Abstract
The recent pneumonia outbreak caused by a novel coronavirus (SARS-CoV-2) is posing a great threat to global public health. Therefore, rapid and accurate identification of pathogenic viruses plays a vital role in selecting appropriate treatments, saving people’s lives and preventing epidemics. It is important to establish a quick standard diagnostic test for the detection of the infectious disease (COVID-19) to prevent subsequent secondary spread. Polymerase chain reaction (PCR) is regarded as a gold standard test for the molecular diagnosis of viral and bacterial infections with high sensitivity and specificity. Isothermal nucleic acid amplification is considered to be a highly promising candidate method due to its fundamental advantage in quick procedure time at constant temperature without thermocycler operation. A variety of improved or new approaches also have been developed. This review summarizes the currently available detection methods for coronavirus nucleic acid. It is anticipated that this will assist researchers and clinicians in developing better techniques for timely and effective detection of coronavirus infection.
Keywords: Coronavirus, Nucleic acid detection, PCR-Based methods, Isothermal nucleic acid amplification-based methods, Microarray-based methods
Graphical abstract
The recent pneumonia outbreak caused by a novel coronavirus (SARS-CoV-2) in China, poses a great threat to global public health. Therefore, rapid and accurate identification of pathogenic viruses plays a vital role in selecting appropriate treatments, saving people’s lives and preventing epidemics. This review summarizes the currently available detection methods for coronavirus nucleic acid. It is anticipated that this will assist researchers and clinicians in developing better techniques for timely and effective detection of coronavirus infection.
Highlights
-
•
This review summarizes the currently available detection methods for coronavirus nucleic acid.
-
•
It will assist researchers in developing better techniques for timely and effective detection of coronavirus infection.
-
•
It will help the establishment of SARS-CoV-2 RNA detection method which is useful for the early diagnosis of COVID-19.
1. Introduction
The fight against infectious diseases caused by viruses remains a challenging and endless task despite the tremendous efforts and significant advances in public healthcare. Infectious diseases pose a significant risk to human health and have led to approximately one-fourth of deaths worldwide [1,2]. Coronaviruses belong to the family of Coronaviridae and may cause respiratory and neurological diseases [3]. Until now, six human coronaviruses (HCoVs) have been identified, namely, HCoV-229E, HCoV-HKU1, HCoV-OC43, HCoV-NL63, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) [4]. In particular, SARS-CoV and MERS-CoV once caused pandemic [5].
Recently, the emergence of novel coronavirus (SARS-CoV-2) has been challenging the global public health community to confront a novel infectious disease (coronavirus disease 2019, COVID-19). As of 22 February 2020, there were 77,816 confirmed cases of COVID-19 and 2012 deaths (https://www.worldometers.info/coronavirus/). In this epidemic, the detections of SARS-CoV-2 RNA were shown to be useful for the early diagnosis of COVID-19, which would be beneficial to controling the sources of infection and help patients to prevent the illness progression. The rapid and accurate detection of coronavirus is therefore becoming increasingly important.
With the advance in molecular biology technology, nucleic acid detection methods have developed rapidly and become a revolutionary technology for virus detection. Especially, the method based on polymerase chain reaction (PCR) is characterized by rapid detection, high sensitivity and specificity, which has been regarded as the "gold standard" for virus detection. Novel PCR-based methods also have been reported to enhance both the specificity and sensitivity of the assay. Besides, several molecular tests which employ non-PCR-based methods, such as isothermal nucleic acid amplification (loop mediated isothermal amplification (LAMP) and nucleic acid sequence-based amplification), were developed for the detection of coronavirus RNA. In this work, we reviewed various approaches currently available for detection of coronavirus. It is anticipated that this will assist researchers to develop rapid and accurate detection techniques.
2. PCR-based methods
PCR is an enzymatic method to produce numerous copies of a gene by separating the two strands of the DNA containing the gene segment, marking its location with a primer, and using a DNA polymerase to assemble a copy alongside each segment and continuously copy the copies. It is widely used to amplify minute quantities of biologic materials so as to provide adequate specimens for laboratory study. Owing to its large range of applications, high sensitivity and high sequence specificity, the PCR-based method has become a routine and reliable technique for detecting coronaviruses [6,7]. Generally, coronavirus RNA is transferred into cDNA by reverse transcription. Afterwards, the PCR is performed and followed by the detection of PCR product through specific detection methods or instruments. Among these, gel visualization and sequencing after PCR are the conventional methods for the detection of coronaviruses [8,9]. However, due to its time-consuming process and high cost, these methods are not commonly used in clinical samples.
Real-time reverse transcriptase-PCR (RT-PCR) detection is currently favored for the detection of coronavirus because of its advantages as a specific, and simple quantitative assay. Moreover, real-time RT-PCR is more sensitive than the conventional RT-PCR assay, which helps much for the diagnosis in early infection [10,11]. Therefore, the real-time RT-PCR assay still is a predominant method to be applied for the detection of all kinds of coronaviruses [12,13], including SARS-CoV-2 [14].
Even so, massive efforts are invested to improve the real-time RT-PCR assay. Since the RT-PCR methods are prone to contamination and require time-consuming sample handling and post-PCR analysis, van Elden et al. [15] described a TaqMan-based real-time RT-PCR that can easily be implemented in the routine diagnostic setting for the detection of HCoV. Moreover, to further improve the sensitivity, Yip et al. [16] designed a real-time quantitative RT-PCR assay for SARS-CoV with the use of 2 TaqMan probes, instead of 1 probe. This simple modification using dual TaqMan probes for quantification has wide applications in areas in which ultrasensitivity is critically required, with the SARS-CoV detection limit of 1 copy RNA per reaction.
In clinical detection, the lack of safe and stable external positive controls (EPC) could become a serious problem in the diagnosis of coronavirus and plenty of attention has been focused to address this problem. However, such problems could be well avoided in real-time RT-PCR assays where EPC is an important component. Yu et al. [17] developed a real-time RT-PCR assay in which the armored RNA was used as EPC to detect the SARS-CoV, with a detection limit of 10 copies/μL. Meanwhile, the rapidly mutating nature of coronaviruses highlights the need for accurate detection of genetically diverse coronaviruses. Therefore, to improve the ability to detect coronavirus precisely and reduce the risk of eliciting false-negative results caused by genome sequence variations, researchers have established multiplex real-time RT-PCR methods with favorable sensitivity for multitarget detection of coronavirus. Hadjinicolaou et al. [18] developed a real-time RT-PCR assay using mismatch-tolerant molecular beacons to distinguish between pathogenic and non-pathogenic strains. The assay incorporated four beacons, targeting four genes in addition to an internal positive control. It was validated using clinical samples, which exhibited target detection ability and specificity with detection limit of 5 copies per reaction.
3. Isothermal nucleic acid amplification-based methods
3.1. Regular LAMP-based methods
The LAMP is a novel isothermal nucleic acid amplification method with high efficiency. It is commonly used for the amplification of DNAs and RNAs, which exhibits great sensitivity and high specificity as a result of its exponential amplification feature and 6 different target sequences identified by 4 different primers simultaneously, respectively [19]. The LAMP assay is rapid and does not require expensive reagents or instruments. Therefore, the application of LAMP test might help to reduce the cost for detection of coronavirus [20]. Herein, a number of LAMP-based coronavirus detection methods have been developed and applied in clinical diagnosis.
Gel electrophoresis is commonly used to analyze the amplified products for an endpoint detection. Poon et al. [21] reported a simple LAMP assay for SARS diagnosis and demonstrated the feasibility of using this technology for detection of SARS-CoV. The ORF1b region of SARS-CoV was chosen for SARS diagnosis and amplified by LAMP reaction in the presence of 6 primers, then the amplified products were analyzed by gel electrophoresis. The detection rates and the sensitivity for SARS-CoV in the LAMP assay are similar to those of conventional PCR-based methods. Pyrc et al. [22] successfully applied LAMP to the detection of HCoV-NL63 which was conducted by agarose gel electrophoresis with favorable sensitivity and specificity in cell cultures and clinical specimens. Notably, the detection limit was found to be 1 copy of RNA template per reaction.
Amplification can be detected as the precipitation of magnesium pyrophosphate or fluorescence dye. This enables the methods to be carried out in real time by monitoring the turbidity of the pyrophosphate or fluorescence, which has effectively overcome the limitations of the endpoint detection [23]. A useful RT-LAMP assay for the diagnosis and epidemiologic surveillance of human MERS-CoV was developed in this way by Shirato et al. [24], which is capable of detecting as few as 3.4 copies of MERS-CoV RNA, and is highly specific, with no cross-reaction with other respiratory viruses. Thai et al. [25] developed a one-step single-tube accelerated real-time quantitative RT-LAMP assay monitored by real-time measurement of turbidity in a photometer for the early and rapid diagnosis of SARS-CoV. In clinical samples, the assay was found to be 100-fold more sensitive than conventional RT-PCR with a detection limit of 0.01 plaque forming units (PFU).
3.2. Sequence-specific LAMP-based methods
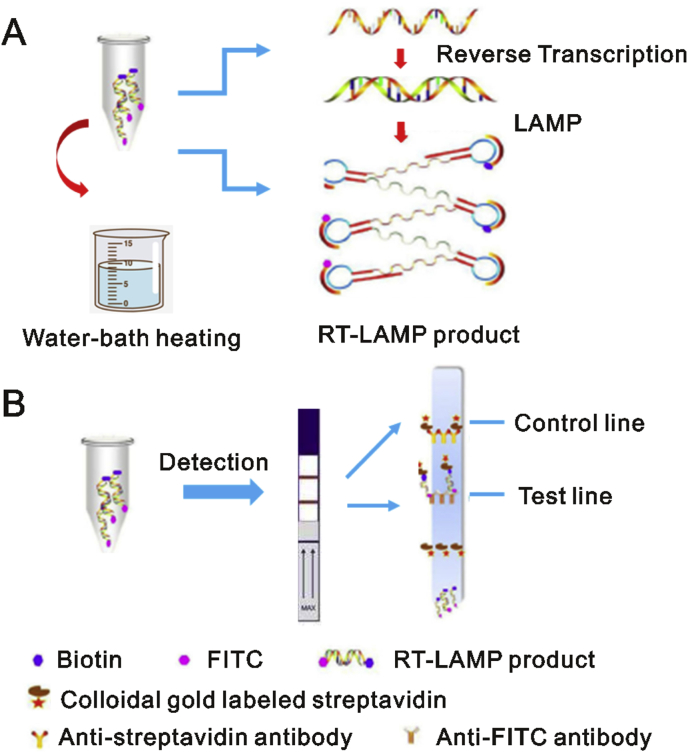
However, if the methods rely on nonspecific signal transduction schemes, such as the fluorescence dyes intercalation into any double-stranded DNA amplicons, or solution turbidity due to the release of pyrophosphates during polymerization, the possibility of unexpected signals derived from primer dimer or non-primer reactions cannot be excluded [26]. A sequence-specific and robust method for monitoring LAMP and other isothermal amplification reactions that can readily separate true signal from nonspecific noise would address this problem. Shirato et al. [27] improved the RT-LAMP assay by using a quenching probe (QProbe) to monitor signal, which has the same performance as the standard real-time RT-PCR assay in the detection of MERS-CoV. Additionally, Huang et al. [4] established a nucleic acid visualization technique that combines RT-LAMP and a vertical flow visualization strip (RT-LAMP-VF) to detect MERS-CoV. As illustrated in Fig. 1A, two loop primers (LF and LB) involved in isothermal amplification are labeled with fluorescein isothiocyanate (FITC) and biotin, respectively. After amplification, the amplicons labeled with biotin can bind to colloidal gold particles conjugated with streptavidin to form a complex which is subsequently captured by an anti-FITC antibody coated on the text line of the strip (Fig. 1B), thus presenting a colored line that is visible to the naked eye. In this instance, MERS-CoV RNA can be detected with a detection limit of 10 copies/μL.
Fig. 1.
Schematic illustration of the RT-LAMP-VF assay. (A) amplification reaction for RT-LAMP and (B) detection on visualization strip. The figure was reproduced with permission from Ref. [4].
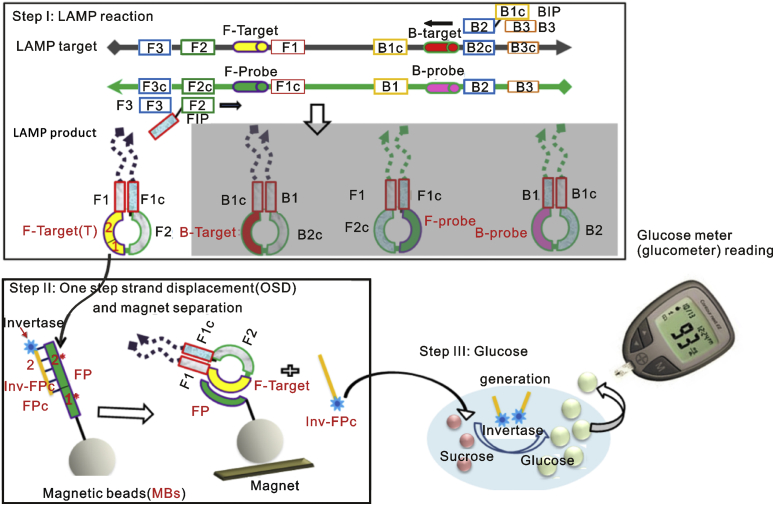
Scientists from Ellington’s group did plenty of work to both improve the specificity of LAMP detection and make readout simpler and more reliable. They replaced the intercalating dye typically used for monitoring in real-time fluorescence with a toehold-mediated strand exchange reaction termed one-step strand displacement (OSD) and applied it for real-time sequence-specific verification of LAMP amplicons [[28], [29], [30]]. The resulting assays could detect 0.02 to 0.2 PFU (5–50 PFU/mL) of MERS-CoV in infected cell culture supernatants within 30–50 min and did not cross-react with common human respiratory pathogens [31]. Other strand exchange signal transductions were developed to make LAMP reaction user-friendly. As shown in Fig. 2, by combining LAMP with a thermostable invertase, Du et al. can directly transduce MERS coronavirus templates into glucose signals, which can be easily read by a commercial glucometer, with a sensitivity as low as 20–100 copies/μL, equating to atto-molar (or low zepto-mole) [32]. The human chorionic gonadotropin (hCG) was also used as the signal as its analyte was already sensitively detected by commercial pregnancy test kits. The hCG was site-specifically conjugated to a DNA oligonucleotide, thereby allowing signal transduction via strand exchange into both capture (signal-off) and release (signal-on) of hCG in the LAMP based virus assay [33]. The incorporation of an engineered hCG reporter protein led to LAMP-to-hCG signal transduction on low-cost, commercially available pregnancy test strips. This assay reliably detected as few as 20 copies of virus templates in both human serum and saliva.
Fig. 2.
Scheme for adapting isothermal amplification to a glucometer. The figure was reproduced with permission from Ref. [32].
Meanwhile, LAMP shows optimal performance at around 65 °C, which always limits its applications. Cai et al. [34] developed a version of LAMP that used phosphorothioated primers (PS-LAMP) to enable more efficient hairpin formation and extension at the termini of growing concatamers, which therefore worked at much lower temperatures. The sensitivity and selectivity for amplicon detection with PS-LAMP at 40 °C were comparable with a regular LAMP reaction at 65 °C.
3.3. Rolling circle amplification-based methods
The rolling circle amplification (RCA) has attracted considerable attention in nucleic acid determination [35,36]. In isothermal conditions, RCA is capable of a 109-fold signal amplification of each circle within 90 min. An efficient assay for the detection of SARS-CoV by RCA was set up in both liquid and solid phases [37], and presented preliminary results on a small number of clinical respiratory specimens. The main advantage of RCA is that it can be performed under isothermal conditions with minimal reagents and avoid the generation of false-positive results, which is frequently encountered in PCR-based assays.
4. Microarray-based methods
The microarray is a detection method with rapid and high throughput. For this method, the coronavirus RNA will first produce cDNA labeled with specific probes through reverse transcription. Then these labeled cDNAs will be loaded into each well and hybridize with solid-phase oligonucleotides fixed on the microarray, followed by a series of washing steps to remove free DNAs. Finally, the coronavirus RNA can be detected by the detection of specific probes. Due to its superiority, the microarray assay has been widely used in the detection of coronavirus [38].
Shi et al. [39] designed a 60mer oligonucleotide microarray according to the sequence of TOR2 and successfully applied it to the detection of SARS coronavirus in clinical samples. However, considering the rapid mutation of SARS-CoV, Guo et al. [40] developed a microarray to detect 24 single nucleotide polymorphism (SNP) mutations among the spike (S) gene of SARS-CoV with 100% accuracy in sample detection. Since coronavirus may lead to a sudden outbreak, it is of great significance that diagnostic assays are able to detect a wide range of coronaviruses and are be deployable at or near the point of care (POC). Therefore, Luna et al. [41] designed a nonfluorescent low-cost, low-density oligonucleotide array for detecting the whole coronavirus genus with a sensitivity equal to that of individual real-time RT-PCR, and Hardick et al. [42] evaluated a novel, portable, and near-POC diagnostic platform based on the microarray chip, the Mobile Analysis Platform (MAP), which has a good performance in identifying virus and acceptable detection limit.
5. Newly developed methods
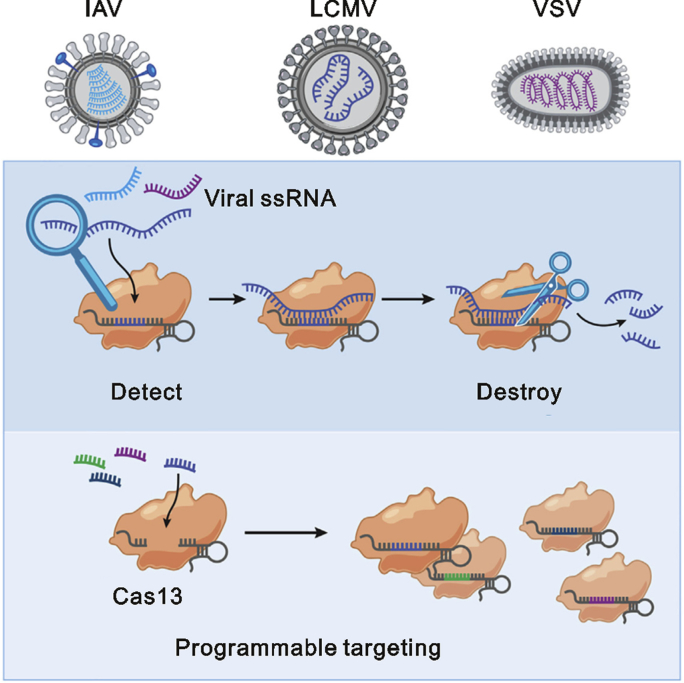
The RNA-targeting CRISPR associated enzyme Cas13 has recently been adapted for rapid and portable sensing of nucleic acids [43,44]. Zhang’s group demonstrated that Cas13 can be programmed to target and destroy the genomes of diverse mammalian single stranded RNA viruses (Fig. 3) [45]. They developed a platform termed SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) that combined isothermal preamplification with Cas13 to detect single molecules of RNA or DNA [46]. It can detect Dengue or Zika virus single-stranded RNA as well as mutations in patient liquid biopsy samples. Their lately protocol for COVID-19 entitled “A protocol for detection of COVID-19 using CRISPR diagnostics” has been reported on website (https://broad.io/sherlockprotocol), which may provide some reference points for researchers interested in further advancing this diagnostics system, highlighting its potential as a multiplexable, portable, rapid, and quantitative detection platform of nucleic acids.
Fig. 3.
Scheme for detection of RNA viruses using Cas13. The figure was reproduced with permission from Ref. [45].
6. Summary and prospect
Currently, the diagnosis of COVID-19 mainly relies on the detection of the coronavirus RNA. It is of great importance to select appropriate detection methods. However, each of the methods described above has its own unique advantages and inevitable disadvantages. PCR is widely used for virus identification with high sensitivity and specificity, but its analysis requires various equipment and educated analysts, which is only possibly accomplished by a well-established laboratory. LAMP is an ultrasensitive nucleic acid amplification method that can often detect small numbers of DNA or RNA templates within roughly an hour, but the requirement for high temperatures still limits its applicability. As for the microarray, the high cost inevitably limits its further application in the detection of coronaviruses. Therefore, it is still necessary to develop more effective and practical methods to overcome the shortcomings of the existing methods in the future.
To date, considerable efforts have been made to improve the detection of coronavirus and a variety of improved or new approaches have been developed. In practical applications, several methods are usually combined to avoid the drawbacks of using single method as much as possible. In short, with the rapid development of new technologies and methods, we believe that there will be more excellent and efficient detection methods developed in the future, which would provide scientists/clinicians with more choices. At the same time, only by balancing the advantages and disadvantages of various detection assays according to specific purposes can we obtain the most economical and optimal option.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We acknowledge financial support from the National Natural Science Foundation of China (Grant 81973281), the Fundamental Research Funds for the Central Universities (2019FZA7017), and Leading Talent of “Ten Thousand Plan”- National High-Level Talents Special Support Plan.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Contributor Information
Su Zeng, Email: zengsu@zju.edu.cn.
Sheng Cai, Email: caisheng@zju.edu.cn.
References
- 1.Morens D.M., Fauci A.S. Emerging infectious diseases: threats to human health and global stability. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim H., Park M., Hwang J. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4:1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- 3.Zhang N., Wang L., Deng X. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92:408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang P., Wang H., Cao Z. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9:1101. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang S.B., Xia S., Ying T.L. A novel coronavirus (2019-nCoV) causing pneumonia-associated respiratory syndrome. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-41020-40372-41424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balboni A., Gallina L., Palladini A. A real-time PCR assay for Bat SARS-like coronavirus detection and its application to Italian Greater Horseshoe Bat Faecal sample surveys. Sci. World J. 2012:989514. doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uhlenhaut C., Cohen J.I., Pavletic S. Use of a novel virus detection assay to identify coronavirus HKU1 in the lungs of a hematopoietic stem cell transplant recipient with fatal pneumonia. Transpl. Infect. Dis. 2012;14:79–85. doi: 10.1111/j.1399-3062.2011.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi D., Johnson G., Draker R. Comprehensive detection and identification of human coronaviruses, including the SARS-associated coronavirus, with a single RT-PCR assay. J. Virol. Methods. 2004;122:29–36. doi: 10.1016/j.jviromet.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setianingsih T.Y., Wiyatno A., Hartono T.S. Detection of multiple viral sequences in the respiratory tract samples of suspected Middle East respiratory syndrome coronavirus patients in Jakarta, Indonesia 2015-2016. Int. J. Infect. Dis. 2019;86:102–107. doi: 10.1016/j.ijid.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Z.Z., Zhang Y.N., He Z.X. A melting curve-based multiplex RT-qPCR assay for simultaneous detection of four human coronaviruses. Int. J. Mol. Sci. 2016;17:1880. doi: 10.3390/ijms17111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noh J.Y., Yoon S.W., Kim D.J. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017;162:1617–1623. doi: 10.1007/s00705-017-3281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman V.M., Eckerle I., Bleicker T. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:3–8. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 13.Lu X.Y., Whitaker B., Sakthivel S.K.K. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J. Clin. Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Elden L.J.R., van Loon A.M., van Alphen F. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J. Infect. Dis. 2004;189:652–657. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yip S.P., To S.S.T., Leung P.H.M. Use of dual TaqMan probes to increase the sensitivity of 1-step quantitative reverse transcription-PCR: application to the detection of SARS coronavirus. Clin. Chem. 2005;51:1885–1888. doi: 10.1373/clinchem.2005.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X.F., Pan J.C., Ye R. Preparation of armored RNA as a control for multiplex real-time reverse transcription-PCR detection of influenza virus and severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2008;46:837–841. doi: 10.1128/JCM.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadjinicolaou A.V., Farcas G.A., Demetriou V.L. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch. Virol. 2011;156:671–680. doi: 10.1007/s00705-010-0906-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notomi T., Okayama H., Masubuchi H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enosawa M., Kageyama S., Sawai K. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 2003;41:4359–4365. doi: 10.1128/JCM.41.9.4359-4365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poon L.L.M., Leung C.S.W., Tashiro M. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin. Chem. 2004;50:1050–1052. doi: 10.1373/clinchem.2004.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyrc K., Milewska A., Potempa J. Development of loop-mediated isothermal amplification assay for detection of human coronavirus-NL63. J. Virol. Methods. 2011;175:133–136. doi: 10.1016/j.jviromet.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori Y., Nagamine K., Tomita N. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 24.Shirato K., Yano T., Senba S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thai H.T.C., Le M.Q., Vuong C.D. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Njiru Z.K. Loop-mediated isothermal amplification technology: towards point of care diagnostics. Plos Neglect. Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirato K., Semba S., El-Kafrawy S.A. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J. Virol. Methods. 2018;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B.L., Chen X., Ellington A.D. Adapting enzyme-free DNA circuits to the detection of loop-mediated isothermal amplification reactions. Anal. Chem. 2012;84:8371–8377. doi: 10.1021/ac301944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y.S., Bhadra S., Li B.L. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem. 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y.S., Stacy A., Whiteley M. Amplicon competition enables end-point quantitation of nucleic acids following isothermal amplification. Chembiochem. 2017;18:1692–1695. doi: 10.1002/cbic.201700317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhadra S., Jiang Y.S., Kumar M.R. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PloS One. 2015;10 doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y., Hughes R.A., Bhadra S. A sweet spot for molecular diagnostics: coupling isothermal amplification and strand exchange circuits to glucometers. Sci. Rep. 2015;5:11039. doi: 10.1038/srep11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Y., Pothukuchy A., Gollihar J.D. Coupling sensitive nucleic acid amplification with commercial pregnancy test strips. Angew. Chem. Int. Ed. 2017;56:992–996. doi: 10.1002/anie.201609108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai S., Jung C., Bhadra S. Phosphorothioated primers lead to loop-mediated isothermal amplification at low temperatures. Anal. Chem. 2018;90:8290–8294. doi: 10.1021/acs.analchem.8b02062. [DOI] [PubMed] [Google Scholar]
- 35.Chapin S.C., Doyle P.S. Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal. Chem. 2011;83:7179–7185. doi: 10.1021/ac201618k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu M.C., Ye J.W., Yang D. Ultrasensitive detection of miRNA via one-step rolling circle-quantitative PCR (RC-qPCR) Anal. Chim. Acta. 2019;1077:208–215. doi: 10.1016/j.aca.2019.05.028. [DOI] [PubMed] [Google Scholar]
- 37.Wang W.K., Fang C.T., Chen H.L. Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J. Clin. Microbiol. 2005;43:962–965. doi: 10.1128/JCM.43.2.962-965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q., Li J., Deng Z.R. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi R., Ma W.L., Wu Q.H. Design and application of 60mer oligonucleotide microarray in SARS coronavirus detection. Chin. Sci. Bull. 2003;48:1165–1169. doi: 10.1007/BF03183928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X., Geng P., Wang Q. Development of a single nucleotide polymorphism DNA microarray for the detection and genotyping of the SARS coronavirus. J. Microbiol. Biotechnol. 2014;24:1445–1454. doi: 10.4014/jmb.1404.04024. [DOI] [PubMed] [Google Scholar]
- 41.Luna L.K.D., Heiser V., Regamey N. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J. Clin. Microbiol. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardick J., Metzgar D., Risen L. Initial performance evaluation of a spotted array Mobile Analysis Platform (MAP) for the detection of influenza A/B, RSV, and MERS coronavirus. Diagn. Microbiol. Infect. Dis. 2018;91:245–247. doi: 10.1016/j.diagmicrobio.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright A.V., Nunez J.K., Doudna J.A. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 44.Gootenberg J.S., Abudayyeh O.O., Lee J.W. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freije C.A., Myhrvold C., Boehm C.K. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gootenberg J.S., Abudayyeh O.O., Kellner M.J. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]