Abstract
Magnesium chelatase (MgCh) is a heterotrimeric enzyme complex, composed of two AAA+ family subunits that can assembly into a double ring structure and a large catalytic subunit. The small AAA+ subunit has ATPase activity and can self‐oligomerize into a ring structure, while the other AAA+ subunit lacks independent ATPase activity. Previous structural studies of the ATPase motor subunit of MgCh from a bacteriochlorophyll‐synthesizing bacterium have identified a unique ATPase clade, but the model of oligomeric assembly is unclear. Here we present the hexameric structure of the MgCh ATPase motor subunit from the chlorophyll‐synthesizing cyanobacterium Synechocystis sp. PCC 6803. This structure reveals details of how the hexameric ring is assembled, and thus provides a basis for further studying the heterotrimeric complex.
Keywords: AAA+ ring hexamer, ATPase, chlorophyll biosynthesis, magnesium chelatase, motor, Synechocystis
1. INTRODUCTION
Magnesium chelatase (MgCh) catalyzes the insertion of Mg into protoporphyrin IX, the first bacteriochlorophyll/chlorophyll specific step of tetrapyrrole biosynthesis in photosynthetic organisms.1, 2, 3 MgCh consists of three subunits, which are BchI, BchD, and BchH in bacteriochlorophyll‐synthesizing organisms, and ChlI, ChlD, and ChlH in chlorophyll‐synthesizing organisms.4, 5, 6, 7 The BchI/ChlI, BchD/ChlD, and BchH/ChlH subunits have respective molecular weights of approximate 40, 70, and 140 kDa. The BchI/ChlI and BchD/ChlD subunits belong to the AAA+ (ATPases associated with diverse cellular activities) protein family, and can assembly into a double ring structure that has been viewed by electron microscopy (EM).8, 9, 10
The double ring BchI–BchD/ChlI–ChlD complex hydrolyzes ATP, which powers the chelation reaction carried out by the largest BchH/ChlH subunit. Mg chelation is a thermodynamically unfavorable reaction, and one Mg chelation reaction requires approximate 15 molecules of ATP.11 The smallest subunit BchI/ChlI has ATPase activity and can self‐associate into a ring structure.12, 13, 14, 15 The crystal structure of BchI from the photosynthetic purple bacterium Rhodobacter capsulatus has been determined in a monomeric state, and has a unique domain arrangement that defines an AAA+ clade.12, 16, 17 Unlike the typical AAA+ proteins, where the C‐terminal α‐helical domain (also called the lid domain) lies at the top of the AAA+ core domain, in BchI, a long helical region (α5) shifts the position of the lid domain from the top to below the core domain. EM maps of R. capsulatus BchI have revealed a hexameric ring structure.8, 10, 15 However, due to limited resolution, it remains largely unknown how the hexamer is assembled. The BchD/ChlD subunit is composed of a C‐terminal integrin I domain preceded by a proline‐rich region and an N‐terminal domain similar to BchI/ChlI, but possesses no ATPase activity.18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 Recently, it has been demonstrated that ChlD links the ATPase activity with the ChlH active site primarily through the integrin I domain.29 Assembly of the BchD/ChlD hexamer is generally assumed to be mediated by the N‐terminal BchI/ChlI‐homologous domain, while contribution from the integrin I domain is unclear. Thus, structural study of the BchI/ChlI hexamer may also help understand the assembly of the BchD/ChlD ring.
The ChlI subunit from the cyanobacterium Synechocystis sp. PCC 6803 has been extensively characterized.11, 14, 18, 20, 22, 26, 27, 28, 29 The ChlI protomers can self‐assemble into a ring structure without ATP. To uncover the interactions stabilizing the assembly, we crystallized and determined its hexameric structure.
2. RESULTS AND DISCUSSION
2.1. Overall structure
We expressed and purified the recombinant Synechocystis ChlI protein, which by itself assembled into oligomeric forms. The predominant form of purified ChlI had a size corresponding to that of a hexamer or heptamer as suggested by size‐exclusion chromatography (Figure S1). The predominant form was crystallized by the vapor diffusion method. The resulting crystals had a hexagonal shape and diffracted to 2.9 Å resolution. The structure was solved by molecular replacement using the R. capsulatus BchI structure (PDB entry: 1G8P) as template,12 and the statistics of data collection and structure refinement are listed in Table 1.
Table 1.
Data collection and structure refinement statistics
| Synechocystis ChlI | |
|---|---|
| Diffraction data | |
| Diffraction source | BL17U1, SSRF |
| Detector | EigerX16M |
| Wavelength (Å) | 0.979 |
| Unit‐cell parameters | |
| a, b, c (Å) | 211.0, 121.1, 119.4 |
| α, β, γ (°) | 90.0, 99.0, 90.0 |
| Space group | C121 |
| Resolution (Å) | 50.00–2.90(3.00–2.90)a |
| Total no. of reflections | 304,832 (30254) |
| No. of unique reflections | 64,295 (6437) |
| Average redundancy | 4.7 (4.7) |
| Mean I/σI | 17.8 (1.8) |
| Completeness (%) | 98.5 (99.7) |
| R merge | 0.076 (0.731) |
| R meas | 0.085 (0.815) |
| CC1/2 | 0.999 (0.853) |
| Refinement | |
| Resolution range (Å) | 49.33–2.90 (3.00–2.90) |
| R work/R free | 0.233/0.252 |
| No. of protein atoms | 11,974 |
| No. of waters | 112 |
| Average B factor (Å2) | 47.49 |
| Protein | 47.60 |
| Water | 36.47 |
| Model quality | |
| RMSZ bond lengths | 0.005 |
| RMSZ bond angles | 0.878 |
| Ramachandran favored (%) | 94.88 |
| Ramachandran allowed (%) | 4.92 |
| Ramachandran outliers (%) | 0.02 |
| PDB code | 6L8D |
Values in parentheses are for highest resolution shell.
The ChlI structure displays a crystallographic pseudo‐hexagonal symmetry (Figure 1a). Its shape is different from the C3‐symmetric BchI model (Figure 1b) reconstructed from the low‐resolution cryo‐EM image.9 It also differs from crystal packing of BchI in the hexagonal P65 space group (Figure 1c), which has a screw axis along the c‐axis. The diameters of the ChlI ring and its center pore are ~120 and ~37 Å, respectively, which is compatible with the size of the BchI hexamer imaged by EM.9, 12, 15 Several fragments in the AAA+ core domain of ChlI are not observed in the electron density map. These correspond to the α2 helix and the three β‐hairpin inserts, α1–β2–β‐hairpin, H2‐insert, and PS‐I insert.16, 17 The amino‐acid sequence of Synechocystis ChlI has 51% identity with that of BchI. Such a high identity is consistent with the r.m.s.d. value of 1.06 Å for the 213 aligned Cα atoms between ChlI (chain A) and BchI, indicating that the protomer structures are highly similar (Figure S2).
Figure 1.
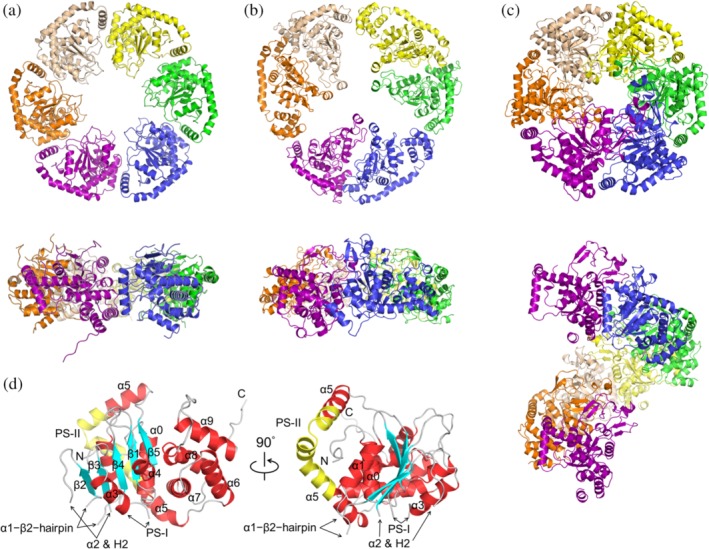
Structure of the MgCh ATPase motor subunit in ribbon representation. (a) ChlI hexamer in top (upper panel) and side (lower panel) views. Six protomers (chains A–F) are colored in purple, orange, beige, yellow, green, and blue, respectively. (b) BchI hexamer with pseudo‐3‐fold symmetry (PDB: 2X31) reconstructed from the cryo‐EM maps. (c) BchI (PDB: 1G8P) in space group P65. The seventh chain along the c‐axis is shown in the same color as the first chain to present the rotational symmetry. (d) ChlI protomer (chain A). The α‐helices (red) and β‐strands (cyan) are labeled following previous conventions for AAA+ proteins.9, 16, 17 The three β‐hairpins, α1–β2–β‐hairpin, H2‐insert (within α2), and PS‐I insert (presensor I insert, between α3 and β4) are denoted by arrows; the PS‐II insert (presensor II insert, within α5) is in yellow; the loops are in gray
For each ChlI protomer, the AAA+ core domain (except the inserts) and the lid domain are well structured. Five α‐helices (α0–α4) and a five‐stranded parallel β‐sheet (β1–β5) constitute the N‐terminal core domain. The C‐terminal lid domain is composed of α6–α9, and the L‐shaped α5 (residues 223–267) acts as a bridge connecting the two domains (Figure 1d). An insertion sequence called the PS‐II insert lies within α5 and bends this helix. This insert defines AAA+ clade 7 that has an unusual arrangement in which the lid domain is repositioned from the top to below the core domain.17
2.2. The ATP‐binding pocket and interprotomer interface
Whereas the domain arrangement of AAA+ clade 7 is unusual, the ATP‐binding pocket at the interprotomer interface resembles the typical configuration found in AAA+ proteins (Figure 2a). The positions of the five key nucleotide interaction motifs (W‐A, W‐B, Arg‐finger, S‐I, and S‐II) are well‐defined in the ChlI hexamer, and cluster in a cleft on the top side of the ring structure. The interprotomer interactions mainly arise from the short arm (residues 223–238) of the L‐shaped α5 and the α6–α7 region of the lid domain. Specifically, the guanidino group of Arg226 within the short arm of α5 forms hydrogen bonds with the carbonyl group of Gly289 and the carboxylic group of Asp293 (Figure 2b). As the six interfaces are not identical, small structural variations exist among protomers. Local interaction networks can be found at the chain B–C/C–D/A–F interface, where the W‐A Arg49 interacts with the aspartate at position 209/286/288 (Figure S3). These interprotomer interactions couple oligomerization to ATP binding and hence possibly motor function.
Figure 2.
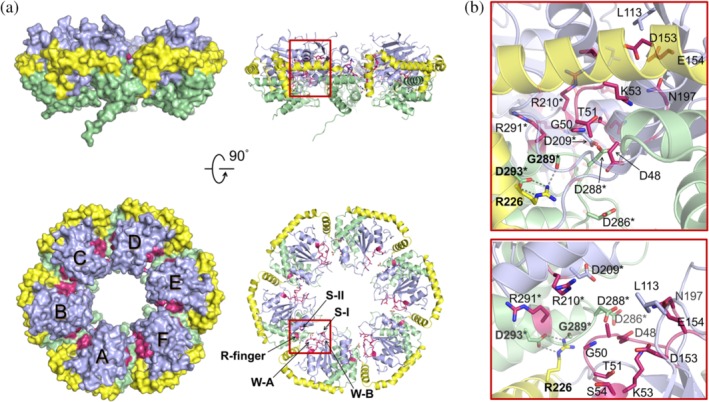
The bipartite ATP‐binding pocket in side (upper panel) and top (lower panel) views. (a) The ATP‐binding motifs in surface (left panel) and ribbon (right panel) representations. The AAA+ core domain is in light blue; the lid domain is in green; α5 is in yellow; the Walker A (W‐A, Gly47–Ser54) and Walker B motifs (W‐B, Asp153–Glu154), the Arg‐finger (Arg210), and the sensor I (S‐I, Asn197) and sensor II motifs (S‐II, Arg291) are in rose. The visible side chains of W‐A, Leu113, W‐B, Asn197, Asp209, Arg210, Arg226, Asp286, Asp288, Arg291, and Asp293, the amide group of Gly50, and the carbonyl group of Gly289 are shown as sticks. (b) Close‐up view the chain A–B interface. The interprotomer hydrogen bonds are shown as dashed lines. Residues of chain B are marked with asterisks
Previous work has shown that four point mutations, Asn269 → Ile of tobacco ChlI, and Leu91 → Phe, Asp187 → Asn, and Arg269 → Lys of barley ChlI, lead to functionally impaired MgCh.30 These conserved residues correspond to Leu113, Asn197 (S‐I), Asp209 and Arg291 (S‐II) in Synechocystis ChlI, and none of them is involved in direct interprotomer interaction (Figure 2b). Leu113 at the apex end of β2 lies on the top of the ATP‐binding cleft, and its phenylalanine mutation could affect the dynamics of nearby residues, which may mediate ATP binding and transduction of conformational change upon ATP hydrolysis. The S‐I asparagine is necessary for ATP hydrolysis,17 and thus mutation of Asn197 to isoleucine could damage the hydrolysis activity. Asp209, preceding the Arg‐finger, mainly participates in the intraprotomer contact with the lid domain and can also interact with neighboring Arg49. The S‐II Arg291 at the base of α7 points away from the W‐A motif of the neighboring protomer, contributing to the intraprotomer contact with the AAA+ core domain. It is possible that during ATP binding, mutations of Asp209 and Arg291 interfere with the trans‐activation (interprotomer communication) of the ChlI hexamer.13, 31, 32
2.3. Implication for the assembly of the ChlI–ChlD double hexamer
A model of the BchI hexamer was built by superimposing individual BchI structures onto the ChlI structure (Figure 3a). The model resembles a 6‐tooth rotor motor, in which each tooth is composed of three β‐hairpins and protrudes from the AAA+ motor (Figure S2). The lid domains and the long arms of the L‐shaped α5 form the base and the outer ring for the motor, respectively. Between adjacent teeth are equally spaced troughs, which allows a zipper dimerization interface with another hexamer in the opposite direction.
Figure 3.
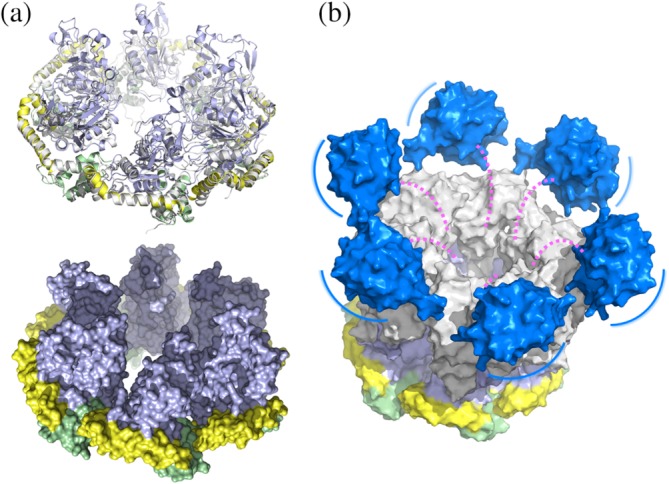
Model of the ChlI–ChlD complex. (a) Superimposition of each BchI protomer (shown in a similar color scheme as in Figure 2) onto the ChlI hexamer (gray) in ribbon (upper panel) and surface (lower panel) representations. The surface of ChlI is not shown. (b) Schematic model of the ChlI–ChlD double hexamer. ChlI is colored as in a. The ChlD N‐terminal domain and C‐terminal integrin I domain, constructed by the SWISS‐MODEL server then aligned onto the planar ChlI, are colored in gray and blue, respectively. The double hexamer model was generated by manually placing the ChlD N‐terminal hexamer atop the ChlI hexamer with maximum interface. The possibly unstructured middle proline‐rich region is represented as dashed magenta lines. The blue lines indicate position movements of the integrin I domains on the periphery of the double ring structure
The apo ChlI structure also provides clues for the assembly of the ATP‐independent ChlD subunit, whose N‐terminal domain is homologous to ChlI. The three key residues (Arg226, Gly289, and Asp293) involved in interprotomer interactions are highly conserved within the BchI/ChlI/ChlD subunits (Figure S4). It is likely that the ChlD subunits assemble in a similar way as the BchI/ChlI subunits. A notable exception is the BchD subunit, which lacks the PS‐II insert and the three residues. Thus, the domain arrangement of BchD seems to be dissimilar from BchI/ChlI/ChlD. This essential difference might explain the discrepancies between BchD and ChlD with respect to the assembly of the BchI–BchD/ChlI–ChlD complex.24, 28
Recently, it has been shown that the ChlD subunit bridges the ATPase activity with the active site at ChlH primarily through its C‐terminal integrin I domain.29 This is structurally reasonable based on our proposed ChlI–ChlD model (Figure 3b), in which the ChlI and ChlD hexamers are able to dimerize through the zipper dimerization interface. The ChlD C‐terminal integrin I domain lies at the top of the double hexamer, and is reminiscent of a propeller blade. As the middle proline‐rich region of ChlD is possibly less structured, the position of integrin I domain could exhibit large adjustments on the periphery of the ChlD hexamer. Upon ATP hydrolysis, the 6‐tooth rotor shaft transmits rotational torque generated by the AAA+ motor to the ChlD subunits, and powers the 6‐blade propeller constituted by the integrin I domains with which the ChlH subunit associates. The overall topology of the ChlI–ChlD double hexamer appears to be similar to that formed by the minichromosome maintenance (MCM) proteins,33 which belong to the AAA+ clade 7 and share mechanistic similarities with BchI.31, 32 While our apo structure lacks information about the nucleotide‐bound state that is required for studying the working mechanism of MgCh, it provides a molecular framework explaining the assembly of the double ring complex.
3. MATERIALS AND METHODS
3.1. Protein expression, purification, and crystallization
The ChlI‐encoding gene slr1030 from Synechocystis sp. PCC 6803 was commercially synthesized (Sangon Biotech, Shanghai, China). The synthetic sequence was amplified by PCR with the primers: 5′‐GGAATTCCATATGATGACTGCCACCCTTG‐3′ (bold: the NdeI restriction site) and 5′‐CCGCTCGAGAGCTTCATCGACAACG‐3′ (bold: the XhoI restriction site). The PCR product was ligated between the NdeI and XhoI restriction sites of the pET‐22b vector (Novagen, Shanghai, China). The resulting vector encodes the full‐length ChlI followed by a C‐terminal His6 tag. The vector was transformed into Escherichia coli BL21(DE3) competent cells for expression. The cells were grown at 37°C till the culture reached an optical density of 0.6 at 600 nm. Then isopropyl β‐d‐thiogalactoside was added to a final concentration of 0.5 mM for induction. The induced cells were grown at 16°C for 20 hr before harvest by centrifugation. The cell pellets were suspended in buffer A (500 mM NaCl and 20 mM Tris‐HCl, pH 7.5) plus 20 mM imidazole, and sonicated in an ice bath. The cell lysate was cleared by centrifugation and incubated with Ni‐NTA agarose (QIAGEN, Shanghai, China) resin at 4°C for 1 hr. Then the resin was packed into an open column (Sangon Biotech, Shanghai, China) and washed with buffer A plus 20 mM imidazole to remove the unbound proteins. The recombinant ChlI was eluted with 200 mM imidazole in buffer A, and concentrated by ultrafiltration through a Millipore 10‐kDa cut‐off filter. The concentrate (2 mL) was loaded onto a 120‐ml HiLoad 16/60 Superdex 200 column (GE Healthcare, Shanghai, China) equilibrated and eluted with buffer A. Fractions containing recombinant ChlI were collected and analyzed by SDS–PAGE. The highly purified fractions were pooled and concentrated to 8 mg ml−1 for crystallization. Crystal trays were set up at 16°C using the vapor diffusion method in a 2‐μl sitting drop containing 1:1 mixture of protein sample and reservoir solution. Crystals appeared in 2 days in the reservoir solution of 0.24 M sodium malonate, pH 7.2, and 18% (w/v) PEG 3350.
3.2. Data collection and structure determination
The crystals were transferred into the reservoir solution plus 20% (v/v) glycerol for cryo‐protection before being flash‐cooled in liquid nitrogen. The diffraction data were collected at a wavelength of 0.9793 Å at 100 K on the BL17U1 beamline of the Shanghai Synchrotron Facility, and processed using the HKL‐3000 program package.34 The initial ChlI model was built by molecular replacement using PHASER in the CCP4 suite,35, 36 and the BchI structure (PDB: 1G8P) was used as the template.12 Further manual corrections and refinements were performed using Coot and the phenix.refine program.37, 38 The final model was evaluated by the MolProbity server.39 All structure figures were prepared with the program PyMOL (Schrödinger LLC, New York, NY).
Supporting information
Figure S1 Purification and crystallization of Synechocystis ChlI.
Figure S2. Structural comparison between Synechocystis ChlI and R. capsulatus BchI.
Figure S3. Top view of the chain B–C interface, chain C–D interface, and chain A–F interface.
Figure S4. Alignment of the N‐terminal regions of ChlI/BchI and ChlD/BchD.
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (2017YFA0503703), the MOE Chang Jiang Scholars Program (Q2017241), and the Anhui Provincial Wanjiang Scholars Program. We thank Ming‐Zhu Wang at Anhui University and the beamline scientists at the Shanghai Synchrotron Radiation Facility for technical support during data collection.
Gao Y‐S, Wang Y‐L, Wang X, Liu L. Hexameric structure of the ATPase motor subunit of magnesium chelatase in chlorophyll biosynthesis. Protein Science. 2020;29:1040–1046. 10.1002/pro.3816
Funding information Anhui Provincial Wanjiang Scholars Program; MOE Chang Jiang Scholars Program, Grant/Award Number: Q2017241; National Key R&D Program of China, Grant/Award Number: 2017YFA0503703
REFERENCES
- 1. Chew AG, Bryant DA. Chlorophyll biosynthesis in bacteria: The origins of structural and functional diversity. Annu Rev Microbiol. 2007;61:113–129. [DOI] [PubMed] [Google Scholar]
- 2. Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. [DOI] [PubMed] [Google Scholar]
- 3. Mochizuki N, Tanaka R, Grimm B, et al. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010;15:488–498. [DOI] [PubMed] [Google Scholar]
- 4. Walker CJ, Willows RD. Mechanism and regulation of Mg‐chelatase. Biochem J. 1997;327:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reid JD, Hunter CN. Current understanding of the function of magnesium chelatase. Biochem Soc Trans. 2002;30:643–645. [DOI] [PubMed] [Google Scholar]
- 6. Masuda T. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth Res. 2008;96:121–143. [DOI] [PubMed] [Google Scholar]
- 7. Al‐Karadaghi S, Franco R, Hansson M, Shelnutt JA, Isaya G, Ferreira GC. Chelatases: Distort to select? Trends Biochem Sci. 2006;31:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elmlund H, Lundqvist J, Al‐Karadaghi S, Hansson M, Hebert H, Lindahl M. A new cryo‐EM single‐particle ab initio reconstruction method visualizes secondary structure elements in an ATP‐fueled AAA+ motor. J Mol Biol. 2008;375:934–947. [DOI] [PubMed] [Google Scholar]
- 9. Lundqvist J, Elmlund H, Wulff RP, et al. ATP‐induced conformational dynamics in the AAA+ motor unit of magnesium chelatase. Structure. 2010;18:354–365. [DOI] [PubMed] [Google Scholar]
- 10. Lundqvist J, Braumann I, Kurowska M, Müller AH, Hansson M. Catalytic turnover triggers exchange of subunits of the magnesium chelatase AAA+ motor unit. J Biol Chem. 2013;288:24012–24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reid JD, Hunter CN. Magnesium‐dependent ATPase activity and cooperativity of magnesium chelatase from Synechocystis sp. PCC6803. J Biol Chem. 2004;279:26893–26899. [DOI] [PubMed] [Google Scholar]
- 12. Fodje MN, Hansson A, Hansson M, et al. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J Mol Biol. 2001;311:111–122. [DOI] [PubMed] [Google Scholar]
- 13. Hansson A, Willows RD, Roberts TH, Hansson M. Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA+ hexamers. Proc Natl Acad Sci U S A. 2002;99:13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reid JD, Siebert CA, Bullough PA, Hunter CN. The ATPase activity of the ChlI subunit of magnesium chelatase and formation of a heptameric AAA+ ring. Biochemistry. 2003;42:6912–6920. [DOI] [PubMed] [Google Scholar]
- 15. Willows RD, Hansson A, Birch D, Al‐Karadaghi S, Hansson M. EM single particle analysis of the ATP‐dependent BchI complex of magnesium chelatase: An AAA+ hexamer. J Struct Biol. 2004;146:227–233. [DOI] [PubMed] [Google Scholar]
- 16. Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. [DOI] [PubMed] [Google Scholar]
- 17. Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. [DOI] [PubMed] [Google Scholar]
- 18. Jensen PE, Gibson LC, Henningsen KW, Hunter CN. Expression of the chlI, chlD, and chlH genes from the cyanobacterium synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium‐protoporphyrin chelatase activity. J Biol Chem. 1996;271:16662–16667. [DOI] [PubMed] [Google Scholar]
- 19. Hansson M, Kannangara CG. ATPases and phosphate exchange activities in magnesium chelatase subunits of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1997;94:13351–13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jensen PE, Gibson LC, Hunter CN. Determinants of catalytic activity with the use of purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from Synechocystis PCC6803. Biochem J. 1998;334:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gräfe S, Saluz HP, Grimm B, Hänel F. Mg‐chelatase of tobacco: The role of the subunit CHL D in the chelation step of protoporphyrin IX. Proc Natl Acad Sci U S A. 1999;96:1941–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jensen PE, Gibson LC, Hunter CN. ATPase activity associated with the magnesium‐protoporphyrin IX chelatase enzyme of Synechocystis PCC6803: Evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP‐dependent interaction of the ChlI and ChlD subunits. Biochem J. 1999;339:127–134. [PMC free article] [PubMed] [Google Scholar]
- 23. Lake V, Olsson U, Willows RD, Hansson M. ATPase activity of magnesium chelatase subunit I is required to maintain subunit D in vivo. Eur J Biochem. 2004;271:2182–2188. [DOI] [PubMed] [Google Scholar]
- 24. Axelsson E, Lundqvist J, Sawicki A, et al. Recessiveness and dominance in barley mutants deficient in Mg‐chelatase subunit D, an AAA protein involved in chlorophyll biosynthesis. Plant Cell. 2006;18:3606–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sawicki A, Willows RD. Kinetic analyses of the magnesium chelatase provide insights into the mechanism, structure, and formation of the complex. J Biol Chem. 2008;283:31294–31302. [DOI] [PubMed] [Google Scholar]
- 26. Adams NB, Reid JD. The allosteric role of the AAA+ domain of ChlD protein from the magnesium chelatase of synechocystis species PCC 6803. J Biol Chem. 2013;288:28727–28732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams NB, Brindley AA, Hunter CN, Reid JD. The catalytic power of magnesium chelatase: A benchmark for the AAA+ ATPases. FEBS Lett. 2016;590:1687–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams NB, Vasilev C, Brindley AA, Hunter CN. Nanomechanical and thermophoretic analyses of the nucleotide‐dependent interactions between the AAA+ subunits of magnesium chelatase. J Am Chem Soc. 2016;138:6591–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farmer DA, Brindley AA, Hitchcock A, et al. The ChlD subunit links the motor and porphyrin binding subunits of magnesium chelatase. Biochem J. 2019;476:1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansson A, Kannangara CG, von Wettstein D, Hansson M. Molecular basis for semidominance of missense mutations in the XANTHA‐H (42‐kDa) subunit of magnesium chelatase. Proc Natl Acad Sci U S A. 1999;96:1744–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. [DOI] [PubMed] [Google Scholar]
- 32. Bae B, Chen YH, Costa A, et al. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17:211–222. [DOI] [PubMed] [Google Scholar]
- 33. Abid Ali F, Douglas ME, Locke J, et al. Cryo‐EM structure of a licensed DNA replication origin. Nat Commun. 2017;8:2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL‐3000: The integration of data reduction and structure solution – From diffraction images to an initial model in minutes. Acta Crystallogr. 2006;D62:859–866. [DOI] [PubMed] [Google Scholar]
- 35. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Afonine PV, Grosse‐Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. 2012;D68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen VB, Arendall WB 3rd, Headd JJ, et al. MolProbity: All‐atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;D66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Purification and crystallization of Synechocystis ChlI.
Figure S2. Structural comparison between Synechocystis ChlI and R. capsulatus BchI.
Figure S3. Top view of the chain B–C interface, chain C–D interface, and chain A–F interface.
Figure S4. Alignment of the N‐terminal regions of ChlI/BchI and ChlD/BchD.


