Highlights
-
•
Non-immunosuppressive derivatives inhibit CoV replication in vitro.
-
•
Coronavirus NL63 and 229E replication depends on CypA.
-
•
Coding non-synonymous SNP variants in the PPIA gene limit HCoV-229E replication.
Abstract
Replication of coronaviruses is inhibited in vitro by cyclosporin A, a well-known immunosuppressive drug which binds to cellular cyclophilins thus inactivating their enzymatic cis-trans peptidyl-prolyl isomerase function. Latter is required for proper folding of cellular proteins and of proteins of several viruses. Here, we summarize present knowledge on the role of cyclophilin A during coronavirus replication. We present data on the effect of cyclophilin A single nucleotide polymorphism mutants on the replication of human CoV-229E demonstrating the requirement of proper cyclophilin A function for virus propagation. Results define cellular cyclophilin A as a host target for inhibition of coronaviruses ranging from relatively mild common cold to highly pathogenic SARS-CoV and MERS-CoV viruses with the perspective of disclosing non-immunosuppressive cyclosporin A analogs to broadly inactivate the coronavirus family.
Current Opinion in Virology 2015, 14:56–61
This review comes from a themed issue on Engineering for viral resistance
Edited by Albrecht von Brunn
For a complete overview see the Issue and the Editorial
Available online 27th August 2015
http://dx.doi.org/10.1016/j.coviro.2015.08.004
1879-6257/© 2015 Elsevier B.V. All rights reserved.
Introduction
Coronaviruses (CoVs) infect a variety of mammalian species including bats, mice, cats, birds and humans causing infection of respiratory and gastrointestinal tracts and the central nervous system [1]. CoVs are enveloped viruses containing the largest known single-stranded RNA genomes (25–32 kb) with positive-sense orientation. They are divided into four genera: Alpha- (HCoV-229E), Beta- (SARS-CoV: lineage B; MERS-CoV: lineage C), Gamma- and Deltacoronavirus [2]. The six human CoVs, namely HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, SARS-CoV and MERS-CoV mainly target the respiratory tract. 15–30% of common colds are caused by HCoVs (229E, OC43, NL63, HKU1) with mostly seasonal occurrence. Whereas 229E and OC43 are known since the mid-1960s, SARS-CoV appeared first in China causing a worldwide outbreak with 8098 cases and 774 deaths in 2002/03 and with enormous socio-economic impact [3]. Arising interest in CoVs led to the discovery of NL63 in 2004 [4] and HKU1 in 2005 [5]. MERS was identified in 2012 in Saudi Arabia. By 31 May 2015 MERS-CoV infections rose to 1154 cases with 431 deaths [6]. In May 2015 a new outbreak occurred in South Korea with over 120 reported cases, 10 deaths as of 11 June [7], and over 2300 individuals placed under quarantine, making it the largest outbreak outside Saudi Arabia.
Until now no effective drug treatment is available neither against the common cold nor the highly pathogenic CoVs. Development of antivirals has concentrated on the development of protease [8, 9] and helicase inhibitors [10, 11]. Great efforts have been made to discover anti-MERS agents by screening defined drug libraries [12, 13, 14]. Although, CoVs display some proofreading activities during replication viral targets are usually prone to develop resistance mutations rather quickly. Therefore, defining cellular co-factors required for viral replication as targets is rather intriguing. Screening 16,671 diverse compounds for anti-229E activity Lundin et al. have identified an inhibitor (K22) specifically targeting membrane-bound coronaviral RNA synthesis at an early step of viral replication [15]. Using unbiased high-throughput protein–protein interaction screening methods we identified cyclophilins as binding factors for CoV proteins and its inhibitor cyclosporin A (CsA) as broad-range anti-coronaviral agent [16••]. CsA binding and inactivation of CypA as cellular co-factor for virus propagation is summarized in the accompanying article in the October issue of Current Opinion in Virology by von Hahn and Ciesek [17]. Here we summarize the inhibitory effect of CsA and non-immunosuppressive derivatives thereof on CypA function during CoV replication. We further describe the effect of the genetic deficiency and of individual CypA SNP mutants on the replication of HCoV-229E indicating the requirement of correctly folded enzymatic groove of CypA.
Cyclophilins and inhibitors
Cyclophilins and FKBPs are members of two ubiquitously distributed PPIase families, collectively called immunophilins [18••]. They are important for a number of cellular processes, for example, protein folding, maturation, trafficking, signal transduction, cell differentiation, apoptosis and infections. CypA and CypB were recognized in 1993 by Y2H techniques to specifically bind to the HIV-1 Gag polyproteins Pr55gag and to capsid p24, but only CypA was demonstrated to be specifically incorporated into HIV-1 virions [reviewed by 19••]. Binding of the amino-terminal domain of HIV-1 capsid to the active groove of CypA was demonstrated by crystal structure [20] and by mutational analysis [21, 22]. Proline-containing peptide substrates bind to this hydrophobic pocket conferring them enzymatic peptidyl-prolyl cis/trans isomerase (PPIases, EC number 5.2.1.8) activity. The isomerase function of Cyp was identified already in 1989 [23•]. It was first shown for HIV-1 that CsA also binds to the groove thus interfering with proper capsid formation and virus replication. In the case of Hepatitis C Virus (HCV) the involvement of Cyps was shown by several groups. Initially conflicting results on which CypA or B was supporting HCV replication could be clarified in favor of CypA [24, 25, 26].
A very important, but from PPIase activity completely independent feature of CsA binding to CypA is the formation of a tri-molecular complex with the cellular phosphatase Calcineurin (Cn). This is a natural coincidence with far-reaching consequences on the immune system: Cn normally dephosphorylates the important immunologic transcription factor NFAT (Nuclear Factor of Activated T-cells), which can then translocate to the nucleus and act as a key regulator of T-cell development and Interleukin-2 production [27•]. CypA/CsA/Cn complexes prevent NFAT dephosphorylation and translocation to the nucleus thus leading to the suppression of the immune system. CsA as a 11mer cyclic peptide displays a ‘surface’ for binding to the PPIase groove of CypA [28] and one for complexing with Cn (Figure 1 ). Intensive efforts were made to separate the PPIase blocking from the immunosuppressive functions of CsA. Modifying side chains of the CsA molecule allowed the development of non-immunosuppressive analogs NIM811 [29, 30], Alisporivir [ALV, Debio-025] [31], SCY-635 [32], sangliferins [33] and a series of newly synthesized CsA position 1-modified compounds [34, 35, 36]. Alisporivir has experienced substantial clinical testing and safety database development with more than 2000 patients treated for up to 48 weeks. NIM811 and SCY-635 have been administered in a very small number (<50 patients) only in short proof-of-concept trials.
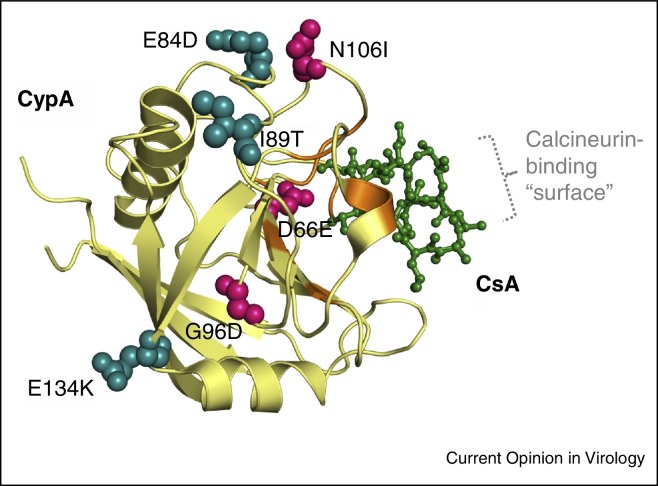
Figure 1.
Crystal structure of human CypA complexed with CsA (1CWA, pdb database, modified) and with coding non-synonymous PPIA gene SNPs. SNPs with accompanying amino acid exchanges introduced in Huh-7.5 PPIA manipulated cell lines [46] are Rs61747111 (D66E), rs17850033 (I89T), rs1059983 (E84D), rs11547706 (G96D), rs17850166 (N106I), rs9769523 (E134K). CypA is shown as a β-sheet structure with the SNP amino acids strongly, or only slightly affecting CoV replication in ball format, colored red and blue, respectively. Active site residues are depicted in orange. CsA binding to the PPIase active pocket of CypA is shown in green. The calcineurin-binding surface of CypA/CsA complex is indicated schematically.
Immunophilins and CoV replication
A first hint on the possible involvement of a cyclophilin, namely CypA, in SARS-CoV replication came from an educated guess finding, which demonstrated interaction of the SARS-CoV nucleocapsid (N) protein with CypA by surface plasmon resonance biosensor technology paralleling the binding of HIV-1 gag to CypA [37]. This finding was supported by a proteomics study which identified CypA as one of a number of cellular proteins incorporated into purified SARS-CoV particles by spectrometric profiling [38]. Inhibitory effects of CsA on CoV replication was reported by several laboratories: (1) using unbiased high throughput Y2H protein–protein interaction screening methods we have noticed the binding of several cyclophilins to SARS-CoV nsp1, and CsA as pan-CoV inhibitor including SARS-CoV, HCoV-229E/-NL63, Feline CoV (FCoV) serotypes I and II [strains Black and 791146], Transmissible Gastroenteritis Virus (TGEV PUR46) and Infectious Bronchitis Virus (IBV Beaudette) [16••]. In a follow-up study it could be demonstrated that, at least for replication of HCoV-NL63 CypA, not CypB is the cyclophilin required for virus replication [36]. As also the immunophilins FKBP1A and FKBP1B showed up as nsp1 interaction partners in the Y2H virus–host protein interaction screens mentioned above, SARS-CoV, HCoV-NL63 and HCoV-229E-GFP/-LUC were tested for sensitivity to FK506. The drug could effectively inhibit replication of these viruses, and HCoV-NL63 did not replicate in FKBP1A/B knockdown CaCo2 cell lines [39]. Thus, PPIase activities of CypA and FKBPs seem to be required for CoV replication but they do not substitute for each other. The current interpretation is that the two classes of PPIases act on different viral proteins. (2) Replication inhibition by CsA up to 4 logs was also shown for GFP-expressing SARS-CoV, 229E and MHV [Mouse Hepatitis Virus] [40] as well as for MERS-CoV [41]. In these studies a specific cyclophilin could not be attributed to SARS-CoV replication. (3) Replication of the highly pathogenic cat CoV Feline Infectious Peritonitis Virus (FIPV) was shown to be sensitive to CsA, but not to FK506 [42]. Rather interestingly, when performing the CsA inhibition experiments with animal CoVs [16••] we also did not find FK506 inhibition of the two FCoV serotypes (Black/791146; unpublished and Heinz-Jürgen Thiel, personal communication), and of TGEV PUR46 and IBV Beaudette strains (unpublished; Christel Schwegmann-Weßels/Georg Herrler, personal communication). (4) Cyclophilin D was shown to play a central role in HCoV-OC43-induced neuronal programmed cell death by CypD knockdown and CsA inhibition. Here, CsA in combination with CypD acted as an inhibitor of the mitochondrial permeabilization transition pore [43].
PPIA gene knockout and coding non-synonymous SNP variants limit HCoV-229E replication
For HCoV-NL63 we have recently shown that replication in CaCo2 cells depends on cyclophilin A (CypA) but not CypB [36]. Human hepatocellular carcinoma cells (Huh-7) and derivatives support the replication of a number of viruses including HCV [44] and HCoV-229E [45]. We utilized Huh-7.5 CypA variant cell lines originally constructed for the analysis of HCV infection [46]. The variants used were Huh-7.5KD-PPIA (CypA knockdowns), Huh-7.5sh-Ctr (non-target controls), Huh-7.5-CypA-KD + wtCypA (CypA knockdowns, reconstituted with CypA) and Huh-7.5-CypA mutants encoding individual, non-synonymous PPIA gene SNPs with amino acid exchanges (D66E, N106I, G96D, E134K, E84D, or I89T; see also Figure 1).
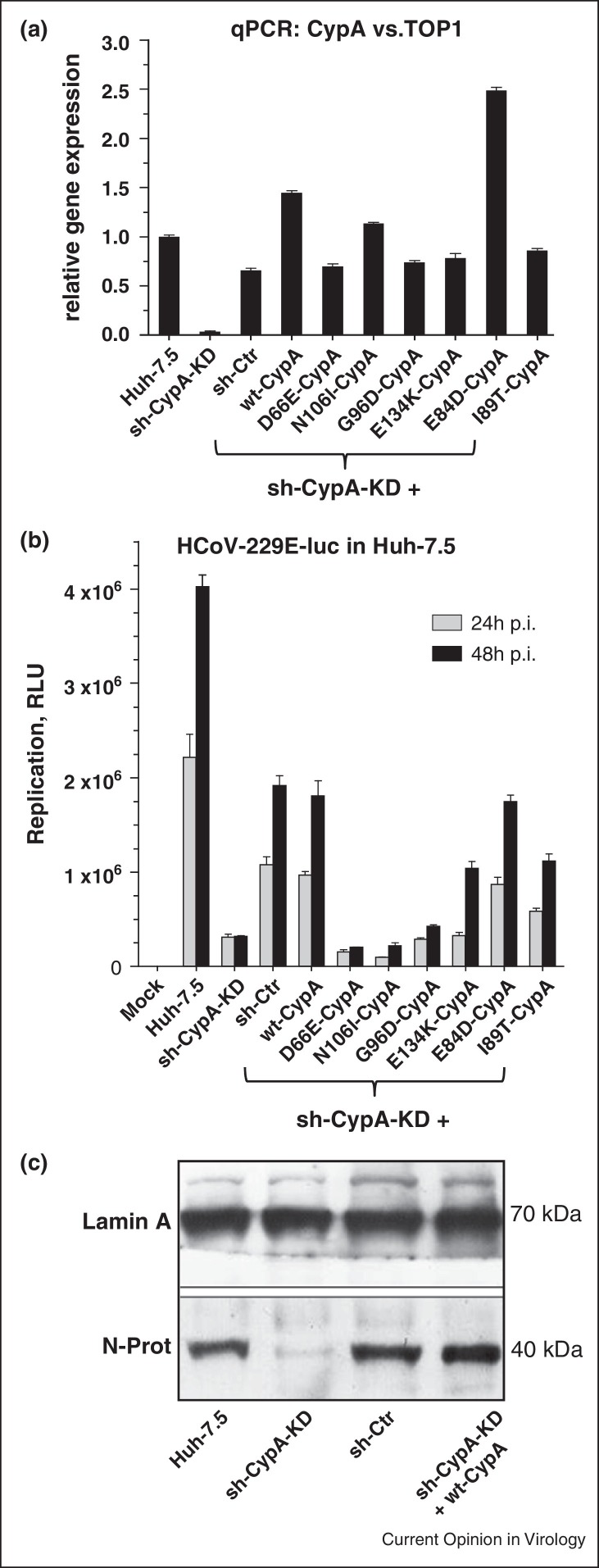
For HCV viral growth behavior was shown to depend on protein stability of the individual SNP variants. Although mRNA levels determined by qPCR analysis were comparable in Huh-7.5wt and CypA variants protein levels of D66E were reduced and G96D and N106I appeared nearly absent [44]. For the infection experiments with HCoV-229E relative CypA mRNA expression levels were analyzed in the various cell lines by qPCR in relation to the house keeping gene hTOP1 with the level in Huh-7.5 set to 1.0 (Figure 2a). CypA mRNA levels in the non-target control (sh-Ctr) and in the D66E, G96D, E134K and I89T variants were slightly decreased. Levels in E84D and in the wtCypA-reconstituted Huh-7.5PPIA-KD (sh-CypA-KD) cells were increased. In Huh-7.5KD-PPIA (sh-CypA-KD) cells PPIA mRNA was close to background confirming that the knockdown was very efficient although not complete.
Figure 2.
Replication analysis of HCoV-229E-LUC in Huh-7.5-KD and single SNP variant mutants. (a) qPCR analysis of CypA expression in sh-CypA-KD, non-target control (sh-Ctr) and with wtCypA reconstituted sh-CypA-KD cells, as well as sh-CypA-KD cells reconstituted with CypA SNP variants carrying amino acid exchanges at D66E, N106I, G96D, E134K, E84D and I89T. Huh-7.5 cells and subclones were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (both Gibco LifeTechnologies), l-glutamine, non-essential amino acids, penicillin, and streptomycin. Cells harboring small hairpin RNA (shRNA) constructs were kept in the presence of blasticidin (5 μg/mL). G418 (750 μg/mL) was additionally added to cells carrying pWPI-encoded CypA variants [46]. hTOP1 was used in qPCR to standardize cyclophilin expression. (b) Replication was measured by determining Renilla Luciferase activity in cell extracts after 24 and 48 h p.I. Values are given as relative light units (RLU). (c) HCoV-229E-LUC N-protein expression as replication measure in Huh-7.5, sh-CypA-KD, non-target control (sh-Ctr) and with wtCypA reconstituted Huh-7.5 cells. qPCR/Western blot methods and materials used are the same as described in [36].
In order to assess growth properties of HCoV-229E-LUC the various cell lines were infected with 0.1 MOI of the virus and grown for 24 and 48 h (Figure 2b). As judged by Luciferase protein expression levels (RLU) virus grew best in Huh-7.5 cells with an about two-fold increase over the sh-CypA-KD + wt-CypA and the non-target sh-Ctr cells. In sh-CypA-KD cells virus replication was decreased by factors of 10 and 20 at 24 h and 48 h timepoints, respectively. In case of the E134K, E84D and I89T mutant virus growth was slightly decreased as compared to the sh-CypA-KD + wt-CypA and the sh-Ctr cells. Very interestingly, in D66E, N106I and G96D, 229E replication was almost completely abolished as it was in the case of sh-CypA-KD cells. To assess viral replication at the level of essential N protein production in HCoV-229E-LUC virus-infected cells (MOI 0.01) Western blot analysis of viral N protein two days p.I. was performed (Figure 2c). N protein levels were similar in Huh-7.5, sh-Ctr and sh-CypA-KD + wt-CypA reconstituted cells. In sh-CypA-KD cells N was almost absent confirming the requirement of CypA for HCoV-229E replication.
Discussion
Genetic variation of host genes involved in virus infection and also in other human diseases [47] is of highest clinical interest as such proteins represent potential molecules for host-targeting (therapeutic) agents (HTAs) with broad-range antiviral activity. Even in the case of rare genetic variants their analysis might give important clues to disease mechanism. In the case of HCV understanding the functional architecture of type III IFN genomic regions and SNPs have improved the knowledge on the pathogenetic mechanism of HCV infection [48]. However, studying the effect of SNP mutations on infection has to be interpreted carefully considering composition of cohorts, differences in disease progression, or duration times of follow-up studies.
For CoVs few host SNP data are available which might give clues on resistance to or promotion of viral infection. For example, SNPs of genes involved in innate immunity were correlated with SARS-CoV load during the initial phase of infection [49]. A 336A > G promoter polymorphism in the gene encoding CD209 (DC-SIGN) was correlated with clinical-pathologic outcomes in 824 serologically confirmed SARS patients [50]. The -336AG/GG genotype was associated with lower standardized lactate-dehydrogenase (LDH) levels compared with the ‘-336AA’ genotype carrying patients with a 60% chance of having a poorer prognosis because of higher LDH levels. In cats, several SNPs have been described in CD209, TNF-α [51] and IFN-γ [52] genes. They were found to be associated with the outcome of FCoV infection, that is, with the susceptibility for or the resistance to Feline Infectious Peritonitis (FIP), an immune-mediated, highly lethal disease without effective therapy and prevention. A genome-wide association study identified 20 SNPs with significant effect for the antibody level against IBV in chicken [53].
By demonstrating the inhibitory potential of CsA and non-immunosuppressive derivatives as effective inhibitors of CoV replication we and others have introduced cyclophilins, the targets of these compounds, as possible host targets for preventing CoV infection. Using CypA knockdown or knockout mutant cell lines we have shown the requirement of the protein for CoV replication.
Most intriguingly, the individual CypA mutations [D66E]/[G96D]/[N106I] did not support 229E-LUC replication as opposed to the [E84D]/[I89T]/[E134K] variants, which is in close agreement with the HCV replication studies in these cell lines [46]. As discussed by these authors the first three functional (with respect to suppression of virus replication) amino acid exchanges are located near the isomerase active site whereas the non-functional second mutation set is located remote (see Figure 1). Destabilization of CypA was identified as the underlying mechanism, resulting in near-complete intracellular CypA depletion. Before infection of the mutant cell lines with HCoV-229E, CypA mRNA levels detected by qPCR analysis were tested and found to be (with the exception of mutant E84D) quite comparable. It is reasonable to assume the similar mechanisms for the reduced and differential replicative behavior of the coronaviruses.
Even though it could be expected that depletion or destabilization of the highly prominent CypA as a house-keeping gene would be detrimental to cell growth it is clear that the mutated Huh-7.5 cell lines proliferate quite normal. Reasons could be either that, as opposed to a knockout the knockdown of CypA is not complete and the activity of the residual molecules suffices for cell growth, or the PPIase functions could be overtaken by other isomerases as was already shown for prolyl isomerases of the Pin1 type [54, 55]. In any case, CypA is a prolyl isomerase required for propagation of HCV and CoVs. From both, the HCV and the HCoV studies it is clear that expression of correctly folded, stable CypA is essential and that blockade of the active groove by CsA or its derivatives blocks enzymatic functions essential for replication of both viruses. For coronaviruses it has to be clarified whether other viral proteins, apart from the CypA binders nsp1 and nucleocapsid, require the proline-directed binding and PPIase activity of CypA and what the underlying mechanism for their involvement in virus replication is.
Due to the fact that CypA SNPs are very rare in the human population and CypA can be effectively inhibited by CsA or non-immunosuppressive derivatives thereof, it is even more intriguing to put efforts into the development of those compounds into broad-spectrum anti-coronaviral drugs.
Conclusions
Several viruses, including coronaviruses and HCV require functional CypA for replication. SNP variants causing amino acid exchanges around the PPIAse active site destabilize the protein and contribute to lower virus replication. Its role during CoV replication needs to be clarified. CypA represents an important host factor whose activity can efficiently be blocked by HTAs like CsA and non-immunosuppressive derivatives thereof.
Conflicts of interest
The authors disclose no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by grants of the German Ministry for Education and Research (BMBF (Zoonosis Network, Consortium on ecology and pathogenesis of SARS (project code 01KI1005F) and Novartis AG, Switzerland, to AvB, and the German Center for Infection Research (DZIF) to SC. We are very grateful to Kristina Lakomek and Karl-Peter Hopfner for valuable help with the Pymol structure display program and to C. Rice for providing the Huh-7.5 cell line.
References
- 1.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z. Middle East Respiratory Syndrome Coronavirus (MERS-CoV); announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS — beginning to understand a new virus. Nat Rev Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert-Koch-Institut Ausbruch von MERS-Coronavirus in Südkorea. Epidemiologisches Bull. 2015 23/2015. [Google Scholar]
- 7.PARK J.-M., YOO C. South Korea cuts rates as MERS clouds outlook; 10th patient dies. Reuters. 2015 http://www.reuters.com/article/2015/06/11/us-health-mers-southkorea-idUSKBN0OQ2U820150611. [Google Scholar]
- 8.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adedeji A.O., Sarafianos S.G. Antiviral drugs specific for coronaviruses in preclinical development. Curr Opin Virol. 2014;8:45–53. doi: 10.1016/j.coviro.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemotherapy. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M. Repurposing of clinically developed drugs for treatment of middle east respiratory syndrome coronavirus infection. Antimicrob Agents Chemotherapy. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaFemina R.L. Alternative screening approaches for discovery of MERS coronavirus inhibitors. Antimicrob Agents Chemotherapy. 2014;58:4251–4252. doi: 10.1128/AAC.03406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundin A., Dijkman R., Bergstrom T., Kann N., Adamiak B., Hannoun C., Kindler E., Jonsdottir H.R., Muth D., Kint J. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10:e1004166. doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall’Armi E., Herzog P., Kallies S. The SARS–coronavirus–host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]; Un-biased high-throughput protein–protein interaction study identifying cyclophilin interaction with CoV proteins and CsA as a pan-CoV inhibitor.
- 17.von Hahn T., Ciesek S. Role of polymorphisms in the cyclophilin A gene PPIA for virus infection. Curr Opin Virol. 2015;Oct. issue doi: 10.1016/j.coviro.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Schiene-Fischer C. Multidomain peptidyl prolyl cis/trans isomerases. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagen.2014.11.012. pii: S0304-4165(14)00389-4. [DOI] [PubMed] [Google Scholar]; Highly informative review on domain organization and function of cyclophilins.
- 19••.Hopkins S., Gallay P.A. The role of immunophilins in viral infection. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagen.2014.11.011. pii: S0304-4165(14)00388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Highly informative review on immunophilins and virus replication.
- 20.Gamble T.R., Vajdos F.F., Yoo S., Worthylake D.K., Houseweart M., Sundquist W.I., Hill C.P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 21.Braaten D., Ansari H., Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorfman T., Weimann A., Borsetti A., Walsh C.T., Göttlinger H.G. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]; The first paper assigning PPIase function to cyclophilins.
- 24.Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lohmann V., Luban J., Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterji U., Bobardt M., Selvarajah S., Yang F., Tang H., Sakamoto N., Vuagniaux G., Parkinson T., Gallay P. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F., Robotham J.M., Nelson H.B., Irsigler A., Kenworthy R., Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]; Highly informative review on NFAT function.
- 28.Mikol V., Kallen J., Pflugl G., Walkinshaw M.D. X-ray structure of a monomeric cyclophilin A-cyclosporin A crystal complex at 2.1 A resolution. J Mol Biol. 1993;234:1119–1130. doi: 10.1006/jmbi.1993.1664. [DOI] [PubMed] [Google Scholar]
- 29.Goto K., Watashi K., Murata T., Hishiki T., Hijikata M., Shimotohno K. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem Biophys Res Commun. 2006;343:879–884. doi: 10.1016/j.bbrc.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 30.Ma S., Boerner J.E., TiongYip C., Weidmann B., Ryder N.S., Cooreman M.P., Lin K. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob Agents Chemother. 2006;50:2976–2982. doi: 10.1128/AAC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paeshuyse J., Kaul A., De Clercq E., Rosenwirth B., Dumont J.M., Scalfaro P., Bartenschlager R., Neyts J. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins S., Scorneaux B., Huang Z., Murray M.G., Wring S., Smitley C., Harris R., Erdmann F., Fischer G., Ribeill Y. SCY-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob Agents Chemother. 2010;54:660–672. doi: 10.1128/AAC.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansson Magnus J., Moss Steven J., Bobardt M., Chatterji U., Coates N., Garcia-Rivera Jose A., Elmér E., Kendrew S., Leyssen P., Neyts J. Bioengineering and semisynthesis of an optimized cyclophilin inhibitor for treatment of chronic viral infection. Chem Biol. 2015;22:285–292. doi: 10.1016/j.chembiol.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malešević M., Gutknecht D., Prell E., Klein C., Schumann M., Nowak R.A., Simon J.C., Schiene-Fischer C., Saalbach A. Anti-inflammatory effects of extracellular cyclosporins are exclusively mediated by CD147. J Med Chem. 2013;56:7302–7311. doi: 10.1021/jm4007577. [DOI] [PubMed] [Google Scholar]
- 35.Prell E., Kahlert V., Rucknagel K.P., Malešević M., Fischer G. Fine tuning the inhibition profile of cyclosporine A by derivatization of the MeBmt residue. Chembiochem. 2013;14:63–65. doi: 10.1002/cbic.201200621. [DOI] [PubMed] [Google Scholar]
- 36.Carbajo-Lozoya J., Ma-Lauer Y., Malešević M., Theuerkorn M., Kahlert V., Prell E., von Brunn B., Muth D., Baumert T.F., Drosten C. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184:44–53. doi: 10.1016/j.virusres.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C., Sun T., He P., Chen J., Shen J. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem Biophys Res Commun. 2004;321:557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., III, Wuthrich K. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carbajo-Lozoya J., Muller M.A., Kallies S., Thiel V., Drosten C., von Brunn A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012;165:112–117. doi: 10.1016/j.virusres.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J Gen Virol. 2011;92:2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W., Posthuma C.C., van der Meer Y., Barcena M., Haagmans B.L. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka Y., Sato Y., Osawa S., Inoue M., Tanaka S., Sasaki T. Suppression of feline coronavirus replication in vitro by cyclosporin A. Vet Res. 2012;43:41. doi: 10.1186/1297-9716-43-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Favreau D.J., Meessen-Pinard M., Desforges M., Talbot P.J. Human coronavirus-induced neuronal programmed cell death is cyclophilin d dependent and potentially caspase dispensable. J Virol. 2012;86:81–93. doi: 10.1128/JVI.06062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartenschlager R., Pietschmann T. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc Natl Acad Sci U S A. 2005;102:9739–9740. doi: 10.1073/pnas.0504296102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A., Baldanti F. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol. 2006;78:938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Hahn T., Schiene-Fischer C., Van N.D., Pfaender S., Karavul B., Steinmann E., Potthoff A., Strassburg C., Hamdi N., Abdelaziz A.I. Hepatocytes that express variants of cyclophilin A are resistant to HCV infection and replication. Gastroenterology. 2012;143:439–447. doi: 10.1053/j.gastro.2012.04.053. e431. [DOI] [PubMed] [Google Scholar]
- 47.Nigro P., Pompilio G., Capogrossi M.C. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]; Highly informative review on CypA in human disease.
- 48.Riva E., Scagnolari C., Turriziani O., Antonelli G. Hepatitis C virus and interferon type III (interferon lambda 3/interleukin 28B and interferon lambda 4): genetic basis of susceptibility to infection and response to antiviral treatment. Clin Microbiol Infect. 2014 doi: 10.1111/1469-0691.12797. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 49.Chen W.J., Yang J.Y., Lin J.H., Fann C.S., Osyetrov V., King C.C., Chen Y.M., Chang H.L., Kuo H.W., Liao F. Nasopharyngeal shedding of severe acute respiratory syndrome-associated coronavirus is associated with genetic polymorphisms. Clin Infect Dis. 2006;42:1561–1569. doi: 10.1086/503843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan K.Y.K., Xu M.-S., Ching J.C.Y., So T.M.K., Lai S.-T., Chu C.-M., Yam L.Y.C., Wong A.T.Y., Chung P.H., Chan V.S.F. CD209 (DC-SIGN) −336A>G promoter polymorphism and severe acute respiratory syndrome in Hong Kong Chinese. Hum Immunol. 2010;71:702–707. doi: 10.1016/j.humimm.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y.T., Hsieh L.E., Dai Y.R., Chueh L.L. Polymorphisms in the feline TNFA and CD209 genes are associated with the outcome of feline coronavirus infection. Vet Res. 2014;45:123. doi: 10.1186/s13567-014-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsieh L.-E., Chueh L.-L. Identification and genotyping of feline infectious peritonitis-associated single nucleotide polymorphisms in the feline interferon-γ gene. Vet Res. 2014;45:57. doi: 10.1186/1297-9716-45-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo C., Qu H., Ma J., Wang J., Hu X., Li N., Shu D. A genome-wide association study identifies major loci affecting the immune response against infectious bronchitis virus in chicken. Infect Genetics Evol. 2014;21:351–358. doi: 10.1016/j.meegid.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Assimes T.L., Holm H., Kathiresan S., Reilly M.P., Thorleifsson G., Voight B.F., Erdmann J., Willenborg C., Vaidya D., Xie C. Lack of association between the Trp719Arg polymorphism in kinesin-like protein-6 and coronary artery disease in 19 case-control studies. J Am Coll Cardiol. 2010;56:1552–1563. doi: 10.1016/j.jacc.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gemmill T.R., Wu X., Hanes S.D. Vanishingly low levels of Ess1 prolyl-isomerase activity are sufficient for growth in Saccharomyces cerevisiae. J Biol Chem. 2005;280:15510–15517. doi: 10.1074/jbc.M412172200. [DOI] [PubMed] [Google Scholar]