Key Points
RNA pseudoknots are structural motifs in RNA that are increasingly recognized in viral and cellular RNAs. They have been shown to have a various roles in virus and cellular gene expression.
Pseudoknots are formed upon base pairing of a single-stranded region of RNA in the loop of a hairpin to a stretch of complementary nucleotides elsewhere in the RNA chain. This simple folding strategy can generate a large number of stable three-dimensional folds, which display a diverse range of highly specific functions.
Pseudoknot function is frequently associated with interactions with ribosomes. The inclusion of pseudoknots in an mRNA can thus confer unusual translational properties.
Many RNA viruses use pseudoknots in the control of viral RNA translation, replication and the switch between the two processes. Some satellite viruses encode ribozymes with active sites that are folded by a pseudoknot.
In cellular RNAs, pseudoknots are associated with all aspects of mRNA function and also ribosome function, as ribosomal RNAs contain numerous pseudoknots. Other essential cellular pseudoknots have been described in telomerase RNA and transfer messenger RNA.
Future research into pseudoknots will focus on structure–function relationships and bioinformatics identification of pseudoknots in genomes. The use of pseudoknots in antiviral applications could also become more widespread.
Supplementary information
The online version of this article (doi:10.1038/nrmicro1704) contains supplementary material, which is available to authorized users.
RNA pseudoknots have been identified in many different viral and cellular RNAs and are known to have various roles in virus and cellular gene expression. Here, Ian Brierley and colleagues review viral pseudoknots and the role of these structural motifs in virus gene expression and genome replication.
Supplementary information
The online version of this article (doi:10.1038/nrmicro1704) contains supplementary material, which is available to authorized users.
Abstract
RNA pseudoknots are structural elements found in almost all classes of RNA. First recognized in the genomes of plant viruses, they are now established as a widespread motif with diverse functions in various biological processes. This Review focuses on viral pseudoknots and their role in virus gene expression and genome replication. Although emphasis is placed on those well defined pseudoknots that are involved in unusual mechanisms of viral translational initiation and elongation, the broader roles of pseudoknots are also discussed, including comparisons with relevant cellular counterparts. The relationship between RNA pseudoknot structure and function is also addressed.
Supplementary information
The online version of this article (doi:10.1038/nrmicro1704) contains supplementary material, which is available to authorized users.
Main
The pioneering work that established pseudoknots as a genuine folding motif in RNA was carried out in the laboratories of Cornelis Pleij, Krijn Rietveld and Leendert Bosch in the early 1980s. These authors were investigating how it was possible for the 3′ ends of some plant virus genomes to possess a number of the functional characteristics of transfer RNAs (tRNAs) yet lack an obvious clover-leaf secondary structure. By applying a 'pseudoknot building principle', it became clear how these viral RNAs could fold into L-shaped structures resembling tRNAs1,2. A seminal paper from the same authors3 subsequently defined the general principles of pseudoknot folding and provided the first examples of pseudoknots in other RNAs — the central pseudoknot of Escherichia coli 16S ribosomal RNA (rRNA) and a pseudoknot present in group I introns.
Since then, many more pseudoknots have been discovered, and they are associated with a remarkably diverse range of biological activities (Supplementary information S1, S2 (tables); reviewed in Refs 4–11). They are especially associated with key roles in the replication cycles of numerous animal and plant viruses, including, in humans, the flavivirus hepatitis C virus (HCV)12, the coronavirus responsible for severe acute respiratory syndrome (SARS-CoV)13, the oncogenic retrovirus T-cell lymphotrophic virus types I and II14 and certain strains of HIV15.
The function of a viral pseudoknot is linked logically to its location in the genome (Fig. 1; Supplementary information S1 (table)). So, in non-coding regions (NCRs) of positive-strand RNA viruses (in which the genomic RNA serves as the mRNA template for translation and then as a template for replication), pseudoknots act in the regulation of initiation of protein synthesis and in template recognition by the viral replicase. By contrast, in coding regions they modulate the elongation and termination steps of translation. Fewer pseudoknots have been documented in the mRNAs of viruses with DNA genomes, but several DNA bacteriophage mRNAs are known to encode pseudoknots16. In the 5′ NCRs, these motifs have roles in the regulation of translation initiation, whereas in the coding region they affect translation elongation. Pseudoknots have also been described in the catalytic RNAs of some RNA satellite viruses, where they have a role in genome replication17.
Figure 1. RNA pseudoknots in virus gene expression.
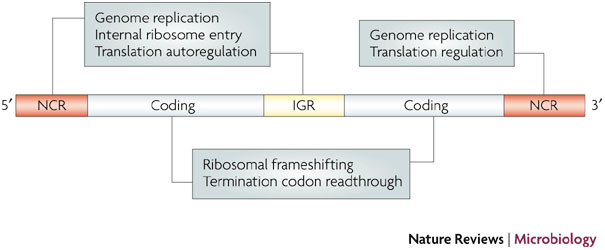
A schematic of a generic RNA virus genome is shown. Viral pseudoknots have been described in the 5′ non-coding region (NCR), the coding region, the intergenic region (IGR) and the 3′ NCR, where they function in various steps of the replication cycle. Although the majority of examples are from positive-strand RNA viruses, pseudoknots also have a role in the replication cycles of certain DNA viruses, satellite RNA viruses and viroids. For simplicity, viral pseudoknots involved in long-range interactions (including virus genome circularization) or possessing catalytic activity are not shown, but are discussed in the text.
Here, we review selected, well characterized examples of pseudoknots in virus genomes — with an emphasis on structure–function relationships — highlighting recent advances in our understanding of pseudoknot conformation at high resolution and exploiting, where relevant, our improved knowledge of ribosome architecture.
What is an RNA pseudoknot?
As defined originally3, a pseudoknot is a structure formed upon base-pairing of a single-stranded region of RNA in the loop of a hairpin to a stretch of complementary nucleotides elsewhere in the RNA chain (Fig. 2). Such pseudoknots, referred to as hairpin type (H-type) pseudoknots, have two base-paired stem regions (S1 and S2) and, depending on the number of loop bases that participate in the pseudoknotting interaction18, two or three single-stranded loops (L1, L2 and L3). In most (>85%; Refs 19, 20) H-type pseudoknots, L2 is absent or very short, and the base-paired stems stack coaxially to form a quasi-continuous helix. In these structures, L1 spans S2 and crosses the deep groove of the helix, whereas L3 spans S1 and crosses the shallow groove (Fig. 2).
Figure 2. RNA pseudoknot structure.
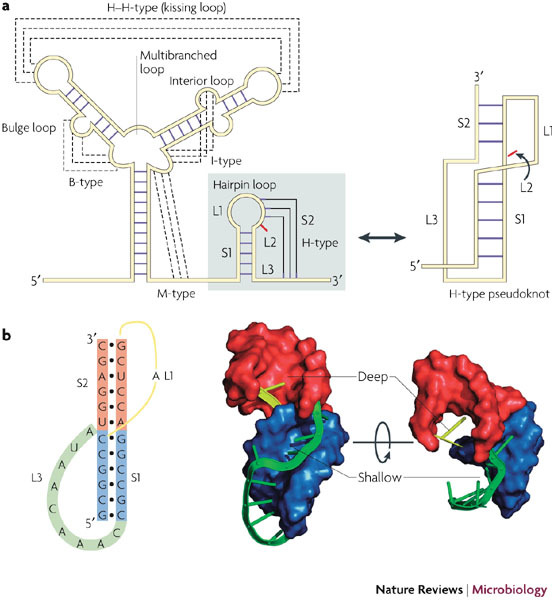
a | Various structural motifs have been described in RNA149. Orthodox secondary structures consist of base-paired regions (stems) connected by single-stranded loops at stem termini (hairpin loop), or in the body of a stem (bulge (B) or interior (I) loop) or at the junction of several stems (multibranched (M) loop). Pseudoknots are considered as a tertiary structure and form when bases in a loop pair with a single-stranded region elsewhere. The hairpin type (H-type) pseudoknot is by far the most common, and this tertiary interaction involves bases in the loop of a hairpin loop. The resultant structure contains two stem regions, S1 and S2, connected by single-stranded loops. In many cases, no unpaired bases are present between the two stems (L2 is zero), and the stems stack coaxially to give a quasi-continuous helix. b | The secondary structure of the pseudoknot of the ribosomal frameshifting signal of simian retrovirus 1 (SRV-1) is shown alongside three dimensional views of the nuclear magnetic resonance model74. The stems are shown as surface representations and the loops as ribbons (all structural images were prepared using PyMol). The polarity and handedness of the double helix leads to inequivalence of the loops, with L1 (yellow) crossing the deep groove and L3 (green) crossing the shallow groove. S1 is blue, S2 is red, L1 is yellow and L3 is green. L2 is not present in the example shown.
The geometry of the pseudoknot is such that when S2 is six or seven base pairs in length, L1 can be as short as a single nucleotide3,21. However, in some pseudoknots, the loops are much longer and include their own secondary structure elements. Pseudoknots are also formed upon base-pairing of single-stranded bulge (B), interior (I) and multibranched (M) loops with complementary regions elsewhere in the RNA (which themselves can be constrained in a secondary structure, for example in intramolecular hairpin–loop–hairpin–loop (H–H or so-called kissing loop) interactions)22. However, unless all of the loop nucleotides are paired, these pseudoknots are generally considered as H-type pseudoknots, as the additional base-pairing interaction is viewed as a substructure within the loop. For this reason, the B, I and M nomenclature (as well as H–H) is not extensively used, and most structures are referred to as H-type pseudoknots and often simply as pseudoknots. An additional issue regarding nomenclature is loop numbering. In the early pseudoknot literature, most examples did not possess unpaired residues between the two stems, so the convention was to name the groove-spanning loops L1 and L2 (now L1 and L3). Here, we have opted for the L1, L2 and L3 nomenclature, which is more generally applicable.
As will be discussed in more detail below, the biological properties of pseudoknots are intimately linked to their structural features4,5,6,7,18,23. For example, the geometry of the junction between the stems and the interactions that can occur between the constituent loops and stems are often of great functional relevance21,24,25,26,27. Indeed, for many viral pseudoknots, much of the primary sequence is unimportant for function, as long as the conformation and overall stability of the structure is maintained. Where precise nucleotide-sequence requirements have been identified, this is likely to reflect a specific structural necessity, although additional roles might be possible (for example, base-specific recognition by proteins).
Pseudoknots and translation
Internal ribosome entry. Most eukaryotic cellular mRNAs are translated in a cap-dependent manner, with the 40S subunit and associated initiation factors scanning along the mRNA until the start codon (AUG) is reached28. Efficient translation also requires mRNA circularization, which is brought about by the interaction of the 3′-end poly(A) tail-binding protein (PABP) with initiation factor 4E (eIF4E), bound to the 5′ cap29. However, the genomes of many positive-strand RNA viruses often lack a cap, a poly(A) tail or both, and translation initiation involves non-standard mechanisms30,31.
One such example is cap-independent internal ribosome entry, in which the ribosome is recruited internally to a structured region of the mRNA (the internal ribosome entry site (IRES), usually located in the 5′ NCR) and often directly to the start codon. Pseudoknots have been identified in a number of IRESs, and their function is best exemplified in the IRESs of the flavivirus HCV and the dicistrovirus cricket paralysis virus (CrPV) (Fig. 3). Unlike the mammalian picornaviral IRESs, which retain a requirement for many or most canonical initiation factors, the mammalian 40S subunit can associate with the HCV IRES in the absence of translation initiation factors, although the formation of the 48S complex (with the initiation codon locked into the mRNA-binding cleft of the small subunit) requires the participation of the eIF2–GTP–Met-tRNAi ternary complex and eIF3 (reviewed in Ref. 32). The HCV IRES forms a defined secondary structure that contains two major hairpins (domains 2 and 3) and an essential pseudoknot structure12 at the base of domain 3 (domain 3e/f) (Fig. 3). IRES function requires domains 2 and 3, but initial binding of the 40S subunit is mediated principally by the basal region of domain 3, including stem–loop 3d, with a modest contribution from the pseudoknot33,34. Comparisons of HCV IRES–40S (Ref. 35) and IRES–80S (Ref. 36) complexes have revealed that the overall appearance of the IRES is similar in the two complexes36. In the 80S complex, the pseudoknot corresponds to an L-shaped density located at the mRNA exit channel in the vicinity of ribosomal protein S5 (rpS5) (Fig. 3; Supplementary information S3 (figure)). The assignment of the pseudoknot to the L-shaped density is based on molecular modelling and is consistent with recent crosslinking data37. The three-dimensional (3D) structure of the HCV pseudoknot is not available, but in essence it is an H-type pseudoknot in which L1 is long and highly structured, and includes the entirety of domains 3a–3e. Why a pseudoknot is present at this key location of the IRES is unclear, but it might be linked to a capacity to bind rpS5 (Ref. 37). This protein is present at the mRNA exit channel and also associates with IRES domain 2 (Refs 36, 38). Boehringer and colleagues propose that the HCV IRES domains function synergistically to position the AUG into the ribosomal peptidyl (P) site, coupled to movement of the pseudoknot36. In this model, a conformational change of the four-way junction (which includes domains 3a and 3c; Fig. 3) pivoted around domain 3d is transmitted to the pseudoknot, which moves towards the mRNA exit channel, positioning the AUG correctly into the ribosomal P site and allowing subunit joining. The movement could be potentiated by eIF3 binding, as this multisubunit initiation factor makes intimate contacts with the HCV IRES39.
Figure 3. Pseudoknots and internal ribosome entry.
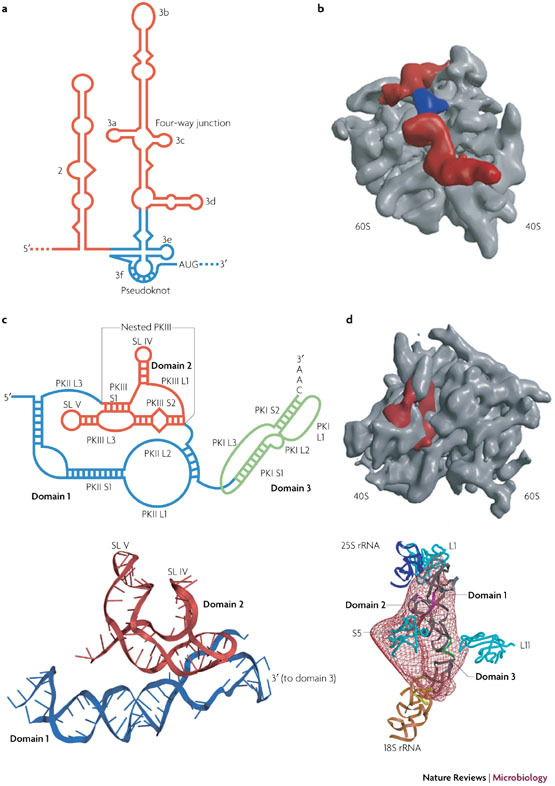
a | A secondary structure representation of the hepatitis C virus (HCV) internal ribosome entry site (IRES) with the pseudoknot shown in blue. b | A surface representation of the human 80S ribosome (grey) in complex with the HCV IRES (red) derived from the cryo-electron microscopy (cryo-EM) structure36. Density corresponding to the pseudoknot is indicated in blue. c | A secondary structure representation of the Plautia stali intestine virus (PSIV) IRES is shown above a ribbon representation of the RNA (domains 1 and 2) derived from the crystal structure27. Domain 3 remains to be solved. The secondary structure of the cricket paralysis virus (CrPV) IRES (not shown) is similar. d | A surface representation of the yeast 80S ribosome (grey) is shown in complex with the CrPV IRES (red) derived from the cryo-EM structure26. Below is a fit of the density to the modelled CrPV IRES showing the interactions that occur between the various domains and ribosomal components. Ribosomal proteins are cyan, 25S ribosomal RNA (rRNA) is purple and 18S rRNA is brown.
Pseudoknots also have an essential role in the function of the intergenic region (IGR) IRES of CrPV40 and other dicistroviruses41. This remarkable IRES, only ∼200 nucleotides in length, has been described as an RNA-based translation factor42 because it recruits ribosomes and activates translation without the involvement of initiation factors or initiator tRNA. The ribosomal 40S and 60S subunits bind directly to the IRES43, which then occupies the ribosomal intersubunit space of the 80S complex and interacts with key components that form the ribosomal aminoacyl (A), P and exit (E) sites.
The three defined domains of this IRES have distinct functional tasks (Fig. 3). Domain 1 contributes to interactions with the 60S subunit in the E- and P-site regions, and domain 2 interacts with the 40S subunit at the E site. Domain 3, which is located predominantly between the A and P sites and is in a similar orientation to ribosome-bound tRNAs, places its most 3′ nucleotide triplet (GCU) into the decoding region of the A site11. Here, it binds the anticodon of tRNAAla, which is brought to the ribosome as part of the ternary complex (elongation factor 1A (eEF1A)–GTP–tRNAAla). Subsequently, tRNAAla is pseudotranslocated (without peptide-bond formation) by eEF2 into the P site, allowing delivery of the next tRNA into the A site and authentic elongation to begin41,43,44.
Modelling and structural analysis of the CrPV IRES and IRESs from related viruses, including Plautia stali intestine virus (PSIV)26,27,45,46, has revealed that the structure is dominated by three H-type pseudoknots (Fig. 3), one per domain. Pseudoknot PKI, which essentially forms all of domain 3, is characterized by the possession of an AU-rich S1 and short loops. The folding of the rest of the IRES is dominated by interactions between pseudoknots PKII and PKIII (Fig. 3). The pseudoknot of domain 2, PKIII, is a nested pseudoknot in that it is entirely contained within L3 of the pseudoknot of domain 1, PKII. Cryo-electron microscopy (cryo-EM) and X-ray crystallography studies26,27 (reviewed in Ref. 11) have indicated a complex folding strategy that forces two small hairpins of PKIII, present as substructures in L1 (stem–loop (SL) IV) and L3 (SL V), to project from a central core and emerge on the same side of the structure to make vital interactions with rpS5 at the E site46,47,48. Pivotal to this folding strategy are pseudoknot loop–helix interactions. In PKIII, for example, an L1–S2 major-groove interaction positions SL IV, and a second interdomain interaction occurs between the minor groove of S2 and four L1 bases of the adjacent pseudoknot PKII, which stabilizes the core. The geometry of the overall fold is such that S2 of PKII stacks on S2 of PKIII to create a wedge-shaped section that occupies the mRNA channel and directs PKI into the decoding site.
Recruitment of the first aminoacyl tRNA to the ribosome requires the participation of eEF2 (Ref. 49). The activation of eEF2 has been linked to an IRES-mediated destabilization of a conserved cellular pseudoknot that is present in helix 18 of the small subunit rRNA (the so-called 530-loop pseudoknot; Supplementary information S2 (table)). This region of rRNA is involved in the enhancement of translational accuracy and tRNA binding50, and its destabilization (probably by the pseudoknot of domain 3) is likely to be crucial to the IRES mechanism.
The structure of the IGR IRESs illustrates how pseudoknots can be used to direct the global folding of an RNA sequence. The pseudoknot motif naturally provides the potential for coaxial stacking of constituent helices, but also the opportunity for additional stacking with helices present in constituent loops. This allows longer helical domains to be generated, a common feature in the organization of global RNA structures51. If the pseudoknot is itself nested in another pseudoknot, additional helical stacking possibilities are created. Superimposed on this is the capacity of the single-stranded loops to interact with constituent stems to add stability, or with other regions of RNA to promote packing of adjacent helices. The compact and complex fold of the IGR IRESs brought about by the nested pseudoknots is a strategy similar to that used to fold certain ribozyme cores, discussed in more detail below. There is no known ribozyme activity associated with IRESs, however, which indicates that this is a general folding strategy that can be used to satisfy different mechanistic requirements.
Autoregulation. One of the first viral pseudoknots to be described16 is encoded by T-even bacteriophages (such as T2, T4 and T6) and functions in translational autoregulation of the gene 32 protein (gp32), a single-stranded DNA-binding protein that mainly functions in replication of the viral double-stranded genomic DNA. The pseudoknot is located some 40 nucleotides upstream of the translation start site (AUG) of the gp32 mRNA and acts as a specific binding site for gp32 itself. At low protein concentrations, pseudoknot-bound gp32 does not overlap the ribosome-binding site, but as protein levels increase, cooperative binding of multiple copies occurs, nucleated at the pseudoknot-bound gp32 (Ref. 52). This assembly blocks access to the Shine–Dalgarno sequence and so represses translation. Unfortunately, molecular details of the gp32–pseudoknot interaction are lacking, but the structure of the pseudoknot isolated from bacteriophage T2 has been solved by nuclear magnetic resonance (NMR)53. It is a classic H-type pseudoknot with short loops and coaxially stacked stems, albeit with S2 rotated by ∼18˚ with respect to S1 to relieve close phosphate–phosphate contacts at the junction while preserving the stabilizing effects of base stacking. Although the T2 pseudoknot possesses only a single nucleotide in L1 (an A), this is stereochemically feasible as the distance between the two phosphates across the deep groove of A-form RNA reaches a minimum when six or seven base pairs of S2 are bridged3,21.
Although the gene 32 system represents the only known viral example of pseudoknot involvement in the autoregulation of translation initiation, there are related examples in cellular mRNAs (Supplementary information S2 (table)). For example, translational repression of the ribosomal S4 α mRNA operon54,55 and autoregulation of ribosomal protein S15 synthesis56 requires specific binding of the respective proteins to pseudoknots in the 5′ untranslated region (UTR) of their own mRNAs.
Frameshifting. RNA pseudoknots in coding regions are principally associated with sites of programmed −1 ribosomal frameshifting. This is a translational mechanism used by many viruses to coordinately express two proteins from a single mRNA at a defined ratio7,57. During elongation, ribosomes decode the mRNA in triplet steps and the reading frame is accurately maintained. However, in frameshifting, the ribosome is forced to shift one nucleotide backwards into an overlapping reading frame and translate an entirely new sequence of amino acids (Fig. 4). In retroviruses, frameshifting at the overlap of the gag and pol open-reading frames (ORFs) allows expression of the viral Gag–Pol polyprotein and sets a defined cytoplasmic Gag:Gag–Pol ratio that is optimized for virion assembly and packaging of reverse transcriptase58. In other RNA viruses, frameshifting allows expression of RNA-dependent RNA polymerases (RdRps)59. Maintaining a precise efficiency of frameshifting has been shown to be crucial to the replication of HIV-1 (Ref. 60) and the retrovirus-like double-stranded RNA virus of yeast, L-A61. Similarly, in other RNA viruses, changing the stoichiometry of non-frameshifted and frameshifted products is also likely to be detrimental. In SARS-CoV, for example, components of the viral replication machinery present in the viral polyproteins pp1a and pp1ab (which are expressed by frameshifting) are predicted to form a heterodimer with a stoichiometry of ∼8:1 (Refs 62, 63), a ratio that is consistent with the natural level of frameshifting64,65. For these reasons, frameshifting has emerged as a potential target for antiviral therapeutics.
Figure 4. Pseudoknots and ribosomal frameshifting.
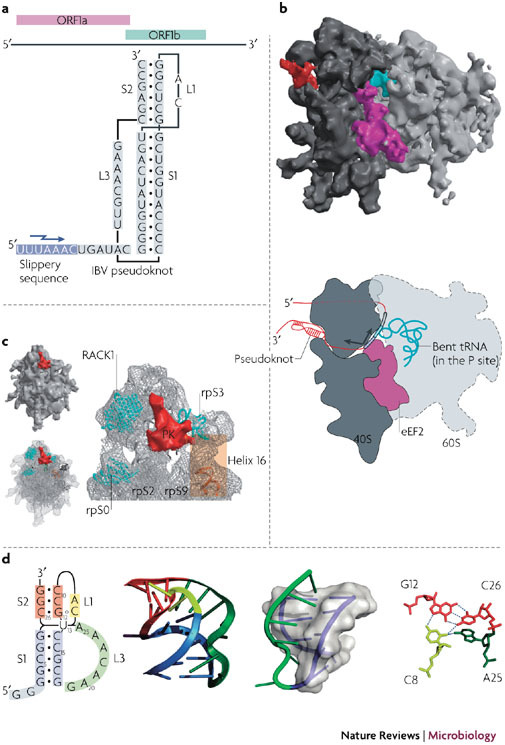
a | The overlapping coding sequences open reading frame 1a (ORF1a) and ORF1b of the genome of the coronavirus infectious bronchitis virus (IBV) are shown above the minimal frameshift-promoting sequences of this virus. The pseudoknot promotes frameshifting at the slippery sequence, indicated by a jagged arrow. b | A representation of the stalled, pseudoknot-engaged rabbit 80S ribosome is shown derived from the cryo-electron microscopy structure71. The 60S subunit is light grey and the 40S subunit is dark grey. The peptidyl (P)-site transfer RNA (tRNA) stalled in the complex is coloured turquoise, the eukaryotic translocase, elongation factor 2 (eEF2), is purple and the pseudoknot structure is red. Below is a schematic of the stalled ribosome. Engagement with the pseudoknot generates a frameshifting intermediate in which the ribosome is stalled during translocation with eEF2 bound, generating tension in the mRNA that bends the P-site tRNA in a(+) sense direction. As a result, the anticodon–codon interaction breaks over the slippery sequence, allowing a spring-like relaxation of the tRNA in a (−) sense direction. c | A close-up view of the pseudoknot (PK) in the stalled complex, with the ribosomal components (rpS0, rpS2, rpS3 and rpS9) in close proximity highlighted. d | The beet western yellows virus (BWYV) pseudoknot73 is illustrated to show examples of features that might confound the ribosomal helicase70. Shown from left to right are: a secondary structure model of the pseudoknot, with U13 drawn to indicate its extrusion form the helix; a ribbon representation of the X-ray structure; the L3–S1 (loop 3–stem 1) triplex interaction, with S1 shown as a transparent surface and the helix within as a purple ribbon; and the BWYV junction quadruple interaction, with hydrogen bonds shown as dashed lines. A triplex also forms at the junction of the two stems (not highlighted). RACK1, receptor for activated C kinase 1. Panels b and c are modified with permission from Ref. 71 © MacMillan Publishers Ltd.
A typical frameshift signal has two essential elements: a heptanucleotide 'slippery' sequence, at which the ribosome-bound tRNAs slip into the −1 frame, and an adjacent mRNA secondary structure, most often an H-type pseudoknot14,66, that stimulates this slippage process (Fig. 4). How the pseudoknot promotes frameshifting is not completely understood, but the mechanism is likely to be linked to the helicase activity of the ribosome, with the pseudoknot presenting an unusual topology that resists unwinding47,67,68,69,70,71. Takyar and colleagues have shown that the prokaryotic 70S ribosome can itself act as a helicase to unwind mRNA secondary structures before decoding70, with the active site located between the head and shoulder of the 30S subunit. Prokaryotic ribosomal proteins S3, S4 and S5 that line the mRNA entry tunnel are implicated in the helicase activity70. Recent cryo-EM images of mammalian 80S ribosomes paused at a coronavirus frameshift-promoting pseudoknot revealed that the ribosomes become stalled during the translocation phase of the elongation cycle with eEF2 bound and in the act of transferring peptidyl-tRNA from the A site to the P site71. Furthermore, the P site tRNA is structurally distorted and in a spring-like conformation. Consistent with its proposed function, the pseudoknot is present at the mRNA entry channel in close proximity to the proteins (rpS3, rpS9 and rpS2) that are likely to form important elements of the mammalian 80S helicase (Fig. 4). Namy and colleagues71 suggest a mechanical model of frameshifting in which the pseudoknot resists unwinding by the helicase, compromising translocation by putting tension on the mRNA, leading to bending of the tRNA anticodon and, ultimately, repositioning the tRNA on the slippery sequence in the −1 reading frame. The pseudoknot is also in close proximity to the ubiquitous ribosomal regulatory protein RACK1 (receptor for activated C kinase 1)72. It is not known whether this protein, or the recruited kinase, has a role in frameshifting, but this could in principle provide a route to regulation of frameshifting during virus infection.
X-ray crystallography and NMR analysis of frameshift-promoting pseudoknots, coupled with functional studies, have revealed features that could account for an intrinsic resistance to unwinding by the ribosomal helicase (Fig. 4). Chief among these is the presence of extensive minor-groove triplex interactions between S1 and the crossing L3, first observed in the pseudoknot of the luteovirus beet western yellows virus (BWYV; Fig. 4)73 and subsequently in related luteoviruses and the gag/pro pseudoknot of simian retrovirus 1 (Ref. 74). The S1–L3 RNA triplex is likely to be the first feature encountered by the elongating ribosome, and the presence of the 'third strand' would conceivably confound unwinding, at least temporarily.
Another feature of functional importance is the architecture of the junction between the constituent pseudoknot helices. Several non-canonical interactions have been described in and around the junction, including base triples, base quadruples and loop–loop Hoogsteen base pairing as well as distortions such as over-rotation of the stems, helical displacement and bending7,75 (Fig. 4). The first NMR study of a frameshift-promoting pseudoknot, from the gag/pro overlap of mouse mammary tumour virus, had indeed indicated a requirement for a specific bent conformation of the pseudoknot, brought about, in part, by the presence of an intercalated unpaired adenosine residue between the two stems76. However, closely related pseudoknots with coaxially stacked stems can clearly stimulate high levels of frameshifting74,77 and it now seems likely that the specific interactions and resultant architectures of the helical junction that are required for frameshifting are strongly context dependent75. Nevertheless, there is no doubt that the junction conformation is crucial to function and might also present a kinetic or thermodynamic barrier to unwinding78,79.
Several cellular homologues of viral frameshift signals have been identified and appear to be derived from endogenous retroviruses80,81,82 (although some recently identified candidates may have a different origin83). The pseudoknots within these genes are fundamentally similar in structure to — and retain the functionality of — their viral progenitors, and have a role in the regulation of gene expression during embryogenesis. Another cellular pseudoknot with close structural similarity to viral frameshift pseudoknots is present in bacterial transfer messenger RNA (tmRNA)84,85,86. This RNA, which contains four pseudoknots (PKI–IV), functions by binding to, and ultimately rescuing, ribosomes stalled on damaged mRNAs by a process termed trans translation86. PKI of tmRNA shows considerable similarity to the BWYV family of pseudoknots, including short stems and loops, extensive base stacking, stabilization of the stem junction by a triplex, exclusion of a uridine at the junction of the two stems and insertion of L3 into the minor groove of S1 (Ref. 84). PKI functions at a different site on the ribosome than frameshift-promoting pseudoknots, however, and its precise role is uncertain. A growing number of pseudoknots have been shown to interact with the ribosome, and it is clear that they can influence function from different sites. This is illustrated in Supplementary information S3 (figure), in which the location on the ribosome of the pseudoknots involved in IRES function, frameshifting and trans translation are compared.
Pseudoknots can also promote +1 ribosomal frameshifting. The best characterized examples come from the cellular gene ornithine decarboxylase antizyme87. However, pseudoknot-dependent +1 frameshifting also appears to be used in the expression of certain structural proteins of the phage of Scott A (PSA) bacteriophage that infects Listeria monocytogenes88, although this remains the only viral example so far.
Stop codon readthrough. In the gammaretroviruses, which are typified by murine leukaemia virus (MuLV), gag and pol are in the same reading frame and are separated by an UAG stop codon. Here, Gag–Pol synthesis requires periodic misreading of the stop codon as a sense (Gln) codon, a process known as termination codon suppression or readthrough. Efficient readthrough is promoted by an H-type pseudoknot spaced precisely eight nucleotides downstream of the stop codon89,90. Retroviral readthrough and frameshift signals are thus similar in terms of overall organization, with the recoding site (a stop codon or a slippery sequence) separated from the pseudoknot by a spacer of similar length. There is little variation in the size of gammaretroviral pseudoknots and considerable primary sequence conservation, especially in the spacer region and L3 of the pseudoknot. Many of these bases are functionally essential89,91,92, but have not yet been linked to an explicit structural role92,93. No high-resolution structures of these pseudoknots are available as yet, and it remains to be seen whether they interact with the ribosome in a manner similar to the pseudoknots involved in frameshifting. However, it is plausible that interactions of the readthrough pseudoknot with the helicase or other ribosomal components could modulate release-factor access or activity and promote misreading of the stop codon by the near-cognate tRNAGln. It has already been shown that sequestration of eRF1, in this case as a result of direct binding by the MuLV reverse transcriptase, can lead to increased readthrough94.
Pseudoknots in the 3′ NCR
Translation and replication. Several positive-strand RNA viruses harbour pseudoknots in the 3′ NCRs (Supplementary information S1 (table)). The best studied examples come from plant viruses and, in particular, the pseudoknot that induces the formation of the tRNA-like structure (TLS) at the end of the genome of turnip yellow mosaic virus1,2 (TYMV; Fig. 5). This short H-type pseudoknot forms part of the aminoacyl acceptor arm of the TLS, stacking against an adjacent hairpin (itself the equivalent of the tRNA T-stem–loop) to generate a quasi-continuous helix that mimics the acceptor arm of tRNA and that can be adenylated and aminoacylated with Val95. NMR studies of this region of the TLS have revealed that the single-stranded loops of the pseudoknot introduce minimal distortion in the A-form helical shape of the molecule. The major groove comfortably accommodates the crossing L1 residues, and L3 is closely anchored to S1 through triplex interactions centred around the adenosine residue in L3. Indeed, this study was the first to reveal the potential for interplay between pseudoknot loops and stem regions21.
Figure 5. Pseudoknots and transfer RNA-like structures.
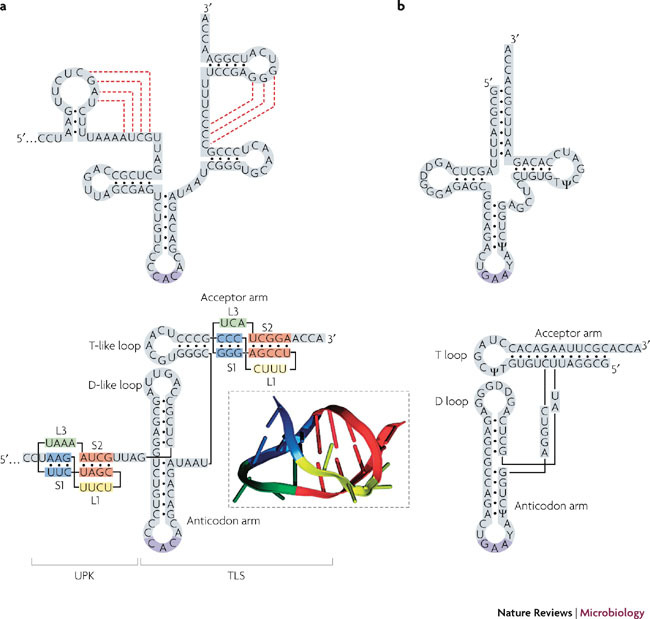
a | The 3′ end of the turnip yellow mosaic virus (TYMV) genomic RNA. In the upper panel, the predicted secondary structure is shown, with pseudoknotting interactions indicated by dashed red lines. Below is a secondary structure representation of the folded molecule, showing the transfer RNA (tRNA)-like structure (TLS) and the pseudoknots in the acceptor arm and upstream of the TLS (UPK). The ribbon representation (boxed) is derived from the nuclear magnetic resonance structure of the acceptor arm pseudoknot21. b | For comparison, secondary structure representations of tRNAPhe are also shown. D, dihydrouridine modified bases; T, ribothymidine base.
The tRNA mimicry accounts for the reactivity of the 3′ end of the viral genome with several enzymes that recognize tRNA. These include the CCA nucleotidyl transferase, which adds a 3′ terminal A to complete the 3′ CCA end of the genome upon infection; valyl tRNA synthetase, which aminoacylates the 3′ end with Val; the elongation factor eEF1A, which binds to the TLS to give a viral RNA–eEF1A–GTP ternary complex; and the viral replicase p69/p206 (Refs 96, 97).
An elegant relationship has been unearthed between the TLS-specific reactivities and the TYMV lifecycle. Upon entry into the cell, the 3′ CCA end is completed and aminoacetylated, at which point translation of the input virus genome, an obligatory step in the production of the virus replicase, is stimulated synergistically by the 5′ cap and the TLS97. The stimulation of translation is maximal when the genome is aminoacetylated and is linked to the formation of a viral RNA–eEF1A–GTP ternary complex97. How eEF1A enhances translation is not known, but the available evidence hints at an unexpected involvement of this elongation factor in the process of translation initiation. It has been suggested98 that binding of the viral RNA ternary complex to the A-site of initiating ribosomes could stimulate initiation at the 5′ end (perhaps in a manner similar to the pseudoknot of the IGR IRES domain 3 discussed above), but recent evidence argues against this specific mechanism99. Nevertheless, close proximity of the virus genome ends during translation (the circularization observed for most cellular mRNAs) could offer an explanation for the translational synergy afforded by the 5′ cap and the 3′ TLS–eEF1A complex.
The TLS of TYMV is also crucial in the switch between translation and replication. As has been elegantly illustrated for bacteriophage Qβ100 and poliovirus101,102, the movement of ribosomes in the 5′→3′ direction is incompatible with negative-strand RNA synthesis, in which the RNA polymerase travels 3′→5′. Thus, following an initial burst of translation, the viral mRNA must be cleared of ribosomes to allow replication from the 3′ end. Negative-strand synthesis in TYMV is initiated from the second C of the 3′ CCA triplet following pseudoknot-dependent binding of the replicase to the TLS103,104. It has been demonstrated recently that this reaction is inhibited by the binding of eEF1A to the valylated TLS105. So, translation is favoured until the levels of viral RdRp are sufficient to compete with eEF1A for binding to the TLS or perhaps until genomes are sequestered into vesicular sites of virus replication, which might be free of competing eEF1A and aminoacyl tRNA synthetases. The TLS might also be involved in genome packaging: in the related brome mosaic virus, viral RNAs lacking the TLS fail to assemble into virions106.
In TYMV, a second pseudoknot is present immediately upstream of the TLS, and this pseudoknot also contributes to translational enhancement, probably by acting as a spacing element to present the functional TLS to enzymes97 (Fig. 5). The presence of such upstream pseudoknots is not uncommon and indeed, tobamo-, hordei-, furo-, pumo- and certain tymoviruses boast clusters of pseudoknots (between two and seven) in this region107, which is termed the upstream pseudoknot domain (UPD). So, in tobacco mosaic virus (TMV), for example, a total of five pseudoknots are present in the 3′ UTR, three of which form the quasi-continuous double helical stalk of the UPD (5′ PKIII, PKII, PKI 3′) and two of which (5′ PKb, PKa 3′) are present in the TLS108. In the TLS, PKa forms part of the acceptor arm and PKb forms the central core, imposing the tRNA-like shape and orienting the UPD109. PKb is a B-type pseudoknot and forms a Y-shaped three-way junction that is proposed from modelling to have some structural similarity to the ribozyme of hepatitis delta virus (HDV; see below), although it displays no catalytic activity109.
Despite the complexity of folding of the TMV TLS, the UPD seems to have usurped its role, at least in terms of translational enhancement110. Although the TLS can be aminoacetylated with His and can bind eEF1A22, the major player is the UPD, in which the highly conserved PKII and PKI can be crosslinked to eEF1A, independently of TLS aminoacylation111, and form an essential element of the promoter for negative-strand synthesis112. The UPD can also bind to the heat-shock chaperone HSP101, although the protein does not appear to be essential for efficient replication of TMV113. It seems that there is a general requirement for the binding of certain cellular proteins to the 3′ end that does not necessarily have to be mediated solely by a TLS.
Another model system used to study the role of viral NCR pseudoknots in facilitating translation, replication and the switch between the two is tomato bushy stunt virus (TBSV)114,115,116,117. The TBSV genome is uncapped and lacks a poly(A) tail. It recruits the translation machinery initially to the 3′ NCR, a process requiring a highly structured 3′ cap-independent translational enhancer (3′ CITE), comprising a Y-shaped hairpin domain upstream of a 3′-end pseudoknot. It is thought that upon infection, the pseudoknot is folded (the closed conformation) and translation factors gather on the 3′ CITE. Translation initiation complexes are then transferred to the vicinity of the 5′ NCR by virtue of genome circularization, mediated, at least in part, by a kissing-loop interaction between a stem–loop in the 3′ CITE and a partner close to the 5′ end of the genome (SL III). Translation complexes then scan to the first AUG and begin protein synthesis. Subsequently, the pseudoknot at the 3′ end of the genome is destabilized, possibly by binding of the accumulating viral RdRp, yielding an 'open' complex that is compatible with negative-strand synthesis117. Another pseudoknot (PK-TD1) in the 5′ NCR is also required for efficient replication, although its mechanism of action is not fully understood.
Pseudoknots in non-coding regions have also been documented in animal RNA viruses, with the vast majority known to be essential for virus replication (Supplementary information S1 (table)). Mostly, they function in translation, genome replication or the switch between the two. However, they are not well characterized structurally (at high resolution) and few details of the molecular interactions in which they participate are available.
Pseudoknots and ribozymes
Hepatitis delta virus. HDV is a satellite RNA virus of humans that replicates in association with a helper virus, hepatitis B virus17. The circular, single-stranded RNA genome of HDV is replicated through an intermediate, the antigenome, by a double rolling-circle mechanism that requires self-cleavage by closely related genomic and antigenomic versions of a ribozyme. The HDV ribozymes have been extensively studied as a model for the mechanism of catalytic RNAs9 and fold into similar structures, characterized by a nested double pseudoknot that helps form the catalytic site and brings great stability to the RNA118 (Supplementary information S4 (figure)). The nested double-pseudoknot motif is also common in cellular ribozymes (Supplementary information S2 (table)), for example, the HDV-like ribozyme present in an intron of the gene that encodes human cytoplasmic polyadenylation binding protein 3 (Ref. 119) and the glmS riboswitch ribozyme present in the 5′ UTR of the gene that encodes glucosamine-6-phosphate synthase in numerous Gram-positive bacteria120,121. The intricate connectivity that results from nesting two pseudoknots within each other makes it possible for these short RNAs to adopt complex and stable 3D folds.
A viral telomerase pseudoknot
Telomerases are ribonucleoprotein complexes responsible for the maintenance of telomeres10. In addition to a specialized reverse transcriptase (TERT), an RNA component (telomerase RNA (TR)) is present and includes the RNA template for telomere addition and a highly conserved pseudoknot necessary for telomerase activity. Like frameshift-promoting pseudoknots, the TR pseudoknot has extensive triplex interactions at the junction of the two stems25 (Supplementary information S5 (figure)). It has been proposed that the pseudoknot region makes contacts with the TERT122 and is needed for telomere repeat-addition processivity123. Indeed, a switch between the pseudoknot and a partially unfolded form might be important in the translocation of telomerase during telomere addition124,125,126. High levels of telomerase activity are detected in a large range of cancers and are closely associated with the immortalization process127,128. Recently, a chicken TR homologue, vTR, has been described in the oncogenic herpesvirus Marek's disease virus129. Deletion of the two copies of vTR from the virus genome led to reduced incidence and severity of lymphoma in infected chickens, indicating that vTR has the attributes of an oncogene130. An understanding of how vTR supports lymphomagenesis in genetic and molecular terms has the potential to yield new insights into telomerase function in cancer131.
The versatile pseudoknot
The development of sophisticated algorithms (Box 1; Table 1) to scan genomic sequences for RNA structural motifs has highlighted the prevalence of the pseudoknot fold in the RNA universe, indicating evolutionary selection. A plausible explanation for this abundance is that pseudoknots offer functional versatility. Some pseudoknots (for example the BWYV frameshift pseudoknot) are extensively stabilized by both Watson–Crick and non-Watson–Crick interactions73 and are more stable than an equivalent hairpin (containing a base-paired stem that is equivalent to S1 plus S2). Thus, a very stable motif can be included in an RNA when space — either in terms of genomic coding capacity or molecular dimensions — is at a premium. At the other end of the spectrum, many pseudoknots are considerably less stable than their equivalent hairpin and could conceivably act as regulatory switches, oscillating between stem–loop and pseudoknot conformations in response to environmental signals132. Pseudoknots also offer binding sites for proteins or single-stranded loops of RNA. The often extensive intra- and intermolecular contacts that pseudoknots engage in provide many targets for such interactions. Indeed, the in vitro selection of RNA aptamers that bind various biomolecules often generates pseudoknotted RNAs (Box 2). Pseudoknotting can also be the most efficient way of folding RNAs in an active conformation (for example, ribozymes).
Table 1.
Examples of pseudoknot prediction programmes
| Task | Programme | Operating system | URL |
|---|---|---|---|
| Pattern matching | RNAmotif | Linux | http://www.scripps.edu/mb/case/casegr-sh-3.5.html |
| Pseudoknot prediction from short sequences | PknotsRG | Linux, OSX, Windows, Web server | http://bibiserv.techfak.uni-bielefeld.de/download/tools/pknotsrg.html |
| Pseudoknot prediction from short sequences | HotKnots | Linux | http://www.cs.ubc.ca/labs/beta/Software/HotKnots |
| Pseudoknot prediction from longer sequences | HPKNOTTER | Web server | http://bioalgorithm.life.nctu.edu.tw/HPKNOTTER |
| Pseudoknot visualization | PseudoViewer | Web server, Windows | http://pseudoviewer.inha.ac.kr |
| Pseudoknot database | PseudoBase | Web server | http://biology.leidenuniv.nl/~batenburg/PKBGetS1.html |
Long-range interactions are also facilitated by this motif to organize global folding (for example, in the ribosome itself; Supplementary information S2 (table)) and to link separate domains of RNA together. In bacteriophage Qβ, although the viral replicase is anchored at a site some 1.2 kb from the 3′ end of the genome where replication initiates, an adjacent pseudoknot forms a long-range interaction that brings the 3′ end to the active site of the polymerase133. Such connections also offer the potential for global regulation of gene expression. In barley yellow dwarf virus, L3 of the frameshift-promoting pseudoknot is over 4 kb in length (of a 5.6-kb genome) and links the frameshift region with the 3′ end of the genome134. It has been suggested that disruption of this long-range contact is a means to regulate ribosome and replicase traffic on the viral genome135.
Especially in RNA viruses, there is often enormous selective pressure, and genomes that are optimized for viral fitness rapidly dominate. Although it is plain that some or all of the features of pseudoknots discussed above could be reproduced in other ways, it might be that this can only be achieved at considerable cost to fitness.
Future perspectives
Although 25 years have passed since the first pseudoknot was described1, much remains to be discovered. Foremost among the tasks ahead is to determine the true frequency of these motifs in natural RNAs. The majority of pseudoknots described in this article were identified as part of the normal process of scientific research, but computational approaches to their identification will be of increasing relevance. To maximize the proportion of genuine candidate pseudoknots identified in such screens, a better understanding of the thermodynamic parameters that govern pseudoknot formation will be invaluable. Additionally, we will need more information about the structure and function of pseudoknots. Until recently, atomic resolution structural information has been restricted to short, often very stable, pseudoknots. Elucidating the cryo-EM and crystal structures of the IGR IRESs of CrPV26 and PSIV27 were huge steps forward, but similar monumental efforts will be required to solve those structures in which the pseudoknot orchestrates the folding of long and complex domains, as is found in many viral NCRs. Several viral pseudoknots remain poorly characterized, both in terms of structure and biological activity. A number of these have been shown to interact with proteins, both viral (including RdRp) and cellular, yet molecular details are lacking. Furthermore, we do not know whether pseudoknots are ever substrates for viral or cellular helicases present in infected cells; such activities could offer an extra level of control of virus gene expression and replication. The use of pseudoknots in antiviral strategies should also be considered more broadly. Antisense oligomers targeting the SARS frameshift-promoting pseudoknot have marked antiviral activity136, and pseudoknotted aptamers have been shown to block HIV replication (Box 2).
The study of viral pseudoknots will continue to excite and challenge researchers. It is unquestionable that many more pseudoknots remain to be discovered, and it is highly likely that some of these will possess novel activities or have unprecedented functions. The field has come a long way since the early experiments of Pleij and his collaborators, yet there is still much to be discovered about these fascinating motifs.
Box 1 | Computational prediction of RNA pseudoknots.
RNA secondary structure prediction is not trivial and is restricted by our incomplete understanding of RNA thermodynamics and folding kinetics, and by long computer processing times137. RNAs can also adopt alternative conformations. Most programmes determine a minimum free-energy structure from the primary sequence, but cannot take into account pseudoknot topology, predominantly owing to the computational complexity138. Indeed, it has been calculated that to find a general method to search sequences for minimum free-energy pseudoknots is an intractable task139. Nevertheless, a substantial proportion (perhaps a quarter) of the pseudoknot literature deals with computational approaches to pseudoknot identification. Most commonly, a heuristic approach is taken where the search is restricted to certain pseudoknot types. However, most programmes are effective only for short sequences, as processing time can increase as the third to sixth power of sequence length, depending on the algorithm used. Another approach that has been used successfully is to perform simulations of RNA folding. These methods do not search for minimum free-energy structures and are therefore computationally more efficient. For longer sequences, a more efficient approach is to use pattern matching to perform a primary screen of a sequence database to gather those sequences with the potential to form pseudoknots of a defined type and subsequently analyse only these sequences. Multiple sequence comparisons are also of use. A combination of approaches is usually needed to have confidence in the outcome of a database search and the pseudoknots predicted within. The capacity of the programme to deal with pseudoknots containing stem regions with non-Watson–Crick pairs or bulged residues is often an issue. Despite these limitations, pseudoknot search programmes are a valuable resource. Examples of commonly used programmes and webservers are provided in Table 1 (see also Ref. 140 for a review).
Box 2 | RNA pseudoknot aptamers.
Aptamers are small nucleic acid ligands that are generated in vitro against various biological molecules. The majority are produced using the SELEX (selective evolution of ligands by exponential enrichment) method141. A pool of randomized RNAs is selectively enriched over repetitive rounds of target binding and subsequent sequence amplification until the surviving pool consists of sequences that bind to the target with high affinity and specificity. Initial experiments with SELEX focused on viral polymerases, confirming the sequence selectivity of T7 DNA polymerase141 and these were followed by the isolation of pseudoknotted aptamers able to bind to HIV-1 reverse transcriptase142. These aptamers have been shown to bind with picomolar affinity143 and can block HIV replication in tissue culture144, inhibiting the polymerase at all stages of genome replication145. The crystal structure of aptamer-bound reverse transcriptase shows how the pseudoknot ligand (coloured strand) is bound along the cleft between the polymerase active site and the RNAse H domain146 (see image; stem 1 (S1) is blue, S2 is red, loop 1 (L1) is yellow, L3 is green and additional non-pseudoknot bases are orange; dark grey is p51 and light grey is p66). The pseudoknot holds the polymerase in a 'closed' conformation, blocking its action by competitive inhibition of the primer–template binding site. Because of their specificity, there is considerable interest in the use of aptamers as therapeutics. However, aptamers cross cell membranes inefficiently owing to their hydrophilicity and are mostly restricted to extracellular applications such as the blocking of viral glycoproteins to prevent cell attachment and viral entry147. To be effective, such aptamers need to be chemically modified so as to resist host ribonucleases. SELEX experiments regularly isolate pseudoknotted ligands148, presumably reflecting the range of binding surfaces that pseudoknots can offer.
Supplementary information
Viral pseudoknots (PDF 111 kb)
Cellular pseudoknots (PDF 113 kb)
Pseudoknot-ribosome interactions. (PDF 487 kb)
The hepatitis delta virus ribozyme. (PDF 230 kb)
The human telomerase RNA pseudoknot. (PDF 173 kb)
Acknowledgements
We are grateful to Stephen Smerdon for critical reading of the manuscript.
Glossary
- Satellite virus
A subviral agent composed of an RNA or DNA molecule that is replicated in association with a helper virus, but encapsidated in its own encoded capsid protein.
- Peptidyl (P) site
The site on the small ribosomal subunit that holds the tRNA molecule that is linked to the growing end of the polypeptide chain.
- Aminoacyl (A) site
The site on the small ribosomal subunit that accepts and proofreads incoming aminoacyl tRNAs prior to peptidyl transfer.
- Exit (E) site
The site on the small ribosomal subunit through which tRNAs pass after they have donated their amino acid to the growing polypeptide chain.
- Ribozyme
An enzyme that has an RNA as it catalytic component.
- ShineDalgarno sequence
(AGGAGG). This sequence is located 5′ of the AUG codon on bacterial mRNAs and functions as the signal for the initiation of protein synthesis.
- Transfer messenger RNA
(tmRNA). An RNA that possesses both tRNA and mRNA characteristics that functions in the recognition and rescue of ribosomes stalled on aberrant mRNAs, the disposal of the causative defective mRNAs and the promotion of the degradation of ribosome-associated protein fragments.
- Rolling-circle replication
A mode of replication that uses a circular molecule as a template to produce concatemers of linear molecules.
- Riboswitch
A conformational switch in an RNA molecule that is induced by a small metabolite, and that leads to a switch in gene-regulatory function.
- Aptamer
An RNA domain, either engineered or natural, that forms a precise 3D structure and selectively binds a target molecule.
Biographies
Dr Ian Brierley graduated in Genetics at Leeds University, UK. Following a PhD in Biochemistry at the University of York, UK, he moved to the Department of Pathology, University of Cambridge, UK, where he is now a senior lecturer in virology. His laboratory studies translational regulation of virus gene expression, focusing on the role of RNA pseudoknots in ribosomal frameshifting.
Dr Simon Pennell is a Leeds University graduate (Microbiology) and did a PhD in virology at Cambridge. Presently, he is an investigator scientist in the Division of Molecular Structure at the National Institute for Medical Research, UK. His principal research interests are the structural aspects of translational control, with an emphasis on ribosomal frameshifting and internal ribosome entry site function.
Dr Robert Gilbert is a graduate of Durham University, UK, with a PhD from the University of Leicester, UK. Following a spell as a postdoctoral research assistant at the University of Oxford, UK, he took up his present post as a Royal Society University Research Fellow in the Division of Structural Biology at Oxford. His research interests include ribosome structure and mechanism during translation initiation and recoding, the structures of pore-forming proteins and of viruses, and the measurement of molecular interactions involving proteins in solution, using cryo-electron microscopy and X-ray crystallography.
Related links
DATABASES
Entrez Genome Project
Entrez Genome
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
Contributor Information
Ian Brierley, Email: ib103@mole.bio.cam.ac.uk.
Simon Pennell, Email: spennel@nimr.mrc.ac.uk.
Robert J. C. Gilbert, Email: gilbert@strubi.ox.ac.uk
References
- 1.Rietveld K, Van Poelgeest R, Pleij CW, Van Boom JH, Bosch L. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982;10:1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rietveld K, Pleij CW, Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pleij CW, Rietveld K, Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pleij CW. Pseudoknots: a new motif in the RNA game. Trends Biochem. Sci. 1990;15:143–147. doi: 10.1016/0968-0004(90)90214-V. [DOI] [PubMed] [Google Scholar]
- 5.ten Dam E, Pleij K, Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992;31:11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- 6.Hilbers CW, Michiels PJ, Heus HA. New developments in structure determination of pseudoknots. Biopolymers. 1998;48:137–153. doi: 10.1002/(SICI)1097-0282(1998)48:2<137::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Giedroc DP, Theimer CA, Nixon PL. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fechter P, Rudinger-Thirion J, Florentz C, Giege R. Novel features in the tRNA-like world of plant viral RNAs. Cell. Mol. Life Sci. 2001;58:1547–1561. doi: 10.1007/PL00000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Been MD. Molecular biology. Versatility of self-cleaving ribozymes. Science. 2006;313:1745–1747. doi: 10.1126/science.1133259. [DOI] [PubMed] [Google Scholar]
- 10.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Hellen CU. Bypassing translation initiation. Structure. 2007;15:4–6. doi: 10.1016/j.str.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel V, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 14.ten Dam EB, Pleij CW, Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baril M, Dulude D, Steinberg SV, Brakier-Gingras L. The frameshift stimulatory signal of human immunodeficiency virus type 1 group O is a pseudoknot. J. Mol. Biol. 2003;331:571–583. doi: 10.1016/S0022-2836(03)00784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPheeters DS, Stormo GD, Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J. Mol. Biol. 1988;201:517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- 17.Taylor JM. Structure and replication of hepatitis delta virus RNA. Curr. Top. Microbiol. Immunol. 2006;307:1–23. doi: 10.1007/3-540-29802-9_1. [DOI] [PubMed] [Google Scholar]
- 18.Westhof E, Jaeger L. RNA pseudoknots. Curr. Opin. Struct. Biol. 1992;2:327–333. doi: 10.1016/0959-440X(92)90221-R. [DOI] [Google Scholar]
- 19.van Batenburg FH, Gultyaev AP, Pleij CW. PseudoBase: structural information on RNA pseudoknots. Nucleic Acids Res. 2001;29:194–195. doi: 10.1093/nar/29.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim N, Shiffeldrim N, Gan HH, Schlick T. Candidates for novel RNA topologies. J. Mol. Biol. 2004;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 21.Kolk MH, et al. NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- 22.Mans RM, Pleij CW, Bosch L. tRNA-like structures. Structure, function and evolutionary significance. Eur. J. Biochem. 1991;201:303–324. doi: 10.1111/j.1432-1033.1991.tb16288.x. [DOI] [PubMed] [Google Scholar]
- 23.Deiman BALM, Pleij CWA. Pseudoknots: a vital feature in viral RNA. Sem. Virol. 1997;8:166–175. doi: 10.1006/smvy.1997.0119. [DOI] [Google Scholar]
- 24.Kim YG, Su L, Maas S, O'Neill A, Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl Acad. Sci. USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nature Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 27.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 29.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/S1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 30.Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res. 2006;119:2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nature Rev. Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 33.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/S1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 36.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Laletina E, et al. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukushi S, et al. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 39.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 40.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/S0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 41.Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the dicistroviridae intergenic region IRESes. J. Mol. Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl Acad. Sci. USA. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/S1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/S0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 47.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 48.Nishiyama T, et al. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J. Biol. Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- 50.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(SICI)1521-3773(19990816)38:16<2326::AID-ANIE2326>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Shamoo Y, Tam A, Konigsberg WH, Williams KR. Translational repression by the bacteriophage T4 gene 32 protein involves specific recognition of an RNA pseudoknot structure. J. Mol. Biol. 1993;232:89–104. doi: 10.1006/jmbi.1993.1372. [DOI] [PubMed] [Google Scholar]
- 53.Holland JA, Hansen MR, Du Z, Hoffman DW. An examination of coaxial stacking of helical stems in a pseudoknot motif: the gene 32 messenger RNA pseudoknot of bacteriophage T2. RNA. 1999;5:257–271. doi: 10.1017/S1355838299981360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang CK, Draper DE. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- 55.Schlax PJ, Xavier KA, Gluick TC, Draper DE. Translational repression of the Escherichia coli α operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem. 2001;276:38494–38501. doi: 10.1074/jbc.M106934200. [DOI] [PubMed] [Google Scholar]
- 56.Mathy N, et al. Specific recognition of rpsO mRNA and 16S rRNA by Escherichia coli ribosomal protein S15 relies on both mimicry and site differentiation. Mol. Microbiol. 2004;52:661–675. doi: 10.1111/j.1365-2958.2004.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brierley I, Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 58.Brierley I, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 60.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag–Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and Gag/Gag–Pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhai Y, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nature Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imbert I, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baranov PV, et al. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plant EP, et al. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/S0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 68.Plant EP, et al. The 9-A solution: how mRNA pseudoknots promote efficient programmed-1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plant EP, Dinman JD. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 71.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su L, Chen L, Egli M, Berger JM, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nature Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michiels PJ, et al. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cornish PV, Stammler SN, Giedroc DP. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen LX, Tinoco I., Jr. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 77.Wang Y, et al. Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: a mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral Gag–Pro frameshift site. RNA. 2002;8:981–996. doi: 10.1017/S1355838202024044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cornish PV, Hennig M, Giedroc DP. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated-1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hansen TM, Reihani SN, Oddershede LB, Sorensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shigemoto K, et al. Identification and characterisation of a developmentally regulated mammalian gene that utilises −1 programmed ribosomal frameshifting. Nucleic Acids Res. 2001;29:4079–4088. doi: 10.1093/nar/29.19.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manktelow E, Shigemoto K, Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wills NM, Moore B, Hammer A, Gesteland RF, Atkins JF. A functional-1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]
- 83.Jacobs JL, Belew AT, Rakauskaite R, Dinman JD. Identification of functional, endogenous programmed-1 ribosomal frameshift signals in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:165–174. doi: 10.1093/nar/gkl1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nonin-Lecomte S, Felden B, Dardel F. NMR structure of the Aquifex aeolicus tmRNA pseudoknot PK1: new insights into the recoding event of the ribosomal trans-translation. Nucleic Acids Res. 2006;34:1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc. Natl Acad. Sci. USA. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 87.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 2003;50:303–317. doi: 10.1046/j.1365-2958.2003.03684.x. [DOI] [PubMed] [Google Scholar]
- 89.Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng YX, Yuan H, Rein A, Levin JG. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J. Virol. 1992;66:5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wills NM, Gesteland RF, Atkins JF. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J. Mol. Biol. 1999;288:837–852. doi: 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- 93.Gluick TC, Wills NM, Gesteland RF, Draper DE. Folding of an mRNA pseudoknot required for stop codon readthrough: effects of mono- and divalent ions on stability. Biochemistry. 1997;36:16173–16186. doi: 10.1021/bi971362v. [DOI] [PubMed] [Google Scholar]
- 94.Orlova M, Yueh A, Leung J, Goff SP. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell. 2003;115:319–331. doi: 10.1016/S0092-8674(03)00805-5. [DOI] [PubMed] [Google Scholar]
- 95.Pinck M, Yot P, Chapeville F, Duranton HM. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature. 1970;226:954–956. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- 96.Giege R. Interplay of tRNA-like structures from plant viral RNAs with partners of the translation and replication machineries. Proc. Natl Acad. Sci. USA. 1996;93:12078–12081. doi: 10.1073/pnas.93.22.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsuda D, Dreher TW. The tRNA-like structure of turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 98.Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. doi: 10.1016/S0092-8674(02)01256-4. [DOI] [PubMed] [Google Scholar]
- 99.Matsuda D, Dreher TW. Cap- and initiator tRNA-dependent initiation of TYMV polyprotein synthesis by ribosomes: evaluation of the Trojan horse model for TYMV RNA translation. RNA. 2007;13:129–137. doi: 10.1261/rna.244407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kolakofsky D, Weissmann C. Possible mechanism for transition of viral RNA from polysome to replication complex. Nature New Biol. 1971;231:42–46. doi: 10.1038/newbio231042a0. [DOI] [PubMed] [Google Scholar]
- 101.Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barton DJ, Morasco BJ, Flanegan JB. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deiman BA, Koenen AK, Verlaan PW, Pleij CW. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J. Virol. 1998;72:3965–3972. doi: 10.1128/jvi.72.5.3965-3972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and –CCR– boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998;4:1083–1095. doi: 10.1017/S1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 106.Choi YG, Rao AL. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 2003;77:9750–9757. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koenig R, et al. Nemesia ring necrosis virus: a new tymovirus with a genomic RNA having a histidylatable tobamovirus-like 3′ end. J. Gen. Virol. 2005;86:1827–1833. doi: 10.1099/vir.0.80916-0. [DOI] [PubMed] [Google Scholar]
- 108.van Belkum A, Abrahams JP, Pleij CW, Bosch L. Five pseudoknots are present at the 204 nucleotides long 3′ noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res. 1985;13:7673–7686. doi: 10.1093/nar/13.21.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Felden B, Florentz C, Giege R, Westhof E. A central pseudoknotted three-way junction imposes tRNA-like mimicry and the orientation of three 5′ upstream pseudoknots in the 3′ terminus of tobacco mosaic virus RNA. RNA. 1996;2:201–212. [PMC free article] [PubMed] [Google Scholar]
- 110.Leathers V, Tanguay R, Kobayashi M, Gallie DR. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell Biol. 1993;13:5331–5347. doi: 10.1128/MCB.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zeenko VV, et al. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 2002;76:5678–5691. doi: 10.1128/JVI.76.11.5678-5691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takamatsu N, Watanabe Y, Meshi T, Okada Y. Mutational analysis of the pseudoknot region in the 3′ noncoding region of tobacco mosaic virus RNA. J. Virol. 1990;64:3686–3693. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Carr T, et al. Tobamovirus infection is independent of HSP101 mRNA induction and protein expression. Virus Res. 2006;121:33–41. doi: 10.1016/j.virusres.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 114.Ray D, Na H, White KA. Structural properties of a multifunctional T-shaped RNA domain that mediate efficient tomato bushy stunt virus RNA replication. J. Virol. 2004;78:10490–10500. doi: 10.1128/JVI.78.19.10490-10500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fabian MR, White KA. 5′-3′ RNA–RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for tombusviridae. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- 116.Fabian MR, White KA. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA–RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Na H, Fabian MR, White KA. Conformational organization of the 3′ untranslated region in the tomato bushy stunt virus genome. RNA. 2006;12:2199–2210. doi: 10.1261/rna.238606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ferre-D'Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 119.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 120.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 121.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc. Natl Acad. Sci. USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. doi: 10.1002/j.1460-2075.1996.tb00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. A molecular switch underlies a human telomerase disease. Proc. Natl Acad. Sci. USA. 2002;99:16998–17003. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Theimer CA, Finger LD, Trantirek L, Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl Acad. Sci. USA. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yingling YG, Shapiro BA. The prediction of the wild-type telomerase RNA pseudoknot structure and the pivotal role of the bulge in its formation. J. Mol. Graph Model. 2006;25:261–274. doi: 10.1016/j.jmgm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 127.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 128.Chen Z, et al. Telomerase activity in Kaposi's sarcoma, squamous cell carcinoma, and basal cell carcinoma. Exp. Biol. Med. 2001;226:753–757. doi: 10.1177/153537020222600807. [DOI] [PubMed] [Google Scholar]
- 129.Fragnet L, Blasco MA, Klapper W, Rasschaert D. The RNA subunit of telomerase is encoded by Marek's disease virus. J. Virol. 2003;77:5985–5996. doi: 10.1128/JVI.77.10.5985-5996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trapp S, et al. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2006;203:1307–1317. doi: 10.1084/jem.20052240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Artandi SE. Telomerase flies the coop: the telomerase RNA component as a viral-encoded oncogene. J. Exp. Med. 2006;203:1143–1145. doi: 10.1084/jem.20060849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wyatt JR, Puglisi JD, Tinoco I., Jr. RNA pseudoknots. Stability and loop size requirements. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 133.Klovins J, van Duin J. A long-range pseudoknot in Qβ RNA is essential for replication. J. Mol. Biol. 1999;294:875–884. doi: 10.1006/jmbi.1999.3274. [DOI] [PubMed] [Google Scholar]
- 134.Paul CP, Barry JK, Dinesh-Kumar SP, Brault V, Miller WA. A sequence required for-1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 2001;310:987–999. doi: 10.1006/jmbi.2001.4801. [DOI] [PubMed] [Google Scholar]
- 135.Barry JK, Miller WA. A-1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl Acad. Sci. USA. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Neuman BW, et al. Inhibition, escape and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 2005;79:9665–9676. doi: 10.1128/JVI.79.15.9665-9676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 138.Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lyngso RB, Pedersen CN. RNA pseudoknot prediction in energy-based models. J. Comput. Biol. 2000;7:409–427. doi: 10.1089/106652700750050862. [DOI] [PubMed] [Google Scholar]
- 140.Shapiro BA, Yingling YG, Kasprzak W. & Bindewald E. Bridging the gap in RNA structure prediction. Curr. Opin. Struct. Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 141.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 142.Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kensch O, et al. HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J. Biol. Chem. 2000;275:18271–18278. doi: 10.1074/jbc.M001309200. [DOI] [PubMed] [Google Scholar]
- 144.Chaloin L, Lehmann MJ, Sczakiel G, Restle T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res. 2002;30:4001–4008. doi: 10.1093/nar/gkf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Held DM, Kissel JD, Saran D, Michalowski D, Burke DH. Differential susceptibility of HIV-1 reverse transcriptase to inhibition by RNA aptamers in enzymatic reactions monitoring specific steps during genome replication. J. Biol. Chem. 2006;281:25712–25722. doi: 10.1074/jbc.M604460200. [DOI] [PubMed] [Google Scholar]
- 146.Jaeger J, Restle T, Steitz TA. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 1998;17:4535–4542. doi: 10.1093/emboj/17.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sayer N, Ibrahim J, Turner K, Tahiri-Alaoui A, James W. Structural characterization of a 2′F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun. 2002;293:924–931. doi: 10.1016/S0006-291X(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 148.Kim MY, Jeong S. RNA aptamers that bind the nucleocapsid protein contain pseudoknots. Mol. Cell. 2003;16:413–417. [PubMed] [Google Scholar]
- 149.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Viral pseudoknots (PDF 111 kb)
Cellular pseudoknots (PDF 113 kb)
Pseudoknot-ribosome interactions. (PDF 487 kb)
The hepatitis delta virus ribozyme. (PDF 230 kb)
The human telomerase RNA pseudoknot. (PDF 173 kb)


