Abstract
Many double-stranded RNA (dsRNA) viruses are capable of transcribing and capping RNA within a stable icosahedral viral capsid. The turret of turreted dsRNA viruses belonging to the family Reoviridae is formed by five copies of the turret protein, which contains domains with both 7-N-methyltransferase and 2′-O-methyltransferase activities, and serves to catalyze the methylation reactions during RNA capping. Cypovirus of the family Reoviridae provides a good model system for studying the methylation reactions in dsRNA viruses. Here, we present the structure of a transcribing cypovirus to a resolution of ~ 3.8 Å by cryo-electron microscopy. The binding sites for both S-adenosyl-l-methionine and RNA in the two methyltransferases of the turret were identified. Structural analysis of the turret in complex with RNA revealed a pathway through which the RNA molecule reaches the active sites of the two methyltransferases before it is released into the cytoplasm. The pathway shows that RNA capping reactions occur in the active sites of different turret protein monomers, suggesting that RNA capping requires concerted efforts by at least three turret protein monomers. Thus, the turret structure provides novel insights into the precise mechanisms of RNA methylation.
Abbreviations: dsRNA, double-stranded RNA; GTase, guanylyltransferase; MTase, methyltransferase; SAM, S-adenosyl-l-methionine; SAH, S-adenosyl-l-homocysteine; ICP, inner capsid particle; cryo-EM, cryo-electron microscopy
Keywords: dsRNA virus, Reoviridae, RNA capping, RNA methyltransferase
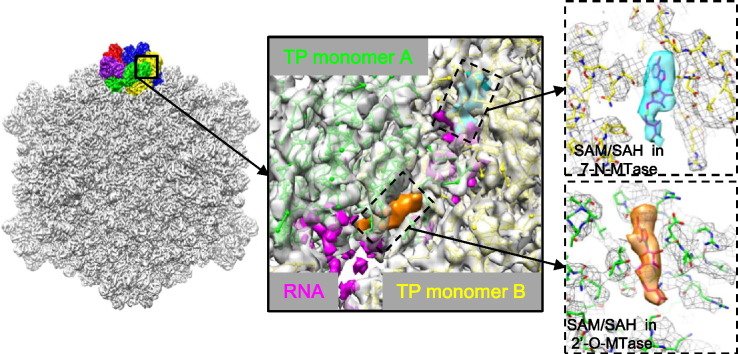
Graphical Abstract
The two active sites in methyltransferases (MTase) in a transcribing dsRNA virus.
Highlights
-
•
Structure of methyltransferases (MTases) and RNA in a transcribing dsRNA virus.
-
•
S-Adenosyl-l-methionine/S-adenosyl-l-homocysteine was observed in the two MTases.
-
•
A pathway was identified through which RNA reaches active sites of the two MTase.
-
•
Methylation reactions require concerted efforts by turret protein monomers.
Introduction
In eukaryotes, capping of the nascent mRNA 5′ end is essential for mRNA splicing, stability, and recognition, as well as export into and translation within the cytoplasm. Many viruses also encode their own enzymes for RNA capping. The mRNA capping process consists of four sequential reactions: (1) an RNA triphosphatase hydrolyzes the 5′ triphosphate of a nascent mRNA to a diphosphate; (2) a guanylyltransferase (GTase) adds a GMP molecule to the RNA 5′ diphosphate to form GpppRNA; (3) a 7-N-methyltransferase (7-N-MTase) transfers a methyl group from S-adenosyl-l-methionine (SAM) to the GpppRNA to form m7GpppRNA and generates S-adenosyl-l-homocysteine (SAH); and (4) a 2′-O-methyltransferase (2′-O-MTase) transfers a methyl group from SAM to the 2′-hydroxyl group of the first nucleotide ribose of the RNA and generates SAH [1].
Double-stranded RNA (dsRNA) viruses are a diverse group of viruses that infect a wide range of hosts including humans, animals, plants, fungi, and bacteria. Most dsRNA viruses have a multilayer spherical capsid. The outer capsid layers of dsRNA viruses present greatest diversity in both sequences and structural organization to adapt to different cell membrane barriers. A common process adopted by many dsRNA viruses is the uncoating of the outer capsid after delivery into the cytoplasm of the host cell, where the inner capsid particles (ICPs) remain intact. In order to protect the viral transcription process from the antiviral defense mechanisms of the host cells, the ICPs contain virally encoded RNA-dependent RNA polymerases and capping enzymes and serve as a stable nano-scale machine for viral mRNA transcription and capping [2].
Cypovirus belongs to the family Reoviridae, which is the largest family of dsRNA viruses causing disease in humans, livestock, insects, and plants [2]. Cypovirus is unique among viruses of the Reoviridae family in that it possesses a single capsid layer. The cypovirus capsid contains transcriptional and capping enzymes, as identified in the ICPs of other multilayer turreted viruses in the Reoviridae family [3]. Previously, we reported the cryo-electron microscopy (cryo-EM) structures of a transcribing cypovirus at ~ 4.1 Å resolution and a non-transcribing cypovirus at ~ 3.9 Å resolution [3], [4]. The turret of cypovirus at each fivefold vertex is formed by five monomers of turret protein VP3, which contains three separate enzymatic domains that catalyze the last three of the four sequential reactions in RNA capping [5]: mRNA GTase, 7-N-MTase, and 2′-O-MTase. In addition to these three enzymatic domains, VP3 contains two other domains: a bridge domain, which bridges the GTase and the two methyltransferases (MTases) according to the domain nomenclature of orthoreovirus turret protein [5], and a brace domain, which serves to brace a spike-like complex at the fivefold vertex in the pentameric turret [3]. The three enzymatic domains of VP3 are topologically identical with those in orthoreovirus [5] and aquareovirus [6], [7]. A GMP moiety, which defines the active site of a GTase domain, was observed in transcribing cypovirus VP3 [4]. However, due to the structural resolution limit of transcribing cypovirus VP3, the active sites of the two RNA MTases, which are defined by positions of SAM/SAH and RNA, were not resolved. How the two RNA MTase activities are facilitated by the turret protein is still unknown.
Besides the Reoviridae family, viral RNA MTases with known structures exist for viruses in three other viral families: protein D1 (7-N-MTase) [8] and VP39 (2′-O-MTase) of vaccinia viruses [9], protein NS5 (has dual functions of 7-N-MTase and 2′-O-MTase) of flaviviruses [10], and nsp16 (2′-O-MTase) of coronaviruses [11]. In vaccinia viruses and coronaviruses, each RNA MTase protein possesses either a 7-N-MTase or a 2′-O-MTase, whereas the flaviviruses NS5 protein only has one MTase domain catalyzing both 7-N-MTase and 2′-O-MTase reactions. In contrast, viruses in the Reoviridae family incorporate the domains of 7-N-MTase and 2′-O-MTase in a single turret protein.
In addition to turreted reoviruses, that is, cypovirus, orthoreovirus, and aquareovirus, a non-turreted virus in the Reoviridae family with known RNA MTase structures is the bluetongue virus. GTase, 2′-O-MTase, and 7-N-MTase of bluetongue virus are all included in a single viral encoded protein VP4 [12]. However, the cypovirus MTase domains are organized differently to those of bluetongue virus. In cypovirus, 7-N-MTase and 2′-O-MTase domains are formed by separated amino acid sequences [3]. In contrast, the bluetongue virus 7-N-MTase is formed by two amino acid sequences (residues 109–174 and residues 378–509), separated by an inserted 2′-O-MTase domain (residues 175–377).
In this study, we have improved the density map of transcribing cypovirus to a resolution of ~ 3.8 Å, which for the first time permits us to observe structures of SAM/SAH molecules and RNA. It represents the first structural analysis of RNA 7-N-MTase and 2′-O-MTase in a transcribing dsRNA virus at near-atomic resolution. Based on our structure of SAM/SAH–turret complex and the distribution of the RNA within the structure, we identified three channels through which the RNA reaches the two active sites of the 7-N-MTase and 2′-O-MTase in a proper order before it is released into the cytoplasm. Our results reveal that the methylation reactions require a concerted effort by at least three turret protein monomers.
Results and Discussion
Cryo-EM is particularly suitable for determining structures of active processes, such as actively transcribing cypoviruses, because such samples can be preserved in their native state by flash freezing, with no crystallization procedures required. The density map of the transcribing cypovirus was improved to ~ 3.8 Å resolution (Fig. 1a–e and Fig. S1). The density maps of turret proteins are of sufficient quality that we can identify nearly all the main chains and approximately 80% of the side chains, permitting us to build an atomic model. The improved density map permitted us to observe new features in the two MTase domains of the turret protein VP3. Our structural analysis of VP3 shows that both MTase domains possess structural characteristics of the class I MTase family [13].
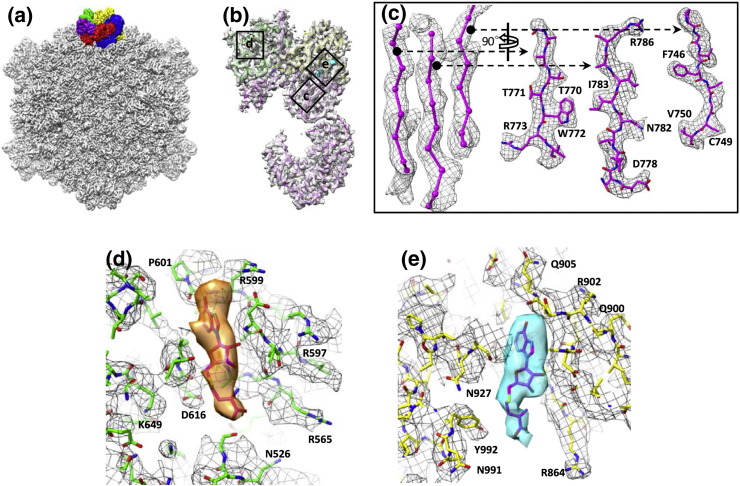
Fig. 1.
Structure of transcribing cypovirus. (a) Overall structure of a transcribing cypovirus filtered at 6 Å resolution along a 2-fold axis. The five VP3 monomers at a fivefold vertex are shown in different colors. (b) Transparent views of the density map of VP3 with its atomic model (only backbone shown) superimposed. The models of the MTase-1 and MTase-2 domains are in green and yellow, respectively. Positions of views labeled c, d, and e are shown in (c), (d), and (e). (c) Zoom-in view of a β-sheet segmented from the structure of VP3 with its atomic model superimposed. (d) Zoom-in view of the SAM/SAH binding site in the MTase-1 domain. An atomic model of SAH is fitted into the density (orange) that is attributed to SAM/SAH molecules located at the MTase-1 domain. Atomic model (sticks) superimposed on its density map (mesh) with some side chains labeled. (e) Zoom-in view of the SAM/SAH binding site of the MTase-2 domain. An atomic model of SAH is fitted in the structure (cyan) that is attributed to SAM/SAH molecules located at the MTase-2 domain. Atomic model (sticks) superimposed on its density map (mesh) with a number of side chains labeled.
The structure of the first MTase (MTase-1) domain (residues 419–461 and 526–725) of cypovirus VP3 remains largely unchanged compared to its counterpart in the non-transcribing cypovirus. However, an additional density feature, which is absent in non-transcribing cypovirus, was observed in the MTase-1 domain of transcribing cypovirus VP3 (Fig. 1d). This additional density cannot be assigned to any main chain or side chain; therefore, we attribute it to either a SAM or a SAH molecule located in the MTase-1 site. However, at the ~ 3.8 Å resolution, it is not possible to discriminate between the SAM and SAH molecules. A conserved catalytic tetrad KDKE (Lys531, Asp616, Lys649, and Glu689), which is thought to be a characteristic of RNA 2′-O-MTases [1], [14], was observed in the MTase-1 site, suggesting that the MTase-1 is 2′-O-MTase (Fig. 2a). An aromatic Tyr620 was observed to interact with the SAM/SAH molecule (Fig. S2), which is thought to be essential for cap methylation [15]. The cypovirus 2′-O-MTase domain is almost topologically identical with other viral 2′-O-MTases for which structures are known, such as the bluetongue virus 2′-O-MTase domain in VP4 [12], the vaccinia virus VP39 [9], and the coronavirus nsp16 [11]. Moreover, when these 2′-O-MTases are structurally aligned, the side chains of the KDKE tetrad in the cypovirus 2′-O-MTase domain closely overlap (Fig. S3a) with those of bluetongue virus 2′-O-MTase domain, vaccinia virus VP39 [9], and coronavirus nsp16 [11]. All side chains of the KDKE tetrad point toward a cleft, which is the putative RNA binding site in the 2′-O-MTase domain (Fig. 2a). According to the crystal structure of vaccinia virus VP39 complexed with a SAH and a 7-N-methylated single-stranded RNA hexamer [16], the Lys531, Lys649, and Glu689 of the conserved tetrad in cypovirus 2′-O-MTase serve to interact with the RNA sugar-phosphate backbone, and Asp616 plays a key role in SAM/SAH binding. In addition to the conserved tetrad, residues Gly562, Asp589, and Val600 around the SAM/SAH structure in the cypovirus 2′-O-MTase are also conserved and overlap closely with those in the bluetongue virus 2′-O-MTase domain of VP4, vaccinia virus VP39, and coronavirus nsp16. In addition, the SAM/SAH structure of cypovirus 2′-O-MTase overlaps closely with those in bluetongue virus 2′-O-MTase domain in VP4, vaccinia virus VP39, or coronavirus nsp16 (Fig. S3b). Side chains around SAM/SAH in these 2′-O-MTases are also similar (Fig. 2b). These results suggest that these 2′-O-MTases are functionally conserved.
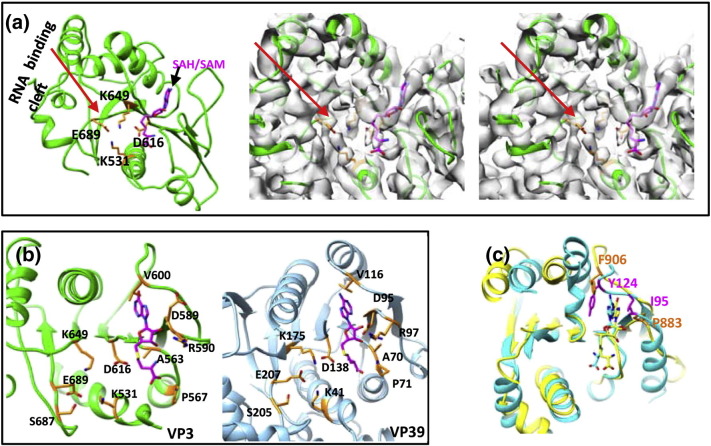
Fig. 2.
The two MTase domains of VP3 and other MTase proteins. (a) Left: Side chains of the KDKE (orange) of the MTase-1 (2′-O-MTase) domain of VP3. The putative RNA binding cleft is indicated by a red arrow. Right: A cross-eye stereo view of the density map (transparent) of the VP3 MTase-1 domain with its atomic model and a SAH molecule superimposed. The side chains of the KDKE and the SAH fit well with the density map (contoured at 1.2σ). The σ is the standard deviation of the density map. (b) Structural comparison between the 2′-O-MTase domain of cypovirus VP3 (left) and that of VP39 (right) of vaccinia virus shows that side chains of the two 2′-O-MTases surrounding SAM/SAH in the two 2′-O-MTases are similar. The tetrad K531, D616, K649, and E689 of 2′-O-MTase domain of VP3 and the tetrad K41, D138, K175, and E207 of VP39 are indicated. (c) Superimposing the 7-N-MTase domains of cypovirus VP3 (yellow) on Ecm1 (cyan). The F906 and P883 in 7-N-MTase of cypovirus are shown in orange, and the Y124 and I95 in Ecm1 are shown in magenta.
Residues 833–1058 form the second MTase (MTase-2) domain of cypovirus VP3. We suggest that this domain is a 7-N-MTase because it lacks a KDKE tetrad. The overall structure of the 7-N-MTase domain is unchanged compared with that in non-transcribing cypovirus [3]. We also observed an additional density feature in the 7-N-MTase domain, which is similar to the SAM/SAH density in the 2′-O-MTase domain of transcribing cypovirus VP3 (Fig. 1e). Thus, we also attribute this density to the presence of a SAM or SAH molecule (Fig. 1e).
A search of the PDB using the Dali server [17] revealed that the cypovirus 7-N-MTase domain most closely resembles Ecm1 (a 7-N-MTase from the microsporidian parasite Encephalitozoon cuniculi) [18] and followed by vaccinia virus 7-N-MTase D1 [8]. Consistent with this, superimposition between the cypovirus 7-N-MTase, Ecm1, and vaccinia virus D1 shows that the SAM/SAH structures in these 7-N-MTases closely overlap. The residues Gly863, Asp868, and Asp882 that interact with the SAM/SAH in the cypovirus 7-N-MTase are conserved and closely overlapped in Ecm1 and vaccinia virus D1. In the cypovirus 7-N-MTase, the adenosine of SAM/SAH is stacked between Phe906 and Pro883, which closely overlap with Tyr124 and Ile95, respectively, in Ecm1 (Fig. 2c). In vaccinia virus D1, the adenosine moiety is stacked between Tyr683 and Arg548. Indeed, mutagenesis experiments have shown that the aromatic nature of the residue is essential for the stacking of SAM adenine rather than the hydroxyl group in tyrosine [15].
In many viruses, the RNA capping reactions are sequential events. In order to reach the active sites of the enzymes efficiently and in proper order, the nascent RNA needs to follow a fixed pathway inside the turret. A previous study of a mammalian orthoreovirus suggested that RNA triphosphatase interacts with RNA-dependent RNA polymerase [19] and therefore is located on the inner surface of the inner capsid adjacent to the fivefold axis [20]. This observation suggests that the first capping reaction (hydrolyzing of 5′ triphosphate) occurs in the reovirus inner capsid before nascent RNA reaches the turret chamber through a peripentonal channel [4] in the capsid shell. Previously, we observed a rod-like density feature in the peripentonal channel in the 4.1-Å-resolution density map of the transcribing cypovirus, which was attributed to a partially resolved RNA [4]. A similar density feature was also observed at nearly the same location of the peripentonal channel in the new 3.8-Å-resolution density map (Fig. S4). In the turret chamber, transfer of GMP to the 5′ diphosphate RNA is followed by N7-guanine and 2′-O methylation [21], [22]. The first enzymatic domain that the nascent RNA reaches is the GTase domain in the turret [4], [20]. The density of the GMP moiety, which is present in the GTase domain in the 4.1-Å-resolution density map [4], is also visible in the GTase domain in the new 3.8-Å-resolution density map (Fig. S5). The GTase domain of one VP3 monomer (e.g., monomer C in Fig. 3a and Movie S1) and the active site of a 7-N-MTase domain of another monomer (monomer A in Fig. 3a and Movie S1) are connected via a channel (denoted as channel-1), which is formed by a bridge domain and a brace domain of monomer A (Fig. 3a and Movie S1) and a brace domain of another monomer (monomer B in Fig. 3a and Movie S1) [3]. Structural analysis of transcribing cypovirus indicates that this channel-1 is approximately 20% wider in the transcribing than that in the non-transcribing cypovirus [4]. We therefore propose that channel-1 guides the RNA molecule, namely, from the GTase domain of monomer C to the active site of the 7-N-MTase of monomer A. In support of this proposal, some fragmented density features approaching the SAM/SAH in 7-N-MTase were observed inside channel-1 (indicated in magenta in Fig. 3a and b). The fragmented density features (indicated in magenta in Fig. S6) have higher density level than noise. Because these density features do not belong to any main chain or side chain of VP3 and were not observed in non-transcribing cypovirus, we assigned them to a part of an RNA molecule present in the channel. The RNA was not resolved, as well as the VP3 structure, because only one RNA molecule is present in the pentameric turret at a time [20] and also due to the fact that the RNA is moving and flexible in the channel during the processes of RNA transcription and capping.
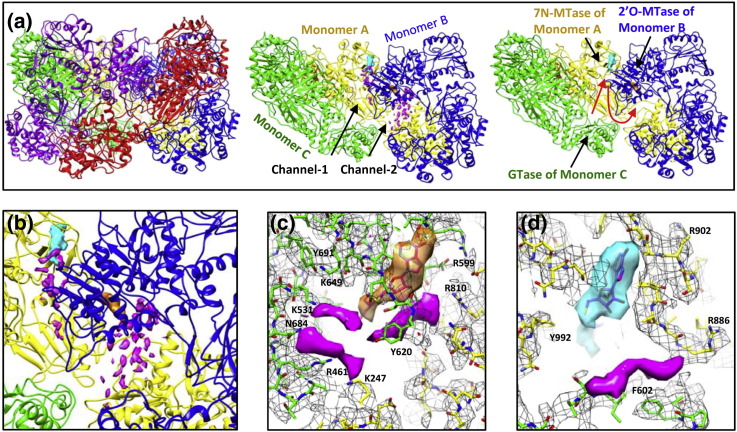
Fig. 3.
Turret structure of a transcribing cypovirus. (a) Atomic model of the turret formed by five VP3 monomers in red, purple, green, yellow, and blue, respectively (left). The two VP3 monomers in red and purple are removed for an inside view of the turret (middle and right). Channel-1 and channel-2 are indicated by two black arrows, and the fragmented density features attributed to RNA are shown in magenta (middle). The RNA pathway is indicated by red arrows, and the enzymatic domains that participate in the capping reactions are indicated by black arrows (right). (b) Zoom-in view of the fragmented density features in channel-1 and channel-2. (c) and (d) are zoom-in views of the fragmented density features of RNA around the SAM/SAH molecules in the domains of 2′-O-MTase and 7-N-MTase (respectively) of cypovirus VP3. The color scheme is the same as that used in Fig. 1d and e, except that the fragmented density features of RNA are in magenta.
Following its N7-guanine methylation, the question remains as to what the pathway could potentially be, along which the RNA reaches the nearest active site of the 2′-O-MTase. Similar to those RNA structures approaching the 7-N-MTase domain, some fragmented density features were also observed in a channel (denoted as channel-2 in Fig. 3a), which is formed by the bridge domain of monomer A, the GTase domain and the 2′-O-MTase domain of monomer B, and which leads to the SAM/SAH molecule and the putative RNA binding cleft of the 2′-O-MTase domain (Fig. 3a–d and Movie S1). We assigned these fragmented density features to RNA as well. The observation of the RNA structures leads us to suggest that the RNA withdraws from the 7-N-MTase domain of one copy of VP3 (monomer A) through channel-1 and approaches the active site of the adjacent 2′-O-MTase domain of another copy of VP3 (monomer B) through channel-2 in the turret (Fig. 3a and b; Fig. S6 and S7 and Movie S1). The process is reminiscent of the two methylations in a flavivirus, for which it was shown that the RNA dissociates from the MTase molecule after N7-guanine methylation and reassociates with another MTase molecule for 2′-O methylation [10].
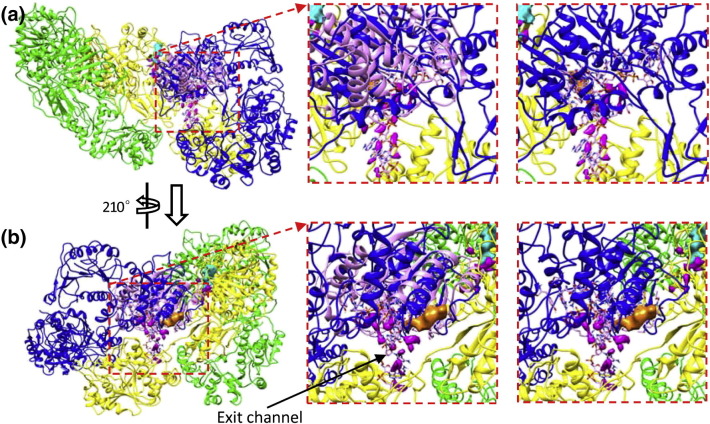
Further evidence on how RNA reaches the 2′-O-MTase site is provided by the crystal structure of vaccinia virus VP39 complexed with a 7-N-methylated, single-stranded RNA hexamer [16]. As described above, the VP39 and cypovirus 2′-O-MTase share a very similar topology, and some key residues interacting with SAM/SAH and RNA are conserved (Fig. 2b). In addition, their SAM/SAH binding sites are superimposed when the cypovirus 2′-O-MTase and the VP39 complex are structurally aligned. These results suggest that the cypovirus 2′-O-MTase binds RNA in a similar manner to VP39. Based on the vaccinia virus VP39 complex structure, we built a model of RNA binding into the cypovirus 2′-O-MTase by superimposing the cypovirus 2′-O-MTase on the VP39 complex. Without any manual position adjustment, the VP39 RNA fitted well into the putative RNA binding cleft in the cypovirus 2′-O-MTase (Fig. 4a and b). Indeed, all these RNA structures approaching the active site of the 2′-O-MTase domain are distributed along the strand of the RNA model built based on the structure of the VP39 complex (Fig. 4b).
Fig. 4.
Fitting the crystal structure of VP39 in complex with a single-stranded RNA hexamer into the 2′-O-MTase domain of VP3. (a) Left: The crystal structure of VP39 in complex with a single-stranded RNA hexamer (pink) is fitted into the 2′-O-MTase domain of a VP3 monomer (blue). Middle: Zoom-in view of the left view. Right: The same view as the middle except that VP39 structure is removed for clarity (only the RNA model is shown). It shows that these fragmented density features of RNA (magenta) approaching the active site of the 2′-O-MTase domain are distributed along the strand of the RNA model built based on the structure of VP39 complex. (b) Left: The structure in (a) is rotated by 210°. Middle: Zoom-in view of the left view. Right: The same view as the middle except that VP39 structure is removed for clarity (only the RNA model is shown). It shows that, after the methylation, the RNA can exit through the channel formed by monomer A (yellow) and monomer B (blue).
We observed that a channel, formed by the 2′-O-MTase domain of monomer B and the GTase domain of monomer A, connects the turret inside and outside (Fig. 4b and Fig. S7). The RNA structures, which are located in the RNA binding cleft of the 2′-O-MTase domain of monomer B, face directly toward the outer end of the channel (Fig. 4b). We also observed some fragmented density features attributable to RNA at the channel outside (Fig. S8). We suggest that the 7-N-methylated and 2′-O-methylated RNA is released from the turret into the cytoplasm through this channel.
Materials and Methods
The details of sample preparation, cryo-EM imaging, and data processing were described previously [4]. In order to confirm that the majority of cypovirus particles were in transcriptional state, we designed two reaction mixtures with different concentrations of GTP, and the reaction mixtures were incubated for 1 h, 3 h, and 6 h, respectively. The first transcription reaction mixture consisted of 70 mM Tris–Ac (pH 8), 10 mM MgAc2, 100 mM NaAc, 4 mM ATP, 2 mM CTP, 2 mM GTP, 2 mM UTP, 20 μCi [α-32P]UTP (specific activity, 3000 Ci/mM), 1 mM SAM, 1 U/μL RNase inhibitor, and purified cypovirus suspension. At 31 °C, a 6-h incubation time of this mixture resulted in darker RNA bands (Fig. S9, lane 6) than 3-h (Fig. S9, lane 3) and 1-h (Fig. S9, lane 2) incubation times, indicating that the reaction continued after 3 h. In the second reaction mixture, the concentration of GTP was decreased to 0.04 mM (the other conditions were identical with those of the first mixture). Different incubation times of this mixture resulted in similar RNA bands (Fig. S9, lanes 1, 5, and 7). We attributed it to the fact that the concentration of GTP was so low that it was exhausted by transcription within 1 h. No band was observed in lane 4 as the mixture did not contain GTP (Fig. S9). From these results, we concluded that cypovirus transcription had occurred, and after 3 h, the majority of the cypovirus particles were still actively transcribing RNA. The transcribing cypovirus particles in the first mixture incubated at 31 °C for 3 h were used for cryo-EM analysis. We collected 3720 micrographs by CCD (Gatan UltraScan4000 model 895) with an FEI 300-kV Titan Krios electron microscope.
Approximately 31,000 particle images were selected from ~ 56,000 particle images and included in the final three-dimensional reconstruction. The orientations and centers of all particle images were determined using an unpublished new software that uses common-line algorithm, as well as message passing interface and open multi-processing parallel technologies. The three-dimensional reconstruction was computed using ISAF algorithm [23], which has been enhanced by fast Fourier-spherical Bessel transformation and parallel computation.
The resolution of the reconstruction was estimated by Fourier shell correlation criterion [24]. The atomic model of VP3 was built based on density map using Coot [25] and then was refined using Rosetta [26]. Density maps of protein subunits were segmented from the capsid density map and visualized using UCSF Chimera [27].
Data deposition
The electron density map and atomic models have been deposited in the Electron Microscopy Data Bank and Protein Data Bank under accession code EMD-5926 and 3J6Q.
The following are the supplementary data related to this article.
Supplementary figures and legends
Atomic model of the turret formed by five VP3 monomers in red, purple, green, yellow, and blue, respectively. The two VP3 monomers in red and purple were for an inside view of the turret. The color scheme in the movie is the same as that in Fig. 3a. The first enzymatic domain that the nascent RNA reaches is the GTase domain of monomer C (green). A channel (denoted as channel-1), which is formed by a bridge domain and a brace domain of monomer A (yellow), as well as a brace domain of monomer B (blue), guides the RNA to the active site of 7-N-MTase of monomer A (yellow). Then, the RNA withdraws from the 7-N-MTase domain of monomer A (yellow) through channel-1 and approaches the active site of the adjacent 2′-O-MTase domain of monomer B (blue) through channel-2, which is formed by bridge domain of the monomer A (yellow), GTase domain, and 2′-O-MTase domain of the monomer B (blue).
Atomic coordinates of turret proteins
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31170697, 31370736, 91230116, and 31230018), the National Basic Research Program of China (2010CB912403), the Program for New Century Excellent Talents in University (NCET-13-0787), Hunan Provincial Natural Science Foundation of China (13JJ1017), and the Scientific Research Foundation for Returned Overseas Chinese Scholars, State Education Ministry. All electron microscopic data were collected at the Center for Bio-imaging, Core Facility for Protein Sciences of Chinese Academy of Sciences. We thank Professor Jingqiang Zhang and Professor Jingchen Sun for providing cypovirus polyhedra.
Conflict of Interest Statement: The authors declare no conflict of interest.
Edited by T. J. Smith
Contributor Information
Hongrong Liu, Email: hrliu@hunnu.edu.cn.
Lingpeng Cheng, Email: lingpengcheng@moon.ibp.ac.cn.
References
- 1.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L., Sun J., Zhang K., Mou Z., Huang X., Ji G. Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc Natl Acad Sci U S A. 2011;108:1373–1378. doi: 10.1073/pnas.1014995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C., Ji G., Liu H., Zhang K., Liu G., Sun F. Cryo-EM structure of a transcribing cypovirus. Proc Natl Acad Sci U S A. 2012;109:6118–6123. doi: 10.1073/pnas.1200206109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinisch K.M., Nibert M., Harrison S.C. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 6.Cheng L., Zhu J., Hui W.H., Zhang X.K., Honig B., Fang Q. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Jin L., Fang Q., Hui W.H., Zhou Z.H. 3.3 Å Cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De la Pena M., Kyrieleis O.J., Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodel A.E., Gershon P.D., Shi X., Quiocho F.A. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85:247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 10.Dong H., Ren S., Zhang B., Zhou Y., Puig-Basagoiti F., Li H. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol. 2008;82:4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutton G., Grimes J.M., Stuart D.I., Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol. 2007;14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 13.Schubert H.L., Blumenthal R.M., Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bujnicki J.M., Rychlewski L. Reassignment of specificities of two cap methyltransferase domains in the reovirus lambda 2 protein. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0038. Published online Aug 23, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao X., Shuman S. Vaccinia virus mRNA (guanine-7-)methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photo-cross-linking of the cap to the methyl acceptor site. Biochemistry. 1996;35:6900–6910. doi: 10.1021/bi960221a. [DOI] [PubMed] [Google Scholar]
- 16.Hodel A.E., Gershon P.D., Quiocho F.A. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 17.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabrega C., Hausmann S., Shen V., Shuman S., Lima C.D. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 19.Kim J., Parker J.S., Murray K.E., Nibert M.L. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor mu2. J Biol Chem. 2004;279:4394–4403. doi: 10.1074/jbc.M308637200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Walker S.B., Chipman P.R., Nibert M.L., Baker T.S. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodel A.E., Gershon P.D., Shi X., Wang S.M., Quiocho F.A. Specific protein recognition of an mRNA cap through its alkylated base. Nat Struct Biol. 1997;4:350–354. doi: 10.1038/nsb0597-350. [DOI] [PubMed] [Google Scholar]
- 22.Hu G., Gershon P.D., Hodel A.E., Quiocho F.A. mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc Natl Acad Sci U S A. 1999;96:7149–7154. doi: 10.1073/pnas.96.13.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H., Cheng L., Zeng S., Cai C., Zhou Z.H., Yang Q. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J Struct Biol. 2008;161:64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal P.B., Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.DiMaio F., Tyka M.D., Baker M.L., Chiu W., Baker D. Refinement of protein structures into low-resolution density maps using Rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and legends
Atomic model of the turret formed by five VP3 monomers in red, purple, green, yellow, and blue, respectively. The two VP3 monomers in red and purple were for an inside view of the turret. The color scheme in the movie is the same as that in Fig. 3a. The first enzymatic domain that the nascent RNA reaches is the GTase domain of monomer C (green). A channel (denoted as channel-1), which is formed by a bridge domain and a brace domain of monomer A (yellow), as well as a brace domain of monomer B (blue), guides the RNA to the active site of 7-N-MTase of monomer A (yellow). Then, the RNA withdraws from the 7-N-MTase domain of monomer A (yellow) through channel-1 and approaches the active site of the adjacent 2′-O-MTase domain of monomer B (blue) through channel-2, which is formed by bridge domain of the monomer A (yellow), GTase domain, and 2′-O-MTase domain of the monomer B (blue).
Atomic coordinates of turret proteins