Abstract
Protein phosphorylation is a key post-translational modification regulating protein function in almost all cellular processes. Although tens of thousands of phosphorylation sites have been identified in human cells, approaches to determine the functional importance of each phosphosite are lacking. Here, we manually curated 112 datasets of phospho-enriched proteins generated from 104 different human cell types or tissues. We reanalyzed the 6,801 proteomics experiments that passed our quality control criteria, creating a reference phosphoproteome containing 119,809 human phosphosites. To prioritize functional sites, we used machine learning to identify 59 features indicative of proteomic, structural, regulatory or evolutionary relevance and integrate them into a single functional score. Our approach identifies regulatory phosphosites across different molecular mechanisms, processes and diseases, and reveals genetic susceptibilities at a genomic scale. Several novel regulatory phosphosites were experimentally validated, including a role in neuronal differentiation for phosphosites in SMARCC2, a member of the SWI/SNF chromatin remodeling complex.
Protein phosphorylation is a post-translational modification (PTM) involved in the regulation of most biological processes and its misregulation has been linked to several human diseases1,2. The full extent of human phosphorylation is still an open question under active investigation through mass spectrometry (MS) approaches3. Notably, an in-depth study of a single cell line identified over 50,000 phosphopeptides and suggested that 75% of the proteome may be phosphorylated4. The aggregation of such studies have led to the identification of over 200,000 phosphosites in resources such as PhosphoSitePlus (PSP)5.
Although analytical challenges remain, the bottleneck in the study of phosphorylation is shifting towards its functional characterization6. Given that phosphorylation can be poorly conserved, it has been suggested that not all phosphorylation is relevant for fitness7–9. Therefore, prioritization strategies are crucial to facilitate the discovery of highly relevant phosphosites10. Different methodologies have been proposed, including identifying phosphosites that are highly conserved11,12, located at interface positions13–15, showing strong regulation, or combinations of such features10,16. Mutational studies have also been used to characterize relevant phosphorylations17, but cannot yet be applied to human phosphorylation at scale.
Machine learning methods remain a poorly explored approach to study the functional relevance of phosphorylation. Here, we generated the largest human phosphoproteome dataset to date, identifying 119,809 human phosphosites. For each phosphosite, we compiled annotations covering 59 features and integrated them into a single score of functional relevance, named here the phosphosite functional score. This score can correctly identify regulatory phosphosites for a diverse set of mechanisms and predict the impact of deleterious mutations.
Results
Mass spectrometry-based proteomics map of the human phosphoproteome
In order to create a comprehensive MS-based definition of the human phosphoproteome, we curated 112 human public phospho-enriched datasets derived from 104 different cell types and/or tissues from the PRIDE database18 (Supplementary Table 1). Using MaxQuant, we jointly re-analyzed the subset of 6,801 human MS experiments passing the quality control criteria, corresponding to 575 days of accumulated instrument time19 (Methods). The joint analysis (deposited in PRIDE, dataset PXD012174) ensured an adequate control of the false discovery rate (FDR) estimated using a target-decoy strategy20 (Methods). FDR was estimated for correct matching to the peptide-spectrum match (PSM), protein and the presence of phosphosite modification(s) and kept at <1% (Methods). The modification localization probability (also called False Localisation Rate) was also estimated, reflecting the confidence of pinpointing which residue carries the phosphorylation. Probabilities above 75% indicate highly confident localizations (Class I sites).
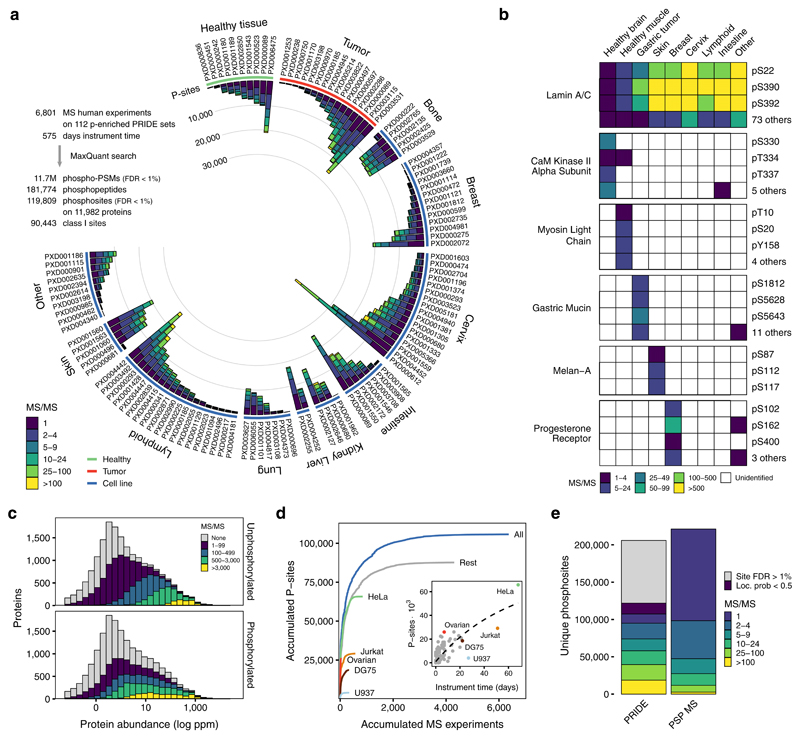
We identified 11.7 million phosphorylated peptide-spectrum matches (PSM-level FDR < 1%), corresponding to 181,774 phosphopeptides spanning 203,930 phosphorylated serines, threonines or tyrosines. Of these, only 119,809 sites passed the 1% site-level FDR correction (59% true positive sites) with 90,443 classified as Class I21. The low true positive percentage suggests that the accumulation of phosphosite identifications from multiple independent searches - as archived in phosphosite databases - might be strongly enriched for potential false positives. The heterogeneity of biological samples analyzed facilitated the identification of phosphosites in proteins expressed in a wide range of tissues from healthy and tumor samples (Figure 1a), allowing us to identify a large number of tissue-specific phosphosites (Figure 1b).
Figure 1. Comprehensive catalog of in-vivo human phosphosites.
a) Number of phosphosites (Localisation probability > 0.5) binned by the number of peptide-spectrum matches coming from the re-analyzed human phospho-enriched datasets curated from PRIDE. b) Examples of broad or tissue-specific phosphosites with spectral count information. c) Phosphopeptide and unphosphorylated peptide MS/MS support for all human proteins binned using the consensus protein abundance from PaxDb d) Cumulative increase in the number of phosphosites for each added MS experiment by biological origin: all samples (blue), top 5 most common cell lines or tissues or remaining samples (grey). MS experiments were sorted by size. Inset - Accumulated instrument time and the total number of phosphosites identified per sample. e) Total number of unique identified phosphosites and MS/MS support in the combined PRIDE analysis and PSP.
To evaluate the phosphoproteome coverage, we studied the proportion of phosphoproteins when stratifying them according to their abundance22. From 14,154 proteins identified, 11,982 (85%) contained at least one FDR-corrected phosphosite. While we observed a bias towards the identification of more abundant proteins, the trend is similar to that of the non-modified peptides present in the samples (Figure 1c). This ratio of phosphoproteins, similar to previous findings4, remained constant regardless of the reported protein abundance in PaxDb (Figure S1). While some cell types, such as HeLa cells, are very over-represented, the identified phosphosites are not strongly biased by common samples. The exclusion of the five most studied cell types (31% of the total instrument time), still results in 83% of the total identified phosphosites (Figure 1d). Together, these results suggest that we have achieved very high coverage of phosphosite identification.
For benchmarking purposes, we compared the identified phosphoproteome against the 221,236 human phosphosites reported by MS in the PSP database (January 2018)5 (Figure 1e). While 11.5% of the high-confidence sites in our study are supported by only 1 MS/MS evidence, 55% of the PSP sites have this level of support. In absolute numbers, we identified 73,973 phosphosites supported by 5 or more MS/MS evidences, while only 47,448 are equally supported in PSP. These results point to the importance of aggregating the growing body of MS phosphoproteomic data while maintaining the statistical reliability.
Prioritizing functional human phosphosites
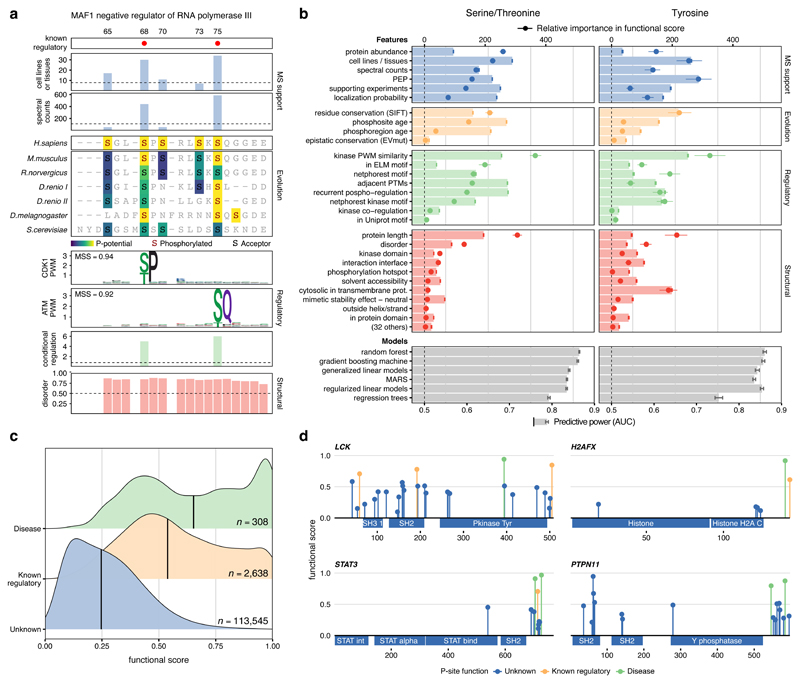
Having identified a comprehensive high confidence human phosphoproteome, we then calculated for every phosphosite a diverse set of features that could indicate importance for fitness (Methods). These properties can be grouped broadly in four categories: the MS-evidence (e.g., spectral counts, localization probability), the phosphosite regulation23 --including the number of conditions regulated or matching to kinase motifs, the structural environment (e.g., at interfaces, surface accessibility), or the evolutionary conservation (Supplementary Note 1). Evolutionary conservation was captured by quantifying residue conservation24, conservation of the phosphorylation within protein domain families11 and the phosphosite evolutionary age, expanding to multicellular species a phylogenetic-based approach previously described for fungal species12. Altogether, we calculated a set of 59 features annotating all phosphosites (Methods, Supplementary Table 2). We illustrate the value of some of these features for identifying regulatory sites in the 65-80 amino acid region of MAF1, a protein involved in the mTORC1 signaling pathway (Figure 2a). While pS68 and pS75 are known to inhibit the MAF1 RNA pol III repression function, three other MS-identified phosphosites in the region have unknown functional roles25. Relative to the other sites, pS68 and pS75 showed: a higher number of spectral counts, higher conservation, a better match to the specificity of kinases, and show condition-specific regulation, including downregulation when treated with the mTOR inhibitors rapamycin or Torin1.
Figure 2. A functional score for human phosphosites.
a) MAF1 phosphosites in the 65-80 region with feature annotations b) Feature discriminative power (AUCs) after repeated cross-validation between phosphosites of known and unknown function (AUC, red, green, yellow and blue bars). Discriminative power (AUCs) after integrating all features using different machine learning algorithms (AUC, grey bars). The contribution of each feature to the final model (point-ranges). c) Distribution of functional scores for all phosphosites (blue), with known regulatory roles (yellow) and associated with human diseases (green). Sample size (n) represents number of phosphosites. d) Functional score and regulatory function for phosphosites in different protein families.
Benefiting from the orthogonal nature of some of these attributes, we next sought to integrate a set of 59 normalized features into a single score that would prioritize phosphosites relevant for fitness. We used 2,638 phosphosites curated by PSP and known to regulate protein function to discriminate, using machine learning, the distinctive properties of functional serine and threonine phosphorylation (S/T) and tyrosine phosphorylation (Y), separately. We first asked how well each of the 59 features independently predicted the known regulatory sites (bars in Figure 2b). The most informative features for function included the number of different cell lines or tissues in which the site had been identified, the phosphosite age, how well it matched a kinase specificity model, how often the phosphosite was regulated in a panel of different perturbations and the presence of neighboring PTMs.
Different machine-learning algorithms were tested for their capacity to integrate the 59 features with most methods displaying similar performances (Methods). The model selected was a gradient boosting machine that achieved an average AUC of 86.1% and 85.7% for ST and Y, respectively26. The contribution of the features included in the model denotes their relevance when combined with the rest of the features (Figure 2b). The performance of each individual feature in isolation and their contribution to the trained models is detailed in Table S2. For example, the consensus protein abundance increased its relevance when integrated with other features, indicating it is not a useful predictor in isolation, but facilitate the interpretation of other features within the model. The integrated predictor displays a much higher performance than any of the features independently.
The final model - named here phosphosite functional score - was applied to generate a score for each of the MS-identified phosphosites, reflecting their importance for organismal fitness (Supplementary Table 3). In Figure 2c, we show how this score ranked known functional phosphosites higher than the overall background and, more interestingly, phosphosites important for human disease - information not included in the model - ranked higher than the other two sets. Our metric very strongly outperforms general-purpose tools such as variant effect predictors when estimating the relevance of functional and disease-associated phosphosites (Figure S2). The functional score correctly distinguished relevant phosphosites across different protein families as illustrated in four different examples, including the LCK kinase, the STAT3 transcription factor, the PTPN11 phosphatase and the H2AFX histone (Figure 2d). In all cases, the phosphosites of known function or associated with diseases were among the top-ranked. Extensive literature search also pointed to highly scored phosphosites that, despite not being included in the true positive set derived from PSP, had supporting evidence of known regulatory function (Figure S3).
Identifying functional phosphosites involved in diverse mechanisms
Phosphorylation modulates protein function via different molecular mechanisms. The distributions of phosphosite functional scores for annotated regulatory sites suggest the method is not strongly biased towards specific mechanisms (Figure S4). Next, we explored the potential of the functional score to prioritize regulatory functions related to protein-protein interactions and transcriptional activity.
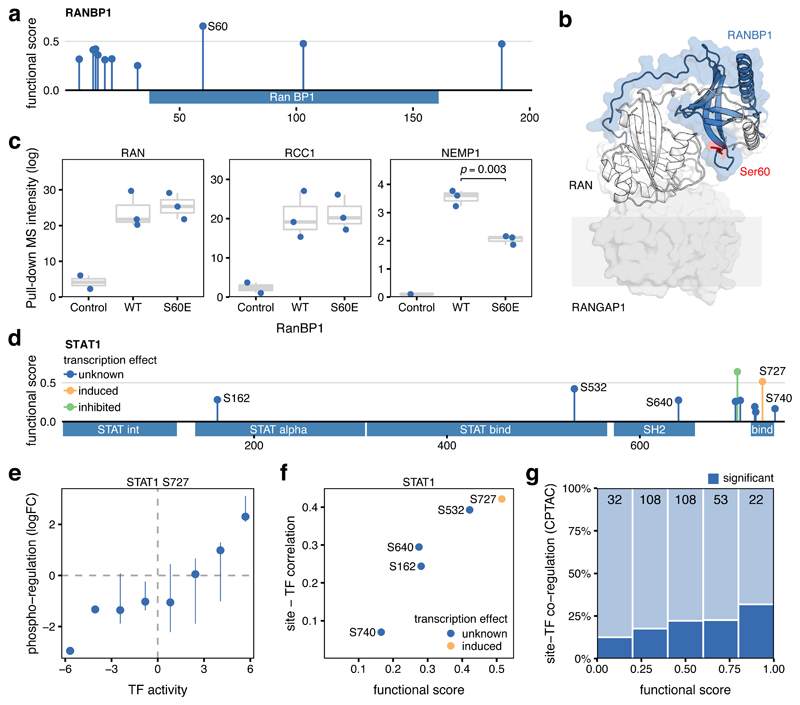
The presence of a phosphosite at a protein interface, one of the used features, is interpreted as being likely to regulate protein-protein interactions (Methods, provided in Supplementary Table 2). For example, the Y34 phosphosite in the Ras Homolog Family Member A (RHOA) had no annotated function in PSP but ranked as a highly functional site (0.56), partly due to its presence in multiple interaction interfaces. Indeed, it has been reported that mutation of Y34 to asparagine27 or phenylalanine28,29 can disrupt certain interactions (Figure S5). To test broadly the value of the functional score for protein interactions, we compiled information on mutational consequences towards measured protein interactions of 394 phosphosite positions30. We observed that phosphosites with a functional score higher than 0.5, had 5 to 10-fold higher chances of the mutation causing changes in interactions (Figure S6).
Independently, we sought to validate as a proof-of-principle a candidate regulatory phosphosite in a protein interface. The PLK1-regulated S60 is the best scoring phosphosite (0.65, Figure 3a) of the RAN Binding Protein 1 (RANBP1)31. This site is upregulated under okadaic acid and it is near the Ran interaction interface and the transmembrane nuclear transporter Ran GTPase Activating Protein 1 (RanGAP) (Figure 3b, PDB:1k5g). To test the impact of pS60 on RanBP1 interactions, we performed an affinity purification experiment comparing the 3xFLAG tagged RANBP1 WT with RANBP1 S60E (Methods, Supplementary Table 4). Among the top scoring interactors, the proteins RAN, RCC1 and NEMP1 were found. The phospho-mutant binding remained similar to the WT in the case of RAN and RCC1 but showed a reproducible decrease (p < 0.003) in binding capability to NEMP1, an interaction partner previously described in other eukaryotes32 (Figure 3c).
Figure 3. Identification of functional sites regulating protein interaction and transcriptional activity.
a) Functional score for 10 phosphosites identified in RANBP1. b) Structural model of RANBP1 in complex with RAN (PDB:1k5g). c) MS binding quantification for RANBP1 interaction partners (RAN, RCC1, and NEMP1) pulling-down the control, the WT, or the S60E RANBP1 mutant. 3 biological replicates between wild type and mutant were compared using a two-sided t test and displayed when significant (p<0.05). Boxes represent Q1-Q3 with a centre in the median value. d) Functional score for 10 phosphosites identified in STAT1 with unknown (blue) known activating (yellow) and inhibitory (green) regulatory activity. e) Pearson correlation between the changes in phosphorylation levels of the known activation site of STAT1 (S727) with the changes in estimated STAT1 transcriptional activity across 74 tumor samples. Pointrange represents the binned median and confidence limits based on non-parametric bootstrap. f) Relationship between STAT1 phosphosite functional score and the Pearson correlation between the TF activity and the phosphorylation changes across 74 tumor samples. g) Fraction of phosphosites in TFs showing significantly correlated changes (Pearson’s correlation p<0.05) with the corresponding TF activity, stratified by their functional score. Data based on 323 transcription factor phosphosites obtained from TCGA and CPTAC consortia (see Methods).
We next explored how the phosphosite prioritization could be used to identify sites implicated in the regulation of transcriptional activity. Changes in the activation state of transcription factors (TFs) across different samples can be approximated by quantifying the changes in expression of their known target genes33. By analyzing 77 breast cancer samples where phosphorylation34 and gene expression35 had been collected, we correlated the changes in phosphosite levels within TFs with changes in estimated TF activity (Methods). We exemplify this approach with phosphosites in STAT1 (Figure 3d), where pS727 is necessary for full transcriptional activation36. We observed, as expected, that increased phosphorylation of S727 was associated with higher estimated TF activities (Figure 3e). Across all STAT1 phosphosites, the functional score stratified all quantified sites based on the correlation between the phosphorylation levels and the TF activity (Figure 3f). We expanded this analysis to the 371 phosphosites in 82 TFs with available paired data and good quality regulons. Although the set of sites with significant site-TF correlation is relatively small (19%), we observed that the functional score was able to prioritize sites in which the changes in phosphorylation strongly correlated with the TF activity (Figure 3g).
Impact of genetic variation on highly functional phosphosites
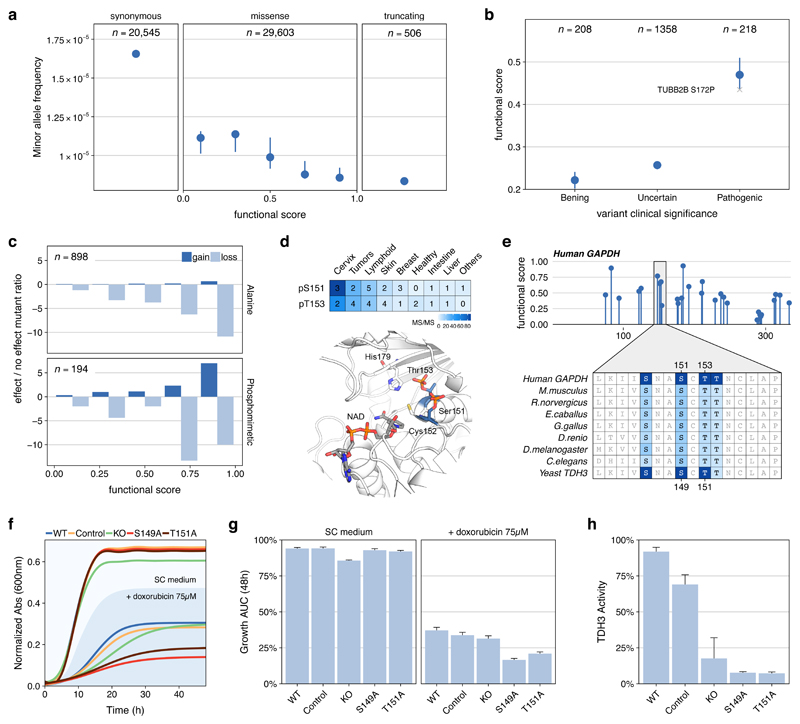
A functional phosphosite is expected to introduce a genetic constraint in the genome and highly functional phosphosites should therefore present a lower tolerance to variation. We analyzed available information on allele frequency of variants in natural populations37 (Figure 4a) and the clinical significance of mutations on human diseases38 (Figure 4b, Methods). In both cases, we observed the expected constraint - mutations in phosphosites with a high functional score were more likely rare in human populations (Figure 4a) and pathogenic (Figure 4b). The annotated phosphoproteome provides an opportunity to further characterize the underlying mechanisms of disease-causing mutations. For example, the S172P mutation in Tubulin, Beta 2B (TUBB2B) has been associated with polymicrogyria, a cortical developmental disease causing poor incorporation of tubulin into microtubules39. The finding of a high functional phosphosite in S172 (0.43, Figure 4b) suggests phospho-regulation as an important disease-related mechanism. Associating disease variants with the functionally annotated phosphoproteome can facilitate the disease interpretation in a signaling context, expanding the possibility for diagnostic and therapeutic strategies. We provide the functional scores for ClinVar variants overlapping phosphorylation sites in Supplementary Table 5.
Figure 4. Consequences of genetic variants for phosphosites with high functional scores.
a) Median (CI > 95%) minor allele frequency for variants sorted by phosphosite functional score and compared with synonymous and stop codon causing variants occurring at phosphosite positions. b) Mean functional score (CI > 95%) for phosphosites at positions with mutations found in patients and having benign, uncertain or pathogenic consequences. The S172P mutation in Tubulin, Beta 2B (TUBB2B) is highlighted as an example - see main text c) Fold ratio of mutations in phosphorylated positions reported in mutagenesis studies having gain/loss of function effects versus no effect and stratified by functional score. d) MS evidence for the phosphorylation of S151 and T153 in GAPDH and their structural context flanking a catalytic cysteine e) Position and functional score for all GAPDH phosphosites and alignment of two human phosphosites (S151 and T153) to the corresponding S. cerevisiae TDH3 (S149 and T151). Color gradient corresponds to supporting evidence of phosphorylation based on ancestral reconstruction f) Consensus growth curves and (g) mean and standard error of the area under the growth curve for wild type (WT), control, GAPDH knockout (KO) and S149 and T151 phospho-deficient mutants in the presence or absence of doxorubicin (75 μM). Every clone is present 4 times in each plate and the experiment repeated 3 times for a total of 12 replicates. h) TDH3 activity as mean and SE measured twice in 3 independent extracts obtained from control and mutant strains.
The introduction of mutations in highly scored phosphosites is also expected to cause important functional consequences. We curated information on protein gain/loss of function as a consequence of 1,092 mutations in phospho-acceptor residues (Methods, Figure 4c). In line with our expectation, the higher the functional score the higher the chances a mutation causes an effect (Figure 4c). Interestingly, mutations to alanine in highly scored acceptors predominantly caused loss-of-function, while mutation to negatively charged residues (i.e. phospho-mimetic) also had an increased chance of causing a gain-of-function effect. To further expand on this idea, we selected to study two highly scored phosphosites of unknown function in glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The two sites - S151 and T153 - flank the catalytic cysteine C152 in a highly conserved region (Figure 4d). Phosphorylation was consistently identified by MS and their phosphosite functional scores (0.63 and 0.70, respectively) indicated a potential regulatory function (Figure 4e). We hypothesized these phosphosites to be important for the regulation of GAPDH enzymatic activity and functionally conserved between human and yeast.
To test this hypothesis, we created two phospho-deficient strains S149 and T151 in the S. cerevisiae main GAPDH gene (TDH3) that corresponds to S151A and T153A in the human GAPDH. For these stains we measured their growth together with the TDH3 gene knockout (KO) in the presence and absence of the topoisomerase inhibitor doxorubicin (Methods). Deletion of TDH3 has been previously reported to cause slow-growth under doxorubicin, a phenotype corroborated in our assay40. Interestingly, more acute growth defects were observed for the 2 phospho-deficient strains under doxorubicin (Figures 4f and 4g). TDH activity assays under no treatment pointed to a depleted enzymatic activity of the 2 phospho-deficient strains (Figure 4h). The growth rate under doxorubicin and the enzyme activity of the point mutants were lower than for the KO strain suggesting that the TDH3 KO may have some compensation via TDH1/TDH2 that is not seen in the mutants. To gain insight into this difference we carried out a thermal proteome profiling experiment41–43 comparing the thermal stability and protein abundance of 2378 proteins between the mutant strains and WT (Methods, Supplementary Table 6). In unstressed conditions, TDH2 protein showed higher abundance in all 3 mutants (KO, S151A and T153A) which is more pronounced in the KO (Figure S7). With doxorubicin, this increase is only significant in the KO (Figure S7, Supplementary Table 6). This increase in TDH2 may partially compensate for the loss of TDH3 activity which would explain the observed differences between the KO and the phospho-mutant strains. In addition, we measured no significant change in stability or abundance of TDH3 itself indicating that the impact of the phospho-mutations is not occurring via the destabilization of the protein. Finally, we found significant enrichment for increased abundance of glycolytic proteins and increased stability of oxidative phosphorylation (Figure S8), indicating that the decrease in activity of TDH3 results in substantial adaptation of the metabolism.
Regulatory phosphosites in the hSWI/SNF remodeling complex member SMARCC2
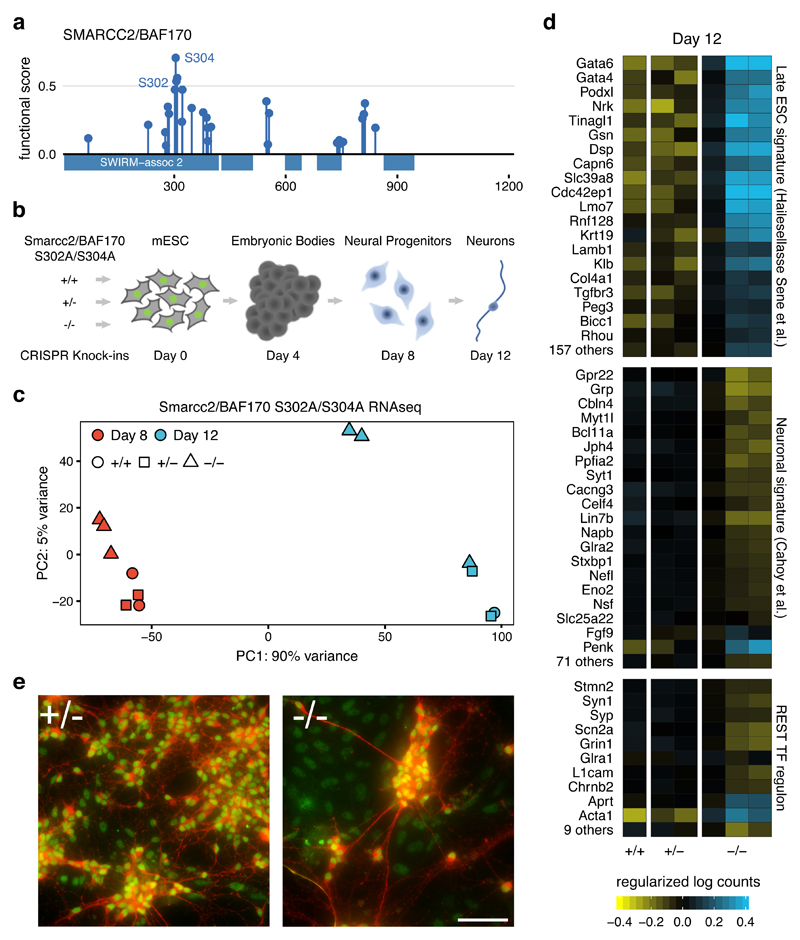
To further exemplify the usefulness of the functional score we studied regulatory sites governing the function of SMARCC2/BAF170 during neuronal differentiation. As part of the SWI/SNF chromatin remodelling complex, SMARCC2 plays an important role in neurogenesis44,45. SMARCC2 expression increases during the differentiation towards the commitment to neuronal precursors. Increased SMARCC2 replaces SMARCC1, composing the neural progenitor-specific hSWI/SNF complex (or npBAF complex)46. This switch recruits the REST (RE1-silencing transcription factor-corepressor) complex an interaction that is important for neurogenesis44. We identified 2 high scoring phosphosites (S302 and S304) in SMARCC2 (Figure 5a) that we hypothesize could play an important regulatory function during neurogenesis.
Figure 5. Smarcc2 S302A/S304A homozygous mutants show delayed neuronal differentiation.
a) Functional score for SMARCC2/BAF170 phosphosites. b) Design for CRISPR Knock-in mutagenesis of control (+/+), heterozygous (+/-) and homozygous (-/-) Smarcc2/Baf170 S302A/S304A mutation followed by expected neuronal differentiation timeline. c) PCA plot based on the normalized RNA-seq levels of 25,411 mouse genes obtained from of the 7 clonal lines at days 8 and 12. d) Normalized RNA-seq log counts of genes contained in signatures of Late ESC, neurons or transcriptionally regulated by REST as measured in 6 independent biological replicates presenting control (+/+), heterozygous (+/-) and homozygous (-/-) backgrounds. e) Merged immunofluorescence images of differentiated cells on day 12 stained with antibodies against neuronal microtubules Map2 (red) and Smarcc2/Baf170 (Green). Scale bar represents 25 μm.
To study the selected phosphosites, we used a murine model of neuronal differentiation and a gene knock-in system (Figure 5b). After verifying the two phosphosites are identified by MS in mouse neuronal tissues47, we used CRISPR-based gene editing in mouse embryonic stem cells (mESC) and independently generated 3 homozygous and 2 heterozygous clones of the double alanine mutant S302A/S304A, as well as control clones (no mutation after CRISPR targeting). Neuronal differentiation was performed over 12 days, with an expected44 increase of Smarcc2 measured by day 12 (Figure S9), concurrent with post-mitotic neuron formation (Figure S10). RNA-seq was performed at days 8 and 12, corresponding to the differentiation to neuronal progenitor cells and post-mitotic neurons. When comparing the mRNA levels using principal component analysis (PCA, Figure 5c), the independent clones showed a strong agreement with the major driver of change (PC1, Figure 5c) being time and the second major driver (PC2, Figure 5c) the mutational status. 2 out of 3 homozygous clones showed a clear separation from the heterozygous and CRISPR controls. A linear model identified 4,776 genes (p < 0.05) showing a gene expression difference that was dependent on the mutational status. This significant transcriptional difference is compatible with a change in the differentiation process as a consequence of the SMARCC2 mutations.
We noted that the homozygous mutants at day 12 displayed transcriptional similarities with the 8-day clones, suggesting a delay in the differentiation. The homozygous mutants showed upregulation of gene signatures associated with murine pluripotency48 and significant downregulation in neuronal differentiation signatures49(Figure 5d). In addition, known REST target genes - a transcriptional repressor of neuronal genes - were significantly down-regulated in the homozygous mutant indicating maintained REST activity (Figure 5d). Neuronal morphology also showed less differentiation in two homozygous clones at 12 days, with observed cell aggregates, fewer neurites and numerous Smarcc2 marked cells without the neuronal marker (e.g., Map2) (Figure 5e). Overall, these results point to a significant delay in the differentiation in the homozygous clones, suggesting that these phosphorylation sites play a role in the regulation neuronal differentiation by SMARCC2.
Discussion
Benefiting from the vast amount of proteomics data deposited by the community in the PRIDE database, we generated a comprehensive human phosphoproteome. The large fraction of phosphosites identified when excluding the most studied cell lines (e.g., HeLa cells) highlights the high coverage achieved. However, some limitations need to be taken into consideration. The difficulties to detect certain phosphopeptides - like those in low abundant proteins - or to accurately localize phosphorylation events, point to a still-incomplete phosphoproteome. Rarefaction curve analysis (Figure S11) also suggests we have not yet reached saturation on the number of identified sites, and the upper limit of phosphosites remains unknown. Our analysis also indicates that the inadequate aggregation of parallel MS searches can lead to very substantial accumulation of false positives, which may be the case for public resources. Our priority has been to provide a high confidence list of phosphosites, implying that some good quality sites might not have made the final list due to the stringent criteria (see Methods). The scale and level of curation of the spectral data analyzed (PRIDE dataset PXD012174) constitute an unprecedented resource to further develop new methods and strategies necessary to understand phosphoproteomics at large scale.
Although adequate methods were in place to prevent a biased scoring system, it is possible the score may be influenced by social bias to better characterize certain functional sites used for training - such as the tendency for researchers to study conserved positions. While we think such biases are possible, we don’t think they dominate the score as it can predict the impact of mutations in phosphorylated residues that were not part of the training. Although we have shown how highly scored phosphorylations are less likely to be mutated or more likely to be involved in disease processes, the full extent of this prioritization has not been explored. Disease variants on functional phosphosites could offer a mechanistic explanation for the disease, possibly indicating a therapeutic strategy perhaps linked to an actionable regulatory kinase.
Despite the accuracy of the functional score, there were several phosphosites of known function that have a low functional score. When stratifying the functional phosphosites by their mechanisms of action (Figure S3) we do not observe a strong difference. As a general trend phosphosites that impact on cellular localization, often overlapping short unstructured motifs, show the lowest performance and those regulating enzymatic activities and conformations – typically within ordered globular regions - show the strongest performance. These trends, although not strong, may point to limitations in the current implementation of the method.
While we integrated a diverse set of features, additional ones may need to be considered in the future. These could include: measuring occupancy for a larger set of phosphosites4, acquiring diverse PTM datasets for different species, paired measurements of phosphoproteomics and protein localization50, and determining the structures of larger set of phosphorylated proteins. Resolving structures of phosphoproteins has been technically very challenging, but recent developments in genetically encoded phosphorylation50,51 should now serve as an opportunity to make progress in this area. Some of the current features may indirectly capture missing signals, such as how spectral counts relate to occupancy or the number of PTMs near a phosphosite may relate to cross-regulation.
Together with the full list of human phosphosites and the spectral data released, the phosphosite functional score and the relevant features associated constitute a systematic resource to understand human signaling on a genomic scale. We have shown examples of how this information can prime the design of experiments in order to identify novel regulatory phosphosites. While some of the features may suggest a possible regulatory mechanism, such as the presence of phosphosites at interface positions, the functional score itself does not predict mechanisms. Moreover, the score might not accurately prioritize regulatory sites acting in groups, despite including features that capture local regulatory effects. Additional experimental and computational approaches need to be developed to study the biochemical consequences of individual and coordinated phosphorylations on a large scale.
Not all residue phosphorylations are likely to contribute equally to organismal fitness and some have speculated that a fraction of phosphosites may serve no purpose in present-day species. Evolutionary studies have suggested that between 35% to 65% sites are constrained and therefore functional9,52. Regardless of the degree of non-functional phosphorylation, there is a clear bottleneck in the characterization of phosphosite function that can be aided by the functional score. The different analyses provide guidance as to how to interpret and make use of the score. Scores above 0.5 were associated with 5 to 10 fold increase in chances of measuring an impact on interactions or phenotypes (Figure 4c and S6). Similarly, pathogenic mutations have an average score of close to 0.5 compared with around 0.2 for benign mutations (Figure 4b). At a 0.5 cut-off around 10% of the phosphoproteome remains with 50% of the true positive phosphosites recovered suggesting that this is useful cut-off for an initial prioritization. The human phosphoproteome and functional score prioritization provided here define a roadmap to study the regulation of proteins involved in almost any cellular process.
Methods
Human phosphoproteome MS search
A list of all 307 human datasets annotated to contain phosphorylations was retrieved from the PRIDE database (June 2017)18. After extensive manual curation of all experimental designs, only untargeted assays that employed phospho-enrichment strategies (e.g. metal oxide affinity chromatography, anti-P-Tyr antibodies, etc.) were included. Datasets with proteomes from more than one species (i.e. infections, xenografts, etc.) were discarded in order to prevent cross-species contaminants. Similarly, datasets with major genetic modifications were also discarded for further analysis. For the remaining 110 PRIDE datasets, additional manual curation was required in order to annotate the biological origin and the search parameters for each raw file. After applying all filters, 6,801 MS raw files remained, corresponding to an accumulated instrument time of 575 days.
All 6,801 MS raw files were jointly searched using MaxQuant 1.6.0.13 (MQ)19 Andromeda engine53 against the UniProt Human Reference Proteome (71,567 sequences, accessed May 2017)54. These were also supplemented with common laboratory contaminants provided by MQ. Based on the labeling strategy or the digestion enzymes, we identified 17 different search parameter groups that were applied to their corresponding raw files. Cysteine carbamidomethylation was set as a fixed modification, while oxidation of methionine (M), protein N-terminal acetylation, and phosphorylation of serine (S), threonine (T) and tyrosine (Y) as variable modifications. Minimum peptide length was set to 7 amino acids, and peptides were allowed to have a maximum of two missed-cleavages. All default parameters were also applied for the Orbitrap instruments, the precursor mass tolerance was set to 20 ppm for the first search and 4.5 for the main search. Fragment mass tolerances were set to 20 ppm and 0.5 Da for FT and IT detectors, respectively. All other mass tolerance settings were kept at default values. The search took 60 days in a Dell PowerEdge R730 rack server configured with Intel(R) Xeon(R) CPU E5-2697 v4 @ 2.30GHz, 64 cores, 256 GB high-capacity DDR4 memory and a 20 TB fast access storage drive.
For the identification of modified peptides, MQ default search parameters were used, including 1% PSM FDR, 1% site-level FDR, a minimum Andromeda score of 40 and a minimum delta score of 6. FDR rates are estimated using a target-decoy strategy. In a standard search, the FDR is controlled on the level of peptide spectrum matches (PSM) and later in the resulting protein groups. In case of modified peptides, the site decoy fraction (SDF) can also be used, and when switched on, it is applied after PSM filtering but separately to protein groups FDR. This filtering step results in a table of PTM sites containing a specified fraction of site decoy hits. The results for phosphorylation sites are contained in the “Phospho (STY)Sites.txt” table. Importantly, since SDF is applied independently of the protein FDR, phosphorylation sites on peptides that are part of proteins that did not pass protein FDR can still appear in this table. In addition to controlling FDR rates, it is often beneficial to include thresholds that ensure that only posttranslationally modified peptide identifications based on spectra of sufficient quality are included. To achieve that, MQ allows to set a minimum Andromeda score and a minimum Andromeda delta score for accepting modified peptides. The Andromeda delta score is the difference between first and second best PSM with a different sequence. Importantly, this filtering happens before site-level FDR correction.
In order to assess the effect of applying false discovery correction at the site level in addition to PSM FDR, two separate searches were performed, one keeping the default 1% site decoy correction and another search removing this filtering. Despite both searches being corrected at a 1% PSM FDR, the 1% site-FDR search identified 119,809 phosphorylated STYs (121,896 when including the 0.98% of decoys), whereas the search performed without the side level correction identified 252,189 STY hits of which 18.48% were decoys. This difference highlights the importance of controlling FDR rates at relevant steps in the identification pipeline of very large PTM datasets. All processing settings and results, including raw and MQ intermediate files, are available in PRIDE under the accession PXD012174.
Functional annotation of the human phosphoproteome
An extended description of the data sources and methods to functionally annotate phosphosites can be found in Supplementary Note 1. The series of features include information about MS supporting evidence, residue conservation, phosphorylation age reconstruction, consesus motif binding, conditional phospho-regulation, 1D structural properties, phosphorylation structural hotspots, structural stability and interfaces and protein topology annotations.
AI phosphosite functional prioritization
An MS-derived reference phosphoproteome was defined based on the 119,809 FDR-corrected phosphosites identified in the UniProt reference proteome54. In order to prevent problems with redundant sequences, we focused our prioritization strategy in the 116,258 sites contained in the subset of 21,009 reviewed proteins within the reference proteome. Every site was annotated based on a comprehensive list containing 85 of the aforementioned features (Supplementary Table 2). The features were filtered as previously described in order to remove correlated features. Continuous variables were centered, scaled, normalized using the Yeo-Johnson power transformation and near zero-variance variables excluded using the R package caret. Only complete features were considered, except for SIFT were a small set of sites from very large proteins required imputation by nearest neighbors. The discrete variables included in the remaining 38 features, were transformed into dummy variables, expanding the list to the final 59 features.
In order to train a machine learning model capable of regressing the functional potential of all phosphosites, we required the phosphoproteome annotated with functional features and a gold standard set of functional phosphosites. From the MS-identified phosphosites, we defined a gold standard using the 2,638 sites with annotated function in the PhosphoSitePlus database (Nov 2017)5 using the remaining sites of unknown function as negatives. A set of regression models were benchmarked using the same strategy. These include random forest, gradient boosting machine, generalized linear models, regularized linear models, Multivariate Adaptive Regression Splines (MARS), and regression trees. The corresponding R packages for each of the models are described in the caret R wrapper. In all cases, a separate predictor for ST and Y residues was built. Nested repeated cross-validation (CV) was used to estimate the generalization error of the underlying model and its (hyper)parameter search. In the inside loop, a 5-fold CV was performed 10 times in order to tune the parameters. In the outside loop, a 3-fold CV was repeated 5 times to quantify the performance of the model using ROC analysis. An extensive search grid was defined for every training/validation/testing cycle and parallelized in a high-performance computing environment.
Despite the small differences in the performance of the best methods, gradient boosting machine was selected as the final model for the functional score, in part due to its more harmonic distribution of scores. The parameters displaying the best results were: 500 trees, interaction depth of 9, 10 minimum observations in a node and shrinkage of 0.0405 for STs and 0.0105 for Tyrosines. Phosphosite functional scores were derived using these parameters as the median values of all scores obtained by 3-fold CV repeated 30 times.
Phospho-deficient RANBP1 pull-down assay
A 3xFLAG tagged fusion of both the WT (UniProt P43487 canonical isoform) and phosphomimetic S60E mutant of RANBP1, were synthesized (GENEWIZ). Lentiviral transduction was used to individually introduce these sequences into HEK293T cells, and RANBP1 expression was controlled under a doxycycline-inducible promoter. Cells from 3 biological replicates of each WT and S60E expression cells were harvested. Additionally, 2 biological replicates of control cells not expressing 3xFLAG RANBP1 were harvested, in which no doxycycline was added to the cells. Protein complexes were purified as previously described55, and analyzed on a Thermo Q-Exactive Plus mass spectrometer. Raw data was searched using MaxQuant19, and high confidence protein-protein interactions were scored using SAINTexpress56.
TF activity inference and phosphosite coregulation
To systematically estimate transcription factor activities and phosphosite co-regulation across a panel of primary tumors, paired basal gene expression and quantitative phosphoproteomic data from the same breast tumors were retrieved from The Cancer Genome Atlas (TCGA)35 and Clinical Proteomic Tumor Analysis Consortium (CPTAC)34, respectively. From the 105 breast tumors with matched phosphoproteomics data, raw counts were downloaded from the Gene Expression Omnibus (GSE62944)57, normalized, processed into counts per million reads and z-transformed. To infer transcription factor (TF) activities from the expression data, 289 good quality TF regulons (A-C) were retrieved from the OmniPath resource58 and an enrichment test per tumor was calculated on them using the analytic Rank-based Enrichment Analysis (aREA) method implemented in Viper59. The Normalized Enrichment Score (NES) was used as an estimate of relative TF activity as described in previous studies33. The changes in phosphorylation for TF sites quantified across the 77 samples passing the CPTAC quality control criteria were correlated with their corresponding TF activities.
Effects of genetic variation in human phosphosites
Variation in natural human populations for 60,706 unrelated individuals was retrieved from the Exome Aggregation Consortium (ExAC)37. The variants with their corresponding adjusted allele frequencies were aligned to UniProt positions using the Needleman-Wunsch global alignment implemented in the Biostrings R package. Only variants observed in at least 10 counts were considered for further analysis. In cases were multiple alleles mapped to the same exact amino acid substitution, the variant with the highest adjusted frequency was preserved. For comparison purposes, Minor Allele Frequency (MAF) for phospho-acceptor residues was calculated as the frequency at which the second most common allele occurs in the population. To further understand the effects in human variation, a total of 159,633 human clinically relevant variants were retrieved from ClinVar38, 1,784 of which match phospho-acceptor residues. The annotated clinical significances were collapsed into benign, uncertain or pathogenic. Finally, the phenotypic consequences of introducing 22,090 missense mutations in 3,022 proteins were retrieved from UniProt54. The 4,764 point-mutations on phospho-acceptor residues were classified into Alanine (A), Phosphomimetic (E or D) or Other depending on the resulting residue. Since the phenotypic consequences are encoded as free text, an in-house parser was required to annotate the effects as gain-of-function, loss-of-function or no effect.
Phenotypic effects of distant phospho-deficient variants
TDH3 phosphomutant construction
Phospho-deficient mutants TDH3 S149A and T151A were constructed by introducing the point mutations into the TDH3 endogenous locus in the Y8205 background strain (MATalpha, his3 1; leu2 0; ura3 0; MET15+; LYS2+; can1::STE2pr-SpHIS5; lyp1::STE3pr-LEU2 + GALpr-SceI-NAT) followed by a URA3 marker after the stop codon. The URA3 marker was flanked by SceI recognition sites and removed by induction of the endonuclease as previously described60. Point mutations were verified by DNA sequencing of the TDH3 ORF.
TDH3 Growth curves description
Yeast strains including wild-type, gene control, TDH3 KO from the yeast gene knockout library61 and the 2 phospho-deficient mutants were inoculated into a final volume of 100 μL SC media with and without 75μM doxorubicin in 96-well plates (initial OD660nm=0.05). 4 biological replicates were inoculated in each plate and the experiment was performed 3 times. All plates were sealed with breathable membranes (Breathe-Easy) and incubated at 30°C in a thermostated incubator (Cytomat 2, Thermo Scientific) with continuous shaking. OD660nm was measured every 30 min for 48h in a Filtermax F5 multimode plate reader (Molecular Devices). Growth curves were estimated by fitting a standard logistic equation included in the Growthcurver R package. Cellular fitness was defined by the area under the curve (AUC).
TDH3 Mutant activity measurement
Yeast strains growing in exponential phase were diluted to the same OD (OD660nm=0.2) in 3 biological replicates and 1ml was collected by centrifugation. Cell lysis was adapted for yeast cells: cell pellets were resuspended in 500μl of cold lysis buffer (20 mM Tris pH8, 15mM EDTA pH8, 15mM EGTA pH8 and 0.1% Triton X-100). Glass beads were added in equal volume (500μl) and cells were lysed by vortexing at 4ºC. Enzymatic activity was quantified twice using the colorimetric Glyceraldehyde-3-Phosphate Dehydrogenase Activity Assay Kit (Abcam) as described by the manufacturer.
Two-dimensional thermal proteome profiling (2D-TPP)
The 2D-TPP protocol43 was adapted to be compatible with S. cerevisiae. Yeast cells were grown overnight at 30°C in YPAD and diluted to OD600 0.1 in 50 ml fresh YPAD media and when OD600 reached ~0.7 cells were treated with or without 75uM Doxirubicin for 2 hours. Cultures were collected by centrifugation at 4000x g for 5 min, immediately frozen in liquid nitrogen, washed with PBS and resuspended to an equivalent of OD660 of 125. 20ul was aliquoted to 10 wells of a PCR plate and the plate was centrifuged at 4.000g for 5 min and was subjected to a temperature gradient for 3 min in a thermocycler (Agilent SureCycler 8800) followed by 3 min at room temperature. Cells were lysed in 30 ul of cold lysis buffer (final concentration: Zymolyase (Amsbio) 0.5mg/ml, 1x protease inhibitor (Roche), 1x phosphatase inhibitor, 250U/ml benzonase and 1mM MgCl2 in PBS for 30 min shaking at 30°C, followed by five freeze-thaw cycles (freezing in liquid nitrogen, followed by 30s at 25°C in a thermocycler and vortexing). Protein digestion, peptide labelling, Mass spectrometry-based proteomics and data analysis were performed as previously described42.
Phenotypic assay of phospho-deficient SMARCC2 during neuronal differentiation
Generation of Smarcc2/BAF170 mutant cell lines
Mouse embryonic stem (ES) cells (129XC57BL/6J) were cultured in media containing Knockout-DMEM (Thermo Fisher) with 15% EmbryoMax FBS (Millipore) and 20 ng/ml leukemia inhibitory factor (LIF, produced by Protein Expression Facility at EMBL Heidelberg), 1% non-essential amino acids, 1% Glutamax, 1% Pen/Strep, 1% of 55mM beta-Mercaptoethanol solution. Cells were maintained at 37°C with 5% CO2. ESCs were routinely tested for mycoplasma absence by PCR.
ES lines with double C to G point mutations encoding S302A and S304A substitutions in Smarcc2/BAF170 were generated using CRISPR-Cas9 mediated knock-in mutation according to the published protocol62. Briefly, oligonucleotides encoding guide sequences were cloned into pSpCas9(BB)-2A-GFP (PX458, Addgene). The resulting plasmid (2 μg Smarcc2_g1 or g2) and 100 μM ssODN repair templates (180 bp, ssODN_Smarcc2_g1 or g2, IDT ultrameres) were nucleofected into ES cells using 4D-nucleofector (Lonza) for gene editing. Two days after nucleofection, individual transfected cells were plated in 96 well plates by FACS. Cells carrying homozygous or heterozygous introduced mutations in Smarcc2/BAF170 were identified by Sanger sequencing. In addition, cells that retained a wild type Smarcc2/BAF170 sequence despite having gone through the CRISPR mutagenesis process were retained as CRISPR controls.
Smarcc2_g1 AGGGGGGCAACTATAAGAAG
Smarcc2_g2 CTCTGGGGTGGGTGAAGGAG
ssODN_Smarcc2_g1
GGGTAAACTGCTGCTCATCCCACGTCCTGTTGTGCAGGTAAACAGCCCAG ATTCAGACAGACGAGACAAGAAGGGGGGCAATTATAAGAAAAGAAAGCGC GCTCCCGCTCCTTCACCCACCCCAGAGGCTAAGAAGAAAAACGCTAAGAA AGGGTAAGCTACCTCCTGTGCCCACACGCG
ssODN_Smarcc2_g2
TCCCACGTCCTGTTGTGCAGGTAAACAGCCCAGATTCAGACAGACGAGAC AAGAAGGGGGGCAACTATAAGAAGAGGAAGCGCGCTCCCGCCCCATCACC CACCCCAGAGGCTAAGAAGAAAAACGCTAAGAAAGGGTAAGCTACCTCCT GTGCCCACACGCGCTGTCCTGATGCCATCTCA
Neuronal Differentiation
ES cells with heterozygous or homozygous Smarcc2 mutations, along with CRISPR control cells, were neuronally differentiated according to Bibel et al63. To start differentiation, ES cells were plated on bacterial Petri dishes in CA media containing DMEM high glucose (Thermo Fisher) with 10% FBS, 1% non-essential amino acids, 1% Glutamax, 1% Pen/Strep, 1% of 55mM beta-Mercaptoethanol solution. After 4 days retinoic acid was added to embryonic bodies at a final concentration of 5μM. For neural culture, after 8 days embryoid bodies were dissociated with trypsin and plated in N2 media composed of regular DMEM supplemented with N2 and B27-VitaminA (Thermo Fisher). Plates were pre-coated with Poly-D-Lysine (Sigma) and Laminin (Roche). Half of the N2 media was changed every two days. Samples of differentiating cells were snap frozen in liquid nitrogen on the day of plating (D0) as well as on the four (D4), eight (D8) and twelve (D12) days following differentiation.
Western Blotting
ES cell-derived neuronal BAF170 protein levels were visualized by western blot. Crude nuclear extracts were prepared from D12 mutant cell lines and CRISPR controls. Extracts were also prepared from D0, D4, D8 and D12 wild type differentiating cells. Extracts were run on SDS/PAGE gels (4-12%), transferred to PVDF membranes and probed with primary antibodies to BAF170 (Active Motif, 1:1000) and Histone H4 (1:6000, Abcam). Membranes were washed, probed with HRP tagged goat anti-rabbit secondary antibodies, and visualized with Immobilon chemiluminescent reagent (Millipore) using a Chemidoc Touch imaging system (Bio-Rad).
Immunofluorescence
Neuronal morphology and SMARCC2/BAF170 protein localization were visualized using indirect immunofluorescence of D12 neuronal cultures. D8 cells were placed on coverslips and fixed on D12 with 2% PFA in PBS for 15 minutes at room temperature. Coverslips were washed 3 times in PBS and permeabilized with 100% methanol for 5 minutes at -20°C. Permeabilized cells were washed twice with wash buffer (0.05% Tween 20 in PBS) and blocked for 30 minutes with 2% BSA in PBS. Cells were then incubated with primary antibodies (1:400 MAP2 (Sigma) and 1:2000 BAF170 (Active Motif)) for 1 hour at room temperature or overnight at 4°C. Coverslips were given 3 washes in wash buffer for 5 minutes each. Secondary antibodies (anti-Mouse Alexa Fluor 594 and anti-Rabbit Alexa Fluor 488) were applied for 30 minutes at room temperature followed by an additional two 5-minute washes in wash buffer. Coverslips were washed briefly in sterile water, inverted and mounted on microscope slides with Prolong Gold antifade reagent. Images were collected at 40x with a Ti-Eclipse Fluorescence Microscope (Nikon). Image Analysis was performed using Fiji and the Cell Counter plugin.
RNA sequencing
Total RNA was extracted from D8 and D12 differentiating cells using the RNeasy RNA isolation kit (Qiagen) followed by DNase digestion using TURBO DNase (Thermo Fisher). mRNAs were isolated from 1 μg of total RNA using a PolyA selection kit (NEB) and sequencing libraries were prepared using the NEBnext Ultra RNA library prep kit for Illumina (NEB). Completed libraries were sequenced on a NextSeq 500 sequencer (Illumina) using a 75 bp single-end run. Reads were aligned to the murine GRCm38 assembly using HISAT2 v2.1.064. Aligned reads were converted to BAM format and sorted using samtools v0.1.1965. Read counts were calculated using htseq-count66 using the murine gene sets annotated in Ensembl v9367. Reads with less than 10 counts were discarded for further analysis. Read counts from all samples were combined using the Rsubread68 and analyzed using DESeq269. Gene set signatures for ESC, neuronal differentiation and TF regulons were downloaded from MSigDB70.
Supplementary Material
ED SUM.
Phosphorylation sites are ranked for functional relevance using a comprehensive, high-quality human phosphoproteome.
Acknowledgements
This study would have been impossible without the selfless deposition of data from hundreds of authors. We would like to extend our gratitude to every one of them. We thank Dr. Jurgen Cox for his insightful advice on the site-decoy multiple testing correction. We would like to thank the members of the Beltrao group for their support collecting features and their relevant comments, as well as to David Ocaña and Andy Cafferkey as part of the EMBL-EBI Technical Services Cluster. We would like to thank Dominic Helm from the EMBL Proteomics core facility for their help with the analysis of thermal proteome profiling samples. This study has been funded by EMBL core funding, and by the Wellcome Trust [grants numbers WT101477MA and 208391/Z/17/Z]. P.B. and D.O. are supported by a Starting Grant Award from the European Research Council (ERC-2014-STG 638884 PhosFunc).
Footnotes
Author contributions
D.O. and P.B. conceived the study, prepared figures and wrote the paper. P.B., A.F.J., D.O. and J.A.V. designed computational analyses. D.O., A.F.J., L.G.-A. and F.S. performed computational analyses. K.-M.N., D.L.S, M.M.S and N.K designed experiments. C.V., M.G., M.S., A.M. A.A.K., A.H. performed experiments. All authors read and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Data and code availability
All MS data - including raw and MQ intermediate- processing settings and results files are available in PRIDE under the accession PXD012174. The functional annotation of the phosphoproteome, as well as the gold standard and resulting functional scores are available in Supplementary Tables. The conditional regulation data used in the feature generation is available in phosphate.com. Code to proceed with the generation of some of the features (e.g. age reconstruction or structural hotspots) is available in their respective repositories as described in Supplementary Notes 1. Further data and code is also available upon request.
References
- 1.Lahiry P, Torkamani A, Schork NJ, Hegele RA. Kinase mutations in human disease: Interpreting genotype-phenotype relationships. Nature Reviews Genetics. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 2.Torkamani A, Kannan N, Taylor SS, Schork NJ. Congenital disease SNPs target lineage specific structural elements in protein kinases. Proc Natl Acad Sci U S A. 2008;105:9011–9016. doi: 10.1073/pnas.0802403105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 4.Sharma K, et al. Ultradeep Human Phosphoproteome Reveals a Distinct Regulatory Nature of Tyr and Ser/Thr-Based Signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 5.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Needham EJ, Parker BL, Burykin T, James DE, Humphrey SJ. Illuminating the dark phosphoproteome. Science Signaling. 2019;12:eaau8645. doi: 10.1126/scisignal.aau8645. [DOI] [PubMed] [Google Scholar]
- 7.Beltrao P, Bork P, Krogan NJ, Van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol. 2013;9:n/a-n/a. doi: 10.1002/msb.201304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanshin E, Bergeron-Sandoval L-P, Isik SS, Thibault P, Michnick SW. A Cell-Signaling Network Temporally Resolves Specific versus Promiscuous Phosphorylation. Cell Rep. 2015;10:1202–1214. doi: 10.1016/j.celrep.2015.01.052. [DOI] [PubMed] [Google Scholar]
- 9.Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends in Genetics. 2009 doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Beltrao P, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strumillo MJ, et al. Conserved phosphorylation hotspots in eukaryotic protein domain families. Nat Commun. 2019;10:1977. doi: 10.1038/s41467-019-09952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studer RA, et al. Evolution of protein phosphorylation across 18 fungal species. Science (80-. ) 2016;354:229–232. doi: 10.1126/science.aaf2144. [DOI] [PubMed] [Google Scholar]
- 13.Betts MJ, et al. Systematic identification of phosphorylation-mediated protein interaction switches. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Šoštarić N, et al. Effects of acetylation and phosphorylation on subunit interactions in three large eukaryotic complexes. Mol Cell Proteomics. 2018;17:2387–2401. doi: 10.1074/mcp.RA118.000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres MP, Dewhurst H, Sundararaman N. Proteome-wide structural analysis of PTM hotspots reveals regulatory elements predicted to impact biological function and disease. Mol Cell Proteomics. 2016;15:3513–3528. doi: 10.1074/mcp.M116.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raguz Nakic Z, Seisenbacher G, Posas F, Sauer U. Untargeted metabolomics unravels functionalities of phosphorylation sites in saccharomyces cerevisiae. BMC Syst Biol. 2016;10:104. doi: 10.1186/s12918-016-0350-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 20.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 21.Olsen JV, et al. Global, In Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochoa D, et al. An atlas of human kinase regulation. Mol Syst Biol. 2016;12:888. doi: 10.15252/msb.20167295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaser R, Adusumalli S, Leng SN, Sikic M. Ng PC SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 25.Michels AA, et al. mTORC1 Directly Phosphorylates and Regulates Human MAF1. Mol Cell Biol. 2010;30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal. 2002;38:367–378. [Google Scholar]
- 27.Houssa B, De Widt J, Kranenburg O, Moolenaar WH, Van Blitterswijk WJ. Diacylglycerol kinase θ binds to and is negatively regulated by active RhoA. J Biol Chem. 1999;274:6820–6822. doi: 10.1074/jbc.274.11.6820. [DOI] [PubMed] [Google Scholar]
- 28.Uezu A, et al. Modified SH2 domain to phototrap and identify phosphotyrosine proteins from subcellular sites within cells. Proc Natl Acad Sci U S A. 2012;109:E2929–E2938. doi: 10.1073/pnas.1207358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worby CA, et al. The Fic Domain: Regulation of Cell Signaling by Adenylylation. Mol Cell. 2009;34:93–103. doi: 10.1016/j.molcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.del-Toro N, et al. Capturing variation impact on molecular interactions in the IMEx Consortium mutations data set. Nat Commun. 2019;10 doi: 10.1038/s41467-018-07709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang HI, Ji JH, Jang YJ. Phosphorylation of ran-binding protein-1 by polo-like kinase-1 is required for interaction with ran and early mitotic progression. J Biol Chem. 2011;286:33012–33020. doi: 10.1074/jbc.M111.255620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibano T, Mamada H, Hakuno F, Takahashi SI, Taira M. The inner nuclear membrane protein Nemp1 is a new type of RanGTP-binding protein in eukaryotes. PLoS One. 2015;10:e0127271. doi: 10.1371/journal.pone.0127271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Alonso L, et al. Transcription factor activities enhance markers of drug sensitivity in cancer. Cancer Res. 2018;78:769–780. doi: 10.1158/0008-5472.CAN-17-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by statl and stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 37.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landrum MJ, et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaglin XH, Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends in Genetics. 2009;25:555–566. doi: 10.1016/j.tig.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Westmoreland TJ, et al. Comparative genome-wide screening identifies a conserved doxorubicin repair network that is diploid specific in Saccharomyces cerevisiae. PLoS One. 2009;4:e5830. doi: 10.1371/journal.pone.0005830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becher I, et al. Pervasive Protein Thermal Stability Variation during the Cell Cycle. Cell. 2018;173:1495–1507.e18. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mateus A, et al. Thermal proteome profiling in bacteria: probing protein state in vivo. Mol Syst Biol. 2018;14:e8242. doi: 10.15252/msb.20188242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savitski MM, et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science (80-. ) 2014;346:1255784. doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 44.Tuoc TC, et al. Chromatin Regulation by BAF170 Controls Cerebral Cortical Size and Thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Devlin B, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–246. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staahl BT, Crabtree GR. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Current Opinion in Neurobiology. 2013;23:903–913. doi: 10.1016/j.conb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu JJ, et al. In vivo brain GPCR signaling elucidated by phosphoproteomics. Science (80-. ) 2018;360:eaao4927. doi: 10.1126/science.aao4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sene K, et al. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krahmer N, et al. Organellar Proteomics and Phospho-Proteomics Reveal Subcellular Reorganization in Diet-Induced Hepatic Steatosis. Dev Cell. 2018;47:205–221.e7. doi: 10.1016/j.devcel.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 51.Rogerson DT, et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray VE, Kumar S. Rampant purifying selection conserves positions with posttranslational modifications in human proteins. Mol Biol Evol. 2011;28:1565–1568. doi: 10.1093/molbev/msr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 54.Bateman A, et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jäger S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teo G, et al. SAINTexpress: Improvements and additional features in Significance Analysis of INTeractome software. J Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Türei D, Korcsmáros T, Saez-Rodriguez J. OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nature Methods. 2016;13:966–967. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- 59.Alvarez MJ, et al. Network-based inference of protein activity helps functionalize the genetic landscape of cancer. Nat Genet. 2016 doi: 10.1038/ng.3593. Network-based. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khmelinskii A, Meurer M, Duishoev N, Delhomme N, Knop M. Seamless gene tagging by endonuclease-driven homologous recombination. PLoS One. 2011;6:e23794. doi: 10.1371/journal.pone.0023794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science (80-. ) 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 62.Ran FAFA, et al. XOne-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bibel M, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 64.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108–e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liberzon A, et al. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.