Abstract
The C-terminal domain (CTD) of the severe acute respiratory syndrome coronavirus (SARS-CoV) nucleocapsid protein (NP) contains a potential RNA-binding region in its N-terminal portion and also serves as a dimerization domain by forming a homodimer with a molecular mass of 28 kDa. So far, the structure determination of the SARS-CoV NP CTD in solution has been impeded by the poor quality of NMR spectra, especially for aromatic resonances. We have recently developed the stereo-array isotope labeling (SAIL) method to overcome the size problem of NMR structure determination by utilizing a protein exclusively composed of stereo- and regio-specifically isotope-labeled amino acids. Here, we employed the SAIL method to determine the high-quality solution structure of the SARS-CoV NP CTD by NMR. The SAIL protein yielded less crowded and better resolved spectra than uniform 13C and 15N labeling, and enabled the homodimeric solution structure of this protein to be determined. The NMR structure is almost identical with the previously solved crystal structure, except for a disordered putative RNA-binding domain at the N-terminus. Studies of the chemical shift perturbations caused by the binding of single-stranded DNA and mutational analyses have identified the disordered region at the N-termini as the prime site for nucleic acid binding. In addition, residues in the β-sheet region also showed significant perturbations. Mapping of the locations of these residues onto the helical model observed in the crystal revealed that these two regions are parts of the interior lining of the positively charged helical groove, supporting the hypothesis that the helical oligomer may form in solution.
Abbreviations: CTD, C-terminal domain containing residues 248–365; SARS-CoV, severe acute respiratory syndrome coronavirus; NP, nucleocapsid protein; SAIL, stereo-array isotope labeling; NTD, N-terminal domain containing residues 45–181; NOE, nuclear Overhauser effect; EMSA, electrophoretic mobility shift assay; HSQC, heteronuclear single-quantum coherence; CT, constant time; UL, uniformly labeled; PDB, Protein Data Bank; CSD, chemical shift displacement; poly-dT, poly-deoxythymine; ssDNA, single-stranded DNA; EDTA, ethylenediaminetetraacetic acid
Keywords: SAIL-NMR, SARS nucleocapsid protein, protein structure determination, nucleocapsid packaging
Introduction
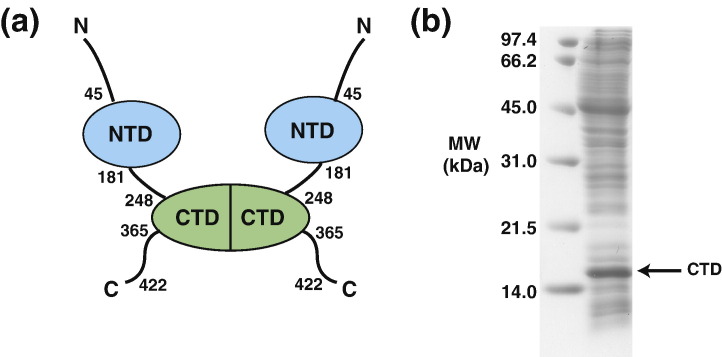
Severe acute respiratory syndrome (SARS) is a recently emergent disease caused by the SARS-associated coronavirus (CoV).1 , 2 The SARS-CoV nucleocapsid protein (NP) packages the viral genomic RNA into a ribonucleoprotein complex and is crucial for the assembly of infectious virus particles. Based on comparative NMR studies of SARS-CoV NP deletion constructs, the protein contains two structural domains: the N-terminal domain (NTD; containing residues 45–181) and the C-terminal domain (CTD; containing residues 248–365),3 flanked by the long, disordered N- and C-termini and the linker sequence (Fig. 1a). The NTD reportedly acts as a putative RNA-binding domain, and the CTD functions as a dimerization domain.3 , 4 Recently, the CTD has also been shown to bind nucleic acids with high affinity.5 Structural investigations of the isolated SARS-CoV NP CTD have been performed by NMR and X-ray crystallography. In a previous NMR study, the topological structure of the isolated SARS-CoV NP CTD as a homodimer was elucidated based on limited intersubunit and intrasubunit nuclear Overhauser effects (NOEs).6 This topology was confirmed by the subsequently reported crystal structures of the SARS-CoV NP CTD and of a shorter construct spanning residues 270–370.5 , 7 The crystal structure of the CTD revealed that residues 248–280 form a positively charged patch, which acts as a putative oligonucleotide-binding region. The patch also participates in intermolecular and intramolecular interactions within the crystal, resulting in the formation of an octameric asymmetric unit. However, previous biochemical and biophysical studies have shown that the CTD exists solely as a dimer in solution.6 These findings motivated us to investigate the nature of the CTD in solution. Initial attempts at the complete structure elucidation of the SARS-CoV NP CTD through NMR were impeded by its short T 2 relaxation times and significant peak overlaps.6
Fig. 1.
Preparation of the SARS-CoV NP CTD. (a) Schematic diagram of the domain architecture of the SARS-CoV NP. (b) SDS-PAGE of the cell-free reaction mixture for the SARS-CoV NP CTD. The left lane shows the molecular weight markers. The band corresponding to the monomer of the SARS-CoV NP CTD is labeled with an arrow.
Recently, we have developed the stereo-array isotope labeling (SAIL) method, which utilizes proteins exclusively composed of stereo- and regio-specifically labeled amino acids.8 Compared to conventional uniform 13C,15N isotopic labeling, the quality of the spectra of the SAIL sample was sufficiently improved, so that a high-resolution solution structure determination could be performed. The sharpened resonance lines and reduced peak overlap with the SAIL method are due to the selective deuteration of many nonlabile protons. The remaining protons with known stereo-specific assignments provide plentiful information about the structure of the protein. In this study, we performed an NMR study of the SARS-CoV NP CTD with the help of the SAIL method. Compared to the protein uniformly labeled with 13C and 15N, the SAIL sample significantly improved the quality of the NMR spectra, to the extent that a high-resolution structure determination could be performed. The tertiary structure obtained by NMR is almost identical with that of the crystal structure, except for a disordered putative RNA-binding domain at the N-terminus. We further applied NMR, mutation analyses, and electrophoretic mobility shift assays (EMSAs) to pinpoint the nucleic-acid-binding site. The active site thus identified agrees well with the helical ribonucleoprotein model suggested by the crystal structure.
Results
Preparation of the SAIL sample of the SARS-CoV NP CTD
The preparation of proteins composed of SAIL amino acids requires cell-free expression to efficiently incorporate the SAIL amino acids into the protein without them being affected by metabolic scrambling in living cells. The expression of the SARS-CoV NP CTD was initially examined in a small-scale reaction (Fig. 1b). Subsequently, the 1H–15N heteronuclear single-quantum coherence (HSQC) of the 15N-labeled SARS-CoV NP CTD produced by cell-free expression was compared with that produced by in vivo expression. These spectra were identical, thus confirming that the structures of the proteins produced by cell-free and in vivo expressions are identical (data not shown). The final sample of the SAIL SARS-CoV NP CTD was produced by using the Escherichia coli cell-free protein synthesis system, with optimizations for the production of labeled NMR samples.9
The SAIL method improves the quality of the NMR spectrum
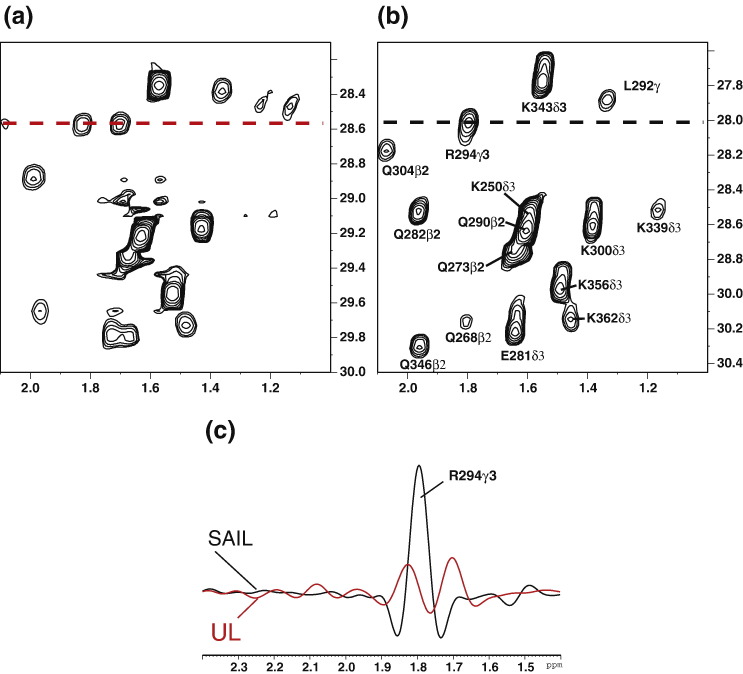
The 1H–13C constant time (CT) HSQC spectra of an aliphatic region were compared between uniformly labeled (UL) and SAIL samples under the same conditions. In the case of the UL sample, the signals were prone to overlap between diastereotopic pairs, and some signals were severely broadened beyond detection in the methylene region (Fig. 2a). In contrast, the corresponding spectrum from the SAIL sample had much better quality than that from the UL sample (Fig. 2b). The signal/noise ratios for the SAIL sample were several times higher than those for the corresponding UL sample, consistent with previous results for calmodulin- and maltodextrin-binding proteins (Fig. 2c).8 For the SARS-CoV NP CTD, the number of peaks to be observed theoretically in the 1H–13C CT HSQC spectra, including methyl, methylene, and methane protons, decreased from 517 for the UL sample to 343 for the SAIL sample, greatly simplifying the analytical process.
Fig. 2.
Comparisons of NMR spectra for the SARS-CoV NP CTD between UL and SAIL in the methylene region. Aliphatic region of 1H–13C CT-HSQC for UL (a) and SAIL (b) SARS-CoV NP CTD. Both spectra were acquired under the same conditions. The sample concentration was 0.5 mM. In (b), assignments for the SAIL sample are labeled. (c) Cross-sections from (a) (red) and (b) (black). The peak scales are identical between the UL and SAIL spectra.
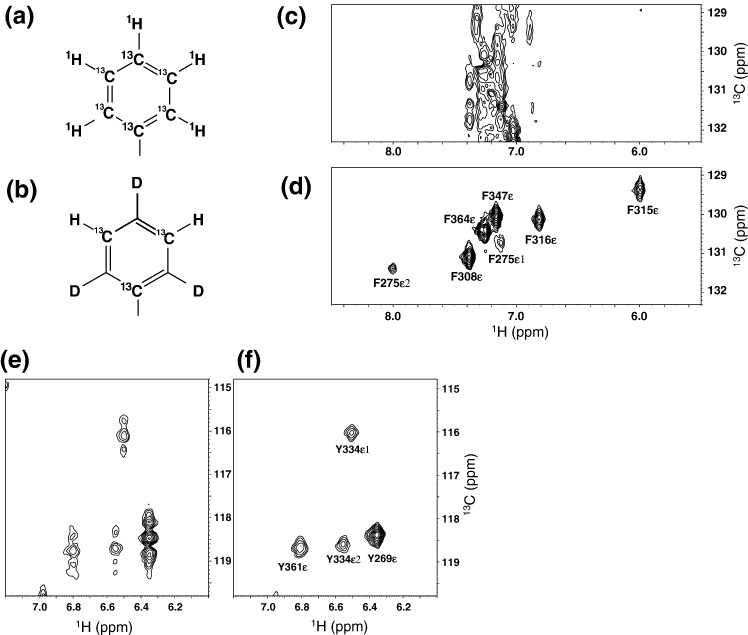
For the aromatic region, improvement due to the use of the SAIL method was even more striking. The aromatic rings of UL Phe and Tyr contain four and five 13C–1H pairs, respectively (Fig. 3a). In the case of SAIL, the six-membered aromatic rings are labeled by alternating 13C–1H and 12C–2H moieties (13C at the ε and γ positions; 12C at the δ and ζ positions) (Fig. 3b).10 In the UL sample, signals for the 1H–13C moieties at the δ, ε, and ζ positions of Phe, and at the δ position of Tyr, are overlapped around 131 ppm in the carbon dimension, thus resulting in severe spectral crowding (Fig. 3c). In contrast, the corresponding region for the SAIL sample was much simpler due to the presence of signals exclusively from the 1H–13C moieties at the ε position of Phe. The short relaxation times led to the detection of resonances that were severely broadened beyond detection with the UL sample (Fig. 3d). Since one-bond 13C–13C couplings in the UL protein did not exist in the SAIL protein, as shown in the Tyr Hε–Cε region (Fig. 3e and f), the CT technique with a long evolution time (∼ 17 ms) is thus not required for the SAIL sample.10
Fig. 3.
Comparisons of NMR spectra for the SARS-CoV NP CTD between UL and SAIL in the aromatic region. Chemical structures of the aromatic rings for UL (a) and SAIL (c) phenylalanine. (b and d) Phenylalanine signals of 1H–13C HSQC for UL (b) and SAIL (d) SARS-CoV NP CTD. (e and f) Tyrosine signals of 1H–13C HSQC for UL (e) and SAIL (f) SARS-CoV NP CTD. To demonstrate the absence of the 1Jcc coupling of aromatic rings for SAIL phenylalanine and tyrosine residues, all 1H–13C HSQC spectra for the aromatic regions were recorded without the CT technique.
With the benefits mentioned above, we were able to acquire a set of NMR spectra with high sensitivity, and the expected chemical shifts were assigned to 91.2% completeness for the SAIL sample.
Solution structure of SARS-CoV NP CTD
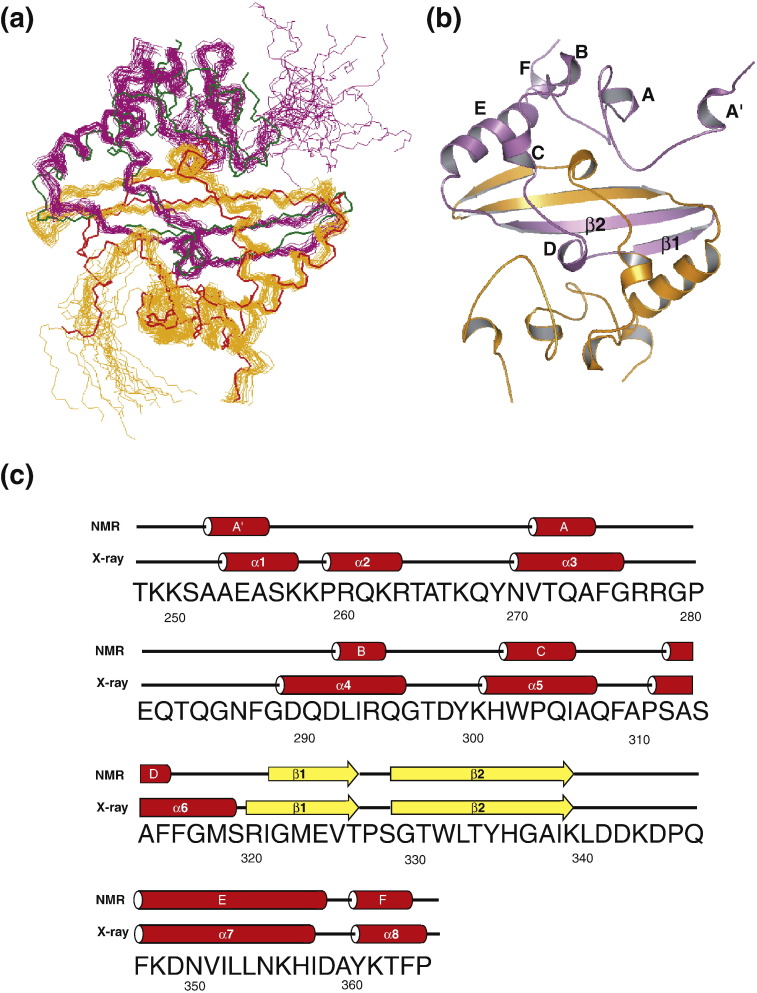
Even though the SARS-CoV NP CTD existed as an octamer in the asymmetric unit in the crystal, abundant evidence suggests that it exists as a homodimer in solution.3 , 6 We manually assigned some of the intersubunit NOE peaks from the SARS-CoV NP CTD based on previous NMR work.6 These manually assigned intersubunit distance restraints were included in the combined automated assignments of the NOE peaks and the structure calculation by CYANA.11 NOE-derived distance restraints totalling 2615 were obtained. Out of 100 structures calculated, the 20 structures with the lowest target function values were selected and energy-refined as the final structures of the SARS-CoV NP CTD. The structural statistics for the NMR structure are summarized in Table 1 . Consistent with the previously reported topological structure of the SARS-CoV NP CTD, the NMR structure of the SARS-CoV NP CTD adopts a domain-swapped homodimer conformation (Fig. 4a and b). The resulting structures exhibited good convergence for both the backbone and the side chains when the regions between residues 260 and 365 of each dimer were superimposed (Fig. 4a). The model has a disordered region spanning residues 248–259 and protruding from the dimer core, which appears to be the result of internal dynamics and prevents the detection of long-range NOEs based on heteronuclear NOE measurements. 15N–{1H} NOE experiments recorded for the SARS-CoV NP CTD showed that the heteronuclear NOE values for this region are smaller than those for the structured region, indicating that the two N-termini are flexible in solution (data not shown). The secondary structural elements of the CTD in solution were defined based on the DSSP algorithm,12 and they corresponded well to those identified in the previous NMR study (Fig. 4b and c).6
Table 1.
Structural statistics for the NMR structure of SARS-CoV CTD
| Parameter | Quantity |
|---|---|
| Completeness of chemical shift assignments (%) | 91.2 |
| Total NOE upper distance bound restraints | 2615 |
| Short range (|i − j| ≤ 1) | 1313 |
| Medium range (1 < |i − j| < 5) | 586 |
| Long range (|i − j| ≤ 5) | 716 |
| Intermolecular | 260 |
| Dihedral angle restraints (φ and ψ) | 236 |
| CYANA target function (Å2) | 2.55 |
| AMBER energy (kcal/mol) | − 6106 |
| Ramachandran plot statistics (%) | |
| Most favored regions | 84.6 |
| Additionally allowed regions | 14.8 |
| Generously allowed regions | 0.6 |
| Disallowed regions | 0.0 |
| Backbone RMSD for residues 260–365 (Å) | 0.77 |
| All heavy atom RMSD for residues 260–365 (Å) | 1.19 |
Fig. 4.
NMR structure of the SARS-CoV NP CTD. (a) Superposition of the 20 lowest-energy NMR structures of the SARS-CoV NP CTD and the corresponding crystal structure spanning residues 248–365.5 The two subunits in each structure are in orange and magenta for the NMR structure, and in red and green for the crystal structure. (b) Ribbon diagram of the solution structure of the SARS-CoV NP CTD. Secondary structure elements are labeled for one subunit. (c) Sequence of the SARS-CoV NP CTD. The secondary structures for the NMR structure and for the crystal structure spanning residues 248–365 are shown above the sequence, with red cylinders for α-helices and yellow arrows for β-strands.
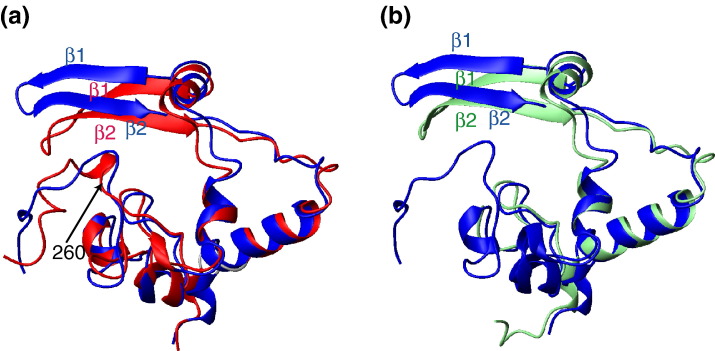
Prior to our NMR investigation, the crystal structures of constructs spanning residues 270–370 [Protein Data Bank (PDB) ID 2gib] and residues 248–365 (PDB ID 2cjr) were elucidated.5 , 7 The overall folds and secondary structure arrangements are very similar between the crystal structures and the NMR structure (Fig. 4a and c). The backbone RMSD between the protomers of the mean NMR structure and the crystal structure of the CTD spanning residues 248–365 is 1.45 Å if residues 260–319 and 333–358 are superimposed. If the NMR structure is superimposed with the crystal structure spanning residues 270–370, then the backbone RMSD between the protomers is 1.26 Å for the regions of residues 274–319 and 333–358. There are two differences, however: first, in both crystal structures, the β-sheet is distorted around residues 320–332 compared to those of the NMR structure (upper left of Fig. 5a and b). Second, the two N-termini (residues 248–259) protruding from the dimer core are disordered in the NMR structure, whereas, in the crystal structure, they are involved in a number of intramonomer and intradimer contacts and are more rigid (Fig. 4a). The disorder of the N-termini in solution was further supported by the analysis of backbone amide-exchange rates (data not shown). We suspect that at least some of these differences are likely to result from the different solvent conditions used for crystallization and/or crystal-packing effects.
Fig. 5.
Superposition of the NMR and crystal structures of a CTD monomer of SARS-CoV NP. The mean NMR structure of a CTD monomer (blue) is superposed on the corresponding crystal structure spanning residues 248–365 (a) (red; PDB code 2cjr)5 and on that encompassing residues 270–370 (b) (light green; PDB code 2gib).7 In (a), the two structures are superposed on the regions of residues 260–319 and 333–358, where the backbone RMSD between them is 1.45 Å. In (b), the two structures are superposed on the regions of residues 274–319 and 333–358, where the backbone RMSD is 1.26 Å.
Nucleic-acid-binding sites of SARS-CoV NP CTD
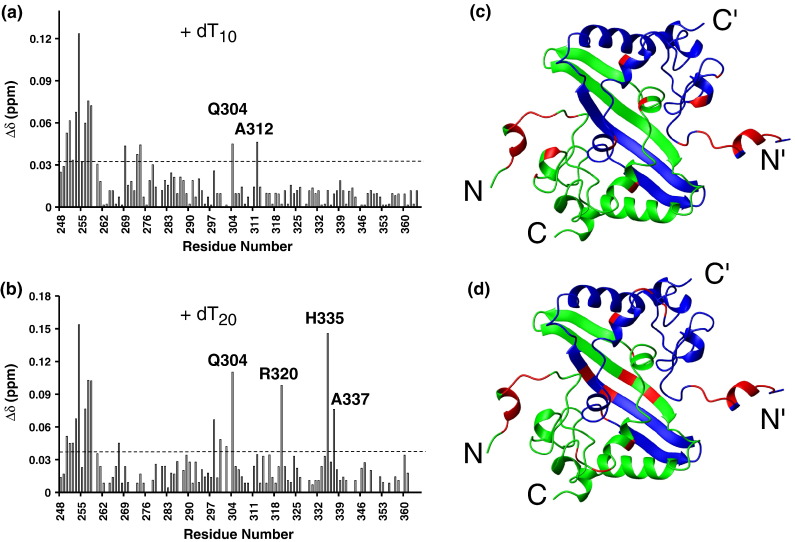
Deletion studies revealed that residues 248–280 are essential for the nucleic-acid-binding activity of the SARS-CoV NP CTD.5 However, the exact residues involved in the binding had not been identified. To identify these residues, we conducted chemical shift displacement (CSD) studies by titrating 10-mer (dT10) or 20-mer (dT20) poly-deoxythymine (poly-dT) single-stranded DNA (ssDNA) into the protein samples. ssDNA were used throughout this study as surrogates of single-stranded RNA. Titration of dT10 or dT20 into the protein sample caused a concentration-dependent gradual shift of some resonances, instead of the appearance of a new set of resonances, suggesting that the binding occurs in the fast-exchange regime, which is indicative of a low-affinity nucleic-acid-binding protein.13 For dT10, significant chemical shift changes were localized primarily in the N-terminal region, particularly K250, E253, A254, S256, K257, and K258 (Fig. 6a and b), while the majority of the other resonances were scarcely affected. This result suggests that dT10 binds to the SARS-CoV NP at the N-terminal flexible segment, without affecting the overall structure of the protein (Fig. 4). The binding constant estimated from the CSD studies at various dT10 concentrations is K d ∼ 30 μM.
Fig. 6.
CSD of SARS-CoV NP CTD titrated with poly-dT ssDNA. Variation of the CSD of SARS-CoV NP CTD titrated with dT10 (a) or dT20 (b). The dashed lines in (a) and (b) represent the cutoff for significant displacements. (c) Spatial locations of residues (red) with CSD values larger than the cutoff value upon titration with dT10 (c) or dT20 (d). The two monomers are in green and blue, respectively.
Similarly, dT20 also generated significant chemical shift changes in the same set of N-terminal residues; however, it also induced CSD of the resonances of R320, H335, and A337, which are located in the β-sheet of the CTD dimer (Fig. 6b and d). Since these residues are not affected when the SARS-CoV NP CTD is bound to dT10 (Fig. 6a and c), we can rule out the effect of long-range structural alterations induced by the binding of oligonucleotides to the N-termini. Our results suggest that R320, H335, and A337 also contribute to nucleic acid binding and could be part of the binding site. This observation is unexpected, as these residues are sequentially and structurally distant from the N-terminal region. It should be noted that we could only add dT20 up to a [dT20]/[CTD monomer] ratio of 1:4. A higher ratio of DNA caused precipitation. As such, we could not obtain a reliable dissociation constant for the complex, and the chemical shift perturbation shown in Fig. 6b may not be the maximum change expected at a saturating dT20 concentration for the complex.
Mutagenesis of the nucleic-acid-binding sites of SARS-CoV NP CTD
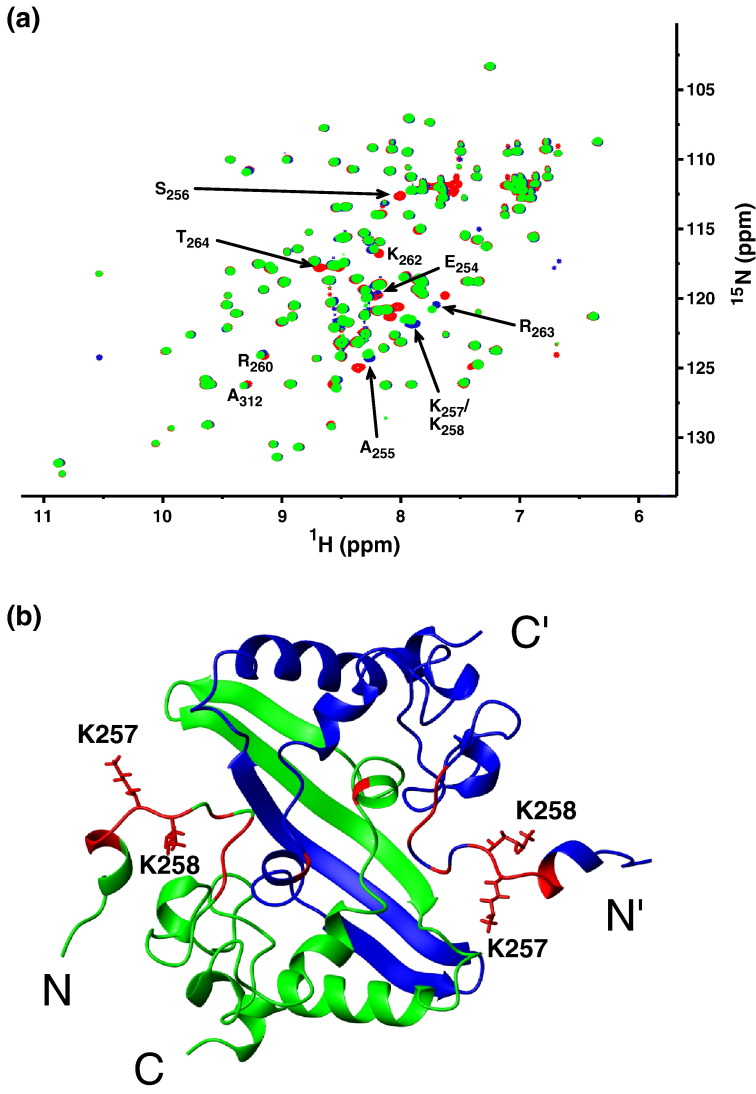
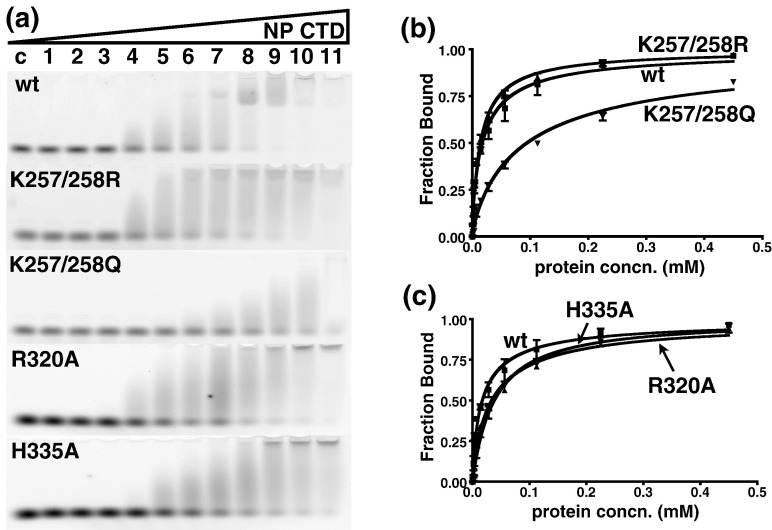
To further quantify the relative contribution of positively charged residues to oligonucleotide binding by the SARS-CoV NP CTD, we produced double mutants targeting K257/K258 and measured the effect of the mutations on the apparent dissociation constant (K d) with fluorescently labeled dT20 through EMSA.14 Since the mutation sites are located in the inherently flexible regions, the mutations did not cause any structural perturbations to other parts of the CTD dimer, as monitored by 15N HSQC spectra (Fig. 7 ). The EMSA results are summarized in Table 2 . We found that the charge-preserving K257R/K258R mutant did not affect the apparent binding affinity, compared to the wild-type construct, whereas the K257Q/K258Q mutant showed a 5-fold reduction in affinity, although binding was not completely abolished (Fig. 8 , Table 2). Our results suggest that the positive charges at positions 257 and 258 of the SARS-CoV NP CTD are significant determinants of its binding affinity towards oligonucleotides.
Fig. 7.
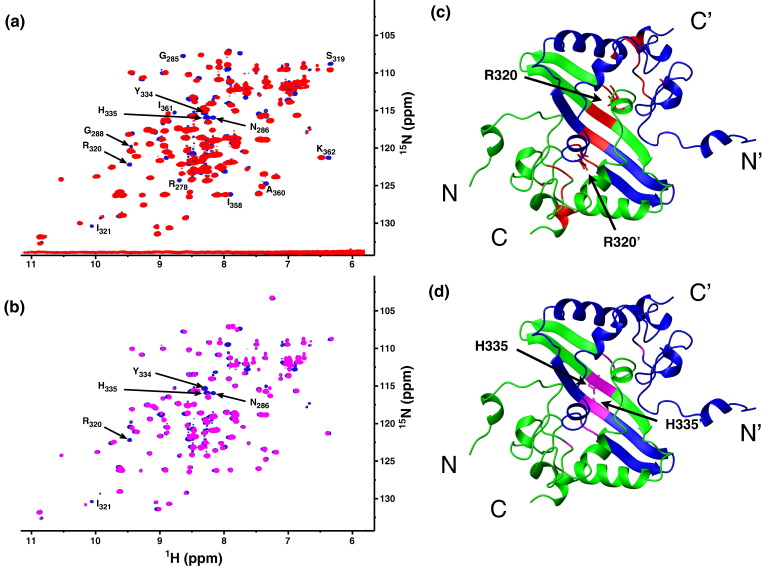
Structure perturbation of K257/K258 double mutants. (a) Overlay of 15N-edited HSQC spectra from wild-type CTD (blue), K257R/K258R (green), and K257Q/K258Q (red) double mutants. Affected resonances are identified by their respective residue types and numbers in the wild-type protein. These are mapped onto the ribbon structure of the CTD dimer in (b).
Table 2.
Binding coefficients for dT20 to SARS-CoV NP CTD
| Protein | Apparent Kd (μM) | Hill coefficient |
|---|---|---|
| Wild type | 17.19 ± 1.51 | 0.82 ± 0.05 |
| K257R/K258R | 13.96 ± 0.92 | 0.92 ± 0.05 |
| K257Q/K258Q | 94.42 ± 5.56 | 0.83 ± 0.04 |
| R320A | 33.81 ± 2.16 | 0.85 ± 0.04 |
| H335A | 38.07 ± 2.25 | 1.01 ± 0.05 |
Fig. 8.
EMSA of SARS-CoV NP CTD mutants. (a) Mobility shift of dT20 bound to wild-type (wt), K257R/K258R (K257/258R), K257Q/K258Q (K257/258Q), R320A, and H335A mutant proteins. The protein concentration was increased by a factor of 2, starting from lane 1 (439 nM) to lane 11 (0.45 mM). Lane C, negative control. (b) Binding curve of the K257/K258 double mutant towards dT20, compared to that of the wild-type protein. (c) Binding curve of the R320A and H335A mutants towards dT20, compared to that of the wild-type protein. Each curve in (b) and (c) represents the best fit from three independent assays. Results are summarized in Table 2.
Similarly, we also generated the R320A and H335A mutants. The 15N HSQC spectra of these mutants revealed that, in both cases, the structural perturbations are mostly limited to the regions adjacent to the Arg320 and His335 mutation sites (Fig. 9 ). However, the R320A mutation had a chemical shift perturbation larger than that of the H335A mutation, probably because the Arg side chain made more contacts with adjacent residues. The changes in the respective apparent dissociation constants were measured by EMSA. Mutations at this secondary binding site lowered the apparent affinity towards dT20 by about twofold (Fig. 8, Table 2). Although the effects were not as remarkable as those of the mutations near the N-terminus, the loss of binding affinity was still measurable. Our results are in agreement with the hypothesis that the β-sheet region of the SARS-CoV NP CTD is part of the nucleic-acid-binding site.
Fig. 9.
Structure perturbation of R320A and H335A mutants. (a) Overlay of 15N-edited HSQC spectra from the wild-type CTD (blue) and the R320A mutant (red). Affected resonances are identified by their respective residue types and numbers in the wild-type protein. (b) Same as in (a), but with the wild-type CTD (blue) and the H335A mutant (magenta). (c) Mapping of residues affected by the R320A mutation (red) in the solution structure of the SARS-CoV NP CTD dimer. The side chains of R320 are shown in a neon representation. (d) Same as in (c), but showing the residues affected by the H335A mutation (in magenta). The side chain of H335 is also shown.
Discussion
SAIL as an emergent tool for solution structure determination
The SAIL method is characterized by a sophisticated labeling pattern that is highly optimized for structure determination, in contrast to other preexisting isotope labeling techniques. Conventional strategies utilizing selective protonation under a predeuterated background lead to a compromise between increased intensity of the labeled protons and loss of information about the deuterated ones. For instance, while the selective protonation of the methyl protons of Ile, Leu, and Val in a deuterated background is very effective for the observation of methyl protons, one cannot obtain any information on the remaining side chains.15 The SAIL method, on the other hand, combines the merits of increased intensity through the deuteration of redundant protons and preservation of the structural information by leaving key protons intact. Of particular interest is the use of SAIL aromatic residues, which was first demonstrated with calmodulin.10 Assignments of chemical shift and NOE peaks involving aromatic signals are indispensable for the high-quality structure determination of proteins, since aromatic residues are often part of the folding core. In the case of the SARS-CoV NP CTD, the aromatic resonances of the UL protein were severely overlapped, making their assignment difficult, if not impossible (Fig. 3b and e). The introduction of aromatic SAIL amino acids in the sample resolved this problem (Fig. 3d and f), ultimately leading to the elucidation of the SARS-CoV NP CTD solution structure. This study is the first case to have demonstrated that the use of the SAIL phenylalanine and tyrosine residues was effective in the NMR spectral analysis of a large protein. This is also the first instance of a homodimeric protein structure to have been solved by the SAIL approach. The SAIL method becomes more effective with increasing molecular weight and allows for the structures of larger proteins with even more intricate features to be solved.8
Differences between the solution structure and the crystal structures of SARS-CoV NP CTD
There are two distinct differences between the structure of the SARS-CoV NP CTD in solution and the structure of the SARS-CoV NP CTD in the crystal. The first is the orientation of the short turn in the β-sheet: in the crystal, the short turn is closer to the N-terminal residues of the same protomer than in the solution structure, resulting in a more compact crystal structure (Fig. 5). This is observed in all of the crystal structures of the SARS-CoV NP CTD solved to date, regardless of the space group and the construct.5 , 7 It is possible that crystal packing is responsible for the compactness. On the other hand, the less compact solution structure of the short hairpin turn could be the result of the more dynamic character of the CTD in an aqueous environment.
The second difference lies in the conformation of the N-termini. The N-termini in the crystal structure of the SARS-CoV NP CTD, spanning residues 248–365, form an ordered conformation anchored by intramolecular and intermolecular interactions between various adjacent residues.5 However, the N-termini in solution are disordered and, in agreement with our previous studies,6 lack a short helix formed by residues 259–263 in the crystal structure (Fig. 4). The CTD is arranged as an octamer within the unit cell of the crystal, and this short helix could be the result of different solvent conditions, crystal packing, and/or the oligomerization process.16 Residues within and adjacent to the short helix participate in intramolecular and intermolecular interactions within the crystal and contribute to the formation of the octamer.5 It is possible that the short helix is selectively stabilized by the formation of new protein–protein contacts in the crystal octamer, similar to those observed for the binding of intrinsically disordered proteins to their targets.17 Transient formation of the short helix is also observed in a few of the conformers within the NMR structural ensemble.
Relevance to ribonucleoprotein packaging
In the crystal structure of SARS-CoV NP248–365, we have previously found that the CTD forms an octamer.5 The packaging of the octamers in the asymmetric unit of the crystal results in two parallel basic helical grooves. Residues 248–280 form a positively charged patch similar to that in the infectious bronchitis virus NP.18 These patches form a large part of the basic helical groove observed in the crystal structure. We postulate that the basic helical groove may serve as the RNA attachment site, and the structure suggests a mechanism for helical RNA packaging in the virus. However, the octamer has not been observed in solution. In our DNA titration study, we found that the spin–spin relaxation time, T 2, of the amide resonances decreases, upon the addition of the DNA oligomer, at a rate faster than that expected for sheer increases in molecular weight. We suspect that the DNA complex forms transient higher-order multimers in solution.
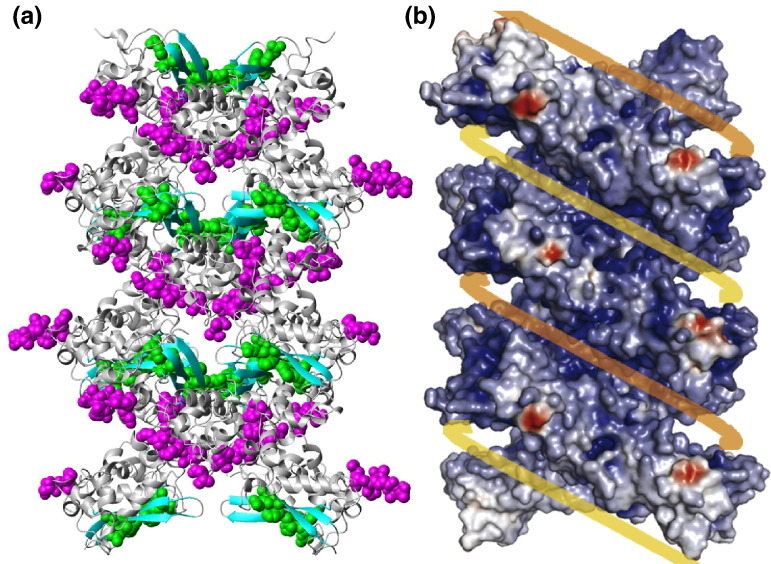
Within the helical oligomer model, one expects nucleic acid binding to stabilize the oligomer structure. To investigate the consistency of our NMR chemical shift perturbation data with the proposed helical model, we mapped the spatial locations of the residues perturbed by dT20 binding onto the helical model proposed by Chen et al. (Fig. 10 ).5 It clearly shows that both the N-terminal residues and the additional perturbed residues in the β-sheet region, namely, R320 and H335, form the interior lining of the positively charged groove. The two N-termini of the dimer reside on the outside edge and in the innermost part of the groove, respectively. The arrangement of the helical structure is such that the interior N-termini are still solvent-accessible. On the other hand, the perturbed β-sheet residues reside in the midregion of the groove. Thus, while the current NMR titration results do not prove the validity of the helical model, they can still be satisfactorily accommodated within this framework. More rigorous data, such as the determination of the structure of the CTD/nucleic acid complex, are necessary to provide definitive proof of the helical model.
Fig. 10.
Spatial locations of the nucleic-acid-binding sites in the helical packing model of the SARS-CoV NP CTD crystal. (a) Binding sites are shown in CPK models, with the N-termini residues in magenta and with the residues on the β-sheet (R320, H335, and A337) in green. The rest of the molecules are shown in a gray ribbon representation, except for the β-sheets, which are in cyan. (b) Surface charge representation of the proposed helical supramolecular complex (adapted from Chen et al.5). The yellow and orange lines represent viral RNA strands. Notice that the binding sites in (a) are located in the positively charged grooves within the supramolecular complex.
Oligonucleotide binding and structural disorder
We have previously shown that the SARS-CoV NP is a modular protein comprising two independent structural domains connected by a 66-residue linker and flanked on each end by the long, disordered N- and C-termini, which comprise 44 and 57 residues, respectively3 (Fig. 1a). The NTD, comprising residues 45–181, has been shown to bind to RNA.4 Here we showed that the CTD also binds to DNA with similar affinity to that of the NTD. We have shown previously that the di-domain fragment NP45-365 binds to DNA and RNA with higher affinity than that of the respective NTD or CTD.15 Taken together, all of these observations suggest that the SARS-CoV NP binds to RNA at multiple sites, and the binding strength is enhanced by the multivalency effect, as multiple binding sites are in contact with the RNA molecule.19 This charge-based nonspecific binding mode works in conjunction with intrinsic disorder to confer two main advantages to the nonspecific binding of oligonucleotides to the CTD. First, because the disordered region is not locked into a single conformation, binding to a variety of partners can occur regardless of the structural features of the partner, as long as the electrostatic interaction provides enough free energy to maintain the bound state. This property allows the CTD to bind to oligonucleotides with different sequences or tertiary structures. This is an important feature of RNA chaperones, of which the SARS-CoV NP is a member, and hints at the possibility that residues 248–270 are involved in the process.20, 21 Second, the unstructured protein molecule can have a greater capture radius for a specific binding site than that of the folded state with its restricted conformational freedom, the so-called “fly-casting mechanism.”22 In this binding scenario, the unfolded state binds weakly at a relatively large distance, and then folds as the protein approaches the binding site. These two advantages could act together to ensure that the CTD is able to bind to a variety of nucleotide sequences with enough affinity to carry out its function, namely, the encapsulation of the viral genome. We envision the extended conformation of the NP molecule as a whole to facilitate its initial contact with the RNA molecule in a fly-casting mechanism. Subsequent rearrangement of the NP molecule in the RNA framework then results in favorable packing of the complex in a helical form.
Materials and Methods
Site-directed mutagenesis
The SARS-CoV NP CTD was cloned from SARS-CoV TW1 strain sequencing vectors (a gift from Dr. P.-J. Chen, National Taiwan University Hospital) as previously described.6 Mutants of the SARS-CoV NP CTD were produced with a QuickChange II kit (Stratagene, La Jolla, CA) on a RoboCycler 96 (Stratagene), in accordance with the manufacturer's recommendations. Primers used for mutagenesis were purchased from Mission Biotech (Taiwan). Mutations were confirmed through DNA sequencing.
Sample preparation
The SARS-CoV NP CTD, encompassing residues 248–365 including an extra MHHHHHHAMG sequence at the N-terminus, was expressed in the E. coli BL21 (DE3) strain for nonlabeled and uniformly labeled samples, as described previously,6 and in a cell-free reaction for the SAIL samples. The production of nonlabeled and uniformly labeled samples by in vivo expression was performed in a conventional manner. The proteins expressed in E. coli were purified in accordance with our previously described protocol.6 The cell-free expression of the SARS-CoV NP CTD was performed as described previously.9 The S30 extract containing minimal residual amino acids was used for the cell-free expression. In the cell-free synthesis of the SARS-CoV NP CTD, the concentration of each SAIL amino acid was set to 0.5 mM, and 2.3 mg of the SAIL-SARS-CoV NP CTD was obtained from a total of 70 mg of SAIL amino mixture. SAIL amino acids were obtained from SAIL Technologies, Inc.‡ The SAIL-SARS-CoV NP CTD thus produced was mainly in soluble form. The SAIL protein was purified by Ni-NTA affinity chromatography in 50 mM sodium phosphate (pH 7.4) and 150 mM NaCl, followed by gel filtration in a buffer containing 50 mM sodium phosphate (pH 7.4), 150 mM NaCl, and 1 mM ethylenediaminetetraacetic acid (EDTA). The eluted SAIL-SARS-CoV NP CTD was then concentrated and exchanged with the NMR buffer.
NMR spectroscopy
The SAIL-SARS-CoV NP CTD sample contained 0.5 mM (10% D2O buffer) and 0.5 mM (100% D2O buffer) of the SAIL SARS-CoV NP CTD in NMR buffer [10 mM sodium phosphate pH 6.0, 50 mM NaCl, 1 mM EDTA, 1 mM 2,2-dimethyl-2-silapentane-5-sulfonate, 0.01% NaN3, 10% D2O, and Complete Mini protease inhibitor mix (Roche)]. SAIL-adapted NMR experiments for the structure determination were performed at 30 °C with Bruker 600-MHz or 800-MHz spectrometers equipped with a TXI triple resonance room-temperature probe or a cryoprobe. 1H–15N HSQC spectra were obtained with a 1-mM 15N-labeled sample in NMR buffer on a Bruker Avance 500-MHz spectrometer equipped with a TXI cryoprobe, using an in-house adaptation of the pulse sequence. For mutant characterization and protein–ssDNA-binding studies in NMR buffer, 15N-labeled samples were prepared in NMR buffer, and spectra were recorded on Bruker Avance 600-MHz or 800-MHz spectrometers equipped with QXI quadruple resonance or TXI probes. The acquired data were processed with the XwinNMR suite (Bruker Biospin, Germany) or with iNMR (Nucleomatica, Italy), and chemical shift assignments were performed. The chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate and deposited in BioMagResBank.
Structure calculation and refinement
The NMR structure calculation of the SARS-CoV NP CTD was started using 28 independent pairs of intersubunit distance restraints obtained from isotope-filtered NOE spectroscopy experiments reported previously.6 Automated NOE cross-peak assignments15 and structure calculations with torsion-angle dynamics16 were performed using a modified version of the program CYANA 2.1, which incorporates SAIL labeling patterns,4 takes the homodimer symmetry explicitly into account for the network anchoring of NOE assignments,15 ensures an identical conformation of the two monomers by imposing torsion-angle difference restraints on all corresponding torsion angles, and maintains a symmetric relative orientation of the two monomers by applying distance difference restraints between symmetry-related intermolecular Cα–Cα distances. Backbone torsion-angle restraints obtained from database searches with the program TALOS17 were incorporated into the structural calculation. Hydrogen-bond restraints were not used. CYANA structure calculations were started from 100 randomized conformers, and simulated annealing with 20,000 torsion-angle dynamics time steps per conformer was performed. The 20 conformers with the lowest final CYANA target function values were subjected to restrained energy refinement in explicit solvent against the AMBER force field.18
CSD studies
A series of 2D 15N-edited HSQC spectra of uniformly 15N-labeled SARS-CoV NP CTD protein (0.5 mM) was recorded in NMR buffer by titrating in different amounts of poly-dT ssDNA (Purigo, Taiwan). The affected amide correlations experienced CSD upon the addition of ssDNA. The unaffected and shifted resonances in uncrowded regions were easily assigned, whereas the shifted resonances in crowded regions were assigned by stepwise titration of the protein with small amounts of ssDNA and by tracing of the changes in the CSD until the desired final concentration is achieved. The final protein/ssDNA ratio was 1:1 for 10-mer and 4:1 for 20-mer. Protein/ssDNA ratios higher than the one presented here resulted in the formation of a precipitate within the sample. The weighted CSD for each residue was calculated with the formula: CSD = (1/2((Δδ1 HN)2 + (Δδ15 N/5)2)1/2, where Δδ denotes the chemical shift difference between the final complex and the free protein resonances. The experimental error in the weighted CSD from the spectral resolution was calculated as: (1/2((SW1 HN/points in 1H)2 + ((SW15 N/points in 15N)/5)2))1/2, where SW denotes the total spectral width of the dimension. Amides with CSD values larger than the average shift of all the peaks plus the experimental error were selected as affected.
EMSA
All experiments were conducted in NMR buffer with 6-aminohexylfluorescein-labeled ssDNA (Purigo). Reactions were set up in 20-μl aliquots each containing 50 nM 6-aminohexylfluorescein-labeled ssDNA. Protein was added to the aliquots starting at a concentration of 500 μM, with each following aliquot containing a 2-fold serial dilution of the protein. A control reaction was set up where only ssDNA and buffer were added. The aliquots were allowed to react at room temperature for 30 min, and then were loaded on a 0.5 × Tris–borate EDTA buffer DNA retardation gel (Invitrogen, Carlsbad, CA). The gel was run at 30 V and 4 °C for 2.5 h, and the bands were visualized with a Typhoon 9410 variable mode imager (Amersham Biosciences, Piscataway, NJ). Quantitation of the free ssDNA band was achieved through the ImageJ software (National Institutes of Health, Bethesda, MA). Bound ssDNA was estimated by subtracting the free ssDNA band of each reaction from that of the control lane. The fraction of bound ssDNA was fitted against the equation: Y = 1/(1 + (K d/X)n), using GraphPad Prism (GraphPad Software, San Diego, CA), where Y is the fraction of ssDNA bound to the protein, X is the protein concentration, K d is the dissociation constant, and n is the Hill coefficient. All experiments were repeated twice.
PDB accession codes
Chemical shift assignments and atomic coordinates have been deposited in BioMagResBank (accession code 15511) and PDB (accession code 2jw8), respectively.
Acknowledgements
This work was supported, in part, by Core Research for Evolutional Science and Technology/Japan Science and Technology Agency (M.K. at Tokyo Metropolitan University), by Technology Development for Protein Analyses from Ministry of Education, Culture, Sports, Science and Technology (M.K. at Nagoya University), by a Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (P.G. at Tokyo Metropolitan University), and by the Volkswagen Foundation (P.G. at J.W. Goethe-University of Frankfurt am Main). This work was also supported by grants NSC 95-2311-B-001-066-MY3 (T.H.) and NSC-95-2113-M-001-035-MY3 (T.H.) from the National Science Council of the Republic of China. Some NMR spectra were obtained at the High-Field Nuclear Magnetic Resonance Center, supported by the National Research Program for Genomic Medicine, Taiwan, ROC.
Edited by M. F. Summers
Footnotes
References
- 1.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek T., Erdman D., Goldsmith C., Zaki S., Peret T., Emery S., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Chang C., Sue S., Yu T., Hsieh C., Tsai C., Chiang Y., et al. Modular organization of SARS coronavirus nucleocapsid protein. Biomed. Sci. 2005;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., et al. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 5.Chen C., Chang C., Chang Y., Sue S., Bai H., Riang L., et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C., Sue S., Yu T., Hsieh C., Tsai C., Chiang Y., et al. The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Lett. 2005;579:5663–5668. doi: 10.1016/j.febslet.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the SARS coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between Corona- and Arteri-viridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kainosho M., Torizawa T., Iwashita Y., Terauchi T., Mei Ono A., Guntert P. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 9.Torizawa T., Shimizu M., Taoka M., Miyano H., Kainosho M. Efficient production of isotopically labeled proteins by cell-free synthesis: a practical protocol. J. Biomol. NMR. 2004;30:311–325. doi: 10.1007/s10858-004-3534-2. [DOI] [PubMed] [Google Scholar]
- 10.Torizawa T., Ono A.M., Terauchi T., Kainosho M. NMR assignment methods for the aromatic ring resonances of phenylalanine and tyrosine residues in proteins. J. Am. Chem. Soc. 2005;127:12620–12626. doi: 10.1021/ja051386m. [DOI] [PubMed] [Google Scholar]
- 11.Güntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 12.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez C., Allain F. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–3645. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garner M.M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tugarinov V., Kay L.E. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 16.Andrec M., Snyder D.A., Zhou Z., Young J., Montelione G.T., Levy R.M. A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins: Struct. Funct. Genet. 2007;69:449–465. doi: 10.1002/prot.21507. [DOI] [PubMed] [Google Scholar]
- 17.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 18.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., et al. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunde B.M., Moore C., Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev., Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zúñiga S., Sola I., Moreno J., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tompa P., Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 22.Shoemaker B.A., Portman J.J., Wolynes P.G. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl Acad. Sci. USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]