Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) infection causes an acute respiratory illness and is associated with a high case fatality rate; however, the pathogenesis of severe and fatal MERS-CoV infection is unknown. We describe the histopathologic, immunohistochemical, and ultrastructural findings from the first autopsy performed on a fatal case of MERS-CoV in the world, which was related to a hospital outbreak in the United Arab Emirates in April 2014. The main histopathologic finding in the lungs was diffuse alveolar damage. Evidence of chronic disease, including severe peripheral vascular disease, patchy cardiac fibrosis, and hepatic steatosis, was noted in the other organs. Double staining immunoassays that used anti–MERS-CoV antibodies paired with immunohistochemistry for cytokeratin and surfactant identified pneumocytes and epithelial syncytial cells as important targets of MERS-CoV antigen; double immunostaining with dipeptidyl peptidase 4 showed colocalization in scattered pneumocytes and syncytial cells. No evidence of extrapulmonary MERS-CoV antigens were detected, including the kidney. These results provide critical insights into the pathogenesis of MERS-CoV in humans.
Middle East respiratory syndrome coronavirus (MERS-CoV) was initially isolated from a sputum specimen of a patient who died of respiratory and renal failure in Saudi Arabia in 2012.1 Recent data have indicated that dromedary camels are likely to transmit MERS-CoV to humans.2, 3 Human-to-human MERS-CoV transmission is well documented, particularly in the setting of nosocomial outbreaks.4, 5
As of July 7, 2015, the World Health Organization (WHO) was notified of 1368 laboratory-confirmed cases of MERS-CoV infection with 487 reported deaths (35.6%) from 26 countries, primarily in men with a median age of 50 years.6 Most cases (>75%) were reported from the Kingdom of Saudi Arabia.6 MERS-CoV infection can result in a wide clinical spectrum from asymptomatic infection, upper respiratory tract illness, to severe pneumonia and multiorgan failure.7, 8, 9 MERS-CoV binds to dipeptidyl peptidase 4 (DPP4) receptors that are primarily in the lower respiratory tract but also distributed in other tissues.10 Although there have been numerous cases and fatalities, the pathologic changes and viral distribution in humans associated with severe MERS-CoV illness are unknown, and knowledge of pathogenesis remains limited.
This report provides the first autopsy, clinicopathologic, immunohistochemical (IHC), and ultrastructural description of a fatal case of MERS-CoV. This patient, who had multiple close contacts, was identified as part of an epidemiologic investigation of a large cluster of MERS-CoV infections at hospital in the United Arab Emirates (UAE) in April 2014. This outbreak occurred in the context of a surge of cases in the Arabian Peninsula in which 515 MERS-CoV cases were identified in Saudi Arabia from April 11 to June 9, 2014 (World Health Organization, http://www.who.int/csr/don/2014_06_13_mers/en, last accessed October 9, 2015).
Materials and Methods
Histopathologic and IHC Studies
Tissues obtained at autopsy were fixed in 10% buffered formalin, paraffin-embedded, sectioned at 4 μm, and stained by routine hematoxylin and eosin. An immunostaining protocol was applied with use of several mouse antibodies and one human antibody to MERS-CoV as described previously.11
IHC assays that used mouse anti-MERS antibodies (Table 1 ) were performed with the use of a polymer-based indirect immunoalkaline phosphatase detection system with colorimetric detection of antibody/polymer complex with Fast Red Chromogen (Thermo Fisher Scientific, Runcom, Cheshire, UK, or Biocare Medical, Concord, CA). IHC assays with the use of the human, anti–MERS-CoV antibody were performed with the Klear Human AP-Polymer Detection Kit with GBI Permanent Red Chromogen (GBI Labs, Bothell, WA). Double-stained assays used the Envision G/2 Double Stain System, Rabbit/Mouse (DAB + Permanent Red) from Dako (Glostrup, Denmark). Antibodies against CD68 (Dako), cytokeratin (Dako), surfactant apoprotein A (Dako), and DPP4 (Origene Technologies, Rockville, MD), followed by the anti–MERS-CoV antibodies [CDC (Atlanta, GA) and University of North Carolina (Chapel Hill, NC)], were used. All assays were performed according to the manufacturer's guidelines. MERS-CoV–infected and –noninfected Vero cells and tissues from non–MERS-CoV cases and normal mouse sera in place of primary antibodies were used as positive and negative controls, respectively.
Table 1.
Primary Antibodies Used for IHC
| Ab No. | Primary Ab | Host | Dilution | Pretreatment | Ab source | Detection | Comments |
|---|---|---|---|---|---|---|---|
| 1510/1513 | MERS-CoV | Mouse hyperimmune serum | 1:200 | PK | CDC, DVD | (no. 1253) UVLP | MERS-CoV_JordanN3 |
| 1511 | EMC S | Mouse hyperimmune serum | 1:500 | PK | Ralph Baric, UNC | (no. 1253) UVLP | |
| 1512 | HKU5.2 N | Mouse hyperimmune serum | 1:500 | AR | CDC, DVD | (no. 1253) UVLP | Bat coronavirus |
| 1514 | γ-Irradiated IN case serum | Human | 1:1000 | AR | CDC, DVD | (no. 1518) Klear | |
| 0274 | CD68 (KP1) | MAB | 1:100 | AR | Dako | (no. 1371) Envision Doublestain | |
| 0304 | Cytokeratin (AE1 and AE3) | MAB | 1:100 | AR | Dako | (no. 1371) Envision Doublestain | |
| 1037 | Surfactant apoprotein A (PE10) | HMAF | 1:100 | AR | Dako | (no. 1371) Envision Doublestain | |
| 1547 | DPP4 (clone OTI11D7) | MAB | 1:100 | PK | Origene Technologies | (no. 1371) Envision Doublestain |
Ab, antibody; AR, antigen retrieval; DPP4, dipeptidyl peptidase 4; DVD, Division of Viral Diseases (CDC); EMC S, spike protein of human coronavirus Erasmus Medical Center; Envision Doublestain, Envision G/2 Double Stain System, Rabbit/Mouse (DAB + Permanent Red); HKU5.2 N, bat coronavirus HKU5.2 nucleoprotein; HMAF, hyperimmune mouse ascitic fluid; Klear, Klear Human AP-Polymer Detection Kit with GBI Permanent Red Chromogen; MAB, monoclonal antibody; MERS-CoV, Middle East respiratory syndrome coronavirus; PK, proteinase K; UNC, University of North Carolina; UVLP, UltraVision Labeled Polymer Detection Kit.
Ultrastructural Studies
Lung tissue samples were excised from a paraffin block with the use of a 2-mm punch and processed for electron microscopy examination as previously described.12
Molecular Analysis
Full genome sequencing by the Sanger method with the use of direct genome walking PCR and next-generation sequencing (Illumina MiSeq; Illumina, San Diego, CA) were performed as previously described on confirmed positive lung tissue,13 named as Abu Dhabi_UAE_8_2014 and deposited into GenBank (http://www.ncbi.nlm.nih.gov; GenBank accession number KP209306). ClustalW was used to align the full genome with the respective available complete or near complete MERS-CoV genomes in GenBank.14 Phylogenetic reconstructions were performed with MrBayes version 3.2 (MrBayes: Bayesian Inference of Phylogeny, http://mrbayes.sourceforge.net, last accessed June 22, 2015) under a General-Time-Reversible model of nucleotide substitution with four categories of γ-distributed rate heterogeneity and a proportion of invariant sites (GTR + 4 + I).15
Results
Clinical History and Findings
The patient was a 45-year-old, nonsmoking, obese (body mass index, 30.5 kg/m2) Filipino man with no relevant past medical history, recent travel, or exposure to sick contacts or farm animals. He worked in a storage room at a paramedic station with no patient care duties. He shared housing with five roommates from the paramedic department.
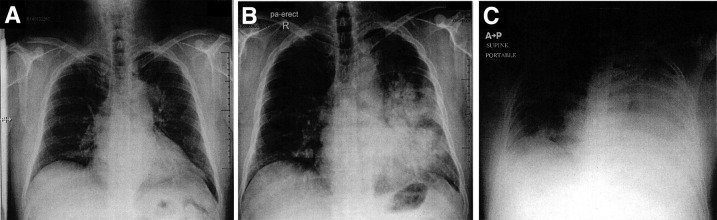
He presented to an emergency department in Abu Dhabi, UAE, on April 2, 2014, with a 4-day history of fever, rhinorrhea, and productive cough. At evaluation, he was afebrile with an oxygen saturation of 99% on room air, and a chest X-ray showed a left-sided opacity (Figure 1 A). The patient was diagnosed as acute bronchitis, prescribed 20 mg prednisolone daily for 5 days and paracetamol, and discharged. He returned to the emergency department on April 6, 2014, with persistent cough and shortness of breath; a chest X-ray showed a left-sided opacity and air bronchograms, and the patient was diagnosed with pneumonia, given a prescription for levofloxacin, and discharged (Figure 1B). He returned to the emergency department the same day, with worsening shortness of breath and was admitted. He had a temperature of 38.7°C, pulse of 113 beats per minute, a respiratory rate of 24 breaths per minute, blood pressure of 123/73, an oxygen saturation of 93% on room air, and left basal crackles and rales. An arterial blood gas on room air showed pH of 7.49, partial pressure of carbon dioxide of 27.6 mm Hg, partial pressure of oxygen of 52.4 mm Hg, and bicarbonate of 20.7 mEq/L, consistent with respiratory alkalosis. Laboratory evaluation showed a normal white blood cell count with lymphopenia (0.5 K/μL) and creatinine of 0.9 mg/dL (Supplemental Table S1). Blood, sputum, and urine cultures were obtained, and a nasopharyngeal swab specimen was sent for MERS-CoV testing. On April 7, day 1 of admission, oseltamivir, ceftriaxone, and azithromycin were started empirically. The day after admission he was transferred to the intensive care unit for tachypnea and respiratory distress and was intubated for mechanical ventilation; a portable chest X-ray showed multiple patchy airspace opacities (Figure 1C). The patient became hypotensive, requiring inotropic support, and developed acute kidney injury and renal failure, requiring dialysis. On April 8, RT-PCR assay of the nasopharyngeal swab specimen collected on April 7 was reported to be positive for MERS-CoV for UpE and ORF1a gene targets.16 Bacterial cultures of blood, sputum, and urine specimens obtained at admission after antibiotic administration were negative. On April 9, he was given 100 mg hydrocortisone intravenously every 8 hours and started on nitric oxide at 16 ppm. The patient's condition continued to deteriorate, and he died April 10.
Figure 1.
CXR images. A: CXR at initial ED visit shows small left-sided opacity. B: CXR at second ED visit, with increase of left-sided opacity. C: Portable CXR 1 day after admission with substantial progression of the opacity. CXR, chest X-ray; ED, emergency department.
Autopsy
The body was kept refrigerated at 4°C, and an autopsy was performed 10 days after death. Notable findings included massive pleural effusion (5 L), substantial pericardial effusion (150 mL), and abdominal effusion; edematous and consolidated lungs; and generalized congestion.
Histopathology, Immunohistochemistry, and Electron Microscopy
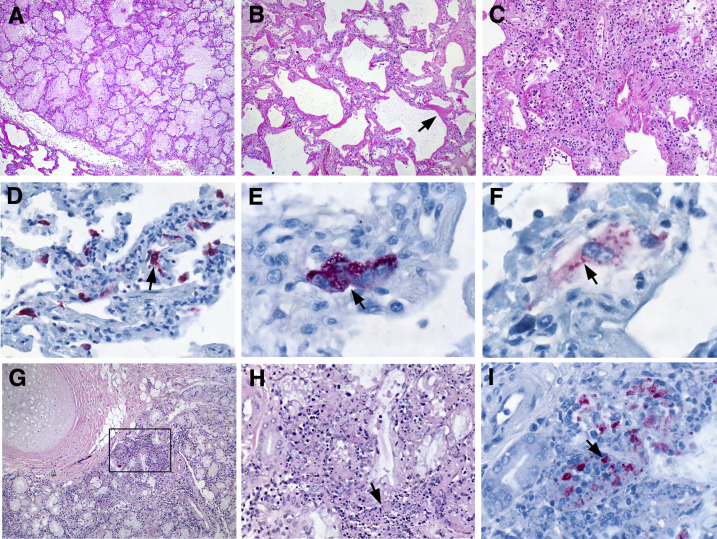
The predominant pulmonary histologic pattern was exudative-phase diffuse alveolar damage with denuding of bronchiolar epithelium, prominent hyaline membranes, alveolar fibrin deposits, type 2 pneumocyte hyperplasia, rare multinucleated syncytial cells, and alveolar septa involved by edema and lymphocytes with fewer plasma cells, neutrophils, and macrophages (Figure 2 , A–C). Dispersed foci of necrotic debris were seen both subpleurally and within alveoli. No viral inclusions were seen, and moderate anthracosis was present. All four antibodies (1510/1513, 1511, 1512, and 1514) highlighted multiple foci of MERS-CoV antigens, predominantly localized within the cytoplasm of pneumocytes and syncytial cells (Figure 2, D–F). Sections of trachea and bronchi showed mild-to-moderate lymphocytic mucosal and submucosal inflammation with a few neutrophils and plasma cells (Figure 2G). Occasional clusters of noninvasive and extracellular candida yeast and hyphae were seen in septa, alveolar spaces, and overlying respiratory epithelium, suggesting postmortem overgrowth. Bronchial submucosal glands were focally necrotic (Figure 2H). Immunostaining for MERS-CoV antigens was identified in both unremarkable and necrotic bronchial submucosal glands (Figure 2I).
Figure 2.
Histopathology of lung from MERS-CoV patient. A: Pulmonary edema. B: Diffuse alveolar damage, including prominent hyaline membrane formation (arrow). C: Alveolar fibrin deposits, type 2 pneumocyte hyperplasia, and thickened alveolar septa involved by edema and a mixed inflammatory infiltrate. D–F: Immunostaining of MERS-CoV antigen in pneumocytes (Ab 1511; D, arrow), a multinucleated syncytial cell (Ab 1511; E, arrow), and a binucleated cell (Ab 1514; F, arrow). G: Moderate lymphocytic inflammation of the submucosal glands. H: Magnified from the boxed area in G. Submucosal glands with focal areas of necrosis (arrow). I: Immunostaining of MERS-CoV antigen in necrotic foci of submucosal glands (arrow; Ab 1512). Original magnification: ×5 (A); ×20 (B–D); ×75 (E); ×100 (F); ×10 (G); ×40 (H); ×63 (I). Ab, antibody; MERS-CoV, Middle East respiratory syndrome coronavirus.
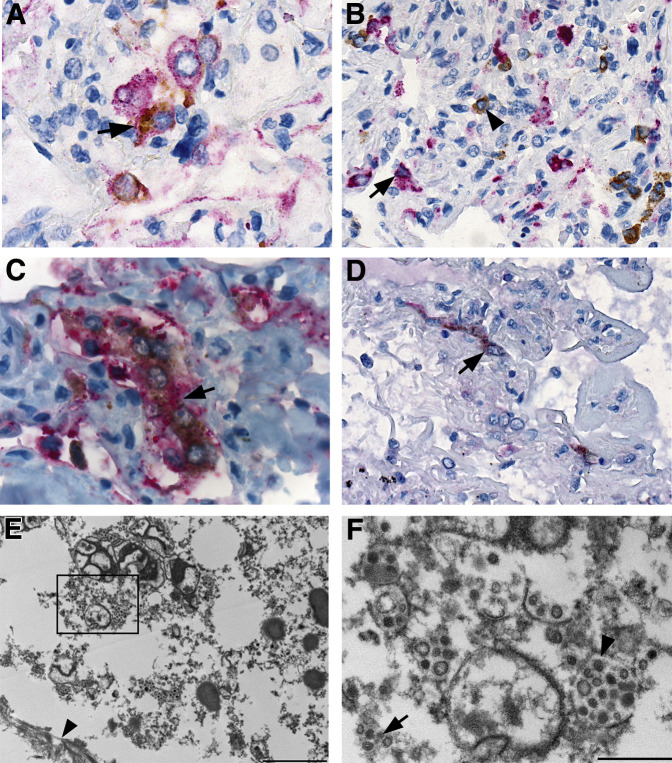
Multiple double immunostaining immunoassays were performed to assess for colocalization of MERS-CoV and cells labeling for cytokeratin, CD68, surfactant, and DPP4. Double staining with cytokeratin confirmed the presence of viral antigens within pneumocytes and epithelial syncytial cells, whereas double staining with CD68 showed two distinct populations with no colocalization of MERS-CoV and macrophages (Figure 3 , A and B). Surfactant double staining revealed type 2 pneumocytes and syncytial cells with intracytoplasmic viral antigens (Figure 3C). DPP4 antigens were detected in pneumocytes, syncytial cells, mononuclear leukocytes, and vascular endothelium, although colocalization of MERS-CoV and DPP4 was observed in scattered pneumocytes and syncytial cells (Figure 3D). Electron microscopy showed infected and degenerated pneumocytes encased between hyaline membranes, composed of fibrin and basement membranes (Figure 3E). Clusters or individualized, predominately spherical, 50 to 150 nm in diameter, viral particles were observed in membrane-bound vesicles (Figure 3F).
Figure 3.
Immunohistochemical and ultrastructural localization of MERS-CoV and associated histologic findings. A: MERS-CoV and cytokeratin antigens in pneumocytes (arrow); red stain, MERS-CoV; brown stain, cytokeratin. B: MERS-CoV antigens in pneumocytes (arrowhead) and CD68 antigens in macrophages (arrow); red stain, CD68; brown stain, MERS-CoV. C: MERS-CoV and surfactant antigens in type 2 pneumocytes (arrow); red stain, surfactant; brown stain, MERS-CoV. D: MERS-CoV and DPP4 in pneumocytes (arrow); red stain, DPP4; brown stain, MERS-CoV. E: Fragmented pneumocyte infected with MERS-CoV, hyaline membrane (arrowhead) present. F: Magnified from the boxed area in E. MERS-CoV virions dispersed as single particles (arrow) or in clusters within membrane-bound vesicles (arrowhead). Spherical and pleomorphic particles ranged in size from 50 to 150 nm diameter. Scale bars: 2 μm (E); 500 nm (F). Original magnification: ×100 (A and C); ×63 (B); ×75 (D). DPP4, dipeptidyl peptidase 4; MERS-CoV, Middle East respiratory syndrome coronavirus.
The kidney showed increased globally sclerotic glomeruli (5% to 10% of glomeruli), thickened Bowman capsules, severe atherosclerosis and hyaline arteriolosclerosis, patchy interstitial inflammation, and intratubular proteinaceous and granular casts. Diminished lymphoid follicles and a robust interfollicular proliferation of pleomorphic immunoblasts intermixed with a polymorphous population of reactive lymphocytes were seen in multiple lymph nodes; the spleen also contained numerous immunoblasts and reactive lymphocytes. The bone marrow was normocellular with maturing trilineage hematopoiesis and left-shifted granulopoiesis. The heart revealed diffuse myocyte hypertrophy, moderate coronary atherosclerosis, and patchy fibrosis. Moderate steatosis, scattered calcifications, and mild portal tract and lobular lymphocytic inflammation were identified in the liver. The sections of the cerebrum and cerebellum were unremarkable. MERS-CoV IHC was negative in multiple specimens from different organs, including kidney, liver, spleen, several lymph nodes, bone marrow, small intestine, and colon.
Overall the histology was well-preserved; although the sections of brain showed minimal-to-mild autolysis and kidney showed moderate autolysis.
Molecular Assays
The genome sequence is similar (>99%) to other known MERS-CoVs and clustered closely with camel-derived MERS-CoV strains (KJ650295 to KJ650297) obtained in Al-Hasa, Saudi Arabia, in 2013, suggesting recent origin in camels, although the patient had no known camel exposures. As indicated from the phylogenetic tree (Supplemental Figure S1), the sequences from this case and close contacts cluster most closely in the same clade, further supporting their transmission link between these cases.
Discussion
This report provides invaluable insights as the first description in the published literature of the clinicopathologic, IHC, and ultrastructural findings of a fatal case of MERS-CoV infection. The histopathologic pattern observed in the lungs was diffuse alveolar damage. MERS-CoV IHC and double staining techniques showed viral antigens were predominantly localized to type 2 pneumocytes and epithelial syncytial cells. Although the pathogenesis of severe and fatal MERS-CoV infection is unknown, these postmortem findings provide critical insights, including evidence that pneumocytes are important targets, suggesting that direct cytopathic effects contribute to MERS-CoV respiratory symptoms. An ex vivo study that examined MERS-CoV–infected human lung tissue found evidence of pneumocyte damage by electron microscopy, including detachment of type 2 pneumocytes, and membrane blebbing, suggestive of apoptosis.17 However, IHC staining for MERS-CoV was patchy, implying that other causes such as immune dysfunction may be relevant. Acute renal failure is commonly observed in critically ill MERS-CoV patients7, 8, 9, 18 and MERS-CoV RNA was detected in urine18, 19; however, no evidence of extrapulmonary MERS-CoV dissemination was observed, suggesting that acute renal failure in this patient was not caused by direct renal infection but likely by other factors, such as hypoperfusion or cytokine dysregulation. The presence of MERS-CoV antigens in submucosal glands provides a mechanism by which the virus enters respiratory secretions and becomes transmissible.
The pathologic features of MERS-CoV are shared by other similar respiratory illnesses, such as severe acute respiratory syndrome (SARS)-CoV. Several reports that evaluated fatal cases of SARS-CoV describe diffuse alveolar damage in various stages as the most characteristic feature,20 with IHC staining of SARS-CoV antigen primarily in alveolar epithelial cells.20 Syncytial cells may also be seen with other coronavirus infections, including SARS-CoV and paramyxovirus infections.20 MERS-CoV entry is mediated by DPP4, which is expressed throughout the lower respiratory tract17 but also numerous other organs, including the kidney.10 Double staining with MERS-CoV and DPP4 IHC was seen in pneumocytes and syncytial cells, consistent with the identification of these cells as targets of MERS-CoV infection. In addition, ex vivo models have detected MERS-CoV antigen in bronchial epithelial cells, pneumocytes, and endothelium and DPP4 in bronchiolar epithelium, endothelium, pneumocytes, and alveolar macrophages,17, 21 correlating with our findings. In contrast, the functional receptor for SARS-CoV is angiotensin-converting enzyme 2.22 Similar to DDP4, angiotensin-converting enzyme 2 has a broad distribution, with heavy expression in alveolar epithelial cells, enterocytes, and endothelial cells.23 Although evidence shows that SARS-CoV also infects type 2 pneumocytes,24, 25 SARS-CoV was detected in several extrapulmonary sites by in situ hybridization and electron microscopy, including circulating lymphocytes, lymphoid tissues, renal distal tubules, and small intestinal mucosa.26 Because MERS-CoV RNA was detected in blood,19 and DPP4 is widely distributed in different tissues,10 extrapulmonary dissemination could be possible, but we did not detect any evidence of MERS-CoV spread outside the respiratory tract.
Pulmonary findings of consolidation, diffuse alveolar damage, and pleural effusions with no evidence of bacterial pneumonia are consistent with the clinical features reported in other critically ill adults with MERS-CoV infection.7, 8, 9 Pathologic evidence of several chronic diseases in this patient, including myocardial fibrosis, atherosclerosis, and hepatic steatosis, correlates with reports of individuals with underlying comorbidities, such as end-stage renal disease, diabetes mellitus, hypertension, or cardiac disease, having an increased risk of developing severe MERS-CoV infection.4, 7 The impact of low-dose oral prednisolone before admission and intravenous hydrocortisone on the clinical course and fatal outcome of this MERS-CoV patient is unknown. Follicular depletion in lymph nodes, consistent with corticosteroid use, was noted, and no bacterial coinfections were identified. The hydrocortisone dosing of 100 mg every 8 hours in the intensive care unit before death was higher than recommended for refractory septic shock by the Surviving Sepsis Campaign guidance,27 but the patient only received a few doses. A systematic review of high-dose corticosteroids for treatment of SARS-CoV reported evidence of harm without survival benefit.28 In patients with influenza A(H1N1)pdm09 virus infection, corticosteroid treatment was associated with increased mortality,29 although further studies are needed. Therefore, until further evidence is available, high-dose corticosteroid treatment for MERS-CoV pulmonary disease should be avoided.30 Current clinical management of MERS-CoV patients is based on supportive care of complications and adherence to recommended infection prevention and control measures (CDC, http://www.cdc.gov/coronavirus/mers/infection-prevention-control.html, last accessed October 9, 2015).
Our findings provide invaluable and unprecedented insights into the histologic changes, pathogenesis, and viral dissemination of MERS-CoV in humans. Further studies, including additional postmortem examinations, evaluation of the host immune response, and identification of sites of viral replication, are necessary to strengthen knowledge on pathogenesis, transmission patterns, and effective treatment strategies for optimal clinical management and infection control practices.
Acknowledgments
We thank Cynthia Goldsmith for reviewing the electron microscopy and Azaibi Tamin for providing the MERS-CoV–infected Vero cell cultures.
Footnotes
See related Commentary on page 507
Supported by CDC operational funds.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Disclosures: None reported.
Current address of M.G.M., National Cancer Institute–Shady Grove, Rockville, MD.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2015.10.024.
Supplemental Data
Phylogenetic analysis of the Abu Dhabi_UAE_8_2014 sequence. The sequence alignment was generated with 76 nearly complete genome sequences published in GenBank (excluding identical sequences). The Abu Dhabi_UAE_8_2014 discussed in this study is noted by red solid circle and the other cases from the UAE cluster are noted by black solid circles. The camel MERS-CoV sequences are labeled with a camel symbol. Posterior probabilities are shown for nodes with a support >0.8. The scale bar indicates the genetic distance, in substitutions/site.
References
- 1.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 3.Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D., AlHajri M.M., Koopmans M.P. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A., KSA MERS-CoV Investigation Team Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Abdallat M.M., Payne D.C., Alqasrawi S., Rha B., Tohme R.A., Abedi G.R., Al Nsour M., Iblan I., Jarour N., Farag N.H., Haddadin A., Al-Sanouri T., Tamin A., Harcourt J.L., Kuhar D.T., Swerdlow D.L., Erdman D.D., Pallansch M.A., Haynes L.M., Gerber S.I., Jordan MERS-CoV Investigation Team Hospital-associated outbreak of Middle East respiratory syndrome coronavirus: a serologic, epidemiologic, and clinical description. Clin Infect Dis. 2014;59:1225–1233. doi: 10.1093/cid/ciu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Middle East respiratory syndrome coronavirus (MERS-CoV–Summary of Current Situation. Lit Update Risk Assess. 7 July 2015 http://www.who.int/csr/disease/coronavirus_infections/risk-assessment-7july2015/en Available at: [Google Scholar]
- 7.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., Makhdoom H.Q., Zumla A.I., Memish Z.A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saad M., Omrani A.S., Baig K., Bahloul A., Elzein F., Matin M.A., Selim M.A., Al Mutairi M., Al Nakhli D., Al Aidaroos A.Y., Al Sherbeeni N., Al-Khashan H.I., Memish Z.A., Albarrak A.M. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int J Infect Dis. 2014;29:301–306. doi: 10.1016/j.ijid.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arabi Y.M., Arifi A.A., Balkhy H.H., Najm H., Aldawood A.S., Ghabashi A., Hawa H., Alothman A., Khaldi A., Al Raiy B. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160:389–397. doi: 10.7326/M13-2486. [DOI] [PubMed] [Google Scholar]
- 10.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel R.M., Morris M., Henning T., Ritter J.M., Jones T.L., Dietz S., Ayers J., Vishwanathan S.A., Jenkins L., Zaki S., Wildemeersch D., Garber D., Powell N., Hendry R.M., McNicholl J., Kersh E.N. Evaluation of pigtail macaques as a model for the effects of copper intrauterine devices on HIV infection. J Med Primatol. 2014;43:349–359. doi: 10.1111/jmp.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paddock C.D., Sanden G.N., Cherry J.D., Gal A.A., Langston C., Tatti K.M., Wu K.H., Goldsmith C.S., Greer P.W., Montague J.L., Eliason M.T., Holman R.C., Guarner J., Shieh W.J., Zaki S.R. Pathology and pathogenesis of fatal Bordetella pertussis infection in infants. Clin Infect Dis. 2008;47:328–338. doi: 10.1086/589753. [DOI] [PubMed] [Google Scholar]
- 13.Cotten M., Lam T.T., Watson S.J., Palser A.L., Petrova V., Grant P., Pybus O.G., Rambaut A., Guan Y., Pillay D., Kellam P., Nastouli E. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis. 2013;19:736–742B. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. ClustalW and ClustalX version 2 (2007) Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 16.Lu X., Whitaker B., Sakthivel S.K., Kamili S., Rose L.E., Lowe L., Mohareb E., Elassal E.M., Al-sanouri T., Haddadin A., Erdman D.D. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hocke A.C., Becher A., Knepper J., Peter A., Holland G., Tonnies M., Bauer T.T., Schneider P., Neudecker J., Muth D., Wendtner C.M., Ruckert J.C., Drosten C., Gruber A.D., Laue M., Suttorp N., Hippenstiel S., Wolff T. Emerging human middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am J Respir Crit Care Med. 2013;188:882–886. doi: 10.1164/rccm.201305-0954LE. [DOI] [PubMed] [Google Scholar]
- 18.Poissy J., Goffard A., Parmentier-Decrucq E., Favory R., Kauv M., Kipnis E., Mathieu D., van der Werf S., Guery B., MERS-CoV Biology Group Kinetics and pattern of viral excretion in biological specimens of two MERS-CoV cases. J Clin Virol. 2014;61:275–278. doi: 10.1016/j.jcv.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D., Junglen S., Müller M.A., Haas W., Guberina H., Röhnisch T., Schmid-Wendtner M., Aldabbagh S., Dittmer U., Gold H., Graf P., Bonin F., Rambaut A., Wendtner C. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholls J.M., Butany J., Poon L.L., Chan K.H., Beh S.L., Poutanen S., Peiris J.S., Wong M. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 2006;3:e27. doi: 10.1371/journal.pmed.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan R.W., Hemida M.G., Kayali G., Chu D.K., Poon L.L., Alnaeem A., Ali M.A., Tao K.P., Ng H.Y., Chan M.C., Guan Y., Nicholls J.M., Peiris J.S. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir Med. 2014;2:813–822. doi: 10.1016/S2213-2600(14)70158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossel E.C., Wang J., Jeffers S., Edeen K.E., Wang S., Cosgrove G.P., Funk C.J., Manzer R., Miura T.A., Pearson L.D., Holmes K.V., Mason R.J. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology. 2008;372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shieh W.J., Hsiao C.H., Paddock C.D., Guarner J., Goldsmith C.S., Tatti K., Packard M., Mueller L., Wu M.Z., Rollin P., Su I.J., Zaki S.R. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum Pathol. 2005;36:303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dellinger R.P., Levy M.M., Rhodes A., Annane D., Gerlach H., Opal S.M., Sevransky J.E., Sprung C.L., Douglas I.S., Jaeschke R., Osborn T.M., Nunnally M.E., Townsend S.R., Reinhart K., Kleinpell R.M., Angus D.C., Deutschman C.S., Machado F.R., Rubenfeld G.D., Webb S., Beale R.J., Vincent J.L., Moreno R., Surviving Sepsis Campaign Guidelines Committee including The Pediatric Subgroup Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Sun W., Svendsen E.R., Tang S., MacIntyre R.C., Yang P., Zhang D., Wang Q. Do corticosteroids reduce the mortality of influenza A (H1N1) infection? A meta-analysis. Crit Care. 2015;19:46. doi: 10.1186/s13054-015-0764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization Clinical management of severe acute respiratory infection when Middle East respiratory syndrome coronavirus (MERS-CoV) infection is suspected. Interim Guidance. 2 July 2015 http://www.who.int/csr/disease/coronavirus_infections/case-management-ipc/en Available at: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of the Abu Dhabi_UAE_8_2014 sequence. The sequence alignment was generated with 76 nearly complete genome sequences published in GenBank (excluding identical sequences). The Abu Dhabi_UAE_8_2014 discussed in this study is noted by red solid circle and the other cases from the UAE cluster are noted by black solid circles. The camel MERS-CoV sequences are labeled with a camel symbol. Posterior probabilities are shown for nodes with a support >0.8. The scale bar indicates the genetic distance, in substitutions/site.