Abstract
Group 2 innate lymphoid cells (ILC2s) are a critical innate source of type 2 cytokines in allergic inflammation. Although ILC2s are recognized as a critical cell population in the allergic inflammation, the regulatory mechanism(s) of ILC2s are less well understood. Here, we show that Regnase-1, an immune regulatory RNAse that degrades inflammatory mRNAs, negatively regulates ILC2 function and that IκB kinase (IKK) complex–mediated Regnase-1 degradation is essential for IL-33– and IL-25–induced ILC2 activation. ILC2s from Regnase-1AA/AA mice expressing a Regnase-1 S435A/S439A mutant resistant to IKK complex–mediated degradation accumulated Regnase-1 protein in response to IL-33 and IL-25. IL-33– and IL-25–stimulated Regnase-1AA/AA ILC2s showed reduced cell proliferation and type 2 cytokine (IL-5, IL-9, and IL-13) production and increased cell death. In addition, Il2ra and Il1rl1, but not Il5, Il9, or Il13, mRNAs were destabilized in IL-33–stimulated Regnase-1AA/AA ILC2s. In vivo, Regnase-1AA/AA mice showed attenuated acute type 2 pulmonary inflammation induced by the instillation of IL-33, IL-25, or papain. Furthermore, the expulsion of Nippostrongylus brasiliensis was significantly delayed in Regnase-1AA/AA mice. These results demonstrate that IKK complex–mediated Regnase-1 degradation is essential for ILC2-mediated type 2 responses both in vitro and in vivo. Therefore, controlling Regnase-1 degradation is a potential therapeutic target for ILC2-contributed allergic disorders.
Keywords: Immunology
Keywords: Allergy, Cytokines, Innate immunity
Targeted mutation of IKK target sites in Regnase-1 attenuates IL-33- and IL-25-induced group-2 innate lymphoid cell activation.
Introduction
Group 2 innate lymphoid cells (ILC2s) produce a large quantity of type 2 cytokines, including IL-5, IL-9, and IL-13, and thus are considered an innate counterpart of Th2 cells (1–4). ILC2s do not express a specific antigen receptor but quickly respond to epithelial cell–derived cytokines, such as IL-33 and IL-25 (also known as IL-17E) (4). This epithelial cytokine-mediated ILC2 activation is sufficient to induce allergic pulmonary inflammation evoked by the instillation of papain (5, 6), house dust mite (7, 8), or Alternaria alternata (9, 10) in mice. ILC2s are also essential for early host protection against helminth infections, including Nippostrongylus brasiliensis (N. brasiliensis) (3, 11). In addition to acute type 2 inflammation, ILC2s contribute to the chronic phase of type 2 inflammation by activating Th2 cells (12, 13) and by acquiring a memory-like phenotype (14). Indeed, ILC2s are implicated in human chronic type 2 disorders, including chronic rhinosinusitis (15, 16) and allergic asthma (17–19). Thus, a deeper understanding of the regulatory mechanisms of ILC2 activation is essential to develop therapeutic measures for type 2 immunity-associated disorders.
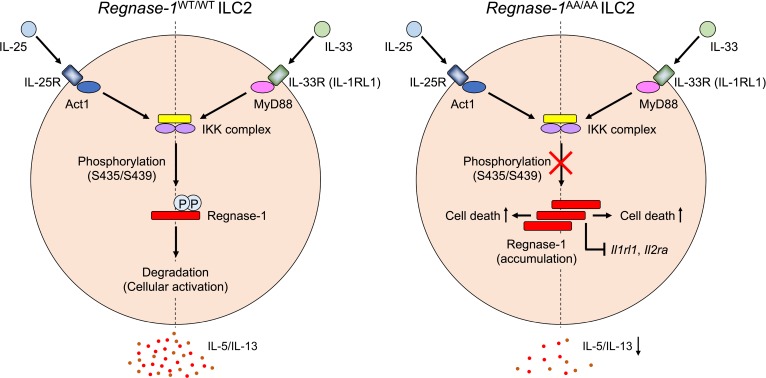
Regnase-1 (also known as Zc3h12a or Mcpip1) is a PIN-like RNAse that destabilizes mRNAs of a set of inflammatory genes (20). Regnase-1–/– mice spontaneously develop autoinflammatory disease caused by aberrant activation of macrophages and CD4+ T cells (20, 21). Regnase-1 is constitutively expressed in macrophages and fibroblasts to prevent “leaky” cytokine production in the resting state (22). Upon stimulation via TLRs or IL-1R, the IκB kinase (IKK) complex, composed of IKKα, IKKβ, and IKKγ (NEMO), phosphorylates Regnase-1, resulting in ubiquitin proteasome–dependent degradation that allows cells to rapidly upregulate mRNAs of inflammatory genes (22). Regnase-1 harbors a canonical IKK target motif, DSGxxS, and a targeted mutation in this motif (S435A/S439A) confers Regnase-1 resistance to IKK complex–mediated degradation and stronger decay activity against Il6 (22). In addition, the S435/S439 motif is required for Regnase-1 degradation downstream of IL-17R signaling (23).
Because IL-33R (composed of ST2 and IL-1RAcP) shares its signaling molecules with TLR/IL-1R, stimulation of cells with IL-33 activates myeloid differentiation factor 88–dependent (MyD88-dependent) signaling (24, 25). IL-1R–associated kinase 4 (IRAK4), IRAK1, and TNF receptor–associated factor 6 (TRAF6) are activated downstream of MyD88, and, in turn, IKK complex phosphorylates IκBα to activate the transcription factor NF-κB (25). Another epithelial cytokine, IL-25, signals through IL-17RA and IL-17RB (26), which recruit Act1 as an adaptor molecule (27, 28), indicating that IL-25 shares its signaling molecules with IL-17 (29). Act1 associates with TRAF6 to induce NF-κB activation through the IKK complex–mediated degradation of IκBα (30, 31). Therefore, Regnase-1 degradation might be controlled by the IKK complex downstream of IL-33 and IL-25. Alternatively, Regnase-1 may contribute to the regulation of IL-33– and IL-25–induced type 2 responses. Although Regnase-1 is initially considered a critical negative regulator of Th1/Th17 responses (20, 21, 32, 33), Regnase-1 also controls Th2 development (34), and the expression of Th2-related genes, including Il5, Il9, Il13, and Gata3, was increased in Regnase-1–/– CD4+ T cells (21, 34, 35). These observations prompted us to investigate the role of Regnase-1 in the activation of ILC2s by IL-33 and IL-25.
Because Regnase-1–/– mice spontaneously develop Th1/Th17 inflammation and die within a few months after birth (20, 21, 35), we took advantage of Regnase-1AA/AA mutant mice where Regnase-1 alleles are mutated to encode Regnase-1 S435A/S439A protein that is resistant to IKK complex–mediated degradation (23). In this study, we show that Regnase-1 undergoes S435/S439 motif–dependent degradation downstream of IL-33 and IL-25 and that Regnase-1 degradation is crucial for IL-33– and IL-25–induced ILC2 activation both in vitro and in vivo.
Results
IL-33 and IL-25 induce Regnase-1 accumulation in Regnase-1AA/AA ILC2s.
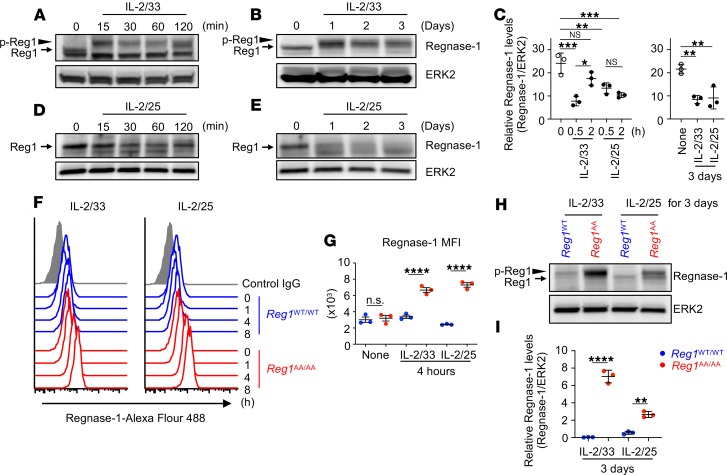
To examine whether Regnase-1 protein is expressed in ILC2s and is controlled downstream of IL-33 and IL-25 signaling, we used in vitro–expanded BM ILC2s. ILC2s (CD45+Lin–CD90.2+CD25+Sca-1+) sorted from BM of WT mice were c-Kit– as previously described for BM ILC2s (refs. 36, 37 and Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131480DS1) and in vitro expanded by IL-2, IL-33, and IL-25. After expansion and rest, ILC2s were stimulated with IL-2 and IL-33 (IL-2/33) or IL-2 and IL-25 (IL-2/25) for varying periods, and Regnase-1 expression was examined by immunoblotting. IL-2/33 stimulation induced a slowly migrating band within 15 minutes (Figure 1A), indicating Regnase-1 phosphorylation at S494/S513 by IRAK1 (22, 23). Then, Regnase-1 expression slightly decreased starting at 30 minutes and recovered by 120 minutes after stimulation (Figure 1, A and C). This is similar to that of LPS-stimulated macrophages, although Regnase-1 levels showed greater dynamic change in macrophages (22). Regnase-1 phosphorylation was maintained for 3 days in IL-2/33–stimulated ILC2s, and the Regnase-1 level gradually decreased over time (Figure 1, B and C). When ILC2s were stimulated with IL-2/25, Regnase-1 expression decreased by 30 minutes, and Regnase-1 maintained a low level of expression level over time (Figure 1, C and D). Although Regnase-1 is phosphorylated at S494/S513 by TANK-binding kinase 1 (TBK1) and inducible IKK (IKKi) downstream of IL-17 (23), slowly migrating Regnase-1 was not detected in IL-2/25–stimulated ILC2s. Regnase-1 gradually decreased upon long-term stimulation with IL-2/25 and had almost disappeared by day 3 (Figure 1, C and E). To examine whether the IKK target motif controls Regnase-1 levels downstream of IL-33 and IL-25, we used BM ILC2s from Regnase-1WT/WT and Regnase-1AA/AA mice. Because Regnase-1AA/AA ILC2s did not expand well in response to IL-33 or IL-25 (discussed below), we only used freshly isolated BM ILC2s for comparing ILC2s from Regnase-1WT/WT and Regnase-1AA/AA mice. Freshly isolated Regnase-1WT/WT and Regnase-1AA/AA ILC2s expressed comparable levels of canonical ILC2s markers (Supplemental Figure 1C). As shown in Figure 1, F and G, intracellular Regnase-1 staining showed that stimulation of both IL-2/33 and IL-2/25 induced a slight decrease in Regnase-1 at 1 hour, which subsequently recovered in WT ILC2s. In Regnase-1AA/AA ILC2s, Regnase-1 was not decreased in response to IL-2/33 or IL-2/25 and accumulated over time. After 3 days of culture, high levels of Regnase-1 accumulated in IL-2/33– and IL-2/25–stimulated Regnase-1AA/AA ILC2s (Figure 1, H and I). Compared with IL-2/25, IL-2/33 induced markedly higher Regnase-1 expression in Regnase-1AA/AA ILC2s. Although phosphorylated Regnase-1 was not observed in IL-2/25–stimulated WT ILC2s, slowly migrating Regnase-1 accumulated in IL-2/25–stimulated Regnase-1AA/AA ILC2s, suggesting that IL-25, similar to IL-17, induces TBK/IKKi-dependent Regnase-1 phosphorylation. Therefore, IKK complex controlled Regnase-1 degradation downstream of IL-33 and IL-25 in ILC2s similar to that downstream of TLR/IL-1 and IL-17, respectively.
Figure 1. IκB kinase complex controls Regnase-1 degradation in ILC2s downstream of IL-33 and IL-25.
(A–E) In vitro–expanded BM ILC2s were stimulated with IL-2 and IL-33 (IL-2/33; A–C) or IL-2 plus IL-25 (IL-2/25; C–E) for the indicated periods. The expression levels of Regnase-1 and ERK2 were determined by immunoblotting. Representative immunoblotting images (A, B, D, and E) and densitometry quantification of Regnase-1 levels (mean [n = 3] ± SD) (C) are shown. (F–I) Freshly isolated BM ILC2s from Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were stimulated with IL-2/33 or IL-2/25 for the indicated periods. (F and G) Intracellular Regnase-1 levels were analyzed by FACS. Representative flow cytometry plots (F) and the quantified graph (mean [n = 3] ± SD) (G) are shown. (H and I) The expression levels of Regnase-1 and ERK2 were determined by immunoblotting. Representative immunoblotting images (H) and densitometry quantification of Regnase-1 levels (mean [n = 3] ± SD) (I) are shown. Data are representative of 2 or 3 independent experiments. Significance was determined by 1-way ANOVA followed by Tukey’s test. **P < 0.01; ***P < 0.001; ****P < 0.0001. Arrows indicate Regnase-1 (Reg1), arrowheads indicate phosphorylated Regnase-1 (p-Reg1). MFI, mean fluorescence intensity.
Regnase-1 degradation is essential for IL-33– and IL-25–induced ILC2 activation.
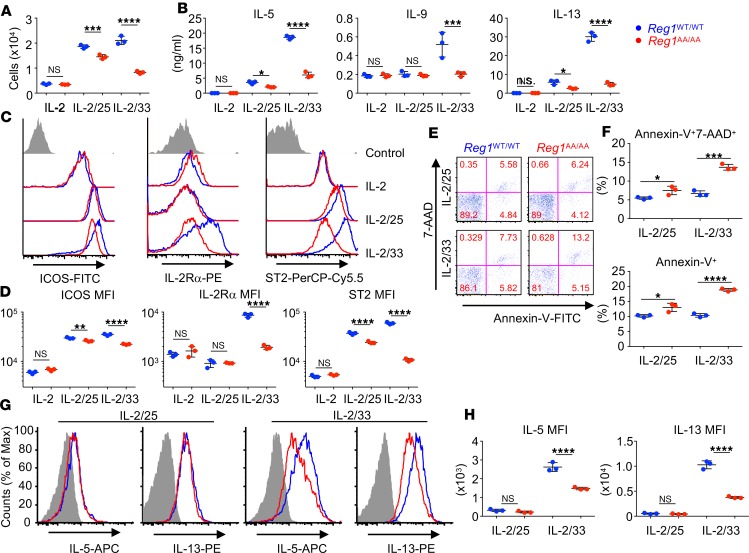
Next, we explored the role of IKK target motif–dependent Regnase-1 degradation in ILC2s. Compared with IL-2 alone, IL-2/33 stimulation induced robust cell proliferation (Figure 2A) and production of type 2 cytokines, IL-5, IL-9, and IL-13 (Figure 2B) from BM ILC2s. IL-2/25 stimulation also induced cell proliferation (Figure 2A) and IL-5 and IL-13, but not IL-9, production (Figure 2B). Regnase-1AA/AA ILC2s showed severely attenuated cell proliferation (Figure 2A) and cytokine production (Figure 2B) in response to IL-2/33. Cell proliferation (Figure 2A) and IL-5 and IL-13 production (Figure 2B) were mildly but significantly decreased in IL-2/25–stimulated Regnase-1AA/AA ILC2s. We also observed defective IL-5 and IL-13 production from Regnase-1AA/AA ILC2s when we used TSLP, another STAT5-activating cytokine, instead of IL-2, suggesting the importance of Regnase-1 downstream of IL-33 and IL-25 rather than downstream of IL-2 signaling (Supplemental Figure 2A). When ILC2s were stimulated with PMA/ionomycin, which bypasses the activation of IRAKs or Act1, Regnase-1WT/WT and Regnase-1AA/AA ILC2s produced comparable amounts of the cytokines (Supplemental Figure 2A). Furthermore, we showed that TPCA-1, an IKKβ inhibitor that suppresses Regnase-1 degradation in IL-1β–stimulated fibroblasts (22), suppressed IL-33– and IL-25–induced IL-5 and IL-13 production from WT ILC2s (Supplemental Figure 2B). Therefore, the defective cytokine production in IL-33– or IL-25–stimulated Regnase-1AA/AA ILC2s should be due to the accumulation of Regnase-1 rather than nonspecific abnormality caused by the mutation.
Figure 2. IκB kinase complex–mediated Regnase-1 degradation is essential for IL-33– and IL-25–induced ILC2 activation.
BM ILC2s from Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were stimulated with IL-2 alone, IL-2 and IL-33 (IL-2/33), or IL-2 and IL-25 (IL-2/25) for 3 days. (A) Live cell number was quantified by FACS. (B) The concentrations of IL-5, IL-9, and IL-13 in the culture supernatants were determined by ELISA. (C and D) Cell surface expressions of ICOS, IL-2Rα, and ST2 were analyzed by FACS. The gray histogram indicates unstained control. (E and F) Cells were stained with annexin V and 7-AAD, and the percentage of dead cells was quantified by FACS. (G and H) Golgi-Stop was added at the final 4 hours of a 3-day culture, and intracellular cytokine levels were analyzed by FACS. Gray histogram indicates unstained control. Data are representative of more than 3 independent experiments. Representative flow cytometry plots (C, E, and G) and mean (n = 3) ± SD (A, B, D, F, and H) are shown. MFI, mean fluorescence intensity. Significance was determined by 1-way ANOVA followed by Tukey’s test (A, B, and D) or 2-tailed Student’s t test (F and H). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
IL-2Rα (38) and ST2 (also known as IL-1RL1 or IL-33Rα) (39) are critical components of the IL-2 and IL-33 receptors, respectively. The costimulatory molecule ICOS is essential for ILC2 activation (40–42). Compared with IL-2 alone, IL-2/33 stimulation upregulated all 3 molecules in WT ILC2s; however, this upregulation was significantly attenuated in Regnase-1AA/AA ILC2s (Figure 2, C and D). ICOS and ST2 levels were slightly but significantly downregulated in IL-2/25–stimulated Regnase-1AA/AA ILC2s. IL-2Rα was not upregulated by IL-2/25 stimulation, even in WT cells. Because forced expression of Regnase-1 induces cellular apoptosis (43, 44), we examined ILC2 cell death after cytokine stimulation. IL-2/33 and IL-2/25 both induced higher cell death (7-aminoactinomycin-D+ [7-AAD+] and/or annexin V+ cells) in Regnase-1AA/AA ILC2s compared with WT cells (Figure 2, E and F). Intracellular IL-5 and IL-13 staining showed decreased cytokine staining in IL-2/33– but not IL-2/25–stimulated Regnase-1AA/AA ILC2s (Figure 2, G and H). IL-9 production promotes ILC2 survival and cytokine production in an autocrine manner (45, 46). However, exogenous IL-9 did not rescue the reduced IL-5/IL-13 production from Regnase-1AA/AA ILC2s (Supplemental Figure 2, C and D).
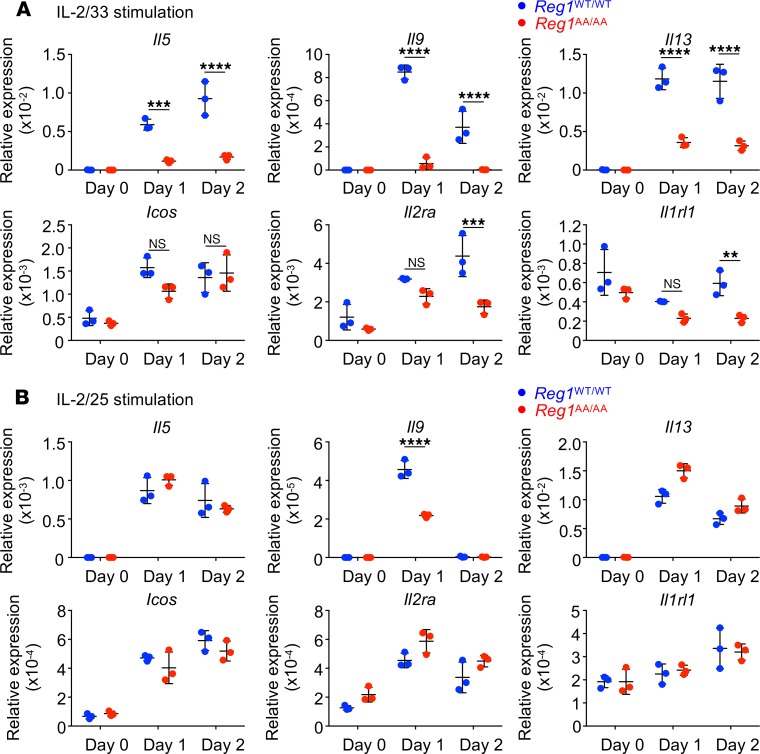
To assess whether decreased cytokine production and surface molecule expression in Regnase-1AA/AA ILC2s occurred at the mRNA level, Regnase-1WT/WT and Regnase-1AA/AA BM ILC2s were stimulated with IL-2/33 or IL-2/25 for 1 or 2 days. The expression of Il5, Il9, Il13, Icos, and Il2ra was upregulated by IL-2/33 stimulation, and Regnase-1AA/AA ILC2s showed significantly decreased expression of these mRNAs, with the exception of Icos (Figure 3A). Although Il1rl1 (which encodes ST2) was not upregulated by IL-2/33 stimulation in WT cells, IL-2/33 stimulation over time decreased Il1rl1 levels in Regnase-1AA/AA ILC2s, and the difference between Regnase-1WT/WT and Regnase-1AA/AA ILC2s was statistically significant on day 2. IL-2/25 stimulation also induced the upregulation of Il5, Il9, Il13, Icos, and Il2ra in ILC2s (Figure 3B). However, the levels of these and Il1rl1 mRNAs were comparable between Regnase-1WT/WT and Regnase-1AA/AA ILC2s, with the exception of Il9, on day 1. Although Gata3, the critical transcription factor for ILC2 functions (47), was a Regnase-1 target gene in CD4+ T cells (21, 34), Gata3 levels were not decreased in Regnase-1AA/AA ILC2s (Supplemental Figure 3). In addition, levels of Il17ra and Il17rb, components of IL-25R, were comparable between Regnase-1WT/WT and Regnase-1AA/AA ILC2s.
Figure 3. Reduced IL-33–inducible mRNA expression in Regnase-1AA/AA ILC2s.
Bone marrow ILC2s from Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were stimulated with IL-2 plus IL-33 (IL-2/33; A) or IL-2 plus IL-25 (IL-2/25; B) for 1 or 2 days. mRNA levels for Il5, Il9, Il13, Icos, Il2ra, and Il1rl1 were determined by quantitative PCR. Data are representative of 2 independent experiments. Mean (n = 3) ± SD are shown. Significance was determined by 2-way ANOVA followed by Bonferroni’s test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
These data demonstrate that the IKK-mediated control of Regnase-1 degradation is essential for IL-33– and IL-25–induced ILC2 activation. Defective IKK-mediated Regnase-1 degradation in ILC2s induces enhanced cell death by IL-33 and IL-25 stimulation and reduces the mRNA levels of IL-33– but not IL-25–inducible genes.
Regnase-1 accumulation in IL-33–stimulated ILC2s destabilizes Il2ra and Il1rl1.
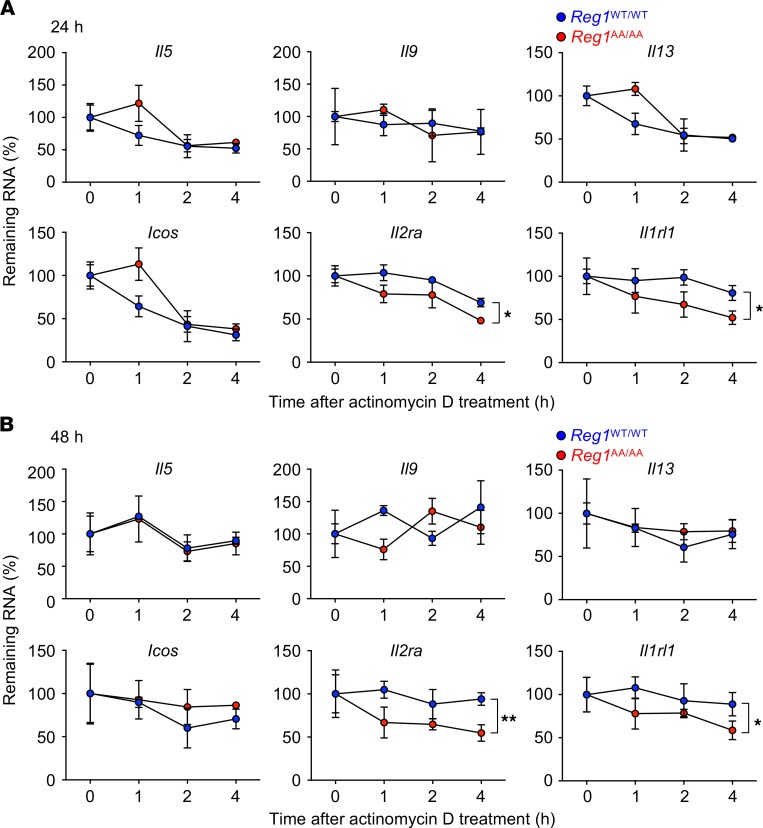
Because several mRNA levels were downregulated in IL-33–stimulated Regnase-1AA/AA ILC2s, we next sought to identify direct target mRNA(s) for Regnase-1 downstream of IL-33 in ILC2s. Regnase-1WT/WT and Regnase-1AA/AA BM ILC2s were stimulated for 24 hours (Figure 4A) or 48 hours (Figure 4B) with IL-2/33 followed by actinomycin D treatment to stop mRNA transcription. Then, the mRNA half-lives within the cells were examined. Il2ra and Il1rl1 were more rapidly degraded in Regnase-1AA/AA ILC2s compared with WT cells after 24 hours (Figure 4A) and 48 hours (Figure 4B) stimulations, while Il5, Il9, Il13, and Icos showed comparable half-lives between Regnase-1WT/WT and Regnase-1AA/AA ILC2s. Regnase-1 started to accumulate after a 4-hour stimulation with IL-2/33 in Regnase-1AA/AA ILC2s (Figure 1, F and G). However, the mRNA half-lives for Il2ra and Il1rl1 were comparable between Regnase-1WT/WT and Regnase-1AA/AA ILC2s at this time point (Supplemental Figure 4), suggesting that relatively high-level Regnase-1 accumulation is required to destabilize Il2ra and Il1rl1.
Figure 4. Il2ra and Il1rl1 are destabilized in IL-33–stimulated Regnase-1AA/AA ILC2s.
BM ILC2s from Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were stimulated with IL-2 plus IL-33 for 24 (A) or 48 hours (B) and then treated with actinomycin D for the indicated periods. mRNA levels for Il5, Il9, Il13, Icos, Il2ra, and Il1rl1 were determined by quantitative PCR, and remaining mRNAs after actinomycin D treatment (relative to 0 hours) were calculated. Data are representative of 2 independent experiments. Mean (n = 3) ± SD are shown. Significance was determined by 2-way ANOVA followed by Bonferroni’s test. *P < 0.05; **P < 0.01.
These results suggest that Regnase-1 accumulation in IL-33–stimulated Regnase-1AA/AA ILC2s destabilizes Il2ra and Il1rl1 mRNAs. Destabilization of the mRNAs may contribute to reduced IL-2R and IL-33R expression and reduced response to the cytokines in IL-33–stimulated Regnase-1AA/AA ILC2s.
Regnase-1AA/AA mice show attenuated IL-33–, IL-25–, or papain-induced acute allergic pulmonary inflammation.
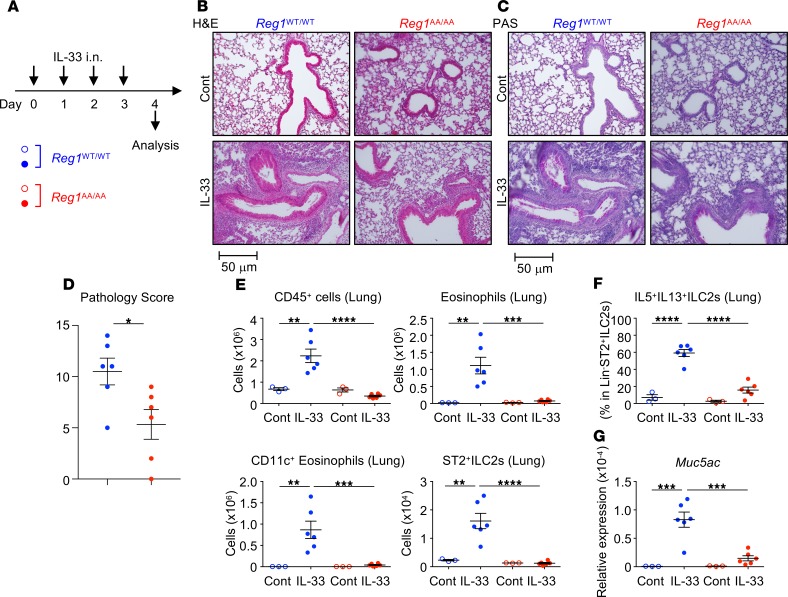
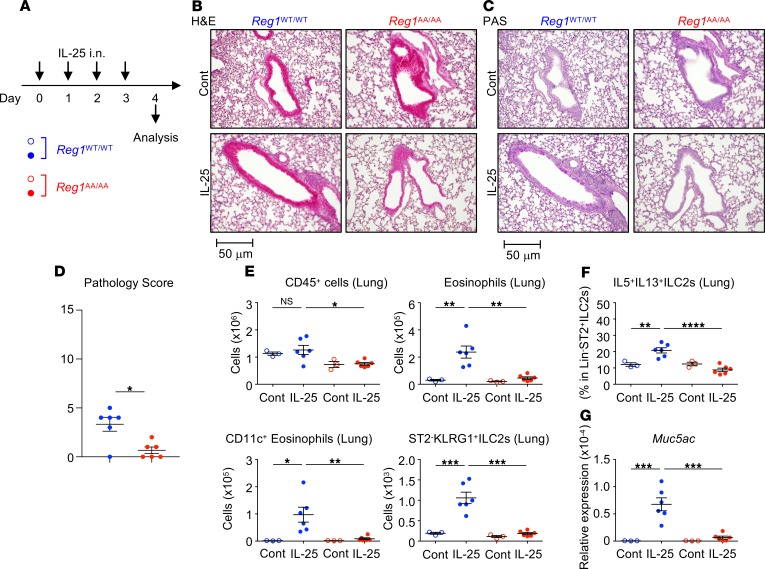
Next, we investigated the in vivo role of Regnase-1 degradation. Regnase-1WT/WT and Regnase-1AA/AA mice were i.n. administered IL-33 for 4 consecutive days, and lung inflammation was analyzed 24 hours after the final challenge (Figure 5A). IL-33–induced lung inflammation was attenuated in Regnase-1AA/AA mice (Figure 5, B–D). Although IL-33 increased the numbers of CD45+ hematopoietic cells and eosinophils in the lungs (Figure 5E and Supplemental Figure 5, A–D) and bronchoalveolar lavages (BALs) (Supplemental Figure 6, A–D) of WT mice, these cells did not accumulate in Regnase-1AA/AA mice. Lung CD11c+ eosinophils represent activated eosinophils (48) and were accumulated in the lungs of IL-33–administered Regnase-1WT/WT but not Regnase-1AA/AA mice (Figure 5E and Supplemental Figure 5D). Numbers of ST2+ ILC2s (CD45+Lin–ST2+CD90.2+Sca-1+ cells) (Figure 5E and Supplemental Figure 5, E and F) and percentages of IL-5/IL-13–producing ILC2s (within CD45+Lin–ST2+ cells) (Figure 5F and Supplemental Figure 5G) in the lungs were markedly lower in IL-33–administered Regnase-1AA/AA mice compared with WT mice. Concomitantly, IL-5 and IL-13 concentrations in BAL fluids (BALFs) were significantly lower in IL-33–administered Regnase-1AA/AA mice (Supplemental Figure 6E). IL-13 induces lung mucin production (49). We found that periodic acid–Schiff+ (PAS+) cells (Figure 5C) and the expression of Muc5ac, which encodes a mucin protein (Figure 5G), were decreased in the lungs of Regnase-1AA/AA mice administered IL-33. Similar to IL-33, Regnase-1AA/AA mice showed decreased IL-25–induced (Figure 6A) pulmonary inflammation (Figure 6, B–D). Eosinophil accumulation in lungs (Figure 6E and Supplemental Figure 7, A–C) was abrogated in Regnase-1AA/AA mice. Eosinophil counts in BALs (Supplemental Figure 8, A–C) also showed a decreased trend in Regnase-1AA/AA mice. As previously reported (50, 51), lung instillation of IL-25 induced ST2–KLRG-1+ rather than ST2+ ILC2s (Figure 6E and Supplemental Figure 7, D–F). Numbers of ST2–KLRG-1+ ILC2s (CD45+Lin–CD90.2+ST2–KLRG-1+ cells) (Figure 6E and Supplemental Figure 7D) and percentages of IL-5/IL-13–producing ILC2s (within CD45+Lin–CD90.2+ cells) (Figure 6F and Supplemental Figure 7E) in the lungs were significantly lower in Regnase-1AA/AA mice compared with WT mice. BALF IL-5 levels showed a decreased trend in Regnase-1AA/AA mice (Supplemental Figure 8D). PAS+ cells (Figure 6C) and Muc5ac expression (Figure 6G) were decreased in the lungs of IL-25–administered Regnase-1AA/AA mice. Although lung administration of IL-25 upregulated the expression of Il33 in WT mice, probably through the induction of inflammation, it was abrogated in Regnase-1AA/AA mice (Supplemental Figure 7G).
Figure 5. Attenuated IL-33–induced pulmonary inflammation in Regnase-1AA/AA mice.
(A) Experimental schema. Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were i.n. administered with IL-33 (100 ng/dose) or PBS (control; Cont) for 4 consecutive days. (B–D) Histological examination. Representative H&E staining (B) and Periodic acid–Schiff staining (PAS) (C) of lung left lobes (scale bar: 50 mm) and the quantified pathology scores of IL-33–treated mice (D) are shown. (E) Total numbers of CD45+ cells (CD45+ singlet cells), eosinophils (CD45+autofluorescence–CD11b+Siglec-F+), CD11c+eosinophils (CD45+autofluorescence–CD11b+Siglec-F+CD11c+), and ST2+ ILC2s (CD45+Lin–ST2+CD90.2+Sca-1+) in the lung left lobes were quantified by FACS. (F) Lung cells were stimulated with PMA and ionomycin for 4 hours. The percentage of IL-5– and IL-13–producing cells within CD45+Lin–ST2+ ILC2s was quantified by intracellular cytokine staining and FACS. (G) mRNA levels for Muc5ac in the lungs were determined by quantitative PCR. Data are representative of 3 independent experiments. Representative images (B and C) and mean (n = 3 for control and n = 6 for IL-33) ± SEM (D–G) are shown. Significance was determined by 2-tailed Student’s t test (D) or 1-way ANOVA followed by Tukey’s test (E–G). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Figure 6. Attenuated IL-25–induced pulmonary inflammation in Regnase-1AA/AA mice.
(A) Experimental schema. Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were i.n. administered with IL-25 (100 ng/dose) or PBS (control; Cont) for 4 consecutive days. (B–D) Histological examination. Representative H&E staining (B) and Periodic acid–Schiff staining (PAS) (C) of lung left lobes (scale bar: 50 mm) and the quantified pathology scores of IL-25–treated mice (D) are shown. (E) Total numbers of CD45+ cells (CD45+ singlet cells), eosinophils (CD45+autofluorescence–CD11b+Siglec-F+), CD11c+eosinophils (CD45+autofluorescence–CD11b+Siglec-F+CD11c+), and ST2–KLRG+ ILC2s (CD45+Lin–CD90.2+ST2–KLRG-1+) in the lung left lobes were quantified by FACS. (F) Lung cells were stimulated with PMA and ionomycin for 4 hours. The percentage of IL-5– and IL-13–producing cells within CD45+Lin–CD90.2+ ILC2s was quantified by intracellular cytokine staining and FACS. (G) mRNA levels for Muc5ac in the lungs were determined by quantitative PCR. Data are representative of 2 independent experiments. Representative images (B and C) and mean (n = 3 for control and n = 6) ± SEM (D–G) are shown. Significance was determined by 2-tailed Student’s t test (D) or 1-way ANOVA followed by Tukey’s test (E–G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
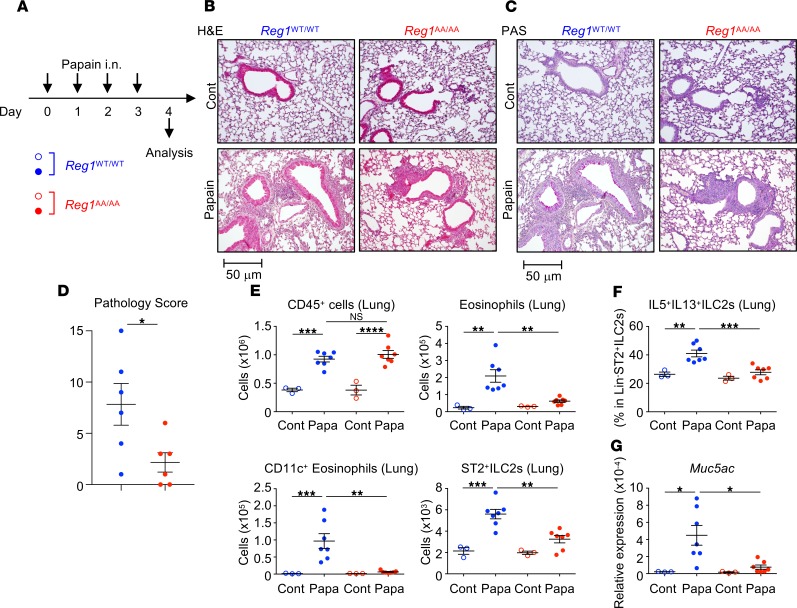
To examine a more physiologically relevant model, we used papain, an allergen that induces ILC2-mediated type 2 pulmonary inflammation (5, 6). Regnase-1WT/WT and Regnase-1AA/AA mice were i.n. administered papain for 4 consecutive days and analyzed 24 hours after the final challenge (Figure 7A). Papain induced lung inflammatory cell infiltration, even in Regnase-1AA/AA mice (Figure 7, B–D). Substantial numbers of CD45+ hematopoietic cells accumulated in the lungs (Figure 7E and Supplemental Figure 9A) and BALs (Supplemental Figure 10, A and B) in Regnase-1AA/AA mice. However, the accumulation of eosinophils in lungs (Figure 7E and Supplemental Figure 9, A–C) and BALs (Supplemental Figure 10, A–C) was severely abrogated in Regnase-1AA/AA mice. Noneosinophil inflammatory cells, including monocytes, neutrophils, macrophages, and lymphocytes, were accumulated in the lungs and BALs of Regnase-1WT/WT and Regnase-1AA/AA mice (Supplemental Figure 9G and Supplemental Figure 10E). This phenotype is similar to that observed for ST2-deficient mice (52). Numbers of ST2+ ILC2s (Figure 7E and Supplemental Figure 9, D and E) and percentages of IL-5/IL-13–producing ILC2s (Figure 7F and Supplemental Figure 9F) in the lungs and BALF IL-5 and IL-13 concentrations (Supplemental Figure 10D) were significantly lower in papain-administered Regnase-1AA/AA mice. PAS+ cells (Figure 7C) and Muc5ac expression (Figure 7G) were decreased in the lungs of papain-administered Regnase-1AA/AA mice. Overall, Regnase-1AA/AA mice showed milder papain-induced lung pathology scores compared with WT mice (Figure 7D). Il33 was upregulated by a single papain administration in WT mice, as previously reported (52), and it was not abrogated in Regnase-1AA/AA mice (Supplemental Figure 9H). Therefore, the defective response to IL-33 may explain the phenotype observed in papain administered Regnase-1AA/AA mice.
Figure 7. Attenuated papain-induced pulmonary inflammation in Regnase-1AA/AA mice.
(A) Experimental schema. Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were i.n. administered with papain (Papa; 10 mg/dose) or PBS (control; Cont) for 4 consecutive days. (B–D) Histological examination. Representative H&E staining (B) and Periodic acid–Schiff staining (PAS) (C) of lung left lobes (scale bar: 50 mm) and the quantified pathology scores of papain–treated mice (D) are shown. (E) Total numbers of CD45+ cells (CD45+ singlet cells), eosinophils (CD45+autofluorescence–CD11b+Siglec-F+), CD11c+eosinophils (CD45+autofluorescence–CD11b+Siglec-F+CD11c+), and ST2+ ILC2s (CD45+Lin–ST2+CD90.2+Sca-1+) in the lung left lobes were quantified by FACS. (F) Lung cells were stimulated with PMA and ionomycin for 4 hours. The percentage of IL-5– and IL-13–producing cells within CD45+Lin–ST2+ ILC2s was quantified by intracellular cytokine staining and FACS. (G) mRNA levels of Muc5ac in the lungs were determined by quantitative PCR. Data are representative of 2 independent experiments. Representative images (B and C) and mean (n = 3 for control and n = 7 for papain) ± SEM (D–G) are shown. Significance was determined by 2-tailed Student’s t test (D) or 1-way ANOVA followed by Tukey’s test (E–G). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
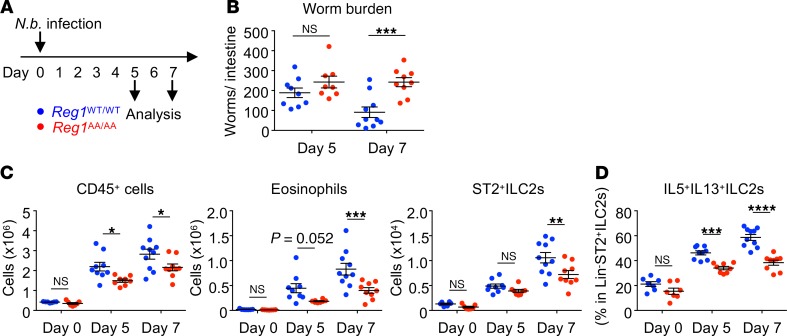
Regnase-1AA/AA mice are susceptible to helminth infection.
Finally, we assessed the role of Regnase-1 degradation in N. brasiliensis infection. Regnase-1WT/WT and Regnase-1AA/AA mice were infected with L3 larvae of N. brasiliensis (Figure 8A). Intestinal worm burden was higher in Regnase-1AA/AA mice than WT mice on day 7 but not on day 5 after infection (Figure 8B), indicating delayed worm expulsion in Regnase-1AA/AA mice. Concomitantly, lung infiltration of CD45+ hematopoietic cells and eosinophils was significantly lower in Regnase-1AA/AA mice (Figure 8C). Although the difference of ST2+ ILC2 numbers between Regnase-1WT/WT and Regnase-1AA/AA mice was relatively small and only statistically significant on day 7 (Figure 8C), the percentage of IL-5/IL-13–producing ILC2s was lower in Regnase-1AA/AA mice on days 5 and 7 (Figure 8D).
Figure 8. Regnase-1AA/AA mice are susceptible to Nippostrongylus brasiliensis infection.
(A) Experimental schema. Regnase-1WT/WT (blue) and Regnase-1AA/AA (red) mice were subcutaneously infected with Nippostrongylus brasiliensis (N.b.). The mice were analyzed at day 5 and 7 after infection. (B) The numbers of worms recovered from small intestines were counted. (C) Total numbers of CD45+ cells (CD45+ singlet cells), eosinophils (CD45+autofluorescence–CD11b+Siglec-F+), and ST2+ ILC2s (CD45+Lin–ST2+CD90.2+Sca-1+) in the lung left lobes were quantified by FACS. (D) Lung cells were stimulated with PMA and ionomycin for 4 hours. The percentage of IL-5– and IL-13–producing cells within CD45+Lin–ST2+ ILC2s was quantified by intracellular cytokine staining and FACS. Pooled data from 2 independent experiments are shown (mean ± SEM). n = 7 (day 0 WT and AA); n = 9 (day 5 WT); n = 8 (day 5 AA); n = 10 (day 7 WT); n = 9 (day 7 AA). Significance was determined by 2-way ANOVA followed by Bonferroni’s test. P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Taken together, these observations demonstrate that Regnase-1 degradation plays a critical role in activating ILC2s in vivo and in acute type 2 pulmonary inflammation.
Discussion
Regnase-1 has been described as a negative regulator of Th1/Th17 inflammatory responses by controlling mRNA stability (20, 21, 32, 33). In this study, we show that Regnase-1 also contributed to type 2 immune responses by regulating IL-33– and IL-25–mediated ILC2 activation.
Although Regnase-1 was identified as an LPS-inducible gene (20), macrophages and fibroblasts constitutively expressed Regnase-1 protein (22). Once cells are activated through TLR/IL-1R, Regnase-1 undergoes degradation, leading to a temporal decrease of Regnase-1, followed by its de novo synthesis. The canonical IKK target motif (DSGxxS) in Regnase-1 (S435/S439) is essential for TLR/IL-1R–induced Regnase-1 degradation. This study demonstrated that the IKK target motif in Regnase-1 was also essential for Regnase-1 degradation in IL-33–stimulated ILC2s, as Regnase-1 accumulated in IL-2/33–stimulated Regnase-1AA/AA ILC2s. In addition to IKKs, IRAK1 phosphorylates Regnase-1 at S494/S513 and forms a slowly migrating Regnase-1, as shown by immunoblotting; this promotes the subcellular translocation of Regnase-1 from the endoplasmic reticulum to the cytoplasm, in which the IKK complex engages with phosphorylated Regnase-1 to facilitate its degradation (22, 23). Upon stimulation of ILC2s with IL-2/33, a slowly migrating Regnase-1 signal was observed within 15 minutes by immunoblotting. Therefore, because IL-33R shares signaling molecules with TLR/IL-1R (25), in IL-2/33–stimulated ILC2s, Regnase-1 can undergo IRAK1-mediated quick phosphorylation followed by IKK-mediated degradation as in TLR/IL-1R–stimulated cells.
Regnase-1 is also involved in IL-17–induced immune responses (32, 33, 53). Regnase-1 S435/S439 controls Regnase-1 degradation in IL-17–stimulated fibroblasts, and TBK1/IKKi are substitutes for the role of IRAK1 (23). Here, we showed that Regnase-1 accumulated in IL-2/25–stimulated Regnase-1AA/AA ILC2s. Although a slowly migrating Regnase-1 was barely detectable in IL-2/25–stimulated WT ILC2s, it accumulated in Regnase-1AA/AA ILC2s. Thus, TBK1/IKKi-mediated Regnase-1 phosphorylation exists downstream of IL-25, but phosphorylated Regnase-1 might be quickly degraded in WT cells. Therefore, because IL-17 and IL-25 belong to the same cytokine family and share Act1 as an adaptor molecule, which interacts with Regnase-1 to induce TBK1/IKKi-mediated phosphorylation (23, 29), Regnase-1 may be controlled downstream of IL-25 in ILC2s by a similar mechanism in IL-17–stimulated cells.
IL-33 induced greater Regnase-1 accumulation in Regnase-1AA/AA ILC2s compared with IL-25. IL-33 was reported to have a greater ILC2-activating capacity compared with IL-25 both in vitro and in vivo (7, 54). This stronger ILC2 activation by IL-33 may explain the higher accumulation of Regnase-1 in IL-2/33–stimulated Regnase-1AA/AA ILC2s. Because of the higher Regnase-1 accumulation, IL-2/33–stimulated Regnase-1AA/AA ILC2s might show a greater decrease in cell proliferation and cytokine production and increase in cell death compared with IL-2/25–stimulated cells. Forced Regnase-1 expression induced cellular apoptosis (44), indicating Regnase-1 accumulation in IL-2/33– or IL-2/25–stimulated Regnase-1AA/AA ILC2s might increase cell death. Intracellular IL-5/IL-13 staining was comparable between IL-2/25–stimulated Regnase-1WT/WT and Regnase-1AA/AA ILC2s, and IL-25–inducible mRNA levels were not decreased in Regnase-1AA/AA ILC2s. Thus, increased ILC2 cell death might decrease cytokine production from IL-2/25–stimulated Regnase-1AA/AA ILC2s. In addition to increased cell death, IL-2/33 stimulation decreased cytokine production on a per-cell-basis and destabilized Il2ra and Il1rl1 mRNAs in Regnase-1AA/AA ILC2s. Although the expression of Il5, Il9, and Il13 mRNAs was decreased in IL-2/33–stimulated Regnase-1AA/AA ILC2s, their stabilities were comparable between Regnase-1WT/WT and Regnase-1AA/AA ILC2s. Therefore, Il5, Il9, and Il13 mRNA might not be direct targets for Regnase-1 in ILC2s, and their reduced expression in IL-2/33–stimulated Regnase-1AA/AA ILC2s might be a secondary effect of decreased signaling from IL-2R and IL-33R. Taken together, the decreased expression of IL-2R and IL-33R and increased cell death might explain decreased cytokine production from IL-2/33–stimulated Regnase-1AA/AA ILC2s.
Because ILC2s are largely tissue-resident (55), the BM ILC2s used for in vitro studies might not share the same characteristics as ILC2s residing in the lungs. However, the results from in vivo IL-33 and IL-25 administration models were similar to those of in vitro experiments with BM ILC2s. IL-33– and IL-25–induced pulmonary inflammation was almost completely abrogated in Regnase-1AA/AA mice. Thus, the role of Regnase-1 degradation might be universal to several ILC2 populations present in different organs. Because IL-33– and IL-25–mediated ILC2 activation plays a central role in acute type 2 pulmonary inflammation induced by allergens (5, 6) or helminth infection (3, 11, 56), defective ILC2 responses should explain attenuated papain-induced lung inflammation and reduced early resistance to N. brasiliensis infection in Regnase-1AA/AA mice. In addition to ILC2s, defects in other innate and adaptive cell populations might be involved in the overall phenotype of Regnase-1AA/AA mice observed in the current study, as Regnase-1 also controls Th2 activation downstream of T cell receptor signaling (34).
Il2ra and Il1rl1 mRNAs were destabilized in IL-33–stimulated but not in IL-25–stimulated Regnase-1AA/AA ILC2s. This might be explained by the amount of Regnase-1 accumulated in ILC2s. Il2ra and Il1rl1 half-lives were comparable between Regnase-1WT/WT and Regnase-1AA/AA ILC2s at 4 hours after IL-2/33 stimulation but were significantly decreased in Regnase-1AA/AA ILC2s at 24 and 48 hours after IL-2/33 stimulation. Therefore, relatively high-level Regnase-1 accumulation might be required to destabilize mRNAs, and Regnase-1 accumulated in IL-2/25–stimulated Regnase-1AA/AA ILC2s might not be sufficient to destabilize the mRNAs. Although the defect in cytokine production in IL-25–stimulated Regnase-1AA/AA ILC2s was milder than that observed in IL-33–stimulated cells, the in vivo response to IL-25 administration was almost completely abrogated in Regnase-1AA/AA mice. Because IL-25 administration induced the upregulation of Il33 in Regnase-1WT/WT but not in Regnase-1AA/AA mice, the phenotype observed in IL-25–administered Regnase-1AA/AA mice should be the sum of the reduced response of the mice to both IL-25 and IL-33. Further study is required to clarify the exact differential role of Regnase-1 and its degradation downstream of IL-33 and IL-25.
The stability of Regnase-1 targets found in CD4+ T cells, such as Icos and Gata3 (21, 34), was not different between Regnase-1WT/WT and Regnase-1AA/AA ILC2s. Regnase-1 might control mRNAs in a cell type–specific manner by interacting with specific partner(s) in the cell population. Alternatively, although Icos and Gata3 mRNAs were stabilized in Regnase-1–deficient CD4+ T cells, excessive Regnase-1 expression in Regnase-1AA/AA cells might not affect the stability.
Because Regnase-1AA/AA ILC2s expressed unusually high levels of Regnase-1 after IL-33 or IL-25 stimulation, the role of Regnase-1 discovered in this study might be specific because of its “overexpression.” Even so, our study demonstrated the potential therapeutic use of Regnase-1 degradation inhibitor(s) for treating allergic disorders. For instance, IKKβ inhibitor(s) are candidate antiasthmatic therapeutics due to their inhibition of NF-κB (57, 58). In addition to suppressing NF-κB, our results suggest that IKKβ inhibitor(s) decrease ILC2 activity by increasing cellular Regnase-1 levels. Because ILC2s and their activation are implicated in the pathogenesis of severe and treatment-resistant asthma (18, 19, 59), Regnase-1 degradation inhibitor(s) might be beneficial for treating severe/refractory asthma.
In summary, we showed that Regnase-1 degradation was essential for ILC2-mediated type 2 responses both in vitro and in vivo. Although we only studied acute type 2 inflammation models, ILC2s might be also involved in chronic and treatment-resistant allergic disorders (15–19). Therefore, controlling Regnase-1 activity is a potential therapeutic target for severe and/or uncontrolled allergic disorders.
Methods
Mice.
C57BL/6 WT mice were purchased from Oriental Yeast. C57BL/6 background Regnase-1AA/AA mice were generated as described previously (23). Mice were maintained under specific pathogen–free conditions at the animal facilities of Hyogo College of Medicine.
Reagents.
Rabbit anti–mouse Regnase-1 mAb (23) and recombinant human IL-33 (60) were prepared as described. Recombinant human IL-2 and mouse IL-9 were purchased from PeproTech. Recombinant and mouse IL-25 and mouse TSLP was purchased from R&D Systems. Actinomycin-D, anti-DNP mouse IgE (SPE-7), ionomycin (A23187), papain from papaya latex, and PMA were from Sigma-Aldrich Japan. TPCA-1 was from abcam. Anti–mouse-IgE mAb (23G3) was purchased from Southern Biotechnology Associates Inc. Anti–mouse-ST2 mAb (DJ8) was purchased from MD Bioproducts. Anti–mouse-ERK2 mAb (C-14) was purchased from Santa Cruz. ECL anti–rabbit IgG HRP-Linked and ECL Prime Western Blotting Detection Reagent were purchased from GE Healthcare. Biotin anti–rabbit IgG was purchased from Vector Laboratories. Annexin V and 7-AAD were purchased from BD Biosciences. Alexa Fluor 488 streptavidin was purchased from Invitrogen. Collagenase and recombinant bovine DNAse-I were purchased from Wako Pure Chemicals. mAbs specific for mouse B220 (RA3-6B2) FITC-conjugated, CD4 (RM4-5) FITC-conjugated, CD11b (M1/70) FITC-conjugated, CD11c (N418) FITC- and biotin-conjugated, CD49b (DX5) FITC-conjugated, CD90.2 (53-2.1) PE-conjugated, and Siglec-F (E50-2440) AlexaFluor647-conjugated were purchased from BD Bioscience. mAbs specific for mouse CD3ε (145-2C11) FITC-conjugated, CD8 (53-6.7) FITC-conjugated, CD11b (M1/70) PE-conjugated, IL-2Rα (PC61) FITC-conjugated, CD45 (30F11) Pacific Blue-conjugated, ICOS (CD15-F9) PE-conjugated, IL-5 (TRFK5) APC-conjugated, KLRG-1 (2F1/KLRG1) Brilliant Violet 510-conjugated, Gr-1 (RB6-8C5) FITC-conjugated, and Sca-1 (D7) APC-conjugated were purchased from BioLegend. mAbs specific for mouse c-Kit (2B8) FITC-conjugated and IL-13 (eBio13A) PE-conjugated were purchased from eBioscience.
Mouse models.
Mice were anesthetized by i.p. injection of Somnopentyl (51.8 μg/g as sodium pentobarbital; Kyoritsu Seiyaku) and IL-33 (100 ng/50 μL/dose), IL-25 (100 ng/50 μL/dose), papain (10 μg/50 μL/dose), or PBS (50 μL/dose as a control) was i.n. administered for 4 consecutive days. Mice were euthanized 24 hours after the final i.n. challenge and analyzed. For N. brasiliensis infection, mice were subcutaneously inoculated with 500 L3 larvae of N. brasiliensis. Worm burden in the small intestine and lung inflammation were analyzed on days 5 and 7 after infection. N. brasiliensis was maintained in male SD rats as previously described (61).
Preparation of ILC2s and in vitro culture.
Lineage– cells were enriched from BM cells using a Lineage Cell Depletion Kit and AutoMACS Separator (Miltenyi Biotech). Then, cells were Fc-blocked with anti-CD16/32, incubated with biotin anti–IL-2Rα antibody and anti–DNP-IgE, and stained using FITC-anti-B220, -CD3, -CD4, -CD8, -CD11b, -CD11c, -CD49b, and -IgE antibodies (lineage markers) as well as PE-anti-CD90.2, PerCP-Cy5.5-streptavidin, APC anti-CD45, and Pacific Blue anti–Sca-1. CD45+Lin–CD90.2+IL-2Rα+Sca-1+ cells were sorted as BM ILC2s using FACS Aria III (BD Biosciences) (Supplemental Figure 1A). In some experiments, ILC2s were expanded for 10 days in complete medium containing IL-2 (3 ng/mL), IL-33 (50 ng/mL), and IL-25 (50 ng/mL). Then, cells were rested in medium only containing IL-2 (3 ng/mL) for 2–3 days before used for experiments. ILC2s were cultured in 96-well palates at 1 × 103 cells/200 μL/well in complete medium containing the combinations of IL-2 (3 ng/mL), TSLP (10 ng/mL), IL-9 (50 ng/mL), IL-33 (100 ng/mL), or IL-25 (100 ng/mL) for varying periods. In some experiments, ILC2s were stimulated with PMA (30 ng/mL) and ionomycin (500 ng/mL). For cytokine production, cell proliferation, or surface molecule expression, cells were cultured for 3 days. Then, the supernatant was harvested and analyzed by ELISA, and cells were subjected to FACS analysis. For intracellular cytokine staining, Golgi-Stop (BD Biosciences) was added for the last 4 hours of culture and cells were subjected to FACS analysis. To analyze mRNA half-life, ILC2s were stimulated with IL-2 (3 ng/mL) and IL-33 (100 ng/mL) for 4, 24, or 48 hours. Then, actinomycin D (2 μg/mL) was added to culture medium to stop transcription. Cells were further cultured for 0, 1, 2, and 4 hours and were collected for quantitative PCR analysis.
Histological examination.
Mice were perfused via the right ventricle with 10 mL PBS. Lung left lobes were dissected from mice and were immediately fixed in 4% paraformaldehyde for 24 hours at 4°C. Samples were embedded in paraffin, and then 4-μm coronal sections were cut with a microtome TU-213 (Yamato Kohki Industrial) and stained with H&E or PAS. Samples were evaluated under an AX80/DP72 microscope (Olympus Life Science). The lung pathology scores were determined by an investigator blinded in terms of bronchial and interstitial cellular infiltration and mucus production (all 0–5 points, thus the maximum potential score is 15).
Preparation of lung and BAL cells.
To prepare lung cells, lung left lobes were dissected from mice and infused with complete medium (RPMI 1640 medium supplemented with 10% fetal bovine serum, 2-ME [50 μM], L-glutamine [2 mM], penicillin [100 U/mL], and streptomycin [100 μg/mL]) containing collagenase (150 U/mL) and DNAse I (10 μg/mL). Then, the samples were minced and digested for 50 minutes at 37°C. Cell suspensions were filtered using a cell strainer and red blood cells were lysed. For FACS analysis, cells were suspended in ice-cold staining buffer (1% BSA in PBS). For intracellular cytokine staining, lung cells were cultured in complete medium containing PMA (30 ng/mL) and ionomycin (500 μg/mL) in the presence of Golgi-Stop (BD Biosciences) for 4 hours. BAL was performed with 3 aliquots of 1 mL PBS per mouse. After centrifugation, BALFs were collected and kept at −80°C for ELISA, and cells were suspended in ice-cold staining buffer for FACS analysis.
FACS analysis.
Cells prepared from in vitro culture, lungs, or BALs were washed in ice-cold staining buffer, Fc blocked with anti-CD16/32 antibody, incubated with each antibody (1:100) for 15 minutes, and washed twice with staining buffer. Intracellular cytokine and Regnase-1 staining were performed using the BD Cytofix/Cytoperm Plus Fixation/permeabilization Kit (BD Biosciences). For intracellular Regnase-1 staining, anti–mouse Regnase-1 mAb and IgG from normal rabbit sera (as a control) (19 μg/mL) were used as primary antibodies. Biotin-conjugated anti–rabbit-IgG mAb (1:500) was used as a secondary antibody, and Alexa Fluor 488 streptavidin (1:500) was used as a tertiary reagent. Data were acquired using a FACS Canto II flow cytometer (BD Biosciences) and analyzed using FlowJo software (version 7.6.1, Tree Star Inc.). To investigate lung inflammatory cells, lung total hematopoietic cells were defined as CD45+ singlet cells (Supplemental Figure 5A). From the CD45+ population, eosinophils (autofluorescence–CD11b+Siglec-F+), monocytes/neutrophils (autofluorescence–CD11b+CD11cintSiglec-F–), macrophages (autofluorescence+CD11chiSiglec-Fhi), lymphocytes (autofluorescence–CD11b–FSCloSSClo), and ILC2s (Lin–ST2+CD90.2+Sca-1+) were defined (Supplemental Figure 9, A–E). In IL-25–administered mice, ILC2s were divided into ST2+ (Lin–CD90.2+ST2+) and ST2–KLRG-1+ (Lin–CD90.2+ST2–KLRG-1+) ILC2s (Supplemental Figure 7D). BAL total hematopoietic cells were defined as CD45+ singlet cells. From the CD45+ population, eosinophils (CD11b+CD11c–Siglec-F+), neutrophils (CD11b+CD11c–Gr-1+Siglec-F–), macrophages (CD11chiSiglec-Fhi), and lymphocytes (CD11b–CD11c–FSCloSSClo) were defined (Supplemental Figure 10, B and C).
ELISA.
Cytokine concentrations in culture supernatants and BALFs were analyzed with mouse IL-5 and IL-13 ELISA DuoSet ELISA Kits (R&D Systems) and the mouse IL-9 ELISA MAX Deluxe Kit (eBioLegend).
Quantitative PCR.
Total RNAs from in vitro–cultured cells and lungs were isolated using the RNeasy Mini Kit (Qiagen) and Sepasol RNA-I Super G (Nakarai), respectively. cDNA was synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo). Gene expression levels were quantified using TaqMan Gene Expression Assays (Applied Biosystems), Premix Ex Taq (Perfect Real Time), and the Thermal cycler dice RT-PCR system (Takara Bio Inc.), with an initial denature at 95°C for 30 seconds, followed by 40–45 cycles of denaturing at 95°C for 5 seconds and annealing and extension at 60°C for 30 seconds. The TaqMan probes used in this study were Il5 (Mm00439646_m1), Il9 (Mm00434305_m1), Il13 (Mm00434204_m1), Il33 (Mm00505403_m1), Icos (Mm00497600_m1), Il2ra (Mm01340213_m1), Il1rl1 (Mm00516117_m1), Il17ra (Mm00434214_m1), Il17rb (Mm00444709_m1), Gata3 (Mm00484683_m1), and 18S rRNA (Applied Biosystems). Results are shown relative to eukaryotic 18S rRNA levels, as determined by the 2-ΔΔC(t) method.
Immunoblotting.
Cells were lysed in lysis buffer (1.0% Nonidet-P40, 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA) containing a Complete Mini protease inhibitor cocktail (Roche). After removing debris by centrifugation, the cell lysates were separated by SDS-PAGE and were transferred onto an Immobilon Membrane (Millipore). Proteins on membranes were visualized with ECL Prime Western Blotting Detection Reagent and ImageQuant LAS4000mini (GE Healthcare). Rabbit anti-mouse Regnase-1 mAb (1.9 μg/mL) and rabbit anti–mouse ERK2 mAb (1:1000) were used as primary antibodies, and ECL anti–rabbit IgG HRP-linked antibody (1:4000) was used as the detection antibody. The quantification of the bands was performed using ImageQuant TL (ver8.1; GE Healthcare). The Regnase-1 levels were shown as relative to ERK2 levels.
Statistics.
Two-tailed Student’s t test (to compare 2 groups), one-way ANOVA followed by Tukey’s test (to compare more than 2 groups), and two-way ANOVA followed by Bonferroni’s test (to analyze time-course experiments) were performed using Prism 6 (version 6.0h, GraphPad Software). P values of less than 0.05 were considered statistically significant.
Study approval.
All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care Committee of Hyogo College of Medicine, which approved these studies (14-007, 16-006, 16-066, and 19-015).
Author contributions
KM designed the study, performed experiments, analyzed data, and wrote the manuscript. HT generated Regnase-1AA/AA mice, provided critical experimental materials, performed experiments, and analyzed data. KY performed N. brasiliensis infection experiments and analyzed data. TA, AF, S. Akasaki, AK, and EK performed experiments and analyzed data. S. Akira generated Regnase-1AA/AA mice, provided critical experimental materials, and supervised the study. TY analyzed data and supervised the study. All authors discussed the results and revised the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science KAKENHI Grant-in-Aid Scientific Research C (16K09927), the Takeda Science Foundation, the Naito Foundation, the MSD Life Science Foundation’s Public Interest Incorporated Foundation (to KM), and the Ministry of Education, Culture, Sports, Science and Technology Grant-in-Aid for Scientific Research’s Strategic Research Foundation at Private Universities (S1001055 to TY). We thank all our colleagues in the Yoshimoto and Akira laboratories for assistance and discussion, C. Minemoto for secretarial assistance, and Y. Miyata and M. Hitomi for technical assistance.
Version 1. 01/28/2020
In-Press Preview
Version 2. 02/27/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(4):e131480.https://doi.org/10.1172/jci.insight.131480.
Contributor Information
Kazufumi Matsushita, Email: mkazu@hyo-med.ac.jp.
Hiroki Tanaka, Email: htanaka@ifrec.osaka-u.ac.jp.
Koubun Yasuda, Email: koubun@hyo-med.ac.jp.
Takumi Adachi, Email: ta-adachi@hyo-med.ac.jp.
Ayumi Fukuoka, Email: Ayumi.Suzuki@glasgow.ac.uk.
Shoko Akasaki, Email: shoko.fufufu-888@docomo.ne.jp.
Atsuhide Koida, Email: atsu-koi@koto.kpu-m.ac.jp.
Etsushi Kuroda, Email: kuroetu@ifrec.osaka-u.ac.jp.
Shizuo Akira, Email: sakira@biken.osaka-u.ac.jp.
Tomohiro Yoshimoto, Email: cytokine.il4@gmail.com.
References
- 1.Moro K, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463(7280):540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 2.Neill DR, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464(7293):1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price AE, et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc Natl Acad Sci USA. 2010;107(25):11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13(2):145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 5.Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012;36(3):451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 6.Morita H, et al. An interleukin-33-mast cell-interleukin-2 axis suppresses papain-induced allergic inflammation by promoting regulatory T cell numbers. Immunity. 2015;43(1):175–186. doi: 10.1016/j.immuni.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein Wolterink RG, et al. Pulmonary innate lymphoid cells are major producers of IL-5 and IL-13 in murine models of allergic asthma. Eur J Immunol. 2012;42(5):1106–1116. doi: 10.1002/eji.201142018. [DOI] [PubMed] [Google Scholar]
- 8.De Grove KC, et al. Dysregulation of type 2 innate lymphoid cells and TH2 cells impairs pollutant-induced allergic airway responses. J Allergy Clin Immunol. 2017;139(1):246–257.e4. doi: 10.1016/j.jaci.2016.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartemes KR, Iijima K, Kobayashi T, Kephart GM, McKenzie AN, Kita H. IL-33-responsive lineage- CD25+ CD44(hi) lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J Immunol. 2012;188(3):1503–1513. doi: 10.4049/jimmunol.1102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013;132(1):205–213. doi: 10.1016/j.jaci.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung LY, et al. IL-33 drives biphasic IL-13 production for noncanonical type 2 immunity against hookworms. Proc Natl Acad Sci USA. 2013;110(1):282–287. doi: 10.1073/pnas.1206587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halim TY, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40(3):425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold MJ, et al. Group 2 innate lymphoid cells facilitate sensitization to local, but not systemic, TH2-inducing allergen exposures. J Allergy Clin Immunol. 2014;133(4):1142–1148. doi: 10.1016/j.jaci.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Gonzalez I, Mathä L, Steer CA, Ghaedi M, Poon GF, Takei F. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity. 2016;45(1):198–208. doi: 10.1016/j.immuni.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Mjösberg JM, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12(11):1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 16.Miljkovic D, et al. Association between group 2 innate lymphoid cells enrichment, nasal polyps and allergy in chronic rhinosinusitis. Allergy. 2014;69(9):1154–1161. doi: 10.1111/all.12440. [DOI] [PubMed] [Google Scholar]
- 17.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134(3):671–678.e4. doi: 10.1016/j.jaci.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SG, et al. Increased numbers of activated group 2 innate lymphoid cells in the airways of patients with severe asthma and persistent airway eosinophilia. J Allergy Clin Immunol. 2016;137(1):75–86.e8. doi: 10.1016/j.jaci.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 19.Nagakumar P, Denney L, Fleming L, Bush A, Lloyd CM, Saglani S. Type 2 innate lymphoid cells in induced sputum from children with severe asthma. J Allergy Clin Immunol. 2016;137(2):624–626.e6. doi: 10.1016/j.jaci.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458(7242):1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 21.Uehata T, et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153(5):1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 22.Iwasaki H, et al. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12(12):1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka H, et al. Phosphorylation-dependent Regnase-1 release from endoplasmic reticulum is critical in IL-17 response. J Exp Med. 2019;216(6):1431–1449. doi: 10.1084/jem.20181078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Liew FY, Girard JP, Turnquist HR. Interleukin-33 in health and disease. Nat Rev Immunol. 2016;16(11):676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- 26.Rickel EA, et al. Identification of functional roles for both IL-17RB and IL-17RA in mediating IL-25-induced activities. J Immunol. 2008;181(6):4299–4310. doi: 10.4049/jimmunol.181.6.4299. [DOI] [PubMed] [Google Scholar]
- 27.Swaidani S, et al. The critical role of epithelial-derived Act1 in IL-17- and IL-25-mediated pulmonary inflammation. J Immunol. 2009;182(3):1631–1640. doi: 10.4049/jimmunol.182.3.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claudio E, et al. The adaptor protein CIKS/Act1 is essential for IL-25-mediated allergic airway inflammation. J Immunol. 2009;182(3):1617–1630. doi: 10.4049/jimmunol.182.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu C, Wu L, Li X. IL-17 family: cytokines, receptors and signaling. Cytokine. 2013;64(2):477–485. doi: 10.1016/j.cyto.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maezawa Y, et al. Involvement of TNF receptor-associated factor 6 in IL-25 receptor signaling. J Immunol. 2006;176(2):1013–1018. doi: 10.4049/jimmunol.176.2.1013. [DOI] [PubMed] [Google Scholar]
- 31.Huang F, Kao CY, Wachi S, Thai P, Ryu J, Wu R. Requirement for both JAK-mediated PI3K signaling and ACT1/TRAF6/TAK1-dependent NF-kappaB activation by IL-17A in enhancing cytokine expression in human airway epithelial cells. J Immunol. 2007;179(10):6504–6513. doi: 10.4049/jimmunol.179.10.6504. [DOI] [PubMed] [Google Scholar]
- 32.Jeltsch KM, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol. 2014;15(11):1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 33.Garg AV, et al. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity. 2015;43(3):475–487. doi: 10.1016/j.immuni.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng H, et al. Monocyte chemotactic protein-induced protein 1 controls allergic airway inflammation by suppressing IL-5-producing TH2 cells through the Notch/Gata3 pathway. J Allergy Clin Immunol. 2018;142(2):582–594.e10. doi: 10.1016/j.jaci.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao R, et al. Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol. 2013;91(5):368–376. doi: 10.1038/icb.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brickshawana A, Shapiro VS, Kita H, Pease LR. Lineage(-)Sca1+c-Kit(-)CD25+ cells are IL-33-responsive type 2 innate cells in the mouse bone marrow. J Immunol. 2011;187(11):5795–5804. doi: 10.4049/jimmunol.1102242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duerr CU, et al. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat Immunol. 2016;17(1):65–75. doi: 10.1038/ni.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldmann TA. The interleukin-2 receptor. J Biol Chem. 1991;266(5):2681–2684. [PubMed] [Google Scholar]
- 39.Schmitz J, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Maazi H, et al. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 2015;42(3):538–551. doi: 10.1016/j.immuni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paclik D, Stehle C, Lahmann A, Hutloff A, Romagnani C. ICOS regulates the pool of group 2 innate lymphoid cells under homeostatic and inflammatory conditions in mice. Eur J Immunol. 2015;45(10):2766–2772. doi: 10.1002/eji.201545635. [DOI] [PubMed] [Google Scholar]
- 42.Kamachi F, Isshiki T, Harada N, Akiba H, Miyake S. ICOS promotes group 2 innate lymphoid cell activation in lungs. Biochem Biophys Res Commun. 2015;463(4):739–745. doi: 10.1016/j.bbrc.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Zhou L, et al. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res. 2006;98(9):1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu W, et al. MCPIP1 selectively destabilizes transcripts associated with an antiapoptotic gene expression program in breast cancer cells that can elicit complete tumor regression. Cancer Res. 2016;76(6):1429–1440. doi: 10.1158/0008-5472.CAN-15-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner JE, et al. IL-9-mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J Exp Med. 2013;210(13):2951–2965. doi: 10.1084/jem.20130071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohapatra A, Van Dyken SJ, Schneider C, Nussbaum JC, Liang HE, Locksley RM. Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis. Mucosal Immunol. 2016;9(1):275–286. doi: 10.1038/mi.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoyler T, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37(4):634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motomura Y, et al. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity. 2014;40(5):758–771. doi: 10.1016/j.immuni.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Grünig G, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282(5397):2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y, et al. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat Immunol. 2015;16(2):161–169. doi: 10.1038/ni.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallrapp A, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature. 2017;549(7672):351–356. doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agoro R, et al. IL-1R1-MyD88 axis elicits papain-induced lung inflammation. Eur J Immunol. 2016;46(11):2531–2541. doi: 10.1002/eji.201646366. [DOI] [PubMed] [Google Scholar]
- 53.Monin L, et al. MCPIP1/Regnase-1 restricts IL-17A- and IL-17C-dependent skin inflammation. J Immunol. 2017;198(2):767–775. doi: 10.4049/jimmunol.1601551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barlow JL, et al. IL-33 is more potent than IL-25 in provoking IL-13-producing nuocytes (type 2 innate lymphoid cells) and airway contraction. J Allergy Clin Immunol. 2013;132(4):933–941. doi: 10.1016/j.jaci.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Moro K, et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol. 2016;17(1):76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 56.Yasuda K, et al. Contribution of IL-33-activated type II innate lymphoid cells to pulmonary eosinophilia in intestinal nematode-infected mice. Proc Natl Acad Sci USA. 2012;109(9):3451–3456. doi: 10.1073/pnas.1201042109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Birrell MA, et al. Ikappa-B kinase-2 inhibitor blocks inflammation in human airway smooth muscle and a rat model of asthma. Am J Respir Crit Care Med. 2005;172(8):962–971. doi: 10.1164/rccm.200412-1647OC. [DOI] [PubMed] [Google Scholar]
- 58.Ogawa H, et al. IκB kinase β inhibitor IMD-0354 suppresses airway remodelling in a Dermatophagoides pteronyssinus-sensitized mouse model of chronic asthma. Clin Exp Allergy. 2011;41(1):104–115. doi: 10.1111/j.1365-2222.2010.03564.x. [DOI] [PubMed] [Google Scholar]
- 59.Kabata H, et al. Thymic stromal lymphopoietin induces corticosteroid resistance in natural helper cells during airway inflammation. Nat Commun. 2013;4:2675. doi: 10.1038/ncomms3675. [DOI] [PubMed] [Google Scholar]
- 60.Kondo Y, et al. Administration of IL-33 induces airway hyperresponsiveness and goblet cell hyperplasia in the lungs in the absence of adaptive immune system. Int Immunol. 2008;20(6):791–800. doi: 10.1093/intimm/dxn037. [DOI] [PubMed] [Google Scholar]
- 61.Matsumoto M, et al. IgG and IgE collaboratively accelerate expulsion of Strongyloides venezuelensis in a primary infection. Infect Immun. 2013;81(7):2518–2527. doi: 10.1128/IAI.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.