ABSTRACT
How temporal cues combine with spatial inputs to control gene expression during development is poorly understood. Here, we test the hypothesis that the Drosophila transcription factor E93 controls temporal gene expression by regulating chromatin accessibility. Precocious expression of E93 early in wing development reveals that it can simultaneously activate and deactivate different target enhancers. Notably, the precocious patterns of enhancer activity resemble the wild-type patterns that occur later in development, suggesting that expression of E93 alters the competence of enhancers to respond to spatial cues. Genomic profiling reveals that precocious E93 expression is sufficient to regulate chromatin accessibility at a subset of its targets. These accessibility changes mimic those that normally occur later in development, indicating that precocious E93 accelerates the wild-type developmental program. Further, we find that target enhancers that do not respond to precocious E93 in early wings become responsive after a developmental transition, suggesting that parallel temporal pathways work alongside E93. These findings support a model wherein E93 expression functions as an instructive cue that defines a broad window of developmental time through control of chromatin accessibility.
KEY WORDS: Temporal transcription factor, Chromatin accessibility, Enhancer, Gene regulation, Ecdysone, Drosophila
Summary: The temporal transcription factor E93 controls both activation and deactivation of different target enhancers in the same cells to respond to spatial cues through regulation of chromatin accessibility.
INTRODUCTION
Cis-regulatory regions such as enhancers and promoters interpret multiple types of inputs to control gene expression during development. These inputs come in the form of both spatial and temporal cues. Spatial cues are often provided by transcription factors, which can contribute information on cell identity (e.g. MyoD), organ identity (e.g. Pha-4) and regional identity (e.g. Hox factors). Additional spatial cues are provided by the activity of signaling pathways such as the Wnt, BMP and EGFR families, which contribute information on distance relative to the source of the signal through their downstream transcriptional effectors. Remarkably, many of these spatial cues are used reiteratively over the course of development, often with different effects on target gene expression. For example, the Hox factor Ubx controls different sets of target genes at different times in Drosophila appendage development, as does the intestine-specifying factor CDX2 during gut development in mouse and humans (Kumar et al., 2019; Pavlopoulos and Akam, 2011). Similarly, EGFR signaling promotes wing vein formation early in Drosophila larval development, whereas later in pupal stages, EGFR represses vein formation and instead promotes differentiation of the complementary intervein cells (Martín-Blanco et al., 1999). Thus, spatial inputs alone are insufficient to account for the sequence of gene expression and cell state changes that occur during development.
Temporal cues provide an additional axis of information that can increase the range of gene expression responses to spatial inputs. Some temporal cues come in the form of post-transcriptional regulators, such as lin-4, lin-28 and let-7 in C. elegans, which control transitions between developmental stages through regulation of RNA stability and translation efficiency (Pasquinelli and Ruvkun, 2002). Other temporal cues come in the form of developmentally restricted expression of transcription factors. For example, in mammals and in Drosophila, the diversity of cell types found in the adult nervous system depends on a temporal cascade of transcription factor expression in neural progenitor cells (Doe, 2017; Holguera and Desplan, 2018). Yet another means of temporal gene regulation involves systemically secreted signals that coordinate the timing of gene expression programs between distant tissues. Ecdysone signaling in insects and thyroid hormone-dependent metamorphosis in amphibians are classic examples of systemic signals that trigger temporal-specific gene expression changes during development.
Although it is clear that both spatial and temporal inputs are necessary for proper gene regulation during development, the mechanisms by which these inputs combine to control target enhancer activity are poorly understood. One potential mechanism for control of the responsiveness of enhancers to transcriptional inputs is regulation of chromatin accessibility. In vivo, the genome is packaged into chromatin. DNA that is wrapped around histone proteins to form nucleosomes is less accessible to transcription factor binding relative to free DNA. For many transcription factors to access their target sequences, nucleosomes must be depleted or remodeled. In principle, the relative accessibility of an enhancer could determine whether it is competent to respond to transcription factor input and thereby help to explain how transcription factors can be reutilized during development with different transcriptional outcomes.
Recently, support for the role of chromatin accessibility in the integration of spatial and temporal factor inputs has emerged from examination of tissues at two different stages of development in Drosophila: neural diversification in the embryo and specification of appendage cell fates in the pupa (Sen et al., 2019; Uyehara et al., 2017). In the embryo, distinct neural stem cell lineages are determined by differential expression of spatial transcription factors. Within a given lineage, neural stem cells utilize sequential expression of temporal transcription factors to produce progeny with distinct identities over time. Importantly, different neural lineages use the same set of temporal transcription factors to specify progeny identities. Using a lineage-specific method of generating genome-wide DNA binding profiles, the temporal transcription factor Hunchback was found to bind different target sites in different neural lineages. Moreover, these target sites correspond to lineage-specific patterns of open chromatin (Sen et al., 2019). These findings indicate that the temporal factor Hunchback does not control open chromatin profiles and does not determine where it binds in the genome. Instead, they suggest that the spatial transcription factors expressed in neuroblasts control open chromatin profiles to drive lineage-specific binding of temporal transcription factors.
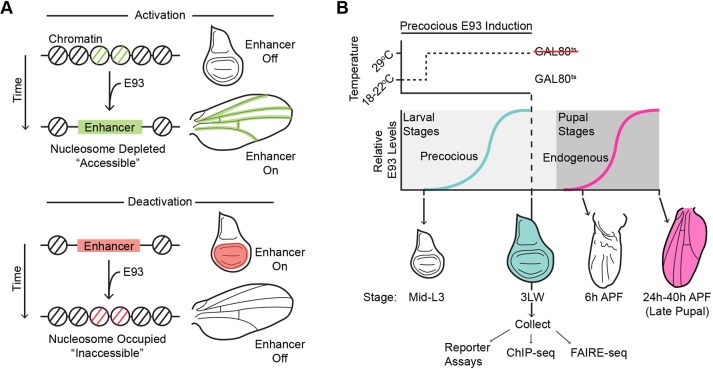
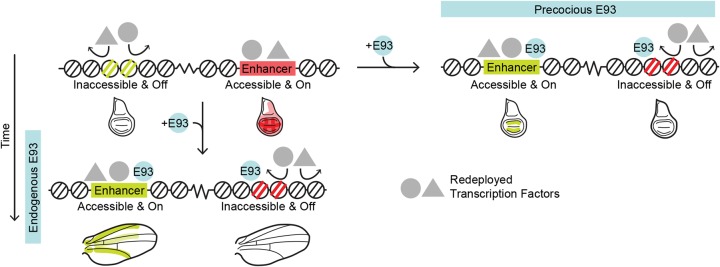
The ecdysone-induced transcription factor E93 (also known as Eip93F) provides a contrasting example of temporal transcription factor function. Similar to Hunchback's role in the embryonic nervous system, E93 functions as a temporal identity factor. E93 is activated during the transition from prepupal to pupal stages of metamorphosis, and E93 loss-of-function mutations exhibit defects in cell fates that are specified during this time (Baehrecke and Thummel, 1995; Mou et al., 2012). Also similar to Hunchback, E93 combines with spatial cues to pattern cell fates. During specification of the pigmented bract cells during pupal leg development, E93 expression makes the Distal-less gene competent to respond to EGFR signaling (Mou et al., 2012). However, in contrast to Hunchback, recent work from pupal wings suggests an essential role for E93 in regulating chromatin accessibility (Uyehara et al., 2017). During metamorphosis, the wing undergoes dramatic morphological, cell fate and gene expression changes to form the notum (back), hinge and wing blade of the adult. Gene expression and chromatin accessibility profiling of larval wing imaginal discs and pupal wings 24 h and 44 h after puparium formation (APF) revealed that these changes coincide with sequential changes in open chromatin genome-wide (Guo et al., 2016; Uyehara et al., 2017). These chromatin accessibility changes strongly correlate with enhancer activity. Sites that open with time correspond to late-acting enhancers switching on, and sites that close with time correspond to early acting enhancers switching off. E93 binds many temporally dynamic open chromatin sites in pupal wings. Moreover, chromatin accessibility profiling of E93 mutants determined that E93 is required for temporal changes in chromatin accessibility and enhancer activity. In the absence of E93, early acting target enhancers fail to close and fail to turn off. Conversely, late-acting target enhancers fail to open and fail to turn on (Fig. 1A). These findings support a model in which E93 functions as a temporal identity factor by acting as a gatekeeper to the genome. In this model, E93 makes late-acting enhancers competent to respond to spatial inputs by increasing their accessibility, whereas it makes early acting enhancers refractory to spatial inputs by decreasing their accessibility, thus allowing for reutilization of spatial inputs.
Fig. 1.
Schematics of E93-mediated enhancer regulation and experimental design. (A) Illustrations of larval wing imaginal discs and late pupal wings showing that E93-dependent changes in chromatin accessibility correlate with temporal changes in the activity of two target enhancers (expression patterns indicated in green and red). (B) Schematic of E93 induction and relative timing of precocious E93 expression (teal) versus endogenous E93 expression (magenta). GAL4 drivers in combination with GAL80ts were used to initiate precocious E93 expression in mid-third instar larvae (L3) for subsequent dissection in wandering third instar larvae (3LW).
In this study, we sought to address two major issues regarding E93-dependent control of enhancer activity. First, E93 appears to simultaneously coordinate the opening and activation of certain target enhancers, whereas it closes and deactivates others; however, the determinants of this context-specific activity are unknown. Second, we sought to determine the sufficiency of E93 expression to regulate target enhancers. Although E93 is required for sequential changes in chromatin accessibility, it is not known whether E93 simply maintains accessibility changes initiated by other factors, or whether it initiates these changes itself.
We took advantage of the temporal sequence of Drosophila wing development to investigate the limits of E93 function by expressing it at an earlier stage of wing development, before the endogenous E93 expression window. We found that precocious E93 expression alters the activity and accessibility of target enhancers, and that it can simultaneously trigger the activation and deactivation of different enhancers in the same cells. Genome-wide profiling demonstrated that these findings are generalizable, and that precocious E93 expression accelerates the wild-type chromatin accessibility program. Finally, we found that not all E93 target enhancers respond to premature E93 expression in wing imaginal discs, even after prolonged exposure. However, these target enhancers become responsive to premature E93 later in prepupal wings, suggesting the requirement of additional temporal inputs that are independent of E93. Together, this work supports a model in which E93 expression defines a broad temporal window, providing competence of genes to respond to inductive signals by regulating chromatin accessibility at target enhancers.
RESULTS
To help define the limits of E93's abilities to regulate enhancer activity and chromatin accessibility we expressed E93 outside of its normal developmental context. In wild-type animals E93 expression is temporally regulated. E93 is not expressed early in wing development, including third instar larvae. It is not until later, during pupal stages, that ecdysone signaling induces E93, with transcript levels peaking by 24 h after the larval-to-pupal transition (24 h APF) (Fig. 1B; Uyehara et al., 2017). Using tissue-specific GAL4 drivers in combination with a UAS-E93 transgene and a ubiquitously expressed temperature-sensitive GAL4 repressor (GAL80ts), we induced E93 in the wing imaginal disc of third instar wandering larvae (3rd Larval Wandering; 3LW), before the time that E93 is normally expressed (Fig. 1B, Fig. S1; Bischof et al., 2013). By switching between the permissive (22°C) and restrictive temperatures (29°C) for GAL80ts, we limited the duration of exogenous E93 expression to 15-24 h at the end of larval development. We refer to this as ‘precocious’ or ‘premature’ E93 expression. Immunofluorescence experiments with E93 antibodies indicated that precocious E93 levels in 3LW wing imaginal discs are ∼twofold greater than endogenous E93 levels in pupal wings (Fig. S1). Thus, this experimental design allows us to determine the impact of near-physiological levels of E93 on enhancer activity and chromatin accessibility.
Precocious E93 expression is sufficient to deactivate a target enhancer
We first examined whether premature expression of E93 is capable of regulating target enhancer activity using a previously identified E93-bound enhancer from the broad (br) locus, which encodes a zinc-finger transcription factor. In wild-type larvae, the brdisc enhancer is active throughout the wing imaginal disc epithelium, with stronger activity in the pouch (Fig. 2A). Later in pupal wings (24-40 h APF), brdisc has turned off (Fig. 2A). In E93 mutant wings the brdisc enhancer fails to turn off (Uyehara et al., 2017). To test the impact of precocious E93 expression on brdisc activity, we expressed E93 in the anterior half of the wing imaginal disc with ci-GAL4. We found that brdisc activity is strongly reduced in E93-expressing cells relative to the wild-type posterior half of 3LW discs (Fig. 2B). Control wing discs from larvae subjected to identical experimental conditions, but lacking the UAS-E93 transgene, showed no change in brdisc activity (Fig. 2B). Similarly, discs from larvae of the experimental genotype, but not subjected to a 29°C shift, showed no induction of the E93 transgene or change in enhancer activity (Fig. S2). We reasoned that E93-dependent repression of brdisc could result either from E93 blocking the initial activation of brdisc, or from E93 turning off brdisc after its initial activation. To discriminate between these possibilities, we assessed enhancer activity shortly after E93 induction. After only 5 h at 29°C we observed induction of E93 in the anterior half of the disc, but no change in brdisc activity in E93-expressing cells relative to E93-nonexpressing cells (Fig. 2B). Thus, precocious E93 triggers brdisc enhancer deactivation instead of simply blocking its activation. En-GAL4-driven E93 expression in the posterior wing compartment resulted in similar brdisc deactivation (Fig. S2). Together, these findings demonstrate that E93 is capable of deactivating a target enhancer even when expressed at an earlier developmental stage.
Fig. 2.
Precocious E93 provides an instructive cue to deactivate and activate specific enhancers. (A,C,E,G) Schematic of spatiotemporal enhancer activity alongside immunofluorescence examples. (A) brdisc is active in wing discs and inactive in pupal wings. (B) Confocal image depicting brdisc activity (green) in control genotypes lacking the UAS-E93 transgene (left panels). brdisc remains active (green) after a 5 h E93 (magenta) induction (middle panels). brdisc activity (green) is lost in E93-expressing cells (magenta) after a 24 h induction (right panels). Green arrows indicate comparable locations in the anterior wing after 5 h and 24 h of E93 expression. (C) tncblade (cyan) is inactive in wing discs but is active in pupal wings in the body wall (arrows) marked by Tsh (yellow), and in intervein cells of the blade. (D) tncblade is inactive in control genotypes lacking the UAS-E93 transgene (left panels). ci-GAL4 pattern is indicated in yellow (GFP). tncblade is active (green) in E93-expressing cells (left of red dashed line) also expressing Tsh (magenta) (right panels). Green arrows indicate colocalization of Tsh expression and precocious activation of tncblade. (E) tncwv (cyan) is inactive in wing discs but is active in pupal wings along longitudinal veins marked by pMad (yellow). Cyan arrows indicate high level tncwv activity in the marginal and L5 veins. Yellow arrows indicate high pMad levels in the marginal and L5 veins. (F) tncwv is inactive in control genotypes lacking the UAS-E93 transgene (left panels). ci-GAL4 pattern is indicated in yellow (GFP). tncwv is active (green) in E93-expressing cells (left of dashed red line) with high pMad levels (magenta) (right panels). Green arrows indicate colocalization of high pMad signal and precocious activation of tncwv. (G) nubvein (green) is inactive in wing discs, but is active in pupal wings along the longitudinal veins. E93 expression is shown in magenta. (H) nubvein (green) is inactive in wing discs regardless of E93 expression (magenta). Dashed white lines outline wing tissue. Red dashed lines indicate the boundary of ci-GAL4 activity, as determined by GFP expression. Scale bars: 100 µm.
Precocious E93 expression is sufficient to activate target enhancers
In addition to brdisc, we examined two E93-bound enhancers from the tenectin (tnc) locus that depend on E93 for proper activation in pupal wings (Uyehara et al., 2017). Tnc encodes a constituent of the extracellular matrix that binds integrins (Fraichard et al., 2010). In wild-type flies, the tncblade enhancer is inactive in larval wing imaginal discs. Later in pupal wings, tncblade is active in the tissue between the developing longitudinal wing veins (Fig. 2C). It is also active at high levels in the body wall adjacent to the wing hinge. Tncblade activity co-localizes with expression of the proximal patterning factor Teashirt (Tsh) in these cells (Fig. 2C). Precocious expression of E93 with ci-GAL4 resulted in premature activation of tncblade in 3LW wing discs in a cluster of cells perpendicular to the anterior-posterior (AP) axis outside the pouch (Fig. 2D). Staining for Tsh and Wingless (Wg) revealed that cells with premature tncblade activity are located in the proximal hinge and neighboring notum (Fig. 2D, Fig. S2; Zirin and Mann, 2007). Similar activation of tncblade occurred in the posterior wing compartment upon precocious E93 expression using en-GAL4 (Fig. S2). Control wing discs lacking the UAS-E93 transgene showed no change in tncblade activity despite being shifted to 29°C for 24 h (Fig. 2D). Discs with the UAS-E93 transgene, but not shifted to 29°C, likewise showed no change in enhancer activity (Fig. S2). Thus, premature E93 expression leads to activation of the tncblade enhancer in the presumptive proximal hinge and notum of wing imaginal discs. Notably, the pattern of precocious tncblade activation resembles its wild-type pattern in pupal wings, indicating that the spatial inputs controlling tncblade in its normal developmental context are similar to those that control the enhancer when E93 is prematurely expressed.
We observed similar outcomes with a second enhancer from the tnc locus, the tncwv enhancer. In wild-type pupal wings, tncwv is active in 10-20 cells surrounding the developing veins, as indicated by phosphorylated Mothers Against Decapentaplegic (pMad) staining, a marker of active Dpp signaling (Fig. 2E; de Celis, 1997). Like tncblade, tncwv is also dependent on E93 for activation (Uyehara et al., 2017). Precocious expression of E93 with ci-GAL4 resulted in premature activation of tncwv in E93-expressing cells flanking the dorsal-ventral (DV) boundary, adjacent to the AP boundary in the pouch of 3LW wing imaginal discs (Fig. 2F). Notably, the pattern of precociously activated tncwv showed a high degree of overlap with strong pMad signal in these discs, similar to the overlap of tncwv with pMad in wild-type pupal wings (Fig. 2F). Using en-GAL4 to drive precocious E93 expression in the posterior compartment resulted in a similar pattern of precocious tncwv activity and similar overlap with active Dpp signaling (Fig. S2). Control larvae subjected to the same 29°C shift, but without the UAS-E93 transgene, showed no change in enhancer activity; neither did larvae with the transgene that were kept at 22°C (Fig. 2F, Fig. S2). Thus, similar to our observations with the tncblade enhancer, these findings demonstrate that precocious E93 expression prematurely switches on late-acting pupal wing enhancers. Moreover, the pattern of premature enhancer activity appears to be guided by similar spatial signals as the wild-type pattern of activity observed later in development, consistent with the proposed role of E93 as a temporal competence factor.
Precocious E93 expression is not sufficient for activation of the nubvein enhancer
Our observations with the br and tnc enhancers suggest that premature expression of E93 accelerates the developmental program active in pupal wings. However, not all E93 target enhancers are sensitive to precocious E93 expression. We previously identified the E93-bound nubvein enhancer, which is normally inactive in early wings and becomes active in 24 h pupal wings in an E93-dependent manner (Fig. 2G; Uyehara et al., 2017). However, in contrast to the tnc enhancers, precocious expression of E93 with en-GAL4 does not activate nubvein in wing imaginal discs (Fig. 2H). We observed no difference in reporter activity in E93-expressing cells relative to their wild-type counterparts in the anterior compartment. Thus, for a subset of target enhancers, E93 expression can support activation outside of their normal developmental context. However, other target enhancers require regulatory inputs in addition to E93 for premature activation.
Precocious E93 binds late targets genome wide
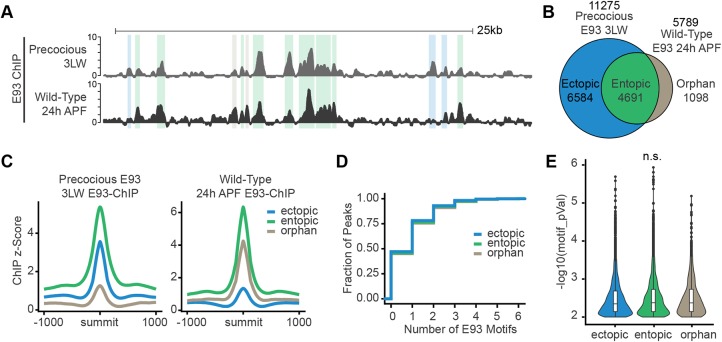
To expand our understanding of the ability of E93 to regulate target enhancers outside of its normal developmental context, we next performed a series of genome-wide profiling experiments in which E93 was prematurely expressed throughout the 3LW wing imaginal disc (Fig. S1). We first performed ChIP-seq to define the DNA binding profiles of precocious E93. Comparison of ChIP-seq profiles for precocious E93 in early wings and endogenous E93 in 24 h pupal wings (late E93 targets) revealed three distinct binding site categories: precocious E93 binding sites that overlap late E93 targets (entopic sites), precocious E93 binding sites that do not overlap late E93 targets (ectopic sites), and late E93 binding sites that do not overlap precocious E93 targets (orphan sites) (Fig. 3A). Of endogenous late targets, 81% are bound by precocious E93, suggesting that the ability of E93 to recognize and bind most of its target sites is not dependent on a late-wing developmental context (Fig. 3B). Notably, the brdisc, tncblade and tncwv enhancers are all bound by precocious E93, consistent with their responsiveness in reporter assays (Fig. 4D,E). By contrast, the nubvein enhancer exhibits low-level binding of precocious E93, indicating that its failure to respond may be due to the inability of E93 to bind nubvein in early wings (Fig. 5E).
Fig. 3.
Precocious E93 recapitulates wild-type late binding profiles. (A) Browser shot of ChIP-seq data for wild-type and precocious E93 wings. Colored highlights correspond to ectopic (blue), entopic (green) and orphan (brown) sites. (B) Venn diagram of peak overlaps between wild-type and precocious E93 ChIP-seq datasets. (C) Average signal plots of ChIP-seq z-score within each binding category for wild-type and precocious E93 ChIP. (D) Cumulative distribution of the number of E93 motifs within 20 bp of the summit for each binding category. (E) Violin plots depicting motif quality (–log10 P-value) for each E93 motif within 20 bp of E93 ChIP peak summits for each binding category. Box plots denote median and interquartile range (IQR), whiskers denote the smallest and largest values within 1.5xIQR for each peak set. Points denote outliers beyond whiskers. n.s., not significant. P>0.05, one-way analysis of means, not assuming equal variance. 3LW, wandering third instar larvae.
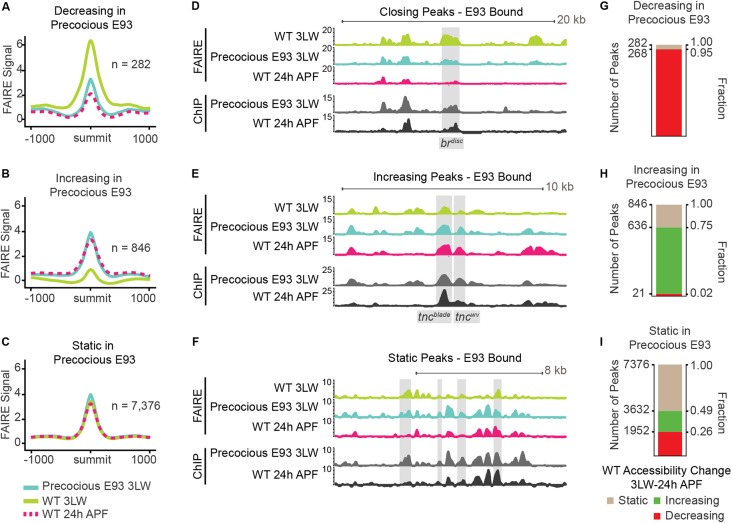
Fig. 4.
Precocious E93 accelerates the wild-type chromatin accessibility program. (A-C) Average FAIRE-seq signal (z-score) from wild-type wandering third instar larvae (3LW) wings (green), wild-type 24 h APF wings (dashed red) and precocious E93 3LW wings (teal) at precocious E93 binding sites that decrease accessibility (A), increase accessibility (B) or remain static (C) in response to precocious E93 expression. (D,E) Browser shots of FAIRE-seq and ChIP-seq signal (z-scores) at the brdisc enhancer (D) and the tncwv and tncblade enhancers (E). (F) Browser shot of static sites bound by precocious E93. (G-I) Stacked bar charts indicating the changes in chromatin accessibility that occur in wild-type development for each of the three E93 binding site categories.
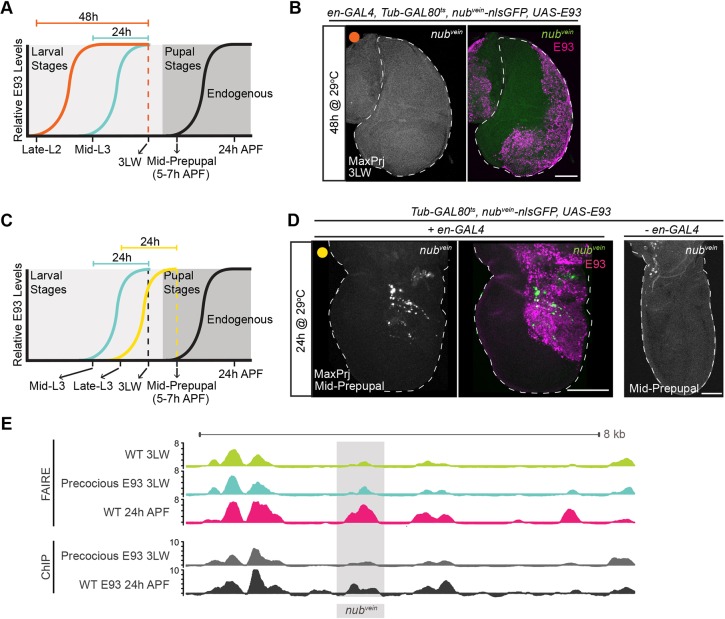
Fig. 5.
nubvein acquires competence to respond to precocious E93 by mid-prepupal stage. (A) Schematic depicting timing of prolonged E93 induction (orange), prior precocious E93-induction (teal) and endogenous E93 expression (black). Dashed vertical line indicates time of dissection. (B) Confocal image of nubvein activity (green) in wandering third instar larvae (3LW) wing disc precociously expressing E93 (magenta) for 48 h. Orange circle indicates the timing of precocious E93 induction from panel A. (C) Schematic depicting timing of E93 induction for mid-prepupal wings (yellow), prior precocious E93-induction (teal) and wild-type E93 expression (black). (D) Confocal images of nubvein activity (green) in mid-prepupal wings precociously expressing E93 (magenta) for 24 h (left panels). Control wing lacking en-GAL4 is shown on the right. Yellow circle indicates the timing of precocious E93 induction from panel C. (E) Browser shot showing FAIRE-seq and ChIP-seq signal (z-scores) at the nubvein enhancer (shaded region). Dashed white lines indicate outline of wing tissue. Scale bars: 100 µm.
The presence of ectopic and orphan binding sites suggests E93 binds to these sites in a context-specific manner. We first compared the levels of E93 ChIP-seq signal between these peak categories. Precocious E93 exhibits increased signal at entopic sites relative to ectopic sites in 3LW wing imaginal discs (Fig. 3C). In addition, endogenous E93 exhibits decreased signal at orphan sites relative to entopic sites in 24 h pupal wings. We therefore sought to define the features that may explain these differences in E93 binding. We observed no difference in the distribution of E93 peaks between binding site categories, demonstrating that localization to specific genomic regions is not a feature that discriminates orphan and ectopic binding sites from entopic sites (Fig. S3A). By contrast, examination of chromatin accessibility at these sites during wild-type development revealed differences between peak categories. Orphan sites are less accessible in early wings but increase in accessibility in late wings (Fig. S3B,C), suggesting that low accessibility may prevent E93 binding in 3LW wing discs. However, many ectopic sites also exhibit low accessibility in 3LW wing discs. Thus, chromatin accessibility may explain some, but not all of the observed differences in E93 binding.
We next sought to determine whether the different E93 binding categories reflect differences in DNA sequence. First, we examined the E93 motif itself and detected no differences in E93 motif enrichment, quality or positioning in ectopic and orphan sites relative to entopic sites (Fig. 3D,E, Fig. S4). Consistent with these findings, position-weight matrices (PWMs) derived from E93 motifs within each binding category were nearly identical to each other (Pearson's R>0.98) (Fig. S4B,C). We conclude that the E93 motif is not the primary determinant of the observed differences in binding. We also performed both de novo and directed motif analyses to determine whether other DNA sequence motifs distinguish ectopic, entopic and orphan categories (Fig. S5). We observed very few differences in motif content between E93 peak categories. Orphan peaks exhibit modest enrichment for homeodomain factor motifs, ectopic peaks are weakly enriched for motifs for the paralogous transcription factors Nub and Pdm2, and entopic peaks exhibit weak enrichment for the zinc-finger factors Crol and Pad. Although the functional significance of these motifs is unclear, the overall assessment is that DNA sequences within each E93 binding site category are highly similar. Only a small amount of differential E93 binding can be explained by the presence of particular transcription factor motifs. Other reasons for the existence of ectopic and orphan peaks include the use of different antibodies in the precocious E93 experiments relative to the endogenous E93 ChIP-seq performed in 24 h pupal wings, and the higher levels of E93 expression in the precocious experimental system.
Precocious E93 expression is sufficient to regulate chromatin accessibility
The ChIP-seq data described above demonstrated that a large majority of targets bound by endogenous E93 in late wings are also bound by precocious E93 in early wings. We next sought to determine the impact of precocious E93 binding on chromatin accessibility. Our previous findings from E93 mutant wings suggested that E93 may function as a competence factor by controlling chromatin accessibility at target enhancers. To further test this model, we performed FAIRE-seq to generate genome-wide open chromatin profiles in wing imaginal discs precociously expressing E93. Comparison of these profiles with wild-type wing imaginal disc FAIRE-seq profiles revealed extensive changes in open chromatin. Using conservative thresholds to define differentially accessible sites bound by E93 (DESeq2 adjusted P value <0.05 and log2 fold change ≥1), we identified 282 sites that decrease in accessibility, 846 sites that increase in accessibility and 7376 sites that remain static in response to premature E93 expression (Fig. 4A-C). Notably, the ratio of sites that open relative to those that close in precocious E93 early wings is similar to the ratio of sites that depend on E93 for opening and closing in wild-type late wings previously identified in E93 loss-of-function experiments (Uyehara et al., 2017). This indicates that the ability of precociously expressed E93 to open chromatin relative to its ability to close chromatin is similar to the abilities of endogenous E93 to regulate chromatin accessibility, despite being expressed outside of its normal developmental context.
To determine the impact of the observed changes in chromatin accessibility induced by premature E93 expression on transcriptional regulation, we examined FAIRE-seq profiles at E93 target enhancers described above. Accessibility of the brdisc enhancer strongly decreases in precocious E93 wing discs (Fig. 4D), consistent with its deactivation in transgenic reporter assays. The brdisc enhancer normally closes between L3 and 24 h pupae, raising the question as to whether any of the other 281 sites that decrease in accessibility in response to premature E93 expression also close over time in wild-type wings. Remarkably, 95% of sites that decrease in accessibility in precocious E93 wing discs also decrease in accessibility during wild-type development (Fig. 4G), suggesting that premature E93 expression recapitulates the normal sequence of enhancer closing. Examination of FAIRE-seq profiles at the tnc enhancers revealed changes in chromatin accessibility that were also consistent with the effects of precocious E93 expression on transgenic reporter activity. Tncwv, and to a lesser extent tncblade, increase in accessibility in response to precocious E93, consistent with the activation of both enhancers in transgenic reporter assays (Fig. 4E). At the genome-wide level, 73% of the sites that increase in response to precocious E93 expression also increase in accessibility during wild-type development (Fig. 4H). Thus, the directionality of chromatin accessibility changes in wings prematurely expressing E93 is preserved relative to the sequential changes in accessibility that normally occur in wild-type wings. This indicates that E93 expression functions as an instructive cue that triggers a response in enhancer accessibility. However, the directionality of this response is not determined by E93.
DNA sequence partially explains differential effects on chromatin accessibility
Although E93 expression is sufficient to change chromatin accessibility at a subset of its target sites (hereafter, E93 ‘sensitive’ sites), E93 does not determine whether target sites increase or decrease in accessibility. The concordance in accessibility changes between precocious E93 and wild-type development suggests the accessibility determinant is either pre-existing on the chromatin or is encoded in the DNA sequence of target enhancers. To test for pre-existing regulatory information, we examined histone post-translational modifications (PTMs) from wild-type 3LW wings (Schertel et al., 2015). We found that specific histone PTMs do not closely correlate with chromatin accessibility changes at E93 target sites (Fig. S6). E93-sensitive sites that decrease in accessibility (‘decreasing E93-sensitive’ sites) exhibit modestly higher average levels of histone PTMs correlated with active transcription (Fig. S6A), such as H3K4me1 and H3K27ac, consistent with these sites being open and active regulatory elements in 3LW wings. However, many decreasing E93-sensitive sites do not exhibit high levels of these PTMs (Fig. S6), indicating that active histone PTMs are not required for E93 to close chromatin. Conversely, high levels of active histone PTMs are found at many E93 binding sites that do not change in accessibility in precocious E93 wings (E93 ‘insensitive’ sites), indicating that the presence of active histone PTMs does not necessitate closing of chromatin upon E93 binding. Sites that increase in accessibility upon E93 binding (‘increasing E93-sensitive’ sites) likewise do not exhibit a clear correlation with histone PTMs. Most increasing E93-sensitive sites lack enrichment of histone PTMs, indicating they are not pre-marked for activation at this stage. However, the absence of histone PTMs does not necessitate opening of chromatin upon E93 binding because many decreasing E93-sensitive and insensitive sites also lack histone PTM enrichment. We conclude that histone PTMs are not the main determinants controlling the differential effects of E93 on chromatin accessibility. Instead, histone PTMs likely reflect the regulatory state of the DNA.
We next examined the DNA sequence of E93-sensitive sites relative to E93-insensitive sites by de novo motif discovery. Decreasing E93-sensitive sites are enriched >twofold for E93 binding site motifs relative to E93-insensitive sites (Fig. S7). Targeted E93 motif analysis revealed that decreasing sites exhibit both greater motif quality and a greater number of E93 motifs relative to increasing E93-sensitive sites or E93-insensitive sites (Fig. S8). We do not detect enrichment of the E93 motif within increasing E93-sensitive sites relative to E93-insensitive sites, likely because of equal enrichment of the E93 motif observed between increasing and static sites (Fig. S8A). Instead, increasing E93-sensitive sites are enriched for motifs matching the zinc-finger transcription factors Br-Z2 (1.4-fold enriched) and Crol (1.3-fold enriched) (Fig. S9). Both br and crol are ecdysone target genes with essential roles in wing development (D'Avino and Thummel, 2000; Schubiger et al., 2005). Br expression is high in larval wings, when these sites exhibit low accessibility, and it decreases during the larval-to-pupal transition, when these sites increase in accessibility in wild-type wings (Guo et al., 2016). Thus, over-representation of Br motifs in increasing E93-sensitive sites suggests a role for Br in keeping pupal regulatory element chromatin inaccessible in larval wings. Crol is expressed at similar levels in both 3LW and pupal wings, and the enrichment of Crol motifs in both entopic E93 sites and increasing E93-sensitive sites suggests Crol may work with E93 to bind DNA (Figs S5, S9, S10; Uyehara et al., 2017). Supporting a potential combinatorial role for E93 and Crol in pupal gene regulation, E93 and crol mutants exhibit similar wing defects, including loss of adhesion and abnormal venation (D'Avino and Thummel, 2000; Mou et al., 2012). Together, these analyses indicate that the differential effects of precocious E93 on chromatin accessibility are at least partially explained by differences in DNA sequence composition of E93 target sites.
Developmental context informs the response of nubvein to precocious E93
Although ∼1100 sites change in accessibility in response to premature E93 expression, the majority of E93-bound sites do not change in accessibility in early wings even though many are dynamic during wild-type development (Fig. 4C,F,I). The nubvein enhancer is representative of this category in that it depends on E93 for opening in pupal wings, but it fails to activate or open in response to precocious E93 expression in larval wing discs (Fig. 2G and Fig. 5E; Uyehara et al., 2017). We considered the possibility that nubvein requires prolonged E93 exposure, relative to E93-sensitive enhancers such as tncblade, in order to become responsive. Prolonged exposure might allow time for E93-initiated events to occur, such as induction of a co-regulator. To test this hypothesis, we doubled the duration of nubvein exposure to E93 (from 24 h to 48 h) by inducing E93 expression earlier in wing development and then dissecting at the same developmental stage as before (3LW) (Fig. 5A). Despite the prolonged exposure to E93, we still observed no change in nubvein reporter activity (Fig. 5B). We next examined the possibility that E93 may require additional developmental inputs in order to activate the nubvein enhancer. To test this hypothesis, we precociously expressed E93 for the same duration as in our initial experiments (24 h), but instead of dissecting at 3LW, we dissected 12 h later (mid-prepupal wings at 5-7 h APF, Fig. 5C). Using this experimental design, we detected clear activation of the nubvein reporter in a subset of E93-expressing cells (Fig. 5D). Thus, the ability of the nubvein enhancer to respond to precocious E93 is dependent on developmental context. It does not respond to E93 in third instar larvae regardless of the duration of E93 expression. However, it does respond to E93 in prepupal wings, suggesting a change in the regulatory environment during the larval-to-prepupal transition makes nubvein competent to respond to E93.
Temporal dynamics of chromatin accessibility indicate context-dependent roles for E93
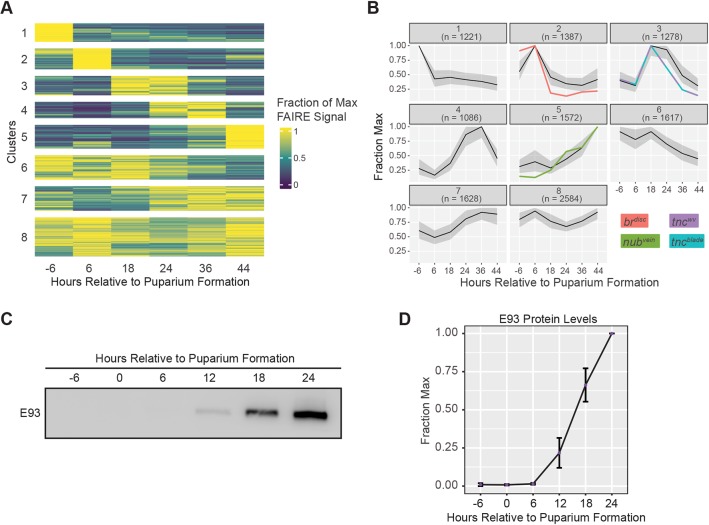
The findings described above indicate that precocious E93 expression controls accessibility and activity of some target sites, but other targets require additional developmentally regulated inputs in order to respond to E93. To gain insight into the extent to which developmental context influences E93 target site responsivity, we examined the timing of chromatin accessibility changes in wild-type wings. Clustering of FAIRE-seq data for E93-bound sites across six time points in wild-type wing development revealed eight distinct temporal chromatin accessibility profiles (Fig. 6A). Notably, the brdisc, nubvein and tnc enhancers fall into different clusters. Brdisc falls into cluster 2 with other E93 targets that close between 6 and 18 h APF (Fig. 6B). The tncblade and tncwv enhancers fall into cluster 3 with other E93 targets that open between 6 and 18 h APF (Fig. 6B). Finally, the nubvein enhancer falls into cluster 5 with other E93 targets that open even later in pupal wing development (Fig. 6B). As each of these enhancers is a bona fide E93 target, their separation into different clusters suggests that E93 regulates target enhancers over a relatively wide range of prepupal and pupal wing development. Supporting this interpretation, western blotting of wild-type wings at 6 h intervals surrounding the larval-to-pupal transition demonstrates that E93 expression overlaps the time points that exhibit changes in chromatin accessibility (Fig. 6C,D). These findings indicate that E93 functions over a broad window of development to control enhancer activity and accessibility, and that this broad window is subdivided into narrower windows through interactions with other developmentally regulated factors.
Fig. 6.
Chromatin accessibility at E93 targets changes over a broad developmental window. (A) Heatmap of chromatin accessibility over time within E93 ChIP peaks represented as fraction of maximum accessibility clustered using k-means (k=8). (B) Line plots depicting FAIRE signal for each cluster in A. Black lines, median fraction of max FAIRE signal; gray area, interquartile range. Accessibility within ChIP-peaks overlapping enhancers is plotted in color indicated by inset labels. (C) Western blot of E93 levels in wild-type wings over time. (D) Quantification of E93 protein levels. Error bars represent s.e.m.
To identify potential co-regulators that subdivide E93 activity during wild-type development, we looked for enrichment of transcription factor motifs in each temporally dynamic cluster (clusters 1-5) relative to all other dynamic clusters. Targeted motif scanning identified motifs with differential enrichment across clusters (Fig. S10). Several of these transcription factors have documented roles in controlling developmental timing. For example, Br motifs are enriched in clusters 3, 4 and 5, which contain E93 targets that open at sequential times after the larval-to-pupal transition. As discussed above, br mRNA levels drop during the larval-to-pupal transition, supporting a potential role for Br in keeping pupal regulatory DNA inaccessible in larval and prepupal wings. Differential motif enrichment for other transcription factors involved in coordinating developmental timing include Ultraspiracle, E74 (Eip74EF) and Abrupt. We also detected differential enrichment of motifs for transcription factors downstream of signaling pathways, including Enhancer of Split, Pointed and Mad, as well as wing patterning factors such as Mirror, Nubbin, Scalloped and Rotund. Finally, we identified strong motif enrichment for Zelda (Zld) in cluster 1, which contains sites that are accessible only in larval wing discs. Zld is a putative pioneer factor required for chromatin accessibility in early Drosophila embryos (Schulz et al., 2015). Intriguingly, Zld is also expressed in the larval wing, and its transcript levels drop eightfold during the larval-to-prepupal transition (Fig. S11; Hamm et al., 2017). The coincident decrease in accessibility of cluster 1 peaks suggests Zld may also have a role in wing development. Thus, a combination of temporal and spatial transcription factors may work with E93 to control accessibility and activity of target enhancers at distinct stages of wing development.
DISCUSSION
Temporal transcription factors as determinants of developmental competence
Spatial cues are iteratively used during development to produce distinct transcriptional outcomes. Many of these spatial inputs come in the form of transcription factors that are expressed at multiple stages of development. However, it is unclear how these factors regulate their given targets only at select times. The findings presented in this study indicate that E93 expression provides competence for target enhancers to respond to spatially restricted inputs. Premature expression of E93 in larval wings switches on the tncblade enhancer in Tsh-expressing cells of the proximal hinge, similar to its wild-type activity pattern in the hinge later in pupal wings. Likewise, larval E93 expression switches on the tncwv enhancer in cells with high pMad levels, similar to its wild-type pattern in pupal wing veins. Notably, neither of these enhancers becomes active in all cells that precociously express E93. Instead, precocious E93 expression activates these enhancers only in populations of cells that appear to receive similar spatial inputs as those in which they normally become active later in development. This suggests that E93 is the limiting factor that enables these enhancers to respond to spatial cues that are used at multiple stages of development. This interpretation is consistent with a previous demonstration that E93 expression makes the Distal-less gene competent to respond to EGFR signaling in the leg (Mou et al., 2012). Importantly, like the spatial cues that regulate the tnc enhancers, the EGFR pathway is active in both early and late legs, and yet EGFR is only capable of activating Distal-less in the presence of E93. Thus, the spatial cues present before E93 expression are insufficient to activate their target enhancers, indicating that E93 is the key determinant for unlocking their activities.
The findings presented here provide new insight into the means by which E93 controls enhancer competence. ChIP-seq demonstrates that E93 binds directly to target enhancers. FAIRE-seq in wings precociously expressing E93 reveals that E93 binding results in chromatin accessibility changes. Together, these findings support a model in which E93 functions as a temporal cue by binding target enhancers and triggering local changes to the chromatin accessibility landscape. Importantly, we observe that E93 initiates distinct effects on chromatin accessibility depending on the target enhancer. At a subset of targets, E93 expression results in increased chromatin accessibility, which may enable binding of other transcription factors that control the spatial pattern of enhancer activity. However, at a different subset of enhancers that is already accessible, E93 expression results in decreased chromatin accessibility. Loss of accessibility may make these enhancers refractory to transcription factor binding and enable redeployment of spatial inputs to other targets. Thus, by controlling the accessibility and consequently the competence of cis-regulatory elements to respond to spatial inputs, temporal transcription factors such as E93 help to control the sequence of gene expression changes that drive development forward in time.
Does E93 control chromatin accessibility on its own, or in combination with other factors?
Although E93 binds directly to target enhancers, this does not mean that E93 controls chromatin accessibility independently of co-regulators. The findings presented in this study suggest a model in which other transcription factors influence the ability of E93 to regulate target enhancer accessibility. Several lines of evidence support this interpretation. First, only a fraction of E93-bound sites exhibit a change in accessibility in response to precocious E93 expression, even though many of them exhibit temporal changes in accessibility that are dependent on E93 later in development. Our motif analyses raise the intriguing possibility that some of the effects of E93 on target chromatin may be limited by other transcription factors in the ecdysone cascade. Motifs for the temporal transcription factor Br are enriched in E93 targets that open during pupal stages. In wild-type wings, br is induced by ecdysone to high levels during larval stages when these sites are inaccessible. Br levels subsequently drop as pupal development progresses, coincident with these sites increasing in accessibility (Guo et al., 2016). Thus, it is possible that Br antagonizes E93 function by maintaining pupal enhancers in an inaccessible chromatin state during larval stages. As E93 deactivates the brdisc enhancer, E93-mediated repression may contribute to the drop in Br levels in prepupal wings. Examples of cross-regulation between ecdysone-induced transcription factors have been previously reported (Mao et al., 2019; Ureña et al., 2016). Thus, some effects of E93 on chromatin accessibility may be indirectly mediated by cross-regulatory interactions between temporal transcription factors.
A second observation supporting a role for co-regulators in E93-dependent control of chromatin accessibility is that the nubvein enhancer only responds to E93 after the larval-to-prepupal transition. Although nubvein depends on E93 for opening and activation in wild-type pupal wings, precocious E93 expression in larval wings does not result in nubvein activation or in increased chromatin accessibility. Nubvein remains inactive even with prolonged exposure to E93 at larval stages, suggesting that its activation is not dependent on a downstream effector of E93 activity. Instead, nubvein exhibits precocious activity only after progression through the larval-to-prepupal transition. This switch in responsivity of nubvein as a function of developmental stage rather than duration of E93 exposure indicates that there is a change in the trans-regulatory environment that occurs independent of E93 activity. One potential trans-regulatory change is fluctuating titers of ecdysone. In Bombyx, E93 binds the ecdysone hormone receptor, an EcR/Usp complex, through its LXXLL nuclear receptor interaction motif (Liu et al., 2015). Hormone binding triggers conformational changes in nuclear receptors that result in differential recruitment of co-regulatory proteins (Glass and Rosenfeld, 2000), and decreasing ecdysone levels during the larval-to-prepupal transition could cause differential association of E93 with EcR/Usp complexes, thus making target enhancers such as nubvein dependent on circulating ecdysone titers.
An additional observation suggesting E93 works with other factors to control chromatin accessibility is the finding that E93 targets do not all experience changes in accessibility at the same time. Clustering of FAIRE-seq data at E93 binding sites from six stages of wild-type wing development revealed distinct temporal patterns of accessibility change. Moreover, these temporal clusters exhibit differential enrichment of transcription factor DNA binding motifs that correspond to transcription factors with varied spatial and temporal functions. This suggests that E93 works in combination with a diverse and dynamic set of co-regulators during pupal wing development to trigger multiple phases of chromatin accessibility regulation. Overall, we envision a model wherein E93 expression functions as a temporal cue that makes target enhancers competent to respond to spatial gene regulatory inputs; other transcription factors that bind with E93 at target enhancers dictate the effect E93 has on chromatin accessibility (Fig. 7).
Fig. 7.
Model of E93 function at target enhancers. E93 controls the competence of transcriptional enhancers to respond to spatial patterning cues and regulates chromatin accessibility genome wide. This suggests that E93 expression initiates a global shift in DNA binding by the transcription factors involved in spatial patterning, referred to here as redeployment. We propose that control of access to enhancers by temporal transcription factors such as E93 allows a limited number of spatial patterning factors to generate a diverse set of transcriptional outputs over time.
Activation and deactivation by E93
Although it is clear that transcription factors often possess both activating and repressing roles, the determinants of this context-specific function are poorly understood. In this study, we find that E93 expression both activates and represses different target enhancers in the same cells at the same time. The brdisc enhancer is active across larval wing discs. This enhancer is closed and deactivated in response to precocious E93 expression. Conversely, the tncblade and tncwv enhancers are opened and activated in response to precocious E93 expression. The pattern of brdisc overlaps the activity pattern of both tnc enhancers, thus strongly indicating that E93 expression is able to enact two opposing transcriptional outputs (activation and deactivation) simultaneously during development. Chromatin accessibility profiling indicates that E93 opens and closes chromatin at hundreds of loci when expressed in larval wings. Thus, the E93-mediated cue to increase or decrease accessibility at target enhancers is not exclusively because of stage-specific expression of co-regulators or a temporally regulated modification of E93 that makes it a dedicated activator or repressor. Instead, how a site responds to E93 is target specific. This is supported by the observation that sites which open or close in response to precocious E93 largely replicate the accessibility changes they normally exhibit during wild-type development. Thus, premature expression of E93 accelerates a regulatory program that is encoded in the DNA sequence of target enhancers. To gain insight into how E93 differentially regulates enhancer accessibility, we examined the DNA sequence of E93-sensitive sites. This analysis revealed that sites that decrease in accessibility in response to E93 binding contain higher quality as well as a greater number of E93 motifs relative to increasing E93-sensitive sites or E93-insensitive sites. Characteristics such as motif quality and quantity can determine whether a transcription factor activates or represses target enhancers (Parker et al., 2011; Scully, 2000; White et al., 2016). Thus, differential E93 motif composition could serve as a key determinant for the opposing effects E93 has on target chromatin. Increased E93 motif content in decreasing E93-sensitive sites could indicate that control of chromatin accessibility at these sites occurs independently of other transcription factors. However, we disfavor this hypothesis because it predicts that decreasing sites would be disproportionately enriched relative to increasing sites among E93-sensitive regions. Comparing the ratio of decreasing to increasing sites reveals no differences in E93-sensitive sites relative to E93-dependent sites. Thus, E93 is no more likely to close chromatin as it is to open it, suggesting both types of target are equally dependent on co-regulators. Further studies are necessary to identify the co-regulator proteins used by E93 to differentially control chromatin accessibility. Identifying these factors will help reveal the mechanisms controlling enhancer competence in development.
MATERIALS AND METHODS
Drosophila culture and genetics
Either ci-GAL4 or en-GAL4 lines were used for enhancer experiments with similar effects on reporter activity. Crosses were raised at 22°C and vials were shifted to 29°C for 24 h to induce E93 expression, unless otherwise indicated. For the experiments presented in Fig. 2B (Early L3), larvae were dissected 5 h after the shift to 29°C. For the experiments presented in Fig. 5B, larvae were dissected 48 h after the shift to 29°C. 3LW were dissected for all experiments, except for the experiments presented in Fig. 5D, in which mid-prepupae (5-7 h APF) were collected after 24 h of E93 induction. The vg-GAL4, Tub>CD2>GAL4, UAS-GFP, UAS-FLP; GAL80ts driver was used for FAIRE-seq and ChIP-seq. Embryos were collected for 6 h on apple juice-agar plates at 25°C and then transferred to 29°C for 36 h. GFP-positive larvae then were picked, transferred to vials and raised for 4.5 days at 18°C. Vials were then switched back to 29°C for 15 h to induce E93 expression. 3LW were dissected.
Fly stocks used were: w; Tub-GAL80ts; tm2/tm6b (Bloomington Drosophila Stock Center, 7108), yw; UAS-E93-3xHA (FlyORF, F000587; Bischof et al., 2013), yw; vg-GAL4, UAS-FLP, UAS-GFP, Tub>CD2>GAL4/CyO (Crickmore and Mann, 2006), yw; en-GAL4 (gift of Greg Matera, University of North Carolina at Chapel Hill, NC, USA), yw; ci-GAL4 (gift of Robert Duronio, University of North Carolina at Chapel Hill, NC, USA), yw; broaddisc-tdTomato (Uyehara et al., 2017), yw; nubvein-nlsGFP (Uyehara et al., 2017), yw; tncwv-tdTomato (Uyehara et al., 2017), yw; tncblade-tdTomato (Uyehara et al., 2017).
Immunofluorescence and image analysis
Immunofluorescence experiments and confocal imaging were performed as previously described (McKay and Lieb, 2013). The following antibodies were used: rabbit anti-E93 (1:2500; Uyehara et al., 2017), mouse anti-HA (1:1000, Sigma-Aldrich, H3663), rabbit anti-HA (1:500, Abcam, ab9110), guinea pig anti-Teashirt (1:1000; Zirin and Mann, 2007), mouse anti-Wingless (1:25, Developmental Studies Hybridoma Bank, 4D4), rabbit anti-Smad3 (phospho S423+S425) (pMad, 1:200, Abcam, ab52903). Alexa Fluor secondary antibodies (Invitrogen, A11032, A11037, A21052, A21071, A21450) were used at 1:1000. Precociously expressed E93 and endogenous E93 at ∼30 h APF were quantified by immunofluorescence using anti-E93 and Alexa 633 secondary antibodies. Fluorescent intensity was measured using ImageJ (Schindelin et al., 2012). Wild-type 30 h APF pupal wings were combined with 3LW wing imaginal discs precociously expressing E93 in the same tube for antibody incubations, then mounted on the same slide and imaged with identical settings (Leica Confocal SP5). E93 levels were quantified by measuring mean gray value in 25×25 pixel selections (10 selections per wing and three wings each). E93 signal was normalized by dividing by the mean background, which was calculated from nine 25×25 pixel selections in E93-negative portions of tissue in each experiment.
High throughput sequencing & data analysis
FAIRE-seq and ChIP-seq were performed as previously described (Uyehara et al., 2017). Briefly, ChIP experiments were performed in duplicate using a minimum of 200 wings for each replicate. Control genotypes contained the GAL4 driver but lacked the UAS-E93-3xHA transgene. Immunoprecipitation was performed using 5 µg of rabbit anti-HA (Abcam, ab9110). FAIRE-seq in precocious E93-expressing wings was performed using 45-60 wings in duplicate. Reads were aligned to the dm3 reference genome with Bowtie2 (Langmead and Salzberg, 2012). ChIP peaks were called with MACS2 (Zhang et al., 2008) on each replicate using background reads from the control genotype (precocious E93 experiments) or from a sonicated genomic DNA library (wild-type 24 h APF E93). ChIP peaks that overlapped between biological replicates were used for analysis. E93 binding categories were identified by intersecting the resulting peak lists from precocious E93 ChIP and wild-type E93 ChIP using the ChIPpeakAnno package from Bioconductor (Gentleman et al., 2004; R Core Team, 2017). Summits from the resulting union ChIP peak list were recomputed from aligned reads from pooled replicates using FunChip (setting d=125) (v1.0.0) (Parodi et al., 2017). Summits from entopic and orphan sites were computed from wild-type late E93 ChIP-seq, whereas summits from ectopic sites were computed from precocious E93 ChIP-seq. Chromatin accessibility differences within precocious E93 ChIP peaks were identified by counting FAIRE-seq reads within the union set of E93 ChIP peaks using featureCounts (setting allowMultioverlap=T) from Rsubread and testing for differential accessibility with DESeq2 using an adjusted P value <0.05 and an absolute log2FoldChange >1 (Liao et al., 2019; Love et al., 2014). Concordance of precocious chromatin accessibility changes with wild-type chromatin accessibility changes was determined using DESeq2, using an adjusted P value <0.05. Average signal line plots were generated using seqplots and ggplot2 from z-score normalized bigwig files at 10 base-pair resolution (Stempor and Ahringer, 2016; Wickham, 2009). Signal tracks were rendered in R with Gviz and cowplot (Hahne and Ivanek, 2016; Wilke, 2017). Overlap of ChIP peaks with genomic feature annotations was performed with ChIPseeker (v1.5.1), using the TxDb.Dmelanogaster.UCSC.dm3.ensGene annotation package from Bioconductor (https://bioconductor.org/packages/release/data/annotation/html/TxDb.Dmelanogaster.UCSC.dm3.ensGene.html).
Motif scanning
The dm3 assembly of the Drosophila melanogaster genome was scanned for the E93 motif from the FlyFactor Survey database using FIMO v4.12.0 (setting –thresh 0.01 –max-strand –text –skip-matched-sequence) (Grant et al., 2011; Zhu et al., 2011). Motifs overlapping a 20-base pair window around ChIP peak summits were identified using GenomicRanges findOverlaps (Lawrence et al., 2013). Motif number per window was quantified by directly counting these overlaps. For each peak category, motif quality within these windows was compared by using the ‘oneway.test’ function in R. Motif centrality within peaks was compared by computing the distance from each peak summit to the nearest E93 motif, then comparing the distribution of distances between binding categories using the ks.test function in R. PWMs of matched E93 motifs from within binding categories were derived by returning the DNA sequence matching each E93 motif detected within each peak. Sequences were converted to position frequency matrices (PFMs) using the ‘PWM’ function from Biostrings, then converted to PFMs using the toPFM function from PWMEnrich (v4.10.0) (https://bioconductor.org/packages/release/bioc/html/Biostrings.html; https://www.bioconductor.org/packages/release/bioc/html/PWMEnrich.html). Sequence logos were rendered using ggseqlogo (Wagih, 2017). Similarity of rederived E93 motifs was compared using a version of the TFBSTools PWMPearson internal function modified to accept PFMs (Tan and Lenhard, 2016). Motif similarity heatmaps of re-derived E93 motifs were rendered using ComplexHeatmap (Gu et al., 2016).
De novo motif analysis
DREME (v4.12.0) was used to scan a 100 bp region around each E93 ChIP peak summit within each E93 binding category using shuffled input sequences as background (using: dreme-py3 -dna -e 0.05 -m 10 -mink 3 -maxk 8) (Bailey, 2011). For analysis within E93 sensitive sites, the same analysis was performed within a 200 bp window around peak summits. Discovered motifs were imported into R using the importMatrix function from motifStack (v1.29.8) (Ou et al., 2018). Similarity values for discovered motifs were determined using the motifSimilarity function from PWMEnrich on all pairwise combinations of discovered motifs (https://www.bioconductor.org/packages/release/bioc/html/PWMEnrich.html). Clustering of PWM similarity was also performed by hierarchical clustering of motif distances computed using MotIV (v1.30.0) functions ‘motifDistances’ and ‘motifHclust’ (http://bioconductor.org/packages/release/bioc/html/MotIV.html). Clustered tree was rendered using ggdendro, ggseqlogo and cowplot https://cran.r-project.org/web/packages/ggdendro/index.html; Wagih, 2017; Wilke, 2017). Discovered motifs were matched to motifs from the Fly Factor Survey using TOMTOM (v4.12.0, using: tomtom -no-ssc -min-overlap 5 -dist “pearson” -evalue -thresh 10.0), displayed matches represent those corresponding to the top hit from this analysis (Gupta et al., 2007).
Targeted motif analysis
Directed motif searches were performed using AME (v5.1.0, setting: ame –scoring avg –method fisher –hit-lo-fraction 0.25 –evalue-report-threshold 10) to scan a 200 bp region around E93 binding category summits (using all ChIP peaks as background) and E93 sensitive summits (insensitive sites used as background). Searches within dynamic clusters were performed using a 100 bp window around the summit of each dynamic cluster, using all other dynamic clusters as background. For all analyses PWMs from the entire Fly Factor Survey were used for detection. All results were first filtered to remove any motifs from transcription factors with FPKM values <5 in wings during our wild-type RNAseq timecourse, reasoning that transcription factors passing this threshold are more likely to be functional during wing development (GSE77562).
Analysis of histone modifications
Data from GSE59769 were processed as described above for ChIP-seq (Schertel et al., 2015). Bigwigs were generated at 10 bp resolution and z-score normalized for analysis. Signal within target regions was extracted using seqPlots for heatmaps and average signal plots (Stempor and Ahringer, 2016). Heatmaps were generated using EnrichedHeatmap (Gu et al., 2018).
Western blotting
Wing discs were dissected from E93GFSTF animals at 6 h intervals relative to puparium formation by staging animals as white prepupae (3LW larvae were used as the −6 h timepoint). Western blots were performed as previously described, with the following changes. Twenty wings were collected per timepoint and stored at −80°C. Samples were lysed in hot Laemmli sample buffer (Leatham-Jensen et al., 2019; Uyehara and McKay, 2019). Samples were run for 60 min at 100 V on a 7.5% Bio-Rad stain-free TGX gel. Total protein stains were collected by laying the PAGE gel directly onto a UV transilluminator for 3 min and imaging on an Amersham Imager 600; the gel was kept hydrated with distilled water during all total protein crosslinking and imaging steps. After imaging the total protein stain, protein was transferred to a 0.2 µm nitrocellulose membrane at 100V for 60 min. E93GFSTF protein was detected using rabbit anti-GFP (1:5000, Abcam, ab290) and HRP-conjugated secondary (1:10,000, donkey anti-rabbit-HRP, GE Healthcare #NA934V) antibodies, and an Amersham ECL prime detection kit (GE Healthcare, RPN2232). Blots were imaged on an Amersham Imager 600. Signals were quantified with FIJI. Each of three replicates were scaled first to total protein then relative to the maximum E93 signal (24 h APF) for quantification.
Supplementary Material
Acknowledgements
We thank Daphne Knudsen for assistance with E93 western blots, Mary Leatham-Jensen for help with FAIRE and ChIP experiments, Jill Dowen and Greg Matera for critical reading of the manuscript, and members of the McKay and Dowen labs for useful discussions. The Wg antibody, developed by Steve Cohen, was obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (NIH) and maintained at The University of Iowa, USA. Stocks obtained from the Bloomington Drosophila Stock Center (NIH Grant P40OD018537) and the Zurich ORFeome Project (FlyORF) were used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.L.N., M.J.N., D.J.M.; Methodology: S.L.N., M.J.N., D.J.M.; Software: S.L.N.; Validation: S.L.N., M.J.N., D.J.M.; Formal analysis: S.L.N., M.J.N., D.J.M.; Investigation: S.L.N., M.J.N., D.J.M.; Resources: S.L.N., M.J.N., D.J.M.; Data curation: S.L.N., M.J.N., D.J.M.; Writing - original draft: S.L.N., M.J.N., D.J.M.; Writing - review & editing: S.L.N., M.J.N., D.J.M.; Visualization: S.L.N., M.J.N., D.J.M.; Supervision: D.J.M.; Project administration: D.J.M.; Funding acquisition: D.J.M.
Funding
M.J.N. was supported in part by NIH Grant T32GM007092 and a National Science Foundation Graduate Research Fellowship under Grant Number DGE-1650116. This work was supported in part by Research Scholar Grant RSG-17-164-01-DDC to D.J.M. from the American Cancer Society, and in part by Grant R35-GM128851 to D.J.M. from the National Institute of General Medical Sciences of the NIH. Deposited in PMC for release after 12 months.
Data availability
Data generated from this study can be found using GEO accession number GSE141738. All code used for data processing and analysis can be found at https://doi.org/10.5281/zenodo.3660089.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.181909.supplemental
References
- Baehrecke E. H. and Thummel C. S. (1995). The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev. Biol. 171, 85-97. 10.1006/dbio.1995.1262 [DOI] [PubMed] [Google Scholar]
- Bailey T. L. (2011). DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653-1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J. and Basler K. (2013). A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434-2442. 10.1242/dev.088757 [DOI] [PubMed] [Google Scholar]
- Crickmore M. A. and Mann R. S. (2006). Hox control of organ size by regulation of morphogen production and mobility. Science 313, 63-68. 10.1126/science.1128650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avino P. P. and Thummel C. S. (2000). The ecdysone regulatory pathway controls wing morphogenesis and integrin expression during Drosophila metamorphosis. Dev. Biol. 220, 211-224. 10.1006/dbio.2000.9650 [DOI] [PubMed] [Google Scholar]
- de Celis J. F. (1997). Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development 124, 1007-1018. [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (2017). Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219-240. 10.1146/annurev-cellbio-111315-125210 [DOI] [PubMed] [Google Scholar]
- Fraichard S., Bougé A.-L., Kendall T., Chauvel I., Bouhin H. and Bunch T. A. (2010). Tenectin is a novel αPS2βPS integrin ligand required for wing morphogenesis and male genital looping in Drosophila. Dev. Biol. 340, 504-517. 10.1016/j.ydbio.2010.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K. and Rosenfeld M. G. (2000). The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121-141. [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L. and Noble W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017-1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R. and Schlesner M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847-2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. and Ishaque N. (2018). EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics 19, 234 10.1186/s12864-018-4625-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Flegel K., Kumar J., McKay D. J. and Buttitta L. A. (2016). Ecdysone signaling induces two phases of cell cycle exit in Drosophila cells. Biol. Open 5, 1648-1661. 10.1242/bio.017525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J. A., Bailey T. L. and Noble W. (2007). Quantifying similarity between motifs. Genome Biol. 8, R24 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F. and Ivanek R. (2016). Visualizing genomic data using Gviz and bioconductor. Methods Mol. Biol. 1418, 335-351. 10.1007/978-1-4939-3578-9_16 [DOI] [PubMed] [Google Scholar]
- Hamm D. C., Larson E. D., Nevil M., Marshall K. E., Bondra E. R. and Harrison M. M. (2017). A conserved maternal-specific repressive domain in Zelda revealed by Cas9-mediated mutagenesis in Drosophila melanogaster. PLoS Genet. 13, e1007120 10.1371/journal.pgen.1007120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguera I. and Desplan C. (2018). Neuronal specification in space and time. Science 362, 176-180. 10.1126/science.aas9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Tsai Y.-H., Chen L., Zhou A., Banerjee K. K., Saxena M., Huang S., Toke N. H., Xing J., Shivdasani R. A. et al. (2019). The lineage-specific transcription factor CDX2 navigates dynamic chromatin to control distinct stages of intestine development. Development 146, dev172189 10.1242/dev.172189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T. and Carey V. J. (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatham-Jensen M., Uyehara C. M., Strahl B. D., Matera A. G., Duronio R. J. and McKay D. J. (2019). Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLoS Genet. 15, e1007932 10.1371/journal.pgen.1007932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Dai F., Guo E., Li K., Ma L., Tian L., Cao Y., Zhang G., Palli S. R. and Li S. (2015). 20-hydroxyecdysone (20E) primary response gene E93 modulates 20E signaling to promote bombyx larval-pupal metamorphosis. J. Biol. Chem. 290, 27370-27383. 10.1074/jbc.M115.687293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Li Y., Gao H. and Lin X. (2019). The direct interaction between E93 and Kr-h1 mediated their antagonistic effect on ovary development of the brown planthopper. Int. J. Mol. Sci. 20, 2431 10.3390/ijms20102431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Blanco E., Roch F., Noll E., Baonza A., Duffy J. B. and Perrimon N. (1999). A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development 126, 5739-5747. [DOI] [PubMed] [Google Scholar]
- McKay D. J. and Lieb J. D. (2013). A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27, 306-318. 10.1016/j.devcel.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou X., Duncan D. M., Baehrecke E. H. and Duncan I. (2012). Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc. Natl. Acad. Sci. USA 109, 2949-2954. 10.1073/pnas.1117559109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Wolfe S. A., Brodsky M. H. and Zhu L. J. (2018). motifStack for the analysis of transcription factor binding site evolution. Nat. Methods 15, 8-9. 10.1038/nmeth.4555 [DOI] [PubMed] [Google Scholar]
- Parker D. S., White M. A., Ramos A. I., Cohen B. A. and Barolo S. (2011). The cis-regulatory logic of hedgehog gradient responses: key roles for Gli binding affinity, competition, and cooperativity. Sci. Signal. 4, ra38-ra38. 10.1126/scisignal.2002077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi A. C. L., Sangalli L. M., Vantini S., Amati B., Secchi P. and Morelli M. J. (2017). FunChIP: an R/Bioconductor package for functional classification of ChIP-seq shapes. Bioinformatics 33, 2570-2572. 10.1093/bioinformatics/btx201 [DOI] [PubMed] [Google Scholar]
- Pasquinelli A. E. and Ruvkun G. (2002). Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18, 495-513. 10.1146/annurev.cellbio.18.012502.105832 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A. and Akam M. (2011). Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. Proc. Natl. Acad. Sci. USA 108, 2855-2860. 10.1073/pnas.1015077108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017). R: A Language and Environment for Statistical Computing.
- Schertel C., Albarca M., Rockel-Bauer C., Kelley N. W., Bischof J., Hens K., van Nimwegen E., Basler K. and Deplancke B. (2015). A large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating Drosophila wing development. Genome Res. 25, 514-523. 10.1101/gr.181305.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M., Carré C., Antoniewski C. and Truman J. W. (2005). Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development 132, 5239-5248. 10.1242/dev.02093 [DOI] [PubMed] [Google Scholar]
- Schulz K. N., Bondra E. R., Moshe A., Villalta J. E., Lieb J. D., Kaplan T., McKay D. J. and Harrison M. M. (2015). Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25, 1715-1726. 10.1101/gr.192682.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully K. M. (2000). Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science 290, 1127-1131. 10.1126/science.290.5494.1127 [DOI] [PubMed] [Google Scholar]
- Sen S. Q., Chanchani S., Southall T. D. and Doe C. Q. (2019). Neuroblast-specific open chromatin allows the temporal transcription factor, Hunchback, to bind neuroblast-specific loci. Elife 8, e44036 10.7554/eLife.44036.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempor P. and Ahringer J. (2016). SeqPlots - Interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 1, 14 10.12688/wellcomeopenres.10004.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G. and Lenhard B. (2016). TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32, 1555-1556. 10.1093/bioinformatics/btw024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ureña E., Chafino S., Manjón C., Franch-Marro X. and Martín D. (2016). The occurrence of the holometabolous pupal stage requires the interaction between E93, Krüppel-Homolog 1 and broad-complex. PLoS Genet. 12, e1006020 10.1371/journal.pgen.1006020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyehara C. M. and McKay D. J. (2019). Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Proc. Natl. Acad. Sci. USA 116, 9893-9902. 10.1073/pnas.1900343116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyehara C. M., Nystrom S. L., Niederhuber M. J., Leatham-Jensen M., Ma Y., Buttitta L. A. and McKay D. J. (2017). Hormone-dependent control of developmental timing through regulation of chromatin accessibility. Genes Dev. 31, 862-875. 10.1101/gad.298182.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O. (2017). ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645-3647. 10.1093/bioinformatics/btx469 [DOI] [PubMed] [Google Scholar]
- White M. A., Kwasnieski J. C., Myers C. A., Shen S. Q., Corbo J. C. and Cohen B. A. (2016). A simple grammar defines activating and repressing cis-regulatory elements in photoreceptors. Cell Rep. 17, 1247-1254. 10.1016/j.celrep.2016.09.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. Springer. [Google Scholar]
- Wilke C. O. (2017). cowplot: Streamlined Plot Theme and Plot Annotations for “ggplot2”. R package version 0.9.4. https://CRAN.R-project.org/package=cowplot R Packag. version 0.9.2 https://cran.r–project.org/package=cowplot.
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. J., Christensen R. G., Kazemian M., Hull C. J., Enuameh M. S., Basciotta M. D., Brasefield J. A., Zhu C., Asriyan Y., Lapointe D. S. et al. (2011). FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 39, D111-D117. 10.1093/nar/gkq858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J. D. and Mann R. S. (2007). Nubbin and Teashirt mark barriers to clonal growth along the proximal-distal axis of the Drosophila wing. Dev. Biol. 304, 745-758. 10.1016/j.ydbio.2007.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.