Abstract
This article reviews and highlights the current development of DNA-based bioanalytical microsystems for point-of-care diagnostics and on-site monitoring of food and water. Recent progresses in the miniaturization of various biological processing steps for the sample preparation, DNA amplification (polymerase chain reaction), and product detection are delineated in detail. Product detection approaches utilizing “portable” detection signals and electrochemistry-based methods are emphasized in this work. The strategies and challenges for the integration of individual processing module on the same chip are discussed.
Keywords: DNA, Bioanalytical microsystem, Lab-on-a-chip, Sample preparation, DNA amplification, Electrochemical detection
1. Introduction
In the 21st Century, integrated and automated bioanalytical systems are going to play leading roles in medical, food, agricultural, environmental, and biodefense testing. At present, blood glucose and pregnancy testing along with antibody-based infectious diseases and biological warfare agent detection have a major share in this multi-billion-dollar market. In the coming years, thanks to the success of various genome projects and the advancement of nucleic acid (NA)-based molecular techniques, nucleic acid testing (NAT) will bring revolutionary changes to this rapidly growing biochip sector. These NA-based micro/nanoanalyzers are expected to offer much higher sensitivity and specificity than non-NA-based technologies.
Since the early work of deoxyribonucleic acid (DNA) manipulations in microchips by Northrup et al. [1] and Woolley et al. [2], tremendous research activities have been carried out to miniaturize the conventional DNA analytical procedures in microchip platforms. These microdevices enjoy the miniaturization advantages of small size, low sample, reagent and power consumption, enhanced analytical performance (e.g. shorter assay time), and high level of integration. An ideal microanalyzer should feature sample in result out kind of automated operation, without any human intervention between individual assay steps. There are three essential components in a complete DNA assay protocol, which include sample preparation, target amplification, and product detection. The implementation of these functionalities on-chip has been individually optimized prior to their final integration. For instance, Northrup et al. developed DNA amplification chips [1], [3], whereas Woolley et al. developed capillary electrophoresis (CE) chip [2]. Afterwards, they combined the two modules to form a microfabricated DNA analysis device [4]. Wilding et al. was another example, they studied microchambers for DNA amplification [5], [6], [7] and microfilters for cell separation [8], [9] separately first. Again, these amplification and preparation functionalities were later integrated onto a single microchip [10].
In the past decade, many integrated DNA analyzers have been developed [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. Of utmost importance, some of these technologies have been successfully commercialized, bringing clinical and on-site NAT into a reality. Companies engaged in this business include Affymetrix (GeneChip® Instrument System [21]), Agilent Technologies (2100 Bioanalyzer [22]), Alderon Biosciences (AndCare 100/800/9600 Portable Electrochemical Instruments [23]), Caliper Life Sciences (LabChip 90 Electrophoresis System [24]), Cepheid (GeneXpert® System [25] and SmartCycler® System [26]), eBiochip Systems (Electrical Array Analyzer [27]), Gen-Probe (Direct Tube Sampling System, DTS™ [28]), Idaho Technology (Ruggedized Advanced Pathogen Identification Device, R.A.P.I.D.® [29] and RAZOR Instrument [30]), IQuum (Liat™ System [31]), Nanogen (NanoChip® Molecular Biology Workstation [32]), Nanosphere (Verigene™ Platform [33]), Roche Molecular Diagnostics (COBAS AMPLICOR™ Analyzer [34]), just to name a few. Most of these instruments have already been widely utilized in central, clinical, and research laboratories, but have not been applied to patient's bedside, doctor's office or battlefield settings yet. This is partly due to the large footprint of these systems, which is attributed to the inclusion of supporting equipment such as pump, thermal cycler, optical detection system, etc.
To date, a number of reviews on micro total analysis systems (μTAS) for NAT have been published [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45]. These general reviews put much emphasis on the state-of-the-art integrated devices, in particular, CE and microarray technologies, which are not targeted for point-of-care (POC) diagnostics and on-site testing. If future handheld NAT devices would like to gain the acceptance like that of the glucose meter, additional attention should be given to the simplicity, versatility, and multiplexing capability of the systems. In this review, we focus on technologies that are promising in realizing future portable DNA analyzers for POC testing of infectious diseases (e.g. human immunodeficiency and hepatitis C viruses), on-site food and water monitoring (e.g. severe acute respiratory syndrome (SARS)-associated coronavirus in avian stock and Escherichia coli O157:H7), as well as bioterrorism agents detection (e.g. Bacillus anthracis). The organization of the following contents is based upon the three basic DNA processing modules of sample preparation, target amplification, and product detection. All relevant on-chip techniques will be covered, except that the product analysis is limited only to electrical/electrochemical detection strategies, which are well suited for total system miniaturization. It should be noted that most of the on-chip sample preparation and DNA amplification techniques were developed before year 2001, while many elegant electrochemical detection schemes were developed after 2001. Finally, directions in the future developments of these DNA-based bioanalytical microsystems are discussed.
2. Sample preparation
The purpose of sample preparation in NAT is to obtain nucleic acids, which can be DNA and/or ribonucleic acid (RNA), of sufficient purity and integrity from raw samples for subsequent amplification and detection. In a conventional setting, it consists of quite a number of steps for cell isolation and lysis followed by NA extraction and purification [46]. Depending upon the sample type, NA concentration, and tolerance of the amplification system, different protocols are needed to prepare amplification-ready samples via the simplest and fastest route. A diagram showing these processing flows is given in Fig. 1 . The easiest situation is that the sample can be directly added to the amplification mixture, without any pretreatment step. This is only applicable to “clean” samples having negligible amount of substances that inhibit the enzymatic target sequence amplification. The cell isolation step is used to select or enrich certain cell types from a complex sample and at the same time to eliminate all other interfering substances. This is particularly important when rare cell detection is encountered. Take the analysis of whole blood as an example, it is a routine practice to isolate the white blood cell (WBC) from the red blood cell (RBC) as the latter one inhibits the amplification step. The cell lysis step, which is divided into chemical and physical means, is introduced to break down the cell membrane and free the NA. Chemical lysis makes use of detergent (sodium dodecylsulfate) or chaotropic agent (guanidinium hydrochloride) whereas physical lysis relies on mechanical, thermal, and electrical means. NA extraction and purification steps are required after the chemical lysis step as the reagents used are not compatible with the amplification system.
Fig. 1.
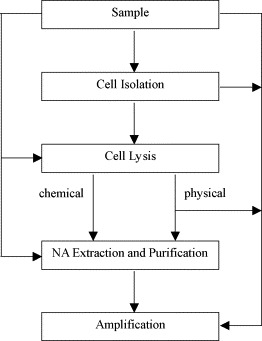
Diagrammatic representation of the processing flows in sample preparation.
The above-mentioned manipulations are very laborious and time-consuming, and the miniaturization of these functionalities on-chip has long been a challenging task [47], [48], [49]. For microchip-based cell isolation, differences in size, charge, and cell-surface properties of different cell types are the driving forces of separation. Wilding et al. demonstrated the use of silicon-based microfilters for the separation of micron-sized particles and cells. A few designs were investigated including arrays of microposts, tortuous channels [8], comb-shaped filters [9], and weir-type filters [10], [15]. A cartoon illustrating the separation of WBC from whole blood with a 3.5 μm-weir-type filter is shown in Fig. 2A. The size of WBC is too large to pass the microfilter, while RBC can easily penetrate through the gap. In addition, this isolation chamber was used as the amplification chamber, simplifying the complexity of the design of the integrated chip. It should be noted that for certain applications and cell types (e.g. WBC and E. coli), the isolated cells could be immediately introduced to the amplification mixture, in which they are thermally lysed [10], [12], [19]. Another well-studied microscale cell separation technique is based on dielectrophoresis (DEP), which deals with the electrical polarization effects applied to different cell types in an inhomogeneous alternating current (a.c.) field [50]. The migration directions of different cell types are determined by the dielectric properties of the cells including applied a.c. field, morphology, structural architecture, electrical double layer associated with surface charge, and many other factors. DEP has been employed for the on-chip separation of a wide variety of biological cells like bacteria [51], [52], cancer cells [53], [54], stem cells [55], and leukocyte subpopulations [56]. An example of DEP-based separation of E. coli from a cell mixture is shown in Fig. 2B. Effort has also been made to integrate DEP-based sample preparation unit with amplification unit [57]. Yet another cell isolation strategy is based on the use of paramagnetic particles [58], [59]. The idea is to coat the surface of the paramagnetic particle with antibody against specific cell surface antigen. The isolation of specific cell types can simply be achieved by mixing the antibody-coated magnetic beads with the sample, followed by a magnetic separation step and adequate washing. Liu et al. reported E. coli cell isolation from whole blood using the immunomagnetic bead approach in an integrated polycarbonate microdevice [20]. In this plastic chip, the cell isolation chamber is also used as the amplification chamber. Recently, Landers and co-workers reported a novel scheme for the separation of sperm and epithelial cells in a microfabricated device. This separation utilized the differential physical properties of the cells, resulting in settlement of the epithelial cells to the bottom of the inlet reservoir and subsequent adherence to the glass substrate. Thus, the sperms cells were separated from the epithelial cell-containing biological mixture at low flow rates.
Fig. 2.
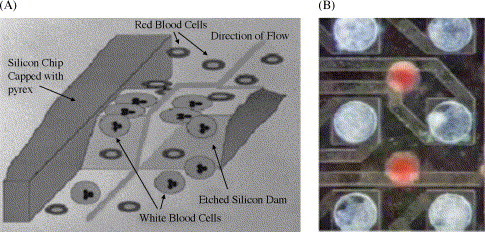
(A) Schematic of weir-type filter. A 3.5-μm gap between the top of the etched silicon dam and the Pyrex glass cover provides active filtration of cells based on size. Adapted from ref. [10]. (B) The separation result of E. coli from a mixture containing human blood cells by means of dielectrophoresis on a microfabricated bioelectronic chip. Adapted from ref. [51].
A number of schemes have been developed for cell disruption in conventional formats, which include chemical, mechanical, thermal, and electrical formats. Some of these schemes have been directly miniaturized for chip-based cell lysis. For example a minisonicator was built by Belgrader et al. to rapidly disrupt bacterial spores mechanically [60], [61]. Thermal lysis can be easily adapted to microfabricated amplification systems as the initial high-temperature step (∼95 °C) employed to denature the double-stranded (ds) DNA template is powerful enough to open up the cell membranes [10], [12], [19]. Electrical lysis of isolated cell on microelectrode was achieved by applying a series of high-voltage pulses [51]. Kellogg et al. developed chip-based chemical lysis procedure with sodium hydroxide, the lysate of which was neutralized prior to the amplification step [62]. Another common chemical lysis protocol with the use of chaotropic agent has also been implemented on microfabricated devices. This step must be followed by NA extraction and purification steps to eliminate the chaotropic agent, which will be discussed in the next paragraph. Besides miniaturizing the conventional formats, other novel schemes have been devised to improve the cell disruption process. One such example was based on a microfluidic filter with nanostructured barbs created using a modified deep reactive ion-etching process, permitting a highly effective yet reagentless mechanical lysis platform [63].
Many NA purification kits are now commercially available for isolating DNA from cell lysate. These kits typically utilize some form of silica gel, glass matrix, or membrane as the NA capture medium. For silica-based method, NA adsorbs to its surface at high concentration of chaotropic salt, while molecules such as protein and polysaccharide do not adsorb and are removed. The adsorbed NA is then eluted under low-salt condition and is ready for amplification. Attempts have been made to implement these strategies on-chip by confining cellulose [13], silica bead [64], silica bead/sol–gel hybrid [65] in microchannels. Micromachined pillar structures of silicon dioxide [66], [67] and photoactivated polycarbonate [68] surfaces have also been proved to be efficient capture media for NA extraction and purification.
3. Target amplification
The amount of NA obtained after the sample preparation step or directly from the raw sample is usually too minute for immediate identification and quantification. Typically, an enzymatic amplification step is carried out to replicate defined target sequence in vitro. The most common one is the polymerase chain reaction (PCR) [69], which was invented by Mullis et al. [70] in 1986. It consists of three basic steps. The first step is denaturation, in which a dsDNA molecule is heat separated to two single strands at a temperature of ∼94 °C. Then, the temperature is cooled to 55–65 °C, at which two oligonucleotide primers (∼20 nucleotides), which flank the target region, bind in a complementary fashion to the two single strands (annealing). After that, the temperature is increased to 72 °C for optimum extension of the primers (successive additions of deoxynucleotide triphosphates, dNTPs) with Thermus aquaticus (Taq) DNA polymerase. At the end of each thermal cycle, the amount of target sequence doubles. In a typical 30-cycle PCR, the target sequence can theoretically be copied for a billion times.
The PCR is basically the core of NAT, and was the first DNA assay component being conducted on-chip. In 1993, Northrup et al. developed a microfabricated silicon-based PCR reaction chamber [1]. Since then, this research area has attracted a great deal of attention, and has already blossomed into a mature stage [71], [72]. Early work on silicon–glass PCR microchips revealed the surface biocompatibility issue. Unlike the conventional polypropylene tubes, the PCR reagents (e.g. Taq polymerase, DNA template, primers, dNTPs, and metal ions) may adsorb to the microchamber surfaces, thereby greatly reducing the amplification efficiency. In view of this, various methodologies have been developed to passivate the surfaces. They can be classified into static and dynamic passivations. In static passivation, the chamber surface is coated with a PCR friendly substance prior to the introduction of the PCR master mix. Silanization has been proven to be an effective way to passivate glass substrates [73]. In cases with silicon–glass hybrid microchambers, a polymer coating is needed after the silanization step [6]. Another commonly used static passivation for silicon substrates is silicon dioxide (SiO2) coating [6], [7], which is compatible with clean-room processes and gives very consistent amplifications. Moreover, Teflon [74] and parylene-C [13] coatings were found to improve the performance of silicon dioxide- and polycarbonate-based microdevices, respectively. In dynamic passivation, the passivation agent is included in the PCR reaction mixture. Bovine serum albumin (BSA) [75], polyethylene glycol (PEG), and polyvinylpyrrolidone (PVP) [73], [76] exhibited significant surface passivation effect in native glass and silicon–glass microchips. It is important to note that these two passivation methods are not mutually exclusive. For instance, silanization–BSA [77], [78], SiO2–BSA [77], [79], SiO2–PEG/PVP [76] as well as BSA–BSA [14] have been demonstrated.
Thermal control is another essential parameter in achieving successful PCR. There are two basic formats of PCR microchip according to its heating algorithm: stationary and flow types. In the former format, similar to the conventional thermal cycler kind of operation, the PCR mixture is enclosed within a microchamber and is heat cycled to the denaturation, annealing, and extension temperatures [3], [5], [7], [11], [77], [79], [80], [81], [82], [83], [84], [85]. An example of such is elucidated in Fig. 3A; while in the latter format, the mixture is pumped through thermostated temperature zones on a microchip [78], [86], [87], [88], [89], [90], as illustrated in Fig. 3B. The main advantage of the flow format is its high speed as there is no need to heat up and cool down the microchip. Nonetheless, it suffers from lower flexibility in terms of cycle number and duration selection as compared to stationary format. In fact, the overall amplification speed can be tuned by the flowrate, but the relative duration of each step is fixed by the microchannel design. The thermal characteristics of the microchips are best evaluated by heating and cooling rates, power consumption, and temperature uniformity. Taking advantage of the small thermal mass of the microchip, high temperature ramp rates and small power consumption can easily be achieved by patterning thin-film heater onto the microchip [3], [77], [78], [82], [83], [84], [87], [90]. Special thermal isolation structures have been fabricated on silicon-based microchambers, resulting in fast thermal cycling (heating and cooling rates of 36–90 and 22–74 °C/s, respectively [82], [83], [84]) with small power requirement (∼1 W).
Fig. 3.
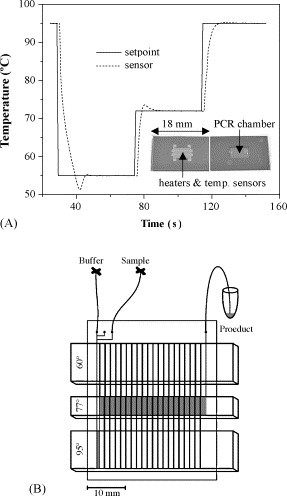
(A) Temperature profile of silicon–glass microchamber. Inset: photographs of the PCR microchip showing the integrated Pt heaters and temperature sensors (left) and the 8 μl reaction chamber (right). (B) Layout of a continuous-flow PCR chip. Adapted from ref. [86].
Material selection deserves great attention when designing one's own amplification unit. The majority of PCR microchips were formed by bonding (anodic [6], [74], [77], [78], [79], [84], thermal [80], [89], or glue [82]) two substrates (silicon, glass, or plastic) together, except those using a mineral oil layer to prevent reagent evaporation during PCR [83], [91]. Depending on the application or some other criteria such as detection method, different combinations of materials have been demonstrated. Among them, silicon–glass hybrid is by far the most common one [5], [74], [77], [78], [79], [82], [83], [84]. If the PCR microchamber/microchannel is etched in a silicon substrate, then the glass substrate acts as a cover plate, and vice versa. Thin-film heaters and temperature sensors are usually patterned on the silicon substrate to ensure good temperature uniformity as a result of its high thermal conductivity. Silicon–silicon microchip has also been developed with a dual-heater reaction chamber design [1]. For PCR–CE monolithic microchips, glass–glass assembly is the desired choice due to the high voltage used in the electrophoretic separation of DNA fragments [81], [92]. Considering the fabrication of disposable devices, polymer has the advantage of low cost compared to silicon and glass. Polycarbonate [13], [80] and polyimide [93] microreactors as well as polydimethylsiloxane–glass hybrid microchip [94] have been successfully utilized for the PCR amplification.
Some work has been done to improve throughput with microchamber array [79], [95], [96], while others focused on the assay sensitivity and demonstrated single copy amplification [75], [91]. Besides the PCR, other DNA amplification strategies [97], [98], including ligase chain reaction, strand displacement amplification, and rolling-circle amplification, can be carried out in microfluidic chip environment. If the target is RNA, then reverse transcription-PCR (RT-PCR) or nucleic acid sequence-based amplification (NASBA) can be used instead.
4. Product detection
Nowadays, methods to detect NA are many and varied [99]. DNA sensor that converts the biorecognition event into an electrical signal is a crucial component of any bioanalytical system for sequence-specific NAT. The sensing protocol basically involves the immobilization of an oligonucleotide onto a transducer surface, and upon the hybridization of complementary target sequence, the binding event is detected by optical, microgravimetric (mass-sensitive), or electrochemical methods. Among them, optical method, in particular, fluorescence-based technique is the most sensitive one and has received the greatest attention in the conventional setting. Nonetheless, the integration of the entire optical system (e.g., laser diode, photodiode, and filter) onto a monolithic chip requires sophisticated fabrication processes, and thereby very costly. In view of this, the electrochemistry-based approach is more suitable for POC and on-site testing with portable analyzers due to its inherent advantages of high speed, low cost, simple instrumentation, and ease of miniaturization. In recent years, a few comprehensive reviews on the electrochemical DNA sensors have been reported [100], [101], [102], [103], [104]. Herein, the focus is on microchip-based/compatible methodologies and the detection of real samples/PCR amplicons is highlighted.
Electrochemical transduction of the hybridization event can be classified into two categories: label-based and label-free approaches. The label-based approach can be further subdivided into intercalator/groove binder, non-intercalating marker, and nanoparticle. On the other hand, the label-free approach is based on the intrinsic electroactivity of the DNA purine bases or the change in interfacial properties (e.g. capacitance and electron transfer resistance) upon hybridization.
The first electrochemical DNA biosensor was reported by Millan and Mikkelsen [105] in 1993, which was based on hybridization indicators of tris(2,2′-bipyridyl)cobalt(III) (Co(bpy)3 3+) or tris(1,10-phenanthroline)cobalt(III) (Co(phen)3 3+). These groove binders or intercalators have higher affinities with dsDNA (probe–target hybrid) than with single-stranded (ss) one (unhybridized probe), leading to the accumulation of redox indicator at the hybrid-formed electrode surface and thus an increased current signal during voltammetric scan [106]. Electroactive intercalators and groove binders other than metal complexes have been extensively studied since Hashimoto et al. published their work on the voltammetric characterization of some commercially available indicators [107]. Hoechst 33258 [108], [109] and daunomycin [110], which behave in a similar fashion as Co(bpy)3 3+ and Co(phen)3 3+, have been successfully applied for real sample or PCR product detection using microfabricated electrodes (gold or screen-printed graphite). Methylene blue, on the other hand, was shown to interact with expose guanine base by Ozsoz and co-workers, thus there was a decrease in peak current upon hybrid formation [111], [112]. The intercalator/groove binder-based approach suffers from low sensitivity as a result of the low binding constant ratio of dsDNA to ssDNA (typical value between 1 and 2). A special redox threading intercalator (ferrocenylnaphthalene diimide), which dissociates very slowly from dsDNA (80 times slower than ssDNA) and exerts little stabilizing effect on the complex with ssDNA (it binds four times more strongly to dsDNA than to ssDNA), was synthesized by Takenaka et al. to lower the detection limit [113], [114].
One way to ensure that the hybridization indicator binds exclusively to either ssDNA or dsDNA is to label the probe or the target with a redox marker. Motorola's Clinical Micro Sensors Division developed a novel sandwich hybridization assay with a ferrocene-modified reporter probe [115], as illustrated in Fig. 4A. A gold electrode was coated with a self-assembled monolayer (SAM) containing a DNA capture probe. Unlabeled target was bound to the surface of the SAM through hybridization with the capture probe. The ferrocene-labeled reporter probe, which was complementary to the target in the region next to the capture probe-binding site, was then held in close proximity to the SAM. The core feature of this detection platform was the ability of the SAM to have electron transfer between the immobilized ferrocene and the gold, while insulating the electrode from the unbound reporter probes. This eSensor™, with its high selectivity, has been widely employed for real clinical samples [116]. Two other ferrocene-labeled capture probe-based formats were developed by Fan et al. [117] (Fig. 4B) and Immoos et al. [118] (Fig. 4C) for the reagentless hybridization detection. The former one was based on molecular beacon type of probe with the hybridization event induced a large conformational change in the stem–loop structure, and thus a decrease in the electron transfer efficiency. In the latter scheme, the capture sequence and the probe sequence (end modified with ferrocene) were linked together via a flexible poly(ethylene glycol) spacer, forming a tri-block macromolecule that was immobilized on a gold electrode surface. The hybridization of a target molecule brought the ferrocene closer to the electrode surface and resulted in an increase in current. An electroactive bead-based amplification strategy with ferrocene compound was reported by Wang et al. [119]. In their work, polystyrene microsphere was internally loaded with ferrocenecarboxaldehyde marker (5 × 1011 molecules/sphere) and was externally modified with a target sequence. Magnetic microparticle immobilized with a probe sequence was used for the selective capture of the target. Such high redox signal per binding event gave rise to a detection limit of 31,000 target molecules.
Fig. 4.
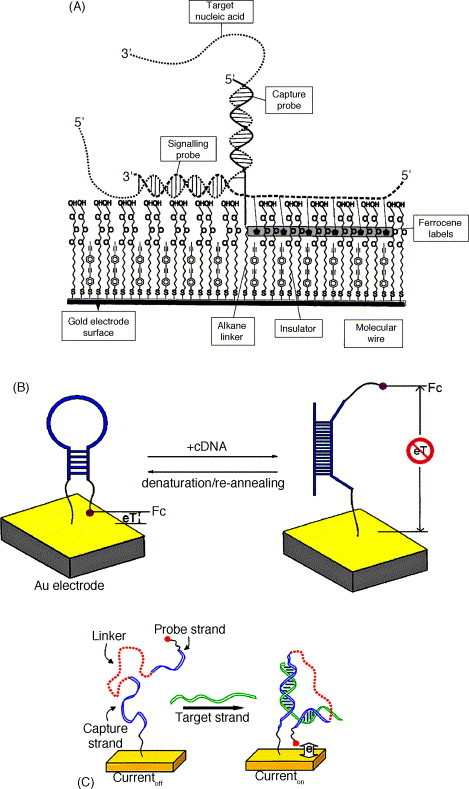
(A) Schematic diagram showing eSensor™'s sandwich hybridization assay with ferrocene-labeled reporter/signaling probe. Adapted from ref. [115]. (B). Pictorial representation of the working principle of the molecular beacon-type capture probe labeled with ferrocene group for the reagentless sequence-specific DNA detection. Adapted from ref. [117]. (C) Electrochemical detection of target DNA sequence using a ferrocene-labeled oligonucleotide-poly(ethylene glycol) triblock macromolecule. Adapted from ref. [118].
Another important class of hybridization indicators that has intrinsic signal amplification capability is redox enzyme. The majority of enzyme-amplified electrochemical DNA hybridization assays utilized horseradish peroxidase (HRP). There have been two main schemes for HRP-based signal transduction. Heller and co-workers pioneered the amperometric sandwich format [120], [121], [122]. The test involved the immobilization of a capture probe onto a redox polymer-coated carbon-based electrode. Then, cohybridization of target and HRP-label reporter probe occurred at the capture probe-modified surface. The redox polymer electrically wired HRP reaction center upon the sandwich hybridization and the whole structure became a catalyst for hydrogen peroxide (H2O2) electroreduction. Target sequence as few as 3000 copies was detected with this technique [122]. Willner and co-workers developed a novel method based on the enzyme-catalyzed precipitation of an insoluble product coupled with Faradaic impedance measurement [123], [124], [125]. In their approach, a capture probe-modified gold electrode was hybridized with a target-biotinylated reported probe, followed by the attachment of avidin–HRP. The enzyme catalyzed the oxidation of 4-chloro-1-naphthol by H2O2, forming an insoluble precipitate on the conductive support and blocking the interfacial electron transfer. Other redox enzymes such as alkaline phosphatase (ALP) [126], glucose oxidase [127], and bilirubin oxidase [128] have also been exploited. Moreover, a number of research groups have devoted themselves to the development of microelectrode array for multiplexed detection [129], [130], [131]. Additional amplification with multiple labels per binding event with carbon nanotube (CNT), 9600 ALP/CNT, was demonstrated by Wang et al. [132].
There has been continuous search for better hybridization indicators in terms of sensitivity and specificity. Since Mirkin and co-workers published their work on the nanoparticle probe-based scanometric DNA detection with extraordinary high selectivity in 2000 [133], there has been considerable interest in utilizing the gold nanoparticle as an electrochemical hybridization indicator. Authier et al. reported the sensitive quantification of an amplified 406 bp human cytomegalovirus DNA sequence with gold nanoparticle probe [134]. The assay relied on (1) immobilization of the amplified sequence (in denatured form) onto a polystyrene microwell; (2) hybridization of a colloidal gold-labeled probe to the immobilized target; (3) release of the gold metal atoms by oxidative metal dissolution (with acidic bromine–bromide solution); and (4) determination of the solubilized Au3+ ions by anodic stripping voltammetry at a screen-printed microband electrode. Later on, other similar schemes were reported [135], [136]. To further increase the sensitivity of the gold nanoparticle-based assay, a signal amplification step coined silver enhancement can be carried out [137], [138], [139], [140]. In this process, the gold nanoparticle catalyzes the reduction of silver ions to silver metal at the nanoparticle surface in the presence of hydroquinone, lowering the detection limit by ∼100 times. It should be noted that the chemical deposition process requires extremely precise time and temperature control. Aiming at a microchip-compatible strategy, Hsing and co-workers developed an electrodeposition means to substitute the chemical one. The protocol commenced with the immobilization of a capture probe onto an electroconductive polymer-coated indium tin oxide (ITO) electrode. Subsequently, in the presence of a biotinylated target, streptavidin–gold was bound to the electrode surface. The onset silver deposition potential for the gold nanoparticle bound sensor surface (with the target) is more positive than the surface without the gold nanoparticle (without the target). With proper selection of the deposition potential, silver deposition occurred preferentially at the target-bound surface.
Mirkin and co-workers presented a promising electrical hybridization detection with the silver–gold (chemical enhancement) approach. The binding event localized gold nanoparticles in an electrodes gap, and the silver enhancement on these nanoparticles bridged the gap, leading to readily measurable conductivity changes. In addition to gold nanoparticle, Wang et al. demonstrated the use of inorganic nanocrystal tracers (zinc sulfide, cadmium sulfide, and lead sulfide) for the simultaneous detection of multiple DNA targets [141].
Indicator-free approaches are emerging technologies in the electrochemical DNA biosensors for simple and fast transduction of the hybridization events. One approach is based on the electroactivity of DNA bases, in particular, the guanine base. This type of sensor requires the immobilization of an inosine-substituted (guanine-free) capture probe onto a solid support (electrode or magnetic bead). The inosine moiety can base-pair with cytosine, but its electroactivity is three orders of magnitude lower than that of guanine [142]. Therefore, the duplex formation can be monitored by the appearance of the guanine oxidation peak [143], [144], [145], [146]. Besides direct oxidation of the guanine base, electrocatalytic oxidation by a redox mediator, Ru(bpy)3 3+ (bpy = 2,2′-bipyridine), has been demonstrated [147], [148].
Another approach of the label-free electrochemical transduction is based on changes in interfacial properties. One such example was an electroactive polypyrrole functionalized with an oligonucleotide capture probe [149]. Specific hybridization of this grafted oligonucleotide with its target induced a significant change in the electrochemical response (voltammetric signal) of the polypyrrole and enabled a sensitive electrical reading of the recognition process. Different signal transduction methods like chronoamperometry [150], [151] and impedance [152] can also be used to probe the hybridization event. Another interfacial properties-related example was ion-channel sensor. In a pH 7 buffer, a gold electrode modified with SAM of peptide nucleic acid (PNA) and 8-amino-1-octanethiol was positively charged due to the protonated amino group. Thus, the electron transfer reaction between an electroactive marker [Ru(NH3)6]3+ and the electrode was hindered because of the electrostatic repulsion between them. Nonetheless, binding of a target sequence to the PNA probe provided an excess negative charge at the surface, thereby facilitating the access of [Ru(NH3)6]3+ to the electrode surface and its redox reaction. This was indicated by the increase in [Ru(NH3)6]3+ reduction current during square-wave voltammetric scan [153].
Recently, there has been a sudden increase in the number of publications on the direct electrochemical/electrical detection of the hybridization event on semiconductor [154], [155] or electrode [156] or field effect transistor (FET) [157], [158], [159] surfaces. These integrated circuit (IC)-compatible sensors, when combined with the state-of-the-art nanotechnology (e.g. silicon nanowire [160]), hold great promise for direct ultrasensitive electrical detection of specific DNA sequences.
5. Future directions and conclusions
After many years of tremendous research efforts in the area of lab-on-a-chip for DNA analysis, almost all individual analytical processes of sample preparation, target amplification, and target detection have reached a mature stage. Nonetheless, portable bioanalytical systems for POC and on-site testing are still in its infancy. To realize handheld DNA analyzers, all the functionalities have to be integrated and the entire operation be automated. One key element to address these issues is microfluidic control. It consists of sample/reagent introduction (macro- to micro-environment interfaces); liquid handling (e.g. mixing); sample/reagent transfer from one module to another one; and on-chip valving/pumping [11]. It should be noted that individual processing steps do not have to be spatially separated. The implementation of two or more functions in the same microvolume reduces the complexity of the design and fabrication as well as cost of the device. For instance, a microchamber in polycarbonate was used for target cell capture and preconcentration together with subsequent cell lysis and PCR (Fig. 5A) [20], whereas a silicon–glass microchamber was used for PCR and electrochemical detection (Fig. 5B) [161].
Fig. 5.
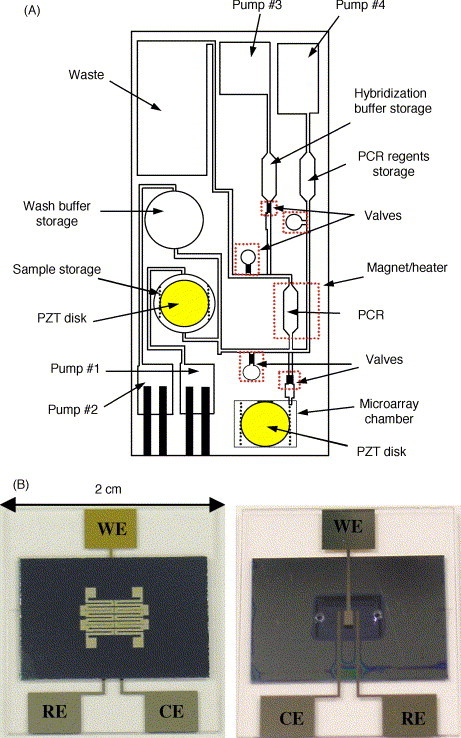
(A) Schematic of the polycarbonate fluidic chip developed by Motorola Labs. Adapted from ref. [20]. (B) Photos showing the top (left) and bottom (right) views of the integrated PCR–electrochemical chip. WE: gold working electrode; CE: Pt counter electrode; and RE: Pt pseudo reference electrode.
There has been keen debate on the need for the PCR if we can accurately detect very low copies of specific DNA sequences (thousands down to the single copy limit). Meanwhile, people argue that there is no need to push the detection limit of the sensor given that the PCR amplicon concentration usually reaches a plateau after 35 cycles irrespective of the initial target amount. It makes sense that it is redundant to perform the PCR if one can detect the sample directly. In reality, the sample contains trace amount of target sequence along with a plethora of unwanted NA even after the sample preparation step. These unwanted NA may interfere with the specific detection of the target sequence. In view of this, the PCR functions to enrich the target sequence so as to minimize the interference. Regarding the detection limit issue, the ability to detect lower DNA concentration at low cycle number allows accurate quantification of the target sequence, which is not possible with the PCR end-point detection (yes or no answer only).
Nanotechnology is going to play a leading role in the development of the disposable DNA analytical chips, especially the electrochemical/electrical detection module. An ideal sensor should be fast, simple, specific, sensitive, and multiplexed. Just like the test strip in glucose and pregnancy tests, the future DNA analytical systems most likely rely on disposable and inexpensive assay chips/cartridges, so as to minimize the potential cross-contamination. The successful development of a DNA analyzer in everyone's hand will have an unprecedented impact on our healthcare, and is anticipated to come true soon.
Acknowledgement
The authors thank the funding support from the Research Grants Council of the Hong Kong Special Administrative Region Government (Project No.: 601504).
References
- 1.M.A. Northrup, M.T. Ching, R.M. White, R.T. Watson, Transducers ’93, Seventh International Conference on Solid State Sensors and Actuators, 1993, p. 924.
- 2.Woolley A.T., Mathies R.A. Proc. Natl. Acad. Sci. USA. 1994;91:11348. doi: 10.1073/pnas.91.24.11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M.A. Northrup, C. Gonzalez, D. Hadley, R.F. Hills, P. Landre, S. Lehew, R. Saiki, J.J. Sninsky, R. Watson, R. Watson Jr., Transducers ’95, Eighth International Conference on Solid-State Sensors and Actuators, 1995, p. 764.
- 4.Woolley A.T., Hadley D., Landre P., deMello A.J., Mathies R.A., Northrup M.A. Anal. Chem. 1996;68:4081. doi: 10.1021/ac960718q. [DOI] [PubMed] [Google Scholar]
- 5.Wilding P., Shoffner M.A., Kricka L.J. Clin. Chem. 1994;40:1815. [PubMed] [Google Scholar]
- 6.Shoffner M.A., Cheng J., Hvichia G.E., Kricka L.J., Wilding P. Nucleic Acids Res. 1996;24:375. doi: 10.1093/nar/24.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J., Shoffner M.A., Hvichia G.E., Kricka L.J., Wilding P. Nucleic Acids Res. 1996;24:380. doi: 10.1093/nar/24.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilding P., Pfahler J., Bau H.H., Zemel J.N., Kricka L.J. Clin. Chem. 1994;40:43. [PubMed] [Google Scholar]
- 9.Cheng J., Fortina P., Surrey S., Kricka L.J., Wilding P. Mol. Diagn. 1996;1:183. doi: 10.1016/s1084-8592(96)70004-8. [DOI] [PubMed] [Google Scholar]
- 10.Wilding P., Kricka L.J., Cheng J., Hvichia G., Shoffner M.A., Fortina P. Anal. Biochem. 1998;257:95. doi: 10.1006/abio.1997.2530. [DOI] [PubMed] [Google Scholar]
- 11.Burns M.A., Johnson B.N., Brahmasandra S.N., Handique K., Webster J.R., Krishnan M., Sammarco T.S., Man P.M., Jones D., Heldsinger D., Mastrangelo C.H., Burke D.T. Science. 1998;282:484. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N., Tan H., Yeung E.S. Anal. Chem. 1999;71:1138. doi: 10.1021/ac981139j. [DOI] [PubMed] [Google Scholar]
- 13.Anderson R.C., Su X., Bogdan G.J., Fenton J. Nucleic Acids Res. 2000;28:e60. doi: 10.1093/nar/28.12.e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khandurina J., McKnight T.E., Jacobson S.C., Waters L.C., Foote R.S., Ramsey J.M. Anal. Chem. 2000;72:2995. doi: 10.1021/ac991471a. [DOI] [PubMed] [Google Scholar]
- 15.Yuen P.K., Kricka L.J., Fortina P., Panaro N.J., Sakazume T., Wilding P. Genome Res. 2001;11:405. doi: 10.1101/gr.155301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belgrader P., Young S., Yuan B., Primeau M., Lee C.A., Pourahmadi F., Northrup M.A. Anal. Chem. 2001;73:286. doi: 10.1021/ac000905v. [DOI] [PubMed] [Google Scholar]
- 17.Higgins J.A., Nasarabadi S., Karns J.S., Shelton D.R., Cooper M., Gbakima A., Koopman R.P. Biosens. Bioelectron. 2003;18:1115. doi: 10.1016/s0956-5663(02)00252-x. [DOI] [PubMed] [Google Scholar]
- 18.Ferrance J.P., Wu Q., Giordano B., Hernandez C., Kwok Y., Snow K., Thibodeau S., Landers J.P. Anal. Chim. Acta. 2003;500:233. [Google Scholar]
- 19.Lagally E.T., Scherer J.R., Blazej R.G., Toriello N.M., Diep B.A., Ramchandani M., Sensabaugh G.F., Riley L.W., Mathies R.A. Anal. Chem. 2004;76:3162. doi: 10.1021/ac035310p. [DOI] [PubMed] [Google Scholar]
- 20.Liu R.H., Yang J., Lenigk R., Bonanno J., Grodzinski P. Anal. Chem. 2004;76:1824. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 21.http://www.affymetrix.com/products/instruments/index.affx.
- 22.http://www.chem.agilent.com/Scripts/PDS.asp?lPage=51.
- 23.http://www.alderonbiosciences.com/prodcat1.html.
- 24.http://www.caliperls.com/products/labchip90.shtml.
- 25.http://www.cepheid.com/Sites/cepheid/content.cfm?id=158.
- 26.http://www.cepheid.com/Sites/cepheid/content.cfm?id=159.
- 27.http://www.ebiochip.com/pdf/ArrayAnalyzer.pdf.
- 28.http://www.gen-probe.com/prod_serv/inst_dts.asp#dts400.
- 29.http://www.idahotech.com/rapid/index.html.
- 30.http://www.idahotech.com/razor/index.html.
- 31.http://www.iquum.com/products/productsdescr.shtml.
- 32.http://www.nanogen.com/products/workstation.htm.
- 33.http://www.nanosphere-inc.com/2_tech/index.html.
- 34.http://www.roche-diagnostics.com/ba_rmd/rmd_products_automated_pcr_systems23.html.
- 35.Mastrangelo C.H., Burns M.A., Burke D.T. Proc. IEEE. 1998;86:1769. [Google Scholar]
- 36.Sanders G.H.W., Manz A. Trends Anal. Chem. 2000;19:364. [Google Scholar]
- 37.Khandurina J., Guttman A. J. Chromatogr. A. 2002;943:159. doi: 10.1016/s0021-9673(01)01451-0. [DOI] [PubMed] [Google Scholar]
- 38.Selvaganapathy P.R., Carlen E.T., Mastrangelo C.H. Proc. IEEE. 2003;91:954. [Google Scholar]
- 39.Ivnitski D., O’Neil D.J., Gattuso A., Schlicht R., Calidonna M., Fisher R. BioTechniques. 2003;35:862. doi: 10.2144/03354ss03. [DOI] [PubMed] [Google Scholar]
- 40.Ferrari M., Stenirri S., Bonini P., Cremones L. Clin. Chem. Lab. Med. 2003;41:462. doi: 10.1515/CCLM.2003.069. [DOI] [PubMed] [Google Scholar]
- 41.Lee S.J., Lee S.Y. Appl. Microbiol. Biotechnol. 2004;64:289. doi: 10.1007/s00253-003-1515-0. [DOI] [PubMed] [Google Scholar]
- 42.Erickson D., Li D. Anal. Chim. Acta. 2004;507:11. [Google Scholar]
- 43.Auroux P.A., Koc Y., deMello A., Manz A., Day P.J.R. Lab Chip. 2004;4:534. doi: 10.1039/b408850f. [DOI] [PubMed] [Google Scholar]
- 44.Obeid P.J., Christopoulos T.K. Crit. Rev. Clin. Lab. Sci. 2004;41:429. doi: 10.1080/10408360490497492. [DOI] [PubMed] [Google Scholar]
- 45.Campas M., Katakis I. Trends Anal. Chem. 2004;23:49. [Google Scholar]
- 46.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Siedman J.G., Smith J.A., Struhl K. John Wiley & Sons; New York: 1992. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. [Google Scholar]
- 47.Cheng J., Kricka L.J., Sheldon E.L., Wilding P. Top. Curr. Chem. 1998;194:215. [Google Scholar]
- 48.Huang Y., Mather E.L., Bell J.L., Madou M. Anal. Bioanal. Chem. 2002;372:49. doi: 10.1007/s00216-001-1191-9. [DOI] [PubMed] [Google Scholar]
- 49.Lichtenberg J., de Rooij N.F., Verpoorte E. Talanta. 2002;56:233. doi: 10.1016/s0039-9140(01)00593-8. [DOI] [PubMed] [Google Scholar]
- 50.Wang X.B., Cheng J. Harwood Academic Publisher; 2001. Biochip Technology. (Chapter 7) [Google Scholar]
- 51.Cheng J., Sheldon E.L., Wu L., Uribe A., Gerrue L.O., Carrino J., Heller M.J., O’Connell J.P. Nature Biotechnol. 1998;16:541. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y., Ewalt K.L., Tirado M., Haigis R., Forster A., Ackley D., Heller M.J., O’Connell J.P., Krihak M. Anal. Chem. 2001;73:1549. doi: 10.1021/ac001109s. [DOI] [PubMed] [Google Scholar]
- 53.Becker F.F., Wang X.B., Huang Y., Pethig R., Vykoukal J., Gascoyne P.R.C. Proc. Natl. Acad. Sci. USA. 1995;92:860. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng J., Sheldon E.L., Wu L., Heller M.J., O’Connell J.A.P. Anal. Chem. 1998;70:2321. doi: 10.1021/ac971274g. [DOI] [PubMed] [Google Scholar]
- 55.Stephens M., Talary M.S., Pethig R., Burnett A.K., Mills K.I. Bone Marrow Transpl. 1996;18:777. [PubMed] [Google Scholar]
- 56.Yang J., Huang Y., Wang X.B., Becker F.F., Gascoyne P.R.C. Biophys. J. 2000;78:2680. doi: 10.1016/S0006-3495(00)76812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. El-Ali, I.R. Perch-Nielsen, C.R. Poulsen, M. Jensen, P. Telleman, A. Wolff, Transducers ’03, The 12th International Conference on Solid State Sensors, Actuators and Microsystems, 2003, p. 214.
- 58.http://www.dynalbiotech.com/.
- 59.Gu H., Ho P.L., Tsang K.W.T., Wang L., Xu B. J. Am. Chem. Soc. 2003;125:15702. doi: 10.1021/ja0359310. [DOI] [PubMed] [Google Scholar]
- 60.Belgrader P., Hansford D., Kovacs G.T.A., Venkateswaran K., Mariella R., Jr., Milanovich F., Nasarabadi S., Okuzumim M., Pourahmadi F., Northrup M.A. Anal. Chem. 1999;71:4232. doi: 10.1021/ac990347o. [DOI] [PubMed] [Google Scholar]
- 61.Belgrader P., Okuzumim M., Pourahmadi F., Borkholder D.A., Northrup M.A. Biosens. Bioelectron. 2000;14:849. doi: 10.1016/s0956-5663(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 62.Kellogg G.J., Arnold T.E., Carvalho B.L., Duffy D.C., Sheppard N.F., Jr. Micro Total Anal. Syst. 2000:239. [Google Scholar]
- 63.Di Carlo D., Jeong K.H., Lee L.P. Lab Chip. 2003;3:287. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- 64.Chen X., Shen K., Liu P., Guo M., Cheng J., Zhou Y. Tsinghua Sci. Technol. 2004;9:379. [Google Scholar]
- 65.Breadmore M.C., Wolfe K.A., Arcibal I.G., Leung W.K., Dichson D., Giordano B.C., Power M.E., Ferrance J.P., Feldman S., Norris P.M., Landers J.P. Anal. Chem. 2003;75:1880. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- 66.Christel L.A., Petersen K., McMillan W., Northrup M.A. J. Biomech. Eng. 1999;121:22. doi: 10.1115/1.2798037. [DOI] [PubMed] [Google Scholar]
- 67.Cady N.C., Stelick S., Batt C.A. Biosens. Bioelectron. 2003;19:59. doi: 10.1016/s0956-5663(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 68.Xu Y., Vaidya B., Patel A.B., Ford S.M., McCarley R.L., Soper S.A. Anal. Chem. 2003;75:2975. doi: 10.1021/ac030031n. [DOI] [PubMed] [Google Scholar]
- 69.Mullis K.B., Ferre F., Gibbs R.A. Birkhauser; Boston: 1994. The Polymerase Chain Reaction. [Google Scholar]
- 70.Mullis K.B., Faloona F., Scharf S.J., Saiki R.K., Horn G.T., Erlich H.A. Cold Spring Harbor. Symp. Quant. Biol. 1986;51:263. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 71.Kricka L.J., Wilding P. Anal. Bioanal. Chem. 2003;377:820. doi: 10.1007/s00216-003-2144-2. [DOI] [PubMed] [Google Scholar]
- 72.Lee J.Y., Kim J.J., Park T.H. Biotechnol. Bioprocess. Eng. 2003;8:213. [Google Scholar]
- 73.Giordano B.C., Copeland E.R., Landers J.P. Electrophoresis. 2001;22:334. doi: 10.1002/1522-2683(200101)22:2<334::AID-ELPS334>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 74.Krishnan M., Burke D.T., Burns M.A. Anal. Chem. 2004;76:6588. doi: 10.1021/ac0488356. [DOI] [PubMed] [Google Scholar]
- 75.Lagally E.T., Medintz I., Mathies R.A. Anal. Chem. 2001;73:565. doi: 10.1021/ac001026b. [DOI] [PubMed] [Google Scholar]
- 76.Lou X.J., Panaro N.J., Wilding P., Fortina P., Kricka L.J. BioTechniques. 2004;36:248. doi: 10.2144/04362ST01. [DOI] [PubMed] [Google Scholar]
- 77.Lee T.M.H., Hsing I.M., Lao A.I.K., Carles M.C. Anal. Chem. 2000;72:4242. doi: 10.1021/ac000384b. [DOI] [PubMed] [Google Scholar]
- 78.Schneegab I., Brautigam R., Kohler J.M. Lab Chip. 2001;1:42. doi: 10.1039/b103846j. [DOI] [PubMed] [Google Scholar]
- 79.Taylor T.B., Winn-Deen E.S., Picozza E., Woudenberg T.M., Albin M. Nucleic Acids Res. 1997;25:3164. doi: 10.1093/nar/25.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang J., Liu Y., Rauch C.B., Stevens R.L., Liu R.H., Lenigk R., Grodzinski P. Lab Chip. 2002;2:179. doi: 10.1039/b208405h. [DOI] [PubMed] [Google Scholar]
- 81.Lagally E.T., Simpson P.C., Mathies R.A. Sens. Actuators B. 2000;63:138. [Google Scholar]
- 82.Poser S., Schulz T., Dillner U., Baier V., Kohler J.M., Schimkat D., Mayer G., Siebert A. Sens. Actuators A. 1997;62:672. [Google Scholar]
- 83.Daniel J.H., Iqbal S., Millington R.B., Moore D.F., Lowe C.R., Leslie D.L., Lee M.A., Pearce M.J. Sens. Actuators A. 1998;71:81. [Google Scholar]
- 84.Yoon D.S., Lee Y.S., Lee Y., Cho H.J., Sung S.W., Oh K.W., Cha J., Lim G. J. Micromech. Microeng. 2002;12:813. [Google Scholar]
- 85.Nagai H., Murakami Y., Morita Y., Yokoyama K., Tamiya E. Anal. Chem. 2001;73:1043. doi: 10.1021/ac000648u. [DOI] [PubMed] [Google Scholar]
- 86.Kopp M.U., de Mello A.J., Manz A. Science. 1998;280:1046. doi: 10.1126/science.280.5366.1046. [DOI] [PubMed] [Google Scholar]
- 87.Sun K., Yamaguchi A., Ishida Y., Matsuo S., Misawa H. Sens. Actuators B. 2002;84:283. [Google Scholar]
- 88.Curcio M., Roeraade J. Anal. Chem. 2003;75:1. doi: 10.1021/ac0204146. [DOI] [PubMed] [Google Scholar]
- 89.Obeid P., Christopoulos T.K., Crabtree H.J., Backhouse C.J. Anal. Chem. 2003;75:288. doi: 10.1021/ac0260239. [DOI] [PubMed] [Google Scholar]
- 90.Fukuba T., Yamamoto T., Naganuma T., Fujii T. Chem. Eng. J. 2004;101:151. [Google Scholar]
- 91.Matsubara Y., Kerman K., Kobayashi M., Yamamura S., Morita Y., Tamiya E. Biosens. Bioelectron. 2005;20:1482. doi: 10.1016/j.bios.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 92.Waters L.C., Jacobson S.C., Kroutchinina N., Khandurina J., Foote R.S., Ramsey J.M. Anal. Chem. 1998;70:158. doi: 10.1021/ac970642d. [DOI] [PubMed] [Google Scholar]
- 93.Giordano B.C., Ferrance J., Swedberg S., Huhmer A.F.R., Landers J.P. Anal. Biochem. 2001;291:124. doi: 10.1006/abio.2000.4974. [DOI] [PubMed] [Google Scholar]
- 94.Hong J.W., Fujii T., Seki M., Yamamoto T., Endo I. Electrophoresis. 2001;22:328. doi: 10.1002/1522-2683(200101)22:2<328::AID-ELPS328>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 95.Trau D., Lee T.M.H., Lao A.I.K., Lenigk R., Hsing I.M., Ip N.Y., Carles M.C., Sucher N.J. Anal. Chem. 2002;74:3168. doi: 10.1021/ac020053u. [DOI] [PubMed] [Google Scholar]
- 96.Nagai H., Murakami Y., Yokoyama K., Tamiya E. Biosens. Bioelectron. 2001;16:1015. doi: 10.1016/s0956-5663(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 97.Coleman W.B., Tsongalis G.J. Humana Press; Totowa: 1997. Molecular Diagnostics for the Clinical Laboratorian. [Google Scholar]
- 98.Demidov V.V., Broude N.E. Horizon Bioscience; Wymondham: 2004. DNA Amplification: Current Technologies and Applications. [Google Scholar]
- 99.Kricka L.J. Ann. Clin. Biochem. 2002;39:114. doi: 10.1258/0004563021901865. [DOI] [PubMed] [Google Scholar]
- 100.Drummond T.G., Hill M.G., Barton J.K. Nature Biotechnol. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 101.Kerman K., Kobayashi M., Tamiya E. Meas. Sci. Technol. 2004;15:R1. [Google Scholar]
- 102.Gooding J.J. Electroanalysis. 2002;14:1149. [Google Scholar]
- 103.Wang J. Anal. Chim. Acta. 2002;469:63. [Google Scholar]
- 104.Wang J. Analyst. 2005;130:421. doi: 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]
- 105.Millan K.M., Mikkelsen S.R. Anal. Chem. 1993;65:2317. doi: 10.1021/ac00065a025. [DOI] [PubMed] [Google Scholar]
- 106.Wang J., Rivas G., Cai X., Dontha N., Shiraishi H., Luo D., Valera F.S. Anal. Chim. Acta. 1997;337:41. [Google Scholar]
- 107.Hashimoto K., Ito K., Ishimori Y. Anal. Chim. Acta. 1994;286:219. [Google Scholar]
- 108.Hashimoto K., Ito K., Ishimori Y. Anal. Chem. 1994;66:3830. doi: 10.1021/ac00093a045. [DOI] [PubMed] [Google Scholar]
- 109.Takahashi M., Okada J., Ito K., Hashimoto M., Hashimoto K., Yoshida Y., Furuichi Y., Ohta Y., Mishiro S., Gemma N. Analyst. 2005;130:687. doi: 10.1039/b414030n. [DOI] [PubMed] [Google Scholar]
- 110.Marrazza G., Chianella I., Mascini M. Biosens. Bioelectron. 1999;14:43. doi: 10.1016/s0956-5663(98)00102-x. [DOI] [PubMed] [Google Scholar]
- 111.Kerman K., Ozkan D., Kara P., Meric B., Gooding J.J., Ozsoz M. Anal. Chim. Acta. 2002;462:39. [Google Scholar]
- 112.Meric B., Kerman K., Ozkan D., Kara P., Erensoy S., Akarca U.S., Mascini M., Ozsoz M. Talanta. 2002;56:837. doi: 10.1016/s0039-9140(01)00650-6. [DOI] [PubMed] [Google Scholar]
- 113.Takenaka S., Yamashita K., Takagi M., Uto Y., Kondo H. Anal. Chem. 2000;72:1334. doi: 10.1021/ac991031j. [DOI] [PubMed] [Google Scholar]
- 114.Yamashita K., Takagi A., Takagi M., Kondo H., Ikeda Y., Takenaka S. Bioconjugate Chem. 2002;13:1193. doi: 10.1021/bc025519u. [DOI] [PubMed] [Google Scholar]
- 115.Umek R.M., Lin S.W., Vielmetter J., Terbrueggen R.H., Irvine B., Yu C.J., Kayyem J.F., Yowanto H., Blackburn G.F., Farkas D.H., Chen Y.P. J. Mol. Diag. 2001;3:74. doi: 10.1016/S1525-1578(10)60655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vernon S.D., Farkas D.H., Unger E.R., Chan V., Miller D.L., Chen Y.P., Blackburn G.F., Reeves W.C. BMC Infect. Dis. 2003;3:12. doi: 10.1186/1471-2334-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan C., Plaxco K.W., Heeger A.J. Proc. Natl. Acad. Sci. USA. 2003;100:9134. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Immoos C.E., Lee S.J., Grinstaff M.W. J. Am. Chem. Soc. 2004;126:10814. doi: 10.1021/ja046634d. [DOI] [PubMed] [Google Scholar]
- 119.Wang J., Polsky R., Merkoci A., Turner K.L. Langmuir. 2003;19:989. [Google Scholar]
- 120.Campbell C.N., Gal D., Cristler N., Branditrat C., Heller A. Anal. Chem. 2002;74:158. doi: 10.1021/ac015602v. [DOI] [PubMed] [Google Scholar]
- 121.Dequaire M., Heller A. Anal. Chem. 2002;74:4370. doi: 10.1021/ac025541g. [DOI] [PubMed] [Google Scholar]
- 122.Zhang Y., Kim H.H., Heller A. Anal. Chem. 2003;75:3267. doi: 10.1021/ac034445s. [DOI] [PubMed] [Google Scholar]
- 123.Patolsky F., Katz E., Bardea A., Willner I. Langmuir. 1999;15:3703. [Google Scholar]
- 124.Alfonta L., Singh A.K., Willner I. Anal. Chem. 2001;73:91. doi: 10.1021/ac000819v. [DOI] [PubMed] [Google Scholar]
- 125.Patolsky F., Lichtenstein A., Willner I. Chem. Eur. J. 2003;9:1137. doi: 10.1002/chem.200390131. [DOI] [PubMed] [Google Scholar]
- 126.Aguilar Z., Fritsch I. Anal. Chem. 2003;75:3890. doi: 10.1021/ac026211z. [DOI] [PubMed] [Google Scholar]
- 127.Xie H., Yu Y.H., Mao P.L., Gao Z. Nucleic Acids Res. 2004;32:e15. doi: 10.1093/nar/gnh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y., Pothukuchy A., Shin W., Kim Y., Heller A. Anal. Chem. 2004;76:4093. doi: 10.1021/ac0495034. [DOI] [PubMed] [Google Scholar]
- 129.Gau J.J., Lan E.H., Dunn B., Ho C.M., Woo J.C.S. Biosens. Bioelectron. 2001;16:745. doi: 10.1016/s0956-5663(01)00216-0. [DOI] [PubMed] [Google Scholar]
- 130.Nebling E., Grunwald T., Albers J., Schafer P., Hintsche R. Anal. Chem. 2004;76:689. doi: 10.1021/ac0348773. [DOI] [PubMed] [Google Scholar]
- 131.Dill K., Montgomery D.D., Ghindilis A.L., Schwarzkopf K.R. J. Biochem. Biophys. Methods. 2004;59:181. doi: 10.1016/j.jbbm.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 132.Wang J., Liu G., Jan M.R. J. Am. Chem. Soc. 2004;126:3010. doi: 10.1021/ja031723w. [DOI] [PubMed] [Google Scholar]
- 133.Taton T.A., Mirkin C.A., Letsinger R.L. Science. 2000;289:1757. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 134.Authier L., Grossiord C., Brossier P. Anal. Chem. 2001;73:4450. doi: 10.1021/ac0103221. [DOI] [PubMed] [Google Scholar]
- 135.Wang J., Xu D., Kawde A.N., Polsky R. Anal. Chem. 2001;73:5576. doi: 10.1021/ac0107148. [DOI] [PubMed] [Google Scholar]
- 136.Ozsoz M., Erdem A., Kerman K., Ozkan D., Tugrul B., Topcuoglu N., Ekren H., Taylan M. Anal. Chem. 2003;75:2181. doi: 10.1021/ac026212r. [DOI] [PubMed] [Google Scholar]
- 137.Wang J., Polsky R., Xu D. Langmuir. 2001;17:5739. [Google Scholar]
- 138.Cai H., Wang Y., He P., Fang Y. Anal. Chim. Acta. 2002;469:165. [Google Scholar]
- 139.Lee T.M.H., Li L.L., Hsing I.M. Langmuir. 2003;19:4338. [Google Scholar]
- 140.Li L.L., Cai H., Lee T.M.H., Barford J., Hsing I.M. Electroanalysis. 2004;16:81. [Google Scholar]
- 141.Wang J., Liu G., Merkoci A. J. Am. Chem. Soc. 2003;125:3214. doi: 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- 142.Thorp H.H. Trends Biotechnol. 1998;16:117. doi: 10.1016/j.tibtech.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 143.Wang J., Rivas G., Fernandes J.R., Paz J.L.L., Jiang M., Waymire R. Anal. Chim. Acta. 1998;375:197. [Google Scholar]
- 144.Ozkan D., Erdem A., Kara P., Kerman K., Meric B., Hassmann J., Ozsoz M. Anal. Chem. 2002;74:5931. doi: 10.1021/ac0257905. [DOI] [PubMed] [Google Scholar]
- 145.Kerman K., Morita Y., Takamura Y., Tamiya E. Electrochem. Commun. 2003;5:887. [Google Scholar]
- 146.Lucarelli F., Marrazza G., Palchetti I., Cesaretti S., Mascini M. Anal. Chim. Acta. 2002;469:93. doi: 10.1016/j.aca.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 147.Napier M.E., Loomis C.R., Sistare M.F., Kim J., Eckhardt A.E., Thorp H.H. Bioconjugate Chem. 1997;8:906. doi: 10.1021/bc9701149. [DOI] [PubMed] [Google Scholar]
- 148.Koehne J., Chen H., Li J., Cassell A.M., Ye Q., Ng H.T., Han J., Meyyappan M. Nanotechnology. 2003;14:1239. doi: 10.1088/0957-4484/14/12/001. [DOI] [PubMed] [Google Scholar]
- 149.Korri-Youssoufi H., Garnier F., Srivastava P., Godillot P., Yassar A. J. Am. Chem. Soc. 1997;119:7388. [Google Scholar]
- 150.Komarova E., Aldissi M., Bogomolova A. Biosens. Bioelectron. 2005;21:182. doi: 10.1016/j.bios.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 151.Ramanaviciene A., Ramanavicius A. Anal. Bioanal. Chem. 2004;379:287. doi: 10.1007/s00216-004-2573-6. [DOI] [PubMed] [Google Scholar]
- 152.Lee T.Y., Shim Y.B. Anal. Chem. 2001;73:5629. doi: 10.1021/ac015572w. [DOI] [PubMed] [Google Scholar]
- 153.Aoki H., Umezawa Y. Electroanalysis. 2002;14:1405. [Google Scholar]
- 154.Macanovic A., Marquette C., Polychronakos C., Lawrence M.F. Nucleic Acids Res. 2004;32:e20. doi: 10.1093/nar/gnh003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cai W., Peck J.R., van der Weide D.W., Hamers R.J. Biosens. Bioelectron. 2004;19:1013. doi: 10.1016/j.bios.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 156.Guiducci C., Stagni C., Zuccheri G., Bogliolo A., Benini L., Samori B., Ricco B. Biosens. Bioelectron. 2004;19:781. doi: 10.1016/s0956-5663(03)00266-5. [DOI] [PubMed] [Google Scholar]
- 157.Kim D.S., Jeong Y.T., Park H.J., Shin J.K., Choi P., Lee J.H., Lim G. Biosens. Bioelectron. 2004;20:69. doi: 10.1016/j.bios.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 158.Uslu F., Ingebrandt S., Mayer D., Bocker-Meffert S., Odenthal M., Offenhausser A. Biosens. Bioelectron. 2004;19:1723. doi: 10.1016/j.bios.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 159.Fritz J., Cooper E.B., Gaudet S., Sorger P.K., Manalis S.R. PNAS. 2002;99:14142. doi: 10.1073/pnas.232276699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hahm J.I., Lieber C.M. Nano Lett. 2004;4:51. [Google Scholar]
- 161.Lee T.M.H., Carles C.M., Hsing I.M. Lab Chip. 2003;3:100. doi: 10.1039/b300799e. [DOI] [PubMed] [Google Scholar]


