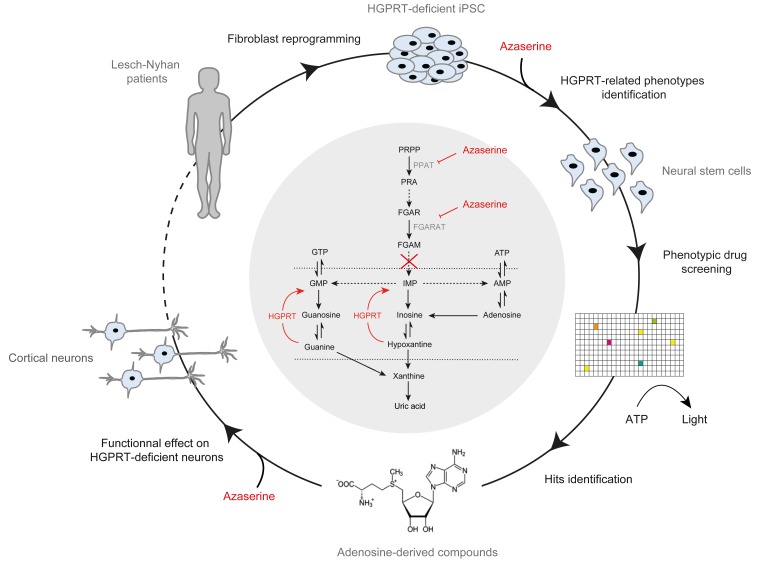
Abstract
Lesch-Nyhan disease (LND) is a rare monogenic disease caused by deficiency of the salvage pathway enzyme hypoxanthine-guanine phosphoribosyltransferase (HGPRT). LND is characterized by severe neuropsychiatric symptoms that currently cannot be treated. Predictive in vivo models are lacking for screening and evaluating candidate drugs because LND-associated neurological symptoms are not recapitulated in HGPRT-deficient animals. Here, we used human neural stem cells and neurons derived from induced pluripotent stem cells (iPSCs) of children affected with LND to identify neural phenotypes of interest associated with HGPRT deficiency to develop a target-agnostic–based drug screening system. We screened more than 3000 molecules and identified 6 pharmacological compounds, all possessing an adenosine moiety, that corrected HGPRT deficiency–associated neuronal phenotypes by promoting metabolism compensations in an HGPRT-independent manner. This included S-adenosylmethionine, a compound that had already been used as a compassionate approach to ease the neuropsychiatric symptoms in LND. Interestingly, these compounds compensate abnormal metabolism in a manner complementary to the gold standard allopurinol and can be provided to patients with LND via simple food supplementation. This experimental paradigm can be easily adapted to other metabolic disorders affecting normal brain development and functioning in the absence of a relevant animal model.
Keywords: Stem cells
Keywords: Drug screens
Human neural stem cells and neurons derived from iPSCs of Lesch-Nyhan disease patients were used to develop a target-agnostic-based drug screening system.
Introduction
Purines are both essential molecules for nucleic acid assembly and the most common carriers of chemical energy in mammalian cells. The cellular pool of purines is tightly maintained by the balance among their de novo synthesis, recycling, and degradation (1). Mutations in enzymes of the synthesis or recycling pathways are accompanied by devastating neurological symptoms (2). One of the most described purine-associated pediatric disorders is Lesch-Nyhan disease (LND), which is caused by deficiency of the salvage pathway enzyme hypoxanthine-guanine phosphoribosyltransferase (HGPRT), an X chromosome–encoded protein (3, 4). As a direct consequence, the recycling of guanine and hypoxanthine into GMP and inosine monophosphate (IMP) is inefficient, leading to uric acid accumulation, formation of uric acid–containing kidney stones, and finally renal failure. LND is also characterized by severe neuropsychiatric disorders, which present with choreoathetosis, dystonia, aggression, and self-injurious behavior (5, 6). Hyperuricemia and the formation of kidney stones can be controlled by the xanthine oxidase inhibitor 1,2-dihydro-4H-pyrazolo, pyrimidin-4-one (allopurinol, ref. 7).
Unfortunately, allopurinol treatment has no positive effect on neuropsychiatric symptoms (8), and new pharmacological approaches are needed to control neurological symptoms, specifically those that are associated with abnormal purine metabolism. However, the links between the metabolic derangements recorded in LND and the resulting neurological manifestations in children are still unknown. Neurological symptoms are not reproduced in genetic animal models of LND, while metabolic hallmarks are; this indicates that either rodent brain cells develop alternative metabolic strategies that are not present in human cells or that some purine-dependent processes are not present during rodent brain development (9, 10). In addition to these purely metabolic actions, purines activate a plethora of cell signalling pathways via membrane receptors that can act additively, synergistically, or antagonistically (2). Related to this mechanistic complexity, the precise measurement of purine intermediate concentrations requires complex techniques, such as HPLC, that are not easily amenable to “hard to access” tissues, such as the CNS. Consequently, most information that is collected is from peripheral cells. However, purine metabolism in the brain, most particularly that during development, appears to be different than that in other organs; it mainly relies upon recycling, with only reduced de novo synthesis activity recorded (11, 12). From a genetic point of view, more than 400 mutations leading to a clinical phenotype have been identified, with a high degree of heterogeneity regarding their effect upon the enzyme activity. It remains, therefore, unclear whether the clinical phenotypes result from a reduced enzymatic activity toward hypoxanthine, guanine, or both (13, 14). Together, this mechanistic complexity ultimately leads to abnormal brain function in LND, but the complexity compromises the development of pharmacological approaches using classical target-based strategies. In contrast, phenotype-based assays monitor the existence of morphological and functional cellular phenotypes of interest that can discriminate between 2 experimental conditions in a target-agnostic manner. Phenotype-based assays enable exploration of a much larger biological space by simultaneously investigating the consequences of multiple and complex modifications of cellular events in their native environment and at endogenous levels. Here, we have used a phenotypic assay approach based on human induced pluripotent stem cells (iPSCs, ref. 15) that are differentiated into neurons to study the effect of HGPRT deficiency. We previously showed that such neurons may be instrumental to identify potentially therapeutic pharmacological compounds using automated high-throughput drug screening coupled with image-based high-content analysis (16–18). In the current study, we show that LND cells demonstrate pathological changes during key steps of the differentiation of human pluripotent stem cell–derived (PSC-derived) neural stem cells (NSC) into cortical neurons. Using this in vitro model, 6 compounds were identified from a collection of over 3000 tested compounds, suggesting paths for clinical development.
Results
To develop a relevant model of LND amenable to drug screening, we used iPSCs reprogrammed from fibroblasts derived from children with LND to obtain HGPRT-deficient NSCs and neurons as a starting material. Primary cultures of fibroblasts isolated from 2 individuals with LND were reprogrammed in parallel with 2 controls using 4 transcription factors: Oct4, Sox2, c-Myc, and KLF4 (ref. 15 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.132094DS1). All iPSC lines self-renewed as undifferentiated colonies with typical PSC morphology (Supplemental Figure 1) and expressed the pluripotency markers Oct-4 and SSEA-4 (Supplemental Figure 1) and TRA 1-81 and SSEA3 (Supplemental Figure 1). The iPSC lines were differentiated into cortical NSCs, as described in Boissart et al. (17), and proliferated at a similar rate and homogeneously expressed the 2 neural markers, SOX2 and Nestin (Supplemental Figure 2). Western blot analysis of HGPRT protein content confirmed a complete absence of HGPRT expression in the 2 donors with LND (Figure 1A). Accordingly, the enzyme was active after reprogramming in the control cells and was inactive in cells derived from the 2 individuals with LND (Supplemental Figure 2). This indicated that HGPRT loss-of-function mutations did not compromise fibroblast reprogramming or iPSC commitment to the neural lineage when phenocopying the enzymatic defect associated with LND.
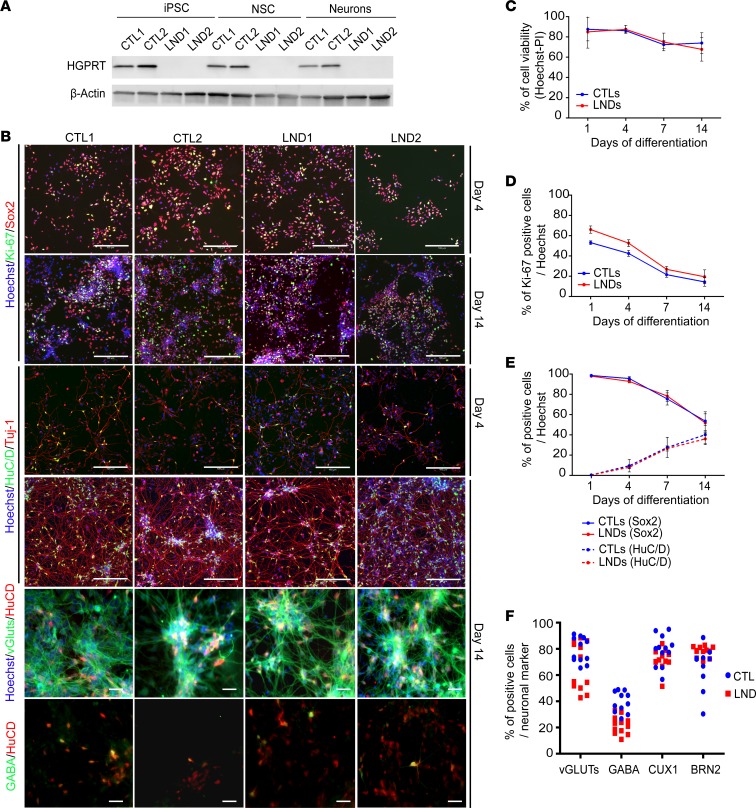
Figure 1. Characterization and phenotypical analysis of iPSCs, NSCs, and cortical neurons derived from control donors and donors with Lesch-Nyhan.
(A) Western blot analysis of HGPRT protein expression in control (CTL) and Lesch-Nyhan disease (LND) iPSCs, NSCs, and cortical neurons. β-Actin was used as a loading control. (B) Representative images of Ki-67 (green) and Sox2 (red) NSC markers and HuC/D (green) and Tuj-1 (red) neuronal markers in control and LND cells on days 4 and 14 of differentiation. Scale bar: 200 μm. vGluts and GABA labeling (green) were combined with HuC/D labeling (red) to identify neuronal subtypes. Scale bar: 20 μm. (C) Quantification of cell viability using a combination of Hoechst and propidium iodide (PI) staining during the process of control (blue lines) or LND (red lines) NSC differentiation. (D and E) Kinetic quantification of the percentage of Ki-67+ cells (D) or HuC/D– and Sox2+ cells (E) during the process of control (blue lines) or LND (red lines) NSC differentiation into postmitotic neurons. (F) Neurons expressing the subtype markers vGLUTs, GABA, CUX1 and BRN2 after 14 days of differentiation in control (blue dots) and LND (red squares) cells. All quantitative results are expressed as the mean ± SD of the 2 control and 2 LND cell lines, with 3 biological replicates per cell line.
Phenotypic characterization of differentiated neurons derived from HGPRT-deficient NSCs.
The consequences of HGPRT deficiency during the differentiation of NSCs into fully differentiated postmitotic neurons were analyzed. Neuronal differentiation was initiated from HGPRT-competent (CTL1 and CTL2, control cell lines) and HGPRT-deficient (LND1 and LND2 cell lines) NSCs in medium that contained N2 and B27 (N2B27, ref. 19); multiparametric assays were conducted during the progression of differentiation from day 4 to day 14. Quantification of Ki67 antigen indicated a progressive decrease of cell proliferation between day 0 (seeding) and day 14, associated with a reduction of the percentage of SOX2+ undifferentiated NSCs (Figure 1, B and D) in control cells. This was paralleled by an increase of the HuC/D+ postmitotic neuronal cell fraction (Figure 1, B and E). At day 14, the percentage of undifferentiated NSCs ranged from 50% to 60% and the percentage of neurons ranged from 40% to 50%. No difference was detected in HGPRT-deficient cells regarding the kinetics or the rate of differentiation. An analysis of the neuronal subtypes indicated that the cultures were mainly composed of glutamatergic and GABAergic neurons expressing superficial cortical layer markers, CUX1 and Brn2. HGPRT deficiency did not modify the neuronal subtypes or the proportion of the different types of neurons in the cultures (Figure 1, B and F, and Supplemental Figure 2).
Very little cell death was measured during the differentiation process, with no difference observed between HGPRT-competent and HGPRT-deficient cells (Figure 1C). In these differentiation conditions, after 14 days, HGPRT-deficient cells produced a global neuronal network that was morphologically indistinguishable from that obtained with HGPRT-competent NSCs (Figure 1B).
We hypothesized that our culture conditions would mask dependency on the purine salvage pathway by providing different carbon resources compared with the physiologically available resources, thus allowing increased de novo synthesis. The hypoxanthine concentrations, measured using HPLC, were elevated in NSCs derived from the 2 donors with LND and were compared with their control counterparts, suggesting a high rate of de novo synthesis not compensated for by downstream salvage (Supplemental Figure 3). Consequently, to reveal phenotypes that are dependent on purine recycling while excluding any potential bias introduced by the overactivation of de novo synthesis, the culture conditions were modified to include low-level inhibition of the de novo pathway. Azaserine, a potent glutamine amidotransferase inhibitor that blocks 2 limiting reactions in the de novo synthesis pathway, was added to the neuronal differentiation medium at either 0.5 or 2 μM, with 80 μM hypoxanthine provided as the principle substrate to the cells to stimulate HGPRT-dependent purine salvage (Supplemental Figure 3). Azaserine was first added to the medium on day 1 of differentiation and was included thereafter with each medium change every other day. The amount of cell death was measured on days 3, 6 and 10 after initiating the treatment, corresponding to days 4, 7, and 11 of differentiation. At these concentrations, azaserine did not compromise the survival of control NSCs with efficient HGPRT activity. In contrast, it induced massive cell death in cultures of HGPRT-deficient NSCs at the 2 concentrations tested. After 10 days of azaserine treatment, only 24.89% (±8.02%) of surviving cells were detected at 0.5 μM and 27.55% (±8.42%) were detected at 2 μM (Figure 2A). This result indicated that inhibition of the de novo pathway was sufficient to reveal an HGPRT-dependent phenotype. However, treating the NSCs from day 1 precluded further evaluation of other differentiation parameters, such as NSC proliferation and engagement into postmitotic neurons. Azaserine treatment was therefore delayed and started only after 5 days of differentiation, a time point at which the NSCs were already committed to becoming postmitotic neurons (Figure 2, B–G). The cells were also analyzed on days 2, 6, 11, and 13 after initiating the treatment, corresponding to days 7, 11, 14, and 18 of differentiation. HGPRT-competent NSCs survived, proliferated, and differentiated into neurons similarly — regardless of whether azaserine was added. In this delayed pattern of treatment, only the 2 μM concentration of azaserine induced cell death in HGPRT-deficient NSCs as they differentiated into neurons but to a lesser extent, with 55.12% (±7.04%) living cells still detected after 11 days of treatment (Figure 2B). The proliferation of HGPRT-deficient cells was also dose-dependently affected by azaserine, as revealed by Ki67 immunoreactivity. Azaserine treatment (2 μM) reduced cell proliferation to 66.10% (±16.18%) compared with that of DMSO-treated cells after 2 days of treatment and to 2% (±1.91%) after 13 days (Figure 2, C and G). Together, azaserine-induced cytotoxicity and cytostaticity led to fewer cells in HGPRT-deficient cultures as the differentiation into neurons proceeded (Figure 2, D and G). After 2 μM azaserine treatment, HuC/D-immunoreactive postmitotic neurons were reduced to 68.96% (±20.79%) of the DMSO-treated cells after 13 days of azaserine treatment (Figure 2, E and G). The sum of modifications induced by azaserine treatment when started on day 5 of differentiation led to a decreased number of neurons produced by HGPRT-deficient NSCs at the 2 concentrations (Figure 2, F and G). An analysis of the neuronal subtypes indicated that azaserine did not specifically affect one neuronal subtype but rather demonstrated toxicity toward both glutamatergic and GABAergic neurons (Supplemental Figure 3).
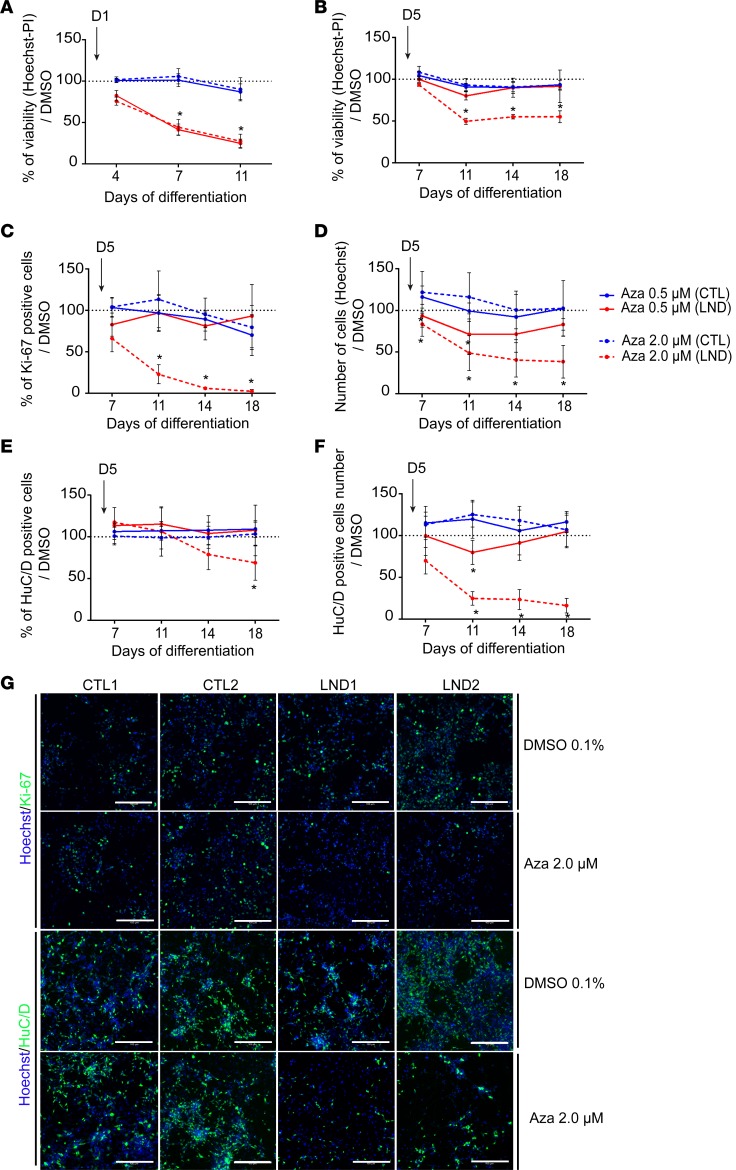
Figure 2. Effect of azaserine treatment on NSCs derived from control individuals or individuals with Lesch-Nyhan disease.
(A) Percentage of cell viability quantified using Hoechst-PI staining after treatment with azaserine starting on day 1 of differentiation (D1) compared with cells exposed to 0.1% DMSO. (B) Percentage of cell viability after azaserine treatment starting on day 5 of differentiation (D5) compared with cells exposed to 0.1% DMSO. (C) Quantification of the percentage of Ki-67+ cells after treatment with azaserine started on day 5 of differentiation compared with cells exposed to 0.1% DMSO. (D) Quantification of the total number of cells quantified using Hoechst nuclear staining after treatment with azaserine started on day 5 of differentiation compared with cells exposed to 0.1% DMSO. (E) Quantification of the percentage of HuC/D+ neurons after treatment with azaserine started on day 5 of differentiation compared with cells exposed to 0.1% DMSO. (F) Quantification of the total number of HuC/D+ neurons after treatment with azaserine started on day 5 of differentiation compared with cells exposed to 0.1% DMSO. (G) Representative images of immunocytochemistry for Ki-67 on day 7 of differentiation and HuC/D on day 14 of differentiation in control (CTL) and Lesch-Nyhan disease (LND) cells exposed to 0.1% DMSO or 2.0 μM azaserine from day 5. Scale bar: 100 μm. All quantitative results are expressed as the mean ± SD of the 2 control (blue lines) and 2 LND (red lines) cell lines, with 4 technical replicates. Azaserine was added at 0.5 (solid lines) or 2.0 μM (dotted lines). *P < 0.01 Holm-Sidak post hoc test.
Identification of LND-relevant pharmacological compounds using target-agnostic high-throughput screening.
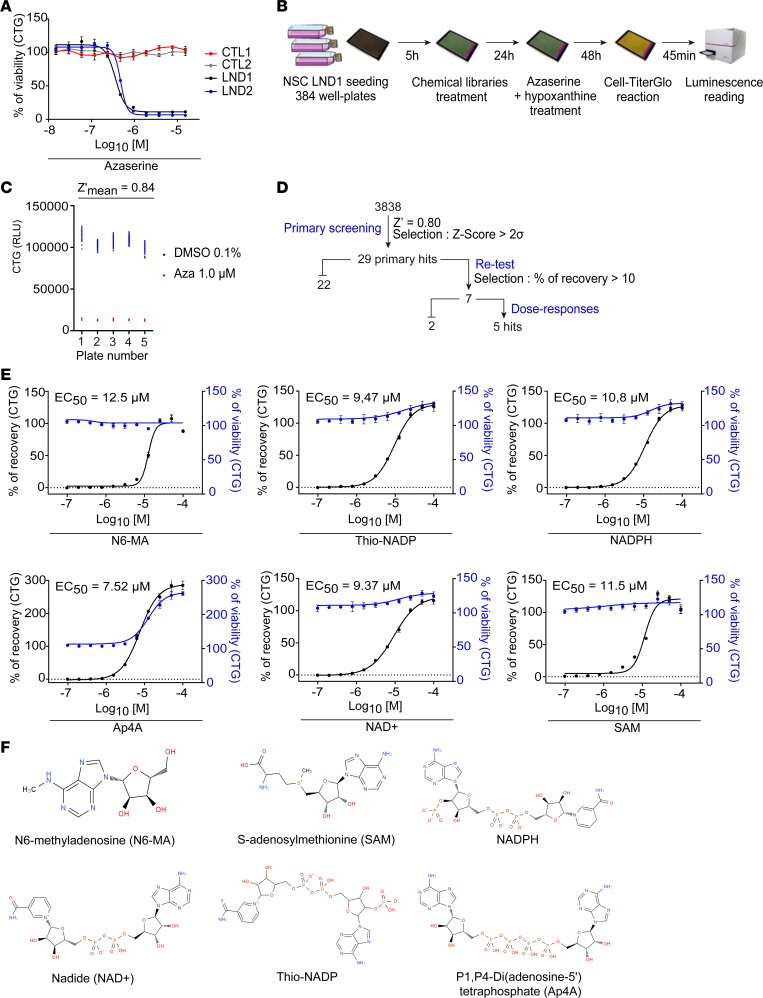
Target-agonistic high-throughput screening (HTS) was used to generate an assay to discover pharmacological compounds that could rescue phenotypes related to HGPRT deficiency. The selected drug screening readout was the cell viability after treating NSCs with azaserine from day 1, i.e., the most dramatic phenotype. The immunofluorescence-based Hoechst/propidium iodide (Hoechst/PI) used thus far was replaced by a luminescence-based method Cell-TiterGlo (CTG) that quantifies ATP content and is an integrated measure of both metabolism and cell death. Dose-response experiments were performed to determine the optimal azaserine concentration to be used (Figure 3A). One micromolar azaserine was chosen because it induced a strong decrease in the ATP content in the 2 LND NSC cultures but had no effect on the control cells. The screening workflow is summarized in Figure 3B. NSCs from LND1 were seeded into 384-well plates and treated with chemical compounds after 5 hours. The next day, 1 μM azaserine was added to the medium. After 48 hours, the ATP content was quantified using CTG. The Z′ factor was calculated using 5 sister plates with cells treated with DMSO as a positive control (maximum value) and cells treated with azaserine as a negative control (minimum value). The mean Z′ factor value was 0.84, which indicates an excellent screening assay (Figure 3C). The Z′ factor was –1.04 when Hoechst/IP was used as a readout, which confirmed the better sensitivity of the CTG method in this screening paradigm. A total of 3838 pharmacological compounds were screened, and a Z score > 2σ was chosen as a selection criteria. After the primary screening, 29 compounds had a Z score > 2σ at 5 or 10 μM (Figure 3D and Supplemental Table 3). After retesting in technical replicates, 7 compounds were selected that consistently induced an increase of more than 10% of CTG signal compared with nontreated cells. Five of those compounds were finally selected because they exhibited a dose-dependent effect with an of EC50 below 13 μM (Figure 3E). All of these compounds shared a common structural core that contained an adenosine moiety (Figure 3F).
Figure 3. Primary high-throughput screening of pharmacological compounds.
(A) Cell-TiterGlo (CTG) estimation of cell viability after treating control and LND cell lines with increasing concentrations of azaserine. The x axis represents azaserine concentration expressed as log10 (concentration), and the concentration is expressed in molarity (M). The results are expressed as the mean ± SD of 4 technical replicates (2 biological replicates). (B) Screening workflow. (C) Z′ factor calculation in 5 sister 384-well plates containing LND1 NSCs treated with 0.1% DMSO or 1.0 μM azaserine. RLU, relative luminescence unit. (D) Primary screening cascade. (E) Dose-response analysis of the 6 hit compounds. The black curve corresponds to the percentage of recovery after treatment of LND1 NSCs with 1.0 μM azaserine. The blue line shows the percentage of viability after treatment with the drug alone without azaserine and represents the toxicity of the compound itself. The results are expressed as the mean ± SD of 4 technical replication (2 biological replicates). (F) Chemical structures of the 6 hit compounds obtained from PubChem (US National Library of Medicine).
To investigate further, we tested S-adenosylmethionine (SAM), a structural homolog of N6-methyladenosine. As CTG signal generally quantifies ATP cellular content, dose-response experiments for the 6 compounds were performed using Hoechst as a readout in order to assess the capacity of the molecules to rescue cell viability. We confirmed the efficiency of all compounds in restoring cellular viability in LND1 NSCs in the presence of 1 μM azaserine in a dose-dependent manner and that they were within the same EC50 range (Supplemental Figure 4).
Adenosine-like compound specificity.
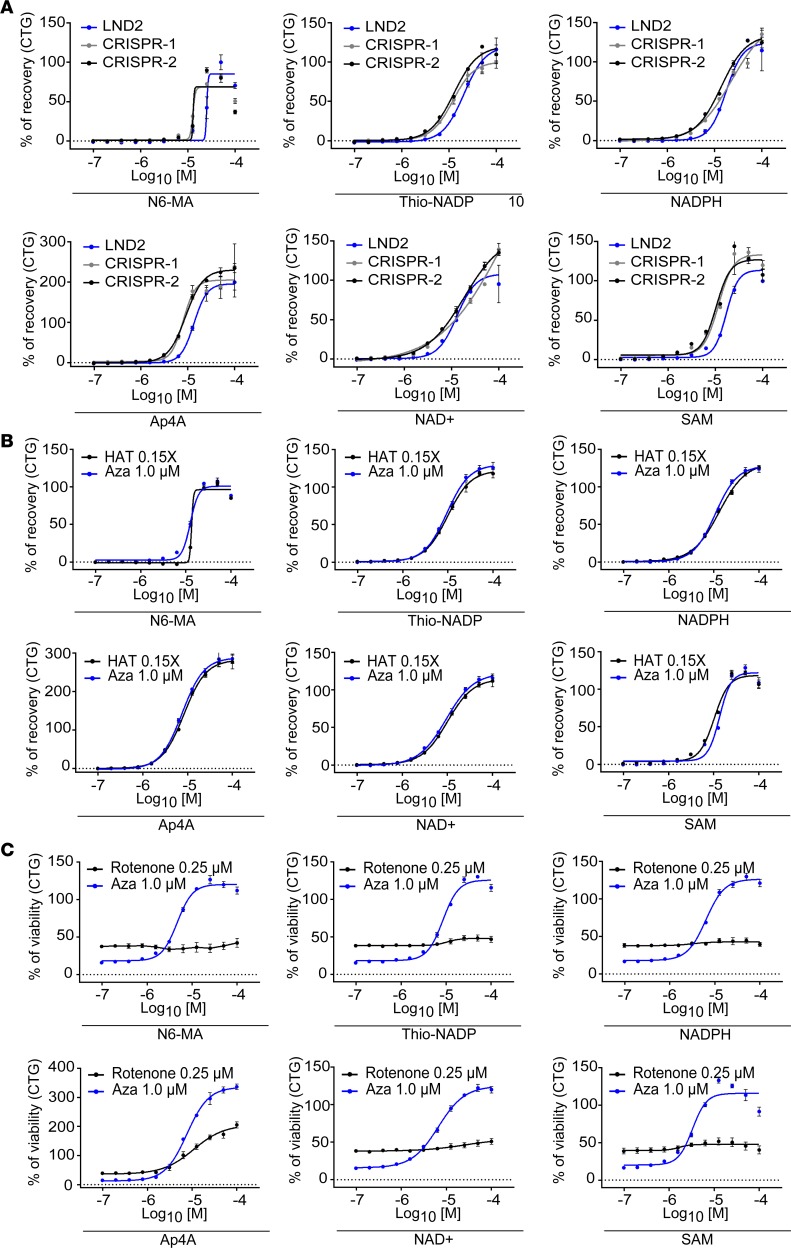
The 6 compounds were retested in cells with other genetic backgrounds. The same efficiency was found for LND2 NSCs as well as for 2 different clones of the hESC line SA001 that had been HGPRT-deleted using CRISPR/Cas9 technology (Figure 4A and Supplemental Figure 5). The assay was also repeated using a selection medium that contained aminopterin instead of azaserine as a de novo pathway inhibitor (HAT medium) to exclude any artefacts due to interactions of the compounds with azaserine. Aminopterin selectively induced cell death in LND NSCs in a dose-dependent manner (Supplemental Figure 5), and a dose of 0.06 μM aminopterin was used to further test compound specificity. The 6 compounds demonstrated a similar rescue efficiency as when using azaserine (Figure 4B). To exclude the possibility that the 6 compounds were solely nonspecific inhibitors of apoptosis, their efficiency was tested using rotenone, an inhibitor of mitochondria complex I that affects NSC survival independently of HGPRT status (Supplemental Figure 5). None of the 6 compounds exhibited neuroprotective activity in that experimental set up (Figure 4C).
Figure 4. Specificity evaluation of the 6 hit compounds.
(A) Evaluation of hit compounds’ dose-dependent activity after treatment with 1 μM azaserine. NSCs were obtained from the LND2 donor (blue line) and from 2 clones derived from HGPRT-competent human embryonic stem cells with HGPRT deletion using the CRISPR/Cas9 system (CRISPR-1 in gray and CRISPR-2 in black). The x axis represents the compound concentration (in molarity) expressed as log10. The results are expressed as the mean ± SD of 4 technical replicates and 2 biological replicates. (B) Evaluation of hit compounds’ dose-dependent activity after treatment of LND NSCs with 1 μM azaserine (blue line) or ×0.15 HAT (black line). The x axis represents the compound concentration (in molarity) expressed as log10. The results are expressed as the mean ± SD of 4 technical replicates and 2 biological replicates. (C) Evaluation of hit compounds’ dose-dependent activity after treatment of LND NSCs with 0.25 μM rotenone (black line) or 1 μM azaserine (blue line). The x axis represents the compound concentration (in molarity) expressed as log10. The results are expressed as the mean ± SD of 4 technical replicates and 2 biological replicates. Aza, azaserine.
Mode of action of the adenosine-like compounds.
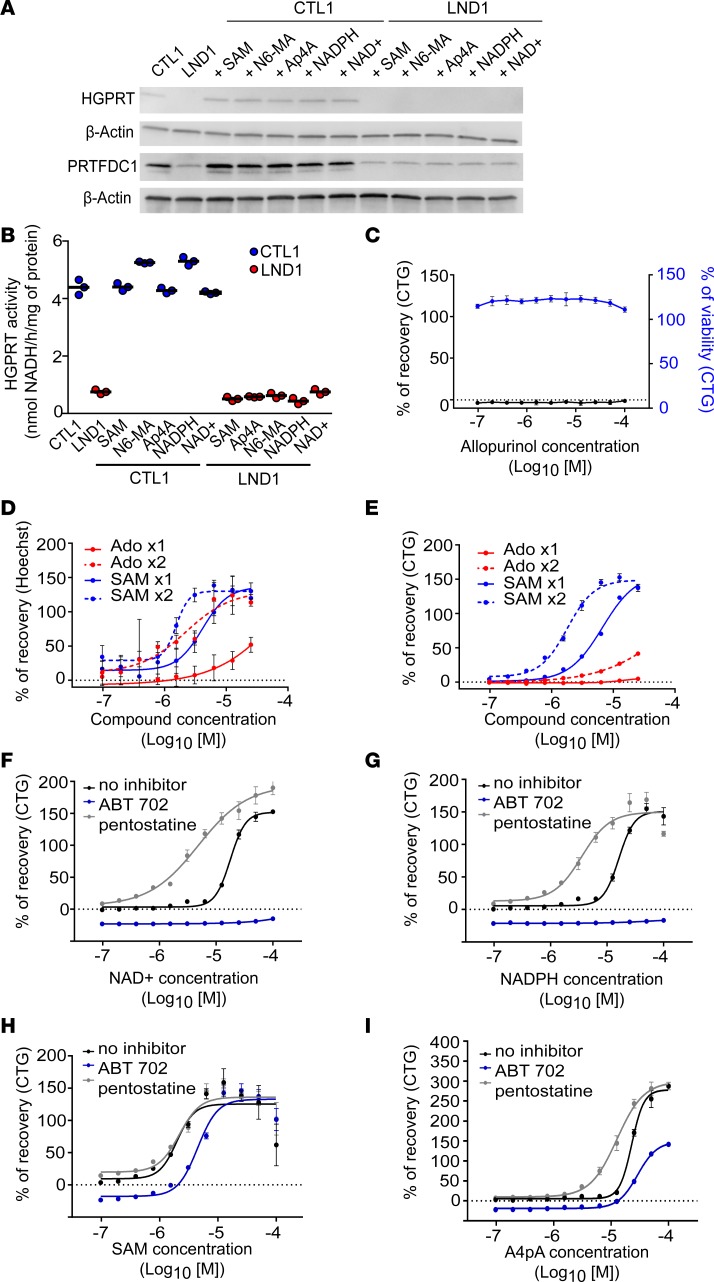
Next, we explored the mechanism of action of the adenosine-like compounds. One hypothesis was that these compounds directly restored an HGPRT-like activity. The expression of HGPRT and of the HGPRT pseudogene PRTFDC1, as well as HGPRT-like enzymatic activity, was quantified after treating LND NSCs with each compound for 72 hours. Neither HGPRT nor PRTFDC1 expression was modified (Figure 5A) nor did the compounds induce any detectable HGPRT-like enzymatic activity (Figure 5B).
Figure 5. Mechanism of action of the hit compounds.
(A) Western blot analysis of HGPRT and PRTFDC1 protein expression in CTL1 and LND1 NSCs treated with SAM (20 μM), N6-MA (20 μM), Ap4A (12.5 μM), NADPH (25 μM), and NAD+ (25 μM) for 72 hours. β-Actin was used as a loading control. (B) HGPRT enzymatic activity in CTL1 (blue) and LND1 (red) NSCs treated with the hit compounds. The bars represent the mean of 3 biological replicates. (C) Allopurinol efficiency in LND1 NSCs treated with 1.0 μM azaserine. CTG was used as the readout (black line). The toxicity of allopurinol alone was also quantified (blue line). The results are expressed as the mean ± SD of 4 technical replicates and 2 biological replicates. (D and E) Hoechst staining (D) or Cell-TiterGlo (CTG) (E) quantification of SAM (blue lines) or adenosine (Ado, red lines) dose-dependent activity after treating LND1 NSCs with 1 μM azaserine. The x axis represents the compound concentration (in molarity) expressed as log10. The results are expressed as the mean ± SD of 4 technical replicates. x1, 1 treatment with the compound (solid line); x2, 2 sequential treatments with the compound (dotted lines). (F–I) CTG-based quantification of NAD+ (F), NADPH (G), SAM (H), and A4pA (I) dose-dependent activity after treating LND1 NSCs with 1 μM azaserine in the presence of 10 μM ABT-702 (adenosine kinase inhibitor) or pentostatin (adenosine deaminase inhibitor). The x axis represents compound concentration (in molarity) expressed as log10. The results are expressed as the mean ± SD of 4 technical replicates.
Allopurinol was then tested to evaluate the possible contribution of xanthine oxidase inhibition. This was not the case, since allopurinol did not rescue azaserine-treated LND NSCs (Figure 5C).
We then evaluated the contribution of the adenosine moiety to the compounds’ mechanism of action. Adenosine was added 5 hours after seeding the NSCs, and azaserine was added 24 hours after seeding. Cell survival was quantified 72 hours after seeding using Hoechst. SAM was used as a reference adenosine-like compound. In these conditions, adenosine-induced recovery was poor compared with SAM. The efficiency of recovery at maximal concentration for adenosine was 51.79% (±10.97%), compared with more than 130% ± 11.92% for SAM. In addition, the EC90 for adenosine was 2.3 × 10–3, whereas the EC90 for SAM was 1.19 × 10–5. A second treatment with adenosine was added at the same time as azaserine in order to increase its bioavailability. Adenosine remained less efficient than SAM (EC90 adenosine, 1.43 × 10–5 vs. EC90 SAM, 2.6 × 10–6, Figure 5D). The same lack of efficiency was recorded when CTG was used as a readout (EC90 adenosine, 1.4 × 10–3 vs. EC90 SAM, 5.4 × 10–6, Figure 5E). Considering that the bioavailability of adenosine in vitro may not have been optimal, the contribution of the adenosine moiety of each compound to act as a source of AMP or inosine was assessed using an inhibitor of the adenosine kinase (ABT-702) or of the adenosine deaminase (pentostatin). ABT-702 totally blocked NAD+ and NADPH, while pentostatin increased the efficiency of NAD+ and NADPH (Figure 5, F and G). Blocking the conversion of adenosine to inosine had no effect on SAM efficiency, while inhibiting its conversion into AMP decreased (but did not suppress) compound efficiency (Figure 5H). ABT-702 only had a partial blocking effect, while pentostatin increased A4pA efficiency (Figure 5I).
Correction of LND-related neuronal phenotypes by adenosine-like compounds.
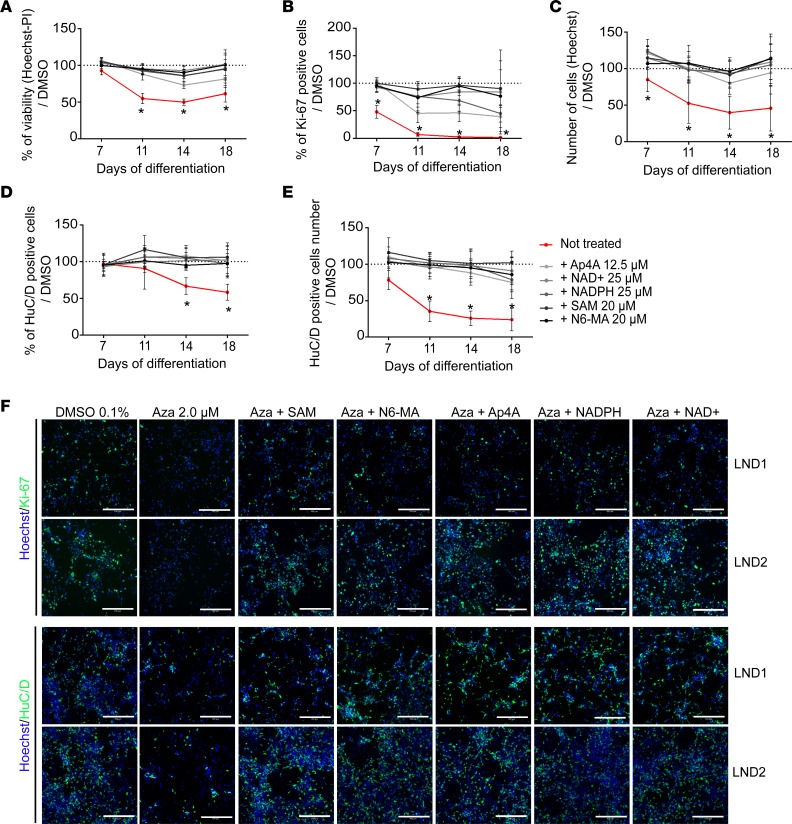
To evaluate whether the compounds had a global positive effect on the different neural phenotypes identified as dependent on the HGPRT-dependent salvage pathway, differentiating NSCs were treated from day 5 with azaserine, and each compound added on days 4, 8, 12, and 15. The cell cultures were stopped and analyzed on days 7, 11, 14, and 18 (Figure 6). The optimal compound concentrations were extrapolated from previous dose-response experiments. All the compounds demonstrated neuroprotective effects against azaserine-induced cell death in developing LND1 and LND2 neurons (Figure 6A). All compounds restored Ki67 expression, although with different efficiencies (Figure 6, B and F), and increased the number of cells in azaserine-treated cultures (Figure 6, C and F). The percentage of HuC/D+ cells and the total number of neurons produced was also increased, reaching levels similar to those observed for cells not exposed to azaserine (Figure 6, D–F). Analysis of neuronal subtypes in the culture treated with SAM suggested that both glutamatergic and GABAergic neurons were rescued by compound treatment (Supplemental Figure 6). Together, these findings indicated that, by promoting adaptive metabolic strategies, all adenosine-like compounds ameliorated the cell survival, proliferation, and differentiation rate of LND NSCs and globally ameliorated neurogenesis in HGPRT-deficient cells (Figure 6F).
Figure 6. Adenosine-like compounds restore LND-relevant neuronal phenotypes.
(A–C) Evaluation of hit compound efficacy upon cell viability (A, Hoechst-PI staining readout), percentage of Ki-67+ cells (B), and total cell number (C) after treating LND neural cells repeatedly with 2.0 μM azaserine from day 5 of differentiation. The results represent the mean ± SD of LND1 and LND2 lines, with 4 technical replicates/donor, compared with 0.1% DMSO-treated cells. (D and E) Evaluation of hit compound efficacy, as assessed through the percentage (E) and total number (F) of HuC/D+ neurons after treating LND neural cells repeatedly with 2.0 μM azaserine from day 5 of differentiation. The results represent the mean ± SD of LND1 and LND2 lines, with 4 technical replicates/donor, compared with 0.1% DMSO-treated cells. (F) Representative images of immunocytochemistry for Hoechst (blue), Ki-67, and HuC/D staining (green) on day 7 of differentiation. Scale bar: 100 μm. Aza, azaserine.
Discussion
The main finding of this study is the establishment of a human model allowing the study of neurodevelopmental defects associated with metabolic disorders. This model enabled the high-throughput screening–based identification of pharmacological compounds with potential therapeutic action on LND neuropsychiatric symptoms. All identified compounds were adenosine donors, although the adenosine metabolic pathway only partially explained their efficiency at rescuing LND cells. This finding was obtained through a target-agnostic–based drug screening system thanks to the use of a pathological model based on neural cells differentiated from iPSC lines derived from patients affected by LND, along with an experimental paradigm that exploited the functional defects revealed by a specific culture medium.
The next step of this work would be to test the efficiency of the hit compounds in animal models of LND. HGPRT-knockout rodent models exhibit metabolic abnormalities consistent with deficient purine salvage (10, 20, 21). To some extend, reduction of dopamine concentration in the caudate-putamen nuclei was also reported both in mice and rats (20, 22). Unfortunately, the rodent model failed to recapitulate the neuropsychiatric characteristics of LND, i.e., dystonia and self-injurious behavior (10, 20). This lack of spontaneous neurobehavioral phenotype renders these models unsuitable for testing the efficacy of the compounds at a functional and integrated level. Evidence of efficiency may therefore rely on human case reports. Indeed, the effect of administration of SAM as a dietary supplement has been documented in some patients with LND (23–26). Due to these case reports, SAM supplementation was evaluated in 21 patients, including 18 with the full spectrum of LND-associated neuropsychiatric disorders. In most cases, supplementation with SAM led to a significant reduction in self-mutilation events and good clinical outcomes. However, Dolcetta and collaborators described aggravation of the symptoms or important adverse effects that some patients could not tolerate, leading the discontinuation of SAM. Although encouraging, these reports described very different situations, with high heterogeneity regarding the age of the patients (mainly adolescents and adults), the duration of the treatment, the dosage of SAM, the type of cotreatment, and, finally, the type of benefit assessment. By demonstrating that SAM actually improved neuronal phenotypes that were directly associated with HGPRT deficiency, our work advocates for a formal reassessment of supplementation with SAM as a beneficial complement in LND. Interestingly, 2 other compounds can be easily assessed in patients with LND, because NAD and NADP can also be provided as food supplements.
Interestingly, all of the identified compounds possess at least one adenosine moiety in their structure. One hypothesis regarding their mode of action is that the release of adenosine can act to replenish adenosine in the cells, which can then be transformed into AMP by adenosine kinase and then into ADP and ATP (27). In addition, IMP can be formed from AMP by AMP deaminase and can then be converted to GMP in 2 reactions, mediated by IMP dehydrogenase, followed by GMP synthase. This constitutes a starting point to replenish GDP and GTP. Alternatively, adenosine can also serve as a source of inosine via the action of adenosine deaminase (28, 29). We investigated the contribution of these 2 adenosine-dependent pathways using ABT-702 or pentostatin to block the conversion of adenosine into AMP and inosine, respectively. In our experimental setup, ABT-702 totally or partially abolished the efficacy of all the tested compounds in rescue of LND cells from azaserine treatment in a dose-dependent manner. In contrast, pentostatin increased the action of NAD, NADPH, and A4p4 but had no effect on SAM and N6M. Together, this demonstrated that the conversion of adenosine into AMP is one of the main modes of action of the compounds, with no contribution of the conversion into inosine. Interestingly, when tested in our paradigm, allopurinol did not provide any protection for azaserine-treated NSCs, indicating a mechanism of action of the adenosine-like compound that was different (and therefore complementary) to the as-yet gold standard treatment used to decrease uricemia in LND.
This study focused primarily on modeling LND neuronal phenotypes using the neural progeny of patient-derived iPSCs. We chose to differentiate iPSCs into cortical progenitors and neurons, because this brain area is reported as functionally and anatomically abnormal in LND (30, 31) and because we previously showed that these cell types are fully amenable to high-throughput drug screening (16). LND is considered a neurodevelopmental disorder with no signs of secondary neurodegeneration (6). The conversion of NSCs into neurons represents a critical developmental period of vulnerability for the emergence of neurodevelopmental disorders (32); therefore, we chose to study the effect of purine deficiency from the very early stage of NSC conversion into neurons rather than studying mature neurons only. We first differentiated the cells using the previously described neural medium N2B27, which did not reveal any alterations associated with HGPRT deficiency at the morphological level. HGPRT deficiency–associated phenotypes were only identified when azaserine, an inhibitor of HGPRT de novo synthesis, was added to the culture medium at concentrations that did not affect HGPRT-competent neural cells. This is in agreement with findings of McKeran and colleagues in vitro that demonstrated the necessity of inhibiting the de novo pathway in cells to study HGPRT deficiency in order to re-create a “brain-like environment” (33). In vitro, cells tend to increase the de novo synthesis to compensate for a recycling deficit, a situation that is not physiologically possible in the brain, where the de novo pathway enzymes are only poorly active (12). This highlights the intrinsic ability of different cell types to mobilize alternative energy supplies to cope with bioenergetics stress — a property termed “metabolic flexibility” (34, 35). However, 1 caveat of our study is that it mainly relied upon morphological observations of the neuronal network. It would be of interest to compare LND and control cells with or without azaserine at a more functional level, for example by using microelectrode arrays or optical assays.
Overall, our study identified metabolic pathways in LND neural cells that can be relevant therapeutic targets to ease the devastating neuropsychiatric symptoms associated with this pathology. Interestingly, these pathways can be activated in patients with LND via simple food supplementation. This also opens the door for larger screening studies of FDA-approved drugs or of collections of novel chemical entities. This experimental paradigm can also be easily adapted to other enzyme-associated neurological disorders that affect normal brain development.
Methods
Neural cell differentiation.
Four iPSC lines, reprogrammed from male fibroblasts obtained from the Coriell Biorepository (Coriell Institute for Medical Research), consisting of 2 control and 2 LND cell lines, were used in this study. The 2 LND lines were reprogrammed from fibroblasts of LND-affected children (National Institute of General Medical Sciences Repository GM02227 and GM23784), both presenting self-injurious behaviors. All cell line details are summarized in Supplemental Table 1. For clarity, the control and LND cells were noted as CTL and LND, respectively, in Supplemental Table 1. Somatic cell reprogramming was performed using the 4 human genes OCT4, SOX2, c-Myc, and KLF4 cloned in Sendai viruses (Invitrogen) and pluripotency characterized according to Nakagawa et al. (36). In addition to iPSCs, a control hESC line was obtained (SA001 hESC, Cellartis) and used under the supervision of the French Bioethics Agency (agreement NOR AFSB 12002 43S). PSC lines were maintained in StemMACS iPS-Brew XF medium (Miltenyi Biotec), supplemented with 1:100 penicillin/streptomycin (Gibco) and vitronectin (Invitrogen) as a matrix. The medium was changed every day, and the cells were manually passaged once per week. Cell cultures were incubated in a 37°C humidified atmosphere with 5% CO2. The differentiation protocol into NSCs was previously published (17). NSCs were cultured in N2B27 medium supplemented with recombinant human fibroblast growth factor 2 (10 ng/mL), epidermal growth factor (10 ng/mL), and brain-derived neurotrophic factor (20 ng/mL) (all from PeproTech). Cell cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C, and the medium was changed every other day. Terminal neuronal differentiation was induced by plating the cells in poly-l-ornithine/laminin-treated 384-well plates in N2B27 medium without growth factors. The medium was changed twice per week with the addition of laminin (1:500, Gibco) to avoid neuronal clumping. Neuronal differentiation was maintained for at least 14 days and up to 18 days to obtain well-characterized cortical neurons.
Western blotting.
Protein expression was quantified by Western blot analysis. Whole-cell pellets were collected and lysed in RIPA lysis buffer (Sigma-Aldrich) containing protease and phosphatase inhibitor cocktail (Thermo Scientific). After centrifugation at 4°C and 10000 g, the supernatants were collected, and the protein concentration was measured using the Pierce BCA Protein Assay kit (Thermo Scientific), following the manufacturer’s instructions. Proteins (20–40 μg) were reduced using dithiothreitol (MilliporeSigma) and Laemmli buffer (Bio-Rad) before being loaded and separated by electrophoresis on 4%–15% SDS-polyacrylamide gels (Criterion Tris-Glycine eXtended Gels, Bio-Rad). The proteins were then transferred to nitrocellulose membranes (Trans-Blot Turbo Nitrocellulose Transfer, Bio-Rad). The membranes were blocked in PBS-Tween-20 0.1% (PBS-T, VWR) containing 5% nonfat dry milk (MilliporeSigma) for 45 minutes at room temperature and then incubated overnight at 4°C with primary antibodies diluted in blocking buffer (Supplemental Table 2). The membranes were then washed 3 times with PBS-T and incubated for 1 hour at room temperature with HRP-labeled secondary antibodies (GE-Healthcare) at 1:10,000 in blocking buffer. The signal was visualized by enhanced chemiluminescence using an Amersham ECL Plus Western Blotting Detection kit (GE Healthcare). β-Actin at 1:50,000 was used as a loading control.
HGPRT enzymatic activity.
HGPRT activity was measured using a PRECICE HPRT Assay kit (Novocib). HGPRT activity was detected as the rate of production of IMP in the presence of α-D-5-phosphoribosyl-1-pyrophosphate (PRPP), followed by IMP oxidation with the reduction of NAD+ to NADH, measurable by the absorbance at 340 nm. PBS-washed cell pellets were resuspended in ice-cold water containing NP-40 lysis buffer (Thermo Scientific) and a protease inhibitor cocktail (Thermo Scientific); the samples were then sonicated at 4°C. After centrifugation, the supernatants were collected to measure HGPRT activity, following the manufacturer’s instructions. The total protein content was determined using a Pierce BCA Protein Assay kit (Thermo Scientific). HGPRT kinetic activity in the cell lysates was measured over 3 hours by reading the absorbance at 340 nm, and enzymatic activity (nmol/mg of protein/hr) was calculated using a recombinant HGPRT enzyme as a standard.
High-throughput drug screening assay.
To conduct high-throughput compound screening, NSCs were plated into poly-l-ornithine/laminin–treated 384-well plates at 5000 cells per well in N2B27 medium without antibiotics using the Biocel 1800 platform (Agilent Technologies). Five hours after plating, the cells were treated with the following chemical libraries: LOPAC 1280 (10 μM, MilliporeSigma, 1280 compounds), Prestwick Chemical Library (5 μM, Prestwick Chemical, 1280 compounds), Prestwick Phytochemical (10 μM, Prestwick Chemical, 320 compounds), and Selleckchem custom library (5 μM, Selleckchem, 958 compounds). A solution of 0.1% DMSO (MilliporeSigma) was used as a control. After 24 hours of treatment, 1 μM azaserine (MilliporeSigma, A1164) and 80 μM hypoxanthine (MilliporeSigma, H9636) were added to the media. After 48 hours of azaserine treatment, the cell survival was quantified using the CellTiter-Glo luminescent cell viability assay (Promega), following the manufacturer’s instructions.
Immunostaining and high-content image analysis.
Cells were fixed using 4% PFA (Electron Microscopy Sciences) and incubated overnight at 4°C with primary antibodies (Supplemental Table 2) diluted in a blocking buffer composed of 1X PBS, 1% BSA (MilliporeSigma) and 0.3% Triton X-100 (MilliporeSigma). The cells were washed 3 times in PBS and incubated for 45 minutes at room temperature with Alexa Fluor–conjugated secondary antibodies (Thermo Scientific). Finally, the cells were washed 3 times in PBS. In addition, Hoechst 33228 (1:3000, MilliporeSigma) nuclear staining was performed during the secondary labeling. The fluorescence signal was detected using the high-content analysis system ImageXpress (Molecular Devices). The apoptotic cell ratio during neuronal differentiation was determined using a Hoechst-PI staining assay (Thermo Fisher Scientific). Living cells were treated with a mixture of Hoechst 33228 (6 μg/mL, MilliporeSigma) and PI (0.4 μg/mL, MilliporeSigma) for 30 minutes at 37°C. The fluorescence signal was detected using an ImageXpress system (Molecular Devices).
Purine measurement by HPLC.
NSCs were dissociated in trypsin (Gibco) and then washed once in PBS. After centrifugation, whole-cell pellets were collected and snap-frozen in liquid nitrogen. They were then resuspended in cold water and sonicated at 4°C. The total protein concentration was measured using a Pierce BCA Protein Assay kit (Thermo Scientific). The cell lysates were treated with 1 M perchloric acid (MilliporeSigma) and incubated for 5 minutes at 40°C, followed by centrifugation at 8000 g at room temperature. The supernatants were collected and stored at –20°C.
Genome editing using the CRISPR/Cas9 system.
HGPRT gene–knockout mutants were generated from the male hESC control line SA001 using CRISPR/Cas9-mediated genome editing. Alt-R CRISPR/Cas9 HPRT crRNA and tracrRNA (Integrated DNA Technologies) were resuspended in Nuclease-Free Duplex Buffer (IDT) to a final concentration of 100 μM and mixed in equimolar concentrations to form RNA-oligo complexes. After incubating for 10 minutes at room temperature, 30 μM Cas9 protein (TacGene) was added to sgRNA at a 1:1.25 ratio and incubated for 15 minutes at room temperature to allow the formation of RNP. At 1–2 hours before transfection, 10 μM ROCK inhibitor (Y27632, StemCell Technologies) was added to the cultured cells. The cells were dissociated with Accutase (StemCell Technologies), and 200,000 cells were added to a 1.5-mL conical tube and gently pelleted. The cells were resuspended in 20 μl of P3 Primary Cell Nucleofector Solution and Supplement (Lonza), following the manufacturer’s instructions. The RNP complex solution was mixed with the cell suspension, and the samples were electroporated using the 4D-Nuclofector system (Lonza). The cells were then plated in 12-well plates containing StemMACS iPS-Brew XF medium (Miltenyi Biotec). A negative Alt-R CRISPR/Cas9 crRNA was used as a control. HGPRT-edited clones were selected using 6-thioguanine medium (A4660, MilliporeSigma) at a final concentration of 1×. Positive clones were amplified and differentiated into NSCs, as described above, and then frozen in a 1:10 mix of 100% DMSO and FBS (MilliporeSigma).
Statistics.
To evaluate assay and automation robustness, the Z′ factor was calculated in 5 sister plates as follows: Z′ factor = 1 – ([3 × σ max + 3 × σ min]/|μmax – μmin|), where σ is the SD of positive controls (maximum) or negative controls (minimum) and μ is the mean signal value of positive and negative controls (37). In the survival experiments, NSCs treated with DMSO were considered as positive controls, and azaserine-treated NSCs served as negative controls.
The percentage of recovery induced by each pharmacological compound was calculated as follows: ([luminescence value of compound-treated well – median of azaserine only–treated wells]/[median value of DMSO only–treated wells – median of azaserine only–treated wells]) × 100.
For the selection of positive compounds in primary screening, the Z score (38) was calculated and was defined as Z = (× − μ)/σ, where × is the relative luciferase activity of an individual compound, μ is the mean luciferase activity of all test compounds, and σ is the SD of all test compounds. Hit compounds were defined as those with a minimum Z score of 2.0, meaning that their luciferase activity was 2 SDs above that of the mean.
ANOVA followed by the Holm-Sidak post hoc test was applied for intergroup multiple comparisons.
For dose-response experiments, nonlinear regression and calculation of the EC50 and EC90 values were performed using PRISM software (GraphPad).
Study approval.
This study has been approved by the French Bioethics Agency (agreement number NOR AFSB 12002 43S) for the use of human pluripotent stem cells including human embryonic stem cells.
Author contributions
VR, JT, and GM conducted high throughput screening. VR, ML, and CB performed secondary compound validation and study the mode of action. LC and CB reprogrammed iPSC, differentiated NSC, and performed all the required quality control. MC conducted CRISPR/Cas-9 based editing. VR, JT, CB, LC, and AB analyzed the data. VR, MP, and AB wrote and edited the manuscript. AB designed the research and supervised the project. All the authors analyzed the data and performed statistical analysis.
Supplementary Material
Acknowledgments
The authors would like to thank Pauline Poydenot, Ronan Crépin, and Cécile Denis for their help in preparing the biological resources and for development of the screening assay as well as Odile Boespflug-Tanguy, Guillaume Pinna, and Jerôme Denis for scientific discussions and evaluation of this work. VR received a PhD grant from the French Ministry of Science and Technology and a grant from the patient and family association Lesch-Nyhan Action. I-Stem is part of the Biotherapies Institute for Rare Diseases and is supported by the Association Française contre les Myopathies (AFM-Téléthon). This study was funded in part by grants from “Investissements d’Avenir” (ANR-11-INBS-0009, INGESTEM; ANR-11-INBS-0011-NeurATRIS). This study used samples from the National Institute of Neurological Disorders and Stroke (NINDS) Human Genetics Resource Center DNA and Cell Line Repository as well as clinical data. This study used data from the NINDS repository (ID numbers GM01869, GM02227, GM23784, and GM04603).
Version 1. 01/28/2020
In-Press Preview
Version 2. 02/27/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(4):e132094.https://doi.org/10.1172/jci.insight.132094.
Contributor Information
Valentin Ruillier, Email: val.ruillier@gmail.com.
Johana Tournois, Email: jtournois@istem.fr.
Claire Boissart, Email: cboissart@istem.fr.
Marie Lasbareilles, Email: mlasbareilles@istem.fr.
Gurvan Mahé, Email: gurvan.mahe987@gmail.com.
Laure Chatrousse, Email: lchatrousse@istem.fr.
Michel Cailleret, Email: mcailleret@istem.fr.
Marc Peschanski, Email: mpeschanski@istem.fr.
Alexandra Benchoua, Email: abenchoua@istem.fr.
References
- 1.Baresova V, et al. Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Hum Mol Genet. 2012;21(7):1534–1543. doi: 10.1093/hmg/ddr591. [DOI] [PubMed] [Google Scholar]
- 2.Fumagalli M, Lecca D, Abbracchio MP, Ceruti S. Pathophysiological role of purines and pyrimidines in neurodevelopment: unveiling new pharmacological approaches to congenital brain diseases. Front Pharmacol. 2017;8:941. doi: 10.3389/fphar.2017.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lesch M, Nyhan WL. A familial disorder of uric acid metabolism and central nervous system function. Am J Med. 1964;36:561–570. doi: 10.1016/0002-9343(64)90104-4. [DOI] [PubMed] [Google Scholar]
- 4.Fu R, et al. Genotype-phenotype correlations in neurogenetics: Lesch-Nyhan disease as a model disorder. Brain. 2014;137(Pt 5):1282–1303. doi: 10.1093/brain/awt202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nyhan WL, O’Neill JP, Jinnah HA, Harris JC. Lesch-Nyhan Syndrome. In: Adam MP, et al. eds. GeneReviews. Seattle, Washington, USA: University of Washington, Seattle; 1993. http://www.ncbi.nlm.nih.gov/books/NBK1149 Updated May 15, 2014. Accessed January 10, 2020.
- 6.Jinnah HA, et al. Delineation of the motor disorder of Lesch-Nyhan disease. Brain. 2006;129(Pt 5):1201–1217. doi: 10.1093/brain/awl056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres RJ, Puig JG. Hypoxanthine-guanine phosophoribosyltransferase (HPRT) deficiency: Lesch-Nyhan syndrome. Orphanet J Rare Dis. 2007;2:48. doi: 10.1186/1750-1172-2-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres RJ, Prior C, Puig JG. Efficacy and safety of allopurinol in patients with the Lesch-Nyhan syndrome and partial hypoxanthine- phosphoribosyltransferase deficiency: a follow-up study of 18 Spanish patients. Nucleosides Nucleotides Nucleic Acids. 2006;25(9-11):1077–1082. doi: 10.1080/15257770600893974. [DOI] [PubMed] [Google Scholar]
- 9.Finger S, Heavens RP, Sirinathsinghji DJ, Kuehn MR, Dunnett SB. Behavioral and neurochemical evaluation of a transgenic mouse model of Lesch-Nyhan syndrome. J Neurol Sci. 1988;86(2-3):203–213. doi: 10.1016/0022-510X(88)90099-8. [DOI] [PubMed] [Google Scholar]
- 10.Engle SJ, et al. HPRT-APRT-deficient mice are not a model for lesch-nyhan syndrome. Hum Mol Genet. 1996;5(10):1607–1610. doi: 10.1093/hmg/5.10.1607. [DOI] [PubMed] [Google Scholar]
- 11.Howard WJ, Kerson LA, Appel SH. Synthesis de novo of purines in slices of rat brain and liver. J Neurochem. 1970;17(1):121–123. doi: 10.1111/j.1471-4159.1970.tb00509.x. [DOI] [PubMed] [Google Scholar]
- 12.Allsop J, Watts RW. Activities of amidophosphoribosyltransferase and purine phosphoribosyltransferases in developing rat brain. Adv Exp Med Biol. 1980;122A:361–366. doi: 10.1007/978-1-4615-9140-5_59. [DOI] [PubMed] [Google Scholar]
- 13.Jinnah HA, Harris JC, Nyhan WL, O’Neill JP. The spectrum of mutations causing HPRT deficiency: an update. Nucleosides Nucleotides Nucleic Acids. 2004;23(8-9):1153–1160. doi: 10.1081/NCN-200027400. [DOI] [PubMed] [Google Scholar]
- 14.Fu R, Jinnah HA. Genotype-phenotype correlations in Lesch-Nyhan disease: moving beyond the gene. J Biol Chem. 2012;287(5):2997–3008. doi: 10.1074/jbc.M111.317701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Darville H, et al. Human pluripotent stem cell-derived cortical neurons for high throughput medication screening in autism: A proof of concept study in SHANK3 haploinsufficiency syndrome. EBioMedicine. 2016;9:293–305. doi: 10.1016/j.ebiom.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boissart C, et al. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl Psychiatry. 2013;3:e294. doi: 10.1038/tp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georges P, Boissart C, Poulet A, Peschanski M, Benchoua A. Protein kinase-A inhibition is sufficient to support human neural stem cells self-renewal. Stem Cells. 2015;33(12):3666–3672. doi: 10.1002/stem.2194. [DOI] [PubMed] [Google Scholar]
- 19.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21(2):183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 20.Meek S, et al. Reduced levels of dopamine and altered metabolism in brains of HPRT knock-out rats: a new rodent model of Lesch-Nyhan disease. Sci Rep. 2016;6:25592. doi: 10.1038/srep25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tschirner SK, Gutzki F, Schneider EH, Seifert R, Kaever V. Neurotransmitter and their metabolite concentrations in different areas of the HPRT knockout mouse brain. J Neurol Sci. 2016;365:169–174. doi: 10.1016/j.jns.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 22.Jinnah HA, et al. Dopamine deficiency in a genetic mouse model of Lesch-Nyhan disease. J Neurosci. 1994;14(3 Pt 1):1164–1175. doi: 10.1523/JNEUROSCI.14-03-01164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolcetta D, et al. Quantitative evaluation of the clinical effects of S-adenosylmethionine on mood and behavior in Lesch-Nyhan patients. Nucleosides Nucleotides Nucleic Acids. 2013;32(4):174–188. doi: 10.1080/15257770.2013.774012. [DOI] [PubMed] [Google Scholar]
- 24.Chen BC, et al. Treatment of Lesch-Nyhan disease with S-adenosylmethionine: experience with five young Malaysians, including a girl. Brain Dev. 2014;36(7):593–600. doi: 10.1016/j.braindev.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Glick N. Dramatic reduction in self-injury in Lesch-Nyhan disease following S-adenosylmethionine administration. J Inherit Metab Dis. 2006;29(5):687. doi: 10.1007/s10545-006-0229-8. [DOI] [PubMed] [Google Scholar]
- 26.Lauber M, Plecko B, Pfiffner M, Nuoffer JM, Häberle J. The effect of S-adenosylmethionine on self-mutilation in a patient with Lesch-Nyhan disease. JIMD Rep. 2017;32:51–57. doi: 10.1007/8904_2016_571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boison D. Adenosine kinase: exploitation for therapeutic gain. Pharmacol Rev. 2013;65(3):906–943. doi: 10.1124/pr.112.006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ipata PL. Origin, utilization, and recycling of nucleosides in the central nervous system. Adv Physiol Educ. 2011;35(4):342–346. doi: 10.1152/advan.00068.2011. [DOI] [PubMed] [Google Scholar]
- 29.Ipata PL, Camici M, Micheli V, Tozz MG. Metabolic network of nucleosides in the brain. Curr Top Med Chem. 2011;11(8):909–922. doi: 10.2174/156802611795347555. [DOI] [PubMed] [Google Scholar]
- 30.Puig JG, Torres RJ. Enzyme activity and brain anatomy: lessons from HPRT deficiency. Lancet Neurol. 2013;12(12):1129–1131. doi: 10.1016/S1474-4422(13)70247-3. [DOI] [PubMed] [Google Scholar]
- 31.Schretlen DJ, et al. Regional brain volume abnormalities in Lesch-Nyhan disease and its variants: a cross-sectional study. Lancet Neurol. 2013;12(12):1151–1158. doi: 10.1016/S1474-4422(13)70238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80(3):588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 33.McKeran RO, Howell A, Andrews TM, Watts RW, Arlett CF. Observations on the growth in vitro of myeloid progenitor cells and fibroblasts from hemizygotes and heterozygotes for “complete” and “partial” hypoxanthine-guanine phosphoribosyltransferase (HGPRT) deficiency, and their relevance to the pathogenesis of brain damage in the Lesch-Nyhan syndrome. J Neurol Sci. 1974;22(2):183–195. doi: 10.1016/0022-510X(74)90245-7. [DOI] [PubMed] [Google Scholar]
- 34.Micheli V, et al. NAD metabolism in HPRT-deficient mice. Metab Brain Dis. 2009;24(2):311–319. doi: 10.1007/s11011-009-9134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen SP, et al. Astrocyte adenosine deaminase loss increases motor neuron toxicity in amyotrophic lateral sclerosis. Brain. 2019;142(3):586–605. doi: 10.1093/brain/awy353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 38.Curtis AE, Smith TA, Ziganshin BA, Elefteriades JA. The mystery of the Z-score. Aorta (Stamford) 2016;4(4):124–130. doi: 10.12945/j.aorta.2016.16.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.