Abstract
Polycomb repressive complex 2 (PRC2) is a histone methyltransferase that methylates histone H3 at Lysine 27. PRC2 is critical for epigenetic gene silencing, cellular differentiation and the formation of facultative heterochromatin. It can also promote or inhibit oncogenesis. Despite this importance, the molecular mechanisms by which PRC2 compacts chromatin are relatively understudied. Here, we visualized the binding of PRC2 to naked DNA in liquid at the single-molecule level using atomic force microscopy. Analysis of the resulting images showed PRC2, consisting of five subunits (EZH2, EED, SUZ12, AEBP2 and RBBP4), bound to a 2.5-kb DNA with an apparent dissociation constant (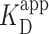 ) of 150 ± 12 nM. PRC2 did not show sequence-specific binding to a region of high GC content (76%) derived from a CpG island embedded in such a long DNA substrate. At higher concentrations, PRC2 compacted DNA by forming DNA loops typically anchored by two or more PRC2 molecules. Additionally, PRC2 binding led to a 3-fold increase in the local bending of DNA’s helical backbone without evidence of DNA wrapping around the protein. We suggest that the bending and looping of DNA by PRC2, independent of PRC2’s methylation activity, may contribute to heterochromatin formation and therefore epigenetic gene silencing.
) of 150 ± 12 nM. PRC2 did not show sequence-specific binding to a region of high GC content (76%) derived from a CpG island embedded in such a long DNA substrate. At higher concentrations, PRC2 compacted DNA by forming DNA loops typically anchored by two or more PRC2 molecules. Additionally, PRC2 binding led to a 3-fold increase in the local bending of DNA’s helical backbone without evidence of DNA wrapping around the protein. We suggest that the bending and looping of DNA by PRC2, independent of PRC2’s methylation activity, may contribute to heterochromatin formation and therefore epigenetic gene silencing.
INTRODUCTION
The Polycomb group (PcG) proteins, including Polycomb repressive complexes 1 and 2 (PRC1 and PRC2), were originally discovered when PcG mutants and knockouts disrupted normal body segmentation in Drosophila melanogaster (1,2). PRC2 is a histone methyltransferase that successively mono-, di- and tri-methylates Lysine 27 of histone H3 (i.e. H3K27me3) (3–6). Disrupting normal PRC2 expression in mice is lethal due to improper embryonic development (3,7). The methyltransferase activity of PRC2 resides in the ‘enhancer of zeste homolog 2’ subunit (EZH2), which forms a core complex with two other PRC2 subunits: the H3K27me3-binding subunit ‘embryonic ectoderm development’ (EED) and the ‘suppressor of zeste 12’ subunit (SUZ12). The ‘retinoblastoma-binding protein 4’ (RBBP4) is shared among all PRC2 complexes, whereas other non-core subunits such as ‘adipocyte enhancer-binding protein 2’ (AEBP2) are only present in some PRC2 subcomplexes (3,8–12). Binding to DNA linkers of nucleosomes or to nucleosome-free regions of the genome with some DNA sequence specificity appears to be important for the recruitment of PRC2 to chromatin (11,13–15). Nuclear pre-mRNA and long noncoding RNAs bind promiscuously to PRC2 and are thought to recruit and/or evict PRC2 from chromatin (9,13,16–18). Finally, binding of the EED subunit to pre-existing H3K27me3 marks is critical for allosteric activation of PRC2 once it is bound (19–22) but may not contribute to initial recruitment of the complex (13).
In contrast to the extensive effort investigating PRC2 recruitment to its sites of action, the key question of how heterochromatin forms once PRC2 arrives remains relatively unexplored. One prominent model is that deposition of H3K27me3 by PRC2 can recruit PRC1 (23–26). Indeed, PRC1 contains the same EED subunit as PRC2, and EED binds H3K27me3 marks (19,27,28). [Note that PRC1 binding can either follow or precede PRC2 (29–31)]. PRC1 has chromatin-compaction activity, which, at least in vitro, is independent of its enzymatic activity in the ubiquitination of H2A (4,32,33). Whether PRC2 itself might have any activities related to chromatin compaction, independent of its H3 methyltransferase activity, has remained unknown and is explored herein.
Atomic force microscopy (AFM) is a powerful tool for investigating the binding of proteins to DNA in a liquid environment (Figure 1A, B) (34–37). In tapping-mode AFM (38), a microscopic cantilever with a nanoscopically sharp tip oscillates above a surface, transiently contacting the surface and measuring the 3D surface topography via detection of a laser reflected off the back of the cantilever. Since AFM operates in a wide variety of buffers without requiring molecular labeling, it provides a versatile platform for investigating biological systems in near-physiological conditions. Recent advances have enabled depositing protein–DNA complexes at biologically relevant ionic conditions on mica that yielded images of DNA in liquid with its native physical properties [persistence length, rise per base pair, width, and helical pitch (39)], in contrast to the still widely used protocol where protein–DNA complexes are imaged in air after rinsing with ultrapure water (40–42). Moreover, achieving a 2D equilibrated DNA conformation on mica yields a more extended molecular configuration than prior deposition protocols for liquid imaging, which in turn facilitates distinguishing protein binding from intramolecular strand crossing (i.e. looped structures).
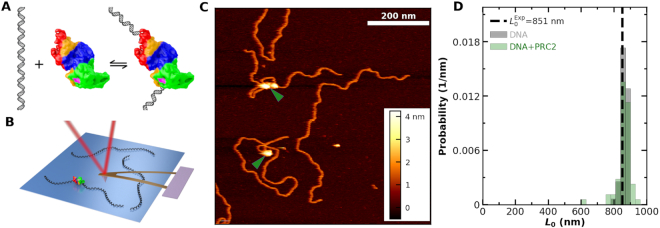
Figure 1.
Imaging PRC2 bound to DNA in liquid. (A) Scheme illustrating DNA reversibly binding to PRC2. (B) Cartoon of atomic force microscopy cantilever imaging of DNA and DNA bound to PRC2 on a mica surface (not to scale). (C) A 700 × 700 μm2 image containing several DNA molecules with and without bound PRC2. Green arrows denote PRC2. (D) A distribution of contour length (L0) for DNA alone (gray, Nmolecules = 52) and uncompacted DNA with PRC2 bound (green, Nmolecules = 59) agrees with the predicted L0 (851 nm; dashed line).
Here, we leverage these advances to image PRC2 bound to DNA as a function of DNA sequence and PRC2 subunit composition. In particular, AFM imaging is used to take a snapshot of the distribution of protein–DNA configurations found in solution by capturing these molecular configurations on the surface. PRC2 was found to compact the DNA by loop formation and to increase the local flexibility of the DNA. We suggest that these two newly characterized properties of bending and looping of DNA by PRC2 could contribute to the ability of PRC2 to form compacted heterochromatin in vivo.
MATERIALS AND METHODS
Purification of PRC2
Human PRC2-5mer complex, comprising of EZH2, EED, SUZ12, RBBP4 and AEBP2 (UniProKB entry isoform sequences Q15910-2, Q15022, O75530-1, Q09028-1 and Q6ZN18-1, respectively), were expressed in insect cells. In brief, standard Bac-to-Bac baculovirus expression system (Expression System) was used to generate baculovirus stocks based on standard protocol. Gp64 detection was used for titering each baculovirus stock (Expression System). Sf9 cells (Invitrogen) were grown to a density of 2 × 106 cells/ml, followed by infection with equal amounts of baculovirus for each subunit. The cells were incubated for an additional 72 h (27°C, 130 rpm), harvested, and snap-frozen with liquid nitrogen for later purification.
A three-column purification scheme was used to purify PRC2 5-mer complexes as previously described (43). Briefly, insect cells were lysed in lysis buffer [10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, 1 mM TCEP (pH 7)] and cell lysate was bound to the amylose resin and washed thoroughly. (Note, all pH values measured at room temperature.) The protein was eluted with 10 mM maltose in 10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM TCEP (pH 7), followed by concentrating to ∼15 mg/ml. PreScission protease was used to digest eluted protein at a mass ratio of 1:100 protease:protein. After overnight incubation at 4°C, cleavage efficiency was checked by SDS-PAGE. The cleaved protein was subject to 5 ml Hi-Trap Heparin column (GE, 17-0407-03) with a gradient over 35 column volumes from Buffer A [10 mM Tris–HCl (pH 7.5), 150 mM NaCl and 1 mM TCEP (pH 7)] to Buffer B [10 mM Tris–HCl (pH 7.5), 2 M NaCl and 1 mM TCEP (pH 7)], with a 1.5 ml/min flow rate. Fractions were checked by SDS-PAGE, and the PRC2 fractions were pooled and concentrated. The concentrated protein was subject to the final sizing column: Superose 6 increase 10/300 GL (GE, 29091596) with running buffer [25 mM NaCl, 10 mM Tris–HCl (pH 7.5), 1 mM TCEP (pH 7)] with a flow rate of 0.5 ml/min. PRC2 peak fractions were checked with SDS-PAGE (Supplementary Figure S1). The correct fractions were pooled and concentrated, as above. Final protein concentration was calculated by Nanodrop (UV absorbance at 280 nm). The ratio of absorbance at 260 nm/280 nm < 0.7 was observed, suggesting no nucleic-acid contamination. The same protocol was used for the purification of the regulatory moiety of PRC2 (SUZ12ΔVEFS, AEBP2, and RBBP4). The catalytic moiety (EZH2, the VEFS domain of SUZ12, and EED with the 81 N-terminal amino acids deleted) was purified as described in Long et al. (44). A three dimensional representation of PRC2 is shown in Supplementary Figure S2.
Purification of DNA substrates
pUC19 plasmid containing twelve Widom 601 positioning sequences was purified using GigaPrep (Qiagen 12191) and cut with EcoRV (NEB R3195M). Efficiency of cutting was determined using a 1% agarose gel. Then, the DNA was adjusted to ∼1 mg/ml and purified via Mono Q column (GE,17-5167-01). Fractions containing the nucleosome template were identified by agarose gel, pooled, and concentrated by ethanol precipitation. DNA was then dissolved in TE buffer. A 200-bp GC-rich (76%) sequence from a CpG island at the Zfpm2 locus, which was suggested to initiate de novo recruitment of PRC2 in embryonic stem cells (45), was designed to be flanked by GC-poor (28%) sequences from lambda DNA. A GC-poor DNA control where the CpG island sequence was substituted by GC-poor lambda DNA sequence was also designed. The DNA substrates were purchased as gblocks (Integrated DNA Technologies) and were cloned into pUC19 plasmid. Purification of the CpG island and GC-poor DNAs were the same as the above protocol. Detailed plots of the GC content of all three constructs are shown Supplementary Figure S3.
Preparation of protein-DNA sample
We first mixed PRC2 at the specified concentration with 20 nM DNA into a final buffer of 25 mM KCl, 38 mM HEPES (pH 7.5), 0.75 mM ZnCl2 and 1 mM TCEP. After a 30 min incubation at 30°C, the sample was diluted ten-fold in deposition buffer [25 mM KCl, 10 mM MgCl2, 10 mM HEPES (pH 7.5)]. We then deposited the sample onto mica using a recently developed protocol (39). Note, this 30 min incubation was much longer than the reported ∼100 s residency time of PRC2 on double-stranded DNA in solution (13), and thus the deposited protein-DNA complex is expected to be equilibrated at the time it is deposited onto the substrate.
As a brief outline of the process, 10-mm-diameter mica disks (Ted Pella, 50) were fixed to a metal disk via epoxy (Ted Pella, 16218), cleaved with masking tape, exposed to unbuffered 100 mM NiCl2 (Sigma 654507), rinsed with copious water, and dried (Whatman 1002-042). We then deposited 20 μl of the protein–DNA mixture onto the surface, waited 2 s, and then serially rinsed the surface with 200 μl of deposition buffer for a total ∼10 ml. Finally, we rinsed the sample with 2 ml of imaging buffer [25 mM KCl, 10 mM NiCl2, 10 mM HEPES (pH 7.5)]. We emphasize that the protein-DNA mixture was never deweted after being deposited. Negative controls with DNA but no PRC2 were prepared as described above, except omitting PRC2.
In order to verify the robustness of our deposition process to extended imaging, a time series assay was performed in both deposition and imaging buffers. In particular, a series of AFM images over ∼70 min confirmed minimal motion of both the unbound DNA and the PRC2-DNA complexes in the imaging buffer (containing NiCl2). Such minimal motion was also seen when imaging in the deposition buffer (containing MgCl2) (Supplementary Figure S4). We expect that proteins remained fixed at the location of initial binding and the DNA absorbed to the mica was capable of nanoscopic rearrangement, but the toplogy of the DNA–PRC2 complex remained unchanged after the exchange from MgCl2 to NiCl2 (Supplementary Figure S5). Surface equilibration of the DNA with respect to the 2D mica surface occurred during the series of gentle rinses in deposition buffer and the subsequent 30 min settling after the sample was loaded into the AFM. Finally, we note that protein or protein complexes bound stably to the mica surface over the time scale of imaging [e.g. stable over ∼1 h of continuous imaging (Supplementary Figure S4)]. Indeed, almost two decades ago, imaging monomeric and dimeric protein complexes absorbed to mica via AFM was introduced as a novel method to determine a protein-protein dissociation constant KD (46).
Experiments using BspMI, a type IIs restriction enzyme, as a DNA localization control were performed as described previously (39). Briefly, DNA from λ bacteriophage (New England Biolabs N3011S) was PCR amplified from positions 9887 to 11785, resulting in a 1899 bp piece of DNA with a BspMI recognition site (5′-ACCTGC-3′) at its center. We next purified the resulting DNA with a PCR purification kit (Qiaquick) and then ran it on a gel. The correct length band was excised and then extracted (Bio-Rad 7326165), concentrated (Millipore UFC501024), and purified a final time (Qiaquick). Experiments then proceeded as described elsewhere (39), except incubation occurred with 100 U/ml of BspMI (New England Biolabs, R0502S) in the buffer, instead of PRC2; MgCl2 was replaced by CaCl2 to prevent DNA cleavage by BspMI.
AFM imaging
We imaged all samples on a commercial AFM (Cypher ES, Asylum Research) featuring a temperature-controlled (19°C), closed-fluidic sample holder. After loading, the sample and cantilever settled for at least 30 min prior to imaging. Images were obtained in tapping mode with a typical set point amplitude of ∼2 nm and a free amplitude of 150% of the set point. We chose the drive frequency as the closest peak of the drive transfer function to the thermal resonance when measured ∼1 μm above the surface. Images of 512 × 512 pixels were acquired at a 2-Hz scan rate. The standard 2 × 2 μm2 images were obtained using a Bruker SNL-10A cantilever (rnom ≈ 2 nm; ktyp = 350 pN/nm) with a 16-kHz resonance in liquid. We used an Olympus BioLever Mini cantilever (rnom = 8 nm; ktyp = 90 pN/nm) when acquiring smaller sized images (e.g. 700 × 700 nm2). The height of the DNA was measured from these smaller sized images and yielded 1.9 ± 0.3 nm (mean ± std. dev, N = 52), in agreement with the expected value of 2.0 nm (47) (Supplementary Figure S6).
Image analysis
All lines scans were flattened with a linear fit to the background of each line, similar to previous AFM image analysis (36,48). Images were analysed using a semi-automated algorithm (39). Binding of PRC2 was associated with a distinct increase in measured height along the axis of DNA. Occasionally, DNA adopted a configuration where the helical backbone crossed itself in the absence of protein. The frequency of such looped structures was ∼8-fold lower when using our deposition protocol that yielded equilibrated DNA with a more extended molecular configuration than standard deposition protocols that yielded kinetically trapped DNA configurations when imaging in liquid (39). Notwithstanding this lower frequency of false positives, we acquired negative controls (i.e. images of DNA without protein). When higher-resolution images were acquired so an individual molecule almost filled the full range of the image (typically 600 × 600 nm2), looped structures were distinguishable from protein bound to the DNA both by height and a volumetric analysis (see section: monomers and multimers of PRC2 distinguished).
Volumetric analysis is the standard analysis for determing the molecular weight and/or the multimeric state of complexes (46,49–51), with an understanding that the resulting deduced volumes inherently include a convolution of the AFM tip radius. As detailed in Supplementary Figure S7, volume analysis was performed by first bounding the region of interest (DNA loop or protein-DNA complex), then fitting a 2D, freely rotating, elliptical Gaussian function to the height as a function of x and y. The volume was measured by integrating the Gaussian over the region of interest. To improve the volume estimation due to inter-image variability in AFM cantilever radius, the DNA in an image was used as a volumetric calibration for that image, conceptually similar to prior work that used naked DNA as a volume standard (52). In our implementation, the DNA height perpendicular to the DNA contour was measured at least 50 nm in contour length from loops or protein. Fitting and then integrating a 1D Gaussian to each height profile along a DNA contour length yielded a DNA volume per unit contour length (Supplementary Figure S7D, E). On a per-molecule basis, the volume of the DNA loop or DNA–PRC2 complex was multiplied by the ratio of the expected to the measured DNA volume per contour length. For simplicity, we assumed the DNA volume per contour length was πr2 based on a rod with a radius of 1 nm. Effectively, this procedure yields the volume of the DNA loop or PRC2 complex, where the volume for each molecule is scaled based on the expected volume per unit contour length of the DNA. We note that omitting our deconvolution procedure did not substantially change the overall shape of the volume distribution or interpretation of the data (Supplementary Figure S8).
To demonstrate that the DNA molecules were equilibrated in 2D on the mica surface, we quantified the DNA’s persistence length (p). A DNA molecule was defined as equilibrated if analysis of its 2D conformation with a 2D WLC model yielded the correct value of p, a definition consistent with prior DNA imaging studies (36,39,53). To do so, we measured the angle θ between two tangent vectors separated by arc length s, a standard analysis (36,53). For this analysis, we selected interpretable DNA molecules, defined as molecules with a configuration containing two or fewer strand crossings. The tangent vector was determined by fitting a third-order, least-squared polynomial spline through user-defined points spaced ∼10 nm along the DNA molecules, excluding looped segments. We then fit the resulting data to
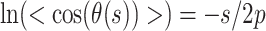 |
(1) |
which is appropriate for analyzing a molecule in 2D (53) (Supplementary Figure S9). This analysis yielded p = 49.1 ± 0.4 nm (mean ± std. dev.; N = 640) consistent with DNA’s known persistence length of 50 nm (54).
Electrophoretic mobility shift assay (EMSA)
EMSA is a well-established technique for determining the apparent binding constant 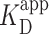 of proteins to DNA (55). The protocol used here followed that of Wang et al. (13), with PRC2 and 32 P-labeled DNA pre-equilibrated 1 h at 30°C in 50 mM Tris (pH 7.5), 100 mM KCl, 0.1 mM ZnCl2, 0.05% NP40, 0.5 mM EDTA and 5% glycerol. Yeast tRNA competitor was omitted, as in the AFM experiments. Dried gels were quantified using a PhosphorImager. Broad bands at intermediate protein concentrations are attributed to some protein dissociation during the 1.5 h electrophoresis time, so all the DNA migrating above the position of free DNA is considered to be bound. The data were then fit to the Hill equation:
of proteins to DNA (55). The protocol used here followed that of Wang et al. (13), with PRC2 and 32 P-labeled DNA pre-equilibrated 1 h at 30°C in 50 mM Tris (pH 7.5), 100 mM KCl, 0.1 mM ZnCl2, 0.05% NP40, 0.5 mM EDTA and 5% glycerol. Yeast tRNA competitor was omitted, as in the AFM experiments. Dried gels were quantified using a PhosphorImager. Broad bands at intermediate protein concentrations are attributed to some protein dissociation during the 1.5 h electrophoresis time, so all the DNA migrating above the position of free DNA is considered to be bound. The data were then fit to the Hill equation:
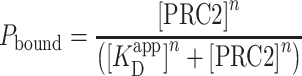 |
(2) |
where Pbound is the fraction of bound DNA, [PRC2] is the concentration of PRC2, n is the Hill coefficient, and 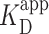 is the apparent dissociation constant.
is the apparent dissociation constant.
RESULTS AND DISCUSSION
High-quality images of PRC2–DNA complexes in liquid
To characterize PRC2 binding to DNA (Figure 1A), we used a recently developed protocol (39) that allows protein-DNA complexes to be gently deposited onto nickel-treated mica under biochemically relevant ionic conditions (25 mM KCl, 10 mM MgCl2). We used the PRC2 complex consisting of EZH2, EED, SUZ12, AEBP2 and RBBP4 and a 2.5-kb DNA substrate consisting of twelve tandem repeats copies of the Widom 601 sequence [12 × 601, unless otherwise noted (see Materials and Methods)]. We then imaged in tapping mode (38) by raster scanning the oscillating AFM tip across the surface (Figure 1B). Figure 1C shows a high signal-to-noise ratio image containing individual DNA molecules bound by zero, one, and multiple PRC2 complexes, where the molecule bound by two PRC2 complexes formed a looped structure.
We considered the possibility that DNA wrapped around the PRC2 complex, akin to DNA wrapping around a histone octamer (56). To look for such putative wrapping, we analyzed the contour length (L0) of unlooped DNA molecules bound by a single PRC2 (see below) and compared that result to DNA molecules imaged with no added PRC2. These two sets of molecules had indistinguishable contour lengths [861 ± 6 nm (mean ± SEM, N = 52) versus 864 ± 3 nm (mean ± SEM, N = 64), respectively] (Figure 1D). Moreover, these measured lengths were within 2% the expected contour length [851 nm (2504 bp)] based on the known rise per base pair for double stranded DNA (0.34 nm/bp). Hence, within the estimated resolution limit of the experiment [≈7 nm (20 bp)], we found no evidence for substantial DNA wrapping around PRC2 and thereby shortening the observed DNA length (Supplementary Figure S10).
Although our deposition and imaging process is based on a protocol that yielded images of DNA on mica in liquid with its native physical properties (persistence length, rise per base pair, width, and helical pitch) (39), we verified that the images collected in the present study also showed the correct persistence length [P = 49.1 ± 0.4 nm (mean ± fitting std. dev.; N = 640)] (Supplementary Figure S9). Moreover, we also showed that imaging in MgCl2 versus NiCl2 yielded similar images of PRC2–DNA complexes (albeit technically more challenging) and that DNA and protein–DNA configurations were stable under both ionic conditions over 1 h of continuous imaging (Supplementary Figure S4). Such image fidelity emphasizes our gentle imaging condition by placing an upper limit on the tip-sample force [≤40 pN (57)], since we report the correct height of the DNA duplex [1.9 ± 0.3 versus 2.0 nm (47), Supplementary Figure S6] in contrast to all but a small set of AFM studies [see for example (39,57)]. In addition, imaging the same individual looped PRC2–DNA complex first in MgCl2 and then in NiCl2 revealed that the looped complex's molecular configuration remained intact and well-bound to the surface during the exchange of buffer (Supplementary Figure S5). Hence, the molecular topology observed in the imaging buffer (i.e. KCl + NiCl2) reflected the topology when the protein–DNA complex was bound by the surface in deposition buffer (i.e. KCl + MgCl2).
Quantifying PRC2 binding by AFM
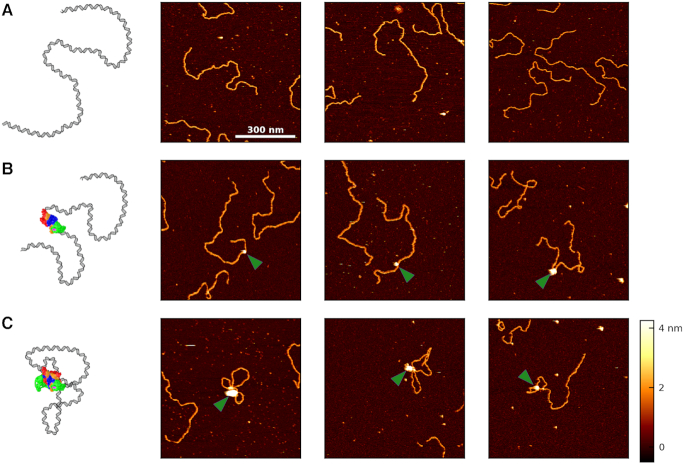
We next sought to establish that the diversity of observed molecular configurations was reproducible and to quantify PRC2 binding to the DNA as a function of protein concentration. Figure 2 provides a gallery of the three general classes of molecular configurations: naked DNA, DNA bound by but not looped by PRC2, and DNA bound and looped by PRC2. The first column in Figure 2 gives a schematic interpretation of the AFM images (not to scale).
Figure 2.
Images of unbound, bound, and looped DNA. (A) A cartoon and three representative images of surface-bound DNA imaged without any bound PRC2. (B) A cartoon and three images showing PRC2 bound to but not looping the DNA. The green arrow denotes PRC2. (C) A cartoon and three images depicting PRC2 bound to and compacting the DNA via looping. All images are 700 × 700 nm2, acquired in liquid at [PRC2] = 50 nM, and use the same color scale shown in the bottom right.
Prior work introduced AFM imaging as a means for determining the dissociation constant KD of protein complexes (46), where the AFM takes a snapshot of the distribution of complexes in the deposited solution. As used in other contexts [e.g. electromobility shift assays (13)], we prefer the use of apparent dissociation constant (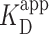 ), as the AFM assay is not strictly reversible. To determine
), as the AFM assay is not strictly reversible. To determine 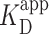 of PRC2 to the 2.5-kb DNA, we varied the concentration of PRC2 at fixed DNA concentration. For each PRC2 concentration, we acquired a series of 2 × 2 μm2 images with a minimum of 70 molecules per concentration and classified the molecular configurations into one of the three general molecular configurations. As expected, increasing the PRC2 concentration while keeping the DNA concentration constant led to an increase in the probability of observing PRC2 bound to DNA (Figure 3A, green points). Representative images at different PRC2 concentrations are shown in Figure 3B–D. In addition, as the PRC2 concentration was increased, the fraction of PRC2-DNA complexes in a compacted, looped state also increased (Figure 3A, brown points). Overall, this analysis included a total of 1152 individual DNA molecules.
of PRC2 to the 2.5-kb DNA, we varied the concentration of PRC2 at fixed DNA concentration. For each PRC2 concentration, we acquired a series of 2 × 2 μm2 images with a minimum of 70 molecules per concentration and classified the molecular configurations into one of the three general molecular configurations. As expected, increasing the PRC2 concentration while keeping the DNA concentration constant led to an increase in the probability of observing PRC2 bound to DNA (Figure 3A, green points). Representative images at different PRC2 concentrations are shown in Figure 3B–D. In addition, as the PRC2 concentration was increased, the fraction of PRC2-DNA complexes in a compacted, looped state also increased (Figure 3A, brown points). Overall, this analysis included a total of 1152 individual DNA molecules.
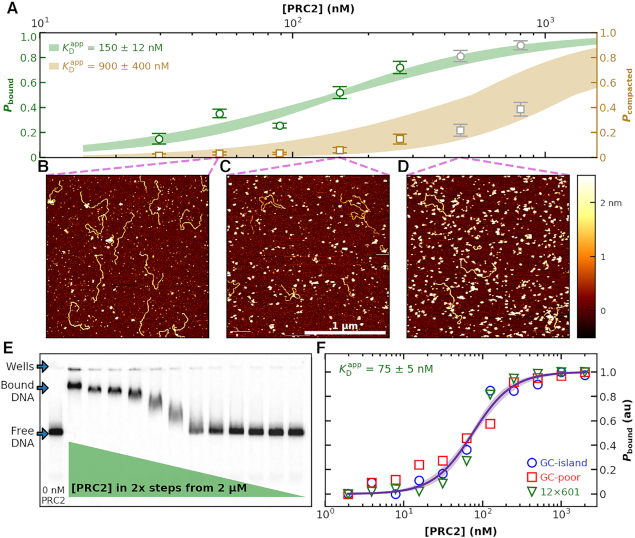
Figure 3.
Binding of PRC2 to DNA as a function of protein concentration. (A) Probability of PRC2 to form a protein–DNA complex [left axis: (green)] and to form a compacted protein–DNA complex [right axis (brown)] plotted as a function of PRC2 concentration. The apparent dissociation constant (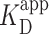 ) was determined by fitting the Hill equation (Equation 2) to the raw data [
) was determined by fitting the Hill equation (Equation 2) to the raw data [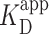 = 150 ± 12 nM (best fit value ± fit std. dev) and n = 1.1 ± 0.1; Nmolecules > 70 per concentration]. The color shaded areas around the markers represent the standard deviation of fitting Equation (2) to the markers. Data from the two highest concentrations (gray) were not used for fitting due to high protein background [though this exclusion did not statistically change the value of
= 150 ± 12 nM (best fit value ± fit std. dev) and n = 1.1 ± 0.1; Nmolecules > 70 per concentration]. The color shaded areas around the markers represent the standard deviation of fitting Equation (2) to the markers. Data from the two highest concentrations (gray) were not used for fitting due to high protein background [though this exclusion did not statistically change the value of 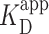 (Supplementary Figure S12)]. (B–D) Representative 2 × 2 μm2 images at concentrations along the binding curve. (E) Gel of an electrophoretic mobility shift assay (EMSA) for PRC2 binding to the same DNA substrate as in panels A–D. (F) Quantification of the EMSA assay shown in panel E (green triangles) and the analogous EMSA quantifications for the 2.5-kb DNA substrates with a GC-poor and GC-rich center (blue circle and red boxes, respectively), as described in the text and Figure 4A. The purple line is a best fit to all three EMSA quantifications, yielding
(Supplementary Figure S12)]. (B–D) Representative 2 × 2 μm2 images at concentrations along the binding curve. (E) Gel of an electrophoretic mobility shift assay (EMSA) for PRC2 binding to the same DNA substrate as in panels A–D. (F) Quantification of the EMSA assay shown in panel E (green triangles) and the analogous EMSA quantifications for the 2.5-kb DNA substrates with a GC-poor and GC-rich center (blue circle and red boxes, respectively), as described in the text and Figure 4A. The purple line is a best fit to all three EMSA quantifications, yielding 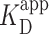 = 75 ± 5 nM (mean ± std. dev.) and a Hill coefficient of 1.7 ± 0.2.
= 75 ± 5 nM (mean ± std. dev.) and a Hill coefficient of 1.7 ± 0.2.
We computed 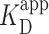 of the PRC2 to the DNA using Equation 2 model, yielding
of the PRC2 to the DNA using Equation 2 model, yielding 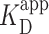 = 150 ± 12 nM (best fit value ± fit std. dev.) and n = 1.1 ± 0.1. The confidence interval associated with one standard deviation around the mean of the fit is plotted (Figure 3A, green shaded curve). For deducing this value of
= 150 ± 12 nM (best fit value ± fit std. dev.) and n = 1.1 ± 0.1. The confidence interval associated with one standard deviation around the mean of the fit is plotted (Figure 3A, green shaded curve). For deducing this value of 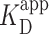 , the two highest concentrations analyzed were excluded due to increasing background of PRC2 on the mica surface potentially confounding the quantification (Supplementary Figure S11). We note, however, that fitting all of the data points yielded a statistically indistinguishable result [140 ± 8 nM (Supplementary Figure S12)]. A similar analysis of compacted DNA defined by the presence of a looped configuration yielded
, the two highest concentrations analyzed were excluded due to increasing background of PRC2 on the mica surface potentially confounding the quantification (Supplementary Figure S11). We note, however, that fitting all of the data points yielded a statistically indistinguishable result [140 ± 8 nM (Supplementary Figure S12)]. A similar analysis of compacted DNA defined by the presence of a looped configuration yielded 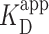 = 900 ± 400 nM and n = 1.4 ± 0.3. (Figure 3A, brown shaded curve). Thus, looping of DNA required substantially higher PRC2 concentrations than simple binding.
= 900 ± 400 nM and n = 1.4 ± 0.3. (Figure 3A, brown shaded curve). Thus, looping of DNA required substantially higher PRC2 concentrations than simple binding.
Similar 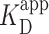 deduced by AFM and ensemble assays
deduced by AFM and ensemble assays
To look for a potential artifact on the stability of the PRC2–DNA complex arising from the surface binding, we measured the 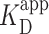 of PRC2 using the same DNA and buffer conditions used in the AFM assay but now with an electrophoretic mobility shift assay (EMSA; see Materials and Methods), an ensemble measurement done entirely in solution (Figure 3E). Quantification of the probability for measuring a DNA molecule bound by PRC2 (Pbound) showed
of PRC2 using the same DNA and buffer conditions used in the AFM assay but now with an electrophoretic mobility shift assay (EMSA; see Materials and Methods), an ensemble measurement done entirely in solution (Figure 3E). Quantification of the probability for measuring a DNA molecule bound by PRC2 (Pbound) showed 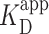 ∼100 nM (Figure 3F), very similar to the
∼100 nM (Figure 3F), very similar to the 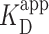 deduced by AFM given the substantial differences in the ensemble and single-molecule assays. Furthermore, at the higher PRC2 concentrations, the additional upward shift of the band indicates multiple PRC2 molecules bound and/or more complex structures with lower electrophoretic mobility, consistent with the structures observed by AFM.
deduced by AFM given the substantial differences in the ensemble and single-molecule assays. Furthermore, at the higher PRC2 concentrations, the additional upward shift of the band indicates multiple PRC2 molecules bound and/or more complex structures with lower electrophoretic mobility, consistent with the structures observed by AFM.
As prior ensemble studies showed a 50-fold difference in 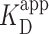 for short 60-bp DNA with 0 versus 100% GC content (13), we also determined the ensemble
for short 60-bp DNA with 0 versus 100% GC content (13), we also determined the ensemble 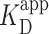 for two other long DNA molecules, with 200-bp segments of either low-GC content (28%) or high-GC (76%) embedded in the longer, low-GC sequence (Figure 4A). Both of these molecules had a lower overall GC content than the 12 × 601 construct (53%). Unexpectedly, EMSA showed that PRC2 bound these three long DNA sequences with similar affinity (Figure 3F). More quantitatively, the combined data from all three sequences were well fit to a cooperative binding model with
for two other long DNA molecules, with 200-bp segments of either low-GC content (28%) or high-GC (76%) embedded in the longer, low-GC sequence (Figure 4A). Both of these molecules had a lower overall GC content than the 12 × 601 construct (53%). Unexpectedly, EMSA showed that PRC2 bound these three long DNA sequences with similar affinity (Figure 3F). More quantitatively, the combined data from all three sequences were well fit to a cooperative binding model with 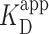 of 75 ± 5 nM (fit ± std. dev.) and a Hill coefficient of 1.7 ± 0.2. Hence, when PRC2 interacts with longer DNA molecules containing a range of natural-like sequences—as opposed to the extremes of 0 and 100% GC content—PRC2 did not exhibit a strong enough GC preference to be detected by EMSA.
of 75 ± 5 nM (fit ± std. dev.) and a Hill coefficient of 1.7 ± 0.2. Hence, when PRC2 interacts with longer DNA molecules containing a range of natural-like sequences—as opposed to the extremes of 0 and 100% GC content—PRC2 did not exhibit a strong enough GC preference to be detected by EMSA.
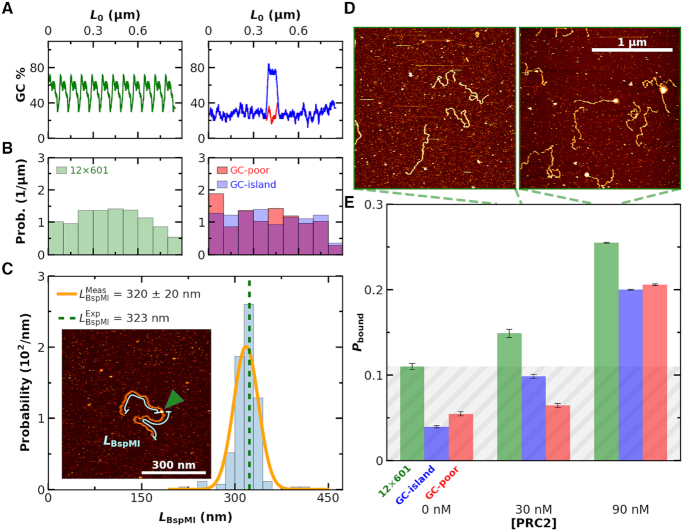
Figure 4.
Binding of PRC2 to 2.5-kb DNA is not strongly sequence dependent. (A) GC content of a DNA construct containing twelve copies of the Widom 601 sequence (left panel, green), a GC-poor construct (right panel, red) and the GC-poor construct containing a 200-bp GC-rich island (right panel, blue). (B) Histogram of distances from PRC2 to both DNA ends for the same sequences as panel A. Note that this analysis does not rely upon knowing the polarity of the DNA relative to the protein. (C) The distribution of binding locations of the type IIs restriction enzyme BspMI to a 646-nm DNA molecule (1899 bp) containing a BspM1 recognition sequence positioned at its center (Nmolecules = 91). The orange line represented a Gaussian fit to the underlying individual measurements of LBspMI. The expected length is indicated by the green dashed line. (Inset) An AFM image showing BspMI (indicated by green arrow) bound to the DNA. Cyan lines illustrate the contour length (LBspMI) from each end of the DNA to the BspMI. (D) Representative 2 × 2 μm2 images at 30 and 90 nM concentrations of PRC2. (E) Probability of PRC2 and DNA complexes (Pbound) as a function of PRC2 concentration for DNA sequences consisting of 12 copies of the Widom 601 sequence (green), a low GC-content (28%) sequence with a GC-rich island (76%) at its center (blue), and a low GC-content sequence (red). The average Nmolecules per DNA substrate type at 0, 30 and 90 nM were greater than 90, 90 and 400, respectively, and the error bars represent the standard error in the mean. Grey shading indicates the false positive rate, based on the highest no added PRC2 control.
PRC2 binds promiscuously along DNA
High-resolution AFM studies provide the ability to measure not only the binding of PRC2 but also to localize that binding along the DNA. We therefore sought to determine if PRC2 would preferentially bind to the GC-rich sequence from the CpG island at the Zfpm2 locus (45) embedded in the otherwise low-GC construct. The low-GC and the 12 × 601 constructs served as controls (Figure 4A). We determined the location of each PRC2 molecule relative to both ends of the DNA and plotted a histogram of those lengths. For the symmetric high-GC island construct, this analysis should have produced a peak at the DNA molecule's center if PRC2 exhibited strong sequence-depending binding. Contrary to this expectation, we saw no spatial preference in PRC2’s binding location, as measured by the distance from the protein to either end of the DNA (Figure 4B) or the distance from the protein to the DNA’s center (Supplementary Figure S13). This result is consistent with the above ensemble measurement of 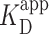 that showed no significant variation with GC content on a 2.5-kb DNA substrate.
that showed no significant variation with GC content on a 2.5-kb DNA substrate.
An immediate question arose: does our single-molecule assay have sufficient spatial resolution to measure sequence-specific localization? To address this concern, we imaged BspMI, a type IIs restriction enzyme, bound to a 646-nm DNA construct containing a single BspMI binding site at its center (Figure 4C). Note, the binding buffer contained Ca2+ in lieu of Mg2+ to allow for site-specific binding without enzyme-induced cleavage. Analysis of these images used the same analysis as for localizing PRC2. The resulting histogram yielded a sharp peak (Figure 4C). The location of the peak quantitatively agreed with the expected location [320 ± 2 nm (mean ± SEM; N = 91) versus 323 nm, respectively]. Thus, the accurate localization of the restriction enzyme, coupled with the lack of a clear binding site for PRC2, suggests that if PRC2 has a specific binding motif, it must occur with similar frequency all along the 2.5-kb DNA substrates, including one containing a CpG island.
In conjunction with the spatial location of PRC2 by AFM, we also quantified the binding of PRC2 to these three DNA constructs at 30 and 90 nM PRC2 (Figure 4D, E), values slightly below the 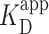 Images of each DNA construct without added PRC2 were used as a control. Overall, for each construct, the increase in Pbound between the two concentrations was similar, though the background level varied for each construct. The increase of Pbound is consistent with the similarity in
Images of each DNA construct without added PRC2 were used as a control. Overall, for each construct, the increase in Pbound between the two concentrations was similar, though the background level varied for each construct. The increase of Pbound is consistent with the similarity in 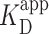 for the different DNA constructs deduced by EMSA (Figure 3F). For simplicity, we use the largest value of the control images as the overall background (Figure 4E, gray shaded region). Future studies requiring higher precision via a lower basal false positive could use shorter DNA molecules that exhibit less spontaneous looping or a smaller field of view than 2 × 2 μm2. We note high-resolution studies of individual molecules are time intensive given the slow acquisition rate of standard AFM imaging (2-Hz line scan) and the total number of molecules in this analysis (N = 2081).
for the different DNA constructs deduced by EMSA (Figure 3F). For simplicity, we use the largest value of the control images as the overall background (Figure 4E, gray shaded region). Future studies requiring higher precision via a lower basal false positive could use shorter DNA molecules that exhibit less spontaneous looping or a smaller field of view than 2 × 2 μm2. We note high-resolution studies of individual molecules are time intensive given the slow acquisition rate of standard AFM imaging (2-Hz line scan) and the total number of molecules in this analysis (N = 2081).
Binding of PRC2 depends on complex composition
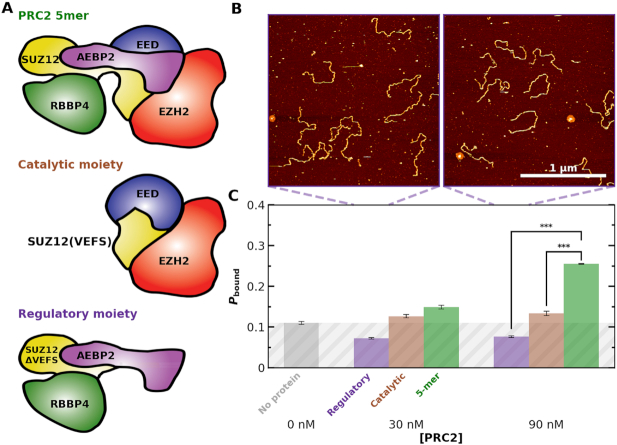
Non-core subunits of PRC2 have a substantial effect on PRC2 binding to chromatin (11,14,15,58) and one current model attributes the contribution of non-core subunits including AEBP2 to their direct binding of DNA. We therefore tested DNA binding with three variants of the PRC2 complex: the full PRC2 complex, a catalytic moiety (EZH2, the VEFS domain of SUZ12, and EED with the 81 N-terminal amino acids deleted) and a regulatory moiety (SUZ12ΔVEFS, AEBP2 and RBBP4) (Figure 5A, Supplementary Figure S1). For each PRC2 variant, we acquired 2 × 2 μm2 images at 30 and 90 nM PRC2 with a minimum of 70 individual DNA molecules per condition (Figure 5B). These concentrations were chosen to enhance the sensitivity to changes in binding affinity, since they were below the single-molecule determined 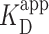 . Analysis of these images showed robust binding of the full PRC2 to the 12 × 601 DNA construct while both PRC2 variants showed binding consistent with background (Figure 5C). Therefore, our single-molecule experiments highlight the importance of PRC2 composition on PRC2 binding to and compacting bare DNA; more specifically, they suggest that the AEBP2 subunit, which contains three zinc-finger domains, enhances the ability of the PRC2 core to bind DNA.
. Analysis of these images showed robust binding of the full PRC2 to the 12 × 601 DNA construct while both PRC2 variants showed binding consistent with background (Figure 5C). Therefore, our single-molecule experiments highlight the importance of PRC2 composition on PRC2 binding to and compacting bare DNA; more specifically, they suggest that the AEBP2 subunit, which contains three zinc-finger domains, enhances the ability of the PRC2 core to bind DNA.
Figure 5.
PRC2 binding depends on PRC2 complex composition. (A) Cartoons illustrating the measured PRC2 moieties. (top) The ‘5mer’ complex consists of EZH2 (red), EED (blue), SUZ12 (yellow), AEBP2 (purple) and RBBP4 (green). (middle) The catalytic moiety of PRC2 consists of EZH2, the VEFS domain of SUZ12, and EED with the 81 N-terminal amino acids deleted. (bottom) The regulatory moiety of PRC2 consists of SUZ12ΔVEFS, AEBP2, and RBBP4. (B) Representative 2 × 2 μm2 images at 30 and 90 nM concentrations of the regulatory moiety of PRC2. (C) Probability of observing a PRC2-DNA complex (Pbound) as a function of PRC2 concentration for the regulatory (purple), catalytic (gray) and 5-mer (green) moieties of PRC2. Concentrations below the 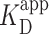 were used to enhance the signal. The triple asterisk represents a statistical significance of P < 0.001. The average Nmolecules at 0, 30 and 90 nM were greater than 80, 90 and 200, respectively, and the error bars represent the standard error in the mean. Gray shading indicates the false positive rate, based on the no added PRC2 control.
were used to enhance the signal. The triple asterisk represents a statistical significance of P < 0.001. The average Nmolecules at 0, 30 and 90 nM were greater than 80, 90 and 200, respectively, and the error bars represent the standard error in the mean. Gray shading indicates the false positive rate, based on the no added PRC2 control.
PRC2-induced DNA compaction via DNA bending
In addition to anchoring DNA loops (Figure 2C), PRC2 might compact DNA by bending DNA’s stiff helical backbone. To look for protein-induced bends, we computed the angle (θbend) between tangent vector of the DNA backbone 10 nm on either side of a bound PRC2 molecule (Figure 6A), akin to earlier work that measured RNA polymerase-induced bends in template DNA (59). The data set for this analysis consisted of all 2 × 2 μm2 images at PRC2 concentrations of 300 nM or less and for PRC2–DNA complexes not involved in a compacted structure (Nprotein = 331) and, as a control, a set of naked DNA molecules (Nmolecules = 70). The distribution of bending angles for unbound DNA is made up of ≈2800 independent segments, since there are many segments separated by 20 nm in a long DNA. As an indication of the underlying data quality, we verified that the measured distribution of the bend angle for the naked DNA quantitatively matched the prediction of the bend angle distribution when using a 2D worm-like chain model and the known persistence length of DNA [P = 50 nm (54)] (Figure 6B, gray bars and dashed line, respectively).
Figure 6.
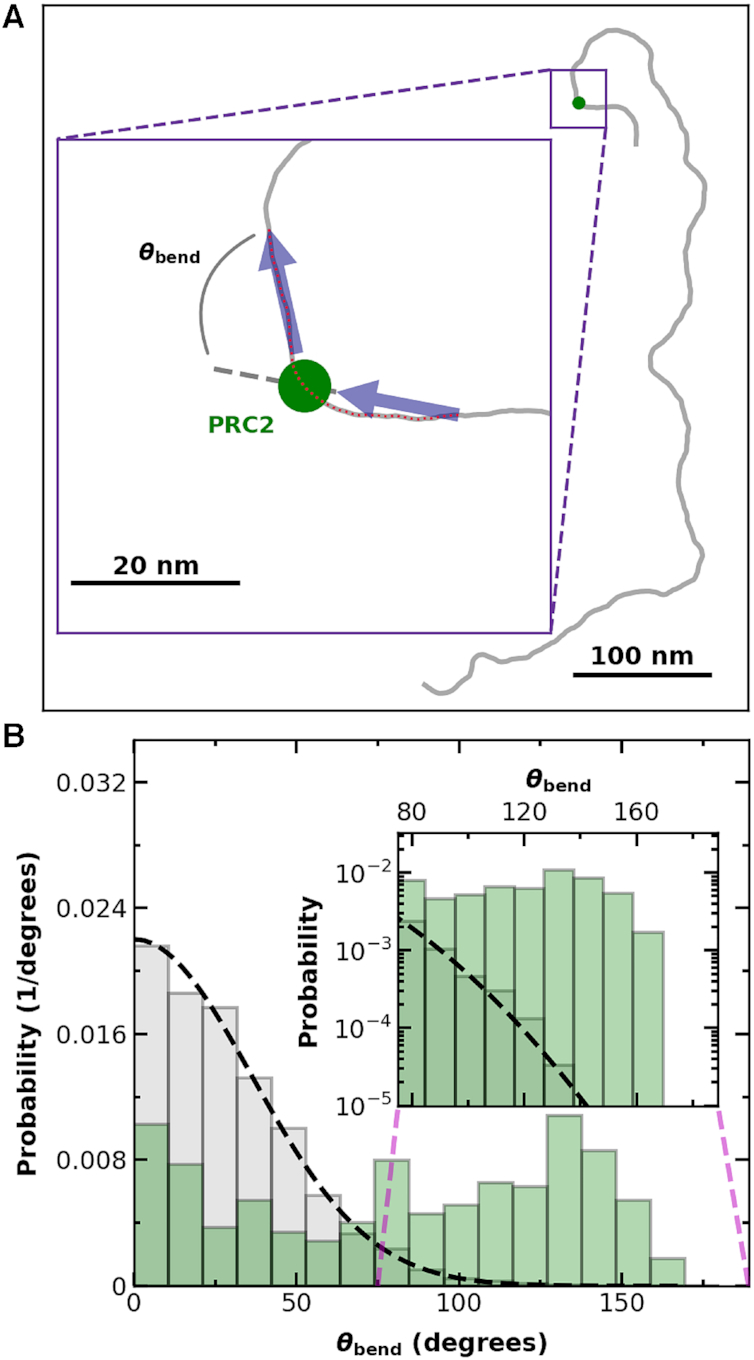
PRC2 binding is associated with an increased probability of large bends in the DNA backbone. (A) A spline denoting a 12 × 601 Widom DNA molecule (gray) bound by PRC2 (green dot). (Inset) Illustrative definition of the local bending angle (θbend) between two tangent vectors (black arrows). (B) Bending angle histogram for DNA (gray bars, Nmolecules = 70) and DNA with PRC2 bound (green bars, Nprotein = 331), where all tangent vectors were spaced 20 nm apart in contour length, and each tangent vector for the PRC2-bound DNA either started or ended at the PRC2, as shown in the cartoon in panel A. Dashed black line is the theoretically predicted bend angle distribution for tangent vectors separated by 20 nm along the DNA with a persistence length of 50 nm (54) equilibrated in two dimensions onto a surface. (Inset) Detailed distribution of large bend angle shows an excess of high angle bends for PRC2-DNA complexes in comparison to experimental and theoretical distributions of unbound DNA. Note, a single 851-nm DNA molecule gives many independent measurements of θbend.
PRC2 binding led to a broad, roughly evenly distributed set of bend angles (Figure 6B, green). Hence, PRC2 does not induce a unique angle in the backbone of DNA, in contrast to the TATA-box binding protein (60,61). On the other hand, PRC2 binding did lead to a 3-fold increase in the average of the absolute value of the bending angle over that of naked DNA [84 ± 3° (mean ± SEM; Nprotein = 331) versus 29.3 ± 0.4° (mean ± SEM; Nsegments = 2844; expected value is 28.9°)]. This enhanced bending appears to occur without substantial wrapping of the DNA around PRC2 that would lead to a shorter observed DNA length correlated with a higher bend angle (Figure 1D, Supplementary Figure S10). Increased bend angles reduce the energetic cost for distal parts of the stiff DNA backbone to interact. These increased bend angles are then a mechanism to facilitate protein-induced loop stabilization (as seen in Figure 2C) and/or association of distal regions of DNA.
Monomers and multimers of PRC2 distinguished
In addition to the mobility shift assay that shows multiple PRC2 molecules bound to individual DNAs (Figure 3E), PRC2 under some conditions binds ssRNA as a dimer (62). Hence, we sought to establish the ability to quantitatively distinguish monomeric from multimeric assembles of PRC2 by AFM. The standard analysis for such quantification is volume (46,49–51), with an understanding that the resulting deduced volumes inherently include a convolution of the tip radius. Initial volume analysis of PRC2-DNA complexes from 2 × 2 μm2 images was limited by the large pixel size = 4 nm. We therefore acquired a set of high-resolution images of PRC2 bound to DNA (pixel size = 1.2 nm) after incubating at 90 nM PRC2 so the background concentration of PRC2 adhered to the mica was not too high (Figure 7A). For this analysis, we selected for molecules where both ends of the DNA were clearly resolved and the contour length could be traced. As a control, looped DNA molecules in the absence of PRC2 were imaged. After a simple analysis to help decrease tip convolution (rnom = 8 nm, see Supplementary Figure S7), a histogram of the resulting PRC2 data showed two distinct peaks consistent with monomers and dimers of PRC2, along with a few larger complexes (N = 63; Figure 7B, green). The peak associated with monomeric PRC2 is clearly distinct from looped naked DNA (N = 63; Figure 7B, gray).
Figure 7.
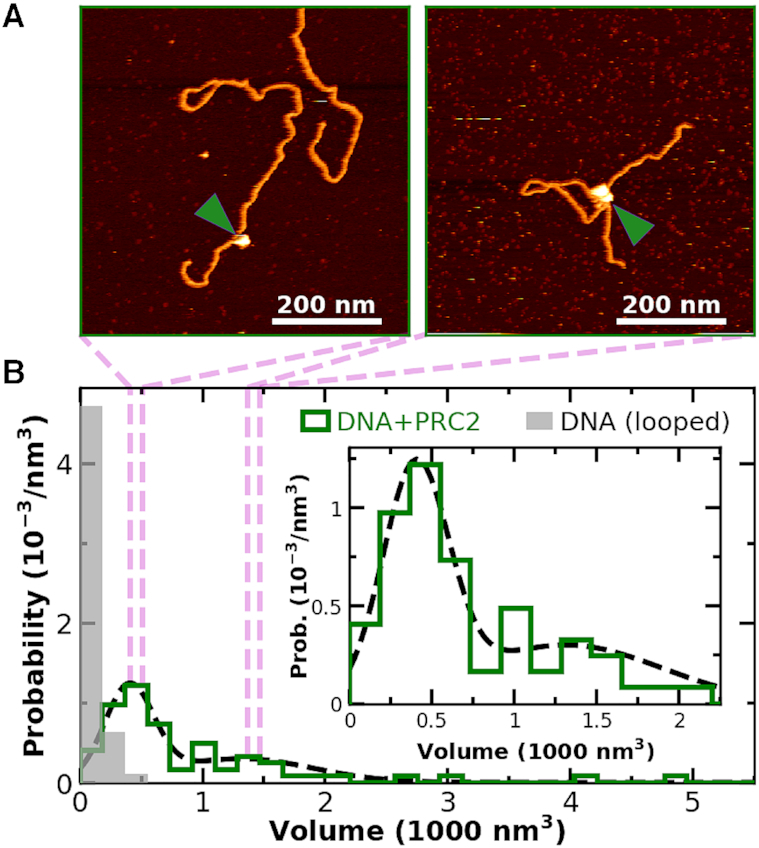
PRC2 binds to individual DNA molecules as monomers and multimeric complexes. (A) Two 600 × 600 nm2 images showing PRC2 bound to DNA, where volumetric analysis suggests monomeric and dimeric (or possibly trimeric) PRC2 complexes. Green arrows denote PRC2. (B) A histogram of the probability of measuring a particular volume at locations where the DNA backbone crosses itself [gray, N = 52] and where DNA is bound to PRC2 [green, N = 64]. Dotted black line is a fit of the sum of two Gaussians (means ± std. dev. are 400 ± 200 and 1300 ± 600 nm3) to the volume distribution of PRC2 bound to DNA. The two bound molecules with the highest volumes, not included in the 64 shown, were excluded for clarity and represent higher multimeric complexes. (Inset) Detailed plot of small volume distribution of PRC2 bound to DNA.
We note though the volume of the second peak is not exactly 2-fold larger, probably arising from three effects: the volume of the looped DNA in the case of compacted DNA (Figure 7A, right panel), tip-convolution artifacts, and the presence of trimeric or higher order complexes (Supplementary Figure S8). Indeed, imaging PRC2 in the absence of DNA indicated dimeric PRC2 complexes at high protein concentrations (Supplementary Figure S14), providing further evidence for multimeric PRC2 despite the absence of DNA. Future studies with sharper tips and shorter DNA molecules can refine these initial results. Such studies would likely benefit from photothermal actuation of the cantilever (63) or peak-force tapping (64) to ensure minimal forces exerted on the DNA.
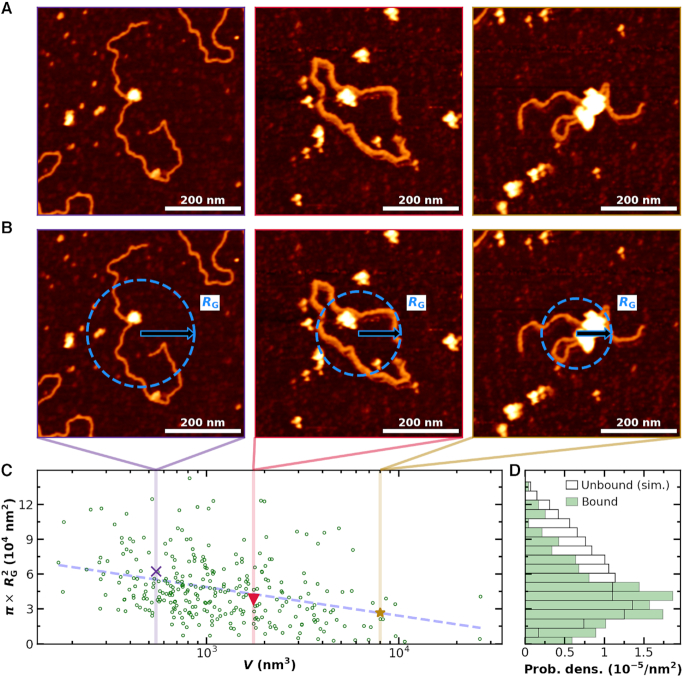
Higher DNA compaction with increasing bound PRC2
Different individual PRC2-DNA complexes displayed different levels of compaction (Figure 8A). A simple metric to quantify compaction on a 2D surface is to compute the mass-weighted area occupied by a DNA molecule (Figure 8B). To do so, we computed the area, defined by 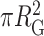 , where RG is radius of gyration, as a function of molecular volume of the PRC2 complex bound to each individual DNA molecule. To analyse a sufficiently large number of molecules (N = 300), we used the 2 × 2 μm2 images at the cost of precision in determining individual molecular volumes. The area shows a clear decrease with increasing molecular volume (Figure 8C). Note the images shown in Figure 8A, B were chosen to fall along the best fit line to the data shown in Figure 8C. The distribution of the areas is skewed towards more compacted structures as compared to the expected distribution for unbound DNA (Figure 8D). Hence, the degree of compaction increased with the number of PRC2 molecules bound to an individual DNA, and thus the degree and frequency of compaction via bending and looping increased with PRC2 concentration.
, where RG is radius of gyration, as a function of molecular volume of the PRC2 complex bound to each individual DNA molecule. To analyse a sufficiently large number of molecules (N = 300), we used the 2 × 2 μm2 images at the cost of precision in determining individual molecular volumes. The area shows a clear decrease with increasing molecular volume (Figure 8C). Note the images shown in Figure 8A, B were chosen to fall along the best fit line to the data shown in Figure 8C. The distribution of the areas is skewed towards more compacted structures as compared to the expected distribution for unbound DNA (Figure 8D). Hence, the degree of compaction increased with the number of PRC2 molecules bound to an individual DNA, and thus the degree and frequency of compaction via bending and looping increased with PRC2 concentration.
Figure 8.
Binding of multiple PRC2 compacts DNA. (A) Three representative AFM images of DNA compacted by PRC2. For clarity, the values between pixels were interpolated using a bicubic polynomial. (B) The same images as in panel A with a superimposed visual definition of the radius of gyration (RG) assuming mass is proportional to height. (C) Compaction area, defined as 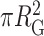 , as a function of PRC2–DNA complex volume (V), as defined in Materials and Methods (green circles, N = 300). Note that for DNA molecules bound by two or more well-separated PRC2 molecules (N = 58), we report the sum of the complex volumes, and for 43 compacted complexes without well-defined contours, as in the rightmost column of panel A, the volumes were corrected as described, except the average correction factor was used. Colored markers and dashed lines indicate the areas and volumes of the example images in panels A and B. A dashed line is plotted to guide the eye to the mean value as a function of V. (D) Distribution of area for simulated DNA with P = 50 nm and L0 = 851 nm (black open bars, N = 4000) and observed PRC2–DNA complexes (green bars). Simulation was performed as in (39). The data shown in panel C were derived from all 2 × 2 μm2 images used in Figure 3A with a deposition concentration of 300 nM PRC2 or less, and the example images in panels A and B all used a PRC2 concentration of 90 nM or less.
, as a function of PRC2–DNA complex volume (V), as defined in Materials and Methods (green circles, N = 300). Note that for DNA molecules bound by two or more well-separated PRC2 molecules (N = 58), we report the sum of the complex volumes, and for 43 compacted complexes without well-defined contours, as in the rightmost column of panel A, the volumes were corrected as described, except the average correction factor was used. Colored markers and dashed lines indicate the areas and volumes of the example images in panels A and B. A dashed line is plotted to guide the eye to the mean value as a function of V. (D) Distribution of area for simulated DNA with P = 50 nm and L0 = 851 nm (black open bars, N = 4000) and observed PRC2–DNA complexes (green bars). Simulation was performed as in (39). The data shown in panel C were derived from all 2 × 2 μm2 images used in Figure 3A with a deposition concentration of 300 nM PRC2 or less, and the example images in panels A and B all used a PRC2 concentration of 90 nM or less.
CONCLUSIONS
PRC2 recruitment to chromatin may depend on multiple interactions of shouvarying affinity, including interactions with histone marks, RNAs, protein factors and DNA. Here, we developed a single-molecule assay for imaging PRC2 binding to DNA in liquid using AFM. The PRC2–DNA complexes were deposited under biochemically relevant ionic conditions by applying a recently developed deposition protocol (39) that builds upon prior work (53,65,66), Notably, this deposition protocol preserved the physical properties of the DNA when bound to the mica substrate. Since surface-bound PRC2–DNA complexes did not exhibit a change in molecular topology during rinsing (Supplementary Figure S5) and were stable for over 1 h of continuous imaging (Supplementary Figure S4) in both imaging and deposition buffer, our sample preparation protocol took an effective ‘snapshot’ of the PRC2–DNA complexes in solution, a long standing interpretation of AFM imaging of protein complexes absorbed to mica (46). We anticipate leveraging this technique for high-resolution studies of PRC2-induced looping and compaction, similar to past studies of other DNA–protein complexes (41,67,68).
The resulting AFM images showed PRC2 binding to DNA and, at higher concentrations, compacting the DNA via intramolecular loops and protein-induced bending of DNA’s stiff helical backbone. Ensemble and single-molecule measurements yielded a similar 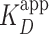 ≈ 100 nM, showing minimal perturbation to the protein-DNA complex during binding. At higher concentrations, PRC2 compacted the DNA via intramolecular loops and protein-induced bending of DNA’s stiff helical backbone. PRC2-induced looping and bending are both unanticipated results, leading us to suggest that PRC2 may directly contribute to heterochromatin formation independent of the H3K27me3 mark that it deposits. The DNA looping involved PRC2 dimers (or possibly multimers), while the bending was induced by a single PRC2 complex. Our findings that dimeric (or multimeric) PRC2 binds to one double-stranded DNA molecule mirrors prior work on RNA (62). Binding depended on the subunit composition of the PRC2. Among the PRC2 complexes tested, only the full PRC2 complex (EZH2, EED, SUZ12, AEBP2, and RBBP4) showed tight binding, consistent with prior work (11). PRC2 binding to long 2.5-kb sequences did not show strong localization to a region of high GC-content (76%), which can be compared to prior work using short 60-bp DNAs where 100% GC content bound 50-fold tighter than 0% GC (13). This result suggests that PRC2 binds tightly but promiscuously along more native-like DNA sequences. In summary, PRC2’s tight, promiscuous binding coupled with its ability to compact DNA via bending and looping suggests a direct role for PRC2 in compacting chromatin and therefore epigenetic silencing.
≈ 100 nM, showing minimal perturbation to the protein-DNA complex during binding. At higher concentrations, PRC2 compacted the DNA via intramolecular loops and protein-induced bending of DNA’s stiff helical backbone. PRC2-induced looping and bending are both unanticipated results, leading us to suggest that PRC2 may directly contribute to heterochromatin formation independent of the H3K27me3 mark that it deposits. The DNA looping involved PRC2 dimers (or possibly multimers), while the bending was induced by a single PRC2 complex. Our findings that dimeric (or multimeric) PRC2 binds to one double-stranded DNA molecule mirrors prior work on RNA (62). Binding depended on the subunit composition of the PRC2. Among the PRC2 complexes tested, only the full PRC2 complex (EZH2, EED, SUZ12, AEBP2, and RBBP4) showed tight binding, consistent with prior work (11). PRC2 binding to long 2.5-kb sequences did not show strong localization to a region of high GC-content (76%), which can be compared to prior work using short 60-bp DNAs where 100% GC content bound 50-fold tighter than 0% GC (13). This result suggests that PRC2 binds tightly but promiscuously along more native-like DNA sequences. In summary, PRC2’s tight, promiscuous binding coupled with its ability to compact DNA via bending and looping suggests a direct role for PRC2 in compacting chromatin and therefore epigenetic silencing.
DATA AVAILABILITY
The AFM images, PAGE images, and DNA sequences are available on Dryad (doi:10.5061/dryad.gqnk98shq). These data are available at the following link: https://datadryad.org/stash/dataset/doi:10.5061/dryad.gqnk98shq.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Steven Burroughs for help in figure production. Mention of commercial products is for information only; it does not imply NIST’s recommendation or endorsement. T.T.P. is a staff member of NIST’s quantum physics division.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB-1716033]; National Institute of Standards and Technology (NIST); T.R.C. is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: National Institute of Standards and Technology.
Conflict of interest statement. T.R.C. is on the board of directors of Merck, Inc., and a scientific advisor to Storm Therapeutics.
REFERENCES
- 1. Lewis E.B. A gene complex controlling segmentation in Drosophila. Nature. 1978; 276:565–570. [DOI] [PubMed] [Google Scholar]
- 2. Jurgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985; 316:153–155. [Google Scholar]
- 3. Margueron R., Reinberg D.. The Polycomb complex PRC2 and its mark in life. Nature. 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simon J.A., Kingston R.E.. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013; 49:808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blackledge N.P., Rose N.R., Klose R.J.. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 2015; 16:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G.. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 2017; 171:34–57. [DOI] [PubMed] [Google Scholar]
- 7. van der Lugt N.M., Domen J., Linders K., van Roon M., Robanus-Maandag E., te Riele H., van der Valk M., Deschamps J., Sofroniew M., van Lohuizen M. et al.. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994; 8:757–769. [DOI] [PubMed] [Google Scholar]
- 8. Davidovich C., Cech T.R.. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015; 21:2007–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaneko S., Bonasio R., Saldana-Meyer R., Yoshida T., Son J., Nishino K., Umezawa A., Reinberg D.. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014; 53:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holoch D., Margueron R.. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 2017; 42:531–542. [DOI] [PubMed] [Google Scholar]
- 11. Youmans D.T., Schmidt J.C., Cech T.R.. Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 2018; 32:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kasinath V., Faini M., Poepsel S., Reif D., Feng X.A., Stjepanovic G., Aebersold R., Nogales E.. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018; 359:940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Paucek R.D., Gooding A.R., Brown Z.Z., Ge E.J., Muir T.W., Cech T.R.. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 2017; 24:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi J., Bachmann A.L., Tauscher K., Benda C., Fierz B., Muller J.. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat. Struct. Mol. Biol. 2017; 24:1039–1047. [DOI] [PubMed] [Google Scholar]
- 15. da Rocha S.T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N.R., Sanulli S., Chow J., Schulz E., Picard C. et al.. Jarid2 Is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell. 2014; 53:301–316. [DOI] [PubMed] [Google Scholar]
- 16. Davidovich C., Zheng L., Goodrich K.J., Cech T.R.. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013; 20:1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Betancur J.G., Tomari Y.. Cryptic RNA-binding by PRC2 components EZH2 and SUZ12. RNA Biol. 2015; 12:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beltran M., Yates C.M., Skalska L., Dawson M., Reis F.P., Viiri K., Fisher C.L., Sibley C.R., Foster B.M., Bartke T. et al.. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016; 26:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Margueron R., Justin N., Ohno K., Sharpe M.L., Son J., Drury W.J. 3rd, Voigt P., Martin S.R., Taylor W.R., De Marco V. et al.. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009; 461:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiao L., Liu X.. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015; 350:aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brooun A., Gajiwala K.S., Deng Y.L., Liu W., Bolanos B., Bingham P., He Y.A., Diehl W., Grable N., Kung P.P. et al.. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 2016; 7:11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Justin N., Zhang Y., Tarricone C., Martin S.R., Chen S., Underwood E., De Marco V., Haire L.F., Walker P.A., Reinberg D. et al.. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016; 7:11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y.. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 24. Wang L., Brown J.L., Cao R., Zhang Y., Kassis J.A., Jones R.S.. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 2004; 14:637–646. [DOI] [PubMed] [Google Scholar]
- 25. Fursova N.A., Blackledge N.P., Nakayama M., Ito S., Koseki Y., Farcas A.M., King H.W., Koseki H., Klose R.J.. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol. Cell. 2019; 74:1020–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Healy E., Mucha M., Glancy E., Fitzpatrick D.J., Conway E., Neikes H.K., Monger C., Van Mierlo G., Baltissen M.P., Koseki Y. et al.. PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol. Cell. 2019; 76:437–452. [DOI] [PubMed] [Google Scholar]
- 27. Xu C., Bian C., Yang W., Galka M., Ouyang H., Chen C., Qiu W., Liu H., Jones A.E., MacKenzie F. et al.. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19266–19271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cao Q., Wang X., Zhao M., Yang R., Malik R., Qiao Y., Poliakov A., Yocum A.K., Li Y., Chen W. et al.. The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 2014; 5:3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H. et al.. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012; 148:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N.P., De Marco V., Elderkin S., Koseki H., Klose R. et al.. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014; 7:1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blackledge N.P., Farcas A.M., Kondo T., King H.W., McGouran J.F., Hanssen L.L., Ito S., Cooper S., Kondo K., Koseki Y. et al.. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014; 157:1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simon J.A., Kingston R.E.. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009; 10:697–708. [DOI] [PubMed] [Google Scholar]
- 33. Kundu S., Ji F., Sunwoo H., Jain G., Lee J.T., Sadreyev R.I., Dekker J., Kingston R.E.. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell. 2017; 65:432–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guthold M., Bezanilla M., Erie D.A., Jenkins B., Hansma H.G., Bustamante C.. Following the assembly of RNA polymerase-DNA complexes in aqueous solutions with the scanning force microscope. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:12927–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shlyakhtenko L.S., Lushnikov A.Y., Lyubchenko Y.L.. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009; 48:7842–7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murugesapillai D., Bouaziz S., Maher L.J., Israeloff N.E., Cameron C.E., Williams M.C.. Accurate nanoscale flexibility measurement of DNA and DNA-protein complexes by atomic force microscopy in liquid. Nanoscale. 2017; 9:11327–11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beckwitt E.C., Kong M., Van Houten B.. Studying protein-DNA interactions using atomic force microscopy. Semin. Cell Dev. Biol. 2018; 73:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hansma P.K., Cleveland J.P., Radmacher M., Walters D.A., Hillner P.E., Bezanilla M., Fritz M., Vie D., Hansma H.G., Prater C.B. et al.. Tapping mode atomic-force microscopy in liquids. Appl. Phys. Lett. 1994; 64:1738–1740. [Google Scholar]
- 39. Heenan P.R., Perkins T.T.. Imaging DNA equilibrated onto mica in liquid using biochemically relevant deposition conditions. ACS Nano. 2019; 13:4220–4229. [DOI] [PubMed] [Google Scholar]
- 40. Wang Y., Ran S., Yang G.. Single molecular investigation of DNA looping and aggregation by restriction endonuclease BspMI. Sci. Rep. 2014; 4:5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murugesapillai D., McCauley M.J., Huo R., Nelson Holte M.H., Stepanyants A., Maher L.J. 3rd, Israeloff N.E., Williams M.C.. DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin. Nucleic Acids Res. 2014; 42:8996–9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sukhanova M.V., Abrakhi S., Joshi V., Pastre D., Kutuzov M.M., Anarbaev R.O., Curmi P.A., Hamon L., Lavrik O.I.. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 2016; 44:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X., Goodrich K.J., Gooding A.R., Naeem H., Archer S., Paucek R.D., Youmans D.T., Cech T.R., Davidovich C.. Targeting of Polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell. 2017; 65:1056–1067. [DOI] [PubMed] [Google Scholar]
- 44. Long Y., Bolanos B., Gong L., Liu W., Goodrich K.J., Yang X., Chen S., Gooding A.R., Maegley K.A., Gajiwala K.S. et al.. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife. 2017; 6:31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mendenhall E.M., Koche R.P., Truong T., Zhou V.W., Issac B., Chi A.S., Ku M., Bernstein B.E.. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010; 6:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ratcliff G.C., Erie D.A.. A novel single-molecule study to determine protein–protein association constants. J. Am. Chem. Soc. 2001; 123:5632–5635. [DOI] [PubMed] [Google Scholar]
- 47. Sinden R.R. DNA Structure and Function. 1994; San Diego: Academic Press. [Google Scholar]
- 48. Hansma H.G., Bezanilla M., Zenhausern F., Adrian M., Sinsheimer R.L.. Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 1993; 21:505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wyman C., Rombel I., North A.K., Bustamante C., Kustu S.. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997; 275:1658–1661. [DOI] [PubMed] [Google Scholar]
- 50. Schneider S.W., Larmer J., Henderson R.M., Oberleithner H.. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflug. Arch. Eur. J. Phys. 1998; 435:362–367. [DOI] [PubMed] [Google Scholar]
- 51. Sanganna Gari R.R., Chattrakun K., Marsh B.P., Mao C., Chada N., Randall L.L., King G.M.. Direct visualization of the E. coli Sec translocase engaging precursor proteins in lipid bilayers. Sci. Adv. 2019; 5:eaav9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fuentes-Perez M.E., Gwynn E.J., Dillingham M.S., Moreno-Herrero F.. Using DNA as a fiducial marker to study SMC complex interactions with the atomic force microscope. Biophys. J. 2012; 102:839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rivetti C., Guthold M., Bustamante C.. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996; 264:919–932. [DOI] [PubMed] [Google Scholar]
- 54. Hagerman P.J. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 1988; 17:265–286. [DOI] [PubMed] [Google Scholar]
- 55. Fried M., Crothers D.M.. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981; 9:6505–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 57. Pyne A., Thompson R., Leung C., Roy D., Hoogenboom B.W.. Single-molecule reconstruction of oligonucleotide secondary structure by atomic force microscopy. Small. 2014; 10:3257–3261. [DOI] [PubMed] [Google Scholar]
- 58. Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C.L., Dynlacht B.D., Reinberg D.. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008; 32:503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rippe K., Guthold M., von Hippel P.H., Bustamante C.. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase · σ54 holoenzyme by scanning force microscopy. J. Mol. Biol. 1997; 270:125–138. [DOI] [PubMed] [Google Scholar]
- 60. Kim J.L., Nikolov D.B., Burley S.K.. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993; 365:520–527. [DOI] [PubMed] [Google Scholar]
- 61. Kim Y., Geiger J.H., Hahn S., Sigler P.B.. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993; 365:512–520. [DOI] [PubMed] [Google Scholar]
- 62. Davidovich C., Goodrich K.J., Gooding A.R., Cech T.R.. A dimeric state for PRC2. Nucleic Acids Res. 2014; 42:9236–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Labuda A., Kobayashi K., Miyahara Y., Grutter P.. Retrofitting an atomic force microscope with photothermal excitation for a clean cantilever response in low Q environments. Rev. Sci. Instrum. 2012; 83:053703. [DOI] [PubMed] [Google Scholar]
- 64. Adamcik J., Berquand A., Mezzenga R.. Single-step direct measurement of amyloid fibrils stiffness by peak force quantitative nanomechanical atomic force microscopy. Appl. Phys. Lett. 2011; 98:193701. [Google Scholar]
- 65. Hansma H.G., Laney D.E.. DNA binding to mica correlates with cationic radius: assay by atomic force microscopy. Biophys. J. 1996; 70:1933–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lyubchenko Y.L., Shlyakhtenko L.S., Ando T.. Imaging of nucleic acids with atomic force microscopy. Methods. 2011; 54:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Malabirade A., Jiang K., Kubiak K., Diaz-Mendoza A., Liu F., van Kan J.A., Berret J.F., Arluison V., van der Maarel J.R.C.. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017; 45:7299–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krzemien K.M., Beckers M., Quack S., Michaelis J.. Atomic force microscopy of chromatin arrays reveal non-monotonic salt dependence of array compaction in solution. PLoS One. 2017; 12:e0173459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AFM images, PAGE images, and DNA sequences are available on Dryad (doi:10.5061/dryad.gqnk98shq). These data are available at the following link: https://datadryad.org/stash/dataset/doi:10.5061/dryad.gqnk98shq.