Abstract
Customizable nanostructures built through the DNA-origami technique hold tremendous promise in nanomaterial fabrication and biotechnology. Despite the cutting-edge tools for DNA-origami design and preparation, it remains challenging to separate structural components of an architecture built from — thus held together by — a continuous scaffold strand, which in turn limits the modularity and function of the DNA-origami devices. To address this challenge, here we present an enzymatic method to clean up and reconfigure DNA-origami structures. We target single-stranded (ss) regions of DNA-origami structures and remove them with CRISPR-Cas12a, a hyper-active ssDNA endonuclease without sequence specificity. We demonstrate the utility of this facile, selective post-processing method on DNA structures with various geometrical and mechanical properties, realizing intricate structures and structural transformations that were previously difficult to engineer. Given the biocompatibility of Cas12a-like enzymes, this versatile tool may be programmed in the future to operate functional nanodevices in cells.
Keywords: DNA nanotechnology, DNA origami, CRISPR-Cas, Molecular devices, Self-assembly
Graphical Abstract
Free at last: DNA-origami nanostructures are set in motion by trans-activated Cas12a that non-specifically chips away single-stranded loops, handles, and tension-loaded tethers. This post-assembly modification method greatly simplifies the design and fabrication of reconfigurable DNA structures, opening up opportunities to operate DNA nanodevices in cells.

Structural DNA nanotechnology generates programmable shapes, patterns, and motions at length scales from a few to thousands of nanometers.[1] Through DNA-directed assembly, it provides an engineering platform to organize guest molecules with spatial and temporal control.[2] DNA origami, which folds a kilobase long single-stranded DNA (ssDNA, termed scaffold strand) with many synthetic oligonucleotides (termed staple strands), has become a popular design strategy, mainly because using enzymatically produced scaffold strands reduces the self-assembly error and production cost.[3] However, these benefits come at the cost of modularity: with the entirety of a DNA structure tethered by a continuous strand, releasing or swapping components becomes challenging. For example, although dynamic, multi-component DNA-origami structures have been constructed — some by exploiting the programmable tension of a partially unstructured scaffold strand,[4] the covalently linked chain of DNA scaffold constrains component mobility, hence limiting potential device functions. Even in static structures, an often-encountered issue is that the unstructured scaffold strand could draw undesired interactions and cloud microscopy studies. Therefore, there remains a need for methods that break scaffolds of pre-folded DNA-origami structures at specific locations while preserving the structures’ integrity and chemical modifications.
Towards this goal, we searched for ssDNA-specific nucleases that function under the ionic and thermal conditions necessary for the stability of DNA-origami structures. We considered known endonucleases from the S1-P1 family, including P1, S1 and mung bean nucleases,[5] as well as the CRISPR-Cas12a (Cpf1), which can be activated by its sequence-specific double-stranded DNA (dsDNA) target (ssDNA activators are less efficient) for nonspecific ssDNA trans-cleavage.[6] To evaluate the nucleases’ DNA-origami post-processing capabilities, we adapted the “force clamp” structure designed by the Liedl group.[7] The structure has a tensioned scaffold strand suspended on a C-clamp. Cleavage of this 87-nt to 276-nt long ssDNA segment can be detected via the release of a pre-hybridized, Cy5-labeled ssDNA probe (Figure 1a, top). By adjusting the length of the suspended scaffold DNA, we built 3 versions of the force clamp containing ssDNA under tensions of 2, 6, and 12 pN (Figure S1–S3).
Figure 1.
Searching for the appropriate nuclease to process ssDNA of a DNA-origami structure. a) Schematics of DNA-origami force clamps used for testing ssDNA scaffold digestion by nucleases (top) and deoxyribozymes (bottom). U-shaped block: DNA-origami force clamp, strand with a star: Cy5-labeled probe, dark ribbon: unhybridized scaffold strand. Inset: 2 deoxyribozymes in tandem (triangles: cleavage sites). b) ssDNA cleavage of 2 pN force clamps by different nucleases at various temperatures after 30 minutes. Top: agarose gel images showing the nuclease-treated force clamps in Cy5 and ethidium bromide (EtBr) channels. Bottom: electron micrographs of the gel-extracted force clamps. Scale bar: 100 nm. c) Kinetics of Cas12a cleavage on 2, 6 and 12 pN force clamps. d) Kinetics of deoxyribozyme self-cleavage on 2, 6 and 12 pN force clamps. Error bars in (c) and (d): standard deviations (n=3).
We first tested the nucleases with the 2 pN force clamp under various temperatures. All nucleases liberated the Cy5 probe bound to the ssDNA in 30 minutes in a temperature-dependent manner (Figure 1b). However, band shifts in agarose gel electrophoresis (AGE) and structures captured by transmission electron microscopy (TEM) indicated that S1 and mung bean nucleases caused damages to the main body of the C-clamp. In contrast, the C-clamps largely maintained structural integrity after being processed by the trans-activated LbCas12a (hereon referred to as Cas12a) and P1 nucleases (Figure S4–S8). The structural damages caused by the S1 and mung bean nucleases are consistent with their reported weak activities of dsDNA digestion, and may be exacerbated by the acidic conditions optimal for their performance.[5] We then tested selected nucleases with the 6 and 12 pN versions of the force clamp. With slower kinetics, Cas12a cut these ssDNA segments without significantly deforming the clamps (Figure 1c & S9–S16). The reduced Cas12a activities at higher tensions could be attributed to difficulties in threading highly-stretched ssDNA through the enzyme’s active site. Likewise, P1 nuclease cleaved ssDNA under 12 pN while preserving the clamp body (Figure S17–S20).
Instead of protein nucleases, one could incorporate deoxyribozyme sequences[8] into the DNA-origami scaffold strand and trigger their self-cleavage by 1–2 mM Zn2+.[3e, 9] We folded the force clamps with such a deoxyribozyme-containing scaffold to study the Zn2+-dependent cleavage of tensioned ssDNA. Here, 2 identical Class I deoxyribozymes flank a Cy5-probe binding region, which releases from the structure upon cleavage (Figure 1a, bottom). Similar to Cas12a and P1 nucleases, deoxyribozymes cleaved the suspended ssDNA tensioned at 2 or 6 pN, albeit slower than the self-cleavage of untensioned deoxyribozymes (Figure 1d & S21). Interestingly, the increase from 2 to 6 pN marginally accelerated Cy5-probe release. We interpret this as the result of reduced cross-hybridization of the 2 sequence-homologous hairpin structures under moderate stretching. However, 12 pN of tension completely diminished the activity of deoxyribozyme (Figure 1d & S22–S23), likely resulting from the force-induced unzipping of its hairpin.[10]
Though requiring crRNA and activator DNA strands for trans-ssDNA cleavage,[6] Cas12a’s independency of Zn2+ or specific nucleic acid sequence lends itself to a wider range of future in vivo applications than P1 nuclease[11] and deoxyribonucleases. Additionally, Cas12a’s crRNA can be engineered to integrate molecular logic circuits,[12] allowing potential regulation of nuclease activity in response to nucleic acid inputs. Given these advantages, we chose Cas12a for the rest of this study.
We started by establishing that Cas12a can trim off an unfolded 1.9 kb scaffold loop of a DNA-origami semicircle within 30 minutes, despite the loop’s greater length than the stretched scaffolds’ in force clamps (Figure 2a & S24–26). Considering DNA-origami structures usually display staple extensions (termed handles) for subsequent chemical modifications (e.g., fluorophores, lipids, and proteins), we reasoned such ssDNA handles must be pre-hybridized to evade Cas12a cleavage. To confirm this, we constructed DNA-origami circles[13] with 8 outward-facing handles. As expected, when premixed with Cy5-labeled complementary ssDNA (anti-handles), the circle retained its fluorophores after 30 minutes of Cas12a digestion (Figure 2b & S27–S30). Conversely, circles directly treated by Cas12a could not bind to the Cy5 anti-handles because of handle losses. These results indicate that the chemical modifications introduced to DNA-origami structures via handle/anti-handle hybridization are preserved after Cas12a post-processing.
Figure 2.
Cas12a activities on DNA-origami structures. Agarose gel images, representative electron micrographs, and cartoon models are shown from left to right. Structures in electron micrographs are extracted from the corresponding bands in agarose gels. a) Cas12a-mediated removal of an unfolded scaffold segment from a DNA-origami semicircle. Digestion time: 30 minutes. Scale bar: 50 nm. b) Selective digestion of unhybridized handles of a DNA-origmai circle by Cas12a. Cas12a, if present before the addition of Cy5 anti-handles, was removed by electrophoresis. Gel images are shown in Cy5 and EtBr channels (1st/2nd: order of reagent addition. NA: not added). Digestion time: 30 minutes. Scale bar: 50 nm. c) Selective release of DNA-origami circles through Cas12a-cleavage of their single-stranded tethers to a DNA rod. Tether protection is achieved by prehybridization. Digestion time: 4 hours. Scale bar: 100 nm.
We then attempted to controllably release structural modules from a multi-component DNA assembly by selective Cas12a cleavage of tethering scaffold strand. For a proof-of-concept demonstration, we designed an origami structure with a rod threading through 3 circles, each tethered to the rod via 3 pairs of 21-nt ssDNA scaffold (tensioned at ~5 pN, Figure S31). Like with staple extensions, here we expect Cas12a to digest only unhybridized scaffold tethers but leave pre-hybridized scaffold strands intact, regardless of their sequences or positions in the DNA structure. Therefore, by selectively protecting the ssDNA tethers via pre-hybridization, we could release specific circles from the rod (Figure 2c & S32–S36). For instance, when all scaffold tethers were protected, both the AGE band mobility and TEM images of Cas12a-treated structures resembled those of undigested controls. However, when a terminal circle’s tethers were unprotected, Cas12a generated 2 major degradation products: 2 circles attached to a rod, and a free circle. We note that even after 4 hours of incubation, a fraction of the structures appeared unprocessed. This could be attributed to the limited accessibility of the tethers caused by incorrect folding of the DNA-origami structure or low clearance between the rod and circles. Intriguingly, protecting only the terminal circles’ tethers led to a mobile circle (macrocycle) trapped between 2 stationary circles (stoppers) on the rod (shaft) to form a rotaxane after Cas12a digestion.
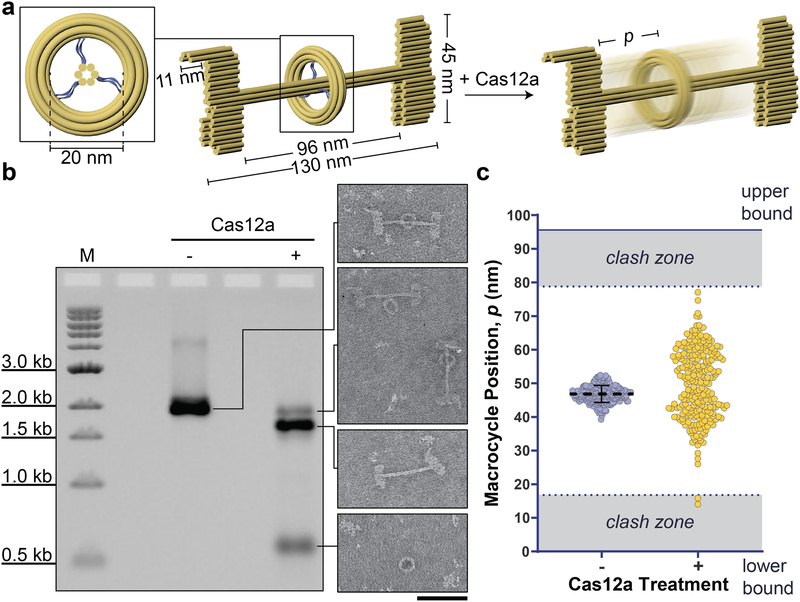
To further investigate the self-assembly and enzymatic releasing efficiencies in synthesizing nanoscopic rotaxanes, a class of mechanically interlocked structures useful for generating molecular switches,[14] we designed a DNA-origami rotaxane with asymmetric stoppers and a longer distance between them. Past attempts to build DNA-origami rotaxanes began with multi-step assembly of components constructed from separate scaffold strands, followed by strand-displacement-mediated release of the macrocycle.[15] In our design, the rotaxane modules — macrocycle, stoppers, and shaft — were first assembled from a single scaffold in one pot, with the macrocycle tethered to the shaft via 6 ssDNA segments (Figure 3a & S37). Subsequently, the tethers were digested by Cas12a for 4 hours to mobilize the macrocycle for free diffusion along the shaft between the stoppers. The one-pot assembly yielded pseudo-rotaxanes (rotaxanes with tethered macrocycles). Upon Cas12a cleavage, ~20% of the stopper-bearing shafts (dumbbells) retained macrocycles and ran as a distinct band after AGE (Figure 3b & S38). The remaining dumbbells separated from the macrocycle, probably because of improper macrocycle threading in initially misfolded pseudo-rotaxanes. After extracting the macrocycle-bearing dumbbells from gel, we identified ~50% of the structures (n=508) as rotaxanes in TEM images, showing a drastic redistribution of macrocycle positions between the stoppers (Figure 3c & S39–S41). The other half of the population within this band appeared as dumbbells with a dethreaded macrocycle attached to the center, which we interpret as a result of the incomplete digestion of initially misfolded pseudo-rotaxanes. Therefore, our overall yield of rotaxane averages ~10% across 3 trials, which is comparable to those generated through multi-step assembly pathways, but without the time-consuming procedures of purifying structural components and sequentially putting them together (a few days to a week). With future optimization of the pseudo-rotaxane design, for example by tuning the positioning and tensions of tethers, we envision such nuclease-mediated reconfiguration methods to enable syntheses of rotaxane-like devices at higher efficiencies.
Figure 3.
Cas12a-facilitated construction of a DNA-origami rotaxane. a) Schematic of the pseudo-rotaxane (left), and the resulting rotaxane (right) after Cas12a removes the 6 ssDNA tethers (blue) joining the macrocycle to the shaft (inset). b) Cas12a-mediated macrocycle release. Left: An EtBr-stained agarose gel image showing the pseudo-rotaxane before and after 4 hours of Cas12a treatment. Right: electron micrographs of the structures within the corresponding gel bands. Scale bar: 100 nm. c) Macrocycle positions measured from electron micrographs of pseudo-rotaxanes (left: n = 208) and rotaxanes (right: n = 251). Clash zones are where the macrocycle would overlap with the terminal stoppers when lying flat on a TEM grid. Upper and lower boundaries are defined by the distance between the stoppers.
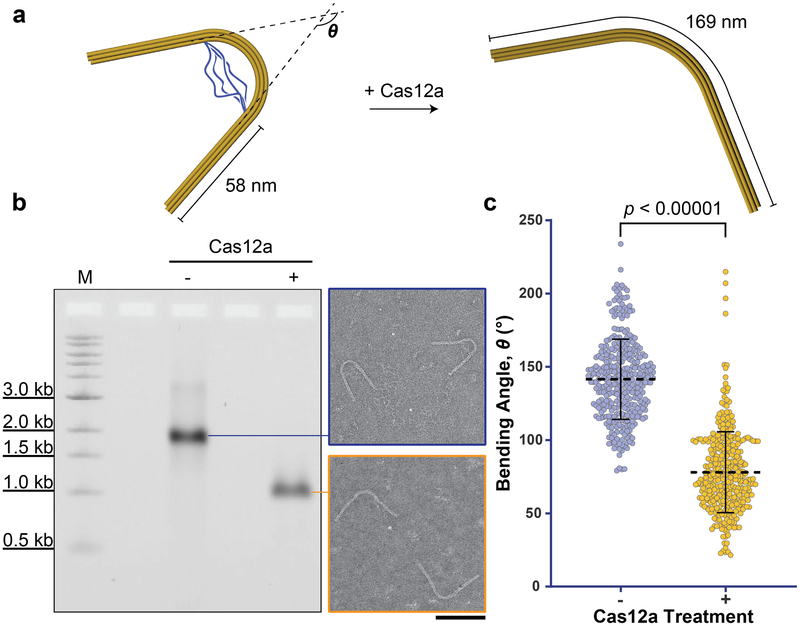
Next, we designed a prestressed elastic beam structure that upon Cas12a-processing, releases its elastic potential energy and changes conformation (Figure 4a & S42). The beam is a 169-nm 6-helix bundle comprising 2 straight arms (each 58 nm) flanking an arc bent by base insertions and deletions[16] as well as 4 tethering ssDNA loops. The combined effect is a bending angle of 142±27 degrees (Figure 4b–4c & S43–S44), consistent with theoretical calculation[17] based on a worm-like chain model[18] for the tethers and a toy model[16] for the beam (see Supporting Information: Materials and Methods). Cas12a should digest the ssDNA tethers and alleviate the initial stress, resulting in the beam’s adoption of a smaller bending angle dictated only by the insertions and deletions within the arc. Indeed, judging from the AGE data, the reconfiguration was efficient (i.e., no detectable side products). Almost all structures treated by Cas12a for 4 hours changed gel mobility owing to tether removal (Figure 4b), which collectively exhibited a down-shifted distribution of bending angles as expected (Figure 4c, S45–S47). In contrast, a beam of the same dimensions containing a bent domain without ssDNA tethers maintained its bending angle after Cas12a treatment (Figure S48–S53), confirming the structural switching is driven by endonuclease cleavage of the pre-tensioned tethers.
Figure 4.
Cas12a-catalyzed reconfiguration of a nanoscopic elastic beam. a) Schematic of the elastic beam undergoing a change in bending angle after Cas12a removes the ssDNA scaffold (blue) tethering its 2 extended arms. b) Cas12a-catalyzed beam reconfiguration characterized by agarose gel electrophoresis (left) and negative-stain TEM (right). Digestion time: 4 hours. Scale bar: 100 nm. c) Quantification of the beams’ bending angles without (left: n = 372) and with (right: n = 338) Cas12a treatment.
In conclusion, we demonstrated that ssDNA-specific nucleases, represented here by Cas12a, are versatile tools for DNA-origami structure modification via selective removal of unpaired scaffold or staple strands. This includes seemingly trivial but otherwise hard-to-achieve tasks like trimming excess scaffold for “pristine” structures. Moreover, this method could trigger conformational changes of a “unibody” DNA-origami structure, potentially enabling greater engineering intricacies without the complexities of multicomponent, stepwise self-assembly. For example, we envision incorporating the elastic beam’s prestressed, switchable design into DNA “nanosprings”[19], thereby enabling membrane tubule constriction on demand. A possible limitation of this technique is the enzyme’s compatibility with detergent, used in constructing DNA-templated bilayers or membrane-protein assemblies[20]. To address this, we verified the potency of Cas12a in up to 5% octyl β-D-glucopyranoside, a commonly used non-ionic detergent for membrane reconstitution (Figure S54). Additionally, force clamps did not aggregate after Cas12a digestion (Figure S9–S13), suggesting their short poly-dT ends remained[21]. Other constraints, including dsDNA damages with excessive enzyme concentration or reaction time (Figure S55–S56) and enzyme accessibility to short, recessed ssDNA (Figure S57–S58), need to be considered when applying this method. Because Cas12a digestion does not require toehold or protospacer adjacent motifs on ssDNA substrates, it is compatible with DNA structures built from virtually any sequences and uniquely suitable for post-processing of single-stranded DNA origami[22]. Further, Cas12a with switchable activities has promising applications in vivo, such as targeted activation of functional nanodevices in response to nucleic acid inputs in different cellular states.
Supplementary Material
Acknowledgements
We thank H. Gu for helpful discussions on deoxyribozyme sequence design. This work is supported by a National Institutes of Health (NIH) Director’s New Innovator Award (GM114830), an NIH grant (GM132114), and a Yale University faculty startup fund to C.L., an Agency for Science, Technology and Research Graduate Scholarship (Singapore) to Q.X., a China Scholarship Council fellowship to C.X., and a National Science Foundation Graduate Fellowship to J.T.P.
Footnotes
Supporting information for this article is available on the WWW under ___________________________.
References
- [1].a) Pinheiro AV, Han D, Shih WM, Yan H, Nat Nanotechnol 2011, 6, 763–772; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Seeman NC, Sleiman HF, Nat Rev Mater 2017, 3, 17068. [Google Scholar]
- [2].a) Aldaye FA, Palmer AL, Sleiman HF, Science 2008, 321, 1795–1799; [DOI] [PubMed] [Google Scholar]; b) Yang YR, Liu Y, Yan H, Bioconjugate Chem 2015, 26, 1381–1395; [DOI] [PubMed] [Google Scholar]; c) Ramezani H, Dietz H, Nat Rev Genet 2019, doi: 10.1038/s41576-019-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Rothemund PW, Nature 2006, 440, 297–302; [DOI] [PubMed] [Google Scholar]; b) Shih WM, Lin C, Curr Opin Struct Biol 2010, 20, 276–282; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tørring T, Voigt NV, Nangreave J, Yan H, Gothelf KV, Chem Soc Rev 2011, 40, 5636–5646; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Hong F, Zhang F, Liu Y, Yan H, Chem Rev 2017, 117, 12584–12640; [DOI] [PubMed] [Google Scholar]; e) Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D, Dietz H, Nature 2017, 552, 84–87. [DOI] [PubMed] [Google Scholar]
- [4].a) Liedl T, Högberg B, Tytell J, Ingber DE, Shih WM, Nat Nanotechnol 2010, 5, 520–524; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Marras AE, Zhou L, Su HJ, Castro CE, Proc Natl Acad Sci U S A 2015, 112, 713–718; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang Z, Yang Y, Pincet F, Llaguno MC, Lin C, Nat Chem 2017, 9, 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Koval T, Dohnálek J, Biotechnol Adv 2018, 36, 603–612. [DOI] [PubMed] [Google Scholar]
- [6].Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA, Science 2018, 360, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nickels PC, Wünsch B, Holzmeister P, Bae W, Kneer LM, Grohmann D, Tinnefeld P, Liedl T, Science 2016, 354, 305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Gu H, Furukawa K, Weinberg Z, Berenson DF, Breaker RR, J Am Chem Soc 2013, 135, 9121–9129; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Du X, Zhong X, Li W, Li H, Gu H, ACS Catal 2018, 8, 5996–6005. [Google Scholar]
- [9].Engelhardt FAS, Praetorius F, Wachauf CH, Brüggenthies G, Kohler F, Kick B, Kadletz KL, Pham PN, Behler KL, Gerling T, Dietz H, ACS Nano 2019, 13, 5015–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Rief M, Clausen-Schaumann H, Gaub HE, Nat Struct Biol 1999, 6, 346–349; [DOI] [PubMed] [Google Scholar]; b) Bockelmann U, Essevaz-Roulet B, Heslot F, Phys Rev Lett 1997, 79, 4489–4492. [Google Scholar]
- [11].Maret W, Int J Mol Sci 2017, 18, 2285. [Google Scholar]
- [12].Oesinghaus L, Simmel FC, Nat Commun 2019, 10, 2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fisher PDE, Shen Q, Akpinar B, Davis LK, Chung KKH, Baddeley D, Šarić A, Melia TJ, Hoogenboom BW, Lin C, Lusk CP, ACS Nano 2018, 12, 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Valero J, Lohmann F, Famulok M, Curr Opin Biotechnol 2017, 48, 159–167; [DOI] [PubMed] [Google Scholar]; b) Jester SS, Famulok M, Acc Chem Res 2014, 47, 1700–1709; [DOI] [PubMed] [Google Scholar]; c) Lu CH, Cecconello A, Willner I, J Am Chem Soc 2016, 138, 5172–5185. [DOI] [PubMed] [Google Scholar]
- [15].a) Powell JT, Akhuetie-Oni BO, Zhang Z, Lin C, Angew Chem Int Ed Engl 2016, 55, 11412–11416; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) List J, Falgenhauer E, Kopperger E, Pardatscher G, Simmel FC, Nat Commun 2016, 7, 12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dietz H, Douglas SM, Shih WM, Science 2009, 325, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhou L, Marras AE, Su HJ, Castro CE, ACS Nano 2014, 8, 27–34. [DOI] [PubMed] [Google Scholar]
- [18].Marko JF, Siggia ED, Macromolecules 1995, 28, 8759–8770. [Google Scholar]
- [19].Grome MW, Zhang Z, Pincet F, Lin C, Angew Chem Int Ed Engl 2018, 57, 5330–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) Perrault SD, Shih WM, ACS Nano 2014, 8, 5132–5140; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yang Y, Wang J, Shigematsu H, Xu W, Shih WM, Rothman JE, Lin C, Nat Chem 2016, 8, 476–483; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dong Y, Chen S, Zhang S, Sodroski J, Yang Z, Liu D, Mao Y, Angew Chem Int Ed Engl 2018, 57, 2072–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhao Z, Zhang M, Hogle JM, Shih WM, Wagner G, Nasr ML, J Am Chem Soc 2018, 140, 10639–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li S, Cheng Q, Liu J, Nie X, Zhao G, Wang J, Cell Res 2018, 28, 491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Han D, Qi X, Myhrvold C, Wang B, Dai M, Jiang S, Bates M, Liu Y, An B, Zhang F, Yan H, Yin P, Science 2017, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.