Abstract
There are many peptides known that inhibit the entry of enveloped viruses into cells, including one peptide that is successfully being used in the clinic as a drug. In this review, we discuss the discovery, antiviral activity and mechanism of action of such peptides. While peptide entry inhibitors have been discovered by a wide variety of approaches (structure-based, accidental, intentional, rational and brute force) we show here that they share a common physical chemical property: they are at least somewhat hydrophobic and/or amphipathic and have a propensity to interact with membrane interfaces. We propose that this propensity drives a shared mechanism of action for many peptide entry inhibitors, involving direct interactions with viral and cellular membranes, as well as interactions with the complex hydrophobic protein/lipid interfaces that are exposed, at least transiently, during virus–cell fusion. By interacting simultaneously with the membrane interfaces and other critical hydrophobic surfaces, we hypothesize that peptide entry inhibitors can act by changing the physical chemistry of the membranes, and the fusion protein interfaces bridging them, and by doing so interfere with the fusion of cellular and viral membranes. Based on this idea, we propose that an approach that focuses on the interfacial hydrophobicity of putative entry inhibitors could lead to the efficient discovery of novel, broad-spectrum viral entry inhibitors. This article is part of a Special Issue entitled: Interfacially Active Peptides and Proteins. Guest Editors: William C. Wimley and Kalina Hristova.
Keywords: Enveloped virus, Entry inhibitor, Fusion protein, Peptide, Interfacial hydrophobicity
Graphical abstract
Highlights
-
•
Enveloped virus fusion and entry are underutilized therapeutic targets.
-
•
Many peptides inhibit enveloped virus entry into cells.
-
•
Many peptide entry inhibitors have a propensity to interact with membrane interfaces.
-
•
Many peptide entry inhibitors are active against multiple, unrelated viruses.
-
•
A proposed model emphasizes the role interfacial hydrophobicity on entry inhibition.
1. Introduction
Enveloped viruses are an ancient and ubiquitous class of human pathogen with infection rates and mortality rates that are often expressed as measurable percentages of the entire human population [1], [2]. This class includes many well-known viruses such as influenza, human immunodeficiency, hepatitis C, small pox, chicken pox, yellow fever, herpes, measles and many more. It also includes tropical pathogens of significant and growing global public health concern (e.g. dengue, lassa and chikungunya viruses), viruses that have recently elicited fears of novel and deadly global pandemics (e.g. avian influenza, SARS and MERS viruses) and viruses with significant biothreat potential (e.g. ebola, hantaviruses, Rift Valley fever virus, and most of the viruses mentioned above). In this review, we discuss the discovery, development, characterization and mechanism of action of peptides that inhibit entry of enveloped viruses into host cells.
Over the past few decades peptides have steadily gained importance in drug design and delivery. Increasingly, focus is shifting toward the development and refinement of techniques for identifying synthetic peptides as drug candidates. Bioactive peptides have been discovered by the use of naturally occurring peptides, through rational engineering, through high-throughput screening, or by structure-based design using sequences from known regions of proteins [3]. These factors are responsible for the emergence of peptides as a growing market in the pharmaceutical industry. Currently there are about 100 peptide based drugs on the market [4], constituting about 10% of the entire drug market [5]. As we describe below, one effective peptide entry inhibitor of an enveloped virus is approved for use in humans [6] and more are in clinical trials [7], [8]. Many other peptide entry inhibitors of enveloped viruses have been described in the scientific literature. In this review we discuss the surprising observation that the majority of known peptide entry inhibitors, which have been discovered by an extraordinary diversity of approaches, share a common physical chemistry: they are somewhat hydrophobic and amphipathic with a propensity for binding to lipid membranes. We hypothesize that their shared physical chemistry could result in a shared mechanism of action; one that enables a generic approach to the discovery of broad-spectrum peptide entry inhibitors for enveloped viruses.
2. Enveloped viruses
2.1. Entry of enveloped viruses into cells
The genomes of all enveloped viruses enter cells through a sequential, multistep process that requires i) virus binding to cell surface receptors and ii) fusion of the viral membrane with a cellular membrane [9]. These essential steps occur by an incompletely understood process in which viral fusion proteins (i.e. envelope glycoproteins, spike proteins, see Fig. 1 ) mediate a temporally and spatially coordinated close contact between the cellular and viral membranes while simultaneously perturbing the continuity of the membranes with hydrophobic segments. For most enveloped virus families these events are triggered by endocytosis of surface-bound virus and subsequent endosomal acidification. For a few viruses, such as HIV, fusion is triggered by conformational changes that occur upon binding to cell surface receptors and co-receptors and is not entirely dependent on endosomal acidification.
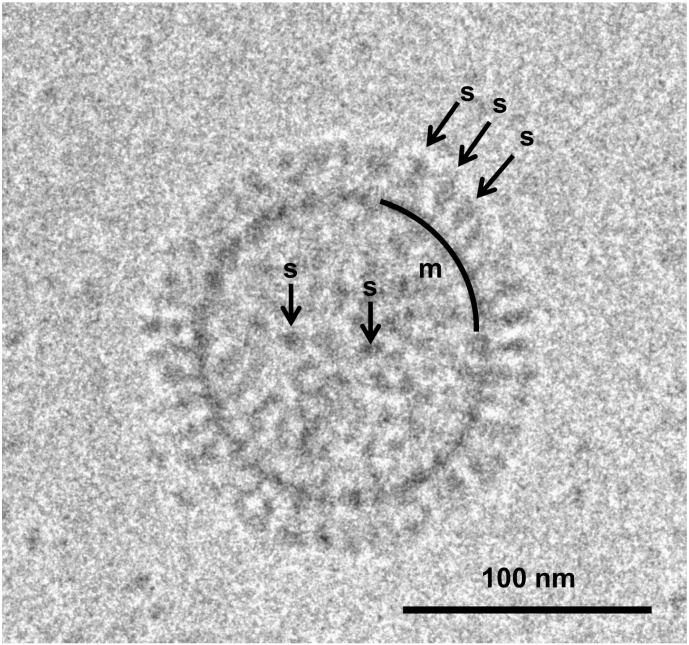
Fig. 1.
Cryo-transmission electron microscopy (CryoTEM) image of an influenza virus particle, showing its classical enveloped virus architecture. The lipid bilayer membrane is bounded on the inner surface by electron-dense matrix proteins (m). The membrane is packed with 16 nm long spikes(s) made of the Class I fusion protein hemagglutinin, here in the pre-fusion (neutral pH) state.
The fusion proteins of enveloped viruses can be grouped into several distinct classes based on the protein secondary and tertiary structure. Class I fusion proteins are predominantly α-helical trimers that fold into an elongated 6-helix bundle [10], [11], [12]. This class includes the well-known “spike” proteins of influenza (hemagglutinin) and HIV (gp160). Class II fusion proteins are defined by their elongated, multi-domain, β-sheet rich structure and are found, for example, in flaviviruses. Class II fusion proteins are dimeric in the pre-fusion configuration, but transition to a trimeric state during low pH-induced fusion [13]. Class III fusion proteins have mixed α/β structure and are found in, for example, the herpes viruses [14].
Despite the structural diversity of their fusion proteins, the entry mechanisms of enveloped viruses share critical biological activities and structural signatures (Fig. 2 ). For all classes, cell surface binding is driven by a specific chain or domain in a fusion protein or protein complex that is distinct from the domain/chain that drives fusion of the viral envelope with the cellular membrane. Membrane fusion is driven by large conformational rearrangements of the fusion protein that expose a hydrophobic fusion peptide or fusion loop while also simultaneously presenting other hydrophobic sequences of the fusion protein to catalyze membrane fusion [15], [16], [17].
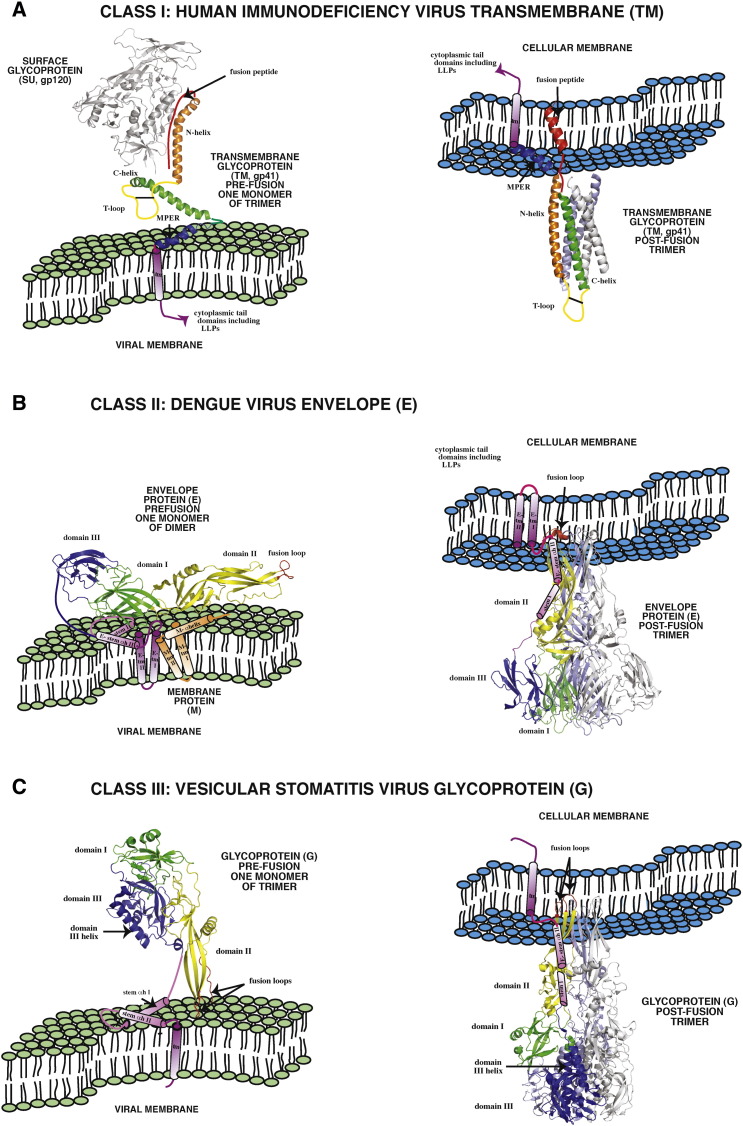
Fig. 2.
Pre- and post-fusion structures of representative viral fusion proteins of Classes I–III. The domain architectures of the three known classes of viral fusion proteins (VFP) of enveloped viruses are shown in the pre-fusion (virion) and post-fusion states. Panel A. The domain architecture of human immunodeficiency virus (family: Retroviridae) transmembrane glycoprotein (TM), a Class I VFP is indicated: red: fusion peptide, orange: amino terminal helix (aka helical region 1, HR1 or NHR), yellow: T loop containing the dicysteine bridge, green: carboxyl terminal helix (aka HR2 or CHR), blue: membrane proximal region (MPER aka aromatic domain), indigo: transmembrane domain (tm). Pre-fusion structure: protein database 4NCO, [135]. Post-fusion structure: 3F4Y, [136]. Fusion peptide in post-fusion state: 1ERF, [137]. MPER structure: 1JAU, [138]. Panel B. The domain architecture of dengue virus (family: Flaviviridae) envelope glycoprotein (E), a Class II VFP is indicated: green (domain I), yellow (domain II) and blue (domain III). The fusion loop, stem and tm helices of E are red, indigo and violet, respectively. The membrane protein (M) is depicted in orange. Pre-fusion structure:1OAN, [139]. Post-fusion structure: 4GSX, [140]. Panel C. The domain architecture of vesicular stomatitis virus (family: Rhabdoviridae) glycoprotein (G), a Class III VFP is indicated: green (domain I), yellow (domain II) and blue (domain III). The fusion loops, stem and tm helices of G are red, indigo and violet, respectively. Pre-fusion structure: 2J6JX, [141]. Post-fusion structure: 2CMZ, [142]. The left portion of Panel B and the depiction of cellular and viral membranes are modified from Fig. 4 of Zhang et al. [143].
In Fig. 2 we show schematic structures of representative Class I, Class II and Class III viral fusion proteins in the pre- and post-fusion states. In addition to the hydrophobic fusion peptide (which is a terminal peptide in Class I proteins, an internal loop in Class II proteins and a pair of fusion loops in Class III fusion proteins) viral fusion proteins also have other hydrophobic sequences that likely interact with membranes and contribute to the fusion process. For example, many have a conserved, hydrophobic, aromatic-rich domain adjacent to the membrane-spanning helix domain. In HIV, this so called “membrane proximal ectodomain region” or MPER is highly conserved and is critical for function [6]. The MPER sequence of HIV gp41 is also the epitope for one of the few broadly neutralizing antibodies against HIV [18]. MPER is very hydrophobic, amphipathic and aromatic [18], [19] which suggests that its function likely involves membrane binding and destabilization [20]. Class I viral fusion proteins also have other internal hydrophobic segments, including leucine zipper like-motifs that lie at the interfaces between helical heptad repeat domains that may also become exposed, at least transiently, during the conformational rearrangements that drive fusion [21]. Class II fusion proteins have similar features. For example, they often have conserved aromatic/hydrophobic juxtamembrane domains, called “stem” domains, that (like MPER) interact strongly with membrane interfaces and can form amphipathic helixes [22]. In addition to the stem domain, Class II fusion proteins have other hydrophobic regions that are likely involved in the fusion process [16], [17]. Interestingly, arenavirus Class II fusion protein complexes include a so called stable signal peptide (SSP) domain, a conserved and required protein which contains two transmembrane helical segments connected by a loop domain [23], [24], [25]. Unlike most signal sequences, arenavirus SSPs remain stably inserted in the viral envelope and associate with the fusion protein in a complex. These hydrophobic SSPs, whose function remains unknown, have the potential to provide additional hydrophobic surface to the protein–membrane interfaces that arise during fusion. Similarly, the architecture of Class III fusion proteins contains hydrophobic patches that may participate in fusion [14] although less is known about Class III fusion proteins, overall.
2.2. Viral entry inhibition: an underutilized therapeutic target
It is reasonable to hypothesize that the function of all viral fusion proteins is similarly dependent on the exposure of multiple hydrophobic segments to form a continuous hydrophobic surface bridging the viral and cellular membranes and allowing for fusion to take place by mixing the lipids in the two membranes. This hypothesis suggests that a broad-spectrum approach to entry inhibition of enveloped viruses may be possible by targeting, through physical chemistry, the hydrophobic surfaces on the membranes and fusion protein that are exposed during fusion. Highlighting the potential vulnerability of the fusion process, it can be inhibited by changes in protein structure [26], by changes in membrane physical properties [27], by changes in the timing of protein conformational changes [28] or by the addition of antibodies, peptides or small molecules that bind the fusion protein [24], [29], [30], [31].
For these reasons, viral entry would seem to be a vulnerable target for antiviral countermeasures. Indeed, there are small molecules that inhibit viral fusion and entry, including inhibitors of influenza [29] and the arenavirus, junin [24]. These compounds function by binding to the fusion protein and inhibiting virus–cell fusion [24], [29]. However, small molecules are not the only possible approach to entry inhibition and may not be ideally suited for interacting with a large conformationally fluid interface between the hydrophobic surfaces that we hypothesize to be critical for inhibition of entry. Larger molecules, such as peptides or other polymers that interact with the hydrophobic membrane–protein interfaces, may also be well suited to be entry inhibitors. For example, it was shown that viral fusion peptides, which are hydrophobic and bind to membranes [32], can inhibit viral entry [33], [34]. Perhaps most importantly, if inhibition occurs due to physical chemical interactions with a transient, hydrophobic protein–membrane interface, then broad-spectrum entry inhibition, without rapid selection for resistance, is possible. In this review we discuss the many known peptide entry inhibitors of enveloped viruses and show that they are, almost always, hydrophobic and/or amphipathic sequences with a propensity for membrane interface binding.
2.3. Enfuvirtide: a peptide entry inhibitor drug against HIV
HIV is a retrovirus that targets and depletes CD4 + T-cells [35], [36], [37] and is responsible for causing AIDS. Globally, tens of millions of people are infected with HIV, leading to 1–2 million deaths per year. The characteristic structural feature of HIV is the presence of protein spikes on its envelope surface [38], [39]. Originally existing as a single chain, gp160, the fusion protein of newly made viruses undergoes proteolytic processing in the Golgi complex by intracellular proteases, producing two subunits, gp120, the distal domain and, gp41, the membrane spanning domain, which is a Class I fusion protein [10]. The collective and co-operative actions of gp120 and gp41 are responsible for promoting viral genome entry into the host cell. The heavily glycosylated gp120 protein is responsible for the recognition and binding of the virus to the host cell, as it binds to the CD4 membrane protein and the co-receptors CCR5 and CXCR4. Binding triggers the conformational changes in gp41 that ultimately lead to fusion of the viral membrane with the host cell membrane [40], [41]. Like other Class I fusion proteins, gp41 is made of a hydrophobic fusion peptide on the N-terminus and an α-helical N-terminal helical heptad repeat (NHR) sequence which is connected to a C-terminal helical heptad repeat (CHR) by a loop domain [10]. Following the CHR is the aromatic/hydrophobic MPER domain, a membrane spanning helical anchor and a C-terminal domain which also contains membrane interacting sequences [20], [42]. During entry, the N-terminal fusion peptide of gp41 becomes exposed and likely interacts with the host cell membrane. At the same time, the N- and C-terminal heptad repeat sequences undergo a major transition from the native state to a more extended helical conformation. The heptad repeat domains drive the assembly of an elongated 6-helix bundle in the trimer. These conformational transitions allow the fusion peptide and other hydrophobic sequences, such as MPER, to destabilize the viral and cell membrane, and also bring the membranes into close proximity, ultimately resulting in mixing of the viral and cell membranes, creating a passage for the release of the viral genome in to the cell [37], [43].
In 1990 Qureshi et al. [44] showed that a peptide named CS3, derived from the loop/C-helix heptad repeat (CHR) domain (referred to as the “fusion initiation region” [45] of the then hypothetical structure of gp41), could block the entry of HIV into cells [38]. The discovery of CS3 catalyzed the search for other gp41-derived peptide entry inhibitors for HIV, which ultimately led to the discovery, development and licensure of enfuvirtide (also known as FUSEON or T20). Enfuvirtide is a peptide entry inhibitor for HIV that was approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for human use in 2003. This injectable antiviral peptide drug is an effective HIV therapeutic belonging to a unique class of entry inhibitor, adding to the already existing classes that include reverse transcriptase inhibitors and HIV protease inhibitors [46], [47].
Enfuvirtide is a 36-residue peptide that is derived from an amphipathic segment of the CHR region of gp41 including the C-terminal end of the CHR and a portion of the aromatic/amphipathic membrane proximal ectodomain region (MPER). The C-terminal half of enfuvirtide is rich with aromatic amino acids and is very hydrophobic. The whole sequence has a strong propensity to form an amphipathic α-helix, which is the structure it forms in the post-fusion 6-helix bundle of gp41. Inhibition of viral infection by enfuvirtide in vitro is through inhibition of entry [6], [46], [47], [48], [49], [50], [51], although the exact molecular mechanism is not entirely clear. Enfuvirtide may bind to multiple regions of gp120/gp41 [6], [46], [47], [48], [49], [50], [51]. Enfuvirtide was shown in one study to inhibit genome entry fusion at a step after some lipid mixing had taken place, but before opening of the fusion pore [52]. Enfuvirtide does not form a stable 6-helix bundle in the presence of NHR sequences, unlike some other CHR-derived peptides [48]. Direct evidence of strong, sequence-specific binding of enfuvirtide to gp41 has not been observed, thus it has been suggested that it interacts with transient fusion intermediate states of gp41. A slightly different variant, DP178, binds strongly to synthetic membranes and was suggested to inhibit fusion pore formation [52] via the effect of direct membrane interactions on gp41 structure and dynamics.
In any case, the success of enfuvirtide in the clinic against a difficult-to-treat, chronic virus demonstrates the potential utility of peptide entry inhibitors as viral therapeutics. In this review we discuss the many peptide entry inhibitors of enveloped viruses that have been identified and characterized since the discovery of enfuvirtide and connect them by hypothesizing an overlapping mechanism of action.
3. Identification of membrane-interacting sequences using the Wimley–White interfacial hydrophobicity scale
Prompted by the discovery and characterization of CS3, enfuvirtide and other hydrophobic/amphipathic peptide entry inhibitors, one of us (R.F.G.) has taken, in recent years, a novel, rational approach to successfully identify numerous peptide entry inhibitors against enveloped viruses with Class I, Class II and Class III fusion proteins. In this approach, potential peptide entry inhibitors are identified by scanning viral fusion proteins for sequences that have a propensity for membrane binding using the Wimley–White interfacial hydrophobicity scale [53], [54] (see next section), followed by direct experimental measurement of antiviral activities of the small number of candidate sequences to identify those with optimal antiviral activity. Later, we discuss each of these peptide entry inhibitor studies and show that this approach is effective, efficient and broadly generalizable.
3.1. The Wimley–White interfacial hydrophobicity scale (WWIHS)
The unique physical–chemical environment at the water–membrane interface drives a unique class of hydrophobic interaction that is dominated by the contribution of aromatic residues [53], [54], [55]. To identify and quantitate interfacial, membrane-binding sequences of peptides and proteins it is appropriate to use the Wimley–White interfacial hydrophobicity scale (WWIHS), an experimentally-determined free energy scale that represents the propensity for individual amino acids, in the context of peptide sequences, to partition from water into a phosphatidylcholine interface [53], [56]. Importantly, the WWIHS is based on whole-residue free energies, and thus has a true zero point that distinguishes peptide sequences that have a propensity to interact with membranes from those that do not. There are two ways that WWIHS scores have been used in the literature and in this review. i) WWIHS is used in the “hydropathy analysis” mode to calculate a sliding window hydrophobicity score along the sequence of a protein. This approach identifies segments of a protein with a propensity to interact with membrane interfaces. ii) WWIHS is used in the “totalizer” mode to calculate the overall interfacial hydrophobicity of a particular peptide sequence. This approach is an accurate predictor of membrane interaction propensity [57]. Here, we calculate WWIHS values by default at low pH (where aspartate and glutamate are uncharged and histidine is charged) to simulate the low pH environment of the endosome (pH 5.0–5.5) where most viral fusion takes place. In support of this assumption, we note that the pKa values of acidic groups on peptides [58], [59] and other molecules [60] are shifted into the range of 6.0 to 7.5 by environmental factors when they are bound to membranes. WWIHS values are calculated assuming random coil peptides partitioned into the bilayer interface. The values are thus minimum possible values, as ΔG can only get more favorable if peptide binding promotes an increase in secondary structure, as is often the case. The possible increase in ΔG with secondary structure (favoring membrane binding) has been estimated to be between 0.3 and 0.6 kcal/mol/residue [58], [59] and thus can be a very large effect.
The interfacial helical hydrophobic moment (iHHM) is another important physical–chemical factor that is relevant to membrane interactions and secondary structure formation by peptides bound to membrane interfaces. The iHHM quantifies the degree to which a peptide sequence would have segregated hydrophobic and hydrophilic faces if it folded into an α-helix [61]. A peptide with a large iHHM can interact strongly with membranes as a helix due to partitioning–folding coupling [58], [59] even when its WWIHS score is not positive overall. Although the exact relationship between partitioning and folding cannot be extracted quantitatively from iHHM, it provides useful landmarks as shown in Table 1 . In this review, we show WWIHS scores for all peptides and include the iHHM of peptide entry inhibitors when the value is large enough to potentially be a factor in membrane interaction (iHHM ≥ 2.0).
Table 1.
Significance of Wimley–White interfacial hydrophobicity scale (WWIHS) score and of interfacial helical hydrophobic moment (iHHM) score. Values have units of kcal/mol and are calculated using the MPEx web utility (http://blanco.biomol.uci.edu/mpex/). See supplemental information for a tutorial on using MPEx to calculate WWIHS and iHHM scores.
| Significance of Wimley–White interfacial hydrophobicity scale (WWIHS) score | |
|---|---|
| Score | Significance |
| > 8 | Very strong membrane partitioning. Free peptide is not readily observable. Binding is likely to drive secondary structure. |
| 6–8 | Strong membrane partitioning. Peptide is mostly bound. |
| 4–6 | Moderate membrane partitioning. Free and bound peptide are both observable. Positive interfacial helical hydrophobic moment (iHHM) will increase binding and helicity. |
| 2–4 | Weak membrane partitioning of random coil peptides. Free peptide is more abundant than bound peptide. Positive iHHM will increase binding and increase helicity of bound peptide. |
| 0–2 | Very weak membrane binding of random coil peptides. Bound peptide is difficult to detect. High helical hydrophobic moment (iHHM) can still drive strong binding. |
| < 0 | No propensity to partition into membranes as a random coil. High iHHM can still drive strong membrane binding as an α-helix. |
| Significance of interfacial helical hydrophobic moment (iHHM) | |
| Score | Significance |
| > 10 | Nearly ideal amphipathicity. Likely to bind strongly to membranes as an α-helix independent of WWIHS score. |
| 6–10 | Very strong amphipathicity. Strong membrane binding and high α-helical secondary structure content are likely. |
| 4–6 | Strong amphipathicity. Strong enhancement of binding is likely. Formation of some α-helical structure is likely. |
| 2–4 | Moderate amphipathicity. Enhancement of membrane binding and helicity is likely for sequences with positive WWIHS score. |
| 0–2 | Weak amphipathicity. Little effect on binding or structure, which will be determined by WWIHS score. |
Wimley–White interfacial hydrophobicity scale scores and helical hydrophobic moments (at neutral or low pH) can be calculated using the “Membrane Protein Explorer” (MPEX) web utility (http://blanco.biomol.uci.edu/mpex/). See supplemental information for a tutorial on using the MPEX utility to calculate WWIHS and iHHM.
3.2. Dengue virus and West Nile virus
The initial proof-of-concept that WWIHS-selected peptides can function as entry inhibitors was by Hrobowski et al. [31]. They identified peptides that successfully inhibited the flaviviruses dengue and West Nile, classified by the US National Institute of Allergy and Infectious Disease (NIAID) as a category A and category B priority pathogens, respectively. The global health crisis caused by dengue virus [62], [63], [64] demonstrates the critical importance of novel therapeutics to public health. Dengue is a vector-borne viral disease, transmitted to humans via infected Aedes aegypti mosquitoes in tropical and sub-tropical regions, with recent expansion to temperate climates. The global incidence of dengue has increased by 30-fold in the past five decades. Despite formidable efforts, no licensed vaccine or approved therapeutic options are available. Dengue infections vary in severity from asymptomatic, to febrile manifestations, to potentially life-threatening dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS). The estimated annual number of infections worldwide stands at 100 million, but this number may be underreported. The annual number of DHS or DSS cases is between 500,000–1 million, with an estimated 22,000 deaths, mainly among children.
West Nile virus is also a flavivirus. The emergence and spread of West Nile virus in North America is a particularly well-documented example of the potential for sudden emergence of public health risk posed by vector borne enveloped viruses. West Nile virus is endemic in Asia, the Middle East and Australia. However, in 1999 West Nile virus infections in humans were identified, and several deaths were reported in Queens, New York, probably after the virus was introduced by an infected bird or animal [65]. The virus spread very rapidly across North America, and then to Central America, rising to about 10,000 identified cases, and about 300 deaths in the US by 2003 [65]. Because the disease is usually mild, the actual number of cases may be as much as 100-fold larger than the reported number [65]. West Nile virus is now endemic across North America. Morbidity and mortality from West Nile virus do occur, usually resulting from viral encephalitis or meningitis [65]. This example illustrates the urgent need for broad-spectrum therapies against enveloped viruses because a future continental or global pandemic might be caused by an enveloped virus that causes high morbidity and mortality.
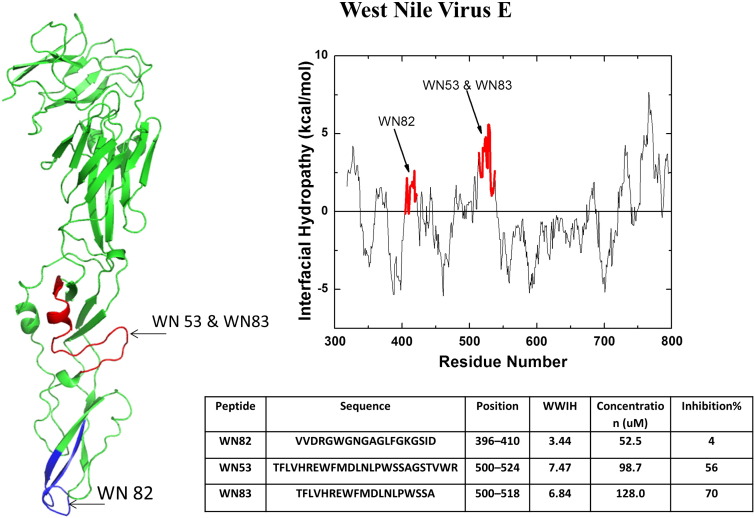
Both of these flaviviruses enter cells via receptor mediated endocytosis [66]. Once internalized, endosomal acidification occurs and the viral fusion protein E, a Class II fusion protein, undergoes a major structural rearrangement which is necessary for the initiation of fusion of the viral and cell membranes. To find putative entry inhibitors, Hrobowski et al. [31] analyzed the fusion proteins of dengue and West Nile virus using WWIHS. A few potential inhibitor candidates were selected based on positive WWIHS scores. Fig. 3, Fig. 4 show the active peptides selected from dengue and West Nile virus surface glycoproteins, respectively. Peptides with positive WWIHS were tested for their ability to inhibit viral plaque formation in vitro. The peptides DN59 from dengue virus (WWIHS = 7.0, iHHM = 5.4) and WN83 from West Nile virus (WWIHS = 7.6, iHHM = 2.3), which are very hydrophobic and amphipathic, proved to be the most effective against dengue and West Nile viruses, respectively with IC50 values around 10 μM [31].
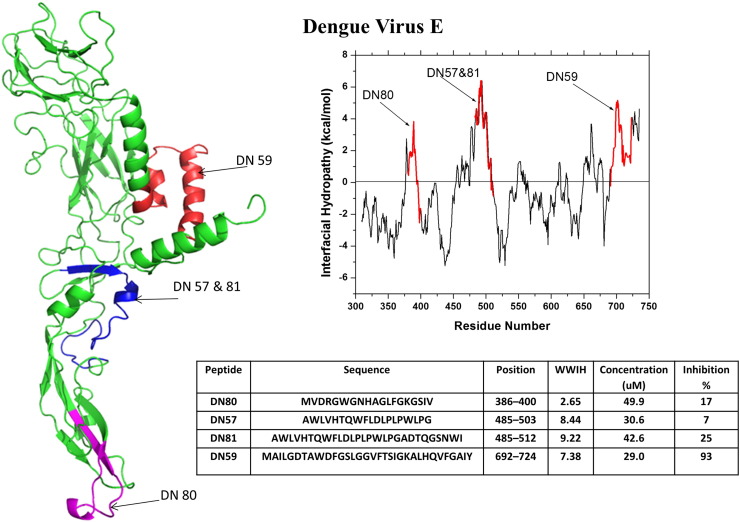
Fig. 3.
Discovery of dengue virus peptide entry inhibitors using WWIHS. The Class II fusion protein, E, of dengue was analyzed using WWIHS to identify putative membrane interacting sequences shown in color. A partial monomeric structure of E [PDB ID: 1OK8] is shown with the WWIHS positive sequences in color. These peptides were tested for inhibition of dengue virus in vitro [31] as shown.
Fig. 4.
Discovery of West Nile virus peptide entry inhibitors using WWIHS. The Class II fusion protein, E, was analyzed using WWIHS to identify putative membrane interacting sequences shown in color. A partial monomeric structure of E [PDB ID: 2I69] is shown with the WWIHS positive sequences in color. These peptides were tested for inhibition of West Nile virus in vitro [31].
DN59 is an aromatic/hydrophobic stem domain peptide that was further studied by Lok et al. [67] who observed a dose-dependent expulsion of the viral genome upon incubation of virus with peptide, indicating a direct virolytic effect. DN59 formed lesions in viral membranes that were observable by cryo transmission electron microscopy. In agreement with the direct virolytic effect, Lok et al. also showed that DN59 binds strongly to virus, to cells and to synthetic membranes and has potent membrane permeabilizing activity in synthetic lipid vesicles. On the other hand, unlike most vesicle-permeabilizing peptides, DN59 has no toxic effect on mammalian cells. Although the active peptides were initially expected to interact specifically with the fusion proteins, the observed non-specific membrane interactions are consistent with the physical chemistry-based identification of potential candidates by WWIHS [53], [67]. Schmidt et al. [68], [69] studied a series of closely related dengue virus stem domain peptides and showed that the peptides bind strongly to viruses prior to attachment and are carried into endosomes along with the virus. Interestingly DN59 and WN83 showed incomplete cross reactivity: DN59 is active against dengue and West Nile viruses, but WN83 is active only against West Nile virus, but not against dengue virus. We discuss below numerous similar cases of incomplete cross-species inhibition by peptide entry inhibitors.
3.3. Severe acute respiratory syndrome corona virus (SARS-CoV) and murine hepatitis virus
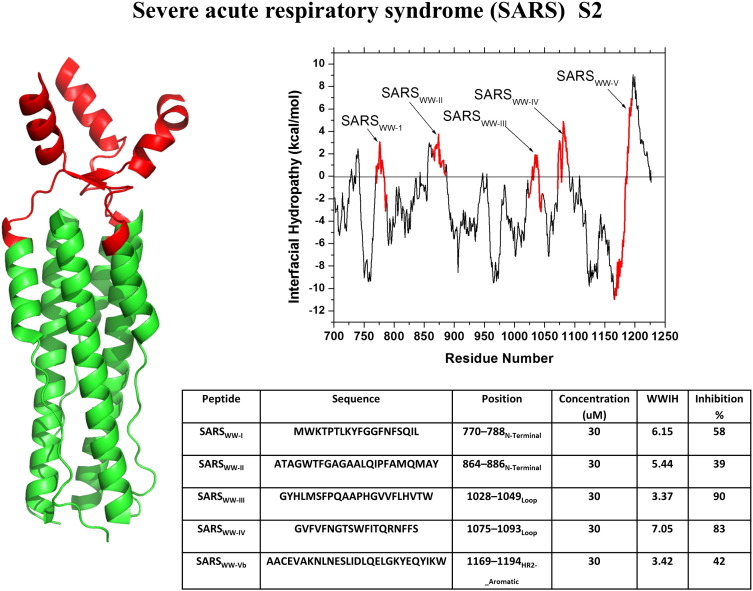
SARS-CoV is a coronavirus that predominantly targets the respiratory tract of humans with symptoms ranging from fever, headache, and cough, to fatal respiratory distress. After its initial outbreak in China, SARS CoV infected at least 8000 individuals in 2003 leading to ~ 700 fatalities [33], [70]. The high mortality rate of SARS elicited fears of a deadly global pandemic [33], [71]. Very recently, a related coronavirus with similarly high mortality, called the Middle East respiratory syndrome coronavirus, or MERS-CoV, has emerged [20], [72]. The rapid rise and spread of MERS once again raised fears of a global pandemic caused by a virus for which there are no known therapeutics. Coronaviruses are enveloped viruses that follow the classical pathway of entry and infection as other enveloped viruses. The surface glycoprotein or fusion protein of the virus, called the S protein, is a typical Class I fusion protein and is divided into two subunits. The S1 subunit is responsible for the binding of the virus to the host cell receptor and the S2 subunit drives fusion of the viral and host membrane [71], [73]. To identify inhibitory peptides against SARS CoV, Sainz et al. [71] analyzed the S2 subunit of the SARS CoV fusion protein using WWIHS. They identified five peptide sequences with high WWIHS scores: SARSWW-I–SARSWW-V (Fig. 5 ). The hydrophobic SARSWW-I (WWIHS = 6.2) sequence is the fusion peptide [22], peptides II, III and IV are found just outside of the heptad-repeat sequences between the N- and C-helical heptad repeat domains. SARSWW-V corresponds to the aromatic-rich juxtamembrane domain, analogous to the stem domain from which DN59 was obtained. Sainz et al. showed that the peptides from all five regions caused measurable inhibition of virus infection, and that SARSWW-III (WWIHS = 5.7) and SARSWW-IV (WWIHS = 7.1) showed the greatest inhibition in vitro with IC50 of 2–4 μM. SARSWW-III and SARSWW-IV were hypothesized to inhibit the conformation change required by the S2 protein during fusion, although the exact mechanism of action remains unknown. In the same paper, Sainz et al. showed that a peptide from the Class I murine hepatitis virus, corresponding to the same hydrophobic segment as SARSWW-IV inhibited MHV and SARS in vitro with IC50 = 4 μM. However, the equivalent SARS peptide did not inhibit MHV.
Fig. 5.
Discovery of SARS coronavirus peptide entry inhibitors using WWIHS. The Class I fusion protein, S2, of SARS was analyzed using WWIHS to identify putative membrane interacting sequences shown in color. A structure for the core trimer of the SARS S2 protein in the prefusion state is shown [PBD ID: 2BEQ]. Only one of the WWIHS positive peptides is included in the structure. These peptides were tested for inhibition of SARS virus in vitro [71].
By successfully using the Wimley–White interfacial hydrophobicity scale to identify peptide entry inhibitors against a second pair of viruses with a different class of fusion protein (Class I vs. Class II), Garry and colleagues demonstrated the power and generality of their approach [33], [71].
3.4. Human cytomegalovirus (HCMV)
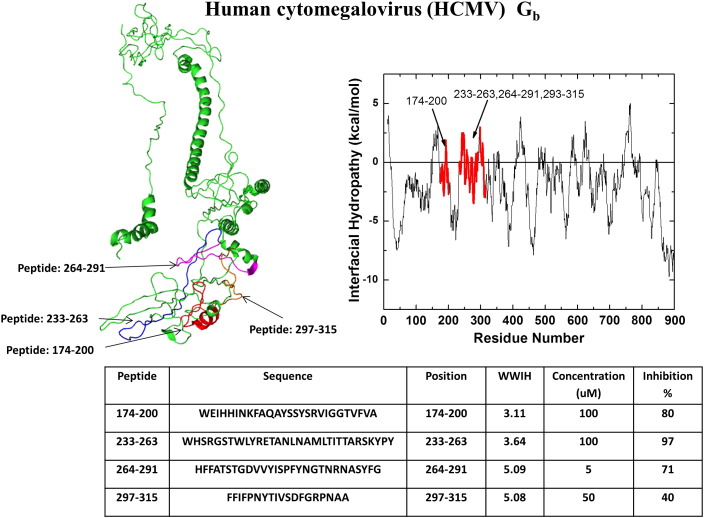
HCMV is a double stranded DNA virus and is a member of the herpes virus family. HCMV, which infects humans through an oral route, is widespread and often asymptomatic. However, infection just before or during pregnancy is associated with high morbidity and mortality for the fetus [74]. Similarly, its widespread abundance leads to high rates of infection and disease in immunocompromised patients, where it can lead to retinitis and blindness. HCMV has a Class III fusion protein in a complex composed of glycoproteins gB, gH and gL. Glycoprotein B (gB) has the primary role in binding and fusion of the virus with the host cell [75], [76]. HCMV can enter many different cell types using an array of envelope glycoproteins, in conjunction with gB. While fusion and entry often occur after endocytosis and acidification, there is also evidence of fusion at neutral pH [76]. Melnik et al. [77] identified nine peptides selected from gB based on their propensity for membrane partitioning as per the WWIHS (Fig. 6 ). Four of these peptides (WWIHS = 3-5) showed ≥ 50% inhibition of HCMV at 1–5 μM peptide [77]. The same peptides also inhibited herpes simplex virus, measles virus and a vesicular stomatitis pseudotype virus.
Fig. 6.
Discovery of human cytomegalovirus peptide entry inhibitors using WWIHS. The Class III fusion protein, Gb, of CMV was analyzed using WWIHS to identify putative membrane interacting sequences shown in color. No homologous three dimensional structure is available for this protein. These peptides were tested for inhibition of CMV virus in vitro [77].
These authors also tested the effect of the same peptide entry inhibitors conjugated to the polycationic tat cell penetrating peptide [78]. They observed that tat conjugation increased antiviral activity significantly against all viruses tested. Importantly, both pre-incubation of cells with tat-conjugated peptides and post-infection treatment provided significant protection. Peptides that were not conjugated to tat had no measurable inhibition when added 24 h post infection while tat conjugated peptides had IC50 values between 1 and 10 μM at 24 h. As tat-cargo conjugates are known to bind to cells and enter by endocytosis [79], this result suggests strongly that entry inhibition takes place in the endosomal compartments which can be readily accessed by tat-conjugated peptides. It is possible that many peptide entry inhibitors, by virtue of their propensity for membrane binding, enter endosomal pathways along with viruses.
The active peptides discovered by Melnik et al. have some overlap with peptides identified in a different study of the related herpes simplex virus. In that study, Akkarawongsa et al. [80] used an entirely different approach to identify entry inhibitor peptides from the Class III fusion protein of HSV (discussed in detail below). Those authors identified a small number of inhibitory peptides, some of which are highly analogous to the ones identified by Melnik et al., and which likewise have positive WWIHS scores.
3.5. Rift Valley fever virus (RVFV)
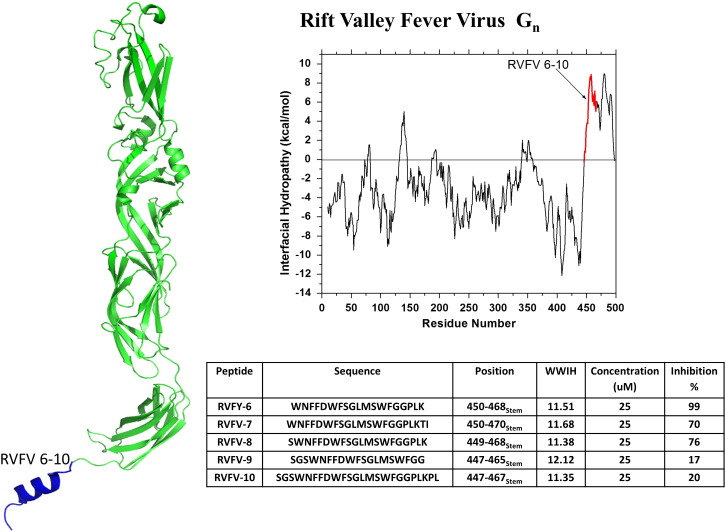
Rift Valley fever virus is an enveloped RNA virus that belongs to the Bunyaviridae family. RVFV infection, which is sometimes accompanied by severe hemorrhagic disease, can be fatal to both humans and livestock [81], [82], [83]. This mosquito-borne virus is endemic in parts of Africa and the Arabian peninsula and has emerged as a significant public health concern [81], [82], [83] and as a potential biothreat agent. As a consequence, RVFV is an NIAID category A priority pathogen, the highest possible priority. The Gn and Gc surface glycoproteins are responsible for the binding and entry of the virus to the host cell. Gn is a Class II fusion protein. The mechanism of fusion for RVFV has not been well studied, however it can be modeled using the well characterized fusion process of other Class II fusion proteins [82], [84]. To identify inhibitory peptides for RVFV, Koehler et al. [82] identified candidates from the sequence of the Gn fusion protein that have positive WWIHS scores (Fig. 7 ). They identified 5 regions of positive WWIHS score, which included the fusion loop peptide and the hydrophobic, aromatic juxtamembrane “stem” domain. Because the stem domain peptides had the best activity, the authors tested a series of truncation and translation variants. The most active peptide, RVFV-6, (WWIHS = 11.5) is very hydrophobic and showed ≥ 95% inhibition of RVFV at 25 μM concentration [82] with an IC50 around 5–10 μM. The authors showed that RVFV-6 binds directly to both cells and virus, and probably inhibits viral entry by inhibiting the fusion step. In support of the somewhat generalizable nature of membrane binding peptide entry inhibitors, RVFP-6 also inhibits an enveloped virus with a Class I fusion proteins (Ebola) and an enveloped virus with Class III fusion proteins (vesicular stomatitis virus). However, it did not inhibit any of three equine encephalitis viruses tested.
Fig. 7.
Discovery of Rift Valley fever virus peptide entry inhibitors using WWIHS. The Class II fusion protein, Gn, of RVFV was analyzed using WWIHS to identify putative membrane interacting sequences shown in color. Variants and truncations were tested to identify the most potent inhibitor. These peptides were tested for inhibition of RVFV and other viruses in vitro. A partial monomer structure of Gn [PDB ID: 4HJC] is shown with the WWIHS positive stem domain shown in color. The stem domain in blue was absent from the crystal structure of Gn. Here it is modeled as an α-helix [82].
Interestingly, one of the WWIHS-positive peptides from the RVFV Gn protein actually increased virus infection at 25 μM, while inhibiting it at 50 μM. A peptide-dependent increase in infection is an effect we also observed in a screen of various membrane binding peptides (unrelated to viral fusion proteins) against a lassa pseudotype virus (AR Hoffmann, WC Wimley and RF Garry, unpublished). Of 14 interfacial binding peptides tested in vitro, 9 inhibited lassa pseudotype virus at ≤ 50 μM, while five increased virus infection at ~ 10 μM, sometimes by more than two-fold, while inhibiting it at 50 μM. Given the hypothesis we are discussing here, it is perhaps not surprising that hydrophobic membrane-interacting peptides have the capacity to promote fusion and viral entry. However, the vast majority of peptides discussed in the literature are inhibitory.
3.6. Influenza virus
Influenza virus is a highly pathogenic enveloped virus with a single stranded RNA genome. It belongs to the Orthomyxoviridae family [85] and is responsible for annual seasonal illnesses as well as occasional global pandemics that occur when new strains arise by recombination. In terms of morbidity and mortality, one of the worst modern pandemics was the “Spanish Flu” outbreak in 1918, caused by an H1N1 strain of influenza A virus which killed as much as 1–3% of the world's population globally [86]. The recently identified H5N1 “avian” influenza virus has a very high mortality rate in humans but is not highly infectious [87]. On the other hand, the so-called “swine flu” or H1N1 pandemic of 2009 infected 10–20% of the global human population (including 30-–40% of school-aged children) in a single season [88]. The US Center for Disease Control (CDC) estimates that up to 575,000 people died [89] during this pandemic, which actually reflects a relatively low mortality rate, given the high infection rate. Every annual flu season raises the possibility of a highly infectious influenza pandemic that also has high mortality, highlighting the urgent need for broad spectrum antiviral therapies.
Influenza virus has two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), which play important roles in the entry of the virus into the host cell and the release of new virus particles from the host cell, respectively. HA was the first viral fusion protein to be characterized and is the archetypal trimeric Class I fusion protein. The precursor HA0 is activated by proteolytic cleavage to give two subunits HA1 and HA2. The globular HA1 is responsible for binding of the virus to sialic acid moieties on protein and lipids on the host cell surface. The membrane-anchored HA2 chain is responsible for promoting membrane fusion. Upon binding, the virus is internalized by clathrin mediated endocytosis. In the late endosomal stage, acidification occurs, triggering a large-scale conformational change in the native HA2 structure that exposes its N-terminal fusion peptide and other hydrophobic sequences and brings the membranes into close proximity for fusion [23], [90], [91].
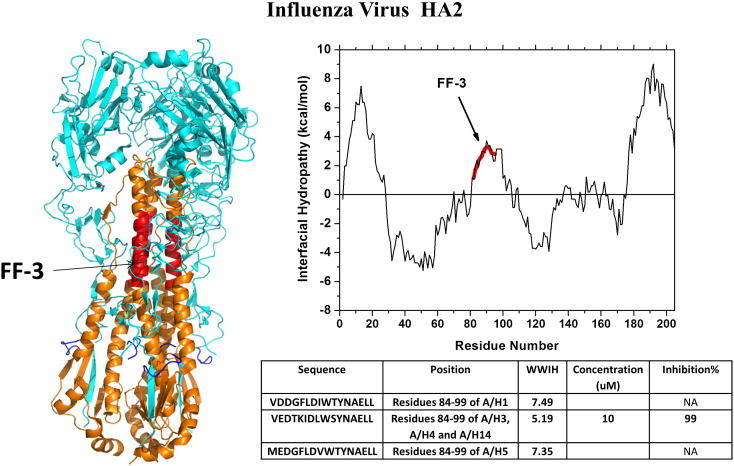
Garry, Wilson and colleagues analyzed the HA2 fusion protein of different strains of influenza A and discovered candidate entry inhibitors that showed robust inhibition in viral plaque assays [92]. The best inhibitory sequences were from the “fusion initiation region” (FIR) of HA2, comprising a hydrophobic segment of the N-helical heptad repeat (NHR) domain of HA2. Several FIR-derived peptides from different strains of influenza were characterized, and the most active 16-residue peptide, named FF-3, was identified. In Fig. 8 we show the peptide sequence from several strains of virus. FF-3, from the H3 strain of influenza, (WWIHS = 5.2) showed the maximum inhibition in viral plaque assays, inhibiting multiple strains of influenza A and B in vivo with IC50 values ≤ 1 μM. FF-3 is currently in phase 1 human clinical trials [7], [8]. Mechanism of action studies suggests that FF-3 is an entry inhibitor despite the fact that it does not interact measurably with influenza hemagglutinin at neutral or low pH (H Badani, RW Wilson, RF Garry and WC Wimley, unpublished).
Fig. 8.
Discovery of influenza virus peptide entry inhibitors using WWIHS. Hemagglutinin, the Class I fusion protein of influenza, was analyzed using WWIHS to identify putative membrane interacting sequences shown in color. A partial structure of an HA1 and HA2 trimer in the prefusion state [PDB ID: 1RD8] is shown with the active WWIHS positive sequence in color. The example shown is for an H3 variant of hemagglutinin. Variants and truncations were tested to identify the most potent inhibitor, FF-3. These peptides were tested for inhibition of influenza in vitro.
3.7. Pichinde virus
Pichinde virus is a model arenavirus, a family that includes multiple NIAID Category A priority pathogens, including lassa, junin and others. The only therapeutic used for arenaviruses is the nucleoside analog ribavirin, which is an off-label use and is not optimal. Thus, as with most classes of enveloped viruses, new classes of inhibitors against arenaviruses are desperately needed. Arenaviruses have envelope glycoprotein complexes comprised of three protein chains: the receptor binding domain, GP1; the Class I fusion protein, GP2; and an unusual “stable signal peptide” (SSP) domain comprised of two transmembrane helices [25] connected by a loop.
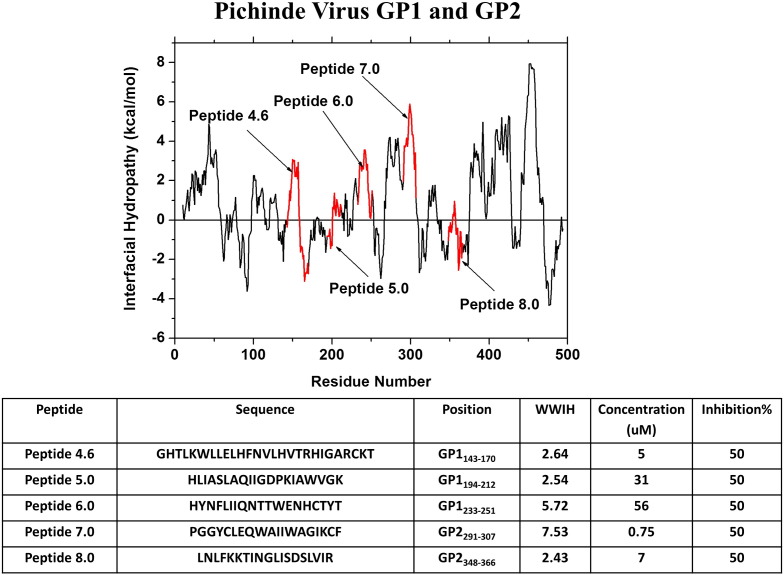
Spence et al. [93] examined the sequences of GP1 and GP2 of pichinde virus using WWIHS and identified 12 peptides from regions with positive WWIHS scores (Fig. 9 ). Five of the peptides had inhibitory activity against pichinde in vitro at concentrations < 100 μM. The most active peptide, corresponds to the fusion peptide of GP2. This peptide, PGGYALEQWAIIWAGIKIF, (WWIHS = 7.2 iHHM = 2.8) is very hydrophobic and inhibits pichinde with IC50 = 0.75 μM [93]. A second highly active peptide AVP-p, LNLFKKTINGLISDSLVIR, (WWIHS = 2.4, iHHM = 3.7) is a somewhat hydrophobic sequence from the N-helix heptad repeat (NHR) domain of GP2 with a strong propensity to form an amphipathic helix (Fig. 8). AVP-p inhibits pichinde with IC50 = 7 μM, and similarly inhibits pseudotype viruses with the envelope glycoproteins of the arenaviruses lassa, junin and machupo. At 7 μM, it did not inhibit vesicular stomatitis, herpes simplex or measles viruses, which belong to other families.
Fig. 9.
Discovery of pichinde virus peptide entry inhibitors using WWIHS. The GP1 and GP2 proteins of the fusion protein complex of pichinde were analyzed using WWIHS to identify putative membrane interacting sequence. No pichinde GP structure is currently available. These sequences were tested to identify the most potent inhibitor against pichinde in vitro [93].
Both of these inhibitory peptides interact directly with synthetic and viral membranes. Interestingly, AVP-p significantly changes the fluidity and organization of the lipids, but it does not bind stably or measurably to the GP2 fusion protein. Despite not interacting with GP2 directly, AVP-p, by cryoTEM and biochemical assays, prematurely triggers the fusion-competent state of GP2, even at neutral pH. Rather than promote infectivity, premature fusion competence is inhibitory, consistent with the proposed critical role of the timing of events for productive fusion. An important conclusion from this work is that a peptide that interacts directly with viral membranes can change their physical properties in a way that affects the structure and function of the fusion protein. A similar conclusion was drawn about the mechanism of action of DP178 against HIV [52]. Because the same experiments have not been done with most other peptide entry inhibitors described here, it remains possible that the mechanism of AVP-p is shared by other peptide entry inhibitors.
3.8. Hepatitis C virus
Hepatitis C virus is an enveloped RNA virus of the flavivirus family that is transmitted by blood-to-blood contact such as by shared needles in iv drug use or improperly sterilized medical equipment. Hepatitis C virus persists chronically in many infected people, frequently leading to hepatitis and liver cancer, both debilitating and costly diseases with high mortality. In fact, deaths from HCV in the U.S. have recently surpassed those caused by HIV [94]. At least several hundred million people are infected with HCV worldwide [94], [95].
HCV has two envelope glycoproteins, E1 and E2, each with its own transmembrane helical anchor. They form a non-covalent dimer in the viral envelope. Their individual roles in cell surface binding and fusion are still not entirely clear. While it had been proposed that E2 is the fusion protein and that it has structural homology to Class II fusion proteins, a recent crystal structure [96] showed that the E2 structure is not homologous to any known class of viral fusion protein. Instead, bioinformatics analyses of the HCV E1 protein suggest that it is a truncated Class II fusion protein (Sabahi A., Dash S, Garry CE, Prabhu, R., Haislip, A.M., Uprichard SL, Wimley WC, McKeating, JA and Garry, RF, in preparation).
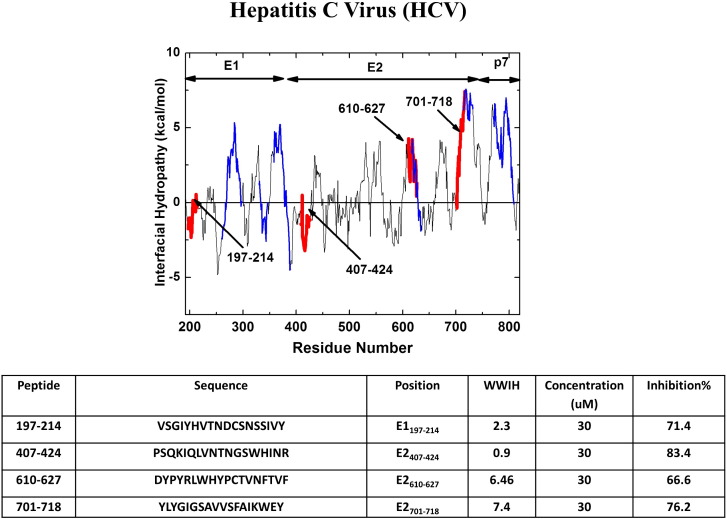
To identify HCV-inhibiting peptides, Sabahi [34] tested overlapping 18-residue peptides from E1 and E2, and also from the viroporin-like p7 protein, for liposome disruption, liposome fusion and viral inhibition. Peptides from five different segments of E1, E2 and p7 showed significant activity against liposomes. All five of these regions correspond to regions of positive WWIHS scores (Fig. 10 ) and include the putative fusion peptide of E1, and the aromatic juxtamembrane stem-like domains of both E1 and E2. A sequence that had been proposed to be the fusion peptide of E2 [97] was not active against liposomes and does not have a positive WWIHS score and therefore is very unlikely to be a fusion peptide [32].
Fig. 10.
Discovery of hepatitis C virus peptide entry inhibitors. The three protein fusion protein complex of HCV containing proteins E1, E2 and p7 was analyzed using WWIHS to identify putative membrane interacting sequences. The colored segments on the plot were peptides from a complete set of overlapping peptides that affected synthetic membranes (blue) or inhibited virus in vitro (red) [34]. The latter sequences are shown in the table.
When the same overlapping peptides were tested in an HCV pseudovirus inhibition assay, four peptides were identified with potent inhibition at μM concentrations. Two of these antiviral peptides correspond to regions of very high WWIHS score and overlap with liposome-active peptides (Fig. 10). The two other active peptides have small, positive WWIHS scores. The same peptides were not active against pseudotype viruses corresponding to murine leukemia virus and vesicular stomatitis virus. Regarding the overarching hypothesis of this review, the HCV data provided by Sabahi et al., are revealing. They directly demonstrate good overlap between predicted membrane binding, experimentally observed membrane perturbation, and antiviral activity. They also show that the overlap is incomplete, similar to the roughly 50% correlation between positive WWIHS scores and entry inhibition in the examples described above. It also agrees with the observations above that cross species inhibition is frequently observed, but is often incomplete. We conclude from these observations that while propensity for membrane binding is a major contributor to peptide entry inhibition, there are other, as yet unknown, factors that also contribute.
4. Other approaches to the discovery of peptide entry inhibitors
4.1. Structure-based identification of peptide entry inhibitors
Peptides such as enfuvirtide and its mimics, derived from the C-helix heptad repeat (CHR) domain of Class I fusion proteins, may be an example of peptide entry inhibitors that act by sequence specific interactions with fusion proteins. In addition to enfuvirtide (WWIHS = − 4.5; iHHM = 5.5 at pH 7) and the closely related DP178 (WWIHS = − 7.0; iHHM = 7.7), there are other peptide entry inhibitors for HIV that are derived from the CHR domain of gp41. One such peptide, called C34 [48], [52] (WWIHS = − 9.6; iHHM = 9.7) inhibits the entry of HIV into cells with IC50 values in the low μM range in vitro [49]. C34 and enfuvirtide have overlapping sequences, yet C34 reportedly has a sequence that enables it to interact specifically with a pocket in gp41, thereby preventing the conformational change required for fusion [48], [52].
In another study, He et al. [98] began with a peptide from the CHR region of gp41 that partially overlaps with enfuvirtide and C34, and subsequently engineered the sequence to be a more ideal amphipathic helix, assuming that this would improve pharmacological properties. In vitro, the peptide they designed, a 32-mer called CP32M, (WWIHS = − 8.7; iHHM = 14.4 at pH 7), is a very potent entry inhibitor against many strains of HIV, including enfuvirtide-resistant strains [98], [99], with IC50 values ranging from nM to pM. CP32M interacts with a protein construct comprised of the NHR sequences of gp41 to form a 6-helix bundle that mimics the native, post fusion 6-helix bundle. Like enfuvirtide and DP178 [48], [52], CP32M was shown to function by inhibition of fusion. Later, both CP32M and the shorter variants were shown by X-ray crystallography to interact with a protein construct containing the core three helix bundle of the N-terminal heptad repeat sequences of gp41. The inhibitors are helical and lie with their hydrophobic face in the hydrophobic groove between the NHR helices [99], [100], just as the CHR helices interact in the post fusion 6-helix bundle of native gp41.
In an additional study [100], the same authors designed overlapping 19–22 residue variants of CP32M (WWIHS − 8 to − 9, iHHM + 15 to + 14) that were shown to have nM IC50 values against all strains of HIV tested. While the WWIHS score for these peptides is negative at neutral pH, their interfacial helical hydrophobic moments are extremely large because the authors made sequence changes intended to create essentially ideal amphipathic helices. Thus, these peptides have a propensity to bind membranes and any other hydrophobic surface by folding into α-helices. In the context of the central hypothesis of this review, this study may be revealing. Some of the engineered sequences are highly active against multiple strains of HIV, yet bear little sequence resemblance to the native sequence of gp41. Compared to CP32M, the active sequences have more than half of their residues changed with dramatically non-conservative substitutions including M → E, I → K, L → K, K → E, I → E, E → I and E → K. One of these sequences was shown to fold into a 6-helix bundle with the NHR sequence of gp41, despite having few native residues in the protein–protein interface, suggesting that the interaction is driven mostly by the alignment of the hydrophobic surface of the amphipathic helix with the hydrophobic groove of the NHR bundle, instead of highly sequence-specific interactions.
Following the early success of enfuvirtide and related peptides against HIV, the same structure-based approach was used to identify entry inhibitors against other enveloped viruses with Class I fusion proteins. For example Rapaport et al. [101] described an inhibitory CHR peptide from the fusion protein of sendai virus. Similarly, Lambert et al. [102] tested enfuvirtide-like CHR sequences from respiratory syncytial virus, parainfluenza virus and measles virus, reporting species-specific inhibition of all three in vitro at sub μM concentrations. Lamb et al. [103] reported that CHR peptides from human T-lymphocyte leukemia virus and bovine leukemia virus inhibit virus entry in a species specific manner. Upon the discovery and identification of the SARS coronavirus in 2003, several groups [33], [71] identified CHR-based peptide entry inhibitors from the SARS Class I fusion protein, S2, using the enfuvirtide/gp41 system as a model. Similarly the discovery of the MERS coronavirus in 2012 has led to the description of peptide inhibitors based on the C-helix heptad repeat sequences of the MERS S2 protein [104]. These authors showed by X-ray crystallography that these peptide inhibitors of MERS co-crystallized as a 6-helix bundle when mixed with a construct containing the N-terminal helical heptad repeat core [104].
The available data suggests that these various CHR-derived peptides may inhibit virus entry by hydrophobic intermolecular interactions with the hydrophobic grooves in the NHR segments of the Class I fusion proteins. Whether sequence specific, native like interactions are essential for activity is not certain. In any case, this interaction, which may occur only with transient intermediate structures, is likely to interfere with the structure and function of the fusion protein during the critical events of fusion. While some CHR entry inhibitors bind stably and specifically to partial gp41 NHR sequences [10], others apparently do not. Despite evidence that these CHR peptides may act by sequence specific interactions, they have one universal feature in common that makes them similar to the other membrane interacting sequences described elsewhere in this review. They are highly amphipathic, with iHHM values between 5 and 15 kcal/mol. This property arises from the fact that, in the native structure, they pack into a very hydrophobic groove in the NHR three helix core that is present in all Class I fusion proteins. Peptides with very large iHHM scores are likely to fold into α-helices and interact with any available hydrophobic surface. Enfuvirtide analogs interact with membranes, thus supporting this hypothesis [48]. The idea that membrane binding may be important is also strengthened by the observation that an inactive enfuvirtide variant with several hydrophobic residues removed from the C-terminus can have its activity restored by the C-terminal addition of an octyl chain [100].
Porotto and colleagues [105] tested a set of “leash” domain peptides from influenza for inhibition. The influenza HA helical bundle contains a linear, non-helical sequence that is similar to the CHR sequences of other Class I fusion proteins in that they pack into grooves in the core N-helix bundle. Unmodified leash domain peptides, including the longest and most hydrophobic variant GTYDHDVYRDEALNNRFQIKGVELKSGYKDW (WWIHS = 2.1; iHHM = 4.7) were inactive against influenza virus in vitro. These authors then attached cholesterol to the amino termini of the leash peptides. This modification by a very hydrophobic cholesterol will effectively tether the peptide to the membrane. Cholesterol-modified leash peptides were found to be potent inhibitors, with EC50 values as low as 0.4 μM. These authors showed that entry inhibition is due to inhibition of fusion, and that multiple strains of influenza virus can be inhibited by cholesterol-conjugated leash peptides. They postulated that cholesterol conjugation insures that the peptide becomes co-encapsulated in the endosome with the virus and can act on the fusion protein when the pH decreases.
4.2. Accidental identification of peptide entry inhibitors
Our main hypothesis is also supported by multiple published reports of other peptides that are entry inhibitors of enveloped viruses. For example, Brandt and colleagues have described the accidental discovery of a peptide with broad-spectrum antiviral activity. This peptide, called EB, was based on a signal sequence, and was initially designed to facilitate cellular entry of an inhibitor of herpes simplex virus [28]. However the “carrier” peptide, EB, was found to be a potent virus inhibitor in pure form. This peptide has an N-terminal solubility sequence, RRKK followed by a hydrophobic, uncharged 16 residue sequence, AAVALLPAVLLALLAP, (WWIHS = 3.5) that has a propensity for membrane interaction. EB inhibits numerous unrelated viruses, including herpes simplex virus [28], influenza virus [106] and vaccinia (small pox) virus [107], consistent with inhibition that is dependent on peptide physical chemistry, rather than on sequence-specific interactions with viral fusion proteins. Inhibition of viral entry by EB occurs at an early stage, likely by inhibition of fusion [108], [109]. Brandt and colleagues showed that EB pre-incubation protected cells from virus, showing that the peptide binds to, or enters, cells [108], [109]. They also showed that EB causes aggregation of both viruses and various proteins, driven by the hydrophobic C-terminal tail of the peptide [108], [109].
In a remarkably similar narrative, Nicol et al. [110] describe the accidental discovery of a very similar peptide with anti-influenza activity. This 12-residue peptide was being studied for its anti-inflammatory properties, but was found to have direct antiviral activity. The initial sequence (called FluPep), WLVFFVIFYFFR (WWIHS = 10.5) is an extremely hydrophobic 12-residue peptide, with physical properties that are very similar to the 16 hydrophobes of EB described in the previous paragraph. The authors increased the apparent solubility of FluPep by adding the same four N-terminal basic residues (RRKK) as found in the peptide EB (above), yielding a peptide they called FluPep4. FluPep4 has IC50 values for inhibition of various influenza viruses in vitro between 135 nM and 30 pM [110]. The authors showed that FluPep4 inhibits virus binding to cells through a direct physical interaction with the virus. Because of its extreme hydrophobicity, FluPep4 will likely interact very strongly with itself and with all cellular and viral membranes.
Graham and colleagues have also reported the accidental discovery of a potent antiviral peptide. Initial information from a yeast 2-hybrid analysis indicated that the fusion protein of respiratory syncytial virus (RSV) could interact with RhoA, a small intracellular GTPase [111]. Following this discovery, the authors identified the linear peptide sequence of RhoA responsible for the interaction, ILMCFSIDSPDSLEN (WWIHS = 4.4). While a physiologically relevant interaction between the fusion protein of RSV and RhoA seemed unlikely, the peptide was nonetheless found to inhibit RSV infection in vitro [21]. The same peptide also inhibits HIV [21]. Subsequent investigation confirmed that viral inhibition was unrelated to any putative interaction with RhoA. Instead the RhoA-derived peptides, which are hydrophobic, interact directly with the virus, and by doing so block cellular binding and entry. In agreement with the hypothesis we are exploring in this review, these authors concluded that “the antiviral activity of RhoA-derived peptides is … a function of the peptides' ‘intrinsic biophysical properties’” and that “the combination of hydrophobicity and negative charge appears to be an effective pattern for many antiviral molecules that target RSV as well as many other enveloped viruses” [21].
4.3. Brute force approaches to the identification of peptide entry inhibitors
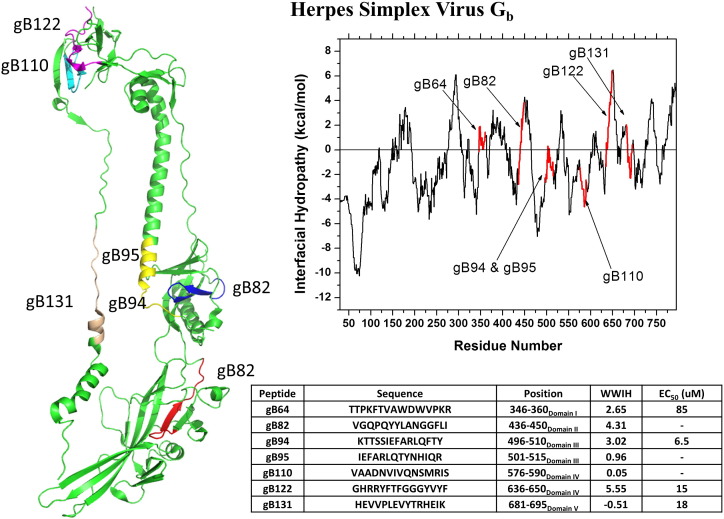
Akkarawongsa et al. generated a comprehensive set of overlapping 15-residue peptides representing the entire surface exposed portion of gB-1, the Class III fusion protein of herpes simplex virus [80]. When the whole set was tested for antiviral activity, seven peptides were identified that inhibit HSV. Three of the peptides were not tested further because they were insoluble (a common problem for the antiviral peptides discussed throughout this review). The three most active peptides had IC50 values less than 20 μM, in vitro. In Fig. 11 , we show the structure and WWIHS score of the gB-1 protein with the positions of the seven inhibitory peptides in red. In very strong support of our overarching hypothesis that membrane interface binding peptides can broadly inhibit viral entry, most of the seven antiviral peptides identified by this non-biased, brute force approach overlap with peaks of positive WWIHS score in the C-terminal portion of the fusion protein. There were also several positive WWIHS segments that did not show activity, a result that is consistent with the success rate reported above for intentionally identifying antiviral peptides using WWIHS. Thus, the efficiency of the approach described earlier of identifying candidates based on WWIHS and then testing the small number of candidates directly for activity is further validated by this work. The authors systematically measured, for each soluble antiviral peptide, direct viral inactivation, inhibition of cell binding, inhibition of entry, and host cell protection. They found that the peptides act mostly through entry inhibition, with a small contribution of direct viral inactivation and host cell protection. No inhibition of cell binding was observed.
Fig. 11.
WWIHS analysis of the putative fusion protein Gb of herpes simplex virus compared with inhibitory peptides identified by Akkarawongsa et al. [80] using a brute force approach based on in vitro testing of a comprehensive set of overlapping peptides from the Gb protein. A partial monomeric structure of Gb [PDB ID: 3NWF] is shown with the active sequences in color. The peptides tested are shown in the table. Those with no EC50 value were not tested due to insolubility.
Similarly, Cheng et al. [27] used a comprehensive set of 441 overlapping 18 residue peptides against the entire polyprotein of hepatitis C virus. They identified 11 peptides with significant antiviral activity, many of which have positive WWIHS scores. The most active inhibitory peptide, called C5A, is very hydrophobic and has a strong membrane binding propensity. It is an amphipathic helix from the membrane anchor domain of the nonstructural protein NS5A. C5A, SWLRDIWDWICEVLSDFK, (WWIHS = 9.0, iHHM = 5.2) interacts directly with the hepatitis C viral envelope and is virolytic with EC50 ≤ 1 μM. It blocks infection most actively if incubated with virus during/prior to the initial entry stage. Interestingly, C5A also eliminated virus in chronically infected cells, suggesting that it can enter cells. C5A made from l-amino acids and C5A made from d-amino acids had the same biological activity [112]. In a separate study C5A was shown to also have potent activity against HIV, and its activity was shown to be due to direct disruption of the viral envelope integrity [112]. In this study also, C5A was able to inhibit an already established HIV infection in vitro. However C5A does not inhibit vesicular stomatitis virus, nor does it inhibit a non-enveloped adenovirus. A less amphipathic but equally hydrophobic variant of C5A with two pairs of amino acids swapped in position is not active against HCV or HIV. C5A permeabilized lipid vesicles with high potency like a lytic toxin, which is consistent with its high WWIHS score and high amphipathicity. However, unlike non-specific lytic peptides (e.g. melittin from bee venom [113]) C5A does not disrupt cellular membranes and is not toxic against living mammalian cells at low concentration [112].
The hydrophobicity, secondary structure, helical hydrophobic moment and mechanism of action of C5A are all very similar to the stem-derived, dengue peptide, DN59, described above. Importantly, the d-amino acid enantiomer of C5A has the same activity as the l-enantiomer, showing that sequence specific peptide–protein interactions are not involved in antiviral activity. Li et al. [112] made a series of C5A variants with altered hydrophobicity and helicity. The anti HCV and anti-HIV activities of these variants in vitro were remarkably similar. Of the 14 variants studied against the two viruses, at least 9 had IC50 values between 0.5 and 5 μM, demonstrating, again, that entry inhibition is not a highly sequence dependent activity and that sequence changes do not abrogate antiviral activity if they do not diminish interfacial hydrophobicity or amphipathicity. Only variants of C5A with nonpolar to polar changes were inactive.
In an approach very similar to the discovery of C5A, Si et al. [114] tested all overlapping 18 amino acid peptides from four proteins thought to be important in the entry of hepatitis C virus: CD81, scavenger receptor B1, claudin 1 and occludin. They tested 113 peptides. Two overlapping peptides from claudin 1 were very potent inhibitors of HCV. These peptides, called CL-58 (MANAGLQLLGFILAFLGW, WWIHS = 8.1; iHHM = 2.5) and CL-59 (AFLGWIGAIVSTALPQWR, WWIHS = 6.0; iHHM = 4.3) are uncharged, very hydrophobic and amphipathic sequences. In fact, these sequences are part of the first membrane spanning α-helix in the claudin-1 protein, and thus are presumably buried in the membrane and not accessible to the virus during entry. These inhibitory sequences block HCV pseudovirus entry and whole virus entry with EC50 around 1–3 μM. They also suppress established HCV infections in cell culture. Inhibition of entry does not occur at the binding step, and the effect of peptide can be removed by washing the virus or the cells. A large number of translation and truncation variants were tested. More than half had similar activity to CL-58 and CL-59. While a scrambled peptide is mostly inactive, the d-amino acid CL-58 peptide is fully active, suggesting that sequence specific intermolecular interactions are not involved in the entry inhibition.
Bai et al. used phage display to identify three peptides that adhere to the West Nile virus fusion protein E [115]. Two of the three peptides that bound strongly to the fusion protein did not inhibit virus in vitro. However, one peptide, which the authors called p1, inhibited West Nile virus in vitro with IC50 = 67 μM. Peptide p1, DTRACDVIALLCHLNT, (WWIHS = 1.9) is more hydrophobic than the two inactive sequences and has a small, but positive WWIHS score. A series of truncation and insertion peptides were tested to find sequences with increased activity. The most hydrophobic of the variants tested, CDVIALLACHLNT, (WWIHS = 3.0), called P9, was also the most potent inhibitor of West Nile virus, with IC50 = 2.6 μM. The authors also tested the peptides against the related dengue virus, which has a fusion protein that is 47% identical to that of West Nile virus. Interestingly, against dengue, the opposite relative activities of P1 and P9 were found: P1 was highly active while P9 was not active. Finally, the authors showed that P9 inhibited West Nile virus in a mouse model with brain involvement, decreasing viral loads and increasing survival dramatically [115]. Remarkably, the peptide P9 was apparently able to cross the blood brain barrier, an important property for therapeutics intended to treat viruses that replicate in the brain.
Reil and colleagues [116], [117] hypothesized that the long recognized inhibition of HIV in patients co-infected with the common, asymptomatic GB virus C, which is related to hepatitis C, was due to a direct interaction between the surface glycoprotein E2 of the GB virus C and gp41, the Class I fusion protein of HIV. These authors examined peptides from the E2 protein and identified two overlapping E2 peptides, WDRGNVTLLCDCPNGPWVWV, (WWIHS = 6.5, iHHM = 3.5) and LCDCPNGPWVWVPAFCQAVG (WWIHS = 5.4, iHHM = 2.9) that inhibit HIV in vitro. These peptides are rich in aromatic residues and are both hydrophobic and amphipathic, giving them a strong propensity for membrane interaction. They inhibit HIV in cell culture with IC50 values of 2 and 0.2 μM, respectively. The authors showed that the two peptides inhibited HIV entry, and do so at an early step, but one that occurs after cell-surface binding, probably fusion. They also showed that this effect is due to an interaction with the HIV gp41 fusion protein that alters its conformational rearrangement, and may prevent the formation of the 6-helix bundle needed for fusion. These authors concluded that the E2 peptides specifically bind at the gp41–gp120 interface based on competitive binding assays and weak sequence similarity between the E2 peptides and HIV gp41. Based on the fact the physical chemistry of the E2 peptides is very similar to the many peptides discussed here, we hypothesize that their mechanism of action may include non-sequence specific interactions between the peptides and the hydrophobic surfaces of the fusion protein–membrane complex.
4.4. Entry inhibition by membrane-permeabilizing, cationic antimicrobial peptides
Cationic antimicrobial peptides constitute a collection of more than 1000 known peptides that are extraordinarily diverse in secondary and tertiary structure, but have similar antibacterial and antifungal activities. AMPs are found in all classes of life and are a critical part of the innate immune systems of vertebrates and invertebrates [118], [119], [120]. Most vertebrates, including humans, produce many different AMPs that have broad spectrum activity against many types of bacteria and unicellular fungi. The activity of AMPs against bacteria has been studied intensively since their discovery in the 1980s [121], [122]. While there are some variations, a unifying mechanism of antibacterial action has emerged from many studies [118], [123]: AMPs preferentially bind to the anionic plasma membranes of bacteria (both Gram positive and Gram negative) and, by virtue of their interfacial activity [123], disrupt the organization and continuity of bacterial membranes. The degree of membrane disruption increases sequentially [124], [125], [126] from loss of transmembrane potential within seconds, to leakage of small molecules, leakage of macromolecules and finally destruction of overall cellular architecture. The membrane disrupting nature of AMPs arises from the fact that they are amphipathic, with polar and hydrophobic surfaces, but that the hydrophobic surfaces are interrupted by polar/cationic residues. We have described how their “interfacial activity” is dependent on “imperfect amphipathicity” [123].
Antiviral activity has been observed, in vitro, for multiple AMPs and it has been proposed that they have evolved to have antiviral activity as well as antibacterial activity. For example, LL37 is a 37-residue cationic, amphipathic helical AMP that is released from neutrophil granules and epithelial cells and that has broad-spectrum activity against many bacteria and pathogenic fungi. Recent reports [127] have shown that LL37 also has broad-spectrum activity against influenza A viruses, in vitro, by a mechanism that is consistent with entry inhibition. The IC50 of LL37 is in the low μM range. LL37-dependent inhibition of virus-cell binding was not observed. Instead, like the anti-dengue peptide DN59 discussed above, LL37 caused a direct physical disruption of the viral membrane [127]. LL37 does not have a favorable WWIHS score, overall, but interacts with membranes because it folds into an α-helix with a very hydrophobic face. The interfacial helical hydrophobic moment, iHHM, of LL37 is 14.2 at neutral pH, which is an extremely high value (see Table 1).
Similarly, multiple classes of defensins, which are cyclic and/or disulfide-crosslinked cationic/hydrophobic peptides, have also been shown to have anti-viral activity against HIV [128], herpes simplex virus [129], influenza A virus [130], [131], and the SARS coronavirus [132]. Inhibition has been reported to arise from peptide-induced viral aggregation and direct virolytic effects. Doss et al. [131] reported a study of a collection of synthetic theta defensin variants (cyclic 18-residue β-sheet rich neutrophil peptides) from which they identified several sequences with IC50 values against influenza virus that are less than 1 μM. Entry inhibition by theta defensins required that the peptide–virus interaction occur before cell binding and was a result of direct peptide–virus interaction; the theta defensins apparently drive large-scale aggregation of viral particles [131].
5. A shared mechanism of action for peptide entry inhibitors?
In 2013 Kumar and colleagues published the “Antiviral Peptide Database” [133] which describes more than 600 antiviral peptides covering all possible mechanisms of action. A significant proportion of those antiviral peptides are entry inhibitors, the class we have discussed in this review. Most of the peptide entry inhibitors described here were initially expected to inhibit viral entry by providing sequence-specific competitive inhibition of intermolecular interactions, mostly involving the viral fusion proteins. However, in the many studies we examined, there is little evidence of highly sequence specific interactions, other than the evidence provided by some Class I, CHR-derived peptides mimicking enfuvirtide. Instead there is strong evidence of generic activity for most peptide entry inhibitors. Peptides that were tested against other enveloped viruses often show at least some cross-species inhibition, even against unrelated viruses. The d-enantiomers show activity, when tested, as do some truncations, insertions and other sequence variations. Yet, even without blocking specific protein–protein interactions, these many peptides effectively inhibit virus entry into cells in vitro with EC50 values from sub nM to low μM. In the few cases reported, inhibition also occurs in vivo.
The mechanisms of action of these peptide entry inhibitors have been studied in multiple laboratories. Some of the peptides have direct physical effects on the virus particles, causing either physical disruption of the lipid bilayer of the viral envelope (virolysis) or causing virus aggregation. Some have been shown to bind to and affect synthetic membranes. Some inhibitory peptides are reported to inhibit binding of viruses to cells, which has been attributed to direct peptide interactions with the virus particles. Other peptides seem to act at the level of membrane fusion, inhibiting fusion of bound virus to cells. In at least one case, fusion inhibition was due to premature initiation of the fusion protein conformation after peptide binding to the viral envelope membrane. Some peptide entry inhibitors bind directly to cells and interfere with virus binding and/or fusion at the cell surface, which may occur only after endocytosis of bound peptide and virus. Many studies suggest that multiple mechanisms are at play simultaneously.
Here we hypothesize that peptides with a propensity for membrane binding, due to interfacial hydrophobicity or amphipathicity, can interfere with enveloped virus entry by direct physical interaction with the hydrophobic surfaces, on both membranes and fusion proteins, that are critical for fusion and entry. In other words, the peptide entry inhibitors discussed here share overlapping mechanisms of action that are derived from their shared physical chemistry as follows: Essentially all the peptide entry inhibitors that we identified are at least somewhat hydrophobic and/or amphipathic with a propensity to bind to bilayer membrane interfaces and other hydrophobic surfaces. Many of them are very hydrophobic and/or highly amphipathic. Furthermore, many of them have acidic isoelectric points so that the low pH environment of late endosomes, where most viral cell fusion takes place, will trigger loss of charge and increased membrane binding. Except for anti-HIV peptides (because HIV fusion takes place at neutral pH) the WWIHS scores we report here reflect the low pH, membrane-bound state where aspartate and glutamate are effectively uncharged and histidine is charged. It is not known, at this time, whether membrane binding can directly affect viral fusion, or whether interaction with the fusion protein itself is an absolute requirement for entry inhibition. However we note, as did Rapaport et al. 20 years ago [101], that membrane binding of an inhibitory peptide will greatly increase the effective concentration of the peptide in the vicinity of the fusion protein thus interaction with membrane and interaction with the fusion protein may be effectively coupled.
Furthermore, we propose that each fusion protein–membrane complex exposes hydrophobic surfaces with somewhat variable detailed physical characteristics. These characteristics include the size, shape and underlying secondary structure of exposed hydrophobic patches, as well as the nature of the polar or charged residues that are near the hydrophobic surfaces. Thus, on the one hand similarities across all classes of fusion proteins give rise to frequent cross-species inhibition by peptide entry inhibitors. But on the other hand, variation in the physical details of the exposed hydrophobic surfaces causes some peptide entry inhibitors to be inactive against some enveloped viruses.
There may also be indirect ways in which interfacial binding peptides can affect viral entry. For example, the spike proteins of enveloped viruses are very crowded in the viral membrane (see Fig. 1) and may interact laterally, leading to the proposal that the conformational change that drives fusion is a cooperative event involving all of the spikes in a virus particle nearly simultaneously. If this is true, then cooperativity results from lateral interactions between proteins, perhaps including the TM domains. Thus, membrane physical properties could be critically important to the events that drive viral entry and it should be possible for peptides to interfere with the function of viral fusion proteins by changing the physical chemistry of the membrane itself by direct interaction. Evidence for this mechanism was discussed above for several peptide entry inhibitors.
Finally we note here the similarity in physical chemistry between membrane partitioning peptides and membrane translocating peptides [134]. Hydrophobic peptides that partition into membranes should also be able to cross cell membranes and enter cells. They may also cross endothelial layers in vivo, perhaps even including the blood brain barrier [115]. Thus peptides that function as entry inhibitors against viruses in vitro by the mechanism we propose here may inherently also have promising bioavailability characteristics in vivo due to membrane binding and translocation. This idea is supported by the very long half time of enfuvirtide in humans (tens of hours) compared to most peptides (minutes). In support of this possibility, we have shown directly that some peptide entry inhibitors can translocate across plasma membranes and enter cells (H. Badani and W.C. Wimley, not shown).
6. Implications and future directions
Effective therapeutics against enveloped viruses are rare. While a few drugs have been developed against HIV, influenza virus, and a few other viruses, these drugs are not ideal, and viruses often become resistant. For most enveloped viruses, there are no effective therapies. Thus entry inhibition represents an attractive and underutilized target in the search for new therapeutics. The hypothesis we have described here, that peptide entry inhibitors act by physical chemical interaction with hydrophobic surfaces exposed during fusion, suggests a novel approach to the discovery of entry inhibitors, one that focuses on the physical chemistry of putative entry inhibitors. Instead of the mostly brute force, structure based or accidental discovery in the literature to date, we propose that an intentional search for novel hydrophobic/amphipathic peptide entry inhibitors based on their physical chemistry, with an orthogonal selection for other critical properties such as solubility and pH sensitivity, could result in highly optimized and clinically effective peptide inhibitors of viral entry. Furthermore, since native-like, specific intermolecular interactions are probably not important, entry inhibitors can be made using protease resistant non-natural or d-amino acid peptides, using peptide mimetics such as peptide-nucleic acids or beta-peptides, or using polymers unrelated to peptides.
Acknowledgements
The authors are thankful for many insightful conversations with many colleagues and lab members, past and present. We are especially indebted to conversations with Russell Wilson, Peter Kulakosky, Tom Voss and William Gallaher.
Footnotes
This article is part of a Special Issue entitled: Interfacially Active Peptides and Proteins. Guest Editors: William C. Wimley and Kalina Hristova.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamem.2014.04.015.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Viral Hemorrhagic Fevers (VHFs) Centers for Disease Control and Prevention. N.p. June 18 2013. http://www.cdc.gov/vhf/virus-families/ Web.
- 2.Strauss J.H., Strauss E.G. 2nd edition. Elsevier; London, UK: 2008. Viruses and Human Disease. [Google Scholar]
- 3.Sato A.K., Viswanathan M., Kent R.B., Wood C.R. Therapeutic peptides: technological advances driving peptides into development. Curr. Opin. Biotechnol. 2006;17:638–642. doi: 10.1016/j.copbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov. Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Craik D.J., Fairlie D.P., Liras S., Price D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 6.Matthews T., Salgo M., Greenberg M., Chung J., DeMasi R., Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 7.Autoimmune Technologies, Safety Study of Flufirvitide-3 Nasal Spray in Healthy Subjects. 2012. http://clinicaltrials.gov/ct2/show/NCT01313962?term=Flufirvitide&rank=1 Web.
- 8.Autoimmune Technologies, Safety, Tolerability, and PK of Escalating Doses of Flufirvitide-3 Dry Powder for Inhalation in Healthy Subjects. 2013. http://clinicaltrials.gov/ct2/show/NCT01990846?term=Flufirvitide&rank=2 Web.
- 9.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 10.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 12.Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 13.Modis Y. Class II fusion proteins. Adv. Exp. Med. Biol. 2013;790:150–166. doi: 10.1007/978-1-4614-7651-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backovic M., Jardetzky T.S. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dessau M., Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1696–1701. doi: 10.1073/pnas.1217780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guu T.S.Y., Zheng W., Tao Y.J. Bunyavirus: structure and replication. Adv. Exp. Med. Biol. 2012;726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- 17.Stiasny K., Fritz R., Pangerl K., Heinz F.X. Molecular mechanisms of flavivirus membrane fusion. Amino Acids. 2011;41:1159–1163. doi: 10.1007/s00726-009-0370-4. [DOI] [PubMed] [Google Scholar]
- 18.Huarte N., Araujo A., Arranz R., Lorizate M., Quendler H., Kunert R., Valpuesta J.M., Nieva J. Recognition of membrane-bound fusion-peptide/MPER complexes by the HIV-1 neutralizing 2 F5 antibody: implications for anti-2 F5 immunogenicity. PLoS ONE. 2012;7:e52740. doi: 10.1371/journal.pone.0052740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyrychenko A., Freites J., He Alfredo J., Tobias J., Wimley Douglas C., Ladokhin William A.S. Structural plasticity in the topology of the membrane-interacting domain of HIV-1 gp41. Biophys. J. 2014;106:610–620. doi: 10.1016/j.bpj.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Sousa R., Reusken C., Koopmans M. MERS coronavirus: data gaps for laboratory preparedness. J. Clin. Virol. 2013;59:4–11. doi: 10.1016/j.jcv.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budge P.J., Graham B.S. Inhibition of respiratory syncytial virus by RhoA-derived peptides: implications for the development of improved antiviral agents targeting heparin-binding viruses. J. Antimicrob. Chemother. 2004;54:299–302. doi: 10.1093/jac/dkh355. [DOI] [PubMed] [Google Scholar]
- 22.Sainz B., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry. 2005;44:947–958. doi: 10.1021/bi048515g. [DOI] [PubMed] [Google Scholar]
- 23.Carr C.M., Kim P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 24.Thomas C.J., Casquilho-Gray H.E., York J., DeCamp D.L., Dai D., Petrilli E.B., Boger D.L., Slayden R.A., Amberg S.M., Sprang S.R., Nunberg J.H. A specific interaction of small molecule entry inhibitors with the envelope glycoprotein complex of the Junín hemorrhagic fever arenavirus. J. Biol. Chem. 2011;286:6192–6200. doi: 10.1074/jbc.M110.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunberg J.H., York J. The curious case of arenavirus entry, and its inhibition. Viruses. 2012;4:83–101. doi: 10.3390/v4010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maselko M., Ward C., Pastey M. A RhoA-derived peptide inhibits human immunodeficiency virus-1 entry in vitro. Curr. HIV Res. 2011;9:1–5. doi: 10.2174/157016211794582605. [DOI] [PubMed] [Google Scholar]
- 27.Cheng G., Montero A., Gastaminza P., Whitten-Bauer C., Wieland S.F., Isogawa M., Fredericksen B., Selvarajah S., Gallay P.A., Ghadiri M.R., Chisari F.V. A virocidal amphipathic {alpha}-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3088–3093. doi: 10.1073/pnas.0712380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones J.C., Turpin E.A., Bultmann H., Brandt C.R., Schultz-Cherry S. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J. Virol. 2006;80:11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu A., Antanasijevic A., Wang M., Li B., Mills D.M., Ames J.A., Nash P.J., Williams J.D., Peet N.P., Moir D.T., Prichard M.N., Keith K.A., Barnard D.L., Caffrey M., Rong L., Bowlin T.L. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza a virus fusion. J. Virol. 2014;88:1447–1460. doi: 10.1128/JVI.01225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper D.A., Lange J.M.A. Peptide inhibitors of virus–cell fusion: enfuvirtide as a case study in clinical discovery and development. Lancet Infect. Dis. 2004;4:426–436. doi: 10.1016/S1473-3099(04)01058-8. [DOI] [PubMed] [Google Scholar]
- 31.Hrobowski Y.M., Garry R.F., Michael S.F. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieva J.L., Suárez T. Hydrophobic-at-interface regions in viral fusion protein ectodomains. Biosci. Rep. 2000;20:519–533. doi: 10.1023/a:1010458904487. [DOI] [PubMed] [Google Scholar]
- 33.Sainz B., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J. Virol. 2005;79:7195–7206. doi: 10.1128/JVI.79.11.7195-7206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabahi A. Tulane University School of Medicine; 2008. Early Events in Hepatitis C Virus Infection: An Interplay of Viral Entry, Decay and Density. (PhD Dissertation) [Google Scholar]
- 35.Pantaleo G., Fauci A.S. New concepts in the immunopathogenesis of HIV infection. Annu. Rev. Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 36.Fauci A.S., Pantaleo G., Stanley S., Weissman D. Immunopathogenic mechanisms of HIV infection. Ann. Intern. Med. 1996;124:654–663. doi: 10.7326/0003-4819-124-7-199604010-00006. [DOI] [PubMed] [Google Scholar]
- 37.Shaw G.M., Hunter E. HIV transmission. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy J.A. HIV pathogenesis: 25 years of progress and persistent challenges. AIDS. 2009;23:147–160. doi: 10.1097/QAD.0b013e3283217f9f. [DOI] [PubMed] [Google Scholar]
- 39.Levy J.A. Virus-host interactions in HIV pathogenesis: directions for therapy. Adv. Dent. Res. 2011;23:13–18. doi: 10.1177/0022034511398874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardaci S., Soster M., Bussolino F., Marchiò S. The V1/V2 loop of HIV-1 gp120 is necessary for Tat binding and consequent modulation of virus entry. FEBS Lett. 2013;587:2943–2951. doi: 10.1016/j.febslet.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 41.Dragic T., Litwin V., Allaway G.P., Martin S.R., Huang Y., Nagashima K.A., Cayanan C., Maddon P.J., Koup R.A., Moore J.P., Paxton W.A. HIV-1 entry into CD4 + cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 42.Postler T.S., Desrosiers R.C. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J. Virol. 2013;87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaBranche C.C., Galasso G., Moore J.P., Bolognesi D.P., Hirsch M.S., Hammer S.M. HIV fusion and its inhibition. Antiviral Res. 2001;50:95–115. doi: 10.1016/s0166-3542(01)00130-9. [DOI] [PubMed] [Google Scholar]
- 44.Qureshi N.M., Coy D.H., Garry R.F., Henderson L.A. Characterization of a putative cellular receptor for HIV-1 transmembrane glycoprotein using synthetic peptides. AIDS. 1990;4:553–558. doi: 10.1097/00002030-199006000-00009. [DOI] [PubMed] [Google Scholar]
- 45.R. Garry, R. Wilson, US Patent: Fusion initiation region in RNA virus envelope proteins, WO/2005/044992, 2004.
- 46.Fung H.B., Guo Y. Enfuvirtide: a fusion inhibitor for the treatment of HIV infection. Clin. Ther. 2004;26:352–378. doi: 10.1016/s0149-2918(04)90032-x. [DOI] [PubMed] [Google Scholar]
- 47.LaBonte J., Lebbos J., Kirkpatrick P. Enfuvirtide. Nat. Rev. Drug Discov. 2003;2:345–346. doi: 10.1038/nrd1091. [DOI] [PubMed] [Google Scholar]
- 48.Liu S., Lu H., Niu J., Xu Y., Wu S., Jiang S. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 2005;280:11259–11273. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 49.Garg H., Viard M., Jacobs A., Blumenthal R. Targeting HIV-1 gp41-induced fusion and pathogenesis for anti-viral therapy. Curr. Top. Med. Chem. 2011;11:2947–2958. doi: 10.2174/156802611798808479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wild C., Oas T., McDanal C., Bolognesi D., Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild C., Greenwell T., Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell–cell fusion. AIDS Res. Hum. Retrovir. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 52.Kliger Y., Gallo S.A., Peisajovich S.G., Munoz-Barroso I., Avkin S., Blumenthal R., Shai Y. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 2001;276:1391–1397. doi: 10.1074/jbc.M004113200. [DOI] [PubMed] [Google Scholar]
- 53.Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 54.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 55.Yau W.M., Wimley W.C., Gawrisch K., White S.H. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 56.White S.H., Wimley W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 57.Snider C., Jayasinghe S., Hristova K., White S.H. MPEx: a tool for exploring membrane proteins. Protein Sci. 2009;18:2624–2628. doi: 10.1002/pro.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wimley W.C., Hristova K., Ladokhin A.S., Silvestro L., Axelsen P.H., White S.H. Folding of beta-sheet membrane proteins: a hydrophobic hexapeptide model. J. Mol. Biol. 1998;277:1091–1110. doi: 10.1006/jmbi.1998.1640. [DOI] [PubMed] [Google Scholar]
- 59.Ladokhin A.S., White S.H. Folding of amphipathic alpha-helices on membranes: energetics of helix formation by melittin. J. Mol. Biol. 1999;285:1363–1369. doi: 10.1006/jmbi.1998.2346. [DOI] [PubMed] [Google Scholar]
- 60.Small D.M., Cabral D.J., Cistola D.P., Parks J.S., Hamilton J.A. The ionization behavior of fatty acids and bile acids in micelles and membranes. Hepatology. 1984;4:77S–79S. doi: 10.1002/hep.1840040814. [DOI] [PubMed] [Google Scholar]
- 61.Segrest J.P., De Loof H., Dohlman J.G., Brouillette C.G., Anantharamaiah G.M. Amphipathic helix motif: classes and properties. Proteins. 1990;8:103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 62.World Health Organization; 2009. Dengue - Guidelines for diagnosis, treatment, prevention and control. [PubMed] [Google Scholar]
- 63.de Mendoza C., Altisent C., Aznar J.A., Batlle J., Soriano V. Emerging viral infections—a potential threat for blood supply in the 21st century. AIDS Rev. 2012;14:279–289. [PubMed] [Google Scholar]
- 64.Bäck A.T., Lundkvist A. Dengue viruses — an overview. Infect. Ecol. Epidemiol. 2013;3 doi: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.West Nile Virus Centers for Disease Control and Prevention. N.p. June 07 2013. http://www.cdc.gov/westnile/statsMaps/final.html Web.
- 66.Zhang Y., Zhang W., Ogata S., Clements D., Strauss J.H., Baker T.S., Kuhn R.J., Rossmann M.G. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lok S.-M., Costin J.M., Hrobowski Y.M., Hoffmann A.R., Rowe D.K., Kukkaro P., Holdaway H., Chipman P., Fontaine K.A., Holbrook M.R., Garry R.F., Kostyuchenko V., Wimley W.C., Isern S., Rossmann M.G., Michael S.F. Release of dengue virus genome induced by a peptide inhibitor. PLoS ONE. 2012;7:e50995. doi: 10.1371/journal.pone.0050995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt A.G., Yang P.L., Harrison S.C. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 2010;6:e1000851. doi: 10.1371/journal.ppat.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt A.G., Yang P.L., Harrison S.C. Peptide inhibitors of flavivirus entry derived from the E protein stem. J. Virol. 2010;84:12549–12554. doi: 10.1128/JVI.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sainz B., Mossel E.C., Gallaher W.R., Wimley W.C., Peters C.J., Wilson R.B., Garry R.F. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 2006;120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fisman D.N., Tuite A.R. The epidemiology of MERS-CoV. Lancet Infect. Dis. 2014;14:6–7. doi: 10.1016/S1473-3099(13)70283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taguchi F. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 1995;69:7260–7263. doi: 10.1128/jvi.69.11.7260-7263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swanson E.C., Schleiss M.R. Congenital cytomegalovirus infection: new prospects for prevention and therapy. Pediatr. Clin. North Am. 2013;60:335–349. doi: 10.1016/j.pcl.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boyle K.A., Compton T. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J. Virol. 1998;72:1826–1833. doi: 10.1128/jvi.72.3.1826-1833.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spear P.G., Longnecker R. Herpesvirus entry: an update. J. Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melnik L.I., Garry R.F., Morris C.A. Peptide inhibition of human cytomegalovirus infection. Virol. J. 2011;8:76. doi: 10.1186/1743-422X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Melnik L.I. Tulane Iniversity School of Medicine; 2013. Peptide inhibition of human cytomegalovirus infection. (PhD Dissertation) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt N., Mishra A., Lai G.H., Wong G.C.L. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 80.Akkarawongsa R, Pocaro NE, Case G, Kolb AW, Brandt CR. Multiple peptides homologous to herpes simplex virus type 1 glycoprotein B inhibit viral infection. Antimicrob. Agents Chemother. 2009;53:987–996. doi: 10.1128/AAC.00793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adam A.A., Karsany M.S., Adam I. Manifestations of severe Rift Valley fever in Sudan. Int. J. Infect. Dis. 2010;14:e179–e180. doi: 10.1016/j.ijid.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 82.Koehler J.W., Smith J.M., Ripoll D.R., Spik K.W., Taylor S.L., Badger C.V., Grant R.J., Ogg M.M., Wallqvist A., Guttieri M.C., Garry R.F., Schmaljohn C.S. A fusion-inhibiting peptide against Rift Valley fever virus inhibits multiple, diverse viruses. PLoS Negl. Trop. Dis. 2013;7:e2430. doi: 10.1371/journal.pntd.0002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madani T.A., Al-Mazrou Y.Y., Al-Jeffri M.H., Mishkhas A.A., Al-Rabeah A.M., Turkistani A.M., Al-Sayed M.O., Abodahish A.A., Khan A.S., Ksiazek T.G., Shobokshi O. Rift Valley fever epidemic in Saudi Arabia: epidemiological, clinical, and laboratory characteristics. Clin. Infect. Dis. 2003;37:1084–1092. doi: 10.1086/378747. [DOI] [PubMed] [Google Scholar]
- 84.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilton P. Spanish flu outdid WWI in number of lives claimed. CMAJ. 1993;148:2036–2037. [PMC free article] [PubMed] [Google Scholar]
- 86.Taubenberger J.K., Hultin J.V., Morens D.M. Discovery and characterization of the, pandemic influenza virus in historical context. Antivir. Ther. 1918;12(2007):581–591. [PMC free article] [PubMed] [Google Scholar]
- 87.Watanabe Y., Ibrahim M.S., Suzuki Y., Ikuta K. The changing nature of avian influenza A virus (H5N1) Trends Microbiol. 2012;20:11–20. doi: 10.1016/j.tim.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Kelly H., Peck H.A., Laurie K.L., Wu P., Nishiura H., Cowling B.J. The age-specific cumulative incidence of infection with pandemic influenza H1N1 2009 was similar in various countries prior to vaccination. PLoS ONE. 2011;6:e21828. doi: 10.1371/journal.pone.0021828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.First Global Estimates of 2009 H1N1 Pandemic Mortality Released by CDC-Led Collaboration Centers for Disease Control and Prevention. N.p. June 25 2012. http://www.cdc.gov/flu/spotlights/pandemic-global-estimates.htm Web.
- 90.Stegmann T., Delfino J.M., Richards F.M., Helenius A. The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J. Biol. Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 91.Stegmann T. Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion. Traffic. 2000;1:598–604. doi: 10.1034/j.1600-0854.2000.010803.x. [DOI] [PubMed] [Google Scholar]
- 92.R. Garry, R. Wilson, US Patent: Influenza inhibiting compositions and methods, US8222204 B2, 2009.
- 93.Spence J. Tulane University School of Medicine; 2013. Design and Characterization of Glycoprotein-derived Peptide Inhibitors of Arena Virus Infection. (PhD Dissertation) [Google Scholar]
- 94.Klevens R.M., Hu D.J., Jiles R., Holmberg S.D. Evolving epidemiology of hepatitis C virus in the United States. Clin. Infect. Dis. 2012;55(Suppl. 1):S3–S9. doi: 10.1093/cid/cis393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lauer G.M., Walker B.D. Hepatitis C virus infection. N. Engl. J. Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 96.Kong L., Giang E., Nieusma T., Kadam R.U., Cogburn K.E., Hua Y., Dai X., Stanfield R.L., Burton D.R., Ward A.B., Wilson I.A., Law M. Hepatitis C virus e2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krey T., d'Alayer J., Kikuti C.M., Saulnier A., Damier-Piolle L., Petitpas I., Johansson D.X., Tawar R.G., Baron B., Robert B., England P., Persson M.A., Martin A., Rey F.A. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.He Y., Cheng J., Lu H., Li J., Hu J., Qi Z., Liu Z., Jiang S., Dai Q. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao X., Chong H., Zhang C., Qiu Z., Qin B., Han R., Waltersperger S., Wang M., He Y., Cui S. Structural basis of potent and broad HIV-1 fusion inhibitor CP32M. J. Biol. Chem. 2012;287:26618–26629. doi: 10.1074/jbc.M112.381079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chong H., Yao X., Qiu Z., Sun J., Zhang M., Waltersperger S., Wang M., Liu S., Cui S., He Y. Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1. FASEB J. 2013;27:1203–1213. doi: 10.1096/fj.12-222547. [DOI] [PubMed] [Google Scholar]
- 101.Rapaport D., Ovadia M., Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lambert D.M., Barney S., Lambert A.L., Guthrie K., Medinas R., Davis D.E., Bucy T., Erickson J., Merutka G., Petteway S.R. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lamb D., Schüttelkopf A.W., van Aalten D.M.F., Brighty D.W. Highly specific inhibition of leukaemia virus membrane fusion by interaction of peptide antagonists with a conserved region of the coiled coil of envelope. Retrovirology. 2008;5:70. doi: 10.1186/1742-4690-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu L., Liu Q., Zhu Y., Chan K.-H., Qin L., Li Y., Wang Q., Chan J., Du L., Yu F., Ma C., Ye S., Yuen K., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee K.K., Pessi A., Gui L., Santoprete A., Talekar A., Moscona A., Porotto M. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J. Biol. Chem. 2011;286:42141–42149. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jones J.C., Settles E.W., Brandt C.R., Schultz-Cherry S. Identification of the minimal active sequence of an anti-influenza virus peptide. Antimicrob. Agents Chemother. 2011;55:1810–1813. doi: 10.1128/AAC.01428-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Altmann S.E., Jones J.C., Schultz-Cherry S., Brandt C.R. Inhibition of Vaccinia virus entry by a broad spectrum antiviral peptide. Virology. 2009;388:248–259. doi: 10.1016/j.virol.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones J.C., Settles E.W., Brandt C.R., Schultz-Cherry S. Virus aggregating peptide enhances the cell-mediated response to influenza virus vaccine. Vaccine. 2011;29:7696–7703. doi: 10.1016/j.vaccine.2011.07.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bultmann H., Girdaukas G., Kwon G.S., Brandt C.R. The virucidal EB peptide protects host cells from herpes simplex virus type 1 infection in the presence of serum albumin and aggregates proteins in a detergent-like manner. Antimicrob. Agents Chemother. 2010;54:4275–4289. doi: 10.1128/AAC.00495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nicol M.Q., Ligertwood Y., Bacon M.N., Dutia B.M., Nash A.A. A novel family of peptides with potent activity against influenza A viruses. J. Gen. Virol. 2012;93:980–986. doi: 10.1099/vir.0.038679-0. [DOI] [PubMed] [Google Scholar]
- 111.Pastey M.K., Crowe J.E., Graham B.S. RhoA interacts with the fusion glycoprotein of respiratory syncytial virus and facilitates virus-induced syncytium formation. J. Virol. 1999;73:7262–7270. doi: 10.1128/jvi.73.9.7262-7270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li G.-R., He L.-Y., Liu X.-Y., Liu A.-P., Huang Y.-B., Qiu C., Zhang X.-Y., Xu J.-Q., Yang W., Chen Y.-X. Rational design of peptides with anti-HCV/HIV activities and enhanced specificity. Chem. Biol. Drug Des. 2011;78:835–843. doi: 10.1111/j.1747-0285.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- 113.Tosteson M.T., Tosteson D.C. The sting. Melittin forms channels in lipid bilayers. Biophys. J. 1981;36:109–116. doi: 10.1016/S0006-3495(81)84719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Si Y., Liu S., Liu X., Jacobs J.L., Cheng M., Niu Y., Jin Q., Wang T., Yang W. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology. 2012;56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bai F., Town T., Pradhan D., Cox J., Ashish M. Ledizet, Anderson J.F., Flavell R.A., Krueger J.K., Koski R.A., Fikrig E. Antiviral peptides targeting the west nile virus envelope protein. J. Virol. 2007;81:2047–2055. doi: 10.1128/JVI.01840-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eissmann K., Mueller S., Sticht H., Jung S., Zou P., Jiang S., Gross A., Eichler J., Fleckenstein B., Reil H. HIV-1 fusion is blocked through binding of GB Virus C E2D peptides to the HIV-1 gp41 disulfide loop. PLoS ONE. 2013;8:e54452. doi: 10.1371/journal.pone.0054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Koedel Y., Eissmann K., Wend H., Fleckenstein B., Reil H. Peptides derived from a distinct region of GB virus C glycoprotein E2 mediate strain-specific HIV-1 entry inhibition. J. Virol. 2011;85:7037–7047. doi: 10.1128/JVI.02366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wimley W.C., Hristova K. Antimicrobial peptides: successes, challenges and unanswered questions. J. Membr. Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang L., Falla T.J. Antimicrobial peptides: therapeutic potential. Expert. Opin. Pharmacother. 2006;7:653–663. doi: 10.1517/14656566.7.6.653. [DOI] [PubMed] [Google Scholar]
- 120.Jenssen H., Hamill P., Hancock R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patterson-Delafield J., Szklarek D., Martinez R.J., Lehrer R.I. Microbicidal cationic proteins of rabbit alveolar macrophages: amino acid composition and functional attributes. Infect. Immun. 1981;31:723–731. doi: 10.1128/iai.31.2.723-731.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. U. S. A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wimley W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rathinakumar R., Wimley W.C. Biomolecular engineering by combinatorial design and high-throughput screening: small, soluble peptides that permeabilize membranes. J. Am. Chem. Soc. 2008;130:9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rangarajan N., Bakshi S., Weisshaar J.C. Localized permeabilization of E. coli membranes by the antimicrobial peptide Cecropin A. Biochemistry. 2013;52:6584–6594. doi: 10.1021/bi400785j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barns K.J., Weisshaar J.C. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim. Biophys. Acta. 2013;1828:1511–1520. doi: 10.1016/j.bbamem.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tripathi S., Tecle T., Verma A., Crouch E., White M., Hartshorn K.L. The human cathelicidin LL-37 inhibits influenza A viruses through a mechanism distinct from that of surfactant protein D or defensins. J. Gen. Virol. 2013;94:40–49. doi: 10.1099/vir.0.045013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tanabe H., Ouellette A.J., Cocco M.J., Robinson W.E. Differential effects on human immunodeficiency virus type 1 replication by alpha-defensins with comparable bactericidal activities. J. Virol. 2004;78:11622–11631. doi: 10.1128/JVI.78.21.11622-11631.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yasin B., Wang W., Pang M., Cheshenko N., Hong T., Waring A.J., Herold B.C., Wagar E.A., Lehrer R.I. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J. Virol. 2004;78:5147–5156. doi: 10.1128/JVI.78.10.5147-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doss M., White M.R., Tecle T., Gantz D., Crouch E.C., Jung G., Ruchala P., Waring A.J., Lehrer R.I., Hartshorn K.L. Interactions of alpha-, beta-, and theta-defensins with influenza A virus and surfactant protein D. J. Immunol. 2009;182:7878–7887. doi: 10.4049/jimmunol.0804049. [DOI] [PubMed] [Google Scholar]
- 131.Doss M., Ruchala P., Tecle T., Gantz D., Verma A., Hartshorn A., Crouch E.C., Luong H., Micewicz E.D., Lehrer R.I., Hartshorn K.L. Hapivirins and diprovirins: novel θ-defensin analogs with potent activity against influenza A virus. J. Immunol. 2012;188:2759–2768. doi: 10.4049/jimmunol.1101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lehrer R.I., Cole A.M., Selsted M.E. θ-Defensins: cyclic peptides with endless potential. J. Biol. Chem. 2012;287:27014–27019. doi: 10.1074/jbc.R112.346098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qureshi A., Thakur N., Tandon H., Kumar M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014;42:D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.He J., Kauffman W.B., Fuselier T., Naveen S.K., Voss T.G., Hristova K., Wimley W.C. Direct cytosolic delivery of polar cargo to cells by spontaneous membrane-translocating peptides. J. Biol. Chem. 2013;288:29974–29986. doi: 10.1074/jbc.M113.488312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Julien J.-P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P., Burton D.R., Sanders R.W., Moore J.P., Ward A.B., Wilson I.A. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Horne W.S., Johnson L.M., Ketas T.J., Klasse P.J., Lu M., Moore J.P., Gellman S.H. Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gordon L.M., Mobley P.W., Lee W., Eskandari S., Kaznessis Y.N., Sherman M.A., Waring A.J. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in lipid detergent and aqueous environments using 13C-enhanced Fourier transform infrared spectroscopy. Protein Sci. 2004;13:1012–1030. doi: 10.1110/ps.03407704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schibli D.J., Montelaro R.C., Vogel H.J. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 139.Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klein D.E., Choi J.L., Harrison S.C. Structure of a dengue virus envelope protein late-stage fusion intermediate. J. Virol. 2013;87:2287–2293. doi: 10.1128/JVI.02957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 142.Roche S., Bressanelli S., Rey F.A., Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 143.Zhang W., Chipman P.R., Corver J., Johnson P.R., Zhang Y., Mukhopadhyay S., Baker T.S., Strauss J.H., Rossman M.G., Kuhn R.J. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.