Key Points
RNA interference (RNAi) is a powerful approach for reducing expression of endogenously expressed proteins for biological applications, or targeting the expression of pathological proteins for therapy.
Several delivery methods are available to achieve RNAi in ex vivo and in vivo settings for therapeutic results.
The development of RNAi-based therapeutics has advanced sufficiently to allow human clinical trials to begin.
Here we outline the broad range of cell-, tissue- and disease-specific approaches under investigation for RNAi therapeutics.
The barriers posed by certain cells and tissues are described, as are issues with off-target silencing.
Subject terms: RNAi, Gene therapy, Technology, miRNAs, Disease genetics, Nucleic-acid therapeutics
RNA interference can elicit specific gene silencing and so holds great potential for treating infectious or genetic diseases. Several small-RNA-based therapies have now reached clinical trials, but further work is still needed to improve delivery and efficacy.
Abstract
RNA interference (RNAi) is a powerful approach for reducing expression of endogenously expressed proteins. It is widely used for biological applications and is being harnessed to silence mRNAs encoding pathogenic proteins for therapy. Various methods — including delivering RNA oligonucleotides and expressing RNAi triggers from viral vectors — have been developed for successful RNAi in cell culture and in vivo. Recently, RNAi-based gene silencing approaches have been demonstrated in humans, and ongoing clinical trials hold promise for treating fatal disorders or providing alternatives to traditional small molecule therapies. Here we describe the broad range of approaches to achieve targeted gene silencing for therapy, discuss important considerations when developing RNAi triggers for use in humans, and review the current status of clinical trials.
Main
RNA interference (RNAi) is a natural cellular process that regulates gene expression and provides an innate defence mechanism against invading viruses and transposable elements1. The finding that dsRNA initiates RNAi was among the most significant recent contributions to cell biology2, and since the discovery that RNAi can be mediated by 21 nucleotide (nt) duplexes3, researchers have worked to harness their potential for addressing biological questions and treating human disease. Some reagents, such as small interfering RNAs (siRNAs), are applied directly to cells, tissues and organisms; others are engineered to be expressed in cells, such as hairpin structures that provide siRNAs when processed. The basic premise underlying the broad utility of RNAi is that, in theory, we can design siRNAs (or vectors encoding them) to target virtually any gene of interest. RNAi technologies use a cell's natural machinery to move exogenously applied siRNAs to the appropriate cellular compartment, where they encounter the correct mRNA target and induce its degradation.
Initial work on RNAi in flies and worms moved quickly to larger mammals and fuelled excitement for potential clinical applications. However, in a similar way to other developing fields in human therapy, such as gene- and antibody-therapy, early excitement has been tempered as a realistic understanding emerges of the milestones that must be reached before the eventual approval of human therapy. Over recent years various complex barriers to achieving efficient RNAi have become evident. These hurdles include: specificity for the target gene; delivery to the correct cell or tissues; the durability of RNAi activity and the ability to redose (if needed); and considerations of the stability of the target mRNA and encoded protein. We have also become aware of the problems posed by the various platforms used to elicit RNAi. However, with setbacks come opportunities. For example, early work in which siRNAs were applied to mouse airway epithelial cells to reduce viral burdens in vivo elicited inhibition of target gene expression4, but it was later found that the induction of an innate immune response probably contributed to the efficacy in these and other experiments5,6. Altering the chemical make-up of the synthetic RNA diminished the immune response, as did avoiding known pro-inflammatory sequences7,8. This finding also opened investigators' eyes to the possibility of purposefully using immunostimulatory RNAi as a direct therapeutic or adjuvant9.
Although the clinical utility of RNAi has not yet been realized, ongoing patient trials provide opportunities for success. The numbers of RNAi-based preclinical and clinical trials have grown over the past several years and have included studies in retinal degeneration, dominantly inherited brain and skin diseases, viral infections, respiratory disorders, cancer and metabolic diseases (Table 1).
Table 1.
Clinical trials for RNAi therapy*
| Clinical setting | Drug | Indication(s) | Target(s) | Sponsor | Status |
|---|---|---|---|---|---|
| Ocular and retinal disorders | TD101 | Pachyonychia congenita | Keratin 6A N171K mutant | Pachyonychia Congenita Project | Completed, Phase I |
| QPI-1007 | Non-arteritic anterior ischaemic optic neuropathy | Caspase 2 | Quark Pharm., Inc. | Active, Phase I | |
| AGN211745 | Age-related macular degeneration; choroidal neovascularization | VEGFR1 | Sirna Therapeutics, Inc. | Completed, Phase I, II | |
| PF-655 | Diabetic macular oedema (DME); age-related macular degeneration (AMD) | RTP801 | Quark Pharm., Inc. | Active, Phase I | |
| SYL040012 | Glaucoma | β2 adrenergic receptor | Sylentis | Active, Phase I, II | |
| Bevasiranib | Diabetic macular oedema | VEGF | Opko Health, Inc. | Completed, Phase II | |
| Bevasiranib | Macular degeneration | VEGF | Opko Health, Inc. | Completed, Phase II | |
| Cancer | CEQ508 | Familial adenomatous polyposis | β-catenin | MDRNA, Inc. | Active, Phase I |
| ALN-PLK1 | Liver tumours | PLK1 | Alnyam Pharm. | Active, Phase I | |
| FANG | Solid tumours | Furin | Gradalis, Inc. | Active, Phase II | |
| CALAA-01 | Solid tumours | RRM2 | Calando Pharm. | Active, Phase I | |
| SPC2996 | Chronic myeloid leukemia | BCL-2 | Santaris Pharm. | Ongoing, Phase I, II | |
| ALN-VSP02 | Solid tumours | VEGF, kinesin spindle protein | Alnylam Pharm. | Active, Phase I | |
| NCT00672542 | Metastatic melanoma | LMP2, LMP7, and MECL1 | Duke University | Active, Phase I | |
| Atu027 | Advanced, recurrent or metastatic solid malignancies | PKN3 | Silence Therapeutics | Active, Phase I | |
| Kidney disorders | QPI-1002/I5NP | Acute kidney injury | p53 | Quark Pharm., Inc. | Terminated, Phase I |
| QPI-1002/I5NP | Delayed graft function kidney transplant | p53 | Quark Pharm., Inc. | Active, Phase I, II | |
| QPI-1002/I5NP | Kidney injury acute renal failure | p53 | Quark Pharm., Inc. | Completed, Phase I | |
| LDL lowering | TKM-ApoB | Hypercholesterolaemia | APOB | Tekmira Pharm. Corp. | Terminated, Phase I |
| PRO-040,201 | Hypercholesterolaemia | APOB | Tekmira Pharm. Corp. | Terminated, Phase I | |
| Antiviral | SPC3649 | Hepatitis C virus | miR-122 | Santaris Pharm | Active, Phase II |
| pHIV7-shI-TAR-CCR5RZ | HIV | HIV Tat protein, HIV TAR RNA, human CCR5 | City of Hope Medical Center/Benitec | Active, Phase 0 | |
| ALN-RSV01 | RSV in volunteers | RSV nucleocapsid | Alnylam Pharm. | Completed, Phase II | |
| ALN-RSV01 | RSV in lung transplant patients | RSV nucleocapsid | Alnylam Pharm. | Completed, Phase I | |
| ALN-RSV01 | RSV in lung transplant patients | RSV nucleocapsid | Alnylam Pharm. | Active, Phase II | |
| APOB, apolipoprotein B; BCL-2, B-cell CLL/lymphoma 2; CCR5, C-C chemokine receptor type 5; LDL, low-density lipoprotein; LMP2, also known as proteasome subunit beta type 9 (PSMB9); LMP7, also known as proteasome subunit beta type 8 (PSMB8); MECL1, also known as proteasome subunit beta type 10 (PSMB10); Pharm., Pharmaceuticals; PKN3, protein kinase N3; PLK1, polo-like kinase 1; RRM2, ribonucleoside-diphosphate reductase subunit M2; RSV, respiratory syncytial virus; RTP801, also known as DNA damage-inducible transcript 4 protein (DDIT4); VEGF, vascular endothelial growth factor. | |||||
| *From ClinicalTrials.gov. | |||||
Here, we provide an overview of RNAi and discuss strategies to use the pathway for directed gene silencing for therapy. We describe delivery systems that might be suitable for different circumstances, and bring to the reader's attention issues that must be surmounted for widespread use in vivo.
Harnessing small RNA biogenesis
The development of RNAi for therapy is based on our understanding of small RNA biogenesis pathways. The two main types of small RNAs involved in gene silencing are microRNAs (miRNAs) and siRNAs, and their processing and targeting is summarized in Fig. 1 (further details can be found in recent reviews10,11,12).
Figure 1. The miRNA and siRNA pathways of RNAi in mammals.
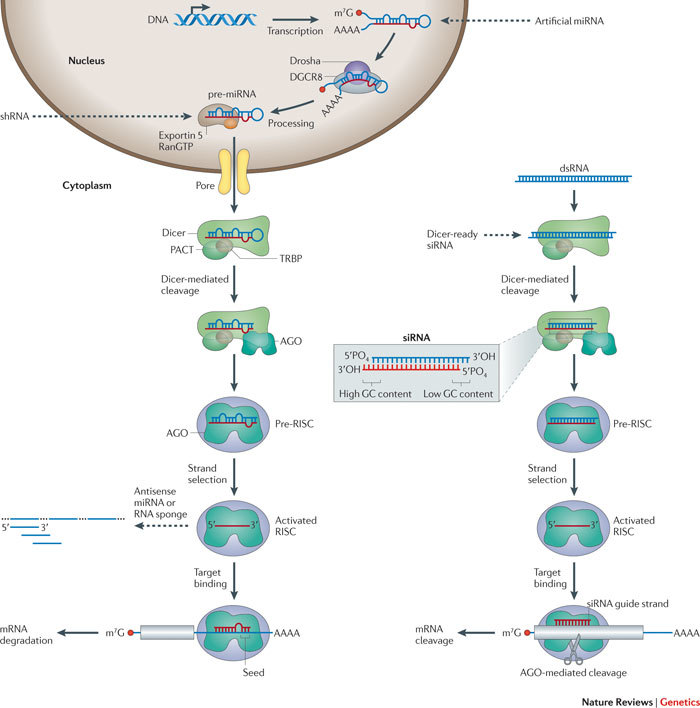
Primary microRNAs (pri-miRNAs) are transcribed by RNA polymerases156,157,158 and are trimmed by the microprocessor complex (comprising Drosha and microprocessor complex subunit DCGR8) into ~70 nucleotide precursors, called pre-miRNAs67,159,160 (left side of the figure). miRNAs can also be processed from spliced short introns (known as mirtrons)161. pre-miRNAs contain a loop and usually have interspersed mismatches along the duplex. pre-miRNAs associate with exportin 5 and are exported to the cytoplasm162,163, where a complex that contains Dicer, TAR RNA-binding protein (TRBP; also known as TARBP2) and PACT (also known as PRKRA) processes the pre-miRNAs into miRNA–miRNA* duplexes116,164,165. The duplex associates with an Argonaute (AGO) protein within the precursor RNAi-induced silencing complex (pre-RISC). One strand of the duplex (the passenger strand) is removed. The mature RISC contains the guide strand, which directs the complex to the target mRNA for post-transcriptional gene silencing. The 'seed' region of an miRNA is indicated; in RNAi trigger design, the off-target potential of this sequence needs to be considered. Long dsRNAs (right side of the figure) are processed by Dicer, TRBP and PACT into small interfering RNAs (siRNAs). siRNAs are 20–24-mer RNAs and harbour 3′OH and 5′ phosphate (PO4) groups, with 3′ dinucleotide overhangs3,166,167. Within the pre-RISC complex, an AGO protein cleaves the passenger siRNA strand. Then, the mature RISC, containing an AGO protein and the guide strand, associates with the target mRNA for cleavage. The inset shows the properties of siRNAs. The thermodynamic stability of the terminal sequences will direct strand loading. Like naturally occurring or artificially engineered miRNAs, the potential 'seed' region can be a source for miRNA-like off-target silencing. shRNA, short hairpin RNA.
miRNAs and siRNAs. miRNAs mediate post-transcriptional gene silencing and are processed from endogenously expressed transcripts (Fig. 1). Either processed strand can mediate post-transcriptional gene silencing, but many miRNAs show asymmetry, primarily loading one strand into the RNA-induced silencing complex (RISC). The small RNA guides RISC to the mRNA target, where the miRNA typically binds to the 3′ UTR. Watson–Crick base pairing between miRNAs and their targets is usually partial, but with high complementarity from bases 2–8 of the miRNA, which is known as the 'seed' region13. Recent data suggest that base pairing can also occur between central miRNA nucleotides and target mRNAs14. Data from several laboratories showed that miRNAs repress the initiation of translation15,16,17, although more recent work indicates that miRNA–mRNA complexes can be transported to cytoplasmic processing bodies18, after which deadenylation and mRNA degradation occurs14,19,20. Interestingly, some miRNA-mediated translational repression is reversible21.
siRNAs are small dsRNAs, 20–24 nt in length, that are processed from longer dsRNAs (Fig. 1). One strand is the 'guide' strand and directs silencing, with the other strand — the 'passenger' — being degraded22,23. Which strand becomes which is determined by the thermodynamic properties of the duplex24,25,26,27. siRNAs generally show full complementarity to their target mRNA, and cleavage occurs 10–12 bases from the 5′ end of the guide strand binding site3,28.
Exogenous inhibitory RNAs. Our understanding of small RNA biogenesis has enabled the development of several strategies for harnessing RNAi pathways for therapy. Recombinant inhibitory RNAs are designed to mimic primary miRNAs (pri-miRNAs) (in the case of artificial miRNAs or exogenous miRNAs) or precursor miRNAs (pre-miRNAs) (in the case of short hairpin RNAs (shRNAs)), whereas chemically synthesized RNA oligonucleotides are designed to mimic Dicer products or substrates. Each class mediates gene silencing but enters the pathway at a different step (Fig. 1). The main differences between exogenously applied oligonucleotide siRNAs and hairpin-based species (shRNA or miRNA shuttles) are the mode of delivery and the duration of gene silencing (Table 2). However, recent advances in non-viral and viral systems are blurring this distinction. In the following sections we describe the main strategies for the design and delivery of inhibitory RNAs.
Table 2.
Methods for the delivery of RNAi triggers to cells and tissues
| Species/formulation | Packaging capacity | Applications and considerations | Refs* |
|---|---|---|---|
| Viral vector | |||
| Adenovirus | Up to ~35 kb, usually <10 kb | dsDNA vector with large packaging capacity, transient expression, highly immunogenic | 76,77 |
| Adeno-associated virus (AAV) | ~4.5 kb | ssDNA vector, small packaging capacity, mildly immunogenic, lasting expression in nondividing cells, capsid pseudotyping/engineering facilitates specific cell-targeting | 82,91,103,108 |
| Lentivirus | Up to 13.5 kb (larger inserts will decrease titre) | RNA vector, integration competent and incompetent forms available, less immunogenic than adenovirus or AAV, envelope pseudotyping facilitates cell targeting, clinical production more difficult than for adenovirus or AAV | 83,84,85,86,87,88,140,155 |
| Herpes simplex virus | 150kb | DNA vector, episomal, lasting expression, immunogenic | 119 |
| Bacterial vector species ‡ | |||
| Escherichia coli, S. Typhymurium§ | Delivery of short hairpin RNA or small interfering RNA to gut tissue | 73,74,75 | |
| Non-viral formulations || | |||
| Nanoparticle | Self-assembling, may target specific receptors, requires technical expertise to prepare | 59 | |
| Stable nucleic acid lipid particle (SNALP) | Stable for systemic delivery, broad cell-type delivery | 51 | |
| Aptamer | Targeting of specific receptors, requires sophisticated screening to develop | 53 | |
| Cholesterol | Stable for systemic delivery, broad cell-type delivery | 46 | |
| *Representative references. | |||
| ‡Bacterial minicells can carry plasmids, short interfering RNAs or drugs. | |||
| §Salmonella enterica subsp. enterica serovar Typhimurium. | |||
| ||The nucleic acids in non-viral carriers can be any size from small oligonucleotides to large artificial chromosomes. | |||
siRNA approaches
The most common method used to harness the RNAi pathway for targeted gene silencing is to transfect 21–22 nt siRNAs into cells. Another option is to use longer, 25–27 nt duplexes that can be processed by Dicer into siRNAs; these are called 'Dicer-ready siRNAs'. In some cases, the silencing potency of Dicer-ready siRNAs can be greater than for siRNAs29,30. For both synthetic triggers, transfection is generally accomplished to high efficiency in cell lines using commercially available transfection reagents. However, as discussed below, alternative packaging is often required for delivery to primary cells and for in vivo applications.
In rational siRNA design it is important to consider the siRNA sequence, the chemical nature of the silencing moiety (for example, RNA with or without modified bases and sugars), the length of the RNA and the nature of the 5′ and 3′ ends. In vitro synthesis of siRNAs using T7 polymerase creates 5′ triphosphates, which can induce type I interferon responses (type I IFN responses)1. Similarly, blunt-ended siRNAs induce cytoplasmic retinoic acid inducible gene 1 protein (RIG1; also known as DDX58) and IFN production. Chemically synthesized siRNAs lacking 5′ triphosphates and containing appropriate 3′ overhangs alleviate these issues.
Many siRNAs, although able to reduce expression of the target gene, are immunostimulatory in a sequence-independent manner because they are recognized by the pattern recognition Toll-like receptors (TLRs)31. TLR3, which can be endosomal or on the cell surface, recognizes dsRNAs and can be activated by uncomplexed 21-mer siRNAs. TLR3 activation inhibits blood and lymphatic vessel growth32,33, which can be advantageous in the setting of corneal vascularization, where inhibition of angiogenesis is desired34. TLR activation can also be advantageous in cancer therapies by stimulating dendritic cells to respond immunologically to cancer cells35. Recently, the generally unwanted stimulation of TLRs by oligonucleotides was used cleverly to achieve gene silencing and immune stimulation for cancer therapy: well-characterized CpG oligonucleotide agonists of TLR9 fused to siRNAs targeting an immune suppressor promoted antitumour immune responses in mice36. This interesting combination of tumour targeting and siRNA immunostimulatory therapy may substantially augment the promising clinical results from the use of TLR9 agonists alone37.
TLR activation would be contraindicated in other settings, such as in attempts to revert or inhibit ischaemia. In addition, endosomal TLR7 and TLR8 recognize ssRNAs and can be activated when siRNAs complexed with carriers are internalized or taken up through receptor targeting. Altering the chemical nature of the siRNA dramatically reduces TLR responses elicited by exogenously applied siRNAs. For example, using 2′-O-methyl-modified purine nucleosides in the passenger strand reduces IFN induction but retains targeting specificity38,39,40. This modification also improves serum stability by reducing susceptibility to RNases41.
siRNA delivery options. Chemically modified siRNAs are most often packaged into carriers for systemic delivery as their negative charge and size prevent cellular penetration. Uncomplexed siRNAs that are delivered systemically are also readily cleared by the kidney and excreted. The array of carriers is vast, and excellent summaries of their chemical make-up and biological properties can be found elsewhere42,43,44,45. Among the most common are lipid-based carriers or cholesterol conjugates to the sense strand of the duplex. Cholesterol-conjugated siRNAs, which are commercially available, enable improved uptake to the liver as they are bound by low-density lipoprotein (LDL) in serum and LDL uptake in the liver is robust46. Lipophilic siRNAs can also bind high-density lipoprotein (HDL); this can target siRNAs to tissues with HDL receptors, such as gut47, kidney47 and vaginal48 epithelial cells and oligodendrocytes in the brain49.
Exciting data in nonhuman primates showed that a single delivery of siRNAs complexed into stable nucleic acid lipid particles (SNALPs) reduced target gene expression for almost 2 weeks50. Recently the same delivery strategy was used successfully to protect nonhuman primates from a lethal challenge of Ebola virus, using siRNAs targeting the expression of three Ebola virus proteins51. Improvements in SNALPs that reduce the doses required for effective silencing in nonhuman primates by ~tenfold will augment the clinical utility of these reagents40.
Complexing siRNAs with carriers also provides opportunities for targeting specific cells or, in the case of cancer, tumour beds. In the first in-human study, nanoparticles designed for enhanced uptake to cancer cells by using transferrin-receptor-targeting ligands showed reduced levels of the target mRNA and evidence for siRNA-mediated cleavage of that target52. As the study continues, it will be interesting to learn the pharmacokinetics of the siRNAs in additional patients and to see the clinical effects of the therapy. Other carriers for tissue-specific targeting include aptamers53,54, antibodies55,56,57,58, peptides and proteins52,59, and oligonucleotide agonists36. Some are synthetically linked to the siRNAs, as in the case of oligonucleotides, peptides and aptamers, whereas others are part of more complex carrier systems (reviewed in Refs 42,60).
Expression systems for RNAi triggers
shRNAs and artificial miRNAs. Expression of shRNAs or artificial miRNAs is achieved by delivering plasmids or by using bacterial or viral vectors. These RNAi triggers are transcribed as sense and antisense sequences connected by a loop of unpaired nucleotides to mimic pre-miRNAs (for shRNAs) or pri-miRNAs (for artificial miRNAs). Following transcription, artificial miRNAs are processed by the Drosha–DGCR8 complex and exported to the cytoplasm, where they engage the pre-RISC complex via the normal miRNA biogenesis pathway (Fig. 1). After cleavage of the passenger (non-guide) strand, the guide strand directs RISC to the mRNA target.
Although the goal for shRNAs is to make a pre-miRNA mimic, shRNA transcripts often do not reflect Drosha cleavage products61,62. If they do not contain the typical 3′ dinucleotide overhang, transport by exportin 5 will be impaired. Reduced shRNA export could cause nuclear accumulation and toxicity63,64. Alternatively, if exportin 5 recognizes the duplexes but shRNA expression is exceptionally high relative to endogenous miRNAs, exportin 5 can become saturated. Overexpression of exportin 5 can alleviate the blockage, but saturation of downstream processing pathways can occur. One method to alleviate saturation of exportin 5 and Argonaute proteins is to overexpress them concomitantly with shRNAs65. However, for therapeutic purposes it might be more appropriate to use weaker promoters61,66 or artificial miRNAs64,67,68,69,70. As miRNA hairpins can be embedded in larger transcripts, the artificial miRNA approach is more suited than shRNAs to RNA polymerase II-based expression systems that provide tissue-specific and inducible gene silencing. The products of RNAi expression systems need to be assessed carefully to ascertain whether the desired strand is loaded into RISC and how well the RNAs are expressed and processed (Fig. 2).
Figure 2. Workflow for testing therapeutic RNAi triggers.
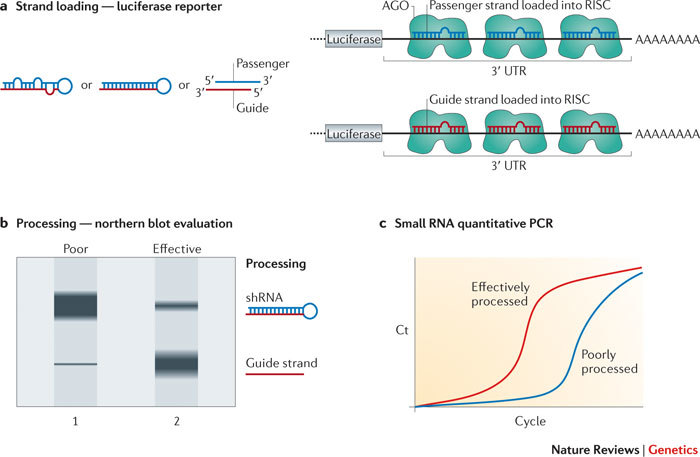
a | Cartoon depicting a luciferase reporter system that is used to confirm that the appropriate strand of small interfering RNAs (siRNAs) or stem–loop platforms from RNAi expression systems is loaded into the RNA-induced silencing complex (RISC). A plasmid with a luciferase reporter that harbours sequences complementary to the guide strand in the 3′ UTR is cotransfected with the RNAi system, and if the appropriate guide strand is loaded, luciferase activity will diminish. When a reporter that contains sequences complementary to the passenger strand is cotransfected, luciferase activity should not be reduced. Because silencing is based on a microRNA (miRNA)-like mechanism, inhibition of luciferase activity will indicate RISC loading, independent of the sequence's ability to induce target cleavage. b | Northern blot analysis can be used to evaluate RNAi triggers expressed from vectors. If the short hairpin RNA (shRNA) or primary miRNA (pri-miRNA) mimics are poorly processed but expressed efficiently, build-up of shRNAs may occur (lane 1). Appropriate processing should yield readily detectable mature, processed siRNAs with minimal levels of unprocessed material (lane 2). Northern blots with probes for the passenger strand can also be used to assess RISC loading of the unintended strand (not shown). c | Small RNA quantitative PCR to quantify the mature product will yield information about overall levels of mature product, which is important to know to understand dosing. The figure shows an example of results obtained from effectively or poorly processed RNA precursors. Cloning and sequencing of the mature small RNAs can be used to assess the silencing RNAs in more detail (not shown). Ct, threshold cycle.
Delivery systems: non-viral vectors. RNAi trigger-expressing plasmids can be packaged into many of the same carriers that can be used for siRNA delivery, although the nature of the particle will change with the different cargo (large DNAs versus small RNAs). Several non-viral platforms for gene delivery are being investigated (reviewed in Ref. 71) and, as for siRNAs, commercial reagents are available for plasmid transfection of cell lines and some primary cells in vitro.
Bacteria can be used as an innovative platform for RNAi delivery; this approach is built on earlier work showing that therapeutic bacteria can enter tumours in patients with cancer72, presumably via a permissive vasculature. The basic premise is that recombinantly engineered Escherichia coli can enter mammalian cells after in vivo delivery and transfer shRNAs. When bacteria containing plasmids that expressed shRNAs targeting β-catenin were fed to mice, β-catenin expression was reduced in the intestinal epithelium73. This platform is now in clinical testing for familial adenomatous polyposis, an inherited form of colon cancer74. A related approach in mice used minicells derived from Salmonella enterica subsp. enterica serovar Typhimurium and targeted them to tumour-cell-surface receptors to deliver siRNAs or shRNAs; this method reduced tumour burden and improved mouse survival75.
Delivery systems: viral vectors. The viral vectors used to deliver shRNAs or artificial miRNAs include murine oncoretroviruses, lentiviruses, adenoviruses, adeno-associated viruses (AAVs), and herpesviruses, among others (Table 2). We refer the reader to recent reviews describing the biology and production of these vector systems76,77,78,79,80. Here we highlight major differences among some of the tools that are relevant to their use for RNAi-based applications, with respect to their tissue tropisms and the fate of their recombinant genomes in host cells.
The genomes of recombinant adenoviruses and AAVs generally remain episomal after the virus has entered the host cell, the viral capsid has been uncoated and the nucleocapsid has been transported to the cell nucleus. An advantage of vector systems with genomes that remain episomal is that insertional mutagenesis is avoided. However, unless only transient expression is desired, their use is limited to cells that divide slowly (for example, some epithelial cells) or not at all (for example, neurons) because the episomal genome will be lost following cell divisions. Naturally occurring differences in capsid structures among adenoviruses or among AAVs affect the ability of the viruses to infect diverse cell types in vitro or specific cells within tissues. Capsid genes can also be manipulated to artificially alter tropism to a cell surface molecule or tissue of choice81,82. Such capsid retargeting takes advantage of the fact that once binding is established, viruses can use secondary receptors for internalization.
A major difference between adenoviruses and AAVs is their packaging capacity (~4.5 kb for AAVs compared with up to 35 kb (although usually less than 10 kb) for adenoviruses)). Another distinguishing point is that all viral genes are removed from AAV vector genomes, whereas recombinant adenoviruses often express many viral genes that may induce immune responses to transduced cells and cause their subsequent elimination. For these reasons, AAVs are generally useful for achieving RNAi in cells that one wants to save, whereas adenoviruses are useful tools for either transient expression or when immune induction is desired. Notably, both adenoviruses and AAVs infect cells at multiple copies per cell, which can be problematic with shRNAs (instead of artificial miRNAs), as dosing may be amplified. Dosing is a function of the copy number and how efficiently the hairpin is expressed and processed62 and, in many cases, higher expression is not necessarily beneficial. If hairpins are inappropriately processed, or expressed at very high levels, toxicity (Box 1) and/or saturation of the RNAi machinery can occur63,64 (see above).
Lentiviruses are another delivery option. If their RNA genomes contain hairpins, they can be cleaved by RNA processing enzymes, which can be a problem during vector production. However, the negative effect this has on vector titres can be rescued by inhibiting the RNAi pathway83. Expression cassette placement is also important for lentiviruses expressing shRNAs or artificial miRNAs, as some configurations negatively affect vector production or shRNA expression84,85,86. Recombinant lentiviruses can transduce dividing and non-dividing cells and generally integrate into transcriptionally active chromatin. A potential problem is that insertion could activate an oncogene or inactivate a tumour suppressor gene. Insertional promiscuity can be dramatically reduced by using integrase-defective lentiviruses87. Like adenoviruses and AAVs, the vector tropism of lentiviruses can be altered, in this case through manipulation of the viral envelope used in vector production88.
The most obvious application for recombinant lentiviruses is for transduction in vitro or, in the case of clinical applications, ex vivo gene transfer to haematopoietic progenitor cells or peripheral blood lymphocytes for treatment of viral infections89. In these settings, recombinant lentiviruses are applied to cells at 1 to 5 vectors per cell, because higher concentrations are often toxic. The final number of integrants per cell is approximately 0.5 to 1 copy per cell. This fact is important when considering lentiviruses for RNAi delivery; if the copy number is low, shRNA expression from strong promoters may be preferred.
Therapeutic applications of RNAi in vivo
Important considerations for therapeutic RNAi are that gene silencing approaches rarely remove 100% of a transcript, that off-target silencing can occur (Box 1) and that each target organ, cell type and target transcript presents unique challenges. In some cases, the goal is to target every cell in an organ, in other instances promiscuous cell tropism is disadvantageous. For example, one might wish to target cancer cells for gene silencing but avoid normal surrounding tissue, or to express the therapeutic RNA in hepatocytes but not Küppfer cells after delivery to the liver. Tissue specificity can be achieved in some cases by incorporating ligands on the carrier that direct transfection or infection to the desired cell, as described above. Alternatively, researchers have taken advantage of the natural tropism90,91 or modified tropism of viral vectors for cell and tissue targeting82, or have used cell-specific promoters to express the silencing RNAs.
Delivery to the respiratory tract. Alterations in gene expression in epithelial cells of the respiratory tract contribute to disease pathogenesis in many disorders including asthma, chronic obstructive airway disease and cystic fibrosis. In addition, these cells are a key site of interaction between the host and the environment and many common viral pathogens replicate in these cells as the initial step in their life cycle, providing opportunities to silence viral gene products or host genes that modify the viral life cycle or the host's response to that virus. These reasons, and the fact that the respiratory tract is an accessible tissue, make the airway epithelium an attractive tissue for exploring RNAi therapies.
The potential of RNAi-based treatments to modify the host response to respiratory virus infections has been extensively studied. In 2004 and 2005 several high-profile papers were published in which synthesized siRNAs or expressed shRNAs were used to inhibit the influenza A virus92,93, severe acute respiratory syndrome (SARS) coronavirus94, respiratory syncytial virus4,94 and parainfluenza virus in animal models4. These studies showed both the promise and potential pitfalls of RNAi as a therapeutic strategy. Although the respiratory tract is readily accessible using topical or aerosol delivery techniques available in the clinic, several years of basic and clinical studies in the field of gene therapy have humbled investigators. These studies used a number of well-conceived viral and non-viral delivery techniques to treat monogenetic disorders such as cystic fibrosis, but the treatments were hindered by the physical barriers posed by the epithelial cells themselves, in addition to their secretions and host defence mechanisms95,96. In principle, delivery of RNAi oligonucleotides might pose a less significant challenge than expression plasmids or other genetic payloads as oligonucleotides need only enter the cytoplasm of surface cells to function. In addition, the mucosal surface of the airways and alveoli are active sites of innate and adaptive immunity, and RNAi delivery vectors or RNAi oligonucleotides may elicit immunologic responses. In the context of anti-infection RNAi strategies, immunostimulation confounded the early preclinical results5,6,32, and possibly the ongoing clinical data97.
In addition to targeting the gene products of respiratory viruses, several groups have used siRNA technology to knock down expression of host gene products or reporter genes in the respiratory tract in animal models. To date, the animal studies have shown little efficacy95 and poor delivery has also been demonstrated in vitro when fully differentiated cell models were studied. In short, effective RNAi activity requires the application of siRNAs before the development of a well-differentiated epithelial barrier96. Thus, efficient delivery remains an important hurdle to overcome as clinical studies are developed.
Antiviral strategies in other tissues. HIV remains an attractive target for drug development, including for therapies based on RNAi89. Examples of strategies include targeting the receptor for the virus and the virus itself55,98. However, the error-prone replication cycles of HIV can be problematic. Combinatorial approaches that include RNAi and other gene silencing approaches have therefore been developed for HIV. An approach undergoing clinical testing uses lentiviral vectors expressing an shRNA targeting an exon shared by HIV tat and rev genes (the tat/rev common exon), combined with two HIV-specific RNA-based inhibitors (a nucleolar-localizing TAR RNA decoy and a C-C chemokine receptor type 5 (CCR5)-targeting hammerhead ribozyme)99. The strategy is to transduce haematopoietic progenitor cells ex vivo and then reinfuse them into patients99. Early data from this Phase I trial show that transduced cells successfully engrafted within 11 days in all four patients treated. Importantly, there were no treatment-related toxicities. Vector expression was documented for up to 24 months in multiple cell lineages, as was expression of the introduced ribozyme and shRNA.
RNAi was recently used to inhibit lethal infection by the filovirus Ebola in a primate model51. A combination of modified siRNAs targeting Ebola L polymerase, viral protein 24 (VP24) and VP35 were SNALP-formulated and delivered intravenously. This strategy protected animals from death, including those that received siRNAs only after the onset of the infection. RNAi-based therapies are also under development for hepatitis B virus (HBV)39,100 and hepatitis C virus (HCV)101. As chronic hepatitis contributes significantly to hepatocellular carcinoma pathogenesis, this further drives interest in new HBV and HCV therapies as a means to reduce disease burden. As with HIV, the hepatitis viruses have a high mutation frequency during viral replication. Therefore, most current antiviral strategies focus on the delivery or expression of more than one RNAi construct to achieve success against chronic hepatitis infection101,102. A current focus of several laboratories is to use miRNA or shRNA expression methods to target more than one viral transcript103,104. Host proteins can also be targeted, for example siRNAs directed to diacylglycerol acyltransferase 1 (DGAT1) can reduce HCV virion production105. Additionally, inhibiting the expression of the host gene product polo-like kinase 1 (PLK1) using siRNA reduces HCV replication106. PLK1 inhibition is also being used in anticancer studies (see below).
Promising results have also been reported using RNAi to modify virus and host gene expression in a mouse model of genital tract herpes simplex virus 2 (HSV-2) infections48,107. Lipid-complexed RNA oligonucleotides inhibited expression of the HSV-2 UL27 and UL29 genes and the host receptor nectin 1 (also known as PVRL1). These approaches showed efficacy in both the prevention and treatment of infection48,107. The manipulation of host miRNAs to inhibit viral expression is also being explored (see further discussion below).
RNAi for neurological disorders. The blood–brain barrier limits access to the central nervous system (CNS) and thus the most practical manner to silence targets in neural cells is through direct injection of the RNAi trigger. As siRNAs have a short half-life, redosing using indwelling catheters would be required for chronic diseases. However, for acute illnesses or delivery to brain tumours, the short half-life of siRNAs may be desirable. By contrast, viral platforms provide lasting expression and may be ideal for chronic disorders. For example, vectors expressing therapeutic RNAi improved disease phenotypes for many months in preclinical studies in rodent models of polyglutamine repeat diseases108,109,110,111,112,113, amyotrophic lateral sclerosis114,115,116, Parkinson's disease117 and Alzheimer's disease118,119. In nonhuman primate brains, viral-vector-based systems are safe120 and, given the encouraging results of AAVs in the human brain and eye121, clinical trials for neurodegenerative diseases with AAVs expressing RNAi triggers are anticipated.
An issue to consider is the delivery to the correct cell type in the brain. Specificity for neurons versus glia has not been achieved for uncomplexed siRNAs, and in fact it has been reported that siRNAs delivered into nonhuman primate brains enter oligodendrocytes49. This might be a suitable method for therapies aimed at treating multiple sclerosis or other white matter diseases. For encapsidated viral vectors, the nature of the protein coat imparts a natural tropism for neurons (in the case of AAV2, for example) or for other cell types91. For enveloped viral vectors, such as those lentiviral systems derived from HIV, the tropism is imparted by the envelope used in vector production in a process known as pseudotyping122. Also, some vectors traffic from the site of injection to other regions of the brain via neuronal connections, but others remain localized. Thus targeting is achieved collectively by the site of injection, the propensity of the virus to infect certain cell types and traffic along or within neuronal axons and neurites, and the promoter used to drive expression of the RNAi trigger.
For chronic, dominantly inherited disorders, it may be preferable to silence only the mutant allele. For some brain diseases there are highly prevalent disease-linked polymorphisms that provide opportunities for allele-specific silencing. Primary dystonia, which is caused by a common GAG deletion in torsin A (TOR1A), is one example123. Other disorders have several disease-linked SNPs present in most patients and these SNPs provide an opportunity for disease allele silencing. However, when targeting a SNP using RNAi, it is critical to consider the potential for off-target binding of the seed sequence in the small RNA (Box 1), as unintended off-target silencing could abrogate any beneficial effects from leaving the wild-type allele intact. Huntington's disease is a case in point. Preclinical work in animal models shows that partial knockdown of both alleles of huntingtin is tolerated and provides clinical benefit124, yet genotyping shows that 4 to 5 SNPs in huntingtin may be present in the majority of patients with Huntington's disease125,126. Small RNAs that are specific to the SNPs may have moderately high off-target potential and therefore should be tested for their long-term tolerability in vivo, as should the safety of partial reductions in expression from both mutant and wild-type alleles.
Targeting metabolic disease and hepatic cancers. One of the first organs tested for the effectiveness of RNAi in vivo was the liver127,128, and RNAi-based treatments for metabolic diseases (such as hypercholesterolaemia), viral infections, cancer6 and liver fibrosis (reviewed in Ref. 129) are in progress. For metabolic diseases, there are preclinical and clinical trials underway for lowering plasma LDLs using siRNAs that target the expression of apolipoprotein B (APOB) and proprotein convertase subtilisin/kexin type 9 (PCSK9). In this work, the siRNAs are complexed to carriers or embedded in liposomal particles (for example, SNALPs). Data from rodents and nonhuman primates50,122 show significant LDL-lowering properties and, in one of the first trials in humans, corporate news releases stated that the SNALP-formulated APOB siRNA was well tolerated at all but the highest dose. Newer formulations that show improved potency in nonhuman primates are under development.
The liver was also one of the first organs targeted in the development of RNAi-based therapies for cancer. One study used SNALPs targeting PLK1, a cell cycle protein that is crucial for the activating phosphorylation of many cell cycle proteins; inhibition of PLK1 induces cell cycle arrest and tumour cell apoptosis130. Mice with hepatic tumours treated with SNALP-formulated PLK1 siRNA showed significant improvements in survival6. In December 2010, this technology advanced to a Phase I trial in humans with liver cancer (Table 1).
Another hepatic cancer application is SNALPs simultaneously delivering siRNAs to kinesin spindle protein (KSP) and vascular endothelial growth factor (VEGF). KSP is required for cell division, and VEGF is required for tumour cell growth. In early 2011, sponsors of a Phase I trial using this approach reported evidence of RNAi activity in biopsied tissue131. These preliminary reports, along with the first report of RNAi activity from exogenously applied siRNA complexes52, are important milestones in the development of RNAi delivery systems as cancer therapeutics.
miRNAs as therapeutic targets. The identification of misregulated miRNAs in cellular transformation and maintenance of the malignant state has profound implications for cancer therapy. As with other misregulated genes, miRNAs can be targets for gene silencing approaches, whether the miRNAs are encoded in the host genome or expressed from oncogenic viruses (reviewed in Ref. 132). Inhibition of oncogenic miRNAs that regulate multiple targets might switch off dozens of cancer-promoting signals. Rather than devising siRNAs to target the misregulated miRNA, researchers have developed miRNA sponges133; these provide alternative binding platforms for the miRNAs and so inhibit their ability to bind and suppress their natural targets (Fig. 1). An early example was the intravenous delivery of antagomirs, which are chemically modified RNA oligonucleotides antisense to the miRNAs134,135. In a primate model of HCV infection, oligonucleotides that sequestered miR-122 inhibited virus replication136. Plasmid- and virus-based approaches are also being used for reducing endogenous miRNA levels133,137. Typically for this approach, strong promoters drive expression of a sequence encoding several miRNA target sites downstream of a reporter. The multiple copies expressed become targets for binding of miRNAs, which are sequestered from targeting their endogenous mRNAs.
Summary and future considerations
In addition to the developments described above, there has been substantial progress in using gene silencing approaches for treating skin138 and retinal diseases139,140. Like the liver and airway, these accessible tissues were early targets for preclinical testing. Exploiting the small RNA biogenesis and gene silencing pathways for heart diseases, either using siRNAs against single targets141 or inhibiting the action of misregulated miRNAs142, has also yielded promising results that are approaching clinical trials.
In addition to its utility as a stand-alone strategy, RNAi may have expanded applications as an adjuvant in multipronged treatment settings. For example, targeting multidrug resistance protein 1 (MDR1; also known as ABCB1) in cancer cells may enhance the activity of chemotherapeutics143, and other host genes have been targeted for similar ends for cancer therapeutics144,145,146. Another RNAi adjuvant strategy is the use of dsRNA oligonucleotides as immunostimulatory agonists alongside vaccines, as in the case of a RIG1 agonist to enhance the activity of a DNA vaccine against influenza9.
RNAi therapy development should consider whether regional delivery and partial knockdown, or global delivery and complete knockdown, is required for a therapeutic result. An example of the former is directed delivery to specific regions of the brain for Parkinson's disease. By contrast, RNAi therapy for cancer may require delivery to all cancer cells. Another important issue that is yet to be resolved is dosing of the therapeutic RNAi. In the case of cancer, pharmacologists must balance the target cells' ability to recover from the exogenously applied siRNAs or anti-miRNA treatments with the practical considerations of patient compliance for repetitive dosing. Will clinical success and eventual cure require treatment for weeks, months or years? Viral vector expression systems for RNAi can overcome this problem by providing sustained expression, but this strategy requires genomic integration of the vector if the target cells are dividing. In order to overcome the potential dangers of genomic integration, methods for integration into genomic 'safe harbours' will be important. With respect to long-term RNAi from viral vectors as therapies for genetic diseases, the question is whether regulated expression is required. To help to answer this question, it is expected that long-term studies in large animals (for example, non-human primates) will yield valuable information regarding chronic application of inhibitory RNAs from various platforms. A further consideration is that although the end points for some trials are obvious, for example lowering blood cholesterol or reducing tumour burden, sensitive and specific end points are not always clear for chronic disorders in which the tissue cannot be easily biopsied or in which biomarkers are not validated.
In slightly more than a decade, we have advanced rapidly from RNAi discovery, to understanding the molecular processes driving small RNA biogenesis and function, to developing reagents that harness the power of the RNAi pathway. Although many hurdles remain for using these technologies for therapy, exciting early clinical results show how far we have come.
Box 1 | Off-target silencing.
When designing therapeutic strategies involving small interfering RNAs (siRNAs) or expression-based systems, it is important to know details about the RNA strand that is incorporated into the RNA-induced silencing complex (RISC) and mediates silencing. Genomically encoded microRNAs (miRNAs) in mammalian cells possess the necessary information within the miRNA duplex for appropriate loading of the miRNA or the miRNA* strand. So, when using chemically synthesized siRNAs or systems that express short hairpin RNAs or artificial miRNAs, the design must take into account what nature has taught us. For example, the designer should consider whether the artificial miRNA has the 5′ and 3′ ends that are necessary for appropriate processing and export, and whether subsequent biochemical processing results in loading of the correct strand into RISC.
The are several methods to determine which strand is loaded into RISC, including northern blots, small RNA PCRs and luciferase-based plasmid systems with targets for the guide strand or the passenger strand placed in the 3′ UTR of the reporter (Fig. 2). Although there are publicly available algorithms that use thermodynamic rules in silencing RNA, it is important to sequence expressed constructs to characterize their termini. Sequencing is necessary to ascertain the relative proportions of the intended mature silencing RNA and RNAs that result when cleavage sites are shifted by one or several nucleotides; such alterations to the RNA could substantially increase off-target silencing. Off-target silencing occurs through the interaction of a seed sequence with transcripts harbouring complementary sequences147,148,149,150,151. siRNAs with high off-target potentials, based on seed complement frequencies in the transcriptome, result in increased silencing of unintended mRNAs and toxicity152. This problem can be reduced by designs that bias RISC loading towards the correct RNA strand and that ensure that processing of silencing RNA precursors only produces the desired small RNA duplex. Incorporating chemical modifications, such as 2′-O-methyl groups, into the guide and passenger strands of chemically synthesized siRNAs reduces indiscriminate effects of ssRNAs and dsRNAs153. Additionally, incorporation of unlocked nucleic acids (acyclic RNA mimics)154 makes the RNAs poor ligands for Toll-like receptors.
Varying amounts of off-target silencing can occur with different silencing platforms because the levels of the silencing RNAs will vary. For example, for expression systems, transfection with plasmids or transduction with adenoviruses or adeno-associated viruses will yield more copies per cell than transduction with lentiviruses. siRNA transfection can also result in abundant off-target silencing if the siRNA has low on-target potency and a moderate or high off-target potential (see above). Less off-target silencing with a lentivirus-based system compared with siRNAs was reported recently, but that study used high doses of siRNAs with high off-target potential155.
Acknowledgements
The authors thank members of the Davidson and McCray laboratories for critical feedback. Work in the authors' laboratories is supported by the Roy J. Carver Trust and the US National Institutes of Health grants HL51670, DK54759 and NS50210.
Glossary
- RNA-induced silencing complex
(RISC). RISC is a group of proteins, including one of the Argonaute proteins, that induces target mRNA cleavage based on loaded small interfering RNA or microRNA guide strands.
- Seed
A sequence of seven bases in a microRNA that is complementary to the mRNA target. This sequence is essential for the initial binding of the microRNA to most targets. Seeds can also exist arbitrarily in small interfering RNAs and processed short hairpin RNAs, causing microRNA-like silencing.
- Primary miRNAs
(pri-miRNAs). The initial transcriptional products of microRNA genes. They are generally >100 nucleotides long and may contain one or more microRNA stem loops that are processed by the microRNA biogenesis pathway.
- Precursor miRNAs
(pre-miRNAs). Hairpin precursors of microRNAs formed by the cleavage of primary microRNAs by DCGR8 and Drosha.
- Dicer
A member of the RNase III family of ribonucleases that cleaves dsRNAs into small interfering RNAs, and precursor microRNAs and mirtrons into microRNAs.
- Type I interferon response
An innate immune response to dsRNA, ssRNA, CpG DNA and other stimuli that triggers a protective antiviral response in host cells. Signalling elicits α- and β-interferon release, which activate multiple components of innate and adaptive immunity.
- Toll-like receptors
(TLRs). A family of receptors that recognize pathogen-associated molecular patterns (PAMPs), including some DNA and RNA molecules.
- CpG oligonucleotides
Immunostimulatory dinucleotide motifs that interact with Toll-like receptor 9.
- Stable nucleic acid lipid particle
(SNALP). A lipid nanoparticle formulation for the systemic delivery of small interfering RNAs to tissues.
- Minicells
Bacteria-derived cells that have no chromosomes and are non-living. They can be loaded with drugs, plasmids or small interfering RNAs.
- Adeno-associated virus
(AAV). A member of the genus Dependovirus. These viruses have small, ssDNA genomes and are not known to cause disease in humans. AAVs are commonly used as recombinant vectors in gene therapy applications.
- Episome
A dsDNA segment that can persist independently of chromosomal DNA.
- Capsid
The outer protein coat of a virus, such as adeno-associated virus or adenovirus.
- miRNA*
The precursor microRNA (pre-miRNA) processed by Dicer generates an miRNA duplex containing the miRNA strand and the miRNA* strand, one of which is loaded into the RNA-induced silencing complex (RISC). The ratio of one strand to the other being loaded into RISC to mediate silencing activity can vary among species, tissues, and disease or developmental settings.
- Off-target effects
Any detectible phenotypic change that is triggered by the RNAi treatment, other than those that are derived directly or indirectly from silencing the targeted mRNA.
- Küppfer cell
A macrophage-like cell that is resident in the liver and is involved in antigen presentation.
- Antagomirs
RNA oligonucleotides that are antisense to endogenous microRNAs. They are used for inhibiting microRNA–mRNA interactions.
Biographies
Beverly Davidson received her Ph.D. from the University of Michigan, USA, in biological chemistry followed by a postdoctoral fellowship in molecular genetics. She is the Vice Chair for Research in Internal Medicine, Associate Director of the Center for Gene Therapy, and holds the Roy J. Carver Biomedical Research Chair at the University of Iowa, USA. Her research interests are focused on molecular therapies for neurogenetic diseases, including Batten disease and the polyglutamine repeat disorders (for example, Huntington's disease), and the role of non-coding RNAs in neurodevelopment and disease.
Paul McCray Jr received his M.D. from the University of Iowa, USA, followed by a paediatric residency and pulmonary fellowship. He is currently a Professor of Pediatrics, Microbiology, and Internal Medicine at the University of Iowa, USA, where he is the Vice Chair for Research in Pediatrics, and holds the Roy J. Carver Chair in Pulmonary Research. His research interests include pulmonary innate immunity, the pathogenesis and treatment of cystic fibrosis, and the development of nucleic acid-based therapies for monogenetic diseases.
Related links
FURTHER INFORMATION
Competing interests
B.L.D. serves on the Scientific Advisory Board of Marina Biotech. P.B.M. declares no competing financial interests.
References
- 1.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nature Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 5.Robbins M, et al. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum. Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 6.Judge AD, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J. Clin. Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornung V, et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 8.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotech. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 9.Luke JM, et al. Coexpressed RIG-I agonist enhances humoral immune response to influenza virus DNA vaccine. J. Virol. 2011;85:1370–1383. doi: 10.1128/JVI.01250-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nature Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 14.Shin C, et al. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiriakidou M, et al. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 17.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 18.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nature Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 19.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 22.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 23.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 25.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 26.Czech B, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakurai K, et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 2010;39:1510–1525. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nature Biotech. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 30.Siolas D, et al. Synthetic shRNAs as potent RNAi triggers. Nature Biotech. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 31.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 32.Kleinman ME, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho WG, et al. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. Proc. Natl Acad. Sci. USA. 2009;106:7137–7142. doi: 10.1073/pnas.0812317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang XL, Sun JF, Wang XY, Du LL, Liu P. Blocking neuropilin-2 enhances corneal allograft survival by selectively inhibiting lymphangiogenesis on vascularized beds. Mol. Vis. 2010;16:2354–2361. [PMC free article] [PubMed] [Google Scholar]
- 35.Cubillos-Ruiz JR, et al. Polyethylenimine-based siRNA nanocomplexes reprogram tumor-associated dendritic cells via TLR5 to elicit therapeutic antitumor immunity. J. Clin. Invest. 2009;119:2231–2244. doi: 10.1172/JCI37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kortylewski M, et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nature Biotech. 2009;27:925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brody JD, et al. In situ vaccination with a TLR9 agonist induces systemic lymphoma regression: a phase I/II study. J. Clin. Oncol. 2010;28:4324–4332. doi: 10.1200/JCO.2010.28.9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Morrissey DV, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature Biotech. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 40.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nature Biotech. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 41.Czauderna F, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 44.Weinstein S, Peer D. RNAi nanomedicines: challenges and opportunities within the immune system. Nanotechnology. 2010;21:232001. doi: 10.1088/0957-4484/21/23/232001. [DOI] [PubMed] [Google Scholar]
- 45.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 47.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature Biotech. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 48.Wu Y, et al. Durable protection from Herpes Simplex Virus-2 transmission following intravaginal application of siRNAs targeting both a viral and host gene. Cell Host Microbe. 2009;5:84–94. doi: 10.1016/j.chom.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Q, et al. Lipophilic siRNAs mediate efficient gene silencing in oligodendrocytes with direct CNS delivery. J. Control. Release. 2010;144:227–232. doi: 10.1016/j.jconrel.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 50.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 51.Geisbert TW, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dassie JP, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nature Biotech. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar P, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SS, et al. RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice. Mol. Ther. 2010;18:370–376. doi: 10.1038/mt.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng X, et al. A novel in vivo siRNA delivery system specifically targeting dendritic cells and silencing CD40 genes for immunomodulation. Blood. 2009;113:2646–2654. doi: 10.1182/blood-2008-04-151191. [DOI] [PubMed] [Google Scholar]
- 59.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 60.Peer D. Induction of therapeutic gene silencing in leukocyte-implicated diseases by targeted and stabilized nanoparticles: a mini-review. J. Control. Release. 2010;148:63–68. doi: 10.1016/j.jconrel.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 61.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 64.McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimm D, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 67.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung KH, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boden D, et al. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 71.Seow Y, Wood MJ. Biological gene delivery vehicles: beyond viral vectors. Mol. Ther. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacDiarmid JA, et al. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell. 2007;11:431–445. doi: 10.1016/j.ccr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 73.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature Biotech. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 74.Keates AC, Fruehauf JH, Xiang S, Parker PD, Li CJ. Cequent Pharmaceuticals, Inc.: the biological pitcher for RNAi therapeutics. Pharmacogenomics. 2007;8:867–871. doi: 10.2217/14622416.8.7.867. [DOI] [PubMed] [Google Scholar]
- 75.MacDiarmid JA, et al. Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nature Biotech. 2009;27:643–651. doi: 10.1038/nbt.1547. [DOI] [PubMed] [Google Scholar]
- 76.Heilbronn R, Weger S. Viral vectors for gene transfer: current status of gene therapeutics. Handb. Exp. Pharmacol. 2010;197:143–170. doi: 10.1007/978-3-642-00477-3_5. [DOI] [PubMed] [Google Scholar]
- 77.Hall K, Blair Zajdel ME, Blair GE. Unity and diversity in the human adenoviruses: exploiting alternative entry pathways for gene therapy. Biochem. J. 2010;431:321–336. doi: 10.1042/BJ20100766. [DOI] [PubMed] [Google Scholar]
- 78.Matrai J, Chuah MK, VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol. Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Overbaugh J, Miller AD, Eiden MV. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 2001;65:371–389. doi: 10.1128/MMBR.65.3.371-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marconi P, Argnani R, Epstein AL, Manservigi R. HSV as a vector in vaccine development and gene therapy. Adv. Exp. Med. Biol. 2009;655:118–144. doi: 10.1007/978-1-4419-1132-2_10. [DOI] [PubMed] [Google Scholar]
- 81.Hajitou A, et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 82.Chen YH, Chang M, Davidson BL. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nature Med. 2009;15:1215–1218. doi: 10.1038/nm.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu YP, et al. Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies. RNA. 2010;16:1328–1339. doi: 10.1261/rna.1887910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harper SQ, et al. Optimization of feline immunodeficiency virus vectors for RNA interference. J. Virol. 2006;80:9371–9380. doi: 10.1128/JVI.00958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amendola M, et al. Regulated and multiple miRNA and siRNA delivery into primary cells by a lentiviral platform. Mol. Ther. 2009;17:1039–1052. doi: 10.1038/mt.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang J, Rossi JJ. Strategies in designing multigene expression units to downregulate HIV-1. Methods Mol. Biol. 2010;623:123–136. doi: 10.1007/978-1-60761-588-0_8. [DOI] [PubMed] [Google Scholar]
- 87.Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nature Biotech. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 88.Anliker B, et al. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nature Methods. 2010;7:929–935. doi: 10.1038/nmeth.1514. [DOI] [PubMed] [Google Scholar]
- 89.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nature Biotech. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stein CS, Martins I, Davidson BL. The lymphocytic choriomeningitis virus envelope glycoprotein targets lentiviral gene transfer vector to neural progenitors in the murine brain. Mol. Ther. 2005;11:382–389. doi: 10.1016/j.ymthe.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 91.Davidson BL, et al. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl Acad. Sci. USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl Acad. Sci. USA. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ge Q, et al. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl Acad. Sci. USA. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li BJ, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nature Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Griesenbach U, et al. Inefficient cationic lipid-mediated siRNA and antisense oligonucleotide transfer to airway epithelial cells in vivo. Respir. Res. 2006;7:26. doi: 10.1186/1465-9921-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caci E, et al. Epithelial sodium channel inhibition in primary human bronchial epithelia by transfected siRNA. Am. J. Respir. Cell. Mol. Biol. 2009;40:211–216. doi: 10.1165/rcmb.2007-0456OC. [DOI] [PubMed] [Google Scholar]
- 97.DeVincenzo J, et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc. Natl Acad. Sci. USA. 2010;107:8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu YP, et al. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol. Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DiGiusto DL, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34+ cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McCaffrey AP, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nature Biotech. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 101.Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl Acad. Sci. USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ely A, Naidoo T, Arbuthnot P. Efficient silencing of gene expression with modular trimeric Pol II expression cassettes comprising microRNA shuttles. Nucleic Acids Res. 2009;37:e91. doi: 10.1093/nar/gkp446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang X, Haurigot V, Zhou S, Luo G, Couto LB. Inhibition of hepatitis C virus replication using adeno-associated virus vector delivery of an exogenous anti-hepatitis C virus microRNA cluster. Hepatology. 2010;52:1877–1887. doi: 10.1002/hep.23908. [DOI] [PubMed] [Google Scholar]
- 104.Shin D, Lee H, Kim SI, Yoon Y, Kim M. Optimization of linear double-stranded RNA for the production of multiple siRNAs targeting hepatitis C virus. RNA. 2009;15:898–910. doi: 10.1261/rna.1268209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herker E, et al. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nature Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen YC, et al. Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J. Virol. 2010;84:7983–7993. doi: 10.1128/JVI.00068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Palliser D, et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 108.Xia H, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nature Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 109.Harper SQ, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol. Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Machida Y, et al. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem. Biophys. Res. Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 112.Alves S, et al. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS ONE. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Drouet V, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 114.Ralph GS, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nature Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 115.Raoul C, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 116.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sapru MK, et al. Silencing of human α-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp. Neurol. 2006;198:382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 118.Singer O, et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nature Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 119.Hong CS, Goins WF, Goss JR, Burton EA, Glorioso JC. Herpes simplex virus RNAi and neprilysin gene transfer vectors reduce accumulation of Alzheimer's disease-related amyloid-β peptide in vivo. Gene Ther. 2006;13:1068–1079. doi: 10.1038/sj.gt.3302719. [DOI] [PubMed] [Google Scholar]
- 120.Hadaczek P, et al. Eight years of clinical improvement in MPTP-lesioned primates after gene therapy with AAV2-hAADC. Mol. Ther. 2010;18:1458–1461. doi: 10.1038/mt.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nature Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 122.Frank-Kamenetsky M, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc. Natl Acad. Sci. USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gonzalez-Alegre P, Bode N, Davidson BL, Paulson HL. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J. Neurosci. 2005;25:10502–10509. doi: 10.1523/JNEUROSCI.3016-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boudreau RL, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pfister EL, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Warby SC, et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nature Biotech. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 128.McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 129.Hu PF, Xie WF. Targeted RNA interference for hepatic fibrosis. Expert Opin. Biol. Ther. 2009;9:1305–1312. doi: 10.1517/14712590903213677. [DOI] [PubMed] [Google Scholar]
- 130.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 131.Alnylam Pharmaceuticals, Inc. Alnylam demonstrates RNAi in man with systemically delivered RNAi therapeutics. Alnylam Pharmaceuticals[online], (2011).
- 132.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nature Rev. Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 134.Pedersen IM, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krutzfeldt J, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 136.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Geusens B, Sanders N, Prow T, Van Gele M, Lambert J. Cutaneous short-interfering RNA therapy. Expert Opin. Drug Deliv. 2009;6:1333–1349. doi: 10.1517/17425240903304032. [DOI] [PubMed] [Google Scholar]
- 139.Chadderton N, et al. Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol. Ther. 2009;17:593–599. doi: 10.1038/mt.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yuan MK, Tao Y, Yu WZ, Kai W, Jiang YR. Lentivirus-mediated RNA interference of vascular endothelial growth factor in monkey eyes with iris neovascularization. Mol. Vis. 2010;16:1743–1753. [PMC free article] [PubMed] [Google Scholar]
- 141.Poller W, Hajjar R, Schultheiss HP, Fechner H. Cardiac-targeted delivery of regulatory RNA molecules and genes for the treatment of heart failure. Cardiovasc. Res. 2010;86:353–364. doi: 10.1093/cvr/cvq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Susa M, et al. Inhibition of ABCB1 (MDR1) expression by an siRNA nanoparticulate delivery system to overcome drug resistance in osteosarcoma. PLoS ONE. 2010;5:e10764. doi: 10.1371/journal.pone.0010764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chang JT, et al. Highly potent and specific siRNAs against E6 or E7 genes of HPV16- or HPV18-infected cervical cancers. Cancer Gene Ther. 2010;17:827–836. doi: 10.1038/cgt.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Herrmann A, et al. Targeting Stat3 in the myeloid compartment drastically improves the in vivo antitumor functions of adoptively transferred T cells. Cancer Res. 2010;70:7455–7464. doi: 10.1158/0008-5472.CAN-10-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu C, et al. Novel biodegradable lipid nano complex for siRNA delivery significantly improving the chemosensitivity of human colon cancer stem cells to paclitaxel. J. Control. Release. 2009;140:277–283. doi: 10.1016/j.jconrel.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 147.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotech. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 148.Jackson AL, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aleman LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Burchard J, et al. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 152.Anderson EM, et al. Experimental validation of the importance of seed complement frequency to siRNA specificity. RNA. 2008;14:853–861. doi: 10.1261/rna.704708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jackson AL, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bramsen JB, et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Klinghoffer RA, et al. Reduced seed region-based off-target activity with lentivirus-mediated RNAi. RNA. 2010;16:879–884. doi: 10.1261/rna.1977810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 157.Monteys AM, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 160.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 161.Chung WJ, et al. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2010;21:286–300. doi: 10.1101/gr.113050.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 163.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Provost P, et al. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 167.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]


