Abstract
Protein semisynthesis – defined herein as the assembly of a protein from a combination of synthetic and recombinant fragments - is a burgeoning field of chemical biology that has impacted many areas in the life sciences. In this review, we provide a comprehensive survey of this area. We begin by discussing the various chemical and enzymatic methods now available for the manufacture of custom proteins containing non-coded elements. This section begins with a discussion of methods that are more chemical in origin and ends with those that employ biocatalysts. We also illustrate the commonalities that exist between these seemingly disparate methods and show how this is allowing for the development of integrated chemoenzymatic methods. This methodology discussion provides the technical foundation for the second part of the review where we cover the great many biological problems that have now been addressed using these tools. Finally, we end the piece with a short discussion on the frontiers of the field and the opportunities available for the future.
1. INTRODUCTION
Nearly twenty years after the completion of the human genome project,1 we have an ever-increasing understanding of the ~ 20,000 human genes and the roles of the proteins which they encode. At the time the catalogue was first announced, it represented a modest number of entries considering the ostensible complexity of the human organism. Today it is apparent that the total size of the human proteome vastly exceeds this genomic tally, a discrepancy explained by an explosion in the molecular diversity arising through spliced isoforms and post-translational modifications (PTMs). The nature of each of these unique proteoforms and how they contribute to cellular biology is the subject of extensive research,2 one that increasingly relies on our ability to manipulate the chemical structure of this class of biomolecules both in vitro and in vivo.
The primary sequence of recombinant proteins may be modified through site-directed mutagenesis, genetically mutating a residue for any of the other 19-common proteinogenic amino acids. Mutagenesis of proteins has been used extensively to characterize structure–function relationships, and as a means to augment biological activity. A key challenge in the modification of protein structure, however, is the ability to introduce chemical functionality that is not normally genetically encoded. In doing so, the effects of protein dynamics, PTMs and protein-protein interactions may be laid bare through the introduction of biophysical probes, amino acid modifications and chemical crosslinkers respectively. The direct chemical labeling of reactive amino acid side-chains, such as cysteine, is widely used to modify proteins with various chemical functionalities.3 Alternatively, the ribosomal incorporation of unnatural amino acids using synthetic biology tools is a powerful way to install chemically unique side-chains into a protein.4 However, engineering of polypeptides using the tools of organic chemistry and chemical peptide synthesis provides the most selective and flexible means to make modified protein sequences, able to not only modify amino acid side chains, but to also alter the chemistry and chirality of the underlying peptide backbone.5 Merging the chemical flexibility of peptide synthesis with the power of recombinant protein engineering is therefore an incredibly attractive route to deciphering how protein structure dictates function.
Approaches to the synthesis of peptides and proteins have historically centered on the chemical condensation of amino acids and peptide precursors into longer polypeptide chains. The earliest such example by Curtius in 1882 is the chemical synthesis of the achiral diglycine molecule from acid chlorides and amines (Figure 1a).6 In the >130 years since, the condensation of amines and carboxylic acids remains the dominant strategy of amide bond synthesis, although it has been greatly refined. Indeed, a practitioner of medicinal or peptide chemistry has a choice of dozens of chemical ‘coupling reagents’ that facilitate the reaction of carboxylic acids with amines to yield amides in high yield.7 In conjunction with the development of protecting groups that mask the amino acid side-chains from the reactive coupling conditions,8 the ability to assemble peptides on a solid support through iterative coupling steps (Solid-Phase Peptide Synthesis – SPPS)9 makes it possible to chemically assemble polypeptides in a user-defined manner reminiscent of ribosomal synthesis (Figure 1b). Despite the flexibility and efficiency of contemporary peptide synthesis, it remains difficult to routinely prepare peptides longer than ~50 amino acids in length, far below the average size of globular protein domains.10 Accessing synthetic polypeptides and proteins beyond this length requires convergent assembly from smaller synthetic fragments. Condensation reactions between fully protected synthetic peptides may be carried out using peptide coupling reagents (i.e. classic fragment condensation11), however such building blocks are notoriously difficult to manipulate in both aqueous and organic solvents, a property that limits this convergent synthetic approach.
Figure 1.
Pioneering examples of peptide bond construction through a) condensation between activated acyl donors and amines, b) Solid-Phase Peptide Synthesis (SPPS), and c) Native Chemical Ligation (NCL – see Figure 3 for additional mechanistic detail).
This problem of how to perform fragment condensation in an operationally simple manner prompted a shift in thought towards how to ligate purified unprotected peptides in aqueous solvent through selective amide bond synthesis.12 This eventually led to the introduction in 1994 of the Native Chemical Ligation (NCL) method by Kent and coworkers who showed that peptides bearing C-terminal thioesters (α-thioesters) and N-terminal cysteine residues respectively can condense under aqueous conditions in high yield (Figure 1c).13 Due to the efficiency of NCL and extensions of this reaction, totally synthetic proteins well in excess of 100 amino acids containing all manner of backbone or side-chain chemical modification may be assembled through iterative ligations of synthetic peptides. Generally speaking, however, the size and number of chemical modifications that one might wish to install into a protein represents a minor proportion of its overall structure. For example, it might be desirable to change just a few residues in an enzyme active site, while leaving all other positions (i.e. most of the molecule) unmodified. Consequently, when synthesizing modified proteins through ligation chemistry, the majority of effort and resources are directed towards the stepwise chemical assembly of peptides containing the standard 20 amino acids. A semisynthetic approach to protein synthesis offers an expedient solution to this problem, wherein a protein is assembled from both synthetic and recombinant fragments, the latter derived from ribosomal protein synthesis, which is both cost effective and unburdened by protein-length limitations.
The synthesis of proteins requires chemoselectivity emblematic of bio-orthogonal chemistry: the ability to forge an amide bond selectively in the presence of the cloud of reactive functionalities commonly present within a polypeptide. This can be achieved by installing chemically orthogonal reactive handles during peptide chemical synthesis. Clearly, when dealing with recombinant proteins, this reactivity must be somehow genetically encoded. Chemical ligation methods that use the reactivity of proteinogenic amino acids, such as cysteine (for NCL) or serine/threonine (Serine/Threonine Ligation – STL),14 are amenable to such extension. However, the full implementation of the semisynthetic strategy requires the ability to install functionalities into recombinant proteins that are not directly incorporated by the ribosome. These include the high-energy α-thioester functionality needed for NCL. This turns out to be a less prosaic protein-engineering problem, one whose solution requires the repurposing of various biocatalytic facilitators. For example, a class of proteins known as inteins emerged as ideal tools for the installation of α-thioesters into recombinant proteins.15 In parallel, approaches for amide bond generation through enzymatic transpeptidation have also appeared, using modified protease16 or transpeptidase17 enzymes (Figure 2). Each of these biocatalysts promote biochemical transformations that are very similar to those that occur during NCL, cleaving and reassembling amide bonds through a series of N-to-S/O acyl shift reactions.18 The innate activity of these biocatalysts therefore makes them especially suited for the semisynthetic manipulation of proteins.
Figure 2.
Timeline of major developments in contemporary protein semisynthesis.
Contemporary protein semisynthesis resembles a manner of biomolecular-orthopedics. The polypeptide backbone, fittingly named, can be altered with high precision, introducing artificial groups throughout. In this review, we inspect each of the ways that enzymes have been used in the semisynthesis of proteins from synthetic and recombinant building blocks. In doing so we hope to harmonize these distinct methods, which share overlapping chemical transformations. We aim to highlight how this chemistry can be leveraged to forge backbone amide bonds on recombinant proteins, whose reactive functionalities are exposed and must be negotiated. In particular, we wish to highlight the power of more recently developed methods employing enzymes and autoprocessing split-intein domains that have been co-opted to serve as biorthogonal protein-based ‘coupling reagents’. We then describe how the protein semisynthesis strategy has been applied to the biochemical and biophysical analysis of proteins. Finally, we offer a perspective on the role these technologies have yet to play for hypothesis-driven biochemistry, particularly in the native cellular context. The total chemical synthesis of proteins (i.e. through the ligation of two or more polypeptides derived exclusively from chemical peptide synthesis), for which there are many excellent recent reviews,5, 19, 20 falls outside the scope of this monograph. Similarly, methods for bioconjugation using the innate reactivity of amino acid side-chains,3, 21 will not be discussed, nor will the remarkable advances that have been made in the last two decades allowing the ribosomal incorporation of many unnatural amino acids.4, 22
2. CHEMICAL LIGATION OF EXPRESSED PROTEINS
Protein semisynthesis merges the fields of protein biotechnology and chemical peptide synthesis. The former area has seen remarkable advances thanks to the tools of molecular and synthetic biology, while the latter has been continually refined over the course of the century to the point of routine automation (at least for small to modest sized peptides). Thus, bringing these two powerful capabilities together in a single manifold offers an appealing route to manipulate the chemical structure of very large proteins. In a sense the best of both worlds can be achieved within a semisynthetic – i.e. part recombinant part chemical – protein framework. This tantalizing prospect, recognized for decades,23, 24 has fueled tremendous technological advances in the last two decades. In the following sections, we discuss how NCL, which is without question the dominant approach for the total chemical synthesis of proteins, has been successfully extended into the realm of protein semisynthesis. In so doing, we highlight how biocatalytic transformations have been instrumental in furnishing recombinant proteins with the necessary functionalities for chemical ligation.
2.1. Native Chemical Ligation
Native chemical ligation is a highly selective amide-bond forming reaction that has been used broadly for the total synthesis and semisynthesis of proteins. The reaction condenses two completely unprotected polypeptide fragments in neutral aqueous conditions, one reaction partner functionalized as an N-terminal cysteine (or reactive equivalent) and the other as an α-thioester (Figure 3). Peptide ligation is initiated by a dynamic trans-thioesterification reaction between the sulfhydryl group of the N-terminal cysteine and the α-thioester moiety, a process that has been generally observed between thiols and acyl-donors including thioesters,25 the rate of which depends on the pKa values of incoming and outgoing nucleophiles. The resulting transthioesterified intermediate then spontaneously rearranges through an S-to-N acyl shift with the pendant α-amine through a 5-membered ring intermediate, creating an amide bond at the ligation junction. The reaction cascade central to NCL was first demonstrated in 1953 by Wieland and coworkers,26 who successfully condensed cysteine and valinyl-thioester. It took over 40 years for this reaction to be applied to the ligation of synthetic polypeptides,13, 27 but then only two years subsequent to be extended to expressed proteins.28
Figure 3.
The mechanism of native chemical ligation (NCL).
The chemistry underlying NCL capitalizes on the unique reactivity of the native cysteine residue. At physiological pH, the sulfhydryl functionality on the cysteine side-chain is in equilibrium with the nucleophilic thiolate form (pKa ~8.5).29 In the cell, this potent nucleophile is utilized in the active sites of the eponymous class of cysteine protease enzymes.30 Processes involving thioester formation and acyl migration are central to several biological processes, including protein ubiquitination,31, 32 transglutamination,33 in the biosynthesis of non-ribosomal peptides,34 intein splicing35, 36 and the covalent modification of bacterial cell-surface proteins.37, 38 (See sections 2.2: “Inteins and Expressed Protein Ligation”, 3.1 “Artificially split inteins”, and 4.1 “Sortase A” for details regarding these last two mechanisms and how they have been co-opted for biotechnology.) Clearly, the biocompatibility of the underlying chemistry of NCL implies the extension of this approach to biomolecules such as recombinant proteins – a capability that was noted in the initial disclosure of the method.13
The rate-determining step in NCL is the initial trans-thioesterification reaction. This bimolecular reaction is not templated, and therefore requires sufficient concentrations of reactants to outcompete hydrolysis of labile thioester reactants. Kent and coworkers investigated the importance of the nature of the α-thioesters used in the reaction, observing accelerated ligations in the presence of aryl thioesters.39 Nonetheless, a reaction between a cysteinyl peptide and a preformed alanyl α-thiophenylester is significantly less rapid (second order rate constant of 0.26 M−1s−1)40 than the commonly used thiol-maleimide bioconjugation reaction (second order rate constant ~300-800 M−1s−1).40, 41 The kinetics of the NCL reaction is also dependent on the nature of the C-terminal amino acid within the α-thioester fragment. Smaller amino side-chains afford more rapid reactions, whereas β-branched amino acids such as valine and isoleucine are more sluggish to react.42 Typical NCL reactions require therefore that at least one of the reactants be in millimolar concentration for efficient conversion to products.
2.1.1. Chemical synthesis of N-terminal cysteinyl peptides
Synthesis of N-terminal cysteinyl peptides for ligation reactions is routine in peptide chemistry, owing to the range of protecting groups available for masking this reactive side-chain.8 These reagents may be prepared by either the Boc or Fmoc-strategy for SPPS (Figure 4), where the sidechain sulfhydryl is protected from acylation, alkylation or oxidation by either benzyl (Bn), p-methylbenzyl (4-MeBn), p-methoxybenzyl (4-MeOBn) or triphenylmethyl (Trt) protecting groups respectively.43–45 Deprotection of the sulfhydryl group then occurs concomitantly with the global deprotection and cleavage with either hydrogen fluoride (Boc-strategy) or concentrated trifluoroacetic acid (Fmoc-strategy) in the presence of carbocation-scavenging reagents. Alternatively, protecting groups orthogonal to the cleavage conditions, such as commonly used S-acetamidomethyl (Acm) and thiazolidine allow the cysteine sulfhydryl to remain protected after peptide purification.46, 47 This strategy, in addition to orthogonal protection of the cysteine α-amine,48 has allowed for sequential multi-step peptide and protein assemblies.47, 49
Figure 4.
Synthesis of N-terminal cysteinyl peptides using either Boc or Fmoc-strategy solid-phase peptide synthesis. (PG = protecting group).
2.1.2. Chemical synthesis of peptide α-thioesters
Compared to N-terminal cysteinyl peptides, peptide α-thioesters require a greater degree of synthetic manipulation to prepare, due to the relative instability of this functionality to basic conditions. (We direct the reader to the following reviews that focus on this topic.20, 50) Through Boc-strategy SPPS, peptide α-thioesters may be synthesized directly on the solid support and remain stable to both the TFA and HF-based deprotection and cleavage conditions (Figure 5a).42, 51, 52 This strategy is incompatible with Fmoc-strategy SPPS due to the instability of thioesters to the repeated exposure to piperidine required for Fmoc-deblocking. Fully protected peptides synthesized using Fmoc-chemistry may be converted to α-thioesters under highly controlled conditions that minimize epimerization of the C-terminal amino acid.53 Alternatively, methods that capitalize on late-stage activation of stable linkers have proven to be robust and flexible routes to α-thioesters (Figure 5b–d). An early example of this involved the use of an alkanesulfonamide “safety-catch” linker that could be activated for α-thioester installation following Fmoc-SPPS.54 Comparable to the NCL reaction cascade in reverse, peptides bearing C-terminal 2-mercapto glycolate esters55, 56 and 2-mercapto alkylamides57–59 can rearrange through acid-catalyzed O/N-to-S acyl shift to form α-thioesters that can be trapped by external thiols to form stable α-thioesters. Peptides functionalized with C-terminal diaminobenzoyl (Dbz) linkers may be converted to N-acyl ureas through on-resin acylation,60, 61 or to N-acyl benzotriazoles through treatment with acidic sodium nitrite solution,62 and subsequently thiolyzed with aryl or alkyl thiols. The use of N-acyl hydrazides, one of the first methods of acyl activation, has been reimagined as a thioester precursor for NCL reactions.63, 64 Chemoselective conversion of this comparatively stable moiety to an α-thioester can be carried out in situ prior to ligation reactions, a method that has been widely adopted for both protein total synthesis and semisynthesis.50 Importantly, resins pre-functionalized with thioester precursors are now commercially available (including the Dbz, bis(2-sulfanylethyl)amino (SEA),59 and N-acyl hydrazide linkers) significantly reducing the barrier to adoption of these powerful technologies.
Figure 5.
Common strategies for the synthesis of peptide thioesters by a) Boc and b-d) Fmoc strategy SPPS.
2.1.3. Expression of N-terminal cysteinyl proteins.
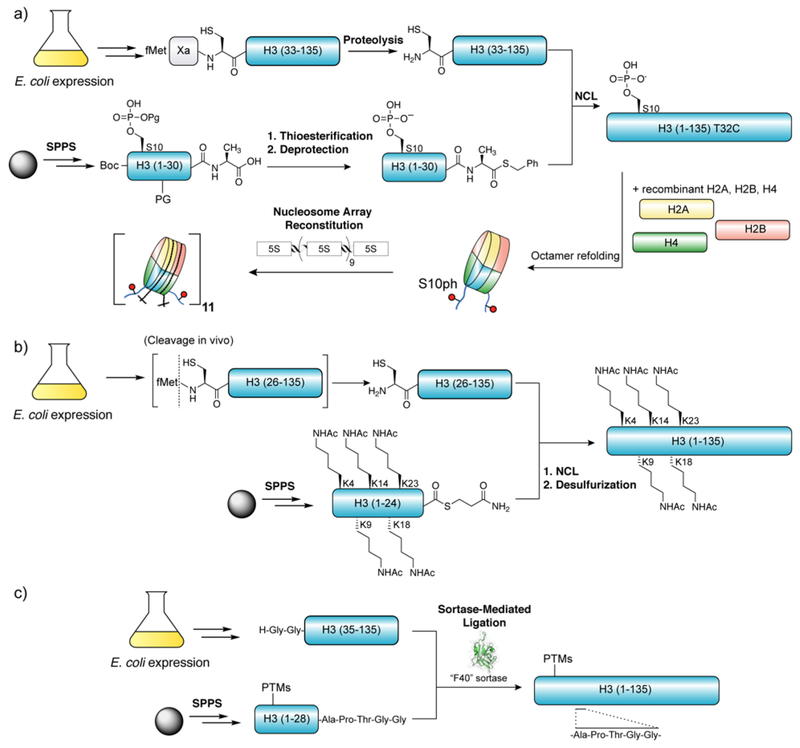
Accessing recombinant protein substrates for NCL has typically been achieved through expression of either full-length or truncated proteins in heterologous host organisms. As for all ribosomally synthesized proteins, genes encoding these proteins must contain an initiator methionine residue at its first position. Various methods, chemical and enzymatic, have been explored in order to transform these constructs into the necessary N-terminal Cys-containing proteins for NCL. The most direct method to install an N-terminal Cys into a recombinant protein is to rely on the endogenous methionine aminopeptidase to remove the initiator methionine from a precursor containing Met-Cys at the N-terminus (Figure 6).65 The kinetics of this processing reaction is highly dependent on the nature of the second amino acid, the enzyme preferring smaller amino acids including Cys as well as Gly, Ala, Ser, Pro, Thr, Val.66 After processing of the N-terminal Met residue, the free N-terminal Cys is prone to conversion in vivo to a thiazolidine through reaction with endogenous electrophilic metabolites such as pyruvate.65 Regeneration of the N-terminal Cys must therefore be carried out prior to NCL, for example, through treatment with methoxylamine.67 Hauser and Ryan employed a similar approach in their semisynthesis of a fusion lipoprotein, expressing the Cys-bearing protein of interested in E. coli as a fusion with the pelB leader peptide sequence (Figure 6).68 This sequence directed the expressed protein to the periplasmic space where an endogenous signal peptidase liberated the N-terminal cysteine-containing protein from the fusion tag.
Figure 6.
Production of recombinant N-terminal cysteinyl proteins through in vivo proteolysis.
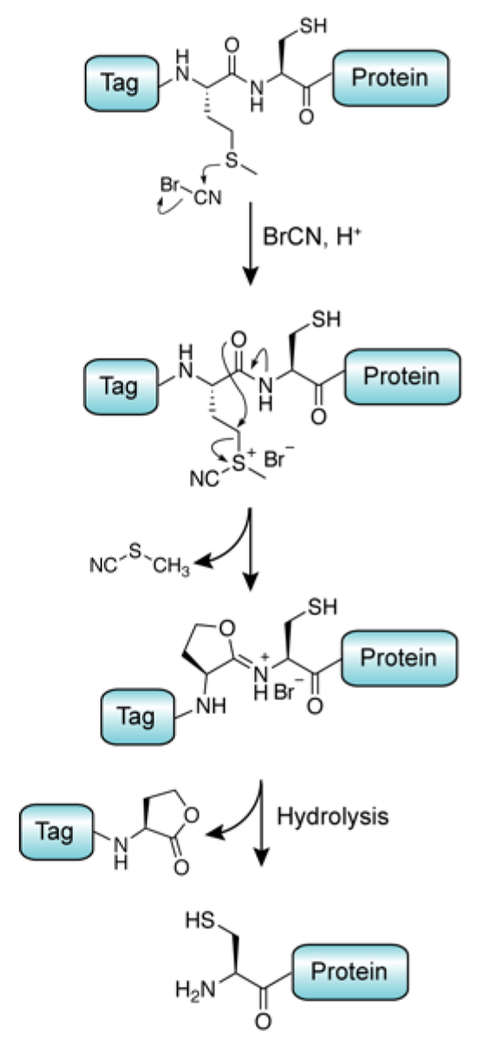
In vitro chemical treatment of proteins with cyanogen bromide has been widely used for the selective cleavage of amide bonds following methionine.69 The reaction of this potent electrophile with the Met thioether side-chain generates an unstable iminolactone, which may be subsequently hydrolyzed (Figure 7). This method has been investigated as a chemical route to isolate fragments of erythropoietin for downstream ligation chemistry, requiring mutation of native methionine residues to avoid off-target cleavage.70
Figure 7.
Mechanism for the chemical cleavage of Met-Cys bonds using cyanogen bromide.
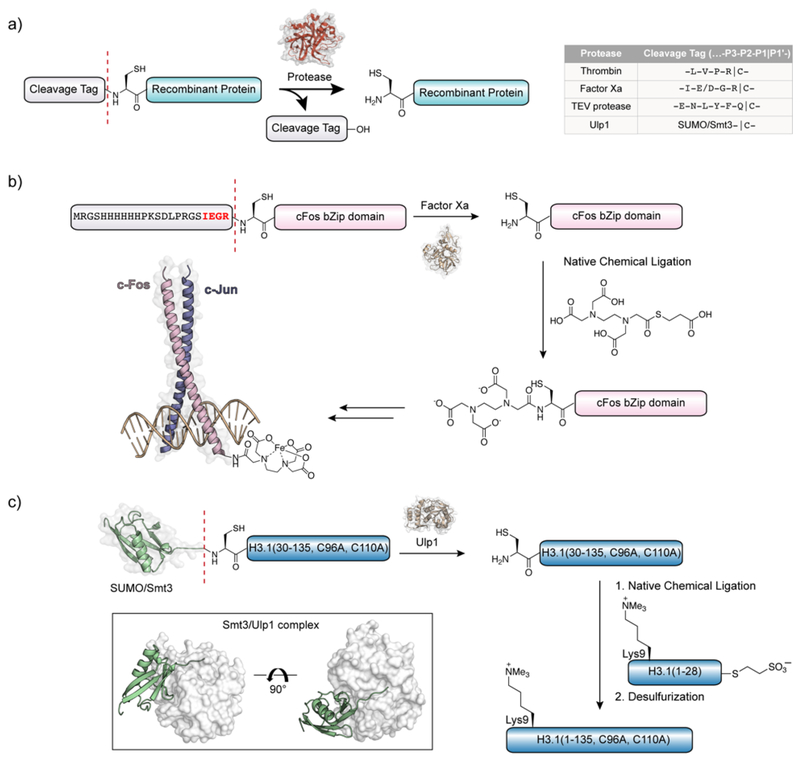
The use of sequence specific proteases to liberate cysteinyl proteins in vitro provides an alternative to relying on the endogenous processing machinery of the cell (Figure 8a). The proteases Factor Xa28, 71, 72 and thrombin73 have been widely used for this purpose due to their tolerance for cysteine at the P1’ positions of their recognition sequences. In the first reported use of NCL on recombinant proteins, Verdine and coworkers expressed the two members of AP-1 transcription factor complex (c-Jun and c-Fos) as N-terminal cysteinyl proteins, genetically fused to purification tags by a Factor Xa recognition sequence (Figure 8b).28 Protease treatment liberated the N-terminal Cys proteins prior to NCL reactions with synthetic thioesters, in this case the small molecule chelator EDTA. Cleavage by both Factor Xa and thrombin requires careful optimization to avoid proteolysis of secondary sites. Such considerations motivated Tolbert and Wong to investigate the use of the highly specific Tobacco Etch Virus (TEV) protease as a way to liberate cysteinyl proteins.74 Besides the increased sequence-specificity relative to Factor Xa and thrombin, this protease may also be overexpressed in E. coli, making this a more accessible and cost effective option.75 Additionally, TEV protease remains active in low concentrations of additives such as detergents and denaturants, conditions that are favorable when working with otherwise insoluble protein fragments.76
Figure 8.
Production of recombinant N-terminal cysteinyl proteins through proteolysis of fusion proteins in vitro. a) Commonly used proteases for the generation of N-terminal cysteinyl proteins for NCL. b) Preparation of recombinant cFos leucine zipper domain with an N-terminal cysteine for NCL (Erlanson et al.).28 c) Preparation of recombinant N-terminal cysteinyl truncated H3 for the semisynthesis of H3K9me3 (Nguyen et al.).77 Inset: Structure of the Smt3/Ulp1 complex (PDB ID code 1EUV).
The small ubiquitin-like modifier (SUMO) protein, often used to boost soluble protein expression, may be removed selectively from fusion proteins through treatment with specific SUMO proteases.78 The Saccharomyces cerevisiae SUMO protein Smt3 and the corresponding protease Ulp1 are the most commonly employed pairing, although other combinations exist.79 Unlike the proteases described previously, SUMO-proteases recognize both the tertiary fold of SUMO as well as the cleavage sequence at the C-terminus (Xaa-Gly-Gly|Xaa), providing highly specific proteolysis (Figure 8c). Like TEV protease, Ulp1 retains activity under mildly denaturing conditions that are often necessary for the purification of aggregation-prone proteins such as histones.80 These features make the Smt3/Ulp1 system especially well-suited for the preparation of N-terminal Cys proteins for use in NCL. Indeed, this SUMO-fusion strategy has emerged as perhaps the ‘go to’ method for the generation of this class of ligation-ready recombinant building block.
2.1.4. Chemical routes to recombinant protein α-thioesters.
As discussed above, many methods for the preparation of synthetic peptide α-thioesters are available, relying on the ability to control chemical manipulations on specific carbonyl groups during peptide synthesis. Such precise control is facile in a chemically defined synthetic peptide, but exceedingly challenging on a recombinant polypeptide. Nonetheless, there has been some success in exploiting certain amino acid sequence motifs embedded within a protein precursor as a way to directly chemically install α-thioesters. For example, MacMillan and coworkers showed that C-terminal cysteines are prone to an N-to-S acyl shift rearrangement under acidic pH, a feature that has been exploited to generate α-thioesters.57 The method has been adapted to afford recombinant protein-hydrazides, which were subsequently transformed into α-thioesters for use in protein ligation applications.81–83 In an elegant example, Otaka and coworkers showed that Ni(II) ions promote the N-to-O acyl shift of Ser-Xaa-His motifs, generating peptide oxo-ester intermediates that can be converted to acyl hydrazides through treatment with hydrazine.84 The related use of this Ni(II)-mediated N-O shift reaction for cleavable protein-tags suggests that this reaction could be extended to the preparation of recombinant protein hydrazides for use in NCL.85
While these chemical cleavage strategies have utility in specific contexts, they do not offer a general solution to the problem of recombinant protein α-thioester generation. Fortunately, a biocatalytic solution to this problem has been developed and, interestingly, this also involves targeted cleavage of a specific peptide bond, albeit in this case facilitated by a remarkable family of auto-processing proteins known as inteins.
2.2. Inteins and Expressed Protein Ligation
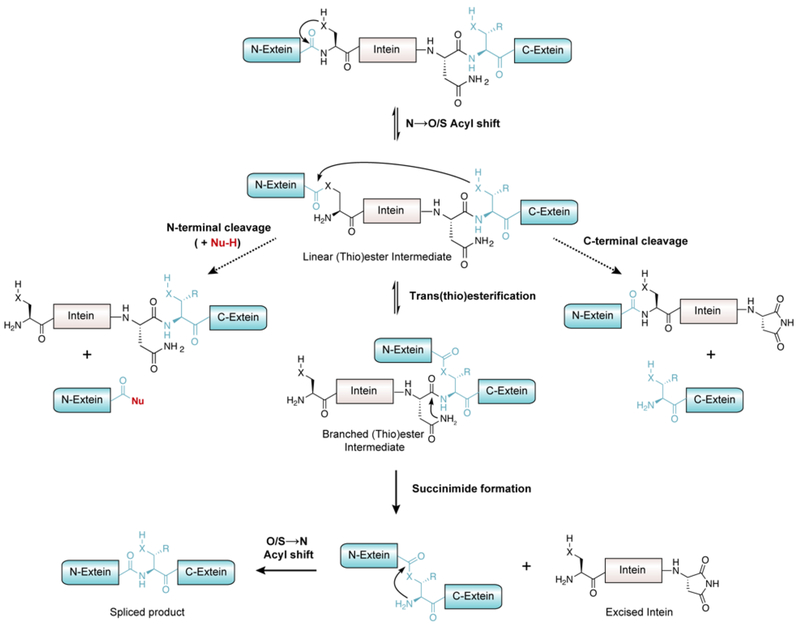
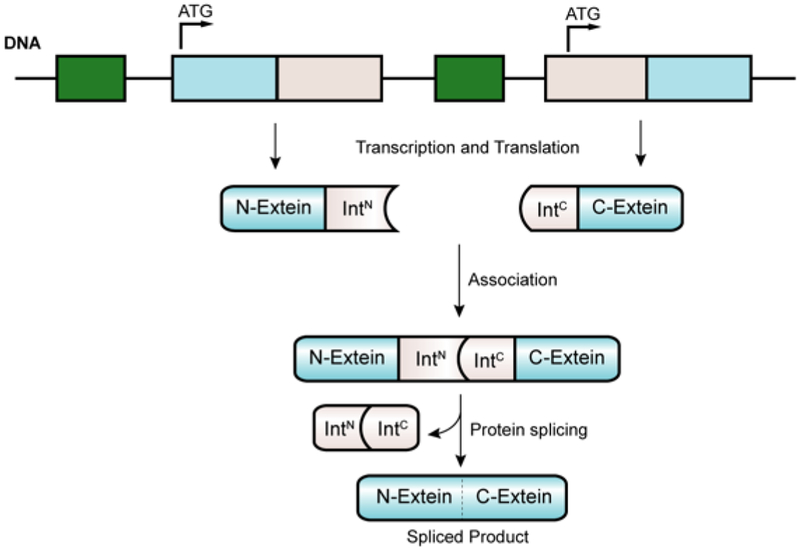
Many proteins become processed through peptide bond cleavage as part of their natural maturation and biological regulation.86 Typically this is achieved in trans through the action of dedicated protease enzymes that catalyze the hydrolysis of a specific amide bond in the precursor polypeptide. However, processing may also occur in cis through the action of auto-processing proteins. Discovered in 1990,87 inteins are such a class of protein domain that catalyze a biochemical process known as protein splicing. These domains, genetically embedded in larger protein-coding genes, undergo splicing post-translationally, which involves the cleavage and re-ligation of the two peptide-bonds immediately flanking the intein domain (Figure 9). Thus, the appended protein domains flanking the intein, referred to as the N- and C-exteins, become ligated together without any traces of the excised intein. Once regarded as curiosities restricted to a small number of microorganisms, we now know, thanks to modern genomic and metagenomics efforts, that inteins are in fact widespread in nature, found in all three branches of life and in many viruses.88, 89 Indeed, these genomic efforts have revealed astonishing functional diversity within the intein family, and as we shall see in the coming sections, this has opened up tremendous opportunities in the semisynthesis area including, but certainly not restricted to, providing a straightforward solution to α-thioester installation in recombinant proteins.
Figure 9.
Post-translational modification of proteins through intein-mediated protein splicing.
2.2.1. Mechanism of intein-mediated protein splicing
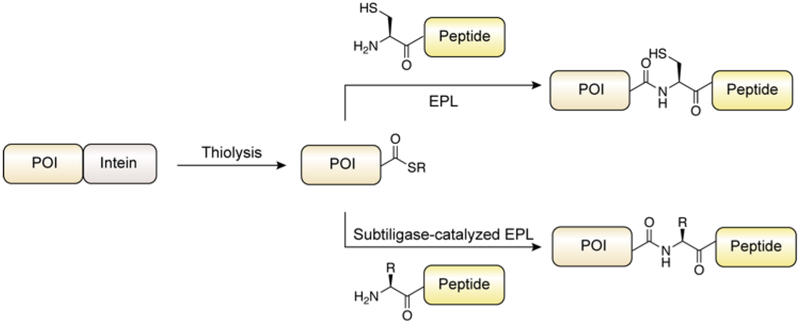
The basic chemical mechanism of protein splicing was elucidated in 1993 by Perler and coworkers.90 Subsequent in vitro biochemical and structural analyses by several research groups mean that we understand the process in great detail.91 Protein splicing follows a series of acyl-shift reactions highly reminiscent of transformations that occur in the NCL reaction cascade (Figure 10). In the canonical mechanism, the first residue of the intein domain (Cys or Ser) undergoes nucleophilic attack on the carbonyl of the preceding amide-bond which is activated by conserved catalytic residues. The resulting oxy(thio)ester linkage then undergoes reversible trans-(thio)esterification with the first residue of the C-extein (exclusively Cys, Ser or Thr) to form a branched oxy(thio)ester intermediate. Resolution of this intermediate then occurs through cyclization of the conserved C-terminal Asn residue of the intein to form a succinimide. This step liberates the intein domain and triggers a spontaneous O/S-to-N acyl shift rearrangement resulting in the N- and C-exteins being joined by a normal peptide bond. Divergence from this mechanism of protein splicing occurs in a small minority of inteins that lack a nucleophilic N-terminal residue, and that instead form the branched oxy(thio)ester intermediate directly.92 However, mutation of this highly conserved feature generally redirects the splicing pathway of the intein towards a cleavage byproduct, where cyclization of the C-terminal asparagine residue occurs prior to forming the branched intermediate, leading to autoproteolysis of the C-extein from the intein. Alternatively, mutation of the C-terminal asparagine of the intein can abolish the ability to undergo branched-intermediate resolution, causing a build-up of activated oxy(thio)ester which can hydrolyze to cleave the N-extein from the intein.92 This autoproteolytic activity makes inteins attractive tools in biotechnology as traceless protein purification tags.93–95 Critically, exogenous thiols may partake in nucleophilic trapping of the acyl-intein thioester, thus liberating the N-extein as an α-thioester,96 which can be isolated and subjected to chemical ligation reactions with synthetic (or recombinant) cysteinyl polypeptides. This semisynthetic extension of NCL using recombinant protein α-thioesters has come to be known as Expressed Protein Ligation (EPL),97 or less commonly Intein-mediated Protein Ligation.98
Figure 10.
Biochemical mechanism of protein splicing and associated N- and C-terminal cleavage pathways.
2.2.2. Production of recombinant protein α-thioesters using inteins.
The protein splicing activity of an intein is, to the first approximation, independent of the nature of the appended N- and C-exteins.91 This biochemical promiscuity is key to the generality of the EPL approach and accounts for the very wide range of systems that have been accessed using the technology. Foundational work in this field employed the vacuolar ATPase subunit intein from Saccharomyces cerevisiae (Sce VMA intein). Indeed, in one of the first applications of EPL, Muir, Sondhi and Cole used this intein in the semisynthesis of the 450-residue protein tyrosine kinase C-terminal Src kinase (Csk), a target beyond the synthetic limits of practical protein chemical synthesis (Figure 11).97 Recombinantly expressed C-terminally truncated Csk, fused to the Sce VMA intein, was treated with synthetic peptides corresponding to the C-terminus of Csk in the presence of thiophenol, which served to liberate a reactive protein α-thioester from the intein. Similarly, Xu and coworkers found that the use of thiophenol and mercaptoethanesulfonate (MESNa) were effective thiol catalysts for both the liberation of protein α-thioesters from both the Sce VMA and Mycobacterium xenopi DNA gyrase A (Mxe GyrA) inteins, and subsequent ligations with synthetic peptides.98 Notable, β-mercaptoethanol and dithiothreitol, while effective for promoting N-terminal extein cleavage, were found to be poor catalysts for the subsequent ligation reaction, presumably due to the known instability of thioesters derived from these reagents.95, 99 In addition to thiolysis, activated protein of interest (POI)-intein fusions can be cleaved by hydrazine to generate protein acyl-hydrazides.63, 100 Liu and coworkers found that cleavage of a fusion between microtubule-associated protein light chain 3 (LC3) and Mxe GyrA could be induced by hydrazine instead of the commonly used thiol MESNa.63 Moreover, cleavage was more efficient with hydrazine and the hydrazide product is chemically more stable than the corresponding α-thioester. Considering the widespread adoption of synthetic peptide acyl-hydrazides as thioester precursors for ligation reactions, it is tempting to predict increased use of intein hydrazinolysis compared to thiolysis in future.
Figure 11.
Expressed protein ligation (EPL) for the semisynthesis of modified Csk.97
Mxe GyrA has been the most widely used intein for the generation of recombinant protein α-thioesters (see Table 1 for a comprehensive listing of EPL applications). The reasons for this are partly historical and partly practical. This intein was among the first to be discovered and made available to the biomedical community.101 However, the small size of this intein relative to other inteins (198 aa), combined with its ability to be refolded from a denatured state, make it attractive when generating protein α-thioesters from insoluble or aggregation prone proteins.102 The Mxe GyrA intein has also been amenable to engineering to increase expression of fusion proteins.103–105 Premature cleavage of the POI-GyrA fusion in vivo can be largely suppressed through a Thr-to-Cys mutation at position 3 of the intein, which temporarily inactivates the catalytic Cys1 residue through the formation of a disulfide.105 Strömgaard and coworkers demonstrated that the GyrA sequence can be reduced by up to 25% through the replacement of superfluous structural elements with unstructured linkers, affording higher yields of protein but without compromising thiol-induced cleavage activity.104 In addition to rational engineering efforts, Shusta and coworkers demonstrated that directed evolution of the Mxe GyrA intein can be used to afford more highly expressing POI-intein fusions (including antibody fragments).103 These studies demonstrate the intrinsic malleability of certain inteins, in this case Mxe GyrA, in order to maximize the production of protein α-thioesters.
Table 1.
Applications of NCL and EPL to protein semisynthesis.a
| Protein | Location of synthetic modification | Modification |
|---|---|---|
| Abl tyrosine kinase | Central | Fluorophore labeling106 |
| n/a | Segmental Isotopic Labeling107 | |
| Adapter protein crk-II | n/a | Segmental isotopic labeling108, 109 |
| Akt1 serine-threonine kinase | C-terminal | Ser473Ph110, Ser477Ph110, Ser479Ph110 |
| β-Amyloid | C-terminal | Unmodified111 |
| Androgenic Gland Hormone | C-terminal | N-linked glycosylation112 |
| Anthrax Lethal Factor | N-terminal | N-terminal acetylation113 |
| Apolipoprotein E (ApoE) | n/a | Segmental Isotopic Labeling114 |
| Arylalkylamine N-acetyltransferase (AANAT) | N-terminal | Thr31Ph, 115 Thr31Pma116 |
| C-terminal | Ser205Ph,115, 117 Ser205Pfa115, 117 | |
| Aspartate transporter GltPh | C-terminal | Arg397Citrulline118 |
| Autophagy-related protein 3 (Atg3) | N-terminal | Lys19Ac119 |
| Central | Lys48Ac119 | |
| Azurin | C-terminal | Cys112Sec120, Met121Nle121, Met121Sec122, Met121Sem121, Met121Hcy123 |
| Casein kinase II (CK2) | C-terminal | Thr344Ph124, Ser347GlcNAc124 |
| Chemotaxis protein CheA | n/a | Segmental Isotopic Labeling125 |
| Checkpoint kinase 2 (CHK2) | N-terminal | Thr68Ph126 |
| Chorismate mutase | C-terminal | Arg90Cit,127 βArg9128 |
| Chaperone protein ClpB | n/a | Segmental isotopic labeling129 |
| N-/C-terminal | Fluorophore labeling130 | |
| Chaperone protein DnaK | n/a | Segmental Isotopic Labeling131 |
| Chemokine (C-X-C motif) ligand 8 (CXCL8) | C-terminal | C-terminal truncation132, fluorescein133, benzoylphenylalanine134, β-peptide mimetics135, segmental isotopic labeling136 |
| Chemokine (C-X-C motif) ligand 12 (CXCL12) | C-terminal | 6-Nitroveratryl (Nvoc) photocages137, 138, fluorescein139 |
| γD-Crystallin | n/a | Segmental isotopic labeling140 |
| 5-Enolpyruvylshikimate-3-phosphate (EPSP) synthase | n/a | Unmodified141 |
| Eph receptor tyrosine kinase | C-terminal | Unmodified142, 143 |
| Erythropoietin (EPO) | N-terminal | N-linked oligosaccharide,144, 145 alkyne tag146 |
| Eukaryotic initiation factor-4A (elF4A) | C-terminal | Ser209Ph147 |
| Glutamyl-prolyl-tRNA synthetase | n/a | Segmental Isotopic Labeling148 |
| Glutaredoxin | N-terminal | Cys11Sec149 |
| Glycosylation-dependent cell adhesion molecule-1 (GlyCAM-1) | N-terminal | Thr/SerGalNAc150 |
| C-terminal | Thr/SerGalNAc150 | |
| Glycogen synthase | C-terminal | Thr668Ph151 |
| α-Hemolysin | Central | Thr117Pra152, Thr117LysAcac153 |
| Heterogeneous nuclear ribonucleoprotein L (hnRNPL) | n/a | Segmental isotopic labeling154 |
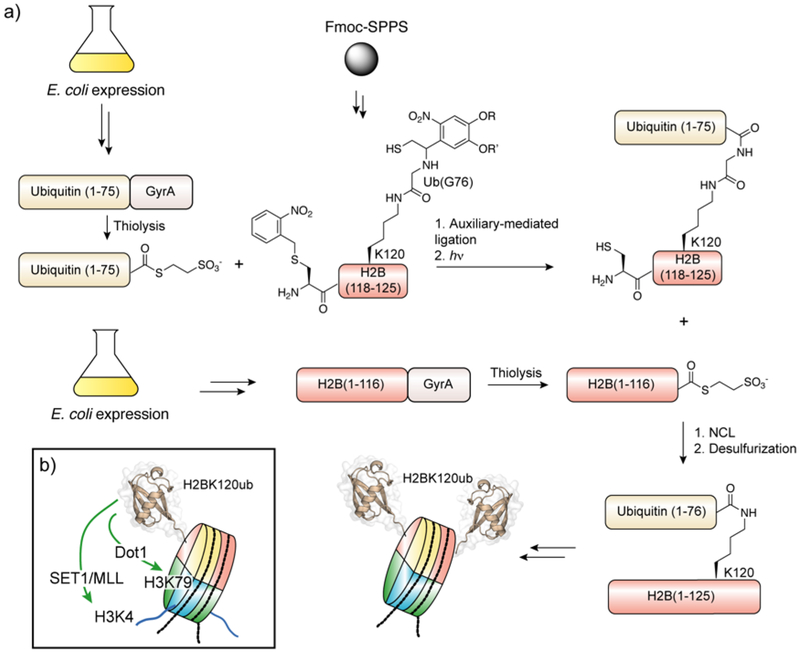
| Histone H2A | N-terminal | Lys5Ac155, Lys9Ac155, Lys13Ac155, Lys13Ub81, Lys15Ac155, Lys15Ub81, Lys15Ub (H2A.X)156 |
| C-terminal | Ser139ph (H2A.X)156, Lys118Ac155 Lys119Ub157,81, 158 | |
| Histone H2B | N-terminal | Ser14ph,67 Lys5Ac,67 Lys11Ac,67 Lys12Ac,67 Lys15Ac67, Lys16Ac155, Lys20Ac155, Lys34Ub81 |
| C-terminal | Lys108Ac155, Ser112GlcNAc155, Lys116Ac155, Lys120Ac155, Lys120Ub159, Lys120Su160, Lys125Ac155 | |
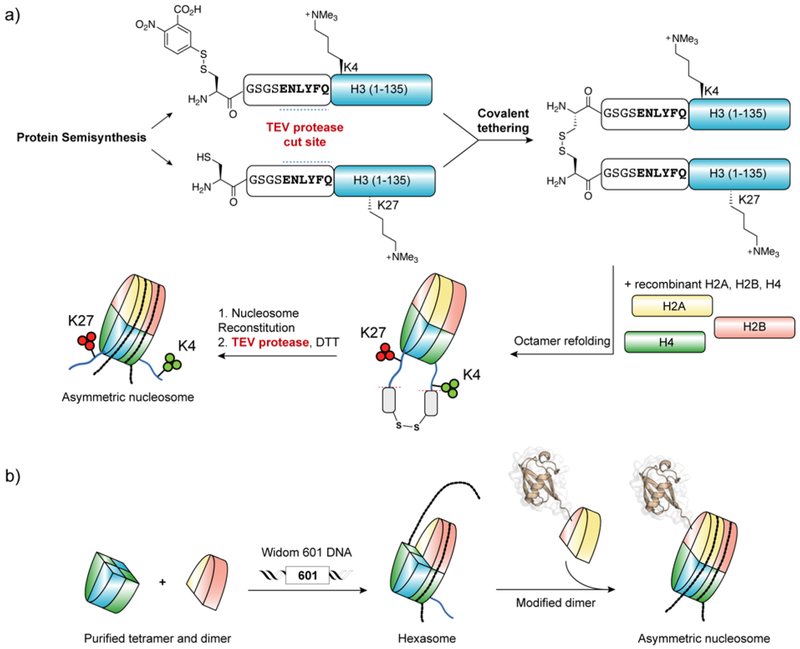
| Histone H3 | N-terminal | Arg2me2155, Lys4Ac161, Lys4me3162–164, Gln5Sero165, Lys9me3161–164, 166–169, Lys9Ac77, 161, Lys14Ac77, 161, Lys18Ac77, 161, 170, Lys18Cr171, Lys23Ac161, 170, Lys27me3162–164, 172, Lys23Ac170, Lys36me3155, 164 |
| Central | Lys79me2/3164, Tyr41Ph155, Arg42me2173 | |
| C-terminal | Lys115Ac174, Lys122Ac174 | |
| Histone H4 | N-terminal | Arg3me2155, Lys5Ac,77, 161, 170 Lys5Cr171, Lys8Ac77, 161, 170, Lys8Cr171, Lys12Ac77, 161, 175 Lys12Cr171, Lys12Su176, Lys16Cr171, Lys16Ac,77, 175, 177 His18pTza178, Lys20Ac,77 Lys20Cr171, Lys20me3168 |
| C-terminal | Lys77Ac155, Lys79Ac155 | |
| Heterochromatin protein 1α (HP1α) | C-terminal | Atto532 labeling179,166 fusion with shigoshin peptide179 |
| Heat shock protein 27 (Hsp27) | C-terminal | Arg188Apy180 |
| Heat shock protein 90 (Hsp90) | n/a | Unmodified181 |
| Huntingtin | N-terminal | Thr3Ph182, 183, Lys6Ac183, Ser13Ph184, Ser16Ph184 |
| Immunity protein Im7 | N-terminal | N-linked glycosylation185 |
| Interferon Response Factor 3 (IRF3) | n/a | Segmental isotop[ic labeling186 |
| Interleukin 6 | Central | N-linked glycosylation187 |
| Interleukin 13 | Central | N-linked glycosylation188 |
| Jarid2 | N-terminal | Lys116me3189, 190, Ser120Ph190, Ser124Ph190, Ser126Ph190 |
| KcsA potassium channel | C-terminal | Unmodified102, 191, Gly77d-Ala192–194, Gly79ester193 |
| Central | Trp68Bta195, Val76ester196, Val76(13C18O)197, Gly77ester,196 Gly77(13C18O)197, Gly79ester,196 Gly79(13C18O)197, Gly77d-Ala193, 196 | |
| KH-type splicing regulatory protein | n/a | Segmental Isotopic Labeling198 |
| KvAP potassium channel | Central | G198dA,192 |
| Lacticin 481 precursor (LctA) | C-terminal | Unmodified199, sequence variants200, Thr33Ph201, 202, Cys38Sec203, Asn39Hse204, Asn39Nva204, Asn39Cba204, Asn39dAsn204, Met40Pra204, Met40Nle204, Met40Nva204, Met40dMet204, Asn41Cba204, Trp43Nal204, Ser28dCys204, Gly29Sar204, Ile31Sar204, His32Sar204, Ile34Sar204, Glu37Sar204 |
| Low molecular weight protein tyrosine phosphatase (LML-PTP) | C-terminal | Tyr131Pmp,205 Tyr132Pmp205 |
| Microcin B17 precursor (McbA) | C-terminal | Sequence variants206, Cys41Hcy |
| Microtubule-associated protein light chain 3 (LC3) | C-terminal | Unmodified63, lipidation207–210 |
| Mitogen-inducible gene 6 (Mig6) | C-terminal | Tyr394Ph211 |
| Mxe GyrA intein | N-terminal | Isotopic labeling212 |
| C-terminal | Isotopic labeling213, Branched isopeptide intermediate,214 Val182hVal214, Thr+1Dap213, 214, His187ThA213, 214, Asn198Nva213 | |
| NaK ion channel | C-terminal | Asp66Csa,215 Asp66Hse215 |
| NEDD4-like E3 ubiquitin-protein ligase WWP2 | N-terminal | Tyr369Ph216 |
| Neurosecretory protein GM | C-terminal | Unmodified217 |
| Notch (V1711 – α-secretase cleavage product) | N-terminal | Nα-Ac218 |
| Nucleolar protein 3 (NPL3) | n/a | Segmental isotopic labeling154 |
| Npu DnaE split intein | n/a | Segmental isotopic labeling219 |
| cGMP phosphodiesterase | C-terminal | Crosslinker (BPA)220 |
| p300 HAT domain (WT and circular permutation) | C-terminal | Unmodified,221 Lys1546Ac,222 Lys1549Ac,222 Lys1551Ac,222 Lys1554Ac,222 Lys1558Ac,222 Lys1560Ac,222 |
| p38α MAP kinase | C-terminal | Thr323Ph223 |
| Paxillin | N-terminal | Tyr31Ph,224 Tyr31Ph(caged)224 |
| Peroxiredoxin-1 | C-terminal | Lys197Ac225 |
| Phosphatase and tensin homolog (PTEN) | C-terminal | Ser380Ph,226–228 Thr382Ph, 226–228 Thr383Ph,226–228 Thr385Ph,226–228 |
| Postsynaptic density protein 95 (PSD-95) | N-terminal | Backbone ester mutants229, 230, Ser73Phos (PDZ1)231 |
| C-terminal | Tyr236Ph (PDZ2)231, Tyr240Ph (PDZ2)231, Y397ph (PDZ3)231 | |
| Polypyrimidine tract binding protein | n/a | Segmental Isotopic Labeling148, 232 |
| Prion Protein | C-terminal | Unmodified,233 GPI anchor234, 235, Palmitoyl anchor234, 236, PEGylation,237 |
| Prochlorosin precursor (ProcA) | C-terminal | [2H]Ser13238 |
| Protein Kinase A | C-terminal | ATP-conjugate239 |
| Rab7 GTPase | C-terminal | Fluorophore labeling240, Cys(Ger)-OMe241–244, diprenylation242 |
| Rad53 serine-threonine kinase | N-terminal | Thr5Ph,245 Thr8Ph,245 Thr12Ph,245 Thr15Ph245 |
| H-Ras GTPase | Central | Gln61GluOMe246, palmitoylation247 |
| K-Ras4B GTPase | C-terminal | Cys(Ger)-OMe248–250, Ser181Ph248 |
| Rheb GTPase | C-terminal | Cys(Ger)-OMe249 |
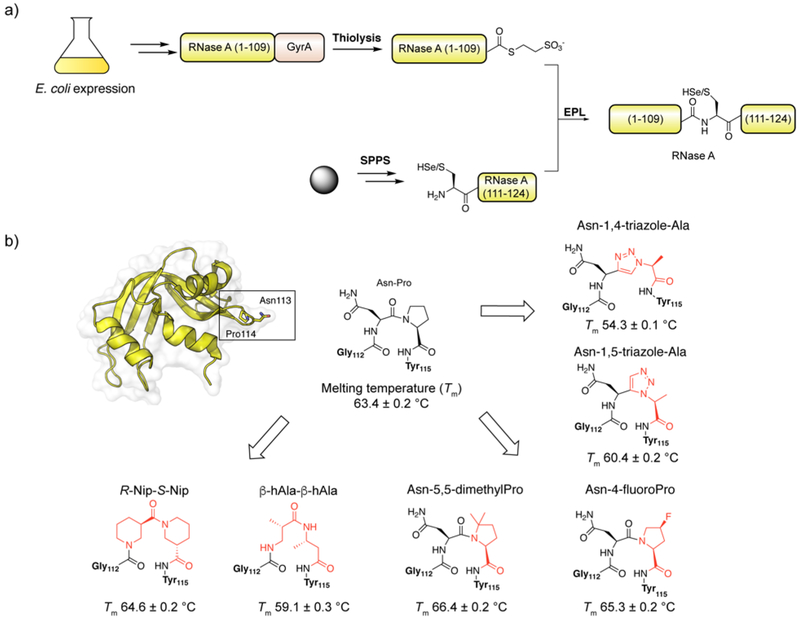
| Ribonuclease A (RNase A) | C-terminal | Unmodified98, Cys110Sec251, β-turn mimics252–256, N-linked glycosylation257, 258 |
| Ribonucleotide reductase (RNR) | C-terminal | Tyr365Ni259, fluorinated Tyr365260, Tyr365Dopa261 |
| S-Adenosylhomocysteine hydrolase | C-terminal | Lys401Ac262, Lys408Ac262 |
| Selenoprotein M (SelM) | n/a | Unmodified (Sec-containing)263 |
| Akt1 serine-threonine kinase | C-terminal | Ser473Ph110, Ser477Ph110, Ser479Ph110 |
| SHP-1 phosphatase | C-terminal | Tyr536Pmp/Pfp264, Tyr580Pmp/Pfp264 |
| SHP-2 phosphatase | C-terminal | Tyr542Ph,265 Tyr542Pmp/Pfp,265 Tyr580Ph,265 Tyr580Pmp/Pfp,265, 266 |
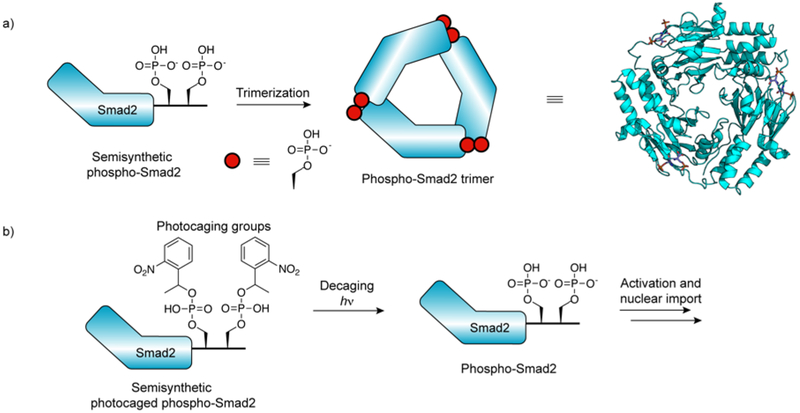
| Smad2 | C-terminal | Ser465Ph267, 268, Ser467Ph267, 268, diazirine photocrosslinker269, photocages270–272 |
| Sortase A | C-terminal | Cys184Sec273, Cys184Hcy273, Arg197Cit274 |
| Src tyrosine kinase | C-terminal | ATP-conjugate275, Tyr527(2-MeTyr)276, Tyr527(b-MeTyr)276, Tyr527Hty276, Tyr527(2,6-F2Tyr)276 |
| Central | Lys57Orn277, Lys57Dab277, Lys57Dap277 | |
| Signal Transducer And Activator Of Transcription 6 (STAT6) | C-terminal | Tyr641Ph (+photocage)278 |
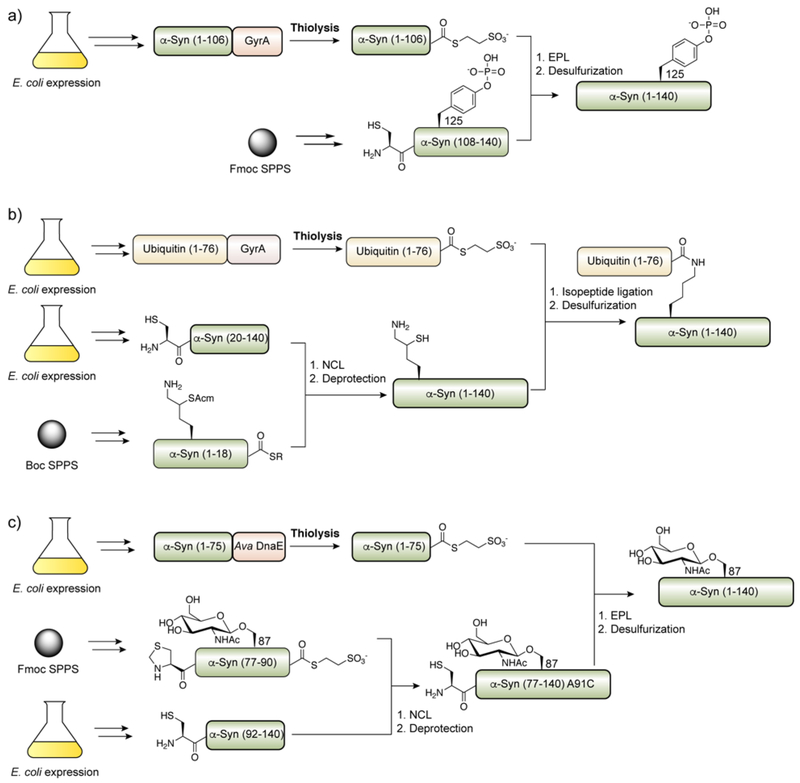
| α-Synuclein | N-terminal | Backbone thioamide279, 280, Nα-Ac281, Lys6Ub282 Lys12Ub4283 |
| C-terminal | Tyr125Ph284, Tyr125Ni285, Ser129Ph286, 287 | |
| Central | Tyr39Ph288, Thr65GlcNAc289, Thr72GlcNAc289, 290, Thr75GlcNAc290, Ser81GlcNAc290, Ser87GlcNAc289–291 | |
| N-/C-terminal | Fluorophore labeling292 | |
| Tau protein | C-terminal | Unmodified293, Ser396Ph49, Ser400GlcNAc294, Ser404Ph49, 295, Ser422Ph296, |
| Central | Lys280Ac49, Lys294Cm297, Ser293Ph297, Ser305Ph297, Tyr310Ph49, | |
| Thioredoxin reductase | C-Terminal | Unmodified (Sec-containing)298, Cys-to-Hcy and -Sec mutants299–305 |
| Thymidine monophosphate kinase | N-terminal | Lys25NBD-Dap306 |
| Transforming growth factor beta (TGFβ) receptor 1 | C-terminal | Thr185Ph,307, 308 Ser187Ph,307, 308 Ser189Ph,307, 308 Ser191Ph307, 308 |
| Trypsin | N-terminal | Unmodified309 |
| Tyrosine-protein kinase Csk | C-terminal | TyrPh97 |
| Ubiquitinb | C-terminal | Diazirine crosslinker310, K6Ub83, K48Ub311, Ser65Ph312, non-hydrolyzable AMP-ester mimic313, 314 |
| Uracil DNA glycosylase | N-terminal | Thr6Ph315, Tyr8Ph315 |
| Zif268 Zinc finger transcription factor | C-terminal | Arg70IDAO,316 Arg70Pra,316 His85TACN,316 Lys89Fl316 |
| β-Xylanase | N-terminal | Fluorinated Glu78317 |
| Yeast α-factor mating receptor | n/a | Segmental Isotopic Labeling318 |
| Ypt1 GTPase | C-terminal | prenylation319, 320 |
| Zinc finger protein (QNK-QDK-RHR) | C-terminal | Arg78Cit321 |
Ac, acetyl; Acac, acetatoacetyl; Apy, argpyrimidine; Bta, β-(3-benzothienyl)-alanine; Cba, cyano β-alanine, Cit, citrulline; Cm, carboxymethyl; Csa, cysteine sulfonic acid; Dab, diaminobutyrate; Dap, diaminopropionate; Fl, fluorescein; GalNAc, N-acetylgalactosaminyl; Ger, geranyl; GlcNAc, N-acetylglucosaminyl; Hcy, homocysteine; Hse, homoserine; Hty, homotyrosine; hVal, hydroxylvaleric acid; IDAO, iminodiacetic acid ornithine; Nal, napthyl alanine; NBD-Dap, N3-nitrobenzofurazane-l-1,3-diaminopropionic acid; Ni, nitro; Nle, norleucine; Nva, norvaline; Orn, ornithine; Pfa, phosphonodifluoromethylenealanine; Pfp, phosphonomethylenephenylalanine; Pmp, phosphonomethylenephenylalanine; Pra, propargylglycine; Ph, phosphoryl; Pma, phosphonomethylenealanine; pTza, phosphoryltriazoylalanine; Sar, sarcosine; Sec, selenocysteine; Sem, selenomethionine; Sero, serotonyl; Su, sumoyl; TACN, triazacyclononane; ThA, 2-thienylalanine; Ub, ubiquityl.
Including the assembly of protein sequences from recombinant and synthetic peptide and protein fragments. Does not include the use of NCL/EPL as a general bioconjugation strategy, e.g. for the generation of antibody conjugates.
Semisynthesis of the protein sequence of ubiquitin itself. Examples of semisyntheses of ubiquitin-modified proteins are listed under the protein of interest.
2.3. Extensions and Alternatives to Native Chemical Ligation
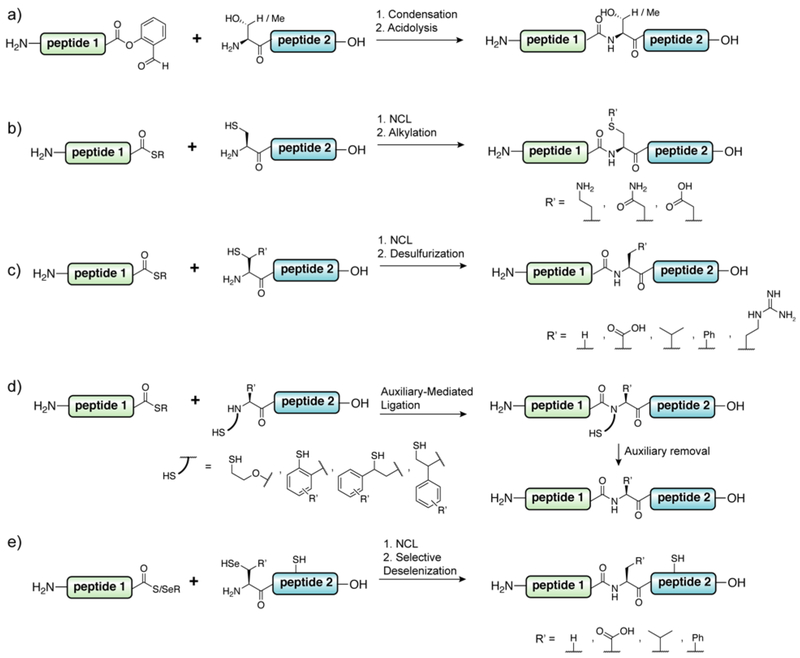
The only strict requirement for a potential native chemical ligation junction within a protein target is that it contains a cysteine residue. While this might not seem like too much of a constraint, the low abundance of cysteine within the proteome (1.1%)322 makes the chances of a suitably located native ligation junction actually quite low. Of course, one can simply introduce a non-native cysteine residue into the target through either mutation or insertion. This commonly employed solution does come with attendant risks given the nucleophilicity and redox activity of the sulfhydryl group. This problem has fueled a great deal of effort geared to towards expanding the number of potential disconnection sites for protein total- and semi-synthesis (Figure 12).
Figure 12.
Extending ligation sites beyond Cys through a) Ser/Thr ligation, b) NCL followed by Cys alkylation, c) NCL followed by desulfurization (not limited to examples of β-thiol amino acids shown – we direct the reader to the following reviews323, 324), d) traceless-NCL using removable ligation auxiliaries,20 e) NCL at selenocysteine followed by selective deselenization (not limited to examples shown – we direct the reader to the following reviews324, 325).
Chemical ligation may be performed between a peptide bearing an N-terminal serine or threonine and a peptide α-ester derived from a suitable hydroxy aldehyde, such as glycolaldehyde or salicaldehyde.326, 327 Selective condensation between the two functionalities occurs through reversible oxazolidine formation, which then rearranges through an O-to-N acyl shift of the proximal ester to form the ligated product (Figure 12a). Salicaldehyde ester auxiliaries can then be removed through acidolytic treatment, generating a native peptide. By virtue of the abundance of serine and threonine residues in the proteome (6.6% and 5.5% respectively)322 the use of Ser/Thr ligation (STL) in a semisynthetic context offers many potential ligation junctions to disconnect the target. In a study by Kirshenbaum and coworkers, the STL reaction was extended from the ligation of exclusively synthetic constructs to the recombinant polypeptides Ribonuclease S-protein and parathyroid hormone (1-34).328 Recombinant proteins bearing N-terminal serines can be obtained through proteolytic liberation strategies akin to those discussed above for NCL, however installation of the required α-ester group into a recombinant fragment is theoretically less straightforward, somewhat limiting the generality of the STL strategy in the semisynthesis arena.
The structure and function of many proteins is unaffected by the introduction of unnatural cysteine residues for the purposes of NCL/EPL reactions (Table 1). However, for proteins that are not amenable to such alteration, there have been considerable methodological developments that expand the approach beyond this rare amino acid. The simplest of these strategies involves elaborating the sulfhydryl group of the cysteine post-ligation (Figure 12b). For example, alkylation can be used to form a range of thioether-containing mimics of proteinogenic side-chains, including lysine,165, 329 glutamine,330 and glutamate.331 Notably, this strategy does not discriminate between the cysteine at the ligation junction and any other cysteines present in the protein. The use of homocysteine as a ligation partner also provides access to methionine residues through methylation after the ligation reaction.332, 333 While not one of the standard 20-amino acids, this residue in protected form may be ribosomally incorporated into recombinant proteins through unnatural amino acid incorporation.334 Alternatively, Petersson and coworkers demonstrated that disulfide-protected homocysteine can be enzymatically installed on the N-terminus of proteins through the activity of a bacterial aminoacyl transferase, a method the authors used for a semisynthesis of α-synuclein.335
Cysteine residues may be formally mutated to alanine through chemical desulfurization reactions (Figure 12c), which expands potential ligation junctions to this highly abundant amino acid.336, 337 As for the use of alkylation reactions post-ligation, such methods are incompatible with endogenous cysteine residues, which will also be chemically modified. As a consequence, these strategies have been extensively applied to the semisynthesis of cysteine-free proteins, such as ubiquitin, α-synuclein and histones (histone H3 contains a single conserved cysteine that may be mutated to alanine) (Table 1). Alternatively, non-participating cysteine residues in recombinant proteins may be masked selectively, e.g with phenacyl groups,180 which can then be unmasked after the ligation and desulfurization reactions. To expand to further ligation junctions, derivatives of proteinogenic amino acids bearing sufhydryls at the β- or γ-position have been synthesized and incorporated into synthetic peptides for use in ligation reactions.323, 324 These hybrid amino acids partake in ligation reactions with synthetic or recombinant polypeptide α-thioesters through the NCL/EPL reaction pathway and may be subsequently converted to the native residue through desulfurization. Of particular note is the use of β-mercapto aspartate by Becker and coworkers in the semisynthesis of pegylated forms of full-length prion protein (PrP).237 The perturbed electronics of β-mercapto aspartate338 allowed this residue to be selectively desulfurized in the presence of two native cysteine residues that are necessary for stabilization of the protein through disulfide formation. Thiolated lysine residues have also been used in the synthesis and semisynthesis of ubiquitinated proteins.339–341 In this case, the non-native sulfydryl reacts with synthetic or recombinant ubiquitin (or potentially SUMO) α-thioesters and direct acyl-shift to the to the ε−amine of Lys to form an isopeptide linkage. An important extension of this strategy is the ability to incorporate δ-mercapto lysine site-specifically into recombinant proteins through unnatural amino acid incorporation,277 work that improved upon an earlier, related approach involving incorporation of cysteinyl-ε-lysine into recombinant proteins.342 An alternative to using non-native amino acids is instead the use of traceless acyl-transfer auxiliaries appended directly to the reactive amine, bearing appropriately positioned sulfhydryl groups (Figure 12d).343 Such auxiliaries are much more sensitive to the steric load surrounding the ligation junction and have been used most effectively at unhindered ligation junctions (e.g. Gly-Gly), for example in the semisynthesis of ubiquitinated proteins.81, 344, 345
Native chemical ligation can also be extended to cysteine’s chalcogen cousin, selenocysteine.251, 346, 347 The selenol group of selenocysteine is substantially more acidic than that of cysteine, existing predominantly as the nucleophilic selenolate at physiological pH, although oxidation to the corresponding diselenide or mixed selenylsulfides is highly facile.325, 348 Akin to the NCL pathway, trans-thio/selenoesterification with polypeptide α-thioesters generates the corresponding Se-linked intermediate that subsequently rearranges through an Se-to-N acyl shift. Raines and coworkers demonstrated that this chemistry is amenable for use in protein semisynthesis, through the reaction with a recombinant α-thioester fragment of ribonuclease A generated through thiolysis of a Mxe GyrA intein fusion.251 Although a genetically encoded amino acid, selenocysteine is not easily introduced into recombinant proteins, which has limited the use of recombinant N-terminal selenocysteinyl proteins as ligation partners. Rozovsky and coworkers were able to express selenocysteine-containing recombinant proteins for ligation through supplementation of the growth medium with selenocystine, leading to the misloading of the cysteinyl-tRNA.263 The resulting selenocysteine-containing proteins were liberated using TEV protease to generate the N-terminal selenocysteinyl protein for ligation. Notably, this strategy will also result in replacement of any internal cysteines in the recombinant fragment with selenocysteine. Thus, in cases where site-specific selenocysteine incorporation is desirable, other approaches are necessary, including unnatural amino acid incorporation349–351 or enzymatic installation using aminoacyl transferase enzymes.352
Initially used as a way to prepare natural selenoproteins, the use of selenocysteine in protein semisynthesis has recently been directed towards the selective chemistry that may be conducted at this amino acid. In particular, selenocysteine can be converted to alanine or serine through phosphine-mediated deselenization under reductive or oxidative conditions, respectively.353, 354 Importantly, reductive selenocysteine-to-alanine conversion may be carried out in the presence of cysteine residues (Figure 12e), offering an alternative to ligation-desulfurization for proteins containing endogenous cysteine residues. In line with this approach, selenated derivatives of proteinogenic amino acids have been prepared for use in ligation-deselenization chemistry.324 Additionally, use of α-selenoesters have been used as acyl-donors that are more reactive than the corresponding α-thioesters with both N-terminal cysteinyl and selenocysteinyl peptides,355, 356 allowing peptide ligations at junctions that may be otherwise intractable (e.g. C-terminal proline).357 While this class of reagents has yet to be extended to protein semisynthesis, this would seem to be a highly productive line of enquiry for the future. Analogous to the generation of α-thioesters, the use of inteins bearing a catalytic selenocysteine residue could provide access to protein α-selenoesters through trans-selenoesterification to extend this chemistry to expressed proteins.350
3. PROTEIN TRANS-SPLICING
The discovery and characterization of protein splicing in cis by contiguous inteins was a clear demonstration that the acyl-shift chemistry utilized in protein ligation chemistry also occurs in nature to alter the primary structure of proteins. However, the development and discovery of trans-splicing split inteins cemented the portrayal of these proteins as nature’s protein ligases. In this case, the fragmented intein pieces associate before auto-catalyzing protein splicing between two separate polypeptides (Figure 13). As one or both of these coupling partners can be synthetic, split inteins offer a chemoenzymatic approach for semisynthetic protein engineering. In this section we will examine the development of protein trans-splicing applications.
Figure 13.
Protein trans-splicing (PTS) mediated by split inteins.
3.1. Artificially split inteins
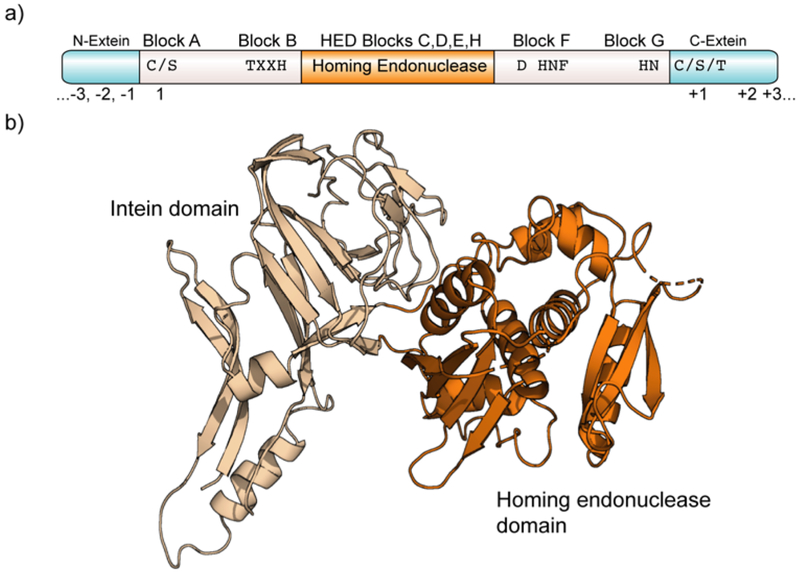
Many contiguous inteins contain a large homing endonuclease domain (HED) embedded within their sequence (Figure 14a). The presence of the HED is thought to allow the horizontal spread of inteins (along with the embedded HED) into host genomes.358 However, the HED is functionally separate from the protein splicing activity of the intein, and there are several examples where this insert has been genetically removed to generate fully active cis-splicing mini-inteins.359, 360 A remarkable feature of HED-containing inteins is the ability of the surrounding protein splicing element to assume its topologically intertwined fold, despite having a very large protein disrupting its primary sequence (Figure 14b). This suggested that it might be possible to reconstitute the intein fold in trans, i.e. by complementation of two separate polypeptide fragments (herein referred to as IntN and IntC) that would normally flank the HED. Indeed, in 1998 it was independently demonstrated in several studies that functional inteins could be reconstituted by refolding N- and C- fragments of the protein (Figure 15a).361–364 Moreover, it was quickly realized that the protein trans-splicing (PTS) phenomenon could be used as a protein ligase system, through trans-splicing between an expressed protein IntN fusion and a synthetic IntC peptide.365
Figure 14.
Intein domain architecture. a) Conserved sequence motifs in inteins relative to intervening homing endonuclease domain. b) Structure of the Sce Vma intein (PDB ID: 1VDE).
Figure 15.
Strategies for converting contiguous inteins into trans-splicing split inteins through a) refolding,361–364 b) induced-proximity,366 and c) reconstitution under native conditions.371
Several other contiguous inteins have been artificially split at the conserved HED insertion site within their sequence (Table 2). In most cases, a refolding step is required to obtain PTS activity, analogous to the Mtu RecA intein. However, there are some exceptions to this rule. For example, the artificially split Sce VMA intein can undergo PTS without a refolding step if (and only if) the two fragments, VMAN and VMAC, are brought into proximity using a two- or three-hybrid paradigm (Figure 15b).366–370 In another example, Mootz and coworkers demonstrated that the artificially split Ssp DnaB intein can effect PTS under native conditions, albeit with reduced efficiency compared to the parental contiguous intein (Figure 15c).371
Table 2.
Artificially and naturally split inteins.
| Intein name | Type | ksplice (s−1)a | t1/2b | Fragment lengthsc (IntN/IntC) | Reference |
|---|---|---|---|---|---|
| Sce VMA | Artificially split | 1.2 x 10−3 | 10 min | 184/64 | 366, 371 |
| Ssp GyrB | Artificially split | 6.9 x 10−5 | 170 min | 150/6 | 373 |
| Ssp DnaB | Artificially split | 9.9 x 10−4 | 12 min | 104/47 | 371 |
| Ssp DnaB | Artificially split | 4.0 x 10−5 | 290 min | 11/142 | 377, 378 |
| Ssp DnaB (M86) | Artificially split | 2.5 x 10−3 | 5 min | 11/142 | 378 |
| Ssp DnaX | Artificially split | 1.9 x 10−4 | 60 min | 144/6 | 375 |
| Ssp DnaE | Naturally split | 6.6 x 10−5 | 175 min | 123/35 | 379, 380 |
| Npu DnaE | Naturally split | 3.7 x 10−2 | 19 s | 102/35 | 380, 381 |
| Ava DnaE | Naturally split | 3.1 x 10−2 | 23 s | 102/35 | 380 |
| Cra DnaE | Naturally split | 1.2 x 10−2 | 58 s | 118/35 | 380 |
| Csp DnaE | Naturally split | 1.8 x 10−2 | 39 s | 99/35 | 380 |
| Cwa DnaE | Naturally split | 5.0 x 10−3 | 140 s | 106/35 | 380 |
| Mcht DnaE | Naturally split | 2.4 x 10−2 | 29 s | 104/35 | 380 |
| Oli DnaE | Naturally split | 1.6 x 10−2 | 43 s | 112/35 | 380 |
| Ter DnaE | Naturally split | 8.5 x 10−3 | 82 s | 101/35 | 380 |
| gp-41–1 | Naturally split | 1.8 x 10−1 | 4 s | 88/36 | 382 |
| gp41–8 | Naturally split | 4.5 x 10−2 | 15 s | 91/44 | 382 |
| NrdJ-1 | Naturally split | 9.8 x 10−2 | 7 s | 105/39 | 382 |
| IMPDH-1 | Naturally split | 8.7 x 10−2 | 8 s | 101/41 | 382 |
| AceL TerL | Naturally split | 1.7 x 10−3 | 7 min | 25/104 | 383 |
| GOS TerL | Naturally split | 1.1 x 10−2 | 65 s | 37/152 | 384, 385 |
In vitro splicing rates under optimal conditions.
Calculated from reported first order rate constants.
Not including extein or initiator methionine residues.
Beyond splitting a contiguous intein at the canonical HED insertion site, considerable effort has been made towards finding alternative split sites with the goal of reducing the size of one of the intein fragments such that it becomes more accessible to SPPS (Table 2, Figure 16).372 The canonical intein split site affords a large IntN fragment (~90-100 aa) and shorter IntC (~30-40 aa) – the former is well beyond the reach to standard SPPS, while latter is close to the limit of synthetic accessibility, thereby allowing only short C-extein cargoes to be appended. With this in mind, the Ssp GyrB mini-intein was artificially split close to the C-terminal splice junction, affording a 6-residue IntC fragment that splices to ~80% completion (albeit with a 10-fold excess of the complementary IntN component).373 The short IntC sequence is easily accessible to peptide synthesis, and was subsequently used for C-terminal labeling of recombinant proteins.374, 375 Notably, this split site has been extended to other systems, specifically the Ssp DnaX and the Ter DnaE-3 mini-inteins.375 Alternatively, artificially split inteins can be generated through fragmenting the primary sequence proximal to the N-terminus, creating a short, synthetically accessible IntN fragment (Figure 16). Accordingly, Mootz and coworkers showed that the Ssp DnaB intein could be artificially split to generate a short IntN (11 aa), which was used to N-terminally label recombinant proteins in vitro.376 This split site was extended to the Rma DnaB intein, which was able to undergo moderately fast PTS (reaction half-life of 22 min) with a small IntN (12 aa) affording >80% spliced product.375 Remarkably, Liu and coworkers demonstrated that PTS can occur in vivo with a DnaB intein split into 3 fragments, comprising a 49 aa C-terminal fragment, an 11 aa N-terminal fragment and a central 94 aa fragment.377
Figure 16.
Common sites for splitting inteins. a) Commonly explored sites for splitting the intein domain in the IntN (S1, S2, S3) and IntC (S10, S11) domains as well as the location of the endonuclease domain (EN/S0). β-strands (β1-β12) are indicated by grey arrows. Split site numbered according to Sun et al.377 b) Split sites mapped onto the structure of the Ssp DnaB mini-intein (PBD ID: 1MI8). The canonical IntN and IntC subdomains (i.e. the S0 split site) are colored light brown and orange respectively.
3.2. Naturally split inteins
In 1998 the first naturally split intein was discovered within the DnaE protein from the Synechocystis sp. strain of cyanobacteria (Figure 17).379 This split intein, named Ssp DnaE, consists of a 123-aa IntN fragment and a 36-aa IntC fragment (including initiator methionine), and likely evolved from a contiguous intein that underwent genomic rearrangement.386 In vitro characterization of this split intein revealed slow protein trans-splicing activity (first-order rate constant of 6.6 (± 1.3) x 10−5 s−1),387 despite the two fragments associating extremely rapidly with nanomolar affinity (Kd < 50 nM).368 A homologue of Ssp DnaE from the Nostoc punctiforme (Npu) cyanobacterium was subsequently cloned and exhibited highly efficient splicing activity in vivo,381 with greater tolerance to local extein sequence variation compared with Ssp. In vitro, Npu DnaE undergoes remarkably fast protein trans-splicing, exhibiting a reaction half-life of less than a minute, compared with > 60 min for Ssp DnaE.388 Further characterization of other DnaE split intein homologues revealed that ultra-fast splicing kinetics are common to this large family of split inteins.380 Indeed, the explosion of genomic sequencing data over the last decade has led to the identification of numerous naturally split inteins, many of which have been shown to have remarkable PTS activities. As a case in point, Mootz and coworkers characterized four non-allelic split inteins discovered through bioinformatic analysis of metagenomic sequence data.382, 389 These inteins (named gp41-1, gp41-8, NrdJ-1 and IMPDH-1) exhibited ultra-fast splicing kinetics, reacting to near-completion with half-lives less than 20s at low micromolar concentration.382 Interestingly, these split inteins share only 40-50% sequence identity with Npu DnaE, supporting the notion that ultrafast splicing kinetics may be a conserved evolutionary feature amongst the majority of allellic and non-allellic split inteins. (In this regard, it is rather ironic that the first naturally split intein characterized, Ssp DnaE, supports comparatively slow PTS kinetics.) Indeed, biochemical analyses suggest that these unrelated splicing elements share a common folding mechanism,390 whereby strong electrostatic interactions drive the association of the split intein fragments, followed by an disorder-to-order “collapse” into the topologically intertwined intein fold to carry out protein splicing.219 It should be noted, however, that the Npu DnaE, gp41-1, gp41-8, NrdJ-1 and IMPDH-1 split intein pairs do not cross-react, i.e. they are functionally orthogonal.390 This attractive feature allows for multiplexing-type applications.390
Figure 17.
A naturally split intein from Synechocystis sp. (Ssp DnaE). a) Schematic for splicing between the naturally split Ssp DnaE intein. b) Structure of the reconstituted Ssp DnaE split intein domain. The SspN and SspC subdomains are colored red and blue respectively (PDB ID: 1ZDE).
The discovery of naturally split inteins with atypical split sites further reinforces nature’s broad use of the PTS paradigm.383, 384 Two of these split inteins, both embedded in the large subunit of the T4-bacteriophage-type DNA-packaging terminase (TerL), originate from the environmentally disparate Ace Lake (AceL) in Antarctica, and the Punta Cormorant lagoon in the Galapagos (GOS). Both AceL-TerL and GOS-TerL have large IntC fragments (104 aa and 152 aa, respectively), and correspondingly short IntN fragments (25 aa and 37 aa, respectively) that are within the realm of synthetic accessibility. Interestingly, the AceL TerL split intein was most active at low temperatures (around 90% splicing yield with a half-life of ~7 minutes at 8 °C)383 while GOS TerL was most active at higher temperatures (around 90% yield with a half-life of between 1-3 min at 30 °C)384, 385 reflecting the evolutionary pressures imparted on both inteins. Two other recently discovered natural atypically split inteins, VidaL T4Lh-1 and VidaL UvsX-2, possess IntN fragments that are even shorter than the TerL split inteins at 15-aa and 16-aa, respectively,391 although the biochemical activity of these systems has yet to be fully explored. In contrast to the canonically split inteins, atypically split inteins appear to associate through hydrophobically driven association,392 followed by disorder-to-order transitions upon complexation. Notably, while the folding mechanisms of canonically and atypically split inteins differ in detail, in both cases the pathways appear to have evolved such that autocatalytic side-reactions are avoided prior to fragment association, a problem that is often observed in artificially split inteins.372
3.3. Engineering of split inteins
While many naturally split inteins have highly favorable biochemical activities, at least relative to their artificially split intein counterparts, there is always room for improvement, whether it be increasing thermal stability or relieving functional constraints imposed by sequence restrictions at the splice junctions. Consequently, split inteins have been the focus of several protein engineering initiatives that have led to enhanced biochemical properties. Consensus protein engineering, wherein multiple sequence alignments are used to design an optimized sequence,393 has been used to improve the chemical and thermal stability of split inteins for PTS applications.392, 394 The application of this strategy to the DnaE family of split inteins, using 73 members predicted to possess fast-splicing kinetics,394 afforded a consensus-fast (Cfa) DnaE intein that possesses superior physicochemical properties and splicing kinetics relative to Npu DnaE, with which it shares 82% sequence homology (Figure 18). Strikingly, the splicing efficiency of Cfa DnaE was largely unaffected up to 8 M urea. The consensus design strategy was also applied to the atypically split TerL inteins, affording an consensus atypically split intein (Cat) with increased activity and stability,392 suggesting that inteins are particularly amenable to this protein engineering strategy.
Figure 18.
Consensus designed DnaE split intein (Cfa DnaE).394 a) Sequence alignment of Npu DnaE and Cfa DnaE split intein. Differences are shown in grey. b) Sequence differences (blue sticks) mapped onto the structure of Npu (PDB ID: 4KL5).
The efficiency of PTS is somewhat context dependent and, as a general rule, all inteins show optimal splicing when the extein sequences at the immediate splice junctions (typically the first 2 or 3 positions of the N- and C-exteins) mimic those of the protein in which they are naturally embedded (see figure 14a for conventional numbering of extein residues common to cis- and trans-splicing inteins). The specifics and stringency of this dependency vary from case to case. For example, the Npu DnaE split intein requires a bulky aromatic residue at the +2 position of the C-extein; structural and functional studies have shown this residue stabilizes nearby catalytic residues in the intein.395 This so-called extein dependence has important implications for the use of split-inteins as tools for traceless protein semisynthesis, where splicing is required to occur in non-native and potentially unfavorable sequence contexts. Strategies employing directed evolution378, 396 and structure-based targeted mutagenesis397 have both been used successfully to relieve this extein dependence. Using PTS to assemble a kanamycin resistance gene in vivo provided a way to select for mutants of a DnaE split intein able to accept the -SGV- sequence at the Int-C-extein junction instead of the native -CFN-.396 This mutant intein was subsequently used to assemble the native sequence of the multidomain Crk-II protein. Of the four mutations responsible for this increased promiscuity, one substitution within a loop proximal to a catalytic His residue was itself capable of stimulating PTS by Npu DnaE when an unfavorable +2 Ala extein residue is present.395 Targeted engineering of this loop through saturation mutagenesis, again coupled to a cell-based selection system, generated an intein with a greatly relieved extein dependence that was subsequently used for the semisynthesis of a modified form of histone H3 (H3K27me3) in isolated nuclei.397 Liu, Mootz and coworkers similarly used directed evolution of the artificially split Ssp DnaB intein to promote more promiscuous splicing.378 Through sequential insertion of the contiguous intein into 10 different locations (with different local extein sequences) within the kanamycin resistance gene, the authors were able to select for inteins with an improved tolerance for non-native exteins. The split form of the best clone (“M86”) exhibited increased fragment affinity relative to the original split intein, as well as greater efficiency in PTS with both native and non-native exteins. It is important to consider that throughout known cis and trans-splicing inteins, each of the 20 common proteinaceous amino acids is sampled at both −1 and +2 extein positions.398
The discovery and engineering efforts described above are providing the protein engineer with an ever-expanding repertoire of split intein tools. From a practical perspective, these reagents are already able to catalyze protein trans-splicing at rates at or beyond the requirements of most protein semisynthesis applications, at least in vitro. Additionally, non-homologous inteins react orthogonally to one another, as the acyl-shift chemistry central to PTS is templated by the intein structure. This allows for multiple PTS reactions to occur simultaneously on the same368, 399 or separate390 polypeptides. The bio-orthogonality of the PTS reaction also makes it especially well suited to semisynthesis applications in living cells, in this case through the cytosolic delivery of synthetic peptides into mammalian cells expressing the complementary split intein fusion.400–404 Notably, this same bio-orthogonality makes PTS an incredibly powerful tool in a host of other biomedical areas, including gene therapy405, 406 and synthetic biology.407 These exciting applications, as well as the many others discussed elsewhere in this review, will surely benefit from the continued discovery and subsequent engineering of split inteins with optimized properties tailored toward the system in question.
4. TRANSPEPTIDATION
Transpeptidation is the transformation of one peptide bond into another through the transacylation of amines. Enzymes that catalyze this reaction, known as transpeptidases, are used in nature to alter the primary sequence of proteins. These enzymes recognize and cleave specific motifs of amino acids, transferring the peptide acyl group to a suitable amine acceptor. In this section we will discuss the discovery and engineering of two families of transpeptidases, sortases and peptide asparaginyl ligases, which have been used extensively by the protein engineering community.
4.1. Sortase A
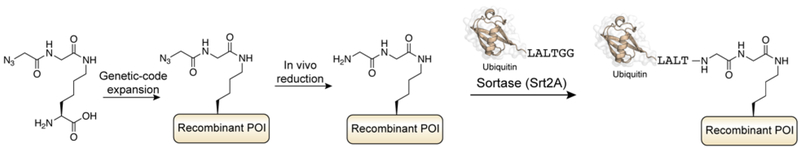
Gram-positive bacteria select a range of proteins for display on their cell surface through covalent attachment to the cell wall envelope.408 In S. aureus, each of these proteins destined for display contain a pentapeptide ‘sorting’ motif, LPxTG (x is variable). In 1999, a mutational screen in S. aureus identified a transpeptidase enzyme, a 206-residue protein containing a single cysteine residue, responsible for recognition of the sorting motif.409 This family of enzymes, dubbed sortases, is present in other gram-positive bacteria including B. subtilis, E. faecalis, S. pnuemoniae and S. pyogenes,410 and most bacterial species contain multiple sortase enzymes (A through F), performing different roles in vivo, such as pilus polymerization. While the number of characterized and hypothetical sortase enzymes continues to grow,411 the most widely studied member of the family remains Sortase A from S. aureus (SaSrtA).
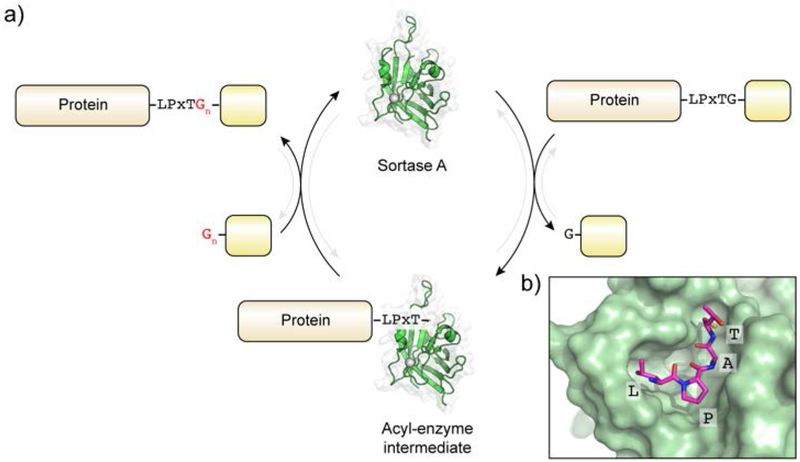
The enzymatic reaction carried out by sortases, and in particular Sortase A, has been widely studied in vitro and is well understood.412, 413 The ~145 residue transpeptidase domain of Sortase A (lacking the N-terminal signal peptide/membrane anchoring sequence) recognizes substrates containing the LPxTG sorting-motif (Figure 19a). The enzyme cleaves the amide bond between the Thr and Gly residues through the catalytic activity of the conserved nucleophilic cysteine, with base catalysis provided by a nearby histidine residue, generating an acyl-enzyme thioester intermediate. In the presence of glycine-terminated peptide nucleophiles, this intermediate is resolved through nucleophilic attack of the α-amine, resulting in a transpeptidation product. In vivo, this nucleophile corresponds to the oligoglycine chain in peptidoglycan cross-bridges. In vitro, peptide substrates containing a minimum of one N-terminal Gly residue are substrates for transpeptidation, although two or more glycines are preferred by the enzyme.412 The specificity for the pentapeptide motif has been illuminated through biochemical and structural analysis of the recombinant enzyme. Kinetic analysis of SaSrtA activity on peptide libraries demonstrated that only changes to position three of the LPxTG motif are tolerated by the enzyme.414 An NMR structure of a covalent complex of SaSrtA and a substrate peptide mimic illuminated how this specificity of SaSrtA for the LPxTG sorting sequence is achieved, specifically through binding to a hydrophobic pocket adjacent to the active site (Figure 19b).415
Figure 19.
Transpeptidation by sortase A. a) Catalytic cycle of SrtA-mediated transpeptidation. b) Close up of the SrtA-substrate complex (PDB ID: 2KID). Substrate is depicted in magenta.
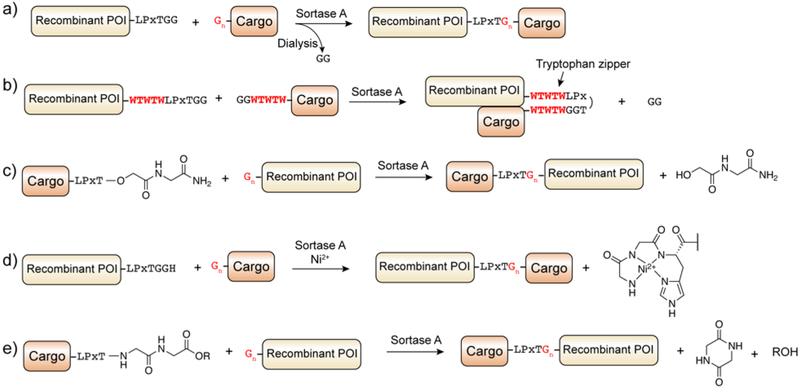
The minimal substrate requirements of SaSrtA suggested that the catalytic activity of this enzyme could be used for enzymatic ligation of peptides and proteins. In a pioneering study, Mao and coworkers used the enzyme to conjugate a range of synthetic peptides to the C-terminus of the green fluorescent protein (GFP), including peptides containing modified amino acids.17 This method of protein conjugation, known as Sortase-Mediated Ligation (SML) and more simply “sortagging”,416 offered a way to modify recombinantly expressed proteins in a controlled manner (Figure 20). The substrate requirements for a protein are minimal, namely that the protein contains either the short pentapeptide motif embedded within an accessible region of the protein (for C-terminal modification), or a N-terminal glycyl-acceptor sequence for (N-terminal modification). These substrates can be easily accessed through recombinant protein expression,417 or through the action of SaSrtA itself.418 Furthermore, cognate synthetic peptide substrates can be accessed through routine SPPS using standard amino acids. The simplicity of the reaction has led to sortagging being widely used to label proteins with a synthetic cargo.419, 420
Figure 20.
Sortase-mediated ligation (sortagging) for the a) C-terminal or b) N-terminal modification of proteins of interest (POI).
Sortase-mediated transpeptidation reactions are inherently reversible, as the product (unavoidably containing a regenerated LPxTG motif) and the glycyl-leaving group are both substrates for the enzyme. In order to drive the reaction to completion, a large excess of the synthetic peptide is typically used to ensure that the reaction equilibrium is pushed towards the product side.17 As this is not always feasible (for example, the peptide may have limited solubility or be difficult to prepare in large amounts), several ways have been found to push the reaction towards completion. This has been achieved most simply through the removal of the small glycyl-leaving groups through dialysis (Figure 21a).421–423 Alternatively, the introduction of a stable β-hairpin structure within the LPxTG motif, through the use of a tryptophan zipper, was found to increase the sortase-mediated ligation between two proteins (MBP and Trx), presumably forcing the sorting sequence in the product into a conformation that is not recognized by the enzyme (Figure 21b).424 Strategies that replace the pro-scissile Thr-Gly amide bond with a methyl ester425 or depsipeptide426 have been shown to increase the efficiency of the reaction, as these substrates generate methanol or hydroxylacetyl leaving groups that are not substrates for the reverse reaction (Figure 21c). Similarly, deactivation of the glycyl-leaving group through diketopiperazine formation,427 or through chelation in a nickel complex428 can prevent the leaving group from re-entering the catalytic cycle (Figure 21d,e). The use of metal chelation by Antos and coworkers is particularly convenient as it relies on native amino acids and can thus be used for peptide- or protein-bound sortase recognition motifs.428 In a conceptually distinct approach, Pentelute and coworkers found that by carrying the reaction out in a flow-based system, the concentration of acyl-acceptor could be decreased by an order of magnitude (300 μM in batch 10-20 μM in flow) while achieving comparable yields and fewer byproducts.429
Figure 21.
Strategies for improving the yield of sortagging reactions.
The low turnover rates of SaSrtA have motivated the engineering and evolution of this enzyme in order to increase the efficiency of transpeptidation (Figure 22). In an elegant application of directed protein evolution, Liu and coworkers used yeast display to anchor randomly mutated SaSrtA proteins to the cell surface, in proximity to one of the transpeptidation substrates.430 Iterative rounds of selection in the presence of decreasing concentrations of the cognate substrate afforded mutant sortase enzymes with up to 140-fold increase in catalytic efficiency compared with the wild type enzyme. Mutations to a loop flanking the LPxTG binding groove, conferred a ~30 fold reduction in Km for the sorting sequence.430 One of these mutants served as a starting point for additional improvements by Chen and coworkers, who used high throughput screening in vitro to identify additional beneficial mutations for enzymatic activity.431
Figure 22.
Evolving the transpeptidase activity of sortase A. a) Schematic for the evolution of sortase activity using yeast display.431 b) Locations of activating mutations mapped onto the structure of WT sortase A (PDB ID: 2KID). LPAT substrate shown in purple.
Using directed protein evolution, SaSrtA has also been reprogrammed to recognize sorting sequences other than the canonical -LPxTG- sequence.432 This has important implications for protein semisynthesis applications, where the -LPxTG- scar in the product may affect the structure and function of the protein. Using their previously developed yeast display method for sortase evolution, Liu and coworkers were able to identify SaSrtA mutants, “eSrtA(4S)” and “eSrtA(2A)”, that recognize -LPxSG- and -LAxTG- sequences in preference to LPXTG.432 The authors then demonstrate that the eSrtA(4S) enzyme is able to modify the native sequence of the protein fetuin A in human plasma. Schwarzer and coworkers used phage display to isolate SaSrtA mutants with relaxed substrate preference at the first position of the sorting motif.433 One such mutant (“F40”) preferred Ala, Asp, Ser, Pro and Gly over the native Leu, albeit at the expense of lower activity relative to the WT enzyme. However, screening of a second-generation library led to the identification of mutants with significantly improved activity for APXTG and FPXTG sequences.434 Importantly, the F40 sortase has found utility for the semisynthesis of N-terminally modified histone H3 through ligation at the native APATG sequence.435–437
The transpeptidation activity of SaSrtA requires Ca2+, which stabilizes the structure of the enzyme.415 The generation of Ca2+-independent mutants is of interest in order to extend the use of the enzyme inside living cells where Ca2+ levels are highly regulated. Sortase A from S. pyogenes (SpSrtA) is Ca2+-independent and has been used by Ploegh and coworkers for intra and intermolecular ligations in the cytosol of S. cerevisiae and HEK293T cells.438 Comparison of the Sortase A sequences from S. aureus and S. pyogenes revealed that calcium binding residues in SaSrtA can be mutated to the corresponding residues of SpSrtA (E105K, E108A/Q) to relieve Ca2+-dependence, although this does lead to a reduction in overall catalytic activity.439 However, combining these mutations with known activating mutations created SaSrtA variants with both increased activity and calcium independence.440–442 These engineered sortases have been shown to be active in living cells, allowing for protein labeling in E. coli443 and 293T cells,444 and even in C. elegans.445 In a particularly impressive recent application, Lang and coworkers used a Ca2+-independent version of a re-programmed eSrt2A enzyme to ligate ubiquitin and SUMO to target proteins in live cells.444 Ribosomal incorporation of unnatural lysine residues, bearing isopeptide-linked diglycines as acyl-acceptors, allowed the generation of SUMOylated and ubiquitinated proteins in a site-specific manner, catalyzed by a calcium-independent, sequence-reprogrammed sortase (Figure 23).
Figure 23.
Sortase-mediated ubiquitination.444
Despite the widespread use of sortase-mediated transpeptidation for the conjugation of biochemical and biophysical probes to proteins,420, 446 its use in traceless protein semisynthesis has been limited compared to NCL/EPL. This is likely due to the sortagging ‘scars’ left behind by the process, which place a significant constraint on the design of traceless (or near traceless) semisynthetic schemes. Nonetheless, the continued use of sophisticated protein evolution methods to these enzymes suggests that these constraints will continue to be loosened, allowing sortagging to be increasingly employed in this area.
4.2. Peptide Asparaginyl Ligases
In plants, the formation of the cyclotide family of cyclic peptide natural products relies on enzymatic cyclization of the peptide backbone. This process is carried out by peptide asparaginyl ligase (PAL) enzymes,447 which may be considered a subset of ligase-type asparaginyl endopeptidases (AEP).448 Whereas AEPs primarily proteolyze Asx-Xaa bonds, PALs catalyze the transpeptidation of Asx-Xaa bonds to form intra- or intermolecularly ligated products. Two such PALs, butelase 1 and OaAEP1b, have been explored for use as peptide and protein transpeptidation catalysts.
4.2.1. Butelase 1
Tam and coworkers isolated a PAL enzyme from the butterfly pea C. ternatea that they named butelase 1.449 Unlike other asparaginyl endopeptidase enzymes, butelase 1 displays a preference for transpeptidation rather than hydrolysis of Asx-containing substrates (Figure 24), suggesting evolutionary specialization as a cyclase or ligase. Indeed, the ~38 kDa active enzyme was able to cyclize many non-native targets including kalata B1, SFTI, histatin-3 and thanatin in yields typically greater than 90% in a 1:400 ratio of enzyme-to-substrate. Importantly, butelase 1 was also able to catalyze transpeptidation in trans, between two small model peptides.
Figure 24.
Transpeptidation by butelase 1. a) Catalytic cycle of butelase 1-mediated transpeptidation. b) Close up of the butelase 1 active site complex (PDB ID: 6DHI).
Compared to sortase A, butelase 1 exhibits greater promiscuity in the sequences it is able to process. Biochemically, the enzyme processes the tripeptide motif –NHV- C-terminally to the asparagine residue, through the activity of a Cys-His-Asn catalytic triad (Figure 24b). The acyl-enzyme intermediate generated from this amidolytic reaction is then resolved through inter/intramolecular attack from an acyl-acceptor. Interestingly, attempts to intercept the acyl-enzyme intermediate with non-peptidic nucleophiles including thiols and hydrazine only afford products of hydrolysis instead of the corresponding thioester or hydrazide products.449 In an intermolecular context, a broad range of amino acids can be accepted as the first residue of the incoming acyl acceptor (i.e. the N-terminus), excluding acidic residues and proline.449 However, the enzyme has a strong preference for hydrophobic amino acids (Ile/Leu/Val) or Cys at the second position of the acyl acceptor peptide. In the case of intramolecular attack, specifically cyclization, these rules are much looser, and the enzyme is able to accept all amino acids at both the first and second positions, at least in certain combinations. Specifically, if either the first residue is Gly or the second residue is Leu, the adjacent position can be any of the 20 common amino acids. Surprisingly this sequence promiscuity even extends to many d-amino acids,450 which was used for the synthesis of analogues of cyclic peptide natural products consisting of d-amino acids except for the Asn-residue at the ligation site.
As per the reversibility of all known transpeptidation processes, butelase-mediated reactions liberate His-Val dipeptides that can serve as a competitive acyl acceptor, requiring excess substrate to drive the reaction to completion.449 To circumvent this problem, Tam and coworkers investigated the use of depsipeptide and thiodepsipeptide substrates for butelase 1.451 The thiodepsipeptide substrates, releasing thiols unable to compete in the transpeptidation reaction, afforded ligations between peptide substrates in >90% yield after 1 h (Figure 25). This strategy allowed for the N-terminal labeling of recombinant GFP and ubiquitin proteins, as well as the synthesis of complex antimicrobial peptide dendrimers.452 In the case of ubiquitin, where the accessibility of the N-terminus was limiting, adding a short linker afforded a labeling yield of ~95% with 0.001 equiv. of enzyme and only four equivalents of thiodepsipeptide reactant.
Figure 25.
Butelase 1-mediated labeling of ubiquitin using a thiodepsipeptide substrate.451
4.2.2. OaAEP1b
OaAEP1b is a PAL enzyme that was isolated from the Oldenlandia affinis plant,453 the source of the cyclotide kalata B1.454 This enzyme can be expressed in E. coli (this is currently not possible for butelase 1) and is able to catalyze the cyclization of non-cyclotide polypeptide precursors,453 as well as the ligation of protein fragments.455 Like butelase 1, OaAEP1b recognizes and processes an -NXX- motif, where XX is typically ‘GL’ (the processed sequence within kalata B1), although ‘AL’ and ‘CL’ sequences considerably improved protein cyclization.455 The catalytic efficiency of this enzyme (~200 M−1s−1) is significantly lower than that of butelase 1 (~100,000 M−1s−1).456 Analysis of a crystal structure of recombinant OaAEP1b identified a Cys residue performing a gate-keeper role near the substrate channel of the enzyme, impeding the catalytic cyclization and ligase activity of the enzyme.456 Mutation of this residue to Ala afforded a PAL enzyme with significantly increased catalytic efficiency (~35 000 M−1s−1) for peptide cyclization, and that was able to catalyze efficient transpeptidation of ubiquitin to short synthetic peptides and to proteins.
Additional identification and characterization of PAL enzymes such as OaAEP1b and butelase 1 will continue to shed light on the factors that favor transpeptidation over hydrolytic activity, including the effects of so-called gate-keeper residues.447, 457 Such information will be critical for the further development of this fascinating class of enzymes, through directed evolution or rational design efforts, for a host of protein chemistry applications.
5. PEPTIDE LIGASES
It is well established that protease enzymes can catalyze amide bond formation through reverse proteolysis between amines and carboxylic acids under certain conditions. In order to capitalize on this activity, extensive efforts have been made to engineer proteases to carry out efficient peptide ligation reactions for the purposes of protein engineering, including semisynthesis. In this section we will describe how proteases have thus been converted into so-called ‘peptide ligases’.
5.1. Subtiligase
In one of the first examples of protein semisynthesis, the serine protease subtilisin BPN’ was used to reassemble RNase A from a complex of two proteolyzed fragments.458 This thermodynamically unfavorable reaction is promoted by the presence of organic co-solvents. To control reverse proteolysis reactions kinetically, activated ester substrates have been used as substrates in the place of C-terminal carboxylic acids.459, 460 In such cases, the preference for hydrolysis of acyl-enzyme intermediates in both cases is greater than the subsequent aminolysis by a suitable acyl acceptor. To avoid such back reactivity, Kaiser and coworkers explored enzymatic coupling of acyl-esters with thiolsubtilisin,461 a chemically “damaged” mutant of subtilisin containing a Ser-to-Cys mutation in the active site (Figure 26a). 462, 463 Thiolsubtilisin, and indeed selenolsubtilisin,464 have greater preference for aminolysis over hydrolysis of acyl-enzyme intermediates, however it should noted that the initial esterase activity of the variants is below that of the WT enzyme.
Figure 26.
Peptide ligation using subtilisin variants. a) Synthesis of short peptides using thiolsubtilisin.461 b) Total synthesis of ribonuclease A containing unnatural catalytic residues using subtiligase.465
In order to improve the catalytic activity of thiolsubtilisin, Wells and coworkers found that an additional mutation to the variant (P225A) was able to relieve steric crowing in the active site, improving the esterase efficiency of the enzyme, which they named ‘subtiligase’.466 In a landmark example of protein synthesis, Wells and coworkers used subtiligase to assemble ribonuclease A from seven synthetic fragments, incorporating unnatural fluorohistidine residues in the active site (Figure 26b).465 Acyl-donor peptides were suitably functionalized with C-terminal glycolate-phenylalanyl amide esters as leaving groups for the ligation reactions, which averaged 70%. Five additional mutations made to subtiligase afforded a variant named “stabiligase” that retained nearly 50% activity in 4M guanidine hydrochloride.16 This allowed enzyme-mediated ligation of a synthetic peptide glycolate ester to a recombinant version of human growth hormone (hGH) protein in the presence of denaturants.
Like the protease from which it is derived, subtiligase engages substrates in an extended conformation, recognizing four amino acids N-terminal to the cleavage/ligation site, and three residues to the C-terminal side.467 Unlike transpeptidase enzymes such as sortase A, subtiligase displays flexibility in the sequences that it can process in the P and P’ sites. On the N-terminal side of the ligation site, the P4 and P1 positions dictate much of the substrate specificity, preferring large hydrophobic residues.466, 468 At the P1’ and P2’ sites, C-terminal to the ligation site, subtiligase accepts most residues except for acidic or β-branched residues at the P1’ position.16 Weeks and Wells recently used a mass-spectrometry-based approach to further profile the specificity of subtiligase for the P1’ and P2’ sites (Figure 27).469 Peptide libraries derived from proteolytically digested E. coli proteome, containing nearly each of the 400 P1’-P2’ dipeptide combinations, were subjected to subtiligase-catalyzed ligation to a probe peptide and analyzed for labeling efficiency. The results indicated that the best substrates had a small amino acid, Met or Arg at the P1’ position and a large hydrophobic or aromatic residue at the P2’ position. Saturation mutagenesis of hot spots that contribute to this specificity led to the identification of subtiligase mutants with a greater substrate tolerance at these key positions. The broadened substrate scope of this library of subtiligase mutants extends the utility of the technology for the tagging and enrichment of neo N-termini in native proteomes,470 as well as for the traceless assembly of semisynthetic proteins. With respect to the latter, the Y217K mutant identified in this screen has been used for cysteine-free semisynthesis of PTEN,471 (See section 6.3 Enzyme-Catalyzed Expressed Protein Ligation). As a further illustration of the tolerance of the subtilisin family to protein engineering efforts, Janssen and coworkers introduced S212C and P216A mutations into a hyperstable, calcium independent variant of subtilisin BPN’, affording a peptide ligase termed, “peptiligase”.472 Further refinements of this system generated a variant, “omniligase”, with broad tolerance for both the P1’ and P2’ positions.473 In light of the recent use of subtiligase for the catalysis of cysteine-free EPL (see Section 6.3 Enzyme-Catalyzed Expressed Protein Ligation), improved subtilisin-derived peptide ligases will continue to find use as catalysts for traceless protein semisynthesis.
Figure 27.
Comprehensive characterization of subtiligase specificity by mass spectrometry.469
5.2. Trypsiligase
Trypsin has long been known to catalyze amide-bond formation under certain conditions.474 Through engineering of the structure of trypsin, Stubbs, Bordusa and coworkers have created a peptide ligase named ‘trypsiligase’ which is able to carry out both transpeptidation and ligation of peptides and peptide esters (Figure 28).475 This version of trypsin possesses four mutations that convert the protease into a transpeptidase enzyme recognizing the tri-peptide motif (-YRH-). In this case, the enzyme cleaves this motif C-terminally to the Tyr residue to form an acyl-enzyme intermediate, that may be intercepted by peptides bearing Arg-His N-terminal dipeptide sequences. This method has been used for the C-terminal labeling of the Fab region of the anti-Her2 antibody with a synthetic peptide,476 although the yield of the reaction suffered from the reversibility and hydrolysis observed for all transpeptidation reactions. Alternatively, trypsiligase accepts substrate mimetics as acyl donors, such as 4-guanidinophenyl (OGp) esters (Figure 28b),477 which can be ligated to acyl acceptors.475 Using this method, proteins including cyclophilin 18, human epidermal growth factor and parvulin 10 were modified N-terminally with various synthetic cargoes, including biotin, PEG and fluorescent dyes.475 While not yet applied beyond labeling, it will be interesting to see if this method is extended to the semisynthesis of proteins, especially those modified within their N-terminal regions, generating only a small -RH- scar. In instances where this motif is contained within the native sequence (e.g. histone H4), trypsiligase could provide one-step traceless protein semisynthesis.
Figure 28.
Trypsiligase catalyzed modification of proteins. a) C-terminal transpeptidation using trypsiligase. B) N-terminal modification of proteins using 4-guanidinophenyl esters.478
6. HYBRID CHEMOENZYMATIC LIGATION TECHNOLOGIES
The technologies described in the preceding sections are usually considered to be separate tools for protein engineering, despite sharing underlying chemistry to promote amide-bond formation, namely, enthalpic activation of the acyl donor, often as a high energy thioester intermediate, and the use of a template (i.e. the substrate binding pocket of an enzyme) to direct amide coupling. Since, as discussed, each of these activation modes comes with associated benefits and weaknesses, researchers have investigated how mixing and matching aspects of these technologies, to create what we will refer to as hybrid ligation methods, might lead to improved routes to semisynthetic proteins. Here we will briefly describe examples of these strategies.
6.1. Transpeptidase-mediated synthesis of protein thioesters
The predominant strategy for the preparation of protein α-thioesters used in semisynthesis schemes is the thiol-mediated cleavage of protein-intein fusions. As an alternative route to these reactive building blocks, the groups of Pentelute and Tam each capitalized on sequence specific transamidation reactions, affording protein α-thioesters that were used in downstream NCL reactions.479, 480 Pentelute and coworkers used sortase-mediated transpeptidation to install an α-thioester group into recombinant proteins bearing C-terminal sorting sequences (Figure 29a).479 Of note, the authors used this approach to insert a synthetic d-peptide sequence between two recombinant protein domains, namely the lethal factor N-terminal domain (LFN) from anthrax toxin and the diptheria toxin α-chain (DTA). The resulting construct that was able to translocate into CHO-K1 cells and inhibit protein synthesis. Tam and coworkers used butelase 1 to create α-thioesters through high yielding transpeptidation between proteins bearing C-terminal NHV tags and glycyl-thioesters.480 This strategy was integrated into a technically impressive sequential modification process (also featuring NCL and sortase-mediated ligation steps) that allowed dual labeling of the N- and C-termini of a recombinant ubiquitin protein (Figure 29b).
Figure 29.
Transpeptidase-mediated assembly of protein thioesters in multistep ligations. a) Sortase-mediated installation of an α-thioester into a recombinant protein for use in NCL.479 LFN, lethal factor N-terminal domain from anthrax toxin; DTA the diptheria toxin α-chain. b) Dual labelling of ubiquitin through EPL of a butelase-assembled protein a-thioester with a synthetic biotinylated peptide, followed by N-terminal sortase-mediated ligation with a fluorescein labeled peptide.480
6.2. Streamlined Expressed Protein Ligation
Split inteins have been used as a way to access protein α-thioesters in a manner that is more controlled than using their contiguous counterparts. This method, named ‘streamlined expressed protein ligation’ (SEPL), capitalizes on the ability to trigger the acyl-shift reactions in the protein splicing cascade through user-controlled reconstitution of the split intein domain.481 In doing so, the premature cleavage of protein-intein fusions that can occur in vivo and during purification can be altogether avoided. Fusion proteins bearing the IntN fragment of the Npu DnaE split intein were converted to α-thioesters through incubation with the thiol reagent MESNa and the cognate IntC fragment, mutated at both the C-terminal asparagine (N137A) and the C-terminal extein (C+1A) residues (Figure 30).481 The strong and specific interaction between the intein fragments allowed this to be performed using an immobilized IntC fragment as a purification step. Importantly, protein α-thioesters bearing each of the 20 amino acids at the C-terminus could be isolated, with the exception of asparagine. This method has been used in the semisynthesis of histones bearing a variety of PTMs,155, 156, 481 the generation of antibody drug conjugates,481–483 and to install biophysical probes into proteins.166, 179 Notably, the SEPL method is likely to be a direct beneficiary of the ongoing efforts to discover and engineer split inteins with improved physicochemical and biochemical properties (see section 3.3 Engineering of split inteins).
Figure 30.
Streamlined EPL using split inteins.481
6.3. Enzyme-Catalyzed Expressed Protein Ligation
The successful use of NCL/EPL for protein semisynthesis is very much dependent upon the process being near traceless. However, the reaction invariably requires a cysteine or related acyl acceptor to facilitate amide-bond formation through proximity-driven entropic activation. To remove this limitation, Cole, Wells and coworkers have used the templating ability of subtiligase to enable ligation reactions between synthetic non-cysteinyl peptides and recombinant protein thioesters generated from intein-fusions (Figure 31).471 Critically, the authors used this approach to generate semisynthetic PTEN in a traceless manner through the ligase-catalyzed condensation between a recombinant fragment containing a C-terminal tyrosine α-thioester and a peptide bearing an N-terminal RY dipeptide sequence.227, 471 Semisynthesis of PTEN using EPL previously required the installation of a non-native Cys residue for the ligation reaction, a mutation which has been found to affect the behavior of the protein in cells.471, 484 Using the enzyme-templated approach, Cole, and coworkers generated both monophosphorylated227 and tetraphosphorylated471 forms of PTEN, which is autoinhibited by these PTMs (see section 7.5.3 for further details). It is anticipated that advances in the engineering of subtiligase and related variants (described earlier) will directly benefit this approach to protein semisynthesis, increasing both the reaction efficiency and sequence tolerance.
Figure 31.
Enzyme-catalyzed expressed protein ligation. Scheme showing intein-mediated generation of a recombinant protein α-thioester, followed by EPL with a cysteinyl peptide (upper) or subtiligase-catalyzed EPL with a non-cysteinyl peptide (lower).471
6.4. Transpeptidase-Assisted Intein Ligation
Protein trans-splicing using ultra-fast split inteins offers an efficient way to re-assemble proteins from recombinant and synthetic fragments at low concentrations. However, the length of the split intein fragments, one of which must be accessed synthetically, can render this approach impractical for protein semisynthesis. Recently, it was demonstrated that the individual split intein fragments could be assembled through a prior transpeptidase mediated ligation reaction.485 Specifically, variants of the enhanced Cfa split intein, mutated to contain recognition sequences for the evolved eSrt2A sortase,432 are assembled from synthetic and recombinant fragments through initial enzyme-mediated transamidation, with the resulting product then undergoing efficient PTS in situ with a complementary intein fragment fused to a protein of interest (Figure 32). A key advantage of this tandem transamidation process is that it reduces the size of the appended intein sequence to a short 7 amino acid peptide tag, which is easily accessible using standard SPPS. This strategy was used for C- and N-terminal labeling of proteins, such as Cas9,486 as well as the traceless semisynthesis of phosphorylated forms of the chromatin architectural proteins linker histone H1 and MeCP2. Further, the successful use of the methodology for the labeling of histones in chromatin in nucleo shows that it is compatible with complex cellular environments. As with other hybrid approaches, this semisynthesis technology will profit from ongoing efforts to engineer improved split inteins and transpeptidases.
Figure 32.
Generation of a C-terminally modified semisynthetic protein using transpeptidase-assisted intein ligation (TAIL).485
7. APPLICATIONS OF PROTEIN SEMISYNTHESIS
The development of new and improved strategies for protein semisynthesis has been driven by the need to understand protein structure and function. Many areas of biological inquiry have benefited greatly from the precision generation of modified proteins, using both chemical and enzymatic approaches. In this section we highlight some key areas where protein semisynthesis methods have helped fuel biological discovery, with emphasis placed on developments that have occurred in the last decade.
7.1. Chromatin modifications
Eukaryotic genomes are packaged into a dense nucleoprotein complex known as chromatin. Like beads on a string, the DNA is spooled into small repeating units known as nucleosomes.487 Each nucleosome contains around 147 bp DNA wound around an octameric complex of histone proteins (two copies each of H2A, H2B, H3 and H4). The disordered N- and C-terminal tails of histones that protrude from the nucleosome core have long been known to serve as hubs for a plethora of PTMs (Figure 33), which have been correlated with the transcriptional state of the underlying genomic locus.488 Accordingly, unraveling the complex biochemical and biophysical implications of histone modifications (colloquially referred to as ‘marks’) has become a dynamic and cross-disciplinary area of inquiry, in which peptide and protein synthesis have played pivotal roles (for detailed accounts that emphasize the important role of chemistry in the area, we direct the reader to the following reviews489–494).
Figure 33.
Histone modifications. a) Structure of a mononucleosome (PDB ID: 1KX5). b) Primary structure of histone tails with important modifications annotated. Ac, acetyl; Cit, citrullyl; Me, methyl; Ph, phosphoryl; Ub, ubiquityl.491
7.1.1. Histones
The generation of modified histone proteins by protein semisynthesis has been especially fruitful for many reasons, the proximity of most PTMs to the N- and C-termini, the ability of histones to be refolded (allowing the use of chaotropes in assembly reactions), and the general lack of endogenous cysteine residues. This last feature has allowed cysteine-specific chemistry to be used in conjunction with amide-bond formation (such as radical desulfurization,337 sulfhydryl-alkylation165, 495 or disulfide formation496, 497) to create completely native sequences or close mimics thereof. In a pioneering study that employed an experimental workflow broadly adopted in the many studies that followed (Figure 34a), Peterson and coworkers used semisynthesis to generate ‘designer’ chromatin containing a specific histone PTM, in this case histone H3 phosphorylated at position S10 (H3S10p), and then used this reagent to gauge how the mark impacted downstream chromatin biochemistry.498 The desired semisynthetic protein was assembled from a synthetic peptide α-thioester spanning residues 1-31 of histone H3 and a recombinant truncated histone spanning residues 32-135, bearing a Thr32Cys mutation necessary for the ligation reaction. The purified semisynthetic protein was refolded into histone octamers with recombinant histones H2A, H2B and H4, followed by deposition onto a DNA fragment containing a tandem array of nucleosome positioning sequences. These nucleosome arrays were then assayed for phosphorylation-dependent effects on chromatin remodeling by the SWI-SNF complex, as well as effects on the acetyltransferase activity of the Gcn5-containing SAGA complex. In contrast to previous studies that employed histone peptide substrates,499, 500 the acetyltransferase activity of the SAGA complex was not stimulated by phosphorylation in a chromatin context, highlighting the importance of using these more physiologically relevant substrates. In the same year, McCafferty and coworkers used a similar NCL-based strategy to prepare polyacetylated (K4Ac, K9Ac, K14Ac, K18Ac and K23Ac) and methylated (K9me3) forms of histone H3, as well as polyacetylated (K5Ac, K8Ac and K12Ac) histone H4.161 Importantly, the authors were able to achieve completely traceless semisynthesis of these proteins through a post-ligation Cys-to-Ala conversion at the ligation junction by means of hydrogenolytic desulfurization (Figure 34b).336 In the years following these first studies, the combination of chemical ligation and cysteine desulfurization (typically now by radical means)337 has been extensively used for the traceless semisynthesis of N-terminally (and C-terminally) modified forms of all four histones (see Table 1). Indeed, histones are without question the most heavily studied proteins using the semisynthesis paradigm.
Figure 34.
Semisynthesis applied to histone H3. a) Semisynthesis of H3Ser10ph through NCL, and incorporation into nucleosome arrays (Shogren-Knaak et al.).498 b) Traceless semisynthesis of polyacetylated H3 using NCL and desulfurization (He et al.).161 c) Traceless semisynthesis of H3 by sortase-mediated ligation using the F40 sortase.
Sortase-mediated ligation has also been used to access semisynthetic histone H3.433, 436, 437 The engineered “F40” sortase recognizes residues 29-33 of histone H3 (APATG) thereby allowing assembly of the native protein from a synthetic peptide, spanning residues 1-33 of histone H3 (containing the non-canonical sortase sequence), and a truncated recombinantly expressed form of histone H3 (33-135) (Figure 34c). Cole and workers used this strategy to prepare nucleosomes containing semisynthetic H3K9Ac as substrates for the CoREST transcriptional repressor in an effort to develop dual action inhibitors against the HDAC and demethylase activities within this multi-protein complex.437 In order to increase the yield of the reaction, this same group subsequently used synthetic glycolate-containing depsipeptides as a way of overcoming the reversible nature of the transpeptidation reaction.435 The increased efficiency associated with this adjustment allowed for the production of various semisynthetic H3 proteins, combinatorially modified with dimethylation at K4 and acetylation at K9, K14 and/or K18. These modified proteins were subsequently used to interrogate the rates of demethylation and deacetylation by the CoREST complex. Although sortase-mediated semisynthesis of H3 cannot currently provide direct access to modifications beyond residue 33, this method has many benefits over other strategies including NCL and desulfurization, by providing access to the native protein sequence in a single step.
Semisynthetic histones bearing C-terminal modifications have been accessed using a combination of EPL and desulfurization chemistry.155, 157, 159, 174, 501 In the first synthesis of ubiquitinated H2B, a synthetic peptide spanning residues 118-125 of the protein was ligated through the side-chain of K120 using an auxiliary-mediated ligation, to a recombinant ubiquitin α-thioester produced from thiolysis of an intein fusion (Figure 35a).345 Photolytic removal of the auxiliary and a protecting group masking the N-terminal cysteine afforded an ubiquitinated peptide that was then ligated to a recombinant protein α-thioester spanning residues 1-116 of the histone, accessed through intein thiolysis. Desulfurization of the non-native Cys residue afforded the native ubiquitinated protein. Subsequent biochemical assays with mononucleosomes containing H2BK120ub demonstrated that this modification stimulates the methyltransferase activity of the catalytic domains of Dot1159 and Set1502–504 (Figure 35b). A revised semisynthesis of ubiquitinated H2B replaced the use of a thiol auxiliary with a Cys residue, effectively creating a G76C mutation of ubiquitin upon ligation that, upon desulfurization, afforded a G76A scar.505 More facile access to ubiquitylated histone H2B, either through this method or through disulfide mimetics, has allowed detailed studies into the role of the PTM in methyltransferase stimulation,310, 496, 506 as well as its impact on chromatin stability and structure.157, 497, 507 This streamlined synthetic approach was also adopted for the semisynthesis of ubiquitinated H2A (H2AK119ub),157 following a similar strategy of dual EPL reactions and subsequent desulfurization, although penicillamine was used as a surrogate for Cys in the second ligation reaction, which was converted to a native Val through desulfurization. Subsequently, auxiliary-directed ligation approaches have been revisited to achieve traceless semisyntheses of H2AK119ub,158 as well as the DNA damage associated H2AK13ub and H2AK15ub.81 It should be stressed that this histone ubiquitination work has stimulated a tremendous amount of peptide and protein chemistry research, studies that are further motivated by the multifarious roles of ubiquitin in eukaryotic biology and the need to access ubiquitinated polypeptides for biochemical studies. Indeed, there have been several highly impactful methodological advances in this area, which have collectively advanced the ubiquitin field greatly. The interested reader is referred to several excellent reviews on this topic.508–511
Figure 35.
Semisynthesis applied to ubiquitinated histone H2B. a) Scheme for the semisynthesis of ubiquininated histone H2B from three fragments.159 b) PTM crosstalks biochemically verified using semisynthetic ubiquitinated histone H2B.
An additional improvement to the generation of modified histones came courtesy of the streamlined EPL protocol,481 which as discussed earlier uses ultrafast split inteins for the generation of protein α-thioesters. This method was used in the preparation of an α-thioester of H2B (residues 1-116) subsequently employed in the semisynthesis of H2BK120Ac. Fierz and coworkers adopted the SEPL approach for the impressive semisynthesis of the histone variant H2A.X phosphorylated at serine 139 (known as γH2A.X) and ubiquitinated at K15 (Figure 36),156 a combination of histone PTMs that is important for the DNA-damage response.512 The remote positions of these modifications, on both the N- and C-termini, required that the authors assemble the histone sequence from three pieces, a synthetic C-terminal cysteinyl peptide spanning residues 135-142 (containing S139ph), a recombinant central portion spanning residues 21-134, and a K15 ubiquitinated N-terminal fragment containing residues 1-20 (generated itself through EPL between a recombinant ubiquitin α-thioester and a synthetic peptide). In order to mask reactive functionalities, the authors expressed the central recombinant portion as an N-terminal SUMO fusion and a C-terminal fusion with the NpuN DnaE split intein. The ligation of this construct to the C-terminal fragment was first achieved through co-incubation in the presence of the complementary NpuC fragment, and additional thiols MESNa and methylthioglycolate, which formed the reactive α-thioester in situ. Following the ligation, treatment with Ulp1 protease unmasked the γH2A.X (21-142) bearing an N-terminal cysteine for the second ligation reaction with the ubiquitinated N-terminal semisynthetic peptide hydrazide. Following ligation and desulfurization, the purified proteins were incorporated into chromatin carrying FRET pairs, and the conformation analyzed on the single-fiber level through total internal reflection fluorescence (TIRF) imaging. Accordingly, it was found that while S139ph does not directly alter chromatin structure, the presence of K15 ubiquitination impairs the formation of higher order chromatin folding.
Figure 36.
Semisynthesis of ubiquitinated and phosphorylated histone H2A.X.156
The discovery of many non-canonical histone modifications has driven the development of new chemistry to access modified nucleosomes. While long known to be ubiquitinated, histone proteins are also modified through SUMOylation,513 with H4K12 sumoylation (H4K12su) being associated with gene repression.514 Chatterjee and coworkers generated H4K12su semisynthetically through a sequence of auxiliary-mediated EPL reactions between a synthetic peptide α-hydrazide and a recombinant SUMO-3 α-thioester (generated from a Mxe GyrA intein fusion), the product of which was then ligated to recombinantly expressed truncated H4.176 Reduction of both the 2-(aminooxy)ethanethiol ligation auxiliary344 and the reactive cysteine (through desulfurization) at various points in the synthesis generated the native sumoylated protein. Interestingly, nucleosomes containing both H4K12su and H3K4me2 were preferred substrates for demethylation by the CoREST complex, dependent on a functional SUMO-interacting motif in CoREST that is proposed to recruit the complex proximal to the H3 tail.176 The synthetic strategy used for the synthesis of H4K12su has been similarly applied to the semisynthesis of sumoylated H2B, although the functional consequences of this modification are yet to be uncovered.160
The covalent attachment of the neurotransmitter serotonin was recently found to occur on Gln5 of histone H3,165 catalyzed by the enzyme tissue transglutaminase 2 (TGM2). In order to test whether Q5 serotonylation can affect the deposition of methyl marks at the proximal H3K4 site, semisynthetic serotonylated H3 was generated through NCL (Figure 37). A synthetic peptide α-thioester spanning residues 1-13 of H3 was synthesized through the thioester-generating SEA-linker,59 and serotonin coupled to an orthogonally protected glutamate residue at position 5 to generate the serotonylated glutamine. Cleavage from the resin afforded a crude serotonylated SEA-functionalized peptide that was converted to a mercaptopropionyl thioester through N-to-S acyl shift and transthioesterification. This peptide was subjected to NCL with a recombinantly produced H3(14-135) bearing an N-terminal cysteine, followed by cysteine alkylation with bromoethylamine329 to give the ligated product with a pseudolysine mimic in place of the native lysine at position 14. Unexpectedly, the presence of the bulky serotonin moiety at H3Q5 did not affect methylation of H3K4 in nucleosomes by the MLL1 methyltransferase complex. Likewise, nucleosomes either unmethylated or trimethylated at H3K4 were equally modified with serotonin by TGM2, shedding light on the observed coexistence of these marks in cells.165
Figure 37.
Semisynthesis of serotonylated histone H3 using NCL followed by cysteine alkylation.165
7.1.2. Asymmetrically Modified Nucleosomes
Nucleosomes have intrinsic two-fold pseudo-symmetry, reflecting the fact that there are two copies of each histone in the nucleoprotein complex.487 In vitro reconstitution of nucleosomes from recombinant and/or semisynthetic histones typically results in retention of this symmetry, since the two copies of each histone in a nucleosome will contain the same modification(s). This is a property that is not altogether shared with nucleosomes in vivo, for example, so-called bivalent domains at developmentally regulated promotors contain nucleosomes bearing both activating (H3K4me3) and repressive (H3K27me3) marks on separate tails.515 Accordingly, efforts have recently been directed to the generation of asymmetrically modified nucleosomes to inform on biochemical cross-talk,515–518 or asymmetric processing mechanisms by remodeling enzymes.519, 520 Initial efforts to generate asymmetric nucleosomes involved refolding of mixtures of modified histones into octamers, followed by two-step affinity purification through non-native tags to generate asymmetrically modified octamers that could be reconstituted onto DNA templates.515 More recently, Fierz and coworkers designed a method to produce nucleosomes containing asymmetrically modified histone H3 through the use of traceless chemically tethered precursors (Figure 38a).516, 518 Semisynthetic H3 proteins, trimethylated at K4, K27 or K36 were fused as asymmetric disulfides through short cysteine containing N-terminal linker sequences. Nucleosomes formed from refolded histone octamers containing these tethered constructs could then be converted in a traceless manner into the native asymmetric nucleosomes through reduction of the disulfide tether and liberation of the linkers with TEV protease. Importantly, these native bivalent nucleosomes validated previous studies suggesting that stimulation of the H3K27 methyltransferase activity of the enzyme PRC2 could be achieved intranucleosomally,515 and that interference of H3K27 methylation by pre-existing H3K4 or K36 methyl marks can only occur on the same histone tail. Notably, these reagents have also proven extremely useful for the development of multivalent probes that specifically sense bivalent nucleosomes in cells.162 This tethering strategy for generating asymmetrically modified nucleosomes has been extended to histone H4, in this case using tethering sequences linked to a lysine side-chain in order to access the natively N-terminally acetylated protein.517 In this way, the authors were able to observe the distributive mechanism of H4K20 methylation by the methyltransferase Set8. Bowman and coworkers have generated asymmetrically modified nucleosomes through the stepwise association of H2A/H2B dimers onto the asymmetric Widom 601 DNA sequence, which is commonly used in nucleosome constitutions (Figure 38b).521 Observing preferential association between H2A/H2B dimers and the TA-rich half of the 601-sequence, the authors were able to purify hexasomal species containing one H2A/H2B dimer, to which another dimer could then be added to complete the nucleosomal structure.519 By generating highly oriented nucleosomes containing a single ubiquitinated H2B, the authors were able to show that this modification can stimulate the activity of the chromatin remodeler Chd1, but only when present on the entry side of the nucleosome (i.e. the side positioned to bind DNA that is pulled onto the nucleosome through remodeling activity). This strategy has also proven useful for the asymmetric labeling of nucleosomes with fluorescent tags, allowing single-molecule FRET analysis of the nucleosome remodeling process.522 We expect that both of these complementary approaches will find further utility in the dissection of the fine molecular mechanisms underpinning the asymmetric sensing of modified nucleosomes and nucleosome arrays by chromatin factors.
Figure 38.
Strategies for the synthesis of asymmetrically modified nucleosomes. a) Reconstitution of asymmetric nucleosomes containing bivalent H3K4me3 and H3K27me3 modifications using traceless disulfide-tethering of semisynthetic H3 proteins.516, 518 b) Stepwise incorporation of histone tetramers and dimers onto the Widom 601 nucleosome positioning DNA sequence.521
Methodological advances have greatly simplified the preparation of modified histone proteins, however elucidating the biochemical effects of each modification remains a daunting task. Moreover, the compounding effects of post-translational modifications, histone variants, disease-associated mutations,523 not to mention DNA modifications,524, 525 creates an overwhelming number of nucleosomal substrates to test individually. The use of DNA-barcoding has allowed for such biochemical experiments to be performed simultaneously on a library of uniquely modified nucleosomes, each identifiable using DNA-sequencing (Figure 39).77 This nucleosome library was first used to profile binding specificity of histone reader proteins, stimulation of p300 acetyltransferase activity in vitro, and multiple PTM cross-talk relationships on a systems level through incubation with an active nuclear extract. Understanding the substrate preference of the ISWI family of chromatin remodeling enzymes was achieved using an expanded 115-member library of modified mononucleosomes.155 Using a previously established assay for chromatin remodeling that is coupled to DNA cleavage by a restriction enzyme, the kinetics of remodeling were measured for 6 unique complexes, which were shown to display differing preferences for the modification state of a nucleosome. Critically, the nucleosome acidic patch, a binding site used by many chromatin binding proteins enzymes,487 was found to be an important regulatory epitope for each of the remodelers. Substrate-profiling using this library has been subsequently used for histone ADP-ribosylation by poly(ADP-ribose) polymerase (PARP) enzymes526 and ubiquitination of H2B by the UBE2A:RNF20/40 ligase machinery.501 In both cases, novel modes of enzyme regulation were revealed, specifically an inhibitory role for H3K9 acetylation in the case of PARP-mediated H3S10 ADP-ribosylation and a stimulatory role of for H2A tail in the case of H2B ubiquitination.
Figure 39.
DNA-barcoded nucleosome library for high-throughput chromatin biochemistry.77 Next-generation DNA sequencing allows for experiment multiplexing and quantitative read-out of individual nucleosome library members.
7.1.3. Chromatin-Associated Proteins
Protein semisynthesis has been used in the study of chromatin-associated proteins other than histones (Table 1). As an example, Fierz and co workers used SEPL to assemble obligate dimers of HP1α,179 a chromatin effector that binds H3K9me3 and induces chromatin compaction.527 This was achieved using an affinity-directed dual EPL approach that resulted in the C-terminal chromo-shadow domain in each copy of the protein being linked via a peptide adaptor derived from the protein shugoshin,528 which has high affinity for the HP1α dimer. The authors found that this covalent dimer had both prolonged binding to chromatin containing semisynthetic H3K9 methylation, as well as increased binding frequency as measured by single molecule TIRF microscopy, indicating that multivalency of HP1α is an important feature for fast and efficient binding to H3K9me3 containing heterochromatin domains.179 In other work, Margueron and coworkers used NCL to generate a methylated form of Jarid2, a cofactor of the PRC2 methyltransferase complex that is itself modified by this enzyme.189 A synthetic peptide α-thioester containing residues 109-123 (containing K116me3) was ligated to a recombinant fragment spanning residues 124-450 to give a semisynthetic fragment of the Jarid2 protein. Interestingly, methylation at this position was shown to bind the EED subunit of PRC2 and stimulate the activity of the catalytic EZH2 methyltransferase subunit of the complex. A recent study, also using semisynthetic Jarid2 fragments, has shown that this stimulation of PRC2 activity on chromatin can be modulated by the presence of specific phosphoserine residues near the site of Jarid2 methylation.190 As another example in the area of chromatin-associated proteins, split DnaE inteins have been used to attach various synthetic cargoes to a nuclease dead version of CRISPR-Cas9 (dCas9) which were subsequently delivered into living cells in complex with guide RNAs for genomic-targeting.486 This system was used to recruit various epigenetic factors to user-defined loci, taking advantage of the availability of small molecule binders to component domains.
7.1.4. Semisynthetic Modification of Native Chromatin
With the help of ultra-fast split inteins, the semisynthesis of histones has been extended to a native chromatin setting.397, 401, 506 In a semisynthesis of H2B ubiquitinated on K120, histone H2B (1-116) was expressed in HEK 293T cells as a fusion to the IntN fragment of the Ava DnaE split intein (Figure 40).401 Isolated nuclei from these cells were then treated with a semisynthetic ubiquitinated H2B peptide (117-125) fused to a complementary IntC fragment, generating the semisynthetic protein through PTS on chromatin. The presence of this mark was sufficient to induce the upregulation of H3K79 methylation, a crosstalk that has been well-characterized in vitro.159 Notably, the ability to manufacture ubiquitinated H2B in nucleo proved instrumental in a subsequent mechanistic dissection of this crosstalk that led to the identification of a functional epitope on the ubiquitin surface required for stimulation of the H3K79 methyltransferase Dot1.506 Access to N-terminally modified histones by PTS in nucleo has also been achieved through the use of the promiscuous CfaGEP DnaE split intein.397 Histone H3 containing the K27me3 heterochromatin mark was generated through the treatment of cell nuclei expressing histone H3 with the IntC fragment embedded in the N-terminal tail with a semisynthetic construct corresponding to the IntN fragment fused to H3 (1-28) containing K27me3.
Figure 40.
Semisynthesis of ubiquitinated histone H2B in isolated nuclei.401
The ability to modify chromatin in nucleo can be considered an intermediate step between in vitro reconstitution of designer chromatin, which is now a fairly established field,489 and the ultimate goal of chemically customizing chromatin in a locus specific manner in living systems.529 While there has been some progress in performed histone semisynthesis in living cells, by delivering split intein fusions into transfected cell lines,401 this does not provide control over genomic location. Efforts to achieve this, perhaps involving the genomic targeting capabilities of dCas9, will certainly test the ingenuity of chemical biologists in the future. However, the great promise such strategies hold for validating causal relationships attendant to epigenetic control of gene expression (and other genomic transactions) argues for this being a worthy undertaking.
7.2. Toxic Proteins in Neurodegenerative Disease
A broad range of neurodegenerative disorders, including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis and prion diseases, share a common feature of accumulation of toxic protein aggregates. The molecular mechanisms behind the aggregation of specific proteins that are linked to these diseases are not altogether understood. Of particular interest are the roles of the various PTMs that decorate these proteins, a question that is, in principle, well suited to the protein semisynthesis approach. In this section, we discuss how semisynthesis has been deployed in this area, allowing the generation of modified forms of these proteins in order to understand the molecular mechanisms behind the neurotoxicity.
7.2.1. α-Synuclein
α-Synuclein is a 140 amino acid protein that is abundantly expressed in the nervous system.530 The aggregation of α-synuclein into intracellular inclusions is a hallmark of Parkinson’s disease (PD) and related neurodegenerative “synucleinopathies”.531, 532 A number of PTMs have been discovered on α-synuclein aggregates isolated from PD patients, including phosphorylation,533 glycosylation,534 ubiquitination535 and nitration.536 The small, unfolded nature of α-synuclein makes total synthesis and semisynthesis useful for the study of this protein (further reviewed elsewhere537). In particular, semisynthetic approaches centered on the use of NCL and EPL have been used to access phosphorylated,284, 286–288 ubiquitinated,282,283 glycosylated,289–291, 538 nitrated285 and Nα-acetylated281 forms of this protein, as well as α-synuclein containing a backbone thioamide which served as a fluorescent quenching probe to study conformational changes during aggregation.280
To prepare C-terminally modified α-synuclein, Lashuel and coworkers used EPL coupled with cysteine desulfurization to install a phospho-tyrosine at position 125 (Y125ph) of the protein (Figure 41a).284 A protein α-thioester spanning residues 1-106 of α-synuclein, generated from thiolysis of a recombinant fusion with the Mxe GyrA intein, was ligated to a synthetic peptide spanning residues 107-140 bearing the phosphotyrosine. Conversion of the reactive Cys107 residue to the native Ala107 was subsequently achieved by radical desulfurization, affording the Y125ph modified protein, which displayed no significant difference in aggregation properties relative to the unmodified protein. By contrast, semisynthetic α-synuclein containing phosphoserine at position 129, generated through the same strategy, was more prone to aggregation and exhibited decreased membrane binding, supporting a role for this modification in the pathogenesis of PD.286, 287
Figure 41.
Semisyntheses of modified forms α-synuclein, including a) C-terminal,284 b) N-terminal,282 and c) central modifications.289
The role of ubiquitination, and the related SUMOylation, in regulating α-synuclein aggregation has been heavily studied using both protein bioconjugation and semisynthesis approaches. In an example of the latter, Lashuel, Brik and coworkers used a combination of NCL and EPL to assemble an N-terminally ubiquitinated version of α-synuclein from three-fragments (Figure 41b).282 In this case, truncated α-synuclein (residues 19-140) containing an N-terminal Cys, accessed directly from recombinant expression in E coli, was ligated to a synthetic peptide α-thioester generated through Boc-strategy SPPS. A δ-mercaptolysine building block was incorporated at position 6 to direct ubiquitination through EPL with a recombinantly expressed ubiquitin α-thioester. Radical desulfurization then afforded the native K6 ubiquitinated α-synuclein. Interestingly, the presence of ubiquitin at this position inhibited α-synuclein aggregation in vitro without altering the membrane-binding affinity, suggesting a protective role for this modification. Impressively, this synthetic strategy has been applied to the semisynthesis of both di- and tetraubiquitinated α-synuclein at K12,283 work that highlights the extraordinary capabilities of modern protein ligation technologies. Other studies in this area, involving the use of streamlined cysteine-based ubiquitination/SUMOylation conjugation strategies,496, 539 have allowed systematic analyses of these modifications as a function of attachment site on α-synuclein.540–542 This has revealed that the effect of ubiquitination on the aggregation and proteosomal stability of the protein is strongly position-dependent, underscoring the nuanced roles of PTMs in regulating protein function.540–542
In another impressive body of work, Pratt and coworkers used semisynthesis to generate a series of α-synuclein variants containing the O-linked monosaccharide, N-acetyl glucosamine (GlcNAc).289–291, 538 These modifications are located within the center of the α-synuclein and require a combination of NCL and EPL reactions to assemble the protein sequence from three fragments (Figure 41c). The strategy taken is illustrated through the assembly of α-synuclein containing GlcNAc at serine 87, in which the protein was assembled from a synthetic peptide α-thioester containing a preinstalled Ser-GlcNAc residue, and two recombinant fragments spanning residues 1-75 and 92-140 respectively. Following sequential ligation reactions, desulfurization of the reactive cysteines afforded the native protein. Using this approach, it was found that attachment of the sugar at S87 and T72 inhibits protein aggregation, without affecting its endogenous membrane binding affinity. Access to a panel of glycosylated forms of α-synuclein has allowed this group to delve deeply into the roles of this modification in regulating the biochemical and biophysical behavior of the protein.
7.2.2. Tau
Tau is a microtubule-associated protein responsible for the stabilization of axonal filaments in neurons.543 Hyperphosphorylation of tau leads to dissociation from microtubules and the formation of insoluble aggregates that are key features of Alzheimer’s disease and other tauopathies.544 Schwarzer, Hackenberger and coworkers used EPL to access tau bearing a C-terminal phosphorylated serine residue (S404ph) (Figure 42a).295 The authors ligated a synthetic phosphopeptide spanning the C-terminal 52 amino acids of tau to a recombinant α-thioester fragment comprising residues 1-389 of the protein, generated through thiolysis of a Mxe GyrA intein fusion. In this case, an Ala390Cys mutation was required for the ligation reaction, which did not affect the function of the semisynthetic proteins based on the ability to promote polymerization of tubulin in vitro. Semisynthetic tau bearing S422ph was accessed in a similar fashion, the aggregation properties of which could be neutralized by an antibody against this known pathological PTM.296 Difficulties in the purification of the semisynthetic protein prompted Hackenberger and coworkers to use a photocleavable biotinylated linker as a traceless purification handle,293 which was employed in the semisynthesis of tau carrying a Ser400GlcNAc modification.294
Figure 42.
Semisyntheses of modified versions of the tau protein.
Lashuel and Haj-Yahya developed a multi-step semisynthetic route to tau amenable to accessing all modifications within the central microtubule-binding domain (Figure 42b).49 The N-terminal fragment, covering residues 2-245, was accessed through recombinant expression and the standard intein thiolysis workflow. Four synthetic fragments were accessed through Fmoc-strategy SPPS, covering residues 246-441. Assembly of the five fragments was achieved through convergent EPL and NCL reactions, incorporating strategic desulfurization steps to selectively convert non-native cysteine residues to the native alanines. The authors used this strategy to synthesize tau proteins bearing an acetyl-lysine at K280, a phosphotyrosine at Y310, and dual phosphoserines at S396 and S404. Tau bearing K280Ac was shown to have enhanced aggregation kinetics relative to the unmodified protein. Most recently, Becker and coworkers developed a semisynthetic route to tau that also allowed access to microtubule binding region of the protein.297 In this case, a three-piece strategy was employed featuring two flanking recombinantly derived fragments and a central synthetic peptide insert (Figure 42c). This route allowed access to variants of the protein containing phosphoserines and an unusual carboxymethyllysine modification. The latter was found to inhibit tubulin polymerization without affecting aggregation, whereas phosphorylation was found to inhibit aggregation without affecting tubulin binding. Collectively, these semisynthetic strategies provide good coverage of the entire C-terminal half of the tau protein and, as a consequence, will greatly aid biochemical and biophysical investigations of this important protein.
7.2.3. Prion Protein
Semisynthesis has been used to generate modified forms of the cellular prion protein (PrP), misfolding of which causes neurological diseases.545, 546 In early work, Becker and coworkers explored two semisynthetic routes to PrP bearing C-terminal palmitoyl groups as mimics of the native glycosylphosphatidylinositol (GPI) anchor.236 The authors first used EPL to assemble the semisynthetic protein, ligating a recombinant PrP α-thioester (residues 90-232, accessed from intein thiolysis) to synthetic palmitoylated peptides (Figure 43a). In the same study, the authors also investigated a PTS-based approach to assemble modified PrP from a recombinant PrP-fused to the IntN fragment of the Ssp DnaE split intein and a synthetic peptide containing the cognate IntC fragment and a short palmitoylated peptide sequence.236 This initial work was soon followed by the semisynthesis of the native GPI-anchored protein (Figure 43b).235 This collaborative work between the Becker and Seeberger groups stands as milestone in the field since it combines cutting-edge approaches in protein semisynthesis and carbohydrate synthesis. Access to these variously lipidated versions of PrP yielded a number of insights into the role of the modification, including how it alters the interaction of the protein with membranes leading to a disruption of membrane integrity.234
Figure 43.
Semisynthesis of the prion protein (PrP). a) Strategy for semisynthesis of C-terminally palmitoylated PrP.236 b) Structure of semisynthetic PrP bearing a native GPI anchor.235 c) Semisynthesis of PrP bearing glycan mimics at positions 181 and 197.338
Li and coworkers reported a different strategy for the semisynthesis of unmodified murine PrP from three fragments.233 An N-terminal recombinant protein α-hydrazide was generated through hydrazinolysis of a chemically cyanylated Cys-containing precursor.547 This recombinant α-hydrazide was converted to an α-thioester in situ and ligated to a synthetic sequence spanning residues 178-230 of mPrP (itself generate from two smaller synthetic fragments). This semisynthetic strategy makes use of the two native Cys residues in the PrP sequence for ligation reactions. However, as one of these junctions comprises a -DC- dipeptide sequence (requiring the use of an unstable Asp α-thioester), Li and coworkers were obliged to mutate this Asp residue to Ser.233 In an alternative approach, Araman and coworkers instead disconnected the protein at an -HD- dipeptide junction,237 which could be reassembled using a combination of EPL and selective desulfurization from a recombinant protein α-thioester and a peptide bearing a β-mercapto Asp building block as a Cys surrogate (Figure 43c).338 The recombinant PrP sequence containing residues 23-177, thiolyzed from a Mxe GyrA intein fusion, was ligated to synthetic peptides spanning residues 178-231 bearing the N-terminal β-mercapto Asp as well as pegylated Asn residues 181 and 197 as mimics of large N-linked glycans at these positions. Selectively desulfurized ligation products (with the native cysteine residues intact) displayed less α-helicity when modified at both Asn181 and Asn197, and all modified PrP variants were able to inhibit the in vitro formation of wild type aggregates at sub-stoichiometric levels.
7.2.4. Huntingtin
The huntingtin protein has been heavily implicated in Huntington’s disease, an inherited disorder characterized by progressive neuronal degeneration.548 The precise physiological function of the protein is still unclear, however, individuals with the disease contain a genetic polymorphism in the huntingtin gene that results in a repeat expansion of glutamine residues in the N-terminal region of the protein.549 Lashuel and coworkers have used NCL to gain semisynthetic access to this biomedically important protein (Figure 44).182 To this end, a recombinant N-terminally truncated fragment (residues 10-90) was expressed with an Ala10 to Cys mutation and ligated to a synthetic peptide bearing a phosphorylated threonine at position 3 and a C-terminal N-acyl urea as a thioester equivalent. Following ligation, the full-length protein was desulfurized in order to access the native sequence at the ligation junction. Interestingly, the presence of phosphorylated threonine significantly slowed the oligomerization of huntingtin into fibrils. This same strategy was then adapted to the semisynthesis of a mutant huntingtin protein, containing an expanded polyQ tract (43Q).183 Similar to the wild-type protein, Thr3 phosphorylation strongly inhibited the aggregation and fibril formation of the semisynthetic mutant huntingtin, although this inhibitory effect could be reversed by the presence of acetylation at Lys6. In addition, subsequent work from Lashuel and coworkers have shown that phosphorylation at Ser13 and Ser16 can also inhibit the aggregation of mutant huntingtin.184 This suggests that crosstalk between these and possible other modifications on the huntingtin N-terminus may influence the structure and aggregation properties of this protein.
Figure 44.
Semisynthesis of phosphorylated huntingtin protein (Htt).182
7.3. Integral membrane proteins
As many as 30% of the proteins encoded in the human genome are predicted to be embedded in cellular membranes.550–552 The biomedical importance of these proteins can hardly be overstated, fully 60% of clinically prescribed drugs target membrane proteins such as G-protein coupled receptors (GPCRs), receptor kinases and ion channels,553, 554 It is equally hard to overstate the technical challenges associated with studying this class of protein – as but one measure of this, only 3.6% of structures in the protein data bank555 as of June 2019 were membrane proteins (although we note that this situation is improving due to technical advances in crystallography and electron microscopy). Extending the range of protein semisynthesis into this arena is enormously challenging due to difficulties associated with preparing and manipulating hydrophobic membrane protein fragments, not to mention the undertaking required to fold these polypeptides into a native structure embedded in a membrane. Nonetheless, there have been a number of remarkable successes in this area, that have yielded important insights into membrane protein function, and that hopefully serve as a beachhead for more work in this area.
7.3.1. Ion Channels
Ion-channels are integral membrane proteins that serve as conduits for ions to pass into and out of cells. This large family of proteins is diversified by the type of ion they conduct and by the regulatory inputs that open and close their conductive pores. Potassium channels exhibit an extremely high degree of selectivity for the K+ ion over other cations, dictated by a stretch of conserved amino acids that form an ion-binding region known as the “selectivity filter”.556 While high-resolution structural studies indicate a key role for the protein backbone in the function of the filter, testing these predictions using conventional mutagenesis is not straightforward. This motivated the development of a semisynthetic route to the archetypical bacterial potassium channel, KcsA, an undertaking that required solving several difficult technical problems along the way, including how to fold the semisynthetic protein into the active homo-tetramer.102, 191
Critically, the optimized semisynthesis that was developed provided chemical access to the selectivity filter (Figure 45a–b), allowing backbone engineering of this region, namely replacement of a key amide with an ester moiety193 and the substitution of a glycine with a D-Ala.194, 557 In both cases, high-resolution crystal structures of the semisynthetic proteins were obtained to complement the functional data (Figure 45c–d). The work on the D-Ala containing KcsA analog is especially noteworthy, since it established a key role for conformational changes in the filter in ion selectivity. In a related study, Valiyaveetil and coworkers assembled the 282-residue archaebacterial voltage-dependent K+ channel, KvAP, from three fragments, two recombinant and one synthetic (Figure 46).192 This involved two sequential ligation reactions, again allowing synthetic access to the selectivity filter. In this case, incorporation of a D-Ala in place of a glycine offered insights into the structural basis of so-called C-type inactivation of the channel. More recently, a semisynthetic version of KcsA was prepared containing site-specific heavy isotopes in the selectivity filter.197, 558 This allowed two-dimensional infrared spectroscopy to be applied to the system, providing insights into the conformational transitions that occur in the filter during multi-ion conduction.
Figure 45.
Semisynthesis applied to the KcsA K+ channel. a) Scheme for the semisynthesis of unmodified KcsA using EPL.191 Two opposite subunits of the refolded tetrameric K+ channel are shown. b) Structure of the selectivity filter of a recombinant KcsA K+ channel (PDB ID code 1K4C). c) Structure of the selectivity filter of a semisynthetic KcsA K+ channel containing a D-Ala mutation (PDB ID code 2IH1).557 c) Structure of the selectivity filter of a semisynthetic KcsA K+ channel a backbone amide to ester mutation (PDB ID code 2H8P).193 Arrows indicate the unnatural ester linkage.
Figure 46.
Semisynthesis of the KvAP voltage-dependent K+ channel using EPL and NCL.192
In another example of semisynthesis as applied to a membrane protein, the Valiyaveetil laboratory used EPL to assemble the Na+-coupled aspartate transporter, GltPh.118 This 418-amino acid protein was assembled using an N-terminal protein α-thioester (residues 1-384) and a synthetic C-terminal 33-residue peptide. Folding of the semisynthetic protein afforded a transporter with a Kd value for aspartate binding similar to a native GltPh control.118 Substituting the highly conserved Arg397 residue in the aspartate binding pocket with the isosteric citrulline demonstrated that this residue is critical for Na+-coupled binding of aspartate and vesicular uptake of the amino acid in vitro.
7.3.2. G-Protein Coupled Receptors
GPCRs are the largest family of cell surface receptors in the human genome and are the targets of over 30% of current drugs on the market. Activated GPCRs are phosphorylated on their C-terminal cytosolic regions by GPCR kinases, a modification that leads to the binding of β-arrestin, triggering several downsteam processes including desensitization, internalization and signaling.559, 560 Consequently, access to phosphorylated versions of GPCRs could help further our understanding of these regulatory processes. With this in mind, Lefkowitz and coworkers employed sortase-mediated ligation to generate a hyperphosphorylated version of the β2-adrenergic receptor (β2AR).561 An evolved sortase430 was used to ligate a synthetic polyphosphorylated peptide to a recombinant version of the receptor equipped with a C-terminal LPETG sortase recognition sequence (Figure 47a). The semisynthetic protein was then reconstituted into high-density lipoprotein particles (nanodiscs).562 Significantly, polyphosphorylation of the GPCR led to recruitment of β-arrestin, allosterically enhancing the affinity of cyanopindolol, an inverse agonist, for the receptor.
Figure 47.
Semisynthesis of the β2-adrenergic receptor (β2AR). a) Semisynthesis of C-terminally phosphorylated β2AR using sortase-mediated ligation followed by reconstitution into lipid nanodiscs.561 b) Preparation of segmentally labeled β2AR using PTS.563 c) Model of conformational change of the β2AR C-terminus following agonist bind and phosphorylation, triggering modulation of transmembrane domain structure and arrestin binding.563
In related work, Shimada and coworkers performed NMR analysis on a segmentally labeled β2AR prepared in vitro using PTS (Figure 47b).563 To observe only resonances corresponding to the C-terminus of the receptor, the authors expressed a C-terminally truncated form of the protein (residues 1-348) in insect cells, fused to the IntN fragment of the Npu DnaE split intein. The remainder of β2AR was expressed in E. coli (in 13C/15N containing media) fused to the IntC fragment, and subsequently spliced onto the β2AR-core. The segmentally labeled ligation product was then reconstituted into nanodiscs, and analyzed by NMR spectroscopy in unmodified form or phosphorylated by GRK2. Interestingly, the receptor C-terminus showed conformational changes upon phosphorylation, potentially caused by differential engagement with the transmembrane region of the GPCR.
7.4. Signaling Proteins
Cellular signaling events require complex networks of signal transduction pathways to interpret and respond to extracellular stimuli. These pathways are typically composed of multiple highly choreographed protein-protein interactions, often driven by post-translational modifications such as protein phosphorylation, that connect extracellular stimuli to a transcriptional output. In this section we will discuss how protein semisynthesis has been applied to proteins involved in cell signaling and signal transduction, including secreted signaling proteins, scaffold proteins, small GTPases and transcription factors.
7.4.1. Secreted Signaling Proteins
Cells secrete a wide range of small proteins, including those historically classed as growth factors, cytokines and hormones, in order to communicate among one another. These couriers of biological messages carry out their biological assignment through engagement with dedicated transmembrane receptors on target cells, which in turn initiate intracellular signaling cascades terminating in a transcriptional output. The activity of these secreted messenger proteins is tuned through extensive modifications that occur during passage through the endoplasmic reticulum and golgi apparatus. Protein chemistry has been used to access these modified proteins, aided by their (typically) small size and ability to be refolded in vitro.
The class of chemotactic cytokines known as chemokines have been accessed using both totally synthetic13, 564–573 or semisynthetic132–139, 188, 574, 575 protein chemistry. Typically, these proteins are only 70-80 residues and have conserved cysteine residues that are conveniently located for synthetic manipulation. Indeed, the chemokine CXCL8, also known as interleukin 8 (IL-8), was the first protein to be prepared using the native chemical ligation strategy.13 EPL has also been used extensively in the preparation of semisynthetic forms of two chemokines, CXCL8132–136, 574 and CXCL12137–139, 575 (stromal cell-derived factor 1). Beck-Sickinger and coworkers have developed robust semisynthetic routes to these chemokines (Figure 48a), allowing the introduction of fluorescent probes,133, 139 photocrosslinking groups,134 photocages,138, 139 isotope labels,136 and β-peptide mimics of the CXCL8 C-terminal helix.135 In this last example, semisynthetic CXCL8 containing an oligo β3-amino acid sequence was prepared, projecting the native functionalities from an artificial secondary structure element in an otherwise native protein scaffold (Figure 48b).135 Remarkably, these chimeric molecules retained activity in CXCR1 stimulation assays.
Figure 48.
Semisynthesis of chemokines. a) General strategy for the semisynthesis of modified CXCL8 and CXCL12, and examples of modifications incorporated. b) Semisynthesis of CXCL8 with a β-peptide sequence at the C-terminus.135
Interleukins are immunomodulatory cytokines, and have provided an arena for the development and implementation of semisynthetic protein chemistry as applied to glycoproteins.187, 188, 576 The heterogeneity of protein glycosylation in vivo makes chemical preparation of such proteins with chemically defined glycans an attractive option with the added appeal of conquering such synthetically demanding molecules, even through total synthesis577, 578. Unverzagt and coworkers used an impressive three-piece ligation strategy to assemble interleukin 6 (IL-6) with an N-linked nonasaccharide.187 N- and C-terminal fragments were expressed recombinantly, the former using a tripartite fusion with two contiguous inteins (Ssp DnaB and Mxe GyrA), each undergoing autoprocessing to liberate the desired IL-6 fragment as an α-thioester with a free N-terminus (Figure 49). Extensive chemical expertise was required to troubleshoot and optimize the synthesis, including the efficient access to the synthetic central fragment, a hexapeptide α-thioester/hydrazide bearing the N-linked nonasaccharide. Nonetheless, the campaign was successful, and gave access to glycoforms of IL-6, bearing N-linked mono- or nonasaccharides, that were correctly folded and were as potent as recombinantly expressed IL-6 in stimulating the proliferation of IL-6 responsive pro B cells (Ba/F3).187, 579 To prepare a glycosylated form of interleukin 13 (IL-13), Kajihara and coworkers used a four-fragment assembly, including two central synthetic peptides and two flanking recombinant fragments (Figure 50).188 The authors used an innovative method to access both N- and C-terminal fragments from a single recombinant protein, employing a combination of chemical cleavage events, including cyanogen bromide cleavage and the so-called “guanidine method”188 of activation and transformation of cysteinyl-peptides into α-acylguanidines – α-thioester equivalents. Ultimately, the authors were able to assemble the 112-amino acid cytokine, bearing an N-linked nonasaccharide at Asn52.
Figure 49.
Semisynthesis of glycosylated interleukin 6.187
Figure 50.
Semisynthesis of glycosylated interleukin 13.188
Of all the glycoproteins studied in the biomedical fields, none have attracted the attention of synthetic protein chemists like erythropoietin (EPO), a hormone that stimulates the production of red blood cells.580, 581 Much of the gravitation pull of this 166-residue glycoprotein stems from its proven medical value – the recombinant version of the protein is used clinically in the treatment of anemia. EPO is natively N- and O-glycosylated at four positions, modifications which are critical for prolonged circulation in vivo,582 although this glycosylation is heterogeneous. This factor opened the door to protein chemistry as a path to homogeneous EPO proteins, using both totally synthetic331, 583–587 and semisynthetic144–146 strategies, to then be used to understand the effects of the carbohydrates on EPO’s pharmacokinetics. Collectively, these studies showcase the incredible power and versatility of modern protein and carbohydrate chemistries – the recent disclosure by the Kajihara group of the total synthesis of five EPO glycoforms provides an especially stunning example of these capabilities.587 While the preparation of native glycoforms represents a major achievement, the considerable synthetic efforts associated with these synthetic campaigns has motivated researchers to consider neoglycoprotein surrogates in order to expedite downstream biological study. Macmillan and coworkers used NCL to prepare semisynthetic EPO with an alkynylated side-chain at position 24, a handle to install a glycan mimic using click chemistry,57 from a recombinant C-terminal protein fragment and an N-terminal peptide α-thioester. Kajihara and coworkers used a similar neoglycoprotein strategy to prepare EPO with two145 and three144 complex sialyloligosaccharides. More recently, Rubini and coworkers reported an amber suppression strategy to site-specifically install a propargylated-lysine residue at various positions in fully recombinant EPO.588 Subsequent, click chemistry using N-glycan azides, yielded several glysosylated EPO conjugates that were shown to be more stable to aggregation relative to the non-glycosylated version of the protein. These and other syntheses of EPO have helped confirm the importance of glycosylation, in particular the degree of sialylation, for the activity of this protein hormone in vivo. More generally, these studies have broken through many of the technical barriers associated with gaining access to homogenous glycoproteins.
7.4.2. Scaffold Proteins
Scaffold proteins play important roles in cellular signaling, serving as hubs for the coordination of signaling cascades through the localization and activation of specific proteins.589 These scaffolds are structurally diverse, but are all typically composed of a modular assembly of discrete binding domains specific for certain motifs present within binding partners. The complex circuitry that these molecular adapters template, further regulated by modifications to the proteins themselves, have made the study of scaffold proteins an important area of investigation. The Crk family of adaptor proteins are involved in many cellular processes including proliferation. These proteins are composed of modular Src Homology 2 (SH2) and Src Homology 3 (SH3) domains that serve as docking sites for protein-protein interactions. Phosphorylation of Tyr221, by the kinases c-Abl and EGFR, leads to intramolecular association between this residue and the N-terminal SH2 domain, forming a closed autoinhibited conformation. These conformational changes have been observed on semisynthetic Crk-II using FRET- and NMR-based methods.108, 109, 130, 590, 591 Strategic placement of donor and acceptor fluorophores onto the N- and C-termini of a Crk-II protein afforded a fluorescent reporter for phosphorylation at Tyr221, providing a direct readout of Abl kinase activity.130, 590 Using sequential protein ligations, the individual domains of Crk-II were also assembled109 is a fashion allowing NMR analysis of the modular protein through segmental isotopic labelling.364, 592 Indeed, NMR-analysis of the segmentally labelled C-terminal SH3 domain of Crk-II, helped to provide a model for Crk-II activation and autoinhibition, where destabilizing structural deformation of this domain counteracts the stabilizing interdomain interactions.108
Protein semisynthesis has been used to study the postsynaptic density protein 95 (PSD-95), an abundant scaffolding protein in the post synaptic density (PSD), a dense network of proteins at the membrane of post synaptic neurons.593 PSD-95 contains three PDZ domains that mediate protein-protein interactions with the C-termini of a range of proteins including neuronal receptors, interactions that concentrate proteins to the PSD.593 In order to understand how these modules engage their ligands, Stromgaard and coworkers used both NCL and EPL to generate semisynthetic forms of the three PDZ domains of PSD-95, bearing either phosphorylation231 or backbone amide-to-ester229 chemical modifications (Figure 51). Phosphorylation of four residues throughout the three PDZ domains (S73ph, Y236ph, Y240ph and Y397ph) generally led to decreased ligand binding, with the exception of stargazin, an auxiliary subunit of the APMA receptor, which displayed increased binding to PDZ3 (Y397ph). This result points to finer regulation of this interaction through post-translational modification of both PSD-95 and stargazin.594 Semisynthetic PDZ domains bearing amide-to-ester mutations within the conserved carboxylate-binding-site showed decreased binding affinity for PDZ-ligands, confirming the importance of H-bonding interactions in the selectivity of binding partners.229 This work has contributed greatly to our understanding of PDZ-ligand engagement generally, and has helped fuel ongoing research into the development of pharmacological agents that disrupt PDZ-ligand interactions for the treatment of various diseases.595
Figure 51.
Semisynthesis of PSD-95 PDZ domains. a) Semisyntheses of phosphorylated PDZ1–3. b) Semisynthesis of PSD-95 PDZ2 with amide-to-ester mutants at the substrate binding site. Right: Important H-bonding interactions in the PDZ-substrate complex backbone, mapped onto a canonical PDZ-substrate structure (PDB ID: 1BE9).229
7.4.3. Small GTPases
Small GTPases are a family of small GTP-binding proteins that play critical roles in the cell, including signal transduction, nuclear import/export, and membrane trafficking. Semisynthesis has been extensively used in the preparation of Rab241–244, 319, 320 and Ras248–250 GTPase proteins in C-terminally prenylated forms. This modification is necessary for association of this soluble protein to the cytoplasmic leaflet of the plasma membrane.596, 597 In early work in this area, Alexandrov and coworkers used EPL to generate a semisynthetic Rab GTPase containing a C-terminal fluorophore and used this to study the interaction with a Rab geranylgeranyl transferase.240 This group further developed their semisynthetic approach in order to chemically install a C-terminal prenyl modification onto Rab7 (Figure 52), using EPL to ligate a recombinant Rab7 α-thioester to a C-terminally prenylated synthetic peptide.244 This basic strategy was subsequently applied to the semisynthesis of the Rab GTPase Ypt1, both in monoprenylated319 and diprenylated320 forms. Impressively, both the singly and doubly prenylated Ypt1 proteins were crystallized in complex with the Rab GDP-dissociation inhibitor protein (GDI), which is responsible for the extraction of GDP-bound Rabs from membranes.319, 320 These structures revealed the binding modes of the two prenyl groups on the Ypt1 C-terminus, one buried in a hydrophobic cavity of GDI and the second laying adjacent to this cavity on the surface of the chaperone protein.320 The binding strength between GDI and prenylated Rab proteins have been subsequently measured in vitro, revealing that both mono and diprenylated GDP-bound Rab7 are bound to GDI with high affinity (Kd 1-5 nM), whereas unprenylated Rab7 is bound weakly (Kd >50 uM).243 However, this high affinity interaction is abolished when the Rab7 protein is bound to a GTP-analog.241 These structural and biochemical experiments have provided key insights into how the Rab:GDI interaction is regulated, helping shape a model for how Rab GTPases are targeted to membranes.
Figure 52.
Semisynthesis of prenylated Rab GTPases. a) Scheme for the semisynthesis of prenylated Rab7 GTPase.244 b) Co-crystal structure of the semisynthetic doubly prenylated Rab GTPase Ypt1 (green cartoon) and GDP dissociation inhibitor (GDI, white surface).320 Unstructured C-terminus is shown as dashed green line. Expansion shows a zoomed view of the two geranylgeranyl moieties. (PDB ID: 2BCG)
7.4.4. TGF-β signaling pathway
Protein semisynthesis has helped our understanding of discrete steps in transforming growth factor-β (TGF-β) signaling, a pathway involved in many processes including cell proliferation, differentiation and migration.598, 599 TGF-β receptors are heterodimeric transmembrane serine/threonine kinases composed of type I and type II TGF-β receptor subunits (TBR-I and TBR-II) that undergo transphosphorylation upon ligand binding. Phosphorylation of TBR-I leads to the activation of this receptor kinase and phosphorylation of a set of transcription factors known as R-Smads. Semisynthesis of the cytoplasmic region of TBR-I illuminated how the receptor becomes activated upon phosphorylation.307, 308, 600 Using NCL, a tetraphosphorylated form of TBR-1 was generated semisynthetically, requiring a challenging synthesis of a peptide α-thioester bearing four phosphorylated serine and threonine residues.307, 308 Through access to these proteins, it was found that phosphorylation of TBR-I plays a duel role in the activation of the receptor, by simultaneously decreasing the affinity for the inhibitory protein FKBP12 as well as increasing the affinity for a substrate R-Smad protein (Smad2).600 Subsequently, semisynthesis of Smad2 itself was undertaken in order to understand the structural effects of C-terminal phosphorylation on this transcription factor.267 Indeed, semisynthetic Smad-2 formed heterotrimers in a phosphorylation dependent manner (Figure 53a). X-ray crystallography revealed this conformational switch is driven by the interaction the phosphoserine residues on the C-terminus of one Smad2 molecule and the mad homology 2 (MH2) domain of a separate molecule.267 Further analysis of phosphorylated variants of Smad2 indicated that both phosphorylation events were required for stable formation of this homotrimer.268 This interaction was captured through covalent UV-crosslinking through a synthetically installed diazirine-containing amino acid.269 Additionally, the incorporation of UV-labile ‘photocages’ onto these key phosphates afforded temporal control over this association, blocking trimerization until deprotected through UV irradiation (Figure 53b).270 Note that in an extension of this caging idea, a semisynthetic version of Smad2 was generated that allowed simultaneous triggering of activity and fluorescence, allowing for tracking of the active protein in live cells.271, 272 The insights gained from these various studies have proved to be significant for the molecular level understanding of modification-driven regulatory events along the entire TGF-β signaling pathway.
Figure 53.
Control of Smad2 activity using semisynthesis. a) Association of semisynthetic phosphorylated Smad2 into trimers (PDB ID code 1KHX). b) Photocaging of Smad2 association and activation.270
7.5. Enzymes
Enzymes have been targets of protein semisynthesis (and, of course, total synthesis) since the earliest days.97, 465, 601–603 The intricacies of enzyme catalysis coupled with the complex modes of allosteric regulation Nature employs to control activity, including through PTMs, means that standard mutagenesis is sometimes a rather blunt instrument when it comes to mechanistic dissection. Semisynthesis, by contrast, offers far greater flexibility in terms of what can be incorporated into an enzyme active site, in principle, making it a more precise tool. In this section, we illustrate this point using select examples from what is a very active area.
7.5.1. Ribonuclease A
Ribonuclease A (RNase A) is a particularly resilient enzyme, and has thus served as a valuable model for the use of protein chemistry to study enzyme structure and function.604 Of note, RNase A was first synthesized in 1969 via two routes, stepwise solid-phase peptide synthesis by Gutte and Merrifield,601, 602 and convergent synthesis by Hirschmann and coworkers.603 Indeed, RNase A has consistently featured in landmark studies in the peptide and protein chemistry fields; the reader is reminded that the enzyme was assembled from a series of synthetic peptide fragments using subtiligase-mediated ligation (see section 5.1). RNase A was also one of the first proteins to be accessed using intein chemistry, assembled semisynthetically by Xu and coworkers as a means to access the cytotoxic enzyme from catalytically inactive recombinant and synthetic fragments (Figure 54a).98 Raines and coworkers used a similar strategy to incorporate a selenocysteine at position 110 using Sec-mediated EPL.251 In both cases, semisynthetic RNase A had equivalent activity to the wild type enzyme, showing that these enzymes were properly folded even when containing a Cys110Sec mutation. Extending the ligation junction to Cys95, Raines and coworkers installed dipeptidyl turn mimics,252, 253 based on the β-amino acids 1 (Nip) or homoalanine (hAla), in order to study the folding and stability of RNase A (Figure 54b). While both of these backbone mutants displayed unchanged enzymatic activity relative to the WT enzyme, the conformational stabilities were affected, negatively in the case of the β-hAla modules but positively in the case of the more rigid Nip-based turns. Subsequent NMR analysis of semisynthetic RNase A, labeled with 15N an 13C isotopes exclusively at Pro-114, illuminated a cis-amide conformational preference at this position.605 The replacement of Pro-114 with a 5,5-dimethylproline (dmPro) residue afforded RNase A with enhanced conformational stability, due to the increased propensity to adopt a cis-peptide bond (Figure 54b).256 Variants containing 1,5-triazole mimics of Pro-114 were also able to support this conformation.254 Interestingly, replacement of Pro-114 with (2S,4S)-4-fluoroproline (a single atom replacement) can even promote this conformational preference through a weakened n→π* interaction, which ordinarily favors the trans isomer.255 Unverzagt and coworkers developed a semisynthetic route to RNase bearing a complex N-linked glycan at Asn34.257, 258 Initial plans to assemble the protein from a recombinant C-terminal fragment and a synthetic peptide bearing a large biantennary glycan were stymied by inefficient peptide synthesis caused by the large nonasaccharide moiety. This necessitated a three-piece ligation strategy between two synthetic peptides (comprising residues 1-25 and 26-39) and a recombinantly expressed fragment from residues 40-124. As is the case for all synthetic preparations of homogeneous glycoproteins, this successful semisynthesis of glycosylated RNase is a tour-de-force, requiring the integration of many tools of carbohydrate, peptide and protein chemistry.
Figure 54.
Semisynthesis of Ribonuclease A (RNase A). a) Scheme for the semisyntheses of RNase A using EPL at Cys or Sec.98, 251 b) Dipeptidyl turn mimics installed in RNase A using EPL. Melting temperatures from themally induced unfolding experiments are shown.
7.5.2. Protein Kinases
Understanding how protein kinase activity is regulated has been an intense area of research for decades, spurred on by the intimate relationship between abnormal signaling and cancer.606 The first report of EPL involved the semisynthesis of a phosphorylated form of the tyrosine kinase Csk, a non-receptor tyrosine kinase that phosphorylates the tail of the Src family of kinases.97 Since this pioneering work, protein semisynthesis has been used to access various kinase enzymes (Table 1), including the TGF-β receptor kinase (also see section 7.4.4),307, 308, 600 Eph tyrosine kinases,142, 143 protein kinase A,239 Rad53,245 p38α MAPK,223 casein kinase II,124 Src,275 and PDK1.607 The kind of questions that these studies have helped address is well illustrated by the work of Cole and coworkers on casein kinase II.124 In this case, EPL was used to install two different modifications into the protein, phosphothreonine at position 344 and O-GlcNAc at Ser347. While the modified proteins showed little to no difference in terms of kinase activity for a substrate peptide, assays on human protein microarrays showed that kinase substrate specificity can be modulated by these two PTMs. Additionally, Thr344 phosphorylation leads to increased cellular stability of casein kinase II, as assessed by microinjection of a semisynthetic protein containing a non-hydrolyzable PTM mimic into cells.124
Protein semisynthesis has illuminated distinct mechanisms of Akt1 kinase activation by phosphorylation.110 Site-specifically phosphorylated Akt1 kinases were prepared through EPL employing recombinant Akt1 α-thioesters (residues 1-459) and synthetic phosphopeptides modified at Ser473, Ser477 and/or Thr479 (residues 460-480) (Figure 55). Fortuitously, the native residue of Akt1 at position 460 is Cys, allowing a traceless semisynthesis to be achieved without the need for any desulfurization steps. In order to install phosphorylation on Thr308, which resides within the kinase activation loop, the Akt1 α-thioester fragment was treated with the protein kinase PDK1. Thus, the final semisynthetic proteins containing both enzymatically and chemically installed phosphorylated amino acids. The authors went on to determine that activation of kinase activity by phosphorylation at Ser473 operates through interaction with the linker region between the kinase domain and the N-terminal pleckstrin homology (PH) domain, thereby relieving autoinhibition.110 Conversely, activation by Ser477/Thr479 phosphorylation appears to function through direct interaction with the activation loop that reduces autoinhibition by the PH domain.
Figure 55.
Semisynthesis of Akt1 kinase. a) Scheme for the semisynthesis of phosphorylated Akt1 kinase using EPL.110 b) Model for the modulation of Akt1 activity through phosphorylation.
7.5.3. Phosphatase enzymes
Protein phosphatase enzymes can themselves be regulated by phosphorylation. Protein semisynthesis has allowed these regulatory mechanisms to be revealed. Cole and coworkers used EPL in order to understand how tyrosine phosphorylation of SHP-2 can affect the activity of this protein tyrosine phosphatase.265, 266 Due to the tendency to autodephosphorylate, the authors prepared semisynthetic SHP-2 containing a non-hydrolyzable phosphotyrosine analogue through EPL between a recombinant protein α-thioester and synthetic peptides containing either phosphonomethylene-phenylalanine or difluorophosphonomethylene-phenylalanine residues in place of phosphotyrosines (Figure 56a). The authors were able to demonstrate that these phosphonotyrosine modifications stimulated the phosphatase activity of SHP-2 through intramolecular interactions with the SH2 domains (Figure 56b). Further, microinjection of semisynthetic SHP-2 bearing phosphonotyrosine at Tyr542 into cells showed that this protein was able to stimulate the MAP kinase pathway more potently than the unmodified protein.
Figure 56.
Semisyntheses of SHP-2 phosphatase. a) Scheme for the semisynthesis of SHP-2 using EPL.265, 266 b) Model for the activation of SHP-2 phosphatase activity through tyrosine phosphorylation.
Originally thought to be a protein tyrosine phosphatase, the phosphatase and tensin homolog (PTEN) is a lipid phosphatase that is regulated by multiple PTMs, including phosphorylation of C-terminal Ser and Thr residues. Semisynthetic PTEN has been generated in order to understand how these modifications affect protein activity.226–228, 471 An initial semisynthesis of the protein involved EPL between a recombinantly produced PTEN α-thioester (residues 1-378) and a tetraphosphorylated synthetic peptide (residues 379-403).228 Notably, mutation of the native Tyr379 to Cys residue was necessary for the ligation reaction, a mutation that has since been shown not to be silent.484 As noted in section 6.3, this issue was subsequently overcome via subtiligase-templated ligation between a PTEN α-thioester (residues 1-377) and a tetraphosphorylated synthetic peptide (residues 378-403) bearing an N-terminal Arg residue (Figure 57a).471 The phosphorylated PTEN proteins displayed reduced catalytic efficiency relative to the unmodified protein, as well as reduced affinity for lipid vesicles.228, 471 Phosphorylation of the C-terminus was shown to promote a more compact form of PTEN, through interactions between the phosphorylated tail and the core that were identified through photocrosslinking and mass-spectrometry (Figure 57b).226 This compact phosphorylated form of PTEN was subsequently demonstrated to be a poorer substrate for ubiquitination by the E3 ligase WWP2,608 suggesting that phosphorylation both regulates the activity and the stability of PTEN. Of note, WWP2 is itself regulated by phosphorylation of an autoinhibitory linker region, a mechanism also illuminated through protein semisynthesis.216
Figure 57.
Semisyntheses of PTEN. a) Scheme for the semisynthesis of phosphorylated PTEN using subtiligase-catalyzed EPL.471 b) Model for the modulation of PTEN structure through phosphorylation.228
7.5.4. p300
The transcriptional coactivator p300 is a histone acetyltransferase (HAT) that is critical for regulating gene expression in mammalian cells.609 Efforts to recombinantly express the p300 HAT domain in E. coli were hampered by autoacetylation of the protein in vivo, as well as the potential to indiscriminately acetylate E. coli host proteins.221 Cole and coworkers found that this protein could be accessed efficiently through semisynthetic assembly of a recombinant α-thioester (spanning residues 1287-1652 of the full length protein), and a 14 amino acid synthetic peptide.221, 222, 610, 611 Importantly, the C-terminally truncated sequence was almost devoid of catalytic activity compared to the semisynthetic product, allowing the authors to study the hypoacetylated form of the enzyme.221 Interestingly, catalysis by the semisynthetic enzyme was stimulated after allowing for autoacetylation to take place. Acetylated residues important for p300 autoactivation were found to reside on a proteolytically sensitive loop region thought to autoinhibit p300 HAT activity. Further studies demonstrated that autoacetylation primarily occurs in trans on up to 17 lysine residues.612 To assess the contributions of individual lysine acetylation events to p300 activity, Cole and coworkers engineered a circularly permuted form of the protein (cp-p300), relocating the autoactivation loop from the middle of the HAT domain to the C-terminus, which could then be modified semisynthetically (Figure 58).222 A series of synthetic 38-mer peptides containing the autoactivation loop and modified with varying numbers of acetyllysine residues, were ligated to a recombinant α-thioester containing remainder of the cp-p300 domain. Under conditions where autoacetylation is minimal, these semisynthetic proteins displayed increasing levels of HAT activity with increasing loop acetylation. Indeed, the cp-p300 domain with 6 preinstalled acetylation sites was equally active as the p300 HAT domain with this loop deleted. Not only do these studies shed light on the regulation of this important coactivator enzyme, they suggest an alternative way to study the internal region of a protein through semisynthesis of a circularly permuted analogue.
Figure 58.
Circular permutation of the p300 HAT domain (PDB ID: 3BIY), which was accessed semisynthetically in order to install acetylated lysine residues.222
7.5.5. Serotonin N-Acetyltransferase
Serotonin N-acetyltransferase [arylalkylamine N-acetyltransferase (AANAT)] is a key regulator of circadian rhythms, responsible for the conversion of serotonin into N-acetyl serotonin, the precursor to melatonin. Phosphorylation of AANAT by PKA at positions Thr31 and Ser205 leads to stabilization of the enzyme, a mechanism that had been presumed to occur through enhanced interaction with two sites of the 14-3-3 protein. This interaction was studied using semisynthetic AANAT, bearing modifications at Thr31 and/or Ser205 as phosphates or the non-hydrolyzable analogues phosphonomethylenealanine (Pma) and phosphonodifluoromethylenealanine (Pfa).116, 117 Pulldown experiments showed that AANAT modified at these positions (Thr31ph/Pma or Ser205ph/Pfa) showed greater affinity for the 14-3-3 protein in vitro than unmodified AANAT, or a Thr31Glu phospho-mimic. Furthermore, these two modifications were able to confer stability to AANAT that was microinjected into cells. Interestingly, phosphorylation at Thr31 was shown to activate the catalytic activity of AANAT through an interaction with 14-3-3ζ that decreases the Km for the substrate tryptamine, although this activation is abolished in the case of the dually modified protein.115 Notably, this enzyme activation was not observed using a serine-to-glutamate mutation as a mimic of phosphoserine, a strategy often applied in cell biology experiments. Analogous results in other systems highlight important limitations of this mutagenesis strategy to study protein phosphorylation, specifically for cases where molecular recognition is strongly dependent on the geometry and charge state of the of phosphoserine/threonine side chains, properties which are often poorly simulated by the glutamate side chain carboxylate.613
7.5.6. Sortase A
Protein chemistry has been used to study the structure and activity of sortase A, itself a valuable tool for protein chemistry. Indeed, refolded totally synthetic forms of the catalytic domain of sortase A are at least as active as the recombinant protein.614 In an attempt to resolve conflicting views regarding the role of the conserved basic Arg197 residue in transpeptidation by the enzyme, McCafferty and coworkers used semisynthesis to incorporate the isosteric and non-ionizable citrulline residue at this position (Figure 59a).274 In this case, a sortase A (25-183) α-thioester was ligated to a synthetic peptide comprising residues 184-206, bearing an N-terminal cysteine and a citrulline at position 197. The citrulline mutation resulted in only modest decrease (less than 3-fold) in catalytic efficiency of the enzyme, arguing for a model of transition-state stabilization through hydrogen-bonding rather than electrostatic interactions mediated by the Arg side-chain (Figure 59b). Schwarzer and coworkers used a semisynthetic approach in order to probe the effects of mutating the catalytic cysteine residue (the only cysteine present in the transpeptidase domain) to either homocysteine or selenocysteine.273 Interestingly, the pH-profile of the Sec-sortase was shifted towards more acidic conditions, although this enzyme displayed a 2-3-fold reduction in activity relative to the WT sequence.
Figure 59.
Semisynthesis of the catalytic domain of sortase A (SrtA). a) Scheme for the semisynthesis of citrullinated SrtA using EPL.274 b) Molecular mechanism of transpeptidation facilitated by H-bonding between the sorting motif and Arg/Cit.
7.5.7. Inteins
As is hopefully clear by this point, the field of protein chemistry has benefitted greatly from exploiting the protein splicing activity of inteins. In turn, these enabling technologies have fed forward into the study of inteins themselves, providing a better understanding of the autocatalyzed protein splicing mechanism. In an early example of this, NCL was used to study the first step in the splicing cascade of the Mxe GyrA intein, namely the N-to-S acyl shift at the N-terminal splice junction (see Figure 10). Specifically, NMR active 15N and 13C isotopes were selectively incorporated at the scissile amide bond by ligating a synthetic peptide α-thioester, containing a single 13C atom at the C’ position of the C-terminal residue, to a uniformly 15N-labeled version of the intein.212 Analysis of the one-bond dipolar coupling between these atoms indicated that this scissile amide is significantly distorted in a manner dependent on the catalytic His75 residue, supporting a role for ground-state destabilization in the initiation of the splicing cascade.
Semisynthesis of the Mxe GyrA protein has also provided insight into the penultimate step of protein splicing, the resolution of the branched intermediate through succinimide formation.213, 214 To generate this branched protein, a recombinant α-thioester fragment corresponding to residues 1-184 of the Mxe GyrA intein was ligated to a synthetic peptide containing the native N- and C- exteins branched through the side-chain of the catalytic +1Thr residue (Figure 60a). Ironically, this semisynthesis actually employed an intein to make an intein (i.e to install the α-thioester into the Mxe GyrA fragment). The refolded semisynthetic, branched intermediate was found to undergo C-terminal asparagine cyclization an order of magnitude faster than a linear intein unable to form the branched intermediate due to a C1A mutation. Additionally, the necessity of histidines 187 and 197 as catalysts for the asparagine cyclization reaction was established through the replacement of these residues with isosteric β-thienylalanines. This insight allowed the x-ray crystal structure of a ‘trapped’ semisynthetic branched intermediate to be solved,214 revealing subtle rearrangements of amino acid side-chains in the active site, such that the C-terminal asparagine residue is positioned for nucleophilic attack on the +1 amide bond, further activated by hydrogen-bonding to a backbone amide bond (Figure 60b).
Figure 60.
Semisynthesis of inteins. a) Scheme for the semisynthesis of the Mxe GyrA branched intermediate using EPL.214 b) Close-up of the active site of the Mxe GyrA (extein residues are colored green and the intein is colored pink). Apparent H-bonds are indicated with dashed lines. c) Folding pathway for the Npu DnaE split intein.219
The fragmented form of split inteins permits the study of catalytic mechanisms without the need to perform formal protein semisynthesis – the split intein complex can simply be constituted from synthetic and recombinant intein fragments.615 However, protein semisynthesis has been used to study the structure of split intein fragments.219 Using EPL, the 102-residue IntN fragment of the Npu DnaE split intein was segmentally labeled with 15N isotopes for NMR spectroscopy. The intein fragment displayed a bipartite structure, where the N-terminal portion is partly folded while the remainder is unstructured in the absence of the cognate IntC fragment. This insight proved instrumental in piecing together the overall folding pathway of Npu DnaE, which involves an initial electrostatic capture step, involving the disordered regions of the IntN and IntC fragments, followed by a hydrophobically driven collapse into the final native fold (Figure 60c). A notable feature of this folding mechanism is that it provides an explanation for why the isolated IntN and IntC fragments have no residual splicing activity, namely the catalytic residues are in disordered regions and only adopt a catalytic configuration upon fragment association. This basic paradigm is also true for the atypically split intein, Cat (Consensus atypical) TerL, even though the details of the folding pathway differ.392
8. SUMMARY AND FUTURE PERSPECTIVES
The marriage of chemical peptide synthesis and recombinant protein production, two vastly different methods of generating polypeptides, has provided controlled access to molecules that could not be generated otherwise. From an engineering point of view, it is quite remarkable that such large biomolecules can be stitched together in a test-tube, often without surgical scars that would give away any hint of non-biological origins. In assembling this review, our motives were two-fold: firstly, to provide an inspection of the available semisynthetic methods of targeted amide bond formation, and secondly to demonstrate the broad use of this enabling protein engineering strategy for the study of many classes of proteins.
The various semisynthetic methods discussed in this review range from those considered to be chemical in origin, such as NCL, to those that are more biologically inspired, including enzyme-catalyzed transpeptidation. These methods have often been considered separate, yet they are unified by common themes of selective acyl-shift chemistry to both break and make selective amide bonds. Enzymes and biocatalysts such as inteins, transpeptidases and proteases are evolved for such precision protein engineering in vivo and have been repurposed by protein chemists to also carry out these tasks in vitro. Currently, each semisynthesis method brings inherent strengths and weaknesses to a particular application, and no one strategy in its current form can truly carry out the role of an all-purpose protein assembly tool. Nonetheless, considerable progress continues to be made in broadening the scope of these strategies, whether it be through the development of new chemistries, the design or evolution of more agile protein ligases or the development of hybrid chemoenzymatic approaches that blend beneficial features of parental methods. Moreover, the field in general is likely to benefit from increased implementation of powerful advances in directed protein evolution.616, 617
While it is remarkable just how far protein semisynthesis has come, both in terms of methodological advances and the biological questions that are now being addressed, it would be wrong to conclude that the field has matured to the point where any biological problem, any protein system, can now be reasonably approached. This is simply not the case. For example, most integral membrane proteins, despite their enormous biological importance, continue to be beyond the reach of current capabilities. Equally, many if not most proteins exist as part of large multi-subunit complexes, a situation that greatly complicates semisynthetic access to these systems, at least if one wishes to study the native complex. Thus, there are plenty of opportunities for profound methodological advancement in the field. Below, we discuss just two of these, recognizing that there are many others.
1. One-step semisynthesis applied to internal regions:
In theory, the size of a protein per se does not dictate whether it is an attractive target for semisynthesis through amide bond construction. Instead, it is the location of the desired modification site within the protein sequence that most often determines whether semisynthesis is a viable strategy. Semisynthetic targets where a modified fragment is installed at the N- or C-termini are the most facile, as these applications only require a single ligation to append a modified peptide onto the end of a truncated protein. On the other hand, modification of an internal region of a protein typically requires that a multistep ligation strategy (i.e. involving the assembly of three or more fragments) be adopted. While there are many impressive examples of this, several of which are described in this review, multistep semisyntheses can be technically quite demanding and are hence less appealing to the non-specialist. In principle, the ability to insert a polypeptide sequence within an expressed protein in a single step would lower the barrier to manipulating internal regions of a protein through semisynthesis. The use of orthogonal transpeptidase enzymes, for example butelase 1 and sortase A, could provide a solution to this problem, through the reversible metathesis of a recombinant sequence with a synthetic sequence through transpeptidation. Alternatively, one could also envision the use of PTS using orthogonal split inteins for such an application, where two templated splicing events irreversibly exchange the recombinant internal sequence with one derived synthetically. In this scenario, synthetic access to peptides bearing two split intein fragments would be required. Accordingly, these reactions would require the use of split inteins with atypically short fragments, or the in situ assembly of split intein fragments as used in TAIL.
2. Protein semisynthesis in the cellular milieu:
The frontier of protein semisynthesis appears, perhaps, not to be the refinement of existing technologies for in vitro applications. Instead, a true challenge is the literal transduction of these technologies into the cellular setting, where biological implications of biochemical and biophysical behavior become apparent. As mentioned previously, semisynthetic proteins may be delivered to cells, for example through microinjection, in order to observe the effects of a given chemical modification. However, in many cases it is preferable to reassemble the protein sequence within the cell itself, especially in cases where the protein target exists in larger complexes with other proteins or, in the case of histone proteins, genomic DNA. Accordingly, the vessel of the semisynthetic reaction is no longer the test tube, but the living cell. Somewhat cruelly, two of the principle tools for protein semisynthesis in vitro, NCL and EPL, are obstructed by the high concentration of reactive thiols in cellular environments. Biocatalytic methods, however, are viable alternatives, as they have been subject to evolutionary pressure in order to function on the surface of and inside living cells. Thankfully, tools capable of carrying out user-defined semisynthesis in cells are now becoming available, achieving a level of proficiency required for cellular applications. Among these, split inteins and transpeptidases have been used to modify proteins in living cells (as described earlier), and it is anticipated that use of these tools will be further explored to generate modified proteins in cells, presumably relying on advanced methods for the delivery of synthetic polypeptides across the plasma membrane, which we note is a very active area of study in chemical biology.618–620 While the challenges associated of this undertaking can hardly be overstated, one could argue that these are no less daunting than those faced by the peptide chemistry community some thirty years ago when it considered protein semisynthesis. Thus, we feel there is some room of optimism here.
In conclusion, protein semisynthesis occupies a unique methodological space at the intersection of synthetic chemistry and molecular biology and has been fortunate enough to benefit from advances in both fields. The range of biological questions answered using semisynthetic proteins is testament to this synergy. This strategy will continue to provide innovative and accessible ways to modify the structure of proteins for molecular-level interrogation, driven by the constant appearance of challenging biological questions and the need to address these in a physiologic context.
ACKNOWLEDGMENT
We thank The Charles H. Revson Foundation (R.E.T.) and the National Institutes of Health (NIH, R37 GM086868 and P01 CA196539) for financial support. We also thank members of the Muir lab for helpful discussions.
ABBREVIATIONS
- AA, aa
amino acid
- A, Ala
alanine
- C, Cys
cysteine
- D, Asp
aspartic acid
- E, Glu
glutamic acid
- F, Phe
phenylalanine
- G, Gly
glycine
- H, His
histidine
- I, Ile
isoleucine
- K, Lys
lysine
- L, Leu
leucine
- M, Met
methionine
- N, Asn
asparagine
- P, Pro
proline
- Q, Gln
glutamine
- R, Arg
arginine
- S, Ser
serine
- T, Thr
threonine
- U, Sec
selenocysteine
- V, Val
valine
- W, Trp
tryptophan
- X, Xaa
any amino acid
- Yaa
any amino acid
- AANAT
arylalkylamine N-acetyltransferase, also known as serotonin N-acetyl transferase
- AceL TerL
atypically split intein in the T4-bacteriophage-type DNA-packaging terminase from the Ace Lake in Antarctica
- Acm
acetamidomethyl
- ADP
adenosine diphosphate
- Akt1
Akt serine/threonine kinase 1, also known as protein kinase B
- AP-1
activator protein 1
- APMA
μ-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- Ava DnaE
naturally split intein in the DnaE protein from Anabaena variabilis
- b2AR
b2-adrenergic receptor
- Bn
benzyl
- Boc
tert-butoxycarbonyl
- Cas9
CRISPR associated protein 9
- Cfa DnaE
consensus designed split intein from the fast-splicing DnaE split inteins
- CfaGEP DnaE
Cfa DnaE with a loop mutation conferring increased extein tolerance
- Chd1
chromodomain-helicase-DNA-binding protein 1, chromatin remodeler, component of the SAGA complex
- CHO-K1
subclone of chinese hampster ovary cell line
- CoREST
RE1 silencing transcription factor, a transcriptional corepressor
- cp-p300
circularly permutated p300 HAT domain
- CRISPR
clustered regularly interspaced short palindromic repeats
- Csk
C-terminal Src kinase
- CXCL12
chemokine (C-X-C motif) ligand 12, also known as stromal cell-derived factor 1
- CXCL8
chemokine (C-X-C motif) ligand 8, also known as interleukin 8
- Dbz
diaminobenzoyl
- dCas9
nuclease dead mutant of Cas9
- dmPro
5,5-dimethylproline
- DnaE
catalytic subunit of bacterial DNA polymerase III
- Dot1
disruptor of telomeric silencing, H3K79 methyltransferase
- DTA
diptheria toxin a-chain
- EDTA
Ethylenediaminetetraacetate
- EED
embryonic ectoderm development, member of the PRC2 complex
- EGFR
epidermal growth factor receptor
- EPL
expressed protein ligation
- EPO
erythropoietin
- eSrtA(2A)
evolved SaSrtA recognizing LAxTG sorting sequence
- eSrtA(4S)
evolved SaSrtA recognizing LPxSG sorting sequence
- ExtC
C-terminal extein sequence
- ExtN
N-terminal extein sequence
- EZH2
enhancer of zeste homolog 2, member of the PRC2 complex
- F40
evolved SaSrtA recognizing yPxTG sorting sequences
- FKBP12
FK506 binding protein 12
- Fmoc
9-fluorenylmethoxycarbonyl
- FRET
Förster resonance energy transfer
- GDI
GDP-dissociation inhibitor protein
- GFP
green fluorescent protein
- GlcNAc
N-acetyl glucosamine
- GltPh
Na+-coupled aspartate transporter
- GOS TerL
atypically split intein in the T4-bacteriophage-type DNA-packaging terminase from the Punta Cormorant lagoon in the Galapagos
- gp41-1/8
split inteins found in the gp41 DNA helicase from unknown host
- GPCR
G-protein coupled receptors
- GPI
glycosylphosphatidylinositol
- GRK2
G-protein coupled receptor kinase 2
- hAla
homoalanine
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HED
homing endonuclease domain
- HEK293T
Human embryonic kidney 293 cell line expressing a mutant version of the SV40 large T antigen
- HF
hydrogen fluoride
- hGH
human growth hormone
- HP1a
heterochromatin protein 1a, binds H3K9me2/3
- IL-13
interleukin 13
- IL-6
interleukin 6
- IMPDH-1
split intein found in the inosine-5’-monophosphate dehydrogenase from unknown host
- IntC
C-terminal split intein fragment
- IntN
N-terminal split intein fragment
- ISWI
imitation switch, chromatin remodeler family
- Jarid2
jumonji and AT-rich interaction domain containing 2, member of the PRC2 complex
- KcsA
potassium channel from the soil bacteria Streptomyces lividans
- KvAP
voltage-dependent potassium channel from Aeropyrum pernix
- LC3
microtubule-associated protein light chain 3
- LFN
lethal factor N-terminal domain from anthrax toxin
- MBP
maltose-binding protein
- MeBn
methylbenzyl
- MeCP2
methyl-CpG binding protein 2
- MeOBn
methoxylbenzyl
- MESNa
mercaptoethanesulfonate sodium salt
- MH2
mad homology 2
- MLL
myeloid/lymphoid or mixed-lineage leukemia, H3K4-specific methyltransferase
- Mtu RecA
intein in the DNA repair protein RecA of Mycobacterium tuberculosis
- Mxe GyrA
intein in DNA gyrase A protien of Mycobacterium xenopi
- NCL
native chemical ligation
- Nip
nipecotic acid
- Npu DnaE
naturally split intein in the DnaE protein from Nostoc punctiforme
- NrdJ-1
split intein found in the ribonucleotide reductase catalytic subunit from unknown origin
- OaAEPlb
asparginyl endopeptidase from Oldenlandia affinis
- OGp
4-guanidinophenyl
- PAL
peptide asparaginyl ligase
- PARP
poly(ADP-ribose) polymerase enzyme
- PD
Parkinson’s disease
- PDK1
pyruvate dehydrogenase kinase 1
- pelB
pectate lyase B
- Pfa
phosphonodifluoromethylenealanine
- PG
protecting group
- PH
pleckstrin homology
- Pma
phosphonomethylenealanine
- POI
protein of interest
- PRC2
polycomb repressive complex 2, H3K27 methyltransferase
- PrP
prion protein
- PSD
post synaptic density
- PSD-95
postsynaptic density protein 95
- PTEN
phosphatase and tensin homolog
- PTM
post-translational modification
- PTS
protein trans-splicing
- Rma DnaB
intein in the DnaB helicase of Rhodothermus marinus
- RNase A
ribonuclease A
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- SaSrtA
Sortase A from S. aureus
- Sce VMA
intein in the vacuolar ATPase subunit of Saccharomyces cerevisiae
- SEA
bis(2-sulfanylethyl)amino
- SEPL
streamlined expressed protein ligation
- SFTI
sunflower trypsin inhibitor-1
- SH2
Src Homology 2 domain
- SH3
Src Homology 3 domain
- SHP-2
Src homology region 2-containing protein tyrosine phosphatase 2
- Smt3
SUMO family protein
- SPPS
solid-phase peptide synthesis
- SpSrtA
sortase A from S. pyogenes
- Ssp DnaB
intein in the DnaB helicase of cyanobacterium Synechocystis sp., strain PCC 6803
- Ssp DnaE
naturally split intein in the DnaE protein from Synechocystis sp., strain PCC 6803
- Ssp DnaX
intein in the DNA polymerase III subunit tau from Synechocystis sp., strain PCC 6803
- Ssp GyrB
intein in DNA gyrase subunit B from Synechocystis sp., strain PCC 6803
- STL
serine/threonine ligation
- SUMO
small ubiquitin-like modifier
- SWI-SNF
switch/sucrose nonfermentable, a nucleosome remodeling complex
- TBR-I/II
type I/II TGF-P receptor subunits
- Ter DnaE-3
naturally split intein in the DnaE protein from Trichodesmium erythraeum
- TEV
tobacco etch virus
- TFA
trifluoroacetic acid
- TGF-β
transforming growth factor-β
- TGM2
tissue transglutaminase 2
- tRNA
transfer RNA
- Trt
triphenylmethyl
- Trx
thioredoxin
- Ulp1
ubiquitin-like-specific protease
- VidaL T4Lh-1 and UvsX-2
atypically split inteins discovered from metagenomic analysis from Lake Vida Brine Hole 2
- WWP2
WW domain containing protein 2, an E3 ubiquitin ligase
Biographies
Biographical Sketch
Tom W. Muir received his B.Sc in Chemistry from the University of Edinburgh in 1989 and his Ph.D. in Chemistry from the same institute in 1993 under the direction of Professor Robert Ramage. Following postdoc studies with Stephen B.H. Kent at The Scripps Research Institute, Muir joined the faculty of The Rockefeller University in 1996, where he rose through the ranks eventually being appointed the Richard E. Salomon Family Professor and Director of the Pels Center of Chemistry, Biochemistry and Structural Biology. In 2011, Dr. Muir joined the faculty of Princeton University as the Van Zandt Williams Jr. Class of ‘65 Professor of Chemistry. He currently serves as Chair of the Chemistry Department. Dr. Muir has published many articles in the areas of peptide and protein chemistry. His current interests lie principally in the area of epigenetics, where he is interested in how changes to chromatin structure drive different cellular phenotypes.
Robert E. Thompson received his received his Ph.D. in Chemistry from the University of Sydney in 2014. During his graduate studies under the supervision of Professor Richard J. Payne and Professor Katrina A. Jolliffe, Rob developed chemical methods for the synthesis of post-translationally modified bioactive peptides. In 2015, he joined the laboratory of Professor Tom W. Muir at Princeton University where he is developing and applying tools in chemistry and protein engineering to study chromatin.
REFERENCES
- 1.International Human Genome Sequencing, C., Finishing the Euchromatic Sequence of the Human Genome. Nature 2004, 431, 931–945. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R; Agar JN; Amster IJ; Baker MS; Bertozzi CR; Boja ES; Costello CE; Cravatt BF; Fenselau C; Garcia BA, et al. , How Many Human Proteoforms Are There? Nat. Chem. Biol 2018, 14, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutureira O; Bernardes GJL, Advances in Chemical Protein Modification. Chem. Rev 2015, 115, 2174–2195. [DOI] [PubMed] [Google Scholar]
- 4.Lang K; Chin JW, Cellular Incorporation of Unnatural Amino Acids and Bioorthogonal Labeling of Proteins. Chem. Rev 2014, 114, 4764–4806. [DOI] [PubMed] [Google Scholar]
- 5.Kent SBH, Total Chemical Synthesis of Proteins. Chem. Soc. Rev 2009, 38, 338–351. [DOI] [PubMed] [Google Scholar]
- 6.Curtius T, Ueber Einige Neue der Hippursäure Analog Constituirte, Synthetisch Dargestellte Amidosäuren. J. Prakt. Chem 1882, 26, 145–208. [Google Scholar]
- 7.El-Faham A; Albericio F, Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev 2011, 111, 6557–6602. [DOI] [PubMed] [Google Scholar]
- 8.Isidro-Llobet A; Alvarez M; Albericio F, Amino Acid-Protecting Groups. Chem. Rev 2009, 109, 2455–2504. [DOI] [PubMed] [Google Scholar]
- 9.Merrifield RB, Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc 1963, 85, 2149–2154. [Google Scholar]
- 10.Wheelan SJ; Marchler-Bauer A; Bryant SH, Domain Size Distributions Can Predict Domain Boundaries. Bioinformatics 2000, 16, 613–618. [DOI] [PubMed] [Google Scholar]
- 11.Nyfeler R, Peptide Synthesis via Fragment Condensation. Methods Mol. Biol 1995, 35, 303–316. [DOI] [PubMed] [Google Scholar]
- 12.Kemp DS, The Amine Capture Strategy for Peptide Bond Formation—An Outline of Progress. Biopolymers 1981, 20, 1793–1804. [Google Scholar]
- 13.Dawson PE; Muir TW; Clark-Lewis I; Kent SBH, Synthesis of Proteins by Native Chemical Ligation. Science 1994, 266, 776–779. [DOI] [PubMed] [Google Scholar]
- 14.Liu H; Li XC, Serine/Threonine Ligation: Origin, Mechanistic Aspects, and Applications. Acc. Chem. Res 2018, 51, 1643–1655. [DOI] [PubMed] [Google Scholar]
- 15.Shah NH; Muir TW, Inteins: Nature’s Gift to Protein Chemists. Chem. Sci 2014, 5, 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang TK; Jackson DY; Burnier JP; Wells JA, Subtiligase: A Tool for Semisynthesis of Proteins. Proc. Natl. Acad. Sci. U.S.A 1994, 91, 12544–12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao H; Hart SA; Schink A; Pollok BA, Sortase-Mediated Protein Ligation: A New Method for Protein Engineering. J. Am. Chem. Soc 2004, 126, 2670–2671. [DOI] [PubMed] [Google Scholar]
- 18.Burke HM; McSweeney L; Scanlan EM, Exploring Chemoselective S-to-N Acyl Transfer Reactions in Synthesis and Chemical Biology. Nat. Commun 2017, 8, 15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson PE; Kent SBH, Synthesis of Native Proteins by Chemical Ligation. Annu. Rev. Biochem 2000, 69, 923–960. [DOI] [PubMed] [Google Scholar]
- 20.Agouridas V; El Mahdi O; Diemer V; Cargoët M; Monbaliu J-CM; Melnyk O, Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations. Chem. Rev 2019, 119, 7328–7443. [DOI] [PubMed] [Google Scholar]
- 21.Sletten EM; Bertozzi CR, Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. Int. Ed 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CC; Schultz PG, Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem 2010, 79, 413–444. [DOI] [PubMed] [Google Scholar]
- 23.Chaiken IM; Komoriya A, Protein Semisynthesis. Trends in Biochemical Sciences 1978, 3, 269–271. [Google Scholar]
- 24.Wallace CJA, Peptide ligation and semisynthesis. Curr. Opin. Biotechnol 1995, 6, 403–410. [DOI] [PubMed] [Google Scholar]
- 25.Hupe DJ; Jencks WP, Nonlinear Structure-Reactivity Correlations. Acyl Transfer Between Sulfur and Oxygen Nucleophiles. J. Am. Chem. Soc 1977, 99, 451–464. [Google Scholar]
- 26.Wieland T; Bokelmann E; Bauer L; Lang HU; Lau H, Über Peptidsynthesen. 8. Mitteilung Bildung von S-haltigen Peptiden durch intramolekulare Wanderung von Aminoacylresten. Justus Liebigs Ann. Chem 1953, 583, 129–149. [Google Scholar]
- 27.Tam JP; Lu YA; Liu CF; Shao J, Peptide Synthesis Using Unprotected Peptides Through Orthogonal Coupling Methods. Proc. Natl. Acad. Sci. U.S.A 1995, 92, 12485–12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlanson DA; Chytil M; Verdine GL, The Leucine Zipper Domain Controls the Orientation of AP-1 in the NFAT·AP-1·DNA Complex. Chem. Biol 1996, 3, 981–991. [DOI] [PubMed] [Google Scholar]
- 29.Thurlkill RL; Grimsley GR; Scholtz JM; Pace CN, pK Values of the Ionizable Groups of Proteins. Protein Sci. 2006, 15, 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otto HH; Schirmeister T, Cysteine Proteases and Their Inhibitors. Chem. Rev 1997, 97, 133–171. [DOI] [PubMed] [Google Scholar]
- 31.Plechanovova A; Jaffray EG; McMahon SA; Johnson KA; Navratilova I; Naismith JH; Hay RT, Mechanism of Ubiquitylation by Dimeric RING Ligase RNF4. Nat. Struct. Mol. Biol 2011, 18, 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berndsen CE; Wolberger C, New Insights into Ubiquitin E3 Ligase Mechanism. Nat. Struct. Mol. Biol 2014, 21, 301–307. [DOI] [PubMed] [Google Scholar]
- 33.Keillor JW; Clouthier CM; Apperley KYP; Akbar A; Mulani A, Acyl Transfer Mechanisms of Tissue Transglutaminase. Bioorg. Chem 2014, 57, 186–197. [DOI] [PubMed] [Google Scholar]
- 34.Marahiel MA; Stachelhaus T; Mootz HD, Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem. Rev 1997, 97, 2651–2673. [DOI] [PubMed] [Google Scholar]
- 35.Noren CJ; Wang JM; Perler FB, Dissecting the Chemistry of Protein Splicing and Its Applications. Angew. Chem. Int. Ed 2000, 39, 450–466. [PubMed] [Google Scholar]
- 36.Eryilmaz E; Shah NH; Muir TW; Cowburn D, Structural and Dynamical Features of Inteins and Implications on Protein Splicing. J. Biol. Chem 2014, 289, 14506–14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guimaraes CP; Witte MD; Theile CS; Bozkurt G; Kundrat L; Blom AEM; Ploegh HL, Site-Specific C-Terminal and Internal Loop Labeling of Proteins Using Sortase-Mediated Reactions. Nat. Protoc 2013, 8, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrickx APA; Budzik JM; Oh SY; Schneewind O, Architects at the Bacterial Surface – Sortases and the Assembly of Pili With Isopeptide Bonds. Nat. Rev. Microbiol 2011, 9, 166–176. [DOI] [PubMed] [Google Scholar]
- 39.Johnson ECB; Kent SBH, Insights into the Mechanism and Catalysis of the Native Chemical Ligation Reaction. J. Am. Chem. Soc 2006, 128, 6640–6646. [DOI] [PubMed] [Google Scholar]
- 40.Saito F; Noda H; Bode JW, Critical Evaluation and Rate Constants of Chemoselective Ligation Reactions for Stoichiometric Conjugations in Water. ACS Chem. Biol 2015, 10, 1026–1033. [DOI] [PubMed] [Google Scholar]
- 41.Schelté P; Boeckler C; Frisch B; Schuber F, Differential Reactivity of Maleimide and Bromoacetyl Functions with Thiols: Application to the Preparation of Liposomal Diepitope Constructs. Bioconjugate Chemistry 2000, 11, 118–123. [DOI] [PubMed] [Google Scholar]
- 42.Hackeng TM; Griffin JH; Dawson PE, Protein Synthesis by Native Chemical Ligation: Expanded Scope by Using Straightforward Methodology. Proc. Natl. Acad. Sci. U.S.A 1999, 96, 10068–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sifferd RH; du Vigneaud V, A New Synthesis of Carnosine, With Some Observations on the Splitting of the Benzyl Group From Carbobenzoxy Derivatives and From Benzylthio Ethers. J. Biol. Chem 1935, 108, 753–761. [Google Scholar]
- 44.Erickson BW; Merrifield RB, Acid Stability of Several Benzylic Protecting Groups Used in Solid-Phase Peptide Synthesis. Rearrangement of O-Benzyltyrosine to 3-Benzyltyrosine. J. Am. Chem. Soc 1973, 95, 3750–3756. [DOI] [PubMed] [Google Scholar]
- 45.Akabori S; Sakakibara S; Shimonishi Y; Nobuhara Y, A New Method for the Protection of the Sulfhydryl Group during Peptide Synthesis. Bull. Chem. Soc. Jpn 1964, 37, 433–434. [Google Scholar]
- 46.Veber D; Milkowski J; Varga S; Denkewalter R; Hirschmann R, Acetamidomethyl. A Novel Thiol Protecting Group for Cysteine. J. Am. Chem. Soc 1972, 94, 5456–5461. [DOI] [PubMed] [Google Scholar]
- 47.Bang D; Chopra N; Kent SBH, Total Chemical Synthesis of Crambin. J. Am. Chem. Soc 2004, 126, 1377–1383. [DOI] [PubMed] [Google Scholar]
- 48.Jbara M; Maity SK; Seenaiah M; Brik A, Palladium Mediated Rapid Deprotection of N-Terminal Cysteine under Native Chemical Ligation Conditions for the Efficient Preparation of Synthetically Challenging Proteins. J. Am. Chem. Soc 2016, 138, 5069–5075. [DOI] [PubMed] [Google Scholar]
- 49.Haj-Yahya M; Lashuel HA, Protein Semisynthesis Provides Access to Tau Disease-Associated Post-translational Modifications (PTMs) and Paves the Way to Deciphering the Tau PTM Code in Health and Diseased States. J. Am. Chem. Soc 2018, 140, 6611–6621. [DOI] [PubMed] [Google Scholar]
- 50.Yan BJ; Shi WW; Ye LZ; Liu L, Acyl Donors for Native Chemical Ligation. Curr. Opin. Chem. Biol 2018, 46, 33–40. [DOI] [PubMed] [Google Scholar]
- 51.Hojo H; Aimoto S, Polypeptide Synthesis Using the S-Alkyl Thioester of a Partially Protected Peptide Segment. Synthesis of the DNA-Binding Domain of c-Myb Protein (142–193)–NH2. Bull. Chem. Soc. Jpn 1991, 64, 111–117. [Google Scholar]
- 52.Camarero JA; Adeva A; Muir TW, 3-Thiopropionic Acid as a Highly Versatile Multidetachable Thioester Resin Linker. Lett. Pept. Sci 2000, 7, 17–21. [Google Scholar]
- 53.Kajihara Y; Yoshihara A; Hirano K; Yamamoto N, Convenient Synthesis of a Sialylglycopeptide-Thioester Having an Intact and Homogeneous Complex-Type Disialyl-Oligosaccharide. Carbohydr. Res 2006, 341, 1333–1340. [DOI] [PubMed] [Google Scholar]
- 54.Backes BJ; Ellman JA, An Alkanesulfonamide “Safety-Catch” Linker for Solid-Phase Synthesis. J. Org. Chem 1999, 64, 2322–2330. [Google Scholar]
- 55.Botti P; Villain M; Manganiello S; Gaertner H, Native Chemical Ligation through in Situ O to S Acyl Shift. Org. Lett 2004, 6, 4861–4864. [DOI] [PubMed] [Google Scholar]
- 56.Zheng J-S; Xi W-X; Wang F-L; Li J; Guo Q-X, Fmoc-SPPS Chemistry Compatible Approach for the Generation of (Glyco)peptide Aryl Thioesters. Tetrahedron Lett. 2011, 52, 2655–2660. [Google Scholar]
- 57.Richardson JP; Chan C-H; Blanc J; Saadi M; Macmillan D, Exploring Neoglycoprotein Assembly Through Native Chemical Ligation Using Neoglycopeptide Thioesters Prepared via N→S Acyl Transfer. Org. Biomol. Chem 2010, 8, 1351–1360. [DOI] [PubMed] [Google Scholar]
- 58.Hojo H; Onuma Y; Akimoto Y; Nakahara Y; Nakahara Y, N-Alkyl Cysteine-Assisted Thioesterification of Peptides. Tetrahedron Lett. 2007, 48, 25–28. [Google Scholar]
- 59.Ollivier N; Dheur J; Mhidia R; Blanpain A; Melnyk O, Bis(2-sulfanylethyl)amino Native Peptide Ligation. Org. Lett 2010, 12, 5238–5241. [DOI] [PubMed] [Google Scholar]
- 60.Blanco-Canosa JB; Dawson PE, An Efficient Fmoc-SPPS Approach for the Generation of Thioester Peptide Precursors for Use in Native Chemical Ligation. Angew. Chem. Int. Ed 2008, 47, 6851–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanco-Canosa JB; Nardone B; Albericio F; Dawson PE, Chemical Protein Synthesis Using a Second-Generation N-Acylurea Linker for the Preparation of Peptide-Thioester Precursors. J. Am. Chem. Soc 2015, 137, 7197–7209. [DOI] [PubMed] [Google Scholar]
- 62.Wang J-X; Fang G-M; He Y; Qu D-L; Yu M; Hong Z-Y; Liu L, Peptide o-Aminoanilides as Crypto-Thioesters for Protein Chemical Synthesis. Angew. Chem. Int. Ed 2015, 54, 2194–2198. [DOI] [PubMed] [Google Scholar]
- 63.Fang G-M; Li Y-M; Shen F; Huang Y-C; Li J-B; Lin Y; Cui H-K; Liu L, Protein Chemical Synthesis by Ligation of Peptide Hydrazides. Angew. Chem. Int. Ed 2011, 50, 7645–7649. [DOI] [PubMed] [Google Scholar]
- 64.Flood DT; Hintzen JCJ; Bird MJ; Cistrone PA; Chen JS; Dawson PE, Leveraging the Knorr Pyrazole Synthesis for the Facile Generation of Thioester Surrogates for use in Native Chemical Ligation. Angew. Chem. Int. Ed 2018, 57, 11634–11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gentle IE; De Souza DP; Baca M, Direct Production of Proteins With N-Terminal Cysteine for Site-Specific Conjugation. Bioconjugate Chemistry 2004, 15, 658–663. [DOI] [PubMed] [Google Scholar]
- 66.Xiao Q; Zhang FR; Nacev BA; Liu JO; Pei DH, Protein N-Terminal Processing: Substrate Specificity of Escherichia coli and Human Methionine Aminopeptidases. Biochemistry 2010, 49, 5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chiang KP; Jensen MS; McGinty RK; Muir TW, A Semisynthetic Strategy to Generate Phosphorylated and Acetylated Histone H2B. ChemBioChem 2009, 10, 2182–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hauser PS; Ryan RO, Expressed Protein Ligation Using an N-Terminal Cysteine Containing Fragment Generated In Vivo From a pe1B Fusion Protein. Protein Expr. Purif 2007, 54, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gross E; Witkop B, Nonenzymatic Cleavage of Peptide Bonds - Methionine Residues in Bovine Pancreatic Ribonuclease. J. Biol. Chem 1962, 237, 1856–1860. [PubMed] [Google Scholar]
- 70.Macmillan D; Arham L, Cyanogen Bromide Cleavage Generates Fragments Suitable for Expressed Protein and Glycoprotein Ligation. J. Am. Chem. Soc 2004, 126, 9530–9531. [DOI] [PubMed] [Google Scholar]
- 71.Camarero JA; Shekhtman A; Campbell EA; Chlenov M; Gruber TM; Bryant DA; Darst SA; Cowburn D; Muir TW, Autoregulation of a Bacterial Sigma Factor Explored by Using Segmental Isotopic Labeling and NMR. Proc. Natl. Acad. Sci. U.S.A 2002, 99, 8536–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romanelli A; Shekhtman A; Cowburn D; Muir TW, Semisynthesis of a Segmental Isotopically Labeled Protein Splicing Precursor: NMR Evidence for an Unusual Peptide Bond at the N-Extein-Intein Junction. Proc. Natl. Acad. Sci. U.S.A 2004, 101, 6397–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu DS; Xu R; Dutta K; Cowburn D, N-Terminal Cysteinyl Proteins Can Be Prepared Using Thrombin Cleavage. FEBS Lett. 2008, 582, 1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tolbert TJ; Wong CH, New Methods for Proteomic Research: Preparation of Proteins With N-Terminal Cysteines for Labeling and Conjugation. Angew. Chem. Int. Ed 2002, 41, 2171–2174. [DOI] [PubMed] [Google Scholar]
- 75.Tropea JE; Cherry S; Waugh DS, Expression and Purification of Soluble His6-Tagged TEV Protease. Methods Mol. Biol 2009, 498, 297–307. [DOI] [PubMed] [Google Scholar]
- 76.Sun CS; Liang JQ; Shi R; Gao XN; Zhang RJ; Hong FL; Yuan QH; Wang SB, Tobacco Etch Virus Protease Retains Its Activity in Various Buffers and in the Presence of Diverse Additives. Protein Expr. Purif 2012, 82, 226–231. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen UTT; Bittova L; Müller MM; Fierz B; David Y; Houck-Loomis B; Feng V; Dann GP; Muir TW, Accelerated Chromatin Biochemistry Using DNA-Barcoded Nucleosome Libraries. Nat. Methods 2014, 11, 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marblestone JG; Edavettal SC; Lim Y; Lim P; Zuo X; Butt TR, Comparison of SUMO Fusion Technology With Traditional Gene Fusion Systems: Enhanced Expression and Solubility With SUMO. Protein Sci. 2006, 15, 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frey S; Görlich D, A New Set of Highly Efficient, Tag-Cleaving Proteases for Purifying Recombinant Proteins. Journal of Chromatography A 2014, 1337, 95–105. [DOI] [PubMed] [Google Scholar]
- 80.Colomina N; Guasch C; Torres-Rosell J, Analysis of SUMOylation in the RENT Complex by Fusion to a SUMO-Specific Protease Domain. Methods Mol. Biol 2017, 1505, 97–117. [DOI] [PubMed] [Google Scholar]
- 81.Ai H; Guo Y; Sun D; Liu S; Qi Y; Guo J; Qu Q; Gong Q; Zhao S; Li J, et al. , Examination of the Deubiquitylation Site Selectivity of USP51 by Using Chemically Synthesized Ubiquitylated Histones. ChemBioChem 2019, 20, 221–229. [DOI] [PubMed] [Google Scholar]
- 82.Pan M; Zheng Q; Ding S; Zhang L; Qu Q; Wang T; Hong D; Ren Y; Liang L; Chen C, et al. , Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains. Angew. Chem. Int. Ed 2019, 58, 2627–2631. [DOI] [PubMed] [Google Scholar]
- 83.Xu L; Huang J-F; Chen C-C; Qu Q; Shi J; Pan M; Li Y-M, Chemical Synthesis of Natural Polyubiquitin Chains through Auxiliary-Mediated Ligation of an Expressed Ubiquitin Isomer. Org. Lett 2018, 20, 329–332. [DOI] [PubMed] [Google Scholar]
- 84.Tsuda Y; Shigenaga A; Tsuji K; Denda M; Sato K; Kitakaze K; Nakamura T; Inokuma T; Itoh K; Otaka A, Development of a Chemical Methodology for the Preparation of Peptide Thioesters Applicable to Naturally Occurring Peptides Using a Sequential Quadruple Acyl Transfer System. ChemistryOpen 2015, 4, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dang B; Mravic M; Hu H; Schmidt N; Mensa B; DeGrado WF, SNAC-Tag for Sequence-Specific Chemical Protein Cleavage. Nat. Methods 2019, 16, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou A; Webb G; Zhu X; Steiner DF, Proteolytic Processing in the Secretory Pathway. J. Biol. Chem 1999, 274, 20745–20748. [DOI] [PubMed] [Google Scholar]
- 87.Kane PM; Yamashiro CT; Wolczyk DF; Neff N; Goebl M; Stevens TH, Protein Splicing Converts the Yeast Tfp1 Gene-Product to the 69-Kd Subunit of the Vacuolar H+-Adenosine Triphosphatase. Science 1990, 250, 651–657. [DOI] [PubMed] [Google Scholar]
- 88.Novikova O; Jayachandran P; Kelley DS; Morton Z; Merwin S; Topilina NI; Belfort M, Intein Clustering Suggests Functional Importance in Different Domains of Life. Molecular Biology and Evolution 2015, 33, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Novikova O; Topilina N; Belfort M, Enigmatic Distribution, Evolution, and Function of Inteins. J. Biol. Chem 2014, 289, 14490–14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu MQ; Southworth MW; Mersha FB; Hornstra LJ; Perler FB, In Vitro Protein Splicing of Purified Precursor and the Identification of a Branched Intermediate. Cell 1993, 75, 1371–1377. [DOI] [PubMed] [Google Scholar]
- 91.Mills KV; Johnson MA; Perler FB, Protein Splicing: How Inteins Escape from Precursor Proteins. J. Biol. Chem 2014, 289, 14498–14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Southworth MW; Benner J; Perler FB, An Alternative Protein Splicing Mechanism for Inteins Lacking an N-Terminal Nucleophile. EMBO J. 2000, 19, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Southworth MW; Amaya K; Evans TC; Xu MQ; Perler FB, Purification of Proteins Fused to Either the Amino or Carboxy Terminus of the Mycobacterium xenopi Gyrase A Intein. BioTechniques 1999, 27, 110–114. [DOI] [PubMed] [Google Scholar]
- 94.Wood DW; Wu W; Belfort G; Derbyshire V; Belfort M, A Genetic System Yields Self-Cleaving Inteins for Bioseparations. Nat. Biotechnol 1999, 17, 889–892. [DOI] [PubMed] [Google Scholar]
- 95.Batjargal S; Walters CR; Petersson EJ, Inteins as Traceless Purification Tags for Unnatural Amino Acid Proteins. J. Am. Chem. Soc 2015, 137, 1734–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chong S; Mersha FB; Comb DG; Scott ME; Landry D; Vence LM; Perler FB; Benner J; Kucera RB; Hirvonen CA, et al. , Single-Column Purification of Free Recombinant Proteins Using a Self-Cleavable Affinity Tag Derived From a Protein Splicing Element. Gene 1997, 192, 271–281. [DOI] [PubMed] [Google Scholar]
- 97.Muir TW; Sondhi D; Cole PA, Expressed Protein Ligation: A General Method for Protein Engineering. Proc. Natl. Acad. Sci. U.S.A 1998, 95, 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Evans TC; Benner J; Xu MQ, Semisynthesis of Cytotoxic Proteins Using a Modified Protein Splicing Element. Protein Sci. 1998, 7, 2256–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gates ZP; Stephan JR; Lee DJ; Kent SBH, Rapid Formal Hydrolysis of Peptide-αthioesters. Chem. Commun 2013, 49, 786–788. [DOI] [PubMed] [Google Scholar]
- 100.Thom J; Anderson D; McGregor J; Cotton G, Recombinant Protein Hydrazides: Application to Site-Specific Protein PEGylation. Bioconjugate Chemistry 2011, 22, 1017–1020. [DOI] [PubMed] [Google Scholar]
- 101.Telenti A; Southworth M; Alcaide F; Daugelat S; Jacobs WR; Perler FB, The Mycobacterium xenopi GyrA Protein Splicing Element: Characterization of a Minimal Intein. J. Bacteriol 1997, 179, 6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Valiyaveetil FI; MacKinnon R; Muir TW, Semisynthesis and Folding of the Potassium Channel KcsA. J. Am. Chem. Soc 2002, 124, 9113–9120. [DOI] [PubMed] [Google Scholar]
- 103.Marshall CJ; Grosskopf VA; Moehling TJ; Tillotson BJ; Wiepz GJ; Abbott NL; Raines RT; Shusta EV, An Evolved Mxe GyrA Intein for Enhanced Production of Fusion Proteins. ACS Chem. Biol 2015, 10, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Albertsen L; Shaw AC; Norrild JC; Strømgaard K, Recombinant Production of Peptide C-Terminal α-Amides Using an Engineered Intein. Bioconjugate Chemistry 2013, 24, 1883–1894. [DOI] [PubMed] [Google Scholar]
- 105.Cui C; Zhao W; Chen J; Wang J; Li Q, Elimination of In Vivo Cleavage Between Target Protein and Intein in the Intein-Mediated Protein Purification Systems . Protein Expr. Purif 2006, 50, 74–81. [DOI] [PubMed] [Google Scholar]
- 106.Cotton GJ; Ayers B; Xu R; Muir TW, Insertion of a Synthetic Peptide into a Recombinant Protein Framework: A Protein Biosensor. J. Am. Chem. Soc 1999, 121, 1100–1101. [Google Scholar]
- 107.Xu R; Ayers B; Cowburn D; Muir TW, Chemical Ligation of Folded Recombinant Proteins: Segmental Isotopic Labeling of Domains for NMR Studies. Proc. Natl. Acad. Sci. U.S.A 1999, 96, 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cho J-H; Muralidharan V; Vila-Perello M; Raleigh DP; Muir TW; Palmer Iii AG, Tuning Protein Autoinhibition by Domain Destabilization. Nat. Struct. Mol. Biol 2011, 18, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blaschke UK; Cotton GJ; Muir TW, Synthesis of Multi-Domain Proteins Using Expressed Protein Ligation: Strategies for Segmental Isotopic Labeling of Internal Regions. Tetrahedron 2000, 56, 9461–9470. [Google Scholar]
- 110.Chu N; Salguero AL; Liu AZ; Chen Z; Dempsey DR; Ficarro SB; Alexander WM; Marto JA; Li Y; Amzel LM, et al. , Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis. Cell 2018, 174, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bockhorn JJ; Lazar KL; Gasser AJ; Luther LM; Qahwash IM; Chopra N; Meredith SC, Novel Semisynthetic Method for Generating Full Length β-Amyloid Peptides. Pept. Sci 2010, 94, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Katayama H; Hojo H; Ohira T; Ishii A; Nozaki T; Goto K; Nakahara Y; Takahashi T; Hasegawa Y; Nagasawa H, et al. , Correct Disulfide Pairing Is Required for the Biological Activity of Crustacean Androgenic Gland Hormone (AGH): Synthetic Studies of AGH. Biochemistry 2010, 49, 1798–1807. [DOI] [PubMed] [Google Scholar]
- 113.Pentelute BL; Barker AP; Janowiak BE; Kent SBH; Collier RJ, A Semisynthesis Platform for Investigating Structure−Function Relationships in the N-Terminal Domain of the Anthrax Lethal Factor. ACS Chem. Biol 2010, 5, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hauser PS; Raussens V; Yamamoto T; Abdullahi GE; Weers PMM; Sykes BD; Ryan RO, Semisynthesis and Segmental Isotope Labeling of the apoE3 N-Terminal Domain Using Expressed Protein Ligation. J. Lipid Res 2009, 50, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Szewczuk LM; Tarrant MK; Sample V; Drury WJ; Zhang J; Cole PA, Analysis of Serotonin N-Acetyltransferase Regulation in Vitro and in Live Cells Using Protein Semisynthesis. Biochemistry 2008, 47, 10407–10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng W; Zhang Z; Ganguly S; Weller JL; Klein DC; Cole PA, Cellular Stabilization of the Melatonin Rhythm Enzyme Induced by Nonhydrolyzable Phosphonate Incorporation. Nat. Struct. Mol. Biol 2003, 10, 1054–1057. [DOI] [PubMed] [Google Scholar]
- 117.Zheng W; Schwarzer D; LeBeau A; Weller JL; Klein DC; Cole PA, Cellular Stability of Serotonin N-Acetyltransferase Conferred by Phosphonodifluoromethylene Alanine (Pfa) Substitution for Ser-205. J. Biol. Chem 2005, 280, 10462–10467. [DOI] [PubMed] [Google Scholar]
- 118.Focke PJ; Annen AW; Valiyaveetil FI, Engineering the Glutamate Transporter Homologue GltPh Using Protein Semisynthesis. Biochemistry 2015, 54, 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Y-T; Yi C; Chen C-C; Lan H; Pan M; Zhang S-J; Huang Y-C; Guan C-J; Li Y-M; Yu L, et al. , A Semisynthetic Atg3 Reveals that Acetylation Promotes Atg3 Membrane Binding and Atg8 Lipidation. Nat. Commun 2017, 8, 14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berry SM; Gieselman MD; Nilges MJ; van der Donk WA; Lu Y, An Engineered Azurin Variant Containing a Selenocysteine Copper Ligand. J. Am. Chem. Soc 2002, 124, 2084–2085. [DOI] [PubMed] [Google Scholar]
- 121.Berry SM; Ralle M; Low DW; Blackburn NJ; Lu Y, Probing the Role of Axial Methionine in the Blue Copper Center of Azurin with Unnatural Amino Acids. J. Am. Chem. Soc 2003, 125, 8760–8768. [DOI] [PubMed] [Google Scholar]
- 122.Ralle M; Berry SM; Nilges MJ; Gieselman MD; van der Donk WA; Lu Y; Blackburn NJ, The Selenocysteine-Substituted Blue Copper Center: Spectroscopic Investigations of Cys112SeCys Pseudomonas aeruginosa Azurin. J. Am. Chem. Soc 2004, 126, 7244–7256. [DOI] [PubMed] [Google Scholar]
- 123.Clark KM; Yu Y; Marshall NM; Sieracki NA; Nilges MJ; Blackburn NJ; van der Donk WA; Lu Y, Transforming a Blue Copper into a Red Copper Protein: Engineering Cysteine and Homocysteine into the Axial Position of Azurin Using Site-Directed Mutagenesis and Expressed Protein Ligation. J. Am. Chem. Soc 2010, 132, 10093–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tarrant MK; Rho H-S; Xie Z; Jiang YL; Gross C; Culhane JC; Yan G; Qian J; Ichikawa Y; Matsuoka T, et al. , Regulation of CK2 by Phosphorylation and O-GlcNAcylation Revealed by Semisynthesis. Nat. Chem. Biol 2012, 8, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Minato Y; Ueda T; Machiyama A; Shimada I; Iwaï H, Segmental Isotopic Labeling of a 140 kDa Dimeric Multi-Domain Protein CheA From Escherichia coli by Expressed Protein Ligation and Protein Trans-Splicing. J. Biomol. NMR 2012, 53, 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raz R; Burlina F; Ismail M; Downward J; Li J; Smerdon SJ; Quibell M; White PD; Offer J, HF-Free Boc Synthesis of Peptide Thioesters for Ligation and Cyclization. Angew. Chem. Int. Ed 2016, 55, 13174–13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kienhöfer A; Kast P; Hilvert D, Selective Stabilization of the Chorismate Mutase Transition State by a Positively Charged Hydrogen Bond Donor. J. Am. Chem. Soc 2003, 125, 3206–3207. [DOI] [PubMed] [Google Scholar]
- 128.Mayer C; Müller MM; Gellman SH; Hilvert D, Building Proficient Enzymes with Foldamer Prostheses. Angew. Chem. Int. Ed 2014, 53, 6978–6981. [DOI] [PubMed] [Google Scholar]
- 129.Rosenzweig R; Farber P; Velyvis A; Rennella E; Latham MP; Kay LE, ClpB N-Terminal Domain Plays a Regulatory Role in Protein Disaggregation. Proc. Natl. Acad. Sci. U.S.A 2015, 112, E6872–E6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cotton GJ; Muir TW, Generation of a Dual-Labeled Fluorescence Biosensor for Crk-II Phosphorylation Using Solid-Phase Expressed Protein Ligation. Chem. Biol 2000, 7, 253–261. [DOI] [PubMed] [Google Scholar]
- 131.Clerico EM; Zhuravleva A; Smock RG; Gierasch LM, Segmental Isotopic Labeling of the Hsp70 Molecular Chaperone DnaK Using Expressed Protein Ligation. Pept. Sci 2010, 94, 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nordsieck K; Baumann L; Hintze V; Pisabarro MT; Schnabelrauch M; Beck-Sickinger AG; Samsonov SA, The Effect of Interleukin-8 Truncations on Its Interactions With Glycosaminoglycans. Biopolymers 2018, 109, e23103. [DOI] [PubMed] [Google Scholar]
- 133.David R; Machova Z; Beck-Sickinger AG, Semisynthesis and Application of Carboxyfluorescein-Labelled Biologically Active Human Interleukin-8. In Biol. Chem, 2003; Vol. 384, pp 1619–1630. [DOI] [PubMed] [Google Scholar]
- 134.David R; Beck-Sickinger AG, Identification of the Dimerisation Interface of Human Interleukin-8 by IL-8-Variants Containing the Photoactivatable Amino Acid Benzoyl-Phenylalanine. Eur. Biophys. J 2007, 36, 385–391. [DOI] [PubMed] [Google Scholar]
- 135.David R; Günther R; Baumann L; Lühmann T; Seebach D; Hofmann H-J; Beck-Sickinger AG, Artificial Chemokines: Combining Chemistry and Molecular Biology for the Elucidation of Interleukin-8 Functionality. J. Am. Chem. Soc 2008, 130, 15311–15317. [DOI] [PubMed] [Google Scholar]
- 136.Nordsieck K; Pichert A; Samsonov SA; Thomas L; Berger C; Pisabarro MT; Huster D; Beck-Sickinger AG, Residue 75 of Interleukin-8 is Crucial for its Interactions with Glycosaminoglycans. ChemBioChem 2012, 13, 2558–2566. [DOI] [PubMed] [Google Scholar]
- 137.Baumann L; Beck-Sickinger AG, Identification of a Potential Modification Site in Human Stromal Cell-Derived Factor-1. Pept. Sci 2010, 94, 771–778. [DOI] [PubMed] [Google Scholar]
- 138.Baumann L; Beck-Sickinger AG, Photoactivatable Chemokines – Controlling Protein Activity by Light. Angew. Chem. Int. Ed 2013, 52, 9550–9553. [DOI] [PubMed] [Google Scholar]
- 139.Bellmann-Sickert K; Baumann L; Beck-Sickinger AG, Selective Labelling of Stromal Cell-Derived Factor 1α with Carboxyfluorescein to Study Receptor Internalisation. J. Pept. Sci 2010, 16, 568–574. [DOI] [PubMed] [Google Scholar]
- 140.Moran SD; Woys AM; Buchanan LE; Bixby E; Decatur SM; Zanni MT, Two-Dimensional IR Spectroscopy and Segmental 13C Labeling Reveals the Domain Structure of Human γD-Crystallin Amyloid Fibrils. Proc. Natl. Acad. Sci. U.S.A 2012, 109, 3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim HJS, H J; Kim HW; Kang S-H; Kim YT, Expressed Protein Ligation of 5-Enolpyruvylshikimate-3-phosphate (EPSP) Synthase: An Application to a Protein Expressed as an Inclusion Body. Bull. Korean Chem. Soc 2007, 28, 2303–2309. [Google Scholar]
- 142.Singla N; Himanen JP; Muir TW; Nikolov DB, Toward the Semisynthesis of Multidomain Transmembrane Receptors: Modification of Eph Tyrosine Kinases. Protein Sci. 2008, 17, 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Singla N; Erdjument-Bromage H; Himanen Juha P.; Muir Tom W.; Nikolov Dimitar B., A Semisynthetic Eph Receptor Tyrosine Kinase Provides Insight into Ligand- Induced Kinase Activation. Chem. Biol 2011, 18, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hirano K; Izumi M; Macmillan D; Tezuka K; Tsuji T; Kajihara Y, Semisynthesis of Erythropoietin Analog Having Three Oligosaccharides. J. Carbohydr. Chem 2011, 30, 306–319. [Google Scholar]
- 145.Hirano K; Macmillan D; Tezuka K; Tsuji T; Kajihara Y, Design and Synthesis of a Homogeneous Erythropoietin Analogue with Two Human Complex-Type Sialyloligosaccharides: Combined Use of Chemical and Bacterial Protein Expression Methods. Angew. Chem. Int. Ed 2009, 48, 9557–9560. [DOI] [PubMed] [Google Scholar]
- 146.Richardson JP; Macmillan D, Optimisation of Chemical Protein Cleavage for Erythropoietin Semi-Synthesis Using Native Chemical Ligation. Org. Biomol. Chem 2008, 6, 3977–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zuberek J; Wyslouch-Cieszynska A; Niedzwiecka A; Dadlez M; Stepinski J; Augustyniak W; Gingras A-C; Zhang Z; Burley SK; Sonenberg N, et al. , Phosphorylation of eIF4E Attenuates Its Interaction With mRNA 5′ Cap Analogs by Electrostatic Repulsion: Intein-Mediated Protein Ligation Strategy to Obtain Phosphorylated Protein. RNA 2003, 9, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Michel E; Skrisovska L; Wüthrich K; Allain FH-T, Amino Acid-Selective Segmental Isotope Labeling of Multidomain Proteins for Structural Biology. ChemBioChem 2013, 14, 457–466. [DOI] [PubMed] [Google Scholar]
- 149.Casi G; Roelfes G; Hilvert D, Selenoglutaredoxin as a Glutathione Peroxidase Mimic. ChemBioChem 2008, 9, 1623–1631. [DOI] [PubMed] [Google Scholar]
- 150.Macmillan D; Bertozzi CR, Modular Assembly of Glycoproteins: Towards the Synthesis of GlyCAM-1 by Using Expressed Protein Ligation. Angew. Chem. Int. Ed 2004, 43, 1355–1359. [DOI] [PubMed] [Google Scholar]
- 151.Baskaran S; Roach PJ; DePaoli-Roach AA; Hurley TD, Structural Basis for Glucose-6-Phosphate Activation of Glycogen Synthase. Proc. Natl. Acad. Sci. U.S.A 2010, 107, 17563–17568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee J; Bayley H, Semisynthetic Protein Nanoreactor for Single-Molecule Chemistry. Proc. Natl. Acad. Sci. U.S.A 2015, 112, 13768–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee J; Boersma AJ; Boudreau MA; Cheley S; Daltrop O; Li J; Tamagaki H; Bayley H, Semisynthetic Nanoreactor for Reversible Single-Molecule Covalent Chemistry. ACS Nano 2016, 10, 8843–8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Skrisovska L; Allain FHT, Improved Segmental Isotope Labeling Methods for the NMR Study of Multidomain or Large Proteins: Application to the RRMs of Npl3p and hnRNP L. J. Mol. Biol 2008, 375, 151–164. [DOI] [PubMed] [Google Scholar]
- 155.Dann GP; Liszczak GP; Bagert JD; Müller MM; Nguyen UTT; Wojcik F; Brown ZZ; Bos J; Panchenko T; Pihl R, et al. , ISWI Chromatin Remodellers Sense Nucleosome Modifications to Determine Substrate Preference . Nature 2017, 548, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kilic S; Boichenko I; Lechner CC; Fierz B, A Bi-Terminal Protein Ligation Strategy to Probe Chromatin Structure During DNA Damage. Chem. Sci 2018, 9, 3704–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fierz B; Kilic S; Hieb AR; Luger K; Muir TW, Stability of Nucleosomes Containing Homogenously Ubiquitylated H2A and H2B Prepared Using Semisynthesis. J. Am. Chem. Soc 2012, 134, 19548–19551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bi X; Yang R; Feng X; Rhodes D; Liu C-F, Semisynthetic UbH2A Reveals Different Activities of Deubiquitinases and Inhibitory Effects of H2A K119 Ubiquitination on H3K36 Methylation in Mononucleosomes. Org. Biomol. Chem 2016, 14, 835–839. [DOI] [PubMed] [Google Scholar]
- 159.McGinty RK; Kim J; Chatterjee C; Roeder RG; Muir TW, Chemically Ubiquitylated Histone H2B Stimulates hDot1L-Mediated Intranucleosomal Methylation. Nature 2008, 453, 812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weller CE; Dhall A; Ding F; Linares E; Whedon SD; Senger NA; Tyson EL; Bagert JD; Li X; Augusto O, et al. , Aromatic Thiol-Mediated Cleavage of N–O Bonds Enables Chemical Ubiquitylation of Folded Proteins. Nat. Commun 2016, 7, 12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.He S; Bauman D; Davis JS; Loyola A; Nishioka K; Gronlund JL; Reinberg D; Meng F; Kelleher N; McCafferty DG, Facile Synthesis of Site-Specifically Acetylated and Methylated Histone Proteins: Reagents for Evaluation of the Histone Code Hypothesis. Proc. Natl. Acad. Sci. U.S.A 2003, 100, 12033–12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Delachat AMF; Guidotti N; Bachmann AL; Meireles-Filho ACA; Pick H; Lechner CC; Deluz C; Deplancke B; Suter DM; Fierz B, Engineered Multivalent Sensors to Detect Coexisting Histone Modifications in Living Stem Cells. Cell Chem. Biol 2018, 25, 51–56. [DOI] [PubMed] [Google Scholar]
- 163.Bartke T; Vermeulen M; Xhemalce B; Robson SC; Mann M; Kouzarides T, Nucleosome-Interacting Proteins Regulated by DNA and Histone Methylation. Cell 2010, 143, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen Z; Grzybowski AT; Ruthenburg AJ, Traceless Semisynthesis of a Set of Histone 3 Species Bearing Specific Lysine Methylation Marks. ChemBioChem 2014, 15, 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Farrelly LA; Thompson RE; Zhao S; Lepack AE; Lyu Y; Bhanu NV; Zhang B; Loh Y-HE; Ramakrishnan A; Vadodaria KC, et al. , Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 2019, 567, 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bryan LC; Weilandt DR; Bachmann AL; Kilic S; Lechner CC; Odermatt PD; Fantner GE; Georgeon S; Hantschel O; Hatzimanikatis V, et al. , Single-Molecule Kinetic Analysis of HP1-Chromatin Binding Reveals a Dynamic Network of Histone Modification and DNA Interactions. Nucleic Acids Res. 2017, 45, 10504–10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Borgel J; Tyl M; Schiller K; Pusztai Z; Dooley CM; Deng W; Wooding C; White RJ; Warnecke T; Leonhardt H, et al. , KDM2A Integrates DNA and Histone Modification Signals through a CXXC/PHD Module and Direct Interaction with HP1 . Nucleic Acids Res. 2016, 45, 1114–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kawakami T; Yoshikawa R; Fujiyoshi Y; Mishima Y; Hojo H; Tajima S; Suetake I, Synthesis of Histone Proteins by CPE Ligation Using a Recombinant Peptide as the C-Terminal Building Block. J. Biochem 2015, 158, 403–411. [DOI] [PubMed] [Google Scholar]
- 169.Franz H; Mosch K; Soeroes S; Urlaub H; Fischle W, Multimerization and H3K9me3 Binding Are Required for CDYL1b Heterochromatin Association. J. Biol. Chem 2009, 284, 35049–35059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Miller TCR; Simon B; Rybin V; Grötsch H; Curtet S; Khochbin S; Carlomagno T; Müller CW, A Bromodomain–DNA Interaction Facilitates Acetylation-Dependent Bivalent Nucleosome Recognition by the BET Protein BRDT. Nat. Commun 2016, 7, 13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bos J; Muir TW, A Chemical Probe for Protein Crotonylation. J. Am. Chem. Soc 2018, 140, 4757–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim D-H; Tang Z; Shimada M; Fierz B; Houck-Loomis B; Bar-Dagen M; Lee S; Lee S-K; Muir TW; Roeder RG, et al. , Histone H3K27 Trimethylation Inhibits H3 Binding and Function of SET1-Like H3K4 Methyltransferase Complexes. Molecular and Cellular Biology 2013, 33, 4936–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Casadio F; Lu X; Pollock SB; LeRoy G; Garcia BA; Muir TW; Roeder RG; Allis CD, H3R42me2a Is a Histone Modification with Positive Transcriptional Effects. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Manohar M; Mooney AM; North JA; Nakkula RJ; Picking JW; Edon A; Fishel R; Poirier MG; Ottesen JJ, Acetylation of Histone H3 at the Nucleosome Dyad Alters DNA-Histone Binding. J. Biol. Chem 2009, 284, 23312–23321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Allahverdi A; Yang R; Korolev N; Fan Y; Davey CA; Liu C-F; Nordenskiöld L, The Effects of Histone H4 Tail Acetylations on Cation-Induced Chromatin Folding and Self-Association. Nucleic Acids Res. 2010, 39, 1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dhall A; Weller CE; Chu A; Shelton PMM; Chatterjee C, Chemically Sumoylated Histone H4 Stimulates Intranucleosomal Demethylation by the LSD1–CoREST Complex. ACS Chem. Biol 2017, 12, 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shogren-Knaak M; Ishii H; Sun J-M; Pazin MJ; Davie JR; Peterson CL, Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [DOI] [PubMed] [Google Scholar]
- 178.Kee J-M; Villani B; Carpenter LR; Muir TW, Development of Stable Phosphohistidine Analogues. J. Am. Chem. Soc 2010, 132, 14327–14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kilic S; Bachmann AL; Bryan LC; Fierz B, Multivalency Governs HP1α Association Dynamics With the Silent Chromatin State. Nat. Commun 2015, 6, 7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Matveenko M; Hackl S; Becker CFW, Utility of the Phenacyl Protecting Group in Traceless Protein Semisynthesis through Ligation–Desulfurization Chemistry. ChemistryOpen 2018, 7, 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Karagöz GE; Sinnige T; Hsieh O; Rüdiger SGD, Expressed Protein Ligation for a Large Dimeric Protein. Protein Eng. Des. Sel 2011, 24, 495–501. [DOI] [PubMed] [Google Scholar]
- 182.Ansaloni A; Wang Z-M; Jeong JS; Ruggeri FS; Dietler G; Lashuel HA, One-Pot Semisynthesis of Exon 1 of the Huntingtin Protein: New Tools for Elucidating the Role of Posttranslational Modifications in the Pathogenesis of Huntington’s Disease. Angew. Chem. Int. Ed 2014, 53, 1928–1933. [DOI] [PubMed] [Google Scholar]
- 183.Chiki A; DeGuire SM; Ruggeri FS; Sanfelice D; Ansaloni A; Wang Z-M; Cendrowska U; Burai R; Vieweg S; Pastore A, et al. , Mutant Exon1 Huntingtin Aggregation is Regulated by T3 Phosphorylation-Induced Structural Changes and Crosstalk between T3 Phosphorylation and Acetylation at K6. Angew. Chem. Int. Ed 2017, 56, 5202–5207. [DOI] [PubMed] [Google Scholar]
- 184.Deguire SM; Ruggeri FS; Fares M-B; Chiki A; Cendrowska U; Dietler G; Lashuel HA, N-Terminal Huntingtin (Htt) Phosphorylation Is a Molecular Switch Regulating Htt Aggregation, Helical Conformation, Internalization, and Nuclear Targeting. J. Biol. Chem 2018, 293, 18540–18558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hackenberger CPR; Friel CT; Radford SE; Imperiali B, Semisynthesis of a Glycosylated Im7 Analogue for Protein Folding Studies. J. Am. Chem. Soc 2005, 127, 12882–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shukla H; Vaitiekunas P; Majumdar AK; Dragan AI; Dimitriadis EK; Kotova S; Crane-Robinson C; Privalov PL, The Linker of the Interferon Response Factor 3 Transcription Factor Is Not Unfolded. Biochemistry 2012, 51, 6320–6327. [DOI] [PubMed] [Google Scholar]
- 187.Reif A; Siebenhaar S; Tröster A; Schmälzlein M; Lechner C; Velisetty P; Gottwald K; Pöhner C; Boos I; Schubert V, et al. , Semisynthesis of Biologically Active Glycoforms of the Human Cytokine Interleukin 6. Angew. Chem. Int. Ed 2014, 53, 12125–12131. [DOI] [PubMed] [Google Scholar]
- 188.Okamoto R; Kimura M; Ishimizu T; Izumi M; Kajihara Y, Semisynthesis of a Post-Translationally Modified Protein by Using Chemical Cleavage and Activation of an Expressed Fusion Polypeptide . Chem. Euro. J 2014, 20, 10425–10430. [DOI] [PubMed] [Google Scholar]
- 189.Sanulli S; Justin N; Teissandier A; Ancelin K; Portoso M; Caron M; Michaud A; Lombard B; da Rocha Simao T.; Offer J., et al. , Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition During Cell Differentiation. Mol. Cell 2015, 57, 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ge EJ; Jani KS; Diehl KL; Müller MM; Muir TW, Nucleation and Propagation of Heterochromatin by the Histone Methyltransferase PRC2: Geometric Constraints and Impact of the Regulatory Subunit JARID2. J. Am. Chem. Soc 2019, 141, 15029–15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Valiyaveetil FI; Sekedat M; Muir TW; MacKinnon R, Semisynthesis of a Functional K+ Channel. Angew. Chem. Int. Ed 2004, 43, 2504–2507. [DOI] [PubMed] [Google Scholar]
- 192.Devaraneni PK; Komarov AG; Costantino CA; Devereaux JJ; Matulef K; Valiyaveetil FI, Semisynthetic K+ Channels Show That the Constricted Conformation of the Selectivity Filter is Not the C-type Inactivated State. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 15698–15703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Valiyaveetil FI; Sekedat M; MacKinnon R; Muir TW, Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel. J. Am. Chem. Soc 2006, 128, 11591–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Valiyaveetil FI; Sekedat M; MacKinnon R; Muir TW, Glycine as a D-Amino Acid Surrogate in the K+ -Selectivity Filter. Proc. Natl. Acad. Sci. U.S.A 2004, 101, 17045–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Komarov AG; Linn KM; Devereaux JJ; Valiyaveetil FI, Modular Strategy for the Semisynthesis of a K+ Channel: Investigating Interactions of the Pore Helix. ACS Chem. Biol 2009, 4, 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Matulef K; Annen Alvin W.; Nix Jay C.; Valiyaveetil Francis I., Individual Ion Binding Sites in the K+ Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation. Structure 2016, 24, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kratochvil HT; Carr JK; Matulef K; Annen AW; Li H; Maj M; Ostmeyer J; Serrano AL; Raghuraman H; Moran SD, et al. , Instantaneous ion configurations in the K+ ion channel selectivity filter revealed by 2D IR spectroscopy. Science 2016, 353, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gallagher C; Burlina F; Offer J; Ramos A, A Method for the Unbiased and Efficient Segmental Labelling of RNA-Binding Proteins for Structure and Biophysics. Sci. Rep 2017, 7, 14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Paul M; Patton GC; van der Donk WA, Mutants of the Zinc Ligands of Lacticin 481 Synthetase Retain Dehydration Activity but Have Impaired Cyclization Activity. Biochemistry 2007, 46, 6268–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chatterjee C; Patton GC; Cooper L; Paul M; van der Donk WA, Engineering Dehydro Amino Acids and Thioethers into Peptides Using Lacticin 481 Synthetase. Chem. Biol 2006, 13, 1109–1117. [DOI] [PubMed] [Google Scholar]
- 201.Chatterjee C; Miller LM; Leung YL; Xie L; Yi M; Kelleher NL; van der Donk WA, Lacticin 481 Synthetase Phosphorylates its Substrate during Lantibiotic Production. J. Am. Chem. Soc 2005, 127, 15332–15333. [DOI] [PubMed] [Google Scholar]
- 202.You YO; van der Donk WA, Mechanistic Investigations of the Dehydration Reaction of Lacticin 481 Synthetase Using Site-Directed Mutagenesis. Biochemistry 2007, 46, 5991–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Xie L; Miller LM; Chatterjee C; Averin O; Kelleher NL; van der Donk WA, Lacticin 481: In Vitro Reconstitution of Lantibiotic Synthetase Activity. Science 2004, 303, 679–681. [DOI] [PubMed] [Google Scholar]
- 204.Levengood MR; Kerwood CC; Chatterjee C; van der Donk WA, Investigation of the Substrate Specificity of Lacticin 481 Synthetase by Using Nonproteinogenic Amino Acids. ChemBioChem 2009, 10, 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schwarzer D; Zhang Z; Zheng W; Cole PA, Negative Regulation of a Protein Tyrosine Phosphatase by Tyrosine Phosphorylation. J. Am. Chem. Soc 2006, 128, 4192–4193. [DOI] [PubMed] [Google Scholar]
- 206.Roy RS; Allen O; Walsh CT, Expressed Protein Ligation to Probe Regiospecificity of Heterocyclization in the Peptide Antibiotic Microcin B17. Chem. Biol 1999, 6, 789–799. [DOI] [PubMed] [Google Scholar]
- 207.Yang A; Hacheney I; Wu Y-W, Semisynthesis of Autophagy Protein LC3 Conjugates. Biorg. Med. Chem 2017, 25, 4971–4976. [DOI] [PubMed] [Google Scholar]
- 208.Huang Y-C; Li Y-M; Chen Y; Pan M; Li Y-T; Yu L; Guo Q-X; Liu L, Synthesis of Autophagosomal Marker Protein LC3-II under Detergent-Free Conditions. Angew. Chem. Int. Ed 2013, 52, 4858–4862. [DOI] [PubMed] [Google Scholar]
- 209.Yang A; Li Y; Pantoom S; Triola G; Wu Y-W, Semisynthetic Lipidated LC3 Protein Mediates Membrane Fusion. ChemBioChem 2013, 14, 1296–1300. [DOI] [PubMed] [Google Scholar]
- 210.Yang A; Pantoom S; Wu Y-W, Elucidation of the Anti-Autophagy Mechanism of the Legionella Effector RavZ Using Semisynthetic LC3 Proteins. eLife 2017, 6, e23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang Z; Raines LL; Hooy RM; Roberson H; Leahy DJ; Cole PA, Tyrosine Phosphorylation of Mig6 Reduces Its Inhibition of the Epidermal Growth Factor Receptor. ACS Chem. Biol 2013, 8, 2372–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Romanelli A; Shekhtman A; Cowburn D; Muir TW, Semisynthesis of a Segmental Isotopically Labeled Protein Splicing Precursor: NMR Evidence for an Unusual Peptide Bond at the N-Extein–Intein Junction. Proc. Natl. Acad. Sci. U.S.A 2004, 101, 6397–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Frutos S; Goger M; Giovani B; Cowburn D; Muir TW, Branched Intermediate Formation Stimulates Peptide Bond Cleavage in Protein Splicing. Nat. Chem. Biol 2010, 6, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu Z; Frutos S; Bick MJ; Vila-Perelló M; Debelouchina GT; Darst SA; Muir TW, Structure of the Branched Intermediate in Protein Splicing. Proc. Natl. Acad. Sci. U.S.A 2014, 111, 8422–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Linn KM; Derebe MG; Jiang Y; Valiyaveetil FI, Semisynthesis of NaK, a Na+ and K+ Conducting Ion Channel. Biochemistry 2010, 49, 4450–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chen Z; Jiang H; Xu W; Li X; Dempsey DR; Zhang X; Devreotes P; Wolberger C; Amzel LM; Gabelli SB, et al. , A Tunable Brake for HECT Ubiquitin Ligases. Mol. Cell 2017, 66, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Masuda K; Furumitsu M; Ooyama H; Iwakoshi-Ukena E; Ukena K, Synthesis of Neurosecretory Protein GM Composed of 88 Amino Acid Residues by Native Chemical Ligation. Tetrahedron Lett. 2016, 57, 804–807. [Google Scholar]
- 218.Bolduc DM; Montagna DR; Gu Y; Selkoe DJ; Wolfe MS, Nicastrin Functions to Sterically Hinder γ-Secretase–Substrate Interactions Driven by Substrate Transmembrane Domain. Proc. Natl. Acad. Sci. U.S.A 2016, 113, E509–E518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shah NH; Eryilmaz E; Cowburn D; Muir TW, Naturally Split Inteins Assemble through a “Capture and Collapse” Mechanism. J. Am. Chem. Soc 2013, 135, 18673–18681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Grant JE; Guo L-W; Vestling MM; Martemyanov KA; Arshavsky VY; Ruoho AE, The N Terminus of GTPγS-activated Transducin α-Subunit Interacts with the C Terminus of the cGMP Phosphodiesterase γ-Subunit. J. Biol. Chem 2006, 281, 6194–6202. [DOI] [PubMed] [Google Scholar]
- 221.Thompson PR; Wang D; Wang L; Fulco M; Pediconi N; Zhang D; An W; Ge Q; Roeder RG; Wong J, et al. , Regulation of the p300 HAT Domain via a Novel Activation Loop. Nat. Struct. Mol. Biol 2004, 11, 308–315. [DOI] [PubMed] [Google Scholar]
- 222.Karukurichi KR; Wang L; Uzasci L; Manlandro CM; Wang Q; Cole PA, Analysis of p300/CBP Histone Acetyltransferase Regulation Using Circular Permutation and Semisynthesis. J. Am. Chem. Soc 2010, 132, 1222–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mittelstadt PR; Yamaguchi H; Appella E; Ashwell JD, T Cell Receptor-mediated Activation of p38α by Mono-phosphorylation of the Activation Loop Results in Altered Substrate Specificity. J. Biol. Chem 2009, 284, 15469–15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Vogel EM; Imperiali B, Semisynthesis of Unnatural Amino Acid Mutants of Paxillin: Protein Probes for Cell Migration Studies. Protein Sci. 2007, 16, 550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rauh D; Fischer F; Gertz M; Lakshminarasimhan M; Bergbrede T; Aladini F; Kambach C; Becker CFW; Zerweck J; Schutkowski M, et al. , An Acetylome Peptide Microarray Reveals Specificities and Deacetylation Substrates for All Human Sirtuin Isoforms. Nat. Commun 2013, 4, 2327. [DOI] [PubMed] [Google Scholar]
- 226.Chen Z; Dempsey DR; Thomas SN; Hayward D; Bolduc DM; Cole PA, Molecular Features of PTEN Regulation by C-Terminal Phosphorylation. J. Biol. Chem 2016, 291, 14160–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Henager S; Henriquez S; Dempsey D; Cole PA, Analysis of Site-specific Phosphorylation of PTEN using Enzyme-Catalyzed Expressed Protein Ligation. ChemBioChem 2019, 20, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bolduc D; Rahdar M; Tu-Sekine B; Sivakumaren SC; Raben D; Amzel LM; Devreotes P; Gabelli SB; Cole P, Phosphorylation-Mediated PTEN Conformational Closure and Deactivation Revealed with Protein Semisynthesis. eLife 2013, 2, e00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pedersen SW; Pedersen SB; Anker L; Hultqvist G; Kristensen AS; Jemth P; Strømgaard K, Probing Backbone Hydrogen Bonding in PDZ/Ligand Interactions by Protein Amide-to-Ester Mutations. Nat. Commun 2014, 5, 3215. [DOI] [PubMed] [Google Scholar]
- 230.Pedersen SW; Hultqvist G; Strømgaard K; Jemth P, The Role of Backbone Hydrogen Bonds in the Transition State for Protein Folding of a PDZ Domain. PLoS One 2014, 9, e95619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pedersen SW; Albertsen L; Moran GE; Levesque B; Pedersen SB; Bartels L; Wapenaar H; Ye F; Zhang M; Bowen ME, et al. , Site-Specific Phosphorylation of PSD-95 PDZ Domains Reveals Fine-Tuned Regulation of Protein–Protein Interactions. ACS Chem. Biol 2017, 12, 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Vitali F; Henning A; Oberstrass FC; Hargous Y; Auweter SD; Erat M; Allain FHT, Structure of the Two Most C‐Terminal RNA Recognition Motifs of PTB Using Segmental Isotope Labeling. EMBO J 2006, 25, 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Shi L; Chen H; Zhang S-Y; Chu T-T; Zhao Y-F; Chen Y-X; Li Y-M, Semi-Synthesis of Murine Prion Protein by Native Chemical Ligation and Chemical Activation for Preparation of Polypeptide-α-Thioester. J. Pept. Sci 2017, 23, 438–444. [DOI] [PubMed] [Google Scholar]
- 234.Chu NK; Shabbir W; Bove-Fenderson E; Araman C; Lemmens-Gruber R; Harris DA; Becker CFW, A C-Terminal Membrane Anchor Affects the Interactions of Prion Proteins With Lipid Membranes . J. Biol. Chem 2014, 289, 30144–30160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Becker CFW; Liu X; Olschewski D; Castelli R; Seidel R; Seeberger PH, Semisynthesis of a Glycosylphosphatidylinositol-Anchored Prion Protein. Angew. Chem. Int. Ed 2008, 47, 8215–8219. [DOI] [PubMed] [Google Scholar]
- 236.Olschewski D; Seidel R; Miesbauer M; Rambold AS; Oesterhelt D; Winklhofer KF; Tatzelt J; Engelhard M; Becker CFW, Semisynthetic Murine Prion Protein Equipped with a GPI Anchor Mimic Incorporates into Cellular Membranes. Chem. Biol 2007, 14, 994–1006. [DOI] [PubMed] [Google Scholar]
- 237.Araman C; Thompson RE; Wang S; Hackl S; Payne RJ; Becker CFW, Semisynthetic Prion Protein (PrP) Variants Carrying Glycan Mimics at Position 181 and 197 Do Not Form Fibrils. Chem. Sci 2017, 8, 6626–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mukherjee S; van der Donk WA, Mechanistic Studies on the Substrate-Tolerant Lanthipeptide Synthetase ProcM. J. Am. Chem. Soc 2014, 136, 10450–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Pickin KA; Chaudhury S; Dancy BCR; Gray JJ; Cole PA, Analysis of Protein Kinase Autophosphorylation Using Expressed Protein Ligation and Computational Modeling. J. Am. Chem. Soc 2008, 130, 5667–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Iakovenko A; Rostkova E; Merzlyak E; Hillebrand AM; Thomä NH; Goody RS; Alexandrov K, Semi-Synthetic Rab Proteins as Tools for Studying Intermolecular Interactions. FEBS Lett. 2000, 468, 155–158. [DOI] [PubMed] [Google Scholar]
- 241.Wu Y-W; Oesterlin LK; Tan K-T; Waldmann H; Alexandrov K; Goody RS, Membrane Targeting Mechanism of Rab GTPases Elucidated by Semisynthetic Protein Probes. Nat. Chem. Biol 2010, 6, 534–540. [DOI] [PubMed] [Google Scholar]
- 242.Durek T; Alexandrov K; Goody RS; Hildebrand A; Heinemann I; Waldmann H, Synthesis of Fluorescently Labeled Mono- and Diprenylated Rab7 GTPase. J. Am. Chem. Soc 2004, 126, 16368–16378. [DOI] [PubMed] [Google Scholar]
- 243.Wu Y-W; Tan K-T; Waldmann H; Goody RS; Alexandrov K, Interaction Analysis of Prenylated Rab GTPase With Rab Escort Protein and GDP Dissociation Inhibitor Explains the Need for Both Regulators. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 12294–12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Alexandrov K; Heinemann I; Durek T; Sidorovitch V; Goody RS; Waldmann H, Intein-Mediated Synthesis of Geranylgeranylated Rab7 Protein In Vitro. J. Am. Chem. Soc 2002, 124, 5648–5649. [DOI] [PubMed] [Google Scholar]
- 245.Chen ES-W; Weng J-H; Chen Y-H; Wang S-C; Liu X-X; Huang W-C; Matsui T; Kawano Y; Liao J-H; Lim L-H, et al. , Phospho-Priming Confers Functionally Relevant Specificities for Rad53 Kinase Autophosphorylation. Biochemistry 2017, 56, 5112–5124. [DOI] [PubMed] [Google Scholar]
- 246.Milić J; Seidel R; Becker CFW; Goody RS; Engelhard M, Semisynthesis of H-Ras With a Glutamic Acid Methylester at Position 61. Pept. Sci 2008, 90, 399–405. [DOI] [PubMed] [Google Scholar]
- 247.Filchtinski D; Bee C; Savopol T; Engelhard M; Becker CFW; Herrmann C, Probing Ras Effector Interactions on Nanoparticle Supported Lipid Bilayers. Bioconjugate Chemistry 2008, 19, 1938–1944. [DOI] [PubMed] [Google Scholar]
- 248.Zhang S-Y; Sperlich B; Li F-Y; Al-Ayoubi S; Chen H-X; Zhao Y-F; Li Y-M; Weise K; Winter R; Chen Y-X, Phosphorylation Weakens but Does Not Inhibit Membrane Binding and Clustering of K-Ras4B. ACS Chem. Biol 2017, 12, 1703–1710. [DOI] [PubMed] [Google Scholar]
- 249.Chen Y-X; Koch S; Uhlenbrock K; Weise K; Das D; Gremer L; Brunsveld L; Wittinghofer A; Winter R; Triola G, et al. , Synthesis of the Rheb and K-Ras4B GTPases. Angew. Chem. Int. Ed 2010, 49, 6090–6095. [DOI] [PubMed] [Google Scholar]
- 250.Gottlieb D; Grunwald C; Nowak C; Kuhlmann J; Waldmann H, Intein-Mediated In Vitro Synthesis of Lipidated Ras Proteins. Chem. Commun 2006, 21, 260–262. [DOI] [PubMed] [Google Scholar]
- 251.Hondal RJ; Nilsson BL; Raines RT, Selenocysteine in Native Chemical Ligation and Expressed Protein Ligation. J. Am. Chem. Soc 2001, 123, 5140–5141. [DOI] [PubMed] [Google Scholar]
- 252.Arnold U; Hinderaker MP; Nilsson BL; Huck BR; Gellman SH; Raines RT, Protein Prosthesis: A Semisynthetic Enzyme with a β-Peptide Reverse Turn. J. Am. Chem. Soc 2002, 124, 8522–8523. [DOI] [PubMed] [Google Scholar]
- 253.Arnold U; Huck BR; Gellman SH; Raines RT, Protein Prosthesis: β-peptides as Reverse-Turn Surrogates. Protein Sci. 2013, 22, 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tam A; Arnold U; Soellner MB; Raines RT, Protein Prosthesis: 1,5-Disubstituted[1,2, 3]triazoles As Cis-Peptide Bond Surrogates. J. Am. Chem. Soc 2007, 129, 12670–12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Arnold U; Raines RT, Replacing a Single Atom Accelerates the Folding of a Protein and Increases Its Thermostability. Org. Biomol. Chem 2016, 14, 6780–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Arnold U; Hinderaker MP; Koditz J; Golbik R; Ulbrich-Hofmann R; Raines RT, Protein Prosthesis: A Nonnatural Residue Accelerates Folding and Increases Stability. J. Am. Chem. Soc 2003, 125, 7500–7501. [DOI] [PubMed] [Google Scholar]
- 257.Piontek C; Ring P; Harjes O; Heinlein C; Mezzato S; Lombana N; Pöhner C; Püttner M; Varón Silva D; Martin A, et al. , Semisynthesis of a Homogeneous Glycoprotein Enzyme: Ribonuclease C: Part 1. Angew. Chem. Int. Ed 2009, 48, 1936–1940. [DOI] [PubMed] [Google Scholar]
- 258.Piontek C; Varón Silva D; Heinlein C; Pöhner C; Mezzato S; Ring P; Martin A; Schmid FX; Unverzagt C, Semisynthesis of a Homogeneous Glycoprotein Enzyme: Ribonuclease C: Part 2. Angew. Chem. Int. Ed 2009, 48, 1941–1945. [DOI] [PubMed] [Google Scholar]
- 259.Yee CS; Seyedsayamdost MR; Chang MCY; Nocera DG; Stubbe J, Generation of the R2 Subunit of Ribonucleotide Reductase by Intein Chemistry: Insertion of 3-Nitrotyrosine at Residue 356 as a Probe of the Radical Initiation Process. Biochemistry 2003, 42, 14541–14552. [DOI] [PubMed] [Google Scholar]
- 260.Seyedsayamdost MR; Yee CS; Reece SY; Nocera DG; Stubbe J, pH Rate Profiles of FnY356−R2s (n = 2, 3, 4) in Escherichia coli Ribonucleotide Reductase: Evidence that Y356 Is a Redox-Active Amino Acid along the Radical Propagation Pathway. J. Am. Chem. Soc 2006, 128, 1562–1568. [DOI] [PubMed] [Google Scholar]
- 261.Seyedsayamdost MR; Stubbe J, Site-Specific Replacement of Y356 with 3,4-Dihydroxyphenylalanine in the β2 Subunit of E. coli Ribonucleotide Reductase. J. Am. Chem. Soc 2006, 128, 2522–2523. [DOI] [PubMed] [Google Scholar]
- 262.Wang Y; Kavran JM; Chen Z; Karukurichi KR; Leahy DJ; Cole PA, Regulation of S-Adenosylhomocysteine Hydrolase by Lysine Acetylation. J. Biol. Chem 2014, 289, 31361–31372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Liu J; Chen Q; Rozovsky S, Utilizing Selenocysteine for Expressed Protein Ligation and Bioconjugations. J. Am. Chem. Soc 2017, 139, 3430–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhang Z; Shen K; Lu W; Cole PA, The Role of C-terminal Tyrosine Phosphorylation in the Regulation of SHP-1 Explored via Expressed Protein Ligation. J. Biol. Chem 2003, 278, 4668–4674. [DOI] [PubMed] [Google Scholar]
- 265.Lu W; Shen K; Cole PA, Chemical Dissection of the Effects of Tyrosine Phosphorylation of SHP-2. Biochemistry 2003, 42, 5461–5468. [DOI] [PubMed] [Google Scholar]
- 266.Lu W; Gong D; Bar-Sagi D; Cole PA, Site-Specific Incorporation of a Phosphotyrosine Mimetic Reveals a Role for Tyrosine Phosphorylation of SHP-2 in Cell Signaling. Mol. Cell 2001, 8, 759–769. [DOI] [PubMed] [Google Scholar]
- 267.Wu J-W; Hu M; Chai J; Seoane J; Huse M; Li C; Rigotti DJ; Kyin S; Muir TW; Fairman R, et al. , Crystal Structure of a Phosphorylated Smad2: Recognition of Phosphoserine by the MH2 Domain and Insights on Smad Function in TGF-β Signaling. Mol. Cell 2001, 8, 1277–1289. [DOI] [PubMed] [Google Scholar]
- 268.Ottesen JJ; Huse M; Sekedat MD; Muir TW, Semisynthesis of Phosphovariants of Smad2 Reveals a Substrate Preference of the Activated TβRI Kinase. Biochemistry 2004, 43, 5698–5706. [DOI] [PubMed] [Google Scholar]
- 269.Vila-Perelló M; Pratt MR; Tulin F; Muir TW, Covalent Capture of Phospho-Dependent Protein Oligomerization by Site-Specific Incorporation of a Diazirine Photo-Cross-Linker. J. Am. Chem. Soc 2007, 129, 8068–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hahn ME; Muir TW, Photocontrol of Smad2, a Multiphosphorylated Cell-Signaling Protein, through Caging of Activating Phosphoserines. Angew. Chem. Int. Ed 2004, 43, 5800–5803. [DOI] [PubMed] [Google Scholar]
- 271.Pellois J-P; Hahn ME; Muir TW, Simultaneous Triggering of Protein Activity and Fluorescence. J. Am. Chem. Soc 2004, 126, 7170–7171. [DOI] [PubMed] [Google Scholar]
- 272.Hahn ME; Pellois J-P; Vila-Perelló M; Muir TW, Tunable Photoactivation of a Post-translationally Modified Signaling Protein and its Unmodified Counterpart in Live Cells. ChemBioChem 2007, 8, 2100–2105. [DOI] [PubMed] [Google Scholar]
- 273.Schmohl L; Wagner FR; Schumann M; Krause E; Schwarzer D, Semisynthesis and Initial Characterization of Sortase A Mutants Containing Selenocysteine and Homocysteine. Biorg. Med. Chem 2015, 23, 2883–2889. [DOI] [PubMed] [Google Scholar]
- 274.Bentley ML; Lamb EC; McCafferty DG, Mutagenesis Studies of Substrate Recognition and Catalysis in the Sortase A Transpeptidase from Staphylococcus aureus. J. Biol. Chem 2008, 283, 14762–14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Shen K; Cole PA, Conversion of a Tyrosine Kinase Protein Substrate to a High Affinity Ligand by ATP Linkage. J. Am. Chem. Soc 2003, 125, 16172–16173. [DOI] [PubMed] [Google Scholar]
- 276.Wang D; Cole PA, Protein Tyrosine Kinase Csk-Catalyzed Phosphorylation of Src Containing Unnatural Tyrosine Analogues. J. Am. Chem. Soc 2001, 123, 8883–8886. [DOI] [PubMed] [Google Scholar]
- 277.Virdee S; Kapadnis PB; Elliott T; Lang K; Madrzak J; Nguyen DP; Riechmann L; Chin JW, Traceless and Site-Specific Ubiquitination of Recombinant Proteins. J. Am. Chem. Soc 2011, 133, 10708–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lahiri S; Seidel R; Engelhard M; Becker CFW, Photocontrol of STAT6 Dimerization and Translocation. Mol. Biosyst 2010, 6, 2423–2429. [DOI] [PubMed] [Google Scholar]
- 279.Wissner RF; Batjargal S; Fadzen CM; Petersson EJ, Labeling Proteins with Fluorophore/Thioamide Förster Resonant Energy Transfer Pairs by Combining Unnatural Amino Acid Mutagenesis and Native Chemical Ligation. J. Am. Chem. Soc 2013, 135, 6529–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Batjargal S; Wang YJ; Goldberg JM; Wissner RF; Petersson EJ, Native Chemical Ligation of Thioamide-Containing Peptides: Development and Application to the Synthesis of Labeled α-Synuclein for Misfolding Studies. J. Am. Chem. Soc 2012, 134, 9172–9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Fauvet B; Fares M-B; Samuel F; Dikiy I; Tandon A; Eliezer D; Lashuel HA, Characterization of Semisynthetic and Naturally Nα-Acetylated α-Synuclein in Vitro and in Intact Cells: Implications for Aggregation and Cellular Properties of α-Synuclein. J. Biol. Chem 2012, 287, 28243–28262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hejjaoui M; Haj-Yahya M; Kumar KSA; Brik A; Lashuel HA, Towards Elucidation of the Role of Ubiquitination in the Pathogenesis of Parkinson’s Disease with Semisynthetic Ubiquitinated α-Synuclein. Angew. Chem. Int. Ed 2011, 50, 405–409. [DOI] [PubMed] [Google Scholar]
- 283.Haj-Yahya M; Fauvet B; Herman-Bachinsky Y; Hejjaoui M; Bavikar SN; Karthikeyan SV; Ciechanover A; Lashuel HA; Brik A, Synthetic Polyubiquitinated α-Synuclein Reveals Important Insights Into the Roles of the Ubiquitin Chain in Regulating Its Pathophysiology. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 17726–17731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hejjaoui M; Butterfield S; Fauvet B; Vercruysse F; Cui J; Dikiy I; Prudent M; Olschewski D; Zhang Y; Eliezer D, et al. , Elucidating the Role of C-Terminal Post-Translational Modifications Using Protein Semisynthesis Strategies: α-Synuclein Phosphorylation at Tyrosine 125. J. Am. Chem. Soc 2012, 134, 5196–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Burai R; Ait-Bouziad N; Chiki A; Lashuel HA, Elucidating the Role of Site-Specific Nitration of α-Synuclein in the Pathogenesis of Parkinson’s Disease via Protein Semisynthesis and Mutagenesis. J. Am. Chem. Soc 2015, 137, 5041–5052. [DOI] [PubMed] [Google Scholar]
- 286.Fauvet B; Lashuel HA, Semisynthesis and Enzymatic Preparation of Post-Translationally Modified α-Synuclein. Methods Mol. Biol 2016, 1345, 3–20. [DOI] [PubMed] [Google Scholar]
- 287.Ma M-R; Hu Z-W; Zhao Y-F; Chen Y-X; Li Y-M, Phosphorylation Induces Distinct alpha-Synuclein Strain Formation. Sci. Rep 2016, 6, 37130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Dikiy I; Fauvet B; Jovičić A; Mahul-Mellier A-L; Desobry C; El-Turk F; Gitler AD; Lashuel HA; Eliezer D, Semisynthetic and in Vitro Phosphorylation of Alpha-Synuclein at Y39 Promotes Functional Partly Helical Membrane-Bound States Resembling Those Induced by PD Mutations. ACS Chem. Biol 2016, 11, 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Marotta NP; Lin YH; Lewis YE; Ambroso MR; Zaro BW; Roth MT; Arnold DB; Langen R; Pratt MR, O-GlcNAc Modification Blocks the Aggregation and Toxicity of the Protein α-Synuclein Associated With Parkinson’s Disease. Nat. Chem 2015, 7, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Levine PM; Galesic A; Balana AT; Mahul-Mellier A-L; Navarro MX; De Leon CA; Lashuel HA; Pratt MR, α-Synuclein O-GlcNAcylation Alters Aggregation and Toxicity, Revealing Certain Residues as Potential Inhibitors of Parkinson’s Disease. Proc. Natl. Acad. Sci. U.S.A 2019, 116, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lewis YE; Galesic A; Levine PM; De Leon CA; Lamiri N; Brennan CK; Pratt MR, O-GlcNAcylation of α-Synuclein at Serine 87 Reduces Aggregation without Affecting Membrane Binding. ACS Chem. Biol 2017, 12, 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lee TC; Moran CR; Cistrone PA; Dawson PE; Deniz AA, Site-Specific Three-Color Labeling of α-Synuclein via Conjugation to Uniquely Reactive Cysteines during Assembly by Native Chemical Ligation. Cell Chem. Biol 2018, 25, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Reimann O; Smet-Nocca C; Hackenberger CPR, Traceless Purification and Desulfurization of Tau Protein Ligation Products. Angew. Chem. Int. Ed 2015, 54, 306–310. [DOI] [PubMed] [Google Scholar]
- 294.Schwagerus S; Reimann O; Despres C; Smet-Nocca C; Hackenberger CPR, Semi-Synthesis of a Tag-Free O-GlcNAcylated Tau Protein by Sequential Chemoselective Ligation. J. Pept. Sci 2016, 22, 327–333. [DOI] [PubMed] [Google Scholar]
- 295.Broncel M; Krause E; Schwarzer D; Hackenberger CPR, The Alzheimer’s Disease Related Tau Protein as a New Target for Chemical Protein Engineering. Chem. Euro. J 2012, 18, 2488–2492. [DOI] [PubMed] [Google Scholar]
- 296.van Ameijde J; Crespo R; Janson R; Juraszek J; Siregar B; Verveen H; Sprengers I; Nahar T; Hoozemans JJ; Steinbacher S, et al. , Enhancement of Therapeutic Potential of a Naturally Occurring Human Antibody Targeting a Phosphorylated Ser422 Containing Epitope on Pathological Tau. Acta Neuropathol. Commun 2018, 6, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ellmer D; Brehs M; Haj-Yahya M; Lashuel HA; Becker CFW, Single Posttranslational Modifications in the Central Repeat Domains of Tau4 Impact its Aggregation and Tubulin Binding. Angew. Chem. Int. Ed 2019, 58, 1616–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Eckenroth B; Harris K; Turanov AA; Gladyshev VN; Raines RT; Hondal RJ, Semisynthesis and Characterization of Mammalian Thioredoxin Reductase. Biochemistry 2006, 45, 5158–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lothrop AP; Snider GW; Flemer S; Ruggles EL; Davidson RS; Lamb AL; Hondal RJ, Compensating for the Absence of Selenocysteine in High-Molecular Weight Thioredoxin Reductases: The Electrophilic Activation Hypothesis. Biochemistry 2014, 53, 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Lothrop AP; Snider GW; Ruggles EL; Patel AS; Lees WJ; Hondal RJ, Selenium as an Electron Acceptor during the Catalytic Mechanism of Thioredoxin Reductase. Biochemistry 2014, 53, 654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Barber DR; Hondal RJ, Gain of function conferred by selenocysteine: catalytic enhancement of one-electron transfer reactions by thioredoxin reductase. Protein Sci 2019, 28, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.O’Keefe JP; Dustin CM; Barber D; Snider GW; Hondal RJ, A “Seleno Effect” Differentiates the Roles of Redox Active Cysteine Residues in Plasmodium falciparum Thioredoxin Reductase. Biochemistry 2018, 57, 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Snider GW; Dustin CM; Ruggles EL; Hondal RJ, A Mechanistic Investigation of the C-Terminal Redox Motif of Thioredoxin Reductase from Plasmodium falciparum. Biochemistry 2014, 53, 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Snider GW; Ruggles E; Khan N; Hondal RJ, Selenocysteine Confers Resistance to Inactivation by Oxidation in Thioredoxin Reductase: Comparison of Selenium and Sulfur Enzymes. Biochemistry 2013, 52, 5472–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Eckenroth BE; Lacey BM; Lothrop AP; Harris KM; Hondal RJ, Investigation of the C-Terminal Redox Center of High-Mr Thioredoxin Reductase by Protein Engineering and Semisynthesis. Biochemistry 2007, 46, 9472–9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Gogolin L; Seidel R; Engelhard M; Goody RS; Becker CFW, Semisynthesis of Human Thymidine Monophosphate Kinase. Pept. Sci 2010, 94, 433–440. [DOI] [PubMed] [Google Scholar]
- 307.Flavell RR; Huse M; Goger M; Trester-Zedlitz M; Kuriyan J; Muir TW, Efficient Semisynthesis of a Tetraphosphorylated Analogue of the Type I TGFβ Receptor. Org. Lett 2002, 4, 165–168. [DOI] [PubMed] [Google Scholar]
- 308.Huse M; Holford MN; Kuriyan J; Muir TW, Semisynthesis of Hyperphosphorylated Type I TGFβ Receptor: Addressing the Mechanism of Kinase Activation. J. Am. Chem. Soc 2000, 122, 8337–8338. [Google Scholar]
- 309.Pál G; Santamaria F; Kossiakoff AA; Lu W, The First Semi-Synthetic Serine Protease Made by Native Chemical Ligation. Protein Expr. Purif 2003, 29, 185–192. [DOI] [PubMed] [Google Scholar]
- 310.Zhou L; Holt MT; Ohashi N; Zhao A; Müller MM; Wang B; Muir TW, Evidence that Ubiquitylated H2B Corrals hDot1L on the Nucleosomal Surface to Induce H3K79 methylation. Nat. Commun 2016, 7, 10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Yang R; Bi X; Li F; Cao Y; Liu C-F, Native Chemical Ubiquitination Using a Genetically Incorporated Azidonorleucine. Chem. Commun 2014, 50, 7971–7974. [DOI] [PubMed] [Google Scholar]
- 312.Han C; Pao K-C; Kazlauskaite A; Muqit MMK; Virdee S, A Versatile Strategy for the Semisynthetic Production of Ser65 Phosphorylated Ubiquitin and Its Biochemical and Structural Characterisation. ChemBioChem 2015, 16, 1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Lu X; Olsen SK; Capili AD; Cisar JS; Lima CD; Tan DS, Designed Semisynthetic Protein Inhibitors of Ub/Ubl E1 Activating Enzymes. J. Am. Chem. Soc 2010, 132, 1748–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Olsen SK; Capili AD; Lu X; Tan DS; Lima CD, Active Site Remodelling Accompanies Thioester Bond Formation in the SUMO E1. Nature 2010, 463, 906–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Weiser BP; Stivers JT; Cole PA, Investigation of N-Terminal Phospho-Regulation of Uracil DNA Glycosylase Using Protein Semisynthesis. Biophys. J 2017, 113, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Fehr F; Nadler A; Brodhun F; Feussner I; Diederichsen U, Semi-Synthesis and Analysis of Chemically Modified Zif268 Zinc-Finger Domains. ChemistryOpen 2012, 1, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kötzler MP; Robinson K; Chen H-M; Okon M; McIntosh LP; Withers SG, Modulating the Nucleophile of a Glycoside Hydrolase through Site-Specific Incorporation of Fluoroglutamic Acids. J. Am. Chem. Soc 2018, 140, 8268–8276. [DOI] [PubMed] [Google Scholar]
- 318.Balambika R; Inui T; Sargsyan H; Arshava B; Cohen LS; Ding FX; Becker JM; Naider F, Synthesis of a Double Transmembrane Domain Fragment of Ste2p by Native Chemical Ligation. Int. J. Pept. Res. Ther 2007, 13, 251–263. [Google Scholar]
- 319.Rak A; Pylypenko O; Durek T; Watzke A; Kushnir S; Brunsveld L; Waldmann H; Goody RS; Alexandrov K, Structure of Rab GDP-Dissociation Inhibitor in Complex with Prenylated YPT1 GTPase. Science 2003, 302, 646–650. [DOI] [PubMed] [Google Scholar]
- 320.Pylypenko O; Rak A; Durek T; Kushnir S; Dursina BE; Thomae NH; Constantinescu AT; Brunsveld L; Watzke A; Waldmann H, et al. , Structure of Doubly Prenylated Ypt1:GDI Complex and the Mechanism of GDI-Mediated Rab Recycling. EMBO J. 2006, 25, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Jantz D; Berg JM, Expanding the DNA-Recognition Repertoire for Zinc Finger Proteins beyond 20 Amino Acids. J. Am. Chem. Soc 2003, 125, 4960–4961. [DOI] [PubMed] [Google Scholar]
- 322.UniProtKB/TrEMBL - Current Release Stastistics Website online. (release 2019_09) https://www.uniprot.org/statistics/TrEMBL (accessed November 4, 2019).
- 323.Malins LR; Payne RJ, Synthetic Amino Acids for Applications in Peptide Ligation–Desulfurization Chemistry. Aust. J. Chem 2015, 68, 521–537. [Google Scholar]
- 324.Kulkarni SS; Sayers J; Premdjee B; Payne RJ, Rapid and Efficient Protein Synthesis Through Expansion of the Native Chemical Ligation Concept. Nature Reviews Chemistry 2018, 2, 0122. [Google Scholar]
- 325.Malins LR; Mitchell NJ; Payne RJ, Peptide Ligation Chemistry at Selenol Amino Acids. J. Pept. Sci 2014, 20, 64–77. [DOI] [PubMed] [Google Scholar]
- 326.Tam JP; Miao ZW, Stereospecific Pseudoproline Ligation of N-Terminal Serine, Threonine, or Cysteine-Containing Unprotected Peptides. J. Am. Chem. Soc 1999, 121, 9013–9022. [Google Scholar]
- 327.Li XC; Lam HY; Zhang YF; Chan CK, Salicylaldehyde Ester-Induced Chemoselective Peptide Ligations: Enabling Generation of Natural Peptidic Linkages at the Serine/Threonine Sites. Org. Lett 2010, 12, 1724–1727. [DOI] [PubMed] [Google Scholar]
- 328.Levine PM; Craven TW; Bonneau R; Kirshenbaum K, Semisynthesis of Peptoid-Protein Hybrids by Chemical Ligation at Serine. Org. Lett 2014, 16, 512–515. [DOI] [PubMed] [Google Scholar]
- 329.Johnson ECB; Malito E; Shen Y; Pentelute B; Rich D; Florián J; Tang W-J; Kent SBH, Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors. J. Mol. Biol 2007, 373, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Torbeev VY; Kent SBH, Convergent Chemical Synthesis and Crystal Structure of a 203 Amino Acid “Covalent Dimer” HIV-1 Protease Enzyme Molecule. Angew. Chem. Int. Ed 2007, 46, 1667–1670. [DOI] [PubMed] [Google Scholar]
- 331.Kochendoerfer GG; Chen S-Y; Mao F; Cressman S; Traviglia S; Shao H; Hunter CL; Low DW; Cagle EN; Carnevali M, et al. , Design and Chemical Synthesis of a Homogeneous Polymer-Modified Erythropoiesis Protein. Science 2003, 299, 884–887. [DOI] [PubMed] [Google Scholar]
- 332.Tam JP; Yu Q, Methionine Ligation Strategy in the Biomimetic Synthesis of Parathyroid Hormones. Biopolymers 1998, 46, 319–327. [DOI] [PubMed] [Google Scholar]
- 333.Aussedat B; Fasching B; Johnston E; Sane N; Nagorny P; Danishefsky SJ, Total Synthesis of the α-Subunit of Human Glycoprotein Hormones: Toward Fully Synthetic Homogeneous Human Follicle-Stimulating Hormone. J. Am. Chem. Soc 2012, 134, 3532–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Uprety R; Luo J; Liu J; Naro Y; Samanta S; Deiters A, Genetic Encoding of Caged Cysteine and Caged Homocysteine in Bacterial and Mammalian Cells. ChemBioChem 2014, 15, 1793–1799. [DOI] [PubMed] [Google Scholar]
- 335.Tanaka T; Wagner AM; Warner JB; Wang YJ; Petersson EJ, Expressed Protein Ligation at Methionine: N-Terminal Attachment of Homocysteine, Ligation, and Masking. Angew. Chem. Int. Ed 2013, 52, 6210–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Yan LZ; Dawson PE, Synthesis of Peptides and Proteins without Cysteine Residues by Native Chemical Ligation Combined with Desulfurization. J. Am. Chem. Soc 2001, 123, 526–533. [DOI] [PubMed] [Google Scholar]
- 337.Wan Q; Danishefsky SJ, Free-Radical-Based, Specific Desulfurization of Cysteine: A Powerful Advance in the Synthesis of Polypeptides and Glycopolypeptides. Angew. Chem. Int. Ed 2007, 46, 9248–9252. [DOI] [PubMed] [Google Scholar]
- 338.Thompson RE; Chan B; Radom L; Jolliffe KA; Payne RJ, Chemoselective Peptide Ligation–Desulfurization at Aspartate. Angew. Chem. Int. Ed 2013, 52, 9723–9727. [DOI] [PubMed] [Google Scholar]
- 339.El Oualid F; Merkx R; Ekkebus R; Hameed DS; Smit JJ; de Jong A; Hilkmann H; Sixma TK; Ovaa H, Chemical Synthesis of Ubiquitin, Ubiquitin-Based Probes, and Diubiquitin. Angew. Chem. Int. Ed 2010, 49, 10149–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Ajish Kumar, K. S.; Haj-Yahya M; Olschewski D; Lashuel HA; Brik A, Highly Efficient and Chemoselective Peptide Ubiquitylation. Angew. Chem. Int. Ed 2009, 48, 8090–8094. [DOI] [PubMed] [Google Scholar]
- 341.Yang R; Pasunooti KK; Li F; Liu X-W; Liu C-F, Dual Native Chemical Ligation at Lysine. J. Am. Chem. Soc 2009, 131, 13592–13593. [DOI] [PubMed] [Google Scholar]
- 342.Li X; Fekner T; Ottesen JJ; Chan MK, A Pyrrolysine Analogue for Site-Specific Protein Ubiquitination. Angew. Chem. Int. Ed 2009, 48, 9184–9187. [DOI] [PubMed] [Google Scholar]
- 343.Offer J, Native Chemical Ligation With Nα Acyl Transfer Auxiliaries. Pept. Sci 2010, 94, 530–541. [DOI] [PubMed] [Google Scholar]
- 344.Weller CE; Huang W; Chatterjee C, Facile Synthesis of Native and Protease-Resistant Ubiquitylated Peptides. ChemBioChem 2014, 15, 1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Chatterjee C; McGinty RK; Pellois J-P; Muir TW, Auxiliary-Mediated Site-Specific Peptide Ubiquitylation. Angew. Chem. Int. Ed 2007, 46, 2814–2818. [DOI] [PubMed] [Google Scholar]
- 346.Quaderer R; Sewing A; Hilvert D, Selenocysteine-Mediated Native Chemical Ligation. Helv. Chim. Acta 2001, 84, 1197–1206. [Google Scholar]
- 347.Gieselman MD; Xie L; van der Donk WA, Synthesis of a Selenocysteine-Containing Peptide by Native Chemical Ligation. Org. Lett 2001, 3, 1331–1334. [DOI] [PubMed] [Google Scholar]
- 348.Muttenthaler M; Alewood PF, Selenopeptide Chemistry. J. Pept. Sci 2008, 14, 1223–1239. [DOI] [PubMed] [Google Scholar]
- 349.Thyer R; Shroff R; Klein DR; d’Oelsnitz S; Cotham VC; Byrom M; Brodbelt JS; Ellington AD, Custom Selenoprotein Production Enabled by Laboratory Evolution of Recoded Bacterial Strains. Nat. Biotechnol 2018, 36, 624–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Mukai T; Sevostyanova A; Suzuki T; Fu X; Söll D, A Facile Method for Producing Selenocysteine-Containing Proteins. Angew. Chem. Int. Ed 2018, 57, 7215–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Liu J; Zheng F; Cheng R; Li S; Rozovsky S; Wang Q; Wang L, Site-Specific Incorporation of Selenocysteine Using an Expanded Genetic Code and Palladium-Mediated Chemical Deprotection. J. Am. Chem. Soc 2018, 140, 8807–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wang YJ; Szantai-Kis DM; Petersson EJ, Chemoselective Modifications for the Traceless Ligation of Thioamide-Containing Peptides and Proteins. Org. Biomol. Chem 2016, 14, 6262–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Malins LR; Mitchell NJ; McGowan S; Payne RJ, Oxidative Deselenization of Selenocysteine: Applications for Programmed Ligation at Serine. Angew. Chem. Int. Ed 2015, 54, 12716–12721. [DOI] [PubMed] [Google Scholar]
- 354.Dery S; Reddy PS; Dery L; Mousa R; Dardashti RN; Metanis N, Insights Into the Deselenization of Selenocysteine Into Alanine and Serine. Chem. Sci 2015, 6, 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Durek T; Alewood PF, Preformed Selenoesters Enable Rapid Native Chemical Ligation at Intractable Sites. Angew. Chem. Int. Ed 2011, 50, 12042–12045. [DOI] [PubMed] [Google Scholar]
- 356.Mitchell NJ; Malins LR; Liu X; Thompson RE; Chan B; Radom L; Payne RJ, Rapid Additive-Free Selenocystine–Selenoester Peptide Ligation. J. Am. Chem. Soc 2015, 137, 14011–14014. [DOI] [PubMed] [Google Scholar]
- 357.Sayers J; Karpati PMT; Mitchell NJ; Goldys AM; Kwong SM; Firth N; Chan B; Payne RJ, Construction of Challenging Proline–Proline Junctions via Diselenide–Selenoester Ligation Chemistry. J. Am. Chem. Soc 2018, 140, 13327–13334. [DOI] [PubMed] [Google Scholar]
- 358.Pietrokovski S, Intein Spread and Extinction in Evolution. Trends Genet. 2001, 17, 465–472. [DOI] [PubMed] [Google Scholar]
- 359.Chong S; Xu M-Q, Protein Splicing of the Saccharomyces cerevisiae VMA Intein without the Endonuclease Motifs. J. Biol. Chem 1997, 272, 15587–15590. [DOI] [PubMed] [Google Scholar]
- 360.Derbyshire V; Wood DW; Wu W; Dansereau JT; Dalgaard JZ; Belfort M, Genetic Definition of a Protein-Splicing Domain: Functional Mini-Inteins Support Structure Predictions and a Model for Intein Evolution. Proc. Natl. Acad. Sci. U.S.A 1997, 94, 11466–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Mills KV; Lew BM; Jiang SQ; Paulus H, Protein Splicing in Trans by Purified N- and C-Terminal Fragments of the Mycobacterium Tuberculosis RecA Intein. Proc. Natl. Acad. Sci. U.S.A 1998, 95, 3543–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Wu H; Xu M-Q; Liu X-Q, Protein Trans-Splicing and Functional Mini-Inteins of a Cyanobacterial dnaB Intein. Biochim. Biophys. Acta 1998, 1387, 422–432. [DOI] [PubMed] [Google Scholar]
- 363.Southworth MW; Adam E; Panne D; Byer R; Kautz R; Perler FB, Control of Protein Splicing by Intein Fragment Reassembly. EMBO J. 1998, 17, 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Yamazaki T; Otomo T; Oda N; Kyogoku Y; Uegaki K; Ito N; Ishino Y; Nakamura H, Segmental Isotope Labeling for Protein NMR Using Peptide Splicing. J. Am. Chem. Soc 1998, 120, 5591–5592. [Google Scholar]
- 365.Lew BM; Mills KV; Paulus H, Protein Splicing In Vitro With a Semisynthetic Two-Component Minimal Intein. J. Biol. Chem 1998, 273, 15887–15890. [DOI] [PubMed] [Google Scholar]
- 366.Mootz HD; Muir TW, Protein Splicing Triggered by a Small Molecule. J. Am. Chem. Soc 2002, 124, 9044–9045. [DOI] [PubMed] [Google Scholar]
- 367.Mootz HD; Blum ES; Tyszkiewicz AB; Muir TW, Conditional Protein Splicing: A New Tool to Control Protein Structure and Function in Vitro and in Vivo. J. Am. Chem. Soc 2003, 125, 10561–10569. [DOI] [PubMed] [Google Scholar]
- 368.Shi JX; Muir TW, Development of a Tandem Protein Trans-Splicing System Based on Native and Engineered Split Inteins. J. Am. Chem. Soc 2005, 127, 6198–6206. [DOI] [PubMed] [Google Scholar]
- 369.Schwartz EC; Saez L; Young MW; Muir TW, Post-Translational Enzyme Activation in an Animal via Optimized Conditional Protein Splicing. Nat. Chem. Biol 2006, 3, 50–54. [DOI] [PubMed] [Google Scholar]
- 370.Alford SC; O’Sullivan C; Obst J; Christie J; Howard PL, Conditional Protein Splicing of α-Sarcin in Live Cells. Mol. Biosyst 2014, 10, 831–837. [DOI] [PubMed] [Google Scholar]
- 371.Brenzel S; Kurpiers T; Mootz HD, Engineering Artificially Split Inteins for Applications in Protein Chemistry: Biochemical Characterization of the Split Ssp DnaB Intein and Comparison to the Split Sce VMA Intein . Biochemistry 2006, 45, 1571–1578. [DOI] [PubMed] [Google Scholar]
- 372.Aranko AS; Wlodawer A; Iwaï H, Nature’s Recipe for Splitting Inteins. Protein Eng. Des. Sel 2014, 27, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Appleby JH; Zhou KS; Volkmann G; Liu XQ, Novel Split Intein for Trans-Splicing Synthetic Peptide onto C Terminus of Protein. J. Biol. Chem 2009, 284, 6194–6199. [DOI] [PubMed] [Google Scholar]
- 374.Volkmann G; Liu XQ, Protein C-Terminal Labeling and Biotinylation Using Synthetic Peptide and Split-Intein. PLoS One 2009, 4, e8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lin Y; Li M; Song H; Xu L; Meng Q; Liu X-Q, Protein Trans-Splicing of Multiple Atypical Split Inteins Engineered from Natural Inteins. PLoS One 2013, 8, e59516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Ludwig C; Pfeiff M; Linne U; Mootz HD, Ligation of a Synthetic Peptide to the N Terminus of a Recombinant Protein Using Semisynthetic Protein Trans-Splicing. Angew. Chem. Int. Ed 2006, 45, 5218–5221. [DOI] [PubMed] [Google Scholar]
- 377.Sun W; Yang J; Liu X-Q, Synthetic Two-Piece and Three-Piece Split Inteins for Protein Trans-Splicing. J. Biol. Chem 2004, 279, 35281–35286. [DOI] [PubMed] [Google Scholar]
- 378.Appleby-Tagoe JH; Thiel IV; Wang Y; Wang Y; Mootz HD; Liu X-Q, Highly Efficient and More General Cis- and Trans-Splicing Inteins through Sequential Directed Evolution. J. Biol. Chem 2011, 286, 34440–34447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Wu H; Hu ZM; Liu XQ, Protein Trans-Splicing by a Split Intein Encoded in a Split DnaE Gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A 1998, 95, 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Shah NH; Dann GP; Vila-Perello M; Liu ZH; Muir TW, Ultrafast Protein Splicing is Common among Cyanobacterial Split Inteins: Implications for Protein Engineering. J. Am. Chem. Soc 2012, 134, 11338–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Iwai H; Zuger S; Jin J; Tam PH, Highly Efficient Protein Trans-Splicing by a Naturally Split DnaE Intein From Nostoc punctiforme. FEBS Lett. 2006, 580, 1853–1858. [DOI] [PubMed] [Google Scholar]
- 382.Carvajal-Vallejos P; Pallisse R; Mootz HD; Schmidt SR, Unprecedented Rates and Efficiencies Revealed for New Natural Split Inteins From Metagenomic Sources. J. Biol. Chem 2012, 287, 28686–28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Thiel IV; Volkmann G; Pietrokovski S; Mootz HD, An Atypical Naturally Split Intein Engineered for Highly Efficient Protein Labeling. Angew. Chem. Int. Ed 2014, 53, 1306–1310. [DOI] [PubMed] [Google Scholar]
- 384.Bachmann A-L; Mootz HD, An Unprecedented Combination of Serine and Cysteine Nucleophiles in a Split Intein with an Atypical Split Site. J. Biol. Chem 2015, 290, 28792–28804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Bachmann A-L; Mootz HD, N-Terminal Chemical Protein Labeling Using the Naturally Split GOS-TerL Intein. J. Pept. Sci 2017, 23, 624–630. [DOI] [PubMed] [Google Scholar]
- 386.Liu XQ, Protein-Splicing Intein: Genetic Mobility, Origin, and Evolution. Annu. Rev. Genet 2000, 34, 61–76. [DOI] [PubMed] [Google Scholar]
- 387.Martin DD; Xu MQ; Evans TC, Characterization of a Naturally Occurring Trans-Splicing Intein From Synechocystis sp. PCC6803. Biochemistry 2001, 40, 1393–1402. [DOI] [PubMed] [Google Scholar]
- 388.Zettler J; Schutz V; Mootz HD, The Naturally Split Npu DnaE Intein Exhibits an Extraordinarily High Rate in the Protein Trans-Splicing Reaction. FEBS Lett. 2009, 583, 909–914. [DOI] [PubMed] [Google Scholar]
- 389.Dassa B; London N; Stoddard BL; Schueler-Furman O; Pietrokovski S, Fractured Genes: A Novel Genomic Arrangement Involving New Split Inteins and a New Homing Endonuclease Family . Nucleic Acids Res. 2009, 37, 2560–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Gramespacher JA; Stevens AJ; Nguyen DP; Chin JW; Muir TW, Intein Zymogens: Conditional Assembly and Splicing of Split Inteins via Targeted Proteolysis. J. Am. Chem. Soc 2017, 139, 8074–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Neugebauer M; Böcker Jana K; Matern Julian CJ; Pietrokovski S; Mootz Henning D, Development of a Screening System for Inteins Active in Protein Splicing Based on Intein Insertion Into the LacZα-Peptide. Biol. Chem 2017, 398, 57–67. [DOI] [PubMed] [Google Scholar]
- 392.Stevens AJ; Sekar G; Gramespacher JA; Cowburn D; Muir TW, An Atypical Mechanism of Split Intein Molecular Recognition and Folding. J. Am. Chem. Soc 2018, 140, 11791–11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Lehmann M; Pasamontes L; Lassen SF; Wyss M, The Consensus Concept for Thermostability Engineering of Proteins. Biochim. Biophys. Acta 2000, 1543, 408–415. [DOI] [PubMed] [Google Scholar]
- 394.Stevens AJ; Brown ZZ; Shah NH; Sekar G; Cowburn D; Muir TW, Design of a Split Intein with Exceptional Protein Splicing Activity. J. Am. Chem. Soc 2016, 138, 2162–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Shah NH; Eryilmaz E; Cowburn D; Muir TW, Extein Residues Play an Intimate Role in the Rate-Limiting Step of Protein Trans-Splicing. J. Am. Chem. Soc 2013, 135, 5839–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Lockless SW; Muir TW, Traceless Protein Splicing Utilizing Evolved Split Inteins. Proc. Natl. Acad. Sci. U.S.A 2009, 106, 10999–11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Stevens AJ; Sekar G; Shah NH; Mostafavi AZ; Cowburn D; Muir TW, A Promiscuous Split Intein With Expanded Protein Engineering Applications. Proc. Natl. Acad. Sci. U.S.A 2017, 114, 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Perler FB, InBase: the Intein Database. Nucleic Acids Res. 2002, 30, 383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Shah NH; Vila-Perello M; Muir TW, Kinetic Control of One-Pot Trans-Splicing Reactions by Using a Wild-Type and Designed Split Intein. Angew. Chem. Int. Ed 2011, 50, 6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Giriat I; Muir TW, Protein Semi-Synthesis in Living Cells. J. Am. Chem. Soc 2003, 125, 7180–7181. [DOI] [PubMed] [Google Scholar]
- 401.David Y; Vila-Perelló M; Verma S; Muir TW, Chemical Tagging and Customizing of Cellular Chromatin States Using Ultrafast Trans-Splicing Inteins. Nat. Chem 2015, 7, 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Borra R; Dong D; Elnagar AY; Woldemariam GA; Camarero JA, In-Cell Fluorescence Activation and Labeling of Proteins Mediated by FRET-Quenched Split Inteins. J. Am. Chem. Soc 2012, 134, 6344–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Jung D; Sato K; Min K; Shigenaga A; Jung J; Otaka A; Kwon Y, Photo-Triggered Fluorescent Labelling of Recombinant Proteins in Live Cells. Chem. Commun 2015, 51, 9670–9673. [DOI] [PubMed] [Google Scholar]
- 404.Braner M; Kollmannsperger A; Wieneke R; Tampé R, ‘Traceless’ Tracing of Proteins – High-Affinity Trans-Splicing Directed by a Minimal Interaction Pair. Chem. Sci 2016, 7, 2646–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Truong D-JJ; Kühner K; Kühn R; Werfel S; Engelhardt S; Wurst W; Ortiz O, Development of an Intein-Mediated Split–Cas9 System for Gene Therapy. Nucleic Acids Res. 2015, 43, 6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Tornabene P; Trapani I; Minopoli R; Centrulo M; Lupo M; de Simone S; Tiberi P; Dell’Aquila F; Marrocco E; Iodice C, et al. , Intein-Mediated Protein Trans-Splicing Expands Adeno-Associated Virus Transfer Capacity in the Retina. Sci. Transl. Med 2019, 11, eaav4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Dobrin A; Saxena P; Fussenegger M, Synthetic Biology: Applying Biological Circuits Beyond Novel Therapies. Integr. Biol 2015, 8, 409–430. [DOI] [PubMed] [Google Scholar]
- 408.Navarre WW; Schneewind O, Proteolytic Cleavage and Cell-Wall Anchoring at the LPXTG Motif of Surface-Proteins in Gram-Positive Bacteria. Mol. Microbiol 1994, 14, 115–121. [DOI] [PubMed] [Google Scholar]
- 409.Mazmanian SK; Liu G; Ton-That H; Schneewind O, Staphylococcus aureus Sortase, an Enzyme that Anchors Surface Proteins to the Cell Wall. Science 1999, 285, 760–763. [DOI] [PubMed] [Google Scholar]
- 410.Spirig T; Weiner EM; Clubb RT, Sortase Enzymes in Gram-Positive Bacteria. Mol. Microbiol 2011, 82, 1044–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Malik A; Kim SB, A Comprehensive In Silico Analysis of Sortase Superfamily. J. Microbiol 2019, 57, 431–443. [DOI] [PubMed] [Google Scholar]
- 412.Huang XY; Aulabaugh A; Ding WD; Kapoor B; Alksne L; Tabei K; Ellestad G, Kinetic Mechanism of Staphylococcus aureus Sortase SrtA. Biochemistry 2003, 42, 11307–11315. [DOI] [PubMed] [Google Scholar]
- 413.Ilangovan U; Ton-That H; Iwahara J; Schneewind O; Clubb RT, Structure of Sortase, the Transpeptidase That Anchors Proteins to the Cell Wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A 2001, 98, 6056–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Kruger RG; Otvos B; Frankel BA; Bentley M; Dostal P; McCafferty DG, Analysis of the Substrate Specificity of the Staphylococcus aureus Sortase Transpeptidase SrtA. Biochemistry 2004, 43, 1541–1551. [DOI] [PubMed] [Google Scholar]
- 415.Suree N; Liew CK; Villareal VA; Thieu W; Fadeev EA; Clemens JJ; Jung ME; Clubb RT, The Structure of the Staphylococcus aureus Sortase-Substrate Complex Reveals How the Universally Conserved LPXTG Sorting Signal Is Recognized. J. Biol. Chem 2009, 284, 24465–24477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Popp MW; Antos JM; Grotenbreg GM; Spooner E; Ploegh HL, Sortagging: A Versatile Method for Protein Labeling. Nat. Chem. Biol 2007, 3, 707–708. [DOI] [PubMed] [Google Scholar]
- 417.Sarpong K; Bose R, Efficient Sortase-Mediated N-Terminal Labeling of TEV Protease Cleaved Recombinant Proteins. Anal. Biochem 2017, 521, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Yamamoto T; Nagamune T, Expansion of the Sortase-Mediated Labeling Method for Site-Specific N-Terminal Labeling of Cell Surface Proteins on Living Cells. Chem. Commun 2009, 10.1039/b818792d, 1022–1024. [DOI] [PubMed] [Google Scholar]
- 419.Dai X; Böker A; Glebe U, Broadening the Scope of Sortagging. RSC Adv. 2019, 9, 4700–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Popp MW-L; Ploegh HL, Making and Breaking Peptide Bonds: Protein Engineering Using Sortase. Angew. Chem. Int. Ed 2011, 50, 5024–5032. [DOI] [PubMed] [Google Scholar]
- 421.Pritz S; Wolf Y; Kraetke O; Klose J; Bienert M; Beyermann M, Synthesis of Biologically Active Peptide Nucleic Acid−Peptide Conjugates by Sortase-Mediated Ligation. J. Org. Chem 2007, 72, 3909–3912. [DOI] [PubMed] [Google Scholar]
- 422.Kobashigawa Y; Kumeta H; Ogura K; Inagaki F, Attachment of an NMR-Invisible Solubility Enhancement Tag Using a Sortase-Mediated Protein Ligation Method. J. Biomol. NMR 2009, 43, 145–150. [DOI] [PubMed] [Google Scholar]
- 423.Refaei MA; Combs A; Kojetin DJ; Cavanagh J; Caperelli C; Rance M; Sapitro J; Tsang P, Observing Selected Domains in Multi-Domain Proteins via Sortase-Mediated Ligation and NMR Spectroscopy. J. Biomol. NMR 2011, 49, 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Yamamura Y; Hirakawa H; Yamaguchi S; Nagamune T, Enhancement of Sortase A-Mediated Protein Ligation by Inducing a β-Hairpin Structure Around the Ligation Site. Chem. Commun 2011, 47, 4742–4744. [DOI] [PubMed] [Google Scholar]
- 425.Antos JM; Chew G-L; Guimaraes CP; Yoder NC; Grotenbreg GM; Popp MW-L; Ploegh HL, Site-Specific N- and C-Terminal Labeling of a Single Polypeptide Using Sortases of Different Specificity. J. Am. Chem. Soc 2009, 131, 10800–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Williamson DJ; Fascione MA; Webb ME; Turnbull WB, Efficient N-Terminal Labeling of Proteins by Use of Sortase. Angew. Chem. Int. Ed 2012, 51, 9377–9380. [DOI] [PubMed] [Google Scholar]
- 427.Liu F; Luo EY; Flora DB; Mezo AR, Irreversible Sortase A-Mediated Ligation Driven by Diketopiperazine Formation. J. Org. Chem 2014, 79, 487–492. [DOI] [PubMed] [Google Scholar]
- 428.David Row R; Roark TJ; Philip MC; Perkins LL; Antos JM, Enhancing the Efficiency of Sortase–mediated Ligations Through Nickel–peptide Complex Formation. Chem. Commun 2015, 51, 12548–12551. [DOI] [PubMed] [Google Scholar]
- 429.Policarpo RL; Kang H; Liao X; Rabideau AE; Simon MD; Pentelute BL, Flow-Based Enzymatic Ligation by Sortase A. Angew. Chem. Int. Ed 2014, 53, 9203–9208. [DOI] [PubMed] [Google Scholar]
- 430.Chen I; Dorr BM; Liu DR, A General Strategy for the Evolution of Bond-Forming Enzymes Using Yeast Display. Proc. Natl. Acad. Sci. U.S.A 2011, 108, 11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Chen L; Cohen J; Song X; Zhao A; Ye Z; Feulner CJ; Doonan P; Somers W; Lin L; Chen PR, Improved Variants of SrtA for Site-Specific Conjugation on Antibodies and Proteins With High Efficiency. Sci. Rep 2016, 6, 31899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Dorr BM; Ham HO; An C; Chaikof EL; Liu DR, Reprogramming the Specificity of Sortase Enzymes. Proc. Natl. Acad. Sci. U.S.A 2014, 111, 13343–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Piotukh K; Geltinger B; Heinrich N; Gerth F; Beyermann M; Freund C; Schwarzer D, Directed Evolution of Sortase A Mutants with Altered Substrate Selectivity Profiles. J. Am. Chem. Soc 2011, 133, 17536–17539. [DOI] [PubMed] [Google Scholar]
- 434.Schmohl L; Bierlmeier J; Gerth F; Freund C; Schwarzer D, Engineering Sortase a by Screening a Second-Generation Library Using Phage Display. J. Pept. Sci 2017, 23, 631–635. [DOI] [PubMed] [Google Scholar]
- 435.Wu M; Hayward D; Kalin JH; Song Y; Schwabe JWR; Cole PA, Lysine-14 Acetylation of Histone H3 in Chromatin Confers Resistance to the Deacetylase and Demethylase Activities of an Epigenetic Silencing Complex. eLife 2018, 7, e37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Ringel AE; Cieniewicz AM; Taverna SD; Wolberger C, Nucleosome Competition Reveals Processive Acetylation by the SAGA HAT Module. Proc. Natl. Acad. Sci. U.S.A 2015, 112, E5461–E5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Kalin JH; Wu M; Gomez AV; Song Y; Das J; Hayward D; Adejola N; Wu M; Panova I; Chung HJ, et al. , Targeting the CoREST Complex With Dual Histone Deacetylase and Demethylase Inhibitors. Nat. Commun 2018, 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Strijbis K; Spooner E; Ploegh HL, Protein Ligation in Living Cells Using Sortase. Traffic 2012, 13, 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Hirakawa H; Ishikawa S; Nagamune T, Design of Ca2+-Independent Staphylococcus aureus Sortase A Mutants. Biotechnology and Bioengineering 2012, 109, 2955–2961. [DOI] [PubMed] [Google Scholar]
- 440.Wuethrich I; Peeters JGC; Blom AEM; Theile CS; Li Z; Spooner E; Ploegh HL; Guimaraes CP, Site-Specific Chemoenzymatic Labeling of Aerolysin Enables the Identification of New Aerolysin Receptors. PLoS One 2014, 9, e109883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Hirakawa H; Ishikawa S; Nagamune T, Ca2+-Independent Sortase-A Exhibits High Selective Protein Ligation Activity in the Cytoplasm of Escherichia coli. Biotechnol. J 2015, 10, 1487–1492. [DOI] [PubMed] [Google Scholar]
- 442.Jeong H-J; Abhiraman GC; Story CM; Ingram JR; Dougan SK, Generation of Ca2+-Independent Sortase A Mutants With Enhanced Activity for Protein and Cell Surface Labeling. PLoS One 2017, 12, e0189068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Glasgow JE; Salit ML; Cochran JR, In Vivo Site-Specific Protein Tagging with Diverse Amines Using an Engineered Sortase Variant. J. Am. Chem. Soc 2016, 138, 7496–7499. [DOI] [PubMed] [Google Scholar]
- 444.Fottner M; Brunner A-D; Bittl V; Horn-Ghetko D; Jussupow A; Kaila VRI; Bremm A; Lang K, Site-Specific Ubiquitylation and SUMOylation Using Genetic-Code Expansion and Sortase. Nat. Chem. Biol 2019, 15, 276–284. [DOI] [PubMed] [Google Scholar]
- 445.Wu Q; Ploegh HL; Truttmann MC, Hepta-Mutant Staphylococcus aureus Sortase A (SrtA7m) as a Tool for In Vivo Protein Labeling in Caenorhabditis elegans. ACS Chem. Biol 2017, 12, 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Tsukiji S; Nagamune T, Sortase-Mediated Ligation: A Gift from Gram-Positive Bacteria to Protein Engineering. ChemBioChem 2009, 10, 787–798. [DOI] [PubMed] [Google Scholar]
- 447.Hemu X; El Sahili A; Hu S; Wong K; Chen Y; Wong YH; Zhang X; Serra A; Goh BC; Darwis DA, et al. , Structural Determinants for Peptide-Bond Formation by Asparaginyl Ligases. Proc. Natl. Acad. Sci. U.S.A 2019, 116, 11737–11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Jackson MA; Gilding EK; Shafee T; Harris KS; Kaas Q; Poon S; Yap K; Jia H; Guarino R; Chan LY, et al. , Molecular Basis for the Production of Cyclic Peptides by Plant Asparaginyl Endopeptidases. Nat. Commun 2018, 9, 2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Nguyen GKT; Wang S; Qiu Y; Hemu X; Lian Y; Tam JP, Butelase 1 is an Asx-Specific Ligase Enabling Peptide Macrocyclization and Synthesis. Nat. Chem. Biol 2014, 10, 732. [DOI] [PubMed] [Google Scholar]
- 450.Nguyen GKT; Hemu X; Quek J-P; Tam JP, Butelase-Mediated Macrocyclization of d-Amino-Acid-Containing Peptides. Angew. Chem. Int. Ed 2016, 55, 12802–12806. [DOI] [PubMed] [Google Scholar]
- 451.Nguyen GKT; Cao Y; Wang W; Liu CF; Tam JP, Site-Specific N-Terminal Labeling of Peptides and Proteins using Butelase 1 and Thiodepsipeptide. Angew. Chem. Int. Ed 2015, 54, 15694–15698. [DOI] [PubMed] [Google Scholar]
- 452.Cao Y; Nguyen GKT; Chuah S; Tam JP; Liu C-F, Butelase-Mediated Ligation as an Efficient Bioconjugation Method for the Synthesis of Peptide Dendrimers. Bioconjugate Chemistry 2016, 27, 2592–2596. [DOI] [PubMed] [Google Scholar]
- 453.Harris KS; Durek T; Kaas Q; Poth AG; Gilding EK; Conlan BF; Saska I; Daly NL; van der Weerden NL; Craik DJ, et al. , Efficient Backbone Cyclization of Linear Peptides by a Recombinant Asparaginyl Endopeptidase. Nat. Commun 2015, 6, 10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Saether O; Craik DJ; Campbell ID; Sletten K; Juul J; Norman DG, Elucidation of the Primary and Three-Dimensional Structure of the Uterotonic Polypeptide Kalata B1. Biochemistry 1995, 34, 4147–4158. [DOI] [PubMed] [Google Scholar]
- 455.Mikula KM; Tascón I; Tommila JJ; Iwaï H, Segmental Isotopic Labeling of a Single-Domain Globular Protein Without Any Refolding Step by an Asparaginyl Endopeptidase. FEBS Lett. 2017, 591, 1285–1294. [DOI] [PubMed] [Google Scholar]
- 456.Yang R; Wong YH; Nguyen GKT; Tam JP; Lescar J; Wu B, Engineering a Catalytically Efficient Recombinant Protein Ligase. J. Am. Chem. Soc 2017, 139, 5351–5358. [DOI] [PubMed] [Google Scholar]
- 457.James AM; Haywood J; Leroux J; Ignasiak K; Elliott AG; Schmidberger JW; Fisher MF; Nonis SG; Fenske R; Bond CS, et al. , The Macrocyclizing Protease Butelase 1 Remains Autocatalytic and Reveals the Structural Basis for Ligase Activity. Plant J. 2019, 98, 988–999. [DOI] [PubMed] [Google Scholar]
- 458.Homandberg GA; Laskowski M, Enzymic Resynthesis of the Hydrolyzed Peptide Bond(s) in Ribonuclease S. Biochemistry 1979, 18, 586–592. [DOI] [PubMed] [Google Scholar]
- 459.Yazawa K; Numata K, Recent Advances in Chemoenzymatic Peptide Syntheses. Molecules 2014, 19, 13755–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Schellenberger V; Jakubke H-D, Protease-Catalyzed Kinetically Controlled Peptide Synthesis. Angew. Chem. Int. Ed 1991, 30, 1437–1449. [PubMed] [Google Scholar]
- 461.Nakatsuka T; Sasaki T; Kaiser ET, Peptide Segment Synthesis Catalyzed by the Semisynthetic Enzyme Thiolsubtilisin. J. Am. Chem. Soc 1987, 109, 3808–3810. [Google Scholar]
- 462.Polgar L; Bender ML, A New Enzyme Containing a Synthetically Formed Active Site. Thiol-Subtilisin. J. Am. Chem. Soc 1966, 88, 3153–3154. [Google Scholar]
- 463.Neet KE; Koshland DE, The Conversion of Serine at the Active Site of Subtilisin to Cysteine: A “Chemical Mutation”. Proc. Natl. Acad. Sci. U.S.A 1966, 56, 1606–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Wu ZP; Hilvert D, Conversion of a Protease Into an Acyl Transferase: Selenolsubtilisin. J. Am. Chem. Soc 1989, 111, 4513–4514. [Google Scholar]
- 465.Jackson DY; Burnier J; Quan C; Stanley M; Tom J; Wells JA, A Designed Peptide Ligase for Total Synthesis of Ribonuclease A With Unnatural Catalytic Residues. Science 1994, 266, 243–247. [DOI] [PubMed] [Google Scholar]
- 466.Abrahmsen L; Tom J; Burnier J; Butcher KA; Kossiakoff A; Wells JA, Engineering Subtilisin and its Substrates for Efficient Ligation of Peptide Bonds in Aqueous Solution. Biochemistry 1991, 30, 4151–4159. [DOI] [PubMed] [Google Scholar]
- 467.McPhalen CA; James MNG, Structural Comparison of Two Serine Proteinase-Protein Inhibitor Complexes: Eglin-C-Subtilisin Carlsberg and CI-2-Subtilisin Novo. Biochemistry 1988, 27, 6582–6598. [PubMed] [Google Scholar]
- 468.Philipp M; Bender ML, Kinetics of Subtilisin and Thiolsubtilisin. Molecular and Cellular Biochemistry 1983, 51, 5–32. [DOI] [PubMed] [Google Scholar]
- 469.Weeks AM; Wells JA, Engineering Peptide Ligase Specificity by Proteomic Identification of Ligation Sites. Nat. Chem. Biol 2017, 14, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Mahrus S; Trinidad JC; Barkan DT; Sali A; Burlingame AL; Wells JA, Global Sequencing of Proteolytic Cleavage Sites in Apoptosis by Specific Labeling of Protein N Termini. Cell 2008, 134, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Henager SH; Chu N; Chen Z; Bolduc D; Dempsey DR; Hwang Y; Wells J; Cole PA, Enzyme-Catalyzed Expressed Protein Ligation. Nat. Methods 2016, 13, 925–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Toplak A; Nuijens T; Quaedflieg PJLM; Wu B; Janssen DB, Peptiligase, an Enzyme for Efficient Chemoenzymatic Peptide Synthesis and Cyclization in Water. Adv. Synth. Catal 2016, 358, 2140–2147. [Google Scholar]
- 473.Schmidt M; Toplak A; Quaedflieg PJLM; Ippel H; Richelle GJJ; Hackeng TM; van Maarseveen JH; Nuijens T, Omniligase-1: A Powerful Tool for Peptide Head-to-Tail Cyclization. Adv. Synth. Catal 2017, 359, 2050–2055. [Google Scholar]
- 474.Oka T; Morihara K, Trypsin as a Catalyst for Peptide Synthesis. J. Biochem 1977, 82, 1055–1062. [DOI] [PubMed] [Google Scholar]
- 475.Liebscher S; Schöpfel M; Aumüller T; Sharkhuukhen A; Pech A; Höss E; Parthier C; Jahreis G; Stubbs MT; Bordusa F, N-Terminal Protein Modification by Substrate-Activated Reverse Proteolysis. Angew. Chem. Int. Ed 2014, 53, 3024–3028. [DOI] [PubMed] [Google Scholar]
- 476.Liebscher S; Kornberger P; Fink G; Trost-Gross E-M; Höss E; Skerra A; Bordusa F, Derivatization of Antibody Fab Fragments: A Designer Enzyme for Native Protein Modification. ChemBioChem 2014, 15, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 477.Thormann M; Thust S; Hofmann H-J; Bordusa F, Protease-Catalyzed Hydrolysis of Substrate Mimetics (Inverse Substrates): A New Approach Reveals a New Mechanism. Biochemistry 1999, 38, 6056–6062. [DOI] [PubMed] [Google Scholar]
- 478.Xu S; Zhao Z; Zhao J, Recent Advances in Enzyme-Mediated Peptide Ligation. Chin. Chem. Lett 2018, 29, 1009–1016. [Google Scholar]
- 479.Ling JJJ; Policarpo RL; Rabideau AE; Liao XL; Pentelute BL, Protein Thioester Synthesis Enabled by Sortase. J. Am. Chem. Soc 2012, 134, 10749–10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Cao Y; Nguyen GKT; Tam JP; Liu CF, Butelase-Mediated Synthesis of Protein Thioesters and Its Application for Tandem Chemoenzymatic Ligation. Chem. Commun 2015, 51, 17289–17292. [DOI] [PubMed] [Google Scholar]
- 481.Vila-Perelló M; Liu Z; Shah NH; Willis JA; Idoyaga J; Muir TW, Streamlined Expressed Protein Ligation Using Split Inteins. J. Am. Chem. Soc 2013, 135, 286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Frutos S; Jordan JB; Bio MM; Muir TW; Thiel OR; Vila-Perelló M, Access to Site-Specific FC–cRGD Peptide Conjugates Through Streamlined Expressed Protein Ligation. Org. Biomol. Chem 2016, 14, 9549–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Frutos S; Hernández JL; Otero A; Calvis C; Adan J; Mitjans F; Vila-Perelló M, Site-Specific Antibody Drug Conjugates Using Streamlined Expressed Protein Ligation. Bioconjugate Chemistry 2018, 29, 3503–3508. [DOI] [PubMed] [Google Scholar]
- 484.Nguyen HN; Afkari Y; Senoo H; Sesaki H; Devreotes PN; Iijima M, Mechanism of Human PTEN Localization Revealed by Heterologous Expression in Dictyostelium. Oncogene 2013, 33, 5688–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Thompson RE; Stevens AJ; Muir TW, Protein Engineering Through Tandem Transamidation. Nat. Chem 2019, 11, 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Liszczak GP; Brown ZZ; Kim SH; Oslund RC; David Y; Muir TW, Genomic Targeting of Epigenetic Probes Using a Chemically Tailored Cas9 System. Proc. Natl. Acad. Sci. U.S.A 2017, 114, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.McGinty RK; Tan S, Nucleosome Structure and Function. Chem. Rev 2015, 115, 2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Jenuwein T; Allis CD, Translating the Histone Code. Science 2001, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 489.Müller MM; Muir TW, Histones: At the Crossroads of Peptide and Protein Chemistry. Chem. Rev 2015, 115, 2296–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Holt M; Muir T, Application of the Protein Semisynthesis Strategy to the Generation of Modified Chromatin. Annu. Rev. Biochem 2015, 84, 265–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Chatterjee C; Muir TW, Chemical Approaches for Studying Histone Modifications. J. Biol. Chem 2010, 285, 11045–11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Boichenko I; Fierz B, Chemical and Biophysical Methods to Explore Dynamic Mechanisms of Chromatin Silencing. Curr. Opin. Chem. Biol 2019, 51, 1–10. [DOI] [PubMed] [Google Scholar]
- 493.Leonen CJA; Upadhyay E; Chatterjee C, Studies of Biochemical Crosstalk in Chromatin With Semisynthetic Histones. Curr. Opin. Chem. Biol 2018, 45, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Fischle W; Mootz HD; Schwarzer D, Synthetic Histone Code. Curr. Opin. Chem. Biol 2015, 28, 131–140. [DOI] [PubMed] [Google Scholar]
- 495.Simon MD; Chu F; Racki LR; de la Cruz CC; Burlingame AL; Panning B; Narlikar GJ; Shokat KM, The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell 2007, 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Chatterjee C; McGinty RK; Fierz B; Muir TW, Disulfide-Directed Histone Ubiquitylation Reveals Plasticity in hDot1L Activation. Nat. Chem. Biol 2010, 6, 267–269. [DOI] [PubMed] [Google Scholar]
- 497.Debelouchina GT; Gerecht K; Muir TW, Ubiquitin Utilizes an Acidic Surface Patch to Alter Chromatin Structure. Nat. Chem. Biol 2016, 13, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Shogren-Knaak MA; Fry CJ; Peterson CL, A Native Peptide Ligation Strategy for Deciphering Nucleosomal Histone Modifications. J. Biol. Chem 2003, 278, 15744–15748. [DOI] [PubMed] [Google Scholar]
- 499.Cheung P; Tanner KG; Cheung WL; Sassone-Corsi P; Denu JM; Allis CD, Synergistic Coupling of Histone H3 Phosphorylation and Acetylation in Response to Epidermal Growth Factor Stimulation. Mol. Cell 2000, 5, 905–915. [DOI] [PubMed] [Google Scholar]
- 500.Lo W-S; Trievel RC; Rojas JR; Duggan L; Hsu J-Y; Allis CD; Marmorstein R; Berger SL, Phosphorylation of Serine 10 in Histone H3 Is Functionally Linked In Vitro and In Vivo to Gcn5-Mediated Acetylation at Lysine 14. Mol. Cell 2000, 5, 917–926. [DOI] [PubMed] [Google Scholar]
- 501.Wojcik F; Dann GP; Beh LY; Debelouchina GT; Hofmann R; Muir TW, Functional Crosstalk Between Histone H2B Ubiquitylation and H2A Modifications and Variants. Nat. Commun 2018, 9, 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Kim J; Guermah M; McGinty RK; Lee J-S; Tang Z; Milne TA; Shilatifard A; Muir TW; Roeder RG, RAD6-Mediated Transcription-Coupled H2B Ubiquitylation Directly Stimulates H3K4 Methylation in Human Cells. Cell 2009, 137, 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Kim J; Kim J-A; McGinty Robert K.; Nguyen Uyen T. T.; Muir Tom W.; Allis CD; Roeder Robert G., The n-SET Domain of Set1 Regulates H2B Ubiquitylation-Dependent H3K4 Methylation. Mol. Cell 2013, 49, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Jeon J; McGinty RK; Muir TW; Kim J-A; Kim J, Crosstalk Among Set1 Complex Subunits Involved in H2B Ubiquitylation-Dependent H3K4 Methylation. Nucleic Acids Res. 2018, 46, 11129–11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.McGinty RK; Köhn M; Chatterjee C; Chiang KP; Pratt MR; Muir TW, Structure–Activity Analysis of Semisynthetic Nucleosomes: Mechanistic Insights into the Stimulation of Dot1L by Ubiquitylated Histone H2B. ACS Chem. Biol 2009, 4, 958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Holt MT; David Y; Pollock S; Tang Z; Jeon J; Kim J; Roeder RG; Muir TW, Identification of a Functional Hotspot on Ubiquitin Required for Stimulation of Methyltransferase Activity on Chromatin. Proc. Natl. Acad. Sci. U.S.A 2015, 112, 10365–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Fierz B; Chatterjee C; McGinty RK; Bar-Dagan M; Raleigh DP; Muir TW, Histone H2B Ubiquitylation Disrupts Local and Higher-Order Chromatin Compaction. Nat. Chem. Biol 2011, 7, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Gopinath P; Ohayon S; Nawatha M; Brik A, Chemical and Semisynthetic Approaches to Study and Target Deubiquitinases. Chem. Soc. Rev 2016, 45, 4171–4198. [DOI] [PubMed] [Google Scholar]
- 509.Meledin R; Mali SM; Brik A, Pushing the Boundaries of Chemical Protein Synthesis: The Case of Ubiquitin Chains and Polyubiquitinated Peptides and Proteins. Chem. Rec 2016, 16, 509–519. [DOI] [PubMed] [Google Scholar]
- 510.Qi Y-K; Si Y-Y; Du S-S; Liang J; Wang K-W; Zheng J-S, Recent Advances in the Chemical Synthesis and Semi-Synthesis of Poly-Ubiquitin-Based Proteins and Probes. Sci. China Chem 2019, 62, 299–312. [Google Scholar]
- 511.Bi X; Pasunooti KK; Liu C-F, Total Chemical and Semisynthetic Approaches for the Preparation of Ubiquitinated Proteins and Their Applications. Sci. China Chem 2018, 61, 251–265. [Google Scholar]
- 512.Ciccia A; Elledge SJ, The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Shiio Y; Eisenman RN, Histone Sumoylation Is Associated With Transcriptional Repression. Proc. Natl. Acad. Sci. U.S.A 2003, 100, 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Nathan D; Ingvarsdottir K; Sterner DE; Bylebyl GR; Dokmanovic M; Dorsey JA; Whelan KA; Krsmanovic M; Lane WS; Meluh PB, et al. , Histone Sumoylation Is a Negative Regulator in Saccharomyces cerevisiae and Shows Dynamic Interplay With Positive-Acting Histone Modifications. Genes Dev. 2006, 20, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Voigt P; LeRoy G; Drury William J.; Zee, Barry M.; Son J; Beck, David B.; Young Nicolas L.; Garcia Benjamin A.; Reinberg D, Asymmetrically Modified Nucleosomes. Cell 2012, 151, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Guidotti N; Lechner CC; Bachmann AL; Fierz B, A Modular Ligation Strategy for Asymmetric Bivalent Nucleosomes Trimethylated at K36 and K27. ChemBioChem 2019, 20, 1124–1128. [DOI] [PubMed] [Google Scholar]
- 517.Guidotti N; Lechner CC; Fierz B, Controlling the Supramolecular Assembly of Nucleosomes Asymmetrically Modified on H4. Chem. Commun 2017, 53, 10267–10270. [DOI] [PubMed] [Google Scholar]
- 518.Lechner CC; Agashe ND; Fierz B, Traceless Synthesis of Asymmetrically Modified Bivalent Nucleosomes. Angew. Chem. Int. Ed 2016, 55, 2903–2906. [DOI] [PubMed] [Google Scholar]
- 519.Levendosky RF; Sabantsev A; Deindl S; Bowman GD, The Chd1 Chromatin Remodeler Shifts Hexasomes Unidirectionally. eLife 2016, 5, e21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Levendosky RF; Bowman GD, Asymmetry Between the Two Acidic Patches Dictates the Direction of Nucleosome Sliding by the ISWI Chromatin Remodeler. eLife 2019, 8, e45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Lowary PT; Widom J, New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-Directed Nucleosome Positioning. J. Mol. Biol 1998, 276, 19–42. [DOI] [PubMed] [Google Scholar]
- 522.Sabantsev A; Levendosky RF; Zhuang X; Bowman GD; Deindl S, Direct Observation of Coordinated DNA Movements on the Nucleosome During Chromatin Remodelling. Nat. Commun 2019, 10, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Nacev BA; Feng L; Bagert JD; Lemiesz AE; Gao J; Soshnev AA; Kundra R; Schultz N; Muir TW; Allis CD, The Expanding Landscape of ‘Oncohistone’ Mutations in Human Cancers. Nature 2019, 567, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Spruijt Cornelia G.; Gnerlich F; Smits Arne H.; Pfaffeneder T; Jansen Pascal W. T. C.; Bauer C; Münzel M; Wagner M; Müller M; Khan F, et al. , Dynamic Readers for 5-(Hydroxy)Methylcytosine and Its Oxidized Derivatives. Cell 2013, 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 525.Beh LY; Debelouchina GT; Clay DM; Thompson RE; Lindblad KA; Hutton ER; Bracht JR; Sebra RP; Muir TW; Landweber LF, Identification of a DNA N6-Adenine Methyltransferase Complex and Its Impact on Chromatin Organization. Cell 2019, 177, 1781–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Liszczak G; Diehl KL; Dann GP; Muir TW, Acetylation Blocks DNA Damage–Induced Chromatin ADP-Ribosylation. Nat. Chem. Biol 2018, 14, 837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Lachner M; O’Carroll D; Rea S; Mechtler K; Jenuwein T, Methylation of Histone H3 Lysine 9 Creates a Binding Site for HP1 Proteins. Nature 2001, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 528.Kang J; Chaudhary J; Dong H; Kim S; Brautigam CA; Yu H, Mitotic Centromeric Targeting of HP1 and Its Binding to Sgo1 Are Dispensable for Sister-Chromatid Cohesion in Human Cells. Mol. Biol. Cell 2011, 22, 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.David Y; Muir TW, Emerging Chemistry Strategies for Engineering Native Chromatin. J. Am. Chem. Soc 2017, 139, 9090–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Lashuel HA; Overk CR; Oueslati A; Masliah E, The Many Faces of α-Synuclein: From Structure and Toxicity to Therapeutic Target. Nat. Rev. Neurosci 2012, 14, 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.McCann H; Stevens CH; Cartwright H; Halliday GM, α-Synucleinopathy Phenotypes. Parkinsonism Relat. D. 2014, 20, S62–S67. [DOI] [PubMed] [Google Scholar]
- 532.Martí MJ; Tolosa E; Campdelacreu J, Clinical Overview of the Synucleinopathies. Mov. Disord 2003, 18, 21–27. [DOI] [PubMed] [Google Scholar]
- 533.Fujiwara H; Hasegawa M; Dohmae N; Kawashima A; Masliah E; Goldberg MS; Shen J; Takio K; Iwatsubo T, α-Synuclein Is Phosphorylated in Synucleinopathy Lesions. Nat. Cell Biol 2002, 4, 160–164. [DOI] [PubMed] [Google Scholar]
- 534.Wani WY; Ouyang X; Benavides GA; Redmann M; Cofield SS; Shacka JJ; Chatham JC; Darley-Usmar V; Zhang J, O-GlcNAc Regulation of Autophagy and α-Synuclein Homeostasis; Implications for Parkinson’s Disease. Mol. Brain 2017, 10, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Giasson BI; Lee VMY, Are Ubiquitination Pathways Central to Parkinson’s Disease? Cell 2003, 114, 1–8. [DOI] [PubMed] [Google Scholar]
- 536.Giasson BI; Duda JE; Murray IVJ; Chen Q; Souza JM; Hurtig HI; Ischiropoulos H; Trojanowski JQ; Y. Lee VM, Oxidative Damage Linked to Neurodegeneration by Selective α-Synuclein Nitration in Synucleinopathy Lesions. Science 2000, 290, 985–989. [DOI] [PubMed] [Google Scholar]
- 537.Chen H; Zhao Y-F; Chen Y-X; Li Y-M, Exploring the Roles of Post-Translational Modifications in the Pathogenesis of Parkinson’s Disease Using Synthetic and Semisynthetic Modified α-Synuclein. ACS Chem. Neurosci 2019, 10, 910–921. [DOI] [PubMed] [Google Scholar]
- 538.Levine PM; De Leon CA; Galesic A; Balana A; Marotta NP; Lewis YE; Pratt MR, O-GlcNAc Modification Inhibits the Calpain-Mediated Cleavage of α-Synuclein. Biorg. Med. Chem 2017, 25, 4977–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Yin L; Krantz B; Russell NS; Deshpande S; Wilkinson KD, Nonhydrolyzable Diubiquitin Analogues Are Inhibitors of Ubiquitin Conjugation and Deconjugation. Biochemistry 2000, 39, 10001–10010. [DOI] [PubMed] [Google Scholar]
- 540.Meier F; Abeywardana T; Dhall A; Marotta NP; Varkey J; Langen R; Chatterjee C; Pratt MR, Semisynthetic, Site-Specific Ubiquitin Modification of α-Synuclein Reveals Differential Effects on Aggregation. J. Am. Chem. Soc 2012, 134, 5468–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Abeywardana T; Pratt MR, Extent of Inhibition of α-Synuclein Aggregation in Vitro by SUMOylation Is Conjugation Site- and SUMO Isoform-Selective. Biochemistry 2015, 54, 959–961. [DOI] [PubMed] [Google Scholar]
- 542.Lewis YE; Abeywardana T; Lin YH; Galesic A; Pratt MR, Synthesis of a Bis-thio-acetone (BTA) Analogue of the Lysine Isopeptide Bond and its Application to Investigate the Effects of Ubiquitination and SUMOylation on α-Synuclein Aggregation and Toxicity. ACS Chem. Biol 2016, 11, 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Buée L; Bussière T; Buée-Scherrer V; Delacourte A; Hof PR, Tau Protein Isoforms, Phosphorylation and Role in Neurodegenerative Disorders. Brain Res. Rev 2000, 33, 95–130. [DOI] [PubMed] [Google Scholar]
- 544.Lee VMY; Goedert M; Trojanowski JQ, Neurodegenerative Tauopathies. Annu. Rev. Neurosci 2001, 24, 1121–1159. [DOI] [PubMed] [Google Scholar]
- 545.Prusiner SB; Scott MR; DeArmond SJ; Cohen FE, Prion Protein Biology. Cell 1998, 93, 337–348. [DOI] [PubMed] [Google Scholar]
- 546.Horwich AL; Weissman JS, Deadly Conformations—Protein Misfolding in Prion Disease. Cell 1997, 89, 499–510. [DOI] [PubMed] [Google Scholar]
- 547.Kajihara Y; Kanemitsu Y; Nishihara M; Okamoto R; Izumi M, Efficient Synthesis of Polypeptide-α-Thioester by the Method Combining Polypeptide Expression and Chemical Activation for the Semi-Synthesis of Interferon-γ Having Oligosaccharides. J. Pept. Sci 2014, 20, 958–963. [DOI] [PubMed] [Google Scholar]
- 548.MacDonald ME; Gines S; Gusella JF; Wheeler VC, Huntington’s Disease. Neuromolecular Med. 2003, 4, 7–20. [DOI] [PubMed] [Google Scholar]
- 549.McColgan P; Tabrizi SJ, Huntington’s Disease: A Clinical Review. Eur. J. Neurol 2018, 25, 24–34. [DOI] [PubMed] [Google Scholar]
- 550.Uhlén M; Fagerberg L; Hallström BM; Lindskog C; Oksvold P; Mardinoglu A; Sivertsson Å; Kampf C; Sjöstedt E; Asplund A, et al. , Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 551.Almén MS; Nordström KJV; Fredriksson R; Schiöth HB, Mapping the Human Membrane Proteome: a Majority of the Human Membrane Proteins can be Classified According to Function and Evolutionary Origin. BMC Biol. 2009, 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.The Human Protein Atlas https://www.proteinatlas.org/ (accessed November 4, 2019).
- 553.Overington JP; Al-Lazikani B; Hopkins AL, How Many Drug Targets Are There? Nat. Rev. Drug. Discov 2006, 5, 993–996. [DOI] [PubMed] [Google Scholar]
- 554.Yin H; Flynn AD, Drugging Membrane Protein Interactions. Annu. Rev. Biomed. Eng 2016, 18, 51–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Berman HM; Westbrook J; Feng Z; Gilliland G; Bhat TN; Weissig H; Shindyalov IN; Bourne PE, The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Doyle DA; Cabral JM; Pfuetzner RA; Kuo A; Gulbis JM; Cohen SL; Chait BT; MacKinnon R, The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science 1998, 280, 69–77. [DOI] [PubMed] [Google Scholar]
- 557.Valiyaveetil FI; Leonetti M; Muir TW; MacKinnon R, Ion Selectivity in a Semisynthetic K+ Channel Locked in the Conductive Conformation. Science 2006, 314, 1004–1007. [DOI] [PubMed] [Google Scholar]
- 558.Kratochvil HT; Maj M; Matulef K; Annen AW; Ostmeyer J; Perozo E; Roux B; Valiyaveetil FI; Zanni MT, Probing the Effects of Gating on the Ion Occupancy of the K+ Channel Selectivity Filter Using Two-Dimensional Infrared Spectroscopy. J. Am. Chem. Soc 2017, 139, 8837–8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Pitcher JA; Freedman NJ; Lefkowitz RJ, G Protein–Coupled Receptor Kinases. Annu. Rev. Biochem 1998, 67, 653–692. [DOI] [PubMed] [Google Scholar]
- 560.Rosenbaum DM; Rasmussen SGF; Kobilka BK, The Structure and Function of G-Protein-Coupled Receptors. Nature 2009, 459, 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Staus DP; Wingler LM; Choi M; Pani B; Manglik A; Kruse AC; Lefkowitz RJ, Sortase Ligation Enables Homogeneous GPCR Phosphorylation to Reveal Diversity in β-Arrestin Coupling. Proc. Natl. Acad. Sci. U.S.A 2018, 115, 3834–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Denisov IG; Sligar SG, Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev 2017, 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Shiraishi Y; Natsume M; Kofuku Y; Imai S; Nakata K; Mizukoshi T; Ueda T; Iwaï H; Shimada I, Phosphorylation-Induced Conformation of β2-Adrenoceptor Related to Arrestin Recruitment Revealed by NMR. Nat. Commun 2018, 9, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Dawson PE, Synthesis of Chemokines by Native Chemical Ligation. Methods Enzymol. 1997, 287, 34–45. [DOI] [PubMed] [Google Scholar]
- 565.Wilken J; Hoover D; Thompson DA; Barlow PN; McSparron H; Picard L; Wlodawer A; Lubkowski J; Kent SBH, Total Chemical Synthesis and High-Resolution Crystal Structure of the Potent Anti-HIV Protein AOP-RANTES. Chem. Biol 1999, 6, 43–51. [DOI] [PubMed] [Google Scholar]
- 566.Ott TR; Lio FM; Olshefski D; Liu XJ; Struthers RS; Ling N, Determinants of High-Affinity Binding and Receptor Activation in the N-Terminus of CCL-19 (MIP-3β). Biochemistry 2004, 43, 3670–3678. [DOI] [PubMed] [Google Scholar]
- 567.Grygiel TLR; Teplyakov A; Obmolova G; Stowell N; Holland R; Nemeth JF; Pomerantz SC; Kruszynski M; Gilliland GL, Synthesis by Native Chemical Ligation and Crystal Structure of Human CCL2. Pept. Sci 2010, 94, 350–359. [DOI] [PubMed] [Google Scholar]
- 568.Tsuji K; Shigenaga A; Sumikawa Y; Tanegashima K; Sato K; Aihara K; Hara T; Otaka A, Development of an Intein-Mediated split–Cas9 System for Gene Application of N–C- or C–N-Directed Sequential Native Chemical Ligation to the Preparation of CXCL14 Analogs and Their Biological Evaluation. Biorg. Med. Chem 2011, 19, 4014–4020. [DOI] [PubMed] [Google Scholar]
- 569.Naruse N; Ohkawachi K; Inokuma T; Shigenaga A; Otaka A, Resin-Bound Crypto-Thioester for Native Chemical Ligation. Org. Lett 2018, 20, 2449–2453. [DOI] [PubMed] [Google Scholar]
- 570.Paolini-Bertrand M; Cerini F; Martins E; Scurci I; Hartley O, Rapid and Low-Cost Multiplex Synthesis of Chemokine Analogs. J. Biol. Chem 2018, 293, 19092–19100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Marcaurelle LA; Mizoue LS; Wilken J; Oldham L; Kent SBH; Handel TM; Bertozzi CR, Chemical Synthesis of Lymphotactin: A Glycosylated Chemokine with a C-Terminal Mucin-Like Domain. Chem. Euro. J 2001, 7, 1129–1132. [DOI] [PubMed] [Google Scholar]
- 572.Brik A; Keinan E; Dawson PE, Protein Synthesis by Solid-Phase Chemical Ligation Using a Safety Catch Linker. J. Org. Chem 2000, 65, 3829–3835. [DOI] [PubMed] [Google Scholar]
- 573.Bellmann-Sickert K; Beck-Sickinger AG, Palmitoylated SDF1 α Shows Increased Resistance against Proteolytic Degradation in Liver Homogenates. ChemMedChem 2011, 6, 193–200. [DOI] [PubMed] [Google Scholar]
- 574.Richardson RM; Pridgen BC; Haribabu B; Ali H; Snyderman R, Differential Cross-Regulation of the Human Chemokine Receptors CXCR1 and CXCR2: Evidence for Time-Dependent Signal Generation. J. Biol. Chem 1998, 273, 23830–23836. [DOI] [PubMed] [Google Scholar]
- 575.Steinhagen M; Hoffmeister P-G; Nordsieck K; Hötzel R; Baumann L; Hacker MC; Schulz-Siegmund M; Beck-Sickinger AG, Matrix Metalloproteinase 9 (MMP-9) Mediated Release of MMP-9 Resistant Stromal Cell-Derived Factor 1α (SDF-1α) from Surface Modified Polymer Films. ACS Appl. Mater. Inter 2014, 6, 5891–5899. [DOI] [PubMed] [Google Scholar]
- 576.Tolbert TJ; Franke D; Wong C-H, A New Strategy for Glycoprotein Synthesis: Ligation of Synthetic Glycopeptides With Truncated Proteins Expressed in E. coli As TEV Protease Cleavable Fusion Protein. Biorg. Med. Chem 2005, 13, 909–915. [DOI] [PubMed] [Google Scholar]
- 577.Lee CL; Liu H; Wong CTT; Chow HY; Li X, Enabling N-to-C Ser/Thr Ligation for Convergent Protein Synthesis via Combining Chemical Ligation Approaches. J. Am. Chem. Soc 2016, 138, 10477–10484. [DOI] [PubMed] [Google Scholar]
- 578.Asahina Y; Komiya S; Ohagi A; Fujimoto R; Tamagaki H; Nakagawa K; Sato T; Akira S; Takao T; Ishii A, et al. , Chemical Synthesis of O-Glycosylated Human Interleukin-2 by the Reverse Polarity Protection Strategy. Angew. Chem. Int. Ed 2015, 54, 8226–8230. [DOI] [PubMed] [Google Scholar]
- 579.Kallen K-J; Grötzinger J; Lelièvre E; Vollmer P; Aasland D; Renné C; Müllberg J; zum Büschenfelde K-HM; Gascan H; Rose-John S, Receptor recognition sites of cytokines are organized as exchangeable modules. Transfer of the leukemia inhibitory factor receptor-binding site from ciliary neurotrophic factor to interleukin-6. J. Biol. Chem 1999, 274, 11859–11867. [DOI] [PubMed] [Google Scholar]
- 580.Miyake T; Kung CK; Goldwasser E, Purification of Human Erythropoietin. J. Biol. Chem 1977, 252, 5558–5564. [PubMed] [Google Scholar]
- 581.Kent SBH, Bringing the Science of Proteins into the Realm of Organic Chemistry: Total Chemical Synthesis of SEP (Synthetic Erythropoiesis Protein). Angew. Chem. Int. Ed 2013, 52, 11988–11996. [DOI] [PubMed] [Google Scholar]
- 582.Dordal MS; Wang FF; Goldwasser E, The Role of Carbohydrate in Erythropoietin Action. Endocrinology 1985, 116, 2293–2299. [DOI] [PubMed] [Google Scholar]
- 583.Wang P; Dong S; Brailsford JA; Iyer K; Townsend SD; Zhang Q; Hendrickson RC; Shieh J; Moore MAS; Danishefsky SJ, At Last: Erythropoietin as a Single Glycoform. Angew. Chem. Int. Ed 2012, 51, 11576–11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Wang P; Dong S; Shieh J-H; Peguero E; Hendrickson R; Moore MAS; Danishefsky SJ, Erythropoietin Derived by Chemical Synthesis. Science 2013, 342, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Liu S; Pentelute BL; Kent SBH, Convergent Chemical Synthesis of [Lysine24, 38, 83] Human Erythropoietin. Angew. Chem. Int. Ed 2012, 124, 1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Chen S-Y; Cressman S; Mao F; Shao H; Low DW; Beilan HS; Cagle EN; Carnevali M; Gueriguian V; Keogh PJ, et al. , Synthetic Erythropoietic Proteins: Tuning Biological Performance by Site-Specific Polymer Attachment. Chem. Biol 2005, 12, 371–383. [DOI] [PubMed] [Google Scholar]
- 587.Murakami M; Kiuchi T; Nishihara M; Tezuka K; Okamoto R; Izumi M; Kajihara Y, Chemical Synthesis of Erythropoietin Glycoforms for Insights Into the Relationship Between Glycosylation Pattern and Bioactivity. Sci. Adv 2016, 2, e1500678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Streichert K; Seitz C; Hoffmann E; Boos I; Jelkmann W; Brunner T; Unverzagt C; Rubini M, Synthesis of Erythropoietins Site-Specifically Conjugated with Complex-Type N-Glycans. ChemBioChem 2019, 20, 1914–1918. [DOI] [PubMed] [Google Scholar]
- 589.Good MC; Zalatan JG; Lim WA, Scaffold Proteins: Hubs for Controlling the Flow of Cellular Information. Science 2011, 332, 680–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Hofmann RM; Cotton GJ; Chang EJ; Vidal E; Veach D; Bornmann W; Muir TW, Fluorescent Monitoring of Kinase Activity in Real Time: Development of a Robust Fluorescence-Based Assay for Abl Tyrosine Kinase Activity. Bioorg. Med. Chem. Lett 2001, 11, 3091–3094. [DOI] [PubMed] [Google Scholar]
- 591.Muralidharan V; Dutta K; Cho J; Vila-Perello M; Raleigh DP; Cowburn D; Muir TW, Solution Structure and Folding Characteristics of the C-Terminal SH3 Domain of c-Crk-II. Biochemistry 2006, 45, 8874–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Cowburn D; Muir TW, Segmental Isotopic Labeling Using Expressed Protein Ligation. Methods Enzymol. 2001, 339, 41–54. [DOI] [PubMed] [Google Scholar]
- 593.Kennedy MB, Signal-Processing Machines at the Postsynaptic Density. Science 2000, 290, 750–754. [DOI] [PubMed] [Google Scholar]
- 594.Hafner A-S; Penn, Andrew C; Grillo-Bosch D; Retailleau N; Poujol C; Philippat A; Coussen F; Sainlos M; Opazo P; Choquet D, Lengthening of the Stargazin Cytoplasmic Tail Increases Synaptic Transmission by Promoting Interaction to Deeper Domains of PSD-95. Neuron 2015, 86, 475–489. [DOI] [PubMed] [Google Scholar]
- 595.Christensen NR; Čalyševa J; Fernandes EFA; Lüchow S; Clemmensen LS; Haugaard-Kedström LM; Strømgaard K, PDZ Domains as Drug Targets. Adv. Therap 2019, 0, 1800143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Brunsveld L; Kuhlmann J; Alexandrov K; Wittinghofer A; Goody RS; Waldmann H, Lipidated Ras and Rab Peptides and Proteins—Synthesis, Structure, and Function. Angew. Chem. Int. Ed 2006, 45, 6622–6646. [DOI] [PubMed] [Google Scholar]
- 597.Goody RS; M. P;,M; Wu Y-W, Mechanisms of Action of Rab Proteins, Key Regulators of Intracellular Vesicular Transport. Biol. Chem 2017, 398, 565–575. [DOI] [PubMed] [Google Scholar]
- 598.Shi Y; Massagué J, Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 2003, 113, 685–700. [DOI] [PubMed] [Google Scholar]
- 599.Massagué J, TGFβ in Cancer. Cell 2008, 134, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Huse M; Muir TW; Xu L; Chen Y-G; Kuriyan J; Massagué J, The TGFβ Receptor Activation Process: An Inhibitor- to Substrate-Binding Switch. Mol. Cell 2001, 8, 671–682. [DOI] [PubMed] [Google Scholar]
- 601.Gutte B; Merrifield RB, The Synthesis of Ribonuclease A. J. Biol. Chem 1971, 246, 1922–1941. [PubMed] [Google Scholar]
- 602.Gutte B; Merrifield RB, Total Synthesis of an Enzyme With Ribonuclease a Activity. J. Am. Chem. Soc 1969, 91, 501–502. [DOI] [PubMed] [Google Scholar]
- 603.Hirschmann R; Nutt RF; Veber DF; Vitali RA; Varga SL; Jacob TA; Holly FW; Denkewalter RG, Total Synthesis of an Enzyme. V. Preparation of Enzymatically Active Material. J. Am. Chem. Soc 1969, 91, 507–508. [DOI] [PubMed] [Google Scholar]
- 604.Raines RT, Ribonuclease A. Chem. Rev 1998, 98, 1045–1066. [DOI] [PubMed] [Google Scholar]
- 605.Nilsson BL; Hondal RJ; Soellner MB; Raines RT, Protein Assembly by Orthogonal Chemical Ligation Methods. J. Am. Chem. Soc 2003, 125, 5268–5269. [DOI] [PubMed] [Google Scholar]
- 606.Cohen P, Protein Kinases — the Major Drug Targets of the Twenty-First Century? Nat. Rev. Drug. Discov 2002, 1, 309–315. [DOI] [PubMed] [Google Scholar]
- 607.Al-Ali H; Ragan TJ; Gao X; Harris TK, Reconstitution of Modular PDK1 Functions on Trans-Splicing of the Regulatory PH and Catalytic Kinase Domains. Bioconjugate Chemistry 2007, 18, 1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Chen Z; Thomas SN; Bolduc DM; Jiang X; Zhang X; Wolberger C; Cole PA, Enzymatic Analysis of PTEN Ubiquitylation by WWP2 and NEDD4–1 E3 Ligases. Biochemistry 2016, 55, 3658–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Dancy BM; Cole PA, Protein Lysine Acetylation by p300/CBP. Chem. Rev 2015, 115, 2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Dancy BM; Crump NT; Peterson DJ; Mukherjee C; Bowers EM; Ahn Y-H; Yoshida M; Zhang J; Mahadevan LC; Meyers DJ, et al. , Live-Cell Studies of p300/CBP Histone Acetyltransferase Activity and Inhibition. ChemBioChem 2012, 13, 2113–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Zucconi BE; Luef B; Xu W; Henry RA; Nodelman IM; Bowman GD; Andrews AJ; Cole PA, Modulation of p300/CBP Acetylation of Nucleosomes by Bromodomain Ligand I-CBP112. Biochemistry 2016, 55, 3727–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Karanam B; Jiang L; Wang L; Kelleher NL; Cole PA, Kinetic and Mass Spectrometric Analysis of p300 Histone Acetyltransferase Domain Autoacetylation. J. Biol. Chem 2006, 281, 40292–40301. [DOI] [PubMed] [Google Scholar]
- 613.Chen Z; Cole PA, Synthetic Approaches to Protein Phosphorylation. Curr. Opin. Chem. Biol 2015, 28, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Deng F-K; Zhang L; Wang Y-T; Schneewind O; Kent SBH, Total Chemical Synthesis of the Enzyme Sortase AΔN59 with Full Catalytic Activity. Angew. Chem. Int. Ed 2014, 53, 4662–4666. [DOI] [PubMed] [Google Scholar]
- 615.Schwarzer D; Ludwig C; Thiel IV; Mootz HD, Probing Intein-Catalyzed Thioester Formation by Unnatural Amino Acid Substitutions in the Active Site. Biochemistry 2012, 51, 233–242. [DOI] [PubMed] [Google Scholar]
- 616.Packer MS; Liu DR, Methods for the Directed Evolution of Proteins. Nat. Rev. Genet 2015, 16, 379–394. [DOI] [PubMed] [Google Scholar]
- 617.Romero PA; Arnold FH, Exploring Protein Fitness Landscapes by Directed Evolution. Nat. Rev. Mol. Cell Biol 2009, 10, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Bruce VJ; McNaughton BR, Inside Job: Methods for Delivering Proteins to the Interior of Mammalian Cells. Cell Chem. Biol 2017, 24, 924–934. [DOI] [PubMed] [Google Scholar]
- 619.Zhang Y; Røise JJ; Lee K; Li J; Murthy N, Recent Developments in Intracellular Protein Delivery. Curr. Opin. Biotechnol 2018, 52, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Stewart MP; Langer R; Jensen KF, Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev 2018, 118, 7409–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]