association between VLDL lipids and insulin resistance is overproduction of VLDL particles and triglycerides from the liver and decreased hydrolysis of FAs from VLDL and chylomicron triglycerides by lipoprotein lipase in other tissues in an insulin resistant state [49, 50].
Higher fraction of triglycerides compared to total lipids was correlated with lower insulin sensitivity across the HDL subclasses. HDL triglycerides reflect exchange of cholesterol esters from HDL with triglycerides in VLDL and chylomicrons by the cholesteryl ester transfer protein (CETP) [51, 52]. This means that increase in HDL triglycerides is another sign of VLDL triglyceride overproduction. In addition, amount of plasma CETP is positively correlated with liver fat content [53], which associates cholesteryl exchange with hepatic insulin resistance. Lipolysis of the large triglyceride-rich particles result in formation of smaller remnant particles which are rich in triglycerides and cholesteryl esters. These particles are a likely causal link between insulin resistance and cardiovascular disease because they can cross arterial walls themselves or, after further lipolysis, as small dense LDL and promote atherosclerosis [50, 54].
Increased monounsaturated fatty acids (MUFAs) were negatively and the ratio of omega 6-FAs to total FAs positively correlated with insulin sensitivity, which agrees with a larger study where MUFAs were predictors of worse present and future glycemia and omega-6 FAs to total FAs was associated with better glycemic profiles [55]. Surprisingly, we found that omega-3 FAs were predictors of insulin resistance. However, while observational and animal studies have found positive associations between omega-3 FAs and insulin sensitivity, trials using omega-3 FA supplementation in humans have shown no benefit on improving insulin sensitivity, which agrees with our finding [56].
We found negative correlations between insulin sensitivity and BCAAs (leucine, isoleucine, valine) and aromatic amino acids (phenylalanine, tyrosine), whereas glycine and glutamine had positive correlation. BCAAs and aromatic amino acids have been associated with increased T2DM risk and glycine and glutamine with lower T2DM risk [3]. Recent Mendelian randomization studies suggest that increased BCAAs may contribute to etiology of T2DM [57] and that insulin resistance increases BCAA concentrations [58, 59]. Genes implicated in these studies suggest that defective catabolism of BCAAs may cause the increased blood BCAA concentrations. In addition, higher acetoacetate was correlated with lower insulin sensitivity in our study. Higher acetoacetate has been previously shown to associate with lower insulin secretion and predict future T2DM among middle-aged men [60]. Of note, acetoacetate had a nonsignificant positive correlation with insulin sensitivity among persons with IFG in the test group, which may have contributed to the overestimation of Muscle-ISI and WB-ISI in this group.
An important finding in this study is that WB-ISI had higher correlation with the gold-standard measurement of whole-body insulin sensitivity than HOMA-IR and revised QUICKI in the test group. This means that serum metabolomics can provide information about insulin sensitivity beyond fasting glucose and insulin measurements. Similarly, Muscle-ISI had a good correlation with insulin-stimulated muscle GU in the test group, albeit not significantly higher than correlations of muscle GU with HOMA-IR or revised QUICKI. Despite HOMA-IR having only moderate correlation with M-value, there is a considerable amount of literature supporting the usefulness of HOMA-IR when estimating risk of T2DM and cardiovascular disease [61–65]. Thus, WB-ISI and Muscle-ISI should also be able to provide clinically meaningful information about T2DM and cardiovascular disease risk.
High AUCs of the ROC curves in the test population show that Muscle-ISI (AUC 0.85) and WB-ISI (0.90) have good performance in determining insulin resistance, based on the previously published cutoffs for muscle insulin-stimulated GU and M-value [33, 66]. WB-ISI had higher AUC of the ROC curve when compared to HOMA-IR and revised QUICKI in the test group. This means that the newly developed indices may be useful for classifying persons as insulin resistant or sensitive. Moreover, we have provided cutoff points for Muscle-ISI and WB-ISI which can be used for this purpose. It should be noted that these cutoffs are meant to be used as a tool to assist identifying persons at risk and not as definite diagnostic criteria. Ideally an optimal tool to determine cutoffs to find persons at risk would be a population-based follow-up study which evaluates how a certain level of insulin resistance translates to future T2DM and cardiovascular disease risk. An Iranian population-based study found HOMA-IR values of 1.85 in women and 2.17 in men to optimally separate persons who later develop T2DM and those who do not [67], but in general, this type of data is scarce and the cutoffs would need to be determined separately in different populations. More research should be performed in this area to determine the possible benefits for using cutoffs when identifying persons at risk.
Strengths of this study include direct determination of skeletal muscle insulin-stimulated GU and having a study population consisting of subjects of both sexes with wide range of insulin sensitivity, BMI, and age. Direct measurement of skeletal muscle insulin-stimulated GU circumvents the possible confounding to M-value measurement by endogenous glucose production. In addition, the determination of serum metabolites is relatively inexpensive. Even though setting up an advanced NMR-metabolomic platform is an expensive task, cost per sample can be likely reduced to a less than $17 when analyses are performed continuously in large scale [68], which would add only a small cost over that of traditional glucose and insulin measurements. The NMR-metabolomic panel used in this study is commercially available and has been already used to characterize hundreds of thousands of blood samples including samples from various large study cohorts [69].
This study has some limitations. The numbers of NFG, IFG, and T2DM subjects were too small to compare performance between the indices created here, HOMA-IR, and revised QUICKI inside these subgroups, albeit the correlations between the new indices and gold-standard measurement was numerically highest in most comparisons (Supplemental Tables 6–7) [39]. The same is true for the comparison between indices evaluating skeletal muscle insulin sensitivity in the whole group. Because this study was based on previous independent projects, the timing of the clamp study and skeletal muscle scan was different across these projects and scan of quadriceps femoris was not available for 23 subjects in our study cohort. Nevertheless, timing of the clamp or scan time did not appear as significant predictor when added to WB-ISI or Muscle-ISI models and Muscle-ISI showed a significant correlation with muscle GU measured from deltoid among the 23 subjects. In addition, OGTT was not performed for all study participants, therefore we could not compare our models with tests of insulin sensitivity based on OGTT.
It should be noted that lipid and glucose medication could potentially reduce accuracy of insulin sensitivity models based on fasting blood lipid and glucose measures. Nevertheless, the use of these medications did not appear as predictors in the models created here and the models were relatively robust for the use of statins or metformin when tested separately in subgroups using these medications and compared to HOMA-IR and revised QUICKI. Finally, insulin measurement is known to vary according to the used assay. Even so, the associations found between M-value and HOMA-IR or revised QUICKI correspond well with the published literature [70].
We have provided a skeletal muscle insulin sensitivity index validated against the gold-standard measurement. Moreover, the WB-ISI model for whole-body insulin sensitivity created here can identify M-value better than HOMA-IR or revised QUICKI in subjects with varying level of glycemic control. In all, these models could be used as easy and inexpensive tools for identifying insulin sensitivity in large clinical studies or clinical setting where more complex measurements would be too costly or time consuming.
Acknowledgments
Financial Support: The study was conducted within the Finnish Centre of Excellence in Cardiovascular and Metabolic Diseases, supported by the Academy of Finland, the University of Turku, Turku University Hospital, Åbo Akademi University and the University of Eastern Finland (grant number 307402 to PN). This work was supported by the Finnish Cultural Foundation, Varsinais-Suomi Regional fund (grant numbers 85141457 and 85182229), Yrjö Jahnsson Foundation (grant number 20146673), Turku University Foundation (grant numbers 10418 and 13179), the University of Turku, Combined Research Fund; and Oskar Öflunds Stiftelse grants to MJH; the Orion Research Foundation grants to MJH and JCH; the Emil Aaltonen Foundation, the European Foundation for the Study of Diabetes, the Diabetes Research Foundation, and the Hospital District of Southwest Finland grants to JCH. In addition, this work was supported by Academy of Finland (grant numbers 296801 and 310561) grants to LLE.
Clinical Trial Information: ClinicalTrials.gov identifier NCT03310502.
Author Contributions: RK contributed to building of the regression models, statistical analysis and writing the manuscript. MJH contributed to planning of the work, collection of data, statistical analysis and writing the manuscript. VH, MB, ALR, KKK, KAV, JCH and RL collected data. MSV performed replication analysis for the regression models. JCH and PI contributed to interpretation of the results and revised the manuscript. LLE and PN supervised the project, interpreted the results and revised the manuscript.
Glossary
Abbreviations
- AUC
area under the curve
- BCAA
branch-chained amino acid
- BMI
body mass index
- FDG
fluorodeoxyglucose
- FA
fatty acid
- FFA
free fatty acid
- GU
glucose uptake
- HDL
high-density lipoprotein
- HOMA-IR
homeostatic model assessment—insulin resistance
- IDL
intermediate-density lipoprotein
- IFG
impaired fasting glucose
- LDL
low-density lipoprotein
- Muscle-ISI
muscle insulin sensitivity index
- NFG
normal fasting glucose
- NMR
nuclear magnetic resonance
- OGTT
oral glucose tolerance test
- PET
positron emission tomography
- QUICKI
quantitative insulin sensitivity check index
- ROC
receiver operating characteristic
- T2DM
type 2 diabetes mellitus
- VLDL
very-low-density lipoprotein
- WB-ISI
whole-body insulin sensitivity index
Additional Information:
Disclosure summary: Nothing to disclose
Data Availability: Restrictions apply to the availability of data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided.
References
- 1. DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237(3):E214–E223. [DOI] [PubMed] [Google Scholar]
- 2. Defronzo RA. Banting Lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58(4):773–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guasch-Ferré M, Hruby A, Toledo E, et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39(5):833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J, Stančáková A, Soininen P, et al. Lipoprotein subclass profiles in individuals with varying degrees of glucose tolerance: a population-based study of 9399 Finnish men. J Intern Med. 2012;272(6):562–572. [DOI] [PubMed] [Google Scholar]
- 5. Goff DC Jr, D’Agostino RB Jr, Haffner SM, Otvos JD. Insulin resistance and adiposity influence lipoprotein size and subclass concentrations. Results from the Insulin Resistance Atherosclerosis Study. Metabolism. 2005;54(2):264–270. [DOI] [PubMed] [Google Scholar]
- 6. Garvey WT, Kwon S, Zheng D, et al. Effects of insulin resistance and type 2 diabetes on lipoprotein subclass particle size and concentration determined by nuclear magnetic resonance. Diabetes. 2003;52(2):453–462. [DOI] [PubMed] [Google Scholar]
- 7. Cartolano FC, Dias GD, de Freitas MCP, Figueiredo Neto AM, Damasceno NRT. Insulin resistance predicts atherogenic lipoprotein profile in nondiabetic subjects. J Diabetes Res. 2017;2017:1018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacLean PS, Vadlamudi S, MacDonald KG, Pories WJ, Houmard JA, Barakat HA. Impact of insulin resistance on lipoprotein subpopulation distribution in lean and morbidly obese nondiabetic women. Metabolism. 2000;49(3):285–292. [DOI] [PubMed] [Google Scholar]
- 9. Festa A, Williams K, Hanley AJ, et al. Nuclear magnetic resonance lipoprotein abnormalities in prediabetic subjects in the Insulin Resistance Atherosclerosis Study. Circulation. 2005;111(25):3465–3472. [DOI] [PubMed] [Google Scholar]
- 10. Hodge AM, Jenkins AJ, English DR, O’Dea K, Giles GG. NMR-determined lipoprotein subclass profile predicts type 2 diabetes. Diabetes Res Clin Pract. 2009;83(1):132–139. [DOI] [PubMed] [Google Scholar]
- 11. Mora S, Otvos JD, Rosenson RS, Pradhan A, Buring JE, Ridker PM. Lipoprotein particle size and concentration by nuclear magnetic resonance and incident type 2 diabetes in women. Diabetes. 2010;59(5):1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fizelova M, Miilunpohja M, Kangas AJ, et al. Associations of multiple lipoprotein and apolipoprotein measures with worsening of glycemia and incident type 2 diabetes in 6607 non-diabetic Finnish men. Atherosclerosis. 2015;240(1):272–277. [DOI] [PubMed] [Google Scholar]
- 13. Mackey RH, Mora S, Bertoni AG, et al. Lipoprotein particles and incident type 2 diabetes in the multi-ethnic study of atherosclerosis. Diabetes Care. 2015;38(4):628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. [DOI] [PubMed] [Google Scholar]
- 15. Perseghin G, Caumo A, Caloni M, Testolin G, Luzi L. Incorporation of the fasting plasma FFA concentration into QUICKI improves its association with insulin sensitivity in nonobese individuals. J Clin Endocrinol Metab. 2001;86(10):4776–4781. [DOI] [PubMed] [Google Scholar]
- 16. Lautamäki R, Airaksinen KE, Seppänen M, et al. Rosiglitazone improves myocardial glucose uptake in patients with type 2 diabetes and coronary artery disease: a 16-week randomized, double-blind, placebo-controlled study. Diabetes. 2005;54(9):2787–2794. [DOI] [PubMed] [Google Scholar]
- 17. Lindroos MM, Majamaa K, Tura A, et al. m.3243A>G mutation in mitochondrial DNA leads to decreased insulin sensitivity in skeletal muscle and to progressive beta-cell dysfunction. Diabetes. 2009;58(3):543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Latva-Rasku A, Honka MJ, Stančáková A, et al. ; T2D-GENES Consortium A Partial Loss-of-Function Variant in AKT2 Is Associated With Reduced Insulin-Mediated Glucose Uptake in Multiple Insulin-Sensitive Tissues: A Genotype-Based Callback Positron Emission Tomography Study. Diabetes. 2018;67(2):334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bucci M, Huovinen V, Guzzardi MA, et al. Resistance training improves skeletal muscle insulin sensitivity in elderly offspring of overweight and obese mothers. Diabetologia. 2016;59(1):77–86. [DOI] [PubMed] [Google Scholar]
- 20. Hirvonen J, Virtanen KA, Nummenmaa L, et al. Effects of insulin on brain glucose metabolism in impaired glucose tolerance. Diabetes. 2011;60(2):443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Viljanen AP, Lautamäki R, Järvisalo M, et al. Effects of weight loss on visceral and abdominal subcutaneous adipose tissue blood-flow and insulin-mediated glucose uptake in healthy obese subjects. Ann Med. 2009;41(2):152–160. [DOI] [PubMed] [Google Scholar]
- 22. Orava J, Nuutila P, Noponen T, et al. Blunted metabolic responses to cold and insulin stimulation in brown adipose tissue of obese humans. Obesity (Silver Spring). 2013;21(11):2279–2287. [DOI] [PubMed] [Google Scholar]
- 23. Eskelinen JJ, Heinonen I, Löyttyniemi E, et al. Muscle-specific glucose and free fatty acid uptake after sprint interval and moderate-intensity training in healthy middle-aged men. J Appl Physiol (1985). 2015;118(9):1172–1180. [DOI] [PubMed] [Google Scholar]
- 24. Immonen H, Hannukainen JC, Iozzo P, et al. Effect of bariatric surgery on liver glucose metabolism in morbidly obese diabetic and non-diabetic patients. J Hepatol. 2014;60(2):377–383. [DOI] [PubMed] [Google Scholar]
- 25. Dadson P, Landini L, Helmiö M, et al. Effect of Bariatric Surgery on Adipose Tissue Glucose Metabolism in Different Depots in Patients With or Without Type 2 Diabetes. Diabetes Care. 2016;39(2):292–299. [DOI] [PubMed] [Google Scholar]
- 26. American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37(Supplement 1):S81-S90. doi: 10.2337/dc14-S081. [DOI] [PubMed] [Google Scholar]
- 27. Peltoniemi P, Lönnroth P, Laine H, et al. Lumped constant for [(18)F]fluorodeoxyglucose in skeletal muscles of obese and nonobese humans. Am J Physiol Endocrinol Metab. 2000;279(5):E1122–E1130. [DOI] [PubMed] [Google Scholar]
- 28. Orava J, Nuutila P, Lidell ME, et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab. 2011;14(2):272–279. [DOI] [PubMed] [Google Scholar]
- 29. Hamacher K, Coenen HH, Stöcklin G. Efficient stereospecific synthesis of no-carrier-added 2-[18F]-fluoro-2-deoxy-D-glucose using aminopolyether supported nucleophilic substitution. J Nucl Med. 1986;27(2):235–238. [PubMed] [Google Scholar]
- 30. Alenius S, Ruotsalainen U. Bayesian image reconstruction for emission tomography based on median root prior. Eur J Nucl Med. 1997;24(3):258–265. [DOI] [PubMed] [Google Scholar]
- 31. Patlak CS, Blasberg RG. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations. J Cereb Blood Flow Metab. 1985;5(4):584–590. [DOI] [PubMed] [Google Scholar]
- 32. Gjedde A. Calculation of cerebral glucose phosphorylation from brain uptake of glucose analogs in vivo: a re-examination. Brain Res. 1982;257(2):237–274. [DOI] [PubMed] [Google Scholar]
- 33. Honka MJ, Latva-Rasku A, Bucci M, et al. Insulin-stimulated glucose uptake in skeletal muscle, adipose tissue and liver: a positron emission tomography study. Eur J Endocrinol. 2018;178(5):523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inouye M, Kettunen J, Soininen P, et al. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol. 2010;6:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8(1):192–206. [DOI] [PubMed] [Google Scholar]
- 36. Soininen P, Kangas AJ, Würtz P, et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134(9):1781–1785. [DOI] [PubMed] [Google Scholar]
- 37.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B-Methodol. 1996;58(1):267–288.
- 38. Dobbin KK, Simon RM. Optimally splitting cases for training and testing high dimensional classifiers. BMC Med Genomics. 2011;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klén R, Honka MJ, Hannukainen JC, et al. Supplementary data. 2020. doi: 10.6084/m9.figshare.9767609.v2. [DOI] [Google Scholar]
- 40. Greiner M, Pfeiffer D, Smith RD. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med. 2000;45(1-2):23–41. [DOI] [PubMed] [Google Scholar]
- 41. R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 42. Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 43. Warnes GR, Bolker B, Bonebakker L, et al. Gplots: Various R programming tools for plotting data. R package version 3.0.1. The Comprehensive R Archive Network (CRAN);2016. https://CRAN.R-project.org/package=gplots [Google Scholar]
- 44. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilcox R. Introduction to robust estimation and hypothesis testing. 3rd ed. Cambridge, Massachusetts, USA: Academic Press; 2011:608. [Google Scholar]
- 46. Sears B, Perry M. The role of fatty acids in insulin resistance. Lipids Health Dis. 2015;14:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Holmes MV, Lange LA, Palmer T, et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet. 2014;94(2):198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fall T, Hägg S, Ploner A, et al. ; ENGAGE Consortium Age- and sex-specific causal effects of adiposity on cardiovascular risk factors. Diabetes. 2015;64(5):1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Larsson M, Vorrsjö E, Talmud P, Lookene A, Olivecrona G. Apolipoproteins C-I and C-III inhibit lipoprotein lipase activity by displacement of the enzyme from lipid droplets. J Biol Chem. 2013;288(47):33997–34008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taskinen MR, Borén J. New insights into the pathophysiology of dyslipidemia in type 2 diabetes. Atherosclerosis. 2015;239(2):483–495. [DOI] [PubMed] [Google Scholar]
- 51. Oliveira HC, de Faria EC. Cholesteryl ester transfer protein: the controversial relation to atherosclerosis and emerging new biological roles. IUBMB Life. 2011;63(4):248–257. [DOI] [PubMed] [Google Scholar]
- 52. Wang H, Peng DQ. New insights into the mechanism of low high-density lipoprotein cholesterol in obesity. Lipids Health Dis. 2011;10:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Y, van der Tuin S, Tjeerdema N, et al. Plasma cholesteryl ester transfer protein is predominantly derived from Kupffer cells. Hepatology. 2015;62(6):1710–1722. [DOI] [PubMed] [Google Scholar]
- 54. Reiner Ž. Hypertriglyceridaemia and risk of coronary artery disease. Nat Rev Cardiol. 2017;14(7):401–411. [DOI] [PubMed] [Google Scholar]
- 55. Mahendran Y, Cederberg H, Vangipurapu J, et al. Glycerol and fatty acids in serum predict the development of hyperglycemia and type 2 diabetes in Finnish men. Diabetes Care. 2013;36(11):3732–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lalia AZ, Lanza IR. Insulin-sensitizing effects of omega-3 fatty acids: Lost in translation? Nutrients. 2016;8(6):329 10.3390/nu8060329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lotta LA, Scott RA, Sharp SJ, et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a mendelian randomisation analysis. Plos Med. 2016;13(11):e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Q, Holmes MV, Davey Smith G, Ala-Korpela M. Genetic support for a causal role of insulin resistance on circulating branched-chain amino acids and inflammation. Diabetes Care. 2017;40(12):1779–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mahendran Y, Jonsson A, Have CT, et al. Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia. 2017;60(5):873–878. [DOI] [PubMed] [Google Scholar]
- 60. Mahendran Y, Vangipurapu J, Cederberg H, et al. Association of ketone body levels with hyperglycemia and type 2 diabetes in 9,398 Finnish men. Diabetes. 2013;62(10):3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song Y, Manson JE, Tinker L, et al. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women’s Health Initiative Observational Study. Diabetes Care. 2007;30(7):1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Færch K, Witte DR, Tabák AG, et al. Trajectories of cardiometabolic risk factors before diagnosis of three subtypes of type 2 diabetes: a post-hoc analysis of the longitudinal Whitehall II cohort study. Lancet Diabetes Endocrinol. 2013;1(1):43–51. [DOI] [PubMed] [Google Scholar]
- 63. Di Pino A, DeFronzo RA. Insulin resistance and atherosclerosis: implications for insulin-sensitizing agents. Endocr Rev. 2019;40(6):1447–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gast KB, Tjeerdema N, Stijnen T, Smit JW, Dekkers OM. Insulin resistance and risk of incident cardiovascular events in adults without diabetes: meta-analysis. Plos One. 2012;7(12):e52036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang T, Lu J, Shi L, et al. ; China Cardiometabolic Disease and Cancer Cohort Study Group Association of insulin resistance and β-cell dysfunction with incident diabetes among adults in China: a nationwide, population-based, prospective cohort study. Lancet Diabetes Endocrinol. 2020;8(2):115–124. [DOI] [PubMed] [Google Scholar]
- 66. Stern SE, Williams K, Ferrannini E, DeFronzo RA, Bogardus C, Stern MP. Identification of individuals with insulin resistance using routine clinical measurements. Diabetes. 2005;54(2):333–339. [DOI] [PubMed] [Google Scholar]
- 67. Ghasemi A, Tohidi M, Derakhshan A, Hasheminia M, Azizi F, Hadaegh F. Cut-off points of homeostasis model assessment of insulin resistance, beta-cell function, and fasting serum insulin to identify future type 2 diabetes: Tehran Lipid and Glucose Study. Acta Diabetol. 2015;52(5):905–915. [DOI] [PubMed] [Google Scholar]
- 68. Vignoli A, Ghini V, Meoni G, et al. High-throughput metabolomics by 1D NMR. Angew Chem Int Ed Engl. 2019;58(4):968–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am J Epidemiol. 2017;186(9):1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Otten J, Ahrén B, Olsson T. Surrogate measures of insulin sensitivity vs the hyperinsulinaemic-euglycaemic clamp: a meta-analysis. Diabetologia. 2014;57(9):1781–1788. [DOI] [PubMed] [Google Scholar]