Abstract
Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) poses a serious public health hazard. The S2 subunit of the S glycoprotein of SARS-CoV carries out fusion between the virus and the host cells. However, the exact mechanism of the cell fusion process is not well understood. Current model suggests that a conformational transition, upon receptor recognition, of the two heptad core regions of S2 may expose the hydrophobic fusogenic peptide or fusion peptide for membrane insertion. Three regions of the S2 subunit have been proposed to be involved in cell–cell fusion. The N-terminal fusion peptide (FP, residues 770–788), an internal fusion peptide (IFP, residues 873–888) and the pre-transmembrane region (PTM, residues 1185–1202) demonstrated interactions with model lipid membranes and potentially involved in the fusion process. Here, we have determined atomic resolution structures of these three peptides in DPC detergent micelles by solution NMR. FP assumes α-helical conformation with significant distortion at the central Gly residues; enabling a close packing among sidechains of aromatic residues including W, Y and F. The 3-D structure of PMT is characterized by a helix–loop–helix with extensive aromatic interactions within the helices. IFP adopts a rather straight α-helical conformation defined by packing among sidechains of aromatic and aliphatic residues. Paramagnetic spin labeled NMR has demonstrated surface localization of PMT whereas FP and IFP inserted into the micelles. Collectively, data presented in this study will aid in understanding fusion mechanism of SARS-CoV.
Abbreviations: SARS-CoV, severe acute respiratory syndrome-associated coronavirus AMPs; FP, fusion peptide; NMR, nuclear magnetic resonance; NOESY, nuclear Overhauser effect spectroscopy; TOCSY, total correlation spectroscopy; DPC, dodecyl phosphocholine
Keywords: SARS-CoV, Cell fusion, NMR, Structure, Fusion protein, Fusion peptide
Graphical abstract
1. Introduction
Membrane fusion is an important step for a successful infection of the enveloped viruses causing the transfer of viral genetic materials into the host cell. The energy barrier of membrane fusion is overcome by a coordinated process mediated by specialized viral fusion proteins [1], [2], [3], [4]. Most fusion proteins are produced as precursor proteins which are often cleaved by host cell proteases, yielding a metastable complex consisted of a receptor binding subunit and a membrane fusion subunit [1], [2], [3], [4]. Upon binding to the cell surface receptor or being in the acidic environment of endosome, fusion proteins are activated following a dramatic conformational transition which essentially sets free a buried hydrophobic fusion peptide from the protein core [1], [2], [5], [6], [7], [8], [9]. The fusion peptide, once exposed, inserts into the outer-leaflet of the lipid bilayer, initiating the cell fusion process [5], [6], [7], [8], [9]. While, a part of the fusion protein undergoes an irreversible refolding to a stable conformation that releases free energy to overcome the membrane fusion barrier [5], [6], [7], [8], [9]. Consequently, a thorough understanding of the molecular basis of the fusion mechanism has been deemed vital not only for the virus life cycle but also for the rational design of potential therapeutics [10], [11], [12], [13], [14]. Therefore, atomic-resolution structures of fusion proteins are essential for obtaining mechanistic insights of membrane fusion process. Toward this, 3-D structures, using X-ray crystallography, of a number of fusion proteins, full length or fragment, are reported in their pre fusion or post fusion states [2], [4], [15], [16], [17], [18], [19]. As such, these atomic-resolution structures have provided important molecular insights for membrane fusion and drug discovery. However, atomic resolution structures of the full-length fusion proteins in complex with lipid membrane remain a challenging task. In order to infer membrane interactions of fusion processes, fusion peptides are investigated for structural and functional characterization in lipid or membrane mimetic environments [20], [21], [22], [23], [24], [25], [26], [27], [28], [29].
During 2002–2003, SARS-CoV emerged as a global health risk which was speared over 29 countries infecting approximately 9000 people with 774 deaths worldwide [30], [31], [32]. More recently, a new coronavirus, designated hCoV-EMC or Middle East Respiratory Syndrome coronavirus (MERS-CoV), has been identified in humans in Middle Eastern countries and England causing death of several hundred of the infected people [33], [34], [35], [36]. Coronaviruses, including SARS-CoV and MERS-CoV, display a broad host range and capable of causing chronic diseases related to the respiratory, hepatic and gastric systems. Due to its recurrent emergence, rapid and facile transmission, and high mortality, it is essential to gain a better understanding of the pathogenesis of this virus to develop antiviral drugs and vaccines for the cure and prevention of SARS and MERS [37], [38], [39], [40].
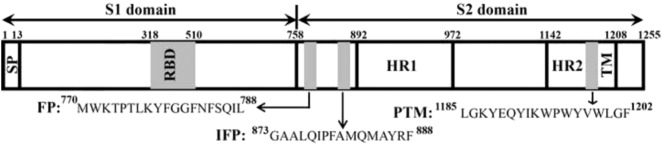
The large positive-stranded RNA genome of SARS-CoV enters into the host through cell fusion mediated by the viral spike (S) glycoprotein [41]. The S1 subunit of the S glycoprotein binds to the cell surface receptor whereas the S2 subunit primarily involves fusion between virus and target cell membrane [42], [43], [44]. The fusion protein or the S2 subunit of SARS-CoV has been designated to the class I type fusion protein [45], [46]. As a member of this class, the S2 subunit of S glycoprotein contains a pair of heptad repeats, HR1 and HR2 and a TM region at the C-terminus. Based on peptides corresponding to HR1 and HR2 it has been demonstrated that HR1 and HR2 forms antiparallel oligomeric complex [45], [46], [47], [48]. Atomic-resolution structures of HR1 and HR2 complex showed formation of a six helix bundle or trimer-of-hairpins structure, a conserved feature of the class I fusion proteins [49], [50], [51], [52]. Three separate regions of the S2 domain of S protein of SARS-CoV have been identified as member active peptides, named as fusion peptide (FP), internal fusion peptide (IFP) and a pre-transmembrane (PTM) domain near the C-terminal TM (Fig. 1 ) (53–56). Vesicle fusions, membrane interactions and biophysical characterization have been carried out for these peptides FP, IFP and PTM, revealing their fusogenic ability, high-affinity to partition into lipid membranes and membrane destabilization [53], [54], [55], [56], [57], [58], [59]. More recently, yet another peptide segment, residues 798 to 815, of S protein has been identified as a potential fusion peptide [60].
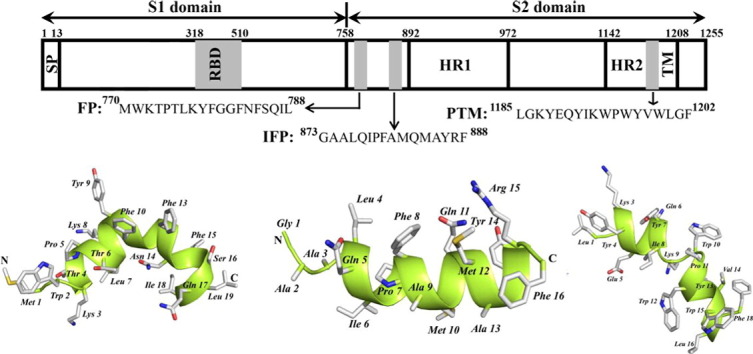
Fig. 1.
Spike glycoprotein of SARS-CoV is composed of two domains S1 and S2 separated by a protease cleavage site. The N-terminal domain has a signal peptide (SP) and a transmembrane domain (TM) is present at the C-terminus. Other regions are receptor binding domain (RBD), two heptad repeat (HR1 and HR2) sequences and putative fusion peptides named as fusion peptide (FP), internal fusion peptide (IFP) and pre transmembrane domain (PTM).
It has been proposed that the membrane fusion in SARS-CoV may involve the HR1, HR2 and these membrane active peptides to form the fusion pore in membrane (53, 56, 59). Notably, synthetic peptides corresponding to these regions demonstrated inhibition of SARS-CoV infection, presumably destabilizing cell fusion process [61]. Atomic resolution structure of any of these fusion peptides of SARS-CoV is yet to be reported. In order to gain insights into membrane fusion mechanism, here we have determined 3-D structures of the three fusion peptides of SARS-CoV in DPC detergent micelles. Our current results would help in better understating of the virus and host cell fusions and may aid in antiviral drug development.
2. Materials and methods
2.1. Peptides and chemicals
Synthetic peptides were purchased commercially from GL-Biochem (China) with purity of > 98%. Deuterated compounds (DPC-d38 and D2O) and 4, 4-dimethyl-4-silapentane-1-sulfonic acid (DSS) were obtained from Cambridge Isotope Laboratories, Inc. (Massachusetts, USA). Paramagnetic lipids, 5-DSA and 16-DSA (doxyl stearic acid) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2. NMR spectroscopy and data processing
NMR spectra were acquired in a Bruker AVANCE II 600 MHz spectrometer equipped with a cryoprobe. Lyophilized peptide samples (0.5 mM) were dissolved in 90% H2O/10%D2O, pH 4.7, containing 125 mM perdeuterated DPC. Typically, two-dimensional (2-D) 1H–1H TOCSY (mixing time 80 ms with 32 scans) and NOESY (mixing time 200 ms with 88 scans) spectra were acquired using spectral width of 13 ppm with 2 K × 512 complex points and the States-TPPI for quadrature detection at the t1 dimension. All NMR experiments were performed at a temperature of 315 K. Water signal was suppressed using WATERGATE. Natural abundance 13C–1H HSQC spectra (for 13Cα chemical shifts) were acquired by dissolving lyophilized peptides (0.5 mM) in D2O containing 125 mM perdeuterated DPC. HSQC spectra were obtained using spectral width of 12 and 80 ppm for 1H and 13C, respectively, with 300 scans. Chemical shifts were referenced with respect to DSS as an internal reference. For probing localization and insertion of peptides in DPC micelles, PRE (paramagnetic relaxation enhancement) studies were conducted by acquiring 2-D TOCSY spectra either in the absence or in the presence of 1 mM of 5-DSA and 16-DSA. Stock solutions of 5-DSA and 16-DSA were made in deuterated methanol. The intensities of the CαH/NH cross-peaks of amino acids in TOCSY spectra were estimated before and after addition of the paramagnetic probes and the % attenuation in intensity for both 5-DSA and 16-DSA were calculated. NMR data were processed using TopSpin 3.0 (Bruker). After zero filling along the t1 dimension, 2 K (t2) × 1 K (t1) data matrices were obtained. Spectral analysis and peak picking were done using SPARKY 3.113.
2.3. Structure calculations
The assigned NOE peaks were classified as strong, medium or weak and translated to an upper bound distance restraints: 2.8, 3.5 and 5.0 Ǻ, respectively. Backbone dihedral angles ϕ and ψ, were calculated using Preditor using 1Hα and 13Cα chemical shifts [62] and used for angular constraints. The DPC-bound structures fusion peptides were calculated using CYANA (version 2.1) [63]. The structure calculation was done in a stepwise manner to get the final ensemble with low RMSD and target function.
3. Results
3.1. NMR analyses of SARS-CoV of FP, IFP and PTM peptides
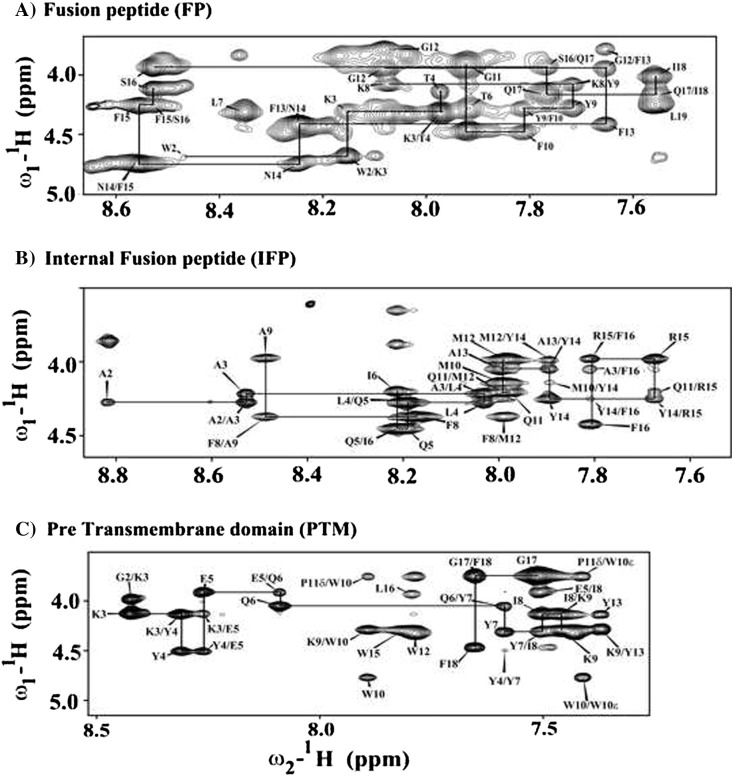
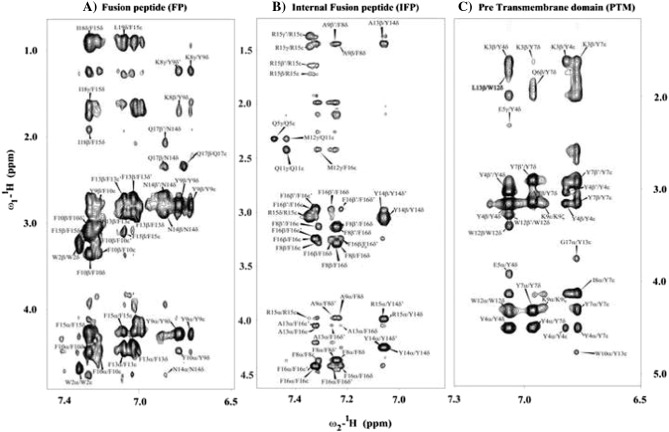
High-quality NMR spectra of SARS fusion peptides were obtained in solution containing DPC micelles. Sequence-specific resonance assignments of fusion peptides were achieved by analyses of two-dimensional 1H–1H TOCSY and NOESY spectra [64]. Fig. 2 shows sequential connectivity in the NOESY spectra of FP (panel A), IFP (panel B) and PTM (panel C). As can be seen, most of the resonances of FP, IFP and PTM are unambiguously identified. Peptide–micelle complexes were found to be stable over the period of time with no new resonances or line-width changes observed, suggesting absence of any potential miceller fusions by these peptides. Analyses of NOESY spectra reveal backbone/backbone backbone/sidechain and sidechain/sidechain NOE contacts for these fusion peptides. FP, IFP and PTM peptides are rich in aromatic amino acids; a number of NOEs could be identified involving aromatic ring protons with the backbone and sidechain protons of aliphatic residues (Fig. 3 ). Fig. 4 summarizes NOE contacts detected for FP (panel A), IFP (panel B) and PTM (panel C) peptides. NOE connectivities are characterized by sequential and medium range for all three fusion peptides (Fig. 4). Noticeably, aromatic residues of FP, IFP and PTM have delineated more number of NOE contacts compared to other residues. Further, residues Y9 and F10 of FP, residues F8, Y14 and F15 and residues Y4 and Y7 of PTM exhibited over 20, 24 and 30 NOEs, respectively (Fig. 4). The amino acid sequence of PTM fusion peptide contains as many as six aromatic residues; most of these residues are found to be involved in a number of NOE contacts (Fig. 4C). Taken together, the large number of NOE contacts observed for FP, IFP and PTM peptides indicate that these fusion peptides assume well-defined conformations in DPC micelles.
Fig. 2.
Fingerprint regions of 1H–1H 2-D NOESY spectra of SARS-CoV fusion peptides showing sequence specific resonance assignments of FP (panel A), IFP (panel B) and PTM (panel C) in DPC micelles.
Fig. 3.
Sections of 1H–1H 2-D NOESY spectra of FP (panel A), IFP (panel B) and PTM (panel C) showing NOEs contacts of low-field shifted aromatic ring proton resonances (6.5–7.5 ppm) with up-field shifted aliphatic resonances (4.5–0.8 ppm).
Fig. 4.
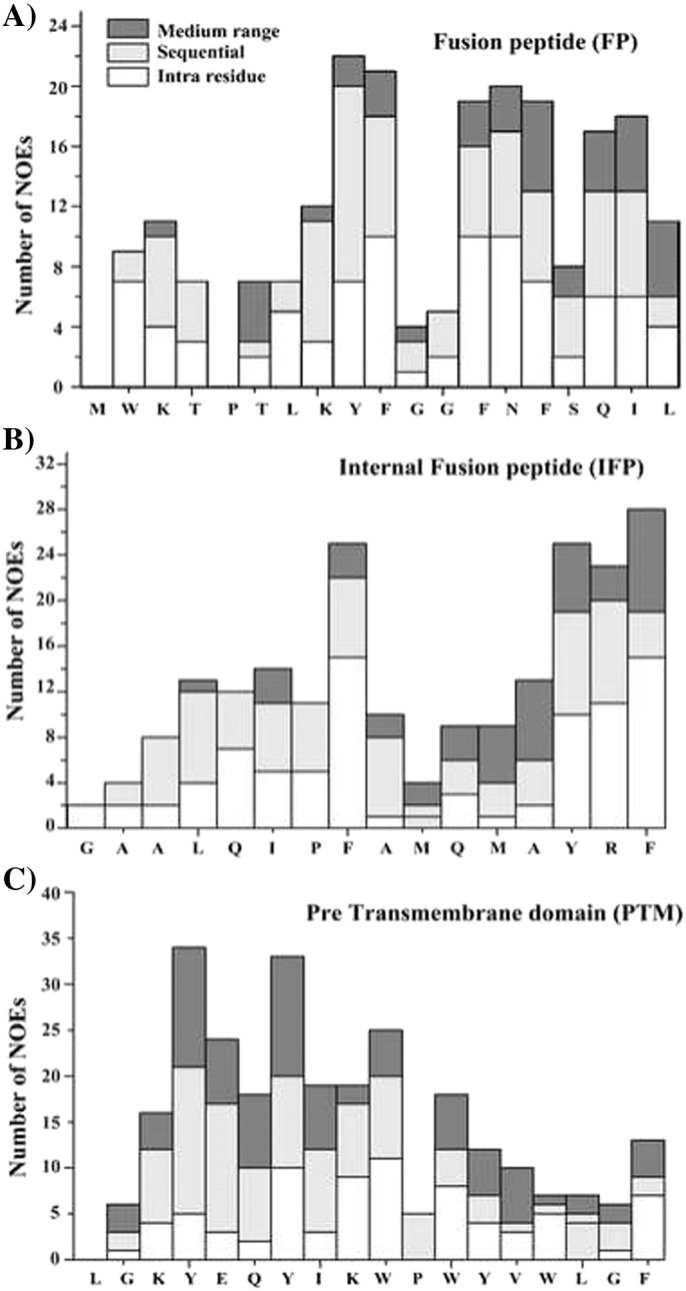
Bar diagram summarizing number and types of NOEs for FP (panel A), IFP (panel B) and PTM (panel C). Intra-residue, sequential and medium range NOEs are marked as white, light gray and dark bars, respectively.
3.2. Atomic-resolution structures of FP, IFP and PTM peptides
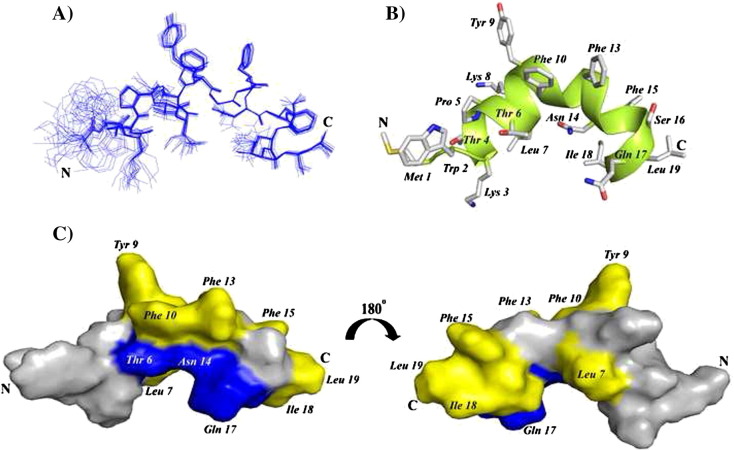
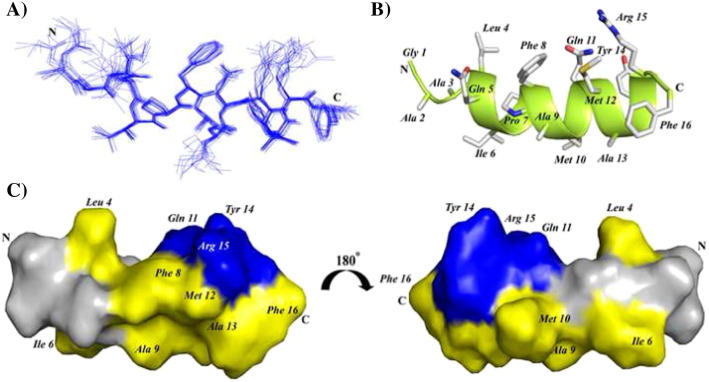
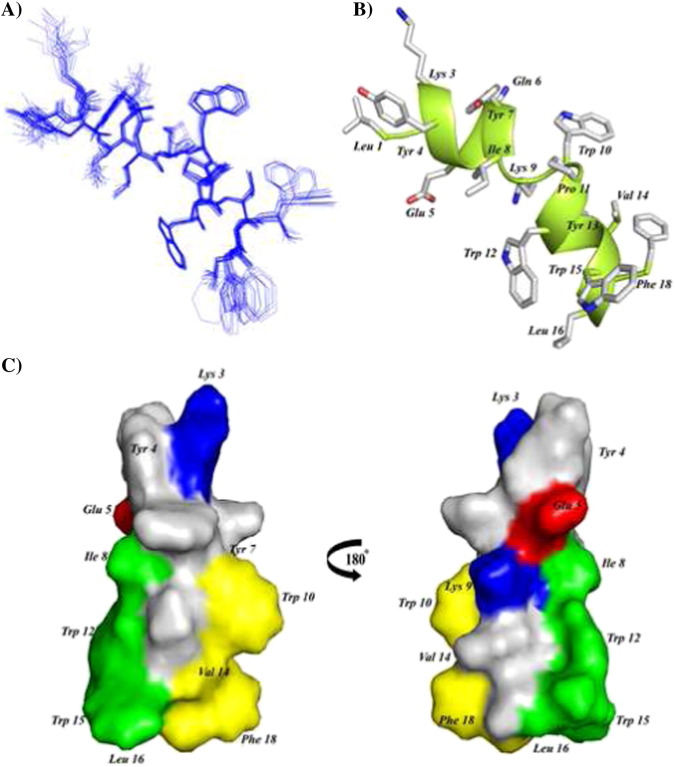
3-D structures of FP, IFP and PTM peptide are determined using CYANA based on NOE-driven distance and backbone dihedral angle (ϕ,ψ) constrains. A total of 153, 148 and 182 distance constrains are used for FP, IFP and PTM, respectively, to obtain the structural ensembles (Table 1 ). Fig. 5A shows superposition of twenty lowest energy structures of FP. The backbone and all heavy atoms RMSD values from the mean structure of FP are estimated to be 0.38 Å and 0.98 Å, respectively (Table 1). As can be seen, backbone atoms of residues T4-L19 and also sidechains of residues Y9, F10, F13, F15 L7, I18 and L19 demonstrate a close superposition whereas residues M1–W2–L3 are loosely superposed in the structural ensemble of FP (Fig. 5A). These observations indicate that a rather rigid structure for most of the residues in T4–L19 segment of FP, although residues at the N-terminus of FP, M1–W2–L3, lacks well-defined conformations. FP assumes a ‘V-shaped’ bend helical conformations for residues P5–I18, while N-terminal residues W2–T4 are found to be largely non-helical or in extended conformations (Fig. 5B). The V-shaped helical conformation of FP appears to be resulting at the centrally located two Gly residues G11–G12 including the residue F10 (Fig. 5B). There are two hydrophobic patches in the bend helical conformations of FP consisted of the sidechains of residues L7, Y9, F10, and F13 and the sidechains of residues F15, I18, and L19 (Fig. 5C). Sidechains of residues T6, N14, and Q17 are positioned along the same face of the bend helix of FP, indicating a potential polar surface (Fig. 5C). Fig. 6A shows twenty lowest energy superposed structure of the IFP. The backbone and all heavy atom RMSDs from the mean structure are found to be 0.43 Å and 0.82 Å, respectively (Table 1). A close superposition can be seen for all residues, except for the first three N-terminal residues G1–A2–A3 (Fig. 6A). The IFP adopts a canonical α-helix encompassing residues L4–R15 (Fig. 6B). One face of the helix is predominantly hydrophobic containing sidechains of residues I6, A9, M10, A13 and F16, whereas the opposite surface of the helix displays sidechains of non-polar and also polar residues (Fig. 6B). In particular, non-polar packing interactions may be sustained by the sidechains of residues L4, F8 and M12 (Fig. 6B). By contrast, a close proximity of sidechains of residues Q11, Y14, and R15 may render polar interactions in the helix of IFP (Fig. 6B). Fig. 7A represents superposition of twenty lowest energy structures of the PTM peptide. The backbone and all heavy atoms RMSD values from the mean structure are restricted to 0.15 Å and 0.80 Å, respectively (Table 1). The primary structure of PTM is rich in aromatic amino acids including one Phe, three Tyr and three Trp residues. The sidechains of these aromatic residues are well defined, except for the terminal residues W15 and F18 (Fig. 7A). The PTM peptide assumes a helix–loop–helix type conformation, whereby residues at the N-terminal K3–Y7 and residues at the N-terminal W10–W15 adopt helical conformations (Fig. 7B). These two short helices are connected by a loop comprised of residues I8 and K9 (Fig. 7B). The C-terminal helix displays an extended hydrophobic patch made of by sidechains of residues W10, V14 and F18 at one face of the helix and residues I8, W12, L16 and W15 located at the opposite face of the helix (Fig. 7C). The N-terminal helix and the loop of PTM structure are largely polar containing basic residues K3 and K9, negatively charged, E5 and polar residues. Sidechains of the residue E5 and the residue K9 are found to be in a close proximity ~ 6 Å, indicating potential salt bridge and/or hydrogen bond interactions (Fig. 7C).
Table 1.
Structural statistics of twenty lowest energy structures of FP, IFP and PTM in DPC micelles.
| Distance constraints | FP | IFP | PTM |
|---|---|---|---|
| Intra residue (i = j) | 89 | 86 | 80 |
| Sequential (i–j = 1) | 44 | 37 | 56 |
| Medium range [1 < |i–j| ≤ 4] | 20 | 25 | 46 |
| Total | 153 | 148 | 182 |
| Angle constraints | |||
| ϕ, ψ constraints | 34 | 28 | 30 |
| Deviation from mean structure | |||
| All backbone atoms | 0.38 | 0.43 | 0.15 |
| All heavy atoms | 0.98 | 0.82 | 0.80 |
| aRamachandran plot for the mean structure | |||
| % residues in the most favored region | 92.9 | 100 | 84.6 |
| % residues in the additionally allowed region | 7.1 | 0.0 | 15.4 |
| % residues in the generously allowed region | 0.0 | 0.0 | 0.0 |
| % residues in the disallowed region | 0.0 | 0.0 | 0.0 |
Calculated using Procheck.
Fig. 5.
(A) Backbone superposition of twenty lowest energy structures of FP in the presence of DPC micelles. (B) A representative structure of the micelle bound FP showing backbone folding as ribbon and sidechain disposition as stick. (C) A surface representation of FP showing localization of polar amino acids (blue color) and hydrophobic residues (yellow). Figures were generated using molecular visualization software Molmol and PyMol.
Fig. 6.
(A) Backbone superposition of 20 lowest energy structures of IFP in DPC micelles. (B) A representative structure of the micelle bound IFP showing backbone folding as ribbon and sidechain disposition as stick. (C) A surface representation of IFP showing localization of polar amino acids (blue color) and hydrophobic residues (yellow). Figures were generated using molecular visualization softwares Molmol and PyMol.
Fig. 7.
(A) Backbone superposition of 20 lowest energy structures of PTM in DPC micelles. (B) A representative structure of the micelle bound PTM showing backbone folding as ribbon and sidechain disposition as stick. (C) A surface representation of PTM showing localization of polar amino acids (blue and red color) and hydrophobic residues (yellow and green). Figures were generated using molecular visualization softwares Molmol and PyMol.
3.3. Localization of FP, IFP and PTM peptides in DPC micelles
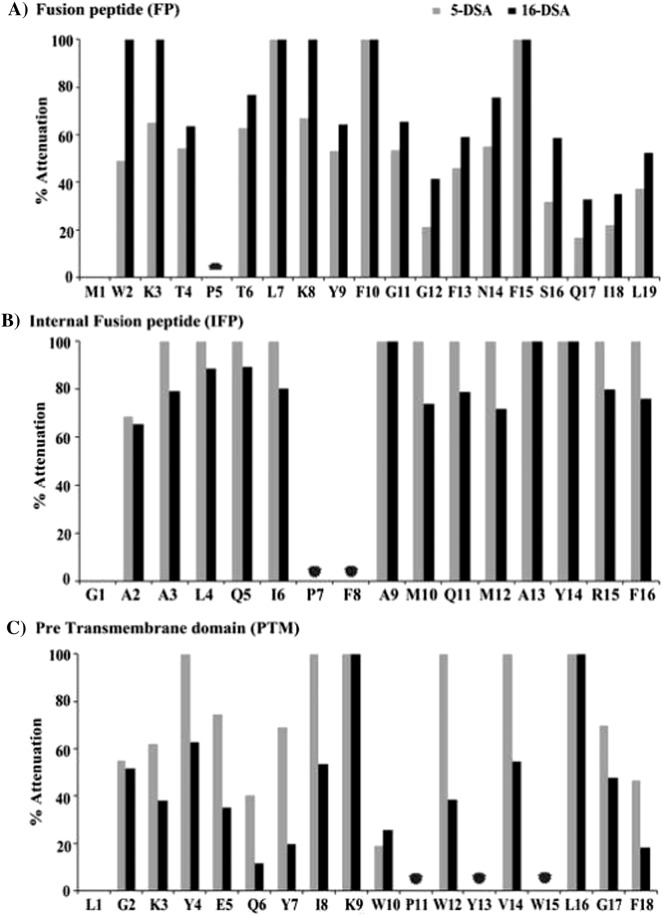
Paramagnetic relaxation enhancement (PRE) NMR studies, by spin labeled doxyl lipids, have been utilized to obtain micelle association and depth of insertion of the three fusion peptides. Two-dimensional TOCSY spectra of the three peptides were acquired either in the absence or in the presence of 1 mM spin labeled lipids, 5-doxyl-stearic acid (5-DSA) and 16-doxyl-stearic acid (16-DSA). Fig. 8 shows %attenuation of αH/NH cross-peak intensity, caused by the PRE probes, of TCOSY spectra for FP (panel A), IFP (panel B) and PTM (panel C). As can be seen, most of residues of three fusion peptides have experienced PRE mediated signal attenuation, indicating localization into micelles. However, three fusion peptides delineate differential signal attenuation in 5-DSA and 16-DSA. For example, residues of IFP appear to show a similar % of attenuation in 5-DSA and 16-DSA (Fig. 8B), whereas a significantly large number of residues of PTM are found to be perturbed by 5-DSA compared to 16-DSA probe (Fig. 8C). By contrast, residues of FP exhibit disparate signal attenuation in 5-DSA and 16-DSA (Fig. 8A). These observations indicate that the three fusion peptides might encompass a different depth of insertion into DPC micelles. It may be noted that a preferential signal attenuation detected in 16-DSA in comparison to that of 5-DSA is indicative of a deeper insertion of the residue into the core of micelle, whereas a residue may be located closer to the micelle head group in case of higher attenuation caused by 5-DSA probe [65], [66]. As can be seen, residues W2, K3 and K8 of FP demonstrate ~ 100% signal attenuation in 16-DSA, 5-DSA has caused lower signal attenuation for these residues, indicating a deep penetration of these residues of the FP into the micelle core (Fig. 8A). Residues L7, F10 and F15 also delineate 100% signal attenuation in the presence of both the doxyl probes (Fig. 8A). Other residues, T4, T6, T9, G11, F13, N14 and S16 show similar degree of perturbation in both probes indicating that these residues of FP are likely to be located toward the center of the micelle with a more surface penetration. The C-terminal residues Q17, I18 L19 and residue G12 exhibit lower perturbation, compared to other residues, in both doxyl probes (Fig. 8A). Thus, these residues may reside closer to the water/head group surface of the micelle. In particular, PRE data suggest that the unstructured segment of the N-terminus and a significant region, except the bend residue G12 and C-terminal end residues, of the helical structure of FP are embedded into the DCP micelle. In case of IFP, PRE studies reveal a nearly 100% signal attenuation, caused by 16-DSA, for almost all the residues, except for A2 (Fig. 8B). 5-DSA has also produced ≥ 80% signal attenuation for these residues of IFP (Fig. 8B). These observations indicate that the most of the residues of IFP helical structure are positioned toward the micelle center. In the case of PTM peptide, an overwhelming number of residues including K3, Y4, E5, Y7, I8, W12, V14, G17 and F18 turns out to be more perturbed by 5-DSA in comparison to 16-DSA probe (Fig. 8C). Whereas, limited signal attenuation can be observed for residues G2, Q6 and W10. Therefore, the helix–loop–helix structure of PTM is largely occupying water/micelle interfacial region. Collectively, PRE studies of the three fusion peptides of SARS-CoV delineate that all three peptides, FP, IFP and PTM assume a parallel orientation in the DPC micelles, whereby, FP and IFP may have a deeper micelle penetration whereas PTM peptide is predominantly surface localized.
Fig. 8.
Bar diagrams showing the % attenuation of CαH/NH cross peaks upon additions of paramagnetic probes, 5-DSA and 16-DSA, for FP (panel A), IFP (panel B) and PTM (panel C). Signal attenuation cannot be obtained for few residues; these are indicated by a filled circle.
4. Discussion
Virus–host membrane fusion mechanism involves sequence of different steps including apposition of membranes, formation of a stalk resulting from hemifusion of the outer leaflets, the generation of a fusion pore through amalgamation of both bilayers at the stalk point, and lastly, enlargement of the fusion pore and mixing [1], [2], [3], [4]. Investigation of SARS-CoV fusion mechanism would contribute to the understating of virus pathology and designing drugs and vaccines. The S protein of SARS-CoV is known to be causing the fusion between the virion envelop and cellular membrane [41], [42], [43], [44]. As a prototype of type I fusion protein, the HR1 and HR2 regions of the S protein of SARS-CoV form trimers of coiled-coil six helix bundle structure [49], [50], [51], [52]. Analogous to the other class I fusion proteins, several trimers of the S protein are involved in membrane juxtaposition, destabilization and fusion pore formation. In addition, a large number of studies have pointed out that multiple regions in the fusion proteins, both in type I and type II are decisive in the membrane fusion process [6], [67], [68]. These segments of fusion proteins bind and destabilize lipid membrane toward the fusion of biological membranes. Three regions, FP, IFP and PTM, of the S protein of SARS-CoV have been identified as plausible membrane fusogenic element (Fig. 1). Synthetic peptides corresponding to FP, IFP and PTM delineate binding to model membranes or liposomes, lipid mixing, destabilization of lipid bilayers and characteristics of typical fusion peptides [53], [54], [55], [56], [57], [58], [59], [60]. However, conformational characteristics of these fusogenic peptides of SARS-CoV were investigated only by CD and FT-IR assessing global conformational preferences in membrane mimicking environments. The current study elucidates the first atomic resolution structures of FP, IFP and PTM peptide in solution containing DPC detergent. It may be noted that 3-D structures of a number of fusion peptides from type I and type II fusion proteins have been obtained in solutions of micelles either DPC or SDS, mimicking membrane environment [23], [26], [28], [29], [69], [70], [71], [72]. Zwitterionic DPC micelles perhaps act as a better mimic of mammalian cell membrane and support tertiary and quaternary packing in short peptides [73], [74]. Our results demonstrate that the three fusion active peptides of SARS-CoV fold into predominantly helical conformations also containing residues with extended or loop like conformations in DPC micelles. In particular, the 3-D structure of FP has been defined by a V-shaped kinked helical structure encompassing residues P5–I18. The N-terminal segment of FP consisting of residues W2–K3–T4 is found to be more flexible. The kinked structure of FP appears to be rather compact and sustained by potential inter-sidechain packing. The surface of the FP structure contains a narrow polar region and extended hydrophobic sections. PRE studies, using spin labeled doxyl lipids, indicate that the residues, e.g. L7, Y9, F10, and F15 at the hydrophobic patches along with structurally flexible residues W2-K3 of the N-terminal of FP are inserted into DPC micelles whereas C-terminal residues are plausibly located at the micelle surface. Previously, intrinsic tryptophan fluorescence studies of FP have also demonstrated insertion of residue W2 in model membranes with restricted solvent accessibility [53], [58]. Notably, the fusion peptide of influenza virus has been found to adopt similar V-shaped conformations in micelles and in lipid bilayers [25], [75].
Most of the residues of IFP, except for the two N-terminal residues, assume helical structure without any bend or kink. The IFP helix is highly hydrophobic, compared to FP and PTM helices, and most of the residues of IFP appear to be mostly inserted into the non-polar region of the DPC micelles. Previous biophysical studies of IFP in model membranes demonstrated incorporation of IFP into the non-polar core and disruption of the acyl chain packing of the lipid bilayers [55]. Taken together, the long hydrophobic patch delineated in the helical structure of IFP may be responsible for such higher portioning of IFP into lipidic environment and perturbation of membrane structures. The aromatic rich PTM peptide of SARS-CoV adopts a helix–loop–helix structure in DPC micelles. The C-terminal helix of PTM is non-polar and contains aromatic packing interactions between residues W12 and W15, whereas the short N-terminal helix is characterized by polar and ionic residues. PRE studies indicate that the PTM helix is more surface localized in comparison to FP and IFP. It is noteworthy that a straight helical structure of a 19-residue long PTM peptide of HIV fusion protein has been determined in DPC micelle solution [76]. The aromatic rich region close to the transmembrane domain is a conserved feature among type I fusion proteins and potentially involved in membrane fusion [56]. Virus–host membrane fusion mechanism is a complex process whereby multiple regions of the fusion protein might be involved in membrane interactions [5], [20], [22]. Some of these regions of the fusion protein can cause membrane fusion and other segments may be required for the membrane anchoring function.
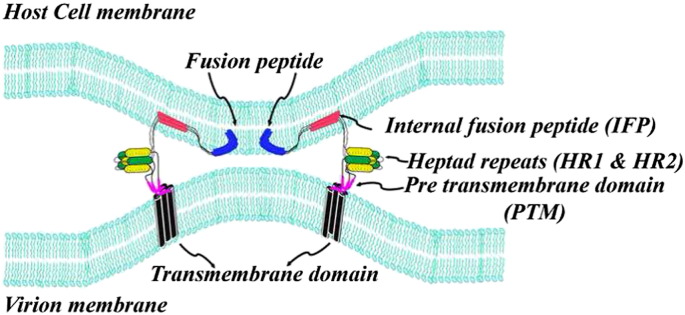
Our results can be used to further refine the current model of membrane fusion of SARS-CoV (Fig. 9 ). Whereby, helical structures of FP, IFP and PTM, along with the TM anchor, insert into the membrane and may align with the six-helix bundle structure of HR1 and HR2 resulting in possible juxtapositioning of the host and viral membranes. The high hydrophobicity of the fusion active structure made of by these helices of the fusion protein would cause membrane mixing and eventually formation of the fusion pore. In addition, the emergence of highly infectious coronavirus termed MERS has prompted discovery of anti-SARS drugs. The fusion peptides of SARS-CoV have been found to be active against viral infection possibly due to their interactions with membranes [10], [60]. Thus, 3-D structures of fusion peptides and membrane fusion mechanism may be further utilized to develop drug against SARS.
Fig. 9.
A hypothetical model of membrane fusion between SARS-CoV and host cells showing involvement of the three putative fusion peptides, FP, IFP and PTM. Upon activation by receptor binding of the S1 domain, the FP (blue color) and IFP (red color) of the S2 domain of the fusion protein insert into host cells causing folding back of the HR1 (green color) and HR2 (yellow color) into six helix bundle. The aromatic residue rich PTM domain (magenta color) and the TM domain residing on the viral membrane provide a contiguous track of hydrophobic helical structures with HR domains and fusion peptides. Such an arrangement may facilitate the mixing of lipid molecules between the two membranes leading to the fusion of the viral and host cells.
Acknowledgment
This work has been supported by the research grants, RG49/10 and RG11/12, from the Ministry of Education (MOE), Singapore. The coordinates and associated NMR parameters are deposited to BMRB data bank with accession numbers: 2rum (FP), 2ruo (IFP) and 2run (PTM).
References
- 1.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 3.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epand R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 6.Peisajovich S.G., Shai Y. Viral fusion proteins: multiple regions contribute to membrane fusion. Biochim. Biophys. Acta. 2003;1614:122–129. doi: 10.1016/s0005-2736(03)00170-6. [DOI] [PubMed] [Google Scholar]
- 7.Tamm L.K. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- 8.Apellaniz B., Huarte N., Largo E., Nieva J.L. The three lives of viral fusion peptides. Chem. Phys. Lipids. 2014;181:40–55. doi: 10.1016/j.chemphyslip.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal R., Durell S., Viard M. HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badani H., Garry R.F., Wimley W.C. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim. Biophys. Acta. 2014;1838:2180–2197. doi: 10.1016/j.bbamem.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews T., Salgo M., Greenberg M., Chung J., DeMasi R., Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 12.Thomas C.J., Casquilho-Gray H.E., York J., DeCamp D.L., Dai D., Petrilli E.B., Boger D.L., Slayden R.A., Amberg S.M., Sprang S.R., Nunberg J.H. A specific interaction of small molecule entry inhibitors with the envelope glycoprotein complex of the Junin hemorrhagic fever arenavirus. J. Biol. Chem. 2011;286:6192–6200. doi: 10.1074/jbc.M110.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper D.A., Lange J.M. Peptide inhibitors of virus-cell fusion: enfuvirtide as a case study in clinical discovery and development. Lancet Infect. Dis. 2004;4:426–436. doi: 10.1016/S1473-3099(04)01058-8. [DOI] [PubMed] [Google Scholar]
- 14.Hrobowski Y.M., Garry R.F., Michael S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissenhorn W., Hinz A., Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- 18.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien J.P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., Ward A.B., Wilson I.A. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peisajovich S.G., Epand R.F., Pritsker M., Shai Y., Epand R.M. The polar region consecutive to the HIV fusion peptide participates in membrane fusion. Biochemistry. 2000;39:1826–1833. doi: 10.1021/bi991887i. [DOI] [PubMed] [Google Scholar]
- 21.Klug Y.A., Ashkenazi A., Viard M., Porat Z., Blumenthal R., Shai Y. Early and late HIV-1 membrane fusion events are impaired by sphinganine lipidated peptides that target the fusion site. Biochem. J. 2014;461:213–222. doi: 10.1042/BJ20140189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh J.K., Peisajovich S.G., Shai Y. Sendai virus internal fusion peptide: structural and functional characterization and a plausible mode of viral entry inhibition. Biochemistry. 2000;39:11581–11592. doi: 10.1021/bi0005963. [DOI] [PubMed] [Google Scholar]
- 23.Han X., Tamm L.K. A host–guest system to study structure–function relationships of membrane fusion peptides. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinhauer D.A., Wharton S.A., Skehel J.J., Wiley D.C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J. Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 26.Lorieau J.L., Louis J.M., Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sackett K., Nethercott M.J., Zheng Z., Weliky D.P. Solid-state NMR spectroscopy of the HIV gp41 membrane fusion protein supports intermolecular antiparallel beta sheet fusion peptide structure in the final six-helix bundle state. J. Mol. Biol. 2014;426:1077–1094. doi: 10.1016/j.jmb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory S.M., Harada E., Liang B., Delos S.E., White J.M., Tamm L.K. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory S.M., Larsson P., Nelson E.A., Kasson P.M., White J.M., Tamm L.K. Ebolavirus entry requires a compact hydrophobic fist at the tip of the fusion loop. J. Virol. 2014;88:6636–6649. doi: 10.1128/JVI.00396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes K.V. Structural biology. Adaptation of SARS coronavirus to humans. Science. 2005;309:1822–1823. doi: 10.1126/science.1118817. [DOI] [PubMed] [Google Scholar]
- 32.Peiris J.S., Yuen K.Y., Osterhaus A.D., Stohr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 33.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 34.Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D., AlHajri M.M., Koopmans M.P. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coleman C.M., Frieman M.B. Coronaviruses: important emerging human pathogens. J. Virol. 2014;88:5209–5212. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L., Liu Q., Wang L., Feng Y., Wang Y., Zheng B.J., Yuen K.Y., Jiang S., Dimitrov D.S. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adedeji A.O., Sarafianos S.G. Antiviral drugs specific for coronaviruses in preclinical development. Curr. Opin. Virol. 2014;8C:45–53. doi: 10.1016/j.coviro.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar V., Jung Y.S., Liang P.H. Anti-SARS coronavirus agents: a patent review (2008–present) Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- 41.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(Pt 7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- 43.Taguchi F. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 1995;69:7260–7263. doi: 10.1128/jvi.69.11.7260-7263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taguchi F., Shimazaki Y.K. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 2000;81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- 45.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosch B.J., Martina B.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J., De Groot R., Osterhaus A.D., Rottier P.J. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng Y., Liu J., Zheng Q., Yong W., Lu M. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure. 2006;14:889–899. doi: 10.1016/j.str.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hakansson-McReynolds S., Jiang S., Rong L., Caffrey M. Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state. J. Biol. Chem. 2006;281:11965–11971. doi: 10.1074/jbc.M601174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sainz B., Jr., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol. 2005;79:7195–7206. doi: 10.1128/JVI.79.11.7195-7206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guillen J., Perez-Berna A.J., Moreno M.R., Villalain J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 2005;79:1743–1752. doi: 10.1128/JVI.79.3.1743-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guillen J., Perez-Berna A.J., Moreno M.R., Villalain J. A second SARS-CoV S2 glycoprotein internal membrane-active peptide. Biophysical characterization and membrane interaction. Biochemistry. 2008;47:8214–8224. doi: 10.1021/bi800814q. [DOI] [PubMed] [Google Scholar]
- 56.Sainz B., Jr., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry. 2005;44:947–958. doi: 10.1021/bi048515g. [DOI] [PubMed] [Google Scholar]
- 57.Guillen J., Moreno M.R., Perez-Berna A.J., Bernabeu A., Villalain J. Interaction of a peptide from the pre-transmembrane domain of the severe acute respiratory syndrome coronavirus spike protein with phospholipid membranes. J. Phys. Chem. B. 2007;111:13714–13725. doi: 10.1021/jp073675y. [DOI] [PubMed] [Google Scholar]
- 58.Guillen J., de Almeida R.F., Prieto M., Villalain J. Structural and dynamic characterization of the interaction of the putative fusion peptide of the S2 SARS-CoV virus protein with lipid membranes. J. Phys. Chem. B. 2008;112:6997–7007. doi: 10.1021/jp7118229. [DOI] [PubMed] [Google Scholar]
- 59.Guillen J., Kinnunen P.K., Villalain J. Membrane insertion of the three main membranotropic sequences from SARS-CoV S2 glycoprotein. Biochim. Biophys. Acta. 2008;1778:2765–2774. doi: 10.1016/j.bbamem.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sainz B., Jr., Mossel E.C., Gallaher W.R., Wimley W.C., Peters C.J., Wilson R.B., Garry R.F. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 2006;120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berjanskii M.V., Neal S., Wishart D.S. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guntert P., Mumenthaler C., Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 64.Wuthrich K. John Wiley and Sons; 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 65.Damberg P., Jarvet J., Graslund A. Micellar systems as solvents in peptide and protein structure determination. Methods Enzymol. 2001;339:271–285. doi: 10.1016/s0076-6879(01)39318-7. [DOI] [PubMed] [Google Scholar]
- 66.Hilty C., Wider G., Fernandez C., Wuthrich K. Membrane protein–lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. Chembiochem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- 67.Samuel O., Shai Y. Participation of two fusion peptides in measles virus-induced membrane fusion: emerging similarity with other paramyxoviruses. Biochemistry. 2001;40:1340–1349. doi: 10.1021/bi001533n. [DOI] [PubMed] [Google Scholar]
- 68.Moreno M.R., Pascual R., Villalain J. Identification of membrane-active regions of the HIV-1 envelope glycoprotein gp41 using a 15-mer gp41-peptide scan. Biochim. Biophys. Acta. 2004;1661:97–105. doi: 10.1016/j.bbamem.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 69.Melo M.N., Sousa F.J., Carneiro F.A., Castanho M.A., Valente A.P., Almeida F.C., Da Poian A.T., Mohana-Borges R. Interaction of the Dengue virus fusion peptide with membranes assessed by NMR: the essential role of the envelope protein Trp101 for membrane fusion. J. Mol. Biol. 2009;392:736–746. doi: 10.1016/j.jmb.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang D.K., Cheng S.F., Deo Trivedi V., Yang S.H. The amino-terminal region of the fusion peptide of influenza virus hemagglutinin HA2 inserts into sodium dodecyl sulfate micelle with residues 16–18 at the aqueous boundary at acidic pH. Oligomerization and the conformational flexibility. J. Biol. Chem. 2000;275:19150–19158. doi: 10.1074/jbc.M907148199. [DOI] [PubMed] [Google Scholar]
- 71.Tan Y., Jiang L., Wang M., Yin F., Deng F., Liu M., Hu Z., Wang H. Mutagenesis and nuclear magnetic resonance analyses of the fusion peptide of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus F protein. J. Virol. 2008;82:8138–8148. doi: 10.1128/JVI.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohanram H., Nip A., Domadia P.N., Bhunia A., Bhattacharjya S. NMR structure, localization, and vesicle fusion of Chikungunya virus fusion peptide. Biochemistry. 2012;51:7863–7872. doi: 10.1021/bi300901f. [DOI] [PubMed] [Google Scholar]
- 73.Porcelli F., Buck-Koehntop B.A., Thennarasu S., Ramamoorthy A., Veglia G. Structures of the dimeric and monomeric variants of magainin antimicrobial peptides (MSI-78 and MSI-594) in micelles and bilayers, determined by NMR spectroscopy. Biochemistry. 2006;45:5793–5799. doi: 10.1021/bi0601813. [DOI] [PubMed] [Google Scholar]
- 74.Saravanan R., Bhattacharjya S. Oligomeric structure of a cathelicidin antimicrobial peptide in dodecylphosphocholine micelle determined by NMR spectroscopy. Biochim. Biophys. Acta. 2011;1808:369–381. doi: 10.1016/j.bbamem.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Vaccaro L., Cross K.J., Kleinjung J., Straus S.K., Thomas D.J., Wharton S.A., Skehel J.J., Fraternali F. Plasticity of influenza haemagglutinin fusion peptides and their interaction with lipid bilayers. Biophys. J. 2005;88:25–36. doi: 10.1529/biophysj.104.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schibli D.J., Montelaro R.C., Vogel H.J. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]