Abstract
Background and Aim:
Houseflies (Musca domestica) are synanthropic insects which serve as biological or mechanical vectors for spreading multidrug-resistant bacteria responsible for many infectious diseases. This study aimed to detect antibiotic-resistant bacteria from houseflies, and to examine their resistance genes.
Materials and Methods:
A total of 140 houseflies were captured using sterile nylon net from seven places of Mymensingh city, Bangladesh. Immediately after collection, flies were transferred to a sterile zipper bag and brought to microbiology laboratory within 1 h. Three bacterial species were isolated from houseflies, based on cultural and molecular tests. After that, the isolates were subjected to antimicrobial susceptibility testing against commonly used antibiotics, by the disk diffusion method. Finally, the detection of antibiotic resistance genes tetA, tetB, mcr-3, mecA, and mecC was performed by a polymerase chain reaction.
Results:
The most common isolates were Staphylococcus aureus (78.6%), Salmonella spp., (66.4%), and Escherichia coli (51.4%). These species of bacteria were recovered from 78.3% of isolates from the Mymensingh Medical College Hospital areas. Most of the isolates of the three bacterial species were resistant to erythromycin, tetracycline, penicillin and amoxicillin and were sensitive to ciprofloxacin, ceftriaxone, chloramphenicol, gentamicin, and azithromycin. Five antibiotic resistance genes of three bacteria were detected: tetA, tetB, mcr-3, and mecA were found in 37%, 20%, 20%, and 14% isolates, respectively, and no isolates were positive for mecC gene.
Conclusion:
S. aureus, Salmonella spp., and E. coli with genetically-mediated multiple antibiotic resistance are carried in houseflies in the Mymensingh region. Flies may, therefore, represent an important means of transmission of these antibiotic-resistant bacteria, with consequent risks to human and animal health.
Keywords: antibiotic resistance genes, antibiotic-resistant bacteria, houseflies
Introduction
Vector-borne diseases have become a global public health concern that is directly related to human health and with the propensity to significantly affect the economy of a country. Several vectors, such as houseflies, mosquitoes, ticks, sand flies, mites, lice, snails carry bacteria, viruses, and parasites, are capable of causing vector-borne illness in humans [1]. According to the WHO [1], major vector-borne diseases are responsible for around 17% of all infectious diseases globally per year, with a particularly high prevalence in tropical and subtropical areas.
The housefly (Musca domestica) of Muscidae family and Diptera order, is known as one of the most abundant and important vectors for devastating diseases of humans and animals [2]. It is frequently and persistently found in human and animal habitats, i.e., animal manure, bedding materials, household kitchens, restaurants, hospitals, dust bins, canteen, and decaying vegetable matter where they reproduce and develop their life cycle [3,4]. The fly usually carries pathogenic micro-organisms by attaching them in their mouth, wings, foot, and body surface, and also through regurgitation of gut contents [5]. Several species of bacteria have been recovered from houseflies, including Staphylococcus aureus, Salmonella spp., Escherichia coli, Shigella spp., Campylobacter spp., Pseudomonas spp., Vibrio spp., Bacillus spp., and Enterococcus faecalis [6-11]. Apart from annoying the animal or human, the presence of these bacteria in flies has been implicated in spreading associated diseases, such as enteric fever, anthrax, shigellosis, cholera, tuberculosis, diarrhea, from human to human, human to animal, and animal to human [12].
Antimicrobial resistance has become a serious issue over recent years throughout the world, and day by day, new resistance mechanisms are emerging and being discovered within the micro-organisms [13]. Antibiotics of the β-lactamase group are most commonly used in staphylococcal infection in humans and animals but, due to a high level of resistance to these and other antibiotics, it becomes increasingly difficult to treat such infections [14]. Methicillin-resistant S. aureus (MRSA), currently recognized as a “superbug,” is resistant to almost every available antibiotic [15]. MRSA is increasingly recognized as a problematic pathogen in environmental settings, carry antibiotic resistance genes such as mecA and mecC [16]; and MRSA with resistance genes such as mecA and nuc is frequently isolated from human, animal, environmental, and food samples in different parts of Bangladesh [17-20]. Similarly, colistin is frequently used in poultry in Bangladesh, although it is the reserved group of antibiotics [21]. Colistin resistance genes (mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5), which appear to be newly developed antibiotic resistance genes, are frequently found in bacteria in the environment [22] and, recently, have been recovered from various environmental samples such as poultry, houseflies, pond water, and sludge samples in Bangladesh [23,24]. Furthermore, in Bangladesh, bacteria with the tetracycline resistance genes tetA, tetB, tetC, and tetD are frequently isolated from the gut of humans, as well as from dairy farms and environmental settings [23,25].
Marshall et al. [16] first showed that houseflies can disseminate antibiotic resistance genes among animals. Houseflies can easily pick up antibiotic-resistant bacteria and transmit them to and between humans and animals [2]. Several recent studies have been undertaken to examine the role of houseflies in relation to the dissemination of antibiotic resistance [26-28].
Houseflies are very common insects and frequently found in Bangladesh as the weather permits favorable conditions for their survival [29]. Several studies have been conducted in Bangladesh related to isolation and antibiogram pattern of bacteria from flies [29-31].
However, there is very limited information on the presence of antibiotic resistance genes of bacteria carried by houseflies in Bangladesh. This study aimed to detect antibiotic-resistant bacteria and their resistance genes from housefly present in Mymensingh city, Bangladesh.
Materials and Methods
Ethical approval and informed consent
No ethical approval was required since the research does not contain any studies with human and animal subjects. However, verbal permission was taken from the concerned authorities during the collection of samples.
Sample collection and processing
A total of 140 houseflies were collected independently using sterile nylon nets from seven different locations, including households, restaurants, the university canteen, veterinary teaching hospital, poultry farms, dairy farms, and Mymensingh Medical College Hospital (MMCH) during the period from July to December 2018, 20 for each location. All the study locations were situated in and around Bangladesh Agricultural University campus and Mymensingh Sadar areas, Mymensingh, Bangladesh (24.45°N, 90.24°E). These places were selected on the basis of fly abundances, condition favorable for their survival, and persistent human movements. Immediately after collection, flies were transferred to a sterile zipper bag from the capture nylon net and brought to the microbiology laboratory within 1 h. Flies were then stored in these zipper bags at −20°C freezer until further processing. Then, flies were identified morphologically using stereo-microscope to ensure that they were M. domestica [32]. Each fly was placed, using sterile forceps, in a 15 ml Falcon tube containing phosphate-buffered saline (PBS) solution and vigorously agitated. Thereafter, 1 ml PBS solution was transferred in a test tube containing nutrient broth, after which it was incubated for 6-8 h at 37°C for enrichment.
Isolation and identification of bacteria
For isolation, a sterile loop was used to inoculate the culture broth onto mannitol salt (MS) agar, xylose lysine deoxycholate (XLD) agar, and eosin methylene blue (EMB) agar (HiMedia, Mumbai, India) culture media. After incubation at 37°C for 24-48 h, colonies that were golden yellow, black center, or a metallic sheen in MS, XLD, and EMB agar, respectively, were identified as presumptive S. aureus, Salmonella spp., and E. coli [33]. Further morphological identification was done through Gram’s staining and biochemical confirmation (sugar fermentation, methyl red, Voges–Proskauer, catalase, and coagulase test) [34]. Presumptive isolates were finally confirmed by polymerase chain reaction (PCR) using previously published genus-specific oligonucleotide primers [35-37].
Antimicrobial susceptibility test
All the isolates of the three bacteria under investigation were tested for antimicrobial susceptibility test by disk diffusion test [38], using ciprofloxacin (5 µg), ceftriaxone (30 µg), chloramphenicol (30 µg), gentamycin (10 µg), azithromycin (30 µg), amoxicillin (30 µg), nalidixic acid (30 µg), streptomycin (10 µg), erythromycin (5 µg), tetracycline (30 µg), penicillin (10 µg), and colistin (10 µg) (HiMedia, Mumbai, India). In all bacterial isolates, 0.5 McFarland suspensions were used as a standard to equalize the turbidity. The zone of growth inhibition in Mueller-Hinton agar media for each isolate was measured and compared with the standards as recommended by the Clinical and Laboratory Standards Institute [39].
Molecular detection of antibiotic resistance genes
Isolates of S. aureus showing phenotypic resistance to amoxicillin and penicillin, Salmonella spp. to tetracycline, and E. coli to colistin were further tested for the detection of antibiotic resistance genes, namely, mecA, mecC, and tetA, tetB, and mcr-3, respectively. Antibiotic resistance genes were determined by PCR using established primers [40-43], as illustrated in Table-1.
Table-1.
List of primers used in this study with sequences.
| Target genes | Primer sequences (5’-3’) | Amplicon size (bp) | References |
|---|---|---|---|
| Staphylococcus aureus | F-GGAGGAAGGTGGGGATGACG R-ATGGTGTGACGGGC GGTGTG |
241 | [35] |
| Salmonella spp. 16S rRNA | F-ACTGGCGTTATCCCTTTCTCTGGTG R-ATGTTGTCCTGCCCCTGGTAAGAGA |
496 | [36] |
| Escherichia coli 16S rRNA | F-AATTGAAGAGTTTGATCATG R-CTCTACGCATTTCACCGCTAC |
704 | [37] |
| mecA | F-AAAATCGATGGTAAAGGTTGG R-AGTTCTGGCACTACCGGATTTTGC |
533 | [40] |
| mecC | F-GAAAAAAAGGCTTAGAACGCCTC R-GAAGATCTTTTCCGTTTTCAGC |
138 | [41] |
| tetA | F-GAAGATCTTTTCCGTTTTCAGC R-CTGTCCGACAAGTTGCATGA |
577 | [42] |
| tetB | F-CCTCAGCTTCTCAACGCGTG R-GCACCTTGCTGATGACTCTT |
634 | [42] |
| mcr-3 | F- TTGGCACTGTATTTTGCATTT R- TTAACGAAATTGGCTGGAACA |
542 | [43] |
Results
Prevalence of bacteria based on molecular identification
A total of 110 (78.6%) isolates of S. aureus, 93 (66.4%) isolates of Salmonella spp., and 72 (51.4%) isolates of E. coli were recovered from 140 houseflies (Table-2 and Figure-1). Among the seven sampling areas, the highest percentages of bacteria were isolated from MMCH (78.3%) and the lowest from households’ flies (48.3%).
Table-2.
Number of bacterial isolates from houseflies.
| Sampling area (n=sample size) | Number of positive bacteria | Total (%) | ||
|---|---|---|---|---|
| Staphylococcus aureus | Salmonella spp. | Escherichia coli | ||
| Households (n=20) | 12 | 9 | 8 | 29 (48.3) |
| Restaurants (n=20) | 17 | 16 | 13 | 46 (76.7) |
| University Canteens (n=20) | 16 | 12 | 12 | 40 (66.7) |
| VTH (n=20) | 18 | 16 | 8 | 42 (70) |
| Poultry farms (n=20) | 12 | 18 | 8 | 40 (66.7) |
| Dairy farms (n=20) | 15 | 8 | 8 | 31 (51.7) |
| MMCH (n=20) | 18 | 14 | 15 | 47 (78.3) |
| Total (n=140) (%) | 110 (78.6) | 93 (66.4) | 72 (51.4) | |
VTH=Veterinary Teaching Hospital; MMCH=Mymensingh Medical College Hospital
Figure-1.
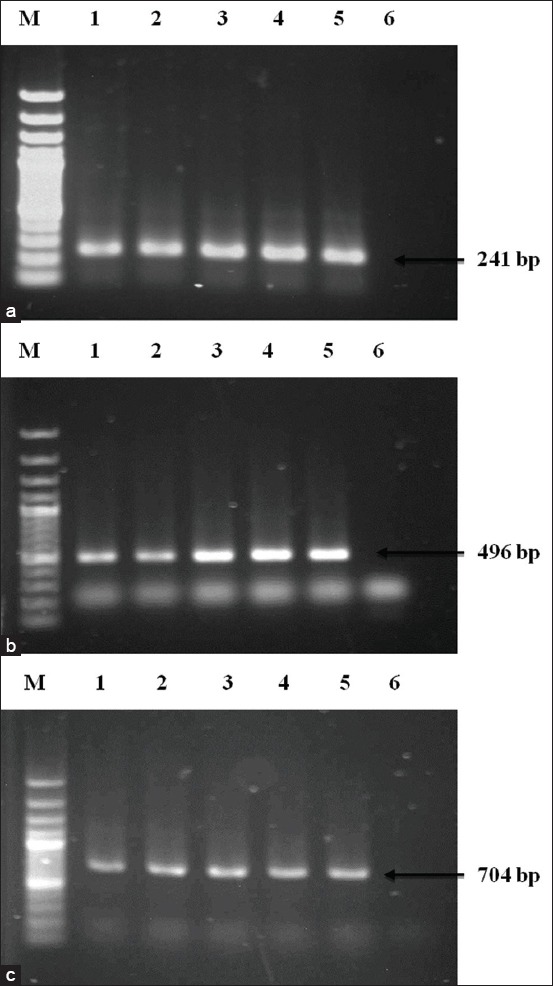
Polymerase chain reaction (PCR) amplification of 16S rRNA of Staphylococcus aureus, Salmonella spp., and Escherichia coli (a) PCR amplification of S. aureus. Lane M: 100 bp DNA Marker, 1-4: Representative S. aureus isolates, 5: Positive control, 6: Negative control. (b) PCR amplification of Salmonella spp. Lane M: 100 bp DNA Marker, 1-4: Representative Salmonella spp. isolates, 5: Positive control, 6: Negative control. (c) PCR amplification of E. coli. Lane M: 100 bp DNA Marker, 1-4: Representative E. coli isolates, 5: Positive control, 6: Negative control.
Antibiotic susceptibility test
Antibiotic resistance pattern of isolated S. aureus, Salmonella spp., and E. coli is shown in Tables-3-5. Isolates of S. aureus were all (100%) resistant to penicillin, followed by 94% resistant to amoxicillin and streptomycin, 93% to erythromycin, and 84% to tetracycline (Table-3). Conversely, 83% were sensitive to chloramphenicol, 80% to ceftriaxone, 78% to ciprofloxacin, and 70% to gentamycin, respectively. For Salmonella spp. (Table-4), isolates were resistant to erythromycin (97%), streptomycin (93%), tetracycline (90%), amoxicillin (88%), and nalidixic acid (62%), but were sensitive to chloramphenicol, ciprofloxacin, gentamycin, ceftriaxone, and azithromycin. Finally, isolates of E. coli were 97% resistant to erythromycin and tetracycline, 88% to streptomycin, and 85% to amoxicillin (Table-5). Fewer isolates were resistant to chloramphenicol (28%), gentamycin (28%), colistin (33%), and ciprofloxacin (38%), respectively.
Table-3.
Antibiotic resistance pattern of Staphylococcus aureus isolated from houseflies.
| Locations | Number of resistant isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CTR | C | GEN | AZM | AMX | NA | S | E | T | P | |
| Households | 2 | 3 | 0 | 2 | 7 | 12 | 10 | 12 | 12 | 11 | 12 |
| Restaurants | 1 | 2 | 1 | 2 | 4 | 16 | 10 | 12 | 16 | 17 | 17 |
| University Canteens | 2 | 1 | 3 | 4 | 8 | 16 | 12 | 16 | 16 | 12 | 16 |
| VTH | 6 | 4 | 2 | 4 | 4 | 16 | 10 | 16 | 16 | 12 | 18 |
| Poultry farms | 4 | 0 | 2 | 3 | 2 | 14 | 8 | 14 | 11 | 13 | 14 |
| Dairy farms | 3 | 5 | 2 | 6 | 6 | 13 | 7 | 15 | 13 | 9 | 15 |
| MMCH | 6 | 7 | 8 | 12 | 8 | 16 | 13 | 18 | 18 | 18 | 18 |
| Total (n=110) (%) | 24 (22) | 22 (20) | 18 (16) | 33 (30) | 39 (36) | 103 (94) | 70 (67) | 103 (94) | 102 (93) | 92 (84) | 110 (100) |
CIP=Ciprofloxacin, CTR=Ceftriaxone, C=Chloramphenicol, GEN=Gentamycin, AZM=Azithromycin, NA=Nalidixic acid, S=Streptomycin, E=Erythromycin, T=Tetracycline, P=Penicillin, VTH=Veterinary Teaching Hospital, MMCH=Mymensingh Medical College Hospital
Table-5.
Antibiotic resistance pattern of Escherichia coli isolated from houseflies.
| Locations | Number of resistant isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CTR | C | GEN | AZM | AMX | NA | S | E | T | CL | |
| Households | 1 | 2 | 0 | 1 | 2 | 8 | 2 | 6 | 6 | 8 | 0 |
| Restaurants | 2 | 1 | 0 | 3 | 8 | 10 | 5 | 13 | 13 | 13 | 4 |
| University Canteens | 4 | 3 | 1 | 3 | 2 | 10 | 6 | 9 | 12 | 12 | 3 |
| VTH | 2 | 2 | 3 | 2 | 4 | 8 | 4 | 8 | 8 | 6 | 3 |
| Poultry farms | 4 | 5 | 6 | 2 | 7 | 5 | 6 | 8 | 8 | 8 | 4 |
| Dairy farms | 4 | 3 | 4 | 3 | 2 | 8 | 4 | 4 | 8 | 8 | 4 |
| MMCH | 10 | 12 | 6 | 6 | 10 | 12 | 8 | 15 | 15 | 15 | 6 |
| Total (n=72) (%) | 27 (38) | 28 (39) | 20 (28) | 20 (28) | 35 (49) | 61 (85) | 34 (47) | 63 (88) | 70 (97) | 70 (97) | 24 (33) |
CIP=Ciprofloxacin, CTR=Ceftriaxone, C=Chloramphenicol, GEN=Gentamycin, AZM=Azithromycin, NA=Nalidixic acid, S=Streptomycin, E=Erythromycin, T=Tetracycline, CL=Colistin, VTH=Veterinary Teaching Hospital, MMCH=Mymensingh Medical College Hospital
Table-4.
Antibiotic resistance pattern of Salmonella spp. isolated from houseflies.
| Locations | Number of resistant isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CTR | C | GEN | AZM | AMX | NA | S | E | T | |
| Households | 1 | 2 | 1 | 1 | 3 | 7 | 4 | 6 | 6 | 7 |
| Restaurants | 2 | 4 | 0 | 2 | 4 | 16 | 2 | 16 | 16 | 16 |
| University Canteens | 1 | 2 | 2 | 3 | 4 | 12 | 6 | 10 | 12 | 9 |
| VTH | 4 | 3 | 2 | 4 | 2 | 12 | 10 | 16 | 16 | 12 |
| Poultry farms | 7 | 8 | 8 | 3 | 12 | 18 | 16 | 18 | 18 | 18 |
| Dairy farms | 2 | 3 | 2 | 4 | 4 | 8 | 8 | 6 | 8 | 8 |
| MMCH | 4 | 10 | 5 | 8 | 3 | 9 | 12 | 14 | 14 | 14 |
| Total (n=93) (%) | 21 (23) | 32 (34) | 20 (22) | 25 (27) | 32 (34) | 82 (88) | 58 (62) | 86 (93) | 90 (97) | 84 (90) |
CIP=Ciprofloxacin, CTR=Ceftriaxone, C=Chloramphenicol, GEN=Gentamycin, AZM=Azithromycin, NA=Nalidixic acid, S=Streptomycin, E=Erythromycin, T=Tetracycline, VTH=Veterinary Teaching Hospital, MMCH=Mymensingh Medical College Hospital
Distribution of antibiotic resistance genes
Among the 110 isolates of S. aureus, 103 that were resistant to both penicillin and amoxicillin and were further investigated for the presence of mecA and mecC genes. Of 103 resistant isolates, 14% were positive for mecA, but all isolates were negative for mecC. In the case of tetracycline-resistant Salmonella, tetA was the most prevalent (44%) resistance gene, compare to tetB (20%). Out of 24 colistin-resistant E. coli isolates, 5 (20%) were positive for mcr-3 gene (Table-6 and Figure-2).
Table-6.
Distribution of antibiotic resistance genes of Staphylococcus aureus, Salmonella spp. and E. coli.
| Locations | Number of penicillin and amoxicillin-resistant Staphylococcus aureus isolates (n=103) | Number of tetracycline-resistant Salmonella spp. isolates (n=84) | Number of colistin-resistant E. coli isolates (n=24) | ||
|---|---|---|---|---|---|
| mecA | mecC | tetA | tetB | mcr-3 | |
| Households | 0 | 0 | 3 | 1 | 0 |
| Restaurants | 1 | 0 | 4 | 2 | 0 |
| University Canteens | 1 | 0 | 3 | 0 | 0 |
| VTH | 2 | 0 | 6 | 2 | 1 |
| Poultry farms | 2 | 0 | 10 | 6 | 1 |
| Dairy farms | 3 | 0 | 3 | 2 | 1 |
| MMCH | 5 | 0 | 8 | 4 | 2 |
| Total (%) | 14 (14) | 0 (0) | 37 (44) | 17 (20) | 5 (20) |
VTH=Veterinary Teaching Hospital, MMCH=Mymensingh Medical College Hospital, E. coli=Escherichia coli
Figure-2.
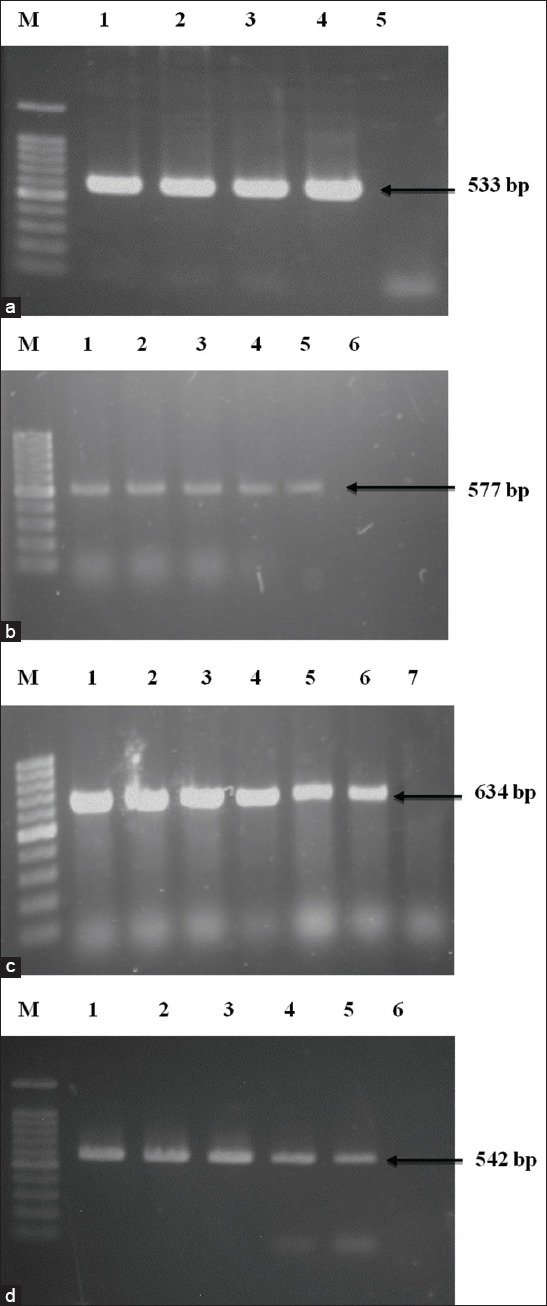
Polymerase chain reaction (PCR) amplification of antibiotic-resistant genes Staphylococcus aureus, Salmonella spp., and Escherichia coli (a) PCR amplification of mecA gene of penicillin and amoxicillin-resistant S. aureus. Lane M: 100 bp DNA Marker, 1-3: Representative S. aureus isolates, 4: Positive control, 5: Negative control. (b) PCR amplification of tetA gene of tetracycline-resistant Salmonella spp. Lane M: 100 bp DNA Marker, 1-4: Representative Salmonella spp. isolates, 5: Positive control, 6: Negative control. (c) PCR amplification of tetB gene of tetracycline-resistant Salmonella spp. Lane M: 100 bp DNA Marker, 1-5: Representative Salmonella spp. isolates, 6: Positive control, 7: Negative control. (d) PCR amplification of mcr3 gene of colistin-resistant E. coli. Lane M: 100 bp DNA Marker, 1-4: Representative E. coli isolates, 5: Positive control, 6: Negative control.
Discussion
The ecology and biology of the housefly (M. domestica) make it a potential mechanical vector for animal and human micro-organisms [4] and, indeed, it can transmit a wide range of pathogenic agents such as bacteria, virus, and fungi [44,45]. Thus, bacteria have frequently been isolated from the external surface of the fly body, as they pick up the pathogens through the mouth, legs, wings, and other body parts during the feeding process, which they carry back the pathogens to animal and human, where they complete their life cycle [46]. In the present study, three bacteria were specifically identified in houseflies, namely, S. aurues, Salmonella spp., and E. coli. Moreover, each fly sample carried at least one pathogen, presumably in association linked with poor hygiene and sanitation in the environment form where they are captured. Previous studies have similarly isolated Salmonella Typhimurium, E. coli, S. aureus, Klebsiella, and Shigella from external surfaces of houseflies circulating in a University Canteen of Dhaka, Bangladesh [29]. Seven places were included in this research from where fly samples were collected. Among them, the highest numbers of bacteria were recovered from the human hospital area (78.3%). The present findings are also comparable with the results of Nazari et al. [2] where higher numbers of bacterial strains were isolated from flies in hospital environments in Hamadan, Iran. In Brazil, Almeida et al. [47] also recovered Staphylococcus spp., Salmonella spp., and E. coli from both internal and external surfaces of houseflies collected from dairy farms. Ommi et al. [48] conducted a study in Iran on houseflies and recovered a significant number of Salmonella and Campylobacter spp. from flies from cattle farms, an animal hospital and a slaughterhouse, compared to chicken farms and human hospital. Ibrahim et al. [49] captured houseflies from a canteen, restaurants, and indoor food preparation premises and isolated several types of Gram-positive and Gram-negative bacteria. The frequency of E. coli, Salmonella spp., and Staphylococcus spp. isolations were 36.8%, 26.3%, and 42.9%, respectively, only slightly lower than in the present study. Ahmed et al. [50] also collected houseflies from different human and animal habitat and recovered E. coli, S. aureus, S. albus, Pseudomonas aeruginosa, Klebsiella, and Salmonella. Similar work was also done in Sokoto metropolis, Nigeria, isolated E. coli, Bacillus spp., Pseudomonas spp., Staphylococcus spp., Enterobacter spp., Proteus spp., Salmonella spp., and Klebsiella spp. [51]. Clearly, therefore, houseflies can act as a potential vector for transmitting these harmful micro-organisms through their external body surfaces and insufficient quantities [52] to cause human or animal infection.
At present, the world is facing the challenges of antibiotic-resistant bacteria, with resistance reported to most of the presently available antibiotics. It has been estimated that around 10 million people will die per year due to antimicrobial resistance by 2050 and could potentially be a significant existential threat to humans and animals [53]. Recently, several studies have been conducted on the housefly regarding antibiotic resistance and show that it plays a significant role in spreading antibiotic-resistant bacteria [2]. In the present study, micro-organisms isolated from houseflies were resistant to multiple antibiotics, and, as a matter of concern, most of the isolates displayed resistance to more than three antibiotics (namely, penicillin, amoxicillin, erythromycin and tetracycline) that are commonly prescribed in the study areas. From the seven studied areas, antibiotic-resistant bacteria were predominantly found in the human hospital areas, because this area is normally filled with sick patients who carry antibiotic-resistant bacteria, and where most of the biological products used in the hospital are simply discarded into dustbins and open places without proper treatment. Similarly, Nazari et al. [2] noted that organisms isolated from houseflies captured from the hospital environment showed higher resistance to antibiotics. In 2016, study of houseflies in the Dhaka district of Bangladesh found all the isolated bacteria were resistant to amoxicillin and cefixime and less resistant to chloramphenicol, gentamycin, and ciprofloxacin [29]. Another report in Chine shows that bacteria isolated from houseflies were resistant to amoxicillin, tetracycline, cefuroxime, and cephalothin [28]. Graham et al. [54] observed that flies captured from poultry farms carried a higher number of antibiotic-resistant bacteria and that houseflies collected from food restaurants carried a large number of antibiotic-resistant enterococci [55]. The findings of these previous studies agree with those of the present study that houseflies carry antibiotic-resistant bacteria [56-59]. An increased number of resistant bacterial strains in houseflies are almost certainly the result of frequent and haphazard use of antibiotics in human and food animals. The present results contribute to this corpus of information that houseflies carry bacteria that are resistant to multiple antibiotics, which might seriously affect human and animal health.
Antibiotic-resistant bacteria carry resistance genes that can genetically transmit to other bacteria [2]. In the present study, five antibiotic resistance genes were detected from three organisms, where tetA was the most common. The gene mcr-3, responsible for the colistin resistance, was found in 20% of E. coli isolates. Colistin is one of the reserve groups of antibiotics that are widely used in veterinary medicine and the agricultural sector both for treatment and prophylactic purposes. In Bangladesh, this drug has been frequently and persistently used by poultry practitioners, with the consequential appearance of resistant microorganisms. In Bangladesh, a recent study conducted by Sobur et al. [21] on poultry farms, houseflies, and pond water samples reported 8% colistin-resistant gene mcr-3. Reports are also available throughout the world for the development of colistin resistance genes. In China, Zhang et al. [60] also recorded the colistin resistance gene (mcr-1, mcr-2, and mcr-3) from houseflies and blowflies. Similarly, in Vietnam, Nguyen et al. [61] reported colistin resistance genes in poultry and pig farm. In the present study, MRSA gene mecA was found higher in number while no isolates were positive for mecC gene. In Bangladesh, penicillin, amoxicillin, and ampicillin are ubiquitously used in human and animal production sectors, as are methicillin and related antibiotics, with the inevitable consequence of the selection of resistant micro-organisms. Several studies in Bangladesh have looked for MRSA and their resistance genes in the dairy sector [17,62,63], but no study yet to be conducted in houseflies related to MRSA resistance genes in Bangladesh. Bacterial containing these antibiotic resistance genes can easily be picked up by houseflies from contaminated places and carried back to humans and animals, in which they can potentially cause severe infection. In addition, medical and veterinary practitioners should follow the guidelines while prescribing the antibiotics.
Conclusion
Houseflies collected from different places carry antibiotic-resistant bacteria and their resistance genes. Therefore, regular surveillance is necessary to fully understand the significance of pathogenic bacteria carried by flies. Flies from hospital areas are contaminated with pathogenic organisms that should be controlled by a hospital authority using proper administrative procedures. In households, a fly net should be used to stop the access of flies into the kitchen. Animal and human waste and other decaying materials should be disposed of properly. Good hygienic and sanitation practices should be mandated in all restaurants and university hall canteens to minimize the fly prevalence.
Authors’ Contributions
SA, AAMS, and ZFH carried out the research, analyzed the data, and wrote the initial draft of the manuscript. MTR contributed to manuscript writing. SS and MAK designed and supervised research work, revised, and finalized the manuscript. All authors read and approved the manuscript before submission.
Acknowledgments
The authors used the laboratory facilities of the Department of Microbiology and Hygiene, Bangladesh Agricultural University, Mymensingh, Bangladesh. The authors are deeply indebted to Dr. T. J. Parkinson, School of Veterinary Science, Massey University, New Zealand, for his useful discussions and critical reading of the manuscript. The research did not receive any specific grant from any funding agency in public, commercial, or not-for-profit organizations.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.World Health Organization. Vector-borne Diseases. 2017. Available from: https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases . Retrieved on 31-10-2017.
- 2.Nazari M, Mehrabi T, Hosseini S.M, Alikhani M.Y. Bacterial contamination of adult house flies (Musca domestica) and sensitivity of these bacteria to various antibiotics, captured from Hamadan city, Iran. J. Clin. Diagn. Res. 2017;11(4):DC04. doi: 10.7860/JCDR/2017/23939.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta A.K, Nayduch D, Verma P, Shah B, Ghate H.V, Patole M.S, Shouche Y.S. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.) FEMS Microbiol. Ecol. 2012;79(3):581–593. doi: 10.1111/j.1574-6941.2011.01248.x. [DOI] [PubMed] [Google Scholar]
- 4.Kassiri H, Akbarzadeh K, Ghaderi A. Isolation of pathogenic bacteria on the house fly Musca domestica L. (Diptera:Muscidae), body surface in Ahwaz hospitals, Southwestern Iran. Asian Pac. J. Trop. Biomed. 2012;2(2):S1116–S1119. [Google Scholar]
- 5.Ugbogu O.C, Nwachukwu N.C, Ogbuagu U.N. Isolation of Salmonella and Shigella species from house flies (Musca domestica L.) in Uturu, Nigeria. Afr. J. Biotechnol. 2006;5(11):1090–1091. [Google Scholar]
- 6.Greenberg B. In Biology and Disease Transmission. Princeton, NJ, USA: Princeton University Press; 1973. Flies and Disease. [Google Scholar]
- 7.Fotedar R, Banerjee U, Samantray J.C, Shriniwas Vector potential of hospital houseflies with special reference to Klebsiella species. Epidemiol. Infect. 1992;109(1):143–147. [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi M, Sasaki T, Saito N, Tamura K, Suzuki K, Watanabe H, Agui N. Houseflies:Not simple mechanical vectors of enterohemorrhagic Escherichia coli O157:H7. Am. J. Trop. Med. Hyg. 1999;61(4):625–629. doi: 10.4269/ajtmh.1999.61.625. [DOI] [PubMed] [Google Scholar]
- 9.Holt P.S, Geden C.J, Moore R.W, Gast R.K. Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Appl. Environ. Microbiol. 2007;73(19):6030–6035. doi: 10.1128/AEM.00803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmad A, Nagaraja T.G, Zurek L. Transmission of Escherichia coli O157:H7 to cattle by house flies. Prev. Vet. Med. 2007;80(1):74–81. doi: 10.1016/j.prevetmed.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Bahrndorff S, De Jonge N, Skovgård H, Nielsen J.L. Bacterial communities associated with houseflies (Musca domestica L.) sampled within and between farms. PLoS One. 2017;12(1):e0169753. doi: 10.1371/journal.pone.0169753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon R.D. Muscid flies (Muscidae) In: Mullen G, Durden L, editors. Medical and Veterinary Entomology. San Diego, CA: Elsevier Science; 2002. pp. 279–301. [Google Scholar]
- 13.World Health Organization. Antibiotic Resistance. 2018. Available from: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance . Retrieved on 15-02-2018.
- 14.Mamza S.A, Egwu G.O, Mshelia G.D. Beta-lactamase Escherichia coli and Staphylococcus aureus isolated from chickens in Nigeria. Vet. Ital. 2010;46(2):155–165. [PubMed] [Google Scholar]
- 15.Stapleton P.D, Taylor P.W. Methicillin-resistant in Staphylococcus aureus . Sci. Prog. 2007;85(1):57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall B, Petrowski D, Levy S.B. Inter-and intraspecies spread of Escherichia coli in a farm environment in the absence of antibiotic usage. Proc. Natl. Acad. Sci. 1990;87(17):6609–6613. doi: 10.1073/pnas.87.17.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haque Z.F, Sabuj A.A.M, Mahmud M.M, Pondit A, Islam M.A, Saha S. Characterization of Staphylococcus aureus from milk and dairy products sold in some local markets of Mymensingh district of Bangladesh. J. Nutr. Food Sci. 2018;8(6):743. [Google Scholar]
- 18.Rahman M.M, Amin K.B, Rahman S.M.M, Khair A, Rahman M, Hossain A, Rahman A.K.M, Parvez M.S, Miura N, Alam M.M. Investigation of methicillin-resistant Staphylococcus aureus among clinical isolates from humans and animals by culture methods and multiplex PCR. BMC Vet. Res. 2018;14(1):300. doi: 10.1186/s12917-018-1611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islam M.A, Parveen S, Rahman M, Huq M, Nabi A, Khan Z.U.M, Ahmed N, Wagenaar J.A. Occurrence and characterization of methicillin-resistant Staphylococcus aureus in processed raw foods and ready-to-eat foods in an urban setting of a developing country. Front. Microbiol. 2019;10:503. doi: 10.3389/fmicb.2019.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das M, Sabuj A.A.M, Haque Z.F, Barua N, Pondit A, Mahmud M.M, Khan M.F.R, Saha S. Characterization of Staphylococcus aureus isolated from human dental infection. Afr. J. Microbiol. Res. 2019;13(14):273–278. [Google Scholar]
- 21.Sobur M.A, Ievy S, Haque Z.F, Nahar A, Zaman S.B, Rahman M.T. Emergence of colistin-resistant Escherichia coli in poultry, house flies, and pond water in Mymensingh, Bangladesh. J. Adv. Vet. Anim. Res. 2019;6(1):50–53. doi: 10.5455/javar.2019.f311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington M.J, de Pinna E, Johnson A.P, Hopkins K.L, Woodford N. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 2016;71(8):2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 23.Sobur M.A, Sabuj A.A.M, Sarker R, Rahman A.T, Kabir S.L, Rahman M.T. Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Vet. World. 2019;12(7):984–993. doi: 10.14202/vetworld.2019.984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam A, Rahman Z, Monira S, Rahman M.A, Camilli A, George C.M, Ahmed N, Alam M. Colistin resistant Escherichia coli carrying mcr-1 in urban sludge samples:Dhaka, Bangladesh. Gut Pathog. 2017;9(1):77. doi: 10.1186/s13099-017-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monira S, Shabnam S.A, Ali S.I, Sadique A, Johura F.T, Rahman K.Z, Alam N.H, Watanabe H, Alam M. Multi-drug resistant pathogenic bacteria in the gut of young children in Bangladesh. Gut Pathog. 2017;9(1):19. doi: 10.1186/s13099-017-0170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahuma N, Ghenghesh K.S, Ben Aissa R, Elamaari A. Carriage by the housefly (Musca domestica) of multiple-antibiotic-resistant bacteria that are potentially pathogenic to humans, in hospital and other urban environments in Misurata, Libya. Ann. Trop. Med. Parasitol. 2005;99(8):795–802. doi: 10.1179/136485905X65134. [DOI] [PubMed] [Google Scholar]
- 27.Macovei L, Miles B, Zurek L. Potential of houseflies to contaminate ready-to-eat food with antibiotic-resistant enterococci. J. Food Prot. 2008;71(2):435–439. doi: 10.4315/0362-028x-71.2.435. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Yang Y, Zhao F, Fan X, Zhong W, Qiao D, Cao Y. Multi-drug resistant gram-negative enteric bacteria isolated from flies at Chengdu airport, China. Southeast Asian J. Trop. Med. Public Health. 2013;44(6):988–996. [PubMed] [Google Scholar]
- 29.Parvez M.A.K, Marzan M, Khatun F, Ahmed M.F, Mahmud S.A, Rahman S.R. Isolation of multidrug-resistant pathogenic bacteria from common flies in Dhaka, Bangladesh. J. Entomol. 2016;13(4):141–147. [Google Scholar]
- 30.Farag T.H, Faruque A.S, Wu Y, Das S.K, Hossain A, Ahmed S, Ahmed D, Nasrin D, Kotoff K.L, Panchilangam S, Nataro J.P, Cohen D, Blackwelder W.C, Levine M.M. Housefly population density correlates with shigellosis among children in Mirzapur, Bangladesh:A time series analysis. PLoS Negl. Trop. Dis. 2013;7(6):e2280. doi: 10.1371/journal.pntd.0002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan M, Mahin A.A, Pramanik M.K, Akter H. Identification of gut bacterial community and their effect on the fecundity of pumpkin fly, bactroceratau (Walker) J. Entomol. 2014;11(2):56–67. [Google Scholar]
- 32.Konemann E.W, Allen S.D, Janda W.M. Mycobacteria. In:Koneman's Color Atlas and Textbook of Diagnostic Microbiology. Philadelphia, PA: Lippincott Williams and Wilkins; 2005. [Google Scholar]
- 33.Bergey D.H, Buchanan R.E, Gibbons N.E American Society for Microbiology. Bergey's Manual of Determinative Bacteriology. Baltimore: Williams and Wilkins Co; 1974. [Google Scholar]
- 34.Cheesbrough M. In:Medical Laboratory Manual for Tropical Countries. 1st ed. London: English Language Book Society; 1985. Microbiology; pp. 400–480. [Google Scholar]
- 35.Stuhlmeier R, Stuhlmeier K.M. Fast, simultaneous, and sensitive detection of staphylococci. J. Clin. Pathol. 2003;56(10):782–785. doi: 10.1136/jcp.56.10.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen N.D, Neibergs H.L, McGruder E.D, Whitford H.W, Behle R.W, Ray P.M, Hargis B.M. Genus-specific detection of salmonellae using the polymerase chain reaction (PCR) J. Vet. Diagn. Invest. 1993;5(3):368–371. doi: 10.1177/104063879300500311. [DOI] [PubMed] [Google Scholar]
- 37.Guan S, Xu R, Chen S, Odumeru J, Gyles C. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 2002;68(6):2690–2698. doi: 10.1128/AEM.68.6.2690-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer A.W, Kirby W.M.M, Sherris J.C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. CLSI supplement M100s. In:Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. Pennsylvania: Clinical and Laboratory Standards Institute, Wayne; 2016. [Google Scholar]
- 40.Lee J.H. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003;69(11):6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stegger Á, Andersen P.S, Kearns A, Pichon B, Holmes M.A, Edwards G, Laurent F, Teale C, Skov R, Larsen A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 2012;18(4):395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 42.Puño-Sarmiento J, Medeiros L, Chiconi C, Martins F, Pelayo J, Rocha S, Blanco J, Blanco M, Zanutto M, Kobayashi R, Nakazato G. Detection of diarrheagenic Escherichia coli strains isolated from dogs and cats in Brazil. Vet. Microbiol. 2013;166(3-4):676–680. doi: 10.1016/j.vetmic.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh T.R, Shen J, Wang Y. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8(3):e00543–17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graczyk T.K, Knight R, Gilman R.H, Cranfield M.R. The role of non-biting flies in the epidemiology of human infectious diseases. Microbes Infect. 2001;3(3):231–235. doi: 10.1016/s1286-4579(01)01371-5. [DOI] [PubMed] [Google Scholar]
- 45.Graczyk T.K, Knight R, Tamang L. Mechanical transmission of human protozoan parasites by insects. Clin. Microbiol. Rev. 2005;18(1):128–132. doi: 10.1128/CMR.18.1.128-132.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khamesipour F, Lankarani K.B, Honarvar B, Kwenti T.E. A systematic review of human pathogens carried by the housefly (Musca domestica L.) BMC Public Health. 2018;18(1):1049. doi: 10.1186/s12889-018-5934-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almeida J.L, Giuffrida R, Andrade R.A.P, Chaves M.P. Muscoid diptera as potential vectors of bacterial agents on dairy farms in the Northern region of Paraná, Brazil. Semin. Cienc. Agrar. 2014;35(6):3127–3138. [Google Scholar]
- 48.Ommi D, Hemmatinezhad B, Hafshejani T.T, Khamesipour F. Incidence and antimicrobial resistance of Campylobacter and Salmonella from houseflies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2016;87(4):1285–1291. [Google Scholar]
- 49.Ibrahim A.W, Ajiboye T.O, Akande T.A, Anibaba O.O. Isolation and identification of pathogenic microorganisms from houseflies. Glob. J. Sci. Front. Res. 2018;18(1):57–64. [Google Scholar]
- 50.Ahmed A.S, Ahmed K.M, Salih S.S. Isolation and identification of bacterial isolates from house flies in Sulaymaniyah city. Eng. Technol. J. 2013;31(1):24–33. [Google Scholar]
- 51.Yalli A.A, Sambo S, Lawal H.M, Tukur U. Study of bacteria on the body surfaces of house flies (Musca domestica) in some homes within Sokoto metropolis. J. Adv. Med. Life Sci. 2017;5(4):1–5. [Google Scholar]
- 52.De Jesús A.J, Olsen A.R, Bryce J.R, Whiting R.C. Quantitative contamination and transfer of Escherichia coli from foods by houseflies Musca domestica L. (Diptera:Muscidae) Int. J. Food Microbiol. 2004;93(2):259–262. doi: 10.1016/j.ijfoodmicro.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 53.de Kraker M.E, Stewardson A.J, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13:11, e1002184. doi: 10.1371/journal.pmed.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graham J.P, Price L.B, Evans S.L, Graczyk T.K, Silbergeld E.K. Antibiotic-resistant enterococci and staphylococci isolated from flies collected near confined poultry feeding operations. Sci. Total Environ. 2009;407(8):2701–2710. doi: 10.1016/j.scitotenv.2008.11.056. [DOI] [PubMed] [Google Scholar]
- 55.Macovei L, Zurek L. Ecology of antibiotic resistance genes:Characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 2006;72(6):4028–4035. doi: 10.1128/AEM.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davari B, Kalantar E, Zahirnia A, Moosa-Kazemi S.H. Frequency of resistance and susceptible bacteria isolated from houseflies. Iran J. Arthropod Borne Dis. 2010;4(2):50. [PMC free article] [PubMed] [Google Scholar]
- 57.Blaak H, Hamidjaja R.A, van Hoek A.H, de Heer L, de Roda Husman A.M, Schets F.M. Detection of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli on flies at poultry farms. Appl. Environ. Microbiol. 2014;80(1):239–246. doi: 10.1128/AEM.02616-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zurek L, Ghosh A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microbiol. 2014;80(12):3562–3567. doi: 10.1128/AEM.00600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell J.M, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh T.R, Shen J. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2(4):16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Wang J, Chen L, Yassin A.K, Kelly P, Butaye P, Li J, Gong J, Cattley R, Qi K, Wang sC. Housefly (Musca domestica) and blowfly (Protophormia terraenovae) as vectors of bacteria carrying colistin resistance genes. Appl. Environ. Microbiol. 2018;84(1):e01736–17. doi: 10.1128/AEM.01736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen N.T, Nguyen H.M, Nguyen C.V, Nguyen T.V, Nguyen M.T, Thai H.Q, Ho M.H, Thwaites G, Ngo H.T, Baker S, Carrique-Mas J. Use of colistin and other critical antimicrobials on pig and chicken farms in Southern Vietnam and its association with resistance in commensal Escherichia coli bacteria. Appl. Environ. Microbiol. 2016;82(13):3727–3735. doi: 10.1128/AEM.00337-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jahan M, Rahman M, Parvej M.S, Chowdhury S.M.Z, Haque M.E, Talukder M.A.K, Ahmed S. Isolation and characterization of Staphylococcus aureus from raw cow milk in Bangladesh. J. Adv. Vet. Anim. Res. 2015;2(1):49–55. [Google Scholar]
- 63.Nusrat J, Ifra T.N, Mrityunjoy A. Detection of methicillin-resistant Staphylococcus aureus rawwithin raw milk and cheese samples. Int. Food Res. J. 2015;22(6):2629–2633. [Google Scholar]


