Abstract
In the constructed habitat in which we spend up to 90% of our time, architectural design influences occupants’ behavioral patterns, interactions with objects, surfaces, rituals, the outside environment, and each other. Within this built environment, human behavior and building design contribute to the accrual and dispersal of microorganisms; it is a collection of fomites that transfer microorganisms; reservoirs that collect biomass; structures that induce human or air movement patterns; and space types that encourage proximity or isolation between humans whose personal microbial clouds disperse cells into buildings. There have been recent calls to incorporate building microbiology into occupant health and exposure research and standards, yet the built environment is largely viewed as a repository for microorganisms which are to be eliminated, instead of a habitat which is inexorably linked to the microbial influences of building inhabitants. Health sectors have re-evaluated the role of microorganisms in health, incorporating microorganisms into prevention and treatment protocols, yet no paradigm shift has occurred with respect to microbiology of the built environment, despite calls to do so. Technological and logistical constraints often preclude our ability to link health outcomes to indoor microbiology, yet sufficient study exists to inform the theory and implementation of the next era of research and intervention in the built environment. This review presents built environment characteristics in relation to human health and disease, explores some of the current experimental strategies and interventions which explore health in the built environment, and discusses an emerging model for fostering indoor microbiology rather than fearing it.
Keywords: Biomonitoring, Dermal exposure, Disease, Environmental monitoring, Epidemiology, Personal exposure
From fear to reluctant acceptance of the microbial world
Long before knowing why it worked, the ill were quarantined to prevent the spread of disease [1]. The idea that microorganisms were solely disease-causing agents to be avoided began with the discovery that basic hand hygiene and aseptic techniques reduced the spread of disease [2, 3], and led to the proposals of Germ Theory in the 1860s [4, 5] and Koch’s postulates in the 1890s [6]. This idea captured our imagination [7], and shaped public policy [8, 9], protocols, best practices, and infection control training—particularly in the healthcare and food service industries. Successive developments in microbiology and microbial ecology, technology, and theory advanced our understanding of microorganism taxonomy, anatomy, physiology, ecology, and infectious potential, and continued to refine our relationship to microorganisms and move us past a strict definition of Germ Theory [4, 10].
In the early-1900s, culture-based investigations revealed the possibility of a commensal microbial community [11]—that which benefits from its association without impacting the host. In the following decades, led by early work from Hungate on protozoa in cattle [12] and McNall-Ngai et al. on squid and Vibrio fischeri [13, 14], the understanding that microorganisms could not only be commensal, but mutualistic with their hosts, began to develop. Investigations into host-microorganism relationships exposed a multitude of microorganism-driven benefits for the host, and even the reliance of some microorganisms or strains on host ecosystems: thus, the abstraction of host-associated microorganisms was solidified [12, 14–16]. Even more nascent is the concept that microbial exposure is integral to our health [17], refuting the paradigm that pathogenic microorganisms are consistently pathogenic, and that some microorganisms could, in fact, be beneficial and essential due to their antagonism with hosts. Our wariness of infectious disease led us to become infatuated with preventing microbial exposure, including those essential microbial-host interactions which were only latently understood. Concurrent to this, urbanized populations saw a rise in allergies, asthma, and inflammation-related chronic diseases, many of which were or seemed to be linked to a lack of microbial exposure; a correlation which was described by the Hygiene Hypothesis [18, 19]. In response, calls propose to redefine “hygiene” from practices that remove all microbiota to simply acting to prevent the spread of pathogenic microorganisms [20]. Yet, not all microorganisms interact with a host in the same way, and not all interactions lead to positive outcomes, and the Hygiene Hypothesis as originally defined did not encompass this nuance [21].
The collective change in ideology regarding microorganisms as beneficial has led to the development of the Old Friends Hypothesis [22]—that early childhood exposure to microorganisms can properly tune the developing immune system such that it will not overreact to future challenges, and aid in establishing a diverse, symbiotic host-associated community. The change in ideology has permeated enough to be implemented into practice. The health sector has redefined its view of the roles that microorganisms play in human health, particularly in establishing an active, yet restrained, immune system and that of a stable and functional microbial community in preventing the overgrowth of certain taxa or activation of pathogenesis [17–21]. Health procedures have even begun to incorporate microorganisms into treatment or prevention regimens, in particular with fecal microbial transplant or probiotics to help stabilize the gut microbiota against disturbance or to recover a diverse community [23–27].
Microorganisms can be found in every environment, including built environments—our habitat of buildings, roads, vehicles, and other human-associated spaces—and even in ultra-clean rooms and space stations [28]. Collectively, these factors mold the built environment’s microbiome [29–31]—all the genetic material of the microbial members present, from which can be derived taxonomic identification and metabolic potential, including activities like nutrient utilization and antimicrobial production [30, 32]. The indoor microbial community is a blend of microorganisms sourced from various ecosystems which seed each particular built environment. Thus, no two homes or two offices share the exact same microbial footprint.
The majority of interest in microbiology of the built environment has focused on microbially-mediated building deterioration or detriment to occupant health from specific microorganisms of interest. Despite advances in awareness of the potential for microbial exposure indoors [33], microorganisms in the built environment are still considered as debris to be removed, instead of members of a microbial ecosystem which is inexorably linked to the influences of building inhabitants [29, 31, 34–37]. Given the propensity for microbial biofilms or overgrowth to pose a threat to the health of occupants, it is not difficult to understand why. Yet, in the same way that a stable host-associated microbial community can resist disturbance that would shift the community and cause harm to the host, indoor microbiologists and architects have pondered the equivalent in the built environment—i.e., a ‘house-associated microbial community’. These so-called ‘healthy buildings’ or ‘bioinformed designs’ would keep microbiology in mind [38–40]. Hypothetically, these designs would acknowledge the impossibility of ridding the built environment of microorganisms indoors and instead learn to carefully use selective pressures and microbial sourcing to cultivate a healthy, symbiotic microbiome [41]. Examples of current practices and supporting research are discussed below.
Insularity or connectivity contributes to microbial transmission between occupants
Architectural design affects human or animal behavior, i.e., their interactions with each other and their surroundings, indoors [42, 43] and how and where we emit and deposit our unique individual microbial communities [44–46]. The occupant-associated proportion of indoor microbial communities can be directly attributed to the individuals who spend sufficient time within that space [43, 47–52] and what household activities are performed, allowing for epidemiological tracking of infectious organisms [53] or microbial forensics [54]. The amount of occupancy in buildings, influenced by building type, occupancy schedule, and indoor activity, facilitates the accrual of human-associated microorganisms [34, 44, 55, 56]. Higher occupant density and increased indoor activity level typically increases social interaction and connectivity, through direct contact, indirect contact with shared surfaces, or via shared air, all of which may facilitate the spread of disease [57–59]. For example, schools and libraries, which have long occupancy schedules, high occupant density, and high turnover of individuals throughout spaces in a day, often have high concentrations of putative human pathogens in the air [60, 61]. Overcrowding in hospitals contributes to the risk of patients contracting a hospital-acquired infection (HAI) during their initial stay, as well as to staff illness [62, 63]. Despite incorporating many ways to isolate occupants, prisons are not designed with infection control in mind [64]; hygiene practices are hampered by resource rationing or location of hygiene facilities. Facilities may be overcrowded, proper infection control training may not be provided to staff, and inmates are under stressful conditions which can modify the host response and their personal microbiome, leaving them vulnerable to infectious disease [64].
The primary avenue of reducing person-to-person microbial transfer is quarantine, achieved indoors by spatial or temporal separation. Quarantine remains an effective way of preventing the spread of infectious agents; however, it is typically applied reactively to control a known infection, and many infectious agents are shed asymptomatically, e.g., Clostridioides difficile [65, 66]. Single-patient rooms in hospitals or assisted-living facilities create a proactive, modified quarantine, which may reduce patient stay durations and reduce errors in care [67]. In work spaces, quarantine may take the form of single-occupancy offices rather than shared or open floor plans, which in survey-based assessments, are correlated with more employee sick leave [68, 69]. However, open floor plans may support aspects of teamwork or increased access to natural light and air which make them desirable or necessary to a space. Likewise, model simulations of social networking and disease transmission indicate that schools that reduce interaction and contact within and between groups of elementary students would be able to reduce influenza transmission more effectively than total school closures [70], which provide the opportunity for out-of-school contact. While effective, quarantine is often at cross purposes with spatial habitation and may be impossible, or only possible for only short periods of time, and in most cases requires significant infrastructure and, therefore, cost.
A contrasting strategy to reducing human-human microbial transmission indoors that has only recently been proposed is simply a return to social practices that embrace positive host-microbial transfers to facilitate a diverse human microbiota. Cohabiting humans share microbiota with each other, the degree of which corresponds to the nature of their relationship and level of intimacy they share [49, 71]. In terms of the built environment, this involves combining space types or otherwise fostering certain human-human interactions. For example, the integration of child-care and aged-care facilities seeks to improve emotional, social, and physical wellbeing [72], and may be inadvertently improving microbial-wellbeing as well. The human microbiome becomes disordered later in life [73–75], which can decrease resistance and resilience to disturbance. Providing social interaction with children, who have a diverse microbiota and whose gastrointestinal tracts still contain beneficial early-colonizer taxa such as bifidobacteria [74, 76], may help older adults recolonize their own microbiota—something like a “two-legged probiotic”. However, microbial transfer in these instances cannot be limited to potentially beneficial microorganisms, and the transference of pathobionts or viruses will pose a risk to elderly adults.
Correspondingly, “host-sourced probiotics” may come in a four-legged version: exposure to dogs, but not cats, are associated with decreased risk of developing asthma or allergies if children are exposed early in life [19], attributed to differences in pet species’ microbial community. In mouse models, early exposure to dust from homes with dogs reduced the immune response to various allergens later in life and altered the gut community [77, 78]. Specifically, exposure to dust from homes with dogs increased the abundance of the bacteria Lactobacillus johnsonii in the gut [77, 78], and even oral administration of the bacteria alone demonstrated similar positive effect to that of the dog-associated dust [77, 78]. Colonization of the gut by L. johnsonii creates favorable conditions for co-colonization of the beneficial bacteria Bacillus fragilis by providing an N-glucan biosynthesis pathway, something which is reduced in the gut of asthmatics [79]. Oral probiotics are often effective in managing respiratory illness symptoms [80], and a small but encouraging amount of evidence exists on aerosolized or inhaled probiotics to similar effect [81, 82]. Intranasal Lactobacillus paracasei reduced inflammation and immune system markers but increased neutropenia in the lungs of mice [81], and aerosolized Lactobacillus rhamnosus protected mice against transplanted lung tumors [82]. Importantly, successful probiotics for humans, in these and other studies, are comprised of lactic-acid bacterial species which are typically found on or near human mucosal surfaces at various body sites, but are not found in the built environment except when sourced from mammalian occupants. Even then, lactobacilli often do not survive the aerobic conditions of buildings.
While the concept of prescribed host-microbial transfer between individuals holds promise, it is also at cross-purposes with instances where quarantine would be prudent. It is important to recognize that in any given public building, be it hospital, school, or office building, there is a diversity of individuals with differing degrees of immune system health and inflammatory exacerbation. Therefore, architectural design and building operation would ideally be flexible and adaptable enough to respond in a manner to best serve differing communities of human inhabitants.
Buildings as microbial reservoirs
Architecture influences building biogeography and patterns of microbial dispersal indoors [44, 46]; the building materials, surfaces, and products we use [43, 83, 84]; the indoor environmental conditions (e.g., temperature, humidity, light, and airflow) [55, 85, 86]; and the connectivity of the indoors to the outdoors and its associated microorganisms [34, 87], all of which can affect the location of microorganisms in the built environment and survival once there. Despite the seeming inhospitality of the built environment [88], many microorganisms can survive indoors for months [89–92], and environmental conditions indoors can facilitate intermittent bacterial and fungal growth [86]. Moreover, there are plenty of nutrient substrates. Human skin squamous cells shed indoors serve as a ready food source for bacteria living in HVAC systems [93], and sufficient nutrients exist in house dust to host a living bacterial community even after 90 days of isolation [85].
The concern for health arises from the interaction between humans, insects, pets, and other occupants and their built environments, which can be described as a bidirectional relationship; humans shed host-associated microbiota [50, 94] as well as track microbiota from their daily lives back to built environments, yet may also reacquire these microorganisms from built environment surroundings [95–97]. For example, hospital patients are more likely to acquire a HAI if the previous room occupant was diagnosed with an HAI [98], and are susceptible to infection by building microorganisms through air, water, and surface or aerosol transmission [99].
Moisture and relative humidity
Moisture is one of the most potent contributors to microbial survival in air and on surfaces [100–103], including resistance to electrostatic charges on surfaces [104], microbial activity [86, 105], and the structure of the microbial community overall as survivors prosper [34, 56, 106]. The ability of microbial cells or spores to become aerosolized from surfaces and be resuspended into air due to occupant traffic or disturbance [107, 108] is increased by a low-relative humidity [109]. Low-relative humidity, such as 20–30%, increases the infection rates of aerosolized particles, such as influenza [110, 111], as well as overall HAI frequencies [112]. This reflects two mechanisms of low relative humidity: (1) the drying of host mucous membranes which increases susceptibility to infection [111, 113], and (2) the increased potential for particles to aerolosize as smaller droplets, stay aloft longer, and travel further distances [111]. Low humidity is also perceived as less comfortable and less fresh [111].
High relative humidity, such as >80%, contributes to microbial survival and activity on surfaces (Fig. 1) [100–103]. It is associated with more mucosal irritation and antigenic potential from fungi [109, 114] and can better facilitate the direct-contact transfer of microorganisms from fomite to host [95]. Microenvironments within surfaces, particularly carpet, can create pockets of higher relative humidity which can further mediate transfer from surfaces (Fig. 1) [115]. Similarly, air thermodynamics creates updrafts and other turbulence which can bring microorganisms into a person’s breathing zone [116], and warm or moist air rises and can create pockets of higher relative humidity when trapped by ceilings. However, by allowing for larger aerosolized particles, high relative humidity can cause particles to settle out of air more quickly. A variable gradient or localization of relative humidity within a building or a space may be required to provide optimal conditions for reducing the dispersal and contraction of potential pathogens in the built environment [111].
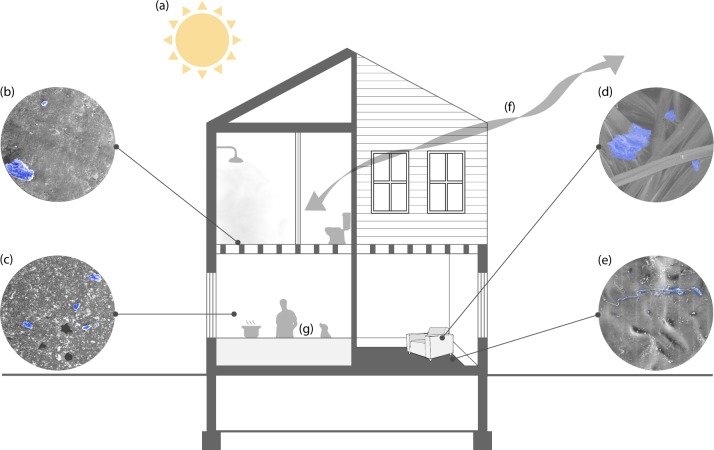
Fig. 1.
Environmental factors and subsequent effects on built environment microorganisms. a Sunlight (both UV light and visible light) have been shown to alter the survival of microorganisms living in the built environment. b Biofilms can form on common built environment surfaces, especially in moist areas such as sinks and showers in bathrooms, facilitating transfer through everyday activities and rituals. c Ambient household conditions constitute a major factor that contributes to the survival and spread of microorganisms. Moisture from daily activities, such as cooking, allows formation of biofilms while and high relative humidity increases the rate of aerosolized microbial cells and spores. Indoor air temperature alters the rate at which built environment inhabitants shed microorganisms. d Frequented household items, such as chairs, can harbor a plentiful supply of nutrients such as human skin squamous cells and other nutrients for microorganisms from built environment inhabitants such as humans, insects, pets, and many others. e Microenvironments within carpet can create pockets of high relative humidity that can aid in the growth, prolonged survival, and transfer of microorganisms from fomite to individual. f Windows provide an access point for microorganisms from the outdoors to access the built environment, contributing to each building’s unique microbial makeup. Ventilation through windows provides air exchange that aids in the reduction of potentially contaminated air. g Humans, insects, pets, and other occupants exchange microorganisms from their own personal microbiome with their built environments contributing to the two-way exchange between hosts and built environment
Dampness or water damage which supports microbial biofilms is especially important for overt microbial growth, especially fungal or mold growth, and biodeterioration of building materials (Fig. 1) [117–121]. Dampness has also been extensively studied in relation to the exacerbation of asthma symptoms [122], and these efforts are not repeated in this review. Motile Escherichia coli are capable of traveling from sink traps back up the drain into the sink, at a rate of 2.5 cm per day, from where they spread to other areas via droplet spray during sink use [123]. Similarly, water heaters host bacterial communities which can spread Mycobacterium via aerosolization at faucets and showerheads [124–126], and Legionella pneumophila can persist in building ventilation systems and infect building occupants [127, 128].
However, development of building systems or cleaning products which intentionally create biofilms have also been utilized to positive effect. Recently, microbial biofilms have been used to reinforce building surfaces instead of decomposing them; namely, through mineralization and deposition of calcium carbonate into concrete [129]. The hydrophobic nature of many bacterial biofilms can impart that protective hydrophobicity onto building surfaces [130], as well. Biofilms have also been used to remediate water and air, especially when other filtration, e.g., reverse osmosis, or sterilization techniques are impractical. Microbial biofilms treat building waste water and even provide non-potable water for reuse within building systems [131]. Similarly, active “green walls” have been used to improve air quality (measured via clean air delivery rate), by utilizing a combination of plant and plant- or soil-associated microorganisms to degrade particulates in conjunction with active airflow [132–134]. However, green walls do have the potential to increase humidity indoors, and poor infrastructure in installations can result in moisture-related structural damage, potentially providing adequate microbial growth conditions. The potential health benefits or infection risks of intentional microbial biofilms indoors have not been thoroughly examined, yet the possibility exists to try and make use of biofilms particularly where they might not interact with occupants.
Temperature
Ambient indoor air temperature has been both positively [34, 114, 135] and inversely [56, 114] correlated with indoor microbial community richness (Fig. 1). It is likely that this correlation has more complex underpinnings, such as the relation of temperature to relative humidity, and with seasonal or local weather conditions and the resultant change in building operations. Most of the studies noting a change in the microbial community correlated with temperature were in buildings utilizing air conditioning [34, 56, 135]. Given that most buildings are maintained at temperatures conducive to microbial growth, it seems less probable that temperature alone is a suitable mark for microbial control indoors, with the exception of viruses which are more sensitive to temperature [103]. However, ambient indoor air temperature does affect animal and human hosts, and the rate at which they shed microorganisms or the degree to which they are susceptible to them. For example, low air temperature (5 °C) did not affect the susceptibility of guinea pigs to acquiring aerosolized influenza virus as the immune system response was not impaired, but this low air temperature did cause infected guinea pigs to produce and shed more virions in oronasal fluids, to produce virions for 40 h longer, and to increase the rate of transmission [110].
Lighting
A great deal of work has focused on the effect of light and architectural design to improve human health, productivity, and comfort [136, 137], specifically pointing to the beneficial effects of full-spectrum sunlight on human physiology. As early as 1845, it was understood that sunlight in buildings was important for human health and there were calls from the medical community to repeal the 100-year old Glass Tax in Britain [138]. The shift towards industrialization, and the reduction of natural daylight indoors, to which the glass tax contributed, was having noticeable health effects for those spending more and more time indoors [136]. In the pre-antimicrobial era, sunlight was used to treat microbial infections such as tuberculosis of the skin [139, 140], and architectural design responded to accommodate health-based infrastructure [136, 140, 141]. In the late 1800s, researchers began studying the bactericidal effects of sunlight [142–144]; quickly realizing the importance of sunlight for controlling pathogenic bacteria, particularly in health care facilities [145–147]. There were even concerns that tall buildings in cities would block the sunlight needed to kill the agent of Typhoid Fever [148]. In cities like New York the skyline still reflects codes related to building setbacks for access to light and air, and the British Law of Ancient Lights, dating back to the Prescription Act of 1189 highlights the understanding of the value of light for health [149].
A large body of research has been conducted on the effect of full-spectrum (sunlight) light [141, 150, 151], ultraviolet light [147, 152–156], and blue or red wavelengths [157–160], on mono-cultured bacterial or fungal survival and activity (Fig. 1). Additionally, light has been shown to reduce virus virulence and increase viron decay [103, 141, 161]. However, microorganisms, particularly bacteria, may easily evolve resistance to UVA and UVB light exposure [153]. UVC light to control microbial growth has recently gained popularity because there is no a priori resistance to it, as it does not penetrate the atmosphere. While effective [155], it is logical to predict that resistance to UVC might develop over time with increased exposure. Only a few studies evaluate the effect of light on whole microbial communities related to the built environment, including those in house dust or on human skin [85, 162, 163]. A recent study demonstrated that different light wavelengths affect the survival of bacteria in complex dust communities differently: visible or UV light reduces the number of living bacteria in dust and results in a less human-associated bacterial community than darkness does [85, 143]. Similarly, indoor surface bacterial communities contained fewer human-associated taxa as the amount of illumination in hospital rooms increased [164]. In terms of light exposure, it appears that daylighting schemes which benefit humans in a myriad of ways also facilitate control of bacterial populations indoors [141], and is likely one of the easiest strategies to support “healthy buildings.” Anecdotally, when architects describe “healthy building” principles, they routinely speak of access to daylight and outside air, and this is supported by the prioritization of daylight in building performance rating systems such as LEED and WELL [165, 166].
Draining or filling the building microbial reservoir
Control of indoor microbial communities typically involves the removal of microbial biomass. New infection-prevention standards have been integrated into healthcare facilities’ architectural designs and clinical procedures [167, 168]. Sophisticated technology exists to monitor hand-hygiene compliance, “time-outs” during surgical procedures for safety checklists, and surface disinfection [169]. Mechanical filtration, non-ionizing UVC, disinfection of building cavities and wall assemblies, room pressurization and laminar ventilation air-flow, have been implemented to control pathogen spread [170], yet, few studies confirm that these interventions decrease HAI [103, 110, 171, 172]. This may be due to the fallibility of typical cleaning behaviors. Common surface cleaning techniques do not remove all microbial biomass from surfaces, where, after just a few days [173], microbial communities are found even in clean rooms or reagents that are considered to be sterile [28, 174–176]. Furthermore, the use of cleaning materials which are themselves contaminated with bacteria simply relocate bacteria between surfaces without removing them [177]. Often times, the use of powerful antimicrobial cleaning solutions can contribute to complacency by cleaning staff [178], and adherence to cleaning protocols may lapse over time [179], leading to the persistence of microorganisms in spaces that are considered clean [176]. However, education on correct and thorough cleaning has been demonstrated to significantly decrease microbial load on frequented surfaces in healthcare environments [180].
Disruption of microbial transfer can be achieved in real-time using ventilation with sufficient air exchanges and/or directed air flow to reduce the impact of potentially contaminated air [57, 58]. This has been widely implemented in health care settings or clean rooms and spaces, and has been proposed for other crowded public spaces [181]. While appropriate ventilation may not effectively reduce all infection transmission, depending on microbial transmission potentials and pathology, it has been demonstrated to reduce the spread of bacterium Mycobacterium tuberculosis, as well as a number of viruses: measles, chickenpox (varicella), influenza, smallpox, and severe acute respiratory syndrome (SARS) [182]. Natural ventilation, which has a lower energy impact and can help mediate relative humidity, has been demonstrated to reduce disease transmission risk [141, 183], and to reduce the human occupant microbial signal in dust by introducing outdoor-associated microorganisms [48, 55, 184]. However, high ventilation rates which cause too much turbulence can negatively affect health by retaining particulate matter in the air [185].
The concept of promoting a dynamic and living indoor microbial community is not widely accepted, and infection control practices continue to view indoor microbiota as a problem that can be rectified given the right cleaning protocols. With the demonstrated failure of current cleaning protocols and mandates, some have investigated the intentional addition of benign microorganisms in the built environment to control pathogenic microorganisms and neutralize the potential negative impact that the microbial world has on our health. In recent years, microbial-based cleaning products (MBCPs) have gained popularity [186] and typically include microorganisms that can outcompete and degrade organic elements of pathogenic microorganisms [187]. The most commonly utilized microorganisms typically belong to the genera Archromobacter, Actinobacter, Alcaligenes, Arthrobacter, Rhodopseudomonas, Rhodobacter, and Lactobacillus [186], the majority of which have been found to degrade xenobiotic compounds [188] and are widely accepted as non-hazardous. Bacillus-based cleaning products have shown promising results in decreasing the viability of pathogenic microorganisms, with continued decreases in pathogenic bacterial load associated with continued cleanings [189]. Further, Bacillus-based cleaning has been shown to reduce antibiotic resistance genes, including those associated with species associated with hospital associated infections [190]. Microbial aerosolizers have also become commercially available to provide a “probiotic for the home”, which are typically bacilli-based and do not appear to contain lactobacilli. Regardless of the bacterial composition, intended use, and health claims, probiotic cleaners generally lack clinical trials at this time.
The lifelong impact of microbial roommates
When contemplating the indoor microbiome, we invariably return to the goal of enhancing the experience and health of its macroscopic occupants. Aside from clear evidence for infection-potential, the role of the built environment on long-term health or chronic conditions is unclear. Occupants may face indirect effects on health or exacerbation of chronic conditions from microbial byproducts, including dozens of bacterial [191] and over 150 fungal allergens [192], as well as microbial volatile organic compounds (mVOCs). These mVOCs have, as yet, only been associated with adverse health effects, such as neurosensory, mucosal membrane, or respiratory reactions [193], which can result in absenteeism or the need for medical intervention [61, 194–196]. Asthma, in particular, is exacerbated by antigens and mVOCs, as well as building material-sourced VOCs [194]. In the United States, an estimated 19.9 million adults suffer from asthma [197], which accounts for more school absences than any other chronic health condition [198], and for approximately $616.9 million USD in losses per year for the United States economy from lost productivity, absenteeism, and emergency medical visits [199].
Different indoor bacterial communities are commonly reported in the homes of dermal or asthmatic atopy or non-atopic asthma sufferers [56, 200] in comparison with otherwise healthy homes. One study found the urban homes of young children with atopic dermatitis or wheezing had lower bacterial richness than homes with healthy children [201] while another study found there was no difference in bacterial richness of asthmatic vs. nonasthmatic homes [200]. Despite the differences in outcome, both studies concluded that the overall community was significantly altered. In addition to reports of decreased abundance of members of the phyla Firmicutes and Bacteroidetes in homes with children who suffer from wheezing or atopy [201], members of the phyla Cyanobacteria and Proteobacteria were increased in asthmatic homes [200]. Many human pathobionts, microorganisms capable of pathogenicity but not inherently infectious [202], belong to the Proteobacteria phylum. Cyanobacteria, typically found in water systems, has been detected in office buildings [203], and were posited to be responsible for the prevalence of allergic reactions to algae [204]. Yet, it has not been demonstrated whether an altered community in dust is a cause of atopic or asthmatic symptoms, or the effect of having a respiratory condition and altering cleaning and ventilation habits, which subsequently alter the indoor community. Moreover, phylum-level changes in a dust community may not be meaningful for all phyla; Firmicutes, Bacteroidetes, and Proteobacteria are found in host-associated and environmental ecosystems, with species or strain-level differences in community composition based on the specific conditions there.
Furthermore, while buildings can exacerbate pre-existing respiratory conditions, the role of the built environment in causing or preventing the development of respiratory conditions remains unclear. Recent research has shown that a diverse and abundant microbial community in house dust, which is often associated with rural farmhouses and homes with pets, is not deleterious to human health [205–208]. Longitudinal studies which point to the presence of pathobionts and antigens in the home during the first few years of life having a long-term protective effect against the development of adverse symptoms [201] lend support to the Old Friends hypothesis. Specifically, exposure to allergens from cockroaches, dogs, and cats were associated with a lower risk of recurrent wheezing [201]. Some of the bacteria of interest were correlated with allergen concentration, including a known cockroach symbiont, and researchers speculated that some of these bacteria in dust may be sourced from cockroaches or mice and that a combination of low bacterial diversity and high antigen concentration may be most efficacious in causing antigen sensitization [201].
Outside of the importance of early microbial and antigen exposure in developing a well-functioning immune system, the potential for positive feedback from the built environment to human health is less well understood. It is possible that a diverse microbial community indoors can facilitate a more diverse host-microbial community, or at least provide a regular source of immune system challenge that can facilitate better host-microbial control. However, environmental microbial communities appear to have short-lived effects on skin microbiota in healthy individuals [163, 209]. In hospital patients, the skin and digestive tract microbial community can be modulated in stays less than 2 weeks, and this disordered host microbiome may allow for colonization from unlikely sources, such as the built environment [210]. Microorganisms found on various surfaces within neonatal intensive care units (NICUs) were subsequently observed in the digestive tract of resident infants, including the same strain of Enterococcus faecalis in the gut of two infants housed in the same ward [211]. The lack of biodiversity within the infants gut at the start of the study provided an ideal environment to track specific strains of microorganisms being transferred between the “sterile” environmental and the human microbiome. Similarly, the Hospital Microbiome Project observed surfaces and patients in a newly opened hospital for the span of a year, revealing more of the intricate relationship between the human microbiome and the built environment using 16S rDNA sequencing [212]. It was observed that when a patient was admitted to a room, they initially acquired taxa that was in the room prior to their stay; however, over several days of occupancy, the patient’s microbial signature began to influence that of the room. Patient skin and the room surfaces were found to be more similar further into the patient’s stay [212].
As in any complex system, a simplified model reveals patterns. In the same way that the transfer of building-sourced microorganisms and the resulting health effects is most easily observed in infants and the immunocompromised, humans living in extreme built environments can model how a simplified indoor microbiome might affect otherwise healthy individuals. In isolated communities such as in the Arctic or Antarctic, heightened social connectedness created by consolidated living conditions can contribute to the spread of human-to-human [213] and building-to-human [214] infections; gastrointestinal disease from water recycling systems is a constant concern [214]. The bacterial community in the gastrointestinal tract of Antarctic visitors decreased in diversity and lost key beneficial members, such as Bifidobacterium [215], both of which can lead to a disrupted intestinal community and risk of disease. Similarly, the sterilization of food and equipment prior to launch, the harsh conditions of space travel, and the isolation from any outside microbial source, contributes to the development of a low-diversity microbial community which is both sparse and tough [28, 216, 217], and does not make for pleasant company [218–220]. The harsh conditions and increase in radiation can contribute to accelerated evolution; a distinct strain of Aspergillus niger was isolated from the International Space Station [221]. Over time, rapid evolution to support hardiness can lead to the development of ‘survivor strains’ which are extremely difficult to kill or eradicate, and recent research has demonstrated this in the built environment [222]. In the same way that HAI are a growing threat to public safety, space-evolved microorganisms and built environments devoid of life may one day hamper our efforts for long-term space travel or residency without radical design changes.
While we are beginning to describe the ways in which the built environment affects health, more research is clearly needed before any accurate conclusions can be reached. The majority of indoor microbiology studies rely on observations and variation within the microbial community, but have not connected these to any potential health outcomes [29, 44, 49, 223, 224]. Similar to the loss of host-associated microorganisms contributing to human immune dysfunction, it has been hypothesized that the loss of macro- and microorganism diversity seen in most urban environments has also reduced our environmental microbial exposures which help train the immune system [225]. However, the loss of ecosystem-scale biodiversity is inexorably tied to increased pollution, as well as changing lifestyle (i.e., activity levels and changes in social engagement), and it is difficult to disentangle these other effects from that of reduced diversity in microbial exposures. Moreover, it has not been conclusively established that a diverse indoor microbiome has any effect on health in otherwise healthy individuals, and the diversity and plasticity of occupants and their respective immune status requires flexibility in the content of that indoor microbiome. Thus, “healthy indoor microbiome” eludes definition, and may only apply to the young or the immunocompromised [210, 211].
Built environment research now and in the future
Epidemiological investigation has shown humans can acquire infectious disease from the built environment [99, 127, 128, 226, 227], but outside of pathogen-based investigations, only a few studies demonstrate microbial transmission from a built environment to a human or animal host [163, 209]. Evaluating the transfer of microorganisms from the built environment to humans has challenges, not the least of which are the technical and logistical challenges of tracking individual cells moving from an area of very low biomass to very high biomass. Additionally, surface material and microorganism characteristics contribute to the likelihood of resuspension or transfer [95, 228, 229], and the complexity of the microbial community of the recipient likely mediates the potential for retention on a host.
As with all microbial communities, elucidation of trends requires both a temporal and spatial resolution of sampling. Microbial transfer from the built environment to humans is often successfully observed using individuals whose microbiota are in a dysbiotic state such as the very young or very sick [127, 128, 210, 211, 230, 231]. Transcriptomics is the sequencing of all an organism’s or community’s RNA transcripts [232], yet it only provides a snapshot of activity. In built environments which may only allow for sporadic activity from microbial communities, or time-dependent activity in the case of communities exposure to daylight, transcriptomics may not yield information on the entire community. Due to the low probability of observing a spontaneous transfer event, an in situ microbial transfer from the built environment to an occupant requires a high resolution of spatial and temporal samples can financially preclude the use of “omics”-technologies which could provide more highly resolved data. Some of these difficulties have been overcome by large-scale, or long-term projects which accumulate sufficiently large datasets to locate infrequent microbial transmissions. Interestingly, several of these utilize 16S rDNA sequencing data, yet with sufficient time or spatial resolution of samples, the inclusion of regular sampling from patients regardless of symptoms, or the examination of low-diversity communities from built environments and hospital patients, the transmission of microorganisms from buildings to bodies was successfully observed [209, 211, 212]. Despite these successes, the logistical difficulty of finding the transmission of a microbial needle in a dust haystack remains a barrier to understanding how microorganisms in buildings affect human health. A great deal of discussion has centered around the development of low-cost, real-time microbial sensing capabilities in buildings [31, 233], which would track total biomass, or detect certain species or antigens, and interface with building automation services to alter operations.
The majority of studies determine the identity of bacteria present by sequencing the 16S ribosomal RNA gene (rDNA). This highly conserved prokaryotic gene provides a useful and reliable, yet inexpensive, means of taxonomic identification. However, the increased use of universal prokaryotic primers favors bacteria over archaea and typically results in underestimated diversity and taxonomic resolution of archaea. Similarly, the difficulty in predicting ecology and host-microorganism interactions using only taxonomic markers (e.g., 16S rDNA, 18S rDNA, Internal Transcribed Spacer (ITS) regions) provides fodder for hypothesis but not mechanism. Another major limiting factor is the lack of strain-specific identification, which obscures evidence that microorganisms from the built environment and those identified as causative agents in hosts are the same microorganisms. In some cases, the specificity of strain is not needed to determine where the microorganism came from, as in the case of Legionella in water systems, due to the specificity of niche location within the built environment that one may find the microorganism. However, many microorganisms spend parts of their lives in different habitats, making it more difficult to determine the origin of that strain. Later research into the microbiomes of the built environment has included the use of metagenome sequencing, which generates data on all the DNA material in an environment, including genomic and plasmidic. Metagenome sequencing has given researchers a holistic view of the indoor microbiome, for example, the ability to determine what genomic characteristics are typical of a specific built environment [46, 222, 234]. Yet, genetic potential does not always translate to activity, and DNA-based technology easily picks up relic DNA from an environment.
An important feature to assess in any microbial community is the determination of which species present are alive and metabolically active [235]; rDNA and rRNA extracted from the same sources resulted in significantly different overall characterization of the microbiota [32, 236]. Analyzing both RNA and DNA in combination, even just rRNA and rDNA, can provide further insight than just one method alone, such as the effect of environmental conditions on which microorganisms thrive while others struggle or perish. There are other methods to select for living microorganisms which may be dormant or only slightly metabolically active, such as amplification inhibitors added prior to nucleic acid extraction. For example, propidium monoazide (PMA) is a membrane-impermeable dye that is only able to enter into cells which have a damaged cell membrane [237]. PMA preferentially covalently binds to double stranded DNA (dsDNA), where it strongly inhibits polymerase chain reaction (PCR) from occurring in downstream laboratory protocols, allowing studies to solely amplify the dsDNA of living cells. By comparing samples from the same community with and without PMA treatment, researchers can begin to understand the current state and recent history of the community in which they are studying [85], respectively. For example, PMA treatment was used to determine how different types of light might select for microbial communities in dust [85]. The detection of living cells is an added technological challenge for real-time microbial monitoring in buildings; however, the detection of living cells may be immaterial in certain circumstances, such as the detection of microbial antigens.
Previous studies looking at health effects have used either correlational studies [191, 238] or mouse models [239]. Mouse models provide a good starting point for epidemiological studies, but often react differentially in comparison to humans when exposed to the same compounds [240], due to differences in metabolic rates, prenatal development and immune system, and interactions related to social structure and use of their environment. Moreover, microbial and chemical exposure may be localized within a building, and the sum of an occupant’s path through the built environment over time leads to tailored exposures from the built environment—recently coined ‘exposomes’ [241, 242]. In recent years, a new technology, ‘organ chips’, has emerged as a potential medium through which to study the built environment. Organ-on-a-chip (OOC) systems replicate human organs by combining microfluidics, three-dimensional tissue constructs, and cultured human cells [243]. OOCs allow researchers to study the effects of differing exposomes on human metabolism, immune response, and toxicity by combining multiple OOCs to create a system of organs, just as in the body [244]. OOC technology could develop into an invaluable tool in the study of the daily exposome on the inhabitants of the built environment.
Yet another challenge in the study of the built environment is determining what effects, if any, may come from our daily exposomes. Perhaps the clearest example of the potential future investigation of the built environment is the recent research that piloted wearable collection devices to track the overall accumulation of microorganisms and abiotic factors over the course of three test subjects’ normal day [242]. A combination of DNA and RNA sequencing, liquid chromatography-coupled spectrometry, and scanning electron microscopy was used to determine the identities of the microorganisms and chemical compounds that the study participants encountered during the sampling period. This study found strong location-dependant relationships between the movement of the study participants and the microorganisms and compounds they were exposed to, alluding to the potential impact that varying built environment compositions could have on microbial exposure. The three individuals were exposed to over 2500 species of microorganisms; however, the authors reported that 43.74% of the analyzed DNA could not be connected to a known species. This demonstrates the tremendous amount of research that needs to be done and is, on its own, a strong reason to invest more time, energy, and money into studying the built environment.
Summary
Overall, our understanding and our views of the relationships we have with microorganisms are evolving [20, 31, 202, 245]. What remains is the ability to know why, when, and how microorganisms transfer from the built environment to occupants, when these interactions matter, under what circumstances these transfers lead to disease, and when these interactions are beneficial to built environment occupants [33]. The variety of occupants and health needs in a space will likely dictate strategies to inform that indoor microbiome, including fostering or inhibiting human to human contact, removing or adding microbial biofilms, and integrating microbial communities into building systems. Similarly, the move towards building automation will need to incorporate technologies designed to monitor and manipulate microorganisms in ways which do not create the harsh selective pressures that may lead to dangerous survivor phenotypes, such as full spectrum sunlight instead of antibiotics or single-ingredient cleaning products. Technological advances such as high-throughput sequencing and transcriptomic analysis have given us new insights into the communities and activities of the microorganisms with which we share our everyday lives. Despite these new insights, we still conceive of our living spaces as places that must protect and isolate us from the world of microorganisms. Using new technology to understand microbial dynamics and host interactions, we continue to evolve past the idea that buildings are isolation chambers to protect occupants from contact with the microbial world and begin to shape salubrious indoor microbial ecosystems through building design.
Acknowledgements
The authors appreciate the efforts of Dr. Roo Vandegrift, Sam Velasquez, Jeff Kline, Fiona Curliss, and Paul Ward in reviewing this manuscript. The authors would also like to thank Mira Zimmerman, Delaney Hetrick, and Julia May for their graphical contributions. This work was funded by grants from the Alfred P. Sloan Foundation to the Biology and the Built Environment Center at the University of Oregon.
Author contributions
SLI conceived of scope, and contributed to writing and editing. PFH and SL contributed to literature review, writing, and editing. GAM, LD, MF, KVDW, and GM contributed to editing.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tognotti E. Lessons from the History of Quarantine, from Plague to Influenza A. Emerging Infectious Dis J. 2013;19:254. doi: 10.3201/eid1902.120312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Best M, Neuhauser D. Ignaz Semmelweis and the birth of infection control. Qual Saf Health Care. 2004;13:233–4. doi: 10.1136/qshc.2004.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister J. On the antiseptic principle in the practice of surgery. Br Med J. 1867;2:246–8. doi: 10.1136/bmj.2.351.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomes NJ. American Attitudes toward the Germ Theory of Disease: Phyllis Allen Richmond Revisited. J Hist Med. 1997;52:17–50. doi: 10.1093/jhmas/52.1.17. [DOI] [PubMed] [Google Scholar]
- 5.Smith KA. Louis pasteur, the father of immunology? Front Immunol. 2012;3:68. doi: 10.3389/fimmu.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans AS. Causation and disease: the Henle-Koch postulates revisited. Yale J Biol Med. 1976;49:175–95. [PMC free article] [PubMed] [Google Scholar]
- 7.Riva MA, Benedetti M, Cesana G. Pandemic fear and literature: observations from Jack London’s The Scarlet Plague. Emerg Infect Dis. 2014;20:1753–7. doi: 10.3201/eid2010.130278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathur P. Hand hygiene: back to the basics of infection control. Indian J Med Res. 2011;134:611–20. doi: 10.4103/0971-5916.90985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teigen PM. Legislating fear and the public health in gilded age Massachusetts. J Hist Med Allied Sci. 2007;62:141–70. doi: 10.1093/jhmas/jrl016. [DOI] [PubMed] [Google Scholar]
- 10.Stewart GT. Limitations of the germ theory. Lancet. 1968;1:1077–81. doi: 10.1016/s0140-6736(68)91425-6. [DOI] [PubMed] [Google Scholar]
- 11.Gause GF. The struggle for existence. Williams & Wilkins Co., Baltimore, Maryland, 1934.
- 12.Hungate RE. Further experiments on cellulose digestion by the protozoa in the rumen of cattle. Biol Bull. 1943;84:157–63. [Google Scholar]
- 13.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–4. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 14.Dunlap PV, McFall-Ngai MJ. Initiation and control of the bioluminescent symbiosis between photobacterium leiognathi and leiognathid fish. Ann N Y Acad Sci. 1987;503:269–83. doi: 10.1111/j.1749-6632.1987.tb40614.x. [DOI] [PubMed] [Google Scholar]
- 15.Adair KL, Douglas AE. Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol. 2017;35:23–9. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Robinson CJ, Bohannan BJM, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–76. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol. 2017;2:eaao1314. doi: 10.1126/sciimmunol.aao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prokopakis E, Vardouniotis A, Kawauchi H, Scadding G, Georgalas C, Hellings P, et al. The pathophysiology of the hygiene hypothesis. Int J Pediatr Otorhinolaryngol. 2013;77:1065–71. doi: 10.1016/j.ijporl.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 19.Liu AH. Revisiting the hygiene hypothesis for allergy and asthma. J Allergy Clin Immunol. 2015;136:860–5. doi: 10.1016/j.jaci.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Vandegrift R, Bateman AC, Siemens KN, Nguyen M, Green JL, Van Den Wymelenberg KG, et al. Cleanliness in context: reconciling hygiene with a modern microbial perspective. Microbiome. 2017;5:76. doi: 10.1186/s40168-017-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scudellari M. News feature: cleaning up the hygiene hypothesis. Proc Natl Acad Sci USA. 2017;114:1433–6. doi: 10.1073/pnas.1700688114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rook GAW, Brunet LR. Microbes, immunoregulation, and the gut. Gut. 2005;54:317–20. doi: 10.1136/gut.2004.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–8. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surawicz CM, McFarland LV, Greenberg RN, Rubin M, Fekety R, Mulligan ME, et al. The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin Infect Dis. 2000;31:1012–7. doi: 10.1086/318130. [DOI] [PubMed] [Google Scholar]
- 25.Na X, Kelly C. Probiotics in Clostridium difficile infection. J Clin Gastroenterol. 2011;45(Suppl):S154–8. doi: 10.1097/MCG.0b013e31822ec787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau CS, Chamberlain RS. Probiotics are effective at preventing Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Int J Gen Med. 2016;9:27–37. doi: 10.2147/IJGM.S98280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mora M, Mahnert A, Koskinen K, Pausan MR, Oberauner-Wappis L, Krause R, et al. Microorganisms in confined habitats: microbial monitoring and control of intensive care units, operating rooms, cleanrooms and the international space station. Front Microbiol. 2016;7:1573. doi: 10.3389/fmicb.2016.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams RI, Bateman AC, Bik HM, Meadow JF. Microbiota of the indoor environment: a meta-analysis. Microbiome. 2015;3:49. doi: 10.1186/s40168-015-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.NASEM. Microbiomes of the Built Environment. Washington, D.C.: National Academies Press; 2017. [PubMed] [Google Scholar]
- 31.Gilbert JA, Stephens B. Microbiology of the built environment. Nat Rev Microbiol. 2018;16:661–70. doi: 10.1038/s41579-018-0065-5. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-Silvan C, Leung MHY, Grue KA, Kaur R, Tong X, Lee PKH, et al. A comparison of methods used to unveil the genetic and metabolic pool in the built environment. Microbiome. 2018;6:71. doi: 10.1186/s40168-018-0453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nazaroff WW. Embracing microbes in exposure science. J Expo Sci Environ Epidemiol. 2019;29:1–10. doi: 10.1038/s41370-018-0075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6:1469–79. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams RI, Lymperopoulou D. Lessons learned when looking for non-neutral ecological processes in the built environment: the bacterial and fungal microbiota of shower tiles. bioRxiv. 2018: 413773. 10.1101/413773.
- 36.Givehchi R, Maestre JP, Bi C, Wylie D, Xu Y, Kinney KA, et al. Quantitative filter forensics with residential HVAC filters to assess indoor concentrations. Indoor Air. 2019;29:390–402. doi: 10.1111/ina.12536. [DOI] [PubMed] [Google Scholar]
- 37.Emerson JB, Keady PB, Clements N, Morgan EE, Awerbuch J, Miller SL, et al. High temporal variability in airborne bacterial diversity and abundance inside single-family residences. Indoor Air. 2017;27:576–86. doi: 10.1111/ina.12347. [DOI] [PubMed] [Google Scholar]
- 38.Green JL. Can bioinformed design promote healthy indoor ecosystems? Indoor Air. 2014;24:113–5. doi: 10.1111/ina.12090. [DOI] [PubMed] [Google Scholar]
- 39.Brown GZ, Kline J, Mhuireach G, Northcutt D, Stenson J. Making microbiology of the built environment relevant to design. Microbiome. 2016;4:6. doi: 10.1186/s40168-016-0152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaler DS. Toward a microbial Neolithic revolution in buildings. Microbiome. 2016;4:14. doi: 10.1186/s40168-016-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yong E. For healthier buildings, just add bacteria? ideas.ted.com. 2017. https://ideas.ted.com/for-healthier-buildings-just-add-bacteria/ (Accessed 19 Feb 2019).
- 42.Purcell AT. The relationship between buildings and behaviour. Build Environ. 1987;22:215–32. [Google Scholar]
- 43.Dunn RR, Fierer N, Henley JB, Leff JW, Menninger HL. Home life: factors structuring the bacterial diversity found within and between homes. PLoS ONE. 2013;8:e64133. doi: 10.1371/journal.pone.0064133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kembel SW, Meadow JF, O’Connor TK, Mhuireach G, Northcutt D, Kline J, et al. Architectural design drives the biogeography of indoor bacterial communities. PLoS ONE. 2014;9:e87093. doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meadow JF, Altrichter AE, Kembel SW, Moriyama M, O’Connor TK, Womack AM, et al. Bacterial communities on classroom surfaces vary with human contact. Microbiome. 2014;2:7. doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu T, Joice R, Vallarino J, Abu-Ali G, Hartmann EM, Shafquat A, et al. Urban transit system microbial communities differ by surface type and interaction with humans and the environment. mSystems 2016; 1. 10.1128/mSystems.00018-16. [DOI] [PMC free article] [PubMed]
- 47.Karkman A, Lehtimäki J, Ruokolainen L. The ecology of human microbiota: dynamics and diversity in health and disease. Ann N Y Acad Sci. 2017;1399:78–92. doi: 10.1111/nyas.13326. [DOI] [PubMed] [Google Scholar]
- 48.Adams RI, Bhangar S, Pasut W, Arens EA, Taylor JW, Lindow SE, et al. Chamber bioaerosol study: outdoor air and human occupants as sources of indoor airborne microbes. PLoS ONE. 2015;10:e0128022. doi: 10.1371/journal.pone.0128022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross AA, Doxey AC, Neufeld JD. The skin microbiome of cohabiting couples. mSystems. 2017;2:e00043–17. doi: 10.1128/mSystems.00043-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meadow JF, Altrichter AE, Bateman AC, Stenson J, Brown GZ, Green JL, et al. Humans differ in their personal microbial cloud. PeerJ. 2015;3:e1258. doi: 10.7717/peerj.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–52. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luongo JC, Barberán A, Hacker-Cary R, Morgan EE, Miller SL, Fierer N. Microbial analyses of airborne dust collected from dormitory rooms predict the sex of occupants. Indoor Air. 2017;27:338–44. doi: 10.1111/ina.12302. [DOI] [PubMed] [Google Scholar]
- 53.Marineli F, Tsoucalas G, Karamanou M, Androutsos G, Mary Mallon (1869-1938) and the history of typhoid fever. Ann Gastroenterol Hepatol. 2013;26:132–4. [PMC free article] [PubMed] [Google Scholar]
- 54.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. 2015;112:E2930–8. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meadow JF, Altrichter AE, Kembel SW, Kline J, Mhuireach G, Moriyama M, et al. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air. 2014;24:41–8. doi: 10.1111/ina.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dannemiller KC, Gent JF, Leaderer BP, Peccia J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air. 2016;26:179–92. doi: 10.1111/ina.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood R, Morrow C, Ginsberg S, Piccoli E, Kalil D, Sassi A, et al. Quantification of shared air: a social and environmental determinant of airborne disease transmission. PLoS ONE. 2014;9:e106622. doi: 10.1371/journal.pone.0106622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrews JR, Morrow C, Walensky RP, Wood R. Integrating social contact and environmental data in evaluating tuberculosis transmission in a South African township. J Infect Dis. 2014;210:597–603. doi: 10.1093/infdis/jiu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salathé M, Kazandjieva M, Lee JW, Levis P, Feldman MW, Jones JH. A high-resolution human contact network for infectious disease transmission. Proc Natl Acad Sci USA. 2010;107:22020–5. doi: 10.1073/pnas.1009094108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borgo B, Mostafavi M. Microbial Air Quality in a 50-year-old Middle School. In: 2007 SACNAS. Society for the Advancement of Chicanos and Native Americans in Science, Kansas City, Missouri, 2007, pp 1–6.
- 61.Hayleeyesus SF, Manaye AM. Microbiological quality of indoor air in university libraries. Asian Pac J Trop Biomed. 2014;4:S312–7. doi: 10.12980/APJTB.4.2014C807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virtanen M, Terho K, Oksanen T, Kurvinen T, Pentti J, Routamaa M, et al. Patients with infectious diseases, overcrowding, and health in hospital staff. Arch Intern Med. 2011;171:1296–8. doi: 10.1001/archinternmed.2011.313. [DOI] [PubMed] [Google Scholar]
- 63.Clements A, Halton K, Graves N, Pettitt A, Morton A, Looke D, et al. Overcrowding and understaffing in modern health-care systems: key determinants in meticillin-resistant Staphylococcus aureus transmission. Lancet Infect Dis. 2008;8:427–34. doi: 10.1016/S1473-3099(08)70151-8. [DOI] [PubMed] [Google Scholar]
- 64.Bick JA. Infection control in jails and prisons. Clin Infect Dis. 2007;45:1047–55. doi: 10.1086/521910. [DOI] [PubMed] [Google Scholar]
- 65.Tschudin-Sutter S, Carroll KC, Tamma PD, Sudekum ML, Frei R, Widmer AF, et al. Impact of toxigenic Clostridium difficile colonization on the risk of subsequent C. difficile Infection in intensive care unit patients. Infect Control Hosp Epidemiol. 2015;36:1324–9. doi: 10.1017/ice.2015.177. [DOI] [PubMed] [Google Scholar]
- 66.Yakob L, Riley TV, Paterson DL, Clements ACA. Clostridium difficile exposure as an insidious source of infection in healthcare settings: an epidemiological model. BMC Infect Dis. 2013;13:376. doi: 10.1186/1471-2334-13-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaudhury H, Mahmood A, Valente M. The use of single patient rooms versus multiple occupancy rooms in acute care environments. coalition for health environments research, 2005. https://www.healthdesign.org/sites/default/files/use_of_single_patient_rooms_v_multiple_occ._rooms-acute_care.pdf.
- 68.Bodin Danielsson C, Chungkham HS, Wulff C, Westerlund H. Office design’s impact on sick leave rates. Ergonomics. 2014;57:139–47. doi: 10.1080/00140139.2013.871064. [DOI] [PubMed] [Google Scholar]
- 69.Pejtersen JH, Feveile H, Christensen KB, Burr H. Sickness absence associated with shared and open-plan offices–a national cross sectional questionnaire survey. Scand J Work Environ Health. 2011;37:376–82. doi: 10.5271/sjweh.3167. [DOI] [PubMed] [Google Scholar]
- 70.Ridenhour BJ, Braun A, Teyrasse T, Goldsman D. Controlling the spread of disease in schools. PLoS ONE. 2011;6:e29640. doi: 10.1371/journal.pone.0029640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dill-McFarland KA, Tang Z-Z, Kemis JH, Kerby RL, Chen G, Palloni A, et al. Close social relationships correlate with human gut microbiota composition. Sci Rep. 2019;9:703. doi: 10.1038/s41598-018-37298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jansen TR. When preschool is in a nursing home. The Atlantic 2016. https://www.theatlantic.com/education/archive/2016/01/the-preschool-inside-a-nursing-home/424827/ Accessed 2 Jan 2019.
- 73.Seidel J, Valenzano DR. The role of the gut microbiome during host ageing. F1000Res. 2018;7:1086. doi: 10.12688/f1000research.15121.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao J-Z, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dimidi E, Christodoulides S, Scott SM, Whelan K. Mechanisms of action of probiotics and the gastrointestinal microbiota on gut motility and constipation. Adv Nutr. 2017;8:484–94. doi: 10.3945/an.116.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol. 2017;8:738. doi: 10.3389/fmicb.2017.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inoue R, Nishio A, Fukushima Y, Ushida K. Oral treatment with probiotic Lactobacillus johnsonii NCC533 (La1) for a specific part of the weaning period prevents the development of atopic dermatitis induced after maturation in model mice, NC/Nga. Br J Dermatol. 2007;156:499–509. doi: 10.1111/j.1365-2133.2006.07695.x. [DOI] [PubMed] [Google Scholar]
- 78.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci USA. 2014;111:805–10. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orsmark-Pietras C, James A, Konradsen JR, Nordlund B, Söderhäll C, Pulkkinen V, et al. Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur Respir J. 2013;42:65–78. doi: 10.1183/09031936.00077712. [DOI] [PubMed] [Google Scholar]
- 80.Mortaz E, Adcock IM, Folkerts G, Barnes PJ, Paul Vos A, Garssen J. Probiotics in the management of lung diseases. Mediators Inflamm. 2013;2013:751068. doi: 10.1155/2013/751068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pellaton C, Nutten S, Thierry A-C, Boudousquié C, Barbier N, Blanchard C, et al. Intragastric and intranasal administration of Lactobacillus paracasei NCC2461 modulates allergic airway inflammation in mice. Int J Inflam. 2012;2012:686739. doi: 10.1155/2012/686739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: a strategy to promote immunosurveillance against lung metastases. Cell Rep. 2018;24:3528–38. doi: 10.1016/j.celrep.2018.08.090. [DOI] [PubMed] [Google Scholar]
- 83.Rutala WA, Weber DJ. Surface disinfection: should we do it? J Hosp Infect. 2001;48(Suppl A):S64–8. doi: 10.1016/s0195-6701(01)90017-9. [DOI] [PubMed] [Google Scholar]
- 84.Vincent M, Hartemann P, Engels-Deutsch M. Antimicrobial applications of copper. Int J Hyg Environ Health. 2016;219:585–91. doi: 10.1016/j.ijheh.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 85.Fahimipour AK, Hartmann EM, Siemens A, Kline J, Levin DA, Wilson H, et al. Daylight exposure modulates bacterial communities associated with household dust. Microbiome. 2018;6:175. doi: 10.1186/s40168-018-0559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dannemiller KC, Weschler CJ, Peccia J. Fungal and bacterial growth in floor dust at elevated relative humidity levels. Indoor Air. 2017;27:354–63. doi: 10.1111/ina.12313. [DOI] [PubMed] [Google Scholar]
- 87.Vandegrift R, Ishaq SL, Kline J, Fahimipour A, Stenson J, Crowley R, et al. Shut the front door: seasonal patterns in window operation drive fungal and bacterial community dissimilarity between indoor and outdoor air. In: The 15th Conference of the International Society of Indoor Air Quality & Climate (ISIAQ). International Society of Indoor Air Quality & Climate, p 2.
- 88.Gibbons SM. The built environment is a microbial wasteland. mSystems. 2016;1:e00033–16. doi: 10.1128/mSystems.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Otter JA, French GL. Survival of nosocomial bacteria and spores on surfaces and inactivation by hydrogen peroxide vapor. J Clin Microbiol. 2009;47:205–7. doi: 10.1128/JCM.02004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis. 2006;6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith SM, Eng RH, Padberg FT., Jr Survival of nosocomial pathogenic bacteria at ambient temperature. J Med. 1996;27:293–302. [PubMed] [Google Scholar]
- 92.Gil F, Lagos-Moraga S, Calderón-Romero P, Pizarro-Guajardo M, Paredes-Sabja D. Updates on Clostridium difficile spore biology. Anaerobe. 2017;45:3–9. doi: 10.1016/j.anaerobe.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 93.Ng TW, Chan PY, Chan TT, Wu H, Lai KM. Skin squames contribute to ammonia and volatile fatty acid production from bacteria colonizing in air-cooling units with odor complaints. Indoor Air. 2017;28:258–65. doi: 10.1111/ina.12439. [DOI] [PubMed] [Google Scholar]
- 94.Hospodsky D, Yamamoto N, Nazaroff WW, Miller D, Gorthala S, Peccia J. Characterizing airborne fungal and bacterial concentrations and emission rates in six occupied children’s classrooms. Indoor Air. 2015;25:641–52. doi: 10.1111/ina.12172. [DOI] [PubMed] [Google Scholar]
- 95.Lopez GU, Gerba CP, Tamimi AH, Kitajima M, Maxwell SL, Rose JB. Transfer efficiency of bacteria and viruses from porous and nonporous fomites to fingers under different relative humidity conditions. Appl Environ Microbiol. 2013;79:5728–34. doi: 10.1128/AEM.01030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marinella MA, Pierson C, Chenoweth C. The stethoscope. A potential source of nosocomial infection? Arch Intern Med. 1997;157:786–90. doi: 10.1001/archinte.157.7.786. [DOI] [PubMed] [Google Scholar]
- 97.Brooks B, Olm MR, Firek BA, Baker R, Geller-McGrath D, Reimer SR, et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome. 2018;6:112. doi: 10.1186/s40168-018-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chemaly RF, Simmons S, Dale C, Jr, Ghantoji SS, Rodriguez M, Gubb J, et al. The role of the healthcare environment in the spread of multidrug-resistant organisms: update on current best practices for containment. Ther Adv Infect Dis. 2014;2:79–90. doi: 10.1177/2049936114543287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Joseph A. The impact of the environment on infections in healthcare facilities. Center for Health Design, Concord, California, 2006.
- 100.Jump RLP, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother. 2007;51:2883–7. doi: 10.1128/AAC.01443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoeksma P, Aarnink A, Ogink N. Effect of temperature and relative humidity on the survival of airborne bacteria. Wageningen UR Livestock Research, 2015. https://library.wur.nl/WebQuery/wurpubs/fulltext/348736.
- 102.Zhao Y, Aarnink AJA, Dijkman R, Fabri T, de Jong MCM, Groot Koerkamp PWG. Effects of temperature, relative humidity, absolute humidity, and evaporation potential on survival of airborne Gumboro vaccine virus. Appl Environ Microbiol. 2012;78:1048–54. doi: 10.1128/AEM.06477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang JW. The effect of environmental parameters on the survival of airborne infectious agents. J R Soc Interface. 2009;6(Suppl 6):S737–46. doi: 10.1098/rsif.2009.0227.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Górny RL, Gołofit-Szymczak M, Cyprowski M, Stobnicka A, Ławniczek-Wałczyk A. Effect of electrical charges on potential of fibers for transport of microbial particles in dry and humid air. J Aerosol Sci. 2018;116:66–82. [Google Scholar]
- 105.Hegarty B, Dannemiller K, Peccia J. Gene expression of indoor fungal communities under damp building conditions: implications for human health. Indoor Air. 2018;28:548–58. doi: 10.1111/ina.12459. [DOI] [PubMed] [Google Scholar]
- 106.Pessi A-M, Suonketo J, Pentti M, Kurkilahti M, Peltola K, Rantio-Lehtimäki A. Microbial growth inside insulated external walls as an indoor air biocontamination source. Appl Environ Microbiol. 2002;68:963–7. doi: 10.1128/AEM.68.2.963-967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu T, Täubel M, Holopainen R, Viitanen A-K, Vainiotalo S, Tuomi T, et al. Infant and adult inhalation exposure to resuspended biological particulate matter. Environ Sci Technol. 2018;52:237–47. doi: 10.1021/acs.est.7b04183. [DOI] [PubMed] [Google Scholar]
- 108.Hyytiäinen HK, Jayaprakash B, Kirjavainen PV, Saari SE, Holopainen R, Keskinen J, et al. Crawling-induced floor dust resuspension affects the microbiota of the infant breathing zone. Microbiome. 2018;6:25. doi: 10.1186/s40168-018-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Frankel M, Hansen EW, Madsen AM. Effect of relative humidity on the aerosolization and total inflammatory potential of fungal particles from dust-inoculated gypsum boards. Indoor Air. 2014;24:16–28. doi: 10.1111/ina.12055. [DOI] [PubMed] [Google Scholar]
- 110.Lowen AC, Mubareka S, Steel J, Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007;3:1470–6. doi: 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wolkoff P. Indoor air humidity, air quality, and health - An overview. Int J Hyg Environ Health. 2018;221:376–90. doi: 10.1016/j.ijheh.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 112.Taylor S, Hugentobler W. Is low indoor humidity a driver for healthcare-associated infections? In: Proceedings, Indoor Air 2016. International Society of Indoor Air Quality and Climate, 2016. https://www.isiaq.org/docs/Papers/Paper340.pdf.
- 113.Davis RE, Dougherty E, McArthur C, Huang QS, Baker MGCold. dry air is associated with influenza and pneumonia mortality in Auckland, New Zealand. Influenza Other Respi Viruses. 2016;10:310–3. doi: 10.1111/irv.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frankel M, Bekö G, Timm M, Gustavsen S, Hansen EW, Madsen AM. Seasonal variations of indoor microbial exposures and their relation to temperature, relative humidity, and air exchange rate. Appl Environ Microbiol. 2012;78:8289–97. doi: 10.1128/AEM.02069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dannemiller KC, Weschler CJ, Peccia J. Fungal and bacterial growth in floor dust at elevated relative humidity levels. Indoor Air. 2017;27:354–63. doi: 10.1111/ina.12313. [DOI] [PubMed] [Google Scholar]
- 116.Khare P, Marr LC. Simulation of vertical concentration gradient of influenza viruses in dust resuspended by walking. Indoor Air. 2015;25:428–40. doi: 10.1111/ina.12156. [DOI] [PubMed] [Google Scholar]
- 117.Ahmed T, Usman M, Scholz M. Biodeterioration of buildings and public health implications caused by indoor air pollution. Indoor Built Environ. 2017;27:752–65. [Google Scholar]
- 118.Pasanen A-L, Juutinen T, Jantunen MJ, Kalliokoski P. Occurrence and moisture requirements of microbial growth in building materials. Int Biodeterior Biodegradation. 1992;30:273–83. [Google Scholar]
- 119.Kuhn DM, Ghannoum MA. Indoor mold, toxigenic fungi, and Stachybotrys chartarum: infectious disease perspective. Clin Microbiol Rev. 2003;16:144–72. doi: 10.1128/CMR.16.1.144-172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kettleson EM, Adhikari A, Vesper S, Coombs K, Indugula R, Reponen T. Key determinants of the fungal and bacterial microbiomes in homes. Environ Res. 2015;138:130–5. doi: 10.1016/j.envres.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mendell MJ, Macher JM, Kumagai K. Measured moisture in buildings and adverse health effects: a review. Indoor Air. 2018;28:488–99. doi: 10.1111/ina.12464. [DOI] [PubMed] [Google Scholar]
- 122.Kanchongkittiphon W, Mendell MJ, Gaffin JM, Wang G, Phipatanakul W. Indoor environmental exposures and exacerbation of asthma: an update to the 2000 review by the Institute of Medicine. Environ Health Perspect. 2015;123:6–20. doi: 10.1289/ehp.1307922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kotay S, Chai W, Guilford W, Barry K, Mathers AJ. Spread from the sink to the patient:in situ study using green fluorescent protein (GFP)-expressing Escherichia coli to model bacterial dispersion from hand-washing sink-trap reservoirs. Appl Environ Microbiol. 2017;83:e03327–16. doi: 10.1128/AEM.03327-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Falkinham JO, 3rd, Iseman MD, de Haas P, van Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6:209–13. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- 125.Thomson R, Tolson C, Carter R, Coulter C, Huygens F, Hargreaves M. Isolation of nontuberculous mycobacteria (NTM) from household water and shower aerosols in patients with pulmonary disease caused by NTM. J Clin Microbiol. 2013;51:3006–11. doi: 10.1128/JCM.00899-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kline S, Cameron S, Streifel A, Yakrus MA, Kairis F, Peacock K, et al. An outbreak of bacteremias associated with Mycobacterium mucogenicum in a hospital water supply. Infect Control Hosp Epidemiol. 2004;25:1042–9. doi: 10.1086/502341. [DOI] [PubMed] [Google Scholar]
- 127.Kool JL, Bergmire-Sweat D, Butler JC, Brown EW, Peabody DJ, Massi DS, et al. Hospital characteristics associated with colonization of water systems by Legionella and risk of nosocomial legionnaires’ disease: a cohort study of 15 hospitals. Infect Control Hosp Epidemiol. 1999;20:798–805. doi: 10.1086/501587. [DOI] [PubMed] [Google Scholar]
- 128.Borella P, Montagna MT, Romano-Spica V, Stampi S, Stancanelli G, Triassi M, et al. Legionella infection risk from domestic hot water. Emerg Infect Dis. 2004;10:457–64. doi: 10.3201/eid1003.020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dade-Robertson M, Keren-Paz A, Zhang M, Kolodkin-Gal I. Architects of nature: growing buildings with bacterial biofilms. Microb Biotechnol. 2017;10:1157–63. doi: 10.1111/1751-7915.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grumbein S, Minev D, Tallawi M, Boettcher K, Prade F, Pfeiffer F, et al. Hydrophobic properties of biofilm-enriched hybrid mortar. Adv Mater. 2016;28:8138–43. doi: 10.1002/adma.201602123. [DOI] [PubMed] [Google Scholar]
- 131.Hesnawi R, Dahmani K, Al-Swayah A, Mohamed S, Mohammed SA. Biodegradation of municipal wastewater with local and commercial bacteria. Procedia Engineering. 2014;70:810–4. [Google Scholar]
- 132.Mikkonen A, Li T, Vesala M, Saarenheimo J, Ahonen V, Kärenlampi S, et al. Biofiltration of airborne VOCs with green wall systems-Microbial and chemical dynamics. Indoor Air. 2018;28:697–707. doi: 10.1111/ina.12473. [DOI] [PubMed] [Google Scholar]
- 133.Russell JA, Hu Y, Chau L, Pauliushchyk M, Anastopoulos I, Anandan S, et al. Indoor-biofilter growth and exposure to airborne chemicals drive similar changes in plant root bacterial communities. Appl Environ Microbiol. 2014;80:4805–13. doi: 10.1128/AEM.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Waring MS. How well do potted plants or bio-walls clean the indoor air of organic gases? October 17–18, 2016. http://nas-sites.org/builtmicrobiome/files/2016/07/Michael-Waring-FOR-POSTING.pdf.
- 135.Ponsoni K, Raddi MSG. Indoor air quality related to occupancy at an airconditioned public building. Braz Arch Biol Technol. 2010;53:99–103. [Google Scholar]
- 136.Sadar John. Through the Healing Glass. 2016. [Google Scholar]
- 137.Himmelfarb P, Scott A, Thayer PS. Bactericidal activity of a broad-spectrum illumination source. Appl Microbiol. 1970;19:1013–4. doi: 10.1128/am.19.6.1013-1014.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.The Duty on Glass. Lancet. 1845;45:214–6.
- 139.Møller KI, Kongshoj B, Philipsen PA, Thomsen VO, Wulf HC. How Finsen’s light cured lupus vulgaris. Photodermatol Photoimmunol Photomed. 2005;21:118–24. doi: 10.1111/j.1600-0781.2005.00159.x. [DOI] [PubMed] [Google Scholar]
- 140.Hobday RA. Sunlight therapy and solar architecture. Med Hist. 1997;41:455–72. doi: 10.1017/s0025727300063043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hobday RA, Dancer SJ. Roles of sunlight and natural ventilation for controlling infection: historical and current perspectives. J Hosp Infect. 2013;84:271–82. doi: 10.1016/j.jhin.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bazzoni CB. The destruction of bacteria through the action of light. Am J Public Health. 1914;4:975–92. doi: 10.2105/ajph.4.11.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ward HM. The action of light on bacteria. Lancet. 1893;141:383. [Google Scholar]
- 144.Downing AMW, Blunt TP. III. Researches on the effect of light upon Bacteria and other organisms. Proc R Soc Lond. 1878;26:488–500. [Google Scholar]
- 145.Medeiros AB, de A, Enders BC, Lira ALBDC. The Florence Nightingale’s Environmental Theory: A Critical. Analysis. Esc Anna Nery. 2015;19:518–24. [Google Scholar]
- 146.Nightingale F. Notes on Hospitals. Longman, Green, Longman, Roberts, and Green, London, 1863.
- 147.Kundsin RB. Architectural design and indoor microbial pollution. Oxford University Press, New York, 1988.
- 148.Stone A (ed.). Proceedings of the Twenty-Seventh Annual Convention of the American Institute of Architects. Inland Architect Press, Chicago, Illinois, 1893.
- 149.Chynoweth P. Progressing the rights to light debate – Part 1: a review of current practice. Structural Survey. 2004;22:131–7. [Google Scholar]
- 150.Hessling M, Spellerberg B, Hoenes K. Photoinactivation of bacteria by endogenous photosensitizers and exposure to visible light of different wavelengths - a review on existing data. FEMS Microbiol Lett. 2017;364:fnw270. doi: 10.1093/femsle/fnw270. [DOI] [PubMed] [Google Scholar]
- 151.Fonseca MJ, Tavares F. The bactericidal effect of sunlight. Am Biol Teach. 2011;73:548–52. [Google Scholar]
- 152.Besaratinia A, Yoon J-I, Schroeder C, Bradforth SE, Cockburn M, Pfeifer GP. Wavelength dependence of ultraviolet radiation-induced DNA damage as determined by laser irradiation suggests that cyclobutane pyrimidine dimers are the principal DNA lesions produced by terrestrial sunlight. FASEB J. 2011;25:3079–91. doi: 10.1096/fj.11-187336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldman RP, Travisano M. Experimental evolution of ultraviolet radiation resistance in Escherichia coli. Evolution. 2011;65:3486–98. doi: 10.1111/j.1558-5646.2011.01438.x. [DOI] [PubMed] [Google Scholar]
- 154.Takada A, Matsushita K, Horioka S, Furuichi Y, Sumi Y. Bactericidal effects of 310 nm ultraviolet light-emitting diode irradiation on oral bacteria. BMC Oral Health. 2017;17:96. doi: 10.1186/s12903-017-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dai T, Vrahas MS, Murray CK, Hamblin MR. Ultraviolet C irradiation: an alternative antimicrobial approach to localized infections? Expert Rev Anti Infect Ther. 2012;10:185–95. doi: 10.1586/eri.11.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Oppezzo OJ. Contribution of UVB radiation to bacterial inactivation by natural sunlight. J Photochem Photobiol B. 2012;115:58–62. doi: 10.1016/j.jphotobiol.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 157.de Sousa DL, Lima RA, Zanin IC, Klein MI, Janal MN, Duarte S. Effect of twice-daily blue light treatment on matrix-rich biofilm development. PLoS ONE. 2015;10:e0131941. doi: 10.1371/journal.pone.0131941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Maclean M, Anderson JG, MacGregor SJ, White T, Atreya CD. A new proof of concept in bacterial reduction: antimicrobial action of violet-blue light (405 nm) in ex vivo stored plasma. J Blood Transfus. 2016;2016:2920514. doi: 10.1155/2016/2920514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Deng Y, Yao J, Wang X, Guo H, Duan D. Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS ONE. 2012;7:e39704. doi: 10.1371/journal.pone.0039704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ondrusch N, Kreft J. Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS ONE. 2011;6:e16151. doi: 10.1371/journal.pone.0016151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sandhu BS, Singh CK. Relationship of sunlight and humidity on the virulence of street rabies virus in saliva. Indian J Anim Sci. 2009;79:24–25. [Google Scholar]
- 162.Patra V, Byrne SN, Wolf P. The skin microbiome: is it affected by uv-induced immune suppression? Front Microbiol. 2016;7:1235. doi: 10.3389/fmicb.2016.01235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Prescott SL, Larcombe D-L, Logan AC, West C, Burks W, Caraballo L, et al. The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ J. 2017;10:29. doi: 10.1186/s40413-017-0160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hartmann B, Benson M, Junger A, Quinzio L, Röhrig R, Fengler B, et al. Computer keyboard and mouse as a reservoir of pathogens in an intensive care unit. J Clin Monit Comput. 2004;18:7–12. doi: 10.1023/b:jocm.0000025279.27084.39. [DOI] [PubMed] [Google Scholar]
- 165.Daylight modeling | WELL Standard. https://standard.wellcertified.com/light/daylight-modeling. Accessed 27 Feb 2019.
- 166.Daylight | U.S. Green Building Council. https://www.usgbc.org/credits/healthcare/v4-draft/eqc-0. Accessed 27 Feb 2019.
- 167.Sehulster L, Chinn RYW, Arduino MJ, Carpenter J, Donlan R, Ashford D, et al. Guidelines for environmental infection control in health-care facilities. Centers for Disease Control and Prevention, 2003. https://francais.cdc.gov/mmwr/preview/mmwrhtml/rr5210a1.htm?mobile=nocontent.
- 168.Lateef F. Hospital design for better infection control. J Emerg Trauma Shock. 2009;2:175–9. doi: 10.4103/0974-2700.55329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, et al. Multistate point-prevalence survey of health care–associated infections. N Engl J Med. 2014;370:1198–208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jacob JT, Kasali A, Steinberg JP, Zimring C, Denham ME. The role of the hospital environment in preventing healthcare-associated infections caused by pathogens transmitted through the air. Health Environ Res. Design J. 2013;7:74–98. [Google Scholar]
- 171.Dancer SJ. Controlling hospital-acquired infection: focus on the role of the environment and new technologies for decontamination. Clin Microbiol Rev. 2014;27:665–90. doi: 10.1128/CMR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Donskey CJ. Does improving surface cleaning and disinfection reduce health care-associated infections? Am J Infect Control. 2013;41:S12–9. doi: 10.1016/j.ajic.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 173.Kwan SE, Shaughnessy RJ, Hegarty B, Haverinen-Shaughnessy U, Peccia J. The reestablishment of microbial communities after surface cleaning in schools. J Appl Microbiol. 2018;125:897–906. doi: 10.1111/jam.13898. [DOI] [PubMed] [Google Scholar]
- 174.La Duc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol. 2007;73:2600–11. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Boyce JM, Havill NL, Dumigan DG, Golebiewski M, Balogun O, Rizvani R. Monitoring the effectiveness of hospital cleaning practices by use of an adenosine triphosphate bioluminescence assay. Infect Control Hosp Epidemiol. 2009;30:678–84. doi: 10.1086/598243. [DOI] [PubMed] [Google Scholar]
- 177.Dharan S, Mourouga P, Copin P, Bessmer G, Tschanz B, Pittet D. Routine disinfection of patients’ environmental surfaces. Myth or reality? J Hosp Infect. 1999;42:113–7. doi: 10.1053/jhin.1999.0567. [DOI] [PubMed] [Google Scholar]
- 178.Dancer SJ. Hospital cleaning in the 21st century. Eur J Clin Microbiol Infect Dis. 2011;30:1473–81. doi: 10.1007/s10096-011-1250-x. [DOI] [PubMed] [Google Scholar]
- 179.Ramphal L, Suzuki S, McCracken IM, Addai A. Improving hospital staff compliance with environmental cleaning behavior. Proc. 2014;27:88–91. doi: 10.1080/08998280.2014.11929065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Garcia dos Santos A, Jr, Menis Ferreira A, Rigotti MA, Ribeiro dos Santos F, Ribeiro Furlan MC, de Andrade D. Efficiency evaluation of the cleaning and disinfection of surfaces in a primary health center. Texto Contexto Enferm. 2018;27:e3720017. [Google Scholar]
- 181.Xu C, Liu L. Personalized ventilation: one possible solution for airborne infection control in highly occupied space? Indoor Built Environ. 2018;27:873–6. [Google Scholar]
- 182.Memarzadeh F. Literature Review: Room Ventilation and Airborne Disease Transmission. Am Soc Healthcare Eng. 2013. http://www.ashe.org/management_monographs/pdfs/mg2013Memarzadeh.pdf.
- 183.Atkinson EBJ, Chartier Y, Pessoa-Silva CL, Jensen P, Li Y, Seto W-H. Natural Ventilation for Infection Control in Health-Care Settings. World Health Organization, 2009. https://www.who.int/water_sanitation_health/publications/natural_ventilation.pdf. [PubMed]
- 184.Adams RI, Miletto M, Taylor JW, Bruns TD. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013;7:1460–1460. doi: 10.1038/ismej.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Carlton EJ, Barton K, Shrestha PM, Humphrey J, Newman LS, Adgate JL, et al. Relationships between home ventilation rates and respiratory health in the Colorado Home Energy Efficiency and Respiratory Health (CHEER) study. Environ Res. 2019;169:297–307. doi: 10.1016/j.envres.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 186.Arvanitakis G, Temmerman R, Spök A. Development and use of microbial-based cleaning products (MBCPs): current issues and knowledge gaps. Food Chem Toxicol. 2018;116:3–9. doi: 10.1016/j.fct.2017.12.032. [DOI] [PubMed] [Google Scholar]
- 187.Spök A, Klade M. Environmental, health and legal aspects of cleaners containing living microbes as active ingredients. 2009.http://www.tb-klade.at/wp-content/uploads/2015/06/IFZ-EWP-3-2010.pdf.
- 188.Pérez-Pantoja D, Donoso R, Junca H, González B, Pieper DH. Phylogenomics of Aerobic Bacterial Degradation ofAromatics. In: Rojo F, editor. Aerobic Utilization of Hydrocarbons, Oils and Lipids. Cham: Springer; 2016. [Google Scholar]
- 189.Vandini A, Temmerman R, Frabetti A, Caselli E, Antonioli P, Balboni PG, et al. Hard surface biocontrol in hospitals using microbial-based cleaning products. PLoS ONE. 2014;9:e108598. doi: 10.1371/journal.pone.0108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Caselli E, D’Accolti M, Vandini A, Lanzoni L, Camerada MT, Coccagna M, et al. Impact of a probiotic-based cleaning intervention on the microbiota ecosystem of the hospital surfaces: focus on the resistome remodulation. PLoS ONE. 2016;11:e0148857. doi: 10.1371/journal.pone.0148857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nordengrün M, Michalik S, Völker U, Bröker BM, Gómez-Gascón L. The quest for bacterial allergens. Int J Med Microbiol. 2018;308:738–50. doi: 10.1016/j.ijmm.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 192.Simon-Nobbe B, Denk U, Pöll V, Rid R, Breitenbach M. The spectrum of fungal allergy. Int. Arch. Allergy Immunol. 2008;145:58–86. doi: 10.1159/000107578. [DOI] [PubMed] [Google Scholar]
- 193.Araki A, Kawai T, Eitaki Y, Kanazawa A, Morimoto K, Nakayama K, et al. Relationship between selected indoor volatile organic compounds, so-called microbial VOC, and the prevalence of mucous membrane symptoms in single family homes. Sci Total Environ. 2010;408:2208–15. doi: 10.1016/j.scitotenv.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 194.Daisey JM, Angell WJ, Apte MG. Indoor air quality, ventilation and health symptoms in schools: an analysis of existing information. Indoor Air. 2003;13:53–64. doi: 10.1034/j.1600-0668.2003.00153.x. [DOI] [PubMed] [Google Scholar]
- 195.Sundell J, Levin H, Nazaroff WW, Cain WS, Fisk WJ, Grimsrud DT, et al. Ventilation rates and health: multidisciplinary review of the scientific literature. Indoor Air. 2011;21:191–204. doi: 10.1111/j.1600-0668.2010.00703.x. [DOI] [PubMed] [Google Scholar]
- 196.Simons E, Hwang S-A, Fitzgerald EF, Kielb C, Lin S. The impact of school building conditions on student absenteeism in Upstate New York. Am J Public Health. 2010;100:1679–86. doi: 10.2105/AJPH.2009.165324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.CDC. Asthma. Centers for Disease Control and Prevention | National Center for Health Statistics. 2019. https://www.cdc.gov/nchs/fastats/asthma.htm. Accessed 6 Feb 2019.
- 198.Hsu J, Qin X, Beavers SF, Mirabelli MC. Asthma-related school absenteeism, morbidity, and modifiable factors. Am J Prev Med. 2016;51:23–32. doi: 10.1016/j.amepre.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nunes C, Pereira AM, Morais-Almeida M. Asthma costs and social impact. Asthma Research and Practice. 2016;3:1. doi: 10.1186/s40733-016-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ciaccio CE, Barnes C, Kennedy K, Chan M, Portnoy J, Rosenwasser L. Home dust microbiota is disordered in homes of low-income asthmatic children. J Asthma. 2015;52:873–80. doi: 10.3109/02770903.2015.1028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134:593–601.e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hooks KB, O’Malley MA. Dysbiosis and its discontents. MBio. 2017;8:01492–01417. doi: 10.1128/mBio.01492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chu W-L, Tneh S-Y, Ambu S. A survey of airborne algae and cyanobacteria within the indoor environment of an office building in Kuala Lumpur, Malaysia. Grana. 2013;52:207–20. [Google Scholar]
- 204.Bernstein IL, Safferman RS. Viable algae in house dust. Nature. 1970;227:851–2. doi: 10.1038/227851a0. [DOI] [PubMed] [Google Scholar]
- 205.Böbel TS, Hackl SB, Langgartner D, Jarczok MN, Rohleder N, Rook GA, et al. Less immune activation following social stress in rural vs. urban participants raised with regular or no animal contact, respectively. Proc Natl Acad Sci USA. 2018;115:5259–64. doi: 10.1073/pnas.1719866115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rintala H, Pitkäranta M, Täubel M. Chapter 4 - Microbial Communities Associated with House Dust. In: Laskin AI, Sariaslani S, Gadd GM (eds). Advances in applied microbiology. Academic Press, 2012, pp 75–120. [DOI] [PubMed]
- 207.Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130:44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 208.Vestergaard DV, Holst GJ, Basinas I, Elholm G, Schlünssen V, Linneberg A, et al. Pig Farmers’ homes harbor more diverse airborne bacterial communities than pig stables or suburban homes. Front Microbiol. 2018;9:870. doi: 10.3389/fmicb.2018.00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cardona C, Lax S, Larsen P, Stephens B, Hampton-Marcell J, Edwardson CF, et al. Environmental sources of bacteria differentially influence host-associated microbial dynamics. mSystems. 2018;3:e00052–18. doi: 10.1128/mSystems.00052-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.McDonald D, Ackermann G, Khailova L, Baird C, Heyland D, Kozar R, et al. Extreme dysbiosis of the microbiome in critical illness. mSphere. 2016;1:e00199–16. doi: 10.1128/mSphere.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Brooks B, Firek BA, Miller CS, Sharon I, Thomas BC, Baker R, et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2014;2:1. doi: 10.1186/2049-2618-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lax S, Sangwan N, Smith D, Larsen P, Handley KM, Richardson M, et al. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med. 2017;9:eaah6500. doi: 10.1126/scitranslmed.aah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Parkinson AJ. The Arctic Human Health Initiative: a legacy of the International Polar Year 2007–9. Int J Circumpolar Health. 2013;72:21655. doi: 10.3402/ijch.v72i0.21655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Barker SF, Packer M, Scales PJ, Gray S, Snape I, Hamilton AJ. Pathogen reduction requirements for direct potable reuse in Antarctica: evaluating human health risks in small communities. Sci Total Environ. 2013;461–2:723–33. doi: 10.1016/j.scitotenv.2013.05.059. [DOI] [PubMed] [Google Scholar]
- 215.Jin J-S, Touyama M, Yamada S, Yamazaki T, Benno Y. Alteration of a human intestinal microbiota under extreme life environment in the Antarctica. Biol Pharm Bull. 2014;37:1899–906. doi: 10.1248/bpb.b14-00397. [DOI] [PubMed] [Google Scholar]
- 216.Venkateswaran K, Vaishampayan P, Cisneros J, Pierson DL, Rogers SO, Perry J. International Space Station environmental microbiome - microbial inventories of ISS filter debris. Appl Microbiol Biotechnol. 2014;98:6453–66. doi: 10.1007/s00253-014-5650-6. [DOI] [PubMed] [Google Scholar]
- 217.Phelan M. Why fungi adapt so well to life in space | Scienceline. Scienceline. 2018. https://scienceline.org/2018/03/fungi-love-to-grow-in-outer-space/. Accessed 10 Feb 2019.
- 218.Crucian BE, Choukèr A, Simpson RJ, Mehta S, Marshall G, Smith SM, et al. Immune system dysregulation during spaceflight: potential countermeasures for deep space exploration missions. Front Immunol. 2018;9:1437. doi: 10.3389/fimmu.2018.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wilson N. A Microbial Hitchhiker’s Guide to the Galaxy: researchers race to understand effects of deep space on microbiome. Bioscience. 2019;69:5–11. [Google Scholar]
- 220.Voorhies AA, Lorenzi HA. The challenge of maintaining a healthy microbiome during long-duration space missions. Front Astronomy Space Stations. 2016;3:23. [Google Scholar]
- 221.Romsdahl J, Blachowicz A, Chiang AJ, Singh N, Stajich JE, Kalkum M, et al. Characterization of Aspergillus niger Isolated from the International Space Station. mSystems. 2018;3:e00112–18. doi: 10.1128/mSystems.00112-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mahnert A, Moissl-Eichinger C, Zojer M, Bogumil D, Mizrahi I, Rattei T, et al. Man-made microbial resistances in built environments. Nat Commun. 2019;10:968. doi: 10.1038/s41467-019-08864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Leung MHY, Wilkins D, Li EKT, Kong FKF, Lee PKH. Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl Environ Microbiol. 2014;80:6760–70. doi: 10.1128/AEM.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Be NA, Thissen JB, Fofanov VY, Allen JE, Rojas M, Golovko G, et al. Metagenomic analysis of the airborne environment in urban spaces. Microb Ecol. 2015;69:346–55. doi: 10.1007/s00248-014-0517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Haahtela T, Holgate S, Pawankar R, Akdis CA, Benjaponpitak S, Caraballo L, et al. The biodiversity hypothesis and allergic disease: world allergy organization position statement. World Allergy Organ J. 2013;6:3. doi: 10.1186/1939-4551-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kim K, DuPont HL, Pickering LK. Outbreaks of diarrhea associated with Clostridium difficile and its toxin in day-care centers: evidence of person-to-person spread. J Pediatr. 1983;102:376–82. doi: 10.1016/s0022-3476(83)80652-0. [DOI] [PubMed] [Google Scholar]
- 227.Delmée M, Verellen G, Avesani V, Francois G. Clostridium difficile in neonates: serogrouping and epidemiology. Eur J Pediatr. 1988;147:36–40. doi: 10.1007/BF00442608. [DOI] [PubMed] [Google Scholar]
- 228.Rusin P, Maxwell S, Gerba C. Comparative surface-to-hand and fingertip-to-mouth transfer efficiency of gram-positive bacteria, gram-negative bacteria, and phage. J Appl Microbiol. 2002;93:585–92. doi: 10.1046/j.1365-2672.2002.01734.x. [DOI] [PubMed] [Google Scholar]
- 229.Ziskind G. Particle resuspension from surfaces: revisited and re-evaluated. Rev Chem Eng. 2006;22:1–123. [Google Scholar]
- 230.Jou J, Ebrahim J, Shofer FS, Hamilton KW, Stern J, Han JH, et al. Environmental transmission of Clostridium difficile: association between hospital room size and C. difficile Infection. Infect Control Hosp Epidemiol. 2015;36:564–8. doi: 10.1017/ice.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Freedberg DE, Salmasian H, Cohen B, Abrams JA, Larson EL. Receipt of antibiotics in hospitalized patients and risk for Clostridium difficile infection in subsequent patients who occupy the same bed. JAMA Intern Med. 2016;176:1801–8. doi: 10.1001/jamainternmed.2016.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–7. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kim H-Y, Estes CR, Duncan AG, Wade BD, Cleary FC, Lloyd CR, et al. Real-time detection of microbial contamination. IEEE Eng Med Biol Mag. 2004;23:122–9. doi: 10.1109/memb.2004.1297183. [DOI] [PubMed] [Google Scholar]
- 234.Fahimipour AK, Ben Mamaar S, McFarland AG, Blaustein RA, Chen J, Glawe AJ, et al. Antimicrobial chemicals associate with microbial function and antibiotic resistance indoors. mSystems. 2018;3:e00200–18. doi: 10.1128/mSystems.00200-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Emerson JB, Adams RI, Román CMB, Brooks B, Coil DA, Dahlhausen K, et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome. 2017;5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Klein AM, Bohannan BJM, Jaffe DA, Levin DA, Green JL. Molecular evidence for metabolically active bacteria in the atmosphere. Front Microbiol. 2016;7:772. doi: 10.3389/fmicb.2016.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nocker A, Cheung C-Y, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods. 2006;67:310–20. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 238.Gao M, Ahern J, Koshland CP. Perceived built environment and health-related quality of life in four types of neighborhoods in Xi’an, China. Health Place. 2016;39:110–5. doi: 10.1016/j.healthplace.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 239.Miller JD, Sun M, Gilyan A, Roy J, Rand TG. Inflammation-associated gene transcription and expression in mouse lungs induced by low molecular weight compounds from fungi from the built environment. Chem Biol Interact. 2010;183:113–24. doi: 10.1016/j.cbi.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 240.Perlman RL. Mouse models of human disease: an evolutionary perspective. Evol Med Public Health. 2016;2016:170–6. doi: 10.1093/emph/eow014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Dai D, Prussin AJ, 2nd, Marr LC, Vikesland PJ, Edwards MA, Pruden A. Factors shaping the human exposome in the built environment: opportunities for engineering control. Environ Sci Technol. 2017;51:7759–74. doi: 10.1021/acs.est.7b01097. [DOI] [PubMed] [Google Scholar]
- 242.Jiang C, Wang X, Li X, Inlora J, Wang T, Liu Q, et al. Dynamic human environmental exposome revealed by longitudinal personal monitoring. Cell. 2018;175:277–91.e31. doi: 10.1016/j.cell.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stucki AO, Stucki JD, Hall SRR, Felder M, Mermoud Y, Schmid RA, et al. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip. 2015;15:1302–10. doi: 10.1039/c4lc01252f. [DOI] [PubMed] [Google Scholar]
- 244.Cho S, Yoon J-Y. Organ-on-a-chip for assessing environmental toxicants. Curr Opin Biotechnol. 2017;45:34–42. doi: 10.1016/j.copbio.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Arnold C. Rethinking sterile: the hospital microbiome. Environ Health Perspect. 2014;122:A182–7. doi: 10.1289/ehp.122-A182. [DOI] [PMC free article] [PubMed] [Google Scholar]