Abstract
Nanopore technology is a promising label-free detection method. However, challenges exist for its further application in sequencing, clinical diagnostics and ultra-sensitive single molecule detection. The development of DNA nanotechnology nonetheless provides possible solutions to current obstacles hindering nanopore sensing technologies. In this review, we summarize recent relevant research contributing to efforts for developing nanopore methods associated with DNA nanotechnology. For example, DNA carriers can capture specific targets at pre-designed sites and escort them from nanopores at suitable speeds, thereby greatly enhancing capability and resolution for the detection of specific target molecules. In addition, DNA origami structures can be constructed to fulfill various design specifications and one-pot assembly reactions, thus serving as functional nanopores. Moreover, based on DNA strand displacement, nanopores can also be utilized to characterize the outputs of DNA computing and to develop programmable smart diagnostic nanodevices. In summary, DNA assembly-based nanopore research can pave the way for the realization of impactful biological detection and diagnostic platforms via single-biomolecule analysis.
INTRODUCTION
As a new platform, nanopore technology presents various advantages over other detection modalities including low cost, high throughput and label-free sample analysis. Accordingly, nanopore analysis has been widely applied to fields such as gene sequencing, personalized medicine, biomedicine, food safety, environmental surveillance, and others. In particular, as a milestone for nanopore detection, in 1996, Deamer et al. proposed DNA sequencing using alpha hemolysin (1). In a typical nanopore experiment, two reservoirs are connected via a single nanometer-sized pore. Upon applying an electric field across the nanopore, electrically charged molecules are driven to translocate the nanopore via an electrophoretic force. When this occurs, conductive ions in the buffer are excluded from the nanopore, resulting in a measurable drop in current. Accordingly, biomolecules of various sizes and charges can be distinguished based on parameters of translocation such as dwell time and current drop amplitude (2). Inherently, nanopore experiments represent a single molecule detection method with ultra-sensitivity and high resolution, capable of discriminating minute differences among single nucleotide bases A, T, G and C. Compared with previous nanopore applications, which mainly focused on gene sequencing (3–6), recent nanopore technologies have also been extended to analyze the physical and chemical properties of various molecules (7) such as nucleic acids (8–12), proteins (13–18), nanoparticles (19–25) and ions (26–32).
Based on materials, nanopores can be divided into two categories: biological nanopore (33) and solid-state nanopore (SS-nanopore) (Figure 1) (34). Biological nanopores comprise channel-bearing proteins such as Phi29 (35–37) and α-hemolysin (38–44). With a narrow channel (general diameter 1 nm–2 nm), protein nanopores can produce sensitive and accurate electrical signals in DNA sequencing due to the similarities they share with the diameter of single-strand DNA (ssDNA). In addition, experimental repeatability is another advantage of biological nanopores, according to which selected portable commercial nanopore devices are being developed. However, issues must be considered when using biological nanopores. For example, it is not easy to readily modify protein nanopores, which limits the scope of their application. Moreover, proteins are more sensitive to external environmental conditions because small variations in temperature or pH will significantly affect biological nanopores’ conformation and activity.
Figure 1.
Schematics of a biological nanopore (A) (33) and a SS nanopore (B) (34). In (A), the target molecule can pass through the pore of α-hemolysin to produce a significant drop in current. The carrier escorting target molecules through SS nanopores induces specific current signals.
SS-nanopores are nanopores are constructed using abiotic materials (45,46). The straightforward fabrication procedures of SS nanopores render diameter control adjustable and scalable, which enables wider sensing ranges to encompass not only small molecules such as DNA, but also large protein targets. However, SS nanopore-based detection faces several challenges. First, the translocation velocity of analytes is generally too fast to allow for consistent identification of smaller biomolecules because of comparatively large nanopore diameters. To solve this problem, various methods including gel substrates (47), molecular modifications on the nanopore (48), blocking the pore with nanobeads (49), applying various high-salt buffers (50) are utilized. However, not all of these methods are satisfactory because of deficiencies such as increased experimental complexity, random systematic noise, and modification difficulties. Additionally, SS nanopore membranes are too thick to acquire high signal-to-noise ratios at a high resolution, compared with biological nanopores in DNA sequencing. In order to improve the resolution and sensitivity of SS nanopores, scientists have combined super-thin two-dimensional materials like molybdenum disulfide (4) and graphene with SS nanopores (51–55). Nevertheless, the preparation of two-dimensional material-based nanopores are complicated, inconvenient, and expensive.
DNA nanotechnology focused on using DNA to construct various self-assembled structures was recently developed. As a versatile technology, DNA assembly is already applied to various nanopore-based analyses. For example, DNA self-assembly structures can be used to capture target biomolecules to form complex spatial arrangements (56), or to serve as possible channels for potential drug delivery (57). Importantly, DNA nanotechnology may also provide practical solutions to the aforementioned challenges in nanopore detection (34,58,59). Several efforts have been made to combine nanopore analysis and DNA nanotechnology in the past decade. For example, scientists directly used DNA nanotechnology to construct assembled nanopores performing target molecule nanopore translocation (60). Notably, in these studies, DNA nanotechnology endowed the nanopore platform with fine-tunable and adjustable properties.
In this review article, we summarize research associating nanopore techniques with DNA nanotechnology. Several categories will be discussed to highlight recent studies combining DNA nanotechnology and nanopore detection. We will primarily discuss recent progress combining DNA nanotechnology and nanopore detection in the following categories: (i) DNA carrier-mediated escort of targets in nanopore translocation; (ii) DNA-assembled nanopores for single- molecule detection with tunable and adjustable properties; (iii) nanopore detection of targets based on the dynamic control of DNA assembly. Combined with DNA nanotechnology, nanopore analysis methods have been greatly improved. To date, several studies have been conducted in this interdisciplinary field. Considering the universal applications of nanopore analysis combined with DNA nanotechnology, this review article highlights related works and inspired ideas to introduce this study area to researchers. Finally, we envision that the association of DNA nanotechnology and nanopores will create many new sensing methods for biodetection, nanoengineering, diagnostics and therapeutics.
DNA NANOTECHNOLOGY
With the recent development of DNA assembly, DNA molecules can serve not only as carriers of genetic information, but also fulfill significant roles in artificial nanosystems through their diversely modified properties, predictable behavior, nanoscale size and programmable features (61). In the 1980s, DNA was characterized as being able to form desired nanostructures via computer-assisted molecular designs, i.e. DNA self-assembly (62). Nadrian Seeman, a pioneer in the field of DNA nanotechnology, constructed various nanostructures based on DNA self-assembly. Utilizing specific DNA molecular recognitions, stable DNA supramolecular structures can spontaneously form via hydrogen bonding, hydrophobic interactions, and van der Waals interactions, among others. There are two main strategies for creating DNA structures: (i) short DNA strands employing DNA tile assembly and (ii) long DNA strands-associated DNA origami assembly. DNA nanotechnology is a promising tool with advantages such as designable nanoscale engineering, high manufacture efficiency, and a convenient preparation process. This method has recently been applied to various areas such as biology, chemistry, medicine, material science, nanoengineering, and molecular computing (63–65). In particular, DNA assembly-based sensing and diagnosis has attracted significant interest. It has been shown that engineered DNA-assembled constructs can facilitate the control of various biomolecules (64), and is an excellent material for complex molecular information processing (65,66).
Tile based self-assembled DNA nanostructures
As the most commonly used primary DNA component, the typical DNA tile generally comprises several short hybridized ssDNA to form a cross-shaped skeleton frame. Then, through cohesive sticky ends, multiple tiles can be linked to assemble more complex structures. As a pioneering DNA assembly method, DNA tiles are generally used to construct a diverse range of nanostructures (67–69).
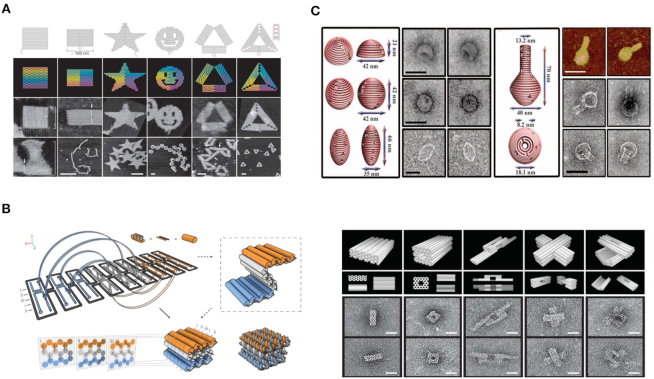
In 1983, DNA tile-based self-assembly was first proposed by Seeman (62). The original naturally occurring tile (Holliday junction) includes four single-stranded DNA. Meanwhile, by rational design, 2D lattices can be constructed using cohesive ends hybridization. Subsequently, to construct more stable and rigid tiles, double crossover (DX) tiles were established including double crossover, antiparallel, odd spacing (DAO) and double crossover, and antiparallel and even spacing (DAE) (67) (Figure 2A). Taking advantage of DX tiles, Hao Yan et al. prepared a 2D rigid grid structure in 1998, where the DNA-assembled structure was imaged by atomic force microscope (AFM), as shown in Figure 2B (69). DNA tiles with high rigidity and more complexity appeared in succession including triple-crossover (TX) (70), paranemic crossover (PX) (71), DX triangle (72), nanoarrays (NAs, shown in Figure 2B and C), nanoroads (NTs) (73) and single-stranded tile (74,75). By controlling the concentration ratios and the lengths of specific DNA strands, Mao et al. manufactured various 2D (76) and 3D structures (77) including tetrahedron, dodecahedron and buckyball arrangements (Figure 2D).
Figure 2.
(A) The structure of 2D DNA tile lattices comprising two and four units (62). The images below represent the structures for DAO and DAE units. (B) The original design and AFM images of nanoribbons (69). (C) Schematic drawings of DNA tiles with multiple DNA bridges (73). (D) Schematic drawings and transmission electron microscope (TEM) images of DNA tile assembled 3D nanostructures (77).
DNA origami-based self-assembly
Compared with minute DNA tiles, DNA origami assembly provides a method for assembling larger DNA nanostructures. DNA origami assembly was developed by Rothemund in 2006, where hundreds of short synthetic oligonucleotides (staple DNA strands) fold a long scaffold strand (typically the M13mp18 genome) into various nanoscale shapes of unprecedented complexity. As shown in Figure 3A, through complementary base-pairing, various 2D DNA structures were assembled including square, rectangle, pentagram, smiling face, triangle and others (78). Consequently, asymmetric shapes such as maps (79), dolphin (80) and characters (81) were also constructed based on DNA origami.
Figure 3.
(A) Schematic illustrations of square, rectangle, star, disk with three holes, triangle with rectangular domains, and sharp triangle with trapezoidal domains (78). (B) Direct self-assembly of DNA into nanoscale three-dimensional shapes and TEM results (86). (C) DNA origami nanostructures with complex 3D curvatures (87).
To construct 3D structures, additional efforts have been made using different design principles: linking 2D self-assembled structures together to obtain 3D aggregations or directly assembling 3D structures. Hollow cubes and prisms were manufactured (82–84) by linking independent 2D structures. Dietz et al. expanded this method by creating DNA nanostructures with controlled curvature and twisting (85). In 2009, Douglas et al. developed a DNA assembly method to directly construct 3D-shaped DNA origami (Figure 3B) (86). In 2011, Hao Yan et al. defined the features of objects via scaffold DNA nanostructures and constructed a connective frame to modify curvature on the surface of 3D DNA origami (Figure 3C) (87).
Dynamic DNA strand displacement
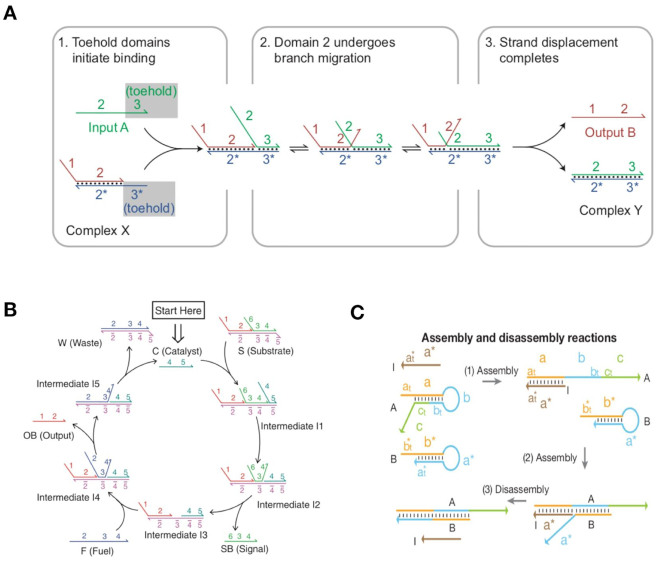
In addition to DNA assembly-based nanostructures, DNA strand displacement, a dynamic DNA regulation method, is another recent development in DNA nanotechnology (88–93). In nature, DNA strand displacement commonly occurs at a very slow reaction rate (88). Toehold-mediated DNA strand displacement reaction (T-SDR) was developed by Yurke et al. in 2000 and greatly accelerates reaction rate by ∼106-fold (89). In a typical T-SDR process, strand displacement initiates from a short single-stranded toehold domain, leading to sequential strand displacement via branch migration (Figure 4A) (90). Importantly, the control of toehold strength enables fine-tuning of T-SDR.
Figure 4.
(A) Schematics of basic DNA strand displacement (90). (B) Catalytic DNA strand displacement circuit where the catalyst DNA can repeatedly participate in multi-cycle reactions (93). (C) Programmable DNA self-assembly pathway based on DNA strand displacement (94).
In recent years, T-SDR has attracted more attention for its easy design and accurate control abilities, and has become a popular area of nanoengineering (91–93). This research context includes the following areas. (i) Regulation of switchable nanodevices and nanostructures. As large-scale DNA molecular systems and origami nanomachines are constructed by DNA molecules, T-SDR- based control can be widely used in DNA-assembled nanosystems by taking advantage of dynamic and precise properties (90). (ii) Molecular signal sensing effected via DNA catalysis (Figure 4B) (93). Since the enzyme-free DNA system can amplify the molecular signal via non-covalent DNA catalysis, catalysis reactions can be used to amplify weak signals (94). (iii) Construction of complex DNA cascading networks and circuits. Relying on specific base-pair recognitions and programmable sequence designs, T-SDR-based DNA regulation was employed to construct various types of molecular motors (89), cascading networks (92–94), and logic circuit operations (95,96). This technique can also be used in areas such as smart molecular sensing and DNA computing (94–98).
THE APPLICATION OF DNA NANOTECHNOLOGY IN NANOPORE SENSING
Nanopore detection is a new and emerging technique for DNA sequencing, single molecule detection, and clinical diagnosis. Although traditional nanopores inherently have features of high sensitivity and label-free detection, several difficulties persist in their use such as uncontrollable translocation and invariable nanopore size or shape. To address these problems, various solutions have been proposed, among which the designable and versatile DNA self-assembly method is particularly attractive.
In reality, DNA nanotechnologies have been widely applied to improve the performance of nanopore detection. There are several primary approaches for the application of DNA nanotechnology in nanopore sensing. (i) DNA-assembled carriers to assist target molecule translocation, where DNA carriers can serve as a position-controllable tool for assisting in the analysis of target molecules. (ii) DNA-assembled nanopores combining DNA structures with SS nanopores or lipid membrane. In this instance, DNA origami channels are designable and programmable, and the shapes and sizes of DNA nanopores are flexible to satisfy various nanopore detection demands. (iii) Dynamic regulations of DNA assembly to perform controllable nanopore sensing. In this method, elegant DNA strand displacement may provide new ideas and schemes for the clinical diagnoses of diseases and single molecule detection. In the past decade, combinations of nanopore analyses and DNA nanotechnology have contributed to the rapid development of new sensing approaches. Herein, we wish to underscore excellent research studies with respect to the development of nanopore analysis combined with DNA nanotechnology.
Nanopore detection of DNA nanostructures
Assembly DNA structures can be created in accurate sizes and shapes, rendering them suitable for analyzing interactions between well-designed DNA structures and nanopores. On the other hand, studies on nanopore-based detections of DNA nanostructures are relatively rare, and many of the interactions involved remain unknown. Therefore, research on nanopore detection of 2D and 3D DNA structures is worth conducting.
In 2014, Plesa et al. assembled linear double-stranded (ds)DNA molecules with branches to survey the velocity of dsDNA during translocation (99). These branches, created using the DNA origami technique, divided the entire dsDNA construct into different segments. As shown in Figure 5A, the relative position of secondary peaks related directly to the location of its corresponding designed branch structure. Thus, through 1D DNA strands and the translocation duration of different segments, the local velocity of translocation could be precisely estimated. In 2016, SS-nanopore analysis of DNA knot structures was conducted in a long dsDNA strand (Figure 5B) (100). In this study, the high-concentration LiCl buffer was used to detect DNA knots. Nanopore analysis found that knotting occurrence became higher alongside increasing DNA strand length. Based on the results, it was estimated that the majority of the DNA knots were tight when passing through the nanopore.
Figure 5.
(A) Schematic illustration of a synthetic DNA construct assembled using a branched DNA structure translocated through a SS-nanopore (99). (B) SS-nanopore analysis of DNA knot structures (100). (C) Schematics for the SS-nanopore detection of DNA cubes and RNA rings (101). (D) Single-state nanopore analysis of DNAzyme cleavage reaction assisted by DNA tetrahedrons (102).
In addition, 2D and 3D DNA structures were also used in nanopore detection. In 2017, Alibakhshi et al. investigated the translocation of nucleic acid nanoparticles (NANPs) such as RNA rings and DNA cubes (Figure 5C) (101). The research detected rings and cubes and analyzed their electrical signatures, respectively. The researchers then mixed the two types of structures and found that the two particles could be reliably distinguished. Inspired by previous work, in 2018, Zhu et al. studied DNAzyme cleavage reaction assisted by DNA tetrahedrons (Figure 5D) (102). The study demonstrated that the signal of dynamic digestion changes on DNA nanostructures can be amplified via the blockade current.
Despite a number of attempts, many related studies still need to be conducted in this area to deepen our understanding of complex DNA structures’ translocation through nanopores. Additionally, DNA nanostructures possess potential application scenarios and values. For example, more interesting DNA 3D nanostructures can be fabricated and characterized directly through nanopores, or act as biomarkers for clinical diagnoses.
DNA assembled carriers escorting cargoes through nanopore
Recently, nanopore studies on the interactions between DNA duplex strands (called ‘DNA carriers’) and target molecules have attracted significant attention. Most of the research conducted in this field is related to protein detection or the characterization of specific DNA structures. In these related works, linear DNA carriers escort target molecules through nanopores in a controlled fashion, which is beneficial for conducting accurate and directional nanopore analysis (103–107).
In DNA carrier escorting nanopore studies, a 1D linear DNA origami carrier is commonly used. Different from 2D or 3D DNA origami structures, which reflect diverse conformational features, linear DNA carriers comprise hybridization of one single-stranded DNA (typically cleaved m13mp18) and hundreds of staple oligonucleotides (56,103,106). One of the advantages of using a linear DNA origami carrier is that the positions of staple DNA strands can be addressable. This means the escorting site can be specifically controlled, which is particularly useful when confronting multiple types and numbers of target molecules on one carrier.
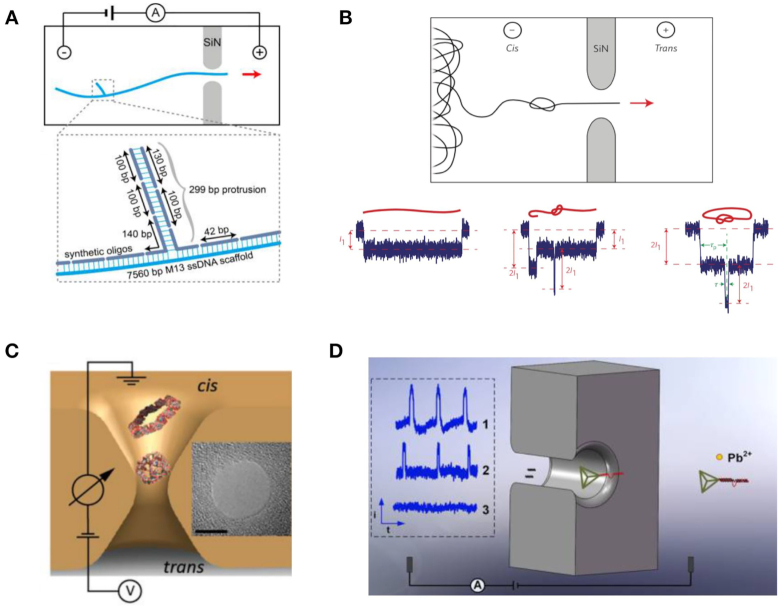
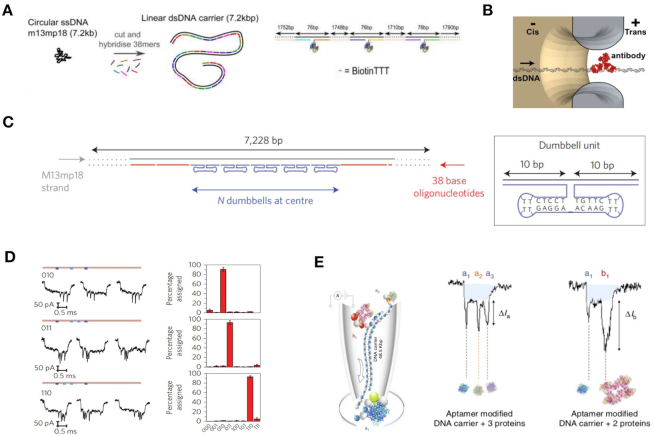
In 2015, Bell et al. synthesized a one-dimensional dsDNA carrier that allowed for the covalent binding of specific proteins at designated positions (Figure 6A) (103). The researchers measured the ionic current signals of DNA carriers with different numbers, as well as the interval and species of binding proteins. In 2016, the ability of DNA carriers to analyze antibody protein detection in the nanomolar range using linear DNA carriers was demonstrated (Figure 6B) (104). The fraction of translocation signals showing a specific target current signal peak of corresponding protein (105). Furthermore, a digitally encoded DNA carrier was also used for multiplexed detection of proteins (Figure 6C) (106). Nanopore signals representing specific barcodes can also be obtained as shown in Figure 6D. In this study, the researchers were able to detect up to four different antibodies with four specific DNA barcodes at nanomolar concentration levels.
Figure 6.
(A) Schematic showing analysis of a linear DNA carrier escorting a target molecule nanopore; a 7.2 kbp DNA carrier escorts proteins with different numbers and positions (103). (B) DNA carrier escorting antibody protein to pass through a SS-nanopore (105). (C) DNA carrier escorting dumbbell DNA structures to produce programmable nanopore signals (106). By controlling the numbers and positions of the dumbbell DNA structures, multiple types of specific nanopore signals can be obtained as shown in (D). (E) Protein nanopore screening in human serum using aptamer-modified DNA carriers (56).
In 2017, Sze et al. also developed an aptamer-binding carrier nanopore method to sense multiple protein targets directly from human serum (56). Employing protein binding with aptamer DNA, unique ionic current signals can be generated to accurately recognize target molecules (Figure 6E). Recently, Chen et al. realized digital data storage and reading based on DNA nanostructures and nanopores. Using a DNA carrier and setting DNA hairpins as encoders, the researchers were able to store digital data arranged in diverse DNA hairpin categories and quantities. The data could also be read by identifying secondary current peaks (107). In areas where the DNA carrier escorted nanopore detection, the work enhanced the aim of realizing a small volume, ultra-sensitive and flexible sensor, or portable digital data storage/reader.
Targets binding with linear DNA carriers has made molecular translocation of nanopores more controllable. Depending on the DNA nanotechnology, binding number, binding position, and type of target, molecules can be designed according to specific demands. Using this method, single target molecules can be recognized and analyzed from complex mixtures through specific binding. For example, physicochemical properties such as reaction kinetic constants between carriers and targets can be speculated based on signal changes during nanopore translocation.
DNA origami blockage to regulating nanopore
Recently, researchers have made significant attempts at directly using DNA origami nanostructures as a nanopore structure associated with lipid membrane or SS nanopores. Through DNA origami assembly, studies involving methods such as DNA origami nanoplates and origami nanopore blockages have been developed, where translocation speeds were able to be controlled by increasing interactions between analytes and DNA scaffold (108–110).
In 2014, Plesa et al. investigated the mechanical properties of DNA origami nanoplates (Figure 7A) (109). Various DNA origami nanoplates were prepared and captured on SS nanopores under certain voltages to test their integral ionic conductance. Different origami nanoplates were designed, with the honeycomb lattice nanoplate providing the best insulation. All nanoplates exhibited a rectification effect, which the authors attributed to structural deformation of the nanoplates (Figure 7 B and C). The nanoplates could be pulled through the pore when using a high electric field force. Similarly, in 2017, Farimani et al. proposed a simulated DNA origami plate-graphene instrument for DNA detection (110). Using molecular dynamic simulations, the researchers computed the ionic conductivity of nanopores on graphene docked with one or two-layered DNA origami. They demonstrated that even the four types of DNA bases could be distinguished according to the blockade current, due to the specific interactions between the DNA origami plate layers and the different DNA bases (Figure 7 D and E).
Figure 7.
(A) Diagram showing a DNA nanoplate and nanopore. (B) Current trace of a nanoplate-nanopore system. (C) A current–voltage (I–V) characteristic curve of a bare pore and the same pore following successful nanoplate docking (109). (D) Schematic of the ionic current simulation system. (E) Theoretically calculated ionic currents for different bases when translocating the origami hybrid nanopore (110).
DNA assembled structures directly serving as nanopores
Biological nanopores and SS-nanopores possess their own strengths and weaknesses, as noted above. In recent years, a combination of nanopore and DNA origami (so-called ‘DNA origami nanopores’) have rapidly been developed. With its strong structural controllability, DNA origami provides a versatile method for assembling designable nanopores with precise and accurate shapes and sizes. It also allows for introducing various addressable modifications at the specific sites of DNA nanopores, thereby endowing nanopores with more powerful functions. In particular, DNA origami nanopores may be suitable in conditions where variable and dynamic nanopore structures are required. In this section, we focus primarily on DNA-assembled structures directly serving as nanopores. Based on the combined-nanopore medium, the DNA-assembled nanopores can be divided into two categories: a DNA-assembled structure combined with SS-nanopores and lipid membrane, respectively.
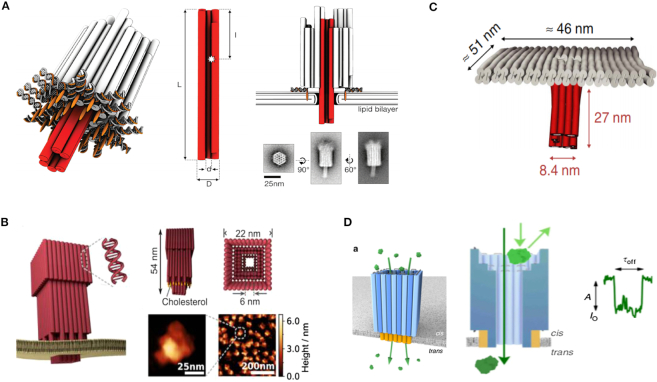
Since SS-nanopores can be fabricated with certain diameters, DNA origami structures can easily be inserted into the SS-nanopore under an electronic field to construct combinational nanopores in specified shapes and sizes (111–116). In 2011, Bell et al. constructed hybrid nanopores comprising DNA origami and SS-nanopores for single molecule sensing (Figure 8A) (112). The researchers inserted DNA origami nanopores into SiN SS-nanopores and demonstrated that the combinational DNA nanopore still allowed target molecules to pass through (Figure 8B). Additionally, a DNA nanoplate nanopore can also be used to created combinational nanopores, where chemical modifications on the nanoplate can improve the geometrical and chemical specifications of the nanopore (Figure 8C) (113). During nanopore analysis, the passage of target molecules translocated through both the SS-nanopore and the nanoplate's nanopore to induce significant signals. Subsequently, glass nanopores were also used to construct hybrid DNA nanopores (Figure 8D) (114). Fluorescently labeled DNA structures and ionic current measurements can demonstrate that the trapping of translocation events occurred during nanopore detection. In addition, the biomolecules modifying DNA nanopores were developed to serve as regulated hybrid DNA nanopores (115,116). For example, in 2018, Ketterer et al. attached a nuclear pore complex (NPC) to a DNA origami ring to study its collective behavior using a SiN nanopore (Figure 8E) (115). Utilizing specific site modifications, the numbers and types of NPCs could be controlled.
Figure 8.
(A) Schematic representation of the DNA origami nanopore (112). (B) Current time curve when a DNA origami nanopore is inserted into a SS-nanopore (112). (C) Schematic of the hybrid nanopore showing the silicon nitride (SiN) membrane (gray) and the DNA nanoplate (red) (113). (D) Typical DNA translocation events for the hybrid nanopore (114). (E) Diagram of the study of a nuclear pore complex based on DNA origami and nanopores (115).
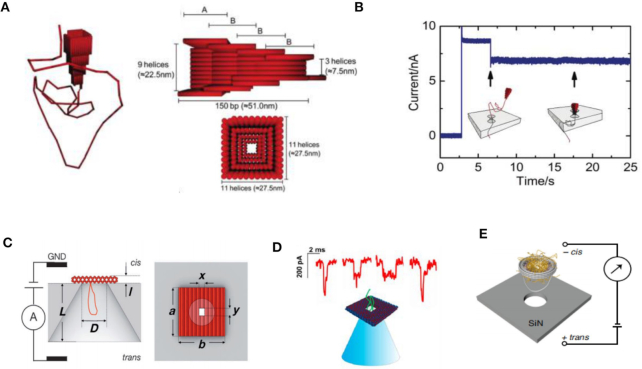
The DNA nanostructures can also serve as nanopores combined with lipid membrane (117–122). Specifically, cholesterol-modified DNA can bind to the membrane via cholesterol group insertion. Then, the DNA origami nanopores can bind to the membrane, allowing target translocation through the nanopores. In 2012, Langecker et al. created an assembled stem using DNA origami for insertion into a lipid membrane via cholesterol moieties (Figure 9A) (120). The properties of this structure were similar to those of natural ion channels. When DNA strand sequence mutations existed in the DNA stem portion, the gating effect was enhanced. This synthetic DNA nanopore can even be used to detect single-stranded DNA molecules. Similarly, another synthetic DNA origami nanopore was established by binding to lipid vesicles (Figure 9B) (121). In 2016, a membrane channel was constructed with a 4 nm diameter nanopore using a DNA origami structure (Figure 9C) (122). This DNA nanopore was able to spontaneously insert itself into lipid bilayers or vesicles. Then, the induced electrical signals of ssDNA and dsDNA were obtained accordingly. Since DNA origami nanopores can be constructed in various diameters to detect DNA molecules, a DNA nanopore with a relatively larger diameter (∼7.5 nm) was constructed to analyze protein molecules (Figure 9D) (60).
Figure 9.
(A) Schematic illustration and TEM images of the transmembrane channel (120). (B) Design and AFM images of DNA origami nanopores (121). (C) Design of the T-shape pore, composed of a double-layered top plate (gray) and a 27 nm-long stem (red) (122). (D) Synthetic protein conductive membrane DNA nanopore (60).
More recently, smaller DNA channels consisting of several DNA strands were synthesized to mimic channel proteins, allowing for spontaneous transport of lipid molecules (123). For example, in 2016, Burns et al. developed an automatic molecular valve made of seven DNA strands, which was able to perform the nanopore open or close functions through DNA strand displacement (Figure 10A) (57). The valve was also sensitive enough to distinguish small molecules that differed by only a single charged group (Figure 10B). Due to its ability to regulate target translocation, the DNA valve can potentially be utilized for drug delivery and synthetic cell or ionic logic circuits. In 2017, Guo et al. established a functional DNA nanopore for uptake by tumor cells (Figure 10C). The DNA nanopore comprised a small DNA tube and was functionalized with Ramos cell aptamers and cell-penetrating peptides (124). Experimental results demonstrated that the DNA nanopores were able to recognize and penetrate Ramos cells with high specificity.
Figure 10.
(A) Schematic illustrations of the DNA nanopore structure with a valve (57). Fluorophore carboxy-fluorescein (CF, red) and sulpho-rhodamine B (SRB, green) are self-quenched molecules. (B) Fluorescence signals of CF and SRB for vesicles with open valves. (C) Schematic illustration of the DNA nanopore's recognition and endocytosis of a tumor cell (124).
Nanopore logic sensing based on dynamic DNA assembly
The fields of smart molecular sensing and molecular computing have rapidly developed in recent years (93–97). Particularly, logic operations based on DNA strand displacement reaction is one of the most common ways through which to achieve intelligent detection and biocomputing. Existing methods for characterizing the output of logic operations are gel and fluorescence arrays (95–97). The superiority of nanopore technology, with features that include single molecule sensing, being label free, and having significant rapidity presents potential detection solutions for addressing particularly smart molecular sensing and biocomputing (125).
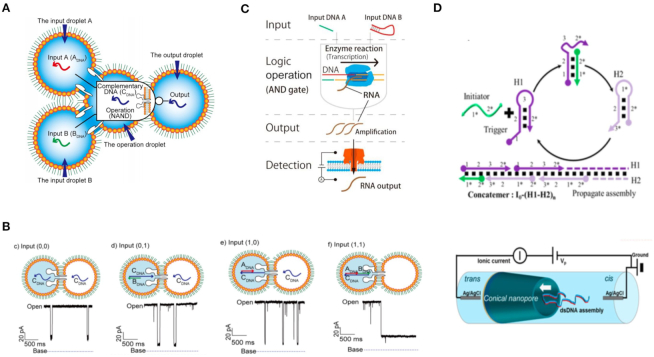
Recently, several research teams used nanopores to characterize the output of DNA logic operations. In 2009, Ali et al. demonstrated that conical nanopores functionalized with polyprotic acid chains showed three levels of conductance based on pH value (126). The researchers utilized the functionalized nanocapillary and different chemical inputs to realize AND and NOT logic gates. In 2016, Yasuga et al. proposed a logic operating system with DNA molecules, droplets, and biological nanopores (127). To realize a NAND operation, they set dsDNA that could not pass through the biological nanopore as ‘0’, while ssDNA that could pass through the pore was set as ‘1’ (Figure 11 A and B). In 2017, Ohara et al. described an AND logic operation using T7 RNA polymerase (Figure 11C) (128). The existence of input DNA A or DNA B represented ‘1’ while the non-existence of input DNA A or DNA B represented ‘0’. Only when both DNA A and DNA B were present, could the electric signal be detected using a biological nanopore. Meanwhile, nanopores could also be used to analyze the complex hairpin DNA structures generated in DNA circuit reactions. For example, Zhu et al. employed the bionanopore technique to characterize complex DNA structures at the single molecule level (129). By analyzing the dwell time and blockade of ionic current signal from DNA structure translocation, information pertaining to hybridization chain reaction could be successfully monitored (Figure 11D).
Figure 11.
Schematics of NAND logic gate operations using biological nanopores (A) and the truth table of a nanopore-based NAND gate (B) (127). (C) Bionanopore detections for performing an AND gate using enzymatic reactions (128). (D) Diagram of using a SS-nanopore to verify organized DNA generated in catalytic hairpin assembly and hybridization chain reaction (HCR) DNA circuit reactions (129).
DNA assembly-based nanopore diagnosis
In recent years, programmable diagnoses based on nanopore methods has attracted significant attention. Combined with DNA assembly, nanopores can also be used in the detection of ultra-sensitive molecules and accurate diagnoses. For example, the DNA strand displacement- assisted nanopore method can be used to sense target molecules at low concentrations, and even in impure clinical samples. Through complex DNA assembly circuit systems, low target signals can be amplified to produce significant nanopore translocation results. This programmable DNA assembly-based nanopore method will undoubtedly contribute to more rapid and accurate diagnoses of diseases in future.
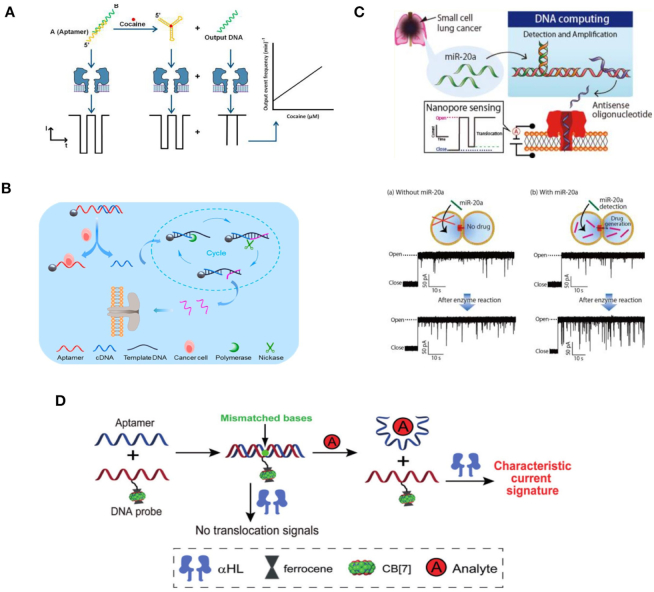
The utilization of nanopores in programmable diagnoses will promote its use in practical clinical applications (130). In 2016, Rauf et al. designed a label-free nanopore biosensor for rapid detection of cocaine in human serum using an aptamer for cocaine (Figure 12A) (131). In the experiment, the aptamer DNA was initially protected by hybridization with a short complementary DNA strand. Then, the greater affinity of the cocaine/aptamer induced displacement of the short complementary DNA strand, thus allowing subsequent biological nanopores’ detection. In 2018, Xi et al. developed a biological nanopore method to detect cancer cells through enzymatic amplification reaction (Figure 12B) (132). Similarly, in 2017, Hiratani et al. described a strategy for cancer diagnoses using microRNA (miRNA) (Figure 12C) (133). The researchers amplified and quantified miRNA from cancer cells through strand displacement and used a biological nanopore to detect targets. In addition, the ultra-sensitive and label-free nanopore method can be used for early diagnoses of various cancers. In 2015, Li et al. reported a series of works about strand displacement-based molecular sensing with biological nanopores. They hybridized aptamers with DNA probes bearing specific biomodification (CB[7]) to form a double-stranded structure that could not translocate through a biological nanopore (Figure 12D) (134). Due to the higher affinity between full complementary DNA strands, hybridization with a receptor can specifically release DNA probes. As a result, a specific ionic current pulse can be observed. Moreover, triplex DNA molecular beacons were also constructed to release DNA probe for diagnosis in 2018 (135). Similarly, via DNA strand displacement, several biological nanopore detection methods were developed for diagnostic applications in the detection of serum and cells (136,137). These works serve as a foundation for the further application of nanopore sensing in practical clinical diagnostics.
Figure 12.
(A) Schematic diagram of an aptamer-based nanopore sensor for cocaine detection (131). (B) Schematic diagram of cancer detection based on biological nanopores (132). (C) Schematic illustration of the nanopore diagnosis system for small cell lung cancer via detection of miR-20a (133). (D) Schematic illustration of a nanopore sensing strategy based on aptamer binding (134).
CONCLUSION
Nanopore technology has attracted interest for use as a single molecule sensor, owing to its advantages of being label-free and fast, with excellent resolution and sensitivity. Although nanopore analysis has undergone significant developed in recent years, several obstacles remain that limit the application of nanopore technology, such as the low signal resolution caused by rapid translocation velocity, invariable nanopore structures, and difficulties related to addressable modification.
Notably, recently developed DNA nanotechnology with its programmable structural design and easy preparation may provide practical solutions to the aforementioned challenges in nanopore detection (54–56). For example, high-speed translocation can be solved by DNA assembly carriers reducing the target passing speed (104–107). In addition, the significant versatility in design afforded by DNA origami can be used to fabricate nanopore channels of any shape and size (120–122). This will allow for performing controlled nanopore analysis according to specific designable selections. In addition, if the DNA assembly method can be used for dynamic control of assembled nanopores, it will be possible to precisely regulate the molecule translocation process in real time. Moreover, it should also be noted that DNA molecules are easy to modify using various chemical groups, which can connect with other materials such as metal nanoparticles and proteins (38,56,104,138). Accordingly, many other materials can be introduced in the construction of hybrid DNA nanopores. More importantly, through DNA nanotechnology, the nanopore can be modified using various materials with sub-nanometer precision. Combining the advantages of DNA nanotechnology and label-free nanopore analysis, DNA assembly-assisted nanopores can be applied to the analysis of proteins, DNA, and other biomolecules. In general, as a promising sensor, the hybrid nanopore analysis can be used to rapidly characterize target molecules and even directly identify the results of complex molecular reaction systems, thus promoting the development of multidisciplinary areas of smart molecular sensing and DNA computing.
On the other hand, critical problems in the DNA hybrid nanopore may still hinder such analysis in further practical applications. Therefore, more efforts are required to solve problems such as current leakage and fluctuations, accurate nanostructure control, and randomly occurring nanopore blockages. Particularly, further research on increasing the stability and controllability of DNA nanopores is required. In addition, although applying the use of DNA nanopores to selected contexts is attractive (such as sequencing and drug delivery), significant developments are required in these areas. Though commercial viability is not yet a reality, it is believed that more significant achievements will be made regarding the combination of nanopores and DNA nanotechnology in the next few years. Taking advantage of recent cross-disciplinary studies on DNA nanotechnology and bioelectronic engineering, the DNA assembly-assisted nanopore method will deliver new opportunities in the areas of single biomolecular analysis, gene sequencing, and clinical diagnosis.
FUNDING
National Key Research and Development Program of China [2017YFE0130600, 2016YFA0501600, 2017YFE0103900]; National Natural Science Foundation of China [61872007, 61320106005, 61772214]; Joint Fund of the Equipment Pre Research Ministry of Education [6141A02033607, 6141A02033608]; Beijing Natural Science Foundation [4182027]; Beijing Municipal Key R&D Project [Z151100003915081]. Funding for open access charge: National Key Research and Development Program of China [2017YFE0130600].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kasianowicz J.J., Brandin E., Branton D., Deamer D.W.. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stein D. Nanopore sequencing: forcing improved resolution. Biophys. J. 2015; 109:2001–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ayub M., Hardwick S.W., Luisi B.F., Bayley H.. Nanopore-based identification of individual nucleotides for direct RNA sequencing. Nano Lett. 2013; 13:6144–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng J., Liu K., Bulushev R.D., Khlybov S., Dumcenco D., Kis A., Radenovic A.. Identification of single nucleotides in MoS2 nanopores. Nat. Nanotech. 2015; 10:1070–1076. [DOI] [PubMed] [Google Scholar]
- 5. Huang S., Romero-Ruiz M., Castell O.K., Bayley H., Wallace M.I.. High-throughput optical sensing of nucleic acids in a nanopore array. Nat. Nanotech. 2015; 10:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fuller C.W., Kumar S., Porel M., Chien M., Bibillo A., Stranges P.B., Dorwart M., Tao C., Li Z., Guo W. et al.. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5233–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu R.J., Ying Y.L., Gao R., Long Y.T.. Confined nanopipette sensing: from single molecules, single nanoparticles, to single cells. Angew. Chem. Int. Ed. 2019; 58:3706–3714. [DOI] [PubMed] [Google Scholar]
- 8. Deamer D.W., Branton D.. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 2002; 35:817–825. [DOI] [PubMed] [Google Scholar]
- 9. Deamer D. Nanopore analysis of nucleic acids bound to exonucleases and polymerases. Annu. Rev. Biophys. 2010; 39:79–90. [DOI] [PubMed] [Google Scholar]
- 10. Ying Y.L., Zhang J., Gao R., Long Y.T.. Nanopore-based sequencing and detection of nucleic acids. Angew. Chem. Int. Ed. 2013; 52:13154–13161. [DOI] [PubMed] [Google Scholar]
- 11. Robert P.J., Aaron M.F., Rukshan T.P., Cynthia J.B., Henry S.W.. Dynamics of a DNA mismatch site held in confinement discriminate epigenetic modifications of cytosine. J. Am. Chem. Soc. 2017; 139:2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang L., Chen X., Zhou S., Roozbahani G.M., Zhang Y., Wang D., Guan X.. Displacement chemistry-based nanopore analysis of nucleic acids in complicated matrices. Chem. Commun. 2018; 54:13977–13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waduge P., Hu R., Bandarkar P., Yamazaki H., Cressiot B., Zhao Q., Whitford P.C., Wanunu M.. Nanopore-based measurements of protein size, fluctuations, and conformational changes. ACS Nano. 2017; 11:5706–5716. [DOI] [PubMed] [Google Scholar]
- 14. Oukhaled G., Mathe J., Biance A.L., Bacri L., Betton J.M., Lairez D., Pelta J., Auvray L.. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 2007; 98:158101. [DOI] [PubMed] [Google Scholar]
- 15. Soskine M., Biesemans A., Moeyaert B., Cheley S., Bayley H., Maglia G.. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 2012; 12:4895–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu D., Bi S., Zhang L., Yang J.. Single-molecule study of proteins by biological nanopore sensors. Sensors. 2014; 14:18211–18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derrington I.M., Craig J.M., Stava E., Laszlo A.H., Ross B.C., Brinkerhoff H., Nova I.C., Doering K., Tickman B.I., Ronaghi M. et al.. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 2015; 33:1073–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yusko E.C., Bruhn B.R., Eggenberger O.M., Houghtaling J., Rollings R.C., Walsh N.C., Nandivada S., Pindrus M., Hall A.R., Sept D. et al.. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2017; 12:360–367. [DOI] [PubMed] [Google Scholar]
- 19. Jubery T.Z., Prabhu A.S., Kim M.J., Dutta P.. Modeling and simulation of nanoparticle separation through a solid-state nanopore. Electrophoresis. 2012; 33:325–333. [DOI] [PubMed] [Google Scholar]
- 20. Tan S., Wang L., Liu H., Wu H., Liu Q.. Single nanoparticle translocation through chemically modified solid nanopore. Nanoscale Res Lett. 2016; 11:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao Y., Lin Y., Qian R.C., Ying Y.L., Si W., Sha J., Chen Y., Long Y.T.. Evidence of single-nanoparticle translocation through a solid-state nanopore by plasmon resonance energy transfer. Chem. Commun. 2016; 52:5230–5233. [DOI] [PubMed] [Google Scholar]
- 22. Darvish A., Goyal G., Aneja R., Sundaram R.V., Lee K., Ahn C.W., Kim K.B., Vlahovska P. M., Kim M.J.. Nanoparticle mechanics: deformation detection via nanopore resistive pulse sensing. Nanoscale. 2016; 8:14420–14431. [DOI] [PubMed] [Google Scholar]
- 23. Roman J., Jarroux N., Patriarche G., Francais O., Pelta J., Le Pioufle B., Bacri L.. Functionalized solid-state nanopore integrated in a reusable microfluidic device for a better stability and nanoparticle detection. ACS Appl. Mater. Interfaces. 2017; 9:41634–41640. [DOI] [PubMed] [Google Scholar]
- 24. Fu K., Han D., Crouch G.M., Kwon S.R., Bohn P.W.. Voltage-gated nanoparticle transport and collisions in attoliter-volume nanopore electrode arrays. Small. 2018; 14:1703248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hulings Z.K., Melnikov D.V., Gracheva M.E.. Brownian dynamics simulations of the ionic current traces for a neutral nanoparticle translocating through a nanopore. Nanotechnology. 2018; 24:445204. [DOI] [PubMed] [Google Scholar]
- 26. Stefureac R.I., Madampage C.A., Andrievskaia O., Lee J.S.. Nanopore analysis of the interaction of metal ions with prion proteins and peptides. Biochem. Cell. Biol. 2010; 88:347–358. [DOI] [PubMed] [Google Scholar]
- 27. Korman C.E., Megens M., Ajo-Franklin C.M., Horsley D.A.. Nanopore-spanning lipid bilayers on silicon nitride membranes that seal and selectively transport ions. Langmuir. 2013; 29:4421–4425. [DOI] [PubMed] [Google Scholar]
- 28. Wang G., Wang L., Han Y., Zhou S., Guan X.. Nanopore detection of copper ions using a polyhistidine probe. Bioelectronics. 2014; 53:453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen L., He H., Xu X., Jin Y.. Single glass nanopore-based regenerable sensing platforms with a non-immobilized polyglutamic acid probe for selective detection of cupric ions. Anal. Chim. Acta. 2015; 889:98–105. [DOI] [PubMed] [Google Scholar]
- 30. Mayne L.J., Christie S.D., Platt M.. A tunable nanopore sensor for the detection of metal ions using translocation velocity and biphasic pulses. Nanoscale. 2016; 8:19139–19147. [DOI] [PubMed] [Google Scholar]
- 31. Roozbahani G.M., Chen X., Zhang Y., Xie R., Ma R., Li D., Li H., Guan X.. Peptide-mediated nanopore detection of uranyl ions in aqueous media. ACS Sensor. 2017; 2:703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roozbahani G.M., Chen X., Zhang Y., Juarez O., Li D., Guan X.. Computation-assisted nanopore detection of thorium ions. Anal. Chem. 2018; 90:5938–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kawano R., Osaki T., Sasaki H., Takinoue M., Yoshizawa S., Takeuchi S.. Rapid detection of a cocaine-binding aptamer using biological nanopores on a chip. J. Am. Chem. Soc. 2011; 133:8474–8477. [DOI] [PubMed] [Google Scholar]
- 34. Keyser U.F. Enhancing nanopore sensing with DNA nanotechnology. Nat. Nanotech. 2016; 11:106–108. [DOI] [PubMed] [Google Scholar]
- 35. Lieberman K.R., Cherf G.M., Doody M.J., Olasagasti F., Kolodji Y., Akeson M.. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J. Am. Chem. Soc. 2010; 132:17961–17972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manrao E.A., Derrington I.M., Laszlo A.H., Langford K.W., Hopper M.K., Gillgren N., Pavlenok M., Niederweis M., Gundlach J.H.. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012; 30:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang S., Haque F., Rychahou P.G., Evers B.M., Guo P.. Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano. 2013; 7:9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campos E., McVey C.E., Carney R.P., Stellacci F., Astier Y., Yates J.. Sensing single mixed-monolayer protected gold nanoparticles by the alpha-hemolysin nanopore. Anal. Chem. 2013; 85:10149–10158. [DOI] [PubMed] [Google Scholar]
- 39. Cabello-Aguilar S., Balme S., Chaaya A.A., Bechelany M., Balanzat E., Janot J.M., Pochat-Bohatier C., Miele P., Dejardin P.. Slow translocation of polynucleotides and their discrimination by alpha-hemolysin inside a single track-etched nanopore designed by atomic layer deposition. Nanoscale. 2013; 5:9582–9586. [DOI] [PubMed] [Google Scholar]
- 40. Zhang X., Price N.E., Fang X., Yang Z., Gu L.Q., Gates K.S.. Characterization of interstrand DNA-DNA cross-links using the alpha-hemolysin protein nanopore. ACS Nano. 2015; 9:11812–11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perera R.T., Fleming A.M., Peterson A.M., Heemstra J.M., Burrows C.J., White H.S.. Unzipping of A-form DNA-RNA, A-form DNA-PNA, and B-form DNA-DNA in the alpha-hemolysin nanopore. Biophys. J. 2016; 110:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ding Y., Kanavarioti A.. Single pyrimidine discrimination during voltage-driven translocation of osmylated oligodeoxynucleotides via the alpha-hemolysin nanopore. Beilstein J. Nanotechnol. 2016; 7:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ding Y., Fleming A.M., Burrows C.J.. Alpha-hemolysin nanopore studies reveal strong interactions between biogenic polyamines and DNA hairpins. Mikrochim Acta. 2016; 183:973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan C.S., Riedl J., Fleming A.M., Burrows C.J., White H.S.. Kinetics of T3-DNA ligase-catalyzed phosphodiester bond formation measured using the alpha-hemolysin nanopore. ACS Nano. 2016; 10:11127–11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Howorka S., Siwy Z.. Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 2009; 38:2360–2384. [DOI] [PubMed] [Google Scholar]
- 46. Kidan L., Kyeong-Beom P., Hyung-Jun K., Jae-Seok Y., Hongsik C., Hyun-Mi K., Ki-Bum K.. Recent progress in solid-state nanopores. Adv. Mater. 2018; 30:1704680. [DOI] [PubMed] [Google Scholar]
- 47. Fujii S., Takeuchi S.. Pesticide vapor sensing using an aptamer, nanopore, and agarose gel on a chip. Lab Chip. 2017; 17:2421–2425. [DOI] [PubMed] [Google Scholar]
- 48. Wei R., Gatterdam V., Wienke R., Tampé R., Rant U.. Stochastic sensing of proteins with receptor modified solid state nanopores. Nat. Nanotech. 2012; 7:257–263. [DOI] [PubMed] [Google Scholar]
- 49. Goto Y., Haga T., Yanagi I., Yokoi T., Takeda K.. Deceleration of single stranded DNA passing through a nanopore using a nanometre sized bead structure. Sci. Rep. 2015; 5:16640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kowalczyk S.W., Wells D.B., Aksimentiev A., Dekker C.. Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett. 2012; 12:1038–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deng Y., Huang Q., Zhao Y., Zhou D., Ying C., Wang D.. Precise fabrication of a 5 nm graphene nanopore with a helium ion microscope for biomolecule detection. Nanotechnology. 2017; 28:045302. [DOI] [PubMed] [Google Scholar]
- 52. Bai Z., Zhang L., Li H., Liu L.. Nanopore creation in graphene by ion beam irradiation: geometry, quality, and efficiency. ACS Appl. Mater. Interfaces. 2016; 8:24803–24809. [DOI] [PubMed] [Google Scholar]
- 53. Kundu S., Karmakar S.N.. Detection of base-pair mismatches in DNA using graphene-based nanopore device. Nanotechnology. 2016; 27:135101. [DOI] [PubMed] [Google Scholar]
- 54. Nam S., Choi I., Fu C.C., Kim K., Hong S., Choi Y., Zettl A., Lee L.P.. Graphene nanopore with a self-integrated optical antenna. Nano Lett. 2014; 14:5584–5589. [DOI] [PubMed] [Google Scholar]
- 55. Qiu W., Skafidas E.. Detection of protein conformational changes with multilayer graphene nanopore sensors. ACS Appl. Mater. Interfaces. 2014; 6:16777–16781. [DOI] [PubMed] [Google Scholar]
- 56. Sze J.Y.Y., Ivanov A.P., Cass A.E.G., Edel J.B.. Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun. 2017; 8:1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burns J.R., Seifert A., Fertig N., Howorka S.. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016; 11:152–156. [DOI] [PubMed] [Google Scholar]
- 58. Bell N.A.W., Keyser U.F.. Nanopores formed by DNA origami: a review. FEBS Lett. 2014; 588:3564–3570. [DOI] [PubMed] [Google Scholar]
- 59. Yong-An R., Han G., Xiangyuan O.. Advances in DNA origami nanopores: fabrication, characterization and applications. Chin. J. Chem. 2018; 36:875–885. [Google Scholar]
- 60. Diederichs T., Pugh G., Dorey A., Xing YZ., Burns J.R., Nguyen Q.H., Tornow M., Tampé R., Howorka S.. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 2019; 10:5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dong Y., Dong C., Wan F., Yang J., Zhang C.. Development of DNA computing and information processing based on DNA-strand displacement. Sci. China Chem. 2015; 10:1515–1523. [Google Scholar]
- 62. Kallenbach N.R., Ma R.I., Seeman N.C.. An immobile nucleic acid junction constructed from oligonucleotides. Nature. 1983; 305:829–831. [Google Scholar]
- 63. Pinheiro A.V., Han D., Shih W.M., Yan H.. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotech. 2011; 6:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen Y.J., Groves B., Muscat R.A., Seelig G.. DNA nanotechnology from the test tube to the cell. Nat. Nanotech. 2015; 10:748–760. [DOI] [PubMed] [Google Scholar]
- 65. Hong F., Zhang F., Liu Y., Yan H.. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 2017; 117:12584–12640. [DOI] [PubMed] [Google Scholar]
- 66. Fu D., Shah S., Song T., Reif J.. DNA-based analog computing. Methods Mol. Biol. 2018; 1772:411–417. [DOI] [PubMed] [Google Scholar]
- 67. Fu T.J., Seeman N.C.. DNA double-crossover molecules. Biochemistry. 1993; 32:3211–3220. [DOI] [PubMed] [Google Scholar]
- 68. Winfree E., Liu F., Wenzler L.A., Seeman N.C.. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998; 394:539–544. [DOI] [PubMed] [Google Scholar]
- 69. Yan H., LaBean T.H., Park S.H., Finkelstein G., Reif J.H.. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003; 301:1882–1884. [DOI] [PubMed] [Google Scholar]
- 70. Labean H.T., Yan H., Kopatsch J., Liu F., Winfree E., Reif J.H., Seeman N.C.. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. JACS. 2000; 122:1848–1860. [Google Scholar]
- 71. Shen Z., Yan H., Wang T., Seeman N.C.. Paranemic crossover DNA: A generalized holliday structure with applications in nanotechnology. JACS. 2004; 126:2324–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ding B., Sha R., Seeman N.C.. Pseudohexagonal 2D DNA crystals from double crossover cohesion. J. Am. Chem. Soc. 2004; 126:10230–10231. [DOI] [PubMed] [Google Scholar]
- 73. Park S.H., Finkelstein G., Labean T.H.. Stepwise self-assembly of DNA tile lattices using dsDNA bridges. J. Am. Chem. Soc. 2008; 130:40–41. [DOI] [PubMed] [Google Scholar]
- 74. Yin P., Hariadi R.F., Sahu S., Choi H.M.T., Park S.H., Labean T.H., Reif J.H.. Programming DNA tube circumferences. Science. 2008; 321:824–826. [DOI] [PubMed] [Google Scholar]
- 75. Wei B., Dai M., Yin P.. Complex shapes self-assembled from single-stranded DNA tiles. Science. 2012; 485:623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. He Y., Chen Y., Liu H., Ribbe A.E., Mao C.. Self-assembly of hexagonal DNA two-dimensional arrays. J. Am. Chem. Soc. 2006; 128:15978–15979.17165718 [Google Scholar]
- 77. He Y., Ye T., Su M., Zhang C., Ribbe A.E., Jiang W., Mao C.. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008; 452:198–201. [DOI] [PubMed] [Google Scholar]
- 78. Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006; 440:297–302. [DOI] [PubMed] [Google Scholar]
- 79. Qian L., Wang Y., Zhang Z., Zhao J., Pan D., Zhang Y., Liu Q., Fan C.. Analogic China map constructed by DNA. Chin. Sci. Bull. 2006; 51:2973–2976. [Google Scholar]
- 80. Andersen E.S., Dong M., Nielsen M.M., Jahn K., Lind-Thomsen A., Mamdouh W., Gothelf K.V., Besenbacher F., Kjems J.. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano. 2008; 2:1213–1218. [DOI] [PubMed] [Google Scholar]
- 81. Pound E., Ashton J.R., Becerril H.A., Woolley A.T.. Polymerase chain reaction based scaffold preparation for the production of thin, branched DNA origami nanostructures of arbitrary sizes. Nano Lett. 2009; 9:4302–4305. [DOI] [PubMed] [Google Scholar]
- 82. Andersen E.S., Dong M., Nielsen M.M., Jahn K., Subramani R., Mamdouh W., Golas M.M., Sander B., Stark H., Oliveira C.L.P. et al.. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009; 459:73–76. [DOI] [PubMed] [Google Scholar]
- 83. Kuzuya A., Komiyama M.. Design and construction of a box-shaped 3D-DNA origami. Chem. Commun. 2009; 28:4182–4184. [DOI] [PubMed] [Google Scholar]
- 84. Endo M., Hidaka K., Kato T., Namba K., Sugiyama H.. DNA prism structures constructed by folding of multiple rectangular arms. J. Am. Chem. Soc. 2009; 131:15570–15571. [DOI] [PubMed] [Google Scholar]
- 85. Dietz H., Douglas S.M., Shish W.M.. Folding DNA into twisted and curved nanoscale shapes. Science. 2009; 325:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M.. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009; 459:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Han D., Pal S., Nangreave J., Deng Z., Liu Y., Yan H.. DNA origami with complex curvatures in three-dimensional space. Science. 2011; 332:342–346. [DOI] [PubMed] [Google Scholar]
- 88. Li Q., Luan G., Guo Q., Liang J.. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002; 30:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yurke B., Turberfield A.J., Mills A.P., Simmel F.C., Neumann J.L.. A DNA-fuelled molecular machine made of DNA. Nature. 2000; 406:605–608. [DOI] [PubMed] [Google Scholar]
- 90. Zhang D., Seelig G.. Dynamic DNA nanotechnology using strand-displacement reactions. Nature Chem. 2011; 3:103–113. [DOI] [PubMed] [Google Scholar]
- 91. Zhang D.Y., Winfree E.. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009; 131:17303–17314. [DOI] [PubMed] [Google Scholar]
- 92. Dirks R.M., Pierce N.A.. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. U.S.A. 2004; 101:15275–15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Seelig G., Soloveichik D., Zhang D.Y., Winfree E.. Enzyme-free nucleic acid logic circuits. Science. 2006; 314:1585–1588. [DOI] [PubMed] [Google Scholar]
- 94. Yin P., Choi H., Calvert C., Pierce N.A.. Programming biomolecular self-assembly pathways. Nature. 2008; 451:318–322. [DOI] [PubMed] [Google Scholar]
- 95. Qian L., Winfree E.. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011; 332:1196–1201. [DOI] [PubMed] [Google Scholar]
- 96. Lopez R., Wang R., Seelig G.. A molecular multi-gene classifier for disease diagnostics. Nature Chem. 2018; 10:746–754. [DOI] [PubMed] [Google Scholar]
- 97. Rothemund W.P., Papadakis N., Winfree E.. Algorithmic self-assembly of DNA tile lattices using dsDNA bridges. J. Am. Chem. Soc. 2008; 130:40–41. [DOI] [PubMed] [Google Scholar]
- 98. Adleman L.M. Molecular computation of solutions to combinatorial problems. Science. 1994; 266:1021–1024. [DOI] [PubMed] [Google Scholar]
- 99. Plesa C., Van Loo N., Ketterer P., Dietz H., Dekker C.. Velocity of DNA during translocation through a solid-state nanopore. Nano Lett. 2015; 15:732–737. [DOI] [PubMed] [Google Scholar]
- 100. Plesa C., Verschueren D., Pud S., van der Torre J., Ruitenberg J.W., Witteveen M.J., Jonsson M.P., Grosberg A.Y., Rabin Y., Dekker C.. Direct observation of DNA knots using a solid-state nanopore. Nat. Nanotech. 2016; 11:1093–1097. [DOI] [PubMed] [Google Scholar]
- 101. Alibakhshi M.A., Halman J.R., Wilson J., Aksimentiev A., Afonin K.A., Wanunu M.. Picomolar fingerprinting of nucleic acid nanoparticles using solid-state nanopores. ACS Nano. 2017; 11:9701–9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhu L., Xu Y., Ali I., Liu L., Wu H., Lu Z., Liu Q.. Solid-state nanopore single-molecule sensing of DNAzyme cleavage reaction assisted with nucleic acid nanostructure. ACS Appl. Mater. Interfaces. 2018; 10:26555–26565. [DOI] [PubMed] [Google Scholar]
- 103. Bell N.A.W., Keyser U.F.. Specific protein detection using designed DNA carriers and nanopores. J. Am. Chem. Soc. 2015; 137:2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kong J., Bell N.A.W., Keyser U.F.. Quantifying nanomolar protein concentrations using designed DNA carriers and solid-state nanopores. Nano Lett. 2016; 16:3557–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Plesa C., Ruitenberg J.W., Witteveen M.J., Dekker C.. Detection of individual proteins bound along DNA using solid-state nanopores. Nano Lett. 2015; 15:3153–3158. [DOI] [PubMed] [Google Scholar]
- 106. Bell N.A.W., Keyser U.F.. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotech. 2016; 11:645–651. [DOI] [PubMed] [Google Scholar]
- 107. Chen KK., Kong JL., Zhu JB., Ermann N., Predki P., Keyser U.F.. Digital data storage using DNA nanostructures and solid-state nanopores. Nano Lett. 2019; 19:1210–1215. [DOI] [PubMed] [Google Scholar]
- 108. Hernández-Ainsa S., Keyser U.F.. DNA origami nanopores: developments, challenges and perspectives. Nanoscale. 2014; 6:14121. [DOI] [PubMed] [Google Scholar]
- 109. Plesa C., Ananth A.N., Linko V., Gülcher C., Katan A.J., Dietz H., Dekker C.. Ionic permeability and mechanical properties of DNA origami nanoplates on solid-state nanopores. ACS Nano. 2014; 8:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Barati Farimani A., Dibaeinia P., Aluru N.R.. DNA origami-graphene hybrid nanopore for DNA detection. ACS Appl. Mater. Interfaces. 2017; 9:92–100. [DOI] [PubMed] [Google Scholar]
- 111. Hernández-Ainsa S., Misiunas K., Thacker V.V., Hemmig E.A., Keyser U.F.. Voltage-dependent properties of DNA origami nanopores. Nano Lett. 2014; 14:1270–1274. [DOI] [PubMed] [Google Scholar]
- 112. Bell N.A.W., Engst C.R., Ablay M., Divitini G., Ducati C., Liedl T., Keyser U.F.. DNA origami nanopores. Nano Lett. 2012; 12:512–517. [DOI] [PubMed] [Google Scholar]
- 113. Wei R., Martin T.G., Rant U., Dietz H.. DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. 2012; 51:4864–4867. [DOI] [PubMed] [Google Scholar]
- 114. Hernández-Ainsa S., Bell N.A.W., Thacker V.V., Gopfrich K., Misiunas K., Fuentes-Perez M.E., Keyser U.F.. DNA origami nanopores for controlling DNA translocation. ACS Nano. 2013; 7:6024–6030. [DOI] [PubMed] [Google Scholar]
- 115. Ketterer P., Ananth A.N., Laman Trip D.S., Mishra A., Bertosin E., Ganji M., van der Torre J., Onck P., Dietz H., Dekker C.. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat. Commun. 2018; 9:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fisher P.D.E., Shen Q., Akpinar B., Davis L.K., Chung K.K.H., Baddeley D., Šarić A., Melia T.J., Hoogenboom B.W., Lin C., Lusk C.P.. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano. 2018; 12:1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Göpfrich K., Zettl T., Meijering A.E.C., Hernández-Ainsa S., Kocabey S., Liedl T., Keyser U.F.. DNA-tile structures induce ionic currents through lipid membranes. Nano Lett. 2015; 15:3134–3138. [DOI] [PubMed] [Google Scholar]
- 118. Burns J.R., Stulz E., Howorka S.. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013; 13:2351–2356. [DOI] [PubMed] [Google Scholar]
- 119. Burns J.R., Al-Juffali N., Janes S.M., Howorka S.. Membrane-spanning DNA nanopores with cytotoxic effect. Angew. Chem. Int. Ed. 2014; 53:12466–12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Langecker M., Arnaut V., Martin T.G., List J., Renner S., Mayer M., Dietz H., Simmel F.C.. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012; 338:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gopfrich K., Li C.Y., Ricci M., Bhamidimarri S.P., Yoo J., Gyenes B., Ohmann A., Winterhalter M., Aksimentiev A., Keyser U.F.. Large-conductance transmembrane porin made from DNA origami. ACS Nano. 2016; 10:8207–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Krishnan S., Ziegler D., Arnaut V., Martin T.G., Kapsner K., Henneberg K., Bausch A.R., Dietz H., Simmel F.C.. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 2016; 7:12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ohmann A., Li C.-Y., Maffeo C., Al Nahas K., Baumann K.N., Göpfrich K., Yoo J., Keyser U.F., Aksimentiev A.. A synthetic enzyme built from DNA flips 107 lipids per second in biological membranes. Nat. Commun. 2018; 9:2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Guo X.L., Yuan D.D., Song T., Li X.M.. DNA nanopore functionalized with aptamer and cell-penetrating peptide for tumor cell recognition. Anal. Bioanal. Chem. 2017; 409:3789–3797. [DOI] [PubMed] [Google Scholar]
- 125. Kawano R. Nanopore decoding of oligonucleotides in DNA computing. Biotechnol. J. 2018; 13:e1800091. [DOI] [PubMed] [Google Scholar]
- 126. Ali M., Mafe S., Ramirez P., Neumann R., Ensinger W.. Logic gates using nanofluidic diodes based on conical nanopores functionalized with polyprotic acid chains. Langmuir. 2009; 25:11993–11997. [DOI] [PubMed] [Google Scholar]
- 127. Yasuga H., Kawano R., Takinoue M., Tsuji Y., Osaki T., Kamiya K., Miki N., Takeuchi S.. Logic gate operation by DNA translocation through biological Nanopores. PLoS One. 2016; 11:e0149667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ohara M., Takinoue M., Kawano R.. Nanopore logic operation with DNA to RNA transcription in a droplet system. ACS Synth. Biol. 2017; 6:1427–1432. [DOI] [PubMed] [Google Scholar]
- 129. Zhu Z.T., Zhou Y., Xu X.L., Wu R.P., Jin Y.D., Li B.L.. Adaption of solid-state nanopore to homogeneous DNA organization verification and label-free molecular analysis without covalent modification. Anal. Chem. 2018; 90:814–820. [DOI] [PubMed] [Google Scholar]
- 130. Liu L., Wu H.C.. DNA-based nanopore sensing. Angew. Chem. Int. Ed. 2016; 55:15216–15222. [DOI] [PubMed] [Google Scholar]
- 131. Rauf S., Zhang L., Ali A., Liu Y., Li J.. Label-free nanopore biosensor for rapid and highly sensitive cocaine detection in complex biological fluids. ACS Sensor. 2017; 2:227–234. [DOI] [PubMed] [Google Scholar]
- 132. Xi D., Li Z., Liu L., Ai S., Zhang S.. Ultrasensitive detection of cancer cells combining enzymatic signal amplification with an aerolysin nanopore. Anal. Chem. 2018; 90:1029–1034. [DOI] [PubMed] [Google Scholar]
- 133. Hiratani M., Ohara M., Kawano R.. Amplification and quantification of an antisense oligonucleotide from target microRNA using programmable DNA and a biological nanopore. Anal. Chem. 2017; 89:2312–2317. [DOI] [PubMed] [Google Scholar]
- 134. Li T., Liu L., Li Y., Xie J., Wu H.C.. A universal strategy for aptamer-based nanopore sensing through host-guest interactions inside alpha-hemolysin. Angew. Chem. Int. Ed. 2015; 54:7568–7571. [DOI] [PubMed] [Google Scholar]
- 135. Guo B., Sheng Y., Zhou K., Liu Q., Liu L., Wu H.C.. Analyte-triggered DNA-probe release from a triplex molecular beacon for nanopore sensing. Angew. Chem. Int. Ed. 2018; 57:3602–3606. [DOI] [PubMed] [Google Scholar]
- 136. Liu L., Li Y., Li T., Xie J., Chen C., Liu Q., Zhang S., Wu H.C.. Selective detection of 8-Oxo-2'-deoxyguanosine in single-stranded DNA via nanopore sensing approach. Anal. Chem. 2016; 88:1073–1077. [DOI] [PubMed] [Google Scholar]
- 137. Liu L., Li T., Zhang S., Song P., Guo B., Zhao Y., Wu H.C.. Simultaneous quantification of multiple cancer biomarkers in blood samples through DNA-assisted nanopore sensing. Angew. Chem. Int. Ed. 2018; 57:11882–11887. [DOI] [PubMed] [Google Scholar]
- 138. Wang F., Zahid O.K., Swain B.E., Parsonage D., Hollis T., Harvey S., Perrino F.W., Kohli R.M., Taylor E.W., Hall A.R.. Solid-state nanopore analysis of diverse DNA base modifications using a modular enzymatic labeling process. Nano Lett. 2017; 17:7110–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]