In Enterococcus faecalis, conjugative plasmids like pCF10 often carry antibiotic resistance genes. With antibiotic treatment, bacteria benefit from plasmid carriage; however, without antibiotic treatment, plasmid gene expression may have a fitness cost. Transfer of pCF10 is mediated by cell-to-cell signaling, which activates the expression of conjugation genes and leads to efficient plasmid transfer. Yet, not all donor cells in induced populations transfer the plasmid. We examined whether induced cells might not be able to functionally conjugate due to premature induced transcript termination. Single-cell analysis showed that most induced cells do, in fact, express all of the genes required for conjugation, suggesting that premature transcription termination within the prgQ operon does not account for failure of induced donor cell gene transfer.
KEYWORDS: conjugation, hybridization chain reaction, plasmid transfer
ABSTRACT
For high-frequency transfer of pCF10 between E. faecalis cells, induced expression of the pCF10 genes encoding conjugative machinery from the prgQ operon is required. This process is initiated by the cCF10 (C) inducer peptide produced by potential recipient cells. The expression timing of prgB, an “early” gene just downstream of the inducible promoter, has been studied extensively in single cells. However, several previous studies suggest that only 1 to 10% of donors induced for early prgQ gene expression actually transfer plasmids to recipients, even at a very high recipient population density. One possible explanation for this is that only a minority of pheromone-induced donors actually transcribe the entire prgQ operon. Such cells would not be able to functionally conjugate but might play another role in the group behavior of donors. Here, we sought to (i) simultaneously assess the presence of RNAs produced from the proximal (early induced transcripts [early Q]) and distal (late Q) portions of the prgQ operon in individual cells, (ii) investigate the prevalence of heterogeneity in induced transcript length, and (iii) evaluate the temporality of induced transcript expression. Using fluorescent in situ hybridization chain reaction (HCR) transcript labeling and single-cell microscopic analysis, we observed that most cells expressing early transcripts (QL, prgB, and prgA) also expressed late transcripts (prgJ, pcfC, and pcfG). These data support the conclusion that, after induction is initiated, transcription likely extends through the end of the conjugation machinery operon for most, if not all, induced cells.
IMPORTANCE In Enterococcus faecalis, conjugative plasmids like pCF10 often carry antibiotic resistance genes. With antibiotic treatment, bacteria benefit from plasmid carriage; however, without antibiotic treatment, plasmid gene expression may have a fitness cost. Transfer of pCF10 is mediated by cell-to-cell signaling, which activates the expression of conjugation genes and leads to efficient plasmid transfer. Yet, not all donor cells in induced populations transfer the plasmid. We examined whether induced cells might not be able to functionally conjugate due to premature induced transcript termination. Single-cell analysis showed that most induced cells do, in fact, express all of the genes required for conjugation, suggesting that premature transcription termination within the prgQ operon does not account for failure of induced donor cell gene transfer.
INTRODUCTION
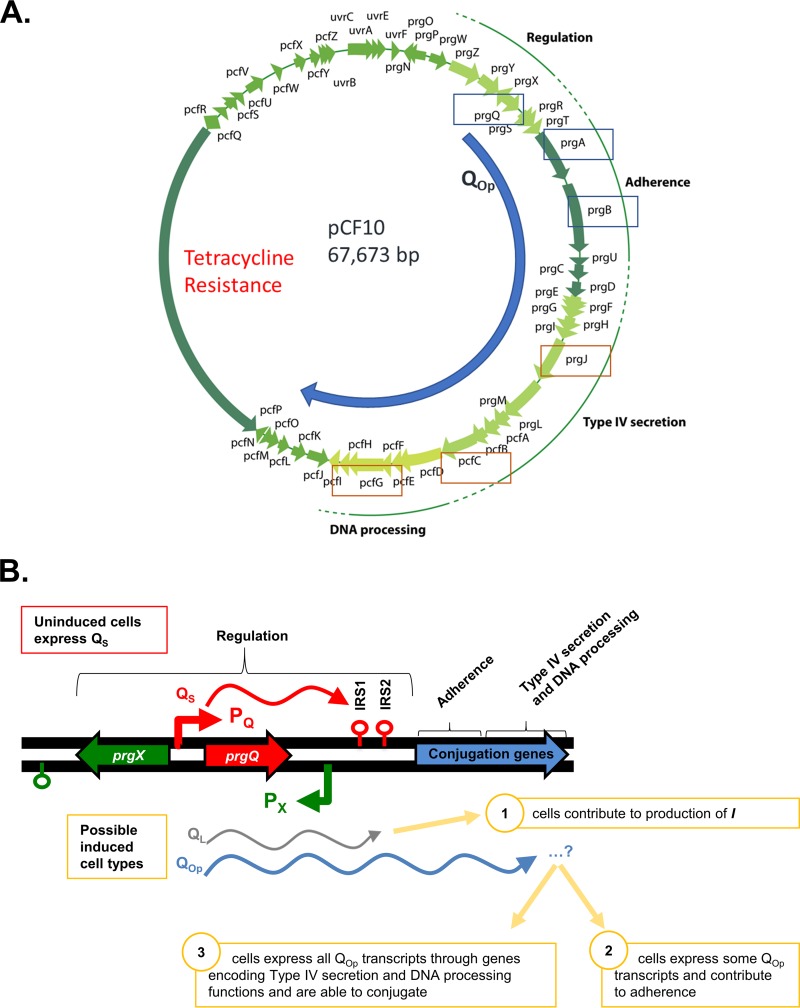
Conjugative transfer of pCF10 in Enterococcus faecalis is enabled by expression of plasmid genes encoding adherence, type IV secretion, and DNA processing machinery (Fig. 1A). Expression and transfer are induced when recipient cells signal donors via the cCF10 (C) peptide (1). Induced expression occurs from promoter PQ, but basal expression also occurs from this promoter in the absence of induction by C (Fig. 1B). Without C, transcripts from PQ terminate at the first inverted repeat sequence (IRS1) and produce QS transcripts (2). Upon induction by C, the frequency of transcription from PQ increases, and transcripts continue past IRS1 and through the entire conjugation operon (3). Here, we considered the possibility that some induced cells are not be able to functionally conjugate due to premature induced transcript termination, resulting in induced transcripts of varied lengths (Fig. 1B). In this work, the term “QL” will be used to refer to induced transcripts terminated at IRS2, and “QOp” (Q operon) will be used to refer to all transcripts extending past IRS2. Furthermore, “early QOp” will be used to discuss downstream genes encoded proximal to IRS2, while “late QOp” will be used to refer to genes encoded distally (15 to 30 kbp downstream). Promoter PQ expression is regulated by the pCF10-encoded PrgX protein (Fig. 1B). The activity of PrgX is modulated by the cCF10 (C) and iCF10 (I) peptide pheromones. PrgX complexed with C allows induced transcription from PQ, while PrgX complexed with I results in increased promoter repression (4). I is produced from pCF10 via translation of prgQ mRNA originating from PQ and generally serves to limit expression of the conjugation machinery in the absence of potential plasmid recipients and to shut down the response following induction.
FIG 1.
Maps of pCF10 and the pCF10 prgX-prgQ regulatory region. Transcripts shown are expressed from promoter PQ, with or without induction by C. (A) Map showing the extended QOp transcript and genes for which HCR transcript-labeling probes were designed. Early QOp transcripts (QL prgA, and prgB) for which HCR probes were designed are boxed in blue, and late QOp transcripts (prgJ, pcfC, and pcfG) for which HCR probes were designed are boxed in orange. (B) In the uninduced state, PQ is modestly active and produces QS transcripts (red), which terminate at IRS1. Upon induction, production of QL (gray) and QOp transcripts (blue) increases dramatically. QL transcripts have are identical to QS transcripts through IRS1 but terminate at IRS2; for this study, we used HCR probes to RNAs produced from the sequences between IRS1 and IRS2, since they are unique to induced cells. QOp transcripts (blue) continue through regions of pCF10 that encode genes required for conjugation. prgX transcripts are generated from the opposite strand and extend from promoter Px, terminating near the 3′ end of prgX (green lollipop). I, iCF10.
Induced expression from PQ has been studied extensively at the population and single-cell levels by a wide variety of methods. Microarrays, reverse transcription-quantitative PCR (RT-qPCR), transcriptome sequencing (RNA-seq), and β-galactosidase reporters have given insight at the population level (5–7). Single-cell responses have been characterized using both fluorescent reporters and hybridization chain reaction (HCR)-based fluorescent transcript labeling (5, 8, 9); clumping and conjugation assays have provided insight into the functional response (1, 2, 10). Together, these methods have revealed that induction is very rapid: induced transcripts can be observed by microarray within 30 min of exogenous C addition (6), and a measurable increase in transconjugants can be quantified within 60 min of mixing donor and recipient cells, even in the absence of exogenous peptide (11). Induction and shutdown have also been shown by RT-qPCR to occur within 15 and 60 min of C addition (11). Furthermore, although single-cell expression analysis methods have supported the timing of early QOp-induced expression and degradation observed by population-level approaches, the former methods have shown that there is a great deal of response variation within populations of cells (9), namely, some cells show induced expression, while others do not. To date, analysis of expression across the entire pCF10 genome—including the full conjugation operon and late QOp genes—has been limited to population averaging methods, as our previous HCR studies focused only on transcripts produced from prgB and upstream loci in the 5′ segment of the prgQ operon. In addition, we have not yet examined multiple induced transcripts within the same cells. The published data are consistent with models where induced expression of a full-length QOp transcript could produce the necessary conjugation machinery from a single initiation event. However, the frequency of transfer from highly induced donor populations is generally below 10−1 (6) (further examined below); this suggests the possibility that only a fraction of the population induced for expression of early QL transcripts actually expresses mRNA from the entire operon. The lack of thorough characterization of downstream gene expression motivated us to further investigate expression and conjugation ability at both the population and single-cell levels.
In this work, we sought to investigate whether (i) all cells induced at promoter PQ are capable of expressing the full QOp transcript or (ii) whether some cells express only partial QOp transcripts terminating before the 3′ end of the operon (Fig. 1B). Note that in the second scenario, we would expect to find substantial numbers of induced donors not expressing 3′ transcripts. This subpopulation of cells could not undergo conjugation, but they might still play important functional roles within a donor population by promoting mating aggregate formation via induced expression of the early QOp gene prgB, (encoding aggregation substance), or by contributing to shutdown of the pheromone response through production of iCF10 (I), which is encoded by the very early gene, prgQ. Early shutdown of the pheromone response could be advantageous to individual cells by preventing expression of the conjugation machinery by all cells in the donor population. Formation of very large aggregates reduces growth rate, and production of excessive levels of PrgB can be lethal (12). Overall, these hypotheses and the open question of QOp transcript length heterogeneity encouraged further investigation of transcription dynamics at the single-cell level.
RESULTS
Most induced donor cells growing in close proximity to a large excess of recipients do not transfer pCF10.
We engineered a conjugation-proficient derivative of pCF10 containing both constitutively expressed (tdTomato) and pheromone-inducible (green fluorescent protein [GFP]) reporter genes. We reasoned that the constitutive reporter should identify all cells carrying the plasmid, while the inducible reporter could be used to assess the induction status of single cells. We used microscopy and flow cytometry to analyze expression of these fluorescent proteins (FPs) in cultures containing mixtures of the engineered donor strain and (for microscopy) a wild-type recipient stained with Hoechst stain or (for flow cytometry) a previously constructed (13) plasmid-free recipient expressing a constitutive cyan fluorescent protein (CFP). The primary objectives were to examine, at the single-cell level, the induction status of donors under different conditions and to detect individual plasmid transfer events. Experiments involving analysis of FP reporter expression in E. faecalis suffer from technical limitations, including sporadic problems tracking single cells over prolonged periods due to localized desiccation of the agarose medium, a substantial lag between transcription and detection of fluorescent signals (likely related to the time required for translation and protein folding in enterococcal cells), excessive processor times and data storage requirements for careful single-cell imaging of large populations, and our inability to use FPs to monitor shutdown of the pheromone response (only stable FPs produced detectable signals in our strains). In spite of these limitations, we did obtain experimental evidence of plasmid transfer from specific donor cells in real time, which has not been described previously in this system. However, we found that only a small fraction of induced cells actually transferred the plasmid under the conditions examined.
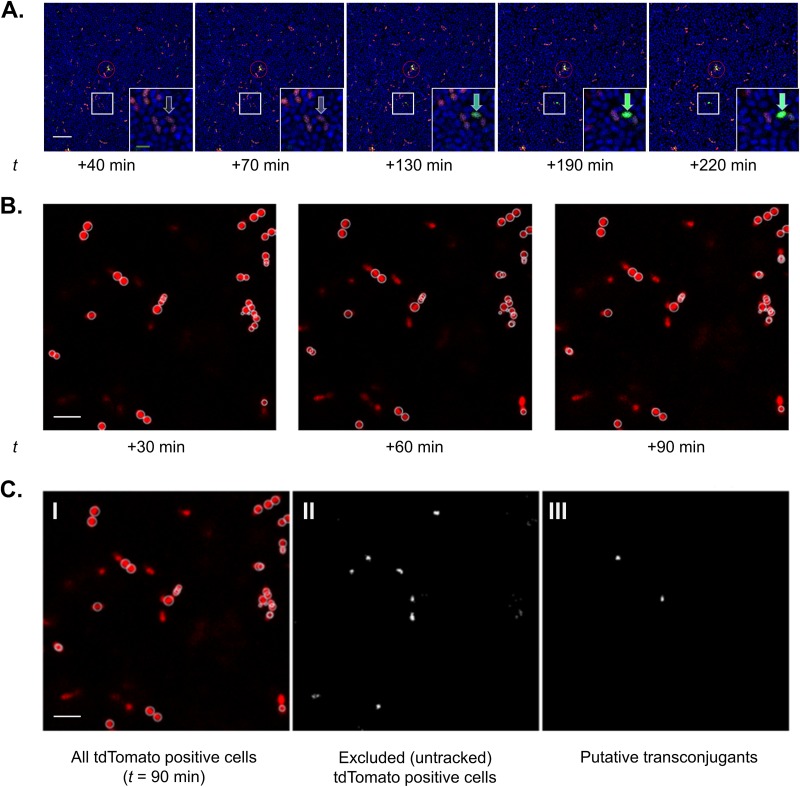
Figure 2 shows representative time lapse microscopic analyses of mixtures of FP-expressing donors and Hoechst-stained recipients growing in close proximity on agarose pads; visual comparisons of the ratios of red to blue cells at the first imaged time point examined (40 min after the mixture was spotted) indicated that both cell types were fairly uniformly distributed, with an apparent ratio of about 1 donor/100 recipients, consistent with the volumes of each cell type (Fig. 2A). Although the donors were given a very short (5-min) treatment with C immediately before mixing with recipients, GFP fluorescence was not detected at the 40-min observation point in the vast majority of the red-fluorescing donor cells; the small number of highly fluorescent cells seen at that time point did not change in appearance over the entire time course and may represent small aggregates or visual artifacts not related to induction (see the circled portion of Fig. 2 for an example). In contrast, a larger proportion of the donor cells (exemplified by the cell depicted in the boxed area magnified in the inset and shown for a much larger field with numerous induced donor cells in Fig. S1 in the supplemental material) showed an increase in signal over the course of the experiment. The slow appearance of induced cells could reflect the lag in GFP folding and also suggests that the donor cells deposited in the agarose were induced by the C produced from surrounding recipient cells during the experiment. By automated periodic imaging of numerous fields over the course of the experiment, we were able to identify recipient cells (blue, Hoechst 33342 positive) that maintained their position in proximity to induced donor cells over the entire course of the experiment but that acquired expression of red fluorescence at later time points (Fig. 2B and C). These cells are believed to represent transconjugants generated in the experiment. From the visual analyses represented in Fig. 2 and Fig. S1, we detected induction of a fraction of donors, but plasmid transfer was only detected from a small minority of these induced donors. It should be noted that pheromone signaling and plasmid transfer in cells embedded in agar pads has not been done before, so these results may not be directly comparable to those from liquid culture experiments or solid-surface matings. Technical issues associated with the long time course of experiments using the agarose pad system also impeded our ability to quantify all induction and transfer events that actually occurred.
FIG 2.
Time-lapse visualization of induction and conjugation via fluorescent reporter protein expression. (A) A dense culture of recipients (blue; Hoechst 33342) and donors (red; constitutive tdTomato expression; exposed to C for 5 min just before mixing) were combined (donor-recipient, 1:100) and sandwiched between an agarose pad and coverslip-bottomed petri dish. Donors express green fluorescent protein (GFP; green) when induced by C. The entire field was tracked every 30 min for 6 h beginning at t =+40 min using time-lapse laser-scanning confocal microscopy; t = +40 to t = +220 min shown here. Induction of a single tdTomato-positive cell (inset). After induction, the cell transcribes the GFP-coupled genes and begins to fluoresce green (arrows). Also, although numerous recipient cells surrounded induced donor cells, induction was not detected for many donors, even over extended time scales. Bars, 10 μm (white) and 2 μm (green, inset). While uncommon, there are rarely donor cells with bright GFP expression even at early (t < 40 min) time points (red circle). Notably, the green fluorescent intensity of these cells was typically static, even over long time periods. (B) Subsequent image analysis allows identification of potential donors (tdTomato positive throughout imaging period) and transconjugants (those cells that become tdTomato positive during imaging). This image shows donor cells that did not change position during the entire experiment. (C) To accurately differentiate potential transconjugants from artifactual signals (nonattached donor cells or autofluorescent debris transiently appearing in the imaging field), individual cells were tracked. White circles indicate cells tracked through all imaging time points during the first 90 min and thus categorized as initial donor cells. Note the rarity of transconjugants. Bar, 2 μm. (C, I) All tdTomato-positive cells observed through the 90-min time point (from panel B). (C, II) The binarized image shows all new red fluorescent signals identified as cells appearing in the field over the 90-min period. (C, III) Among the cells identified in panel C, II, two potential transconjugants met the requirements that potential transconjugants were spatially adjacent to an existing induced donor cell. Other excluded/untracked tdTomato-positive cells were likely secondary to minor deformation of the agarose substrate over time leading to shifts in included focal planes and/or transient, nonattached cells. Bar, 2 μm.
To validate the microscopy results and quantify induction and transfer in mixed cultures, we also analyzed broth mating mixtures by flow cytometry. For these experiments, the donors were preinduced with C for 1 h, prior to mixing with a 10-fold excess of recipients for 2 h. A major complication with these experiments was the tendency of the mixtures to form large clumps—mediated by surface expression of the PrgB adhesin in induced donors (14)—at later time points. Thus, we pretreated the mixtures with EDTA and proteinase K along with sonication to try and disperse clumps prior to analysis by flow cytometry (see methods in the supplemental material for details). These treatments reduced clumping sufficiently to enable to analysis of FP expression levels in populations containing 1 × 105 to 5 × 105 cells for levels of the various FPs. We initially showed that donor cultures not induced with exogenous C contained extremely low numbers of GFP-fluorescent cells (see Fig. S2 in the supplemental material). As depicted in Fig. S3 in the supplemental material, the majority of the donor cells exposed to C for 1 h and then mixed with excess recipients for 2 h were induced, but the putative transconjugants after 2 h of mixed incubation represented <2% of the total population. The relatively low percentage of the donors classified as “induced” that transferred in the above-described experiment could be related to failure of donors (expressing the complete transfer machinery) to contact recipients with sufficient affinity to form a productive mating pair. It is also possible that some apparently induced donors actually did not express all the genes required for a functional conjugation machine. Overall, the lack of transfer of the majority of induced donors observed in these two experiments is in agreement with previous studies, providing justification for the experiments described below.
Single-cell analysis of induced expression of genes throughout the QOp transcript appears similar after induction by high levels of exogenous C.
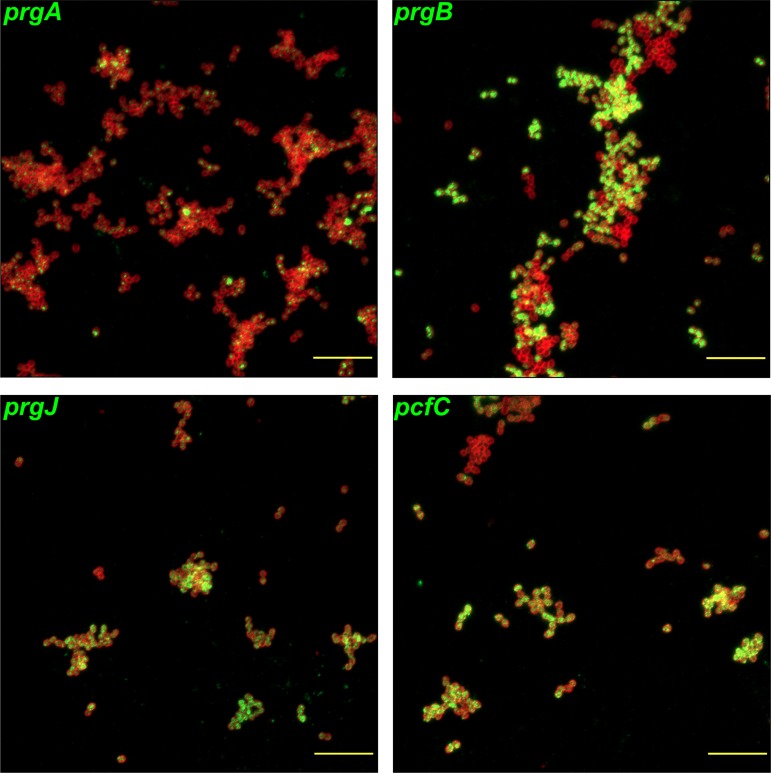
We previously carried out extensive single-cell (9, 15) and population-based (11, 16) analyses of mRNAs produced from the 5′ “early gene” segment of the prgQ operon, as indicators of pheromone induction. To test whether there was significant heterogeneity in QOp transcript length among subpopulations of induced cells, we designed fluorescence in situ hybridization chain reaction (HCR) probes for both early (QL, prgA, and prgB RNAs) and late QOp transcripts (prgJ, pcfC, and pcfG) (Fig. 1A). We examined variation in QOp transcript length in a large population of cells subjected to induction with a high concentration of exogenous C. Cells were harvested before addition of C and at 30 and 120 min after C addition. Expression of induced prgA, prgB, prgJ, pcfC, and pcfG transcripts from aliquots of induced donor cultures were initially visualized via fluorescence in situ HCR labeling with single-transcript probes and microscopy (Fig. 3). Expression of these transcripts was not observed for uninduced cells (harvested before C addition (data not shown, but as reported previously [9]). Expression levels of these transcripts were robust in the majority of cells at the 30-min time point, demonstrating that late QOp transcripts were expressed in a similar percentage of cells as those containing early QOp transcripts (Fig. 3); visual inspection of these images suggests that both early and late prgQ gene expression can be detected by HCR, and in comparable fractions of the induced donor population. Shutdown appeared to be well under way by the 120-min time point (data not shown, but as seen previously [9]).
FIG 3.
HCR labeling of induced genes suggest that induced early and late transcripts appear to be expressed in a similar fraction of cells. E. faecalis OG1RF/pCF10 was harvested for HCR labeling at 30 min after addition of 50 ng · ml−1 C. Induced prgA, prgB, prgJ, and pcfC transcripts were fluorescently labeled by HCR in separate paired samples. Red, cell envelopes labeled with Alexa Fluor 647-wheat germ agglutinin (AF647-WGA) conjugate highlighting the outlines of individual bacterial cells; green, HCR-labeled transcripts (Alexa Fluor 488). Bars, 10 μm.
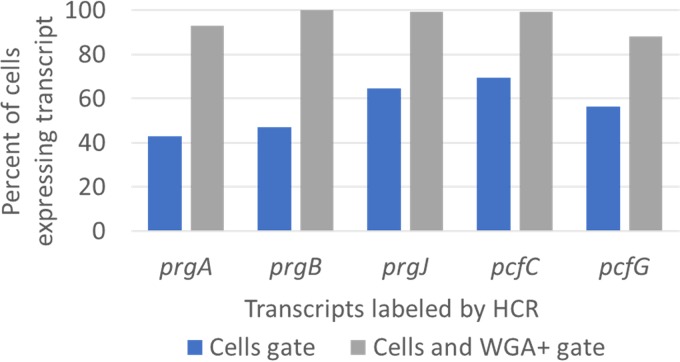
For quantitative analysis of the percentages of cells fluorescently labeled by HCR in larger samples (>1,000 for each sample analyzed) of induced donors, flow cytometry was used (Fig. 4). Cells were labeled with fluorescently tagged wheat germ agglutinin (WGA), which binds the bacterial cell envelope; multiple complementary gating strategies were employed for robustness. Typical forward and side scatter properties of E. faecalis cells were used to gate on probable cells, and staining by WGA was used to eliminate analysis of noncellular particles. Regardless of the gating strategy, the percentage of cells expressing each transcript remained similar for early and late QL transcripts.
FIG 4.
Flow cytometry analysis of HCR labeling of induced genes supports the hypothesis that induced early and late transcripts are expressed in a similar percentage of cells. E. faecalis OG1RF/pCF10 was harvested for HCR labeling at 30 min after addition of 50 ng · ml−1 C. Induced prgA, prgB, prgJ, pcfC, and pcfG transcripts were fluorescently labeled by HCR in separate paired samples. Cells were also labeled with Alexa Fluor 647-wheat germ agglutinin (AF647-WGA) conjugate, which binds cell envelopes. Cells were gated by typical forward and side scatter (FSC and SSC) properties (“Cells gate”), and the percentages of cells with Alexa Fluor 488 HCR staining were quantified. Probable cells based on FSC and SSC properties were also further gated by AF647-WGA staining, and the percentages of cells with Alexa Fluor 488 HCR staining were requantified (“Cells and WGA+ gate”).
In individual cells, induced expression of early QOp genes is similar to that of late QOp genes after induction by low levels of exogenous C.
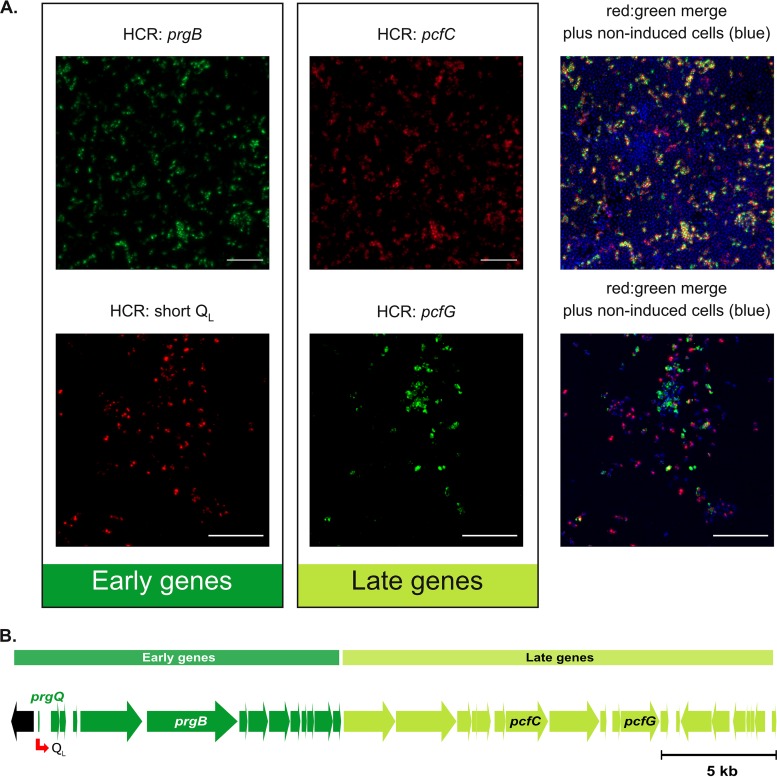
After analysis of early and late QOp transcripts expressed upon addition of high levels of C failed to reveal heterogeneity in late transcript expression (Fig. 3 and 4), we considered whether the lack of heterogeneity could have been due to an artificially high concentration of C. Thus, we next induced with a low level of exogenous C to create more variability in induction and took a more targeted approach, in which we simultaneously labeled representative sets of early and late QOp transcripts in populations of cells harvested 30 min after C addition (Fig. 5). In one instance, we colabeled prgB and pcfC transcripts by HCR. Alternatively, we colabeled a portion of the QOp transcript between IRS1 and IRS2 (QL) and pcfG by HCR. Although this approach did create more heterogeneity in induction (the cultures contained more uninduced cells), visual inspection of the images from both dual-gene HCR labeling experiments strongly indicated that cells induced to express early QOp transcripts also expressed late QOp transcripts. Furthermore, if there was any level of differential labeling of early versus late genes in individual cells, it appeared as though there was actually more expression of the late QOp gene than of the early QOp gene in many cells and that this was not an artifact of the fluorophore used in the HCR labeling. Fluorophore swapping (see Materials and Methods) also allowed us to confirm that these findings were not an artifact of the fluorophore used to label each transcript. Quantification of two-gene expression patterns in single cells, presented below in Fig. 6, confirmed these conclusions.
FIG 5.
Induced E. faecalis cells that express early induced transcripts also express late induced transcripts. (A) E. faecalis OG1RF/pCF10 was harvested for HCR labeling at 30 min after addition of 2.5 ng · ml−1 C. Green and red, transcripts labeled by HCR with Alexa Fluor 488 and Alexa Fluor 647, respectively; blue, nucleic acids (primarily DNA) labeled with Hoechst 33342 highlight individual cells. (Top) Induced prgB (green, early gene) and pcfC (red, late gene) HCR-labeled transcripts. (Bottom) Induced pcfG (green, late gene) and QL (red, early gene) HCR-labeled transcripts. Bars, 10 μm. (B) Schematic showing the distances between the genes whose transcripts were probed using HCR. See Fig. 1 for a more complete map. Not shown is a control experiment in which an identically induced aliquot of donor cells was subjected to HCR analysis for both QL and pcfG expression but where the fluorophores used for detection were swapped; the fractions of positive cells for each transcript were not detectably different from those shown in the images in the lower part of panel A.
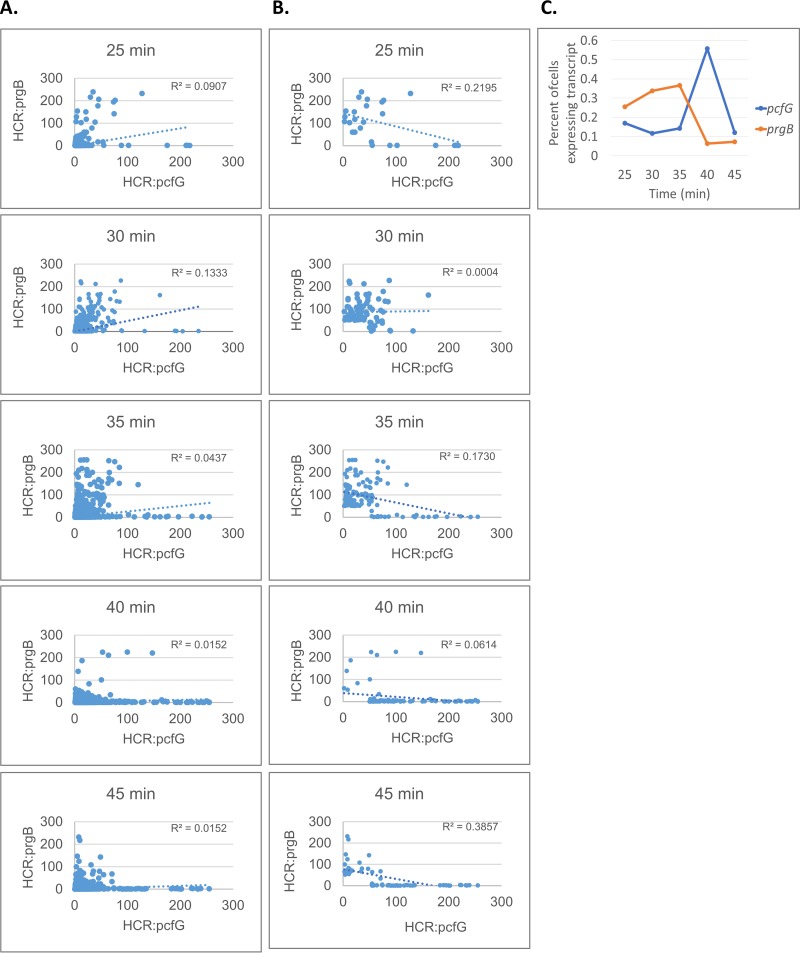
FIG 6.
Analysis of HCR-labeled cells suggests that peak expression of early and late induced genes may be temporally distinct. E. faecalis cells carrying pCF10 were induced as described for Fig. 5 and harvested for HCR labeling at 25, 30, 35, 40, and 45 min after the addition of 2.5 ng · ml−1 C. Induced early (prgB) and late (pcfG) transcripts were fluorescently labeled by HCR and quantified by image analysis, as described above and in Materials and Methods. (A and B) Plotted values reflect relative intensity of the fluorescent HCR labeling in individual cells for each transcript. R2 values reflect the cells plotted on each graph. (A) All analyzed cells are plotted. (B) Only cells over the HCR intensity threshold of 50 for either labeled transcript are plotted. (C) Percentages of cells with HCR staining over the threshold were quantified and plotted for each time point. The total numbers of imaged cells analyzed for each time point are as follows: 7,061 (25 min), 23,114 (30 min), 28,646 (35 min), 15,759 (40 min), and 31,506 (45 min).
Peak expression of early and late induced genes may occur at different times after induction by exogenous C.
To further explore the possibility that late QOp gene expression would be more robust than early QOp gene expression at 30 min after addition of C, we again pursued a dual-gene HCR labeling approach (labeling prgB and pcfG). Cells were harvested at 25, 30, 35, 40, and 45 min postinduction with a low level (2.5 ng · ml−1) of exogenous C (Fig. 6). Fluorescence corresponding to each HCR-labeled transcript was quantified and plotted for every cell observed by microscopy (Fig. 6A) and for only cells above a threshold of induction (Fig. 6B). In both cases, we observed that the occurrence of cells with high fluorescence corresponding to prgB transcripts is greater at earlier time points, while the occurrence of cells with high fluorescence corresponding to pcfG transcripts is greater at later time points. This is also reflected by the slope of the trendlines when cells below the induction threshold are removed (Fig. 6B). Furthermore, quantification of the percentage of cells expressing each transcript at each time point showed that the peaks for induced expression of prgB and pcfG were temporally distinct (Fig. 6C). Assuming an elongation rate of 40 nucleotides/s, there could be a lag of 10 min or more for an elongation complex to traverse the template from prgB to pcfG, which could account for the differences in the timing of peak expression levels. However, the number of cells induced for prgB expression with no detectable pcfG signal was low at all time points, while at the late time points, numerous cells with high levels of pcfG message and undetectable prgB message were apparent. While HCR analysis only captures a snapshot of each cell at a single time point, these data support the dynamics of pCF10 induction that have been observed in the past by population averaging methods (6), where the observed maximum induction and subsequent shutdown occurred temporally in the 5′→3′ direction across the entire QOp transcript.
DISCUSSION
The complex regulatory circuits controlling the enterococcal pheromone response have been studied extensively for pCF10 (2, 17, 18) and for other enterococcal plasmids (14, 19–21). While expression of the entire prgQ operon is induced by C and many details of the pheromone response initiation process have been characterized (2, 22–24), the extent of our knowledge about expression of the downstream genes and how a given induction state is related to functional conjugation ability is limited. We have observed repeatedly that in relatively short matings with preinduced donors (where the transfer frequencies primarily reflect a single round of transfer), only a small fraction, about 1 to 10%, of highly induced donors transfer, even when surrounded by a significant excess of recipients at high densities (11). In recent experiments, we used fluorescent reporters of induction and plasmid transfer to examine induction and transfer frequencies at the single-cell level in real time. When induced donors were mixed with an excess of recipients, both single-cell microscopic analyses (Fig. 2; see Fig. S1 in the supplemental material) and flow cytometric analysis (Fig. S2 and S3) of populations indicated that only a small fraction of induced donors completed conjugation. A potential model to explain these observations postulates premature termination of QOp transcription in large subpopulations of induced donor cells prior to expression of all genes encoding the conjugation machinery. Comparative HCR single-cell analysis of selected pairs of upstream and downstream genes enabled us to rigorously address the possibility that premature termination of transcripts in subpopulations of induced donors takes place and explains the observed discrepancies between induction (as reflected in upregulation of PQ-proximal genes) and transfer ability. Interestingly, our cumulative data strongly suggest that failure of many induced donors to conjugate is not due to lack of transcription for downstream genes; inefficient transfer may thus relate to inefficient formation of stable mating pairs.
Overall, analyses of HCR labeling suggest that cells which initiate induction can complete expression of the full QOp transcript. Furthermore, peak expression of early and late induced genes may be temporally distinct and degradation of transcripts after induced expression is rapid. These new data were the first to explore long QOp gene expression at the single-cell level and show the temporality of induction of genes throughout the QOp transcript. Since the majority of cells that initiate induction appear to complete expression of the full QOp transcript, these data refute the hypothesis that significant subpopulations of cells induced to transcribe early prgQ genes fail to generate full-length transcripts. These conclusions are not inconsistent with earlier microarray and expression data, since these previous studies were not designed to resolve the two models. Spatial constraints could have limited the efficiency of transfer by fully induced cells, accounting for our observations, using fluorescent reporters of induction and conjugation, of low transfer frequencies by induced populations and for those in previous studies (11).
There remain limitations to analysis by HCR transcript labeling. HCR analysis and comparison of gene expression is limited by the fact that the fluorescence signal emitted by HCR-labeled transcripts is more useful for assessing relative levels of transcripts than for absolute values. Absolute numbers of transcripts may be more variable than is reflected by mean HCR intensity values, since HCR labeling functions through an amplification step, in which the HCR amplifiers chain react to increase the fluorescence emitted by a single transcript. Overall, HCR-based analysis is best for judgement of presence versus absence of individual transcripts, including analysis of multiple transcripts within single cells. Additionally, since the HCR technique is designed for detection of transcripts at a single point in time, it is intrinsically unable to track the history of expression within individual cells in its current format.
Given these limitations, it remains possible that there is simply more variation in prgB expression than in pcfG expression. For example, an individual cell may express multiple copies of the prgB transcript (which could be beneficial to individual cells as noted above), while expressing a single copy of a downstream transcript (e.g., pcfG) required for successful conjugation. Such a scenario might result in similar HCR labeling for both transcripts that actually represent different transcript levels that could be reflected by population-level transcript quantification. Slight variation in absolute transcript abundance for early versus late QOp transcripts in individual cells could also be caused by differential stability, very weak terminators, or stochastic termination of transcription due to polymerase fall-off that is known to occur at a low rate for long transcripts (25). However, our data support the conclusion that strong terminators within the long QOp transcript (that might create detectable heterogeneity in transcript length) do not exist.
In conclusion, the data presented here suggest that heterogeneity in induced transcript length in different donor cell subpopulations is not present. Rather, the majority of induced cells go on to express the full QOp transcript encoding all of the functions required for conjugation (induced cell type 3 from Fig. 1B). If strong checkpoint-like terminators in the long QOp transcript were present, we would have expected to find a substantial number of cells that did not express late QOp transcripts (induced cell types 1 and/or 2 from Fig. 1B). While it is still possible that heterogeneity in the exact number of transcripts in individual cells exists at some level, it is unlikely that there are strong checkpoints that create major population subtypes with distinct conjugation potential.
Looking to the overall mechanics of the pCF10 system, this work demonstrates the commitment cells make to undergoing conjugation upon induction. For the propagation of the pCF10 plasmid on an evolutionary time scale, tight inducible control of induction—which reduces superfluous costly production of the conjugation machinery encoded by QOp—may have been essential. Even if a very low percentage of cells may be induced, it is likely that they will express all of the genes required for conjugation and be functionally able to transfer the plasmid. Finally, it would be interesting to define the mechanisms of induced transcript degradation and further develop the HCR technique for temporal use.
MATERIALS AND METHODS
Bacterial growth, induction, and cell harvest for HCR.
E. faecalis strain OG1RF containing the conjugative plasmid pCF10 was used throughout these experiments (26). OG1RF is a derivative of the original OG1 clinical isolate (27) with spontaneous resistance to rifampin and fusidic acid (1, 28). E. faecalis strains were grown statically at 37°C in M9 medium containing 3 g · liter−1 yeast extract, 10 g · liter−1 Casamino Acids, 36 g · liter−1 glucose, 0.12 g · liter−1 MgSO4, and 0.011 g · liter−1 CaCl2. Antibiotics, when used for selection, were at the following concentrations: tetracycline (Tet), 10 μg · ml−11; rifampin (Rif), 200 μg · ml−1; and fusidic acid (Fus), 25 μg · ml−1. Antibiotics were used to verify expected antibiotic resistances and not during experimental growth.
For HCR experiments, overnight cultures were subcultured 1:10 in M9 medium and grown to the early exponential phase (≈2 h to an optical density at 600 nm [OD600] of ≈1.2). Cultures were induced with 2.5 or 50 ng · ml−1 cCF10 (C) peptide, and cells were harvested at 25 to 45 min after C addition. Fixation of cells began immediately upon harvest, when they were mixed 1:1 with electron microscopy (EM)-grade 8% paraformaldehyde (PFA; 4% final concentration) and fixed for >20 h at 4°C. Following fixation, cells were isolated by centrifugation at 13,000 × g for 5 min and subsequently resuspended in phosphate-buffered saline containing potassium (KPBS) with trace RNaseOUT (Invitrogen).
HCR labeling approach.
HCR labeling generally followed our previously published methods (9). Briefly, cells were permeabilized and HCR labeling and counterstaining was done on cells in suspension. To reduce clumps in suspensions analyzed by flow cytometry, we vortexed the cell suspensions in KPBS containing 2 mM EDTA immediately prior to injection into the instrument; gating strategies described in the results section were used to eliminate remaining clumps from the fluorescence analysis. HCR hybridization probes were designed to target genetic loci throughout induced QOp from pCF10, including QL (encoding an RNA; our probes targeted the region between IRS1 and IRS2, encoding the 3′ end of this transcript, which is only expressed in induced cells) and the protein-coding genes prgA, prgB, prgJ, pcfC, and pcfG. Nucleic acid probes and hairpin amplifiers were obtained from Molecular Instruments (for more information, see Tables S1 and S2 in the supplemental material). After cell labeling by HCR and counterstaining by Alexa Fluor 647-wheat germ agglutinin (AF647-WGA) or Hoechst 33342, cells were mounted as previously described (9).
For HCR labeling shown in Fig. 3 and quantified in Fig. 4, different transcripts, including prgA, prgB, prgJ, pcfC, and pcfG, were labeled in paired samples. For detection of these transcripts, HCR amplifiers conjugated to Alexa Fluor 488 were used, and cells were counterstained with AF647-WGAto label cell envelopes.
For HCR labeling shown in Fig. 5, transcripts prgB and pcfC were colabeled in one sample, and transcripts pcfG and “QL” were colabeled in another sample. To confirm the reproducibility of transcript detection in replicate induced cultures and to rule out the possibility that the results could be biased by the fluorophores used to detect specific transcripts, we carried out HCR analysis of pcfG and QL transcripts in an identically induced culture where we swapped fluorophores. We obtained expression patterns indistinguishable from the analysis of these two transcripts shown in Fig. 5A. For these samples, HCR amplifiers conjugated to Alexa Fluor 488 were used to label prgB and pcfG transcripts, while amplifiers conjugated to Alexa Fluor 647 were used to label pcfC and QL transcripts. For both samples, cells were counterstained with Hoechst 33342, which binds nucleic acids.
For HCR labeling quantified in Fig. 6, transcripts prgB and pcfG were colabeled. HCR amplifiers conjugated to Alexa Fluor 488 were used to label pcfG, and HCR amplifiers conjugated to Alexa Fluor 647 were used to label prgB. Cells were counterstained with Hoechst 33342, which binds nucleic acids to facilitate high-throughput image analysis by identifying the HCR fluorescent signals within Hoechst-positive bacterial cells.
HCR microscopy, image processing, and analysis.
For images shown in Fig. 3 and 5 and for image analysis data shown in Fig. 6, images were acquired using an Axio Observer.Z1 confocal microscope equipped with an LSM 800-based Airyscan system in normal confocal mode (Zeiss). Confocal images were acquired using a 100× 1.46-numerical-aperture (NA) objective (Zeiss) with 280-nm z-stacks using Zen software (version 2.1; Zeiss). Image stacks were deconvolved using Huygens (version 19.10.0p1; SVI) and are shown here as maximum intensity projections. Quantification was done via Matlab (version 2015b; Mathworks) as described previously (9). Briefly, the blue fluorescence channel (Hoechst 33342) was used to identify the locations of individual cells, and colocalized fluorescent overlap from HCR labeled transcripts was quantified (Fig. 6A and B). The percentages of cells expressing each transcript were determined (Fig. 6B and C) based on a threshold HCR intensity of 50, which was chosen based on expression in the uninduced population and represents a conservative cutoff for expression (which likely miscategorized some cells with transcripts as cells without transcripts).
HCR analysis by flow cytometry.
For analysis of HCR labeling presented in Fig. 4, flow cytometry was used to characterize HCR labeling. A 10-μl aliquot of HCR-labeled cell samples in suspension were diluted in 1 ml KPBS and analyzed by flow cytometry. Cells were analyzed for fluorescence from AF647-WGA and Alexa Fluor 488 conjugated to the HCR transcript label using a BD Fortessa X-20 flow cytometer and BD FACSDiva software (version 8.0). Data were analyzed using FlowJo software (version 10.2). Flow cytometry settings and the compensation matrix used in this analysis can be found in Table S3 in the supplemental material. Cells were gated by typical forward and side scatter (FSC and SSC) properties (Fig. 4, “Cells gate”), and the percentages of cells with AF488 HCR staining were quantified. Probable cells based on FSC and SSC properties were also further gated by AF647-WGA staining, and the percentages of cells with AF488 HCR staining were requantified (Fig. 4, “Cells and WGA+ gate”).
Fluorescent protein reporter strain construction.
E. faecalis codon-optimized versions of the red fluorescent proteins (RFPs) mCherry and tdTomato (29) were synthesized (DNA2.0). A superfolder GFP construct originally optimized for Streptococcus pneumoniae (30) was also found to work well in E. faecalis. The intergenic region between pcfR and Tn925 was chosen for introduction of the constitutively expressed RFP (tdTomato and mCherry). For the inducible GFP, the intergenic region between prgC and prgD was chosen. Vectors with the homologous recombination sites of pcfR_p23_RestrictionSites:BamHI&SphI_Tn925 and prgC_RestrictionSites:BglII&SphI_prgD were also obtained from DNA2.0. The FPs were restriction digested and ligated in the intergenic regions of the above vectors. Upon successful integrations, the pcfR_p23_RFP_Tn925 and prgC_GFP_prgD were digested and ligated to the pCJK218 vector (containing the pheS counterselectable marker) intended for markerless allelic exchange into the pCF10 backbone (31). The built pCJK218_ prgC_GFP_prgD vector was introduced into E. faecalis by electroporation with selection on brain heart infusion (BHI) agar plates supplemented with chloramphenicol (Cm) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 100 μg · ml−1) at 30°C. Subsequently, the transformants were used to inoculate cultures, grown to an optical density at 600 nm (OD600) of 0.2, and shifted to 42°C for 3 h. Dilutions were spread on BHI supplemented with Cm and X-Gal (100 μg · ml−1) at 42°C. Blue colonies were screened for the integration event by PCR, and the integrant colonies were used to inoculate cultures in BHI and passaged from overnight cultures at 30°C for two successive days in BHI with no selection for the second recombination event to excise the remnants of the vector. Successful integrants were isolated by plating on counterselection medium containing p-Cl-Phe and X-Gal. The resulting white colonies were PCR and sequence verified. This process was repeated for pCJK218_ pcfR_p23_RFP_Tn925 (tdTomato and mCherry) to obtain the final plasmid pCF10_tdTomato_iGFP and pCF10_mCherry_iGFP.
Microscopic analysis of pheromone induction and plasmid transfer using fluorescent protein reporter strains.
Overnight cultures were centrifuged, washed twice with 1 ml KPBS containing 2 mM EDTA, and diluted 1:5 in fresh M9 medium. The bacterial strains for live-cell imaging were grown to the exponential growth phase by incubating the cell suspension for 1 h at 37°C. The recipient cells were counterstained with Hoechst 33342 and washed. Donor cells similarly grown to the exponential phase, exposed to 10 ng/ml C for 5 min, and then immediately washed (but not counterstained) were mixed with the recipients at a ratio of ∼1 donor/100 recipients. The cell suspension was concentrated in 100 μl, and 10 μl of the cell suspension was added onto an agarose pad made by sandwiching molten agarose (M9 growth medium containing 0.6% low melting temperature agarose) in between 2 coverslips. The cells were then “sandwiched” between the agar pad and a glass-bottomed petri dish coated with poly-d-lysine (MatTek Corp.). A 3-ml aliquot of molten (37°C) agar was then added to the dish and left at room temperature for a few minutes to ensure complete solidification of the agar before mounting of the specimen for imaging.
Imaging for monitoring induction and conjugation events was done using a Zeiss LSM800 inverted laser scanning confocal microscope using the 405-nm, 488-nm, and 561-nm lasers for excitation of Hoechst 33342 stain, CFP, GFP, or tdTomato, respectively. Confocal images were acquired through a 63×, 1.40-NA oil immersion objective (Zeiss). Images were acquired of 1/16 of the field in a tile pattern every 30 min, starting 40 min after mixing the donors with recipients and over a total time course of 4 to 6 h. Resulting image stacks were deconvolved and aligned using Huygens Professional (version 19.10.0p1; SVI) using recommended parameters; composite figures were generated with maximum intensity projections using ImageJ (v 1.52q; NIH).
Image analysis for fluorescent protein reporter strains.
Images were imported into ImageJ (version 1.49m; NIH) and subjected to background subtraction with a rolling ball radius of 50 pixels using the internal ImageJ function. The background-subtracted images were flattened using a maximum intensity projection and exported as TIFF images. These preprocessed images were then imported into Mathematica (Wolfram Research) for further analysis, depicted in Fig. 2 and Fig. S1. To identify cell positions, the red channel (corresponding to tdTomato fluorescence used as a reference channel) images from all time points were binarized, and then distance transform and maximum detection functions were applied to create cell location markers. Using the markers previously found as positional information, a watershed transformation was applied to the binarized images, and the SelectComponents function was used to identify any object found by the watershed algorithm. The ComponentsMeasurements function was then used to find the centroid coordinates and the equivalent disk radius of all identified cells. These cell location markers were used to mark all tdTomato-positive (donor plus transconjugant) cells. Using these coordinates as starting points, the algorithm ImageFeatureTrack was applied to all images within the time series to track the tdTomato-positive cells through the images. The tdTomato-positive cells that were tracked though the entire time course were likely to form the initial pool of donor cells in the field. In order to identify new tdTomato-positive cells, these continuously positive cells were excluded from all the time points by using a black disk created using the centroid location and equivalent disk radius. The untracked tdTomato-positive cells consist of the potential new transconjugants and cells which may have drifted in from adjacent lateral locations or z planes. In order to narrow our calls for potential transconjugants, we applied an additional restriction that the untracked tdTomato-positive cells must be adjacent to a tracked preexisting tdTomato-positive/GFP-positive (induced donor) cell.
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
These studies were supported by NIH grant R35GM118079 (G.M.D.). R.J.B.E. was supported in part by predoctoral training grants (T32-GM008347 and T90-DE0227232). A.M.T.B. was supported in part by a postdoctoral training grant (T32-AI055433). S.A.O. was also supported in part by the T32-GM008347 predoctoral traineeship.
Computational resources were provided by the Minnesota Supercomputing Institute (MSI) at the University of Minnesota (http://www.msi.umn.edu).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 3.Bensing BA, Manias DA, Dunny GM. 1997. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol 24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Bandyopadhyay A, Kozlowicz BK, Haemig HAH, Tai A, Hu W-S, Dunny GM. 2017. Mechanisms of peptide sex pheromone regulation of conjugation in Enterococcus faecalis. MicrobiologyOpen 6:e00492. doi: 10.1002/mbo3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandyopadhyay A. 2018. Systems analysis of pheromone signaling and antibiotic resistance transfer in Enterococcus faecalis. PhD thesis University of Minnesota, Minneapolis, MN. [Google Scholar]
- 6.Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol 187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlowicz BK, Dworkin M, Dunny GM. 2006. Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity? Int J Med Microbiol 296:141–147. doi: 10.1016/j.ijmm.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook L, Chatterjee A, Barnes A, Yarwood J, Hu W-S, Dunny G. 2011. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol 81:1499–1510. doi: 10.1111/j.1365-2958.2011.07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breuer RJ, Bandyopadhyay A, O’Brien SA, Barnes AMT, Hunter RC, Hu W-S, Dunny GM. 2017. Stochasticity in the enterococcal sex pheromone response revealed by quantitative analysis of transcription in single cells. PLoS Genet 13:e1006878. doi: 10.1371/journal.pgen.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandyopadhyay A, O’Brien S, Frank KL, Dunny GM, Hu W-S. 2016. Antagonistic donor density effect conserved in multiple enterococcal conjugative plasmids. Appl Environ Microbiol 82:4537–4545. doi: 10.1128/AEM.00363-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee A, Cook L, Shu C-C, Chen Y, Manias DA, Ramkrishna D, Dunny GM, Hu W-S. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110:7086–7090. doi: 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatty M, Camacho MI, Gonzalez-Rivera C, Frank KL, Dale JL, Manias DA, Dunny GM, Christie PJ. 2017. PrgU: a suppressor of sex pheromone toxicity in Enterococcus faecalis. Mol Microbiol 103:398–412. doi: 10.1111/mmi.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes AMT, Dale JL, Chen Y, Manias DA, Greenwood Quaintance KE, Karau MK, Kashyap PC, Patel R, Wells CL, Dunny GM. 2017. Enterococcus faecalis readily colonizes the entire gastrointestinal tract and forms biofilms in a germ-free mouse model. Virulence 8:282–296. doi: 10.1080/21505594.2016.1208890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clewell DB, Dunny GM. 2002. Conjugation and genetic exchange in enterococci, p 265–300. In Gilmore M, Clewell D, Courvalin P, Dunny G, Murray B, Rice L (ed), The enterococci. ASM Press, Washington, DC. [Google Scholar]
- 15.Erickson RJB, Manias DA, Hu W, Dunny GM. 2019. Effects of endogenous levels of master regulator PrgX and peptide pheromones on inducibility of conjugation in the enterococcal pCF10 system. Mol Microbiol 112:1010–1023. doi: 10.1111/mmi.14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny GM, Antiporta MH, Hirt H. 2001. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides 22:1529–1539. doi: 10.1016/s0196-9781(01)00489-2. [DOI] [PubMed] [Google Scholar]
- 17.Dunny GM, Berntsson R. 2016. Enterococcal sex pheromones: evolutionary pathways to complex, two-signal systems. J Bacteriol 198:1556–1562. doi: 10.1128/JB.00128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breuer RJ, Hirt H, Dunny GM. 2018. Mechanistic features of the enterococcal pCF10 sex pheromone response and the biology of Enterococcus faecalis in its natural habitat. J Bacteriol 200:e00733-17. doi: 10.1128/JB.00733-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clewell DB. 2011. Tales of conjugation and sex pheromones: a plasmid and enterococcal odyssey. Mob Genet Elements 1:38–54. doi: 10.4161/mge.1.1.15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. 2013. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70:289–302. doi: 10.1016/j.plasmid.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67:277–301. doi: 10.1128/mmbr.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bensing BA, Meyer BJ, Dunny GM. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci U S A 93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shu CC, Chatterjee A, Dunny G, Hu WS, Ramkrishna D. 2011. Bistability versus bimodal distributions in gene regulatory processes from population balance. PLoS Comput Biol 7:e1002140. doi: 10.1371/journal.pcbi.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee A, Johnson CM, Shu C-C, Kaznessis YN, Ramkrishna D, Dunny GM, Hu W-S. 2011. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci U S A 108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters JM, Vangeloff AD, Landick R. 2011. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol 412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunny G, Funk C, Adsit J. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- 27.Gold OG, Jordan HV, van Houte J. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch Oral Biol 20:473–477. doi: 10.1016/0003-9969(75)90236-8. [DOI] [PubMed] [Google Scholar]
- 28.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 30.Martin B, Granadel C, Campo N, Hénard V, Prudhomme M, Claverys JP. 2010. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol 75:1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x. [DOI] [PubMed] [Google Scholar]
- 31.Vesić D, Kristich CJ. 2013. A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J Bacteriol 195:1815–1824. doi: 10.1128/JB.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.