Abstract
Mass spectrometry analysis of the Ebola virus soluble glycoprotein sGP identified a rare post-translation modification, C-mannosylation, which was found on tryptophan (W) 288. This modification has not been described for any other viral protein; however, many viral transmembrane glycoproteins contain one or more of the recognition motifs (W-x-x-W). Elimination of the C-mannose on sGP did not significantly alter protein biosynthesis, processing or structure. Furthermore, the protective effect of sGP on endothelial barrier function, currently the only known activity of sGP, was unaltered. It is possible that C-mannosylation may be a common post-translational modification of viral transmembrane glycoproteins where it could play a role in particle maturation and/or entry by stabilizing the structure of these proteins. In this regard, C-mannosylation of sGP may be an anomaly resulting from the unique manner in which this protein is generated as the product of unedited transcripts from the glycoprotein gene of Ebola.
Keywords: C-mannosylation, Ebola, Glycoprotein, Post-translational modification, Structure–function
Introduction
Ebola viruses (EBOV) belong to the family Filoviridae in the order Mononegavirales. These viruses cause a severe hemorrhagic fever, termed Ebola hemorrhagic fever (EHF), with fatality rates as high as 90% (Feldmann et al., 2003). The fourth gene of the EBOV genome encodes the precursors of a soluble non-structural glycoprotein (pre-sGP) and the structural transmembrane glycoprotein (pre-GP) (Sanchez et al., 1996, Volchkov et al., 1995). Pre-sGP is the primary product and is processed by signalase and furin cleavage into sGP and Δ-peptide (Volchkova et al., 1999), both of which are secreted. sGP has been detected in the serum of infected individuals (Sanchez et al., 1996) and is hypothesized to play a role in the pathogenesis of EHF (Feldmann et al., 2003). Pre-GP undergoes similar processing to yield the mature viral spike protein GP1,2, which consists of the two disulphide-bonded cleavage fragments GP1 and GP2 (Sanchez et al., 1998, Volchkov et al., 1998). sGP shares the 295 N-terminal amino acids with GP1 but has a unique 28 amino acid carboxyl-terminus, whereas pre-GP is only expressed after RNA editing (Volchkov et al., 1995). Despite the large region of shared primary sequence, these proteins are markedly different in their structure, with sGP forming homodimers (Volchkova et al., 1998, Barrientos et al., 2004, Falzarano et al., 2006), which according to the more recent studies seem to exist in a parallel rather than an anti-parallel orientation, and GP1,2 forming trimers (Sanchez et al., 1998). These proteins also differ in their function; GP1,2 has been shown to mediate virus entry (attachment and fusion) (Feldmann et al., 2001) while sGP, besides additional yet unknown functions, might have an anti-inflammatory role (Wahl-Jensen et al., 2005). As part of our efforts to characterize the structure of sGP (including co- and post-translational modifications) for future functional studies, we recently demonstrated that in contrast to GP1,2, which is heavily N- and O-glycosylated (Feldmann et al., 2001), sGP carries solely N-linked carbohydrates (Falzarano et al., 2006). Here we discuss the discovery of a novel type of glycosylation, termed C-mannosylation, which has yet to be identified for any viral protein. Zaire ebolavirus (ZEBOV) sGP was found to possess a C-mannose on tryptophan (Trp) residue 288 (W288). A mutant sGP lacking this modification was unaltered in its biosynthesis, processing, structure and potential anti-inflammatory function. This raises the question as to whether C-mannosylation of sGP has functional consequences or if it is a fortuitous occurrence due to the parasitic nature of viruses in mammalian cells.
Results
Ebola virus sGP is C-mannosylated
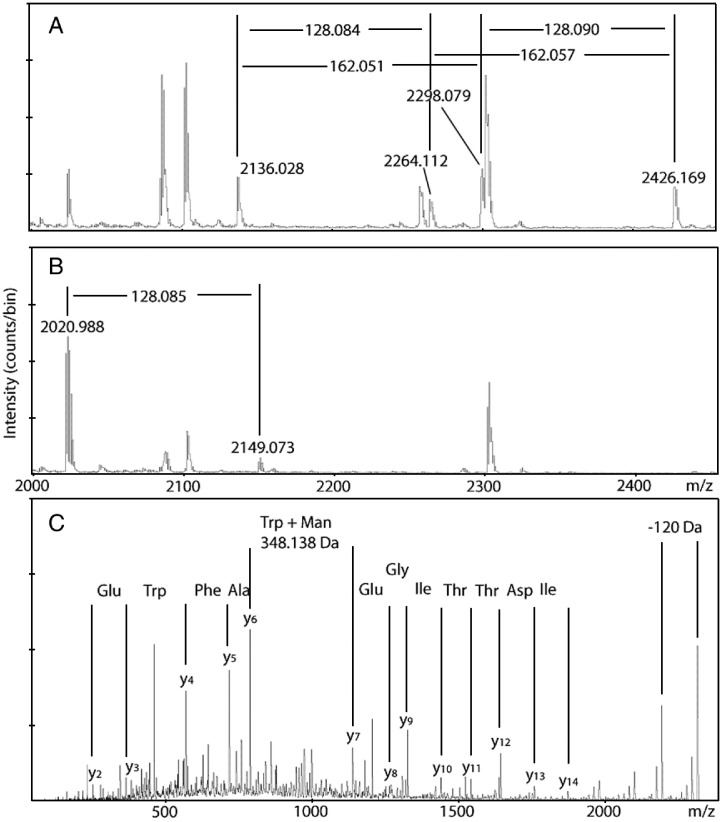
sGP peptide mapping by MALDI MS and HPLC-MALDI MS was performed as previously described following trypsin digestion (Falzarano et al., 2006). Two unmodified tryptic peptides containing amino acids 288 to 291 (WAFW) were found: T26 (VNPEIDTTIGEWAFWETK) and T26 + T27 (VNPEIDTTIGEWAFWETKK) (Fig. 1A). The presence of a peptide with a missed cleavage is typical when there are two adjacent cut sites (KK) and in this case results in two peaks separated by ∼ 128.095 Da. Both peptides showed + 162 Da adducts, consistent with the mass of a hexose residue (162.053 Da, Delta Mass—a database of protein post-translational modifications, http://www.abrf.org/index.cfm/dm.home). A comparison of the relative intensities of the non-modified peptides and their respective + 162 Da adducts (Fig. 1A) suggested that approximately 60% of the peptides were modified. MALDI MS analysis allowed all four of the fragments to be observed simultaneously, while HPLC-MALDI MS analysis introduces an additional retention time shift for the + 162 Da modified species—they elute ∼ 1 min earlier under the chromatographic conditions used. This finding is consistent with the addition of a hydrophilic sugar moiety to a hydrophobic Trp residue. The identification of this post-translational modification was further supported by the presence of the recognition sequence, W-x-x-W, for C-mannosylation at amino acid position 288 to 291 (W-A-F-W) of sGP, which was part of both peptides.
Fig. 1.
Identification of C-mannosylation on sGP by mass spectrometry. (A) A portion of the MALDI/TOF MS spectrum of tryptic digests of purified sGP containing non-modified m/z 2136.028 (VNPEIDTTIGEWAFWETK), 2264.112 (VNPEIDTTIGEWAFWETKK) and C-mannosylated fragments m/z 2298.079 and 2426.169, respectively. The + 162 Da shift resulting from the addition of the mannose is indicated. (B) Spectrum demonstrating that the fragment of sGP-WXXA mutated to prevent C-mannosylation does not contain a peak shift of + 162 Da. Respective peaks of m/z 2020.988 (VNPEIDTTIGEWAFAETK) and 2149.073 (VNPEIDTTIGEWAFAETKK) are separated by 128.085 (one Lys residue). (C) MS/MS spectrum of C-mannosylated peptide m/z 2298. The y-series of ions from the fragmentation of the (277–294) VNPEIDTTIGEWAFWETK peptide are shown as well as the characteristic 120-Da loss from the parent peptide. The difference of 348.138 Da between the y6 and y7 ions corresponds to the 288-tryptophan residue with a mannose attached (186.079 + 162.053 = 348.132).
Final confirmation of W288 C-mannosylation was obtained during low-energy CID MS/MS analysis of the modified peptides. Fig. 1C shows MS/MS spectrum of the + 162 Da adducts of the T26 (VNPEIDTTIGEWAFWETK) peptide (MH+ 2298). A complete series of y ions (y2 to y6) confirms that W291 is unmodified, while all y ions starting at y7 have a mass increase of 162 Da. This is a strong indication that a mannose residue is attached to W288. Furthermore, the characteristic 120 Da loss (Furmanek and Hofsteenge, 2000) that occurs during CID of mannosylated peptides was also observed (Fig. 1C). MS/MS analysis of the second mannosylated peptide T26 + T27 (VNPEIDTTIGEWAFWETKK) (MH+ 2426) showed similar results.
To ensure that this modification was not unique to the specific cell line used, sGP was also purified from transfected Vero E6 and Huh7 cell lines. MALDI MS revealed a similar spectra to that of the 293T cell derived sGP with modified peptides indicating the presence of a mannose residue (Figs. 2A, B). This is not unexpected as others have reported that C-mannosylation occurs in multiple tissue types from diverse species (Krieg et al., 1997, Doucey et al., 1998). In addition, sGP purified from ZEBOV-infected Vero cells was shown to contain the C-mannosylated peptide T26 following HPLC-MALDI MS (Fig. 2C), but in contrast to the transfected cell lines, only C-mannosylated peptides were detected. To confirm this result, the peak corresponding to C-mannosylated T26 was selected for MS/MS which yielded results identical to Fig. 1C (data not shown).
Fig. 2.
C-mannosylation of sGP occurs in multiple cell lines and during virus infection. (A) MALDI/TOF MS spectrum of tryptic digests of purified sGP from Huh7 and (B) Vero E6 cell lines showing non-modified m/z 2136 (VNPEIDTTIGEWAFWETK), 2264 (VNPEIDTTIGEWAFWETKK) and C-mannosylated fragments m/z 2298 and 2426, respectively. The addition of the mannose (+ 162 Da) is indicated. (C) Spectrum of fraction 28 from purified supernatant of Zaire ebolavirus-infected Vero E6 cells indicating C-mannosylated peptide m/z 2298.082.
Alanine mutation of W291 prevents C-mannosylation of Ebola virus sGP
In order to confirm C-mannosylation of W288, a mutant sGP with an altered recognition sequence (sGP-WXXA), which has previously been demonstrated to eliminate C-mannosylation (Krieg et al., 1998), was created (Fig. 3A). sGP-WXXA was expressed at similar levels to sGP and was secreted from the cell as a homodimer with the same molecular mass as sGP (Fig. 3A), indicating that biosynthesis, processing and transport of the mutant was not significantly affected. Interestingly, the mutation obviously destroyed one of the immunogenic epitopes on sGP since monoclonal antibody Z42/3.7 was not able to detect sGP-WXXA by western blot (Fig. 3B). A comparable peptide mapping of sGP-WXXA by MALDI MS and HPLC-MALDI MS resulted in the elimination of the mass shift observed for both peptides T26 and T26 + T27 derived from wild-type sGP (Fig. 1B). All identified post-translational modifications and disulphides bonds for sGP are summarized in Fig. 3C.
Fig. 3.
Characterization of sGP and sGP-WXXA biosynthesis and structure. SDS–PAGE (10%) analysis of sGP and sGP-WXXA under reducing (R) and non-reducing (NR) conditions using monoclonal antibodies (A) P1302H11 (1:100 dilution) and (B) Z42/3.7 (1:4000 dilution). The numbers on the left indicate molecular mass in kDa. Lane 1, sGP-WXXA; lane 2, sGP. (C) Schematic representation of the sGP homodimer (grey) indicating known post-translational modifications, including the site of C-mannosylation (red W) as well as N-glycosylation sites (green N) and all disulphide bonds (blue C). The location of the signal peptidase cleavage site (SP) and the furin cleavage site (FC) is indicated, as is Δ-peptide (brown).
C-mannosylation of sGP does not affect its anti-inflammatory role
We have recently demonstrated that sGP is able to restore the decrease in barrier function of endothelial cells, which can be provoked through TNF-α treatment (Wahl-Jensen et al., 2005). This potential anti-inflammatory role of sGP is structure dependent (Falzarano et al., 2006) and currently the only functional test for this protein. It has been suggested that C-mannosylation may play a role in protein folding, especially involving folding around disulphide bonds. The location of the C-mannosylated W288 is in close proximity to the cysteine residue involved in one of the intermolecular disulphide bonds (C306) important for the formation of sGP homodimers (Barrientos et al., 2004, Falzarano et al., 2006). As observed previously (Wahl-Jensen et al., 2005), treatment of endothelial cells with TNF-α leads to a decrease in transendothelial resistance (TER), which can be partially reversed by sGP (Fig. 4 ). The same rescue effect was also observed when endothelial cells were treated simultaneously with TNF-α and sGP-WXXA (Fig. 4), demonstrating that the lack of C-mannosylation of W288 did not alter this biological function of sGP, although it may still be important for another as of yet unidentified function.
Fig. 4.
Effect of C-mannosylation on sGP rescue of TNF-α-treated endothelial cells under static conditions. Human endothelial cells (HUVEC) were equilibrated for 1 h to generate a baseline TER. TNF-α with and without sGP or sGP-WXXA (10 μg ml− 1) were added to the medium as indicated. Following treatment, the chambers were monitored by impedance spectroscopy. There was a significant difference (P < 0.05) between the TER of cells treated with TNF-α alone and cells treated with TNF-α and sGP or sGP-WXXA at 900 min for sGP (**) and 420 min for sGP-WXXA (*).
C-mannosylation could be widely distributed among viral proteins
A search of the non-redundant NCBI RefSeq viral protein database revealed 2323 proteins that contain the recognition sequence W-x-x-W for C-mannosylation. Only proteins that enter the endoplasmic reticulum are considered to have the potential to be C-mannosylated; therefore, protein sequences containing W-x-x-W were run through the SignalP 3.0 server in both NN and HMM modes to predict whether they could possess a signal peptide. This predictive modeling gives a yes/no (NN) or a predicted signal peptide/non-secretory protein/signal anchor prediction (HMM). From this analysis, 373 viral proteins were predicted to have signal peptides, indicating that they may have the potential to be C-mannosylated. Among these, EBOV GP1,2 contains 3 potential C-mannosylation sites, which are highly conserved in all 4 EBOV species, including one in the same position as sGP, due to the similarity in the primary sequence of the two proteins (see introduction and Feldmann et al., 2001). Further candidate proteins for C-mannosylation can be found among RNA viruses and include the single transmembrane proteins vesicular stomatitis virus (VSV) G, Borna disease virus (BDV) G and Crimean–Congo hemorrhagic fever virus (CCHF) GC, the hemagglutinin and M2 proteins of influenza virus, the E2 protein of SARS-coronavirus, the spike proteins of other human coronaviruses and the envelope proteins of human immunodeficiency virus (HIV)-1 and 2 (Table 1 ). This modification may also be present in glycoproteins of DNA viruses including Hepatitis B virus, Myxoma virus and numerous Herpesviruses.
Table 1.
Proteins from selected viruses that contain the C-mannosylation recognition sequence W-x-x-W and are predicted to have the potential to cross the ER membrane as determined by SignalP 3.0 in both Neural Net (NN) and Hidden Markov Model (HMM) modes
| Family | Virus | Protein | NN | HMM | |
|---|---|---|---|---|---|
| dsDNA | Poxviridae | Myxoma virus | m4.1 | Yes | Signal peptide |
| Molluscum contagiosum virus | MC011L | Yes | Signal peptide | ||
| Herpesviridae | Human herpesvirus 1 | Virion glycoprotein M | No | Signal anchor | |
| Human herpesvirus 2 | Virion glycoprotein M | No | Signal anchor | ||
| Tegument protein | No | Signal peptide | |||
| Hepadnaviridae | Hepatitis B virus | S protein | Yes | Signal peptide | |
| RT | Retroviridae | Friend murine leukemia virus | Envelope protein | Yes | Signal peptide |
| Human immunodeficiency | gp41 | Yes | Signal peptide | ||
| Virus 1/2 | gp160 | Yes | Signal peptide | ||
| Simian immunodeficiency virus | env | Yes | Signal peptide | ||
| dsRNA | Reoviridae | Human rotavirus | VP7 | Yes | Signal anchor |
| (-)ssRNA | Bornaviridae | Borna disease virus | Glycoprotein | Yes | Signal peptide |
| Rhabdoviridae | Vesicular stomatitis virus | Glycoprotein | Yes | Signal peptide | |
| Mokola virus | Transmembrane glycoprotein | Yes | Signal peptide | ||
| Filoviridae | Cote d'Ivoire ebolavirus | Secreted glycoprotein | Yes | Signal peptide | |
| Structural glycoprotein | Yes | Signal peptide | |||
| Reston ebolavirus | Small/secreted non-structural glycoprotein | Yes | Signal peptide | ||
| Structural glycoprotein | Yes | Signal peptide | |||
| Sudan ebolavirus | Secreted glycoprotein | Yes | Signal peptide | ||
| Structural glycoprotein | Yes | Signal peptide | |||
| Zaire ebolavirus | sGP | Yes | Signal peptide | ||
| ssGP | Yes | Signal peptide | |||
| Virion spike glycoprotein precursor | Yes | Signal peptide | |||
| Orthomyxoviridae | Influenza A virus (A/New York/392/2004 (H3N2)) | Hemagglutinin | Yes | Signal peptide | |
| Influenza C virus | CM2 protein | Yes | Signal peptide | ||
| Bunyaviridae | Crimean–Congo hemorrhagic fever virus | Glycoprotein precursor | Yes | Signal peptide | |
| (+)ssRNA | Coronaviridae | Human coronavirus 229E | Surface glycoprotein | Yes | Signal peptide |
| 4a protein | Yes | Signal peptide | |||
| Membrane protein | Yes | Non-secretory | |||
| Murine hepatitis virus | E1 glycoprotein | No | Signal anchor | ||
| E2 glycoprotein precursor | Yes | Signal peptide | |||
| Spike glycoprotein S | Yes | Signal peptide | |||
| Membrane protein M | Yes | Signal anchor | |||
| SARS coronavirus | E2 glycoprotein precursor | Yes | Signal peptide | ||
| Matrix protein | Yes | Signal anchor | |||
| Flaviviradae | Dengue virus type 1 | Non-structural protein 2B | Yes | Signal anchor | |
| West Nile virus | Polyprotein precursor | No | Signal peptide | ||
| Hepatitis C virus | NS2 protein | Yes | Signal anchor | ||
| Togaviridae | Ross River virus | 6K protein | No | Signal anchor | |
| Rubella virus | Glycoprotein E1 | No | Signal peptide | ||
| Parvoviridae | Canine parvovirus | Polyprotein | No | Signal peptide |
NN predicts the presence/absence of a signal peptide (yes/no). HMM predicts the presence of a signal peptide/signal anchor or the lack thereof (non-secretory). The viruses are grouped according to the guidelines of the International Committee on Taxonomy of Viruses (ICTV).
Discussion
C-mannosylation is a relatively unknown and poorly studied post-translation modification that has been implicated in protein folding and/or trafficking (Furmanek and Hofsteenge, 2000). Here we present the first identification of C-mannosylation on a viral protein—ZEBOV sGP. Abolishment of C-mannosylation did not significantly alter the biosynthesis, processing or release of sGP. It also did not affect its anti-inflammatory function but may impact on another as of yet unidentified sGP function.
C-mannosylation involves the covalent attachment of an α-mannopyranosyl residue to the indole C2 carbon atom of Trp via a C–C linkage (Hofsteenge et al., 1994). This modification was first described for W7 of human RNase 2 (Hofsteenge et al., 1994). C-mannosylation has been shown to be part of the normal biosynthetic pathway in Caenorhabditis elegans, amphibians, birds and mammals but does not appear to occur in insect, plant, yeast or bacterial cells (Doucey et al., 1998, Krieg et al., 1997). The linear recognition sequence consists of W-x-x-W/F, in which the first Trp residue becomes mannosylated (Doucey et al., 1998). Replacement of the second Trp with a Phe residue reduces the efficiency of C-mannosylation 3.5-fold (Krieg et al., 1998). The C-mannosylation synthesis pathway, which appears to be enzymatically catalyzed by proteins present in liver microsomes (Doucey et al., 1998, Krieg et al., 1998), has been dissected and is as follows: Mannose (Man) → → GDP-Man → Dolichylphosphate-Man (Dol-P-Man) → (C2-Man-Trp) (Doucey et al., 1998).
A number of different cellular proteins have been identified as having C-mannosylated Trp residues with some of these proteins containing multiple modifications (Hofsteenge et al., 2001). Proteins that contain C-mannosylation serve a variety of cellular functions and include extracellular matrix proteins, axonal guidance proteins, proteins involved in angiogenesis, metalloproteases and a number of proteins from the complement system, including properdin and the terminal complement components C6 to C9. It appears that for RNase2, as well as the six polypeptides from the complement system, the hydrophilic mannose residue fills a cavity formed by the main and side chain atoms of a loop on an exposed region of the protein, thus playing a structural role (Mosimann et al., 1996). It has also been suggested that the C-mannose may be involved in protein transport as mutations in the W-x-x-W motif of the erythropoietin receptor and MP20 (a bovine lens protein that is C-mannosylated at W43 and W61) stall the transport of these proteins in the ER-Golgi interface or the ER, respectively (Hilton et al., 1996, Perez-Vilar et al., 2004). Other studies have identified that mutagenesis of the Trp residues in the W-x-x-W motif abolishes the binding of the ligand for the IL-2 receptor (Miyazaki et al., 1991).
The discovery of protein C-mannosylation in a viral protein is of interest. In many cases, post-translational modifications are essential for the function and/or regulation of a given protein. Previous work has shown that sGP was able to reverse the permeability increasing effect of TNF-α on endothelial cells and partially restore endothelial barrier function (Wahl-Jensen et al., 2005). This finally provided us with a functional assay system for sGP that was recently used to demonstrate that the activity of sGP is critically dependent on its structure as maintained by correct intermolecular disulphide bonding (Falzarano et al., 2006). Here we introduced a point mutation into sGP that is sufficient to abolish C-mannosylation and thus may alter the structure of sGP. However, our investigations did not reveal changes in the biosynthesis, transport, structure or function of the mutant sGP-WXXA compared to wild-type sGP (Fig. 3, Fig. 4). Moreover, the potential anti-inflammatory effect of sGP (Wahl-Jensen et al., 2005) might not be the sole function, as it has been suggested by others that sGP may play a role in immune evasion by acting as a decoy molecule for EBOV-specific humoral immune responses or bind to neutrophils and subsequently inhibit their activation (Ito et al., 2001, Yang et al., 1998). Unfortunately, these proposed functions have been difficult to prove directly.
The vast majority of the viral proteins that contain a W-x-x-W motif and have the potential to be C-mannosylated are surface glycoproteins (Table 1). With this in mind, the modification of sGP may be an anomaly resulting from the unique manner in which this protein is generated as the product of unedited transcripts from the glycoprotein gene of EBOV (Feldmann et al., 2001). While not excluding a role of C-mannosylation on sGP function, it could be that C-mannosylation may impact on the function of the EBOV transmembrane glycoprotein GP1,2. This protein contains multiple potential C-mannosylation sites with two located adjacent to the predicted transmembrane region of GP2 and one on W288 of GP1, which is identical to the one on sGP. The location of a C-mannose on the extracellular portion near the membrane is not unprecedented as bovine lens protein MP20 also contains this modification in a similar location (Perez-Vilar et al., 2004). The location of a C-mannosylation site just inside the transmembrane region of the BDV G and on the extracellular portion of the VSV G may further indicate that this modification is important for the orientation of membrane proteins or trafficking of viral membrane proteins to the cell surface, as has been suggested for C-mannosylated mammalian proteins (Ervin et al., 2005, Hilton et al., 1996). Alternatively, C-mannosylation of viral proteins could also be a chance event due to the parasitic nature of mammalian viruses that highjack the cellular machinery to complete their life cycle; however, we find this unlikely.
Materials and methods
Cloning and site-directed mutagenesis
The region of the GP gene corresponding to the ZEBOV (strain Mayinga) sGP open reading frame lacking the region expressing the signal peptide was cloned into the pDisplay vector (Invitrogen, Burlington, ON), which contains an endogenous signal peptide and an N-terminal HA-tag. Site-directed mutagenesis was performed with the QuikChangeII site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions with specifically altered oligonucleotides resulting in the W288AFW → W288AFA mutation in sGP. The presence of the mutations was verified by sequencing after purification of plasmid DNA using the QIAfilter Plasmid Maxi kit (Qiagen, Mississauga, ON).
Protein expression and purification
The appropriate plasmid was transfected into low passage 293T cells, Vero E6 (both human embryonic kidney cells) and Huh7 (human hepatoma cells) using FuGene6 (Roche Diagnostics, Laval, QC) (6 μl per 1 μg DNA) in OptiMEM (Invitrogen). Seventy-two hours post-transfection, supernatant was collected and centrifuged at 5000×g for 15 min to remove cell debris. The cell-free supernatant was concentrated with a Centricon PL-70 (30000 MWCO) (Millipore, Cambridge, ON) and added to an anti-HA affinity matrix (Roche Diagnostics, Laval, QC) as per the manufacturer's instructions and rotated overnight at 4 °C. The supernatant plus the matrix were added to gravity flow columns and washed extensively with TNE buffer (20 mM Tris–HCl, 100 mM NaCl, 0.1 mM EDTA) containing Tween20 (0.05%). The HA-tagged glycoproteins were eluted with excess HA peptide (American Peptide Company, Sunnyvale, CA) and the HA peptide eliminated by multiple washes with TNE buffer through an Amicon Ultra 4 (30000 MWCO) (Millipore). The presence of the recombinant protein was confirmed by western blot using two monoclonal antibodies directed against ZEBOV GP (Z42/3.7 and P1302H11, courtesy of Ayato Takada, Hokkaido University, Japan and Mavanur Suresh, University of Alberta, respectively).
Virus infection
Vero E6 cells were infected with ZEBOV (strain Mayinga) in the biosafety level 4 (BSL4) laboratory at the National Microbiology Laboratory at an MOI of approximately 0.5. Three days post-infection, the supernatant was removed and centrifuged at 5000×g for 15 min. To remove most of the virus, the supernatant was centrifuged at 21000 RPM in an SW-41 rotor for 30 min. The supernatant was poured off and passed through a 0.22-μm filter. SDS was added to the supernatant to 0.5% and the sample was irradiated (2 MRad). The supernatant was dialyzed extensively against 20 mM Tris–HCl, 500 mM NaCl, pH 7.4 buffer (TN buffer) and then added to a concanavalin A column (GE Healthcare). Unbound protein was washed through with TN buffer. Bound protein was eluted with 0.75 mM methyl-α-d-glucopyranoside in TN buffer. The fractions were concentrated with an Amicon (Millipore) and subjected to western blot. The appropriate fraction was then prepared for mass spectrometry.
Mass spectrometry
Sample preparation for peptide mapping by MALDI MS and HPLC-MALDI MS was performed as previously described (Falzarano et al., 2006). Briefly, samples were reduced (10 mM DTT, 65 °C, 30 min), alkylated (50 mM iodoacetamide, room temperature, 45 min), dialyzed (7,500 MWCO, against 100 mM ammonium bicarbonate) and digested with 1:50 trypsin/substrate ratio (with or without prior deglycosylation with PNGaseF (New England Biolabs, Pickering, ON). The resultant digests were either spotted directly on MALDI targets with a 2,5-dihydroxybenzoic acid matrix solution or directly injected (5 μl) into the micro HPLC system. Both non-separated samples and chromatographic fractions were analyzed by MS and by tandem mass spectrometry (low-energy collision-induced dissociation (CID), MS/MS) in the Manitoba/Sciex prototype MALDI quadrupole/TOF (QqTOF) mass spectrometer. This instrument provides resolving power of 10,000 full width at half maximum (FWHM) and mass accuracy of ∼ 10 ppm in both MS and MS/MS modes.
Endothelial cell culture and impedance spectroscopy
Impedance spectroscopy is a highly sensitive biophysical assay that provides a unique possibility to study the endothelial barrier function. This technique determines the transendothelial electrical resistance (TER) of a cultured endothelial cell monolayer and predominantly reflects changes in paracellular permeability. Human umbilical vein endothelial cells (HUVEC) were isolated and cultured as described elsewhere (Schnittler et al., 1990) and TER was determined as previously described (Seebach et al., 2000). Briefly, an alternating voltage was applied and the impedance magnitude was measured at frequencies between 10 Hz and 1 MHz between the electrode area of the ITO slide and a counterelectrode. The TER was calculated from the resultant spectra. Prior to addition of TNF-α (1 ng ml− 1) (R&D Systems, Minneapolis, MN) and sGP or sGP-WXXA (10 μg ml− 1), cells were equilibrated for 1 to 2 h to establish a baseline TER. All electrical resistance data are presented as normalized to baseline resistance values (TER/TER0). TER data are shown as mean ± standard error. Data were compared by unpaired t-test. Values were considered to be statistically significant when P was < 0.05.
Viral protein database search for C-mannosylation motif
Protein sequences from the non-redundant NCBI RefSeq viral protein database (ftp://ftp.ncbi.nih.gov/refseq/release/viral/viral1.protein.faa.gz) were scanned for the recognition sequence W-x-x-W. These sequences were then truncated to the N-terminal 90 amino acid residues and run through SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) in both Neural Net (NN) and Hidden Markov Model (HMM) modes. These algorithms predict whether a protein is likely to contain a signal peptide.
Acknowledgments
We would like to thank Ayato Takada (Hokkaido University, Sapporo, Japan) and Mavanur Suresh (University of Alberta, Edmonton, Alberta) for supplying the Zaire ebolavirus-specific monoclonal antibodies Z42/3.7 and P1302H11, respectively. We would like to thank John Wilkins (Manitoba Centre for Proteomics, Winnipeg, Manitoba) for helpful discussions and we would further like to thank Allison Groseth, Hideki Ebihara and Ute Ströher for critical review of the manuscript. This work has been supported by a grant from the Canadian Institutes of Health Research (MOP 40321) awarded to H.F., the Public Health Agency of Canada and by the Deutsche Forschungsgemeinschaft within the priority program “ Infection of the endothelium” awarded to H.S.
References
- Barrientos L.G., Martin A.M., Rollin P.E., Sanchez A. Disulfide bond assignment of the Ebola virus secreted glycoprotein SGP. Biochem. Biophys. Res. Commun. 2004;323(2):696–702. doi: 10.1016/j.bbrc.2004.08.148. [DOI] [PubMed] [Google Scholar]
- Doucey M.A., Hess D., Cacan R., Hofsteenge J. Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell. 1998;9(2):291–300. doi: 10.1091/mbc.9.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervin L.A., Ball L.E., Crouch R.K., Schey K.L. Phosphorylation and glycosylation of bovine lens MP20. Invest. Ophthalmol. Visual Sci. 2005;46(2):627–635. doi: 10.1167/iovs.04-0894. [DOI] [PubMed] [Google Scholar]
- Falzarano D., Krokhin O., Wahl-Jensen V., Seebach J., Wolf K., Schnittler H.J., Feldmann H. Structure–function analysis of the soluble glycoprotein, sGP, of Ebola virus. ChemBioChem. 2006;7(10):1605–1611. doi: 10.1002/cbic.200600223. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Volchkov V.E., Volchkova V.A., Stroher U., Klenk H.D. Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 2001;82(Pt 12):2839–2848. doi: 10.1099/0022-1317-82-12-2839. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Jones S., Klenk H.D., Schnittler H.J. Ebola virus: from discovery to vaccine. Nat. Rev., Immunol. 2003;3(8):677–685. doi: 10.1038/nri1154. [DOI] [PubMed] [Google Scholar]
- Furmanek A., Hofsteenge J. Protein C-mannosylation: facts and questions. Acta Biochim. Pol. 2000;47(3):781–789. [PubMed] [Google Scholar]
- Hilton D.J., Watowich S.S., Katz L., Lodish H.F. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J. Biol. Chem. 1996;271(9):4699–4708. doi: 10.1074/jbc.271.9.4699. [DOI] [PubMed] [Google Scholar]
- Hofsteenge J., Muller D.R., de Beer T., Loffler A., Richter W.J., Vliegenthart J.F. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry. 1994;33(46):13524–13530. doi: 10.1021/bi00250a003. [DOI] [PubMed] [Google Scholar]
- Hofsteenge J., Huwiler K.G., Macek B., Hess D., Lawler J., Mosher D.F., Peter-Katalinic J. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 2001;276(9):6485–6498. doi: 10.1074/jbc.M008073200. [DOI] [PubMed] [Google Scholar]
- Ito H., Watanabe S., Takada A., Kawaoka Y. Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 2001;75(3):1576–1580. doi: 10.1128/JVI.75.3.1576-1580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg J., Glasner W., Vicentini A., Doucey M.A., Loffler A., Hess D., Hofsteenge J. C-mannosylation of human RNase 2 is an intracellular process performed by a variety of cultured cells. J. Biol. Chem. 1997;272(42):26687–26692. doi: 10.1074/jbc.272.42.26687. [DOI] [PubMed] [Google Scholar]
- Krieg J., Hartmann S., Vicentini A., Glasner W., Hess D., Hofsteenge J. Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp. Mol. Biol. Cell. 1998;9(2):301–309. doi: 10.1091/mbc.9.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T., Maruyama M., Yamada G., Hatakeyama M., Taniguchi T. The integrity of the conserved ‘WS motif’ common to IL-2 and other cytokine receptors is essential for ligand binding and signal transduction. EMBO J. 1991;10(11):3191–3197. doi: 10.1002/j.1460-2075.1991.tb04881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann S.C., Newton D.L., Youle R.J., James M.N. X-ray crystallographic structure of recombinant eosinophil-derived neurotoxin at 1.83 A resolution. J. Mol. Biol. 1996;260(4):540–552. doi: 10.1006/jmbi.1996.0420. [DOI] [PubMed] [Google Scholar]
- Perez-Vilar J., Randell S.H., Boucher R.C. C-mannosylation of MUC5AC and MUC5B Cys subdomains. Glycobiology. 2004;14(4):325–337. doi: 10.1093/glycob/cwh041. [DOI] [PubMed] [Google Scholar]
- Sanchez A., Trappier S.G., Mahy B.W., Peters C.J., Nichol S.T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad Sci. U. S. A. 1996;93(8):3602–3707. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Yang Z.Y., Xu L., Nabel G.J., Crews T., Peters C.J. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 1998;72(8):6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittler H.J., Wilke A., Gress T., Suttorp N., Drenckhahn D. Role of actin and myosin in the control of paracellular permeability in pig, rat and human vascular endothelium. J. Physiol. 1990;431:379–401. doi: 10.1113/jphysiol.1990.sp018335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebach J., Dieterich P., Luo F., Schillers H., Vestweber D., Oberleithner H., Galla H.J., Schnittler H.J. Endothelial barrier function under laminar fluid shear stress. Lab. Invest. 2000;80(12):1819–1831. doi: 10.1038/labinvest.3780193. [DOI] [PubMed] [Google Scholar]
- Volchkov V.E., Becker S., Volchkova V.A., Ternovoj V.A., Kotov A.N., Netesov S.V., Klenk H.D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214(2):421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- Volchkov V.E., Feldmann H., Volchkova V.A., Klenk H.D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U.S.A. 1998;95(10):5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkova V.A., Feldmann H., Klenk H.D., Volchkov V.E. The nonstructural small glycoprotein sGP of Ebola virus is secreted as an antiparallel-orientated homodimer. Virology. 1998;250(2):408–414. doi: 10.1006/viro.1998.9389. [DOI] [PubMed] [Google Scholar]
- Volchkova V.A., Klenk H.D., Volchkov V.E. Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology. 1999;265(1):164–171. doi: 10.1006/viro.1999.0034. [DOI] [PubMed] [Google Scholar]
- Wahl-Jensen V.M., Afanasieva T.A., Seebach J., Stroher U., Feldmann H., Schnittler H.J. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J. Virol. 2005;79(16):10442–10450. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Delgado R., Xu L., Todd R.F., Nabel E.G., Sanchez A., Nabel G.J. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279(5353):1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]