Abstract
Background
Nanoscale surface roughness has been suggested to have antibacterial and antifouling properties. Several existing models have attempted to explain the antibacterial mechanism of nanoscale rough surfaces without direct observation. Here, conventional and liquid-cell TEM are implemented to observe nanoscale bacteria/surface roughness interaction. The visualization of such interactions enables the inference of possible antibacterial mechanisms.
Methods and Results
Nanotextures are synthesized on biocompatible polymer microparticles (MPs) via plasma etching. Both conventional and liquid-phase transmission electron microscopy observations suggest that these MPs may cause cell lysis via bacterial binding to a single protrusion of the nanotexture. The bacterium/protrusion interaction locally compromises the cell wall, thus causing bacterial death. This study suggests that local mechanical damage and leakage of the cytosol kill the bacteria first, with subsequent degradation of the cell envelope.
Conclusion
Nanoscale surface roughness may act via a penetrative bactericidal mechanism. This insight suggests that future research may focus on optimizing bacterial binding to individual nanoscale projections in addition to stretching bacteria between nanopillars. Further, antibacterial nanotextures may find use in novel applications employing particles in addition to nanotextures on fibers or films.
Keywords: liquid TEM, graphene liquid cell, antibacterial nanopatterns, antibacterial surface topology, antibacterial microparticles
Introduction
Antibacterial treatments are frequently implemented in modern medicine, agriculture, and antimicrobial textiles.1,2 Bacteria are, however, capable of developing resistance to many antibacterial treatments.3,4 Conventional antibiotics typically act by preventing the upkeep of the cell wall, interfering with the bacterial metabolism, or by inhibiting the synthesis of nucleic acids or proteins.5 Antibiotics may also depolarize the cell membrane, allowing ion influx.6 This change in ion concentration can kill bacteria.7 Antibiotic resistance occurs through limiting uptake of the antibiotic, removal of the antibiotic from within the bacteria, inactivation of the antibiotic through chemical modification or hydrolysis, or modification of the antibiotic’s target.8,9 In gram-negative bacteria, the outer plasma membrane provides intrinsic protection against the uptake of many antibiotics.10 Antibiotics primarily depend on the presence of porin proteins in the plasma membrane.11 These porin proteins facilitate the transport of hydrophilic antibiotics across the cell envelope.11 Mutations in the porin proteins may prevent the transportation of antibiotics.11 Bacteria may also reduce the number of porins.11 In contrast, gram-positive bacteria lack the outer plasma membrane and are less likely to limit the uptake of antibiotics.12 Gram-positive bacteria thus primarily depend on removal of the antibiotic, destruction of the antibiotic, or inactivation of the antibiotic via chemical modification.5 Alternatively, bacteria may alter the target of the antibiotic to prevent the antibiotic effects.5
Three general approaches to battle antibiotic-resistant bacteria exist: 1) continued development of conventional antibiotics, to which bacteria continue to adapt, 2) sterilization techniques using chemical agents, such as bleach, other harsh chemicals, or heating and cleaning of surfaces; such approaches have limited medical applications, require continuous reapplication, and may have detrimental environmental ramifications. 3) Novel fundamentally-different antibacterial treatments that rely on silver or specific surface topologies.13–19 To this end, bacteria were shown to develop resistance to silver treatments.20,21 Silver may also present an ecological hazard similar to that posed by chemical or antibiotic treatments.22 Regardless of whether an antibacterial treatment features an antibiotic drug, strong chemical, or silver, introduction of substances toxic to bacteria may be inherently caustic and induce environmental damage.16,23
Antibacterial surface topologies were first observed in nature. Cicada wings and dragonfly wings exhibit nanotextures which induce bacterial death and reduce bacterial fouling.24–27 These antibacterial nanotextures may be synthesized on a variety of biodegradable, non-toxic, and non-pathogenic materials without the presence of any bactericidal chemical or elemental agent.28 In previous works, Serrano et al explored nanotexturing of biomedical sutures,28 which showed reduced bacterial attachment, suggesting the sutures would be less likely to transmit infections prior to implantation.28 However, nanostructured cicada wings were previously shown to kill bacteria with a physical mechanism rather than by preventing bacterial adhesion.26
The bactericidal property of nanopillar surface topologies has been proposed to occur through several mechanisms: Pogodin et al proposed that bacteria bind to the top of nanopillars.26 The bacteria then attempt to bind below the surface and are consequently stretched between the nanopillars.26 This stretching of the cell membrane supposedly causes rupture of the cell wall and induces bacterial death.26 In both gram-negative and gram-positive bacteria, the cell wall includes a layer of peptidoglycan, which provides mechanical support.29 In gram-positive bacteria, the cell envelope includes the outer cell wall, an intermediate periplasmic space, and an interior plasma membrane.29 In gram-negative bacteria, the cell envelope includes an outer lipopolysaccharide and protein layer, an intermediate periplasmic space, a middle cell wall, a second periplasmic space, and an interior plasma membrane.29 Linklater et al proposed another mechanism, in which the bacteria induce mechanical forces on the surface of nanopillars upon binding.30 These forces then deflect the nanopillars, which then induce strain on the bacteria upon relaxation of the pillars.30 This strain damages the cell wall and induces the observed cell death.30 While nanopillar surfaces were experimentally shown via traditional fluorescent light microscopy and scanning electron microcopy (SEM) to induce bacterial death, the mechanism of bactericidal activity was mathematically inferred in both Linklater et al and Pogodin et al rather than demonstrated experimentally.26,30 In another mechanism, Michalska et al proposed that black silicon nanopillars directly penetrate the cell wall of the bacterium.31 However, Linklater et al later suggested that the bacterial death observed in Michalska et al occurred via the strain mechanism.30,31
Additional studies have focused on mathematical modeling of bacteria/nanotexture interaction.32–34 By necessity, these mathematical models included assumptions such that the stiffness of the bacteria may be modeled as a single-layer of peptidoglycan, that deformation of the bacterial cell wall does not induce other metabolic processes (eg, apoptosis) through programmed cell death, and that the bacteria binding to the surface occurs through purely physics-based methods, such as hydrophobic interactions and Van der Waals forces. The initial bacteria/surface contact is indeed dependent on physical factors, such as surface charge and wettability (hydrophilicity/hydrophobicity).35–39 However, bacteria/surface binding occurs in a second stage, wherein adhesion forces far exceed the forces in the initial contact period and are dominated by multiple adhesion proteins and attachment pili.35–39 Bacterial binding of nanotextures is an active process and depends on the metabolic rate, which confirms the active role of protein adhesion in bacteria/nanotexture binding.40 Thus, while mathematical models are insightful and may prove useful for offering predictions, experimental verification of these mathematical models is also essential. Further, multiple bactericidal mechanisms are possible and may not be mutually exclusive.
Observational non-mechanistic studies provide direct evidence of the efficacy of a treatment on a particular bacterial strain. However, bacterial strains vary in size, stiffness, and metabolic processes.41 Due to these bacterial differences, observational studies require testing antibacterial treatments iteratively against multiple strains to produce a generalized inference of the real-world applications of any antibacterial approach.31 Experimental studies of the E.coli/nanotexture interaction mechanisms are thus needed to complement existing observational studies and mathematical models. Here, both conventional transmission electron microscopy (TEM) and liquid-phase TEM are implemented to observe the mechanism of bacterial death. Liquid-phase TEM allows for high-resolution nanoscale observations of the E. coli/nanotexture interactions without requiring preservation of the sample. E. coli K12 is well-characterized in literature as a model organism and was thus chosen for this investigation.42,43 In contrast to previous works on films or larger fibers, here poly(lactic-co-glycolic acid) (PLGA) microparticles (MPs) are nanotextured and are shown to have an antibacterial effect on E. coli in liquid media.26,28,30,31 PLGA is biocompatible, biodegradable, low-cost, FDA-approved, and may be functionalized with other antibacterial treatments to produce a combined antibacterial effect if desired.44,45 Suspensions containing E. coli and nanotextured PLGA MPs were imaged in TEM via both conventional TEM and liquid-phase TEM via graphene liquid cells (GLCs) to observe bacterial death and structural changes in E. coli.
Materials and Methods
Bacteria Culturing
E. coli K12 were commercially purchased (Escherichia coli (Migula) Castellani and Chalmers, ATCC® 29425™). Starter cultures were preserved in 50% glyceraldehyde and 50% phosphate buffered saline (PBS) and frozen at −80°C as previously described.46 E. coli was cultured in 50mL lysogeny broth (LB) at 37.5°C on an orbital shaker at 100RPM as previously described.47
PLGA Microparticle Etching
PLGA MPs were commercially purchased (Degrdex® PLGA microspheres, LG500). The particles varied between several nanometers to several micrometers in diameter. The PLGA MPs were then dispersed in DI water and sonicated for 5 mins. The PLGA MP suspension was then dropcast in 10μL quantities onto approximately 5x5cm copper sheets and allowed to dry completely for at least 12 hrs. The PLGA-covered copper sheets were then plasma etched via the method described by Serrano et al.28 The particles were etched using a South Bay PC150 plasma etcher under 200 ppm oxygen at 100 W in 1 min increments, with 2 mins between each increment to prevent heating of the sample. After etching, the copper sheets were cut into strips and placed in microcentrifuge tubes containing 1 mL sterile PBS and sonicated for 5 mins to remove the PLGA MPs from the copper sheets.
SEM of PLGA MPs
PLGA MPs on copper after plasma etching were gold sputter-coated for 1 min and imaged in SEM via a RAITH100 eLine EBL at 2kV to 3kV with working distances of 13 mm to 20 mm.
AgNP Solution Preparation
Silver NPs (AgNPs) were commercially purchased and added to sterile PBS. The suspensions were then sonicated to facilitate suspension of the AgNPs, and exposed to UV light to sterilize the sample.
Colony Counting
E. coli were cultured in LB broth to the stationary phase of E. coli growth. Ten microliters were then extracted from each E. coli culture to determine viability prior to the addition of antibacterial materials. PLGA MPs and AgNPs were added to the appropriate E. coli samples, which were then immediately returned to the incubator. The extracted samples were diluted to 10−2, 10−4, and 10−6, and plated on LB agar plates, as described previously.48 The colony counting procedure was repeated at 4 hrs and 8 hrs after the addition of the antibacterial or control samples to the E. coli culture to observe bacterial death over time. This procedure was repeated six times to produce statistically valid data. The results were analyzed via Student’s t-test.49
Conventional TEM Imaging
E. coli was cultured to the stationary phase at which point PLGA MPs etched for 2 mins were added, incubated an additional 20 mins, and then fixed for imaging with conventional TEM. The stationary phase refers to the phase of bacterial population growth at which the total population is constant.50 Bacteria and the antibacterial agent were centrifuged to isolate the sample as a pellet, with the LB broth removed. The samples were then preserved in resin via glutaraldehyde and osmium fixing and staining, as per Feng et al.51 This conventional TEM method includes microtoming the samples to produce cross-sectional 2D slices for imaging.52 Some of the rod-shaped, bacilli E. coli may thus appear circular due to the orientation of these bacteria within the 2D slice.53 A JEOL 1220 TEM was used for image acquisition. TEM images were false-colored for clarity. The original, unaltered images are included in the SI.
In situ TEM Imaging
E. coli was cultured to stationary phase in LB broth. PLGA MPs etched for 2 mins were added to the solution and incubated for an additional 20 mins. The LB broth was then removed via centrifugation and the sample rehydrated in PBS. Approximately 1 μL of the solution was placed on a graphene-coated TEM grid and then covered with another graphene-coated grid to produce a graphene sandwich. Textor et al provided a detailed review of this GLC synthesis process.54 The incident electron dose varied between 3×109 Gy/s to 6×1010 Gy/s throughout the experiment. A JEOL 1220 TEM was used for imaging. TEM images were false-colored for clarity. The original, unaltered images are included in the SI.
Results and Discussion
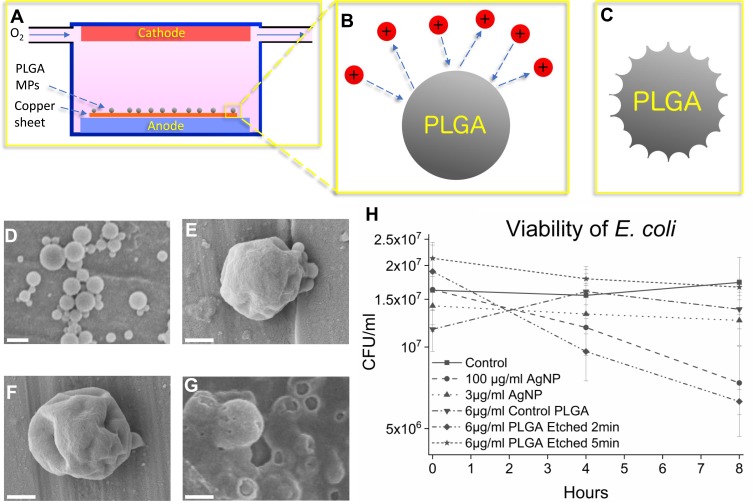
In the first step, the PLGA MPs were plasma-treated to produce nanotextures on their surface. The MPs were plasma etched for 1, 2, and 5 mins to induce different surface roughness (see Material and Methods, Figure 1A–C). The PLGA MPs were imaged via SEM to observe the surface topology (Figure 1D–G). The PLGA MPs show minimal nanotexturing in particles etched for 1 min, while at 2 mins these nanotextures are deeper and more pronounced. At 5 mins of plasma etching, the sample is highly damaged leaving behind damaged, disordered PLGA. Particles etched for 2 mins, with the roughest surface morphology, were selected for antibacterial testing.
Figure 1.
Schematic of plasma etching of PLGA MPs (A-C), SEM imaging of the control PLGA MPs and the plasma-etched PLGA MPs (D-G) and viability graph of E. coli control and in the presence of antibacterial agents (H). In (A) oxygen flows in low concentration (200 ppm volume) through a charged environment to create free radicals (B) which etch the surface, leaving behind rough MPs (C). In (D) unetched particles show smooth surface morphology. In (E) the PLGA MPs show mild dimpling of the surface after 1 min of plasma etching. In (F) the PLGA MPs etched for 2 mins show more pronounced, sharp patterns as opposed to the PLGA MPs shown in (D) and (E). Finally, in (G) 5 mins of etching severely damaged the MPs, reducing the PLGA to primarily amorphous PLGA aggregations. The bactericidal efficacy of PLGA MPs and AgNPs are compared in (H) where control and bacteria treated with 3 μg/mL AgNPs do not show decreased CFU/mL, while the 100 μg/mL AgNPs samples show decreased viability. The error bars in (H) represent the standard error. The unetched PLGA does not exhibit a statistically significant bactericidal effect, nor does the PLGA etched for 5 mins. However, the PLGA etched for 2 mins, which features rough MP surfaces, does exhibit a statistically significant bactericidal effect. The scale bar is 1 µm in (D), and 500 nm in (E), (F), and (G).
In the next step, the bacterial activities of treated and untreated PLGA MPs were studied and compared with AgNPs. Unetched smooth PLGA MPs, PLGA MPs etched for 2 mins, and PLGA MPs etched for 5 mins were added in concentrations of 6 μg/mL to separate E. coli cultures at the stationary growth phase (see Material and Methods, Figure 1H). Additionally, AgNPs, known to have bactericidal effects, were added to separate LB broth solutions in concentrations of 3 μg/mL AgNPs and 100 μg/L AgNPs to create positive controls (see Material and Methods).21 The results show the PLGA MPs etched for 2 mins lowered the viability of E. coli by approximately 64% (p<0.05) as compared to controls, while the 100 μg/mL AgNPs exhibited bactericidal effects which lowered the viability of E. coli by approximately 57% (p<0.10) (Figure 1H). The E. coli sample containing 3 μg/mL AgNP may not have shown a statistically significant antibacterial effect (Figure 1H) due to the large size of the AgNPs used in this work, which may reduce the bactericidal efficiency of AgNPs as compared to smaller AgNPs used in other studies (Figure 1H).55 The control, smooth PLGA MPs did not kill E. coli, in agreement with previous findings that PLGA is non-toxic.44 The highly damaged amorphous PLGA MPs likewise do not exhibit any bactericidal effect, suggesting that the plasma etching process does not alter the chemistry or surface properties of the PLGA MPs to produce bactericidal effects. Further, the increased surface area on the highly damaged PLGA etched for 5 mins would be expected to increase the bactericidal effect if bacterial death was due to a chemical effect rather than the rough surface topology. Only the PLGA MPs etched for 2 mins with the rough nanotextured surface kill bacteria, to which only the nanotextures are unique, thus indicating that it is the nanotextures that kill E. coli.
Direct mechanical damage to the cell wall compromises the cell envelope permeability and induces cell death.26 Metabolically mediated cell wall degradation may occur due to a cessation of metabolic cell wall maintenance or by the production of intracellular autolysins which actively break down the cell wall.56 Further, escape of the bacterial cytosol, the fluid within bacteria, and a decrease in volume are indicative of bacterial death.57 Conventional light microscopy therefore uses fluorescent dyes to determine the integrity of the cell wall regardless of the mechanism of bacterial death.58 However, TEM allows nanoscale observation of the cell envelope and MPs and thus does not require fluorescent dyes since cell-wall damage can be directly deserved.51,59
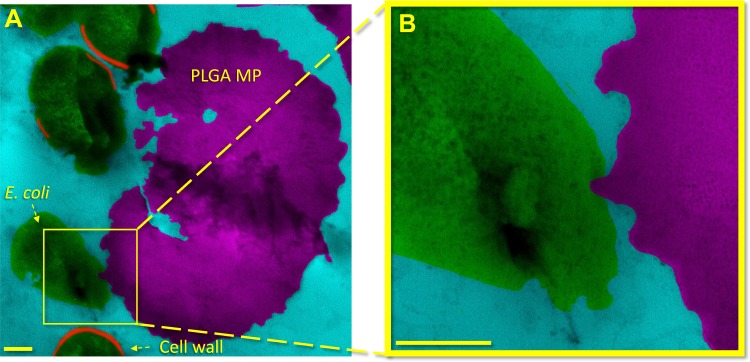
Nanotextured PLGA MP/bacteria surface interactions were examined via conventional TEM as shown in Figure 2 (see Material and Methods) and Figure S1. In this figure, a PLGA MP has rough nanotexture and uneven edges as observed with SEM (Figure 1). As shown in Figure 2A and B (progressive magnification) (also in Figure S1), the MP has a surface protrusion inside an adjacent bacterium. The location of the surface protrusion suggests that the MP sharp features pierced the cell envelope of the bacterium, including the outer membrane, peptidoglycan layer, and inner membrane. Piercing the cell envelope would produce local cell damage and lysis of the bacterium.26,30,31,60 The absence of the bacterial cell envelope in Figure 2B indicates that this damage was sufficient to stop the metabolic maintenance of the cell envelope or produce apoptosis.60 In contrast to mathematical modeling in previous works, in Figure 2 the E. coli is not stretched between nanopillars but rather is in contact with a single sharp protrusion, which appears to have been directly penetrated the cell surface (Figure S1).26,30,32–34
Figure 2.
Bacterial death is induced by rough MP topography as observed in a cross-sectional image obtained via conventional TEM. In (A) and (B) low to high magnifications of surface interaction between an E. coli and a rough PLGA particle etched for 2 mins is shown. A sharp peak on the PLGA particle appears to have penetrated the E. coli bacterium cell envelope. The area bracketed in (A) is shown at higher magnification in (B). The scale bars in (A) and (B) are 200nm.
E. coli and PLGA MPs etched for 2 mins were then encapsulated together in GLCs (Figures 3 and S2). In GLCs, a liquid sample is encapsulated between two layers of graphene (see Material and Methods).61–64 The graphene layers are impermeable to liquid matter, fully encasing the sample, and allow transmission of the electron beam through the sample. The graphene also dissipates energy, which reduces undesired effects of the beam exposure (SI, Liquid-TEM Imaging).57 Previous works have shown that the effects of changes in pressure, temperature, free radical generation, and exposure to the electron beam itself are non-significant under low electron doses for large samples, such as those containing E. coli, as implemented here (SI notes).57,59,61,65–76 Thus, previous works indicate that E. coli 1) maintain their metabolic processes, 2) maintain their cell envelopes, and 3) continue to undergo binary fission within liquid-phase TEM.57,59,61,65–76
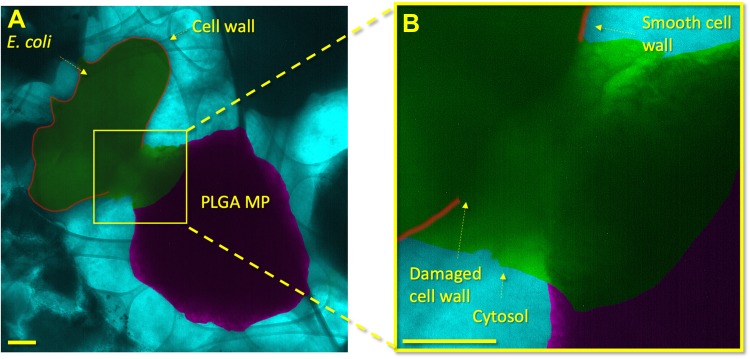
Figure 3.
GLC encapsulation of E. coli and PLGA MPs shows localized damage to the cell envelope of the bacterium. In (A), a lower-magnification image shows an overview of the E. coli and PLGA MP. In (B), a higher magnification shows the PLGA MP and bacterium in close proximity. The cell wall proximal to the PLGA MP shows damage with a shape similar to the adjacent MP. A high contrast liquid indicates the cytosol of the bacterium, which clearly identifies the death of the bacterium. The cytosol has a distinct contrast from the PBS medium due to the proteins, glycans, and other bacterial components within it. Other areas of the E. coli cell wall are smooth and show no damage or degradation. The scale bars in (A) and (B) are 200 nm.
Liquid-phase TEM (Figures 3 and S2) provides further evidence of the penetration of nanoprotrusions in E.coli, as also observed forensically with conventional TEM (Figures 2 and S1). The cell envelope of the bacterium immediately adjacent to a rough PLGA MP is highly damaged and shows escape of the bacterium’s cytosol. In contrast, areas distal to the PLGA MP surface protrusion show smooth, undamaged cell envelopes. The localization of cell damage to the site of bacterium/PLGA MP interaction indicates that the bacterium was not killed by general metabolic factors, which would cause diffuse damage throughout the bacterium cell envelope. Bacterial death thus must have occurred due to the localized damage produced by the E. coli/surface interaction.
Gram-negative bacteria, including E. coli, contain peptidoglycan cell walls approximately 2nm to 8nm in thickness. In contrast, gram-positive bacteria have cell wall thicknesses from approximately 20nm to 35nm.41,77 Peptidoglycan has been considered to provide the structural support and is a primary contributor to the mechanical properties of bacteria.78 The thickness of the peptidoglycan layer might be intuitively expected to reflect the stiffness of bacteria. In contrast to this expectation, initial experimental studies reported that gram-negative bacteria exhibit an average Young modulus of approximately 30MPa while gram-positive bacteria exhibit a Young modulus of 20MPa.41 However, further studies showed that the Young modulus varies significantly depending on the bacterial species, growth medium, preparation method, dry or wet state, and measurement method, with previously reported Young modulus values between 0.05MPa and 769MPa.41,79 This variation in mechanical properties reflects cell wall variations in proteins, phospholipids, teichoic acids, lipoteichoic acids, and differences in peptidoglycan-peptidoglycan binding or interactions.80 Peptidoglycans consist of alternating sugar moieties, β-(1,4) linked N-acetylglucosamine and N-acetylmuramic acid, which are then attached to a peptide chain.81,82 While the sugar moieties are highly conserved between bacterial species, the attached peptide chains are bacterial strain-specific and vary significantly in peptide chain length and amino acid composition.41 Peptidoglycans may be monomers or cross-linked as dimers or trimers, with linkages between a variety of amino acid residues.83,84 The variation in peptide chains and the activity of crosslinking proteins produces unique chain crosslinking.85,86 The degree and type of peptidoglycan cross-linking are thus highly positively correlated with the stiffness of the bacteria.80 Some bacteria may also contain actin-like filaments that form a rudimentary cytoskeleton.87 These actin-like filaments may influence the mechanical properties of bacteria, including their stiffness.87 In short, the thickness of the peptidoglycan layer and the gram-positive/gram-negative classification are not sufficient to establish the mechanical properties of bacteria.41 This suggests that the bactericidal nature of nanoscale surface topologies may also be effective in gram-positive bacteria, since the cell wall of such bacteria may be comparable to E. coli.41
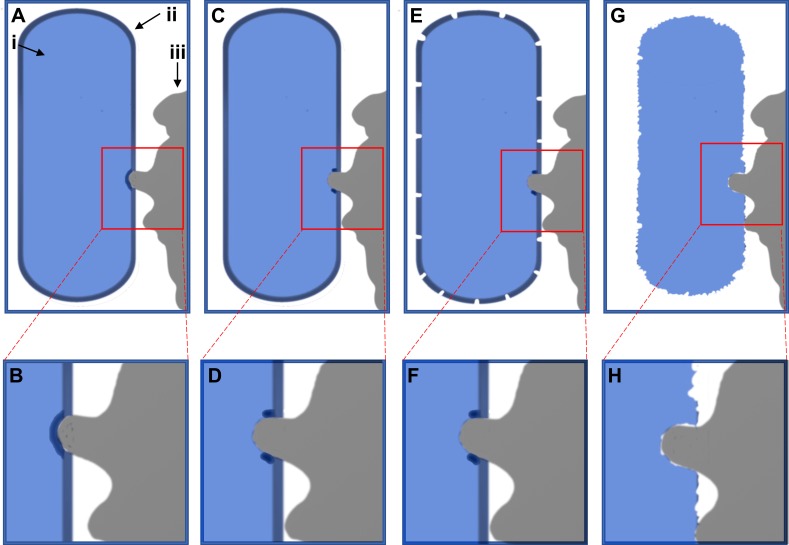
The deformation of the bacterial cell wall thus depends not on simple physical forces and the thickness of the peptidoglycan layer, but on the Young modulus of the bacterial cell wall and the attachment forces of the bacterial adhesion proteins. E. coli was previously shown to exhibit a Young modulus of 22 MPa.88 This suggests the attachment pressure of E. coli exceeds this value to produce strain within the cell wall to produce the damage shown in Figures 2 and 3 (Figures S1 and S2). The bactericidal mechanism of antibacterial nanotextures observed here and supported by literature is illustrated schematically in Figure 4. First, simple physical forces, eg, hydrophobic/hydrophilic interactions, bring the E. coli into contact with the PLGA particle surface.38,39 The adhesion proteins then attach and contract to bind the E. coli to the PLGA surface.38,39 However, the protruding fixture on the PLGA surface exerts a local force opposing the adhesion proteins bound to the non-protruding surface (Figures 2,3 and S1,S2). This causes stretching in the E. coli cell membrane over the PLGA nanopillar, as experimentally observed here in Figures 2 and 3 (Figures S1 and S2) and illustrated in Figure 4A–D, which damages the cell envelope. This damage causes the death of the E. coli and leakage of the cytosol localized to the nanotexture projection, as experimentally observed in Figures 3 and S2. This cell death later results in total degradation of the cell envelope due to reduced metabolic cell envelope maintenance or production of autolysis, as experimentally observed in Figure 2 (Figure S1) and illustrated in Figure 4E–H.56 Here, a mechanism by which nanoscale surface roughness may kill E. coli K12 has been shown.
Figure 4.
Schematic illustration of bacterial death mechanism. The bacterium is shown in blue on the left side of the image (i), whereas the cell envelope is indicated as a dark blue layer on the outer edge of the bacterium (ii). The PLGA particle is shown in grey on the right-hand side (iii). In (A) and (B) the particle has come into contact and deforms the cell envelope, before breaking it in (C) and( D). This damage then causes degradation of the cell envelope in (E) and (F), before the cell wall disintegrates in (G) and (H).
Conclusions
We report rough nanotextured polymer MPs that exhibit antibacterial action. Such MPs have applications distinct from previously designed media, such as antibacterial gels, liquid suspensions, or powders. The nanotextured surface topology of the MPs was shown to be bactericidal via a localized E. coli/surface interaction mechanism. The MP surface protrusions deform bacterial cell walls to induce bacterial death rather than act by damaging bacterial cell walls via stretching between nanopillars. This cell-wall damage suggests that the attachment pressure exceeded the force required for deformation of the cell wall of the bacterium. The deformation of the cell envelope appears to cause cell lysis and death of the E. coli. Future studies are required to explore the nanoscale interaction of rough surface topology with other types of bacteria, and to further exploit the antibacterial mechanism suggested here. One should be aware that the success rates of liquid-phase TEM experiments are very low. The reproducibility of such liquid-phase TEM tests is also very challenging and needs to be revisited. We believe more careful studies are needed to ensure the bacteria are alive in the TEM environment.
Acknowledgments
This work was funded by the Nonwovens Institute (North Carolina State Univ. Grant 16-195). T. Shokuhfar and E. Firlar acknowledge NSF CAREER Award #1564950. Partial support from NSF Award #1803693 is acknowledged. R. Shahbazian-Yassar’s efforts were supported by NSF Award # 1710049. This work made use of instruments in the Electron Microscopy Core of UIC’s Research Resources Center (RRC). The authors would like to thank the University of Illinois at Chicago’s Nanotechnology Core Facility of the College of Engineering and the RRC, Vice Chancellor’s Office for Research, for its services and instrumentation use. Portions of this work were carried out in the Fluorescence Imaging Core (FIC) of the RRC at UIC.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.McArthur JV, Tuckfield RC, Baker-Austin C. Antimicrobial textiles. Berlin, Heidelberg: Springer; 2012:135–152. [DOI] [PubMed] [Google Scholar]
- 2.Witte W. Medical consequences of antibiotics use in agriculture. Science. 1998;379(5353):996–997. doi: 10.1126/science.279.5353.996 [DOI] [PubMed] [Google Scholar]
- 3.Livermore DM, Brown DF. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48:59–64. doi: 10.1093/jac/48.suppl_1.59 [DOI] [PubMed] [Google Scholar]
- 4.Davies J. Origins and evolution of antibiotic resistance. Microbiologia. 1996;74(3):417–433. [PubMed] [Google Scholar]
- 5.Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482–501. doi: 10.3934/microbiol.2018.3.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kepplinger B, Morton-Laing S, Seistrup KH, et al. Mode of action and heterologous expression of the natural product antibiotic vancoresmycin. ACS Chem Biol. 2018;13(1):207–214. doi: 10.1021/acschembio.7b00733 [DOI] [PubMed] [Google Scholar]
- 7.Cadelis MM, Pike EIW, Kang W, et al. Exploration of the antibiotic potentiating activity of indolglyoxylpolyamines. Eur J Med Chem. 2019;183:111708. doi: 10.1016/j.ejmech.2019.111708 [DOI] [PubMed] [Google Scholar]
- 8.Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303(6–7):287–292. doi: 10.1016/j.ijmm.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 9.Chancey ST, Zähner D, Stephens DS. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 2012;7:959–978. doi: 10.2217/fmb.12.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epand RM, Walker C, Epand RF, Magarvey NA. Molecular mechanisms of membrane targeting antibiotics. Biochim Biophys Acta Biomembr. 2016;1858(7):980–987. doi: 10.1016/j.bbamem.2015.10.018 [DOI] [PubMed] [Google Scholar]
- 11.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6(12):893–903. doi: 10.1038/nrmicro1994 [DOI] [PubMed] [Google Scholar]
- 12.Lambert PA. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol Symp Suppl. 2002;92:46S–54S. doi: 10.1046/j.1365-2672.92.5s1.7.x [DOI] [PubMed] [Google Scholar]
- 13.Balaban NQ, Liu J. Evolution under antibiotic treatments: interplay between antibiotic persistence, tolerance, and resistance In: Lewis K, editor. Persister cells and infectious disease. Cham: Springer International Publishing; 2019:1–17. [Google Scholar]
- 14.Durão P, Balbontín R, Gordo I. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol. 2018;26(8):677–691. doi: 10.1016/j.tim.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 15.Silindir M, Özer AY. Sterilization methods and the comparison of E-beam sterilization with gamma radiation sterilization. Fabad J Pharm Sci. 2009;34:43–53. [Google Scholar]
- 16.Alavi M, Rai M. Recent advances in antibacterial applications of metal nanoparticles (MNPs) and metal nanocomposites (MNCs) against multidrug-resistant (MDR) bacteria. Expert Rev Anti Infect Ther. 2019;17(6):419–428. doi: 10.1080/14787210.2019.1614914 [DOI] [PubMed] [Google Scholar]
- 17.Alavi M, Karimi N, Valadbeigi T. Antibacterial, antibiofilm, antiquorum sensing, antimotility, and antioxidant activities of green fabricated Ag, Cu, TiO2, ZnO, and Fe3O4 NPs via protoparmeliopsis muralis lichen aqueous extract against multi-drug-resistant bacteria. ACS Biomater Sci Eng. 2019;5(9):4228–4243. doi: 10.1021/acsbiomaterials.9b00274 [DOI] [PubMed] [Google Scholar]
- 18.Alavi M, Rai M. Recent progress in nanoformulations of silver nanoparticles with cellulose, chitosan, and alginic acid biopolymers for antibacterial applications. Appl Microbiol Biotechnol. 2019;103(21–22):8669–8676. doi: 10.1007/s00253-019-10126-4 [DOI] [PubMed] [Google Scholar]
- 19.Alavi M, Nokhodchi A. An overview on antimicrobial and wound healing properties of ZnO nanobiofilms, hydrogels, and bionanocomposites based on cellulose, chitosan, and alginate polymers. Carbohydr Polym. 2020;227:115349. doi: 10.1016/j.carbpol.2019.115349 [DOI] [PubMed] [Google Scholar]
- 20.Panáček A, Kvítek L, Smékalová M, et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat Nanotechnol. 2018;13:65–71. doi: 10.1038/s41565-017-0013-y [DOI] [PubMed] [Google Scholar]
- 21.Randall CP, Gupta A, Jackson N, Busse D, O’Neill AJ. Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J Antimicrob Chemother. 2015;70(4):1073–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panyala NR, Peña-Méndez EM, Havel J. Silver or silver nanoparticles: a hazardous threat to the environment and human health? J Appl Biomed. 2008;6:117–129. doi: 10.32725/jab.2008.015 [DOI] [Google Scholar]
- 23.Kim S, Choi JE, Choi J, et al. Oxidative stress-dependent toxicity of silver nanoparticles in human hepatoma cells. Toxicol Vitr. 2009;23(6):1076–1084. doi: 10.1016/j.tiv.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 24.Kelleher SM, Habimana O, Lawler J, et al. Cicada wing surface topography: an investigation into the bactericidal properties of nanostructural features. ACS Appl Mater Interfaces. 2016;8(24):14966–14974. doi: 10.1021/acsami.5b08309 [DOI] [PubMed] [Google Scholar]
- 25.Hasan J, Webb HK, Truong VK, et al. Selective bactericidal activity of nanopatterned superhydrophobic cicada Psaltoda claripennis wing surfaces. Appl Microbiol Biotechnol. 2013;97(20):9257–9262. doi: 10.1007/s00253-012-4628-5 [DOI] [PubMed] [Google Scholar]
- 26.Pogodin S, Hasan J, Baulin VA, et al. Biophysical model of bacterial cell interactions with nanopatterned cicada wing surfaces. Biophys J. 2013;104(4):835–840. doi: 10.1016/j.bpj.2012.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandara CD, Singh S, Afara IO, et al. Bactericidal effects of natural nanotopography of dragonfly wing on escherichia coli. ACS Appl Mater Interfaces. 2017;9(8):6746–6760. doi: 10.1021/acsami.6b13666 [DOI] [PubMed] [Google Scholar]
- 28.Serrano C, García-Fernández L, Fernández-Blázquez JP, et al. Nanostructured medical sutures with antibacterial properties. Biomaterials. 2015;52:291–300. doi: 10.1016/j.biomaterials.2015.02.039 [DOI] [PubMed] [Google Scholar]
- 29.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5):a000414. doi: 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linklater DP, De Volder M, Baulin VA, et al. High aspect ratio nanostructures kill bacteria via storage and release of mechanical energy. ACS Nano. 2018;12(7):6657–6667. doi: 10.1021/acsnano.8b01665 [DOI] [PubMed] [Google Scholar]
- 31.Michalska M, Gambacorta F, Divan R, et al. Tuning antimicrobial properties of biomimetic nanopatterned surfaces. Nanoscale. 2018;10:6639–6650. doi: 10.1039/C8NR00439K [DOI] [PubMed] [Google Scholar]
- 32.Xue F, Liu J, Guo L, Zhang L, Li Q. Theoretical study on the bactericidal nature of nanopatterned surfaces. J Theor Biol. 2015;385:1–7. doi: 10.1016/j.jtbi.2015.08.011 [DOI] [PubMed] [Google Scholar]
- 33.Ayazi M, Golshan Ebrahimi N, Jafari Nodoushan E. Bacterial adhesion reduction on the surface with a simulated pattern: an insight into Extrand model. Int J Adhes Adhes. 2019;88:66–73. doi: 10.1016/j.ijadhadh.2018.10.017 [DOI] [Google Scholar]
- 34.Li X. Bactericidal mechanism of nanopatterned surfaces. Phys Chem Chem Phys. 2015;18(2):1311–1316. doi: 10.1039/C5CP05646B [DOI] [PubMed] [Google Scholar]
- 35.An YH, Friedman RJ. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J Biomed Mater Res. 1998;43:338. doi: 10.1002/(ISSN)1097-4636 [DOI] [PubMed] [Google Scholar]
- 36.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12(1):49–62. doi: 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elbourne A, Chapman J, Gelmi A, Cozzolino D, Crawford RJ, Truong VK. Bacterial-nanostructure interactions: the role of cell elasticity and adhesion forces. J Colloid Interface Sci. 2019;546:192–210. doi: 10.1016/j.jcis.2019.03.050 [DOI] [PubMed] [Google Scholar]
- 38.Goulter RM, Gentle IR, Dykes GA. Issues in determining factors influencing bacterial attachment: a review using the attachment of Escherichia coli to abiotic surfaces as an example. Lett Appl Microbiol. 2009;49(1):1–7. doi: 10.1111/j.1472-765X.2009.02591.x [DOI] [PubMed] [Google Scholar]
- 39.Tuson HH, Weibel DB. Bacteria-surface interactions. Soft Matter. 2013;9:4368–4380. doi: 10.1039/c3sm27705d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köller M, Ziegler N, Sengstock C, Schildhauer TA, Ludwig A. Bacterial cell division is involved in the damage of gram-negative bacteria on a nano-pillar titanium surface. Biomed Phys Eng Express. 2018;4:055002. doi: 10.1088/2057-1976/aad2c1 [DOI] [Google Scholar]
- 41.Auer GK, Weibel DB. Bacterial cell mechanics. Biochemistry. 2017;56(29):3710–3724. doi: 10.1021/acs.biochem.7b00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blount ZD. The unexhausted potential of E. coli. Elife. 2015;4:e05826. doi: 10.7554/eLife.05826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keseler IM, Collado-Vides J, Gama-Castro S, et al. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers (Basel). 2011;3(3):1377–1397. doi: 10.3390/polym3031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson JM, Shive MS. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv Drug Deliv Rev. 1997;28(1):5–24. doi: 10.1016/S0169-409X(97)00048-3 [DOI] [PubMed] [Google Scholar]
- 46.Son MS, Taylor RK. Growth and maintenance of Escherichia coli laboratory strains. Curr Protoc Microbiol. 2012;Chapter 5:Unit5A.4. [DOI] [PubMed] [Google Scholar]
- 47.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189(23):8746–8749. doi: 10.1128/JB.01368-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breed RS, Dotterrer WD. The number of colonies allowable on satisfactory agar plates. J Bacteriol. 1916;1(3):321–331. doi: 10.1128/JB.1.3.321-331.1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Winter JCF. Using the student’s t-test with extremely small sample sizes. Pract Assess Res Eval. 2013;18(10):1–12. [Google Scholar]
- 50.Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3:371–394. doi: 10.1146/annurev.mi.03.100149.002103 [DOI] [Google Scholar]
- 51.Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000;52(4):662–668. doi: [DOI] [PubMed] [Google Scholar]
- 52.Winey M, Meehl JB, O’Toole ET, Giddings TH. Conventional transmission electron microscopy. Mol Biol Cell. 2014;25(3):319–323. doi: 10.1091/mbc.e12-12-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Jiang Y, Ding Y, et al. Mechanistic investigation into antibacterial behaviour of suspensions of ZnO nanoparticles against E. coli. J Nanoparticle Res. 2010;12:1625–1636. doi: 10.1007/s11051-009-9711-1 [DOI] [Google Scholar]
- 54.Textor M, De Jonge N. Strategies for preparing graphene liquid cells for transmission electron microscopy. Nano Lett. 2018;18(6):3313–3321. doi: 10.1021/acs.nanolett.8b01366 [DOI] [PubMed] [Google Scholar]
- 55.Lok C-N, Ho C-M, Chen R, et al. Silver nanoparticles: partial oxidation and antibacterial activities. J Biol Inorg Chem. 2007;12(4):527–534. doi: 10.1007/s00775-007-0208-z [DOI] [PubMed] [Google Scholar]
- 56.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berry V. Impermeability of graphene and its applications. Carbon N Y. 2013;62:1–10. doi: 10.1016/j.carbon.2013.05.052 [DOI] [Google Scholar]
- 58.Stiefel P, Schmidt-Emrich S, Maniura-Weber K, Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015;15:36. doi: 10.1186/s12866-015-0376-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohanty N, Fahrenholtz M, Nagaraja A, Boyle D, Berry V. Impermeable graphenic encasement of bacteria. Nano Lett. 2011;11(3):1270–1275. doi: 10.1021/nl104292k [DOI] [PubMed] [Google Scholar]
- 60.Islam MS, Aryasomayajula A, Selvaganapathy PR. A review on macroscale and microscale cell lysis methods. Micromachines. 2017;8(3):83. doi: 10.3390/mi8030083 [DOI] [Google Scholar]
- 61.Firlar E, Ouy M, Bogdanowicz A, et al. Investigation of the magnetosome biomineralization in magnetotactic bacteria using graphene liquid cell-transmission electron microscopy. Nanoscale. 2019;11(2):698–705. doi: 10.1039/C8NR08647H [DOI] [PubMed] [Google Scholar]
- 62.Wang C, Qiao Q, Shokuhfar T, Klie RF. High-resolution electron microscopy and spectroscopy of ferritin in biocompatible graphene liquid cells and graphene sandwiches. Adv Mater. 2014;26(21):3410–3414. doi: 10.1002/adma.v26.21 [DOI] [PubMed] [Google Scholar]
- 63.Ghodsi SM, Anand S, Shahbazian-Yassar R, Shokuhfar T, Megaridis CM. In situ study of molecular structure of water and ice entrapped in graphene nanovessels. ACS Nano. 2019;13(4):4677–4685. doi: 10.1021/acsnano.9b00914 [DOI] [PubMed] [Google Scholar]
- 64.Yuk JM, Park J, Ercius P, et al. High-resolution EM of colloidal nanocrystal growth using graphene liquid cells. Science. 2012;336(6077):61–64. [DOI] [PubMed] [Google Scholar]
- 65.Woehl TJ, Kashyap S, Firlar E, et al. Correlative electron and fluorescence microscopy of magnetotactic bacteria in liquid: toward in vivo imaging. Sci Rep. 2014;4:6854. doi: 10.1038/srep06854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kennedy E, Nelson EM, Tanaka T, Damiano J, Timp G. Live bacterial physiology visualized with 5 nm resolution using scanning transmission electron microscopy. ACS Nano. 2016;10(2):2669–2677. doi: 10.1021/acsnano.5b07697 [DOI] [PubMed] [Google Scholar]
- 67.Kennedy E, Nelson EM, Damiano J, Timp G. Gene expression in electron-beam-irradiated bacteria in reply to “Live cell electron microscopy is probably impossible.” ACS Nano. 2017;11(1):3–7. doi: 10.1021/acsnano.6b06616 [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Chen X, Cao H, Deng C, Cao X, Wang P. A structural study of Escherichia coli cells using an in situ liquid chamber TEM technology. J Anal Methods Chem. 2015;2015:829302. doi: 10.1155/2015/829302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghodsi SM, Megaridis CM, Shahbazian‐Yassar R, Shokuhfar T. Advances in graphene-based liquid cell electron microscopy: working principles, opportunities, and challenges. Small Methods. 2019;3:453001. doi: 10.1002/smtd.v3.5 [DOI] [Google Scholar]
- 70.Shin D, Park JB, Kim Y-J, et al. Growth dynamics and gas transport mechanism of nanobubbles in graphene liquid cells. Nat Commun. 2015;6:6068. doi: 10.1038/ncomms7068 [DOI] [PubMed] [Google Scholar]
- 71.Khestanova E, Guinea F, Fumagalli L, Geim AK, Grigorieva IV. Universal shape and pressure inside bubbles appearing in van der Waals heterostructures. Nat Commun. 2016;7:12587. doi: 10.1038/ncomms12587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gänzle M, Liu Y. Mechanisms of pressure-mediated cell death and injury in Escherichia coli: from fundamentals to food applications. Front Microbiol. 2015;6:599. doi: 10.3389/fmicb.2015.00599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grogan JM, Schneider NM, Ross FM, Bau HH. Bubble and pattern formation in liquid induced by an electron beam. Nano Lett. 2014;14(1):359–364. doi: 10.1021/nl404169a [DOI] [PubMed] [Google Scholar]
- 74.Levanduski L, Jaczynski J. Increased resistance of Escherichia coli O157: h7to electron beam following repetitive irradiation at sub-lethal doses. Int J Food Microbiol. 2008;121(3):328–334. doi: 10.1016/j.ijfoodmicro.2007.11.009 [DOI] [PubMed] [Google Scholar]
- 75.Egerton RF. Control of radiation damage in the TEM. Ultramicroscopy. 2013;127:100–108. doi: 10.1016/j.ultramic.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 76.Egerton RF. Mechanisms of radiation damage in beam-sensitive specimens, for TEM accelerating voltages between 10 and 300 kV. Microsc Res Tech. 2012;75(11):1550–1556. doi: 10.1002/jemt.v75.11 [DOI] [PubMed] [Google Scholar]
- 77.Beeby M, Gumbart JC, Roux B, Jensen GJ. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol. 2013;88(4):664–672. doi: 10.1111/mmi.2013.88.issue-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci. 2008;105(48):18953–18957. doi: 10.1073/pnas.0808035105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuson HH, Auer GK, Renner LD, et al. Measuring the stiffness of bacterial cells from growth rates in hydrogels of tunable elasticity. Mol Microbiol. 2012;84(5):874–891. doi: 10.1111/mmi.2012.84.issue-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loskill P, Pereira PM, Jung P, et al. Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the staphylococcus aureus cell envelope. Biophys J. 2014;107(5):1082–1089. doi: 10.1016/j.bpj.2014.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vollmer W. Structure and biosynthesis of the Murein (Peptidoglycan) sacculus In: Ehrmann M, editor. The periplasm. ASM Press;2007:198–213. [Google Scholar]
- 82.van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11(3):25R–36R. doi: 10.1093/glycob/11.3.25R [DOI] [PubMed] [Google Scholar]
- 83.Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta Biomembr. 2008;1778(9):1714–1734. doi: 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 84.Glauner B, Holtje JV, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263(21):10088–10095. [PubMed] [Google Scholar]
- 85.Snowden MA, Perkins HR. Peptidoglycan cross-linking in Staphylococcus aureus An apparent random polymerisation process. Eur J Biochem. 1990;191(2):373–377. doi: 10.1111/ejb.1990.191.issue-2 [DOI] [PubMed] [Google Scholar]
- 86.Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol. 2008;190(13):4782–4785. doi: 10.1128/JB.00025-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci. 2010;107(20):9182–9185. doi: 10.1073/pnas.0911517107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Si F, Li B, Margolin W, Sun SX. Bacterial growth and form under mechanical compression. Sci Rep. 2015;5:11377. doi: 10.1038/srep11367 [DOI] [PMC free article] [PubMed] [Google Scholar]