Key Points
mRNAs are protected at their 5′ ends by a cap structure consisting of an N7-methylated GTP molecule linked to the first transcribed nucleotide by a 5′–5′ triphosphate bond.
The cap structure is essential for RNA splicing, export and stability, and allows the ribosomal complex to recognize mRNAs and ensure their efficient translation.
Uncapped RNA molecules are degraded in cytoplasmic granular compartments called processing bodies and may be detected as 'non-self' by the host cell, triggering antiviral innate immune responses through the production of interferons.
Conventional RNA capping (that is, of mRNAs from the host cell and from DNA viruses) requires hydrolysis of the 5′ γ-phosphate of RNA by an RNA triphosphatase, transfer of a GMP molecule onto the 5′-end of RNA by a guanylyltransferase, and methylation of this guanosine by an (guanine-N7)-methyltransferase. Subsequent methylations on the first and second transcribed nucleotides by (nucleoside-2′-O)-methyltransferases form cap-1 and cap-2 structures.
Viruses have evolved highly diverse capping mechanisms to acquire cap structures using their own or cellular capping machineries, or by stealing cap structures from cellular mRNAs.
Virally encoded RNA-capping machineries are diverse in terms of their genetic components, protein domain organization, enzyme structures, and reaction mechanisms and pathways, making viral RNA capping an attractive target for antiviral-drug design.
Subject terms: Virology, Cellular microbiology, Pathogens, Innate immunity, Virus-host interactions
Capping the 5′ end of eukaryotic mRNAs with a 7-methylguanosine moiety enables efficient splicing, nuclear export and translation of mRNAs, and also limits their degradation by cellular exonucleases. Here, Canard and colleagues describe how viruses synthesize their own mRNA cap structures or steal them from host mRNAs, allowing efficient synthesis of viral proteins and avoidance of host innate immune responses.
Abstract
In the eukaryotic cell, capping of mRNA 5′ ends is an essential structural modification that allows efficient mRNA translation, directs pre-mRNA splicing and mRNA export from the nucleus, limits mRNA degradation by cellular 5′–3′ exonucleases and allows recognition of foreign RNAs (including viral transcripts) as 'non-self'. However, viruses have evolved mechanisms to protect their RNA 5′ ends with either a covalently attached peptide or a cap moiety (7-methyl-Gppp, in which p is a phosphate group) that is indistinguishable from cellular mRNA cap structures. Viral RNA caps can be stolen from cellular mRNAs or synthesized using either a host- or virus-encoded capping apparatus, and these capping assemblies exhibit a wide diversity in organization, structure and mechanism. Here, we review the strategies used by viruses of eukaryotic cells to produce functional mRNA 5′-caps and escape innate immunity.
Main
The cap structure found at the 5′ end of eukaryotic mRNAs consists of a 7-methylguanosine (m7G) moiety linked to the first nucleotide of the transcript via a 5′–5′ triphosphate bridge1 (Fig. 1a). The cap has several important biological roles, such as protecting mRNA from degradation by 5′ exoribonucleases and directing 'pre-mRNA splicing and mRNA export from the nucleus2. In addition, the cap confers stability to mRNAs and ensures their efficient recognition by eukaryotic translation initiation factor 4E (eIF4E) for translation3,4. Conversely, RNA molecules with unprotected 5′ ends are degraded in cytoplasmic granular compartments called processing bodies (P-bodies)5. Uncapped RNAs, such as nascent viral transcripts, may also be detected as 'non-self' by the host cell, triggering (in mammalian cells) an antiviral innate immune response through the production of interferons6,7.
Figure 1. RNA cap structure and canonical capping mechanisms.
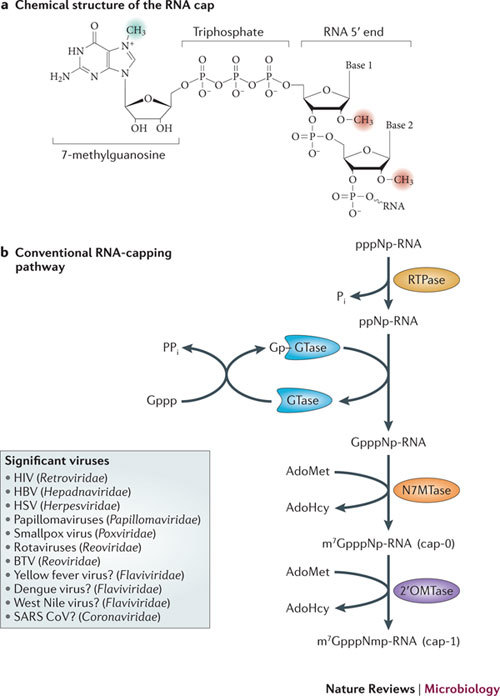
a | The mRNA cap consists of a 7-methylguanosine linked to the 5′ nucleoside of the mRNA chain through a 5′–5′ triphosphate bridge. The methyl group at the N7 position of the guanosine is shaded green, and the 2′-O-methyl groups of the first and second nucleotide residues, forming the cap-1 and the cap-2 structures, respectively, are shaded red. b | The cap-0 structure is formed on nascent RNA chains by the sequential action of three enzymes. First, the RNA triphosphatase (RTPase) hydrolyses the γ-phosphate of the nascent RNA (pppNp-RNA, in which N denotes the first transcribed nucleotide and p denotes a phosphate group) to yield a diphosphate RNA (ppNp-RNA) and inorganic phosphate (Pi). Then, guanylyltransferase (GTase) reacts with the α-phosphate of GTP (Gppp), releasing pyrophosphate (PPi) and forming a covalent enzyme–guanylate intermediate (Gp–GTase). The GTase then transfers the GMP molecule (Gp) to the 5′-diphosphate RNA to create GpppNp-RNA. In the final step, (guanine-N7)-methyltransferase (N7MTase) transfers the methyl group from S-adenosyl-L-methionine (AdoMet) to the cap guanine to form the cap-0 structure, 7-methyl-GpppNp (m7GpppNp), and releases S-adenosyl-L-homocysteine (AdoHcy) as a by-product. The capping reaction is completed by methylation of the ribose-2′-O position of the first nucleotide by the AdoMet-dependent (nucleoside-2′-O)-methyltransferase (2′OMTase), generating the cap-1 structure (m7GpppNm2′-Op). The box contains examples of viruses that acquire their cap structures using the cellular capping machinery or encode their own viral capping machineries that adopt the canonical pathway. Question marks indicate viruses that are likely to follow this conventional pathway. The RNAs capped by viral enzymes are indistinguishable from cellular mRNA and can thus be translated into proteins by the cellular ribosomal machinery. BTV, bluetongue virus; HBV, hepatitis B virus; HSV, herpes simplex viruses; SARS CoV, severe acute respiratory syndrome coronavirus.
During virus–host co-evolution, viral RNA-capping pathways that lead to the same cap structure as that of host mRNAs have been selected for as efficient mechanisms to ensure both escape from detection by the innate immune system and efficient production of viral proteins. Compared with canonical eukaryotic mRNA capping, viral mRNA capping is highly diverse in terms of its genetic components, protein domain organization, enzyme structures and reaction mechanisms. Here, we review what is known about the various mRNA-capping pathways used by viruses that infect eukaryotes, paying particular attention to human pathogens. We also attempt to connect the pathways, machineries, structures and reactions involved in the viral RNA-capping process, and the specific cellular factors that trigger a response from the innate immune system.
Capping, decapping and turnover of host RNA
Nascent cellular mRNAs are generally produced in the nucleus in a 5′-triphosphate form and are modified co-transcriptionally by a set of cap-synthesizing enzymes. These enzymes are recruited by the DNA-dependent RNA polymerase RNA pol II on pausing, once the transcript is approximately 20–25 nucleotides long. Interestingly, two viruses — vaccinia virus (from the Poxviridae family of double-stranded DNA (dsDNA) viruses) and mammalian orthoreovirus (from the dsRNA Reoviridae family) — played a major part in the discovery of the RNA cap1,8,9,10,11, because they produce high levels of capped viral mRNAs and encode their own capping machinery, allowing bona fide RNA cap synthesis in vitro. Mammalian orthoreovirus mRNAs and dsRNA genomes were first shown to be blocked at the 5′ end, preventing phosphorylation by polynucleotide kinase9,10. Purified mammalian orthoreovirus preparations were then demonstrated to be able to methylate mRNA 5′ ends, and the full mRNA cap structures were deciphered for both dsRNA orthoreoviruses9,10,12 (those targeting humans and insects) and the dsDNA species vaccinia virus11, followed by many other viral species1,13,14.
The three canonical capping reactions responsible for the synthesis of the cap structure are outlined in Fig. 1b. This pathway is found in all eukaryotic species, as well as in most DNA viruses and members of the family Reoviridae, and consists of the following reactions: hydrolysis of the γ-phosphate of the primary transcript by an RNA 5′-triphosphatase (RTPase); transfer of GMP to the 5′-diphosphate RNA (ppNp-RNA; in which N is the first transcribed nucleotide and p is a phosphate group) by a guanylyltransferase (GTase) through a 'ping-pong' mechanism, leading to the formation of GpppNp-RNA; and methylation of the guanosine moiety by a cap-specific S-adenosyl-L-methionine (AdoMet)-dependent (guanine-N7)-methyltransferase (N7MTase), providing the minimal RNA cap chemical structure, named cap-0 (m7GpppNp), which is recognized by the translation factor eIF4E15. Further methylation reactions catalysed by (nucleoside-2′-O)-methyltransferases (2′OMTases) can occur on the first and second nucleotides 3′ to the triphosphate bridge, yielding cap-1 (m7GpppNm2′-O), and cap-2 (m7GpppNm2′-OpNm2′-Op) structures, respectively (Fig. 1). Cap-4 structures (m7GpppNm2′-OpNm2′-OpNm2′-OpNm2′-Op) were also identified on parasite mRNAs16.
Following translation, the lifespan of an mRNA molecule is controlled by two main processes in eukaryotic cells: first, the removal of its poly(A) tail and subsequent 3′-to-5′ exonucleolytic degradation, and second, an mRNA-decapping step that allows 5′-to-3′ exonucleolytic degradation (see Refs 17, 18 for a review). Interestingly, decapped RNA may apparently be re-capped by the combined action of an as-yet-unknown 5′-monophosphate kinase19 interacting with a host cell GTase that is also present in minor amounts in the cytoplasm (see Ref. 20 for a review). Cytoplasmic decapping is catalysed by DCPS, a member of the NUDIX hydrolase superfamily, and stimulated by decapping enhancer proteins. Following RNA decapping in P-bodies, transcripts are quickly degraded by 5′-to-3′ exonucleases such as XRN1. Thus, 5′-triphosphate mRNAs are almost completely absent in the cytoplasm. The eukaryotic cell has consequently evolved mechanisms to sense triphosphate RNA as non-self and uses these RNA species to trigger an innate immune response6,7. Viruses are the most common cell invaders to produce cytoplasmic mRNAs and have therefore been under a selective pressure to evolve counteracting mechanisms to conceal their RNA 5′ ends from the innate immunity machineries of the host cell.
Some viruses, such as those in the family Picornaviridae (single-stranded positive-sense RNA viruses (ss(+)RNA viruses); for example, polioviruses and encephalomyocarditis virus (EMCV)) recruit the 43S pre-initiation complex in a 5′-cap-independent manner (Box 1). Other viruses (for example, viruses of the family Caliciviridae, which are also ss(+)RNA viruses) covalently attach their RNA 5′ end to a VPg-like protein, which directly interacts with the cap-binding protein eIF4E and initiates translation of viral mRNAs21. In members of the Picornaviridae, VPg may be lost before translation22,23.
However, by far the most common viral mechanism for ensuring efficient translation of viral proteins and avoiding immune surveillance mechanisms is through the acquisition of a cap structure. The remarkable diversity of mechanisms that lead to an RNA cap structure identical to that of the host cell mRNAs is described below.
Conventional capping of viral RNA
Even when the viral replication cycle includes a nuclear phase, any virus entering a host cell must express its genes in the cytoplasm using the host translation machinery. Viral genomes can be made of single-stranded or double-stranded nucleic acids, either RNA or DNA, and the corresponding strategies used for protecting their RNA transcripts are outlined in Fig. 2.
Figure 2. RNA 5′ ends in the mammalian-virus world.
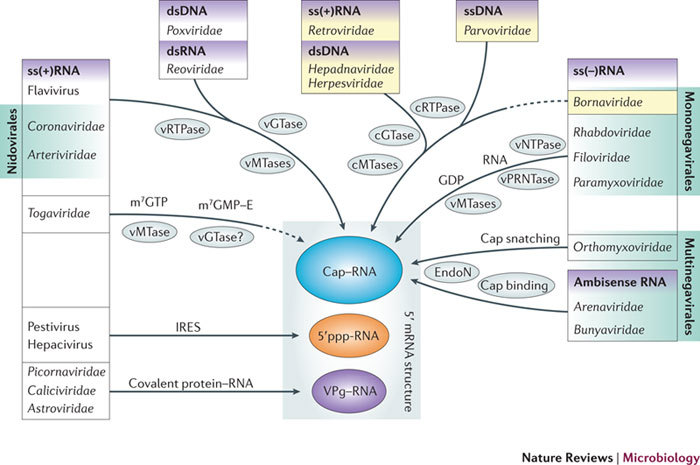
Mammalian viruses, with the exception of those from the single-stranded positive-sense RNA (ss(+)RNA) virus genera Pestivirus and Hepacivirus, use strategies to chemically modify their mRNA 5′ ends through either covalent attachment of a protein (VPg for ss(+)RNA viruses from the families Picornaviridae, Caliciviridae and Astroviridae) or covalent attachment of an RNA cap structure (all other viruses). Arrows indicate the type of RNA 5′ end protection that is used by these viral groups, and the enzymes and mechanisms involved are indicated. Dashed arrows indicate a likely but incompletely demonstrated pathway. Viral and cellular proteins are distinguished with the prefixes v and c, respectively. Yellow shading highlights viral groups for which the life cycle includes a nuclear phase in the host cell. This list of viral taxa is non exhaustive and used as an example only. ds, double-stranded; E, enzyme; EndoN, endonuclease; GTase, guanylyltransferase; IRES, internal ribosome entry site; m7, 7-methyl; MTase, methyltransferase; NTPase, nucleotide 5′-triphosphatase; p, phosphate group; PRNTase, polyribonucleotidyl transferase; RTPase, RNA triphosphatase; ss(−)RNA, single-stranded negative-sense RNA.
Cap structures can be added to viral RNAs by one of the three following mechanisms. In the first mechanism, most viruses that synthesize their mRNA using cellular RNA pol II use the cellular capping machinery (Figs 1b, 2), as exemplified by most DNA viruses (except for those from the dsDNA virus family Poxviridae) and by RNA viruses such as those of the family Retroviridae (ss(+)RNA viruses) and the family Bornaviridae (single-stranded negative-sense RNA viruses (ss(−)RNA viruses)). A second strategy consists of acquiring cap structures from cellular mRNAs by 'cap snatching' (Figs 2, 3). RNA viruses such as members of the families Orthomyxoviridae, Arenaviridae and Bunyaviridae — which are ss(−)RNA viruses, with the latter two families also referred to as ambisense RNA viruses — steal short, capped cellular mRNAs through endonucleolytic cleavage. The stolen short, capped mRNAs are then used by the viral polymerase to prime synthesis of viral RNA. For the third method, many viruses encode their own capping machinery and have evolved a diverse set of dedicated enzymes and mechanisms to carry out capping. Accordingly, most viruses with an ssRNA genome synthesize or acquire the RNA cap using their own set of enzymes (Fig. 2). Within this diversity, the capping of viral mRNA can be classified as either 'conventional', when it follows the mRNA-capping pathways used by eukaryotes and DNA viruses (that is, the sequential action of RTPase, GTase and MTases) (Figs 1b, 2), or 'unconventional', when it does not (see below and Fig. 3).
Figure 3. Unconventional capping pathways.
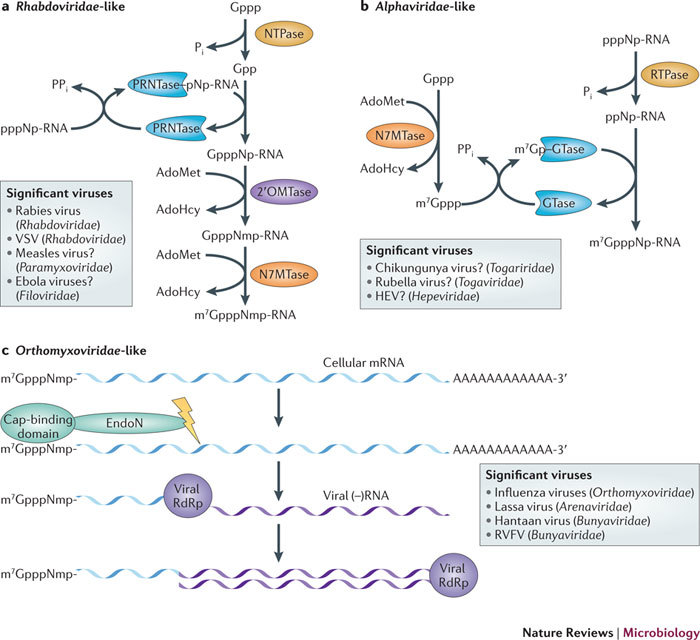
a | The RNA-capping mechanism of negative-sense RNA ((−)RNA) viruses such as those of the family Rhabdoviridae. The NTPase hydrolyses the γ-phosphate of GTP (Gppp) to yield a GDP (Gpp) and inorganic phosphate (Pi). Polyribonucleotidyl transferase (PRNTase) reacts with the nascent viral RNA (pppNp-RNA; in which N denotes the first transcribed nucleotide), releasing pyrophosphate (PPi) and forming a covalent PRNTase–pNp-RNA intermediate. The PRNTase then transfers the RNA molecule to the GDP to create GpppNp-RNA. (Nucleoside-2′-O)-methyltransferase (2′OMTase) transfers the methyl group from S-adenosyl-L-methionine (AdoMet) to the first nucleotide of the RNA. The capping reaction is then completed by methylation of the cap by the AdoMet-dependent (guanine-N7)-methyltransferase (N7MTase). The box lists examples of viruses that acquire their cap structures using such a capping pathway. Question marks indicate viruses that are likely to follow this conventional pathway. b | The RNA-capping mechanism of positive-sense RNA ((+)RNA) viruses such as those of the family Alphaviridae. The RNA triphosphatase (RTPase) hydrolyses the γ-phosphate of the viral RNA to yield a diphosphate RNA (ppNp-RNA) and Pi. A GTP molecule is methylated at its N7 position by the AdoMet-dependent N7MTase. Guanylyltransferase (GTase) then binds the N7-methyl-GTP (m7Gppp), forming a covalent link with a catalytic histidine (m7Gp–GTase) and releasing PPi. The GTase then transfers the m7GMP molecule (m7Gp) to the 5′-diphosphate RNA to create m7GpppNp-RNA. The box indicates viruses that acquire their cap structures using such a capping pathway. c | the RNA-capping mechanism of (–)RNA viruses such as those of the family Orthomyxoviridae; this mechanism is referred to as cap snatching. The PB2 subunit of the viral RNA-dependent RNA polymerase (RdRp) binds to the 5′ end of cellular capped mRNAs (which are enriched in the processing (P)-bodies), and the PA subunit then releases short capped RNAs by using its endonuclease (EndoN) activity. These capped RNAs are used as primers by the viral RdRp in viral transcription to generate viral mRNA using the viral (–)RNA as a template. The RdRp then synthesizes the complementary negative-sense strand. The box provides example of viruses that acquire their cap structures using a similar capping pathway. Note that most of the mRNAs that are capped by viral enzymes are indistinguishable from cellular mRNAs and can be translated into proteins by the cellular ribosomal machinery. AdoHcy, S-adenosyl-L-homocysteine; HEV, hepatitis E virus; RVFV, Rift Valley fever virus; VSV, vesicular stomatitis virus.
The best characterized conventional RNA-capping system is that exemplified by the dsDNA virus vaccinia virus, which expresses a multifunctional mRNA cap-synthesizing enzyme (D1) containing RTPase, GTase and N7MTase domains24. The 5′-triphosphate of the nascent mRNA is first hydrolysed by the RTPase to yield 5′-diphosphate RNA, which is then sequentially transferred to other internal domains25,26 to be capped and methylated, the latter reaction with allosteric stimulation through direct association with viral D12 protein25,26. Cap assembly is completed by the viral VP39, a bifunctional protein that catalyses the 2′-O-methylation of the cap-0 structure and also acts as an elongation factor for poly(A) polymerase27. Viruses from the dsRNA virus family Reoviridae share the same pathway as vaccinia virus. The enzymes remain physically associated in an inner capsid (or 'transcriptionally active core'), which constitutes a molecular machine or 'assembly line' that is able to transcribe the genome, synthesize the cap and inject the resulting mRNA into the cytoplasm of the host cell for translation. In addition, further assignment and characterization of the GTase activity in viruses of the genus Flavivirus and those of the family Coronaviridae may join these viruses, which are ss(+)RNA viruses, to the conventional RNA-capping pathway group.
Conventional viral RNA-cap-synthesizing enzymes
RNA triphosphatases. RTPases catalyse the cleavage of the interphosphate bond between the β-phosphate and the γ-phosphate of 5′-triphosphorylated mRNA (Fig. 1b). A range of enzyme structures exist (Fig. 4a), indicating that during evolution several independent solutions evolved for this initial step of the capping reaction, which is often found in association with RNA-binding and strand separation activities. Most RTPases are also able to hydrolyse NTPs28,29,30.
Figure 4. Structural constituents of viral capping machineries, folds and mechanisms.
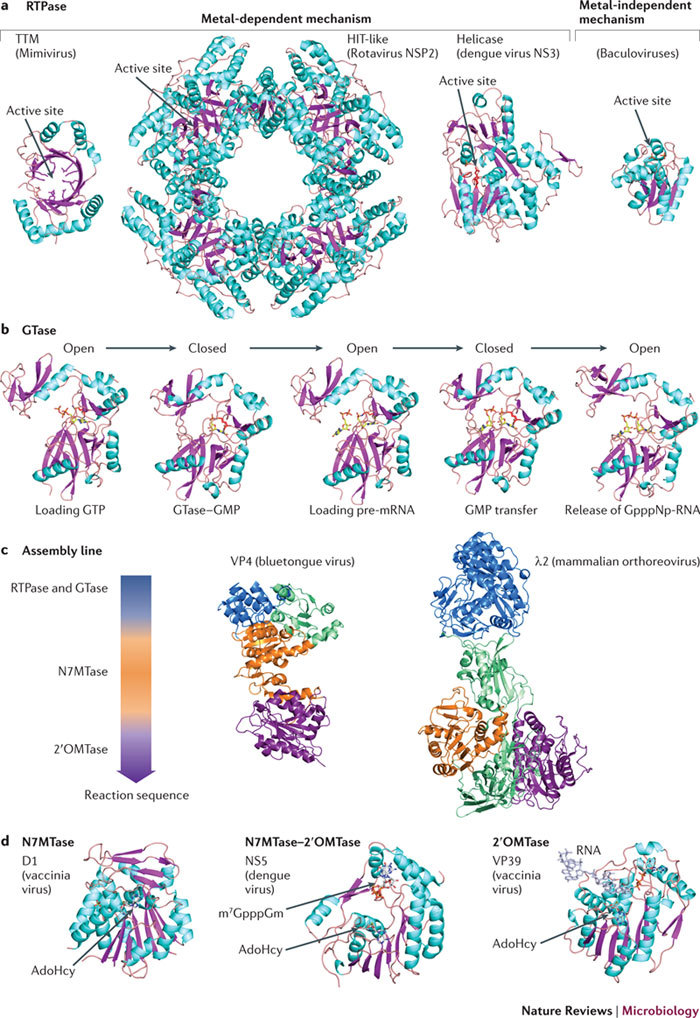
a | Different RNA triphosphatase (RTPase) folds. For the metal-dependent mechanism, the triphosphate tunnel metalloenzyme (TTM) fold is exemplified by the structure of the RTPase (Protein Data Bank (PDB) accession code 2QZE) from the genus Mimivirus, which consists of double-stranded DNA (dsDNA) viruses; the histidine triad (HIT)-like fold of the RTPase from dsRNA rotaviruses (PDB accession code 1L9V) and the helicase fold of the RTPase from the single-stranded positive-sense RNA (ss(+)RNA) virus dengue virus (PDB accession code 2BHR) are also shown. For the metal-independent mechanism, the fold of the RTPase from the dsDNA baculoviruses (PDB accession code 1YN9) is the only viral structure available so far. Structures are coloured cyan for α-helices and pink for β-strands. b | Structure-based model of a guanylyl transfer mediated by the dsDNA virus Paramecium bursaria chlorella virus 1 guanylyltransferase (GTase). Presented are the five stages of the reaction: loading of GTP onto the active site, with the GTase in an open conformation (PDB accession code 1CKO); the GTase in a closed conformation and the creation of covalent intermediate GTase–GMP through a lysine residue (model derived from the structure in PDB accession code 1CKN); the intermediate GTase–GMP reopening to bind a pre-mRNA molecule (model derived from the structure in PDB accession code 1CKO); the GTase closing again to complete the transfer (model derived from the structure in PDB accession code 1CKN); and, transfer completed, the GTase opening to release the GpppNp-RNA (in which p is a phosphate group and N is the first transcribed nucleotide) (PDB accession code 1CKM). Structures are coloured cyan for α-helices and pink for β-strands. c | Assembly line structures. Protein VP4 (PDB accession code 2JHC) of bluetongue virus (a dsRNA virus from the genus Orbivirus) and protein λ2 of mammalian orthoreovirus (a dsRNA virus) (PDB accession code 1EJ6). The colour code correlates with the colour of each domain as represented in the assembly sequence arrow to the left. Domains in green are extra domains that do not take part directly in the capping mechanism. d | Methyltransferase structures. Left: the (guanine-N7)-methyltransferase (N7MTase) domain of protein D1 from the dsDNA virus vaccinia virus (PDB accession code 2VDW) in complex with a molecule of S-adenosyl-L-homocysteine (AdoHcy). Middle: the N7MTase–(nucleoside-2′-O)-methyltransferase (2OMTase) domain of NS5 from dengue virus (ss(+)RNA) in complex with the cap analogue 7-methyl-G (m7G)-pppGm2′-O and AdoHcy (PDB accession code 2P41). Right: VP39, the 2′OMTase of the dsDNA virus vaccinia virus, in complex with a capped RNA and AdoHcy (PDB accession code 1AV6). All figures were prepared using PyMOL.
RTPases can be categorized into four groups, further defined by the metal dependency of the hydrolytic reaction mechanism that they use to cleave the interphosphate bond. Metal-dependent RTPases can be organized into three structural groups: the histidine triad (HIT)-like fold (α–β complex), the triphosphate tunnel metalloenzyme (TTM) and the viral RNA helicase-like fold (Fig. 4a).
The HIT-like fold is found in NSP2, from the dsRNA rotaviruses, and so far it is the only RTPase identified with such a fold. It has an amino-terminal helical domain and a carboxyl terminus with an α–β fold that resembles the ubiquitous cellular HIT group of nucleotidyl hydrolases31. The nucleotide-binding site is located in the cleft between the two domains, which contains a histidine residue involved in binding and in Mg2+-dependent hydrolysis of both NTP and RNA substrates. NSP2 self-assembles into a doughnut-shaped octamer, a quaternary structural organization that creates several RNA-binding sites, which are presumably needed to destabilize RNA duplexes during genome replication and packaging30,32,33.
TTM enzymes hydrolyse NTPs in the presence of Mn2+ or Co2+ and are found in fungi and protozoa, and in most DNA viruses that encode an RTPase, including poxviruses, chlorella virus, baculoviruses and mimiviruses34,35,36,37. The RTPase Cet1 from Saccharomyces cerevisiae is nearly identical to that of mimiviruses and serves as a paradigm for the TTM group. Its structure features a characteristic tunnel lined by eight antiparallel β strands (Fig. 4a) and a double glutamate motif36,38. The mRNA supposedly sits in the tunnel, which harbours several charged and hydrophilic side chains that coordinate Mn2+ and SO2− in the crystal structure. SO2− is thought to indicate the position of the γ-phosphate of mRNA.
The third group comprises proteins with NTPase and helicase activity, belonging to the large helicase superfamilies SF1 and SF2 (Refs 39, 40, 41). Such NTPase–helicase family members are found in flaviviruses (for example, NS3 proteins of dengue virus, yellow fever virus, West Nile virus and Japanese encephalitis virus), coronaviruses (for example, nsp13 of severe acute respiratory syndrome coronavirus (SARS CoV), although for this member of the SF1 helicases the crystal structure is not known), alphaviruses (for example, the protein nsP2) and potexviruses (for example, protein 1A of bamboo mosaic virus), all of which are ss(+)RNA viruses, and also in viruses of the family Reoviridae (for example, the protein λ1), which are dsRNA viruses. The fold adopted by SF2 helicases features two RecA-like subdomains between which a cleft accommodates the nucleotide or 5′-triphosphate RNA substrate42. The RecA subdomains I and II carry the Walker A and B motifs. Residues belonging to the Walker A motif (also named motif I) form the P-loop, which stabilizes the terminal phosphate moiety of the substrate, whereas acidic residues from the Walker B motif (also named motif II or DEXD box) coordinate the Mg2+ needed for hydrolysis42. Structural and biochemical studies revealed that the RTPase and NTPase activities of helicase enzymes from flaviviruses (ss(+)RNA) have a common catalytic site28,42,43,44,45,46,47. Similar biochemical studies have mapped an associated helicase–RTPase activity in other viral families, including alphaviruses (ss(+)RNA)48. However, structural data are needed to better understand how these multifunctional viral proteins coordinate their various activities.
Metal-independent RTPases are found in plants, metazoa and viruses such as baculoviruses (dsDNA)49,50. They belong to the cysteine phosphatases superfamily. The RTPase reaction proceeds in two steps, starting with entry of the 5′-triphosphate RNA into the active-site tunnel and formation of a covalent cysteinyl-S-phosphoester adduct. The catalytic cysteine responsible for the nucleophilic attack on the γ-phosphate of the RNA 5′ end belongs to a P-loop motif (HCXXXXXR(T/S)). The second step releases inorganic phosphate. Cysteine phosphatases adopt a characteristic α–β fold with a central twisted, five-stranded parallel β-sheet flanked by six α-helices49. The catalytic cysteine that specifically recognizes the γ-phosphate of the RNA 5′ end resides at the bottom of the substrate-binding pocket, whereas other conserved residues from the P-loop, together with surrounding residues, form a positively charged channel that can accommodate the α-phosphate and the β-phosphate of the RNA 5′ end. The shape of the binding pocket dictates the selectivity for triphosphate RNA and seems too deep to grant diphosphate RNA access to the active site.
Guanylyltransferases. GTases of DNA viruses contain two domains: a nucleotidyltransferase (NTase) domain that is conserved in capping enzymes, RNA ligases and DNA ligases51, and a C-terminal oligonucleotide-binding domain that is observed in capping enzymes and several DNA ligases. Sequence alignments aided by structural information for several family members identified conserved residues and motifs both in the nucleotide-binding site and in the NTase site51,52. A lysine-containing motif, KXDG(I/L), is conserved among the GTases encoded by several DNA viruses (vaccinia virus, Shope fibroma virus and African swine fever virus) and the yeasts S. cerevisiae and Schizosaccharomyces pombe. The lysine in this motif was shown to be the catalytic residue in the GTase of vaccinia virus53,54,55 and in the yeast capping enzyme56. Moreover, this motif is conserved in the active site of polynucleotide ligases, which, like capping enzymes, catalyse an enzymatic reaction via the formation of a covalent Lys–NMP intermediate57.
The structure of the GTases from Paramecium bursaria chlorella virus 1 (dsDNA), humans58 and the yeast Candida albicans (the protein Cgt1) have been solved individually and, for the chlorella virus GTase and Ctg1, in complexes with cap analogues, GTP or as a covalent GTase–GMP intermediate59,60,61. These structures suggest the following reaction scheme, during which the oligonucleotide-binding and NTase domains, acting as a clamp, undergo a series of large conformational changes. These domains open up to load GTP, close to catalyse the formation of a covalent enzyme–GMP complex through the action of the catalytic lysine, open again to release pyrophosphate and bind the pre-mRNA substrate, close again to catalyse nucleotidyl transfer to the RNA and finally re-open to release the GpppNp-RNA product51,60 (Fig. 4b).
The dsRNA viruses of the family Reoviridae use one multifunctional capsid protein for the capping reaction. λ2 of the dsRNA virus mammalian orthoreovirus62, VP4 of bluetongue virus (in the genus Orbivirus63) and VP3 from cytoplasmic polyhedrosis virus (in the genus Cypovirus64) act as assembly lines, such that the RNA substrate is shuttled from one domain to the next (see below). The GTase domains of these proteins feature different folds (see below and Fig. 4c) to DNA virus GTases. Despite these structural differences, viruses from the genera Orthoreovirus and Aquareovirus use a conserved lysine residue (although not part of the signature sequence found in DNA viruses) to form a covalent intermediate with GMP65,66. In flaviviruses (ss(+)RNA), the 2′OMTase domain has been proposed to act as a GTase67,68. However, the proposed catalytic lysine is not conserved in flaviviruses69.
Methyltransferases. Despite the limited overall amino acid sequence identity in the large family of AdoMet-dependent MTases, most of these enzymes share a common structural core made of seven β-strands flanked by three α-helices on each side of the sheet, similar to the core found in the catechol-O-MTase (a class I AdoMet-dependent MTase70,71). This core catalytic domain has evolved extensions that consist of structurally non-conserved domains and allow these MTases to accommodate a range of methyl acceptors70. The MTases exist either as isolated proteins (for example, nsp16 viruses from the Coronaviridae; ss(+)RNA72), or as domains of larger proteins (such as NS5 of viruses from the ss(+)RNA virus genus Flavivirus69,73 or the multidomain cap assembly lines of dsRNA viruses from the Reoviridae62). In some instances the same protein domain can have dual N7MTase and 2′OMTase activities (for example, the MTase domain in NS5 from flaviviruses74,75), sharing the same cofactor-binding site. When this is the case, repositioning of the RNA substrate must occur.
Both N7MTases and 2′OMTases share the class I family fold (Fig. 4d). N7-methyl transfer is thought to be promoted by optimal positioning of the reacting groups, mediated by several aromatic residues, and by an electrostatic environment that is favourable for the reaction25,76. By contrast, 2′OMTases rely on a conserved catalytic tetrad, Lys-Asp-Lys-Glu69,77,78. The catalytic reaction was deciphered by several structural studies of the 2′OMTase VP39 of vaccinia virus79,80. It was suggested that residues in the vicinity of the catalytic Lys175 (the second lysine in the Lys-Asp-Lys-Glu motif) decrease the pKa value of its ε-amino group. This orientates the 2′-hydroxyl group of the ribose for a nucleophilic in-line sn-2 attack on the AdoMet methyl group81.
Using cap analogues, the molecular basis for recognition of GpppNp-RNA versus cap-0-RNA has also been characterized for VP39 (Refs 80, 82). The m7G is stacked between two aromatic residues, and electron delocalization and electrostatically enhanced stacking owing to N7 methylation favours the recognition of the cap over GTP (Fig. 4d).
One interesting aspect of MTase activity relates to its regulation: in three cases, interfacial activation is achieved either by a cofactor protein (such as the vaccinia virus D12 subunit, which enhances the N7MTase activity of D1 through an allosteric mechanism25,26, and the SARS CoV (ss(+)RNA) metalloprotein nsp10, which acts as a cofactor to activate the 2′OMTase nsp16 but not the N7MTase nsp14 (Ref. 83)) or by binding to lipid membranes (as is the case for nsP1 of the ss(+)RNA alphaviruses).
What determines the sequence of the two methylation steps? For flaviviruses74,75 or coronaviruses83,84, which are ss(+)RNA viruses, the order in which methylations are performed is not encoded in the global protein architecture. Rather, variations in kinetics and affinity may dictate the order in which reactions occur83,85. In the case of flaviviruses, RNA secondary structures also seem to be important: whereas the N7MTase activity of the bifunctional NS5 MTase domain requires a long substrate encompassing a specific stem loop RNA structure, christened stem loop A (SLA), the 2′OMTase activity is able to transfer a methyl group to short RNA acceptors75.
Cap assembly lines. Several dsRNA viruses encode structural proteins that are packaged with their genome in the viral particle and are able to perform the four reactions needed to synthesize a cap-1 structure, much like an assembly line (Fig. 4c). The key molecular components of the RNA-capping machinery in members of the dsRNA virus family Reoviridae are RNA-directed RNA polymerase (named VP1 in orbiviruses and rotaviruses, and λ3 in orthoreoviruses) and a multifunctional cap-synthesizing enzyme (named VP4 in orbiviruses, VP3 in rotaviruses and λ2 in orthoreoviruses). Both λ2 and VP4 are composed of four domains that were identified as RTPase, GTase, N7MTase and 2′OMTase, respectively62,63. The spatial arrangement of the different protein domains reflects the time sequence of the enzymatic reactions that are required for mRNA capping following synthesis by the viral polymerase (Fig. 4c). Although it is unclear how and when RTPase activity occurs, a complete pathway has been proposed in which guanylyl transfer occurs near the base of the pentameric 'turret' (formed by λ2 in orthoreoviruses), followed by N7-methylation and 2′-O-methylation of the mRNA62. Therefore, in dsRNA viruses, the sequence of steps in the cap synthesis pathway should remain identical to the sequence in the capping pathway for cellular mRNAs and for DNA viruses such as vaccinia virus, for which the capping machinery is embedded in a multidomain protein complex.
The GTase domain and both MTase domains of bluetongue virus (a dsRNA virus in the genus Orbivirus) were unambiguously mapped on the VP4 structure, but the position of the RTPase domain remains uncertain63. However, it is believed that both the RTPase and GTase activities reside in the same C-terminal domain of VP4, a unique architecture that is reminiscent of, but distinct from, double-domain RTPase–GTase proteins found in metazoans and plants. The enzymatic activity requires Mg2+ (Refs 63, 86, 87). However, VP4 does not adopt a typical metal-dependent RTPase fold. A cysteine residue is found in a deep cavity similar to that harbouring the catalytic motif of the cysteine phosphatase superfamily63. Thus, these assembly line enzymes seem to have incorporated features from various phylogenetic origins.
Unconventional cap synthesis pathways
The first indication that there are deviations from the conventional RNA-capping pathway for viral mRNAs came in the early 1970s, around the time of the discovery of the RNA cap structure. Since then, is has been demonstrated that the ss(−)RNA virus vesicular stomatitis virus (VSV) and ss(+)RNA alphaviruses (from the family Togaviridae) can synthesize a viral RNA cap that is identical to a cellular RNA cap, albeit constructed through a completely different mechanism. Although alphaviruses do not proceed further than synthesizing a cap-0 structure, the fact that divergent biosynthetic pathways converge to the consensus cap structure indicates that the selective pressure to maintain this structure must be high.
The Mononegavirales RNA-capping pathway. Mononegavirales is a viral order of ss(−)RNA viruses with unsegmented genomes, such as VSV and rabies virus (in the family Rhabdoviridae), measles virus (from the family Paramyxoviridae), bornavirus (from the family Bornaviridae), and Ebola viruses and Marburg viruses (from the family Filoviridae). These viruses encode a multifunctional L protein that carries RNA-dependent RNA polymerase (RdRp) and RNA cap synthesis activities. These enzymes have evolved independently from other known eukaryotic cap-synthesizing enzymes, and the L proteins of VSV88,89, spring viraemia of carp virus90, human respiratory syncytial virus91 and Chandipura virus92 transfer GDP rather than GMP to the RNA 5′ end. Part of a domain in the conserved region V of L protein contains the GDP polyribonucleotidyl transferase (PRNTase) activity, and forms a covalent enzyme–pNp–RNA intermediate (Fig. 3a) with the nascent viral RNA. The covalent bond with RNA involves a conserved histidine residue present in an 'HR' motif instead of the lysine residue used by conventional GTases92. The 5′-monophosphorylated viral mRNA start sequence then receives GDP generated from GTP93 by an as-yet-unknown NTPase. The VSV MTase, present in domain VI of L protein78,94, subsequently methylates the core cap structure at the ribose-2′-O position of the first nucleotide, followed by methylation at the guanine-N7 position, generating GpppAm2′-O-RNA and m7GpppAm2′-O-RNA, respectively95,96,97. The capping reaction seems to be dependent on RNA length, indicating a possible spatial rearrangement in L protein98. Although no crystal structure is available yet, the MTase activities share the AdoMet-binding and active site, which comprises a conserved Lys-Asp-Lys-Glu catalytic tetrad, typical of 2′OMTases.
The RNA-capping pathway of the alphavirus-like togaviruses. Alphaviruses (ss(+)RNA viruses, such as Semliki Forest virus, sindbis virus and chikungunya virus) synthesize a cap-0 structure through a non-conventional mechanism (Fig. 3b). The Semliki Forest virus replicase protein nsP1 has N7MTase and GTase activities, the latter still incompletely characterized, and is presumably involved in the capping of viral mRNA after nsP2-mediated cleavage of the β–γ phosphate bond at the 5′ end99,100. Although the GTase activity of nsP1 has not been fully demonstrated, GTP leads to the formation of a covalent enzyme–GMP complex, albeit only in the presence of AdoMet99,101. Accordingly, mutagenesis and in vitro assays have also revealed that the MTase catalyses the transfer of a methyl group from AdoMet to the N7 position of GTP before the formation of the covalent m7GMP–enzyme complex. The covalent link involves a conserved catalytic histidine residue that is required for the GTase reaction but not for MTase activity. Interestingly, brome mosaic virus replicase protein 1A, bamboo mosaic virus nsp102, tobacco mosaic virus P126 (Ref. 103) and hepatitis E virus p110 (Ref. 104) have properties similar to the N7MTase and GTase of alphaviruses, pointing to a common evolutionary origin for these distantly related viruses of plants and animals. Crystal structures are not yet available for any of these enzymes.
Virus-mediated RNA cap snatching
Among the ss(−)RNA viruses, those of the families Arenaviridae, Bunyaviridae and Orthomyxoviridae have a segmented RNA genome and form the order tentatively named Multinegavirales (Fig. 2). These viruses do not have a cap-synthesizing machinery. However, they have evolved to steal caps from host mRNAs in order to prime their own viral replication, in a process known as cap snatching (Refs 105, 106, 107). Cap snatching involves three steps (Fig. 3c). First, the 5′-methylated cap-1 or cap-2 structure of a host mRNA is bound by a specific site in the viral RdRp (or possibly the N protein108). Then, endonucleolytic cleavage of the cellular mRNA occurs several nucleotides downstream from the cap structure. Finally, this short, capped RNA is used as a primer for the synthesis of viral mRNA by the RdRp. The sequence, length and structure of the mRNA 5′ end that comes with the cap varies from one virus to the other. Most sequences are 15–20 nucleotides long106,109,110,111,112, but arenaviruses, nairoviruses and thogotoviruses use shorter primers113,114,115,116. Following endonucleolytic cleavage, the 'decapped' cellular mRNAs (Box 2) are targeted to the degradation machinery, resulting in the downregulation of cellular mRNAs.
Enzymes from the cap-snatching pathway. Cap snatching was found initially in influenza viruses (ss(−)RNA viruses of the family Orthomyxoviridae), which serve as a model system for the other two viral families known to use snatching (ss(−)RNA viruses of the families Bunyaviridae and Arenaviridae), although differences in the proteins involved and the lengths of snatched sequences are expected, as shown for Thogoto virus117. The influenza virus polymerase is made of three subunits: PA, PB1 and PB2. A cap-binding domain was found in the central region of the PB2 subunit118, and an endonuclease domain at the N terminus of the PA subunit119,120. The structures of both domains shed light on the molecular mechanisms leading to cap snatching (see below). The cap-binding domain has a novel fold, although the mode of m7G binding by aromatic stacking is similar to that used in other cap-binding proteins. By contrast, the endonuclease domain of PA has a fold that is characteristic of the two-metal-dependent PD(D/E)XK nuclease superfamily but has the peculiarity of a metal-ligating histidine residue in the active site, conferring Mn2+ specificity121 (Fig. 5).
Figure 5. Unconventional capping machineries. Endonucleases and cap-binding PB2.
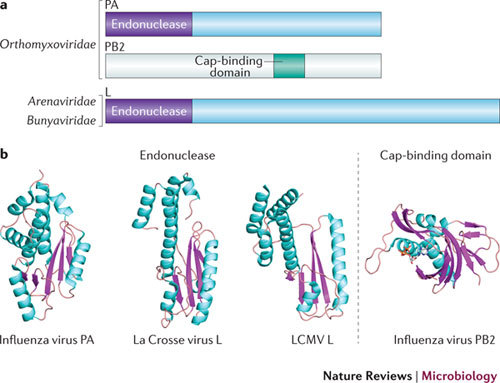
a | Domains involved in the 'cap-snatching' mechanism. The organization of cap-snatching domains of influenza viruses (single-stranded negative-sense RNA (ss(−)RNA) viruses of the family Orthomyxoviridae), and corresponding domains of distantly related viruses of the families Arenaviridae and Bunyaviridae (also ss(−)RNA viruses). Influenza virus polymerase is composed of three proteins of multiple domains: PA, PB1 and PB2. PA and PB2 are involved in cap snatching. PA carries the endonuclease domain in its amino terminus, whereas PB2 has an inner domain responsible for cap binding. Mapping of the domain organization for arenaviruses and bunyaviruses is less advanced; only the endonuclease domain is mapped to the amino terminus of L protein. The cap-binding domain is not clearly identified, as it is thought to be in either L protein or nucleocapsid (N or NP) protein, depending on the virus. b | Structures of the different endonuclease domains of viruses from the families Orthomyxoviridae (influenza viruses), Arenaviridae (lymphocytic choriomeningitis virus (LCMV)) and Bunyaviridae (La Crosse virus) (Protein Data Bank (PDB) accession codes 3HW4, 3JSB and 2XI5, respectively), and of the cap-binding domain from an influenza virus (PDB accession code 2VQZ). Despite having no sequence similarities, the folds of these endonuclease domains are conserved, suggesting a convergent evolution. Structures are coloured cyan for α-helices and pink for β-strands.
In contrast to orthomyxoviruses, both arenaviruses and bunyaviruses have a single protein (L) carrying the polymerase and cap-snatching activities. Recent studies showed that a Mn2+-dependent endonuclease that is homologous to that of orthomyxoviruses exists at the N terminus of arenaviral and bunyaviral L protein122,123(Fig. 5a). Mutational analysis and cell-based replicon assays demonstrated that viral nuclease activities are essential for cap-dependent transcription of viral mRNA124. These domains have a conserved architecture and mechanism, which suggests an evolutionary link between them despite their low sequence identity (Fig. 5b).
A preliminary electron microscopy study of L protein from Machupo virus (an arenavirus) showed a central ring similar to the RdRp of dsRNA viruses but decorated with additional, non-conserved accessory domains. The authors speculate that these extra domains are involved in cap snatching, and that arenaviral and bunyaviral L proteins contain independently folded and functional domains, as does the influenza virus RdRp125,126.
The recent nucleoprotein (NP) structure and enzymatic assay of Lassa virus (ss(−)RNA; Arenaviridae) revealed a second nuclease108, which is probably involved in damping the interferon response127, and a dTTP-binding site, which is proposed to be a cap-binding site involved in cap snatching. The idea that the cap-binding site for cap snatching may be in the nucleoprotein rather than the L protein had already been suggested for hantaviruses (ss(−)RNA; Bunyaviridae)128. In this model, L and N (as the NP protein is called in hantaviruses) cooperate in the process. Meanwhile, the structure of N from Rift Valley fever virus (also a bunyavirus)129 does not present any of these features. These data call for caution before extending the assignation of such a function to related nucleoproteins.
Innate immunity and RNA capping
Mammalian cells have co-evolved with viruses and have developed several mechanisms to detect a viral infection (such as detecting uncapped or partially capped RNA, VPg–RNA, and so on) and induce an antiviral response in neighbouring cells130,131,132. This innate immunity is based on a small number of receptors called pattern recognition receptors (PRRs), which discriminate self from non-self components. Non-self detection depends on the recognition of a limited set of pathogen-associated molecular patterns (PAMPs), which are molecules or components that are characteristic of infectious agents, such as viral nucleic acids133. The presence of non-self nucleic acids is detected through sensors such as Toll-like receptors (TLRs), which recognize DNA or RNA in intracellular compartments that do not usually contain these molecules. Moreover, several PRRs sense the presence of foreign nucleic acids directly in the cytoplasm — namely, the NOD-like receptors (NLRs) and the retinoic acid-inducible gene (RIG)-like receptors (RLRs)134. As host cell RNA is present in the cytosol, PRRs sense uncommon RNA structures that are present in infected cells, such as dsRNA, RNA presenting a 5′-triphosphate, RNA with an incompletely methylated cap structure (cap-0, for mammalian standards) or RNA bearing a protein covalently attached to the 5′ end (such as VPg). The detection of PAMPs by PRRs triggers intracellular signalling events that mainly induce the production of type I interferon (IFN), interleukin-1 (IL-1) and pro-inflammatory cytokines, as well as the establishment of a cellular antiviral state in order to limit viral propagation135,136 (Fig. 6).
Figure 6. Sensing of viral RNA by the innate immune system.
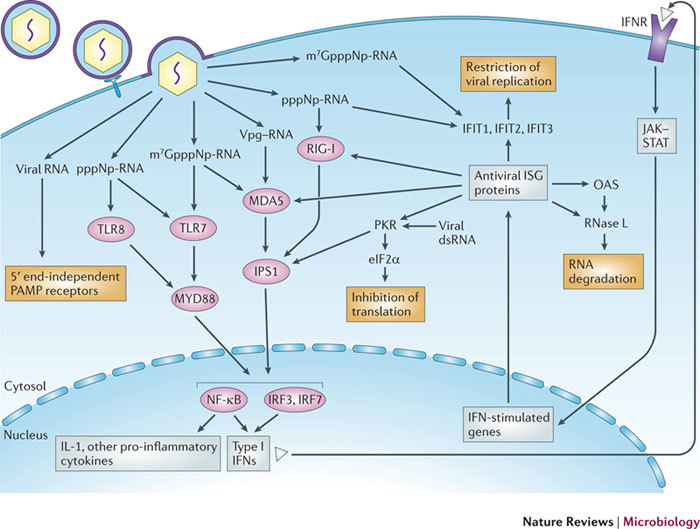
A virus that infects a cell releases viral RNA into the host cytoplasm or endosomes in one of four forms: RNA protected by a 5′ cap-1 structure (7-methyl-Gppp-2′-O-methyl-NP-RNA, depicted here as m7GpppNmp-RNA, in which N is the first transcribed nucleotide and p is a phosphate group), 5′-triphosphate RNA (pppNp-RNA), RNA linked to VPg or RNA carrying a 5′ cap-0 structure (m7GpppNp-RNA). Cap-1 RNA is recognized by pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs), retinoic acid-inducible gene (RIG)-like receptors (RLRs) and NOD-like receptors (NLRs), which in this case recognize, for example, double-stranded RNA (dsRNA). In absence of a cap structure, the 5′-triphosphate of RNA is sensed by the RLR RIG-I, and the VPg–RNA and cap-0 RNA are recognized by another RLR, MDA5. Both RIG-I and MDA5 recruit the mitochondrial-anchored protein interferon-β (IFNβ) promoter stimulator 1 (IPS1), which in turn recruits appropriate inhibitor of NF-κB kinase (IKK) proteins to activate nuclear factor-κB (NF-κB) and IFN regulatory factors (IRFs). This results in the induction of type I IFNs and the production of pro-inflammatory cytokines such as interleukin-1 (IL-1). The endosomal receptor TLR7 can recognize cap-0 RNA and 5′-triphosphate RNA, whereas TLR8 recognizes only 5′-triphosphate RNA. TLR7 recruits the adaptor protein MYD88 through a Toll–IL-1 receptor (TIR)–TIR domain interaction, leading to the activation of NF-κB and IRF3 or IRF7; this results in the induction of type I IFNs and the production of IL-1 and other pro-inflammatory cytokines. Antiviral restriction factors are also stimulated by type I IFN receptor (IFNR) and are known to inhibit the replication of RNA viruses carrying non-capped genomes. Autocrine and paracrine IFN binds IFNR and initiates the JAK–STAT (Janus kinase–signal transducer and activator of transcription) signalling cascade. Among hundreds of proteins encoded by IFN-stimulated genes (ISGs), some antiviral proteins specifically target uncapped viral RNA. The IFIT (IFN-induced protein with tetratricopeptide repeats) proteins specifically sequester 5′-triphosphate RNA (IFIT1) or cap-0 RNA (IFIT2). Protein kinase, RNA activated (PKR) recognizes 5′-triphosphate RNA through an amino-terminal dsRNA-binding domain composed of two binding motifs. Activated PKR phosphorylates eukaryotic translation initiation factor 2α (eIF2α) through its kinase domain and blocks protein translation. RNase L is stimulated by 2′,5′oligo(A) oligonucleotides synthesized by oligoadenylyl synthases (OASs) and is also involved in the degradation of capped and non-capped viral RNA.
Among the PRRs, the TLR family is the best studied innate immunity sensor family. TLRs are transmembrane proteins that are mainly expressed in immune cells, such as macrophages and dendritic cells. They are localized in endosomal compartments or at the cell surface. TLRs contain a leucine-rich repeat motif that recognizes PAMPs, and a Toll–IL-1 receptor (TIR) domain is present in the cytoplasmic part of the protein, ensuring signal transduction through TIR domain interaction with the TIR domains of cytoplasmic adaptor proteins such as MYD88 and TRIF (also known as TICAM1). TLR3, TLR7 and TLR8 are activated by different kinds of RNAs. TLR3 detects dsRNA, whereas TLR7 and TLR8 recognize ssRNA carrying a 5′-triphosphate or a cap-0 structure137.
In contrast to TLRs, the NLRs and RLRs are localized in the cell cytoplasm132 and detect the presence of intracellular invaders. Among the RLRs, RIG-I (also known as DDX58) and MDA5 (also known as IFIH1) seem to discriminate non-self RNA from self RNA on the basis of the RNA 5′ end134. Metazoan self RNA presents a 5′ cap-1 or cap-2 structure. RIG-I protein is specialized in the detection of 5′-triphosphate RNA, whereas MDA5 senses the presence of RNA with a cap-0 structure or linked to a protein such as VPg138. RIG-I and MDA5 consist of two N-terminal caspase-recruitment domains (CARDs), a central DEXH box-containing RNA helicase–ATPase domain and a C-terminal regulatory domain (CTD)139. It is likely that, in the absence of ligands, the CARDs of RIG-I are auto-inhibited by other domains of the protein. For RIG-like proteins, nucleic acid binding to the RNA-binding site of the CTD induces a conformational change resulting in interaction of the CARD with the signalling adaptor molecule IFNβ promoter stimulator 1 (IPS1; also known as CARDIF, MAVS or VISA). IPS1 recruits a signalling complex in order to activate transcription factors such as interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB), leading to the expression of IFNβ and other proteins that drive the antiviral response (Fig. 6).
The molecular basis for RNA recognition with and without 5′ end modification and/or overhanging nucleotides was analysed for RIG-I and MDA5. Initial studies indicated that RIG-I specifically recognizes 5′-triphosphate-containing ssRNA140. It was later found that RIG-I requires base-paired structures in conjunction with a 5′-triphosphate to trigger an antiviral response141. The molecular basis of the specific interaction between the RIG-I CTD and 5′-triphosphate dsRNA was deciphered by a crystallographical study of the CTD in complex with RNA142. This study revealed that 5′-triphosphate dsRNA binds to the CTD of RIG-I more tightly than its single-stranded counterpart. The 5′-triphosphate is sequestered in a lysine-rich cleft of the CTD, with a phenylalanine residue stacked to the terminal base pair. Interestingly, 5′-triphosphate dsRNA methylated at the 2′-O position of its first or second nucleotide is expected to create a steric conflict with the CTD of RIG-I. Accordingly, it does not stimulate the RIG-I pathway.
In contrast to RIG-I, MDA5 is thought to recognize either a viral RNA 5′ end carrying structures that are distinct from a 5′-triphosphate or longer, structured (mesh) RNA that would be generated during the viral life cycle. Indeed, MDA5 was reported to sense both dsRNA and ssRNA bearing a 5′ cap-0 (Refs 138, 143) or linked to VPg. Structural analysis of the MDA5 CTD indicated that its global fold is similar to that of the RIG-I CTD144, with amino acid differences in the domains involved in the recognition of the RNA 5′ end.
The involvement of MDA5 and TLR7 in the detection of cap-0-containing RNA was recently characterized in the antiviral response observed for a coronavirus mutant lacking 2′OMTase activity138. The replication of this mutant virus was dramatically impaired in infected mice. However, the replication of this virus was restored in MDA5-, TLR7- or type I IFN receptor (IFNR)-deficient mice. It has also been suggested that other interferon-stimulated genes (ISGs) restrict the replication of 2′OMTase-deficient viruses145. Accordingly, IFIT1 (IFN-induced protein with tetratricopeptide repeats 1; also known as IFI56) and IFIT2 (also known as IFI54) were reported to limit the replication of West Nile virus (an ss(+)RNA virus of the genus Flavivirus), vaccinia virus (a dsDNA virus of the family Poxviridae) and murine hepatitis virus (an ss(+)RNA virus of the family Coronaviridae) lacking 2′OMTase activity145. IFIT1 was recently shown to bind and sequester 5′-triphosphate RNA into a multiprotein complex containing IFIT2 and IFIT3 (also known as IFI60) in order to exert its antiviral effect146. Therefore, it is likely that 2′-O-methylation of the RNA cap promotes escape from the host innate antiviral response through avoidance of IFIT-mediated suppression.
Conclusions
Since the discovery of 'blocked and methylated' mRNA ends nearly 40 years ago, viruses have played an essential part in deciphering the process of mRNA capping, as well as its relationship with various cellular processes such as transcription, translation and innate immunity. Viral RNA capping is a field that still has a lot of uncharted territory: whether the RNA 5′ ends are protected or not is still unknown for many neglected viral families, and the GTase resists identification even for some studied human pathogens (for example, the ss(+)RNA viruses of the order Nidovirales).
It is likely that, during their co-evolution with their hosts, viruses evolved different adaptation strategies to protect their RNA transcripts. The diversity of mechanisms expressed in nature to add a cap to an RNA 5′ end is larger than that described here. Future research will also aim to elucidate how the fine-tuning between host-mediated decapping of viral RNA, virus-mediated capping of viral RNA and host innate immunity is performed. Moreover, it has been shown that viral antigenomes are not capped147, an observation that has now been extended to many different viruses. Thus, 'no capping' signals probably exist. In addition, the abundance of template RNAs must certainly need to be finely regulated for optimal viral replication. The corresponding signals and regulations are largely unknown.
Finally, owing to its spectacular mechanistic diversity, RNA capping is an attractive field for the design of antiviral drugs. Several molecules have been proposed to act directly or indirectly on viral RNA capping. Ribavirin is a broad-spectrum antiviral agent that is active against several viruses that add or snatch RNA caps, and its pleiotropic mechanism includes targeting the RNA-capping machinery148,149. So far, efforts to design MTase inhibitors have used the AdoMet and AdoHcy (S-adenosyl-L-homocysteine) backbone to synthesize analogues that are specific to viral enzymes150. The increasing knowledge about active-site differences between cellular and viral MTases is expected to provide antiviral selectivity. Last, inhibitors of cap-snatching endonuclease have long been known119,151,152,153. Recently published crystal structures119,122,123 of their targets should inform antiviral-drug design projects.
Box 1 | Getting around the lack of capping.
Some viruses (such as single-stranded positive-sense RNA (ss(+)RNA) viruses from the families Picornaviridae, Caliciviridae and Astroviridae) do not have a cap structure at the 5′ end of their mRNAs or genomic RNAs. Rather, they covalently attach to the 5′ RNA end a protein termed VPg21 and/or carry an internal ribosome entry site (IRES) structure in the 5′ untranslated region154. IRESs have now been found in many different cellular and viral RNAs, including those of ss(+)RNA viruses from the families Picornaviridae and Dicistroviridae, the genus Lentivirus and the Flaviviridae genera Hepacivirus and Pestivirus. In pestiviruses, genomic RNA remains in a 5′-triphosphate form and thus promotes high levels of expression of host interferon-stimulated genes. However, these viruses also trigger several pathways that limit the antiviral response155,156, resulting in a competitive advantage for IRES-dependent translation of viral genes157.
Box 2 | The RNA-decapping pathway of viruses.
Viruses cap and decap RNA, and many viruses regulate the decapping pathway in order to control the ratio of viral and cellular mRNAs.
Decapping of cellular mRNA by Saccharomyces cerevisiae L-A virus (from the Totiviridae family of double-stranded RNA viruses) proceeds through a decapping enzyme carried by the Gag subunit of the capsid; this Gag subunit is responsible for covalently binding cap structures (7-methyl-GpppG (m7GpppG), in which p is a phosphate group) of cellular mRNA158. The decapping activity of Gag aids in the expression of viral RNA, apparently by producing large amounts of cellular RNA decoys that inhibit the S. cerevisiae enzyme 5′–3′ exoribonuclease 1 (Xrn1)159 and compete with degradation of viral RNA. How the viral mRNA is recruited by the eukaryotic translation initiation factor 4E (eIF4E) complex remains to be elucidated. In the case of the family Poxviridae (double-stranded DNA viruses), the decapping enzyme (D10) increases the turnover of host mRNAs and contributes to the shutdown of host protein expression160. Moreover, D10 seems to preferentially degrade m7GpppGm2′-O rather than m7GpppAm2′-O and thereby hydrolyses early-phase viral RNA carrying predominantly m7GpppGm2′-O cap structures161. In other words, this viral pathway benefits from having mRNAs (produced by the capping apparatus) that will be recruited by the eIF4E complex, and simultaneously removes the potential competition from cellular mRNAs for ribosome binding. Viral 'cap-snatching' (see main text) also results in this imbalance, favouring expression of viral genes. Finally, several viruses have been reported to interfere with the cellular RNA-trafficking and decoy machinery. First, a single-stranded positive-sense RNA (ss(+)RNA) enterovirus was shown to inhibit the ability of cells to form stress granules by cleaving RAS·GAP-binding protein (G3BP) family members. The kinetics of poliovirus-induced processing (P)-body disruption correlates with production of viral proteases that induce the degradation of P-body proteins, such as DCP1A and the 3′-deadenylase complex component PAN3. Recently, the ss(+)RNA viruses hepatitis C virus and HIV were also reported to be connected to the cellular decapping machinery and to regulate it (reviewed in Ref. 162).
Acknowledgements
The authors are in debt to all previous and current laboratory members, too numerous to name, for their contributions and involvement in the study of viral RNA replication and capping. Special thanks go to B. Selisko for her tireless dedication to and help with the scientific elaboration of this manuscript. This work was supported in part through funding by the Fondation pour la Recherche Médicale (Programme Aide aux équipes), the French Direction Générale de l'Armement (contrat 07co404), Infectiopôle-Sud and the European Union Seventh Framework Programme (FP7/2007–2013) through the project SILVER (Small inhibitor leads against emerging RNA viruses; grant agreement 260644).
Glossary
- Pre-mRNA splicing
A post-transcriptional modification of pre-mRNA, in which introns are excised and exons are joined in order to form a translationally functional, mature mRNA.
- γ-phosphate
The third phosphate attached at the 5′ end of the ribose moiety of a nucleotide.
- 'Ping-pong' mechanism
A two-step mechanism in which a substrate molecule first forms a (covalent) link with the enzyme and is then transferred to an acceptor molecule to yield a product.
- Poly(A) tail
A string of AMP that is added to the 3′ end of mRNA.
- NUDIX hydrolase superfamily
A family of proteins that hydrolyse a wide range of organic pyrophosphates, including NDPs, NTPs, dinucleoside and diphosphoinositol polyphosphates, nucleotide sugars and RNA caps, with varying degrees of substrate specificity.
- Single-stranded positive-sense RNA viruses
(ss(+)RNA viruses). Viruses that have or produce mRNAs that are co-linear to their genomic RNA.
- 43S pre-initiation complex
A multiprotein complex composed of eukaryotic translation initiation factor 3 (eIF3), eIF4A, eIF4E and eIF4G associated with the small ribosomal subunit. This pre-initiation complex scans the mRNA towards the 'start' codon (typically AUG), where translation is initiated.
- Single-stranded negative-sense RNA viruses
(ss(−)RNA viruses). Viruses that have or produce mRNAs that are complementary to their genomic RNA.
- Ambisense RNA viruses
Viruses (such as members of the families Arenaviridae and Bunyaviridae) that have or produce both mRNAs that are co-linear to and mRNAs that are complementary to their genomic RNA, although most mRNAs are complementary in polarity.
- β-phosphate
The second phosphate attached at the 5′ end of the ribose moiety of a nucleotide.
- Walker A and B motifs
Motifs that are present in nucleotide-binding proteins but also in a range of proteins with widely varying functions, including ATP synthases, myosins, transducins, helicases, kinases and RecA proteins. The Walker A motif contains a phosphate-binding loop (P-loop) motif with the consensus sequence GXXXGK(T/S), and the Walker B motif contains the consensus sequence (R/K)XXXXGXXXXLhhhhD (in which h refers to any hydrophobic residue).
- Nucleophilic attack
Generally, a starting point for a chemical reaction; a doublet of electrons selectively attacks the positive or partially positive charge of the atomic nucleus in order to create a new chemical bond.
- α-phosphate
The first phosphate attached at the 5′ end of the ribose moiety of a nucleotide.
- pKa value
The acid dissociation constant, a quantitative measurement of the strength of a chemical group as an acid in solution. It corresponds to the pH value at which half of the ionizable group is either protonated or deprotonated.
- ε-amino group
A positively charged group found at the extremity of a lysine side chain. The ε-amino group is a primary amine and, owing to its high pKa value, it is reactive and often participates in reactions at the active site of enzymes.
- Stem loop
A hairpin structure in single-stranded RNA or DNA, resulting from intramolecular base-pairing when two regions of the same strand contain partial or perfect anti-complementary nucleotide sequences.
- Cell-based replicon assays
Assays that allow one to follow the replication of a 'minimal viral genome' encoding the viral replication complex but no structural or envelope proteins, which are usually replaced by reporter genes (such as luciferase or chloramphenicol acetyl transferase genes).
- Viral antigenomes
Viral RNAs that are complementary strands to the genome. The antigenome strand is used as a matrix for the synthesis of new viral genomes, and of viral mRNAs in the case of positive-sense RNA viruses.
Biographies
Etienne Decroly obtained a Ph.D. in biochemistry in 1994 from the Université Libre de Bruxelles at the Laboratoire de Chimie Physique des Macromolécules aux Interfaces (LCPMI), Belgium, under Jean Marie Ruyschaert. This was followed by post-doctoral training at the Institut de Recherches Cliniques de Montréal (IRCM), Canada, under Nabil G. Seidah and at the Institut National de la Santé at de la Research Médical (INSERM) U372, Marseilles, France, under Robert Vigne. He is currently a member of the team Viral Replication: Structure, Mechanisms, and Drug Design at the Architecture and Functions of Biological Macromolecules Laboratory, Aix-Marseille Université, France, where he holds a Centre National de la Reserche Scientifique (CNRS) position as Chargé de Recherche. His current interests are the structure and function of enzymes involved in RNA capping in single-stranded, positive-strand RNA viruses.
François Ferron obtained his Ph.D. in biochemistry and bioinformatics in 2005 from the Université de la Méditerranée, Marseilles, at the CNRS, working under Bruno Canard and Sonia Longhi. He undertook postdoctoral training in structural biology at Boston Biomedical Research Institute, Massachusetts, USA, and became a research fellow at the Physiology Department of the University of Pennsylvania, School of Medicine, Philadelphia, USA, working with Roberto Dominguez. In 2008, he joined the CNRS team Proteins from Emergent Viruses and Parasitology in Marseilles, under Julien Lescar, to work on phleboviral genome packaging. Since 2010, he has held a CNRS researcher position in the team Viral Replication: Structure, Mechanisms, and Drug Design at the Architecture and Functions of Biological Macromolecules Laboratory, Aix-Marseille Université. His current interests are focused on the structural assembly of macromolecular complexes that form viral replication machineries.
Julien Lescar obtained a Ph.D. in 1993 from Université Paris-Sud XI, Orsay, working at the Institut Pasteur, Paris, France, under the supervision of Roberto Poljak and Pedro Alzari. After a postdoctoral stay in the laboratory of Gabriel Waksman at Washington University in St Louis, USA, and again at the Institut Pasteur, under Graham Bentley, he worked as a beamline postdoctoral scientist for 3 years at the European Synchrotron Radiation Facility in Grenoble, France. In 2000, he became a member of the CNRS at the Centre de Recherches sur les Macromolecules Vegetales, as Charge de Recherches. In 2002, he took a sabbatical leave from the CNRS to set up his own group in Singapore at the School of Biological Sciences, Nanyang Technological University. He is now working at the Architecture and Function of Biological Macromolecules laboratory, Aix-Marseille Université. His main research interests are in using structural biology tools — mainly X-ray crystallography — to understand the structure of viruses and viral proteins and to apply this knowledge to assist the design of antiviral compounds.
Bruno Canard obtained a Ph.D. in 1991 in microbiology and biochemistry from the Université Paris VII at the Institut Pasteur, under Stewart T. Cole, and then carried out postdoctoral training in 1995–1998 at the Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, working with Charles C. Richardson. He is currently the principal investigator of the team Viral Replication: Structure, Mechanisms, and Drug Design at the Architecture and Functions of Biological Macromolecules Laboratory, Aix-Marseille Université, where he holds a CNRS position as Directeur de Recherche. His current interests are the structure and function of enzymes involved in viral replication and RNA capping, and their use in the design of antiviral drugs.
Related links
DATABASES
Protein Data Bank
FURTHER INFORMATION
Accession codes
Accessions
GenBank/EMBL/DDBJ
Competing interests
The authors declare no competing financial interests.
References
- 1.Shatkin A. Capping of eucaryotic mRNAs. Cell. 1976;9:645. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE., Jr Transcription units for mRNA production in eukaryotic cells and their DNA viruses. Prog. Nucleic Acid Res. Mol. Biol. 1979;22:327–353. doi: 10.1016/S0079-6603(08)60803-X. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, et al. A protein binding the methylated 5′-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc. Natl Acad. Sci. USA. 1976;73:1559–1563. doi: 10.1073/pnas.73.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Perry RP. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res. 1977;4:4133–4149. doi: 10.1093/nar/4.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Kiledjian M. Decapping the message: a beginning or an end. Biochem. Soc. Trans. 2006;34:35–38. doi: 10.1042/BST0340035. [DOI] [PubMed] [Google Scholar]
- 6.Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv. Vir. Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuichi Y, Muthukrishnan S, Shatkin AJ. 5′-Terminal m-7G(5′)ppp(5′)Gmp in vivo: identification in reovirus genome RNA. Proc. Natl Acad. Sci. USA. 1975;72:742–745. doi: 10.1073/pnas.72.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shatkin AJ. Methylated messenger RNA synthesis in vitro by purified reovirus. Proc. Natl Acad. Sci. USA. 1974;71:3204–3207. doi: 10.1073/pnas.71.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei CM, Moss B. Methylated nucleotides block 5′-terminus of vaccinia virus messenger RNA. Proc. Natl Acad. Sci. USA. 1975;72:318–322. doi: 10.1073/pnas.72.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shatkin AJ, Both GW. Reovirus mRNA: transcription translation. Cell. 1976;7:305–313. doi: 10.1016/0092-8674(76)90159-8. [DOI] [PubMed] [Google Scholar]
- 13.Furuichi Y, Morgan M, Muthukrishnan S, Shatkin AJ. Reovirus messenger RNA contains a methylated, blocked 5′-terminal structure: m7G(5′)ppp(5′)GmpCp- Proc. Natl Acad. Sci. USA. 1975;72:362–366. doi: 10.1073/pnas.72.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 15.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1987;89:951–961. doi: 10.1016/S0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 16.Perry KL, Watkins KP, Agabian N. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl Acad. Sci. USA. 1987;84:8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 18.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell. Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenberg DR, Maquat LE. Re-capping the message. Trends Biochem. Sci. 2009;34:435–442. doi: 10.1016/j.tibs.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow I, et al. Calicivirus translation initiation requires an interaction between VPg and eIF4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambros V, Pettersson RF, Baltimore D. An enzymatic activity in uninfected cells that cleaves the linkage between poliovirion RNA and the 5′ terminal protein. Cell. 1978;15:1439–1446. doi: 10.1016/0092-8674(78)90067-3. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V, Baltimore D. Purification and properties of a HeLa cell enzyme able to remove the 5′-terminal protein from poliovirus RNA. J. Biol. Chem. 1980;255:6739–6744. [PubMed] [Google Scholar]
- 24.Cong P, Shuman S. Mutational analysis of mRNA capping enzyme identifies amino acids involved in GTP binding, enzyme-guanylate formation, and GMP transfer to RNA. Mol. Cell. Biol. 1995;15:6222–6231. doi: 10.1128/MCB.15.11.6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De la Peña M, Kyrieleis OJP, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J. Biol. Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 27.Schnierle BS, Gershon PD, Moss B. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc. Natl Acad. Sci. USA. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benarroch D, et al. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328:208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Myette JR, Niles EG. Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleotide triphosphate phosphohydrolase activities demonstrate that both activities are carried out at the same active site. J. Biol. Chem. 1996;271:11945–11952. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- 30.Vasquez-Del Carpio R, Gonzalez-Nilo FD, Riadi G, Taraporewala ZF, Patton JT. Histidine triad-like motif of the rotavirus NSP2 octamer mediates both RTPase and NTPase activities. J. Mol. Biol. 2006;362:539–554. doi: 10.1016/j.jmb.2006.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayaram H, Taraporewala Z, Patton JT, Prasad BVV. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature. 2002;417:311–315. doi: 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- 32.Taraporewala Z, Chen D, Patton JT. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taraporewala ZF, Patton JT. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 2001;75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benarroch D, Smith P, Shuman S. Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure. 2008;16:501–512. doi: 10.1016/j.str.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Gu M, Lima CD. Processing the message: structural insights into capping and decapping mRNA. Curr. Opin. Struc. Biol. 2005;15:99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Lima CD, Wang LK, Shuman S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/S0092-8674(00)81541-X. [DOI] [PubMed] [Google Scholar]
- 37.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 38.Lehman K, Schwer B, Ho CK, Rouzankina I, Shuman S. A conserved domain of yeast RNA triphosphatase flanking the catalytic core regulates self-association and interaction with the guanylyltransferase component of the mRNA capping apparatus. J. Biol. Chem. 1999;274:22668–22678. doi: 10.1074/jbc.274.32.22668. [DOI] [PubMed] [Google Scholar]
- 39.Gorbalenya AE, Koonin EV. One more conserved sequence motif in helicases. Nucleic Acids Res. 1988;16:7734. doi: 10.1093/nar/16.15.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorbalenya AE, Koonin EV. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo D, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl Acad. Sci. USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lescar J, et al. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Luo D, et al. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol. Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo D, et al. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampath A, et al. Structure-based mutational analysis of the NS3 helicase from dengue virus. J. Virol. 2006;80:6686–6690. doi: 10.1128/JVI.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balistreri G, Caldentey J, Kääriäinen L, Ahola T. Enzymatic defects of the nsP2 proteins of Semliki Forest virus temperature-sensitive mutants. J Virol. 2007;81:2849–2860. doi: 10.1128/JVI.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Changela A, Ho CK, Martins A, Shuman S, Mondragón A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 2001;20:2575–2586. doi: 10.1093/emboj/20.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takagi T, Moore CR, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/S0092-8674(00)80272-X. [DOI] [PubMed] [Google Scholar]
- 51.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc. Natl Acad. Sci. USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cong P, Shuman S. Covalent catalysis in nucleotidyl transfer. A KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J. Biol. Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 54.Niles EG, Christen L. Identification of the vaccinia virus mRNA guanyltransferase active site lysine. J. Biol. Chem. 1993;268:24986–24989. [PubMed] [Google Scholar]
- 55.Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme–guanylate intermediate. Proc. Natl Acad. Sci. USA. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwer B, Shuman S. Mutational analysis of yeast mRNA capping enzyme. Proc. Natl Acad. Sci. USA. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindahl T, Barnes DE. Mammalian DNA ligases. Annu. Rev. Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 58.Chu C, et al. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl Acad. Sci. USA. 2011;108:10104–10108. doi: 10.1073/pnas.1106610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2003;11:1549–1561. doi: 10.1016/S1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 60.Håkansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/S0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 61.Håkansson K, Wigley DB. Structure of a complex between a cap analogue and mRNA guanylyl transferase demonstrates the structural chemistry of RNA capping. Proc. Natl Acad. Sci. USA. 1998;95:1505–1510. doi: 10.1073/pnas.95.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 63.Sutton G, Grimes JM, Stuart DI, Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nature Struct. Mol. Biol. 2007;14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 64.Cheng L, et al. Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc. Natl Acad. Sci. USA. 2011;108:1373–1378. doi: 10.1073/pnas.1014995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J, Tao Y, Reinisch KM, Harrison SC, Nibert ML. Orthoreovirus and Aquareovirus core proteins: conserved enzymatic surfaces, but not protein–protein interfaces. Virus Res. 2004;101:15–28. doi: 10.1016/j.virusres.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 66.Luongo CL, Reinisch KM, Harrison SC, Nibert ML. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein λ2. J. Biol. Chem. 2000;275:2804–2810. doi: 10.1074/jbc.275.4.2804. [DOI] [PubMed] [Google Scholar]
- 67.Issur M, et al. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 69.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/S0959-440X(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 71.Schubert H. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Decroly E, et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egloff MP, et al. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J. Mol. Biol. 2007;372:723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 74.Dong H, Ren S, Li H, Shi PY. Separate molecules of West Nile virus methyltransferase can independently catalyze the N7 and 2′-O methylations of viral RNA cap. Virology. 2008;377:1–6. doi: 10.1016/j.virol.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ray D, et al. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/S1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 77.Bujnicki JM, Rychlewski L. Reassignment of specificities of two cap methyltransferase domains in the reovirus λ2 protein. Genome Biol. 2001;2:RESEARCH0038.1–RESEARCH0038.6. doi: 10.1186/gb-2001-2-9-research0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ferron F, Longhi S, Henrissat B, Canard B. Viral RNA-polymerases – a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 2002;27:222–224. doi: 10.1016/S0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 79.Hodel AE, Gershon PD, Shi X, Quiocho FA. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85:247–256. doi: 10.1016/S0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- 80.Lockless SW, Cheng HT, Hodel AE, Quiocho FA, Gershon PD. Recognition of capped RNA substrates by VP39, the vaccinia virus-encoded mRNA cap-specific 2′-O-methyltransferase. Biochemistry. 1998;37:8564–8574. doi: 10.1021/bi980178m. [DOI] [PubMed] [Google Scholar]
- 81.Li C, Xia Y, Gao X, Gershon PD. Mechanism of RNA 2′-O-methylation: evidence that the catalytic lysine acts to steer rather than deprotonate the target nucleophile. Biochemistry. 2004;43:5680–5687. doi: 10.1021/bi0359980. [DOI] [PubMed] [Google Scholar]
- 82.Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol. Cell. 1998;1:443–447. doi: 10.1016/S1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 83.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Decroly E, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramadevi N, Burroughs NJ, Mertens PP, Jones IM, Roy P. Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc. Natl Acad. Sci. USA. 1998;95:13537–13542. doi: 10.1073/pnas.95.23.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ramadevi N, Roy P. Bluetongue virus core protein VP4 has nucleoside triphosphate phosphohydrolase activity. J. Gen. Virol. 1998;79:2475–2480. doi: 10.1099/0022-1317-79-10-2475. [DOI] [PubMed] [Google Scholar]
- 88.Abraham G, Rhodes DP, Banerjee AK. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 89.Abraham G, Rhodes DP, Banerjee AK. Novel initiation of RNA synthesis in vitro by vesicular stomatitis virus. Nature. 1975;255:37–40. doi: 10.1038/255037a0. [DOI] [PubMed] [Google Scholar]
- 90.Gupta KC, Roy P. Alternate capping mechanisms for transcription of spring viremia of carp virus: evidence for independent mRNA initiation. J. Virol. 1980;33:292–303. doi: 10.1128/jvi.33.1.292-303.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barik S. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 1993;74:485–490. doi: 10.1099/0022-1317-74-3-485. [DOI] [PubMed] [Google Scholar]
- 92.Ogino T, Banerjee AK. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen. Virol. 2010;91:1311–1314. doi: 10.1099/vir.0.019307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 94.Bujnicki JM, Rychlewski L. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng. 2002;15:101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- 95.Li J, Fontaine-Rodriguez EC, Whelan SP. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahmeh AA, Li J, Kranzusch PJ, Whelan SPJ. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Testa D, Banerjee AK. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 1977;24:786–793. doi: 10.1128/jvi.24.3.786-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tekes G, Rahmeh AA, Whelan SP. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog. 2011;7:e1002073. doi: 10.1371/journal.ppat.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ahola T, Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl Acad. Sci. USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vasiljeva L, Merits A, Auvinen P, Kääriäinen L. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 2000;275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- 101.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li YI, Chen YJ, Hsu YH, Meng M. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of bamboo mosaic virus replicase. J. Virol. 2001;75:782–788. doi: 10.1128/JVI.75.2.782-788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Merits A, et al. Virus-specific capping of tobacco mosaic virus RNA: methylation of GTP prior to formation of covalent complex p126-m7GMP. FEBS Lett. 1999;455:45–48. doi: 10.1016/S0014-5793(99)00856-X. [DOI] [PubMed] [Google Scholar]
- 104.Magden J, et al. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 2001;75:6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bouloy M, Plotch SJ, Krug RM. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc. Natl Acad. Sci. USA. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caton AJ, Robertson S. Structure of the host-derived sequences present at the 5′ ends of influenza virus mRNA. Nucleic Acids Res. 1980;8:2591–2603. doi: 10.1093/nar/8.12.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Plotch SJ, Bouloy M, Krug RM. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl Acad. Sci. USA. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bishop DHMY, Gay ME, Matsuoko Y. Non Viral heterogeneous sequences are present at the 5′ ends of one species of snowshoe hare bunyavirus S. complementary RNA. Nucleic Acids Res. 1983;11:6409–6418. doi: 10.1093/nar/11.18.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bouloy M, Pardigon N, Vialat P, Gerbaud S, Girard M. Characterization of the 5′ and 3′ ends of viral messenger RNAs isolated from BHK21 cells infected with Germiston virus (Bunyavirus) Virology. 1990;175:50–58. doi: 10.1016/0042-6822(90)90185-T. [DOI] [PubMed] [Google Scholar]
- 111.Eshita Y, Ericson B, Romanowski V, Bishop DH. Analyses of the mRNA transcription processes of snowshoe hare bunyavirus S and M RNA species. J. Virol. 1985;55:681–689. doi: 10.1128/jvi.55.3.681-689.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patterson JL, Holloway B, Kolakofsky D. La Crosse virions contain a primer-stimulated RNA polymerase and a methylated cap-dependent endonuclease. J. Virol. 1984;52:215–222. doi: 10.1128/jvi.52.1.215-222.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garcin D, et al. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jin H, Elliott RM. Non-viral sequences at the 5′ ends of Dugbe nairovirus S mRNAs. J. Gen. Virol. 1993;74:2293–2297. doi: 10.1099/0022-1317-74-10-2293. [DOI] [PubMed] [Google Scholar]
- 115.Raju R, et al. Nontemplated bases at the 5′ends of Tacaribe virus mRNAs. Virology. 1990;174:53–59. doi: 10.1016/0042-6822(90)90053-T. [DOI] [PubMed] [Google Scholar]
- 116.Weber F, Haller O, Kochs G. Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel” cap-stealing” mechanism in tick-borne orthomyxoviruses? J. Virol. 1996;70:8361–8367. doi: 10.1128/jvi.70.12.8361-8367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leahy MB, Dessens JT, Nuttall PA. In vitro polymerase activity of Thogoto virus: evidence for a unique cap-snatching mechanism in a tick-borne orthomyxovirus. J. Virol. 1997;71:8347–8351. doi: 10.1128/jvi.71.11.8347-8351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guilligay D, et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature Struct. Mol. Biol. 2008;15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 119.Dias A, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 120.Yuan P, et al. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature. 2009;458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 121.Crépin T, et al. Mutational and metal binding analysis of the endonuclease domain of the influenza virus polymerase PA subunit. J. Virol. 2010;84:9096–9104. doi: 10.1128/JVI.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010;6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morin B, et al. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010;6:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lelke M, Brunotte L, Busch C, Günther S. An N-terminal region of Lassa virus L protein plays a critical role in transcription but not replication of the virus genome. J. Virol. 2010;84:1934–1944. doi: 10.1128/JVI.01657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kranzusch PJ, et al. Assembly of a functional Machupo virus polymerase complex. Proc. Natl Acad. Sci. USA. 2010;107:20069–20074. doi: 10.1073/pnas.1007152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ruigrok RWH, Crépin T, Hart DJ, Cusack S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 2010;20:104–113. doi: 10.1016/j.sbi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 127.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl Acad. Sci. USA. 2011;108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl Acad. Sci. USA. 2008;105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ferron F, et al. The hexamer structure of the rift valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7:e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koyama S, Ishii KJ, Coban C, Akira S. Innate immune response to viral infection. Cytokine. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 131.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol. Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 132.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 134.Brennan K, Bowie AG. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010;13:503–507. doi: 10.1016/j.mib.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 135.Hansen JD, Vojtech LN, Laing KJ. Sensing disease and danger: a survey of vertebrate PRRs and their origins. Dev. Comp. Immunol. 2011;35:886–897. doi: 10.1016/j.dci.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 136.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 137.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 138.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nature Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-β-inducing potential. Eur. J. Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 140.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 141.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nature Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luthra P, Sun D, Silverman RH, He B. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc. Natl Acad. Sci. USA. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li X, et al. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch. Biochem. Biophys. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 145.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nature Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 147.Garcin D, Kolakofsky D. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J. Virol. 1990;64:6196–6203. doi: 10.1128/jvi.64.12.6196-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hong Z, Cameron CE. Pleiotropic mechanisms of ribavirin antiviral activities. Prog. Drug Res. 2002;59:41–69. doi: 10.1007/978-3-0348-8171-5_2. [DOI] [PubMed] [Google Scholar]
- 149.Magden J, Kääriäinen L, Ahola T. Inhibitors of virus replication: recent developments and prospects. Appl. Microbiol. Biotechnol. 2005;66:612–621. doi: 10.1007/s00253-004-1783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lim SP, et al. Small molecule inhibitors that selectively block dengue virus methyltransferase. J. Biol. Chem. 2011;286:6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kuzuhara T, Iwai Y, Takahashi H, Hatakeyama D, Echigo N. Green tea catechins inhibit the endonuclease activity of influenza A virus RNA polymerase. PLoS Curr. 2009;1:RRN1052. doi: 10.1371/currents.RRN1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Parkes KEB, et al. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem. 2003;46:1153–1164. doi: 10.1021/jm020334u. [DOI] [PubMed] [Google Scholar]
- 153.Tomassini J, et al. Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother. 1994;38:2827–2837. doi: 10.1128/AAC.38.12.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 155.Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- 156.Malmgaard L. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- 157.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Blanc A, Goyer C, Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol. Cell. Biol. 1992;12:3390–3398. doi: 10.1128/MCB.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nature Struct. Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 160.Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc. Natl Acad. Sci. USA. 2007;104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McLennan AG. Decapitation: poxvirus makes RNA lose its head. Trends Biochem. Sci. 2007;32:297–299. doi: 10.1016/j.tibs.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 162.Gaglia MM, Glaunsinger BA. Viruses and the cellular RNA decay machinery. Wiley Interdiscip. Rev. RNA. 2010;1:47–59. doi: 10.1002/wrna.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


