Key Points
Positive-strand RNA ((+)RNA) viruses are major pathogens of humans, animals and plants. These viruses actively reprogramme the host cell metabolism to support the infection process and to allow viruses to escape or suppress host defence mechanisms.
To facilitate RNA replication, (+)RNA viruses interact with numerous host molecules via protein–protein, RNA–protein and protein–lipid interactions. These interactions are crucial for the formation of viral replication organelles, which produce new viral RNA progeny in host cells.
All characterized (+)RNA viruses assemble viral replication complexes, containing both viral and host proteins, on intracellular membranes. The subverted host factors play crucial parts in all steps of (+)RNA virus replication. Therefore, host factors are key determinants of viral pathology as well as viral evolution.
As there can be ∼20,000–30,000 different proteins in a typical eukaryotic cell, identifying all the proteins that are subverted by a given (+)RNA virus is a daunting task. Genome-wide and global proteomics approaches have recently emerged as a powerful means to identify the host factors involved in (+)RNA virus replication.
An emerging theme from the genome-wide screens is that many of the host proteins subverted for (+)RNA virus replication are unique for a given virus. This suggests that (+)RNA viruses have evolved different ways to utilize host cell resources.
In spite of the diverse sets of host factors co-opted by various viruses, functional and mechanistic studies suggest that different host proteins provide similar functions during viral RNA replication.
Common host factors that are recruited by (+)RNA viruses for their replication include: RNA-binding proteins that facilitate viral RNA synthesis; proteins involved in membrane bending that contribute to the formation of membrane-bound replication complexes; lipid synthesis enzymes that affect lipid composition and have a role in making a favourable microenvironment for viral replication; and chaperones and prolyl isomerases that facilitate the proper folding and functions of viral replication proteins during assembly of the viral replication complexes.
Subject terms: Cellular microbiology, Infectious diseases, Pathogens, Virology
The replication of positive-sense RNA ((+)RNA) viruses involves numerous interactions between the RNA and proteins of the virus and proteins, membranes and lipids of the host. Host factors are thus key determinants of viral pathology as well as viral evolution. In this Review, Nagy and Pogany outline our current understanding of the host factors that facilitate the replication of (+)RNA viruses.
Abstract
Positive-sense RNA ((+)RNA) viruses such as hepatitis C virus exploit host cells by subverting host proteins, remodelling subcellular membranes, co-opting and modulating protein and ribonucleoprotein complexes, and altering cellular metabolic pathways during infection. To facilitate RNA replication, (+)RNA viruses interact with numerous host molecules through protein–protein, RNA–protein and protein–lipid interactions. These interactions lead to the formation of viral replication complexes, which produce new viral RNA progeny in host cells. This Review presents the recent progress that has been made in understanding the role of co-opted host proteins and membranes during (+)RNA virus replication, and discusses common themes employed by different viruses.
Main
The success of viruses as pathogens depends on their ability to replicate within host cells. They achieve this by actively reprogramming host cell metabolism to support the infection process and to allow viruses to escape or suppress host defence mechanisms. As a result, virus-infected cells undergo a series of major changes during infection.
Positive-sense RNA ((+)RNA) viruses include the human pathogens hepatitis C virus (HCV), West Nile virus (WNV), dengue virus (DENV) and severe acute respiratory syndrome coronavirus (SARS CoV). Although (+)RNA viruses have limited coding potential, containing between three and ten genes, these viruses can replicate efficiently in infected host cells owing to their use of host proteins, membranes, lipids and metabolites. Using molecular mimicry, a virus can trick host factors into performing novel functions that frequently target host cells. However, (+)RNA viruses can also induce strong antiviral responses. Viral infection, then, often represents a race between the virus and the host to gain control over the resources of the infected cell.
Following viral entry, the viral (+)RNAs are released and then translated to produce viral replication proteins. These proteins recruit the viral (+)RNA for replication, which requires the assembly of viral replication complexes (VRCs) on subcellular membrane surfaces. Each VRC first produces a complementary negative-sense RNA ((−)RNA) using the original (+)RNA as a template. The (−)RNA is then used by the viral replicase to synthesize new (+)RNA molecules that undergo additional rounds of translation and replication to form new viral particles that can egress from cells, or move from cell to cell in the case of plant (+)RNA viruses1,2,3,4 (Fig. 1).
Figure 1. Schematic infection cycle of positive-sense RNA viruses.

Positive-sense RNA ((+)RNA) viruses enter animal cells by endocytosis and plant cells through wounds. When the virus is inside the cell, the (+)RNA genome is released into the cytosol, where it is translated by the host ribosomes. The resulting viral replication proteins then recruit the (+)RNA to subcellular membrane compartments, where functional viral replication complexes (VRCs) are assembled. A small amount of negative-sense RNA ((−)RNA) is synthesized and serves as a template for the synthesis of a large number of (+)RNA progeny. The new (+)RNAs are released from the VRCs, whereas the (−)RNA is retained. The released (+)RNAs start a new cycle of translation and replication, become encapsidated, and then exit the cells (in the case of animal viruses) or move to neighbouring cells through plasmodesmata (in the case of plant viruses).
The replication of (+)RNA viruses involves numerous interactions between components from the virus (RNA and proteins) and the host (proteins, membranes and lipids). All characterized (+)RNA viruses assemble VRCs, which contain both viral and host proteins, on intracellular membranes1,5,6,7,8,9,10. These membranes can be derived from various organelles, such as the endoplasmic reticulum (ER), mitochondria, vacuole, Golgi, chloroplasts and peroxisomes, or from plasma membranes, or they emerge through the formation of novel cytoplasmic vesicular compartments derived from the ER or, possibly, autophagosomal membranes11,12,13,14,15,16,17,18,19,20. Thus, subverted host factors have crucial roles in all steps of (+)RNA virus replication. It follows, therefore, that host factors are key determinants of viral pathology as well as viral evolution4,9,14,21.
This Review outlines our current understanding of the host factors that facilitate the replication of (+)RNA viruses. Recent progress has come from in vitro replicase assays, reverse genetics approaches, intracellular localization studies, genome-wide screens to identify host factors affecting viral replication, and the use of natural or model hosts. The replication of RNA viruses has been described in several recent publications2,3,4,14,20,22. Here, an in-depth description of co-opted host factors is provided for representative (+)RNA viruses, including flaviviruses, coronaviruses, picornaviruses, plant tombusviruses and plant bromoviruses. A better understanding of the functions of the co-opted host factors could lead to the development of more potent antiviral strategies.
The discovery of co-opted host factors
As there can be ∼20,000–30,000 different proteins in a typical eukaryotic cell, identifying all the proteins that are subverted by a given (+)RNA virus is a daunting task. Numerous approaches have been developed in recent years, and in this section we briefly describe some successful strategies.
Genome-wide approaches have emerged as a powerful means to identify the host factors involved in (+)RNA virus replication. For example, genome-wide screens of Saccharomyces cerevisiae (Box 1) led to the identification of ∼130 S. cerevisiae genes that affect the replication of tomato bushy stunt virus (TBSV)23,24, ∼30 genes that affect TBSV RNA recombination25,26,27, and ∼100 genes that influence the replication of brome mosaic virus (BMV)28. Interestingly, only a small subset of S. cerevisiae genes affects replication of both BMV and TBSV, suggesting that these distantly related (+)RNA viruses depend on mostly different host factors and cellular processes23,24,28.
Genome-wide approaches based on RNA interference (RNAi) have been carried out with Drosophila C virus16,29, WNV30, DENV31 and HCV32,33,34,35,36,37 in various animal cell types. This approach led to the identification of several hundred host genes that affect (+)RNA virus replication, but this is probably an underestimation owing to the considerable level of genetic redundancy in animals, the variable efficiency of the protein knockdowns, the diversity of experimental conditions and cell types used, and the possible 'off-target' effects of some small interfering RNAs (that is, unwanted silencing of non-target genes). Further analysis of the functions of the identified genes will be necessary to confirm that they are indeed co-opted for viral replication.
Global proteomic approaches have also been widely used to identify host components that are part of VRCs or interact with viral replication proteins. For example, yeast two-hybrid screens using proteins from HCV and its human host38 revealed that viral non-structural protein 3 (NS3) and NS5A, which are involved in HCV RNA replication, had the largest number of interacting proteins (214 and 96, respectively). These data indicate that NS3 and NS5A might have crucial roles in reprogramming host cell metabolism during HCV infection through altering the cellular functions of the interacting host proteins. Proteomic approaches have also been used to identify host proteins that interact directly with the viral RNA of coronaviruses, alphaviruses and WNV (viral RNA was purified, and bound host proteins were then identified by mass spectrometry9,39,40,41,42,43,44) and with the viral RNA of TBSV and BMV (using a protein microarray)45,46.
Taken together, the results of global screens involving HCV, TBSV, BMV, WNV, DENV and Drosophila C virus confirm that (+)RNA viruses depend on numerous host cell components for robust RNA replication. Moreover, the overlap among the sets of host proteins identified by different screens for the same virus is low, which suggests that the screens have not yet reached saturation levels; one alternative explanation, however, is that some interactions identified in these screens are fortuitous, nonspecific or non-functional. In any case, these studies clearly indicate that multiple approaches are needed to catalogue the host–virus 'interactome' (that is, all the host proteins that specifically interact with viral RNAs and proteins).
The host factors that have been identified through such approaches are likely to have diverse roles during (+)RNA virus replication, including roles as mediators of the intracellular transport of viral proteins and viral RNA; as chaperones that facilitate folding of viral proteins; as helicases or RNA chaperones that assist the folding of viral RNA; as regulators of the switch from translation to replication (by promoting template recognition); and as enzymes involved in lipid metabolism to drive membrane proliferation14,24,28,38,47,48. Notable surprises from the genome-wide screens include the alteration of host lipid metabolism by DENV hijacking of autophagosomes49, the requirement for a host microRNA in HCV replication50, the involvement of membrane-bending host proteins in TBSV and BMV replication51,52, and the effect of a Ca2+,Mn2+-ATPase pump on TBSV RNA replication and recombination53. Interestingly, many of the identified host factors are conserved in eukaryotes, suggesting that (+)RNA viruses selectively target conserved host functions as opposed to host species-specific factors. Such a strategy would help viruses broaden their host range.
Roles of host factors in (+)RNA virus replication
One emerging theme from the genome-wide screens is that many of the host genes that are apparently subverted for (+)RNA virus replication are unique for a given virus. This suggests that (+)RNA viruses have evolved different ways to utilize host cell resources. However, in spite of the diverse sets of host factors that are co-opted by different viruses, functional and mechanistic studies suggest that different host proteins could provide similar functions during replication of viral RNA. The common functions performed by diverse host proteins during various steps of viral replication are discussed below.
Selection and recruitment of viral (+)RNA for replication. The viral (+)RNA participates in several competing processes that are required for successful infection and are also highly regulated and compartmentalized to escape RNA degradation22 (Fig. 1). Therefore, for replication to take place, the viral genomic RNA, together with viral and host factors, must be actively recruited to the appropriate subcellular membrane surfaces.
Viral replication proteins can bind selectively to viral (+)RNA, which probably leads to recruitment of the viral RNA from translation to replication54,55,56,57,58,59. However, host proteins are also involved. For example, in the case of human poliovirus (PV), a central role for host poly(rC)-binding protein 2 (PCBP2) in RNA template recruitment has been suggested (Fig. 2). PCBP2 is an RNA-binding protein involved in mRNA stabilization and the regulation of transcription and translation, and it binds to the internal ribosome entry site (IRES) in PV (+)RNA to promote cap-independent translation. When bound to the viral RNA, PCBP2 is cleaved by the PV-encoded RNA-dependent RNA polymerase (RdRp) precursor (3CD) or protease (3C) proteins and, as a result, loses one of its three RNA-binding sites. However, the cleaved form of PCBP2 retains two RNA-binding sites that allows the protein to bind to the cloverleaf structure at the 5′ untranslated region (UTR) of PV (+)RNA (Fig. 2a). This is crucial for PV RNA replication, as it brings together the 3′ and 5′ ends of the viral RNA through interaction with another host protein family, the poly(A)-binding proteins (PABPs)60,61 (Fig. 2a). The PV-encoded 3CD is also involved in RNA template selection and in switching of the viral RNA from translation to replication by binding to the cloverleaf-like structure at the 5′ UTR and influencing genome circularization54,55,60,62,63.
Figure 2. Remodelling the cellular secretory pathway to support human poliovirus replication.

a | The human poliovirus (PV) viral replication complex (VRC) assembles on the Golgi–trans-Golgi network (TGN) membrane and contains proteolytically cleaved host poly(rC)-binding protein 2 (PCBP2) and the virally encoded RNA-dependent RNA polymerase (RdRp) precursor protein, 3CD, bound to the 5′ cloverleaf-like structure of the positive-sense RNA ((+)RNA), with a host poly(A)-binding protein (PABP) bound to the 3′ poly(A) tail. As a result of the interaction between PCBP2 and PABP, the ends of the PV RNA genome are brought into proximity, facilitating the cleavage of 3CD and the release of the RdRp (3D-POL). 3D-POL then starts synthesis of the negative-sense RNA ((–)RNA) using VPg–UU (di-uridylated VPg protein) as a primer. The newly made double-stranded RNA (dsRNA) replicative form (RF), which consists of hybridized (+)RNA and (−)RNA, is partly unwound by the chaperone activity of the host heterogeneous nuclear ribonucleoprotein C (hnRNPC) and the binding of PCBP2 and 3CD to the 5′ end of the (+)RNA. After cleavage of 3CD, the released 3D-POL starts (+)RNA synthesis, which will go on for many rounds to produce excess amounts of (+)RNA progeny. b | PV begins replicating in the Golgi–TGN compartment. The virally encoded tail-anchored membrane protein 3A binds to the small GTPase ADP-ribosylation factor 1 (ARF1) and promotes the degradation of the host general vesicular transport protein p115, preventing the recruitment of ɛ-COP and β-COP (components of the vesicle coatomer complex 1, which surrounds transport vesicles in the early secretory pathway) to ERGIC. These events slow down vesicular transport from the endoplasmic reticulum (ER) to the Golgi–TGN. The production of additional 3A molecules on the newly made PV (+)RNAs results in remodelling of the Golgi–TGN and ERGIC compartments into viral replication organelles (the VRCs). These organelles provide optimal replication for PV owing to the presence of an increased amount of the lipid phosphatidylinositol 4-phosphate (PtdIns4P), which is produced by the recruited host PtdIns4P synthesis enzyme, PtdIns4-kinase-β (PI4KIIIβ). c | Trafficking between the ER and the Golgi in uninfected cells. The ERGIC is decorated with COP1, which is recruited by ARF1 and its guanine nucleotide exchange factor, GBF1.
Additional host proteins with documented roles in the recruitment of viral (+)RNA from translation to replication are listed in Table 1. Overall, these proteins facilitate the recruitment of different viral (+)RNAs for replication (Fig. 1) by protecting the viral (+)RNA from degradation, preventing a collision between the ribosome and the VRC (which compete for the same viral (+)RNA while progressing in opposite directions) and facilitating the selection of a cognate viral (+)RNA for replication (as VRCs of a given virus do not usually support the replication of RNAs from other viruses or from the host).
Table 1.
Roles of host factors in replication of positive-sense RNA viruses
| Host factors | Viruses | Functions | Refs |
|---|---|---|---|
| RNA-binding proteins | |||
| eEF1α | TBSV, WNV | RNA recruitment and (−)RNA synthesis | 123,125 |
| PCBP2 | PV | RNA recruitment and (−)RNA synthesis | 60,61 |
| PABP | PV | RNA recruitment and (−)RNA synthesis | 62 |
| PTB | HCV, coronaviruses | RNA recruitment and (−)RNA synthesis | 141 |
| LSM1 | BMV, HCV | RNA recruitment | 142,143 |
| hnRNPs | PV, HCV, coronaviruses | RNA recruitment, and (+)RNA and (−)RNA synthesis | 129,130,144,145 |
| TIA1, TIAR | WNV | (+)RNA synthesis | 131 |
| GAPDH (Tdh2 and Tdh3) | TBSV | Asymmetric (+)RNA synthesis | 133,134 |
| Cellular chaperones | |||
| HSP70 | TBSV, FHV | VRC assembly or activation and replication protein recruitment | 75,113,117 |
| HSP90 | FHV, HCV | VRC assembly or activation | 115,120 |
| HSP40 | FHV, BMV, flaviviruses | VRC assembly or activation | 118,119 |
| CYPA | HCV | Polyprotein processing and RdRp NS5B folding | 121 |
| CYPB | HCV | Conformation of the RdRp NS5B | 122 |
| FKBP8 | HCV | Interaction with NS5A and VRC stabilization | 120 |
| Protein targeting | |||
| TOM1 and TOM3 | ToMV | Replication protein targeting, and anchoring to membranes | 65 |
| VAPA | HCV | NS5A and NS5B binding, and anchoring to membranes | 66,67 |
| FBXL2 | HCV | NS5A recruitment to membranes | 69 |
| PEX19 | TBSV | Replication protein recruitment to peroxisome membranes | 73 |
| PI4KIIIβ | PV | Viral RdRp 3D-POL recruitment to membranes | 82 |
| Membrane remodelling and lipid synthesis | |||
| ARF1, GBF1 | PV | Viral replication organelle formation | 82 |
| PI4KIIIβ | PV | PtdIns4P enrichment in membranes co-opted for VRCs | 82 |
| PI4KIIIα | HCV | PtdIns4P enrichment in membranes co-opted for VRCs | 87 |
| EDEM1, OS9 | Coronaviruses | Edemosome formation (possibly containing VRCs) | 90 |
| LC3-I | Coronaviruses | VRC formation | 90 |
| FASN | DENV | Fatty acid synthesis and VRC formation | 91 |
| HLH106 | DCV | Fatty acid synthesis and, possibly, VRC formation | 16 |
| Ole1 | BMV | Fatty acid desaturation and VRC formation | 92 |
| Erg25, SMO1, SMO2 | TBSV | Sterol synthesis and VRC formation | 94 |
| Cellular lipids | |||
| Sterols | TBSV, WNV, HCV | VRC formation | 94,99 |
| Fatty acids | DENV, BMV, DCV, HCV | VRC formation | 92,101 |
| Phospholipids | TBSV, FHV | VRC formation | 104,106 |
| PtdIns4P | HCV, PV | Replication protein binding to membranes, and VRC formation | 82,87 |
| Membrane-shaping proteins | |||
| ESCRT proteins | TBSV | VRC formation | 52 |
| Reticulons | BMV | VRC formation | 51 |
| Other host proteins | |||
| PMR1, ECA1 and ECA3 | TBSV | VRC activity and RNA recombination | 53 |
| ARF1, ADP-ribosylation factor 1; BMV, brome mosaic virus; CYP, cyclophilin; DCV, Drosophila C virus; DENV, dengue virus; ECA, calcium-transporting ATPase, ED type; EDEM1, ER degradation-enhancing α-mannosidase-like 1; eEF1α, elongation factor 1α; ESCRT, endosomal sorting complexes required for transport; FASN, fatty acid synthase; FBXL2, F-box and LRR-repeat protein 2; FHV, Flock House virus; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HCV, hepatitis C virus; hnRNPs, heterogeneous nuclear ribonucleoproteins; HSP, heat shock protein; LC3-I , non-lipidated microtubule-associated proteins 1A and 1B light chain 3A; NS, non-structural protein; PABP, poly(A)-binding protein; PCBP2, poly(rC)-binding protein 2; PI4K, PtdIns 4-kinase; PTB, polypyrimidine tract-binding protein; PtdIns4P, phosphatidylinositol-4-phosphate; PV, human poliovirus; RdRp, RNA-dependent RNA polymerase; (+)RNA, positive-sense RNA; (−)RNA, negative-sense RNA; TBSV, tomato bushy stunt virus; TIAR, TIA1-related protein; ToMV, tomato mosaic virus; VAPA, vesicle-associated membrane protein-associated protein A; VRC, viral replication complex; WNV, West Nile virus. | |||
Targeting viral replication proteins to the replication site. Host proteins participate in targeting the virally encoded replication proteins to their replication sites within the appropriate subcellular compartments. For example, replication of tomato mosaic virus (ToMV) depends on two Arabidopsis thaliana membrane proteins termed TOM1 and TOM3, which interact with the helicase-like ToMV replication protein, 130K64,65. TOM1 and TOM3 are also part of the ToMV VRC, in which they probably act as membrane anchors for the replication proteins64,65. Similarly, the human vesicle-associated membrane protein-associated protein A (VAPA), a SNARE-like protein, might serve as a membrane anchor for HCV replication proteins. Indeed, VAPA interacts with the HCV replication proteins NS5A and NS5B66, an interaction that could be important for the association of NS5A and NS5B with the intracellular ER-derived membranes that constitute the site of HCV replication. Inhibiting the expression of VAPA, which is partially associated with lipid rafts, results in relocation of NS5B from detergent-resistant to detergent-sensitive membranes67. Detergent-resistant lipid rafts — cholesterol- and sphingolipid-rich microdomains — in subcellular membranes could serve as nucleation sites to increase the local concentration of viral replication proteins and facilitate the selective binding of key host factors, ultimately leading to the formation of VRCs67,68. In addition, a geranylgeranylated cellular protein, FBXL2 (F-box and LRR-repeat protein 2; also known as FBL2), might be involved in the recruitment of HCV NS5A to intracellular membranes69.
The transport of potyvirus replication proteins and RNA to the sites of replication occurs in viral protein-induced vesicles that move from the ER to the outer chloroplast membrane. This process seems to rely on the endosomal trafficking pathway and the actomyosin system70. It has been proposed that only a single genomic RNA molecule is transported in each vesicle71. TBSV requires the peroxisomal shuttle protein PEX19, which is necessary for the formation of the peroxisome membrane, and the chaperone heat shock protein 70 (HSP70) to target viral replication proteins to the peroxisome72,73,74,75. However, the peroxisomal membrane is not essential for TBSV replication, as TBSV RNA can replicate in a pexΔ S. cerevisiae strain, (which is defective in peroxisome biogenesis) as efficiently as in a wild-type strain24,76. The finding that TBSV can replicate efficiently on the ER membrane in the absence of peroxisomes suggests that some RNA viruses have the flexibility to use different host membranes for their replication. In addition, TBSV replication proteins can be transported to the sites of replication in the form of multimolecular complexes that include viral RNA and some host factors77. Pre-organization of replication factors into multicomponent complexes in the cytoplasm could facilitate efficient transport and colocalization of these components to the replication sites and, thus, the assembly of functional VRCs.
Assembly of the VRC
The assembly of a functional VRC occurs on intracellular membrane surfaces and appears to be a highly regulated event. Long-range RNA–RNA interactions, cis-acting viral RNA elements and trans-acting viral and host factors contribute to the fidelity and efficiency of VRC assembly. The formation of VRCs probably serves to concentrate crucial viral and host factors, facilitate optimal conditions for viral RNA synthesis, and protect sensitive RNA and protein components from recognition and destruction by the host antiviral surveillance system.
Detailed studies of VRCs are challenging, as these complexes form complicated and fragile structures that are frequently part of large viral replication organelles. The HCV VRC contains approximately one viral (−)RNA molecule and five viral (+)RNA molecules, together with hundreds of viral NS proteins, the viral RdRp and subverted host proteins78. All these components are present within spherules (invaginations of lipid membranes) that are resistant to proteases and nucleases and are formed from ER-derived membrane. Similar structures have been also observed with alphaviruses, Flock House virus, BMV and TBSV11,13,52,79,80.
Cellular membrane remodelling during VRC assembly. (+)RNA viruses subvert selected organellar membranes by dynamically remodelling and deforming them, giving rise to spherules and vesicles, or by inducing the formation of viral replication organelles4,20,52,79,80,81. For example, a recent model proposed that PV initially starts replication on pre-existing Golgi and trans-Golgi network (TGN) membranes followed, at the peak of PV infection, by redistribution of the newly made viral proteins to viral replication organelles82 (Fig. 2b,c). These organelles form close to the ER exit sites and are enriched in various host proteins such as the small RAS-family GTPase ADP-ribosylation factor 1 (ARF1), GBF1 (a guanine nucleotide exchange factor (GEF) for ARF1) and phosphatidylinositol 4-kinase-β (PI4KIIIβ; also known as PI4Kβ), which is involved in phosphatidylinositol-4-phosphate (PtdIns4P) synthesis82. The subversion of PI4KIIIβ leads to enrichment of the membrane compartment with phosphoinositide lipids such as PtdIns4P82 (Fig. 2b). This helps to recruit the PV RdRp, 3D-POL, which binds selectively to PtdIns4P in the membrane. The probable role of GTP-bound ARF1 (the membrane-associated active form) is to recruit other cellular effector proteins to change the membrane curvature, induce the formation of transport vesicles from intracellular organelles and modify the lipid composition of membranes83,84. It is likely that PV proteins 3A and 3CD act synergistically to reorganize the host secretory trafficking pathway in order to facilitate the formation of individual VRCs and replication organelles82,83. Similar replication organelles are probably also formed during flaviviral infections82.
HCV uses a similar strategy to build subcellular vesicles or ER-derived 'membranous webs', in which HCV VRCs are found85. The ER-bound HCV protein NS5A recruits the host kinase PI4KIIIα (also known as PI4Kα), another protein involved in PtdIns4P synthesis37,85,86. NS5A binding stimulates PI4KIIIα kinase activity to enrich the membrane compartment with PtdIns4P87. The PtdIns4P-rich membrane can then facilitate the recruitment of additional host factors, such as oxysterol-binding protein 1 (OSBP1), which is required for HCV replication88. Both PI4KIIIα kinase and PtdIns4P are crucial for the formation and integrity of the membranous web structure of the HCV replication organelle87 (Fig. 3a). Remarkably, both PV and HCV seem to use PtdIns4P enrichment to create a favourable microenvironment for RNA replication, but they co-opt different cellular kinases to do so. One possible explanation for this is the availability of these host kinases in different subcellular compartments. For example, ER-bound NS5A co-opts PI4KIIIα kinase, which is also ER localized and thus likely to be readily available. By contrast, PV co-opts the Golgi-localized PI4KIIIβ, which could be available in the ERGIC compartment (Fig. 2). The mechanisms used by HCV and PV for PtdIns4P enrichment of cellular membranes provide a good illustration of a common theme in the biology of (+)RNA viruses: different host proteins can be co-opted by different viruses to provide mechanistically similar functions during viral replication.
Figure 3. Structures of the flavivirus replication complexes.

a | Hepatitis C virus (HCV) replication takes place at a unique subcellular compartment, the endoplasmic reticulum (ER)-derived membranous web, which consists of clusters of HCV-induced vesicles and lipid droplets (LD) and is the proposed site of viral assembly. The virus-encoded replication protein non-structural protein 5A (NS5A) binds to human vesicle-associated membrane protein-associated protein A (VAPA), which, together with NS4B, seems to be important for the association of NS5A and NS5B with cholesterol-rich lipid rafts in the ER-derived membranes. The membrane-bound NS5A then recruits and activates the host kinase phosphatidylinositol 4-kinase-α (PI4KIIIα), which leads to an enrichment of phosphatidylinositol-4-phosphate (PtdIns4P) that could facilitate recruitment of oxysterol-binding protein 1 (OSBP1) and the formation of the sterol-rich 'membranous web' to support HCV replication. The high concentration of PtdIns4P probably also facilitates the recruitment of additional host factors to the membranous web. The viral protein NS4B is also important for the formation of the membranous web, possibly by inducing membrane curvature (not shown). b | Electron micrograph of the viral replication complexes (VRCs) induced during dengue virus (DENV) infections. The white arrowhead indicates a DENV-induced vesicle in the ER with an opening towards the nuclear envelope; the black arrowhead points to a virus particle in a cisterna close to a Golgi stack. c | Three-dimensional modelling of the VRCs induced during DENV infections. The arrow indicates a putative virus budding site, and red spheres represent virus particles. ARF1, ADP-ribosylation factor 1; NE, nuclear envelope; Ve, vesicle. Parts b,c are reproduced, with permission, from Ref. 140 © (2009) Elsevier.
In addition, coronaviruses induce the formation of interconnected double-membraned vesicles (DMVs)89 by exploiting a regulatory pathway that controls ER-associated protein degradation (ERAD)90. Mouse hepatitis virus (MHV), a coronavirus, seems to prevent degradation of the short-lived ER chaperones ER degradation-enhancing α-mannosidase-like 1 (EDEM1) and OS9, which are crucial regulators of ERAD, by trapping them in structures called edemosomes (Fig. 4). It has been hypothesized that the accumulation of MHV replication proteins in edemosomes, which are decorated with LC3-I (non-lipidated microtubule-associated proteins 1A and 1B light chain 3A; a homologue of yeast Atg8, a ubiquitin-like protein that is required for the formation of autophagosomal membranes), transforms these structures into permanent DMVs90. The coronaviral replication proteins nsp2 and nsp3 are colocalized with LC3-I on the surface of DMVs during the entire course of MHV infection. Indeed, depletion of LC3 (which results in a reduction of both the lipidated form, LC3-II, and the non-lipidated form) by RNAi severely inhibits DMV formation and MHV replication90. The actual function of DMVs has not been fully defined, but they could be the sites of viral replication and virion assembly. The viral content could be released from DMVs by fusion with other membranes or by disintegration of the DMV membranes.
Figure 4. Proposed remodelling of the endoplasmic reticulum-associated protein degradation tuning pathway during coronavirus replication.

a | In normal, uninfected cells, the endoplasmic reticulum-associated protein degradation (ERAD) tuning pathway downregulates the levels of the ER chaperones EDEM1 (ER degradation-enhancing α-mannosidase-like 1) and OS9 in the ER via transport to the lysosome, mediated by LC3-I (non-lipidated microtubule-associated proteins 1A and 1B light chain 3A)-coated edemosomes. b | Coronavirus (CoV) infection transforms the edemosomes into double-membraned vesicles (DMVs) that probably contain viral replication complexes (VRCs). The actual orientation of the CoV VRCs in DMVs is not yet known. c | Electron tomography-based model of CoV DMVs. Narrow connections between several DMVs form a network that is also connected to the ER. Part c is reproduced from Ref. 9.
The role of lipids in VRC assembly. Another major group of host factors that affects VRC assembly is involved in regulating the lipid composition of selected intracellular membranes. Membrane lipids can serve as scaffolds for the assembly of VRCs or can provide crucial lipid cofactors to regulate the function of the viral replicase. For example, the DENV protein NS3 recruits fatty acid synthase (FASN), a major rate-limiting enzyme in fatty acid biosynthesis, to the ER membrane, where DENV replicates91 (Fig. 3b,c). Moreover, the interaction between NS3 and FASN results in increased FASN activity91. This leads to de novo synthesis of fatty acids that then integrate into the ER and DENV VRC membranes, suggesting that the local synthesis of fatty acids facilitates the expansion of the ER and the formation of DENV VRCs in infected cells. DENV also increases the amount of free fatty acids derived from lipid droplets in mammalian cells by inducing autophagy49. The released free fatty acids become available for the assembly of DENV VRCs and lead to an increase in cellular ATP, as they are metabolized by β-oxidation in the mitochondria. Increased ATP is expected to enhance cellular processes and stimulate DENV replication49. Thus, DENV and the closely related HCV seem to rely on the local enrichment of different lipids (fatty acids versus PtdIns4P) to promote the assembly of VRCs. These differences could be explained by the different cell types or hosts infected by these viruses (HCV infects liver cells in humans, whereas DENV not only infects humans and non-human primates but also persists in the mosquito vector), which could dictate the available lipid synthesis pathways.
The generation of new membrane surfaces via fatty acid biosynthesis also has a major effect on Drosophila C virus replication in Drosophila melanogaster cells: depletion of host HLH106, a protein that regulates fatty acid metabolism, blocks the formation of Drosophila C virus-induced vesicular compartments16. In addition, in S. cerevisiae inactivation of the gene encoding Ole1, a fatty acid Δ9-desaturase that participates in the synthesis of unsaturated fatty acids, reduces the activity of the BMV replicase, probably owing to reduced binding of one of the replicase components (BMV replication protein 1a) to membranes with a lower ratio of unsaturated fatty acids92. This results in reduced efficiency of VRC assembly and a low level of replication.
Lipids also seem to affect tombusvirus replication. Electron microscopy images of plant cells infected with tombusviruses show extensive remodelling of membranes and indicate that active lipid biosynthesis occurs52,80,93. Moreover, genome-wide screens in S. cerevisiae identified 14 host genes involved in lipid metabolism that affect tombusvirus replication and recombination23,24,25,27. Detailed studies with Erg25, an important enzyme in the sterol biosynthesis pathway in S. cerevisiae, revealed that sterols are crucial for TBSV replication94. Sterols are ubiquitous and essential membrane components in all eukaryotes, affecting membrane rigidity, fluidity and permeability by interacting with other membrane lipids and proteins95,96. Sterols are also important for the replication of other (+)RNA viruses, such as HCV, WNV, DENV and Norwalk virus97,98,99,100,101. For example, infection of mammalian cells with WNV leads to redistribution of cholesterol from the plasma membrane to the sites of viral replication and also results in reduced antiviral responses99.
Phospholipids are major components of cellular membranes, affecting the size, shape and rigidity of cells and intracellular organelles102, and they affect the replication of several (+)RNA viruses by their effects on the binding of viral replication proteins to membranes103,104,105. For example, reducing phospholipid levels in S. cerevisiae cells affects TBSV replication in various ways, leading to reduced template activity of the viral replicase, and to altered subcellular localization and reduced stability of the viral replication proteins24,106. Interestingly, genes involved in the production of phosphatidylcholine in D. melanogaster are crucial for Flock House virus replication107. Therefore, the amount and ratio of various phospholipids are important for the replication of several (+)RNA viruses.
Membrane-shaping proteins. Other host factors recruited by some (+)RNA viruses for their own replication are involved in bending membranes. Indeed, dynamic remodelling and deforming of membranes to give rise to unique spherule structures seem to require host proteins. For example, tombusviruses recruit ESCRT (endosomal sorting complexes required for transport) proteins24, which have a major role in sorting cargo proteins from the endosomes to multivesicular bodies by membrane invagination and vesicle formation108,109,110,111. Recruitment of ESCRT proteins for TBSV replication might facilitate VRC assembly, including the formation of TBSV-induced spherules (Fig. 5) and vesicles in infected plant cells52. Accordingly, the ubiquitylated TBSV p33 was found to interact with S. cerevisiae vacuolar protein sorting-associated protein 23 (Vps23; an ESCRT-I protein) and Bro1 (an accessory ESCRT factor)52,112, resulting in the recruitment of Vps23 and possibly Bro1 to peroxisomes, the sites of TBSV replication. This is followed by the recruitment of ESCRT-III proteins (Snf7 and Vps24), which could assist optimal assembly of the VRC, facilitate colocalization of the viral replication proteins p33 and p92 within selected areas in the membrane, and promote the formation of viral spherules by deforming the membranes or stabilizing the 'neck' structure (the narrow opening in the spherules that faces the cytosol) (Fig. 5). It seems that protection of the viral RNA is compromised if the VRC assembles in the absence of ESCRT proteins52. Thus, ESCRT proteins appear to affect the quality of VRC assembly by contributing to the protection of viral RNA from ribonucleases.
Figure 5. Functions of host factors during tombusvirus replication in Saccharomyces cerevisiae.
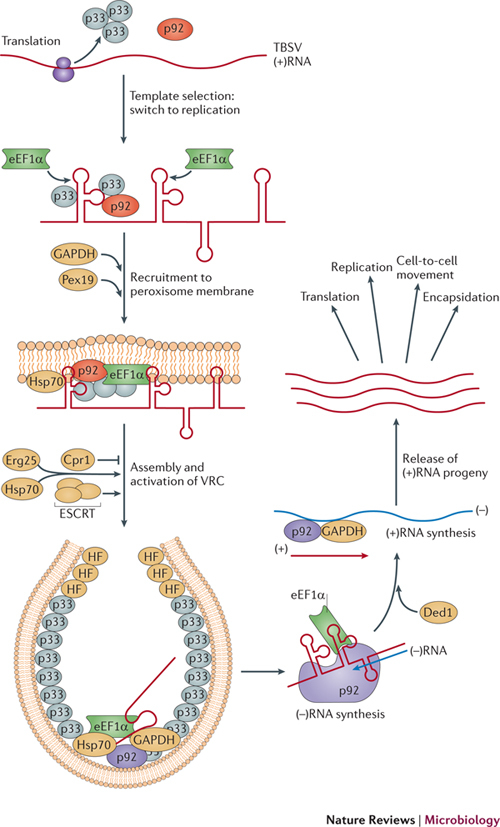
Tombusvirus (such as tomato bushy stunt virus (TBSV)) RNA translation and replication, and the host proteins that affect these processes in a Saccharomyces cerevisiae model host. The TBSV RNA-dependent RNA polymerase (RdRp) precursor p92 is activated during the assembly process (as indicated by the colour change) in an as-yet-unknown way. TBSV p33 is an RNA chaperone involved in viral RNA recruitment and assembly of the viral replication complexes (VRCs). The presence of host proteins (denoted host factor (HF) here) is currently hypothetical, and they could be ESCRT-III (endosomal sorting complexes required for transport III) factors. The known cellular functions for the shown host proteins are: elongation factor 1α (eEF1α), required for translation elongation; Pex19, a cytosolic shuttle protein involved in transporting peroxisomal membrane proteins to the peroxisome; heat shock protein 70 (Hsp70), a molecular chaperone involved in protein folding, mostly in the cytosol; Erg25, an enzyme for sterol biosynthesis; Cpr1 (also called cyclophilin), a cytosolic prolyl isomerase; ESCRT, a large family of proteins involved in membrane bending, protein transport and degradation via the endosomal pathway; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzymes, a ubiquitous protein family that is a key component of cytosolic energy production; Ded1, a DEAD box helicase involved in translation initiation.
Anther group of membrane-shaping proteins, the reticulon homology proteins (Rhps), are involved in assembly of the spherule-like structures during BMV replication in S. cerevisiae51. The BMV helicase-like replication protein 1a recruits Rhps to the perinuclear ER, resulting in spherule formation. In addition, Rhps affect some functions of protein 1a, such as induction of VRC formation or recruitment of viral RNA. Thus, membrane-shaping proteins of the ESCRT and Rhp groups appear to be involved in the formation of spherules, in which (+)RNA replication can be protected from host immune surveillance.
Roles of host chaperones. Cellular chaperones are also involved in VRC assembly. For example, the tombusvirus replicase was completely inactive when assembled in vitro using an S. cerevisiae cell-free extract that lacked heat shock protein 70 (Hsp70)113. The addition of purified recombinant Hsp70 to cell-free extracts was necessary for VRC assembly and replication of the viral RNA113. Therefore, it seems that Hsp70 is necessary for TBSV replication (Fig. 5). In addition, Hsp70 seems to be important for the insertion of tombusvirus replication proteins into intracellular membranes75 and for the folding and stability of the tombusvirus VRC114. Inhibition of the activity or expression of Hsp70 also affects the replication of ToMV and turnip crinkle virus, suggesting that subversion of Hsp70 is common among (+)RNA viruses75.
Another example of the role of cellular chaperones is found in the replication of Flock House virus, which is affected by HSP70 and HSP90 proteins as well as by J-domain HSP40-family chaperones115,116,117. Moreover, Ydj1, a J-domain protein of the HSP40 chaperone family, was proposed to affect the assembly and/or activation of the BMV VRC before (–)RNA synthesis in S. cerevisiae118. Also, the assembly or activity of flaviviral VRCs is affected by DNAJC14, an HSP40 family chaperone in mammalian cells119. Thus, various host chaperones are involved in activation and assembly of several (+)RNA virus VRCs, making these chaperones potential targets for antiviral approaches.
In addition, prolyl isomerases (PPIases) are involved in the assembly of the HCV VRCs. The immunophilin FKBP8 (a PPIase) interacts with the HCV phosphoprotein NS5A and with HSP90, and this interaction may have a role in stabilizing the VRC120. Even more importantly, the cytosolic protein cyclophilin A (CYPA; also known as PPIase A), which has PPIase activity, is crucial for assembly of the HCV VRC, probably because it affects processing of the HCV polyprotein and folding of the RdRp NS5B121. CYPB (also known as PPIase B), another cellular PPIase, also affects the conformation of NS5B, which in turn affects the RNA-binding ability and template activity of this viral protein122.
Synthesis of progeny viral RNA
Another common theme in the replication of (+)RNA viruses is the subversion of various host RBPs to facilitate viral RNA synthesis. Several of the RBPs that are co-opted by different viruses probably have mechanistically similar functions during viral replication, as discussed below.
Host factors that regulate (−)RNA synthesis. Although all (+)RNA viruses encode their own RdRps and usually 1–6 auxiliary replication proteins, host proteins are predicted to participate in each step of RNA synthesis. For example, elongation factor 1α (eEF1α), which is a highly abundant cellular protein that delivers the aminoacyl-tRNA to the elongating ribosome, is a permanent resident of the tombusvirus VRC and binds to the 3′ UTR of the TBSV RNA (in yeast, this is the TEF1 and TEF2 forms of eEF1α, but in plants and mammals the specific form of eEF1α involved is not yet known)46. Mutational analysis of eEF1α, in combination with in vitro replication studies, suggests that eEF1α promotes (−)RNA synthesis by the tombusvirus VRC by binding to both the RdRp and the 3′ UTR of the (+)RNA molecule, thus facilitating the proper positioning of the RdRp on the template RNA for efficient initiation of RNA synthesis123. Similarly, eEF1α binds to the 3′ UTR of the (+)RNA of WNV, and certain mutations introduced at the eEF1α-binding sites of the viral RNA inhibit both (+)RNA–eEF1α binding and (−)RNA synthesis21,124. Moreover, eEF1α colocalizes with the WNV VRC in infected cells, indicating that eEF1α facilitates the interaction between the viral replicase and the WNV RNA 3′ UTR125. The involvement of eEF1α in viral (+)RNA replication might be common, as eEF1α binds to many viral (+)RNAs and replication proteins, and its high abundance in cells might facilitate its recruitment for viral replication. Indeed, the bacterial homologue of eEF1α, termed EF-Tu, is known to affect (−)RNA synthesis for the bacteriophage Qβ126,127, suggesting that subversion of eEF1α-like host proteins by (+)RNA viruses might be an ancient process.
Other host RBPs seem to facilitate proper positioning of the viral RdRp on the RNA template to promote (−)RNA synthesis. For example, co-opted PCBP2 forms a complex with PV 3CD while bound to the cloverleaf structure in the PV (+)RNA 5′ UTR128 (Fig. 2a). After activation of PV RdRp by cleavage of 3CD, the replication initiation complex at the 5′ end is brought to the 3′ end by genome circularization through the interaction of PCBP2 with the PABP bound to the poly(A) sequence in the 3′ UTR62 (Fig. 2a). Interactions between polypyrimidine tract-binding protein (PTB; also known as heterogeneous nuclear ribonucleoprotein I), which is bound to the 5′ UTR of the MHV genome, and hnRNPA1, which is bound to 3′ UTR, might have a comparable role in facilitating (−)RNA synthesis by viral RdRp. Mutations in MHV RNA that impair binding of host factors to the UTRs inhibit replication of the viral RNA129.
Host factors that regulate (+)RNA synthesis. Host RBPs are also important for (+)RNA synthesis. For example, hnRNPC, which binds to the 3′ end of the PV (–)RNA through its RNA recognition motif (RRM) domain (containing an arginine-rich motif), also interacts with the replication protein 3CD, probably recruiting 3CD to the (−)RNA template130. The RNA chaperone activity of hnRNPC could facilitate the folding of (−)RNA or the replicative double-stranded RNA intermediate into a replication-compatible conformation, thus promoting RNA synthesis130 (Fig. 2a).
Other host proteins involved in (+)RNA synthesis are the stress granule protein TIA1 and TIA1-related protein (TIAR), which contain three RRM motifs. TIA1 and TIAR have been proposed to bind to WNV (−)RNA, promoting efficient (+)RNA synthesis42,131. In addition, the sequestration of these proteins for WNV replication might inhibit the formation of stress granules and P-bodies, thus potentially blocking mRNA degradation131.
Regulation of asymmetric RNA synthesis by host factors. During TBSV replication, host glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is involved in regulation of asymmetric RNA synthesis (Fig. 5). This hallmark feature of (+)RNA viruses leads to the generation of 10- to-100-fold more (+)RNA progeny than (−)RNA replication intermediates. GAPDH, which is present in the tombusvirus VRC114, is a highly conserved, abundant and ubiquitous protein that is a key component of cytosolic energy production132. Downregulation of GAPDH inhibits TBSV replication in S. cerevisiae and in plants and results in production of (+)RNAs and (–)RNAs in a 1:1 ratio, instead of asymmetric RNA synthesis133. GAPDH might promote asymmetric RNA synthesis by selectively anchoring the (−)RNA template to the VRC, allowing efficient access of the (−)RNA replication intermediate to viral RdRp, whereas (+)RNA progeny (which are not bound by GAPDH) are released from VRC into the cytosol133. In addition, GAPDH stimulates (+)RNA synthesis in vitro by recruiting the TBSV replication protein p92 to the (−)RNA template within the VRC134.
Other host factors that affect RNA synthesis. In addition to the above RBPs, other host factors can affect viral RNA synthesis by regulating VRC activity. For example, Pmr1, an S. cerevisiae Ca2+,Mn2+-ATPase pump, affects the activity of the tombusvirus replicase. Inactivation of PMR1 increases the rate of TBSV RNA recombination and replication53. It has been proposed that the viral replicase uses the far more abundant Mg2+ over Mn2+ when Pmr1 works efficiently (that is, at low cytosolic Mn2+ concentrations), leading to a normal replication rate and low-frequency RNA recombination53. However, inhibiting Pmr1 increases the level of cytosolic Mn2+, promoting more efficient use of Mn2+ by the viral replicase, resulting in an increased replication rate and high-frequency RNA recombination. Host Ca2+,Mn2+-ATPase pumps might also influence other viral RdRps, as Mn2+ is known to affect the activity of several DNA and RNA polymerases in vitro.
Conclusions and future directions
Several common themes are emerging from studies of the functions of host factors in (+)RNA virus replication. Many of the identified host factors are conserved in eukaryotes, suggesting that these viruses selectively target highly conserved host functions. Moreover, a large fraction of the subverted host proteins is unique for a given virus, indicating that (+)RNA viruses have evolved different ways to exploit host cells. However, although the co-opted host proteins are diverse, they might fulfil similar or related functions during viral RNA replication.
Common host factors that are recruited by (+)RNA viruses for their replication include: RBPs that facilitate viral RNA synthesis; proteins involved in membrane bending that contribute to the formation of membrane-bound replication complexes; lipid synthesis enzymes (such as FASN and PI4KIIIβ) that affect lipid composition and have a role in creating a favourable microenvironment for viral replication; and chaperones and PPIases that facilitate the proper folding and functions of viral replication proteins during VRC assembly.
In spite of these recent advances, our current understanding of (+)RNA virus replication and virus–host interactions is far from complete. Further application of proteomics and genome-wide studies will expand this area of research. Importantly, functional and mechanistic studies based on biochemical approaches (such as the use of cell-free extracts and single-molecule techniques) in combination with live-cell imaging will be needed to test all the candidate proteins identified. In vitro reconstitution of VRCs would be extremely useful for the dissection of unknown functions of the host proteins that are co-opted for viral replication. Super-resolution imaging and electron tomography to determine the position and orientation of host proteins within VRCs will advance our understanding of the mechanisms involved in (+)RNA virus replication. In addition, proteomics can be used to study the possible roles of post-translational modifications of viral replication proteins. These advances should lead to a better understanding of viral replication and the interactions between host factors and (+)RNA viruses, which are key aspects of viral pathogenesis. Characterizing the functions of host factors in (+)RNA virus replication might also help develop novel antiviral strategies and potential biotechnological applications of these viruses.
Box 1 | Saccharomyces cerevisiae as a model host.
The use of Saccharomyces cerevisiae as a model organism to study host–virus interactions has several advantages. Plants and most animals have large genomes with extensive genetic redundancy (functional duplications) in various functions, whereas S. cerevisiae has a small genome with only ∼6,000 genes, more than 60% of which have been characterized to some extent. The small genome size also means that there is a reduced level of redundancy. In addition, less than 7% of the genes carry introns, which greatly simplifies prediction of the encoded proteins. Moreover, toolboxes and gene libraries are available for the controlled expression of selected genes. One advantage to performing genome-wide screens in S. cerevisiae is the availability of collections of knockout mutants, such as the yeast single-gene-knockout (YKO) library, or libraries containing gene sets with regulated expression, such as the yTHC library135. Gene libraries with fluorescent tags or affinity tags for identification of the subcellular localization of proteins and for protein purification, respectively, are also available136. Microarray chips with DNA oligonucleotides for most S. cerevisiae genes, and protein arrays with 4,100 purified proteins from this organism, have been produced137. Finally, the databanks for S. cerevisiae genes are the most complete among those for eukaryotes. For example, a global analysis of protein localization has been carried out, and detailed protein–protein interaction networks and genetic interaction maps have been constructed138,139.
Acknowledgements
The authors thank D. Smith and members of the Nagy lab for discussion. The authors apologize to those colleagues whose research on the replication of (+)RNA viruses is not mentioned in this Review owing to space restrictions. This work was supported by the US National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Glossary
- Peroxisomes
Organelles that are found in virtually all eukaryotic cells and are involved in the catabolism of fatty acids, damino acids and polyamines. Peroxisomes are important for energy metabolism.
- Genome-wide approaches
Studies based on high-throughput screens that include a systematic analysis of all or most of the genes encoded by the genome of a particular host organism.
- RNA interference
(RNAi). A pathway that, in most eukaryotes, controls which genes are active and their relative activities. The pathway is induced by small interfering RNAs (siRNAs), which are small double-stranded RNAs. Using a library of siRNAs, the expression of each gene in a given cell line or organism can artificially be knocked down.
- Proteomic approaches
Global approaches that involve all or most of the proteins encoded by the genome of the particular host.
- Yeast two-hybrid screens
Screens looking for proteinprotein or proteinnucleic acid interactions in Saccharomyces cerevisiae cells. The target protein is screened for physical interaction against a library of full-length or truncated genes that are expressed from a plasmid.
- Protein microarray
A microarray based on purified (host) proteins fixed to the chip surface. These arrays are used to identify proteinprotein or proteinnucleic acid interactions, substrates of proteins or targets of biologically active small molecules.
- Cap-independent translation
A method of translation used by several positive-sense RNA viruses, in which the usual requirement for the interaction of host translation initiation factors with the 5-cap (a special tag bound to the 5 end of an mRNA molecule) is circumvented, and select host initiation factors are recruited by alternative means to participate in translation.
- SNARE-like protein
One of a large superfamily of proteins with a primary role in vesicle fusion.
- Geranylgeranylated cellular protein
A protein that has been modified post-translationally by the attachment of one or two 20carbon lipophilic geranylgeranyl isoprene units (from geranylgeranyl diphosphate) to a cysteine at the carboxyterminus of the protein. Geranylgeranylation is proposed to function as a membrane anchor for proteins.
- Actomyosin
A protein complex composed of actin and myosin.
- Viral replication organelles
Large cellular structures that contain many individual viral replication complexes.
- Trans-Golgi network
A complex network of membranes and associated vesicles in the so-called trans-face of the Golgi, which is involved in sorting and shipping proteins to their intended destinations in the cell.
- ER exit sites
Selected areas within the endoplasmic reticulum (ER) from which transport vesicles carrying newly synthesized proteins and lipids bud off for transport to the Golgi apparatus.
- ERGIC
(Endoplasmic reticulumGolgi intermediate compartment). An organelle in eukaryotic cells that mediates trafficking between the ER and the Golgi, facilitating the sorting of cargo.
- Autophagy
A tightly regulated catabolic process involving the degradation of cellular components through the lysosomal machinery.
- Stress granule
A type of dense proteinRNA aggregation in the cytosol, not surrounded by membrane, that appears when the cell is under stress. Stress granules may store and protect RNAs from harmful conditions under stress, and they serve as decision points for untranslated mRNAs for further storage, degradation or translation re-initiation.
- P-bodies
(Processing bodies). Distinct foci within the cytoplasm, consisting of enzymes involved in mRNA turnover.>
Biographies
Peter D. Nagy is a full professor at the University of Kentucky, Lexington, USA. His research interests are identifying and dissecting the functions of the host factors involved in the replication of RNA viruses and RNA recombination. His laboratory developed tombusvirus replication and recombination systems in the model host Saccharomyces cerevisiae. His laboratory also studies the assembly of the viral replication complex and virus–host interactions that lead to the host cells being reprogrammed into viral replication factories.
Judit Pogany received her Ph.D. in 1997 for studies on the formation of defective RNAs in bromoviruses. She is currently a staff scientist at the University of Kentucky, USA. She has developed a unique cell-free assay based on yeast extracts to support complete replication of tomato bushy stunt virus. She is currently studying the roles of recruited host chaperones in tombusvirus replication.
Related links
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
References
- 1.Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagy PD. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 2008;46:217–242. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- 3.Novoa RR, et al. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nature Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 6.Buck KW. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- 8.Nagy PD, Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344:211–220. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Shi ST, Lai MM. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strauss JH, Strauss EG. Viral RNA replication. With a little help from the host. Science. 1999;283:802–804. doi: 10.1126/science.283.5403.802. [DOI] [PubMed] [Google Scholar]
- 11.Spuul P, et al. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J. Virol. 2011;85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spuul P, Balistreri G, Kaariainen L, Ahola T. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 2010;84:7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy PD, Pogany J. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv. Virus Res. 2010;76:123–177. doi: 10.1016/S0065-3527(10)76004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison MR. Seeking membranes: positive-strand RNA virus replication complexes. PLoS Biol. 2008;6:e270. doi: 10.1371/journal.pbio.0060270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherry S, et al. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger D, Bienz K. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 2005;86:707–718. doi: 10.1099/vir.0.80442-0. [DOI] [PubMed] [Google Scholar]
- 18.Kirkegaard K, Jackson WT. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy. 2005;1:182–184. doi: 10.4161/auto.1.3.2065. [DOI] [PubMed] [Google Scholar]
- 19.Rust RC, et al. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 2001;75:9808–9818. doi: 10.1128/JVI.75.20.9808-9818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinton MA. Host factors involved in West Nile virus replication. Ann. NY Acad. Sci. 2001;951:207–219. doi: 10.1111/j.1749-6632.2001.tb02698.x. [DOI] [PubMed] [Google Scholar]
- 22.Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Serviene E, Gal J, Panavas T, Nagy PD. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 2006;80:7394–7404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panavas T, Serviene E, Brasher J, Nagy PD. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl Acad. Sci. USA. 2005;102:7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serviene E, Jiang Y, Cheng CP, Baker J, Nagy PD. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 2006;80:1231–1241. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng CP, Serviene E, Nagy PD. Suppression of viral RNA recombination by a host exoribonuclease. J. Virol. 2006;80:2631–2640. doi: 10.1128/JVI.80.6.2631-2640.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serviene E, et al. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl Acad. Sci. USA. 2005;102:10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kushner DB, et al. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl Acad. Sci. USA. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherry S, et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng TI, et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413–1421. doi: 10.1002/hep.21608. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl Acad. Sci. USA. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supekova L, et al. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J. Biol. Chem. 2008;283:29–36. doi: 10.1074/jbc.M703988200. [DOI] [PubMed] [Google Scholar]
- 36.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl Acad. Sci. USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaillancourt FH, et al. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5–10. doi: 10.1016/j.virol.2009.02.039. [DOI] [PubMed] [Google Scholar]
- 38.de Chassey B, et al. Hepatitis C virus infection protein network. Mol. Syst. Biol. 2008;4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li HP, Zhang X, Duncan R, Comai L, Lai MM. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl Acad. Sci. USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanda SK, Leibowitz JL. Mitochondrial aconitase binds to the 3′ untranslated region of the mouse hepatitis virus genome. J. Virol. 2001;75:3352–3362. doi: 10.1128/JVI.75.7.3352-3362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spagnolo JF, Hogue BG. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/JVI.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, et al. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007;367:212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Frolova E, et al. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, et al. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl Acad. Sci. USA. 2007;104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, et al. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385:245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger KL, Randall G. Potential roles for cellular cofactors in hepatitis C virus replication complex formation. Commun. Integr. Biol. 2009;2:471–473. doi: 10.4161/cib.2.6.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe. 2009;5:318–328. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 51.Diaz A, Wang X, Ahlquist P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl Acad. Sci. USA. 2010;107:16291–16296. doi: 10.1073/pnas.1011105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barajas D, Jiang Y, Nagy PD. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5:e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaag HM, Pogany J, Nagy PD. A host Ca2+/Mn2+ ion pump is a factor in the emergence of viral RNA recombinants. Cell Host Microbe. 2010;7:74–81. doi: 10.1016/j.chom.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 54.Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamarnik AV, Andino R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 2000;74:2219–2226. doi: 10.1128/JVI.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osman TA, Buck KW. Identification of a region of the tobacco mosaic virus 126- and 183-kilodalton replication proteins which binds specifically to the viral 3′-terminal tRNA-like structure. J. Virol. 2003;77:8669–8675. doi: 10.1128/JVI.77.16.8669-8675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, et al. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 2005;79:13747–13758. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Wynsberghe PM, Ahlquist P. 5′ cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J. Virol. 2009;83:2976–2988. doi: 10.1128/JVI.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79:4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter BL, Parsley TB, Ehrenfeld E, Semler BL. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002;76:12008–12022. doi: 10.1128/JVI.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogt DA, Andino R. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 2010;6:e1000936. doi: 10.1371/journal.ppat.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamanaka T, et al. Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J. Virol. 2002;76:2491–2497. doi: 10.1128/jvi.76.5.2491-2497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamanaka T, et al. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl Acad. Sci. USA. 2000;97:10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tu H, et al. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 67.Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324:450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 69.Wang C, et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Wei T, et al. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 2010;84:799–809. doi: 10.1128/JVI.01824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cotton S, et al. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 2009;83:10460–10471. doi: 10.1128/JVI.00819-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beckham CJ, et al. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pathak KB, Sasvari Z, Nagy PD. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379:294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 74.Wang RY, Stork J, Pogany J, Nagy PD. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology. 2009;394:28–38. doi: 10.1016/j.virol.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang RY, Stork J, Nagy PD. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 2009;83:3276–3287. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jonczyk M, Pathak KB, Sharma M, Nagy PD. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362:320–330. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 77.Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338:81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 78.Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwartz M, et al. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 82.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belov GA, Ehrenfeld E. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle. 2007;6:36–38. doi: 10.4161/cc.6.1.3683. [DOI] [PubMed] [Google Scholar]
- 84.Belov GA, Habbersett C, Franco D, Ehrenfeld E. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J. Virol. 2007;81:9259–9267. doi: 10.1128/JVI.00840-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berger KL, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl Acad. Sci. USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borawski J, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reiss S, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Amako Y, Sarkeshik A, Hotta H, Yates J, 3rd, Siddiqui A. Role of oxysterol binding protein in hepatitis C virus infection. J. Virol. 2009;83:9237–9246. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reggiori F, et al. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heaton NS, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl Acad. Sci. USA. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 2003;77:12819–12828. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navarro B, Russo M, Pantaleo V, Rubino L. Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication. J. Gen. Virol. 2006;87:705–714. doi: 10.1099/vir.0.81325-0. [DOI] [PubMed] [Google Scholar]
- 94.Sharma M, Sasvari Z, Nagy PD. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J. Virol. 2010;84:2270–2281. doi: 10.1128/JVI.02003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bloch K. Sterol molecule: structure, biosynthesis, and function. Steroids. 1992;57:378–383. doi: 10.1016/0039-128X(92)90081-J. [DOI] [PubMed] [Google Scholar]
- 96.Bloch KE. Sterol structure and membrane function. CRC Crit. Rev. Biochem. 1983;14:47–92. doi: 10.3109/10409238309102790. [DOI] [PubMed] [Google Scholar]
- 97.Chang KO. Role of cholesterol pathways in norovirus replication. J. Virol. 2009;83:8587–8595. doi: 10.1128/JVI.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothwell C, et al. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology. 2009;389:8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 99.Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 100.Sagan SM, et al. The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem. Cell Biol. 2006;84:67–79. doi: 10.1139/o05-149. [DOI] [PubMed] [Google Scholar]
- 101.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl Acad. Sci. USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nohturfft A, Zhang SC. Coordination of lipid metabolism in membrane biogenesis. Annu. Rev. Cell Dev. Biol. 2009;25:539–566. doi: 10.1146/annurev.cellbio.24.110707.175344. [DOI] [PubMed] [Google Scholar]
- 103.Palomares-Jerez MF, Villalain J. Membrane interaction of segment H1 (NS4BH1) from hepatitis C virus non-structural protein 4B. Biochim. Biophys. Acta. 2011;1808:1219–1229. doi: 10.1016/j.bbamem.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 104.Stapleford KA, Rapaport D, Miller DJ. Mitochondrion-enriched anionic phospholipids facilitate flock house virus RNA polymerase membrane association. J. Virol. 2009;83:4498–4507. doi: 10.1128/JVI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahola T, Lampio A, Auvinen P, Kaariainen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma M, Sasvari Z, Nagy PD. Inhibition of phospholipid biosynthesis decreases the activity of the tombusvirus replicase and alters the subcellular localization of replication proteins. Virology. 2011;145:141–152. doi: 10.1016/j.virol.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Castorena KM, Stapleford KA, Miller DJ. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics. 2010;11:183. doi: 10.1186/1471-2164-11-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 109.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nature Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 111.Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 112.Barajas D, Nagy PD. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology. 2009;397:358–368. doi: 10.1016/j.virol.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pogany J, Stork J, Li Z, Nagy PD. In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc. Natl Acad. Sci. USA. 2008;105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Serva S, Nagy PD. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 2006;80:2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Castorena KM, Weeks SA, Stapleford KA, Cadwallader AM, Miller DJ. A functional heat shock protein 90 chaperone is essential for efficient Flock House virus RNA polymerase synthesis in Drosophila cells. J. Virol. 2007;81:8412–8420. doi: 10.1128/JVI.00189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kampmueller KM, Miller DJ. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 2005;79:6827–6837. doi: 10.1128/JVI.79.11.6827-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weeks SA, et al. A targeted analysis of cellular chaperones reveals contrasting roles for heat shock protein 70 in flock house virus RNA replication. J. Virol. 2010;84:330–339. doi: 10.1128/JVI.01808-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tomita Y, et al. Mutation of host dnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 2003;77:2990–2997. doi: 10.1128/JVI.77.5.2990-2997.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yi Z, et al. Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS Pathog. 2011;7:e1001255. doi: 10.1371/journal.ppat.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Okamoto T, et al. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kaul A, et al. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watashi K, et al. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 123.Li Z, et al. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 2010;6:e1001175. doi: 10.1371/journal.ppat.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davis WG, Blackwell JL, Shi PY, Brinton MA. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 2007;81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kidmose RT, Vasiliev NN, Chetverin AB, Andersen GR, Knudsen CR. Structure of the Qβ replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl Acad. Sci. USA. 2010;107:10884–10889. doi: 10.1073/pnas.1003015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takeshita D, Tomita K. Assembly of Qβ viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl Acad. Sci. USA. 2010;107:15733–15738. doi: 10.1073/pnas.1006559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barton DJ, O'Donnell BJ, Flanegan JB. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang P, Lai MMC. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brunner JE, et al. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 2005;79:3254–3266. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl Acad. Sci. USA. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta. 1999;1432:159–184. doi: 10.1016/S0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 133.Wang RY, Nagy PD. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe. 2008;3:178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 134.Huang TS, Nagy PD. Direct inhibition of tombusvirus plus-strand RNA synthesis by a dominant-negative mutant of a host metabolic enzyme, GAPDH, in yeast and plants. J. Virol. 2011;85:9090–9102. doi: 10.1128/JVI.00666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 136.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 137.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 138.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 139.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 140.Welsch SS, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host. Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Aizaki H, Choi KS, Liu M, Li YJ, Lai MM. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 2006;13:469–480. doi: 10.1007/s11373-006-9088-4. [DOI] [PubMed] [Google Scholar]
- 142.Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl Acad. Sci. USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Scheller N, et al. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl Acad. Sci. USA. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Choi KS, Mizutani A, Lai MM. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 2004;78:13153–13162. doi: 10.1128/JVI.78.23.13153-13162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim CS, Seol SK, Song OK, Park JH, Jang SK. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J. Virol. 2007;81:3852–3865. doi: 10.1128/JVI.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


