Significance
Many cell types can find their ways in the environment following gradients of external signaling molecules either by migrating (chemotaxis) or growing (chemotropism) toward the source of the signal. In both cases, the proteins that control cytoskeleton dynamics concentrate, by an autocatalytic mechanism, in the membrane facing the uphill direction of the gradient. Because this polarization is self-amplifying, gradient sensing is susceptible to mistakes, especially because cells have intrinsic polarity cues that can compete with external gradients. Here, we show that, in budding yeast, the presence of internal cues used to guide patterns of division can interfere with the ability to track sexual-pheromone gradients used to find mating partners.
Keywords: chemotropism, Rsr1, signal integration, Rax1, cytokinesis
Abstract
Polarity decisions are central to many processes, including mitosis and chemotropism. In Saccharomyces cerevisiae, budding and mating projection (MP) formation use an overlapping system of cortical landmarks that converges on the small G protein Cdc42. However, pheromone-gradient sensing must override the Rsr1-dependent internal polarity cues used for budding. Using this model system, we asked what happens when intrinsic and extrinsic spatial cues are not aligned. Is there competition, or collaboration? By live-cell microscopy and microfluidics techniques, we uncovered three previously overlooked features of this signaling system. First, the cytokinesis-associated polarization patch serves as a polarity landmark independently of all known cues. Second, the Rax1-Rax2 complex functions as a pheromone-promoted polarity cue in the distal pole of the cells. Third, internal cues remain active during pheromone-gradient tracking and can interfere with this process, biasing the location of MPs. Yeast defective in internal-cue utilization align significantly better than wild type with artificially generated pheromone gradients.
Cell polarity is central to all living organisms for proliferation, differentiation, and morphogenesis. Examples of polarity-associated processes are widely distributed during embryo development, chemotaxis of bacteria or immune cells, chemotropism observed during axon guidance, cell migration, epithelial polarity, and asymmetric cell division. Similarly, malfunction of polarity mechanisms is associated with diverse pathological conditions such as neurodegenerative diseases, genetic disorders, and cancer (1–3).
During polarity establishment, cells are able to interpret a variety of cues of different types. Extracellular, spatially distributed cues include gradients of chemoattractants (chemotaxis/chemotropism), tissue stiffness (durotaxis), and substrate adhesion (haptotaxis). Mammalian cells exposed to a uniform chemoattractant concentration become highly polarized (4) in ways that reveal intrinsic polarity biases (5). For example, the position of the centrosome defines the polarity axis in neutrophils stimulated with uniform concentrations of the bacterial chemoattractant peptide fMLP (6). Intrinsic biases can constrain the dynamics of polarity proteins but are thought to be overwhelmed by external gradients (7). This is especially interesting in the cases where input cues are conflicting. Data suggest that overlapping gradients of different chemoattractants compete with each other in the response of cells, suggesting the existence of a hierarchy of chemoattractants (8). However, the dynamics of the competitions between multiple external signals or between external and internal cues is far from understood.
In this complex context, a question that still remains unanswered is how multiple overlapping signals are interpreted, processed, and integrated so that a cell makes an appropriate polarity response. To address this issue, the current study focused on a polarization system controlled by the small G protein Cdc42, a master regulator of cell polarity throughout eukaryotic cells (9, 10), using Saccharomyces cerevisiae as a model system, which is arguably one of the best studied examples of cell polarity (11, 12).
Yeast polarize their growth following both internal cues (during cell division to form a bud) and external cues (in response to sexual pheromone gradients to find a mate). In both cases, a core set of proteins concentrates to form a polarity patch. Cdc42 is locally activated (exchanging GDP for GTP) by its guanine nucleotide exchange factor (GEF) Cdc24, and this interaction is stabilized by the scaffold protein Bem1 (13, 14). In a positive feedback loop, Cdc42-GTP recruits more of the Bem1-Cdc24 complex, helping to activate more Cdc42 (15). Therefore, polarization in this system is a highly self-reinforcing process. Cdc42-GTP then recruits the formins Bni1 and Bnr1, which nucleate linear actin filaments. Bni1 is part of the polarisome, a complex organized by the Spa2 and Pea2 proteins, which acts as the focal point for polymerization of actin monomers into actin cables (16, 17). Transport of membrane vesicles along these cables allows polarized cell growth.
The direction of polarization is controlled by distinct cues during budding and mating. For budding, polarization is directed to specific sites on the cell surface by internal landmark proteins, known as budding cues. During mating, yeast use extracellular cues: haploid cells of the mating types MATa and MATα secrete a- and α-factor pheromones, respectively, forming gradients that the cell of the opposite mating type detects and tracks to form a mating projection (MP) (18). Because budding cues and pheromone gradients use the same core polarity machinery based on Cdc42, there is potential for the distinct cues to compete with each other. Thus, this system provides a good platform to study the integration of spatial information by the Cdc42 module in eukaryotes.
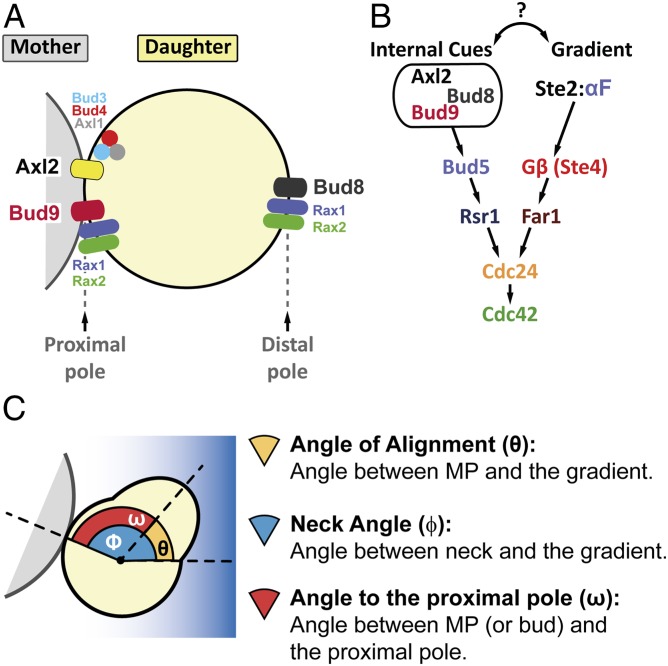
Budding cues are localized at both poles of the cell (Fig. 1A). The proximal pole of a daughter cell is defined by the place where cytokinesis occurred, whereas the distal pole is located opposite to it. Haploid yeast follow an axial budding pattern, in which buds form adjacent to the immediately preceding division site (next to the proximal pole in first-time mothers). In contrast, diploid cells use a bipolar pattern, placing the first bud usually at the distal pole and, in subsequent divisions, at either pole (19, 20). At the molecular level, Axl2/Bud10 and Bud9 are the proximal cues while Bud8 is the distal cue (21). Axl2 is positioned next to the latest cytokinesis ring by a complex formed by Bud3, Bud4, and the haploid-specific protein Axl1 (22). Axl2 dominates over the other cues, causing axial budding in haploids. In some yeast strains, like those from the W303 background, Axl2 is inefficiently positioned due to a mutation in Bud4, and therefore budding follows a mixed axial-bipolar pattern (23). In diploids, yeast use Bud9 and Bud8 to choose either pole. In late G1, these protein landmarks locally activate the small G protein Rsr1/Bud1 by recruiting its GEF Bud5 (24, 25). Active Rsr1-GTP brings Cdc24 to the membrane, which activates Cdc42 to initiate budding (26). Precise spatiotemporal regulation of the activity of Rsr1 is achieved by regulation of the localization of its inhibitor Bud2, a GTPase-activating protein (27). In the absence of Rsr1, the Cdc42 module is activated stochastically at a random place on the cell cortex (28), resulting in randomly placed buds (29, 30).
Fig. 1.
Internal cues and the mating-pheromone-response pathway converge on the activation of Cdc42. (A) The proximal and distal poles of a daughter cell are specified relative to the position of the mother cell. For budding, axial-budding cells utilize the protein Axl2, the axial landmark, which is positioned in every cell cycle adjacent to the last division site (the proximal pole in daughter cells) by the action of Bud3, Bud4, and Axl1. Bipolar budding cells use distal-pole Bud8 and proximal-pole Bud9 to choose the budding site. The Rax1-Rax2 complex participates in the localization of both Bud8 and Bud9. (B) In late G1, Axl2, Bud8, or Bud9 (depending on cell type and condition) can recruit Bud5, which activates Rsr1, which then brings Cdc24, the Cdc42 GEF, to the membrane to generate a concentrated pool of active Cdc42-GTP. During mating in MATa cells, α-factor binds to its receptor Ste2, and the activated receptor dissociates the G-protein trimer into Gα (not illustrated) and Gβ/Ste4. Ste4 recruits the scaffold protein Ste5 (not illustrated) and the adaptor protein Far1 in complex with Cdc24 to the side of the membrane exposed to the higher concentration of α-factor. As in budding, Cdc24 locally activates Cdc42. (C) Definition of the three angles measured in our studies: θ, the angle of alignment; ϕ, the neck angle; and ω, the angle to the proximal pole.
Mating pheromones stimulate cellular responses by binding a G-protein–coupled receptor (Ste2 in MATa and Ste3 in MATα) (Fig. 1B), which then triggers dissociation of a Gαβγ heterotrimer into Gα (Gpa1) and Gβγ (Ste4 + Ste18) (31). Free Gβγ recruits the scaffold protein Ste5 and the adaptor protein Far1 to the plasma membrane. Ste5 activates a MAPK-dependent cascade that induces mating-related genes, stimulates polarization, and arrests the cell cycle in G1 phase (32, 33). Far1 associates with Cdc24 and thus links activated receptors to the Cdc42 polarization module (34, 35). In this way, the pheromone gradient biases the cytoskeleton and cell growth in the direction of the mating partner. If Gβγ is decoupled from the polarity machinery (e.g., by mutations that disrupt Far1-Cdc24 binding), yeast cannot track gradients, and instead MPs form at a default site, defined supposedly by the budding cues (36, 37). Similarly, during isotropic stimulation, yeast form MPs guided to the default site (38). The evidence supporting the use of budding cues as default site comes from experiments with randomly budding cells, such as Δrsr1, that seem to place their MPs randomly as well. However, because in that work (38) the positions of MPs were measured relative to bud scars that were themselves randomly placed, it was not possible to define whether the MPs were positioned randomly relative to the proximal and distal poles. Thus, the question remains as to whether MPs follow budding cues.
The interaction between budding cues and pheromone-gradient tracking is still controversial. Two main models have been proposed for gradient detection in S. cerevisiae: global and local sensing (39). In the first case, polarization would form directly toward the pheromone source as a global reaction of the system. For that to take place, it was thought that proper detection of pheromone gradients involved inactivation of (see ref. 40), or interference with, internal budding cues. The latter might be accomplished, for example, by strongly biasing Cdc24 association toward Far1 instead of Rsr1 (35). In the local-sensing model, an initial “exploratory” polarity patch, established independently of gradient direction, would gradually correct its position through local sampling of the concentration of pheromone. This is possible because the polarity patch is highly dynamic and mobile, especially at low α-factor concentrations (41, 42). In this scenario, inactivation of budding cues might not be necessary. In fact, it was recently suggested that this initial patch forms at budding cues (43, 44).
Considering that budding cues remain active during pheromone stimulation, we decided to revisit this simple, yet unanswered, question: What happens when intrinsic and extrinsic spatial cues are not aligned? Is there competition, so that internal cues interfere with efficient gradient sensing, or collaboration? While in the process of answering this question, we discovered that the default polarization site in response to pheromone is the result of a much more complex system of landmarks, which has, surprisingly, remained unnoticed. Once this system was uncovered, we found that intrinsic polarity sites indeed can interfere with pheromone-gradient sensing and that their removal significantly improves gradient tracking.
Results
Default Polarization Sites Interfere with Gradient Sensing.
To answer the question of how internal and external cues interact, we first analyzed how yeast orient in the pheromone gradients created in our microfluidic devices (45). Unless otherwise indicated, we used, as the parent strain, a MATa Δbar1 strain of the W303 background (SI Appendix, Table S1) (46). Deletion of BAR1, which encodes a secreted α-factor protease, prevents self-induced changes in the α-factor concentration (47). For the gradient assay, we used a derivative with the pheromone-inducible reporter PPRM1-YFP to calibrate the α-factor concentrations in the device. We placed cells in 300-μm-wide chambers and then formed a linear α-factor gradient from 0 to ∼12 nM (SI Appendix, Fig. S1 A–D). After 2 h, we measured the angle of the MP relative to the gradient, θ (Fig. 1C). For quantification, we divided the chamber into eight equal-size regions, from low to high α-factor. At the lowest end of the gradient (SI Appendix, Fig. S1E, regions 1 and 2), yeast did not arrest, and hence there were no MPs. In region 3, with average α-factor concentration between 1 and 4 nM, yeast oriented best [mean cosθ of 0.81 ± 0.03 (a perfectly oriented MP has cos(0°) = 1) and low SD of angles] (SI Appendix, Fig. S1 E and F). At higher α-factor, orientation progressively deteriorated to random (mean cosθ close to zero and a high SD). Previous work obtained similar overall gradient-tracking performance (mean cosθ) and also found that gradient sensing is best at low α-factor (48–50). In all subsequent microfluidic experiments, we used the cosine and the SD of θ to define the region of good alignment.
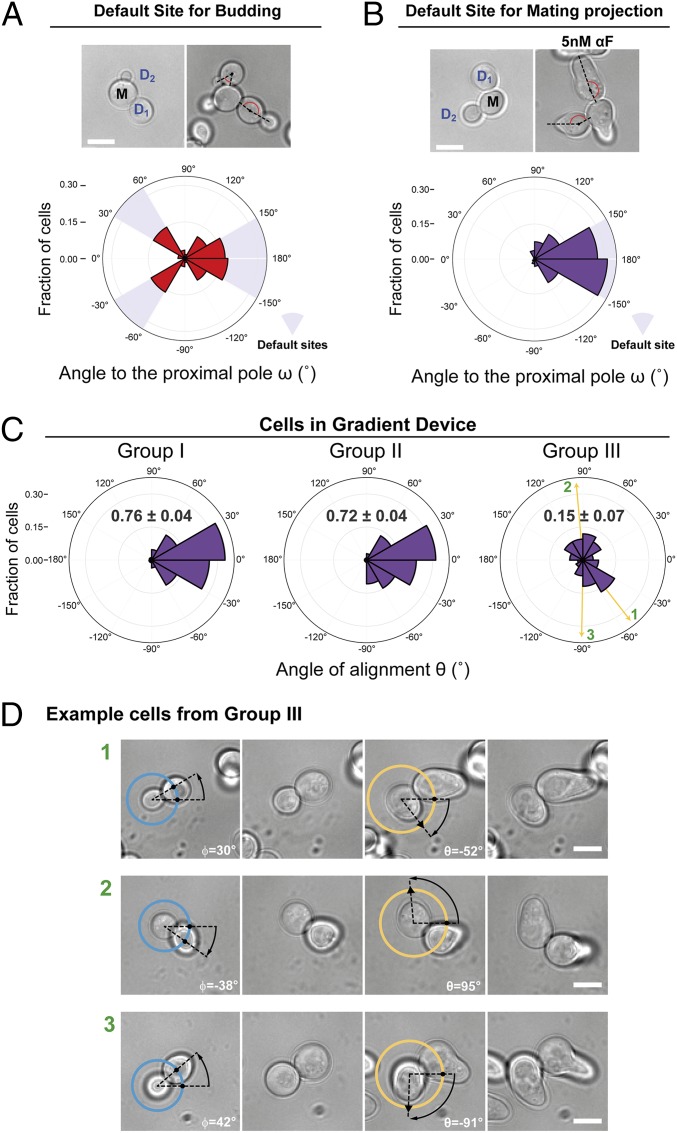
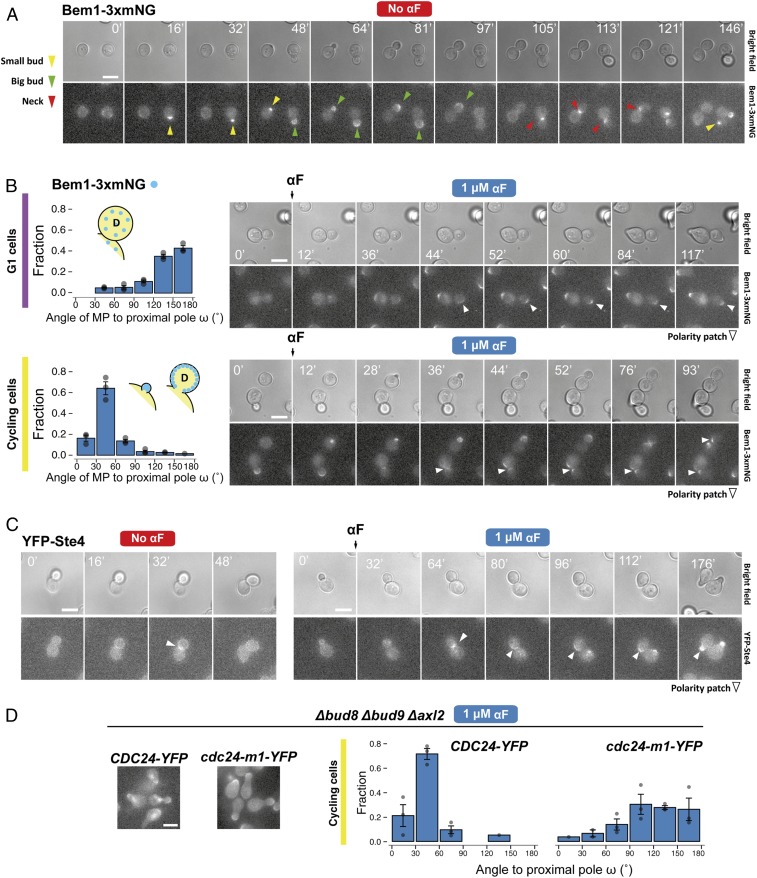
To evaluate if the internal bud-position cues affect MP positioning when yeast are challenged with pheromone gradients, we first determined the default site used by our parental strain. This is important because yeast are immobilized on the glass bottom of the device chamber in random orientations, and thus the orientation of the default site with respect to the pheromone gradient will vary in the cell population. We evaluated the position of the default MP site by stimulating cells with isotropic (no gradient) α-factor at a concentration at which the strain used detected gradient direction well in the device (5 nM). We did this by measuring the angle between the MP and the proximal pole, ω, in daughter cells (Fig. 1C). Nearly all MPs formed at or near the distal pole, indicating that this is the default site for MPs (Fig. 2B). This was surprising, because for budding these cells used both proximal and distal poles (Fig. 2A). This result is an indication that MP and bud-site selection use different rules.
Fig. 2.
Internal polarity cues interfere with gradient sensing. (A and B) Budding and MP default sites used by the MATa Δbar1 strain ACL379. Yeast were imaged during vegetative growth or after stimulation with isotropic (no gradient) 5 nM α-factor (αF). In daughter cells, the angle between the proximal pole and the first bud or MP, ω, was measured as illustrated by the diagram (Fig. 1C). Images show the measurement of ω for two different daughter cells, D1 and D2. (Scale bar, 5 μm.) Polar plots show distributions of ω. Note the “no polarization zone” at angles between −30° and 30°. (C) Δbar1 cells (ACL379) were exposed to a gradient of α-factor in a microfluidic device. Daughter cells were divided into three groups depending on ϕ (Fig. 1C): group I, distal pole facing the gradient (|ϕ| > 120°); group II, distal pole perpendicular to the gradient (60° < |ϕ| < 120°); and group III, distal pole facing away from the gradient (|ϕ| < 60°). Polar plots show the distributions of θ, the angle of alignment, where 0° corresponds to perfect alignment. The mean cosθ ± SEM is indicated in each group. Arrows indicate the angles of alignment of the three cells shown in D. The data from four independent experiments were pooled (total number of cells: 270). (D) Examples of cells from group III. For each cell, ϕ and θ are indicated. The pheromone source is on the right. (Scale bar, 5 μm.)
Next, we performed gradient-sensing experiments. To simplify the analysis, we divided daughter cells into three groups based on the “neck angle” (i.e., between the cell’s proximal-distal axis and the gradient direction), ϕ (Fig. 1C). The results strongly supported the hypothesis that default sites interfere with gradient sensing (Fig. 2C). Cells oriented well when the default site neither faced nor opposed the gradient (group II), even better when the default site faced the gradient (group I), and worse when it opposed the gradient (group III). Note that we measured the angle of the first polarized growth direction, before MPs could reorient to correct their initial MP direction (see example of a reorienting MP in Fig. 2D, cell 3) (49). Remarkably, only a small fraction corrected their gradient alignment (24 ± 7%).
We obtained the same qualitative results using cells of another commonly used genetic background, S288C. These cells used the proximal pole for both budding and MPs (SI Appendix, Fig. S2A). The proximal-pole preference was due to the presence of a functional Bud4 protein since swapping alleles between S288C and W303 largely swapped their budding and MP default sites as well (SI Appendix, Fig. S3). When exposed to an α-factor gradient, S288C cells showed the same biases as W303 cells, but, due to the opposite location of their default site, the behavior in groups I and III was reversed (compare Fig. 2C and SI Appendix, Fig. S2C).
Thus, when gradient and default sites are opposed, the conflict between external and internal cues is such that the cell’s choice is usually incorrect. These findings provide solid evidence that intrinsic polarization landmarks can interfere with gradient sensing.
A Pheromone-Specific Cue Independent of the Mitotic-Landmark System.
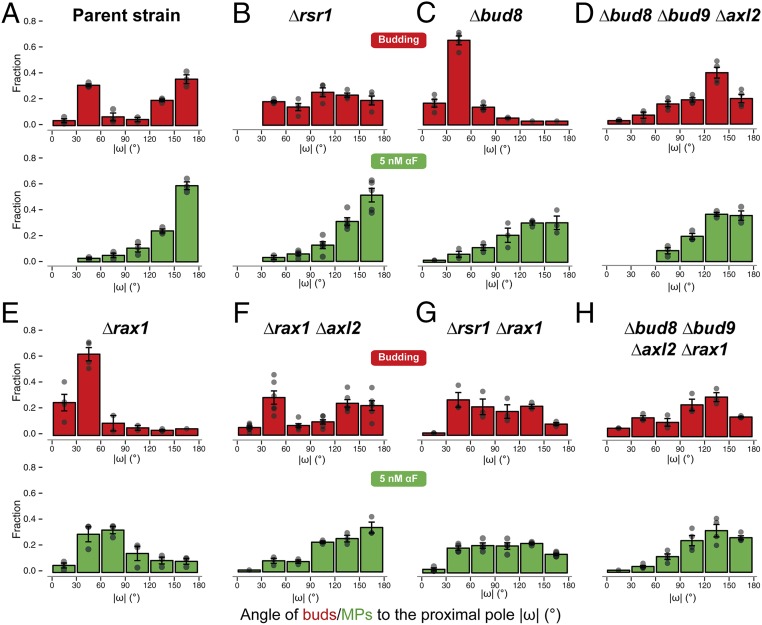
The above results suggest that inactivating the molecular components of internal cues might improve gradient-sensing capabilities. To obtain such cueless strains, we first tried Δrsr1 cells, which, as expected, budded randomly (Fig. 3B, Top) due to the interrupted communication between the Cdc42 module and the landmark proteins Bud8, Bud9, and Axl2 (Fig. 1A). Unexpectedly, in uniform pheromone, Δrsr1 cells formed MPs with a clear distal bias (Fig. 3B, Bottom). Distal MP formation was also seen in the proximal-budding strain Δbud8 and in the triple landmark deletion strain Δbud8 Δbud9 Δaxl2 (Fig. 3 C and D). Taken together, in contrast to previous literature (38), these observations pointed to the existence of a pheromone-specific cue at the distal pole independent of Bud8 (the only budding cue that directs budding to the distal pole) and of Rsr1.
Fig. 3.
Simultaneous deletion of RAX1 and RSR1 results in random MPs in low αF. Distribution of the angle ω in daughter cells for the first bud (red) or the MP in response to uniform 5 nM α-factor (green) in the parent strain (ACL379) (A) or in the indicated deletion strains (B–H). In all cases, the angle distributions were divided into 30° bins, and the fraction of cells in each bin was calculated. Note that the data for strain ACL379 are the same as in Fig. 2A but displayed in a linear histogram (absolute value of ω). Data from at least three to four independent experiments were pooled to calculate the means ± SEMs (bars and whiskers). Strains: parent strain (ACL379); Δrsr1 (YGV5405); Δbud8 (YGV5429); Δbud8 Δbud9 Δaxl2 (YGV5433); Δrax1 (YGV5259); Δrax1Δxl2 (YGV5408); Δrsr1Δrax1 (YGV5838); and Δbud8 Δbud9 Δaxl2 Δrax1 (YGV5839).
We then found evidence indicating that this MP-specific distal cue is the Rax1-Rax2 complex. First, deletion of RAX1 caused MPs to become mostly proximal (Fig. 3E). This bias was Axl2-dependent since it disappeared when we deleted AXL2 or RSR1 in the Δrax1 strain (Fig. 3 F and G). Second, and more importantly, Δrsr1Δrax1 cells made randomly positioned MPs. Interestingly, the quadruple mutant Δaxl2 Δbud8 Δbud9 Δrax1 had a slight residual distal tendency (Fig. 3H), suggesting that Rsr1 might also work as a weak distal cue. Lack of internal landmarks in Δrsr1Δrax1 cells did not affect the shape of MPs or the dynamics of polarity patch formation (SI Appendix, Fig. S4), suggesting that these landmarks’ only role might be to bias polarization to specific locations. The above results were independent of genetic background: Δrsr1 made distal MPs and the Δrsr1Δrax1 made random MPs in S288C cells as well (SI Appendix, Fig. S5). This was expected since in Δrsr1 strains the genetic status of BUD4 should be irrelevant (Fig. 1 A and B). Thus, at low α-factor, Δrsr1Δrax1 yeast are cueless for MP formation.
The Cytokinesis Patch as a Seed for MPs, Independently of Internal Cues.
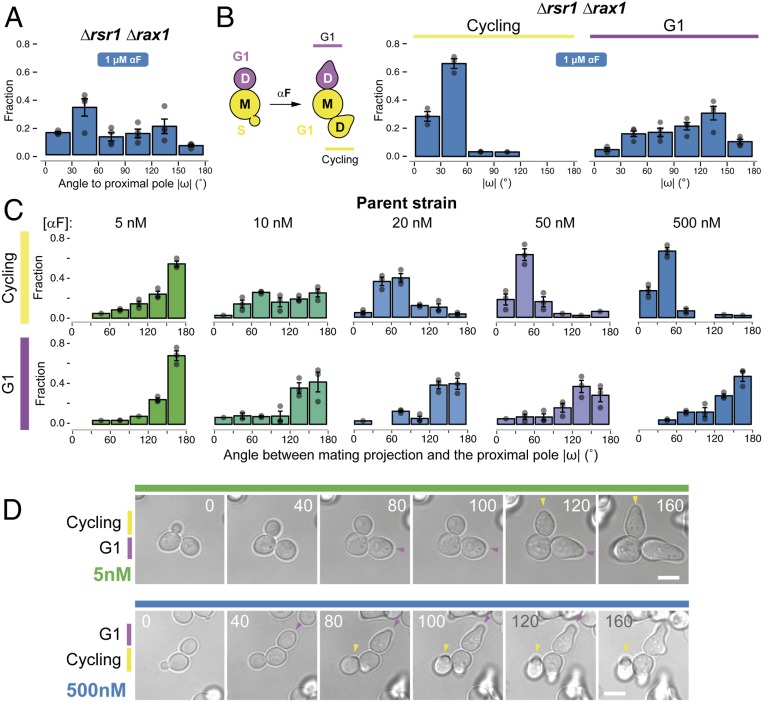
Surprisingly, when we tested the Δrsr1Δrax1 strain in uniformly high (1 μM) α-factor, MP positioning was not random. Instead, a significant fraction of the cells used the proximal pole (Fig. 4A), suggesting the existence of additional regulation. Further investigation revealed that MP positioning in high α-factor depended strongly on the cell-cycle position at the time of stimulation (Fig. 4B). This was true not only in Δrsr1Δrax1 but also in the parent strain. If daughter cells were still buds at the time of pheromone stimulation (“cycling” cells) (Fig. 4B), they uniformly formed MPs around the proximal pole at high α-factor concentration. In contrast, if daughters were already in G1 when we added α-factor, they behaved as when stimulated with low α-factor: the parent strain used the distal pole, whereas the Δrsr1Δrax1 cells placed MPs in random locations (Fig. 4 B–D). Moreover, cycling cells made proximal MPs in all landmark-deletion strains tested, including the triple-cue deletion strain Δaxl2 Δbud8 Δbud9 (SI Appendix, Fig. S6 A–F), as well as in Δrsr1 and Δrsr1 Δrax1 strains of the S288C background (SI Appendix, Fig. S7). Thus, in cycling cells there is an Rsr1- and Rax1-independent mechanism that operates at high α-factor concentrations and directs MPs to the proximal pole. The existence of this mechanism, together with the Rax1/Rax2-dependent default site at the distal pole, explains the behavior of cycling wild-type cells across all α-factor concentrations (Fig. 4C). In contrast, G1 cells selected the distal pole at all pheromone concentrations.
Fig. 4.
Default sites differ between G1 and cycling cells. (A) Exponential cultures of the Δrsr1 Δrax1 strain YGV5838 were subjected to live-cell imaging in the presence of 1 μM α-factor. The angle of the MP relative to the proximal pole (ω) was measured in daughter cells as in Fig. 3. (B, Left) Diagram illustrating classification into G1 and cycling daughter cells at the time of the addition of α-factor. (B, Right) Distribution of angle ω in cycling and G1 Δrsr1Δrax1 cells from A. (C) Distribution of angle ω in cycling and G1 cells of the ACL379 parent strain exposed to the indicated pheromone concentrations. Data from three independent experiments (points) were pooled to calculate the mean ± SEM (bars and whiskers). (D) Time-lapse images of ACL379 cells from C stimulated with 5 or 500 nM αF. Numbers correspond to time in minutes after pheromone addition. Arrowheads indicate the position of the MP. (Scale bar, 5 μm.)
We wondered why cycling and G1 cells behaved so differently. One obvious difference is that cycling cells make an MP only after finishing cytokinesis, whereas G1 cells can immediately execute a chemotropic response. Thus, we monitored the polarity-patch dynamics in cycling and G1 cells using strains expressing Bem1 (see the Introduction) fused to three tandem copies of mNeonGreen (Bem1-3xmNG). In G1 cells, Bem1-3xmNG translocated directly to the distal pole to form a new polarization site (Fig. 5 A and B and Movie S1). In cycling cells, the presence of α-factor seemed to prevent the dispersal of the cytokinesis-related polarity patch, which normally occurs when cells enter G1. Instead, this patch persisted at the neck and appeared to be used directly for MP polarization (Fig. 5B and Movie S2).
Fig. 5.
Polarity-patch dynamics during default MP site selection. (A) Time-lapse fluorescence microscopy of YGV5097 cells expressing Bem1-3xmNG (three copies of monomeric NeonGreen fluorescent protein in tandem) with different bud sizes (a proxy for cell-cycle position). Arrowhead colors indicate the different patterns of Bem1 localization: concentrated during apical growth of incipient and small buds (yellow), in the periphery of big buds during isotropic growth (green), and at the neck during cytokinesis (red). (B, Left) Distribution of MP angle ω in G1 (Top) and cycling (Bottom) Bem1-3xmNG cells. (B, Right) Dynamics of Bem1-3xmNG in typical G1 (Top) and cycling (Bottom) cells exposed to 1 μM α-factor. Arrowheads mark the location of the Bem1 patch. (C) Time-lapse images of cells expressing N-terminally YFP-tagged Ste4 (TCY3064) in vegetative growth (“No αF”) or after exposure to 1 μM αF. In A, B, and C, times of image acquisition are indicated in minutes. (D) Distribution of MP angle ω in cycling Δbud8 Δbud9 Δaxl2 cells expressing Cdc24-YFP (YGV6097) or Cdc24-m1-YFP (YGV6100) after exposure to 1 μM αF. Images illustrate the localization of Cdc24-YFP and Cdc24-m1-YFP after 2 h of exposure to αF. Note the absence of nuclear Cdc24-YFP in the m1 mutant due to the interrupted interaction with Far1. (Scale bars, 5 μm.)
If the above interpretation is correct, it would explain why cycling cells choose the proximal pole even in the absence of Rsr1 and Rax1 at high α-factor concentration (Fig. 4 and SI Appendix, Fig. S7). That is, the prepolarized structure at the neck could hold the pheromone-pathway polarity components at that site. A good candidate is Gβ (Ste4) because it is at the neck during cytokinesis even in the absence of α-factor (51) (Fig. 5 C, Left, and Movie S3) and might be tethered to the patch by the Far1-Cdc24 complex. Monitoring a YFP-Ste4 patch at the neck showed that, indeed, when pheromone was added, Ste4 moved only slightly to the side of the cytokinesis site, where an MP then formed (Fig. 5 C, Right, and Movie S4). Blocking the connection between the patch and Ste4 should abolish proximal MP positioning. To test that, we used a Δbud8 Δbud9 Δaxl2 strain that also expresses Cdc24-m1, a mutant protein with an impaired association with Far1, but normal binding to Rsr1. In this strain, MPs showed a strong distal-pole bias (Fig. 5D), indicating that the proximal-pole choice did involve the connection between the mating pathway and the cytokinesis patch via the Far1-Cdc24 tether.
Dose-Dependent Switch Depends on MP Patch Mobility.
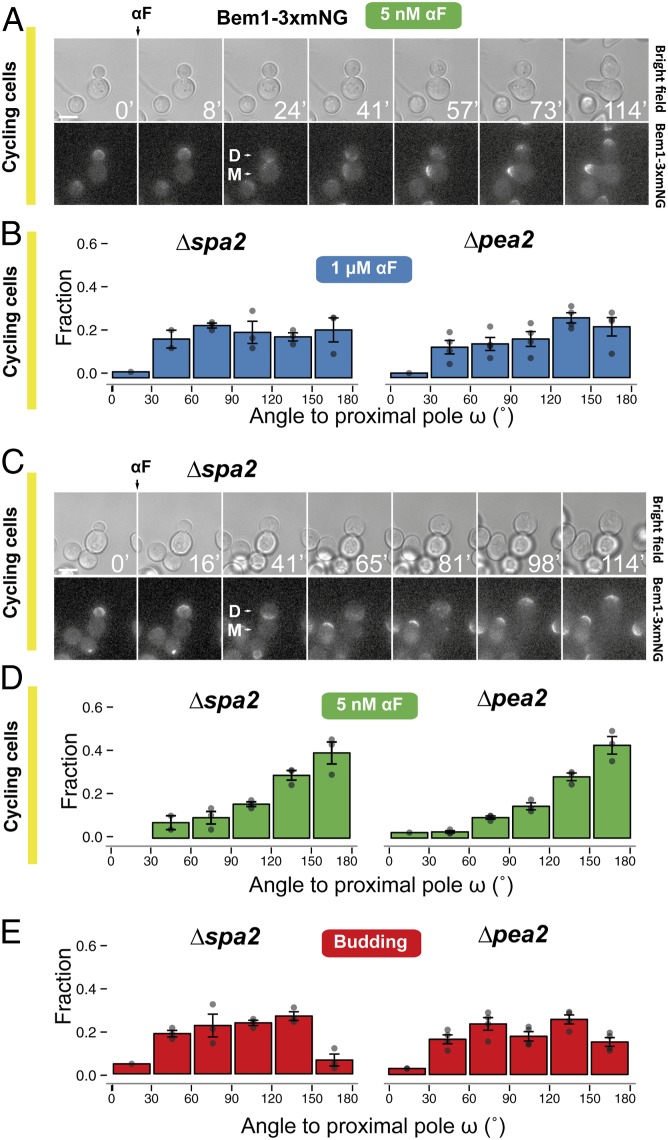
What, then, explains the dose-dependent switch from proximal-pole to distal-pole MP formation in cycling cells (Fig. 4C)? We imagined two alternatives. At low α-factor concentrations, the patch at the neck might either fall apart and reform directly at the distal pole, or instead it might migrate through the plasma membrane. Observations of Bem1-3xmNG in cycling cells exposed to 5 nM α-factor seemed to support the migration alternative because in all cells analyzed we detected a slow movement of a broad Bem1 signal from the neck toward the distal pole (Fig. 6A and Movie S5). Migration could be the result of fast cycles of partial disassembly followed by reassembly in a nearby location, with an overall directional movement toward the distal pole. Patch migration presumably happens at lower α-factor doses because the polarity-patch mobility increases under these conditions (41, 52).
Fig. 6.
Participation of the cytokinesis-related polarity patch in selection of MP sites. (A) Dynamics of Bem1-3xmNG localization followed by live-cell fluorescence microscopy in the parental strain (YGV5097) exposed to 5 nM α-factor. Times are in minutes. “D” marks the daughter and “M” the mother cell. (Scale bar, 5 μm.) (B) Distributions of MP angle ω in cycling Δspa2 (YRB3862) and Δpea2 (YRB3865) daughters exposed to 1 μM α-factor. Data from three independent experiments (points) were pooled to calculate the means ± SEMs (bars and whiskers). (C) As in A, dynamics of Bem1-3xmNG localization in Δspa2 (YGV5836) stimulated with 1 μM α-factor. (D) Distributions of MP angle ω in cycling Δspa2 (YRB3862) and Δpea2 (YRB3865) cells exposed to 5 nM α-factor. (E) Distributions of first-bud angle ω in Δspa2 (YRB3862) and Δpea2 (YRB3865) cells growing vegetatively.
To further test the idea that patch migration plays a role in the dose-dependent switch in MP localization, we artificially increased patch mobility by deleting SPA2 or PEA2, which encode two components of the polarisome. The instability of the patch in these mutants is reflected in the shape of the MPs, which are significantly broader at high α-factor (53). As with lowering the dose of pheromone, these deletions resulted in a reduced frequency of MPs at the proximal pole at high α-factor (Fig. 6B) and migration of the Bem1-3xmNG cap away from the bud neck (Fig. 6C and Movie S6). MPs formed elsewhere, resulting in an essentially random distribution (Fig. 6B), consistent with the hypothesis that movement of the polarity patch allows it to escape the neck prepolarization. Further increasing patch mobility in Δspa2 or Δpea2 cells by also lowering the α-factor concentration resulted in MPs forming mainly at the distal pole (Fig. 6D), indicating that the polarisome is not required for use of the distal landmarks during pheromone stimulation [contrary to what happens during budding, which was random in these strains (54) (Fig. 6E)].
In a reciprocal experiment, we reinforced the stability of the cytokinesis patch and asked if this blocked the switch from proximal-pole to distal-pole MPs. We did this by deleting the neck-localized Cdc42 inhibitor Rga1 (55–57). Indeed, the absence of Rga1 in cells exposed to 5 nM α-factor resulted in proximal MPs in both the parent and Δrsr1Δrax1 strains (SI Appendix, Fig. S8), further supporting the hypothesis that patch migration requires destabilization of the initial cytokinesis patch.
Taken together, these results indicate that the increased patch mobility at low α-factor explains why, under these conditions, cycling cells use similar sites for MPs as do G1 cells by de novo polarization (SI Appendix, Fig. S6).
Improved Gradient-Sensing in Cueless (Δrsr1 Δrax1) Cells.
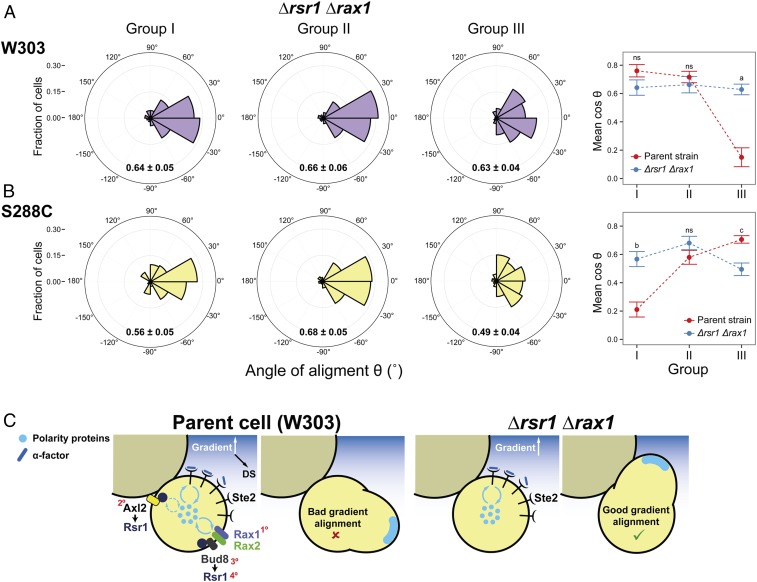
With a better understanding of how internal landmarks govern default-site positioning of MPs, we next evaluated the performance of the landmark-free Δrsr1 Δrax1 mutant in a gradient device, expecting that an improvement might be seen in those daughter cells that had their default polarization sites opposed to the direction of the gradient. To this end, we classified Δrsr1 Δrax1 daughters into the same three groups that we had used for the parent strain (Fig. 2C). We did not discriminate between G1 and cycling daughters because, at the α-factor concentrations used to score gradient sensing, MP positioning in Δrsr1 Δrax1 cells (and in their parent strain) is not influenced by cell-cycle position (Figs. 3G and 4C). As predicted, the Δrsr1 Δrax1 cells tracked gradients well even when the gradient direction was opposed to the default site (group III cells, Fig. 7 A, Left), outperforming in this case the parent strain (Fig. 7 A, Right). Thus, the presence of landmarks that direct MPs to the default site hinders gradient sensing. As in the parent strain, only a few of those cells that oriented incorrectly managed to reorient (18 ± 4%), indicating that the correcting mechanism is independent of internal landmarks.
Fig. 7.
Improved gradient sensing in cueless (Δrsr1 Δrax1) cells. (A and B) Δrsr1 Δrax1 cells in the W303 (YGV5838) (A) and S288C (YDV6164) (B) backgrounds were exposed to an α-factor gradient generated in a microfluidic device, and the angles of the MPs relative to the gradient, θ, were measured. (Left) Data are shown in polar histograms as in Fig. 2C. Number of cells: 311 (YGV5838) and 390 (YDV6164). (Right) The means of cosθ ± SEM, indicated within the polar histograms, are plotted for the parental and Δrsr1 Δrax1 strains in both backgrounds. Data for parent strains are from Fig. 2C and SI Appendix, Fig. S2C. Statistical differences between parent and Δrsr1 Δrax1 were calculated by Kolmogorov–Smirnov test (P values: a, 3.5 × 10−7; b, 1.9 × 10−9; c, 0.049; ns, nonstatistical differences). (C) Model for the competition between internal cues and gradient decoding. In RSR1 RAX1 cells, the presence of internal cues can compete for a shared pool of polarity proteins, thereby affecting the cell’s ability to track the gradient. In the scheme, only competition for cytosolic components (blue circles) is represented, but the same argument can be applied for membrane components (Discussion). In Δrsr1 Δrax1 cells, the absence of internal cues removes the competition, improving the detection of gradient direction. In the parental W303 strain, the order of strength of the different landmarks is depicted by red numbers: Rax1-Rax2Distal > Axl2Proximal ≥ Bud8Distal > Rsr1(itself)Distal. (In S228C-background cells, Axl2 is stronger than Rax1-Rax2; see also SI Appendix, Table S2.)
Like W303-background cells, S288C-background Δrsr1Δrax1 cells tracked gradients better than their parental strain when exposed to α-factor gradients, as evident in the group I cells in which the default site of the parent cells opposed the direction of the gradient (Fig. 7B).
Discussion
In this work, we revisited a matter studied since the early 1990s: the interaction between different polarization cues in yeast. The external cue provided by a pheromone gradient during mating must override the internal polarity cues used for budding. Using live-cell microscopy and microfluidics techniques, we uncovered three previously overlooked features of this signaling system. First, the cytokinesis-related polarization patch can serve as a polarity cue independently of all known landmark proteins. Second, the Rax1-Rax2 complex functions as a pheromone-specific polarity cue at the distal pole. Finally, internal cues remain active during pheromone-gradient tracking and interfere with this process, thus biasing the location of MPs (Fig. 7C).
Interaction Between Gradient Sensing and Intrinsic Polarity Cues.
On the question of local- vs. global-sensing modes of gradient tracking, our results suggest that yeast cells do both, depending on the preexistence of a polarity patch. Cycling cells, starting from the cytokinesis patch, use local sensing, whereas G1 cells may use global sensing, directly polarizing at the site of maximum extracellular signal. Consistent with our findings, recent work by others, in which the analysis was restricted to cycling cells, found support for local sensing (43, 44). Moreover, work using mating mixtures showed that G1 cells can form de novo polarizations that are aligned with a mating partner, consistent with a global-sensing response (58). For either sensing mode, we showed that internal landmarks are not necessary. Quite the opposite, yeast devoid of classical cues track gradients better than their parental counterparts.
How do intrinsic cues interfere with the localization or stabilization of the polarity patch that tracks pheromone concentration? Our observations are compatible with a competition for membrane-associated polarity components, facilitated, for example, by the presence of Gβγ at the membrane. The presence of a fixed focus (the default site) may partly deplete an incipient pheromone-induced Gβγ patch facing the gradient, thereby destabilizing it. Then, through positive feedback, one particular position may become fixed. This idea is reminiscent of the recent proposal that patches may exist in an immature, indecisive (mobile) state and then evolve into a committed, less mobile state (58).
As we have shown in yeast, mammalian cells can also polarize in ways that show the influence of internal cues (5). In a remarkable example, neutrophils stimulated with uniform concentrations of the bacterial chemoattractant peptide fMLP usually polarize to the left of a vector drawn from the center of the nucleus to the centrosome (6). This chiral behavior can be annulled or even reversed (rightward bias) by genetic and chemical interventions, indicating its mechanistic basis. However, in the presence of external gradients, these internal biasing forces are believed to be overcome (7). In the same way that internal and external signals compete in yeast, neutrophils exposed to simultaneous gradients show antagonism in which some attractants dominate over others (8). The mechanisms that underlie competition between different cues are still unclear.
A Pheromone-Promoted Role for Rax1 during MP Positioning.
Although at first sight seemingly in contradiction, our results showing that Δrsr1 yeast do not position MPs randomly are actually consistent with the original observations suggesting that they do (38). Time-lapse analysis of single cells led to a different interpretation than in the earlier study because we used the mother-bud axis to score MP positioning instead of bud scars (SI Appendix, Fig. S9). Ours is not the first evidence of distal polarization in daughter cells in the Δrsr1 background. Δrsr1 daughters from diploid cells in general or from haploid cells committed to invasive growth still showed a degree of distal budding (59, 60). This bias is caused by polarisome-dependent delivery of vesicles to the distal pole during apical growth, which promotes positioning of bipolar landmarks (54). However, this mechanism does not seem to promote distal MPs because the polarisome-deficient mutants Δspa2 and Δpea2, which in our hands show random budding (Fig. 6E), still show intact distal bias (by de novo polarization in G1 cells or at low α-factor in cycling cells) (Fig. 6D and SI Appendix, Fig. S6 G and H). Instead, our evidence indicates that the extra distal bias of MPs in ∆rsr1 cells is due to Rax1.
How does Rax1 promote the formation of distal-pole MPs? As we have shown, it is not due to the localization of Bud8; thus, we propose that it is the localization of the Rax1/Rax2 complex itself that concentrates the polarization machinery at the distal pole. We propose that Rax1 directly attracts the G protein-Far1 complex. In fact, Rax1 contains an RGS domain in its cytoplasmic side that binds and may regulate Gα/Gpa1 (61). A role of Rax1 through the G protein itself (instead of downstream, as the classical landmarks do) suggests that Rax1 may determine Cdc24 localization only indirectly, via Far1. Consistent with this hypothesis, the cdc24-m1 Δrsr1 double mutant, in which Far1 cannot interact with Cdc24, fails to stabilize the polarity patch, which instead moves continuously around the cell cortex (41, 42).
It seems that the use of default polarization sites could have been positively selected through evolution. In a natural environment, mating is thought to occur following spore germination in fly frass (62). In fact, tetrad dissolution in insect guts is thought to enhance out-breeding, i.e., the mating of cells from different asci (63). However, a large fraction of germinating spores chooses to bud and form microcolonies instead of committing to mating immediately after germination (62). In homothallic yeast, mothers usually switch mating type, which enables mating with their previous daughters. Thus, microcolony formation should increase selfing at the expense of out-breeding. However, a distal bias for polarization in daughter cells may have been selected to reduce mating with the mother cell located at its proximal pole.
MP Site-Selection Rules Depend on Pheromone Concentration and the Cell-Cycle Stage.
The behavior of G1 and cycling cells is quite different. In the absence of gradients, direct observation of the polarity proteins revealed that G1 cells formed MPs by de novo polarization at the default site irrespective of pheromone dose. In stark contrast, the default localization of MPs in cycling cells is regulated by α-factor concentration. At high pheromone, the cytokinesis-related polarity patch, to which the Ste4-Far1 complex tethers, is used as a cue for MP formation. Only at low-pheromone concentrations is the patch able to detach from this prepolarized starting point. There are at least two possible reasons why patch detachment from the neck might happen at low α-factor concentrations: increased patch mobility and a weaker connecting tether. First, in the context of the “patch-wandering” model, patch mobility increases at low pheromone due to slower ligand:receptor-binding dynamics coupled with vesicle delivery of new unbound receptors (41, 52). In addition, low activation of the MAPK Fus3 at low α-factor might fail to stabilize the patch (43, 64). Second, the connecting tether should be weaker in the low-pheromone range due to lower availability of dissociated Gβγ dimers to form a complex with Far1. Low Gβγ concentrations would also reduce the abundance of the Gβγ-Far1-Cdc24 complex, compromising the ability of Gβγ to connect with the cytokinesis patch at the neck.
Cytokinesis Patch as a Polarity Cue.
It is intriguing that the formation of proximal MPs in response to saturating α-factor even in the absence of protein landmarks has remained unnoticed for so long. The use of the cytokinesis site as a polarity cue may be a feature shared by many polarized cells. For example, in Drosophila melanogaster during nervous-system development, the mitotic-cleavage site determines the axis of the precursor cell and defines where the first neurite forms (65). In epithelial cells, the mitotic midbody helps preserve the apical-basal tissue architecture during proliferation by providing a spatial cue for the formation of the apical daughter-cell interface (66). In yeast, the polarization-proximal bias enables growth of a daughter cell toward its mother after it has switched mating type so that it mates with it; at the same time, the distal bias dependent on Rax1-Rax2 may represent a counterbalance to enable genetic outcrossing.
Final Remarks.
The presence of intrinsic polarity cues is ubiquitous in eukaryotic cells. In neurons, for example, polarity is inherited from the preexisting apical-basal polarity of the neuroepithelial progenitor cell or from the site of cell division (5). These cells also must track gradients of regulatory molecules during axon guidance. Thus, the interaction between internal and external cues is a common situation in cell biology. In this work, we have demonstrated that intrinsic landmarks may bias the polarity system within a cell either positively or negatively depending on the situation. We expect that these results will help our understanding of gradient sensing as a cell behavior that integrates a variety of polarity systems.
Materials and Methods
Strains.
S. cerevisiae strains were derived from MATa Δbar1 parents of the W303 and S288C genetic backgrounds by standard nucleic-acid and yeast-manipulation procedures. More information on strains used and their construction is provided in SI Appendix, Tables S1, S3, and S4.
Time-Lapse Microscopy.
Cultures in exponential growth were sonicated and diluted to a concentration of 2.5 × 105 cells/mL. Polyethylene glycol [average molecular weight (MW) = 3,550, Sigma catalog no. P4338] was added to all media at 0.1% to avoid nonspecific binding of α-factor (67). Then, 20 μL of cell suspension was applied to individual wells of 384-well glass-bottom plates precoated with 1 mg/mL Con A (Sigma catalog no. C7275). Plates were centrifuged to assist the attachment of cells. In the microscope, two to three image fields per well were selected, and the time-lapse imaging was started. After the first time point, 20 μL of α-factor (Anaspec, custom-made) was added to final concentrations of 0 to 1,000 nM, depending on the experiment.
For imaging, a fully motorized Olympus IX-81 microscope with an Olympus UplanSapo objective (63×; numerical aperture = 1.35), coupled with an HQ2 (Roper Scientific) cooled charged-coupled device camera, was used with filter sets for yellow fluorescent protein (YFP), cyan fluorescent protein, and thetramethylrhodamine isothiocyanate (TRITC) (41028, 31044v2, and 41004, Chroma Technologies). For assessing Bem1-3xmNG or YFP-Ste4 localization, Z-stacks were acquired, and maximum-fluorescence projections were used.
Chemotropism Assays in Microfluidic Devices.
Microfluidic devices designed for the generation of stable gradients in open chambers were fabricated using standard protocols for polydimethylsiloxane microfluidic-device construction as described previously (45, 68).
To improve adherence of cells to the glass, the bottom of the chambers was treated with poly-d-lysine (1 mg/mL; Sigma catalog no. P6407) at 4 °C overnight and then incubated with ConA (1 mg/mL; Sigma catalog no. C7275) for 1 h at room temperature. Next, the device was filled with 0.22 μm of sterile, filtered water using a vacuum-assisted method (69). Subsequently, the two ports of the device were connected with tubing and syringes filled with filtered synthetic complete medium alone or with 50 nM αF and either 0.1 mg/mL bromophenol blue or dextran-TRITC (MW = 4,400; Sigma catalog no. T1037) as a tracking dye. All media contained 0.1% wt/vol polyethylene glycol (MW = 3,550; Sigma) to prevent nonspecific binding of α-factor to the container’s surfaces (67). Water hydrostatic pressure drove all flow. Formation of the gradients was monitored by measuring bromophenol blue fluorescence. Finally, the flow was stopped, the chambers were washed with medium, and cells from a mildly sonicated yeast exponential culture were loaded on top of the device. Cells were allowed to settle and bind to the bottom glass before resuming the flow. Imaging was performed as described for glass-bottom plates.
Angle Determinations.
Angles were manually measured using custom-written macros for ImageJ on bright-field time-lapse image stacks. The angle of alignment (θ) was measured between the MP and the gradient. The angle ω was measured between the MP and the mother-daughter neck (i.e., the proximal pole). Cells that rotated or moved led to inexact determination of the proximal pole or the MP and thus were excluded from the analysis.
Data Analysis.
All data analysis was done in R (70). For angle linear histograms, cells were split into six bins, from 0 to 180°, according to the position of the mating projection, and the fraction of cells in each bin in each experiment was calculated. The means ± SEMs of at least three independent assays were plotted. For gradient experiments, first the region of the device where cells could detect gradient direction was established. To do that, the chambers were divided in 50-μm windows parallel to the gradient direction. In each window, the SD and the cosine of the angle of alignment (θ) were calculated. In a region of good gradient sensing, the dispersion of angles drops below the level of the distribution of angles in cell populations exposed to isotropic α-factor. SDs and cosθ values were used to define the region of the devices used for analyses. In these experiments, only the first polarization was considered, and two angles were scored: the angle of the mating projection (θ) and the neck angle (ϕ), both relative to the gradient direction. Based on the second angle, cells were divided into three groups: the distal pole facing the gradient (I, |ϕ|>120°); the distal pole perpendicular to the gradient (II, 60°<|ϕ|<120°); and the proximal pole facing the gradient (III, |ϕ|<60°). In each group, polar histograms with 30°-bins were plotted after pooling cells from independent microfluidic assays.
Statistical differences in cosθ were calculated using the nonparametric Kolmogorov–Smirnov test.
Data Availability Statement.
All data discussed in the paper are available to readers in ref. 71.
Supplementary Material
Acknowledgments
We thank Peter Pryciak, Pablo Aguilar, and Andreas Constantinou for helpful discussions and comments on the manuscript; the Albert Folch laboratory for assistance with microfluidic experiments; Andreas Constantinou for providing plasmid A666; and the editor and the two anonymous reviewers for numerous constructive comments and suggestions. This work was supported by Grants PICT2013-2210, PICT2015-3824, and PICT2016-0949 from the Argentine Agency of Research and Technology (to A.C.-L.) and by Grant 1R01GM097479-01 from the National Institute of General Medical Sciences, NIH (to Roger Brent and A.C.-L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper can be found in Mendeley Data at http://doi.org/10.17632/wpczdh5rnk.1.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912505117/-/DCSupplemental.
References
- 1.Aguilar B. J., Zhu Y., Lu Q., Rho GTPases as therapeutic targets in Alzheimer’s disease. Alzheimers Res. Ther. 9, 97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egorov M. V., Polishchuk R. S., Emerging role of Cdc42-specific guanine nucleotide exchange factors as regulators of membrane trafficking in health and disease. Tissue Cell 49, 157–162 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Haga R. B., Ridley A. J., Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 7, 207–221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Servant G., et al. , Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287, 1037–1040 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yogev S., Shen K., Establishing neuronal polarity with environmental and intrinsic mechanisms. Neuron 96, 638–650 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Xu J., et al. , Polarity reveals intrinsic cell chirality. Proc. Natl. Acad. Sci. U.S.A. 104, 9296–9300 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millius A., Dandekar S. N., Houk A. R., Weiner O. D., Neutrophils establish rapid and robust WAVE complex polarity in an actin-dependent fashion. Curr. Biol. 19, 253–259 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heit B., et al. , PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat. Immunol. 9, 743–752 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Adams A. E. M., Johnson D. I., Longnecker R. M., Sloat B. F., Pringle J. R., CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111, 131–142 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etienne-Manneville S., Cdc42The centre of polarity. J. Cell Sci. 117, 1291–1300 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Goryachev A. B., Leda M., Many roads to symmetry breaking: Molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell 28, 370–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H.-O., Bi E., Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71, 48–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butty A.-C., et al. , A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 21, 1565–1576 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y., Cerione R., Bender A., Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J. Biol. Chem. 269, 2369–2372 (1994). [PubMed] [Google Scholar]
- 15.Kozubowski L., et al. , Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18, 1719–1726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheu Y. J., Santos B., Fortin N., Costigan C., Snyder M., Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053–4069 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chenevert J., Valtz N., Herskowitz I., Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics 136, 1287–1296 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson C. L., Hartwell L. H., Courtship in S. cerevisiae: Both cell types choose mating partners by responding to the strongest pheromone signal. Cell 63, 1039–1051 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Chant J., Pringle J. R., Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751–765 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freifelder D., Bud position in Saccharomyces cerevisiae. J. Bacteriol. 80, 567–568 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi E., Park H.-O., Cell polarization and cytokinesis in budding yeast. Genetics 191, 347–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang P. J., Angerman E., Jung C.-H., Park H.-O., Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J. Cell Sci. 125, 3840–3849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voth W. P., Olsen A. E., Sbia M., Freedman K. H., Stillman D. J., ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot. Cell 4, 1018–1028 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang P. J., Sanson A., Lee B., Park H.-O., A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292, 1376–1378 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krappmann A.-B., Taheri N., Heinrich M., Mösch H.-U., Distinct domains of yeast cortical tag proteins Bud8p and Bud9p confer polar localization and functionality. Mol. Biol. Cell 18, 3323–3339 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y., Bender A., Cerione R. A., Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 270, 626–630 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Park H.-O., Sanson A., Herskowitz I., Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 13, 1912–1917 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wedlich-Soldner R., Altschuler S., Wu L., Li R., Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231–1235 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Bender A., Pringle J. R., Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. U.S.A. 86, 9976–9980 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chant J., Herskowitz I., Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203–1212 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Bardwell L., A walk-through of the yeast mating pheromone response pathway. Peptides 25, 1465–1476 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Dohlman H. G., Thorner J. W., Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu. Rev. Biochem. 70, 703–754 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Pryciak P. M., Huntress F. A., Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12, 2684–2697 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butty A. C., Pryciak P. M., Huang L. S., Herskowitz I., Peter M., The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282, 1511–1516 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Nern A., Arkowitz R. A., A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J. Cell Biol. 144, 1187–1202 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nern A., Arkowitz R. A., A GTP-exchange factor required for cell orientation. Nature 391, 195–198 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Valtz N., Peter M., Herskowitz I., FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 131, 863–873 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madden K., Snyder M., Specification of sites for polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol. Biol. Cell 3, 1025–1035 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegemann B., Peter M., Local sampling paints a global picture: Local concentration measurements sense direction in complex chemical gradients. BioEssays 39, 1600134 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Roemer T., Madden K., Chang J., Snyder M., Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 10, 777–793 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Dyer J. M., et al. , Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23, 32–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nern A., Arkowitz R. A., G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell 5, 853–864 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Hegemann B., et al. , A cellular system for spatial signal decoding in chemical gradients. Dev. Cell 35, 458–470 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Wang X., et al. , Mating yeast cells use an intrinsic polarity site to assemble a pheromone-gradient tracking machine. J. Cell Biol. 218, 3730–3752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura A. C., et al. , Utilization of extracellular information before ligand-receptor binding reaches equilibrium expands and shifts the input dynamic range. Proc. Natl. Acad. Sci. U.S.A. 111, E3860–E3869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colman-Lerner A., et al. , Regulated cell-to-cell variation in a cell-fate decision system. Nature 437, 699–706 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Sprague G. F. Jr, Herskowitz I., Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J. Mol. Biol. 153, 305–321 (1981). [DOI] [PubMed] [Google Scholar]
- 48.Muller N., et al. , A predictive model for yeast cell polarization in pheromone gradients. PLOS Comput. Biol. 12, e1004795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallier L. G., Segall J. E., Snyder M., The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton 53, 251–266 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Moore T. I., Chou C.-S., Nie Q., Jeon N. L., Yi T.-M., Robust spatial sensing of mating pheromone gradients by yeast cells. PLoS One 3, e3865 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suchkov D. V., et al. , Polarization of the yeast pheromone receptor requires its internalization but not actin-dependent secretion. Mol. Biol. Cell 21, 1737–1752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClure A. W., et al. , Role of polarized G protein signaling in tracking pheromone gradients. Dev. Cell 35, 471–482 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valtz N., Herskowitz I., Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol. 135, 725–739 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheu Y. J., Barral Y., Snyder M., Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235–5247 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong Z., et al. , Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 179, 1375–1384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meitinger F., et al. , A memory system of negative polarity cues prevents replicative aging. Cell 159, 1056–1069 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Miller K. E., Lo W.-C., Lee M. E., Kang P. J., Park H.-O., Fine-tuning the orientation of the polarity axis by Rga1, a Cdc42 GTPase-activating protein. Mol. Biol. Cell 28, 3773–3788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henderson N. T., et al. , Ratiometric GPCR signaling enables directional sensing in yeast. PLoS Biol. 17, e3000484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cullen P. J., Sprague G. F. Jr, The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell 13, 2990–3004 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michelitch M., Chant J., A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr. Biol. 6, 446–454 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Chasse S. A., et al. , Genome-scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 5, 330–346 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McClure A. W., Jacobs K. C., Zyla T. R., Lew D. J., Mating in wild yeast: Delayed interest in sex after spore germination. Mol. Biol. Cell 29, 3119–3127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reuter M., Bell G., Greig D., Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 17, R81–R83 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Matheos D., Metodiev M., Muller E., Stone D., Rose M. D., Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 165, 99–109 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pollarolo G., Schulz J. G., Munck S., Dotti C. G., Cytokinesis remnants define first neuronal asymmetry in vivo. Nat. Neurosci. 14, 1525–1533 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Herszterg S., Pinheiro D., Bellaïche Y., A multicellular view of cytokinesis in epithelial tissue. Trends Cell Biol. 24, 285–293 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Liu B., et al. , Parts-per-million of polyethylene glycol as a non-interfering blocking agent for homogeneous biosensor development. Anal. Chem. 85, 10045–10050 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Keenan T. M., Hsu C.-H., Folch A., Microfluidic “jets” for generating steady-state gradients of soluble molecules on open surfaces. Appl. Phys. Lett. 89, 114103 (2006). [Google Scholar]
- 69.Monahan J., Gewirth A. A., Nuzzo R. G., A method for filling complex polymeric microfluidic devices and arrays. Anal. Chem. 73, 3193–3197 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Bush A., Chernomoretz A., Yu R., Gordon A., Colman-Lerner A., Using Cell-ID 1.4 with R for microscope-based cytometry. Curr. Protoc. Mol. Biol. Chapter 14, Unit 14.18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vasen G., Colman-Lerner A., Supporting Data: Mitotic and pheromone-specific intrinsic polarization cues interfere with gradient sensing in S. cerevisiae. Mendeley Data. 10.17632/wpczdh5rnk.1 (17 February 2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available to readers in ref. 71.