Abstract
This brief report serves as an introduction to a supplement of the Journal of Infectious Diseases entitled “Next-Generation Sequencing (NGS) Technologies to Advance Global Infectious Disease Research.” We briefly discuss the history of NGS technologies and describe how the techniques developed during the past 40 years have impacted our understanding of infectious diseases. Our focus is on the application of NGS in the context of pathogen genomics. Beyond obvious clinical and public health applications, we also discuss the challenges that still remain within this rapidly evolving field.
Keywords: infectious diseases, next-generation sequencing, pathogen genomics
Deoxyribonucleic acid (DNA) was first discovered and isolated in 1869 by Friedrich Miescher [1]. Nevertheless, the significance of this discovery remained unnoticed for decades, because it was generally believed that proteins were the molecules that held the genetic code of life. This all changed in 1944, when DNA was shown to encode hereditary properties, initiating a search to decode the information carried within it [2]. In 1953 came the discovery of the structure of DNA [3], and, since then, we have witnessed an explosion in technological advances that have facilitated both the manipulation and reading of nucleic acid sequences. These include tools such as polymerase chain reaction, molecular cloning, Sanger sequencing, and, more recently, CRISPR/Cas editing and next-generation sequencing (NGS). These techniques, taken together, have enabled major discoveries and breakthroughs in medicine and public health.
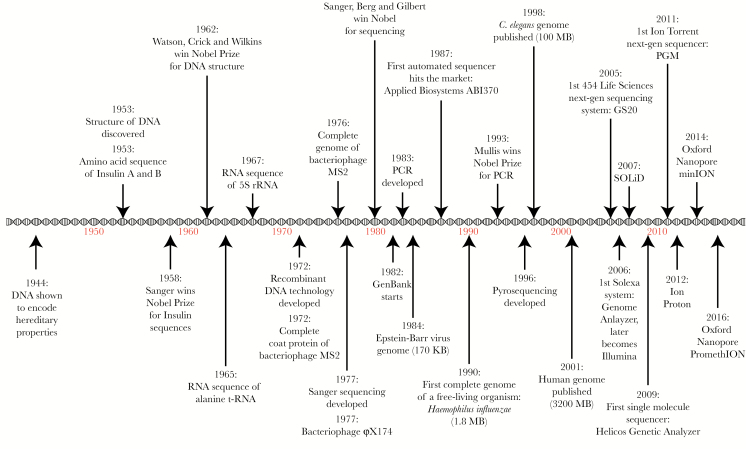
The rapid development of sequencing approaches (Figure 1), together with concomitant innovations in bioinformatics methods and algorithms, have ushered in an era of fast and relatively inexpensive sequencing and big data analysis. The first semi-automated platform for DNA sequencing was reported in 1986 [4], only 10 years after the development of the Sanger method [5] (first-generation sequencing). A year later, in 1987, the first completely automated sequencer hit the market [6]. By 1996, pyrosequencing was reported as the first-ever method for massively parallel DNA sequencing [7] (second-generation sequencing, or NGS). The first commercially available NGS platform arrived in 2005, followed by exponential improvements in sequencing speed and accuracy, and rapid drops in sequencing cost and platform size. As a result, NGS is now more affordable and widely distributed than ever, revolutionizing the way we explore biological questions and enabling applications that were previously unfathomable. Today, we have portable NGS platforms that fit on the palm of the hand and can be run on batteries in remote locations away from the controlled environment of a traditional research laboratory [8]. We are also now entering the era of third-generation sequencing, where additional technological advances facilitate sequencing of extremely long fragments of DNA, several kilobases in length [9].
Figure 1.
Timeline of scientific discoveries and technological advances that have enabled the rapid development of sequencing approaches. DNA, deoxyribonucleic acid; PCR, polymerase chain reaction; rRNA, ribosomal ribonucleic acid.
The developments of the past 40 years have had a very significant positive impact on our understanding of infectious diseases. Next-generation sequencing has been used to study a wide range of medically important viruses, bacteria, parasites, fungi, and other pathogens. In this supplement of the Journal of Infectious Diseases, we present a compilation of reviews and original research articles that highlight a few exciting applications of NGS in the context of pathogen genomics. Although these articles cover just a fraction of the pathogens and pathogen-induced diseases that have been studied by NGS, they complement each other by collectively touching on the major considerations pertinent in moving NGS from the bench to the bedside, from the laboratory to the field, from single pathogen genome characterization to an understanding of disease epidemiology and evolution, and from journal publications to public health and clinical decision making. The supplement has received input from NGS scientists, developers, users, and other stakeholders. Authors include basic and applied scientists, bioinformaticians, evolutionary biologists, clinical microbiologists, physicians, epidemiologists, and those in the public health workforce. Therefore, their perspectives and conclusions encompass individual patient care, laboratory research, local, national and international public health control, and global epidemic response.
The pathogens and infections covered herein include medically important respiratory RNA and DNA viruses and bacteria, enteric viruses and bacteria, cosmopolitan and tropical parasites, mosquito-borne viruses, prosthetic joint infections, undifferentiated meningoencephalitis, and pandemic agents. The scope of studies ranges from the use of NGS to rapidly identify pathogens directly from uncultured clinical and surveillance samples, to understanding the functional and epidemiological patterns of antimicrobial resistance, and to dissecting host genomic responses to infections from a systems biology perspective. In many cases, these are directly relevant to the design and evaluation of vaccines, therapeutics, infection control, and a range of nonmedical public health countermeasures for epidemics. From a technical standpoint, the supplement discusses the application of NGS techniques such as shotgun metagenomics, RNAseq, ribosome profiling, targeted sequencing, and minor variant reconstruction, as well as a broad suite of post-pipeline analyses that unifies various combinations of genomic, clinical, epidemiological, and ecological data. When applied to specific pathogen research, these may require development of additional ad hoc technical solutions or improvements at various levels, including the experimental, sequencing, and bioinformatics realms. As such, the supplement constitutes a platform from which the complexities of these techniques, in regard to their application to pathogen research, can be put forth for analysis, discussion, and improvement.
Beyond the clinical and public health applications, scientific advances, and technical updates described, much of the supplement examines the challenges that still prevent NGS from achieving its full translational impact on patient care and public health. These challenges remain considerable. Among what we estimate to be just a partial list, we have identified the following: the technical demands of conducting NGS-enabled research in public health laboratories around the world; the importance of, and increased need for, training and quality-control mechanisms to conducting, analyzing, and interpreting NGS platform outputs; the complexity of clinical validation and regulatory requirements for microbial diagnostic metagenomic NGS technologies; the integration of pathogen NGS into existing clinical and public health frameworks; and the equitable and timely access to NGS data and analysis during outbreaks and epidemics. Yet, when possible, contributors also explore pathways toward overcoming many of these obstacles. Collectively, we remain optimistic about the current and future prospects for precision medicine and precision public health in this new era of pathogen NGS.
Notes
Disclaimer. The views expressed here are those of the authors and do not reflect the official policy or position of the Department of Defense, USUHS, the NIH, nor the US Government. Several authors are US Government employees or military service members. This work was prepared as part of their official duties. Title 17 USC. § 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 USC. §101 defines US Government work as prepared by military service members or employees of the US Government during official duties.
Financial support. M. L. L. is funded by Pontificia Universidad Católica del Perú and the National Institute of Allergy and Infectious Diseases.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Dahm R. Friedrich Miescher and the discovery of DNA. Dev Biol 2005; 278:274–88. [DOI] [PubMed] [Google Scholar]
- 2. Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus Type III. J Exp Med 1944; 79:137–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watson JD, Crick FH. The structure of DNA. Cold Spring Harb Symp Quant Biol 1953; 18:123–31. [DOI] [PubMed] [Google Scholar]
- 4. Smith LM, Sanders JZ, Kaiser RJ, et al. . Fluorescence detection in automated DNA sequence analysis. Nature 1986; 321:674–9. [DOI] [PubMed] [Google Scholar]
- 5. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977; 74:5463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prober JM, Trainor GL, Dam RJ, et al. . A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science 1987; 238:336–41. [DOI] [PubMed] [Google Scholar]
- 7. Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem 1996; 242:84–9. [DOI] [PubMed] [Google Scholar]
- 8. Quick J, Loman NJ, Duraffour S, et al. . Real-time, portable genome sequencing for Ebola surveillance. Nature 2016; 530:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Batty EM, Chaemchuen S, Blacksell S, et al. . Long-read whole genome sequencing and comparative analysis of six strains of the human pathogen Orientia tsutsugamushi. PLoS Negl Trop Dis 2018; 12:e0006566. [DOI] [PMC free article] [PubMed] [Google Scholar]