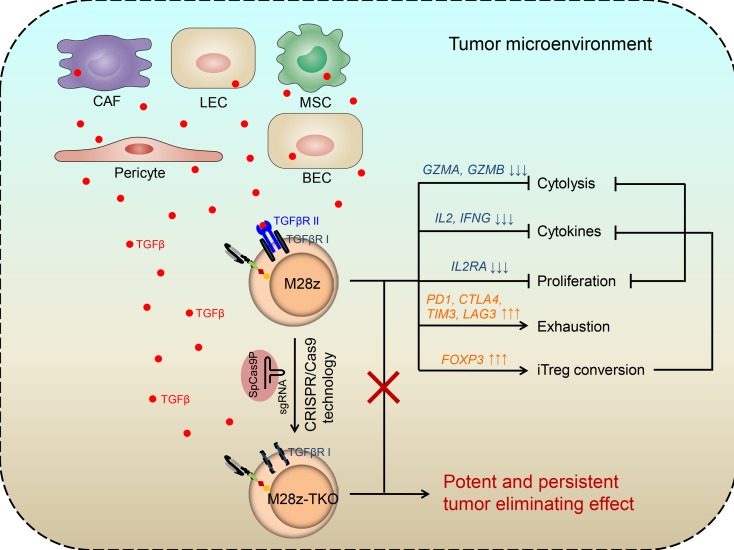
Abstract
In recent years, chimeric antigen receptor–modified T cell (CAR T cell) therapy has proven to be a promising approach against cancer. Nonetheless, this approach still faces multiple challenges in eliminating solid tumors, one of which being the immunosuppressive tumor microenvironment (TME). Here, we demonstrated that knocking out the endogenous TGF-β receptor II (TGFBR2) in CAR T cells with CRISPR/Cas9 technology could reduce the induced Treg conversion and prevent the exhaustion of CAR T ce lls. Meanwhile, TGFBR2-edited CAR T cells had better in vivo tumor elimination efficacy, both in cell line–derived xenograft and patient-derived xenograft solid tumor models, whether administered locally or systemically. In addition, the TGFBR2-edited CAR T cells could eliminate contralaterally reinoculated xenografts in mice effectively, with an increased proportion of memory subsets within circulating CAR T cells of central memory and effector memory subsets. In conclusion, we greatly improved the in vitro and in vivo function of CAR T cells in TGF-β–rich tumor environments by knocking out endogenous TGFBR2 and propose a potentially new method to improve the efficacy of CAR T cell therapy for treating solid tumors.
Keywords: Oncology, Therapeutics
Keywords: Cancer immunotherapy
Inhibition of the immunosuppressive TGFβ signaling by knocking out endogenous TGFBR2 greatly improved the persistence and efficacy of CAR-T cells against solid tumors in mice.
Introduction
In recent years, chimeric antigen receptor–modified T cells (CAR T cells) have emerged as a promising class of cancer therapeutics (1–3). Anti-CD19 CAR T cells have achieved remarkable therapeutic efficacy in treating hematological malignancy, and two products were approved by FDA in 2017 (4, 5). However, despite many attempts, CAR T cells targeting various tumor-associated antigens still fail to yield favorable clinical response in patients with solid tumors in a reproducible manner (2, 6–8). One of the major challenges that CAR T cells face after infiltrating into the solid tumor is the suppressive tumor microenvironment (TME). The solid tumor TME contains numerous cell types in addition to cancer cells as well as extracellular matrix components and inflammatory mediators. In this complex microenvironment, T cells encounter many inhibitory cells and molecules that can impair their survival, activation, proliferation, and effector functions (9, 10).
TGF-β, one of the most important regulators in TME, can be secreted by stromal cells, such as cancer-associated fibroblasts, blood endothelial cells, mesenchymal stem cells, lymphatic epithelial cells, and pericytes (9). There are 3 TGF-β ligands: TGF-β1, TGF-β2, and TGF-β3. TGF-β1 is the most commonly upregulated in tumor cells and the TME (11, 12). The TGF-β ligands signal through type I and type II TGF-β receptors (TGF-βRI and TGF-βRII, respectively). Upon binding to TGF-βRII, TGF-βRI is recruited, phosphorylated, and activated to phosphorylate the downstream mediators, and it then modulates gene transcription (13, 14). TGF-β plays a very important role in tumor initiation and progression. Meanwhile, TGF-β has an adverse effect on antitumor immunity and significantly inhibits host tumor immune surveillance (12, 15). It has been reported that TGF-β markedly suppresses the cytotoxic function of CD8+ T cells through transcriptional downregulation of genes encoding key proteins, such as perforin, granzymes, and cytotoxins (16, 17). TGF-β affects CD4+ T cell differentiation and function by inducing Treg conversion (17, 18). In addition, inhibiting TGF-β signaling with a TGF-β1 antibody (19), TGF-βRI inhibitor (20), or overexpressing dominant-negative TGF-βRII have all been shown to enhance the tumor elimination ability of T cells (21, 22).
In this study, we characterized the effects of TGF-β on CAR T cells and found that CAR T cells could be induced into a Treg-like phenotype that was FOXP3 dependent. In addition, we found that the presence of TGF-β1 accelerated the exhaustion of CAR T cells by upregulating PD1. We completely eliminated the negative effect of TGF-β by knocking out TGFBR2 in CAR T cells using the CRISPR/Cas9 system and observed dramatically improved function both in vitro and in cell line–derived xenograft (CDX) and patient tumor–derived xenograft (PDX) models. Impressively, after PDX tumor was eradicated by TGFBR2-KO CAR T cells, the contralaterally reinoculated patient-derived tumor was again fast cleared, demonstrating the potent and persistent in vivo antitumor efficacy.
Results
TGF-β1 negatively regulates CAR T cell cytotoxic function via TGF-βreceptor. To study CAR T cells targeting solid tumors, we designed antimesothelin CAR. The scFV was derived from an antimesothelin monoclonal antibody P4 (23), and CD28 costimulatory domain and CD3z were used as intercellular signaling components. We generated CAR T cells (M28z) via lentiviral transduction and used the mesothelin+ tumor cell lines CRL5826 and OVCAR-3 as target cells.
We first confirmed the low expression level of endogenous TGF-β1 in CRL5826 cells and generated clonal cell lines expressing TGF-β1 at different levels (CRL5826-TGF-β1-low, -intermediate, -high). Although these cell lines released different amounts of TGF-β1, there was no difference of cell lysis when they were cocultured with M28z CAR T cells (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133977DS1), suggesting that the TGF-β1 released by these cell lines could not inhibit CAR T cell function, likely being the latent form.
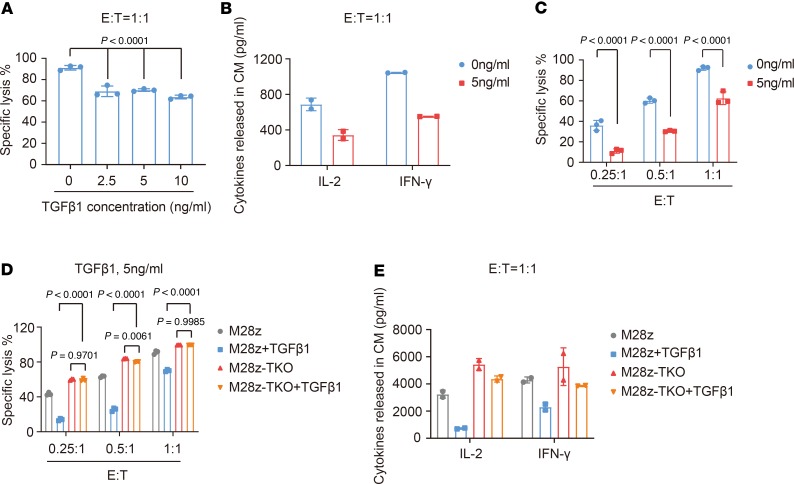
Then, we added different concentrations of human recombinant TGF-β1 to the M28z CAR T and CRL5826 coculture system and observed their effect on the cytotoxic function of CAR T cells. As shown in Figure 1A, the lysis of CRL5826 by M28z CAR T cells at a 1:1 effector-to-target (E/T) ratio was reduced to a similar level when 2.5, 5, or 10 ng/mL TGF-β1 was added. Subsequently, we used 5 ng/mL TGF-β1 in our in vitro experiments. The release of IL-2 and IFN-γ by CAR T cells was also markedly reduced in the presence of TGF-β1 (Figure 1B).
Figure 1. TGF-β1 suppresses cytolysis of CAR T cells and their ability to release cytokines via TGF-β receptor.
(A) Specific lysis of CRL5826 tumor cells after coculture with M28z CAR T cells at a 1:1 effector/target (E/T) ratio, in the presence of 0, 2.5, 5, and 10 ng/mL TGF-β1. (B) IL-2 and IFN-γ secretion after coculture with M28z CAR T cells at a 1:1 E/T ratio in the presence of 5 ng/mL TGF-β1. (C) M28z CAR T cell–mediated tumor lysis in the presence of 5 ng/mL TGF-β1 at 0.25:1, 0.5:1 and 1:1 E/T ratios. (D and E) TGFBR2 KO completely rescues the negative effects of TGF-β1 on CAR T cell-mediated tumor lysis (D) and (E) IL-2 and IFN-γ secretion. M28z-TKO, TGFBR2 KO M28z. Mean ± SD of 3 technical replications per assay. Ordinary 1-way ANOVA and Dunnett’s multiple comparisons test were used in A; 2-way ANOVA and Sidak’s multiple comparisons test were used in C; 2-way ANOVA and Tukey’s multiple comparisons test were used in D. The assays in A, C, and D were repeated more than 3 times and those in B and E were repeated 2 times.
Considering that the number of T cells was much lower than that of the tumor cells upon infiltration into the solid tumor TME, we evaluated the effect of TGF-β1 at a lower E/T ratio. Impressively, as we lowered the E/T ratio, the inhibitory effect became more pronounced. At the E/T ratio of 0.25:1, the CAR T cell–mediated tumor lysis in the presence of TGF-β1 was only about one-quarter of that in the absence of TGF-β1 (Figure 1C). These data indicate that TGF-β1 negatively regulates the cytotoxic function of CAR T cells and that the inhibition level corresponds to the E/T ratio.
To rescue the CAR T cells from this immune suppression effect of TGF-β1, we sought to eliminate TGF-β receptor by knocking out the TGFBR2 gene in CAR T cells. Upon optimization, we achieved KO efficiency of 50%–80% (Supplemental Figure 2). Knocking out TGFBR2 did not affect the proliferation, CAR expression and T cell subtype of M28z CAR T cells (Supplemental Figure 3). Using 3 different E/T ratios, we compared the specific lysis ability of control (M28z) and TGFBR2-KO CAR T (M28z-TKO) cells and found that TGFBR2 KO could completely rescue the negative effect of TGF-β1 on tumor lysis (Figure 1D) and cytokine release (Figure 1E). These results indicate that TGF-β1 inhibits CAR T cell function solely through activating the TGF-β receptor.
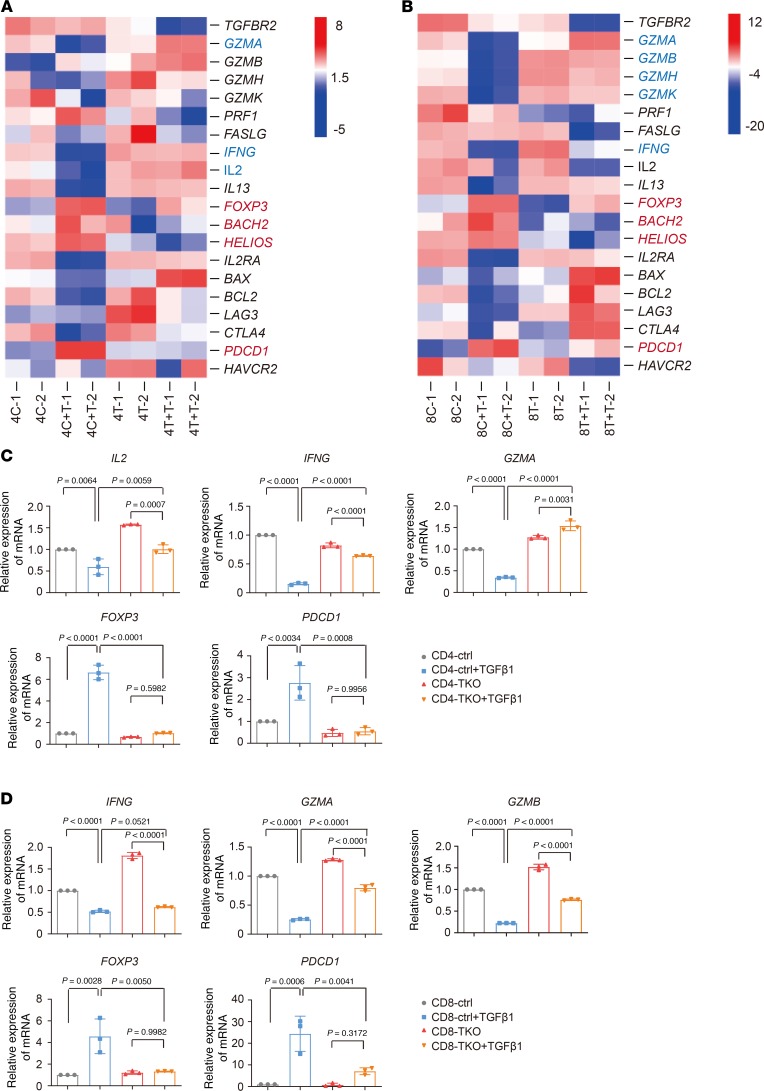
TGF-β1 affects the expression of key genes in CD4 and CD8 CAR T cells. To explore the molecular mechanisms mediating the inhibition of CAR T cell function by TGF-β, after being cocultured with tumor cells in the presence or absence of TGF-β1, M28z and M28z-TKO CAR T cells were sorted into CD4+ and CD8+ populations, and RNA-seq was performed. The expression of cytokine-related genes, such as IFNG and IL2, was markedly downregulated in CD4 cells after addition of TGF-β1 (Figure 2A, 4C + T vs. 4C). In the CD8+ subset, the expression of granzyme-related genes, GZMA, GZMB, GZMH, and GZMK, was decreased significantly after addition of TGF-β1 (Figure 2B, 8C + T vs. 8C). These results were consistent with our observation of the reduction of cytokine release and tumor lysis by CAR T cells in the presence of TGF-β1 (Figure 1, A and B). Furthermore, TGF-β1 induced the upregulation of Treg-related genes, such as FOXP3, BACH2, and HELIOS, both in CD4+ and CD8+ subsets, suggesting a possible conversion of CAR T cells to Treg-like status in the presence of antigen stimulation and TGF-β1. Consistent with a previous study indicating that TGF-β1 induced PD1 expression (24), we observed an evident upregulation of PDCD1 in CAR T cells with TGF-β1 addition. In addition, a good number of exhaustion-related signature genes (25–31) were also upregulated in CAR T cells in the presence of TGF-β1 (Supplemental Figure 4), suggesting that TGF-β1 induces CAR T cell exhaustion. With TGFBR2 removed, the TGF-β1 effects on the expression of above-mentioned genes were largely abolished (Figure 2, A and B, and Supplemental Figure 4; 4T vs. 4T + T and 8T vs. 8T + T). We further confirmed the TGF-β1–induced expression changes of these genes by qPCR (Figure 2, C and D). Therefore, we focused on the TGF-β1–induced Treg (iTreg) phenotype and CAR T cell exhaustion in the following experiments.
Figure 2. TGF-β1 affects the expression of key genes in CD4 and CD8 CAR T cells.
(A and B) Expression changes of key functional genes in CD4 (A) and CD8 (B) cells. (C and D) Expression changes of target genes in CD4 (C) and CD8 (D) cells confirmed by qPCR. 4C, CD4 ctrl; 4C + T, CD4 ctrl + TGF-β1; 8C, CD8 ctrl; 8C + T, CD8 ctrl + TGF-β1; 4T, CD4-TKO; 4T + T, CD4-TKO + TGF-β1; 8T, CD8-TKO; 8T + T, CD8-TKO + TGF-β1. Mean ± SD of 3 technical repetitions in 1 assay. Ordinary 1-way ANOVA and Tukey’s multiple comparisons test were used. The assays in C and D were repeated 2 times.
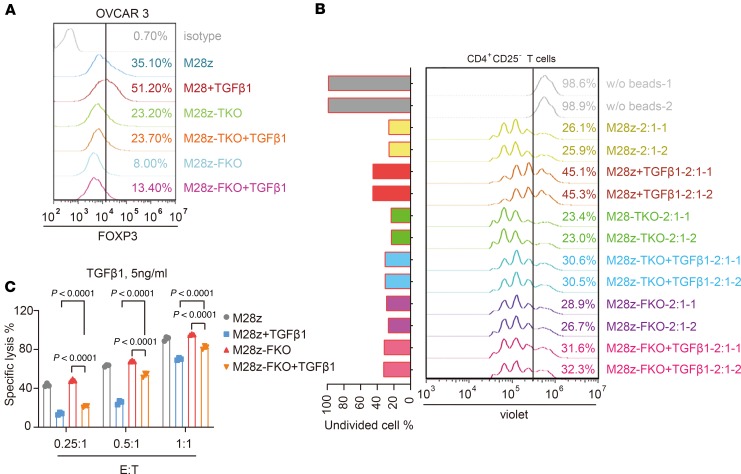
TGF-β1induces a Treg-like phenotype of CAR T cells. Consistent with the upregulation of FOXP3 in RNA-seq and qPCR data (Figure 2), FACS analysis also showed elevated FOXP3 protein levels after CAR T cells were cocultured with either CRL5826 or OVCAR-3 cells in the presence of TGF-β1 (Supplemental Figure 5, B and D). To evaluate whether these FOXP3+ cells can inhibit T cell proliferation, we performed a proliferation suppression assay (Supplemental Figure 5A). The effector cells (CAR T cells) were first cocultured with tumor cells in the presence or absence of TGF-β1 and then were harvested and incubated with responder T cells in normal T cells culture media. Effector cells, after being cocultured with tumor cells in the presence of TGF-β1, suppressed the proliferation of responder cells to a greater extent compared with the TGF-β1 minus control samples (M28z-1:1 and M28z-2:1), while this effect was more prominent when using OVCAR-3 cells (Supplemental Figure 5, C and E). This suggests that a portion of CAR T cells were converted into functional iTreg-like cells under the condition of antigen simulation and TGF-β1 presence.
To confirm that this effect was due to FOXP3 upregulation, we performed a proliferation suppression assay using TGFBR2- and FOXP3-KO (Supplemental Figure 6) CAR T cells. Compared with that in WT cells, expression of FOXP3 was maintained at a lower level in M28z-TKO and FOXP3-KO M28z (M28z-FKO) cells cocultured with tumor cells in the presence of TGF-β1 (Figure 3A). Accordingly, the proliferation suppression abilities of effector cells from the M28z-TKO + TGF-β1 and M28z-FKO + TGF-β1 groups were much lower than that in M28z + TGF-β1 group (Figure 3B). This indicates that FOXP3 played an essential role in the iTreg-like phenotype induced by TGF-β1. In addition, we observed an improvement in the tumor lysis capability of M28z-FKO cells in the presence of TGF-β1; however, FOXP3 editing could only partially rescue the cytotoxic function reduction induced by TGF-β1 (Figure 3C).
Figure 3. TGF-β1 induced FOXP3-dependent iTreg-like cell conversion of CAR T cells.
(A) FOXP3 expression in control and M28z-TKO CAR T cells after incubation with OVCAR3 cells in the presence or absence of 5 ng/mL TGF-β1. (B) Proliferation suppression ability of iTreg-like cells converted from control, M28z-TKO, and M28z-FKO CAR T cells after incubation with OVCAR3 cells in the presence of 5 ng/mL TGF-β1. (C) FOXP3 KO partially rescues the negative effects of TGF-β1 on CAR T cell–mediated tumor lysis. M28z-TKO, TGFBR2-KO M28z; M28z-FKO, FOXP-KO M28z. Mean ± SD of 3 technical repetitions in 1 assay. Two-way ANOVA and Tukey’s multiple comparisons test were used. The assays in A and C were repeated more than 3 times and those in B were repeated 2 times.
These results suggest that the inhibition of TGF-β1–FOXP3 signal improves CAR T cell function by reducing iTreg-like cell conversion. However, this pathway is only partially responsible for the negative effects of TGF-β1 on CAR T cell function.
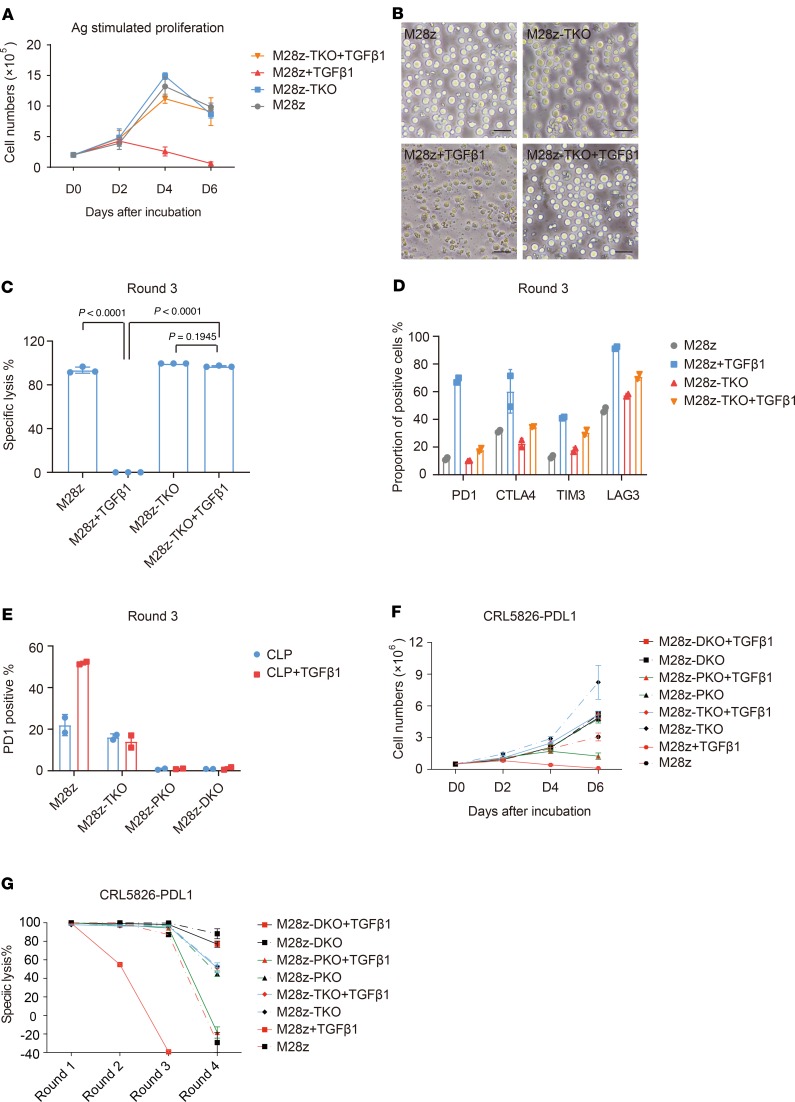
TGF-β1 accelerated CAR T cell exhaustion. Persistent activation of T cell signaling leads to exhaustion, and exhausted T cells have decreased proliferative capacity and effector function as well as overexpression of immune checkpoint genes, such as PDCD1 (32). After observing the significant increase of PDCD1 expression in M28z cells after addition of TGF-β1 (Figure 2), we further explored whether TGF-β1 would accelerate CAR T cell exhaustion using a multiround antigen stimulation assay. We found that TGF-β1 significantly affected the proliferation of CAR T cells upon multiple challenges of tumor cells. At the fourth challenge, almost all CAR T cells died in the presence of TGF-β1, while the control cells survived well. Knocking out TGFBR2 rendered CAR T cells unresponsive to the TGF-β1 effect, leading to survival similar to that of the control group (Figure 4, A and B). In the presence of TGF-β1, the tumor lysis activity of CAR T cells was reduced to nothing after 2 rounds of challenges, while M28z-TKO cells remained active (Figure 4C). Accordingly, addition of TGF-β1 upregulated the expression of major exhaustion markers, such as PD1, TIM3, LAG3, and CTLA4, in CAR T cells after 3 rounds of tumor challenges, and knocking out TGFBR2 reduced this effect (Figure 4D). Using qPCR analysis, we also observed the general upregulation of well-established exhaustion-related genes on CAR T cells after antigen rechallenges in the presence of TGF-β1 (Supplemental Figure 7). It is worth noting that among these markers, PD1 was one of the most upregulated and there were still a considerable number of cells expressing PD1 in M28z-TKO cells, indicating that although they were unresponsive to TGF-β1 signaling, they were still prone to PD1 suppression, as engaged by corresponding ligands in the TME.
Figure 4. TGFBR2 KO rescues CAR T cell exhaustion induced by TGF-β1.
(A) Growth curve and (B) representative images of M28z and M28z-TKO CAR T cells after multiple rounds of CRL5826 stimulation in the presence of 5 ng/mL TGF-β1. Scale bar: 40 μm. (C) Specific tumor lysis by M28z and M28z-TKO cells after 3 rounds of CRL5826 stimulation in the presence of 5 ng/mL TGF-β1. (D) Immune checkpoint gene expression of M28z and M28z-TKO cells after 3 rounds CRL5826 stimulation in the presence of 5 ng/mL TGF-β1. (E) Proportion of PD1+ cells in M28z, M28z-TKO, M28z-PKO, and M28z-DKO cells after 3 rounds of incubation with CRL5826-PDL1 in the presence of 5 ng/mL TGF-β1. (F) Proliferation and (G) tumor lysis capability of M28z, M28z-TKO, M28z-PKO, and M28z-DKO cells during 3 rounds of incubation with CRL5826-PDL1 cells in the presence of TGF-β1. M28z-TKO, TGFBR2-KO M28z; M28z-PKO, PD1-KO M28z; M28z-DKO, TGFBR2 and PD1 double-KO M28z. Mean ± SD of 3 technical repetitions in 1 assay. Ordinary 1-way ANOVA and Tukey’s multiple comparisons test were used in C. The assays in A–C were repeated more than 3 times and those in D–G were repeated 2 times.
To evaluate how PD1 upregulation contributes to the inhibitory effect of TGF-β1, we generated PDCD1-KO (M28z-PKO) and PDCD1/TGFBR2 double-KO (M28z-DKO) CAR T cells, and we overexpressed PDL1 to model a more suppressive TME. Compared with the upregulation of PD1 in M28z cells induced by TGF-β1, the expression of PD1 was reduced to basal levels in M28z-PKO and -DKO cells, indicating efficient gene editing (Figure 4E). Knocking out PDCD1 indeed improved CAR T cell proliferation upon multiple tumor challenges, while it was still worse than TKO and DKO (Figure 4F). In the presence of TGF-β1 and PDL1 overexpression, the tumor lysis capability of M28z cells was reduced at round 2 and completely lost at round 3. M28z-PKO cells were able to achieve over 90% tumor lysis at round 3 and lost efficacy at round 4. In contrast, M28z-TKO cells were able to maintain about 60% tumor lysis capability at round 4 (Figure 4G). All of these data showed that PD1 upregulation partially contributed to the negative regulation effect of TGF-β. On the other hand, TGF-β signaling was only partially responsible for PD1 expression, as a portion of TKO cells could still express PD1 and were therefore subject to PDL1 suppression. The DKO CAR T cells had the best performance, eliminating about 90% of tumor at round 4 (Figure 4G), suggesting that simultaneous blocking both TGF-β and PD1 signaling can further improve the CAR T cell’s resistance to suppressive TME.
TGFBR2 editing enhanced tumor elimination efficacy of CAR T cells in CDX models.
After observing the superior in vitro function of M28z-TKO CAR T cells, we evaluated in vivo efficacy using a CDX model established by grafting CRL5826 cells subcutaneously in NPG mice (VitalStar). The expression of mesothelin of this tumor model was confirmed by IHC (Figure 5A). Despite the low level TGF-β1 secretion of in vitro cultured CRL5826 cells (Supplemental Figure 1), the CDX was rich in TGF-β1 (Figure 5A). We suspected that this TGF-β1 in TME was partially released by the mouse stromal cells. To confirm the effect of mouse TGF-β1, we added it to human CAR T cells and observed an inhibitory effect similar to that of human TGF-β1 (Supplemental Figure 8).
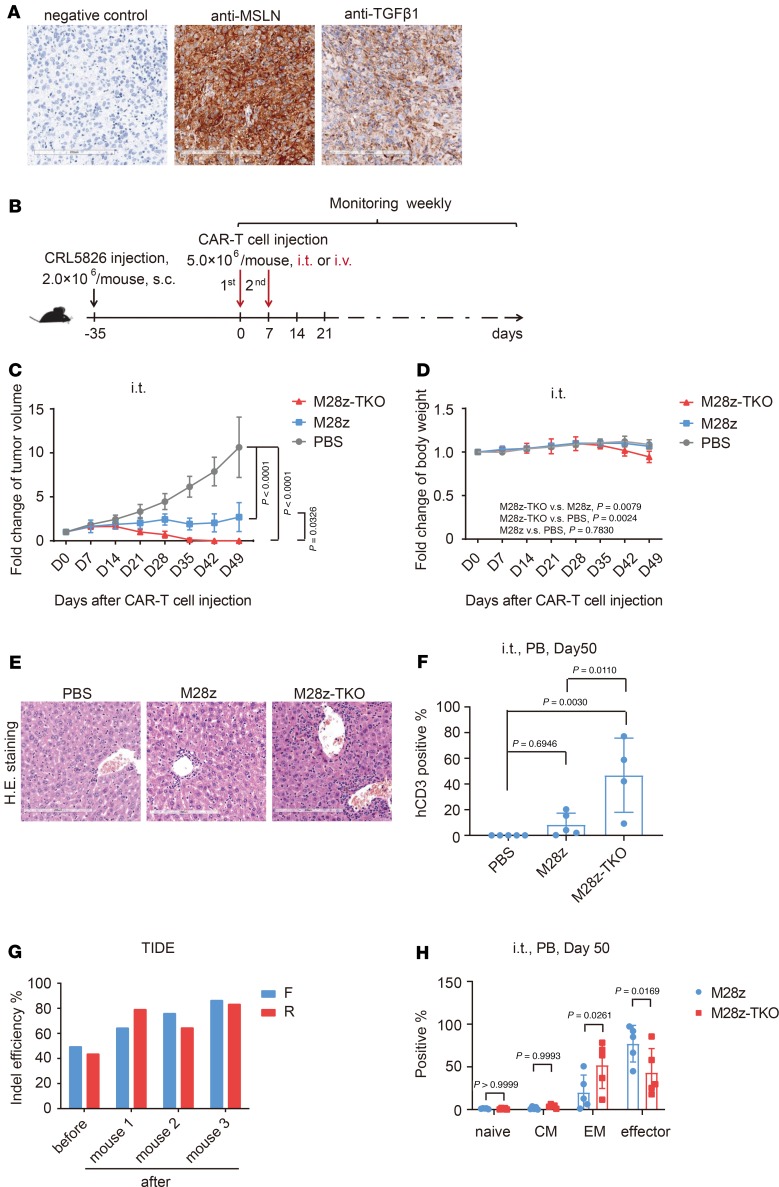
Figure 5. TGFBR2 KO improved in vivo tumor elimination efficacy of CAR T cells in CDX models after local administration.
(A) Mesothelin and TGF-β1 expression in the CDX model. Scale bar: 200 μm. (B) Schematic of the in vivo experimental design using CDX models. (C) Fold changes of tumor volume and (D) mouse body weight after intratumor (i.t.) administration of CAR T cells. (E) Inflammatory cell infiltration of CAR T cells into liver detected by H&E staining in the CDX model. Scale bar: 200 μm. (F) Peripheral blood analysis of the proportion of hCD3+ cells after CAR T cell i.t. administration. (G) Indel frequencies of M28z-TKO cells, before and after administration, determined by TIDE analysis. (H) Analysis of T cell subsets in peripheral blood after CAR T cell i.t. administration. M28z-TKO, TGFBR2-KO M28z; PB, peripheral blood; CM, central memory; EM, effector memory. Mean ± SD, n = 5. Two-way ANOVA and Tukey’s multiple comparisons test was used in C and D. Ordinary 1-way ANOVA and Tukey’s multiple comparisons test was used in F. Two-way ANOVA and Sidak’s multiple comparisons test was used in H.
After the tumor reached a defined size (200–300 mm3), we performed intratumor (i.t.) injections of M28z and M28z-TKO CAR T cells (Figure 5B). Compared with the continuous increase of tumor volume in the PBS group, M28z CAR T cells slowed down tumor growth significantly. However, they failed to clear the tumor up to 49 days after injection. In comparison, the tumors in all 5 mice were completely eliminated 35 days after M28z-TKO injection (Figure 5C and Supplemental Figure 9A). The body weight of M28z-TKO–treated mice decreased gradually (Figure 5D). Meanwhile, severe fur loss and swollen feet were observed in 5 of 5 M28z-TKO–treated mice, and 1 died 49 days after injection. Body dissection showed that the number of inflammatory cells infiltrated into mice liver was increased in M28z-TKO–treated group (Figure 5E). These phenomena indicated that graft-versus-host disease (GvHD) occurred in M28z-TKO–treated mice. Consistent with observed GvHD, a high proportion of human CD3+ (hCD3+) cells was detected in mice peripheral blood. As shown in Figure 5F and Supplemental Figure 9B, nearly 60% of cells were hCD3+ in the blood of M28z-TKO–treated mice, while less than 10% blood cells of M28z-treated mice were hCD3+, showing that CAR T cells with TGF-βR2 editing had a much better proliferation in vivo. Meanwhile, the gene editing efficiency increased substantially in M28z-TKO cells 50 days after injection (Figure 5G), which further confirmed the TGF-βR2–edited cells had a proliferation advantage. In addition, the proportion of effector memory cells (CCR7–CD45RO+) and central memory cells (CCR7+CD45RO+) was notably increased in the peripheral blood of M28z-TKO–treated mice (Figure 5H).
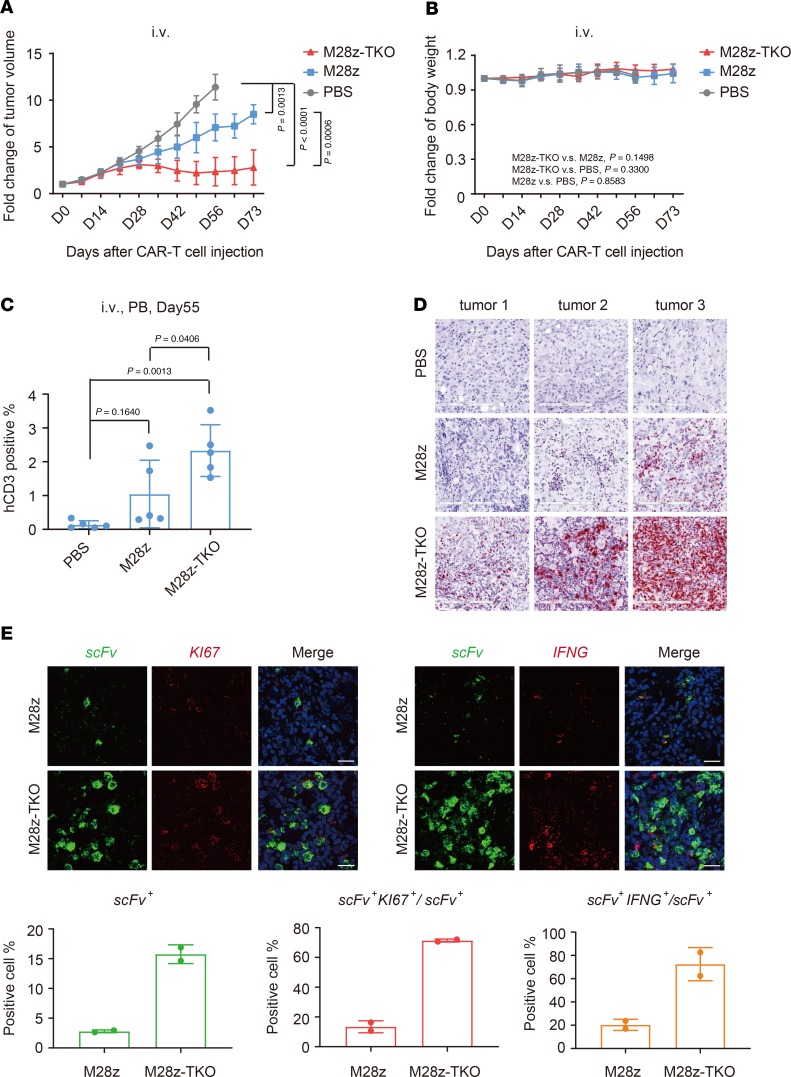
After confirming the robust efficacy of M28z-TKO cells via i.t. injection, we administered CAR T cells to the same CDX model via i.v. route. Compared with i.t. injection, i.v. injection of M28z cells showed less robust function in controlling tumor growth, as observed in previous studies (33). M28z-TKO cells delivered through i.v. injection performed significantly better than M28z cells, controlling tumor growth efficiently while not completely eradicating the tumor (Figure 6A and Supplemental Figure 9C). Symptoms of GvHD were not observed in any of these i.v. injected mice (Figure 6B), and the peripheral blood samples were analyzed 55 days after CAR T injection. The average percentage of hCD3+ cells in the M28z-TKO–treated group was higher than that in the M28z-treated group; however, the percentage was relatively low compared with that of i.t. injected mice (Figure 6C). At day 75 after treatment, we terminated the experiments and harvested the tumors for analysis. Using RNAscope technology, we found that the number of tumor-infiltrating M28z-TKO cells was much higher than M28z cells (Figure 6D) and a marked portion of these M28z-TKO CAR T cells were functional and proliferating, as they were IFNG+ or KI67+ (Figure 6E). These data indicate that blocking the TGF-β pathway in CAR T cells by gene editing can significantly improve their survival and proliferation within the TME.
Figure 6. TGFBR2 KO improved in vivo tumor elimination efficacy of CAR T cells in CDX models after systematic administration.
(A) Fold change of tumor volume and (B) mouse body weight after CAR T cell i.v. administration. (C) Peripheral blood analysis of the proportion of hCD3+ cells after CAR T cell i.v. administration. (D) Detection of tumor-infiltrating CAR T cells by RNAscope. Scale bar: 200 μm. (E) Characterization of the expression of KI67 and IFNG in tumor-infiltrating CAR T cells by RNAscope. Scale bar: 20 μm. M28z-TKO, TGFBR2-KO M28z; PB, peripheral blood. Mean ± SD, n = 5 in A–C, n = 2; 10 pictures were taken randomly per tumor section in E. Two-way ANOVA and Tukey’s multiple comparisons test were used in A and B. Ordinary 1-way ANOVA and Tukey’s multiple comparisons test were used in C.
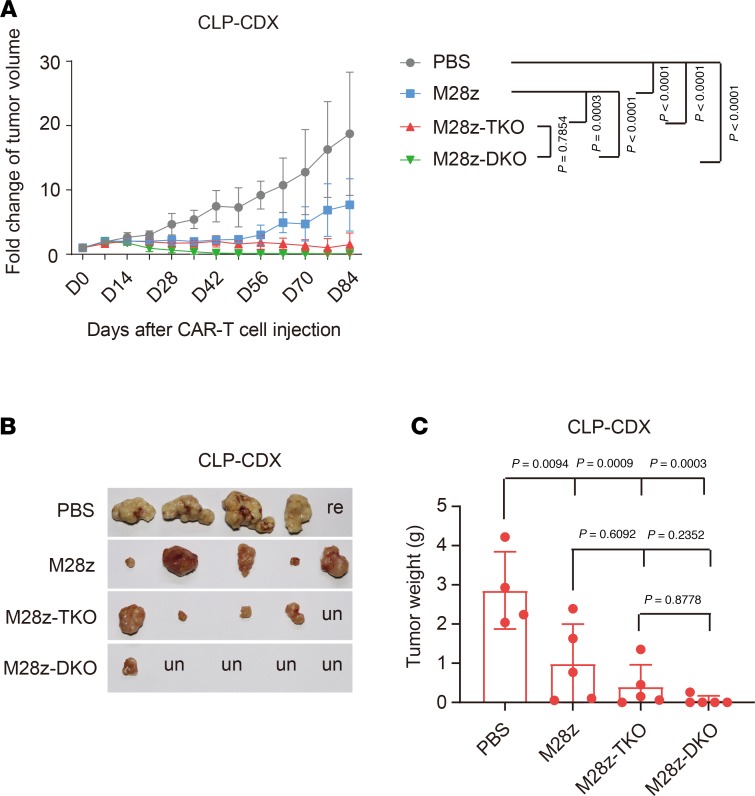
As the TME usually contains multiple inhibitory signals, we further challenged M28z-TKO CAR T cells with a more suppressive TME. We overexpressed PDL1 in CRL5826 cells, generated a CRL5826-PDL1 CDX model, and performed i.t. injections of CAR T cells (Supplemental Figure 10). Compared with the M28z-treated group, M28z-TKO CAR T cells showed better efficacy by controlling the tumor volume at a lower level, indicating that TGF-β signaling blockade could improve CAR T cell function in a TME with high concentrations of TGF-β and PDL1. However, in contrast to the fast eradication of the CRL5826 tumor in all 5 mice with i.t. injection of M28z-TKO cells, the CRL5826-PDL1 tumor was cleared in only 1 of 5 mice by day 84. Next, we administered M28z-DKO cells, in which both TGF-β and PD1 signaling were abolished by gene editing, and observed the elimination of CRL5826-PDL1 tumor in 4 of 5 M28z-DKO–treated mice (Figure 7). These results suggest that, for treating solid tumors, blocking multiple inhibitory pathways in a TME-specific manner might be necessary for CAR T cells to function optimally.
Figure 7. TGFBR2 and PD1 double-KO improved in vivo tumor elimination efficacy of CAR T cells in PDL1 overexpression CDX models.
(A) Fold change of tumor volume after intratumor administration of CAR T cells in a CRL5826-PDL1 CDX model. (B) Tumor sizes and (C) weights at the end of this experiment. M28z-TKO, TGFBR2-KO M28z; M28z-DKO, TGFBR2 and PD1 double-KO M28z; CLP-CDX, CRL5826-PDL1 CDX model; re, reinoculation; un, undetectable. Mean ± SD, n = 5, except n = 4 in PBS group in C. Two-way ANOVA and Tukey’s multiple comparisons test were used in A. Ordinary 1-way ANOVA and Tukey’s multiple comparisons test were used in C.
TGFBR2 editing enhanced tumor elimination efficacy of CAR T cells in pancreatic carcinoma PDX models.
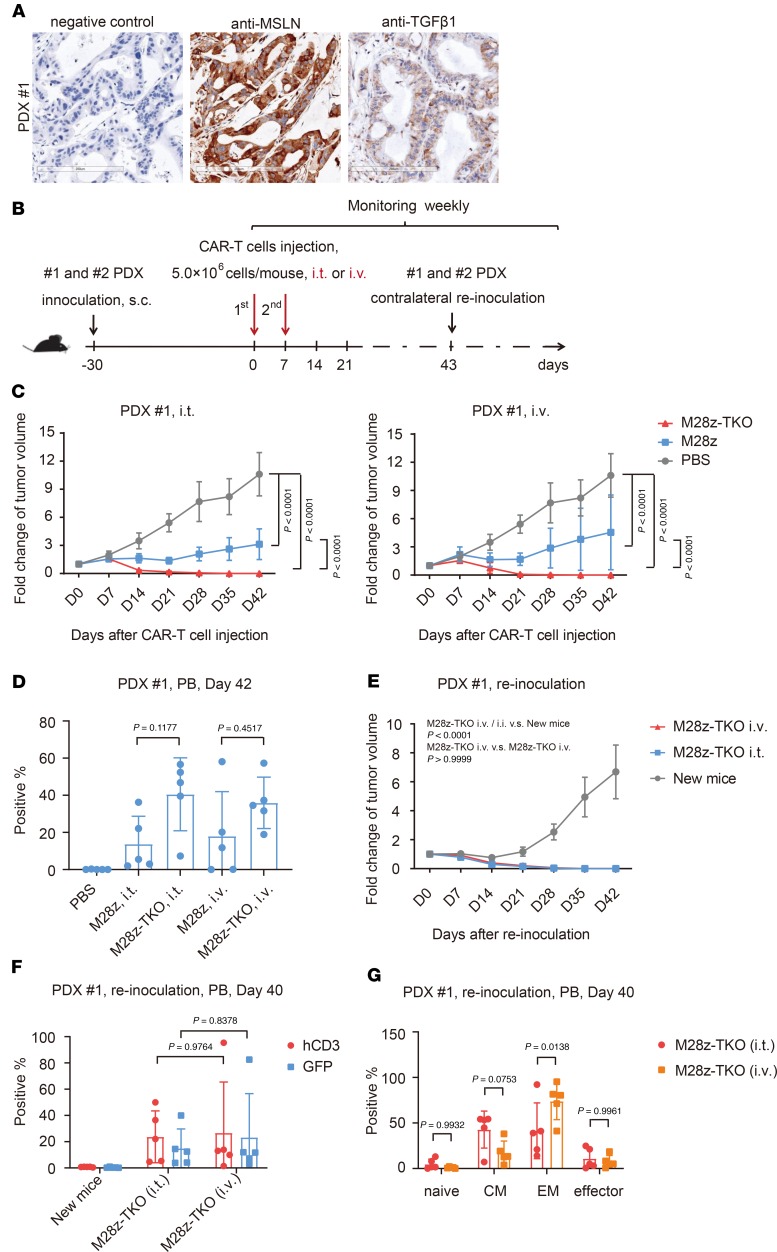
To evaluate CAR T cell efficacy in models more relevant to primary tumor, we established 1 pancreatic carcinoma patient-derived xenograft (PDX) model that expressed mesothelin and TGF-β1 in a relatively uniform manner (Figure 8A). M28z and M28z-TKO CAR T cells were delivered via either i.v. or i.t. injection (Figure 8B). Compared with the fast increase of tumor size in the PBS group, both i.v. and i.t. injection of M28z cells slowed down the tumor growth, although the tumor still grew to about 3 times the initial volume at the time of first injection. In contrast, only 21 days after the first injection, M28z-TKO i.t. and i.v. injection reduced the tumor to less than 10% of initial volume, and the tumors were completely eradicated at day 28 (Figure 8C). The average proportions of hCD3+ cells were about 40% of the periphery blood of M28z-TKO–treated mice, either via i.t. or i.v. injection (Figure 8D). These results were consistent with data from the CDX model, further confirming the superior in vivo proliferation and function of M28z-TKO cells.
Figure 8. TGFBR2-KO CAR T cells have potent and persistent tumor elimination efficacy in PDX tumor 1 model.
(A) Expression of mesothelin and TGF-β1 on PDX tumor 1. Scale bar: 200 μm. (B) Schematic of the in vivo experimental design using PDX models. (C) Fold change of tumor volume after i.t. or i.v. CAR T cell administration in PDX model 1. (D) The proportion of hCD3+ cells in the peripheral blood of PDX tumor 1 model on day 42 after i.t. or i.v. administration of CAR T cells. (E) Fold change of tumor volume after PDX tumor 1 reinoculation. (F) The proportion of hCD3+ and GFP+ cells and (G) T cell subsets in the peripheral blood of PDX model 1 on day 40 after tumor reinoculation. M28z-TKO, TGFBR2-KO M28z; i.t., intratumor; PB, peripheral blood. Mean ± SD, n = 5. Two-way ANOVA and Tukey’s multiple comparisons test were used in C, E, and F. Ordinary 1-way ANOVA and Tukey’s multiple comparisons test were used in D. Two-way ANOVA and Sidak’s multiple comparisons test were used in G.
Since the majority of these hCD3+ cells were effector memory and central memory subsets (Supplemental Figure 11A), we wondered whether M28z-TKO cells have long-lasting antitumor efficacy. At day 43 after treatment, 15 days after the eradication of tumor, we removed the tumor from PBS control mice and reinoculated these tumor-free TKO-treated mice contralaterally. Except for 1 mouse from the M28z-TKO i.v.-treated group, which died from GvHD at the end of this experiment, the reinoculated xenograft tumors were eliminated completely in all mice by day 35 (Figure 8E and Supplemental Figure 11B). The proportion of hCD3+ and GFP+ cells remained at around 20%–40% in the peripheral circulation of these mice (Figure 8F). T cell subset analysis showed that effector memory and central memory cells still accounted for the majority of hCD3+ cells in M28z-TKO group (Figure 8G). The body weights of mice in the M28z-TKO–treated group were lower than those in the M28z-treated group (Supplemental Figure 11, C and D), suggesting that GvHD caused by enhanced T cell proliferation during first and second xenograft challenges.
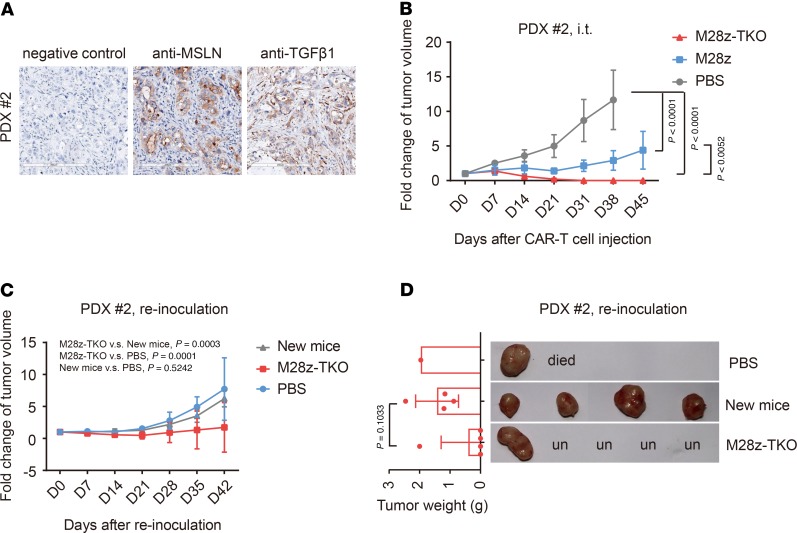
Considering that the antigens recognized by CAR T cells are usually only expressed in partial primary tumor cells, we established another PDX model of pancreatic carcinoma, which has a weaker and more heterogenous expression of mesothelin and a more compact structure (Figure 9A). With only some of the tumor cells expressing the antigen recognized by CAR, we evaluated whether CAR T cells could still completely eliminate the tumor. Impressively, while tumors in the mice injected with M28z cells continued to grow, tumors were completely eradicated from all mice treated with M28z-TKO by day 31 (Figure 9B). At day 40, we reinoculated the same primary tumor sample to 5 M28z-TKO–treated tumor-free mice and observed complete elimination of tumor in 4 of 5 mice by day 35 after tumor reinoculation (Figure 9, C and D). In contrast to the first PDX model, in this experiment, no mice showed severe weight lost or any GvHD phenotype during the initial treatment or the reinoculation of tumor (Supplemental Figure 12A). We examined the peripheral blood of mice at day 34 after initial injection and day 22 after reinoculation of tumor, the percentage of hCD3+ cells was around 5% (Supplemental Figure 12, B and C). These results suggest that PDX tumor 2 does not induce CAR T cell proliferation as efficiently as PDX tumor 1, likely due to its lower and heterogenous mesothelin expression and more concrete structure. However, even with fewer circulating cells, M28z-TKO cells can still efficiently eradicate the tumor, even after a second inoculation, indicating a potent and lasting antitumor effect.
Figure 9. TGFBR2-KO CAR T cells have potent and persistent tumor elimination efficacy in PDX tumor 2 model.
(A) Expression of mesothelin and TGF-β1 on PDX tumor 2. Scale bar: 200 μm. (B) Fold change of tumor volume after i.t. CAR T cell administration in PDX model 2. (C) Fold change of tumor volume after PDX tumor 2 reinoculation. (D) Tumor sizes and weights were detected at the end of the PDX tumor 2 reinoculation experiment. M28z-TKO, TGFBR2-KO M28z; i.t., intratumor; PB, peripheral blood; un, undetectable. Mean ± SD, n = 5, except in D. Two-way ANOVA and Tukey’s multiple comparisons test were used in B and C. Unpaired t test was used in D.
Discussion
TGF-β, one of the most important regulators in the TME, greatly affects immune cell function and tumor progression (9). In this study, we explored the mechanisms of TGF-β effects on CAR T cells and found that TGF-β1 induced FOXP3-dependent iTreg-like cell conversion and PD1-dependent CAR T cell exhaustion in vitro. By knocking out endogenous TGFBR2, we completely rescued the negative effects of TGF-β1. M28z-TKO cells showed superior proliferation and tumor lysis capability after multiple rounds of tumor challenges in vitro. Compared with the strategy of expressing dominant-negative TGF-βRII to compete with the endogenous receptor (34), gene editing blocks TGF-β signaling completely and improves CAR T cell function, despite the increased concentration of TGF-β1 (Supplemental Figure 13).
One of the main reasons for the poor clinical efficacy of CAR T cells in treating solid tumor is the lack of robust in vivo proliferation (34–37). In this study, we demonstrated that knocking out TGFBR2 enables CAR T cells to survive and proliferate better in tumor xenograft mouse models, leading to significantly improved antitumor efficacy. Although different administrative routes and tumor models induced different levels of CAR T proliferation, cells with TGFBR2 KO consistently showed superior proliferation. In some models, the M28z-TKO cells accounted for about 60% of peripheral blood cells 50 days after administration, indicating highly robust in vivo proliferation capability. Within tumor, M28z-TKO cells also existed at higher numbers, expressing both proliferation and functional markers (Figure 6, D and E). More importantly, not only the total number of CAR T cells, but also the proportions of central and effector memory T cells, were increased, leading to sustained antitumor efficacy upon repetitive tumor challenges (Figures 8 and 9). Our results suggest that completely blocking TGF-β signaling by gene editing is a promising strategy to improve the proliferation of CAR T cells when treating solid tumor patients.
Considering that the clinical therapeutic effect of CAR T cells is likely affected by the histopathologic type, and the heterogeneity of tumor-associated antigen expression in solid tumor (37, 38), we established 2 PDX models using different pancreatic carcinoma samples to evaluate the function of TGFBR2-KO CAR T cells. We observed that these 2 xenografts, regardless of loose or compact structure and complete or partial expression of target antigen, were both eradicated efficiently by TGFBR2-KO CAR T cells, and the residual edited CAR T cells could eliminate the reinoculated xenografts effectively. These results suggest that TGFBR2-edited CAR T cells function effectively in different primary tumor environments. However, we also want to point out that TGF-β signals through many cell types at TME to induce T cell suppression, and our CDX and PDX models were only recapitulating some aspects of the TME. It is possible that the lack of TGF-βRII on T cells will allow for greater signaling through other cell subsets, which is an important issue to be addressed using more sophisticated tumor models.
In the complex suppressive TME, different inhibitory cells and molecules inhibit T cell function via multiple pathways (9). In the CRL5826 CDX model, which weakly expressed PDL1 and was enriched for TGF-β1, CAR T cells that had TGF-β signaling inhibited via gene editing eradicated the tumors much more efficiently. In comparison, in a TME highly expressing PDL1 (CRL5826-PDL1 CDX model), although TGFBR2-edited CAR T cells had the tumor-eradicating advantage compared with the unedited cells, they were not able to eradicate tumor in most mice, indicating that blocking only 1 inhibitory pathway was not enough to render the CAR T cells resistant to complex TME. When both PD1 and TGF-β pathways were blocked by PDCD1-TGFBR2 double KO, CAR T cells were able to clear the tumor from 4 of 5 mice. These observations highlight the fact that inhibiting pathways specific to a particular TME could improve CAR T cell function in treating solid tumors and suggest a TME-specific strategy of editing CAR T cell genome for its usage in clinic.
In conclusion, we improved the function of CAR T cells in a TGF-β–rich environment by editing endogenous TGFBR2, proposing a general strategy to improve the efficacy of CAR T cell therapy treating solid tumor. Gene editing of multiple genes in CAR T cells to avoid the combined inhibitory effects of multiple immune factors might provide a more targeted and effective therapy for different cancer patients.
Methods
Cell lines.
Human 293T and HepG2 cells were maintained in DMEM (GIBCO) supplemented with 10% (v/v) FBS (GIBCO), 100 U/mL penicillin (GIBCO), and streptomycin (GIBCO). CRL5826-luciferase cells and CRL5826-luci-PDL1 cells were maintained in RPMI1640 medium (GIBCO) supplemented with 10% (v/v) FBS, 100 U/mL penicillin and streptomycin. OVCAR3 cells were maintained in RPMI1640 medium supplemented with 20% (v/v) FBS, 100 U/mL penicillin, and streptomycin. All cell lines were purchased from ATCC and cultured at 37°C in a 5% CO2 atmosphere.
CD3+ T cell isolation, activation, and expansion.
Fresh umbilical cord blood units were obtained from healthy volunteer donors with informed consent from the Beijing Cord Blood Bank (Beijing, China), and mononuclear cells were separated with human mononuclear cell separation medium 1.007 (Beijing Dongfang Huahui Biomedical Technology Co. Ltd.). T cells were isolated using the EasySep human T cell enrichment kit (Stemcell Technologies) and activated and expanded with anti-CD3/anti-CD28 Dynabeads (Thermo Fisher Scientific) at the ratio of 1:1 according to the manufacturer’s instruction. T cells were cultured in X-VIVO15 medium (Lonza) supplemented with 5% (v/v) heat-inactivated FBS (GIBCO) and 300 IU/mL recombinant human IL-2 (Sino Biological Inc.).
Generation of antimesothelin CAR T cells.
Antimesothelin CAR was constructed, and antimesothelin CAR T cells (M28z) were generated and expanded as previously described (23, 39). Briefly, freshly purified primary CD3+ T cells were activated for 24 hours and then infected with lentivirus harboring antimesothelin CAR. Lentivirus was produced by cotransfecting with packaging plasmids pMD2.G, psPAX2, and M28z-GFP into 293T cells. The virus supernatants were harvested 48 and 72 hours after transfection, and the virus was prepared after concentration (MilliporeSigma). Dominant-negative TGF-β receptor II–overexpressed (DN-overexpressed) M28z CAR T cells were prepared after constructing EF1α-M28z-P2A-DN and EF1α-DN-P2A-M28z plasmids as previously described (40).
Generation of a TGF-β1–overexpressing CRL5826 cell line. EF1-TGF-β1-P2A-tomato plasmid was constructed. Lentivirus was produced by cotransfecting with packaging plasmids pMD2.G, psPAX2, and the expression plasmid into 293T cells. The virus supernatants were harvested 48 and 72 hours after transfection, and the virus was prepared after concentration (MilliporeSigma). Then, tomato+ CRL5826 cells were sorted and cultured after infection with the lentivirus.
In vitro transcription.
In vitro transcription (IVT) single guide RNAs (sgRNAs) were prepared as previously described (39, 41). Briefly, plasmid PX330 (42230) containing sgRNA backbone was used as PCR template. The acquired PCR amplicon containing T7 promoter, 20-bp target sequence, and sgRNA backbone was used as IVT template. The IVT was performed using the MEGAshortscript T7 kit (Thermo Fisher Scientific). For the calf intestine phosphatase (NEB) treatment, 2 U enzyme was added to each μg of in vitro transcribed sgRNA and further incubated at 37°C for 1 hour. sgRNA was then purified with MEGAclear columns (Thermo Fisher Scientific) and eluted with elution buffer.
Electroporation of HepG2 cell lines and CAR T cells.
Cas9 and sgRNA ribonucleoprotein were prepared immediately before electroporation. 6 μg Cas9 protein (provided by Shenzhen Fapon Biological Therapy Co. Ltd.) with 6 μg sgRNA was incubated at room temperature for 20 minutes. 5 × 105 HepG2 or CAR T cells were centrifuged at 200 g for 5 minutes, resuspended in 20 μl transfection buffer containing indicated ribonucleoprotein, and then transferred into the electroporation cuvette. 4D-Nucleofector System N (Lonza), SE cell line 4D-nucleofector kit (V4XC-1032, Lonza), and program EH-100 were used for HepG2 cell line electroporation. 4D-Nucleofector System N (Lonza), P3 Primary Cell 4D-Nucleofector X Kit (V4XP-3024, Lonza), and program EO-115 were used for CAR T cell electroporation. After electroporation, cells were resuspended in prewarmed medium, transferred into culture plate, and incubated at 37°C in an atmosphere of 5% CO2 respectively.
Analysis of gene editing efficiency.
The genomic disruption level of TGFBR2 in HepG2 cell line, TGFBR2 and FOXP3 in CAR T cells was determined by surveyor nuclease assay using the surveyor mutation detection kit (Integrated DNA Technologies Inc.). The percentage of target disruption was quantified by densitometry and calculated as described previously (42). The indel frequencies of TGFBR2 and FOXP3 were also measured by TIDE (Tracking Indels by Decomposition) analysis (43). The PCR primers used for the amplification of target loci are listed in Supplemental Table 1.
Luciferase-based cytolysis assay.
CRL582-luciferase cell-based cytotoxicity was assessed as previously described (39, 41). Briefly, CRL5826-luciferase cells or CRL5826-luciferase-PDL1 cells and effector cells were suspended at a density of 1 × 105 cells/mL in RPMI1640 medium and then seeded in white opaque plate at the indicated ratio and incubated at 37°C in 5% CO2 for 48 hours. 10 μl Steady-Glo luciferase substrate (Promega) was added, and 5 minutes later, luminescence was recorded by PerkinElmer VICTOR X3. The results were reported as percentage of killing based on the luciferase activity in the wells compared with tumor cells alone (percentage killing = 100 – [(RLU from well with effector and target cell coculture)/(RLU from well with target cells) × 100]).
Real-time PCR.
GFP+ effector cells were isolated by fluorescent cell sorting 3 days after coincubation with CRL5826 tumor cells in the absence or presence of 5 ng/mL TGF-β1. Total RNA was extracted using the RNAmini Kit (Qiagen) according to the manufacture’s instructions. cDNA was synthesized by reverse transcription using the TransScript-Uni One-Step gDNA Removal and cDNA Synthesis Supermix Kit (TransGen Biotech). mRNA quantification of IL2, IFNG, GZMA, GZMB, FOXP3, and PDCD1 was performed with the CFX96 real-time detection system (Bio-Rad). The housekeeping gene GAPDH was used as an internal control. The qPCR primers used in this study are listed in Supplemental Table 1.
RNA-seq.
M28z CAR T cells were incubated with CRL5826 tumor cells in the absence or presence of 5 ng/mL TGF-β1. CD4+GFP+ and CD8+GFP+ effector cells were isolated by fluorescent cell sorting after 3-day coincubation. Samples were sent to BIOMARK Technologies. RNA extraction, library building and sequencing, and data analysis were all performed by this company. Raw sequencing data were deposited in the NCBI’s Sequence Read Archive (SRA BioProject PRJNA598415).
Flow cytometry.
CytoFLEX (Beckman Coulter Inc.) was used to perform fluorescence expression analysis. Cells were harvested and prepared according to the manufacture’s protocols. The antibodies used are as follows: TGF-βRII-PE (R&D Systems, FAB2411P); LAG3-PE (BD Biosciences, 565616); and CD25-Percp (302625), FOXP3-APC (17-4776-42), CD3-Pacific Blue (300329), CD4-PE (317410), CD8a-APC (301014), PD1-PE (329906), CTLA-APC (349908), TIM3-APC (345012), CD45RO-PE (304206), and CCR7-Percp (353242) (all from BioLegend).
Cytokine ELISA.
Supernatants of effector cells were harvested 16 hours after coincubation with target tumor cells (CRL5826-luci) at a 1:1 ratio (2 × 104 cells each). Cytokines (IFN-γ and IL-2) produced by effector cells (M28z and M28z-TKO) were evaluated by ELISA Kits (BioLegend) according to the manufacturer’s protocols. To quantify the concentration of TGF-β1 in CRL5826 and CRL5826-TGF-β1 OE cell line culture medium, FBS-free basic medium was changed when the cell confluence was over 80%. 24 hours later, the culture medium was collected, and TGF-β1 released by these cells was evaluated by the ELISA Kit (R&D Systems) according to the manufacturer’s protocols.
Multi-round antigen stimulation assay.
2 × 105 CAR T cells were coincubated with CRL5826 or CRL5826-PDL1 tumor cells at a E/T of 2:1 in the absence or presence of 5 ng/mL TGF-β1. Two days later, the tumor cells were all lysed, and new tumor cells were added to the incubation system after CAR T cell counting. By the analogy, new tumor cells were added every 2 days, at a E/T of 2:1, until the CAR T cells could not lyse anymore. The concentration of TGF-β1 was maintained at 5 ng/mL in the addition group. The CAR T cells were collected in the last round, and expression levels of related checkpoints were detected.
Proliferation suppression assay.
2 × 106 CAR T cells were coincubated with OVCAR3 tumor cells at a E/T of 1:1 in the absence or presence of 5 ng/mL TGF-β1. Three days later, GFP+ CAR T cells, as effector cells, were sorted by flow cytometry. CD4+CD25– T cells, as responder cells, were isolated according to the manufacturer’s instructions (STEMCELL Technology). Responder cells, stained with violet (Life Technology), were incubated with effector cells at a ratio of 1:2 or 1:4 in the medium containing CD3/28 beads (Life Technology) and 100 IU/mL IL-2. Five days later, the proliferation of responder cells were analyzed by flow cytometry.
Animal model and in vivo CAR T cell function detection.
Six-week-old female NOD-Prkdcscid Il2rgnull (NPG) mice (VitalStar) were inoculated subcutaneously with 2 × 106 CRL5826-luci tumor cells. Mice were randomly divided into 3 groups, PBS, M28z, and M28z-TKO (n = 5), when the tumor volumes were between 200 and 300 mm3. 5 × 106 CAR T cells (CAR+ was 50%) were administrated i.t. or i.v. twice with a 1-week interval. Body weight, tumor size, and the onset of GvHD were monitored weekly. Mouse peripheral bloods were collected, and the proportion and subsets of human cells were analyzed. Mice liver and tumor tissues were fixed and embedded in paraffin. Liver sections were stained by H&E (MilliporeSigma) to detect the infiltration of inflammatory cells. Immunohistochemical staining was used to detect the expression of Mesothelin (Abcam) and TGF-β1 (Abcam) in tumor sections. Tumor sections were stained with the RNAscope Red Detection Kit with an antimesothelin antibody scFv probe (ACD Technologies) to detect the infiltration of CAR T cells. Tumor sections were stained with the RNAscope Multi-fluorescent Detection Kit with an antimesothelin antibody scFv, anti-MKI67, and anti-IFNG probes (ACD Technologies) to detect the function of infiltrated CAR T cells.
NPG mice engrafted with pancreatic carcinoma PDX (VitalStar) were also randomly divided into the above-mentioned 3 groups when the tumor sizes were between 200 and 300 mm3. 5 × 106 CAR T cells (CAR+ was 50%) were administrated i.t. or i.v. twice with a one-week-interval. Body weight, tumor size, and the onset of GvHD were monitored weekly. Mouse peripheral bloods were collected, and the proportion and subsets of human cells were analyzed. Contralateral reinoculation of the same PDXs was done when the primary PDX was removed completely. Body weight, tumor size, and the onset of GvHD were also monitored weekly. Mouse peripheral bloods were also collected, and the proportion and subsets of human cells was analyzed.
Statistics.
Data are shown as mean ± SD. All statistical comparisons were evaluated by ordinary 1-way or 2-way ANOVA or unpaired t test (2 tailed). The exact P values were labeled in corresponding figures. P values of less than 0.05 were considered significant.
Study approval.
All experiments involving animals were approved by the Animal Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences.
Author contributions
NT, CC, and HW designed the study and planned the experiments. NT, CC, XZ, MQ, and WM performed the experiments. NT and CC analyzed the data. NL generated the M28z plasmid. NT and HW wrote the manuscript. CC, XFW, and WH edited the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Ruiqi Wang and Wen Sun for editing the manuscript and Yi Yang (Beijing Cord Blood Bank) for her help in preparing the cord blood samples. This work was supported by the National Key Research and Development Program of China (2018YFA0107703), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010503), and the National Natural Science Foundation of China (81773269).
Version 1. 01/30/2020
In-Press Preview
Version 2. 02/27/2020
Electronic publication
Footnotes
Conflict of interest: A PCT patent (application serial no. PCT/CN2019/106109) has been filed base on this work.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(4):e133977.https://doi.org/10.1172/jci.insight.133977.
Contributor Information
Na Tang, Email: tangna@ioz.ac.cn.
Chen Cheng, Email: chengc1113@163.com.
Xingying Zhang, Email: zhangxingying@ioz.ac.cn.
Miaomiao Qiao, Email: 1365924262@qq.com.
Na Li, Email: linawhere@163.com.
Wei Mu, Email: muweicelltherapy@163.com.
Xiao-Fei Wei, Email: xfwei@chinacord.org.
Weidong Han, Email: hanwdrsw@sina.com.
Haoyi Wang, Email: wanghaoyi@ioz.ac.cn.
References
- 1.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim WA, June CH. The principles of engineering immune cells to treat cancer. Cell. 2017;168(4):724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalos M, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95):95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuster SJ, et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N Engl J Med. 2017;377(26):2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neelapu SS, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377(26):2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson LA, June CH. Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 2017;27(1):38–58. doi: 10.1038/cr.2016.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosti P, Maher J, Arnold JN. Perspectives on chimeric antigen receptor T-cell immunotherapy for solid tumors. Front Immunol. 2018;9:1104. doi: 10.3389/fimmu.2018.01104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knochelmann HM, Smith AS, Dwyer CJ, Wyatt MM, Mehrotra S, Paulos CM. CAR T Cells in Solid Tumors: Blueprints for Building Effective Therapies. Front Immunol. 2018;9:1740. doi: 10.3389/fimmu.2018.01740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15(11):669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 10.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 11.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Pang Y, Moses HL. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31(6):220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakao A, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi H, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89(7):1165–1173. doi: 10.1016/S0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 15.Dahmani A, Delisle JS. TGF-β in T cell biology: implications for cancer immunotherapy. Cancers (Basel) 2018;10(6):E194. doi: 10.3390/cancers10060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas DA, Massagué J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8(5):369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Trapani JA. The dual adverse effects of TGF-beta secretion on tumor progression. Cancer Cell. 2005;8(5):349–350. doi: 10.1016/j.ccr.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. J Exp Med. 2001;194(5):629–644. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pu N, et al. Neutralizing TGF-β promotes anti-tumor immunity of dendritic cells against pancreatic cancer by regulating T lymphocytes. Cent Eur J Immunol. 2018;43(2):123–131. doi: 10.5114/ceji.2018.77381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sow HS, Ren J, Camps M, Ossendorp F, Ten Dijke P. Combined inhibition of TGF-β signaling and the PD-L1 immune checkpoint is differentially effective in tumor models. Cells. 2019;8(4):E320. doi: 10.3390/cells8040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster AE, et al. Antitumor activity of EBV-specific T lymphocytes transduced with a dominant negative TGF-beta receptor. J Immunother. 2008;31(5):500–505. doi: 10.1097/CJI.0b013e318177092b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7(10):1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 23.Bergan L, Gross JA, Nevin B, Urban N, Scholler N. Development and in vitro validation of anti-mesothelin biobodies that prevent CA125/Mesothelin-dependent cell attachment. Cancer Lett. 2007;255(2):263–274. doi: 10.1016/j.canlet.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Park BV, et al. TGFβ1-mediated SMAD3 enhances PD-1 expression on antigen-specific T cells in cancer. Cancer Discov. 2016;6(12):1366–1381. doi: 10.1158/2159-8290.CD-15-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 26.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 27.Derré L, et al. BTLA mediates inhibition of human tumor-specific CD8+ T cells that can be partially reversed by vaccination. J Clin Invest. 2010;120(1):157–167. doi: 10.1172/JCI40070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai G, Anumanthan A, Brown JA, Greenfield EA, Zhu B, Freeman GJ. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol. 2008;9(2):176–185. doi: 10.1038/ni1554. [DOI] [PubMed] [Google Scholar]
- 29.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44(5):989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto M, et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med. 2018;69:301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 33.Yu J, et al. Anti-GD2/4-1BB chimeric antigen receptor T cell therapy for the treatment of Chinese melanoma patients. J Hematol Oncol. 2018;11(1):1. doi: 10.1186/s13045-017-0548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloss CC, et al. Dominant-negative TGF-β receptor enhances PSMA-targeted human CAR T cell proliferation and augments prostate cancer eradication. Mol Ther. 2018;26(7):1855–1866. doi: 10.1016/j.ymthe.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeku O, Li X, Brentjens RJ. Adoptive T-cell therapy for solid tumors. Am Soc Clin Oncol Educ Book. 2017;37:193–204. doi: 10.14694/EDBK_180328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraietta JA, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. 2018;24(5):563–571. doi: 10.1038/s41591-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guedan S, Alemany R. CAR-T cells and oncolytic viruses: joining forces to overcome the solid tumor challenge. Front Immunol. 2018;9:2460. doi: 10.3389/fimmu.2018.02460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, et al. Armored inducible expression of IL-12 enhances antitumor activity of glypican-3-targeted chimeric antigen receptor-engineered T cells in hepatocellular carcinoma. J Immunol. 2019;203(1):198–207. doi: 10.4049/jimmunol.1800033. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27(1):154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieser R, Attisano L, Wrana JL, Massagué J. Signaling activity of transforming growth factor beta type II receptors lacking specific domains in the cytoplasmic region. Mol Cell Biol. 1993;13(12):7239–7247. doi: 10.1128/MCB.13.12.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu W, Tang N, Cheng C, Sun W, Wei X, Wang H. In vitro transcribed sgRNA causes cell death by inducing interferon release. Protein Cell. 2019;10(6):461–465. doi: 10.1007/s13238-018-0605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 43.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.