Abstract
How virulence evolves after a virus jumps to a new host species is central to disease emergence. Our current understanding of virulence evolution is based on insights drawn from two perspectives that have developed largely independently: long-standing evolutionary theory based on limited real data examples that often lack a genomic basis, and experimental studies of virulence-determining mutations using cell culture or animal models. A more comprehensive understanding of virulence mutations and their evolution can be achieved by bridging the gap between these two research pathways through the phylogenomic analysis of virus genome sequence data as a guide to experimental study.
Subject terms: Evolutionary genetics, Viral pathogenesis, Phylogenetics, Phylogenomics, Viral genetics, Viral evolution, Viral transmission, Ebola virus, Virus-host interactions, Influenza virus
The virulence of viruses is a major determinant of the health burden of viral infections in humans and other species. In this article, Geoghegan and Holmes discuss how largely disparate research fields — theoretical modelling of virulence evolution and experimental dissection of genetic virulence determinants in laboratory model systems — can be bridged by considering real genomic data of viral evolution in a phylogenetic context. They describe the underlying principles of virulence evolution and how they apply to real-world viral infections and outbreaks of global importance.
Introduction
Despite many advances in our understanding of virus evolution, how virulence evolves in a virus, particularly following a jump to a new host species, continues to be contentious. Will a virus become more or less virulent in a new host? What level of virulence is optimized by natural selection and why? Is there any consistent association between host-jumping and virulence such that predictions can be made about how virulence might evolve following emergence? Not only will the answers to these questions reveal fundamental aspects of virus biology, but they also may assist in infectious disease management and mitigation, particularly as humans, other animals and plants face a continual threat from emerging viruses.
The term ‘virulence’ has different meanings depending on context, can be assessed in a variety of ways and is often only an operational measure1. To be as general as possible, we assume a simple working definition of virulence: the harm caused by pathogen infection, particularly in terms of host morbidity and mortality. Virulence is also a complex trait determined by a combination of pathogen, host and environmental factors. Although it is obviously necessary to understand all three, we focus on the pathogen (virus) component as this is the most tractable, with the small genomes and rapid replication and evolution of viruses facilitating comparative and experimental studies, and because there is strong evidence for heritable virus genetic variation for virulence2. Although we purposely focus on the analysis of virus genomes, genome-wide association studies (GWAS) promise to open up new ways to explore the evolutionary impact of viral infections on host genomes3 and hence the intimate interaction between host and virus4.
Another complexity in studies of virulence evolution is that metagenomics is increasingly showing that mixed (that is, polymicrobial) infections are commonplace5, and even seemingly healthy hosts can carry multiple microorganisms of the kind often thought to be pathogenic6,7. Determining which microorganism is the cause of a particular disease syndrome can be troublesome, and it is possible that overt illness might result from synergistic interactions between multiple microorganisms that overwhelm the host. Hence, the model of one pathogen–one disease that has dominated studies of human infections, and implicitly models of virulence evolution, may be overly simplistic. It is also possible that measurements of relative virulence vary between humans and wildlife populations. For example, whereas dengue is considered an important infectious disease of humans, the virulence of dengue virus is low in terms of overall mortality in humans, and infections of equivalent severity may go unnoticed in wildlife, particularly as there is a strong sampling bias towards the most virulent presentations.
Virulence evolution in viruses has traditionally been studied from one of two separate research paths — the theoretical and the empirical — that have largely been pursued independently. Although theory and empiricism have each generated important and parallel insights, they have each been able to paint only a partial picture of virulence evolution. Few attempts have been made to bridge this divide8,9.
There is now a large body of long-standing evolutionary theory that considers what level of virulence maximizes pathogen fitness under variable conditions, such as differing modes of transmission, levels of co-infection, selection pressures, and both within and between hosts10. Although of great value, a drawback is that this work is unavoidably based on a small number of case studies, the most famous of which is the co-evolution of myxoma virus (MYXV) and European rabbits following the release of MYXV as a biological control11. However, the insights from examples such as MYXV may be insufficient to adequately inform theoretical models confronted with novel, real-world emergence events.
By contrast, empirical studies involve laboratory-based methods to identify the mutations that affect virulence (that is, virulence determinants), usually on the basis of a combination of reverse genetics and cell culture and/or animal models8,9,12–16. These studies are often very successful in pinpointing causal mutations (see Table 1 for illustrative examples) and are commonplace following the emergence of a new disease. However, because the mutations identified through experimental studies are not considered in an evolutionary context, their relevance for general theories of virulence evolution is usually ignored. In addition, in vitro methods may not reflect real-world selection pressures, there may be little consideration about how virulence mutations affect inter-host transmission, and animal models commonly differ from the species infected in the field. For example, virulence determinants in MYXV identified on the basis of in vitro studies and mouse models have often not been upheld in reverse genetic experiments using the natural rabbit host17. Similarly, despite the regularity of their use, there has been a long-standing debate over the validity of ferrets as accurate models for human influenza18.
Table 1.
Examples of virulence determinants in viruses
| Virus | Virulence determinants | Method of detection | Refs | ||
|---|---|---|---|---|---|
| In vitro | In vivo | Phylogenetics | |||
| RNA viruses | |||||
| West Nile virus (WNV) | Threonine to proline at amino acid 249 (T249P) in NS3 helicase protein increases virulence in American crows | – | ✓ | ✓ | 58 |
| Human immunodeficiency virus (HIV) | Can be considered through variation in set point viral load | – | – | ✓ | 104, 113 |
| Ebola virus (EBOV) | A82V in the glycoprotein appeared to increase human mortality | ✓ | – | ✓ | 70, 116 |
| Avian influenza A virus (AIV) H5N1 | L627E in the PB2 protein attenuated the infection outcome in mice | – | ✓ (mice) | – | 85 |
| P42S in the NS1 protein dramatically increased the virulence in mice | – | ✓ (mice) | – | 147 | |
| 1918 H1N1 human influenza A virus | Four C-terminal residues (Glu–Ser–Glu–Val) of the NS1 protein are associated with increased virulence in mice | – | ✓ (mice) | – | 88 |
| 2009 H1N1pdm human influenza A virus | E47K in the HA2 subunit of the stalk region of the HA protein increased infectivity in ferrets | ✓ | ✓ (ferrets) | – | 148 |
| H3N2 human influenza A virus | D189N and V194I in NS1 attenuated virulence in mice | – | ✓ (mice) | – | 87 |
| Influenza B virus | K338R in the polymerase acidic protein | – | ✓ (mice) | – | 84 |
| Zika virus (ZIKV) | S139N in the PrM protein exacerbated symptoms in mice | – | ✓ (mice) | – | 125 |
| Enterovirus A71 (EVA71) | G145E or Q145E in the capsid protein VP1 increased virulence in non-human primates | – | ✓ (non-human primates) | – | 14 |
| Japanese encephalitis virus (JEV) | Multiple amino acid substitutions in the E protein (L107F, E138K, I176V, T177A, E244G, Q264H, K279M, A315V, S366A and K439R) were sufficient for complete attenuation of virulence in mice | – | ✓ (mice) | – | 149 |
| Porcine reproductive and respiratory syndrome virus (PRRSV) | Mutant virus with a spontaneous 88-amino-acid deletion in NSP2 resulted in increased mRNA expression of type I interferon and chemokine genes in host cells in vitro, suggesting an increase in virulence | ✓ | – | – | 150 |
| Newcastle disease virus (NDV) | E402A mutation in the C-terminus of the nucleoprotein resulted in larger viral plaques that appeared more quickly and increased the virulence of NDV | ✓ | – | – | 151 |
| Severe acute respiratory syndrome coronavirus (SARS-CoV) | A recombinant lacking the E protein is attenuated in vitro and in mice | ✓ | ✓ (mice) | – | 13 |
| Infectious pancreatic necrosis virus (IPNV) | Thr217 and Ala221 of VP2 protein are highly virulent in salmon | ✓ | ✓ (salmon) | – | 15, 152 |
| Rabies virus (RABV) | I333R in glycoprotein increases pathogenicity | – | ✓ (mice) | – | 153 |
| DNA viruses | |||||
| Myxoma virus (MYXV) | Wild-type ANK repeat proteins, M148R and M149R, are associated with increased virulence in European rabbits | ✓ | ✓ (rabbits) | 154 | |
| Marek’s disease virus (MDV) | Two different mutant viruses, one bearing a 173-amino-acid deletion in the lipase homologous domain and the other having an alanine point mutation at the serine nucleophile position, resulted in enhanced chicken survival | ✓ | ✓ (chickens) | 100 | |
| Murine polyomavirus (MuPyV) | Y296A in VP1 increased virulence in mice | – | ✓ (mice) | – | 155 |
| Sheep pox virus (SPPV) | A deletion in the SPPV-019 kelch-like gene, ΔKLP, resulted in markedly attenuated virulence | – | ✓ (sheep) | – | 156 |
This table shows examples of mutations that have been proposed to determine virulence in various RNA and DNA viruses and the method used to identify them (in vitro, in vivo or by phylogenetic analysis). ANK, ankyrin; E, envelope; HA, haemagglutinin; NS1, non-structural protein 1; NSP2, non-structural polyprotein 2; PB2, polymerase basic protein 2; pdm, pandemic; PrM, pre-membrane protein.
Bridging the gap between the theoretical and empirical approaches would bring a new impetus to studies of virulence evolution. In this Review, we outline how this can be achieved within a phylogenomics framework. We show that virus phylogenies are being increasingly used to help identify virulence determinants and that the data obtained can be used to test general theories of virulence evolution. Such a phylogenomic approach to studying virulence evolution is timely because of the rapidity with which virus genome sequence data are now being generated, including during ongoing disease outbreaks of emerging viruses19–21, and because of the development of new phylogeny-based methods for studying and visualizing genomic data22–24. However, the success of this approach also requires that phylogenomic data are combined with relevant clinical, epidemiological and experimental metadata so that a direct link can be made between virulence, virus genotype and phenotype, and population fitness.
Disease emergence and virulence evolution
Arguably the most interesting context of virulence evolution is following a host jump as this sits at the heart of virus emergence, and the question of how virulence will evolve is commonly asked following the appearance of a new virus or the emergence of an existing virus with altered host range (Box 1). To make meaningful inferences, it is important to compare virulence in both the reservoir (that is, donor) and novel (that is, recipient) host species. Although this may sound straightforward and is tractable in some cases25, in reality it faces a number of difficulties. In many cases, including common infections such as hepatitis C virus26, as well as emerging infectious diseases such as Zika virus (ZIKV), the reservoir species is unknown or is at best uncertain. Even if a reservoir species is known, we generally know little, if anything, about virulence in that species, and there is likely to be an ascertainment bias towards the most virulent cases. For example, although species of fruit bats appear to be reservoirs for Ebola virus (EBOV)27, little is known about its virulence in these animals28, as is true of many wildlife infections. The identification of host species may also change with better sampling, which is invariably poor in wildlife. For example, it was long thought that the canine parvovirus (CPV) that emerged in dogs in the late 1970s had jumped from cats infected by a closely related virus29. However, more recent sampling of wild carnivore species has shown this to be incorrect, such that the true reservoir species for CPV is unclear30. Therefore, it is crucial to understand disease processes, including virulence, in reservoir hosts under natural conditions, which will require more detailed studies of animal ecology.
Although we currently know little about virulence in reservoir species, comparative data tell us that, on average, low-virulence infections have a greater chance of successfully establishing transmission cycles in humans than viruses with higher mortality31. This greater chance is presumably because high virulence requires a greater supply of susceptible hosts during the early stages of emergence.
Box 1 Cross-species virus transmission and virulence evolution.
There is no clear understanding on whether emerging viruses become more or less virulent following a jump to a new host. Before a host jump, pathogens are likely to have been selected to optimize their virulence for onward transmission in the reservoir host. However, immediately following a host jump, the pathogen is likely to be initially poorly adapted to the new host, which may inhibit successful emergence135, and the host is likely to be poorly adapted to the new pathogen25. Indeed, high host mortality is associated with a reduced likelihood of successful onward transmission in the case of human viruses31.
There are three possible scenarios for virulence evolution immediately following a host jump (the filled nodes in the figure):
Some of the most devastating epidemics have been associated with an increase in virulence following a cross-species transmission event (see the figure, part a). For example, whereas HIV almost invariably progresses to AIDS if untreated in humans, in African non-human primates, the closely related simian immunodeficiency virus (SIV) persists without causing disease136,137 or is associated with less overt disease than in humans138. Similarly, the (deliberate) transfer of myxoma virus (MYXV) from the South American tapeti, where it was largely benign, initially resulted in mortality of ~99% in European rabbits11.
No apparent change in virulence following a host jump can be seen in H3N8 influenza A virus that jumped from horses to dogs139,140 (see the figure, part b). Disease is mild in both cases, and there are minimal biological differences between these viruses141. Similarly, rabies virus (RABV) causes an equally severe, ultimately fatal encephalitis in foxes and dogs and following spillover to humans142.
Cases of decreased virulence may be largely overlooked because avirulent viruses will often persist in a host population without detection. An example of decreased virulence following a host jump may be that of the infectious haematopoietic necrosis virus (IHNV) that spread from sockeye salmon to rainbow trout in North America and Japan (see the figure, part c). Upon the initial cross-species transmission event in the 1950s to 1960s, the virus seemed at first to be avirulent in the new host in contrast to the mortality seen in sockeye salmon. Since then, however, virulence in some genotypes of IHNV in rainbow trout has increased, itself associated with an increase in transmissibility143,144.
For many diseases, however, the extent of changes in virulence upon emergence is not known, in part because little is usually known about disease epidemiology in reservoir hosts, and there is likely to be a sampling bias towards the cases of highest virulence.
Theories of virulence evolution
Evolutionary biologists have had a long fascination with virulence32–37. Because there is a very large literature base on this subject, we necessarily provide only a brief overview here. A straightforward interpretation of virulence evolution is that natural selection will optimize the level of virulence that maximizes pathogen fitness, expressed as the basic reproductive number (R0)1, although in reality fitness is shaped by a complex set of host–pathogen interactions38,39. Current evolutionary theory tells us that when a virus jumps to a new species, its initial virulence can vary from asymptomatic to highly pathogenic, and precisely where it lies on this virulence spectrum is difficult to predict. However, it is possible that the direction of virulence evolution can be anticipated, at least in part, if the key relationship between virulence and transmissibility, and hence fitness, is understood. Importantly, there is also evidence from insect viruses that host phylogeny is able to predict some aspects of virulence evolution following species jumps, with related host species tending to have similar levels of virulence25.
A commonly stated idea is that there is often an evolutionary trade-off between virulence and transmissibility because intra-host virus replication is necessary to facilitate inter-host transmission but may also lead to disease, and it is impossible for natural selection to optimize all traits simultaneously. In the case of MYXV, this trade-off is thought to lead to ‘intermediate’ virulence grades being selectively advantageous: higher virulence may mean that the rabbit host dies before inter-host transmission, whereas lower virulence is selected against because it does not increase virus transmission rates. A similar trade-off model has been proposed to explain the evolution of HIV virulence40. However, many doubts have been raised about the general applicability of the trade-off model35,41–43, virus fitness will be affected by traits other than virulence and transmissibility39,41,44, contrary results have been observed in experimental studies45 and relatively little is known about evolutionary trade-offs in nature. For example, in the case of the second virus released as a biocontrol against European rabbits in Australia — rabbit haemorrhagic disease virus (RHDV) — there is evidence that virulence has increased through time, probably because virus transmission often occurs through blow flies that feed on animal carcasses, making host death selectively favourable46. Similarly, experimental studies of plant RNA viruses have shown that high virulence does not necessarily impede host adaptation47 and, in the case of malaria, higher virulence was shown to provide the Plasmodium parasites with a competitive advantage within hosts48.
Other factors in addition to evolutionary trade-offs can shape the level of virulence in an emerging virus. For example, ‘short-sighted’ virulence evolution within a single host may be detrimental for inter-host transmission49, and newly emerged ‘spillover’ infections that have experienced only a limited number of transmission events are likely to have virulence levels that have not yet been optimized for transmissibility by natural selection50. Accordingly, for spillover infections, ongoing transmission may be largely at the mercy of random drift effects, including the severe population bottlenecks that routinely accompany such events51. Finally, it is possible that virulence may sometimes simply be a coincidental by-product of selection for another trait or selection for transmission in another species.
Theory therefore tells us that natural selection can increase or decrease pathogen virulence, depending on the particular combination between host, virus and environment1,32,33,37,41,52,53. Although providing a useful framework, theory can provide only useful generalities because the relevant factors vary substantially and need to be assessed on a case-by-case basis. Virulence evolution could, however, be better understood if its genomic basis were known.
Phylogenomics for assessing virulence evolution
Phylogenetic studies of viruses, including those that consider whole-genome sequences, are commonplace and are often used to understand a variety of aspects of virus evolution. In particular, virus phylogenies are being increasingly used to understand the evolution of key phenotypic traits such as virulence9 (and see the examples described below). Phylogenomics provides an informative way to help understand virulence evolution and establishes a set of hypotheses that can be tested using appropriate experimental assays8,9. We also believe that phylogenomics provides valuable information on how natural selection acts on virulence and can be used to test general models of virulence evolution, thereby providing a key link between theoretical and empirical approaches to studying virulence evolution. The crux of this approach involves mapping mutations onto phylogenetic trees of viruses sampled within and/or between disease outbreaks and from reservoir and novel hosts. The phylogenetic location of these changes — whether they fall on shallow or deep nodes (branches) and/or singularly or in parallel — makes it possible to infer, at least in broad terms, the selection pressures acting on virulence mutations and from this infer important aspects of virulence evolution (Box 1; Fig. 1).
Fig. 1. Phylogenomics of virulence evolution.
a | A model phylogeny with virulence determinants mapped to a fairly deep node suggesting that higher virulence has increased virus fitness. b | A model phylogeny with virulence traits mapped to shallow nodes suggesting that higher virulence reduced pathogen fitness so that viruses with these mutations are purged from the population or require compensatory mutations. c | A model phylogeny with a high-virulence mutation arising multiple times independently owing to parallel or convergent evolution. The occurrence of parallel/convergent mutations that occur more frequently than by chance8 is likely to reflect adaptive evolution (Fig. 2). d | The relationship between virulence, fitness and host jumps. A virus is assumed to be at a fitness peak (high R0), in this case high virulence, in the reservoir host, so that the mutations determining both virulence and host range are expected to be subject to strong purifying selection (for example, a low value of dN/dS). As the virus emerges in the new recipient host, it will initially be maladapted (that is, reside in a fitness valley) and subject to genetic drift as the population is small. As it adapts to the new host, virulence will be selectively optimized (in this case declining), increasing R0 and resulting in positive selection (for example, dN/dS > 1, although other measures of selection pressure are available). Once the virus becomes adapted to the new host, the virulence determinants are again subject to purifying selection.
The greater the fitness of a virulence determinant, the more rapidly it will spread through the virus population and the deeper it will fall on a virus phylogeny (that is, closer to the root of the tree), including on the branch linking reservoir and novel hosts. Of particular importance are repeated occurrences of the same mutation falling on deep branches across multiple outbreaks, or multiple cross-species transmission events, as both parallel evolution and convergent evolution can be signatures of adaptive evolution8,54–57 (Fig. 1). For example, in the case of West Nile virus (WNV), a single mutation in the virus helicase protein repeatedly evolved in high-mortality outbreaks in birds, which is indicative of a selective advantage58. Similarly, the reversion to virulence in oral polio vaccine (OPV) strains of poliovirus has been associated with extensive parallel evolution8, and parallel evolution was also associated with host-specific adaptation in experimental studies of cross-species transmission involving Drosophila virus C59. Because adaptive evolution has been at play, phylogenies of the sequence in question may have a characteristic shape24, and the sequences associated with selected branches may also contain genomic signatures indicative of positive selection, such as the rapid fixation of amino acid changes or an increased rate of nonsynonymous to synonymous substitutions per site (ratio dN/dS)8,60. In the case of frequent parallel or convergent evolution for specific virulence mutations, it is also possible that the amino acid sites involved will have signatures of positive selection, such as an elevated dN/dS (as was the case in WNV; see below).
Following the same logic, mutations that fall on shallow branches in virus phylogenies (that is, closer to the tips) are present in a smaller proportion of the population and are therefore more likely to be of lower fitness such that they may be removed by purifying selection. Hence, virulence-determining mutations that repeatedly fall on tip branches alone are likely to inhibit some other aspect of pathogen fitness, thereby reducing R0 at the population scale.
Although this approach has a solid theoretical and empirical basis61,62, a complicating factor is that a virulence-determining mutation that has very recently emerged will necessarily fall towards the tips rather than on an internal branch even if it is selectively advantageous. Similarly, although popular, dN/dS measures are less robust over short timescales, such as during outbreaks, because mutations may not have reached fixation by positive selection or had time to be purged by purifying selection, and it can be difficult to detect selected mutations that occur only once63,64. Approaches to detect positive selection that do not rely on dN/dS, such as those based on tree shape24, or tracking mutations that are increasing in frequency compared with those thought to be evolving neutrally64,65, may therefore add analytical power.
The phylogenetic mapping of virulence mutations can proceed in two ways depending on the extent of a priori knowledge. In a ‘top-down’ approach, in which virulence determinants are unknown, a virus phylogeny is inferred, mutations are mapped onto this phylogeny and the mutations on key branches are then identified. Such ‘key branches’ include those directly associated with cross-species transmission events, invasions of new geographic areas, increases in rates of transmission, spikes in morbidity and/or mortality or clear instances of positive selection. The mutations identified in this way are candidates for virulence determinants that can be tested in an appropriate experimental framework8. An example of this approach is shown in Fig. 2. The second, ‘bottom-up’, approach utilizes existing knowledge of virulence determinants, such as that determined by an experimental study. The putative virulence determinant is then mapped onto the phylogeny, and its phylogenetic location (that is, deep or shallow branch, singular or parallel/convergent evolution) is used to infer how it affects virulence evolution, whether it is associated with reciprocal mutations that reflect evolutionary trade-offs and the selection pressures it faces.
Fig. 2. Example of how phylogenomics can guide the experimental analysis of virulence determinants.
The evolution of virulence in strains of oral polio vaccine (OPV)8. OPV is an attenuated form of poliovirus that can occasionally revert to a virulent form and cause outbreaks of poliomyelitis. A | Phylogenetic analysis of OPV strains in nature reveals that some mutations associated with high virulence have experienced more frequent parallel evolution than expected by chance (and occupy well supported nodes) and hence are likely to be seletively favoured8. B | Computational evolutionary analysis then reveals that this parallel evolution for high virulence is associated with a hypothetical threonine-to-proline (T-to-P) amino acid change that is subject to significant adaptive evolution (which can be detected in a variety of ways)60,63. C | The virulence impact of these mutations is then confirmed in both in vitro (cell culture; part Ca) and in vivo (mouse; part Cb) experimental studies. In all cases, the red shading signifies increased virulence.
Although this phylogenomic approach is being increasingly used to identify virulence determinants, and we discuss a number of real data examples below, it can be used to make general statements about the nature of virulence evolution. Specifically, a virulence mutation that falls deep in the phylogeny such that it is inherited in all subsequent branches, and one evolving in parallel or with evidence of positive selection, necessarily implies that virulence is selectively advantageous. Conversely, a virulence determinant that occurs sporadically on shallow branches and is subject to strong purifying (negative) selection suggests that virulence is not directly beneficial, probably because it inhibits some other component of overall fitness. In such cases, each instance of high virulence may represent an independent and transient evolutionary event. If only a single mutation is associated with a change in virus virulence, as in the case of WNV, then this change in virulence is likely to be selectively advantageous without an evolutionary trade-off with transmissibility, as a reduction in transmissibility would probably need to be compensated for by additional reciprocal mutations located elsewhere in the genome. Hence, if multiple mutations fall on a branch associated with a change in virulence, it is possible that some are virulence determinants and the others are associated with evolutionary trade-offs on other traits.
Although we have described it in terms of emerging viruses, this phylogenomic approach can, in theory, be applied to any system in which a phylogeny can be inferred and in which it is possible to experimentally assess the impact of individual mutations on virulence. Similarly, it can be used to study other virological traits associated with disease emergence, particularly host range. For example, the repeated evolution of the same amino acid changes following the cross-species transmission of avian influenza virus to humans strongly suggests that they directly affect host range66, and a similar approach has been used to elucidate the nature of the evolutionary arms race between viruses and their hosts67,68.
Critically, however, the approach described here should also be considered an idealized one that works best when a limited number of genomic mutations act independently to shape virulence. Virulence determinants may be harder to identify when there are more complex interactions between mutations9, which appears to be true of MYXV (Box 2). Although epistasis is likely to be commonplace in RNA viruses69, little is currently known about whether virulence mutations interact epistatically70. Similarly, this approach may work best for RNA viruses because their constrained genome sizes mean that there are probably a limited number of virulence determinants, increasing the likelihood that they are subject to parallel and/or convergent evolution, and rates of recombination (which complicate phylogenetic relationships) are often fairly low within species71.
Box 2 The phylogenomics of virulence evolution in myxoma virus.
The co-evolution of European rabbits (Oryctolagus cuniculus) and the myxoma virus (MYXV) released as a biological control against them is arguably the most famous case study in virulence evolution. Fenner and colleagues93 were able to identify ‘grades’ of MYXV virulence using laboratory assays in European rabbits. The most lethal grade I strains (shaded red in the figure) were characterized by almost 100% mortality, whereas the most ‘attenuated’ grade V strains had mortality <50% and longer survival times. Grade I strains were released in both Australia (starting strain, SLS) and Europe (starting strain, Lausanne) in the early 1950s, and soon after lower virulence viruses began to appear in parallel in both continents, with intermediate virulence grades (for example, grade III) becoming the most common in the field.
Although of large importance, the studies of Fenner were necessarily limited in that the genetic basis of virulence evolution was unknown. That the trajectory of virulence evolution was the same in both Australia and Europe meant that the same virulence determinants might be involved on both continents. Therefore, it was a surprise when the first large-scale genome comparisons of MYXV revealed that virulence grades changed frequently across the virus phylogeny, as depicted in the phylogeny, and that different mutations appeared in the Australian and European epidemics (depicted as different filled shapes at branch tips), with no parallel evolution of possible virulence determinants within Australia and Europe94,145. For example, there were no mutations that were unique to the most attenuated grade V viruses, nor to the highest virulence grade I viruses. Hence, there are multiple genetic routes to attenuation or virulence in MYXV, such that there has been convergent evolution for phenotype but not genotype, and both attenuating and virulence-restoring mutations have been fixed in Australian MYXV. Therefore, there is no simple way to predict virulence evolution from genome-scale comparisons in the case of MYXV and no parallel or convergent evolution to guide experimentation. Indeed, experimentally verified virulence determinants in rabbits remain elusive17.
The ongoing evolution of MYXV virulence reflects both mutations acquired in the virus and resistance evolution in the rabbit host11. This co-evolutionary process has also resulted in a marked increase in MYXV virulence146. Unlike viruses sampled in the 1950s before the evolution of any host resistance, most virus isolates sampled in the 1990s that have evolved in the face of host resistance induce a lethal immune collapse syndrome with similarities to septic shock146.
Examples of virulence evolution in nature
To illustrate how a phylogenomic approach can shed light on the evolution of virus virulence, we now briefly outline a number of cases in which it can be or has been applied. We begin by considering cases in which virulence determinants have been successfully mapped (WNV and avian influenza A virus (AIV)), move on to those in which revealing the mutations that underpin changes in virulence has been more complex (MYXV, Marek’s disease virus (MDV) and HIV) and end by examining virulence evolution in two recent disease outbreaks (EBOV and ZIKV). When possible, we also outline what the phylogenomic analysis in each case has told us about the evolution of virulence in general.
West Nile virus
In 1999, a new lineage of WNV became the leading cause of arthropod-borne viral encephalitis in humans and horses in North America, spreading from east to west across the continent72 and causing severe mortality in many bird species, particularly the American crow73. Phylogenomic analysis revealed that a single Thr249Pro (T249P) amino acid substitution in the virus NS3 helicase protein was associated with high-virulence WNV outbreaks in corvids on multiple continents. Experimental analysis in captive crows then showed that this mutation was sufficient to explain the high fatality rates in American crows, perhaps because it increased the rate of virus replication58. WNV therefore provides an important example of where a single genetic switch controls virulence, which is obviously the easiest scenario to detect using a phylogenomic approach. Of more general importance was that T249P evolved in parallel and experienced an elevated rate of nonsynonymous change, suggesting that high virulence was selectively favoured in the absence of an evolutionary trade-off as no reciprocal mutations were observed elsewhere in the viral genome58. However, because WNV infects a variety of bird species, it is possible that the repeated appearance of T249P in fact reflects aspects of viral evolution in different hosts. In particular, American robins may have been responsible for a substantial proportion for the cross-continent spread of the virus74, in which case T249P may have been selected to increase replication (transmissibility) in that species, a coincidental by-product of which was heightened virulence in crows.
Avian influenza A virus
Ever since the emergence of the highly pathogenic H5N1 subtype of influenza virus, there has been concern over whether this influenza virus could establish sustained transmission in humans, in which it causes only sporadic spillover infections at present75,76. More recently, highly pathogenic H7N9 has sporadically infected humans77 and continues to spread through poultry populations in China78, evolving from a low-virulence ancestor79. Although the true number of human cases, and hence accurate mortality, is difficult to ascertain, it is clear that both H5N1 and H7N9 cause fairly high mortality in humans and could have serious consequences were they to trigger a large-scale human epidemic. This concern has led to attempts to use genomic data to help in pandemic risk assessment80.
At the virus subtype level, the presence of a run of polybasic amino acids in the hinge region between the HA1 and HA2 subunits that make up the haemagglutinin (HA) protein of influenza virus helps it establish a systemic, and subsequently more serious, infection and thereby acts as a useful marker of high-virulence strains of the H5 and H7 AIV subtypes79,81,82. This marker makes it relatively easy to distinguish between potentially low-virulence and high-virulence AIVs, although what triggers the evolution of the high-pathogenicity variants in these subtypes is unclear83. Other individual amino acid changes, affecting a variety of gene functions, have also been proposed as specific virulence determinants for H5N1 (refs84–86) as well as in those viruses that circulate in human populations such as seasonal H3N2 (ref.87), the H1N1 virus responsible for the global pandemic of 1918–1919 (ref.88) (in which host inflammatory and cell death responses to infection appear to play a key role)89, and influenza B virus90 (Table 1).
The key unresolved question is how natural selection will shape both virulence and transmissibility if an AIV-like H5N1 or H7N9 virus is eventually able to develop sustained transmission in humans. An added complexity is that phylogenomic analyses reveal a consistent set of mutations that distinguish human and avian influenza viruses, although whether these affect host range alone, or both host range and virulence, is unclear66,84.
Myxoma virus
The canonical study of virulence evolution following a species jump is MYXV in European rabbits, with a body of classic work undertaken by Fenner and colleagues91–93 (Box 2). In both Australia and Europe, highly virulent strains of MYXV were used as a biological control against the European rabbit population, with releases beginning in the early 1950s. In both continents, the same trajectory of virulence evolution was observed: virulence declined from the highly virulent (that is, grade I) release strains to encompass a far wider range of virulence grades, including the most attenuated grade V strains, with strains of ‘intermediate’ virulence the most commonly sampled in the field. This pattern, reflecting a combination of the virus evolving more attenuated strains and the host developing resistance, fuelled the idea of a trade-off between virulence and transmissibility.
Sixty years after the initial release of MYXV, the first large-scale genomic studies of its spread were performed (Box 2). Phylogenomic analysis revealed that the virulence phenotype has changed on a regular basis94. However, a major surprise was that each change in virulence was associated with a different set of mutations across multiple genes94,95. Although which mutations had the greatest impact on virulence is still unclear and requires further experimental analysis, such a phylogenomic pattern indicates that there are multiple routes to achieving the same levels of virulence, including attenuation, such that there has been convergent evolution for phenotype but not genotype. It is likely that this evolutionary flexibility in part reflects the fairly large genome size of MYXV (a double-stranded DNA virus of ~160,000 bp), which may mean that there is a large number of potential virulence determinants that can interact through epistasis96, in turn complicating any phylogenomic analysis.
Marek’s disease virus
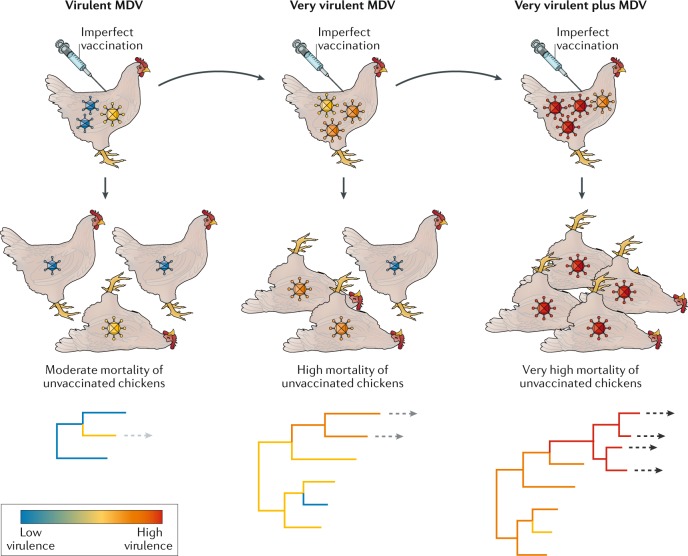
Whether ‘imperfect’ (that is, ‘leaky’) vaccination against infectious disease, in which disease symptoms are reduced but there is less impact on virus replication and transmission, will change the selection pressures acting on the pathogen and affect virulence evolution has been the source of debate97,98. Although still contentious, particularly in the case of human disease, there is good evidence that imperfect vaccination has increased virulence in the case of MDV, a DNA herpesvirus that poses a major problem to the poultry industry99. In the 1960s, the appearance of virulent MDV strains forced the development of the first generation of Marek’s disease vaccines. However, because these vaccines were imperfect, ‘very virulent’ MDV began to appear within 10 years, necessitating a second-generation vaccine. This very virulent MDV was followed, more rapidly, by the appearance of ‘very virulent plus’ MDV, requiring a third-generation vaccine (Fig. 3). Imperfect MDV vaccines enhance virulence by elongating the infectious periods and hence transmission potential of virulent strains that would have been removed by natural selection before transmission in the absence of vaccination99. Although the genomic basis to MDV virulence evolution is currently uncertain, with some causative amino acid changes proposed100, initial phylogenomic studies suggest that, as in the case of MYXV, there are multiple genetic pathways to high virulence101 (and which again may reflect the fairly large size of the viral genome). Not only does virulence evolution in MDV have important implications for vaccination strategies against other diseases in which vaccine efficacy is fairly low102, but it also shows that in some circumstances increased virulence can be selectively advantageous.
Fig. 3. Evolution of virulence in the context of imperfect vaccination.
In the 1960s, a vaccine was developed for Marek’s disease virus (MDV) of chickens present on poultry farms. This imperfect vaccine reduced disease symptoms but did not prevent virus replication, thereby extending the infectious periods, and hence potential for transmission, of virulent strains that would have been removed by natural selection before transmission to a new host in the pre-vaccine era99. Because of this, ‘very virulent’ MDV began to appear within 10 years, necessitating the development of a second-generation vaccine that was also imperfect. This was followed, in an even shorter period, by the appearance of ‘very virulent plus’ MDV, requiring a third-generation vaccine. Although the genomic basis of MDV virulence is currently unknown, the phylogenies at the bottom of the figure hypothetically assign virulence to multiple causative mutations (as in the case of myxoma virus). The dashed arrows indicate the evolution of viruses to the next virulence grade.
HIV
Given the importance of HIV to human health and that it ignited much of the research on disease emergence, it is no surprise that there has been considerable discussion on the evolution of HIV virulence103–107. Indeed, it is striking that HIV in humans is markedly more virulent than the closely related viruses that naturally infect non-human primates in Africa (Box 1). Although there have been suggestions that HIV has begun to evolve reduced virulence108, discussions of the trajectory of virulence evolution are necessarily complicated by the fact that antiviral therapy has greatly extended life expectancy.
HIV virulence is often approximated as the degree of variation in the set point viral load (SPVL) that is established soon after initial infection104. The higher the SPVL, reflecting greater levels of virus replication, then the more rapidly the patient will progress to AIDS in the absence of antiviral therapy, although other studies have suggested that the replicative capacity of the virus itself is a more informative marker of virulence and is also a direct measure of virus fitness109. Indeed, some ‘controller’ individuals are able to control levels of HIV in the absence of antiviral therapy, and it has been shown that this is in part due to infection with viruses of reduced replicative capacity110.
Importantly, viral genetic variation may play a more important role in shaping HIV virulence than host factors, with approximately one-third of the observed variability in SPVL assigned to virus factors111 and only ~13% seemingly due to the host112. This observation also implies that SPVL, and hence virulence, can be selectively optimized113. In support is evidence that SPVL, and hence virulence, has declined in some African HIV subtypes, even accounting for the use of antiviral therapy, and that this reflects a trade-off between virulence and transmissibility114. Importantly, however, despite many studies into the determinants of HIV virulence, the virus genomic mutations responsible for determining SPVL are still uncertain and multiple genes may be involved104. The difficulty in assigning the genetic determinants of SPVL may be in part due to genetic variation across viral populations111. For example, heritability in SPVL was highest (~60%) between individuals in the Swiss HIV cohort, which also represents the most homogenous viral population113.
Ebola virus
The 2013–2016 outbreak of EBOV (Makona variant) in West Africa was the largest and longest described in humans since the first description of the disease in 1976, with approximately 29,000 cases and some 11,000 deaths. In addition to hindering attempts at disease control, this elongated period of transmission in humans may have resulted in different selection pressures from those faced in the animal reservoir. This outbreak also raised key questions about virulence evolution, particularly whether natural selection would have favoured EBOV variants causing higher or lower human case fatality rates had the virus not been stamped out by public health intervention115.
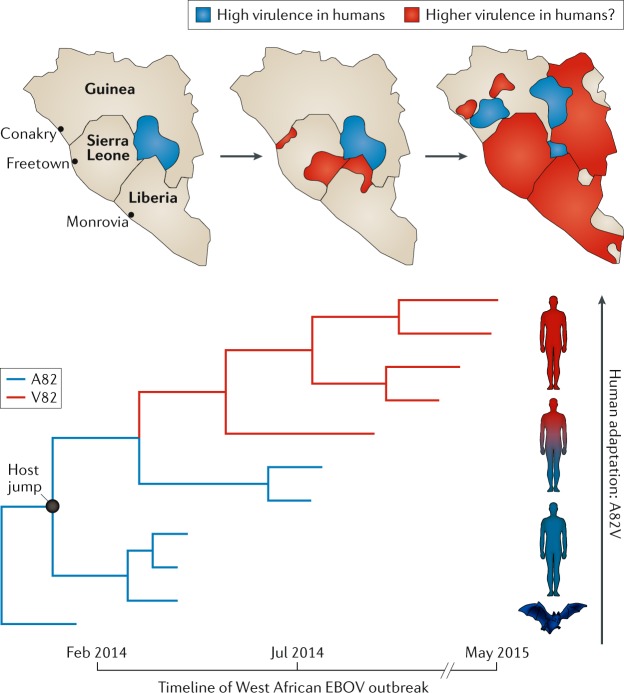
Phylogenetic analysis of EBOV during the 2013–2016 outbreak revealed an Ala82Val (A82V) substitution in the virus glycoprotein to be of particular importance70,116. A82V is notable as it falls on a deep internal branch of the EBOV phylogeny, compatible with adaptive evolution, whereas other amino acid changes are associated with individual or only small clusters of sequences. Moreover, A82V improves binding to the human NPC1 receptor utilized by EBOV117, which would increase infectivity in humans, at the same time reducing infectivity in cells from the bat reservoir species70,116. Intriguingly, the appearance of A82V on the EBOV phylogeny is associated with two key epidemiological features: an increase in case numbers and an increase in mortality (Fig. 4). If there was indeed an increase in both EBOV transmissibility and virulence, then higher virulence is likely to have directly increased viral fitness, and in the absence of an evolutionary trade-off as only a single substitution was identified. However, these apparent changes in phenotype also coincided with the movement of the virus from Guinea to Sierra Leone, such that any change in case numbers and mortality could in fact be due to a change in epidemiological factors (such as access to health care or differing human demographics and/or transmission networks), and recent studies using animal models suggest that A82V has no direct impact on virulence118.
Fig. 4. The relationship between host adaptation and the evolution of virulence in Ebola virus.
A model Ebola virus (EBOV; Makona variant) phylogeny illustrates the evolution of a single amino acid substitution (glycoprotein, A82V) that is associated with viral adaptation to the human host during the West African EBOV outbreak of 2013–2016. A82V improves binding to the human NPC1 receptor utilized by EBOV, increasing infectivity in humans (red) while simultaneously reducing infectivity in cells from the bat reservoir species (blue)70,116. Maps above the phylogeny show the spread of EBOV over the timeline of the outbreak in the three affected countries in West Africa, where blue-shaded regions correspond to the wild-type virus variant (A82) and red-shaded areas correspond to mutated virus variant (V82). It is possible that A82V was also associated with an increase in both EBOV case numbers and mortality (that is, virulence) as the outbreak progressed, such that increased virulence is directly selectively advantageous, although this is confounded by epidemiological factors.
Zika virus
ZIKV is the most recent emerging virus to lead to a major public health scare and is puzzling because a seemingly benign virus suddenly increased in virulence, causing severe neurological disease in humans. Before 2007, there were fewer than 20 human cases of ZIKV reported and all were mild infections restricted to Africa and Asia9,119. Consequently, neither the disease caused by ZIKV nor the molecular determinants of ZIKV virulence were well characterized, and it is likely that there was systematic under-reporting of infections, including those associated with severe disease. In 2007, the Pacific Islands reported the first major outbreaks of ZIKV before the virus spread to the Americas in 2014. Although the majority of human infections range from asymptomatic to mild, the virus was associated with the neurological Guillain–Barré syndrome in French Polynesia in 2013 (ref.120), and those cases from the Americas, particularly Brazil, were linked to more severe diseases, including congenital abnormalities such as microcephaly121. Phylogenetic analysis revealed that the most recent Zika epidemics are due to the Asian lineage of ZIKV, rather than the African lineage122, and that the virus spread cryptically in Brazil for at least a year before its detection20. Although there are multiple amino acid differences between the African and Asian lineages122,123, it has been claimed that those in the Asian lineage that spread through the Americas may be directly linked to both increased infectivity in Aedes aegypti mosquitoes124 and microcephaly, most notably a Ser139Asn (S139N) amino acid change in the PrM protein125. However, there is still considerable uncertainty in this area, with others arguing that viruses from both lineages can cause neurovirulence but that cases often go unreported9,126. Hence, the case of ZIKV highlights the difficulty in assessing virulence evolution within a background of sparse and biased sampling even with phylogenomic data and shows the importance of collecting reliable, real-time epidemiological data even in low-incidence situations.
Future directions and recommendations
Virulence evolution has been one of the longest-standing issues in evolutionary biology. Although a strong body of theory has been developed, there are few cases in which we understand the forces that have shaped particular instances of virulence evolution and even fewer in which we have successfully linked evolutionary theory with individual genomic changes. We believe that a synthesis of experimental studies of virulence determinants and long-standing theory of virulence evolution set within a phylogenomic framework will generate a more comprehensive understanding of virulence evolution. In particular, not only does a phylogenomic approach enable potential virulence determinants to be identified, which is being increasingly used in the case of emerging viruses, but this analysis also sheds light on the models of virulence evolution that have occupied theoreticians for decades.
Recent advances in real-time genomics during disease outbreaks127,128 and the increased demand for precision in public health interventions may help in the development of a new understanding of the evolution of pathogen virulence. We contend that this can be achieved within a phylogenomic framework as long as relevant data are available and strong links are made between genomics, phylogenetics, epidemiology, and experimental studies of virus virulence and fitness. Therefore, it is critically important to collect clinical (that is, disease symptoms and severity) and epidemiological (that is, time and place of sampling) metadata concurrently with the sequencing of virus genomes and to sample across a range of clinical syndromes, not just those associated with severe disease. We also stress the value of gathering concurrent and historical data from likely reservoir species as these will provide a more complete insight into virulence evolution and determining the full range of microorganisms that infect a particular species, as well as their interactions, as assigning disease syndromes to individual pathogens may often be difficult. Thankfully, advances in metagenomics now make the latter task feasible6,7,129. Similarly, there is a marked lack of good virulence grading schemes among viral infections. Although such schemes can sometimes be simplistic, assuming discrete virulence categories that may not exist in nature and incorporating degrees of subjectivity, the case of MYXV shows that they are key to considering the relationship between genotype and phenotype that is essential to understanding virulence evolution.
Finally, it is possible that an increased understanding of virulence evolution drawn from a phylogenomic approach may contribute to new strategies for pathogen control and eradication, and there is a clear potential for this framework to inform and improve the fields of disease management and the biological control of invasive pests. Although predicting where and when a new disease might emerge is clearly unfeasible because of the immense complexities involved54,130, predicting the overall trajectory of the virulence evolution of a virus in a novel host may be more achievable. Once again, biocontrol presents a compelling example. Although controversial131–133, the proposed release of cyprinid herpesvirus 3 (CyHV-3) as a biological control against invasive common carp (Cyprinus carpio L.) in Australia may present a unique opportunity to follow, in real time, the co-evolution between host and virus at both the genotypic and phenotypic scales. Both theory and virus natural history predict that CyHV-3 virulence will decline with time134, and it will be interesting and informative to see how any such virulence evolution is manifest in phylogenomic data.
Acknowledgements
E.C.H. is funded by an Australian Research Council Australian Laureate Fellowship (FL170100022). The authors thank J. Bull for helpful discussions.
Reviewer information
Nature Reviews Genetics thanks S. Alizon and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Glossary
Glossary
- Virulence
Although virulence can be defined in a range of case-specific ways, the simplest definition is the severity or harmfulness of a pathogen.
- Metagenomics
The study of genetic material recovered from microbial (including viral) communities in which genome sequence data are simultaneously generated for all the microorganisms present.
- Biological control
The reduction in numbers or elimination of pest organisms through the introduction of a pathogen.
- Virulence determinants
Mutations in pathogen genomes that directly affect virulence.
- Reverse genetics
Experimental method used to identify gene function by modifying the sequence of a target gene and analysing its phenotypic consequences.
- Phylogenomics
The inference of evolutionary patterns and/or processes through the comparison of whole-genome sequences.
- Phylogeny
Depiction of the evolutionary history of a genetically related group of organisms. Contains branches and nodes.
- Emerging infectious diseases
Infectious diseases that have recently appeared in populations or known diseases that are rapidly increasing in incidence or geographic range.
- Basic reproductive number
(R0). Represents the number of secondary infections caused by an infectious host in an entirely susceptible population.
- Evolutionary trade-off
Occurs when a change in one trait increases fitness but simultaneously reduces fitness because of its impact on another trait, thereby preventing the organism from optimizing both traits.
- Spillover
The initial and sometimes transient appearance of a pathogen in a new species following a host jump.
- Population bottlenecks
Reductions, sometimes drastic, in the size of populations. They often accompany inter-host virus transmission.
- Cross-species transmission
The transmission of a pathogen from one host species to another. Also called host-jumping or host-switching.
- Parallel evolution
An evolutionary process by which two or more separate lineages develop identical characteristics independently.
- Convergent evolution
The descendants of unrelated ancestors that have evolved similar traits independently.
- Positive selection
Natural selection that leads to advantageous mutations spreading through a population. Mutations in coding regions can be either synonymous (which do not change the amino acid; measured as dS) or non-synonymous (which change the amino acid; measured as dN). The dN/dS ratio >1 is sometimes used to infer the occurrence of positive selection, although the accuracy of this measure depends on various factors, including the timescale of sampling.
- Purifying selection
Natural selection that acts to remove low-fitness (including deleterious) mutations from populations. It is the most common form of natural selection and gives a dN/dS < 1. Also called negative selection.
- Epistasis
The interaction among genes at different loci. The function and evolution of one gene may be dependent on the presence of one or more other genes.
- Subtype of influenza virus
Influenza A viruses are categorized into subtypes on the basis of the diversity in two proteins present in the viral envelope: haemagglutinin (H or HA) and neuraminidase (N or NA).
- Heritability
The amount of phenotypic variation in a population that is attributable to individual genetic differences.
Author contributions
Both authors contributed to all aspects of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
National Geographic: leaky vaccines enhance spread of deadlier chicken viruses: http://phenomena.nationalgeographic.com/2015/07/27/leaky-vaccines-enhance-spread-of-deadlier-chicken-viruses/
The Scientist Infographic: evolving virulence: https://www.the-scientist.com/?articles.view/articleNo/50451/title/Infographic--Evolving-Virulence/
References
- 1.Bull JJ, Lauring AS. Theory and empiricism in virulence evolution. PLOS Pathog. 2014;10:e1004387. doi: 10.1371/journal.ppat.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMahon DP, et al. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. Biol. Sci. 2016;283:20160811. doi: 10.1098/rspb.2016.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat. Rev. Genet. 2017;18:41–50. doi: 10.1038/nrg.2016.132. [DOI] [PubMed] [Google Scholar]
- 4.Ansari MA, et al. Genome-to-genome analysis highlights the effect of the human innate and adaptive immune systems on the hepatitis C virus. Nat. Genet. 2017;49:666. doi: 10.1038/ng.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, et al. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J. Clin. Microbiol. 2011;49:3463–3469. doi: 10.1128/JCM.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi M, et al. Redefining the invertebrate virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 7.Shi M, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 8.Stern A, et al. The evolutionary pathway to virulence of an RNA virus. Cell. 2017;169:35–46. doi: 10.1016/j.cell.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossi SL, Ebel GD, Shan C, Shi P-Y, Vasilakis N. Did Zika virus mutate to cause severe outbreaks? Trends Microbiol. 2018 doi: 10.1016/j.tim.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alizon S, Lion S. Within-host parasite cooperation and the evolution of virulence. Proc. Biol. Sci. 2011;278:3738–3747. doi: 10.1098/rspb.2011.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr PJ, et al. Myxoma virus and the leporipoxviruses: an evolutionary paradigm. Viruses. 2015;7:1029–1061. doi: 10.3390/v7031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebihara H, et al. Molecular determinants of Ebola virus virulence in mice. PLOS Pathog. 2006;2:e73. doi: 10.1371/journal.ppat.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jimenez-Guardeño JM, et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLOS Pathog. 2014;10:e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataoka C, et al. The role of VP1 amino acid residue 145 of enterovirus 71 in viral fitness and pathogenesis in a cynomolgus monkey model. PLOS Pathog. 2015;11:e1005033. doi: 10.1371/journal.ppat.1005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song H, Santi N, Evensen O, Vakharia VN. Molecular determinants of infectious pancreatic necrosis virus virulence and cell culture adaptation. J. Virol. 2005;79:10289–10299. doi: 10.1128/JVI.79.16.10289-10299.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uraki R, et al. Virulence determinants of pandemic A(H1N1)2009 influenza virus in a mouse model. J. Virol. 2013;87:2226–2233. doi: 10.1128/JVI.01565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Reverse engineering field isolates of myxoma virus demonstrates that some gene disruptions or loss of function do not explain virulence changes observed in the field. J. Virol. 2017;91:e01289–17. doi: 10.1128/JVI.01289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh DY, Hurt AC. Using ferrets as an animal model for investigating influenza antiviral effectiveness. Front. Microbiol. 2016;7:80. doi: 10.3389/fmicb.2016.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudas G, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faria NR, et al. Epidemic establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes EC, Dudas G, Rambaut A, Andersen KG. The evolution of Ebola virus: insights from the 2013–2016 epidemic. Nature. 2016;358:193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadfield J, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lässig M, Łuksza M. Can we read the future from a tree? eLife. 2014;3:e05060. doi: 10.7554/eLife.05060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neher RA, Russell CA, Shraiman BI. Predicting evolution from the shape of genealogical trees. eLife. 2014;3:e03568. doi: 10.7554/eLife.03568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longdon B, et al. The causes and consequences of changes in virulence following pathogen host shifts. PLOS Pathog. 2015;11:e1004728. doi: 10.1371/journal.ppat.1004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter S, et al. Differential infection patterns and recent evolutionary origins of equine hepaciviruses in donkeys. J. Virol. 2016;91:e01711–e01716. doi: 10.1128/JVI.01711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leroy EM, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–566. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 28.Allocati N, et al. Bat-man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Dis. 2016;2:16048. doi: 10.1038/cddiscovery.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truyen U, Evermann JF, Vieler E, Parrish CR. Evolution of canine parvovirus involved loss and gain of feline host range. Virology. 1996;215:186–189. doi: 10.1006/viro.1996.0021. [DOI] [PubMed] [Google Scholar]
- 30.Allison AB, et al. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLOS Pathog. 2014;10:e1004475. doi: 10.1371/journal.ppat.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geoghegan JL, Senior AM, Di Giallonardo F, Holmes EC. Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl Acad. Sci. USA. 2016;113:4170–4175. doi: 10.1073/pnas.1521582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- 33.Bull JJ. Virulence. Evolution. 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 34.Ebert, D. in Evolution in Health and Disease (ed. Stearns, S. C.) 161–172 (Oxford Univ. Press, Oxford, 1999).
- 35.Ebert D, Bull JJ. Challenging the trade-off model for the evolution of virulence: is virulence management feasible? Trends Microbiol. 2003;11:15–20. doi: 10.1016/s0966-842x(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 36.Ewald PW. Evolution of Infectious Diseases. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 37.Read AF. The evolution of virulence. Trends Microbiol. 1994;2:73–76. doi: 10.1016/0966-842x(94)90537-1. [DOI] [PubMed] [Google Scholar]
- 38.Furió V, et al. Relationship between within-host fitness and virulence in the vesicular stomatitis virus: correlation with partial decoupling. J. Virol. 2012;86:12228–12236. doi: 10.1128/JVI.00755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lion S, Metz JAJ. Beyond R0 maximisation: on pathogen evolution and environmental dimensions. Trends Ecol. Evol. 2018;33:458–473. doi: 10.1016/j.tree.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc. Natl Acad. Sci. USA. 2007;104:17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: history , current state of affairs & the future. J. Evol. Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 42.Alizon S, Michalakis Y. Adaptive virulence evolution: the good old fitness-based approach. Trends Ecol. Evol. 2015;30:248–254. doi: 10.1016/j.tree.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Lipsitch M, Moxon ER. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 1997;5:31–37. doi: 10.1016/S0966-842X(97)81772-6. [DOI] [PubMed] [Google Scholar]
- 44.Novella IS, Elena SF, Moya A, Domingo E, Holland JJ. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J. Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betancourt M, Escriu F, Fraile A, García-Arenal F. Virulence evolution of a generalist plant virus in a heterogeneous host system. Evol. Appl. 2013;6:875–890. doi: 10.1111/eva.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Giallonardo F, Holmes EC. Virus biocontrol: grand experiments in disease emergence and evolution. Trends Microbiol. 2015;23:83–90. doi: 10.1016/j.tim.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willemsen A, Zwart MP, Elena SF. High virulence does not necessarily impede viral adaptation to a new host: a case study using a plant RNA virus. BMC Evol. Biol. 2017;17:25. doi: 10.1186/s12862-017-0881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Roode JC, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl Acad. Sci. USA. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 50.Lloyd-Smith JO, et al. Epidemic dynamics at the human-animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergstrom CT, McElhany P, Real L A. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc. Natl Acad. Sci. USA. 1999;96:5095–5100. doi: 10.1073/pnas.96.9.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolker BM, Nanda A, Shah D. Transient virulence of emerging pathogens. J. R . Soc. Interface. 2010;7:811–822. doi: 10.1098/rsif.2009.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity , and control. Trends Microbiol. 2012;20:336–342. doi: 10.1016/j.tim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolan PT, Whitfield ZJ, Andino R. Mapping the evolutionary potential of RNA viruses. Cell Host Microbe. 2018;23:435–446. doi: 10.1016/j.chom.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sackman AM, et al. Mutation driven parallel evolution during viral adaptation. Mol. Biol. Evol. 2017;34:3243–3253. doi: 10.1093/molbev/msx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart C-B, Schilling JW, Wilson AC. Convergent evolution of lysozyme sequences? Nature. 1988;332:788. doi: 10.1038/332787c0. [DOI] [PubMed] [Google Scholar]
- 57.Frickel J, Feulner PGD, Karakoc E, Becks L. Population size changes and selection drive patterns of parallel evolution in a host-virus system. Nat. Commun. 2018;9:1706. doi: 10.1038/s41467-018-03990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brault AC, et al. A single positively selected West Nile viral mutation confers increased avian virogenesis in American crows. Nat. Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Longdon B, et al. Host shifts result in parallel genetic changes when viruses evolve in closely related species. PLOS Pathog. 2018;14:e1006951. doi: 10.1371/journal.ppat.1006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver S, et al. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLOS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pybus OG, et al. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol. Biol. Evol. 2007;24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 63.Bhatt S, Holmes EC, Pybus OG. The genomic rate of molecular adaptation of the human influenza A virus. Mol. Biol. Evol. 2011;28:2443–2451. doi: 10.1093/molbev/msr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geoghegan JL, Senior AM, Holmes EC. Pathogen population bottlenecks and adaptive landscapes: overcoming the barriers to disease emergence. Proc. Biol. Sci. 2016;283:1837. doi: 10.1098/rspb.2016.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morley VJ, Turner PE. Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change. Evolution. 2017;71:872–883. doi: 10.1111/evo.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taubenberger JK, et al. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 67.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLOS Biol. 2004;2:e275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bedhomme S, Hillung J, Elena SF. Emerging viruses: why they are not jacks of all trades? Curr. Opin. Virol. 2015;10:1–6. doi: 10.1016/j.coviro.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 70.Urbanowicz RA, et al. Human adaptation of Ebola virus during the West African outbreak. Cell. 2016;167:1079–1087. doi: 10.1016/j.cell.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat. Rev. Micro. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Giallonardo F, et al. Fluid spatial dynamics of West Nile virus in the USA: rapid spread in a permissive host environment. J. Virol. 2016;90:862–872. doi: 10.1128/JVI.02305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LaDeau SL, Kilpatrick AM, Marra PP. West Nile virus emergence and large-scale declines of North American bird populations. Nature. 2007;447:710–713. doi: 10.1038/nature05829. [DOI] [PubMed] [Google Scholar]
- 74.Kilpatrick AM. Globalization, land use, and the invasion of West Nile virus. Science. 2011;334:323–327. doi: 10.1126/science.1201010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guan Y, et al. H5N1 influenza: a protean pandemic threat. Proc. Natl Acad. Sci. USA. 2004;101:8156–8161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302:1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 77.Lam TT-Y, et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522:102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 78.Yang L, et al. Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in mainland China. J. Virol. 2017;91:e01277–e01217. doi: 10.1128/JVI.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi W, et al. Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low pathogenic ancestor. J. Virol. 2018;92:e00921–e00917. doi: 10.1128/JVI.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lipsitch M, et al. Viral factors in influenza pandemic risk assessment. eLife. 2016;5:e18491. doi: 10.7554/eLife.18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. Bioessays. 2003;25:657–671. doi: 10.1002/bies.10303. [DOI] [PubMed] [Google Scholar]
- 82.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 83.Monne I, et al. Emergence of a highly pathogenic avian influenza virus from a low pathogenic progenitor. J. Virol. 2014;88:4375–4388. doi: 10.1128/JVI.03181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Russell CA, et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 86.Pearce MB, et al. Enhanced virulence of clade 2.3.2.1 highly pathogenic avian influenza A H5N1 viruses in ferrets. Virology. 2017;502:114–122. doi: 10.1016/j.virol.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. NS1 protein amino acid changes D189N and V194I affect interferon responses, thermosensitivity , and virulence of circulating H3N2 human influenza A viruses. J.Virol. 2017;91:e01930–e01916. doi: 10.1128/JVI.01930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl Acad. Sci. USA. 2008;105:4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kash JC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bae J-Y, et al. A single amino acid in the polymerase acidic protein determines the pathogenicity of influenza B viruses. J. Virol. 2018;92:e00259–e00218. doi: 10.1128/JVI.00259-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fenner F. Biological control as exemplified by smallpox eradication and myxomatosis. Proc. R . Soc. B. 1983;218:259–285. doi: 10.1098/rspb.1983.0039. [DOI] [PubMed] [Google Scholar]
- 92.Fenner F, Marshall ID. A comparison of the virulence for European rabbits (Oryctolagus cuniculus) of strains of myxoma virus recovered in the field in Australia, Europe and America. J. Hyg. 1957;55:149–191. doi: 10.1017/s0022172400037098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fenner F, Ratcliffe FN. Myxomatosis. Cambridge: Cambridge Univ. Press; 1965. [Google Scholar]
- 94.Kerr PJ, et al. Evolutionary history and attenuation of myxoma virus on two continents. PLOS Pathog. 2012;8:e1002950. doi: 10.1371/journal.ppat.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kerr PJ, et al. Genome scale evolution of myxoma virus (MYXV) reveals host-pathogen adaptation and rapid geographic spread. J. Virol. 2013;87:12900–12915. doi: 10.1128/JVI.02060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elena SF, Carrasco P, Daròs JA, Sanjuán R. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 2006;7:168–173. doi: 10.1038/sj.embor.7400636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gandon S, Mackinnon MJ, Nee S, Read AF. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- 98.Smith T. Imperfect vaccines and imperfect models. Trends Ecol. Evol. 2002;17:154–156. [Google Scholar]
- 99.Read AF, et al. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLOS Biol. 2015;13:e1002198. doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kamil JP, et al. vLIP, a viral lipase homologue, is a virulence factor of Marek’s disease virus. J. Virol. 2005;79:6984–6996. doi: 10.1128/JVI.79.11.6984-6996.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trimpert J, et al. A phylogenomic analysis of Marek’s disease virus reveals independent paths to virulence in Eurasia and North America. Evol. Appl. 2017;10:1091–1101. doi: 10.1111/eva.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mackinnon MJ, Read AF. Immunity promotes virulence evolution in a malaria model. PLOS Biol. 2004;2:e230. doi: 10.1371/journal.pbio.0020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carre N, et al. Has the rate of progression to AIDS changed in recent years? AIDS. 1997;11:1611–1618. doi: 10.1097/00002030-199713000-00010. [DOI] [PubMed] [Google Scholar]
- 104.Fraser C, et al. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science. 2014;343:1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Galai N, et al. Temporal trends of initial CD4 cell counts following human immunodeficiency virus seroconversion in Italy , 1985-1992. The human immunodeficiency virus Italian seroconversion study. Am. J. Epidemiol. 1996;143:278–282. doi: 10.1093/oxfordjournals.aje.a008739. [DOI] [PubMed] [Google Scholar]
- 106.Herbeck JT, et al. Is the virulence of HIV changing? A meta-analysis of trends in prognostic markers of HIV disease progression and transmission. AIDS. 2012;26:193–205. doi: 10.1097/QAD.0b013e32834db418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vanhems P, Lambert J, Guerra M, Hirschel B, Allard R. Association between the rate of CD4 + T cell decrease and the year of human immunodeficeny virus (HIV) type 1 seroconversion among persons enrolled in the Swiss HIV cohort study. J. Infect. Dis. 1999;180:1803–1808. doi: 10.1086/315110. [DOI] [PubMed] [Google Scholar]
- 108.Arien KK, Vanham G, Arts EJ. Is HIV-1 evolving to a less virulent form in humans? Nat. Rev. Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Claiborne DT, et al. Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4 + T cells, and disease progression. Proc. Natl Acad. Sci. USA. 2015;112:E1480–E1489. doi: 10.1073/pnas.1421607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miura T, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J. Virol. 2010;84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blanquart F, et al. Viral genetic variation accounts for a third of variability in HIV-1 set-point viral load in Europe. PLOS Biol. 2017;15:e2001855. doi: 10.1371/journal.pbio.2001855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fellay J, et al. Common genetic variation and the control of HIV-1 in humans. PLOS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Alizon S, et al. Phylogenetic approach reveals that virus genotype largely determines HIV set-point viral load. PLOS Pathog. 2010;6:e1001123. doi: 10.1371/journal.ppat.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blanquart F, et al. A transmission-virulence evolutionary trade-off explains attenuation of HIV-1 in Uganda. eLife. 2016;5:e20492. doi: 10.7554/eLife.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sofonea MT, Aldakak L, Boullosa LFVV, Alizon S. Can Ebola virus evolve to be less virulent in humans? J. Evol. Biol. 2018;31:382–392. doi: 10.1111/jeb.13229. [DOI] [PubMed] [Google Scholar]
- 116.Diehl WE, et al. Ebola virus glycoprotein with increased infectivity dominated the 2013-2016 epidemic. Cell. 2016;167:1088–1098. doi: 10.1016/j.cell.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ng M, et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. eLife. 2015;4:e11785. doi: 10.7554/eLife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marzi A, et al. Recently identified mutations in the Ebola virus-Makona genome do not alter pathogenicity in animal models. Cell Rep. 2018;23:1806–1816. doi: 10.1016/j.celrep.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chan JF, Choi GK, Yip CC, Cheng VC, Yuen KY. Zika fever and congenital Zika syndrome: an unexpected emerging arboviral disease. J. Infect. 2016;72:507–524. doi: 10.1016/j.jinf.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cao-Lormeau VM, et al. Guillain–Barré syndrome outbreak associated with Zika virus infection in French Polynesia: a case–control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fauci AS, Morens DM. Zika virus in the Americas — yet another arbovirus threat. N. Engl. J. Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 122.Simonin Y, van Riel D, Van de Perre P, Rockx B, Salinas S. Differential virulence between Asian and African lineages of Zika virus. PLOS Negl. Trop. Dis. 2017;11:e0005821. doi: 10.1371/journal.pntd.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grubaugh ND, Faria NR, Andersen KG, Pybus OG. Genomic insights into Zika virus emergence and spread. Cell. 2018;172:1160–1162. doi: 10.1016/j.cell.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 124.Liu Y, et al. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature. 2017;545:482–486. doi: 10.1038/nature22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yuan L, et al. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science. 2017;358:933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 126.Rosenfeld AB, Doobin DJ, Warren AL, Racaniello VR, Vallee RB. Replication of early and recent Zika virus isolates throughout mouse brain development. Proc. Natl Acad. Sci. USA. 2017;114:12273–12278. doi: 10.1073/pnas.1714624114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Firth C, Lipkin WI. The genomics of emerging pathogens. Annu. Rev. Genom. Hum. Genet. 2013;14:281–300. doi: 10.1146/annurev-genom-091212-153446. [DOI] [PubMed] [Google Scholar]
- 128.Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shi M, Zhang Y-Z, Holmes EC. Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 2018;243:83–90. doi: 10.1016/j.virusres.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Geoghegan JL, Holmes EC. Predicting virus emergence amidst evolutionary noise. Open Biol. 2017;7:170189. doi: 10.1098/rsob.170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lighten J, van Oosterhout C. Biocontrol of common carp in Australia poses risks to biosecurity. Nat. Ecol. Evol. 2017;1:87. doi: 10.1038/s41559-017-0087. [DOI] [PubMed] [Google Scholar]
- 132.Marshall J, et al. Biocontrol of invasive carp: risks abound. Science. 2018;359:877. doi: 10.1126/science.aar7827. [DOI] [PubMed] [Google Scholar]
- 133.McColl KA, Sheppard AW, Barwick M. Safe and effective biocontrol of common carp. Nat. Ecol. Evol. 2017;1:134. doi: 10.1038/s41559-017-0134. [DOI] [PubMed] [Google Scholar]
- 134.McColl KA, Sunarto A, Holmes EC. Cyprinid herpesvirus 3 and its evolutionary future as a biological control agent for carp in Australia. Virol. J. 2016;13:206. doi: 10.1186/s12985-016-0666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andre JB, Hochberg ME. Virulence evolution in emerging infectious diseases. Evolution. 2005;59:1406–1412. [PubMed] [Google Scholar]
- 136.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: Showing AIDS the door. Science. 2012;335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. Understanding the benign nature of SIV infection in natural hosts. J. Clin. Invest. 2007;117:3148–3154. doi: 10.1172/JCI33034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Crawford PC, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 140.Parrish CR, Murcia PR, Holmes EC. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J. Virol. 2015;89:2990–2994. doi: 10.1128/JVI.03146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Feng KH, et al. Equine and canine influenza viruses H3N8 viruses show minimal biological differences despite phylogenetic divergence. J. Virol. 2015;89:6860–6873. doi: 10.1128/JVI.00521-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bourhy H, Dautry-Varsat A, Hotez PJ, Salomon J. Rabies, still neglected after 125 years of vaccination. PLOS Negl Trop. Dis. 2010;4:e839. doi: 10.1371/journal.pntd.0000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garver KA, Batts WN, Kurath G. Virulence comparisons of infectious hematopoietic necrosis virus U and M genogroups in sockeye slamon and rainbow trout. J. Aquat. Anim. Health. 2006;18:232–243. doi: 10.1577/H05-038.1. [DOI] [PubMed] [Google Scholar]
- 144.Penaranda MM, Purcell MK, Kurath G. Differential virulence mechanisms of infectious hematopoietic necrosis virus in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. Gen. J. Virol. 2009;90:2172–2182. doi: 10.1099/vir.0.012286-0. [DOI] [PubMed] [Google Scholar]
- 145.Kerr PJ, et al. The evolution of myxoma virus: genomic and phenotypic characterization of isolates from Great Britain reveals multiple successful evolutionary pathways distinct from those in Australia. PLOS Pathog. 2017;13:e1006252. doi: 10.1371/journal.ppat.1006252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kerr PJ, et al. Next step in the ongoing arms race between myxoma virus and wild rabbits in Australia is a novel disease phenotype. Proc. Natl Acad. Sci. USA. 2017;114:9397–9402. doi: 10.1073/pnas.1710336114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jiao P, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cotter CR, Jin H, Chen Z. A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLOS Pathog. 2014;10:e1003831. doi: 10.1371/journal.ppat.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gromowski GD, Firestone CY, Whitehead SS. Genetic determinants of Japanese encephalitis virus vaccine strain SA14-14-2 that govern attenuation of virulence in mice. J. Virol. 2015;89:6328–6337. doi: 10.1128/JVI.00219-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.He W, et al. Effect of an 88-amino-acid deletion in nsp2 of porcine reproductive and respiratory syndrome virus on virus replication and cytokine responses in vitro. Arch. Virol. 2018;163:1489–1501. doi: 10.1007/s00705-018-3760-7. [DOI] [PubMed] [Google Scholar]
- 151.Yu X, et al. The glutamic residue at position 402 in the C-terminus of Newcastle disease virus nucleoprotein is critical for the virus. Sci. Rep. 2017;7:17471. doi: 10.1038/s41598-017-17803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Panzarin V, et al. Low evolutionary rate of infectious pancreatic necrosis virus (IPNV) in Italy is associated with reduced virulence in trout. Virus Evol. 2018;4:vey019. doi: 10.1093/ve/vey019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dietzschold B, et al. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl Acad. Sci. USA. 1983;80:70–74. doi: 10.1073/pnas.80.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Blanie S, Mortier J, Delverdier M, Bertagnoli S, Camus-Bouclainville C. M148R and M149R are two virulence factors for myxoma virus pathogenesis in the European rabbit. Vet. Res. 2009;40:11. doi: 10.1051/vetres:2008049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bauer PH, et al. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J. Virol. 1995;69:7925–7931. doi: 10.1128/jvi.69.12.7925-7931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Balinsky CA, et al. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J. Virol. 2007;81:11392–11401. doi: 10.1128/JVI.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]